NLRP3 is an innate immune receptor that needs to be tightly regulated to prevent overshooting immune responses. Stutz et al. demonstrate that NLRP3 is phosphorylated as a safeguard against accidental activation, and that dephosphorylation involving PP2A activity is required for NLRP3 activation.
Abstract
NLRP3 is a cytosolic pattern recognition receptor that senses microbes and endogenous danger signals. Upon activation, NLRP3 forms an inflammasome with the adapter ASC, resulting in caspase-1 activation, release of proinflammatory cytokines and cell death. How NLRP3 activation is regulated by transcriptional and posttranslational mechanisms to prevent aberrant activation remains incompletely understood. Here, we identify three conserved phosphorylation sites in NLRP3 and demonstrate that NLRP3 activation is controlled by phosphorylation of its pyrin domain (PYD). Phosphomimetic residues in NLRP3 PYD abrogate inflammasome activation and structural modeling indicates that phosphorylation of the PYD regulates charge–charge interaction between two PYDs that are essential for NLRP3 activation. Phosphatase 2A (PP2A) inhibition or knock-down drastically reduces NLRP3 activation, showing that PP2A can license inflammasome assembly via dephosphorylating NLRP3 PYD. These results propose that the balance between kinases and phosphatases acting on the NLRP3 PYD is critical for NLRP3 activation.
Introduction
NLRP3 is a cytosolic pattern recognition receptor that senses microbes and endogenous danger signals. Upon activation, NLRP3 forms an inflammasome with the adapter ASC, resulting in caspase-1 activation, release of proinflammatory cytokines, and cell death (Latz et al., 2013). The exact activation mechanism of NLRP3 remains unclear, but it is assumed that its leucine-rich repeats (LRRs) have a regulatory function and hinder oligomerization of NLRP3 via its nucleotide-binding oligomerization domain (NOD). Upon activation, NLRP3 oligomerizes and triggers the helical fibrillar assembly of the adapter ASC via pyrin domain (PYD)–PYD interactions (Latz et al., 2013; Lu et al., 2014). ASC fibrils assemble into large structures, called ASC specks (Franklin et al., 2014; Schmidt et al., 2016) and recruit pro-caspase-1, leading to its autoproteolytic activation. Caspase-1 mediates activation and release of highly proinflammatory cytokines of the IL-1 family and triggers pyroptosis of macrophages. Whereas NLRP3 activation is beneficial in host responses to certain pathogens, it can be detrimental under sterile inflammatory conditions, such as atherosclerosis or gout (Strowig et al., 2012). Unsurprisingly, NLRP3 inflammasome assembly is controlled at transcriptional and posttranslational levels. Resting macrophages express insufficient amounts of NLRP3 and require a priming signal before NLRP3 inflammasome activation (Bauernfeind et al., 2009; Franchi et al., 2009). Additionally, a fast, nontranscriptional priming pathway induces NLRP3 deubiquitination (Juliana et al., 2012; Py et al., 2013), thereby allowing NLRP3 to get activated. Furthermore, NLRP3 activity can be blocked by nitrosylation (Hernandez-Cuellar et al., 2012; Mao et al., 2013; Mishra et al., 2013). These studies indicate that NLRP3 activation is tightly regulated by the combined effects of various PTMs.
We were therefore questioning whether NLRP3 was regulated by additional posttranslational modifications. Using mass spectrometry (MS) we identified three phosphorylated serines in NLRP3, all of which were conserved across species. Among these, one phosphorylated serine was located in the PYD of NLRP3 within a charge–charge interaction interface, suggesting that phosphorylation of NLRP3 PYD likely disturbed the assembly and interaction of PYDs. Indeed, phosphomimetic residues in this position inhibited NLRP3-ASC, as well as NLRP3 PYD–PYD interactions. Protein phosphatase PP2A was involved in NLRP3 dephosphorylation, and knockdown of PP2A inhibited NLRP3 inflammasome activation. These data suggest that accidental NLRP3 activation is prevented via phosphorylation of the PYD and that dephosphorylation involving PP2A facilitates NLRP3 activation.
Results
NLRP3 is phosphorylated at multiple serines
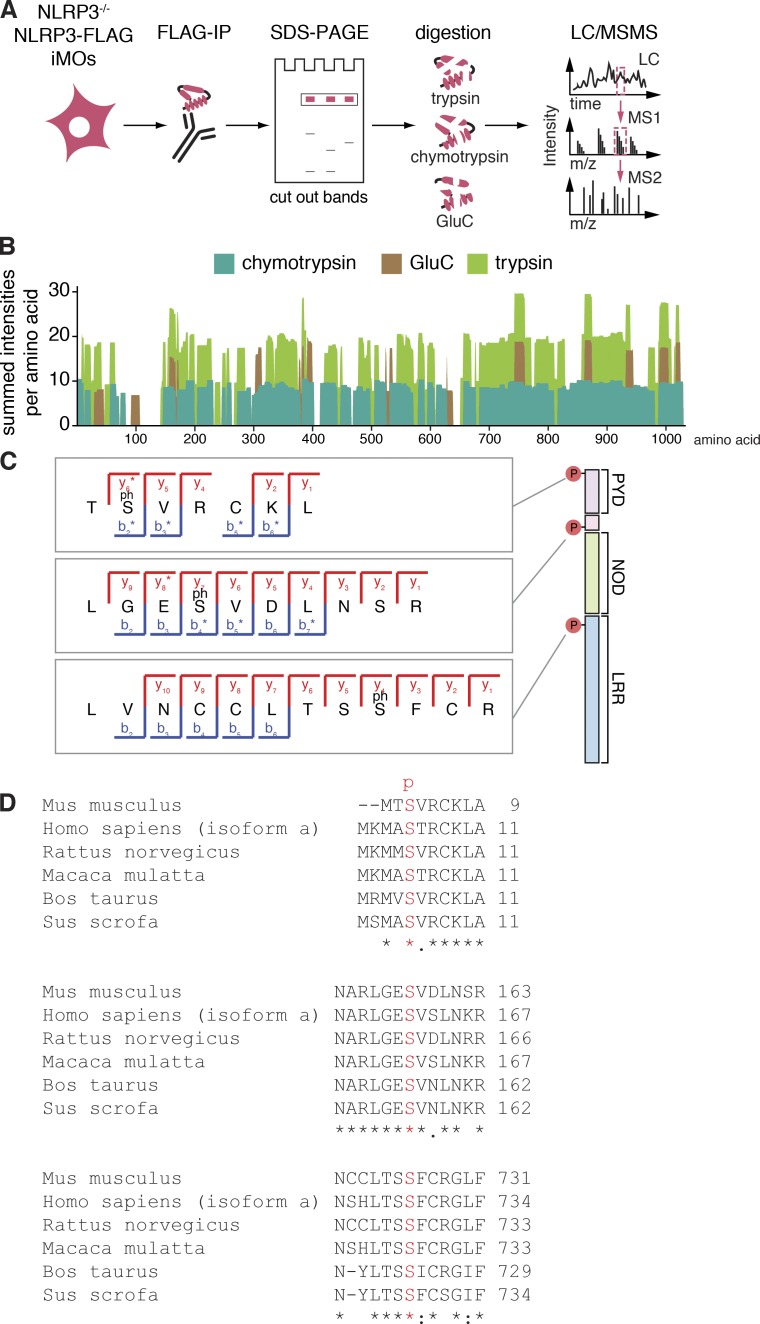
To elucidate whether phosphorylation of NLRP3 can regulate its activity, we used quantitative proteomics (Fig. 1 A). For this, we used a murine cell line retrovirally expressing NLRP3-FLAG, which expresses NLRP3-FLAG to levels of endogenous NLRP3 when primed with LPS (Juliana et al., 2012). NLRP3-FLAG purified by immunoprecipitation and SDS-PAGE was digested by trypsin, chymotrypsin, or Glu-C to achieve diverse proteolytic peptides and maximal NLRP3 sequence coverage. Reverse phase ultra-high performance liquid chromatography, followed by identification via high-resolution MS achieved 92% NLRP3 sequence coverage (Fig. 1 B) and identified three phosphorylation sites. One phosphorylation site was located in the PYD (amino acid S3 in mouse, which corresponds to S5 in human), the second site positioned between the PYD and the NOD (S157 in mouse, corresponding to S161 in human), and a third site was identified in the LRR region (S725 in mouse, S728 in human) (Fig. 1 C and Fig. S1, A to C). All identified phosphosites were conserved across species (Fig. 1 D).
Figure 1.
NLRP3 is phosphorylated. (A) Mass spectrometry workflow for the analysis of NLRP3. (B) Summed peptide intensities per amino acid display quantitative evidences for overlapping peptides. 92% of the NLRP3 sequence is covered by identified and quantified peptides using a combination of trypsin, chymotrypsin, and Glu-C. Representative of at least two different experiments. (C) Identification of phosphorylated NLRP3 peptides by mass spectrometry. MS2 fragmentation spectra illustrating the coverage of y-ion and b-ion series. Phosphorylated amino acids are indicated with (ph). Spectra are representative of at least two different experiments. (D) ClustalW alignment of NLRP3 orthologs surrounding the phosphorylated residues (highlighted in red). "*" indicates fully conserved residue: strongly similar and "." weakly similar properties. See also Fig. S1.
