The innate sensor STING is critical for innate immune activation, but its role in adaptive immunity is unknown. By studying pathogenic mutations in STING found in patients, Cerboni et al. show that active STING inhibits proliferation in T lymphocytes.
Abstract
Activation of the cyclic dinucleotide sensor stimulator of interferon (IFN) genes (STING) is critical for IFN and inflammatory gene expression during innate immune responses. However, the role of STING in adaptive immunity is still unknown. In this study, we show that STING activation reduces the proliferation of T lymphocytes. This activity was independent of TBK1 and IRF3 recruitment and of type I IFN but required a distinct C-terminal domain of STING that activates NF-κB. Inhibition of cell proliferation by STING required its relocalization to the Golgi apparatus and caused mitotic errors. T lymphocytes from patients carrying constitutive active mutations in TMEM173 encoding STING showed impaired proliferation and reduced numbers of memory cells. Endogenous STING inhibited proliferation of mouse T lymphocytes. Therefore, STING, a critical innate sensor, also functions intrinsically in cells of the adaptive immune system to inhibit proliferation.
Introduction
Innate control of adaptive immunity relies on the paradigm that activation of innate sensors in specialized cells leads to extrinsic signals, such as cytokines, that instruct lymphocytes for adaptive immunity (Iwasaki and Medzhitov, 2015). However, innate sensors may adopt distinct activity when they function intrinsically in cells of adaptive immunity, such as T cells. The inflammasome receptor NLRP3 was recently identified as a transcription factor for T helper type 2 cells (Th2 cells), although this activity was not linked to the activation of NLRP3 (Bruchard et al., 2015). Here, we examined the activity adopted by stimulator of IFN genes (STING) in CD4+ T cells.
STING is a receptor for cyclic dinucleotides such as 2′3′-cGAMP (2′3′–cyclic guanosine monophosphate–adenosine monophosphate) produced by cGAS (cGAMP synthase) in response to cytosolic double-stranded DNA (Ishikawa and Barber, 2008; Burdette et al., 2011; Wu et al., 2013). STING activation induces its relocation from the endoplasmic reticulum to the Golgi (Ishikawa et al., 2009). During this process, STING recruits the noncanonical IκB kinase TBK1, which phosphorylates serine 366 in the C-terminal tail (CTT) of STING, generating a platform for IRF3 recruitment and phosphorylation by TBK1 (Liu et al., 2015). STING also activates NF-κB through a poorly resolved mechanism, although TBK1 has also been implicated (Abe and Barber, 2014). Phosphorylated IRF3 and NF-κB subsequently induce type I IFN and inflammatory gene expression. In DCs, STING activation additionally induces expression of co-stimulatory molecules, leading to cell maturation and launching of adaptive immunity (Li et al., 2013).
Monogenic immune dysregulation syndromes have been instrumental in the understanding of the contribution of individual proteins to immunity. Genetic defects in components of the innate nucleic acid–sensing and –signaling pathway leading to an increase in the production of type I IFNs have been identified and grouped as interferonopathies (Crow and Manel, 2015). However, the disease phenotypes associated are broad, affecting several organ systems, and have been classified as autoinflammation (RNASEH2, ADAR, DDX58, and TMEM173), infection (STAT1-GOF, C1Q, C1R, C1S, C4, CYBB, NCF1, and NCF2), autoimmunity (TREX1, PSMB8, IFIH1, and ACP5), and malignancy (SAMHD1; Meyts and Casanova, 2016). This suggests that the implicated proteins could have nonredundant functions distinct from their described role in type I IFN production and nucleic acid sensing.
