Abstract
Prions are self-propagating protein aggregates that act as protein-based elements of inheritance in fungi. Although prevalent in eukaryotes, prions have not been identified in bacteria. Here we found that a bacterial protein, transcription terminator Rho of Clostridium botulinum (Cb-Rho), could form a prion. We identified a candidate prion-forming domain (cPrD) in Cb-Rho and showed that it conferred amyloidogenicity on Cb-Rho and could replace the PrD of a yeast prion-forming protein functionally. Furthermore, its cPrD enabled Cb-Rho to access alternative conformations in E. coli – a soluble form that terminated transcription efficiently and an aggregated, self-propagating prion form that was functionally compromised. The prion form caused genome-wide changes in the transcriptome. Thus, Cb-Rho functions as a protein-based element of inheritance in bacteria, suggesting that the emergence of prions predates the evolutionary split between eukaryotes and bacteria.
Main Text
First described as the protein-based causative agent of the fatal transmissible spongiform encephalopathies (1), prions have also been uncovered in fungi where they act as protein-based elements of inheritance that confer new phenotypes on those cells that harbor them (2, 3). Fungal prions are formed by proteins that can access alternative conformations, including a self-perpetuating amyloid fold (the prion form) that is characteristically heritable (4). At least a dozen prion-forming proteins with diverse functions have been uncovered in budding yeast (4), where prions have been shown to confer growth advantages under specific conditions (3). Nonpathogenic prion-like proteins have also been described in mammals (5), Aplysia (6), Drosophila (6) and, most recently, Arabidopsis (7). Although bacteria have been shown to propagate a yeast prion (8, 9), it is not known if bacterial prions exist.
We used a previously described Hidden Markov Model-based algorithm trained on a set of yeast prion-forming proteins (10, 11) to mine ~60,000 bacterial genomes for proteins containing candidate prion-forming domains (cPrDs) (Table S1). Among the proteins identified by this analysis was the transcription termination factor Rho of Clostridium botulinum E3 str. Alaska E43 (Cb), which contains a 68-residue cPrD (residues 74-141) (Fig S1) (12). Rho is a highly conserved hexameric helicase that loads onto nascent transcripts and couples ATP hydrolysis to RNA translocation, resulting in RNA polymerase transcription termination (13). Phyletic analysis of bacterial Rho orthologs revealed that many Rho proteins contain an N-terminal insertion domain (NID) (14); the Cb-Rho cPrD is located within an NID (Fig 1A). Notably, many Rho orthologs from distantly related bacteria contain similarly situated cPrDs (Fig S2).
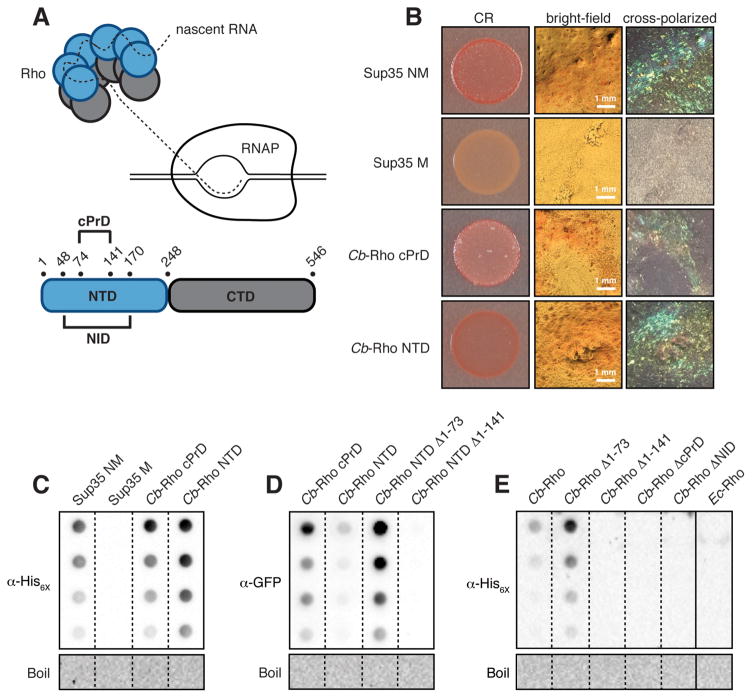
Figure 1.