Phosphorylation at serine 5 inhibits inflammasome activation
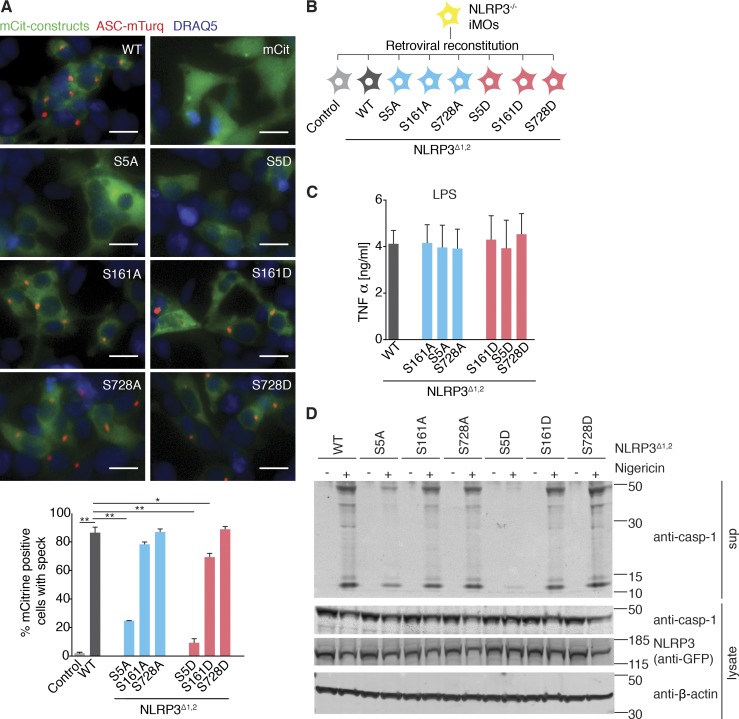
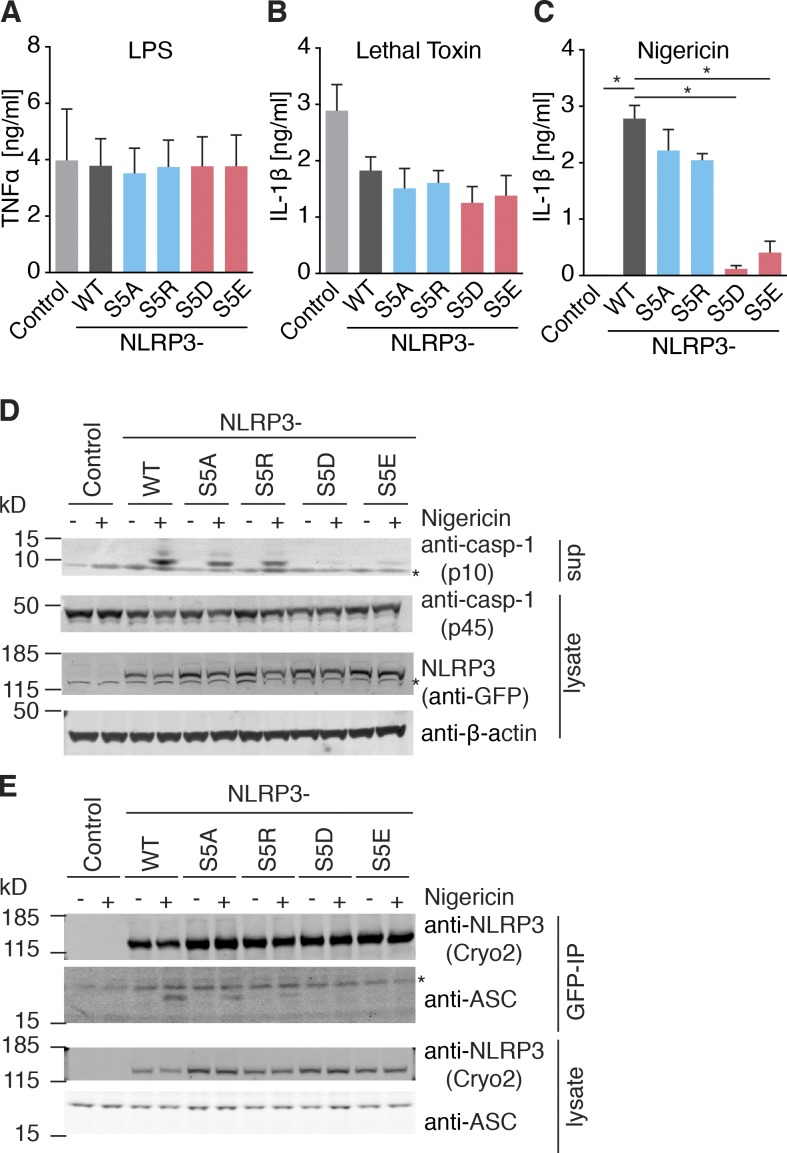
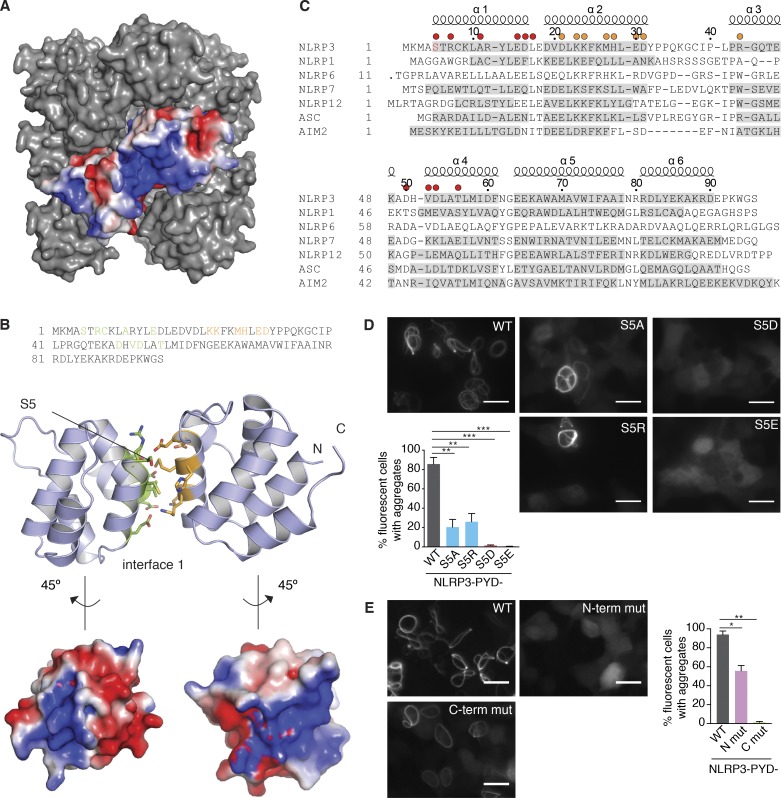
In human NLRP3, two isoforms with alternate start codons are described. The main isoform a is two residues longer than isoform e. Isoform e, however, is used as a reference for the autoinflammatory disease database Infevers (Touitou et al., 2004), and it is referred to here as NLRP3Δ1,2 to obtain consistency with the NCBI Protein RefSeq database. Because the length of the N terminus can influence protein activity (Gibbs et al., 2014), we first tested the function of the phosphosites in the context of the short isoform (NLRP3Δ1,2). We generated mammalian expression constructs of human NLRP3Δ1,2 mutated at the three identified phosphorylatable serines individually to nonphosphorylatable alanines or phospho-mimetic glutamates. Overexpression of NLRP3 is sufficient to cause ASC speck formation in HEK cells stably expressing ASC (Yu et al., 2006), and indeed, we observed ASC speck formation when we transfected the WT form of NLRP3Δ1,2 into HEK-FlpIn cells stably expressing ASC-mTurquoise (Fig. 2 A). However, ASC speck formation was completely abolished when S5 was mutated to the phosphomimetic aspartate (S5D) and strongly reduced when changed to alanine (S5A). Mutation of either S161 or S728 resulted in a comparable activity to the WT form of NLRP3Δ1,2. To investigate the effect of the phosphorylation sites in the more physiological setting of a macrophage and in a stimulation-dependent fashion, we retrovirally reconstituted immortalized NLRP3 deficient murine macrophages with fluorescent WT or mutant NLRP3Δ1,2 and FACS sorted cells to achieve comparable NLRP3 expression levels (Fig. 2 B). TNF production in response to LPS was not influenced by the expression of the different NLRP3Δ1,2 forms (Fig. 2 C). In contrast, NLRP3 inflammasome activation by nigericin was reduced in cells expressing NLRP3Δ1,2 in which mutations were introduced at position S5, but not in cells expressing NLRP3Δ1,2, in which mutations of the serines S161 or S728 were introduced (Fig. 2 D). We found that mutating the serine at position 5 to alanine rendered NLRP3 partially responsive to nigericin, whereas a mutation to the phosphomimetic glutamate completely abolished its function. We next tested the corresponding mutations at position 5 (S5A, S5D) in context of the long isoform (isoform a), which is generally used to study human NLPR3 activation. Since we reasoned that negative charges in this position were involved in regulation of NLRP3 activity, we introduced two additional mutations: The positively charged arginine, which cannot be phosphorylated (S5R), and the phosphomimetic glutamic acid (S5E). TNF responses toward LPS and IL-1β responses toward the NLRP1B activator lethal toxin were similar in reconstituted NLRP3 deficient macrophages expressing the different NLRP3 mutant proteins (Fig. 3, A and B). In contrast, IL-1β release (Fig. 3 C) or caspase-1 cleavage (Fig. 3 D) in response to nigericin treatment were abrogated in reconstituted NLRP3 deficient macrophages expressing NLRP3 phosphomimetic mutations (S5E, S5D), but not in cells expressing nonphosphorylatable mutations (S5A, S5R). The activation of NLRP3 is thought to form the molecular seed that nucleates the formation of helical fibrils of ASC via PYD interactions (Lu et al., 2014). We hypothesized that the introduction of negative charges by phosphorylation at position S5 would disturb the interaction between NLRP3 and ASC. Hence, we coimmunoprecipitated NLRP3 and ASC and found that NLRP3 WT and NLRP3 containing S5A or S5R mutations interacted with ASC in activated cells, whereas S5 phosphomimetic mutants did not (Fig. 3 E). Together, these data suggest that NLRP3 activation can be regulated by the charge of a single amino acid in the PYD.
