Taylor et al. show that PD1 negatively regulates KLRG1+ ILC-2s by attenuating STAT5 activation. Blocking PD1 signaling significantly enhances the protective function of ILC-2s in helminthic infection.
Abstract
Group 2 innate lymphoid cells (ILC-2s) regulate immune responses to pathogens and maintain tissue homeostasis in response to cytokines. Positive regulation of ILC-2s through ICOS has been recently elucidated. We demonstrate here that PD-1 is an important negative regulator of KLRG1+ ILC-2 function in both mice and humans. Increase in KLRG1+ ILC-2 cell numbers was attributed to an intrinsic defect in PD-1 signaling, which resulted in enhanced STAT5 activation. During Nippostrongylus brasiliensis infection, a significant expansion of KLRG1+ ILC-2 subsets occurred in Pdcd1−/− mice and, upon adoptive transfer, Pdcd1−/− KLRG1+ ILC-2s significantly reduced worm burden. Furthermore, blocking PD-1 with an antibody increased KLRG1+ ILC-2 cell number and reduced disease burden. Therefore, PD-1 is required for maintaining the number, and hence function, of KLRG1+ ILC-2s.
Introduction
Group 2 innate lymphoid cells (ILC-2s) lack antigen-specific receptors and primarily render their function via cytokine signaling (Bartemes et al., 2012; Spits and Cupedo, 2012; Huang et al., 2015). ILC-2s were initially described by several groups and designated as natural helper cells (Koyasu et al., 2010; Moro et al., 2010), nuocytes (Neill et al., 2010; Barlow et al., 2012, 2013), or innate helper 2 cells (Price et al., 2010) that respond to tissue-derived signals including IL-25, IL-33 and thymic stromal lymphopoietin (TSLP). ILC-2s express IL-33 receptor (ST2), IL-25 receptor (IL-17RB), KLRG1 and naturally reside in tissue sites such as the lung, small intestine, skin and adipose tissues. ILC-2s initiate immune responses against parasites (Fallon et al., 2006; Huang et al., 2015), participate in inflammatory processes, such as airway hyperactivity (Chang et al., 2011), allergen induced lung inflammation (Motomura et al., 2014), and allergic atopic dermatitis (AD) in humans (Salimi et al., 2013). ILC-2s also contribute toward lung tissue repair (Monticelli et al., 2011), adipose tissue homeostasis (Brestoff et al., 2015; Lee et al., 2015), and cutaneous wound healing (Yin et al., 2013; Rak et al., 2016). Therefore, elucidating immunoregulatory mechanisms that can modulate ILC-2 cell number and function can identify important checkpoints that can be manipulated for controlling type 2–mediated immune responses.
Recent studies on ILC-2s in airway inflammation have identified a positive regulatory axis driven by ICOS signaling (Maazi et al., 2015; Molofsky et al., 2015; Paclik et al., 2015). Studies on negative co-receptor mediated regulation of ILC-2s has been restricted to the role of KLRG1, which has been previously shown to inhibit ILC-2 effector response (Salimi et al., 2013). Here, we have investigated the role of PD-1 in regulating KLRG1+ ILC-2 subsets and demonstrate the downstream signaling mechanism by which PD-1 regulates KLRG1+ILC-2s. PD-1 is related to the CD28 superfamily and is expressed on activated T cells, B cells, monocytes, and macrophages. It has two binding partners, namely PDL-1 (Dong et al., 1999) and PDL-2 (Latchman et al., 2001; Keir et al., 2008; Fife et al., 2009). Co-stimulation of PD-1 by either of these ligands activate inhibitory signals in T cells which either prevent T cell proliferation or render a regulatory phenotype to the T cells (Fife et al., 2009; Francisco et al., 2009; Amarnath et al., 2010, 2011). These varied immune-tolerant signaling cascades occur through SHP1/2 phosphatases, which are recruited to the ITIM and ITSM cytoplasmic domains of the PD-1 receptor (Okazaki et al., 2001; Parry et al., 2005). The recruited SHP1/2 phosphatases dephosphorylate STATs and/or AKT, thereby dampening T helper cell function (Franceschini et al., 2009; Francisco et al., 2009; Amarnath et al., 2011). In particular PD-1 can specifically inhibit STAT5 signaling in T regulatory cells (Franceschini et al., 2009). It is yet to be clarified if such PD-1–mediated tolerance mechanisms occur in ILC subsets.
Tumors (Wang and Chen, 2011), viruses (Barber et al., 2006; Day et al., 2006; Trautmann et al., 2006), and bacteria (Das et al., 2006; Beswick et al., 2007; Barber et al., 2011) manipulate the PD-1 signaling pathway to evade host immune responses. In particular, clinical trials that use PD-1 blocking antibody have shown phenomenal success in cancer immunotherapy (Topalian et al., 2012; Yaqub, 2015). Parasitic worms also exploit the PD-1 pathway to create an immune-suppressive microenvironment by inducing macrophages with suppressor function (Smith et al., 2004; Terrazas et al., 2005). Hence, PD-1–mediated tolerance mechanisms in adaptive and innate immune cells, with respect to tumors and pathogens, have been extensively studied. However, the cellular mechanism by which PD-1 modulates ILC-2 function during disease pathogenesis is still largely unknown.
In this study, we have explored whether PD-1 regulates ILC-2 cells. We demonstrate that PD-1 is a critical negative regulator of KLRG1+ ILC-2 subsets. Disrupting PD-1 signaling either by genetic deletion or by antibody blockade significantly enhanced KLRG1+ ILC-2 cells in both number and function, thereby efficiently clearing Nippostrongylus brasiliensis worms in mice. In humans, we found that PD-1 is exclusively expressed by ILC-2s (and not ILC-1 or ILC-3) and regulates human ILC-2 function.
Results
Pdcd1−/− mice possess enhanced KLRG1+ ILC-2 subsets
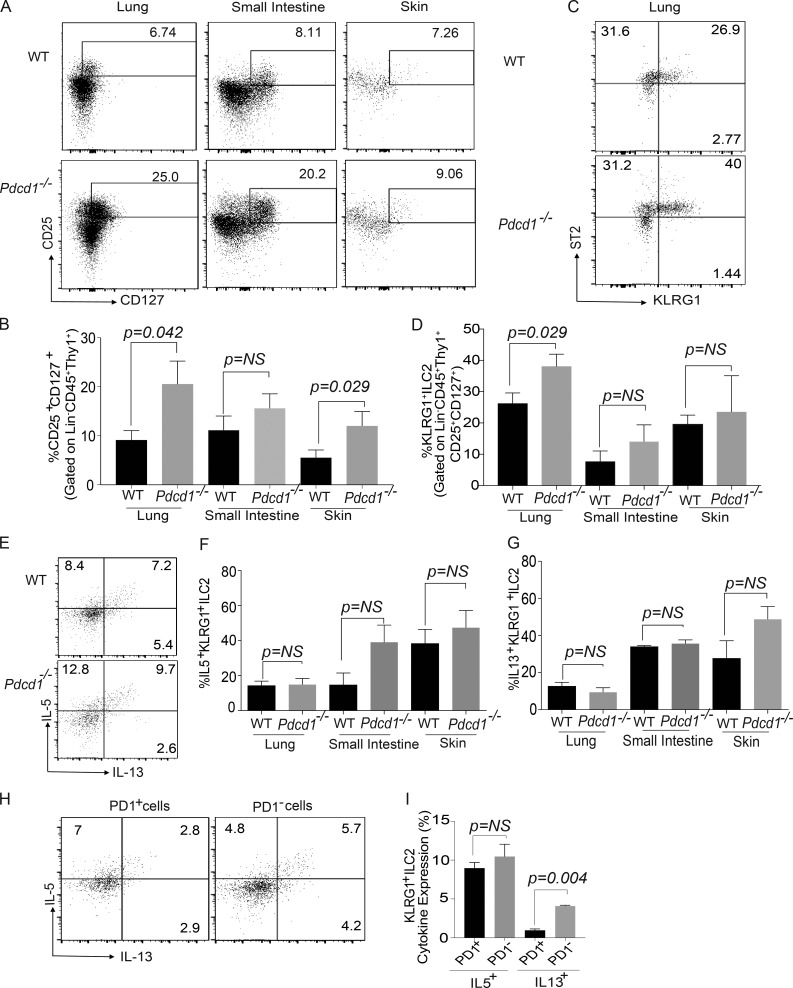
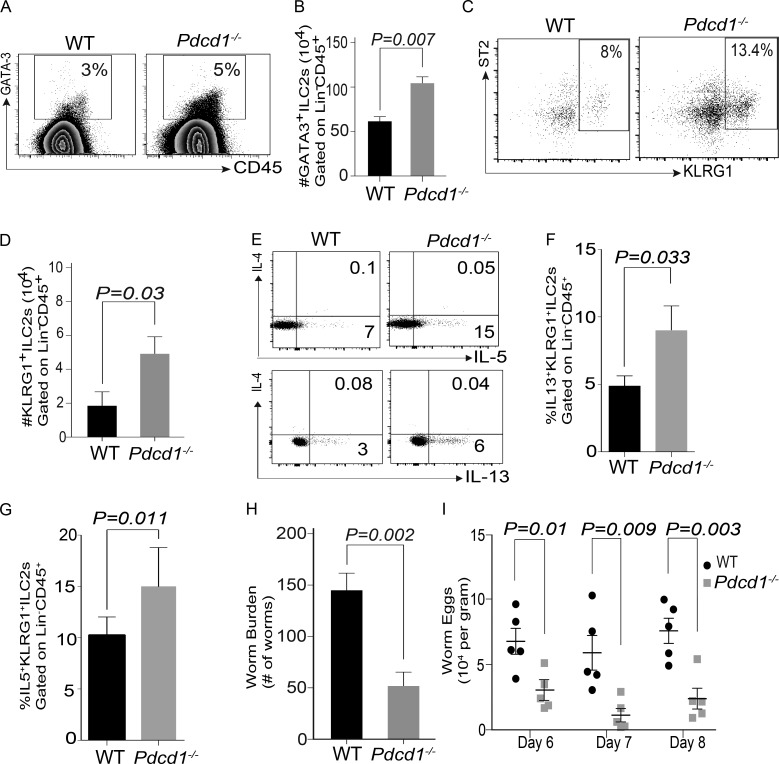
The expression and regulatory function of PD-1 in T cells, B cells, and myeloid cells has been previously characterized (Agata et al., 1996; Okazaki et al., 2002), but its role in ILCs is yet to be defined. Using previously defined gating strategy for ILC-2s (Chang et al., 2011; Monticelli et al., 2011; Halim et al., 2012), we found that Lin−CD45+CD90+CD25+CD127+ KLRG1+ ILC-2s expressed PD-1 in WT mice (Fig. S1, A–C). We next explored the functional consequence of PD-1 expression on KLRG1+ILC-2s by comparing the frequency of these subsets in WT and Pdcd1−/− mice. Initial analysis suggested that WT and Pdcd1−/− KLRG1+ILC-2s showed similar trend in IL-7R and IL-2R surface expression (Fig. S1, D and E). Therefore, we investigated the frequency of ILC-2s in WT and Pdcd1−/− mice. In all organs tested, tissue-resident Lin−CD45+CD90+CD25+CD127+ ILCs were increased and a significant difference was noted in the lungs and skin of Pdcd1−/− mice (Fig. 1, A and B). Next, we evaluated if there was a difference in ILC-2 subsets between WT and Pdcd1−/− mice. A significant increase in KLRG1+ ILC-2s was observed in the lungs of Pdcd1−/− mice (Fig. 1 C). Although there was a small increase in KLRG1+ ILC-2s in the small intestine and skin, a significant statistical difference was not observed (Fig. 1 D). We next evaluated if there was a difference in the cytokine expression within the KLRG1+ ILC-2s. Although both WT and Pdcd1−/− KLRG1+ ILC-2s had similar IL-5 and IL-13 expression by intracellular flow cytometry (Fig. 1, E–G), the cytokine profile of WT KLRG1+ILC-2s that were PD-1− or PD-1+ was altered. In the WT ILC-2s, PD-1− population produced higher amounts of effector cytokines such as IL-13 compared to the PD-1+ subsets (Fig. 1, H and I). These data led us to investigate the kinetics of PD-1 expression on KLRG1+ILC-2s and their potential function in these cells. We found that KLRG1+ILC-2s up-regulated PD-1 expression in response to IL-2 and IL-7 stimulation. PD-1 expression was significantly increased in the presence of IL-33 cytokine, but a similar effect was not noted with IL-25 or TSLP (Fig. S1, F and G). Collectively, these data suggest that PD-1 expression on ILC-2s is regulated by integrative signals from IL-2, IL-7, and IL-33.