Activating mutations in TMEM173 have been described in humans leading to a severe early onset inflammatory disease characterized by interstitial lung disease and vascular skin disease particularly targeting the extremities (Jeremiah et al., 2014; Liu et al., 2014). The reported mutations lie in the dimerization domain and were proposed to mimic the effect of 2′3′-cGAMP binding. STING with activating mutation was reported to be localized in the Golgi at steady state in the absence of ligand stimulation and to induce constitutive type I IFN expression in cell lines. Accordingly, circulating type I IFN and inflammatory cytokines have been detected in these patients. Interestingly, alteration in the immunological phenotype such as lymphopenia and leukopenia in patients with constitutively active STING were also observed (Jeremiah et al., 2014; Liu et al., 2014). Here, we show that patients carrying an active mutation in TMEM173 have a T cell imbalance, and we leverage this finding to show that STING adopts an antiproliferative activity in CD4+ T cells.
Results
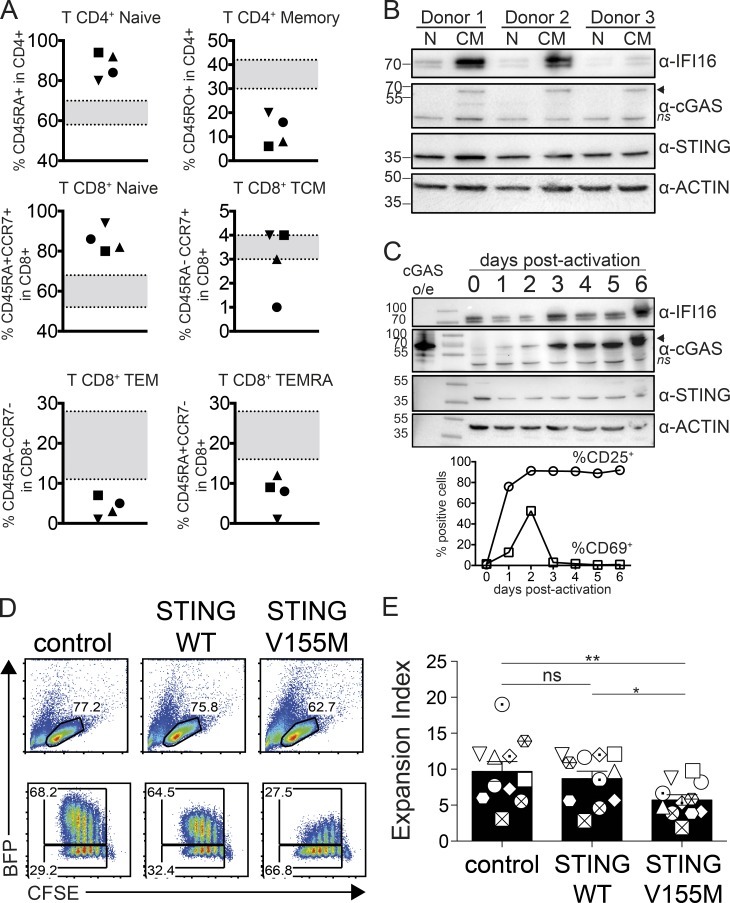
Clinical parameter analysis of patients carrying activating TMEM173 mutations revealed a peripheral T cell compartment imbalance characterized by an increased fraction of naive CD4+ and CD8+ T cells and a reduced fraction of memory cells (Fig. 1 A and Table S1). This raised the possibility that STING may have activities in lymphocytes. We focused on CD4+ T lymphocytes obtained from healthy donors and examined the expression of STING and upstream sensors cGAS and IFI16 at the protein level. STING was expressed at similar levels in resting naive and central memory CD4+ T cells, whereas cGAS and IFI16 were more expressed in memory cells (Fig. 1 B). We followed protein expression during activation of naive CD4+ T cells in vitro. STING expression was maintained over time, whereas cGAS and IFI16 were induced during the first few days of activation (Fig. 1 C). Thus STING, a sensor of innate immunity, is also expressed in cells of adaptive immunity. To examine the impact of WT and mutated active STING on CD4+ T cells, we developed an overexpression approach using BFP-2A lentivectors combined with cell proliferation profile analysis. CD4+ T cells from healthy donors transduced with control vector or STING WT steadily proliferated (Fig. 1, D and E). In contrast, CD4+ T cells transduced with STING carrying the patients’ activating mutation V155M showed reduced expansion (Fig. 1, D and E). Strikingly, the fraction of cells expressing the highest level of STING V155M gradually decayed at each cell cycle. Untransduced BFP-negative cells in the same well expanded normally, indicating that the effect required cell-intrinsic STING V155M activity (Fig. S1).