Figure 2.
Phosphorylated residues between PYD/NOD and in LRR region show little influence on NLRP3 activation. (A) Images and quantification of HEK293 FlpIn cells expressing ASC-mTurquoise transfected with NLRP3-mCitrine WT, the indicated NLRP3 mutants or mCitrine-HA (as a control). Bar, 20 µm. Images are representative of four independent experiments. n = 4 ± SEM. *, P < 0.05; **, P < 0.01 (ANOVA with Holm-Sidak). (B) NLRP3-deficient immortalized macrophages (iMOs) were reconstituted with point-mutated NLRP3-mCitrine. (C) Quantification of TNF by ELISA in NLRP3-deficient iMOs reconstituted with NLRP3-mCitrine WT or indicated mutations after 3-h LPS priming. n = 3 ± SEM. (D) Immunoblot of NLRP3 deficient iMOs reconstituted with NLRP3-mCitrine WT or indicated mutations after stimulation with nigericin (1 h) or left untreated. Immunoblots are representative of two independent experiments.
Figure 3.
Phosphorylation of S5 inhibits NLRP3 activation. (A–C) Quantification of IL-1β and TNF by ELISA in NLRP3-deficient iMOs reconstituted with NLRP3-mCitrine WT or indicated mutations after 3 h LPS priming (for [A] TNF) and stimulated with (B) lethal toxin (6 h) or (C) nigericin (1 h). (A–C) n = 3 ± SEM; (C) *, P < 0.05 (ANOVA with Holm-Sidak). (D) Immunoblot of NLRP3-deficient iMOs reconstituted with NLRP3-mCitrine WT or indicated mutations after stimulation with nigericin (1 h) or left untreated. Immunoblots are representative of three independent experiments. (E) Co-immunoprecipitation (IP) of ASC with NLRP3-mCitrine. NLRP3-deficient iMOs reconstituted with NLRP3-mCitrine WT or indicated mutations were primed with LPS for 2 h and left untreated (none) or stimulated with nigericin (1 h). NLRP3 was immunoprecipitated using anti-GFP antibodies. Asterisk denotes unspecific bands. Immunoblots are representative of two independent experiments.
Phosphorylation interferes with charge–charge interactions in the NLRP3 PYD
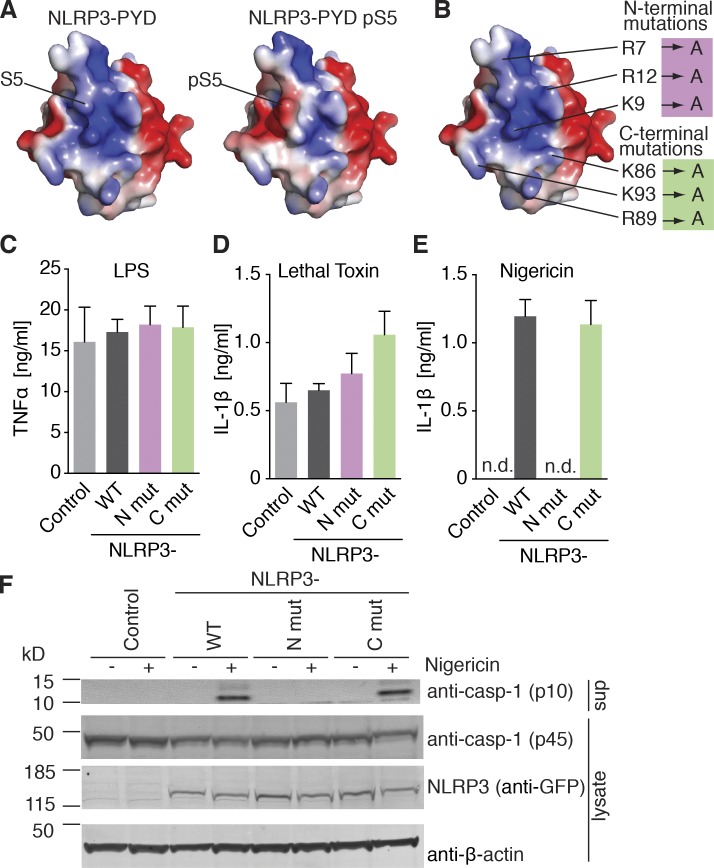
Charge–charge interactions play an important role in death-fold domain interactions as has been shown for the interaction of ASC PYDs (Moriya et al., 2005; Lu et al., 2014). To gain insights into the mechanisms by which the charge switch at position 5 in NLRP3 PYD can regulate inflammasome formation, we used the existing crystal structure of NLRP3 PYD (Bae and Park, 2011) and displayed the electrostatic surface representation of NLRP3 PYD with S5 in the nonphosphorylated state. According to this analysis, S5 appears to be accessible for modification and is located in a polybasic patch formed by three positively charged residues each in helices 1 and 6 (Fig. 4 A, left). Modeling of electrostatic surface charges using phosphorylated S5 showed that the two negative charges introduced by phosphoserine altered the surface charge pattern to near neutralization of the net positive surface charge in this patch (Fig. 4 A, right). To investigate whether the identified positively charged residues that build the charged patch around S5 are critical for NLRP3 activation, we mutated the three residues in helix 1 (R7, K9, and R12) to alanines (N-terminal mutant) or the three residues in helix 6 (K86, R89, and K93) to alanines (C-terminal mutant) (Fig. 4 B). Reconstitution of NLRP3-deficient macrophages with the N- or C-terminal NLRP3 charge change mutants did not affect LPS responsiveness (Fig. 4 C) or NLRP1B activation (Fig. 4 D). However, NLRP3-deficient macrophages expressing the N-terminal NLRP3 mutant completely failed to respond to NLRP3 inflammasome activation with IL-1β secretion (Fig. 4 E) or caspase-1 activation (Fig. 4 F), whereas the respective responses of cells expressing the C-terminal NLRP3 mutant were comparable to NLRP3 WT-expressing cells. Together, these data show the critical function of the positively charged patch within the NLRP3 PYD for NLRP3 inflammasome activation.
Figure 4.
Phosphorylation interferes with a polybasic cluster in the NLRP3 PYD. (A) Electrostatic surface representation of NLRP3 PYD. (left) Nonphosphorylated PYD (PDB accession no. 3QF2); (right) surface charge changes with phosphorylated serine 5 modeled. (B) Mutations introduced in the N/C terminal region of NLRP3 PYD. (C–E) Quantification of IL-1β and TNF by ELISA in NLRP3 deficient iMOs reconstituted with NLRP3-mCitrine WT or PYD-N-terminal (R7A, K9A, and R12A) or PYD-C-terminal (K86A, R89A, and K93A) mutations after 3 h LPS priming (for [C] TNF) and stimulated with (D) lethal toxin (6 h) or (E) nigericin (1 h). (C–E), n = 3 ± SEM. (F) Immunoblot of NLRP3-deficient iMOs reconstituted with NLRP3-mCitrine WT or indicated mutations after 2 h LPS priming and left untreated (none) or stimulated with nigericin (1 h). Immunoblots are representative of two independent experiments.