Cb-Rho contains an amyloidogenic cPrD. (A) (Top) Cartoon of a Rho hexamer engaging RNAP. (Bottom) Domain organization of Cb-Rho highlighting its cPrD and NID. (B) E. coli cells exporting the indicated protein domain were spotted on solid medium containing the amyloid-binding dye Congo Red (CR). The PrD of yeast Sup35 (NM) and its derivative (M) serve as positive and negative controls, respectively. Plate-derived material visualized by bright-field microscopy and between crossed polarizers reveals “apple-green” birefringence characteristically exhibited by CR-bound amyloid aggregates. (C) Amyloid-like aggregates (characterized by their resistance to denaturation in SDS) were detected in plate-derived material from (B) as assessed by filter-retention analysis. Undiluted sample and three two-fold dilutions are shown. Aggregates were no longer detected once boiled (see Supplementary Materials). (D) SDS-stable aggregates were detected in cell lysates containing the indicated cPrD-containing Cb-Rho fragment fused to mYFP, but not a cPrD-lacking variant. (E) SDS-stable aggregates were detected in cell lysates containing hexahistidine-tagged, cPrD-containing Cb-Rho, but not cPrD-lacking variants or Ec-Rho. (C–E) See Fig S3 for immunoblot analyses.
A characteristic of most prion-forming proteins is their ability to assemble as amyloid aggregates (3, 4). Therefore, we tested Cb-Rho for amyloidogenicity using an E. coli-based secretion assay that detects extracellular amyloid (15). Both the 68-residue cPrD of Cb-Rho and a 248-residue Rho fragment encompassing the cPrD in the structurally well-defined Rho N-terminal domain (NTD) (13) had amyloid-forming propensities by this test (Fig 1A, B, C; Fig S3A). These Cb-Rho domains also formed amyloid-like material when fused to monomeric YFP and produced in the E. coli cytoplasm, as did a truncated NTD fragment retaining the complete cPrD (NTD Δ1-73) but not an NTD variant lacking the cPrD (NTD Δ1-141) (Fig 1A, D; Fig S3B). Similarly, full-length Cb-Rho and Cb-Rho Δ1-73 formed amyloid-like material in the E. coli cytoplasm, but excess E. coli Rho or three Cb-Rho variants lacking the cPrD did not (Fig 1A, E; Fig S3C). Thus, the cPrD confers amyloid-forming potential on Cb-Rho.
Next, we asked whether the Cb-Rho cPrD could functionally replace the PrD of the yeast prion-forming protein Sup35, an essential translation release factor. Strains containing Sup35 in its non-prion form ([psi−] strains) are proficient for translation termination, whereas strains containing Sup35 in its prion form ([PSI+] strains) exhibit stop codon-readthrough detectable as a heritable colony-color phenotype (4). Sup35 has an N-terminal PrD (Sup35NM) that can be functionally replaced by heterologous PrDs and a C-terminal moiety (Sup35C) with translation release activity (11). We replaced Sup35NM with several Cb-Rho fragments (cPrD, NTD, or NTD Δ1-73) and constructed yeast strains containing the three resulting Cb-Rho-Sup35C chimeras as the sole source of translation release activity. In each case the cells exhibited a [psi−]-like phenotype that could undergo conversion to a stable [PSI+]-like phenotype (Fig 2A, B), the propagation of which was dependent on the chaperone Hsp104 (Fig 2C), mirroring the Hsp104 dependence of the Sup35 and other yeast prions (4). Cells containing three variants of the Cb-Rho NTD-Sup35C chimera lacking the cPrD exhibited [psi−]-like phenotypes only (Fig 2A, B). Thus, Cb-Rho cPrD can functionally substitute for Sup35NM. Additionally, our finding that the Cb-Rho cPrD-Sup35C chimera exhibited stable [psi−]-like and [PSI+]-like phenotypes in yeast enabled us to demonstrate that Cb-Rho aggregates produced in bacteria were infectious when introduced into yeast cells (Fig S5).