Figure 1.
T cell imbalance induced by constitutive active STING. (A) Frequency of naive and memory CD4+ and CD8+ T cell compartments in patients carrying activating TMEM173 mutations. Each symbol represents an individual donor. n = 4 donors. TCM, T central memory; TEM, T effector memory; TEMRA, T effector memory CD45RA+. Age-matched expected values are indicated in gray. (B) Immunoblot of cGAS, STING, IFI16, and actin expression in resting naive (N) or central memory (CM) CD4+ T cells from blood of healthy donors. n = 3 donors. The arrowhead indicates the specific cGAS band, and ns indicates the non-specific band. (C) Immunoblot of cGAS, STING, IFI16, and actin (top) and surface expression of CD25 and CD69 (bottom) in naive CD4 T cells from day 0 to day 6 after activation with PHA and IL-2. Data are representative of n = 2 donors. o/e, overexpression. (B and C) Molecular mass is shown in kilodaltons. (D) Proliferation profile of naive CD4+ T cells 4 d after transduction with BFP lentivectors coding for control, STING WT, or STING V155M. (E) Expansion index in BFP-positive and proliferating cells as in D. n = 11. Data are mean ± SEM. One-way ANOVA with Tukey’s correction was used. *, P < 0.05; **, P < 0.01.
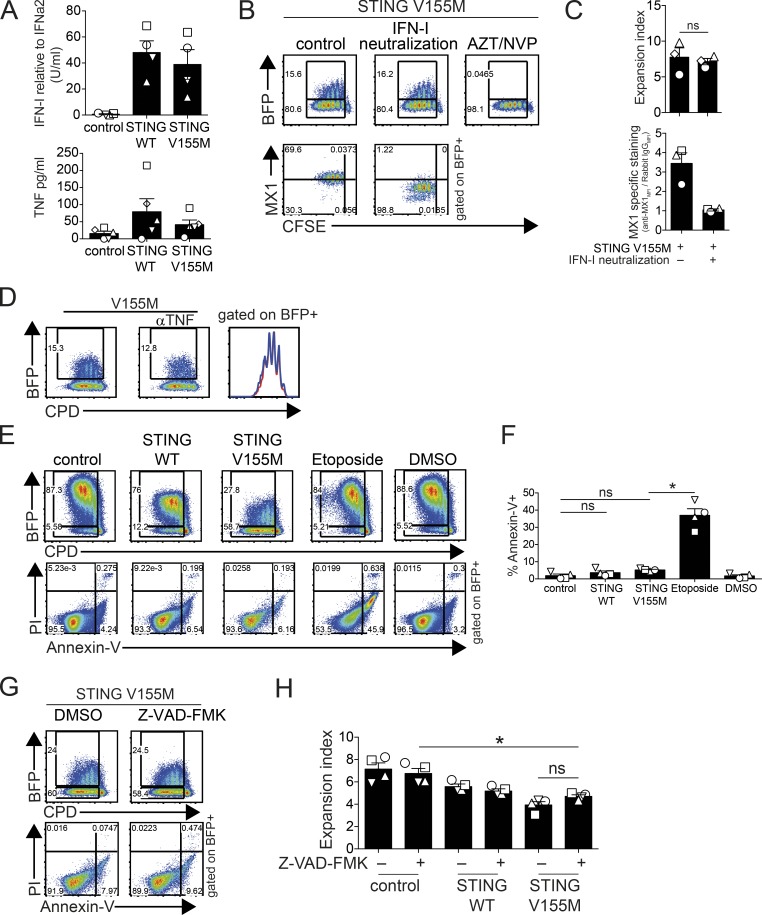
STING activation leads to the expression of type I IFN and TNF (Sun et al., 2013). CD4+ T cells transduced with STING WT and STING V155M produced type I IFN and TNF (Fig. 2 A). The bulk levels of TNF and IFN were similar between WT and V155M samples, whereas the fraction of cells expressing STING was greatly reduced in the case of V155M (Fig. 1 D). Thus, V155M is also a STING-activating mutation in primary CD4+ T cells. Although proliferation of BFP-negative cells was not affected by STING V155M–expressing cells, autocrine TNF or IFN could be required in cells expressing STING to inhibit proliferation. We used neutralizing reagents to assess the role of these cytokines in the reduced expansion of STING V155M cells. To control for type I IFN neutralization, we measured the expression of MX1, an IFN-stimulated gene (ISG; Fig. S2 A). MX1 was induced in STING V155M cells and abrogated when type I IFN was neutralized (Fig. 2, B and C). However, the STING V155M cells conserved their reduced expansion under type I IFN neutralization conditions (Fig. 2, B and C). To assess the TNF neutralization, we used an NF-κB reporter in CD4+ T cells. Recombinant TNF stimulated reporter expression, which was neutralized with anti-TNF/TNFR antibodies (Fig. S2 B). In contrast, parallel