Serine 5 is located in a PYD–PYD interaction interface
To better understand how exactly S5 and the positively charged patch is located to support homotypic interactions between PYDs, we used the structure of NLRP3 PYD (Bae and Park, 2011) to form a NLRP3 PYD/PYD interaction model based on a structure of ASC PYD helical fibrils (Lu et al., 2014) (Fig. 5 A and B). According to this model, S5 is the first residue in helix 1, which participates in forming the most dominant interaction interface 1 between two PYDs (Fig. 5 B). The interface is mainly based on electrostatic interactions, possibly explaining why introducing two negative charges (via phosphorylation) or a negatively charged residue (such as glutamate or aspartate) would have a disruptive effect. Based on this model, it appears likely that phosphomimetic mutations of S5 disrupt PYD–PYD interactions. Notably, a negative charge at this position in helix 1 is unique among PYDs of other inflammasome sensor molecules, which have neutral or positively charged residues at this position in a structure-based alignment, indicating that phosphorylation of NLRP3 PYD could represent a unique mechanism for the regulation of the NLRP3 inflammasome (Fig. 5 C). To determine whether negative charges at position 5 would also interfere with NLRP3 PYD homotypic interactions, we overexpressed the NLRP3 PYD WT or the mutant forms to high levels in HEK cells to force PYD interaction in the absence of the NOD, which regularly drives NLRP3 self-assembly. Indeed, prolonged expression of NLRP3 PYD-mCitrine resulted in filament formation, similar to what had been demonstrated for ASC PYD (Masumoto et al., 2001; Hornung et al., 2009). In contrast, mutations of S5 to phosphomimetic residues ablated the NLRP3 PYD self-assembly into filaments (Fig. 5 D) under identical experimental conditions. Mutation to alanine or arginine reduced, but did not abrogate, filament formation, indicating that a perfect fit at position 5 of the PYD seems crucial for the interaction between NLRP3 PYDs. We also tested whether the positively charged patch at the N-terminus of NLRP3 PYD was required for NLRP3 PYD interaction. Indeed, mutation of the three positively charged residues to alanines in helix 1 (N-terminal mutant) surrounding S5 led to a complete abrogation of filament formation, whereas mutation of the positively charged region in helix 6 of NLRP3 PYD (C-terminal mutant) did not (Fig. 5 E). Together, these data suggest that phosphorylation of serine 5 neutralizes a critical positively charged surface patch within the NLRP3 PYD, which regulates both the interaction of NLRP3 PYD with itself and with ASC to initiate inflammasome signaling.
Figure 5.
S5 is located in a PYD-PYD interaction interface. (A) Model of a NLRP3 PYD filament based on the crystal structure of NLRP3 PYD (PDB 3QF2) and the EM structure of ASC filaments (PDB 3J63). (B, top) Sequence of NLRP3 PYD with residues participating in interface 1a highlighted in green, the residues of interface 1b colored in orange. Interacting residues of the interfaces 1a and 1b are derived from a sequence alignment with ASC PYD in fibrillary state (PDB 3J63). (middle) Ribbon representation of two interacting NLRP3 PYD. Residues are color coded as above. (bottom) Electrostatic surface representation of NLRP3 PYD. (C) Structure-based sequence alignment of inflammasome-forming human PYDs. Helices are indicated above the alignment and shaded in gray in the individual sequences. The residues potentially participating in interface 1a are marked by a red dot above the alignment, the residues of interface 1b are marked by an orange dot. (D) Images and quantification of HEK293T cells transfected with constructs expressing NLRP3-PYD(1–99)-mCitrine WT or the indicated NLRP3 mutants. Bar, 20 µm. Images are representative of five independent experiments. n = 5 ± SEM; **, P < 0.01; ***, P < 0.001 (ANOVA with Holm-Sidak). (E) Images and quantification of HEK293T cells transfected with constructs expressing NLRP3-PYD(1–99)-mCitrine WT or PYD-N-terminal (R7A, K9A, and R12A) or PYD-C-terminal (K86A, R89A, and K93A) mutations. Bar, 20 µm. Images are representative of three independent experiments. n = 3 ± SEM; *, P < 0.05; **, P < 0.01 (ANOVA with Holm-Sidak).
Phosphatase PP2A is involved in dephosphorylation and activation of NLRP3
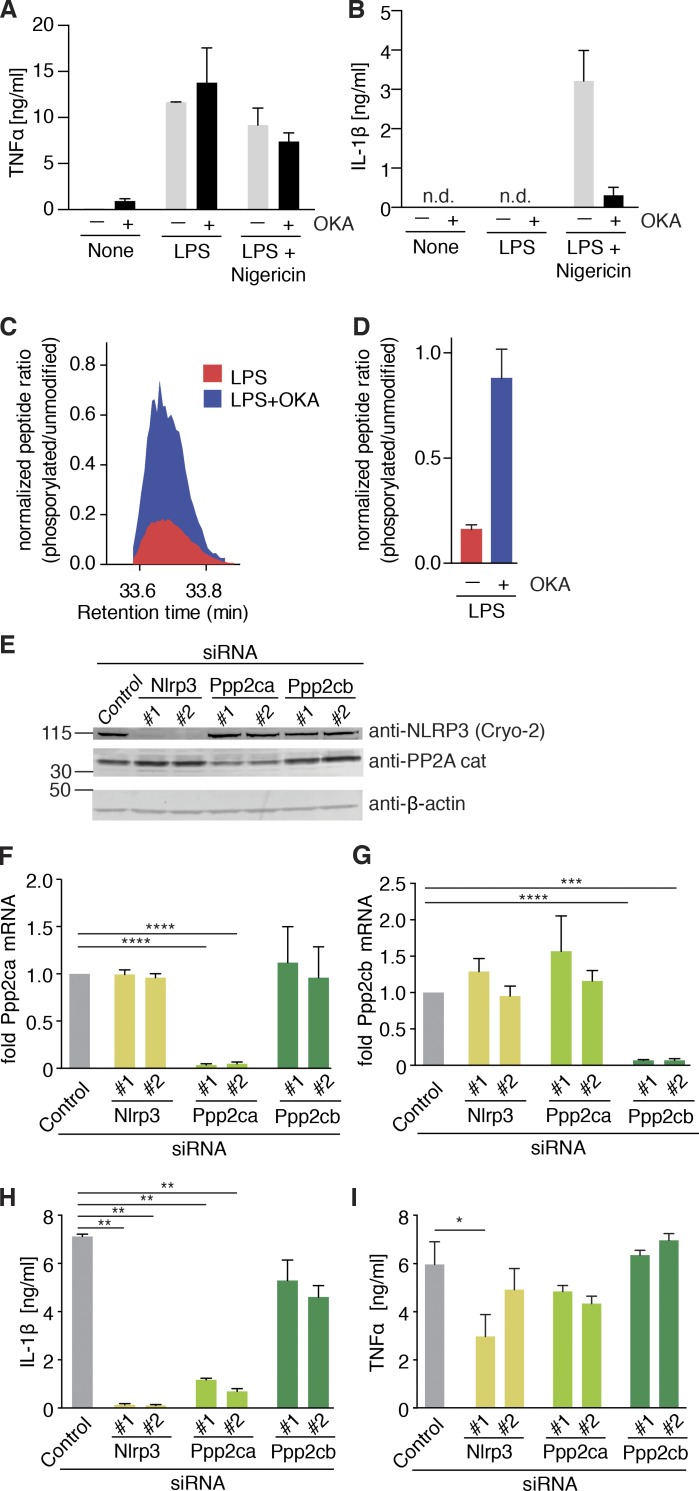
Inhibition of protein phosphatase 2A (PP2A) by okadaic acid (OKA) has been described to inhibit NLRP3 activation (Luheshi et al., 2012). Thus, we investigated whether PP2A played a role in dephosphorylating S5 of NLRP3. OKA pretreatment did not change LPS-induced TNF secretion (Fig. 6 A), but inhibited IL-1β secretion in response to nigericin stimulation (Fig. 6 B). We next used MS to determine whether PP2A inhibition would influence the phosphorylation status of serine 5. Indeed, OKA pretreatment resulted in increased S5 phosphorylation in LPS primed macrophages (Fig. 6, C and D), suggesting that PP2A is involved in NLRP3 dephosphorylation. Next, we tested whether a knockdown of PP2A enzyme levels would inhibit NLRP3 activation. There are two isoforms of the catalytic subunit, Ppp2ca and Ppp2cb. Because PPP2CA and PPP2CB only differ in three amino acids, isoform-specific antibodies are not available and only the knockdown of the dominantly expressed isoform PPP2CA could be visualized by immunoblot (Fig. 6 E). However, both isoforms were knocked down strongly and specifically on mRNA level (Fig. 6 F and G). Only Ppp2ca knockdown strongly inhibited NLRP3 inflammasome activation, whereas Ppp2cb knockdown had no influence (Fig. 6 H). Both Ppp2ca and Ppp2cb did not have an influence on TNF secretion in response to LPS treatment (Fig. 6 I). These data demonstrate a role for PP2A (specifically catalytic subunit isoform α) in the dephosphorylation and activation of NLRP3.