Figure 2.
Cb-Rho behaves as a prion in yeast. (A) Yeast cells (containing native Sup35 or the indicated Cb-Rho fragment fused to Sup35C) were spotted on adenine-lacking medium containing (Gal) or lacking (Raf) galactose to induce transient overproduction of plasmid-encoded Sup35NM-EYFP (in Sup35-containing cells) or Cb-Rho cPrD-EGFP fusion protein (in Sup35 fusion protein-containing cells) (see Fig S4 for fluorescence microscopy and Supplementary Materials). Adenine-lacking medium selects for [PSI+] clones and transient overproduction of its PrD triggers conversion of a prion protein to the prion form (4). (B) Phenotypes of [psi−], [PSI+], [psi−]-like, and [PSI+]-like colonies from (A) restreaked on rich medium. [psi−]-like and [PSI+]-like states are designated [rho-X-C−] and [RHO-X-C+], respectively. (C) Phenotypes of colonies from (B) restreaked on rich medium following passage on GuHCl-containing medium (transient inactivation of Hsp104).
We then asked whether Cb-Rho could interconvert between non-prion and self-perpetuating prion conformations in E. coli cells, with conversion to the prion form causing decreased Rho activity. To detect Rho activity, we used a reporter in which the Rho-dependent terminator tR1 is placed between a phage promoter and lacZ (Fig 3A). Decreased Rho activity in a strain harboring this reporter should result in increased expression of lacZ (detectable as blue colony-color on indicator medium). Although we were unable to replace E. coli rho, an essential gene, with Cb rho (see Supplementary Materials), we could construct a strain encoding a Cb-Rho NTD – E. coli-Rho CTD chimera in place of E. coli Rho. This strain exhibited a slow-growth phenotype, which we could ameliorate by supplementing the chromosomally encoded Rho chimera with excess plasmid-encoded Rho chimera (Cb-Rho NTD Δ1-73-E. coli-Rho CTD) (Fig 3A; Supplementary Materials). Cells containing this plasmid gave rise to both pale blue colonies (henceforth referred to as pale colonies and indicative of high Rho activity) and blue colonies (indicative of low Rho activity) (Fig 3B). This phenotypic heterogeneity suggested that plasmid encoded Rho chimera was capable of accessing alternative conformations: a soluble, non-prion conformation (pale colonies) and an aggregated, prion conformation (blue colonies).
Figure 3.
Cb-Rho exhibits prion behavior in E. coli. (A) Schematic of E. coli reporter strain containing terminator tR1 upstream of lacZ and chromosomally encoded Rho chimera. Reporter strain growth defects were rescued by a plasmid directing the IPTG-inducible synthesis of hexahistidine-tagged Rho chimera Δ1-73 (see Supplementary Materials). (B) A representative inducer-containing indicator plate showing blue and pale colonies of isogenic cells described in (A). (C) SDS-stable aggregates were detected in cell lysates of cultures inoculated with blue colonies (blue dot); those inoculated with pale colonies (pale blue dot) contained little aggregated material and those inoculated with naïve colonies (yellow dot) of reporter strain cells producing plasmid encoded Rho chimera lacking the cPrD (Δ1-141) contained none. We note that production of Rho chimera Δ1-141 was toxic and could not be evaluated for colony-color phenotype. Overproduction of ClpB, but not a catalytically inactive ClpB mutant (ClpB*), under the control of an arabinose (Ara)-inducible promoter cured cells of SDS-stable aggregates (see Fig S6 for immunoblot analyses). (D) A representative example of terminator readthrough in “blue” cells. Rockhopper-normalized RNA-seq counts aligned to sense/antisense (+/−) strands are shown for cells descended from pale colonies (pale blue tracks) or blue colonies (blue tracks). Genes exhibiting statistically significant changes in expression are colored in blue. Solid red line indicates a transcript with its previously inferred Rho-dependent terminator (cross bar) (20). Dashed pink line shows extension of the transcript in “blue” cells.