Figure 1.
Characterization of ILC-2 subsets in WT and Pdcd1−/− mice. Lungs, small intestine, and skin were harvested from WT and Pdcd1−/− mice, and then ILC-2 subsets were characterized by flow cytometry. Lin− was defined as CD3−, CD4−, CD8−, NK1.1−, GR1−, Ter119−, CD5−, CD11b−, CD11c−, F4/80−, CD45R/B220−, and CD19−. Gating strategy for ILCs included an initial gating of Lin−CD45+Thy1+. ILCs were then characterized as Lin−CD45+ Thy1+CD127+CD25+ and ILC-2s as Lin−CD45+ Thy1+CD127+CD25+ST2+KLRG1+. Representative flow plots of ILC subset frequency in the lungs, small intestine, and skin in the WT and Pdcd1−/− mice (A). Summary of ILC frequency in the lungs, small intestine, and skin (B). Representative flow plots of ILC-2 subset frequency in the lungs in the WT and Pdcd1−/− mice (C). Summary of KLRG1+ ILC-2 frequency in the lungs, small intestine, and skin (D). Cytokine expression of ILC-2 subsets were measured by stimulating the cells with cell stimulation cocktail for 4 h, and expression of cytokines such as IL-5 and IL-13 were measured by flow cytometry. The expression of IL-5 and IL-13 by Lin−CD45+ Thy1+CD127+CD25+ST2+KLRG1+ in WT and Pdcd-1−/− mice; representative example in lungs (E) and summary in the various organs (F and G). KLRG1+ILC-2 cells from the lungs of WT mice were gated for PD1− and PD1+, and then the cytokine profile was monitored by intracellular flow cytometry (H and I). Experiments were repeated three times reproducibly, and data shown are from n = 3–5 mice for all panels. Data shown is mean ± SEM. A Student’s t test was performed to determine statistical significance between the various cohorts. P ≤ 0.05 was considered significant. Significant p-values are denoted in the figures.
PD-1 deficiency does not affect ILC-2 development
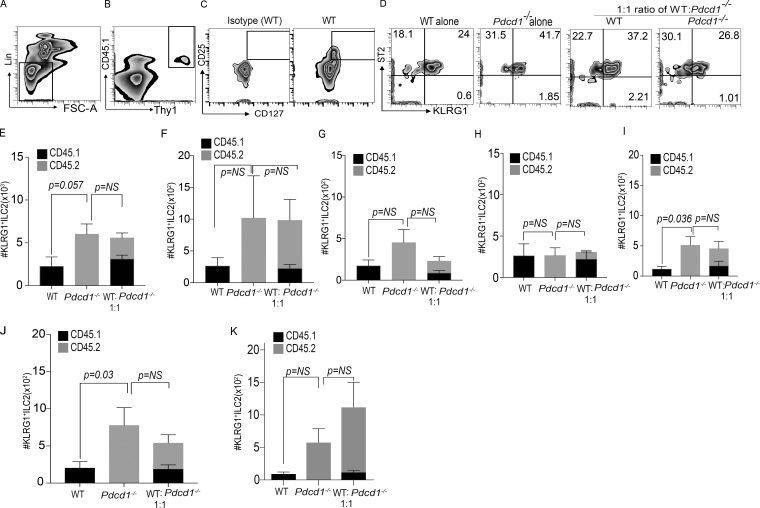
We next used a mixed BM chimera experiment to test two aspects of PD-1 function in KLRG1+ILC-2 biology, namely development and function. Mixed BM chimera experiments were performed in which T cell–depleted BM was adoptively transferred into WT B6 recipients after total body irradiation. 8 wk after transplant, ILC-2 frequency and numbers were quantified. Murine recipients that received WT BM had a tendency toward lower numbers of KLRG1+ ILC-2s in the lungs compared with Pdcd1−/− recipients, but this difference did not reach significance (Fig. 2, A–E). A similar trend was noted in the small intestine and mesenteric LNs (Fig. 2, F and G). We next evaluated if ILC-2 cell development in the Pdcd1−/− mice was altered in the presence of WT BM (i.e., in the 1:1 mixed ratio cohort). Our data suggests that ILC-2 cell numbers in the WT and Pdcd1−/− compartments were not significantly altered in the mixed chimera cohorts (1:1 ratio). In addition, these data further identified an intrinsic function for PD-1 in regulating KLRG1+ ILC-2 numbers. In line with the increase in cell numbers, we also found that the IL-13 cytokine production of KLRG1+ ILC-2s was increased in the lungs in the Pdcd1−/− cohorts but did not reach significance in 1:1 mixed chimera cohorts (Fig. 2, H and I). Therefore, lack of PD-1 did not alter KLRG1+ILC-2 cell development or function. To confirm the role of PD-1 in the development and intrinsic regulation of ILC-2s, experiments were repeated in Rag2−/−γc−/− hosts that lack host derived ILCs. These murine recipients were reconstituted with T depleted BM from WT or Pdcd1−/− mice. A significant increase in ILC-2 numbers were noted in the lungs of Pdcd1−/− BM recipients and a trend toward increase in ILC-2 cell numbers were noted in the small intestine (Fig. 2, J and K). No significant changes were noted in the ILC-2 cell numbers in the mixed chimera cohorts, both in the lungs and small intestine, which may indicate inhibitory signals from the WT compartment that can restrict Pdcd1−/− ILC-2 numbers. In summary the mixed chimera experiments demonstrate that PD-1 deficiency does not inhibit ILC-2 cell development.
Figure 2.
PD-1 deficiency does not alter ILC-2 cell development. Host CD45.1+C57BL6 mice were subjected to total body irradiation (1,050 cGy), and then reconstituted with either CD45.1+ WT BM (10 million) or CD45.2+Pdcd1−/− BM (10 million). Some cohorts received both WT and Pdcd1−/− BM at a 1:1 ratio (5 million WT:5million Pdcd1−/−). At 8 wk after BM transplant, ILC-2 were analyzed by flow cytometry via gating on Lin− CD45+ Thy1+ CD127+ CD25+ ST2+ KLRG1+ (Representative data from lungs; A–D) and cell numbers were characterized in the lungs, small intestine, and mesenteric LNs. Summary of the absolute numbers of KLRG1+ILC-2 cells in the lungs (E), small intestine (F), and mesenteric LNs (G). Single-cell suspensions were stimulated with cytokine stimulation cocktail, and cytokine profile was measured by flow cytometry. Summary of absolute numbers of IL-5 and IL-13 from lung KLRG1+ILC-2s (H and I). Host C57BL6 Rag−/−γc−/− mice were reconstituted with either CD45.1+ WT BM (10 million), or CD45.2+Pdcd1−/− BM (10 million). Some cohorts received both WT and Pdcd1−/− BM at a 1:1 ratio. At 8 wk after BM transplant, the lung and small intestine were harvested from the mice and KLRG1+ILC-2 cell numbers were characterized. Summary of the absolute numbers of KLRG1+ILC-2 cells in the lungs (J) and small intestine (K) in the various cohorts was analyzed by flow cytometry. Animals per cohort was n = 5. Data shown are mean ± SEM. Experiments were repeated twice. A one-way ANOVA analysis followed by a multiple comparison test [Turkey] was performed to determine statistical significance between the various cohorts. P ≤ 0.05 was considered significant. Significant p-values are denoted in the figures.
PD-1 intrinsically inhibits STAT5 signaling in KLRG1+ ILC-2s
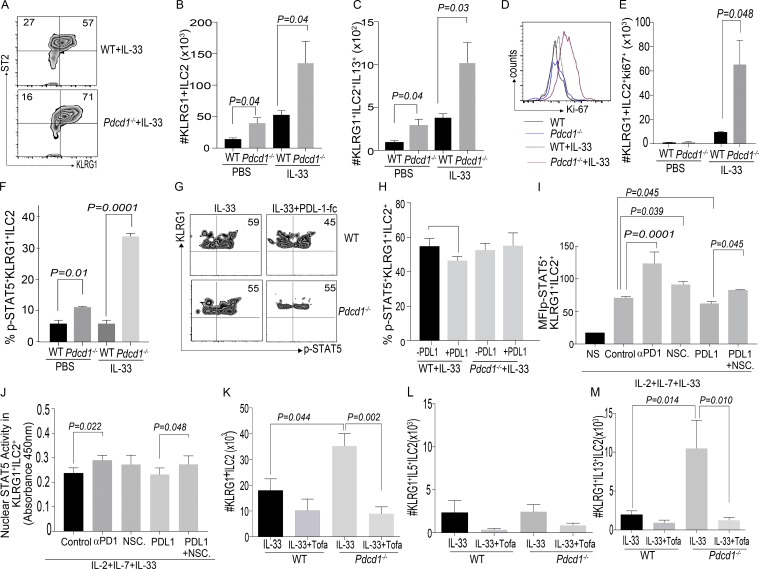
Because our data suggested that PD-1 intrinsically regulated ILC-2 numbers, we next tested if PD-1 altered the proliferative potential of KLRG1+ILC-2s. Lung-resident WT and Pdcd1−/− KLRG1+ ILC-2s were stimulated in vitro with various ILC-2 promoting cytokines, of which IL-33 in combination with IL-2 and IL-7 significantly enhanced the proliferation of Pdcd1−/− ILC-2s compared with WT ILC-2s. No significant proliferative difference was noted with other ILC-2 cytokines, i.e., IL-25 and TSLP (Fig. S2, A and B). We next investigated if the in vitro effect was replicated in vivo on IL-33 priming of WT and Pdcd1−/− murine recipients. Toward this goal, WT and Pdcd1−/− mice were treated with rmIL-33 for 3 d as previously described (Maazi et al., 2015), and then numbers of KLRG1+ ILC-2s were determined. Compared with WT mice, Pdcd1−/− mice had significantly increased numbers of KLRG1+ ILC-2 cells (Fig. 3, A and B). No difference in effector cytokine frequency was noted between WT and Pdcd1−/− recipients (Fig. S2 C), but a significant increase in the absolute numbers of IL-13+KLRG1+ cells was observed (Fig. 3 C). We next investigated if in vivo IL-33–stimulated KLRG1+ ILC-2s had enhanced proliferative potential. Our data demonstrated that KLRG1+ILC-2s from Pdcd1−/− mice had significantly enhanced proliferative capacity (Fig. 3, D and E).
Figure 3.