Figure 6.
NLRP3 PYD gets dephosphorylated with involvement of protein phosphatase 2A. (A and B) Quantification of (A) TNF and (B) IL-1β by ELISA in NLRP3-deficient iMOs reconstituted with NLRP3-FLAG after 15 min pretreatment with okadaic acid (OKA) or left untreated (none), followed by 2 h LPS priming (for TNF) and stimulated with nigericin (1 h). (A and B) n = 2 ± SEM. (C) Ion intensities for the peptide containing the phosphorylated S3 (acTphSVRCKL; m/z = 493.23046; z = 2), normalized to the nonphosphorylated counterpart (acTSVRCKL; m/z = 453.24729; z = 2) are plotted at the indicated retention times for LPS-treated and LPS+OKA-treated samples. Representative of two independent experiments. (D) Ratio of the apex of phosphorylated to nonphosphorylated peptide intensities (mean ± SEM of replicates, representative of two independent experiments). (E) Immunoblot of NLRP3-deficient iMOs reconstituted with NLRP3-FLAG WT and transfected with the indicated siRNAs for 40 h. Immunoblots are representative of three independent experiments. (F and G) Same as E, but showing mRNA levels (fold of control, normalized to Hprt expression) for (F) Ppp2ca and (G) Ppp2cb. (F and G) n = 4 ± SEM. ***, P < 0.001; ****, P < 0.0001 (ANOVA with Holm-Sidak). (H and I) Quantification of IL-1β (H) and TNF (I) by ELISA in NLRP3-deficient iMOs reconstituted with NLRP3-FLAG WT, transfected with the indicated siRNAs for 40 h, and then primed for 2 h with LPS (for TNF) and stimulated with nigericin (1 h). (H and I) n = 3 ± SEM. *, P < 0.05; **, P < 0.01 (ANOVA with Holm-Sidak).
Discussion
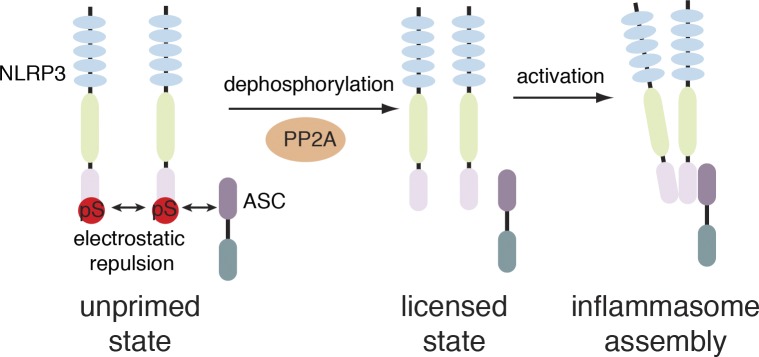
In summary, we identified a critical licensing mechanism of NLRP3 by dephosphorylation of a single amino acid in the PYD. Our data suggest that in an unstimulated state, NLRP3 PYD phosphorylation hinders interaction of NLRP3 PYDs and NLRP3 and ASC PYDs via electrostatic repulsion, thereby preventing accidental assembly. Upon dephosphorylation involving the activity of PP2A, the electrostatic repulsion is removed allowing for downstream signaling to occur. This step functions as a licensing mechanism, but does not directly lead to inflammasome formation (Fig. 7). Instead, additional signals may be required for either the recruitment of ASC or the assembly of ASC helical fibrils.
Figure 7.
Model for licensing through dephosphorylation. In unprimed cells, NLRP3 is phosphorylated at S5, resulting in electrostatic repulsion of pyrin domains. Dephosphorylation involving PP2A licenses NLRP3 for activation and is required before inflammasome assembly can occur.
Together with other findings describing that NLRP3 activation is regulated by ubiquitination and nitrosylation, our data suggest that NLRP3 integrates various signals from multiple upstream pathways. NLRP3 activation appears to be restricted to a defined cellular activation state that allows for NLRP3 seed formation, recruitment of ASC, and initiation of downstream signaling. A recent study found that NLRP3 was phosphorylated at Y861 in the LRR after 12 h of LPS priming and was dephosphorylated during the activation step (Spalinger et al., 2016). In our MS approach, we focused on shorter priming times (2 h) and did not detect this phosphorylation event, despite sequence coverage at this position in the protein. Furthermore, in situations in which PKA is triggered, NLRP3 can be phosphorylated at position 295, leading to its inhibition (Guo et al., 2016; Mortimer et al., 2016). We have profiled NLRP3 phosphorylation without triggers causing PKA activation. Together, these data indicate that NLRP3 regulation through phosphorylation is dynamic and likely depends on the timing of priming and stimulants and the cell type. Similarly to our findings for NLRP3, licensing, but not activation, through dephosphorylation has been described for the nucleic-acid sensing PRRs RIG-I and MDA5 (Wies et al., 2013). Interestingly, RIG-I and MDA5 are phosphorylated by members of the Protein Kinase C family (Maharaj et al., 2012), raising the question whether these kinases may also play a role in NLRP3 regulation. Additionally, the inflammasome sensor molecule pyrin is kept inactive by a phosphorylated serine, and loss of the phosphorylation results in activation of the pyrin inflammasome (Masters et al., 2016). Together with our results, these studies show that the balance between kinase and phosphatase activity in immune cells acts to license the activation of the most upstream signaling events. The introduction of charge changes in death domains by phosphorylation appears to have evolved as an effective mechanism to prevent assembly of large molecular signaling complexes. A better understanding of the different signaling networks that regulate NLRP3 phosphorylation could enable novel strategies for therapeutic intervention in diseases associated with overly active NLRP3 such as atherosclerosis, gout, or type II diabetes.
Materials and methods
Reagents
Ultrapure LPS was obtained from InvivoGen, nigericin was obtained from Invitrogen, okadaic acid was obtained from Calbiochem, lethal factor and protective antigen were obtained from List Biological Laboratories, and Gene Juice was purchased from Merck. DRAQ5 was purchased from eBioscience. The ELISA kits for mouse IL-1β and mouse TNF were obtained from R&D Systems and were used according to the manufacturer’s instructions.
Plasmids, cell lines, and stimulation conditions
Plasmids for NLRP3-mCitrine, NLRP3-tagBFP-FLAG, NLRP3-PYD(1–99), mutants, and control constructs were generated with standard cloning techniques.
HEK293-FlpIn TREx cell line (Invitrogen) with a single insertion of ASC-mTurquoise was generated according to manufacturer’s instructions. NLRP3-deficient (Nlrp3−/−) murine immortalized macrophages (iMOs) and NLRP3 FLAG-overexpressing NLRP3-deficient iMOs have been previously described (Hornung et al., 2008; Juliana et al., 2010). NLRP3-deficient IMOs were retrovirally transduced with constructs for the indicated NLRP3 mutants fused C-terminally to an mCitrine-tag. After retroviral transduction, cells were flow cytometrically sorted to similar levels of mCitrine expression. Cell lines were checked for mycoplasma contamination via PCR.
For stimulation, immortalized macrophages were primed for 2–3 h with 200 ng/ml LPS. For experiments that did not include analysis of IL-1β, the priming step was omitted. Inflammasome stimulation was performed with 10 µM nigericin (60–90 min) or 1 µg/ml lethal toxin (1 µg/ml lethal factor plus 1 µg/ml protective antigen) (6 h). PP2A inhibition was performed 15 min before LPS priming with 1 µM okadaic acid.
RNA interference
SilencerSelect siRNAs (Thermo Fisher Scientific) were used for knockdowns in NLRP3 FLAG-overexpressing NLRP3-deficient iMOs. The following siRNAs were used: Ctrl #1, 4390846; Ctrl #2, s229087; Nlrp3 #1, s103710; Nlrp3 #2, s103711; Ppp2ca #1, s72066; Ppp2ca #2, s72067; Ppp2cb #1, s72069; Ppp2cb #2, s72070. Reverse transfection in 6-well format was performed using 15 µl Lipofectamine RNAiMAX (Thermo Fisher Scientific), 2.4 × 105 cells and siRNAs at 50 nM final concentration. 40 h after transfection, the cells were harvested, counted, and either replated for stimulation or used for qPCR/immunoblots.