treatment of the STING V155M cells with anti-TNF/TNFR antibodies did not rescue the expansion of STING V155M cells (Fig. 2 D and Fig. S2 C). The proliferation of BFP-negative cells was also not affected by IFN or TNF neutralization (Fig. 2, B and D). Thus, inhibition of CD4+ T cell proliferation by STING V155M was independent from autocrine type I IFN and TNF signals. Activation of mouse STING by a synthetic agonist induces apoptosis of malignant mouse B cells (Tang et al., 2016), and STING activation in endothelial cells was shown to induce apoptosis (Liu et al., 2014). Greater than 90% of cells expressing STING V155M remained negative for annexin V, indicating no overt induction of apoptosis (Fig. 2, E and F). To determine whether apoptosis was implicated in the reduced proliferation, we used the pan-caspase inhibitor Z-VAD-FMK. Z-VAD-FMK protected from etoposide-induced apoptosis in T cells, validating the approach (Fig. S2 D), but did not rescue the proliferation of cells expressing STING V155M (Fig. 2, G and H).
Figure 2.
STING has an intrinsic antiproliferative activity in T cells. (A) Type I IFN activity (top; n = 4 donors) and TNF concentration (bottom; n = 5 donors) in supernatants of naive CD4+ T cells 4 d after transduction with control, STING WT, or STING V155M BFP lentivectors. (B) BFP and intracellular MX1 expression and proliferation profile (CFSE) after type I IFN neutralization or azidothymidine (AZT)/nevirapine (NVP) treatment (BFP only) in naive CD4+ T cells transduced with STING V155M BFP lentivectors. (C) Expansion index (top) and MX1-specific intracellular staining (bottom) in proliferating BFP-positive cells as in B. n = 3. MFI, mean fluorescence intensity. (D) BFP expression and proliferation profile (CPD) after TNF neutralization in naive CD4+ T cells transduced with STING V155M BFP lentivectors. (E) Annexin V and PI staining in naive CD4+ T cells transduced with control, STING WT, or STING V155M BFP lentivectors or treated with 25 µM etoposide or DMSO. (F) Frequency of annexin V–positive cells in BFP-positive cells as in E. n = 4. (G) BFP expression, proliferation profile, and annexin V and PI staining in naive CD4+ T cells transduced with STING V155M BFP lentivectors after treatment with 50 µM Z-VAD-FMK or DMSO. (H) Expansion index in BFP-positive naive CD4+ T cells transduced with control, STING WT, or STING V155M BFP lentivectors after treatment with Z-VAD-FMK or DMSO. n = 4. Data are mean ± SEM. One-way ANOVA test with Tukey’s correction was used. *, P < 0.05.