Lack of PD-1 signaling results in a significant increase in KLRG1+ ILC-2 numbers. WT and Pdcd1−/− mice were either treated with PBS or IL-33 (200 ng/mice) for 3 d. ILC-2 frequency and numbers were evaluated by flow cytometry at day 3. Gating strategy include: Lin−CD45+ Thy1+CD127+CD25+ST2+KLRG1+. Representative flow plots of KLRG1+ILC-2 subsets in WT and Pdcd1−/− (A) and summary of absolute numbers of KLRG1+ ILC-2 in WT and Pdcd1−/− lungs were evaluated (B). Cytokine expression of ILC-2 subsets were measured by stimulating the cells with cell stimulation cocktail for 4 h, and expression of cytokines such as IL-5 and IL-13 were measured by flow cytometry. The absolute number of IL-13+ cells within Lin−CD45+ Thy1+CD127+CD25+ST2+KLRG1+ in WT and Pdcd-1−/− mice (C). Ki67 staining in the different cohorts were analyzed by flow cytometry (D and E). WT or Pdcd-1−/− were treated with PBS or IL-33 (200ng/mice) for 1 d. Lung ILC-2 cells were isolated, and then restimulated with IL-2 for 15 min, followed by phospho-STAT5 flow cytometry. Summary of the frequency of p-STAT5 signaling in the various cohorts (F). Experiments were repeated where WT or Pdcd-1−/− mice were treated with PBS or IL-33 (200 ng/mice) for 3 d. Lung ILC-2 cells were restimulated with IL-2 alone or IL-2 along with coated PDL-1fc chimera (5 µg/ml) for 15 min, followed by phospho-STAT-5 flow cytometry. Representative flow plots showing p-STAT5 signaling in WT (G; top) or Pdcd-1−/− (G; bottom). Summary of the frequency of p-STAT5 signaling in the various cohorts (H). Experiments were repeated where WT mice were treated with IL-33 (200 ng/mice) for 3 d, and lung ILC-2s were stimulated for 24 h with IL-2, IL-7, and IL-33. In addition, cohorts were treated with Isotype control antibody, αPD-1 antibody, NSC87877, PDL-1fc, or PDL-1 Fc plus NSC87877. After 24-h stimulation, cells were restimulated with IL-2 (100 ng/ml) and subjected to p-STAT5 flow cytometry. Mean fluorescence intensity of p-STAT5 in KLRG1+ILC-2s from various cohorts (I). Experiments were repeated, and ILC-2s were stimulated in vitro for 3 d, and then nuclear lysates were tested for STAT5a (J). Mechanistic experiments were set up with WT and Pdcd1−/− mice, where cohorts were treated with rmIL-33 + Vehicle (DMSO) or with rmIL-33 + Tofacitinib (15mg/kg/d/mouse). Absolute numbers of KLRG1+ILC-2s were evaluated at day 3 after treatment (K), and functional cytokine expression was monitored in the various cohorts (L and M). Animals per cohort was n = 5. Data shown are mean ± SEM and cumulative of at least two repeats. A one-way ANOVA analysis followed by a multiple comparison test (Tukey) was performed to determine statistical significance between the various cohorts. P ≤ 0.05 was considered significant. Significant p-values are denoted in the figures.
On identifying the in vitro and in vivo effect of IL-33, the influence of IL-25 was investigated. IL-25 treatment did not alter the frequency and numbers of KLRG1+ ILC-2s in the Pdcd1−/− mice (Fig. S2, D and E).
The signaling pathways that resulted in an increase in KLRG1+ ILC-2 cells in Pdcd1−/− mice were next explored. Because ILC-2s depend on STAT5 signaling for proliferation and PD-1 inhibits STAT5 signaling in T cells (Franceschini et al., 2009), we explored if a similar mechanism operated in ILC-2s. To measure STAT5 signaling in the KLRG1+ ILC-2 subsets, WT and Pdcd1−/− mice were either treated with PBS or rmIL-33 for 1 d, as previously described (Maazi et al., 2015). KLRG1+ILC-2s were then ex vivo stimulated with IL-2 for 15 min, and then subjected to phospho-STAT5 flow cytometry. STAT5 signaling was significantly enhanced in the Pdcd1−/− KLRG1+ ILC-2 subsets that were treated with either PBS or rmIL-33 (Fig. 3 F). We next investigated if providing PDL-1 ligand had an effect in inhibiting STAT5 activation in WT KLRG1+ ILC-2 subsets. For these experiments, WT and Pdcd1−/− mice were treated with IL-33 for 3 d, and then KLRG1+ ILC-2 cells were stimulated in the presence of plate-coated PDL-1 fc-chimera plus IL-2 or IL-2 alone for 15 min, and then phospho-STAT5 flow cytometry was performed. In the WT mice treated with rmIL-33, sufficient STAT5 signaling was noted, which was significantly decreased in the presence of PDL-1 (Fig. 3, G and H). No difference in p-STAT5 signaling was detected in the Pdcd1−/− cohorts treated with PDL-1. In addition, these data suggest that ILC-2s from Pdcd1−/− mice exhibit robust STAT5 activation (one dose of in vivo IL-33), and this effect is recapitulated in the WT ILC-2s after three doses of in vivo IL-33. The initial boost in STAT5 activation, along with lack of response to PDL-1–mediated inhibitory effects, may contribute toward significant proliferation in the Pdcd1−/− cohorts.
To further explore this mechanism, we sequentially inhibited PD-1 or its downstream signaling components, namely SHP1/2 in ex vivo culture of KLRG1+ILC-2s. IL-33 was administered to WT and Pdcd1−/− mice for 3 d, and then KLRG1+ILC-2s were stimulated for 24 h in the presence of IL-2, IL-7, and IL-33. Certain cohorts were treated with an isotype control antibody (denoted as control in figure), anti–PD-1 antibody, selective SHP1/2 inhibitor (NSC87877), PDL-1 fc, and PDL-1 fc+SHP1/2 inhibitor. After 24-h stimulation, cells from the various cohorts were restimulated for 15 min with IL-2, and then subjected to phospho-STAT5 flow cytometry. We found that blocking PD-1 or its signaling pathway significantly increased p-STAT5 activation in KLRG1+ ILC-2s. This increase in p-STAT5 was inhibited when the PD-1 pathway was activated instead of blocked (by providing the ligand PDL-1fc). Inhibition of p-STAT5 in the cohorts that were treated with PDL-1 fc was significantly reversed upon blocking SHP1/2 (Fig. 3 I). Collectively, these data suggest that altering PD-1 signaling in KLRG1+ILC-2s can modulate STAT5 activation. The impact of these conditions (PD-1 signaling blockade) on nuclear STAT5a/b content in KLRG1+ILC-2s was investigated. A similar experimental approach was used, but the cells were expanded for 72 h instead of 24 h. After stimulation, cells from the different cohorts were isolated and nuclear extracts were prepared and subjected to a nuclear transcription factor assay. We found that blocking PD-1 significantly increased nuclear STAT5 in KLRG1+ILC-2s. An increase was also noted in the presence of SHP1/2 blocker, but did not reach statistical significance. Providing PDL-1 fc in these long-term cultures did not alter STAT5a nuclear content, but blocking SHP1/2 (in the PDL-1 fc group) did increase the content of nuclear STAT5a in KLRG1+ ILC-2s (Fig. 3 J). The nuclear content of STAT5b showed a similar profile to that of STAT5a (Fig. S2 f). Next, we investigated the reason for the lack of PDL-1 fc effect in long-term cultures. We found that IL-33 stimulation for 72 h up-regulated PDL-1 expression on KLRG1+ILC-2s (Fig. S2 g), suggesting that providing PDL-1 fc as a ligand may be redundant under these stimulating conditions. We finally evaluated the proliferation of KLRG1+ ILC-2s under these various culture conditions. Blocking PD-1 or SHP1/2 significantly enhanced the proliferative potential of KLRG1+ ILC-2s, which was significantly inhibited by PDL-1 fc stimulation. Similar to the p-STAT5 assays, blocking SHP1/2 in the presence of PDL-1 fc reversed the proliferative effect (Fig. S2, Ha and I). Therefore, these data suggest that modulating PD-1 signaling in KLRG1+ ILC-2s can alter STAT5 activation status and can also affect proliferation in long-term cultures.
Next, we explored if PD-1 signaling independently (bypassing STAT5) affected proliferation of KLRG1+ ILC-2s. We found that co-injection of both IL-33 and Tofacitinib (JAK/STAT inhibitor) in Pdcd1−/− mice significantly reduced KLRG1+ ILC-2 cell numbers and IL-13 cytokine production in the lungs (Fig. 3, K–M; and Fig. S2, J and K). These data demonstrate that the PD-1 pathway does not operate independently of STAT5 in inducing proliferation of KLRG1+ ILC-2s.
Pdcd1−/− KLRG1+ILC-2s gene expression pattern indicates enhanced effector function
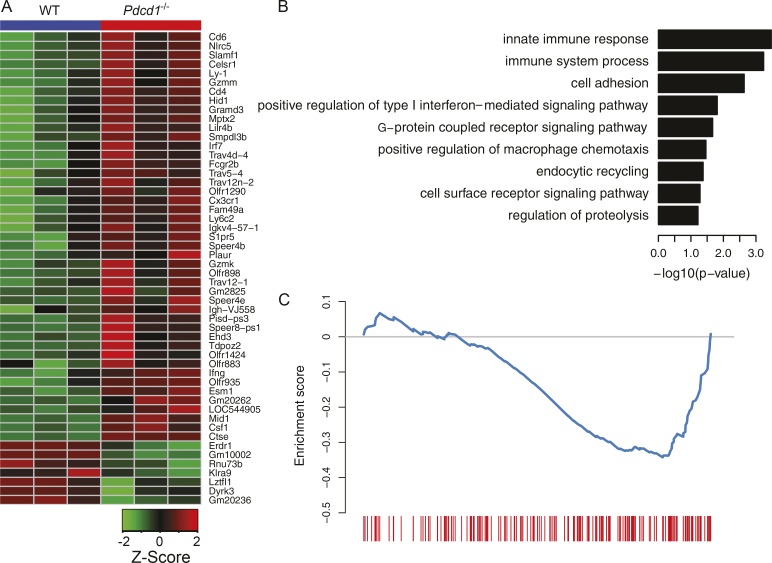
To further explore the molecular mechanism of PD-1 regulation on ILC-2 cells, we performed a microarray assay to compare gene expression patterns of KLRG1+ ILC-2s between WT and Pdcd1−/−. Dozens of differentially expressed genes were identified, with 45 genes up-regulated and 7 down-regulated in Pdcd1−/− (false discovery rate [FDR] ≤ 0.15). Consistently, the expression levels of STAT5 target genes (e.g., slamf1 and csf1) were increased in PD-1–deficient KLRG1+ ILC-2s, whereas cell proliferation or activation-associated genes (e.g., klra9 and lztfl1) were also altered (Fig. 4 A). Gene ontology enrichment analysis of up-regulated genes revealed that the lack of PD-1 affected signaling pathway on ILC-2s, including innate immune responses, macrophage chemotaxis, cell-cell interaction and metabolism (Fig. 4 B). We also performed gene set enrichment analysis against published gene set (Duraiswamy et al., 2011) and the result showed that PD-1 deficiency had similar impact on gene expression patterns between ILC-2s and CD8+ T cells (FDR q-value = 0.036; Fig. 4 C), suggesting that a similar intrinsic regulatory mechanism of PD-1 signal may apply to both innate and adaptive lymphocytes.
Figure 4.