Mass spectrometry analysis
NLRP3-FLAG-overexpressing NLRP3 deficient iMOs were left untreated, stimulated with 200 ng/ml LPS for 2 h, or treated with 1 µM okadaic acid for 15 min before stimulation with 200 ng/ml LPS for 2 h (all in serum-free DMEM). After stimulation, cells were scraped into the medium, pelleted at 2,000 g for 10 min, and lysed in FLAG-lysis buffer (50 mM Tris-HCl, pH 7.4, 150 mM NaCl, 1 mM EDTA, 1% Triton X-100, supplemented with cOmplete protease and PhosSTOP [Roche] inhibitor, 100 µM PMSF, 10 mM iodoacetamide, and 250 U/ml benzonase [Sigma-Aldrich]) for 30 min on ice. Nuclei were removed by centrifugation at 355 g for 10 min. Equal amounts of protein were used for immunoprecipitation using anti-FLAG M2 affinity gel (Sigma-Aldrich) for 1 h at 4°C. The affinity gel was washed three times with FLAG buffer without inhibitors, once with high salt FLAG buffer (50 mM Tris-HCl, pH 7.4, 1 M NaCl,1 mM EDTA, 1% Triton X-100) and once with FLAG buffer without inhibitors. Samples were eluted using 2× SDS sample buffer (40 mM Tris HCl, pH 8.0, 15.7% glycerol, 4% SDS, and 0.017% bromphenol blue), reduced with 10 mM dithiothreitol, followed by alkylation with 55 mM iodoacetamide. The samples were run on 8% Tris-glycine SDS-PAGE gels, fixed, and stained with Coomassie (0.3% Coomassie Brilliant Blue, 45% methanol, and 10% acetic acid in water). After destaining (10% acetic acid, 15% isopropanol in water), gel slices containing NLRP3 were excised and reduced with 10 mM dithiothreitol, followed by alkylation with 55 mM iodoacetamide. We digested the samples with either trypsin, chymotrypsin or GluC and desalted them on C18 material. Peptides were separated with an EASY-nLC (Thermo Fisher Scientific) with in-house packed columns (75 µm inner diam, 20 cm length, 1.9 µm C18 particles from Dr. Maisch GmbH) at 250 nl/min and a column temperature of 45°C using a gradient from 5 to 30% acetonitrile in water containing 0.5% formic acid).
A Quadrupole Orbitrap mass spectrometer (Scheltema et al., 2014; Q Exactive plus and Q Exactive HF; Thermo Fisher Scientific) was coupled to the HPLC system via a nanoelectrospray ion source (Thermo Fisher Scientific). We used a data-dependent acquisition method with a survey scan range of 300 to 1,650 m/z, at a resolution of 60,000 at m/z and the selection of up to five most abundant features with a charge state ≥2 for fragmentation at a normalized collision energy of 27 and a resolution of 15,000 at m/z 200. To limit repeated sequencing, dynamic exclusion of sequenced peptides was set to 20 s. Thresholds for ion injection times and ion target values were set to 20 ms and 3E6 for the survey scans and 120 ms and 1E5 for the MS/MS scans. Data were acquired using the Xcalibur software (Thermo Fisher Scientific). MS raw files were processed with the MaxQuant software (v1.3.0.3) (Cox and Mann, 2008). We used the Andromeda (Cox et al., 2011) to search MS2 spectra against the mouse UniProt database and 247 common potentially contaminating proteins (Cox et al., 2011).
We set enzyme specificity accordingly for trypsin (C-terminal to lysine and arginine excluding proline), chymotrypsin (C-terminal to tryptophan, tyrosine, phenylalanine, leucine, methionine, and histidine with the following exceptions: tryptophan excluding methionine or proline; methionine excluding tyrosine; histidine excluding aspartate, methionine, or tryptophan), and GluC (C-terminal to glutamic acid), and allowed a maximum of two missed cleavages. Phosphorylation of serine, threonine and tyrosine, as well as N-terminal acetylation and methionine oxidation were set as variable modifications, carbamidomethylcystein as fixed modification. A false discovery rate (FDR) cutoff of 5% was applied at the peptide level. Identified and quantified peptides were mapped to NLRP3 and visualized with R (R Development Core Team, 2011). For the quantification of phosphorylated NLRP3 S3, (ac)TphSVRCKL (m/z = 493.23046; z = 2), and (ac)TSVRCKL (m/z = 453.24729; z = 2) were extracted from raw files with 6 ppm tolerance and a minimal signal-to-noise ratio of 5. Peptide intensities of (ac)TpSVRCKL were normalized by the apex of (ac)TSVRCKL and retention times were apex aligned for visualization.
Structural modeling and sequence alignments
Protein sequence alignments for Fig. 1 were done using ClustalW2 (EMBL-EBI web services) (McWilliam et al., 2013). NLRP3 protein sequences are reported in the NCBI Protein RefSeq database with accession nos. NP_001073289.1 (Homo sapiens), NP_665826.1 (Mus musculus), NP_001095689.1 (Bos taurus), NP_001178571.1 (Rattus norvegicus), NP_001243699.1 (Sus scrofa), and NP_001107823.1 (Macaca mulatta). All other protein sequence alignments were performed with the Align web server or with MultAlign (Corpet, 1988) for the alignment of multiple sequences. Human NOD family protein sequences are reported in the UniProt database (UniProt Consortium, 2015) with accession nos. Q96P20 (NLRP3), Q9C000 (NLRP1), P59044 (NLRP6), Q8WX94 (NLRP7), P59046 (NLRP12), Q9ULZ3 (ASC), and O14862 (AIM2). Secondary structure elements were derived from protein structures with Protein Data Bank accession codes 3QF2 (NLRP3), 1PN5 (NLRP1), 2KM6 (NLRP7), 2L6A (NLRP12), 2KN6 (ASC), and 4O7Q (AIM2).
The structure building of a phosphate group to serine 5 of human NLRP3 was made with the graphical program COOT (Emsley and Cowtan, 2004) using the crystal structure 3QF2 (Bae and Park, 2011) (chain A) of the NLRP3 PYD as template. Modeling of the nonmodified NLRP3 PYD into a fibrillary structure was achieved by using the reconstruction of the ASC filament structure 3J63 (Lu et al., 2014) for model building. Superimposition of the NLRP3 PYD 3QF2 (chain A) with the ASC PYD 1UCP exhibited a root-mean-square deviation value of 1.78 Å. Overall, 15 PYD protein chains were mutually exchanged from ASC to NLRP3 to gain a full model of the fibrillary structure. Residues contributing to the molecular interfaces of the highly symmetric PYD fibril model were determined from the structure alignment with ASC. Molecular diagrams of protein structures were drawn with PyMOL v1.7.0.3 (Schrodinger Scientific Inc.) using the plugin APBS (Baker et al., 2001) for electrostatic surface calculations.
ASC speck analysis after NLRP3 overexpression
HEK293-FlpIn TREx ASC-mTurquoise cells were transfected with 200 ng/96 well of the indicated constructs with Lipofectamine 2000 according to manufacturer’s instructions. 24 h after transfection, cells were fixed with 4% formaldehyde and nucleic acids were stained with 2.5 µM DRAQ5. Cells were imaged using a Observer.Z1 epifluorescence microscope, 20× objective (dry, PlanApochromat, NA 0.8; ZEISS), Axiocam 506 mono, and ZEN Blue software (ZEISS).
NLRP3-PYD filament formation
HEK293T cells were transfected with 200 ng/96 well of the indicated constructs with Lipofectamine 2000 according to manufacturer’s instructions. 48 h after transfection, cells were fixed with 4% formaldehyde. Cells were imaged using an Observer.Z1 epifluorescence microscope, 20× objective (dry, PlanApochromat, NA 0.8; ZEISS), Axiocam 506 mono, and ZEN Blue software (ZEISS).
Image analysis
Image analysis of all HEK transfection experiments was done manually using Fiji software (Schindelin et al., 2012) and the Cell Counter plugin. For better visibility, brightness and contrast were adjusted. A minimum of 10 images was analyzed for each condition in each experiment.
Sample preparation for caspase-1 immunoblots
Cells were lysed with NP-40 buffer (20 mM Tris-HCl, pH 7.4, 150 mM NaCl, 1 mM EDTA, 1% Nonidet P-40, 10% glycerol, cOmplete protease, and PhosSTOP [Roche] inhibitor), nuclei were removed by centrifugation (10 min, 355 g) and protein concentration was normalized after measurement with a bichinonic acid assay (BCA; Thermo Fisher Scientific). Protein from supernatants was precipitated by adding an equal volume of methanol and 0.25 volumes of chloroform and centrifugation for 3 min at 20,000 g. The upper phase was discarded, the same volume of methanol of the previous step was added and the sample was centrifuged for 3 min at 20,000 g. The pellet was dried and taken up in LDS-sample buffer containing reducing agent (Invitrogen).
Sample preparation for NLRP3 expression levels
WT immortalized macrophages were either left untreated or primed with 200 ng/ml LPS for either 3 h or 6 h. NLRP3-tagged cells were not treated (their expression is not driven by the endogenous NLRP3 promoter, but instead by the retroviral vector long terminal repeat). The cells were lysed with NP-40 lysis buffer (20 mM Tris-HCl, pH 7.4, 150 mM NaCl, 1 mM EDTA, 1% Nonidet P-40, 10% glycerol, cOmplete protease and PhosSTOP (Roche) inhibitor), nuclei were removed by centrifugation (10 min, 355 g) and protein concentration was normalized after measurement with a bichinonic acid assay (BCA; Thermo Fisher Scientific). Immunoblots were performed as described below. The band intensities for NLRP3 and actin were measured using ImageStudio 3 software (LI-COR Biosciences), the ratio NLRP3/actin was calculated and graphed using GraphPad Prism (version 6.0).