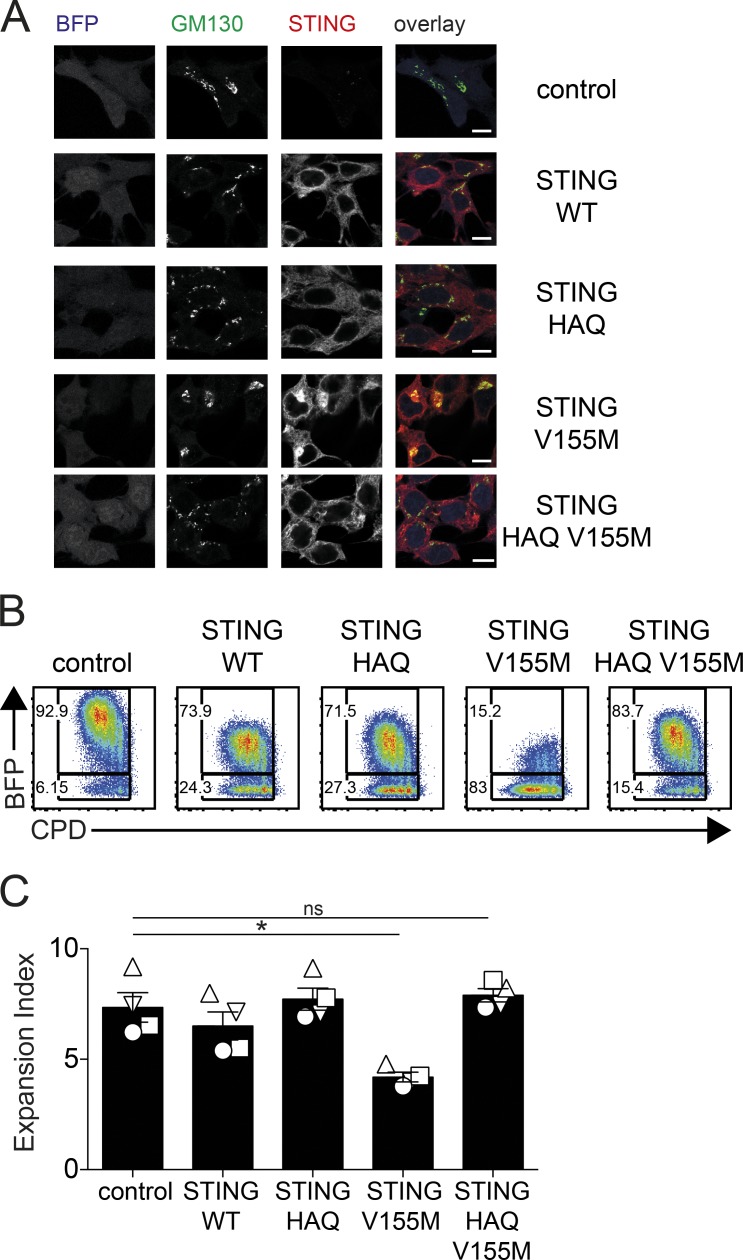
Next, we sought to determine whether the signaling activity of STING V155M was required to inhibit T cell proliferation. To do so, we examined the ability of second-site mutations in STING V155M to interfere with its activity. The natural polymorphism R71H-G230A-R293Q (HAQ) in TMEM173 WT has been proposed to inhibit its response to cyclic dinucleotides (Jin et al., 2011; Yi et al., 2013). We hypothesized that HAQ could function as a genetic inhibitor of STING signaling. We introduced the HAQ polymorphism in STING WT and STING V155M. The presence of HAQ in STING V155M abolished its constitutive activity in a reporter assay for type I IFN transcription (Fig. S2, E and F). Noteworthy, STING HAQ and STING HAQ V155M remained partially responsive to 2′3′-cGAMP stimulation in this assay, indicating that the HAQ polymorphism preferentially impacts the constitutive activity of STING. Whereas STING V155M localized to the Golgi, a hallmark of STING activation, STING HAQ V155M localized to the endoplasmic reticulum similar to unstimulated STING WT (Fig. 3 A). This validated that the HAQ polymorphism functionally inhibited STING signaling by the constitutive active mutant V155M. Strikingly, in CD4+ T cells, the presence of HAQ in STING V155M restored cell proliferation (Fig. 3, B and C). Thus, the activated state of STING V155M is required to inhibit proliferation of T cells.