Comparison of gene expression profiles of KLRG1+ ILC-2s between WT and Pdcd1−/−. Lin−CD90+KLRG1+ ILC-2s were sorted from the lungs of WT or Pdcd1−/− mice that had been treated with IL-33 for 3 d. Total RNA was isolated, and then subjected to microarray analysis. Heat map for the expression patterns of differentially expressed genes (A). Representative GO terms enriched for genes up-regulated in Pdcd1−/−. Only GO terms from the biological process category is shown. The log10-scaled p-value is indicated in the x axis (B). Gene set enrichment analysis of the effect of Pdcd1 knockout for gene expression. The genes are ranked by fold change as evaluated by microarray data, and compared with the gene set available from GEO under accession no. GSE26495 (NAIVE_VS_PD1LOW_CD8_TCELL_DN). The blue curve shows the running enrichment score (ES) for the gene set as the analysis walks down the ranked gene list. The leading edge for the gene set is shown as short bars at the bottom. The normalized enrichment score (NES) is calculated as –1.51, and the false discovery rate FDR is calculated as 0.036 (C). A one-way ANOVA analysis was performed to determine statistical significance. Microarray analysis was performed in triplicates, and each experiment consisted of n = 5 mice per cohort.
KLRG1+ILC-2 cells from Pdcd1−/− mice possess anti-helminth immunity
We next evaluated if PD-1 deficiency modulated ILC-2 function during parasitic helminth infections. To investigate this, WT and Pdcd1−/− mice were infected with N. brasiliensis, and then KLRG1+ ILC-2 subset numbers were evaluated. In the lungs, the frequency and numbers of ILC-2 (GATA3+) or KLRG1+ ILC-2 subsets were similar in both WT and Pdcd1−/− cohorts on day 5 after worm infection (Fig. S3). In the Pdcd1−/− mice, near the site of infection (mesenteric LNs; MLNs), a significant increase in GATA3+ILC-2 frequency (Fig. 5 A) and absolute numbers (Fig. 5 B) was noted. In addition a significant increase in KLRG1+ ILC-2 subset frequency (Fig. 5 C), absolute numbers (Fig. 5 D), and cytokine production (Fig. 5, E–G) was also noted. Next, the function of the Pdcd1−/− KLRG1+ ILC-2 cells independent of adaptive immunity in clearing worm burden was evaluated. For these experiments, Rag2−/−γc-/− mice were infected with N. brasiliensis, followed by adoptive transfer of either WT or Pdcd1−/− KLRG1+ ILC-2 cells. A significant difference in the worm burden (Fig. 5 H) and worm eggs (Fig. 5 I) was noted in recipients that were reconstituted with Pdcd1−/− KLRG1+ ILC-2 subsets.
Figure 5.
KLRG1+ ILC-2 cells from Pdcd1−/−are efficient at clearing N. brasiliensis from Rag−/−γc−/− mice. WT or Pdcd1−/− mice were infected with N. brasiliensis, and then KLRG1+ ILC-2 numbers in the mesenteric LNs were evaluated by flow cytometry. Representative flow plots showing GATA3+ ILC-2 frequency in the WT and Pdcd1−/− (A), summary of absolute numbers of GATA3+ILC-2+ cells (B), representative flow plots showing KLRG1+ILC-2 subset frequency in WT and Pdcd1−/− mice (C), and absolute numbers of KLRG1+ ILC-2 numbers in WT and Pdcd1−/− mice (D). Functional cytokine expression was monitored using intracellular flow cytometry, and both frequency and absolute numbers were measured (E–G). Rag2−/−γc−/− mice were infected with N. brasiliensis, and then reconstituted with either WT or Pdcd1−/− KLRG1+ ILC-2 cells. Worm burden (H) and eggs in the feces (I) of host mice is shown. Black circles represent WT and gray squares represent Pdcd1−/−. Animals per cohort was n = 4–5, and data shown are mean ± SEM. Experiments were performed twice; data shown is from one repeat. A Student’s t test was performed to determine statistical significance between the various cohorts in all the panels. P ≤ 0.05 was considered significant. Significant p-values are denoted in the figures.
Blocking PD-1 in Rag1−/− mice enhances ILC-2 function during parasitic helminth infections
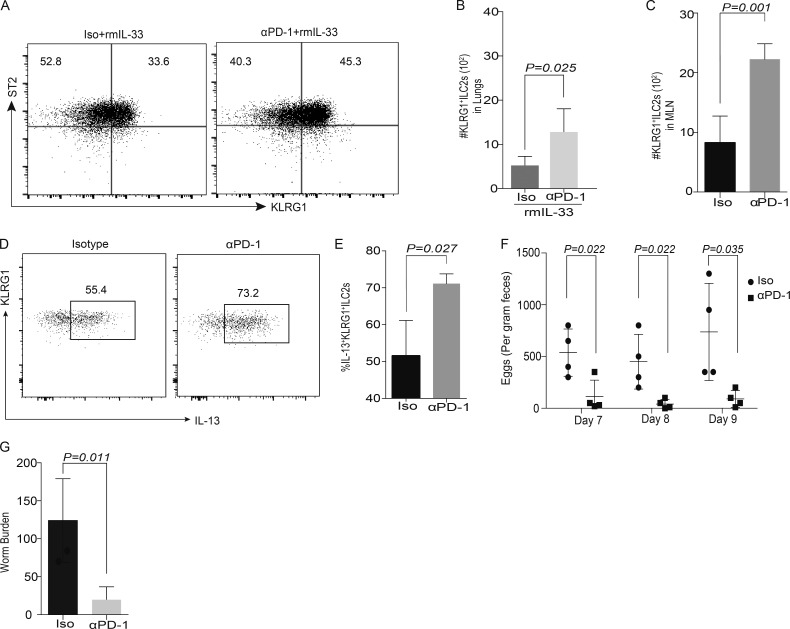
Experiments were performed to identify if exogenous blocking of PD-1 using an antibody would have similar effects on KLRG1+ ILC-2 subsets in vivo. Rag1−/−mice were injected with either isotype or αPD-1 antibody at day 0, and then treated with rm-IL-33 from day 1 to 3. In cohorts that were treated with both αPD-1 and rmIL-33 cytokine, a significant increase in KLRG1+ ILC-2 cell frequency numbers was noted (Fig. 6, A and B). To identify if blocking PD-1 can enhance therapeutic potential via ILC-2 cells, we repeated PD-1 blocking experiments in Rag1−/− mice infected with N. brasiliensis. Blocking PD-1 during ongoing helminth infection significantly enhanced ILC-2 numbers (Fig. 6 C) and cytokine production in MLNs (Fig. 6, D and E) and diminished both worm eggs in the feces (Fig. 6 F) and worm burden (Fig. 6 G). These results identify a potential wider application for anti–PD-1 therapeutics in helminth infections.
Figure 6.
Administration of αPD-1 can significantly diminish worm burden in Rag1−/−recipients. Rag1−/− mice were treated with isotype or αPD-1 antibody (250 µg/mice), followed by rmIL-33 (200 ng/mice), for 3 d. Gating strategy included Lin−CD45+ Thy1+CD127+CD25+ST2+KLRG1+. Representative flow plots of ILC-2 in Isotype and αPD-1 cohorts (A) and summary of absolute numbers of KLRG1+ ILC-2 in Isotype and αPD-1 cohorts’ lungs (B) were evaluated. Rag1−/− mice were infected with 300 Larvae 3 of N. brasiliensis on day 0. Anti–mouse PD-1 antibodies or IgG isotype control were i.p. injected into mice on day 0, 3, 6, and 9, at a dose of 250 µg antibody per mouse each time. Leukocytes were isolated from MLNs on day 14, and the numbers of total ILC-2 cells were analyzed by flow cytometry (C). Cytokine expression was analyzed using intracellular flow cytometry (D and E). Feces were collected on day 7, 8, and 9 from individual mouse, and the worm eggs were counted (F). Adult worms in small intestine were counted on day 14 (G). Animals per cohort was n = 4–5. Data shown are mean ± SEM. Experiments were repeated twice. Data on n = 5 mice is shown (A–C). A Student’s t test was performed to determine statistical significance for the data shown in all the panels. P ≤ 0.05 was considered significant. Significant p-values are denoted in the figures.
PD-1 regulates KLRG1+ ILC-2s in humans
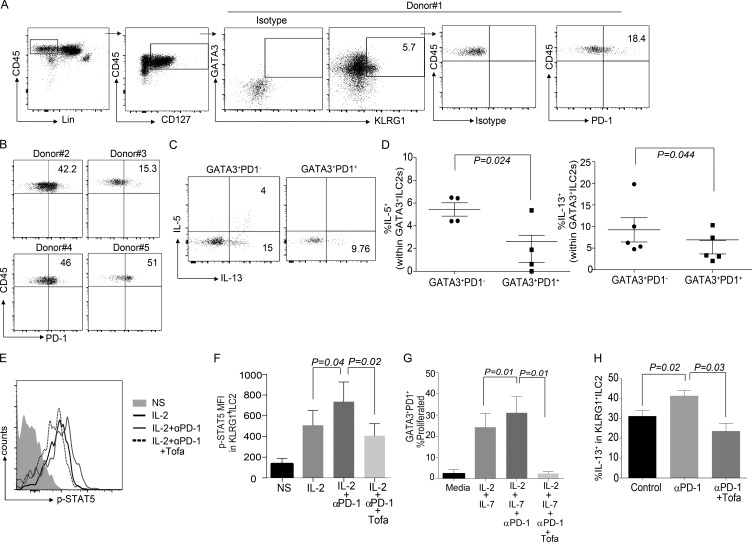
We next explored if PD-1 is expressed on ILC subsets in human PBMCs. PD-1 was almost exclusively expressed on ILC-2s as compared with ILC-1 and ILC-3 subsets (Fig. S4, A and B). On further characterization based on IL-7Rα, KLRG1, and GATA3, we found that human KLRG1 subsets also expressed PD-1 at a similar frequency to murine ILC-2s (Fig. 7, A and B). Similar to murine data, PD-1+ subsets secreted significantly lower amounts of IL-5 and IL-13 compared with PD-1− subsets (Fig. 7, C and D). We next evaluated if PD-1 also intrinsically inhibited STAT5 signaling in human ILC-2s. STAT5 signaling was enhanced when PD-1 was blocked on ILC-2s, and this effect was reversed by the JAK/STAT inhibitor Tofacitinib (Fig. 7, E and F). We next investigated if nuclear STAT5a/b content in KLRG1+ILC-2s is altered when PD-1 pathway is manipulated. Similar to murine data, blocking PD-1 resulted in an increase in STAT5a/b nuclear content in human ILC-2s (Fig. S4, C and D). After this, we evaluated the downstream effects of STAT5 activation on ILC-2 proliferation. PD-1+GATA3+ cells were inherently less proliferative as compared with the PD-1−GATA3+ ILC-2s (Fig. 7 G; and Fig. S4, E and F). On inhibiting PD-1 signaling, an increase in proliferation was noted, which was reversed on inhibiting Jak/STAT signaling through Tofacitinib (Fig. 7 G; and Fig. S4, E and F). In summary, PD-1 intrinsically regulates human KLRG1+ ILC-2s by inhibiting STAT5 phosphorylation.
Figure 7.