Immunoprecipitation assay for interaction with endogenous ASC
Cells were left untreated or stimulated with 10 µM nigericin for 45 min in serum-free medium (SFM). After stimulation, the cells were scraped in SFM and centrifuged at 2,000xg for 10 min at 4°C. The cell pellet was resuspended in hypotonic lysis buffer (20 mM Hepes, pH 7.4, 10 mM KCl, 1 mM EDTA, cOmplete protease and PhosSTOP (Roche) inhibitor) and lysed by syringing. The cell lysate was centrifuged at 350 g for 5 min at 4°C, and the supernatant was diluted 1:1 with 2× immunoprecipitation buffer (100 mM Tris, pH 7.8, 300 mM NaCl, 0.2% Nonidet P-40, and 10 mM EDTA) containing 25 U/ml benzonase (Sigma-Aldrich). The resulting soluble cell lysate was precleared using protein G Dynabeads (Invitrogen). The precleared cell lysate was incubated with anti-GFP antibody (A11122; Invitrogen) at 4°C overnight. Protein G Dynabeads were added to each sample, incubated for 2 h, and then washed three times with immunoprecipitation buffer (50 mM Tris, pH 7.8, 150 mM NaCl, 0.1% Nonidet P-40, and 5 mM EDTA). Bead-bound proteins were eluted with SDS-PAGE sample buffer.
Immunoblots
Proteins were separated by 4–12% SDS-PAGE in precast gels (Novex; Invitrogen) with MOPS buffer (Novex; Invitrogen). Proteins were transferred onto Immobilon-FL PVDF membranes (Millipore) and nonspecific binding was blocked with 3% BSA in Tris-buffered saline for 1 h, followed by overnight incubation with specific primary antibodies in 3% BSA in Tris-buffered saline with 0.1% Tween-20.
Primary antibodies were used as follows: NLRP3 (1:5,000 dilution; Cryo-2) from Adipogen, caspase-1 p45, and p10 (1:200 dilution; sc-514) from Santa Cruz Biotechnology (1:1,000 dilution; A11122) from Invitrogen, FLAG (1:1,000 dilution, clone M2) from Sigma-Aldrich, actin (mouse or rabbit, both 1:1,000 dilution) from LI-COR Biosciences, ASC (1:1,000 dilution; AL177) from Adipogen, PP2A-catalytic subunit (1:40,000 dilution, clone 46) from BD. Membranes were then washed and incubated with the appropriate secondary antibodies (coupled to IRDye 800CW or IRDye 680RD; 1:25,000 dilution; LI-COR Biosciences), washed, and analyzed with an Odyssey CLx imaging system (LI-COR Biosciences) and ImageStudio 3 Software (LI-COR Biosciences).
Quantitative real-time PCR (qPCR)
qPCR quantifications were performed essentially as previously described (De Nardo et al., 2014) with the following changes: 250–500 ng of RNA were used for the RT-PCR and the qPCR was performed using QuantStudio 6 PCR System (Thermo Fisher Scientific). The primer sequences were as follows: Hprt, forward 5′-TGAAGTACTCATTATAGTCAAGGGCA-3′ and reverse 5′-CTGGTGAAAAGGACCTCTCG-3′; Ppp2ca, forward 5′-TCTTCCTCTCACTGCCTTGGT-3′ and reverse 5′-CAGCAAGTCACACATTGGACCC-3′; Ppp2cb, forward 5′-AAGGCGTTCACCAAGGAGCT-3′ and reverse 5′-ACAGCGGACCTCTTGCACAT-3′.
Statistical analysis
Unless otherwise stated, the significance of differences between groups was evaluated by one-way analysis of variance (ANOVA) with repeated measures with Holm-Sidak’s post-comparison test and Geisser-Greenhouse correction. Statistical analysis was performed with GraphPad Prism (version 6.0). Data were considered significant when P < 0.05 (*), 0.01 (**), 0.001(***), or 0.0001(****). Data are graphed as mean, error bars show SEM unless otherwise stated.
Online supplemental material
Fig. S1 shows the annotated MS2 spectra of the experiment performed in Fig. 1 C to identify phosphorylated residues.
Supplementary Material
Acknowledgments
We thank M. Mann for support and critical comments, and S. Dewitz as well as M. Dodel for technical assistance. We thank A. Dolf and P. Wurst (flow cytometry core facility Bonn) for assistance with fluorescence-activated cell sorting. We thank E. Alnemri (Thomas Jefferson University Philadelphia) for the NLRP3 FLAG-overexpressing NLRP3 deficient murine macrophages. B. Franklin, M. Geyer, and E. Latz are members of the excellence cluster ImmunoSensation.
E. Latz is supported by grants from the German Research Foundation (DFG; SFB645, 704, 670, 1123, TRR57, 83) and the European Research Council (InflammAct).
E. Latz and M. Geyer are co-founders and A. Stutz is an employee of IFM Therapeutics. The authors declare no additional competing financial interests.
Author contributions: A. Stutz, F. Meissner, and E. Latz designed the study, A. Stutz, C.-C. Kolbe, G.L. Horvath, B.S. Franklin, O. van Ray, R. Brinkschulte, and F. Meissner performed experiments. A. Stutz, G.L. Horvath, and F. Meissner analyzed data. R. Stahl designed and generated the expression constructs used in this study. M. Geyer created the structural models and provided critical input. A. Stutz, F. Meissner, and E. Latz wrote the manuscript with input from all authors.
Footnotes
Abbreviations used:
- ASC
- apoptosis-associated speck-like protein containing a CARD
- LRR
- leucine-rich repeat
- iMO
- immortalized macrophage
- MS
- mass spectrometry
- NLRP3
- NACHT, LRR and PYD domains-containing protein 3
- NOD
- nucleotide-binding oligomerization domain
- OKA
- okadaic acid
- PP2A
- protein phosphatase 2A
- PYD
- pyrin domain
References
- Bae J.Y., and Park H.H.. 2011. Crystal structure of NALP3 protein pyrin domain (PYD) and its implications in inflammasome assembly. J. Biol. Chem. 286:39528–39536. 10.1074/jbc.M111.278812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker N.A., Sept D., Joseph S., Holst M.J., and McCammon J.A.. 2001. Electrostatics of nanosystems: application to microtubules and the ribosome. Proc. Natl. Acad. Sci. USA. 98:10037–10041. 10.1073/pnas.181342398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauernfeind F.G., Horvath G., Stutz A., Alnemri E.S., MacDonald K., Speert D., Fernandes-Alnemri T., Wu J., Monks B.G., Fitzgerald K.A., et al. . 2009. Cutting edge: NF-kappaB activating pattern recognition and cytokine receptors license NLRP3 inflammasome activation by regulating NLRP3 expression. J. Immunol. 183:787–791. 10.4049/jimmunol.0901363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corpet F. 1988. Multiple sequence alignment with hierarchical clustering. Nucleic Acids Res. 16:10881–10890. 10.1093/nar/16.22.10881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox J., and Mann M.. 2008. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol. 26:1367–1372. 10.1038/nbt.1511 [DOI] [PubMed] [Google Scholar]
- Cox J., Neuhauser N., Michalski A., Scheltema R.A., Olsen J.V., and Mann M.. 2011. Andromeda: a peptide search engine integrated into the MaxQuant environment. J. Proteome Res. 10:1794–1805. 10.1021/pr101065j [DOI] [PubMed] [Google Scholar]
- De Nardo D., Labzin L.I., Kono H., Seki R., Schmidt S.V., Beyer M., Xu D., Zimmer S., Lahrmann C., Schildberg F.A., et al. . 2014. High-density lipoprotein mediates anti-inflammatory reprogramming of macrophages via the transcriptional regulator ATF3. Nat. Immunol. 15:152–160. 10.1038/ni.2784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P., and Cowtan K.. 2004. Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60:2126–2132. 10.1107/S0907444904019158 [DOI] [PubMed] [Google Scholar]
- Franchi L., Eigenbrod T., and Núñez G.. 2009. Cutting edge: TNF-alpha mediates sensitization to ATP and silica via the NLRP3 inflammasome in the absence of microbial stimulation. J. Immunol. 183:792–796. 10.4049/jimmunol.0900173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franklin B.S., Bossaller L., De Nardo D., Ratter J.M., Stutz A., Engels G., Brenker C., Nordhoff M., Mirandola S.R., Al-Amoudi A., et al. . 2014. The adaptor ASC has extracellular and ‘prionoid’ activities that propagate inflammation. Nat. Immunol. 15:727–737. 10.1038/ni.2913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibbs D.J., Bacardit J., Bachmair A., and Holdsworth M.J.. 2014. The eukaryotic N-end rule pathway: conserved mechanisms and diverse functions. Trends Cell Biol. 24:603–611. 10.1016/j.tcb.2014.05.001 [DOI] [PubMed] [Google Scholar]