Additional findings fulfilled key predictions of the hypothesis that alternative protein conformations, including a self-perpetuating prion form, were responsible for the pale and blue colony-color phenotypes: (i) Plasmid DNA originating from either blue or pale colonies revealed no sequence differences within or surrounding the chimeric rho gene and the plasmids behaved indistinguishably when retransformed into naïve reporter strain cells (Fig 4A). (ii) Cell lysates prepared from overnight cultures inoculated with blue colonies contained Rho protein aggregates, but those prepared from cultures inoculated with pale colonies contained little aggregated material (Fig 3C; Fig S6). (iii) “Blue” and “pale” cell cultures produced predominantly blue and pale colonies, respectively, when plated on indicator medium; this bias was even more pronounced when blue and pale colonies were resuspended and replated without intervening liquid growth, a procedure that enabled us to estimate the probability of spontaneous loss (< 0.8% per cell per generation) and appearance (< 0.2% per cell per generation) of the Rho prion (Fig 4A; Supplementary Materials). (iv) The blue colony-color phenotype (i.e. the Rho prion) could be propagated for ≥ 120 generations and its maintenance depended on continued synthesis of the Rho chimera (Fig 4B; Supplementary Materials). (v) Reminiscent of the effect of Hsp104 overproduction on [PSI+] yeast cells (4), the blue colony-color phenotype was “cured” by overproduction of the disaggregase ClpB (the bacterial ortholog of Hsp104) (Fig 3C; Fig 4C). (vi) Transcription profiling on cells descended from either blue or pale parent colonies revealed genome-wide readthrough of Rho-dependent terminators specifically in cells derived from blue colonies (Fig 3D; Fig S7; Fig S8; Table S2) (16). Thus, Cb-Rho can undergo conversion to a self-propagating prion conformation in E. coli cells, eliciting genome-wide changes in the transcriptome.
Figure 4.
Propagation of the Cb-Rho prion occurs over ≥ 120 generations. (A) Naïve, blue, or pale parent colonies of reporter strain cells producing Rho chimera Δ1-73 were either grown overnight in liquid medium (+) or simply resuspended (−), and then replated on inducer-containing indicator plates. (B) The blue colony-color phenotype was propagated over five rounds of replating without intervening growth in liquid medium (see Supplementary Materials). (C) The blue colony-color phenotype was cured upon arabinose (Ara)-inducible overproduction of ClpB, but not a catalytically inactive ClpB mutant (ClpB*). Values represent the mean and standard deviation of three biological replicates (see Supplementary Materials).
We also observed Cb-Rho prion behavior without protein overproduction by constructing a strain encoding Cb-Rho NTD Δ1-73-Ec-Rho CTD at the native chromosomal rho locus. The resulting cells were healthy and yielded both blue and pale colonies when plated on indicator medium (Fig S9). Blue colonies spontaneously gave rise to pale colonies at a low frequency (Fig S9B, C), and the blue colony-color phenotype was cured by transient ClpB overproduction (Fig S9B; see Supplementary Materials). Moreover, pale colonies gave rise to blue colonies upon transient exposure to 5% ethanol, a stress condition known to confer a fitness advantage to E. coli cells carrying a reduced function rho allele (Fig S9B; see Supplementary Materials) (17).
The identification of Cb-Rho as a bacterial prion-forming protein establishes protein-based heredity in the bacterial domain of life, suggesting that the emergence of prion-dependent phenomena predates the divergence of Bacteria and Eukaryota. Moreover, the presence of cPrDs in Rho proteins of bacteria representing at least 6 phyla, including the dominant constituents of the human gut microbiota, suggests that the impact of bacterial prion-based phenomena may be far reaching. Rho and Sup35 prion formation have an intriguing similarity. Whereas formation of the Sup35 prion triggers genome-wide changes in the proteome due to stop codon readthrough (18), formation of the Rho prion triggers genome-wide changes in the transcriptome due to terminator readthrough. Prions may represent a source of epigenetic diversity in bacteria, which can contribute to bacterial fitness in a variety of settings, facilitating immune evasion in the context of infection (19), for example, or enabling antibiotic tolerance in quasi dormant “persister” cells (20). Moreover, because prion formation typically results in a reduced function phenotype, it is notable that adaptive null mutations in bacteria are common, often facilitating survival in response to environmental challenge (21).