PD-1 regulation of KLRG1+ILC-2 is conserved in human PBMCs. Human PBMCs were obtained from normal healthy volunteers and characterized for the expression of PD-1 in the various ILC subsets. Gating strategy included Lin−CD45+CD127+GATA3+KLRG1+ (A). PD-1 expression was analyzed in n = 5 donors (A and B). ILC-2 cells were gated as either GATA3+PD-1− or GATA3+PD-1+, and then characterized for cytokine expression (C and D). PBMCs were stimulated with rhIL-2 (1,000 IU) + Vehicle (DMSO) + Isotype control for 15 min, and then p-STAT5 expression was measured by flow cytometry. Certain cohorts were treated with rhIL-2 + Vehicle + αPD-1 or rhIL-2 + Tofacitinib (0.3 µM) + αPD-1. The representative flowplot and mean fluorescence intensity of pSTAT5 in various cohorts were measured by flow cytometry (E and F; n = 4 donors). PBMCs were labeled with Cell Trace Violet. and then ILC-2s were stimulated with rhIL-2 + rhIL-7 (40 ng/ml) + Vehicle (DMSO) + Isotype control or with rhIL-2 + rhIL-7 + Vehicle + αPD-1 or with rhIL-2 + rhIL-7 + Tofacitinib + αPD-1. Dilution of Cell Trace Violet as a measure of proliferation was performed at day 5 after stimulation (G; n = 4 donors). Experiments were performed in multiple donors as stated, and data are represented as mean + SEM. Human PBMCs (4 million) were adoptively transferred into NSG murine recipients and treated with rhIL-2, rhIL-7, and rhIL-33 for 3 d, along with Vehicle (DMSO) and Isotype control antibody. This cohort is termed as control; certain cohorts received either αPD-1 or αPD-1 and Tofacitinib, in addition to rhIL-2, rhIL-7, and rhIL-33. At day 3 after adoptive transfer, lungs were harvested and stimulated with PMA/ionomycin, and then human ILC-2 subsets were characterized. Human CD45+ cells were gated followed by CD45+Lin− CD127+GATA3+KLRG1+ gating. Cytokine expression of KLRG1+ ILC-2s were evaluated in the various cohorts (H). For in vivo experiments, each cohort had n = 4 mice; data are represented as mean ± SEM. A Student’s t test was performed to determine statistical significance in C and D, and a one-way ANOVA analysis followed by a multiple comparison test [Tukey] was performed to determine statistical significance between the various cohorts in F–H. P ≤ 0.05 was considered significant. Significant p-values are denoted in the figures.
We next investigated if blocking PD-1 by antibody treatment increased human ILC-2 numbers and function in vivo. Initial experiments suggested that similar to our murine data, IL-33 was the most efficient stimuli that induced robust proliferation of PD-1+ ILC-2 cells (Fig. S4 G). NSG recipients were reconstituted with PBMCs and then treated with anti-PD-1 antibody along with rhIL-33 for 3 d. We found that blocking PD-1 significantly enhanced cytokine production in GATA3+KLRG1+ cell subsets (Fig. 7 h). The enhanced cytokine secretion capacity of ILC-2s was inhibited in cohorts that received Tofacitinib in addition to rhIL-33 and αPD-1 antibody treatment. Therefore, in humans PD-1 negatively regulates KLRG1+ ILC-2 cells by modulating the STAT5 activation pathway.
Discussion
Approximately one-third of the world’s population is infected with one or more types of parasitic helminth worms however treatment options for these ubiquitous infections are limited (Hotez et al., 2008). Type 2 immune responses, which are initiated by ILC-2s, are pivotal in clearing helminth worms in adults, but the regulatory pathways that can modulate the function of ILC-2s are still largely unexplored. Emerging literature on ILC-2 populations in mice and humans (Maazi et al., 2015; Molofsky et al., 2015; Paclik et al., 2015; Seillet et al., 2016; Yu et al., 2016), combined with the data presented here, has identified a pivotal role for co-receptor–mediated regulation of ILC-2s. Whereas immune-activating signals that maintain ILC-2s have been extensively studied, there is paucity in understanding the regulatory pathways that modulate ILC-2 cell number and function in vivo.
This report is the first to demonstrate that IL-33–responsive KLRG1+ ILC-2s are regulated by PD-1 in both mice and humans. When ILC-2s are activated by IL-33 (through the IL-33 receptor ST2), they respond by secreting type 2 cytokines and up-regulate KLRG1. Our data indicates that PD-1 selectively inhibits the proliferation of KLRG1+ ILC-2s via modulating STAT5 function. Here, we propose a new regulatory axis, whereby activated mature KLRG1+ ILC-2s up-regulate PD-1 on exposure to IL-33. PD-1 then inhibits uncontrolled expansion of ILC-2 effector population by blocking proliferation. PD-1, therefore, is a critical negative checkpoint and contributes toward maintaining KLRG1+ ILC-2 cell numbers.
Recent studies indicated that PD-1 is a novel maker for ILC precursors (Yu et al., 2016), but whether PD-1 could regulate the development and function of ILCs, particularly ILC-2s, was unclear. We observed that, first, the frequency of mature KLRG1+ ILC-2s was increased in the Pdcd1−/− mice; second, reconstituted Pdcd1−/− ILC-2 cells were capable of secreting functional cytokines; and third, ILC-2 cell development was not compromised when BM from Pdcd1−/− mice was adoptively transferred into immune-deficient Rag2−/−γc−/− mice. Notably, Seillet et al. (2016) observed that Pdcd1−/− mice possess a similar frequency of ILC-2 precursors to WT mice. In addition, PD-1 has been suggested to be a marker on ILC-2 precursors (Yu et al., 2016). Our study is the first to identify a mechanistic role for PD-1 in mature ILC-2 and propose that although PD-1 is expressed on ILC-2 precursors, lack of PD-1 does not alter ILC-2 development or their cytokine production capacity in mixed chimera experiments.
In keeping with most ILC subsets, ILC-2s are responsive to γc cytokines (Bartemes et al., 2012) and explicitly depend on γc cytokines and STAT5 signaling for proliferation and survival. A specific increase in proliferation was noted in the Pdcd1−/− ILC-2s, which then led us to investigate the interplay between PD-1 and STAT5 signaling in these cells. Notably, STAT5 signaling can be modulated by co-receptors such as ICOS in ILC-2s (Maazi et al., 2015), and PD-1 signaling can inhibit T cell function and proliferation by dampening STAT5 (Francisco et al., 2009). In agreement with these previous studies, a significant increase in STAT5 phosphorylation and proliferation was noted in KLRG1+ ILC-2 numbers in Pdcd1−/− recipients, which was conserved in human ILC-2s. In all these studies (Francisco et al., 2009; Maazi et al., 2015), the PD-1 activating ligand that was primarily responsible for inhibiting STAT5 signaling is PDL-1, and hence we used a similar experimental methodology in the current study. We found that PDL-1 binding to PD-1 specifically down-regulated STAT5 activation in KLRG1+ ILC-2s.
Although the importance of PD-1 signaling in inhibiting STAT5 activation has been previously suggested in T cells (Francisco et al., 2009), two caveats remain. First, no direct mechanistic evidence has linked these pathways; second, it is not clear whether similar mechanisms exist in ILC-2s. Our in vitro and in vivo mechanistic data in murine and human ILC-2s has demonstrated that inhibiting or inducing PD1 signaling pathway can alter STAT5 activation on KLRG1+ILC-2s. Importantly, PD-1 does not operate independently of STAT5 in promoting ILC-2 cell proliferation.
In addition to cellular mechanisms, we sought to verify if Pdcd1−/− KLRG1+ ILC-2s exhibited a gene profile that correlated with enhanced effector function. The microarray data suggested that ILC-2s deficient in PD-1 had increased expression of STAT5 target genes such as slamf1 (Wang et al., 2015), and STAT5 activating genes such as csf1 (Novak et al., 1996). Of note, the gene profile of Pdcd1−/− ILC-2s followed a pattern that is found in effector memory CD8+ T cells. In particular, we found that klra9, which identifies exhaustion in CD8+ T cells (Wherry et al., 2007), was down-regulated in Pdcd1−/− ILC-2s. Gene set enrichment analysis further correlated with the cellular mechanistic assays in highlighting the enhanced effector function of Pdcd1−/− KLRG1+ILC-2s.
The function of Pdcd1−/− KLRG1+ILC-2 cells was found to be superior to their WT counterparts in eliciting host-mediated pathogen responses in in vivo worm expulsion experiments. Given that KLRG1+ ILC-2 subsets are important in the expulsion of worms (Huang et al., 2015), the potency of these Pdcd1−/− KLRG1+ILC-2 cells in Rag2−/−γc−/− identifies a new therapeutic role for PD-1 in inducing robust type 2 responses. Furthermore, the differential numbers of KLRG1+ILC-2 cells in the MLNs of Pdcd1−/− mice during infection suggests the importance of PD-1 as a checkpoint modulator that can regulate ILC-2 cell numbers in the MLN during host–pathogen–mediated immune responses. Of note is the lack of difference in ILC-2 cell numbers in the lungs of infected Pdcd1−/− mice. N. brasiliensis life cycle includes migration through the lungs; however, a clear difference in WT and Pdcd1−/− mice with respect to ILC-2 cell numbers was not observed. Hence, alternate regulatory mechanisms independent of PD-1 may inhibit ILC-2s in different organs during parasitic infections. This differential increase in ILC-2 numbers could be important in translating these findings to helminth immunity.
To confirm the potential therapeutic benefits in host–pathogen–mediated immune responses, PD-1 antibody experiments were performed in Rag1−/− recipients that were infected with N. brasiliensis. To date, the data presented here is the first to show that αPD-1 therapy results in an increase in KLRG1+ ILC-2 numbers, which is reflected in a significant decrease in worm burden. These results suggest that blocking PD-1 can skew immune responses to type 2 phenotype, thereby inducing strong antiparasitic responses. In addition, this observation can be extrapolated to the cancer immunotherapy trials (Topalian et al., 2012; Yaqub, 2015), whereby the primary reason for reduced autoimmune colitis in patients who receive PD-1 therapy can be attributed to increased type 2 responses (Ruyssers et al., 2009; Bashi et al., 2015).
Lastly, the murine N. brasiliensis experiments in Rag−/−γc−/− mice demonstrated that PD-1 intrinsically regulates ILC-2 cell function with minimal contribution from bystander immune cells. In the adoptive transfer experiments, murine recipients reconstituted with Pdcd1−/− ILC-2s had significantly lower worm burden than the WT cohorts. These data demonstrate that an intrinsic defect in PD-1 signaling in ILC-2s contribute to robust antiparasitic helminth responses. Indeed, if PD-1 elicits its function through bystander immune cells, no difference between cohorts that received WT and Pdcd1−/− ILC-2s will be noted.
In summary, we have demonstrated that ILC-2 cells are regulated by PD-1, lack of which enhances KLRG1+ ILC-2 cell numbers and effector function. The cellular signaling mechanism by which KLRG1+ ILC-2 cell numbers are increased in Pdcd1−/− mice is through enhanced STAT5 activation, which results in elevated γc cytokine–mediated cell proliferation and cytokine secretion. Collectively, these data represent an ideal translational route to manipulating ILC-2 cell numbers in vivo. Therefore, PD-1 blockade may be beneficial in situations that require significant ILC-2 cell numbers (helminth immunity) and this effect can be controlled by Tofacitinib to prevent deleterious uncontrolled type 2 inflammatory processes.
Materials and methods
Mice
Wild-type C57BL/6 (B6) and B6.Pdcd1−/− littermates were bred and maintained in a specific pathogen–free facility at the National Cancer Institute (NCI; Bethesda, MD). Mice were housed and maintained according to National Institutes of Health guidelines and all experiments were performed in accordance with the NCI Animal Care and Use Approval and institutional guidelines. All mice used in the experiments were age and sex-matched. CD45.1+ C57BL6, NOD-SCID-γc−/− (NSG) and B6. Rag2−/−γc−/− mice were obtained from Jackson Laboratories and maintained at the NCI or at Newcastle University, UK under a home office approved project license. All the experimental procedures performed at Newcastle University incorporated the NC3R guidelines for animal research, and the data were presented according to the ARRIVE guidelines.