- Guo C., Xie S., Chi Z., Zhang J., Liu Y., Zhang L., Zheng M., Zhang X., Xia D., Ke Y., et al. . 2016. Bile Acids Control Inflammation and Metabolic Disorder through Inhibition of NLRP3 Inflammasome. Immunity. 45:802–816. 10.1016/j.immuni.2016.09.008 [DOI] [PubMed] [Google Scholar]
- Hernandez-Cuellar E., Tsuchiya K., Hara H., Fang R., Sakai S., Kawamura I., Akira S., and Mitsuyama M.. 2012. Cutting edge: nitric oxide inhibits the NLRP3 inflammasome. J. Immunol. 189:5113–5117. 10.4049/jimmunol.1202479 [DOI] [PubMed] [Google Scholar]
- Hornung V., Bauernfeind F., Halle A., Samstad E.O., Kono H., Rock K.L., Fitzgerald K.A., and Latz E.. 2008. Silica crystals and aluminum salts activate the NALP3 inflammasome through phagosomal destabilization. Nat. Immunol. 9:847–856. 10.1038/ni.1631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hornung V., Ablasser A., Charrel-Dennis M., Bauernfeind F., Horvath G., Caffrey D.R., Latz E., and Fitzgerald K.A.. 2009. AIM2 recognizes cytosolic dsDNA and forms a caspase-1-activating inflammasome with ASC. Nature. 458:514–518. 10.1038/nature07725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juliana C., Fernandes-Alnemri T., Wu J., Datta P., Solorzano L., Yu J.-W., Meng R., Quong A.A., Latz E., Scott C.P., and Alnemri E.S.. 2010. Anti-inflammatory compounds parthenolide and Bay 11-7082 are direct inhibitors of the inflammasome. J. Biol. Chem. 285:9792–9802. 10.1074/jbc.M109.082305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juliana C., Fernandes-Alnemri T., Kang S., Farias A., Qin F., and Alnemri E.S.. 2012. Non-transcriptional priming and deubiquitination regulate NLRP3 inflammasome activation. J. Biol. Chem. 287:36617–36622. 10.1074/jbc.M112.407130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Latz E., Xiao T.S., and Stutz A.. 2013. Activation and regulation of the inflammasomes. Nat. Rev. Immunol. 13:397–411. 10.1038/nri3452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu A., Magupalli V.G., Ruan J., Yin Q., Atianand M.K., Vos M.R., Schröder G.F., Fitzgerald K.A., Wu H., and Egelman E.H.. 2014. Unified polymerization mechanism for the assembly of ASC-dependent inflammasomes. Cell. 156:1193–1206. 10.1016/j.cell.2014.02.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luheshi N.M., Giles J.A., López-Castejón G., and Brough D.. 2012. Sphingosine regulates the NLRP3-inflammasome and IL-1β release from macrophages. Eur. J. Immunol. 42:716–725. 10.1002/eji.201142079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maharaj N.P., Wies E., Stoll A., and Gack M.U.. 2012. Conventional protein kinase C-α (PKC-α) and PKC-β negatively regulate RIG-I antiviral signal transduction. J. Virol. 86:1358–1371. 10.1128/JVI.06543-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao K., Chen S., Chen M., Ma Y., Wang Y., Huang B., He Z., Zeng Y., Hu Y., Sun S., et al. . 2013. Nitric oxide suppresses NLRP3 inflammasome activation and protects against LPS-induced septic shock. Cell Res. 23:201–212. 10.1038/cr.2013.6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masters S.L., Lagou V., Jéru I., Baker P.J., Van Eyck L., Parry D.A., Lawless D., De Nardo D., Garcia-Perez J.E., Dagley L.F., et al. . 2016. Familial autoinflammation with neutrophilic dermatosis reveals a regulatory mechanism of pyrin activation. Sci. Transl. Med. 8:332ra45 10.1126/scitranslmed.aaf1471 [DOI] [PubMed] [Google Scholar]
- Masumoto J., Taniguchi S., and Sagara J.. 2001. Pyrin N-terminal homology domain- and caspase recruitment domain-dependent oligomerization of ASC. Biochem. Biophys. Res. Commun. 280:652–655. 10.1006/bbrc.2000.4190 [DOI] [PubMed] [Google Scholar]
- McWilliam H., Li W., Uludag M., Squizzato S., Park Y.M., Buso N., Cowley A.P., and Lopez R.. 2013. Analysis Tool Web Services from the EMBL-EBI. Nucleic Acids Res. 41(W1):W597-600 10.1093/nar/gkt376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishra B.B., Rathinam V.A.K., Martens G.W., Martinot A.J., Kornfeld H., Fitzgerald K.A., and Sassetti C.M.. 2013. Nitric oxide controls the immunopathology of tuberculosis by inhibiting NLRP3 inflammasome-dependent processing of IL-1β. Nat. Immunol. 14:52–60. 10.1038/ni.2474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moriya M., Taniguchi S., Wu P., Liepinsh E., Otting G., and Sagara J.. 2005. Role of charged and hydrophobic residues in the oligomerization of the PYRIN domain of ASC. Biochemistry. 44:575–583. 10.1021/bi048374i [DOI] [PubMed] [Google Scholar]
- Mortimer L., Moreau F., MacDonald J.A., and Chadee K.. 2016. NLRP3 inflammasome inhibition is disrupted in a group of auto-inflammatory disease CAPS mutations. Nat. Immunol. 17:1176–1186. 10.1038/ni.3538 [DOI] [PubMed] [Google Scholar]
- Py B.F., Kim M.-S., Vakifahmetoglu-Norberg H., and Yuan J.. 2013. Deubiquitination of NLRP3 by BRCC3 critically regulates inflammasome activity. Mol. Cell. 49:331–338. 10.1016/j.molcel.2012.11.009 [DOI] [PubMed] [Google Scholar]
- R Development Core Team 2011. R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing, Vienna. [Google Scholar]
- Scheltema R.A., Hauschild J.-P., Lange O., Hornburg D., Denisov E., Damoc E., Kuehn A., Makarov A., and Mann M.. 2014. The Q Exactive HF, a Benchtop mass spectrometer with a pre-filter, high-performance quadrupole and an ultra-high-field Orbitrap analyzer. Mol. Cell. Proteomics. 13:3698–3708. 10.1074/mcp.M114.043489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindelin J., Arganda-Carreras I., Frise E., Kaynig V., Longair M., Pietzsch T., Preibisch S., Rueden C., Saalfeld S., Schmid B., et al. . 2012. Fiji: an open-source platform for biological-image analysis. Nat. Methods. 9:676–682. 10.1038/nmeth.2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt F.I., Lu A., Chen J.W., Ruan J., Tang C., Wu H., and Ploegh H.L.. 2016. A single domain antibody fragment that recognizes the adaptor ASC defines the role of ASC domains in inflammasome assembly. J. Exp. Med. 213:771–790. 10.1084/jem.20151790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spalinger M.R., Kasper S., Gottier C., Lang S., Atrott K., Vavricka S.R., Scharl S., Gutte P.M., Grütter M.G., Beer H.-D., et al. . 2016. NLRP3 tyrosine phosphorylation is controlled by protein tyrosine phosphatase PTPN22. J. Clin. Invest. 126:1783–1800. 10.1172/JCI83669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strowig T., Henao-Mejia J., Elinav E., and Flavell R.. 2012. Inflammasomes in health and disease. Nature. 481:278–286. 10.1038/nature10759 [DOI] [PubMed] [Google Scholar]
- Touitou I., Lesage S., McDermott M., Cuisset L., Hoffman H., Dodé C., Shoham N., Aganna E., Hugot J.-P., Wise C., et al. . 2004. Infevers: an evolving mutation database for auto-inflammatory syndromes. Hum. Mutat. 24:194–198. 10.1002/humu.20080 [DOI] [PubMed] [Google Scholar]
- UniProt Consortium 2015. UniProt: a hub for protein information. Nucleic Acids Res. 43(D1):D204–D212. 10.1093/nar/gku989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wies E., Wang M.K., Maharaj N.P., Chen K., Zhou S., Finberg R.W., and Gack M.U.. 2013. Dephosphorylation of the RNA sensors RIG-I and MDA5 by the phosphatase PP1 is essential for innate immune signaling. Immunity. 38:437–449. 10.1016/j.immuni.2012.11.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J.-W., Wu J., Zhang Z., Datta P., Ibrahimi I., Taniguchi S., Sagara J., Fernandes-Alnemri T., and Alnemri E.S.. 2006. Cryopyrin and pyrin activate caspase-1, but not NF-kappaB, via ASC oligomerization. Cell Death Differ. 13:236–249. 10.1038/sj.cdd.4401734 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.