Supplementary Material
Summary.
The identification of a bacterial protein that forms a prion establishes protein-based heredity in the bacterial domain of life.
Acknowledgments
We thank A. Lancaster and the late Susan Lindquist, to whose memory this work is dedicated, for sharing the Hidden Markov Model-based algorithm, R. Washburn and M. Gottesman for the tR1-lacZ reporter, and S. Dove, S. Garrity and B. Nickels for invaluable advice. RNA-seq data were deposited at Gene Expression Omnibus (GEO) under accession number GSE90485. Work was supported by NIH Pioneer Award OD003806 and grant GM115941 (A.H.).
Footnotes
References and Notes
- 1.Aguzzi A, Baumann F, Bremer J. The prion’s elusive reason for being. Annu Rev Neurosci. 2008;31:439–477. doi: 10.1146/annurev.neuro.31.060407.125620. [DOI] [PubMed] [Google Scholar]
- 2.Wickner RB. [URE3] as an altered URE2 protein: evidence for a prion analog in Saccharomyces cerevisiae. Science. 1994;264:566–569. doi: 10.1126/science.7909170. [DOI] [PubMed] [Google Scholar]
- 3.Newby GA, Lindquist S. Blessings in disguise: biological benefits of prion-like mechanisms. Trends Cell Biol. 2013;23:251–259. doi: 10.1016/j.tcb.2013.01.007. [DOI] [PubMed] [Google Scholar]
- 4.Liebman SW, Chernoff YO. Prions in yeast. Genetics. 2012;191:1041–1072. doi: 10.1534/genetics.111.137760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cai X, Chen J, Xu H, Liu S, Jiang QX, Halfmann R, Chen ZJ. Prion-like polymerization underlies signal transduction in antiviral immune defense and inflammasome activation. Cell. 2014;156:1207–1222. doi: 10.1016/j.cell.2014.01.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Si K, Kandel ER. The role of functional prion-like proteins in the persistence of memory. Cold Spring Harb Perspect Biol. 2016 doi: 10.1101/cshperspect.a021774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chakrabortee S, Kaytekin C, Newby GA, Mendillo ML, Lancaster A, Lindquist S. Luminidependens (LD) is an Arabidopsis protein with prion behavior. Proc Natl Acad Sci U S A. 2016;113:6065–6070. doi: 10.1073/pnas.1604478113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Garrity SJ, Sivanathan V, Dong J, Lindquist S, Hochschild A. Conversion of a yeast prion protein to an infectious form in bacteria. Proc Natl Acad Sci U S A. 2010;107:10596–10601. doi: 10.1073/pnas.0913280107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yuan AH, Garrity SJ, Nako E, Hochschild A. Prion propagation can occur in a prokaryote and requires the ClpB chaperone. Elife. 2014;3:e02949. doi: 10.7554/eLife.02949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lancaster AK, Nutter-Upham A, Lindquist S, King OD. PLAAC: a web and command-line application to identify proteins with prion-like amino acid composition. Bioinformatics. 2014;30:2501–2502. doi: 10.1093/bioinformatics/btu310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Alberti S, Halfmann R, King O, Kapila A, Lindquist S. A systematic survey identifies prions and illuminates sequence features of prionogenic proteins. Cell. 2009;137:146–158. doi: 10.1016/j.cell.2009.02.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pallarès I, Iglesias V, Ventura S. The Rho termination factor of Clostridium botulinum contains a prion-like domain with a highly amyloidogenic core. Front Microbiol. 2016;6:1516. doi: 10.3389/fmicb.2015.01516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Skordalakes E, Berger JM. Structure of the Rho transcription terminator: mechanism of mRNA recognition and helicase loading. Cell. 114:135–146. doi: 10.1016/s0092-8674(03)00512-9. [DOI] [PubMed] [Google Scholar]