Isolation of ILC-2 cells and ex vivo stimulation
Spleens and MLNs were mechanically crushed into single-cell suspensions. Lungs and small intestine were digested in Liberase TL (Roche) and DNase (Roche; Asseman et al., 2003), and ILCs were isolated as previously described. For skin ILCs, a small piece of mouse dorsal skin was removed, minced, and incubated with serum-free media containing Liberase TL and DNase at 37°C. Single-cell suspensions were prepared by mashing the tissue slurry through a 100-µm nylon mesh. Cells were sequentially filtered through 70- and 40-µm nylon filters. To assess production of cytokines, both murine and human ILC-2s were stimulated for 4 h in complete RPMI + 10% FBS with cell stimulation cocktail (PMA, ionomycin, Brefeldin A, and monensin; eBioscience) as per manufacturer’s instruction, followed by intracellular staining. Unstimulated controls were used to determine gating strategy for flow cytometric plots in figures. All human-derived materials were collected under an approved ethics plan (Durham and Tees Valley Research Ethics Committee [12/NE/0121], granted 16th April 2012).
BM mixed chimera experiments
BM cells from B6 or B6.Pdcd1−/− mice were harvested and T cell depleted. Congenic recipients B6.CD45.1+ mice were conditioned with total body irradiation (1,050 cGy) and rescued with 10million T-depleted BM (TDBM) cells from B6.CD45.1+ WT or B6 CD45.2+ Pdcd-1−/−mice. 8 wk later, lungs and small intestine were digested and immune cells were isolated followed by ILC-2 subsets characterization. Experiments were repeated in host B6.Rag2−/−γc−/− mice, where mice were reconstituted with either congenic WT BM (CD45.1) or Pdcd1−/−BM (CD45.2).
Flow cytometry
The following fluorochrome-conjugated antibodies were used for flow cytometry. Anti-CD3ε (145-2c11), anti-CD8 (53-6.7), anti-CD19 (6D5), anti–Gr-1 (RB6-8C5), anti-KLRG1 (2F1/KLRG1), anti-CD25 (PL61), anti-CD45.2 (104), anti–PD-1 (RMP1-30), anti-CD4 (GK1.5), anti–IL-5 (TRFK5), anti-CD45.1 (A20), anti-CD127 (A7R34), anti-CD25 (PC61), anti-CD5 (53-7.3), anti-CD11c (N418), anti-CD11b (M170), anti-Ter119, anti-CD45R/B220, and anti-F4/80 (BM8) were obtained from BioLegend. Anti–IL-4 (11B11) and anti-Thy1 (53–2.1) were obtained from BD. Anti-KLRG1 (2F1), anti-Ki67 (SoLA15), anti-Gata3 (TWAJ), and anti–IL-13 (eBio13A) were purchased from eBioscience. Anti-ST2 (DJ8) was obtained from MD Bioproducts. Single-cell suspensions were generated from indicated organs, and then incubated with anti-Fc receptor antibody (2.4G2) and stained with Lin PE-Cy5.5 (CD3+, CD4+, CD8+, NK1.1+, Gr1+, CD5+, Ter119+, CD11b+, CD11c+, F4/80+, CD45R/B220+, and CD19+), CD45 APC-Cy7, KLRG1 BV421 or APC-Cy7, ST2 FITC, GATA3 efluor660, and PD-1 BV605, CD127 BV510 or BV711, CD25 Pe-cy7, PDL-1 BV711, Thy1 PE. For intracellular (IC) cytokine measurement, cells were fixed, permeabilized, and stained with IL-13PE, IL-4 BV605, and IL-5 BV421. For human experiments, PBMCs were stained with Lin BV510 (CD19+, CD56+, CD3+, CD20+, CD16+, CD11c+, CD11b+, CD5+, TCRα/β+, and CD14+), PD-1 APC-Cy7 (clone EH12.2H7) or PD-1 Pe-Cy7 (clone:J10S), GATA3 efluor 660, KLRG1 PerCPefluor710 (clone 13F12F2), CD45 FITC (clone 2D1) or CD45 PeCy7 (clone H130), IL13 FITC (clone: JES10-SA2), IL5 BV421, and ST2 APC (R&D systems). ILCs were detected using the following gating strategy. First cells were gated as Lin−CD45+Thy1+, and then ILCs were defined as Lin−CD45+Thy1+ CD25+CD127+. ILC-2s were defined as Lin−CD45+Thy1+ CD25+CD127+ST2+KLRG1+. In human cells, ILC-2s were defined as Lin−CD45+CD127+GATA3+. Cells were analyzed using LSRII flow cytometer with FACSDiva software (BD) and analysis was performed with FlowJo 10.1 software (Tree Star).
In vivo ILC-2 cell stimulation assays
WT or Pdcd1−/− mice were treated with rmIL-33 (R&D Systems; 200 ng/mice) or PBS for 3 d, and then lungs were isolated for ILC-2 characterization and functional phospho STAT-5 phosphorylation assays. In certain experiments, Rag1−/− mice were treated with either αPD-1 antibody (BioXcell; clone RMP1-14; 250 µg/mice/d) or mouse IgG2a isotype control. After antibody treatment, mice were treated with rm-IL-33 or PBS, and then lungs were isolated for ILC-2 characterization. For human into murine experiments, PBMCs were injected into NSG mice (4 million/mouse), followed by i.p injection of rh-IL-2 (500 ng/mouse/d), rh-IL-7 (500 ng/mouse/d), and rh-IL-33 (200 ng/mouse/d). Certain cohorts were treated with mouse isotype control (250 µg/mouse/day) and DMSO Vehicle or with αPD-1 (250 µg/mouse/d) and Tofacitinib (15 mg/kg/d/mouse) for 3 d. Lungs were harvested at day 5 after adoptive transfer, and then human ILC-2 numbers and function were evaluated.
STAT5 phosphorylation assays
For STAT-5 phosphorylation assays, WT and Pdcd1−/−mice were treated with either PBS or rmIL-33 once, and then lungs were isolated the next day. ILC-2s were stimulated with IL-2 (100 IU) for 15 min, and then cells were stained for p-STAT5 and analyzed by flow cytometry. For PD-1 signaling assays, WT and Pdcd1−/− mice were treated with either PBS or rmIL-33 for 3 d, and then lungs were isolated. ILC-2s were stimulated with IL-2 (100 IU) alone or IL-2 plus plate-coated PDL-1-Fc chimera (5 µg/ml) for 15 min, and then stained with p-STAT5 antibody. For PDL-1-Fc chimera coating, PDL-1-Fc was resuspended in PBS (5 µg/ml), and then added to wells in a 24-well plate. The plate was then incubated for 3 h at 37°C and then washed three times with PBS and then used for cell culture. In a second set of experiments, WT ILC-2 cells were treated with rhIL-2 (40 ng/ml), rmIL-7 (40 ng/ml), and rmIL-33 (40 ng/ml). In addition, cohorts were treated with either isotype control antibody (10 µg/ml) or αPD-1 (10 µg/ml), or NSC87877 (SHP1/2 selective inhibitor; 10 µM resuspended in complete mouse media), PDL-1 fc (coated onto plates, and then washed with PBS three times before cell culture), or PDL1 fc plus NSC87877 for 24 h. ILC-2s were then restimulated with IL-2 for 15 min, and then subjected to p-STAT5 flow cytometry. For nuclear analysis of STAT5a/b, ILC-2s were stimulated for 72 h under similar conditions, and then restimulated with IL-2 for 15 min. Nuclear lysates were prepared according to the manufacturer’s instructions (Abcam), and then subjected to transcription factor ELISA-based assay (Abcam). Similar culture methodology was used for detecting human nuclear STAT5 activity in human ILC-2s. For in vivo STAT5 inhibition experiments, WT or Pdcd1−/− mice were treated with rmIL-33 plus DMSO or rmIL-33 plus Tofacitinib (15 mg/kg/day) i.p for 3 d. Lungs were evaluated for KLRG1+ILC-2 cell number and function after treatment.
N. brasiliensis infection
Mice were given subcutaneous injection of 300 third-stage N. brasiliensis larvae (provided by J.F. Urban Jr., US Department of Agriculture) and cell transfer was performed on the same day if required. Mouse feces were collected from each individual mouse on day 7–9 after infection, and eggs were counted. Worm burden in small intestines was measured on day 14.
Proliferation assays
For cell trace violet proliferation assays, murine ILC-2s and human ILC-2s were stained with cell trace violet in serum-free media as per manufacturer’s instructions. For murine cultures, ILC-2s were stimulated with rhIL-2 (40 ng/ml), rmIL-7 (40 ng/ml), and rmIL-33 (40 ng/ml), or rmIL-25 (40 ng/ml), or TSLP (40 ng/ml), and cultured for 72 h in cell culture media (RPMI, supplemented with 10% FCS, glutamine [2 mM], nonessential amino acids [0.1 mM], 2-mercaptoethanol [50 µM], sodium pyruvate [1 mM], penicillin and streptomycin [100 U/ml]), and then proliferation was measured by flow cytometry. Human ILC-2s were expanded in the presence of rhIL-2 (100 IU), rhIL-7 (40 ng/ml). Certain cohorts were treated with αPD-1 (20 µg/ml) or isotype control (20 µg/ml) alone with either Vehicle (DMSO) or Tofacitinib (0.3 µM). Where indicated, rhIL-33 (40 ng/ml) or rhIL-25 (40 ng/ml), or rhTSLP (40 ng/ml) or rhIL-4 (40 ng/ml), was added to the cultures. Cells were expanded for 5 d, and then proliferation was measured by flow cytometry.
Microarray
For microarray analysis, flow sorted ILC-2s were pelleted, and then resuspended in TRIzol and shipped to Central Biotechnology Services (CBS; Cardiff University, Cardiff, Wales, UK). Total RNA extraction and microarray experiment was performed by the CBS. In brief, total RNA was isolated, and RNA quality was checked on Agilent Bioanalyzer. All samples used for microarray analysis have high quality score (RIN > 9). RNA was reverse transcribed and amplified, and sense strand cDNA was fragmented and labeled using Affymetrix WT Pico terminal labeling kit. Three replicates (each replicate was from five mice) of each group were hybridized to Affymetrix mouse Gene ST 2.0 GeneChip in Affymetrix hybridization oven at 45°C, 60 rpm for 16 h. Washing and staining were performed on Affymetrix Fluidics Station 450 and scanned on Affymetrix GeneChip scanner 3000. Data were collected using Affymetrix AGCC software. The data discussed in this publication have been deposited in NCBI Gene Expression Omnibus (GEO) and are accessible under GEO accession no. GSE94609. The R/Bioconductor package oligo (Carvalho and Irizarry, 2010) was used for preprocessing and normalization of the microarray data using RMA algorithm (Irizarry et al., 2003). The normalized expression values are log2-scaled, and probes were annotated (MacDonald, 2016). After excluding genes with low variance across arrays (<50% quantile) using genefilter (Bourgon et al., 2010) or without gene annotation, differentially expressed genes were identified as those with at least twofold differences and with FDR < 0.15 using limma (Ritchie et al., 2015). Gene Ontology enrichment analysis for up-regulated genes was conducted using DAVID bioinformatics resources (Huang et al., 2009). The GSEA software (Subramanian et al., 2005) was used for gene set enrichment analysis. The gene expression data were compared against the curated Immunological Signatures gene set (C7.all.v5.2), with the permutation type set as gene_set.