Figure 3.
STING signaling activity is required to inhibit T cell proliferation. (A) Staining for STING and GM130 and BFP expression in 293FT cells transduced with control, STING WT, STING HAQ, STING V155M, or STING HAQ V155M lentivectors (portion of one field out of four fields, representative of three independent experiments). Bars, 10 µM. (B) Naive CD4+ T cell proliferation profile (CPD) 4 d after transduction with BFP lentivectors coding for control, STING WT, STING HAQ, STING V155M, or STING HAQ V155M. (C) Expansion index in BFP-positive cells as in B. n = 4. Data are mean ± SEM. One-way ANOVA with Dunnett’s correction was used. *, P < 0.05.
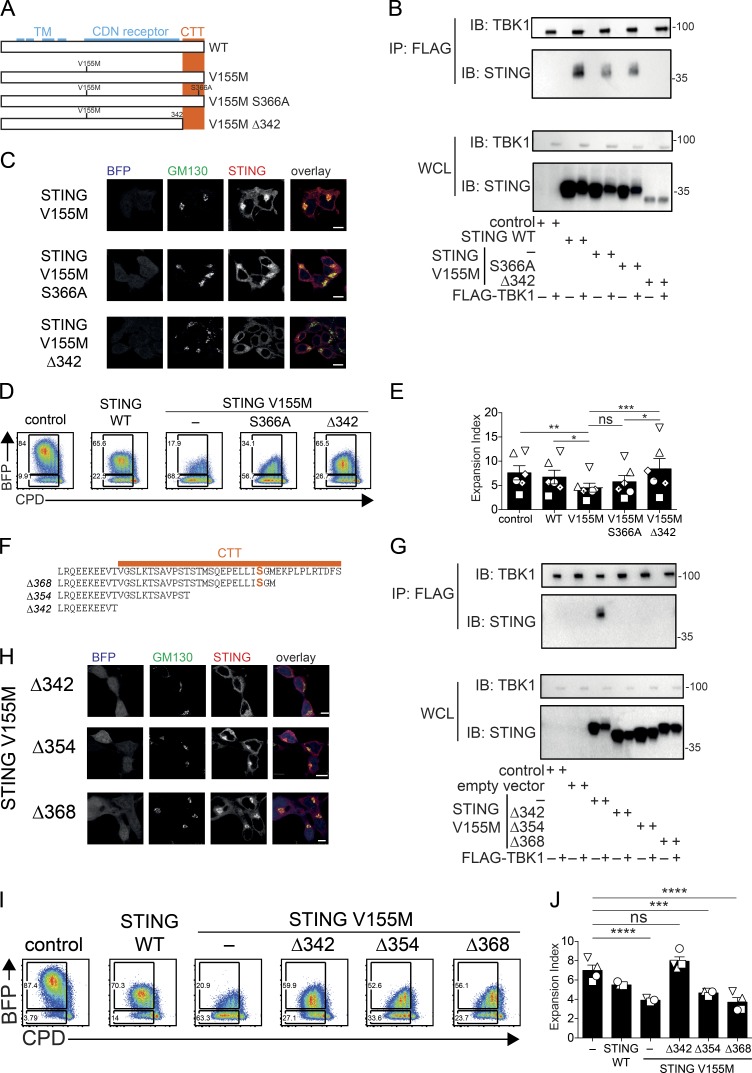
We sought to determine whether STING V155M required IRF3 binding and TBK1 binding to inhibit proliferation. We introduced the S366A mutation in STING V155M, which was shown to prevent IRF3 recruitment after cGAMP-mediated activation (Liu et al., 2015). We also deleted the CTT of STING (Δ342), a domain that was shown to be sufficient for TBK1 binding and proposed to be required in STING for TBK1 binding (Fig. 4 A; Tanaka and Chen, 2012; Kranzusch et al., 2015). We validated that Δ342 in STING V155M but not S366A prevented interaction with TBK1 (Fig. 4 B). The Δ342 mutation also abolished the localization of STING V155M to the Golgi (Fig. 4 C). In T cells, both mutations abolished the production of type I IFN and the up-regulation of MX1 (Fig. S3, A and B). However, only the Δ342 mutation abolished the antiproliferative activity of STING (Fig. 4, D and E). Thus, the antiproliferative activity of STING V155M requires the CTT but not the IRF3 binding site.