- 14.D’Heygère F, Rahbi M, Boudvillain M. Phyletic distribution and conservation of the bacterial transcription termination factor Rho. Microbiology. 2013;159:1423–1436. doi: 10.1099/mic.0.067462-0. [DOI] [PubMed] [Google Scholar]
- 15.Sivanathan V, Hochschild A. Generating extracellular amyloid aggregates using E. coli cells. Genes Dev. 2012;26:2659–2667. doi: 10.1101/gad.205310.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Peters JM, Mooney RA, Grass JA, Jessen ED, Tran F, Landick R. Rho and NusG suppress pervasive antisense transcription in Escherichia coli. Genes Dev. 2012;26:2621–2633. doi: 10.1101/gad.196741.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Freddolino PL, Goodarzi H, Tavazoie S. Fitness landscape transformation through a single amino acid change in the Rho terminator. PLoS Genet. 2012;8:e1002744. doi: 10.1371/journal.pgen.1002744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Baudin-Baillieu A, Legendre R, Kuchly C, Hatin I, Demais S, Mestdagh C, Gautheret D, Namy O. Genome-wide translational changes induced by the prion [PSI+] Cell Rep. 2014;8:439–448. doi: 10.1016/j.celrep.2014.06.036. [DOI] [PubMed] [Google Scholar]
- 19.Stewart MK, Cookson BT. Non-genetic diversity shapes infectious capacity and host resistance. Trends Microbiol. 2012;20:461–466. doi: 10.1016/j.tim.2012.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Balaban NQ, Merrin J, Chait R, Kowalik L, Leibler S. Bacterial persistence as a phenotypic switch. Science. 2004;305:1622–1625. doi: 10.1126/science.1099390. [DOI] [PubMed] [Google Scholar]
- 21.Hottes AK, Freddolino PL, Khare A, Donnell ZN, Liu JC, Tavazoie S. Bacterial adaptation through loss of function. PLoS Genet. 2013;9:e1003617. doi: 10.1371/journal.pgen.1003617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Link AJ, Phillips D, Church GM. Methods for generating precise deletions and insertions in the genome of wild-type Escherichia coli: applications to open reading frame characterization. J Bacteriol. 1997;179:6228–6237. doi: 10.1128/jb.179.20.6228-6237.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.McClure R, Balasubramanian D, Sun Y, Bobrovskyy M, Sumby P, Genco CA, Vanderpool CK, Tjaden B. Computational analysis of bacterial RNA-seq data. Nucleic Acids Res. 2013;41:e140. doi: 10.1093/nar/gkt444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Saldana AJ. Java Treeview: extensible visualization of microarray data. Bioinformatics. 2004;20:3246–3248. doi: 10.1093/bioinformatics/bth349. [DOI] [PubMed] [Google Scholar]
- 25.Letunic I, Bork P. Interactive tree of life (iTOL) v3: an online tool for the display and annotation of phylogenetic and other trees. Nucleic Acids Res. 2016;44:W242–W245. doi: 10.1093/nar/gkw290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kim HC, Washburn RS, Gottesman ME. Role of E. coli NusA in phage HK022 Nun-mediated transcription termination. J Mol Biol. 2006;359:10–21. doi: 10.1016/j.jmb.2006.02.081. [DOI] [PubMed] [Google Scholar]
- 27.Tanaka M, Weissman JS. An efficient protein transformation protocol for introducing prions into yeast. Methods Enzymol. 2006;412:185–200. doi: 10.1016/S0076-6879(06)12012-1. [DOI] [PubMed] [Google Scholar]
- 28.Matsumoto Y, Shigesada K, Hirano M, Imai M. Autogenous regulation of the gene for transcription termination factor Rho in Escherichia coli: localization and function of its attenuators. J Bacteriol. 1986;166:945–958. doi: 10.1128/jb.166.3.945-958.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Martinez A, Opperman T, Richardson JP. Mutational analysis and secondary structure model of the RNP1-like sequence motif of transcription termination factor Rho. J Mol Biol. 1996;257:895–908. doi: 10.1006/jmbi.1996.0210. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.