Statistics
Results are expressed as mean with error bars showing ±SEM unless otherwise indicated. Statistical significance was determined for normally distributed data by using a Student’s t test or a one-way ANOVA, followed by appropriate multiple comparison tests. All analysis were performed using Prism 7.0 (GraphPad Software), and differences were considered significant when P ≤ 0.05.
Online supplemental material
Fig. S1 shows gating strategy for ILC analysis, cytokine receptor expression, and PD-1 expression during homeostasis and cytokine stimulation on ILC-2s. Fig. S2 shows the proliferative effect of ILC-2 driving cytokines on WT and Pdcd1−/− ILC-2s. Effect of in vivo IL-25 administration on ILC-2 cell numbers in WT and Pdcd1−/− ILC-2s, and the importance of PD-1 and STAT5 signaling in WT and Pdcd1−/− ILC-2s. Fig. S3 demonstrates the changes in ILC-2 numbers during helminth infection in WT and Pdcd1−/− mice. Fig. S4 demonstrates the significance of PD-1 in human KLRG1+ ILC-2s.
Supplementary Material
Acknowledgments
We thank J.F. Urban Jr. at the US Department of Agriculture for providing N. brasiliensis larvae. We would like to thank Dr. Amanda Redfern, Central Biotechnology Services, Cardiff University, for performing microarray experiments.
This work was supported by the Intramural Research Program, National Institutes of Health and a Newcastle University Research Fellowship, Newcastle University. N.J. Reynolds’s research is supported by the Newcastle NIHR Biomedical Research Centre and the Newcastle MRC/EPSRC Molecular Pathology Node.
N.J. Reynolds’s research group has received grant and consultancy income from Genentech through Newcastle University. The authors declare no additional competing financial interests.
Author contributions: S. Taylor performed experiments and analyzed data. Y. Huang performed N. brasiliensis experiments and wrote the paper. G. Mallett, C. Stathopolou, E.L. Martin, T.C. Felizardo, E.L. Woodward, J. Scott, and M.S. Elias performed experiments. M.-A. Sun analyzed microarray data. N.J. Reynolds critically read the manuscript and helped write the paper. W.E. Paul provided intellectual input and support for N. brasiliensis experiments. D.H. Fowler reviewed and edited the manuscript. S. Amarnath conceptualized, supervised, and planned experiments and wrote the paper.
Footnotes
Abbreviations used:
- ILC
- innate lymphoid cell
- TSLP
- thymic stromal lymphopoietin
References
- Agata Y., Kawasaki A., Nishimura H., Ishida Y., Tsubata T., Yagita H., and Honjo T.. 1996. Expression of the PD-1 antigen on the surface of stimulated mouse T and B lymphocytes. Int. Immunol. 8:765–772. 10.1093/intimm/8.5.765 [DOI] [PubMed] [Google Scholar]
- Amarnath S., Costanzo C.M., Mariotti J., Ullman J.L., Telford W.G., Kapoor V., Riley J.L., Levine B.L., June C.H., Fong T., et al. 2010. Regulatory T cells and human myeloid dendritic cells promote tolerance via programmed death ligand-1. PLoS Biol. 8:e1000302 10.1371/journal.pbio.1000302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amarnath S., Mangus C.W., Wang J.C., Wei F., He A., Kapoor V., Foley J.E., Massey P.R., Felizardo T.C., Riley J.L., et al. 2011. The PDL1-PD1 axis converts human TH1 cells into regulatory T cells. Sci. Transl. Med. 3:111ra120 10.1126/scitranslmed.3003130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asseman C., Read S., and Powrie F.. 2003. Colitogenic Th1 cells are present in the antigen-experienced T cell pool in normal mice: control by CD4+ regulatory T cells and IL-10. J. Immunol. 171:971–978. 10.4049/jimmunol.171.2.971 [DOI] [PubMed] [Google Scholar]
- Barber D.L., Wherry E.J., Masopust D., Zhu B., Allison J.P., Sharpe A.H., Freeman G.J., and Ahmed R.. 2006. Restoring function in exhausted CD8 T cells during chronic viral infection. Nature. 439:682–687. 10.1038/nature04444 [DOI] [PubMed] [Google Scholar]
- Barber D.L., Mayer-Barber K.D., Feng C.G., Sharpe A.H., and Sher A.. 2011. CD4 T cells promote rather than control tuberculosis in the absence of PD-1-mediated inhibition. J. Immunol. 186:1598–1607. 10.4049/jimmunol.1003304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barlow J.L., Bellosi A., Hardman C.S., Drynan L.F., Wong S.H., Cruickshank J.P., and McKenzie A.N.. 2012. Innate IL-13-producing nuocytes arise during allergic lung inflammation and contribute to airways hyperreactivity. J. Allergy Clin. Immunol. 129:191–8.e1: 4. 10.1016/j.jaci.2011.09.041 [DOI] [PubMed] [Google Scholar]
- Barlow J.L., Peel S., Fox J., Panova V., Hardman C.S., Camelo A., Bucks C., Wu X., Kane C.M., Neill D.R., et al. 2013. IL-33 is more potent than IL-25 in provoking IL-13-producing nuocytes (type 2 innate lymphoid cells) and airway contraction. J. Allergy Clin. Immunol. 132:933–941. 10.1016/j.jaci.2013.05.012 [DOI] [PubMed] [Google Scholar]
- Bartemes K.R., Iijima K., Kobayashi T., Kephart G.M., McKenzie A.N., and Kita H.. 2012. IL-33-responsive lineage- CD25+ CD44(hi) lymphoid cells mediate innate type 2 immunity and allergic inflammation in the lungs. J. Immunol. 188:1503–1513. 10.4049/jimmunol.1102832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bashi T., Bizzaro G., Ben-Ami Shor D., Blank M., and Shoenfeld Y.. 2015. The mechanisms behind helminth’s immunomodulation in autoimmunity. Autoimmun. Rev. 14:98–104. 10.1016/j.autrev.2014.10.004 [DOI] [PubMed] [Google Scholar]
- Beswick E.J., Pinchuk I.V., Das S., Powell D.W., and Reyes V.E.. 2007. Expression of the programmed death ligand 1, B7-H1, on gastric epithelial cells after Helicobacter pylori exposure promotes development of CD4+ CD25+ FoxP3+ regulatory T cells. Infect. Immun. 75:4334–4341. 10.1128/IAI.00553-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourgon R., Gentleman R., and Huber W.. 2010. Independent filtering increases detection power for high-throughput experiments. Proc. Natl. Acad. Sci. USA. 107:9546–9551. 10.1073/pnas.0914005107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brestoff J.R., Kim B.S., Saenz S.A., Stine R.R., Monticelli L.A., Sonnenberg G.F., Thome J.J., Farber D.L., Lutfy K., Seale P., and Artis D.. 2015. Group 2 innate lymphoid cells promote beiging of white adipose tissue and limit obesity. Nature. 519:242–246. 10.1038/nature14115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carvalho B.S., and Irizarry R.A.. 2010. A framework for oligonucleotide microarray preprocessing. Bioinformatics. 26:2363–2367. 10.1093/bioinformatics/btq431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang Y.J., Kim H.Y., Albacker L.A., Baumgarth N., McKenzie A.N., Smith D.E., Dekruyff R.H., and Umetsu D.T.. 2011. Innate lymphoid cells mediate influenza-induced airway hyper-reactivity independently of adaptive immunity. Nat. Immunol. 12:631–638. 10.1038/ni.2045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das S., Suarez G., Beswick E.J., Sierra J.C., Graham D.Y., and Reyes V.E.. 2006. Expression of B7-H1 on gastric epithelial cells: its potential role in regulating T cells during Helicobacter pylori infection. J. Immunol. 176:3000–3009. 10.4049/jimmunol.176.5.3000 [DOI] [PubMed] [Google Scholar]
- Day C.L., Kaufmann D.E., Kiepiela P., Brown J.A., Moodley E.S., Reddy S., Mackey E.W., Miller J.D., Leslie A.J., DePierres C., et al. 2006. PD-1 expression on HIV-specific T cells is associated with T-cell exhaustion and disease progression. Nature. 443:350–354. 10.1038/nature05115 [DOI] [PubMed] [Google Scholar]
- Dong H., Zhu G., Tamada K., and Chen L.. 1999. B7-H1, a third member of the B7 family, co-stimulates T-cell proliferation and interleukin-10 secretion. Nat. Med. 5:1365–1369. 10.1038/70932 [DOI] [PubMed] [Google Scholar]
- Duraiswamy J., Ibegbu C.C., Masopust D., Miller J.D., Araki K., Doho G.H., Tata P., Gupta S., Zilliox M.J., Nakaya H.I., et al. 2011. Phenotype, function, and gene expression profiles of programmed death-1(hi) CD8 T cells in healthy human adults. J. Immunol. 186:4200–4212. 10.4049/jimmunol.1001783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fallon P.G., Ballantyne S.J., Mangan N.E., Barlow J.L., Dasvarma A., Hewett D.R., McIlgorm A., Jolin H.E., and McKenzie A.N.. 2006. Identification of an interleukin (IL)-25-dependent cell population that provides IL-4, IL-5, and IL-13 at the onset of helminth expulsion. J. Exp. Med. 203:1105–1116. 10.1084/jem.20051615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fife B.T., Pauken K.E., Eagar T.N., Obu T., Wu J., Tang Q., Azuma M., Krummel M.F., and Bluestone J.A.. 2009. Interactions between PD-1 and PD-L1 promote tolerance by blocking the TCR-induced stop signal. Nat. Immunol. 10:1185–1192. 10.1038/ni.1790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franceschini D., Paroli M., Francavilla V., Videtta M., Morrone S., Labbadia G., Cerino A., Mondelli M.U., and Barnaba V.. 2009. PD-L1 negatively regulates CD4+CD25+Foxp3+ Tregs by limiting STAT-5 phosphorylation in patients chronically infected with HCV. J. Clin. Invest. 119:551–564. 10.1172/JCI36604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francisco L.M., Salinas V.H., Brown K.E., Vanguri V.K., Freeman G.J., Kuchroo V.K., and Sharpe A.H.. 2009. PD-L1 regulates the development, maintenance, and function of induced regulatory T cells. J. Exp. Med. 206:3015–3029. 10.1084/jem.20090847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halim T.Y., MacLaren A., Romanish M.T., Gold M.J., McNagny K.M., and Takei F.. 2012. Retinoic-acid-receptor-related orphan nuclear receptor alpha is required for natural helper cell development and allergic inflammation. Immunity. 37:463–474. 10.1016/j.immuni.2012.06.012 [DOI] [PubMed] [Google Scholar]
- Hotez P.J., Brindley P.J., Bethony J.M., King C.H., Pearce E.J., and Jacobson J.. 2008. Helminth infections: the great neglected tropical diseases. J. Clin. Invest. 118:1311–1321. 10.1172/JCI34261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang W., Sherman B.T., and Lempicki R.A.. 2009. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 4:44–57. 10.1038/nprot.2008.211 [DOI] [PubMed] [Google Scholar]
- Huang Y., Guo L., Qiu J., Chen X., Hu-Li J., Siebenlist U., Williamson P.R., Urban J.F. Jr., and Paul W.E.. 2015. IL-25-responsive, lineage-negative KLRG1(hi) cells are multipotential ‘inflammatory’ type 2 innate lymphoid cells. Nat. Immunol. 16:161–169. 10.1038/ni.3078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irizarry R.A., Hobbs B., Collin F., Beazer-Barclay Y.D., Antonellis K.J., Scherf U., and Speed T.P.. 2003. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics. 4:249–264. 10.1093/biostatistics/4.2.249 [DOI] [PubMed] [Google Scholar]
- Keir M.E., Butte M.J., Freeman G.J., and Sharpe A.H.. 2008. PD-1 and its ligands in tolerance and immunity. Annu. Rev. Immunol. 26:677–704. 10.1146/annurev.immunol.26.021607.090331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koyasu S., Moro K., Tanabe M., and Takeuchi T.. 2010. Natural helper cells: a new player in the innate immune response against helminth infection. Adv. Immunol. 108:21–44. 10.1016/B978-0-12-380995-7.00002-1 [DOI] [PubMed] [Google Scholar]
- Latchman Y., Wood C.R., Chernova T., Chaudhary D., Borde M., Chernova I., Iwai Y., Long A.J., Brown J.A., Nunes R., et al. 2001. PD-L2 is a second ligand for PD-1 and inhibits T cell activation. Nat. Immunol. 2:261–268. 10.1038/85330 [DOI] [PubMed] [Google Scholar]
- Lee M.W., Odegaard J.I., Mukundan L., Qiu Y., Molofsky A.B., Nussbaum J.C., Yun K., Locksley R.M., and Chawla A.. 2015. Activated type 2 innate lymphoid cells regulate beige fat biogenesis. Cell. 160:74–87. 10.1016/j.cell.2014.12.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maazi H., Patel N., Sankaranarayanan I., Suzuki Y., Rigas D., Soroosh P., Freeman G.J., Sharpe A.H., and Akbari O.. 2015. ICOS:ICOS-ligand interaction is required for type 2 innate lymphoid cell function, homeostasis, and induction of airway hyperreactivity. Immunity. 42:538–551. 10.1016/j.immuni.2015.02.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacDonald J.W.2016. mogene20sttranscriptcluster.db: Affymetrix mogene20 annotation data (chip mogene20sttranscriptcluster). R package version 8.5.0. http://bioconductor.org/packages/clariomdhumanprobeset.db/
- Molofsky A.B., Van Gool F., Liang H.E., Van Dyken S.J., Nussbaum J.C., Lee J., Bluestone J.A., and Locksley R.M.. 2015. Interleukin-33 and interferon-γ counter-regulate group 2 innate lymphoid cell activation during immune perturbation. Immunity. 43:161–174. 10.1016/j.immuni.2015.05.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monticelli L.A., Sonnenberg G.F., Abt M.C., Alenghat T., Ziegler C.G., Doering T.A., Angelosanto J.M., Laidlaw B.J., Yang C.Y., Sathaliyawala T., et al. 2011. Innate lymphoid cells promote lung-tissue homeostasis after infection with influenza virus. Nat. Immunol. 12:1045–1054. 10.1038/ni.2131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moro K., Yamada T., Tanabe M., Takeuchi T., Ikawa T., Kawamoto H., Furusawa J., Ohtani M., Fujii H., and Koyasu S.. 2010. Innate production of TH2 cytokines by adipose tissue-associated c-Kit+Sca-1+ lymphoid cells. Nature. 463:540–544. 10.1038/nature08636 [DOI] [PubMed] [Google Scholar]
- Motomura Y., Morita H., Moro K., Nakae S., Artis D., Endo T.A., Kuroki Y., Ohara O., Koyasu S., and Kubo M.. 2014. Basophil-derived interleukin-4 controls the function of natural helper cells, a member of ILC2s, in lung inflammation. Immunity. 40:758–771. 10.1016/j.immuni.2014.04.013 [DOI] [PubMed] [Google Scholar]
- Neill D.R., Wong S.H., Bellosi A., Flynn R.J., Daly M., Langford T.K., Bucks C., Kane C.M., Fallon P.G., Pannell R., et al. 2010. Nuocytes represent a new innate effector leukocyte that mediates type-2 immunity. Nature. 464:1367–1370. 10.1038/nature08900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novak U., Mui A., Miyajima A., and Paradiso L.. 1996. Formation of STAT5-containing DNA binding complexes in response to colony-stimulating factor-1 and platelet-derived growth factor. J. Biol. Chem. 271:18350–18354. 10.1074/jbc.271.31.18350 [DOI] [PubMed] [Google Scholar]
- Okazaki T., Maeda A., Nishimura H., Kurosaki T., and Honjo T.. 2001. PD-1 immunoreceptor inhibits B cell receptor-mediated signaling by recruiting src homology 2-domain-containing tyrosine phosphatase 2 to phosphotyrosine. Proc. Natl. Acad. Sci. USA. 98:13866–13871. 10.1073/pnas.231486598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okazaki T., Iwai Y., and Honjo T.. 2002. New regulatory co-receptors: inducible co-stimulator and PD-1. Curr. Opin. Immunol. 14:779–782. 10.1016/S0952-7915(02)00398-9 [DOI] [PubMed] [Google Scholar]
- Paclik D., Stehle C., Lahmann A., Hutloff A., and Romagnani C.. 2015. ICOS regulates the pool of group 2 innate lymphoid cells under homeostatic and inflammatory conditions in mice. Eur. J. Immunol. 45:2766–2772. 10.1002/eji.201545635 [DOI] [PubMed] [Google Scholar]
- Parry R.V., Chemnitz J.M., Frauwirth K.A., Lanfranco A.R., Braunstein I., Kobayashi S.V., Linsley P.S., Thompson C.B., and Riley J.L.. 2005. CTLA-4 and PD-1 receptors inhibit T-cell activation by distinct mechanisms. Mol. Cell. Biol. 25:9543–9553. 10.1128/MCB.25.21.9543-9553.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price A.E., Liang H.E., Sullivan B.M., Reinhardt R.L., Eisley C.J., Erle D.J., and Locksley R.M.. 2010. Systemically dispersed innate IL-13-expressing cells in type 2 immunity. Proc. Natl. Acad. Sci. USA. 107:11489–11494. 10.1073/pnas.1003988107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rak G.D., Osborne L.C., Siracusa M.C., Kim B.S., Wang K., Bayat A., Artis D., and Volk S.W.. 2016. IL-33-dependent group 2 innate lymphoid cells promote cutaneous wound healing. J. Invest. Dermatol. 136:487–496. 10.1038/JID.2015.406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritchie M.E., Phipson B., Wu D., Hu Y., Law C.W., Shi W., and Smyth G.K.. 2015. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 43:e47 10.1093/nar/gkv007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruyssers N.E., De Winter B.Y., De Man J.G., Loukas A., Pearson M.S., Weinstock J.V., Van den Bossche R.M., Martinet W., Pelckmans P.A., and Moreels T.G.. 2009. Therapeutic potential of helminth soluble proteins in TNBS-induced colitis in mice. Inflamm. Bowel Dis. 15:491–500. 10.1002/ibd.20787 [DOI] [PubMed] [Google Scholar]
- Salimi M., Barlow J.L., Saunders S.P., Xue L., Gutowska-Owsiak D., Wang X., Huang L.C., Johnson D., Scanlon S.T., McKenzie A.N., et al. 2013. A role for IL-25 and IL-33-driven type-2 innate lymphoid cells in atopic dermatitis. J. Exp. Med. 210:2939–2950. 10.1084/jem.20130351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seillet C., Mielke L.A., Amann-Zalcenstein D.B., Su S., Gao J., Almeida F.F., Shi W., Ritchie M.E., Naik S.H., Huntington N.D., et al. 2016. Deciphering the innate lymphoid cell transcriptional program. Cell Reports. 17:436–447. 10.1016/j.celrep.2016.09.025 [DOI] [PubMed] [Google Scholar]
- Smith P., Walsh C.M., Mangan N.E., Fallon R.E., Sayers J.R., McKenzie A.N., and Fallon P.G.. 2004. Schistosoma mansoni worms induce anergy of T cells via selective up-regulation of programmed death ligand 1 on macrophages. J. Immunol. 173:1240–1248. 10.4049/jimmunol.173.2.1240 [DOI] [PubMed] [Google Scholar]
- Spits H., and Cupedo T.. 2012. Innate lymphoid cells: emerging insights in development, lineage relationships, and function. Annu. Rev. Immunol. 30:647–675. 10.1146/annurev-immunol-020711-075053 [DOI] [PubMed] [Google Scholar]
- Subramanian A., Tamayo P., Mootha V.K., Mukherjee S., Ebert B.L., Gillette M.A., Paulovich A., Pomeroy S.L., Golub T.R., Lander E.S., and Mesirov J.P.. 2005. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA. 102:15545–15550. 10.1073/pnas.0506580102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terrazas L.I., Montero D., Terrazas C.A., Reyes J.L., and Rodríguez-Sosa M.. 2005. Role of the programmed Death-1 pathway in the suppressive activity of alternatively activated macrophages in experimental cysticercosis. Int. J. Parasitol. 35:1349–1358. 10.1016/j.ijpara.2005.06.003 [DOI] [PubMed] [Google Scholar]
- Topalian S.L., Hodi F.S., Brahmer J.R., Gettinger S.N., Smith D.C., McDermott D.F., Powderly J.D., Carvajal R.D., Sosman J.A., Atkins M.B., et al. 2012. Safety, activity, and immune correlates of anti-PD-1 antibody in cancer. N. Engl. J. Med. 366:2443–2454. 10.1056/NEJMoa1200690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trautmann L., Janbazian L., Chomont N., Said E.A., Gimmig S., Bessette B., Boulassel M.R., Delwart E., Sepulveda H., Balderas R.S., et al. 2006. Upregulation of PD-1 expression on HIV-specific CD8+ T cells leads to reversible immune dysfunction. Nat. Med. 12:1198–1202. 10.1038/nm1482 [DOI] [PubMed] [Google Scholar]
- Wang S., and Chen L.. 2011. Immunobiology of cancer therapies targeting CD137 and B7-H1/PD-1 cosignal pathways. Curr. Top. Microbiol. Immunol. 344:245–267. [DOI] [PubMed] [Google Scholar]
- Wang Z., Medrzycki M., Bunting S.T., and Bunting K.D.. 2015. Stat5-deficient hematopoiesis is permissive for Myc-induced B-cell leukemogenesis. Oncotarget. 6:28961–28972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wherry E.J., Ha S.J., Kaech S.M., Haining W.N., Sarkar S., Kalia V., Subramaniam S., Blattman J.N., Barber D.L., and Ahmed R.. 2007. Molecular signature of CD8+ T cell exhaustion during chronic viral infection. Immunity. 27:670–684. 10.1016/j.immuni.2007.09.006 [DOI] [PubMed] [Google Scholar]
- Yaqub F. 2015. Nivolumab for squamous-cell non-small-cell lung cancer. Lancet Oncol. 16:e319 10.1016/S1470-2045(15)00033-9 [DOI] [PubMed] [Google Scholar]
- Yin H., Li X., Hu S., Liu T., Yuan B., Ni Q., Lan F., Luo X., Gu H., and Zheng F.. 2013. IL-33 promotes Staphylococcus aureus-infected wound healing in mice. Int. Immunopharmacol. 17:432–438. 10.1016/j.intimp.2013.07.008 [DOI] [PubMed] [Google Scholar]
- Yu Y., Tsang J.C., Wang C., Clare S., Wang J., Chen X., Brandt C., Kane L., Campos L.S., Lu L., et al. 2016. Single-cell RNA-seq identifies a PD-1(hi) ILC progenitor and defines its development pathway. Nature. 539:102–106. 10.1038/nature20105 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.