Figure 4.
miniCTT, a functional domain of STING distinct from TBK1 and IRF3 binding sites that inhibits T cell proliferation. (A) Schematics of STING S366A and Δ342 mutants. TM denotes the transmembrane domains, CDN denotes the cyclic dinucleotide receptor domain, and the CTT is also shown (Kranzusch et al., 2015). (B) Immunoprecipitation (IP) of STING mutants by TBK1. 293FT cells were transfected with control, STING WT, V155M, V155M S366A, or V155M Δ342 plasmids and FLAG-TBK1 plasmid for 24 h. Cell lysates were prepared and immunoprecipitated with anti-FLAG beads. Input cell lysates and immunoprecipitates were analyzed by Western blot with the indicated antibodies. IB, immunoblot; WCL, whole-cell lysate. Molecular mass is shown in kilodaltons. (C) Staining for STING and GM130 and BFP expression in 293FT cells transduced with STING V155M, V155M S366A, or V155M Δ342 BFP lentivectors (portion of one field out of four fields, representative of three independent experiments). Bars 10 µM. (D) Naive CD4+ T cell proliferation profile (CPD) 4 d after transduction with control, STING WT, V155M, V155M S366A, or V155M Δ342 BFP lentivectors. (E) Expansion index in BFP-positive cells as in D. n = 6. Data are mean ± SEM. One-way ANOVA with Tukey’s correction was used. *, P < 0.05; **, P < 0.01; ***, P < 0.005. (F) Schematics of STING CTT deletion mutants Δ342, Δ354, and Δ368. Serine 366 is indicated in bold orange. (G) Immunoprecipitation of STING mutants by TBK1. 293FT cells were transfected with control, STING V155M, V155M Δ342, V155M Δ354, or V155M Δ368 plasmids and FLAG-TBK1 plasmid for 24 h. Cell lysates were prepared and immunoprecipitated with anti-FLAG beads. Input cell lysates and immunoprecipitates were analyzed by Western blotting with the indicated antibodies. Molecular mass is shown in kilodaltons. (H) Staining for STING and GM130 and BFP expression in 293FT cells transduced with STING V155M Δ342, V155M Δ354, or V155M Δ368 BFP lentivectors (portion of one field out of four fields). Bar, 10 µM. (I) Cell proliferation profile in naive CD4+ T cells transduced with control, STING WT, V155M, V155M Δ342, V155M Δ354, or V155M Δ368 BFP lentivectors. (J) Expansion index in BFP-positive cells as in I. n = 4. Data are mean ± SEM. One-way ANOVA with Dunnett’s correction was used. ***, P < 0.001; ****, P < 0.0001.
To determine whether TBK1 binding was required for the antiproliferative activity of STING V155M, we introduced partial deletions (Δ354 and Δ368) in the CTT of STING V155M (Fig. 4 F). Similar to Δ342, Δ354 and Δ368 lost the ability to bind TBK1 (Fig. 4 G) and to induce type I IFN and ISG expression (Fig. S3, C and D). Unexpectedly, STING V155M Δ354 and Δ368 localized to the Golgi (Fig. 4 H), suggesting that they retained their activated state. Strikingly, STING V155M Δ354 and Δ368 also conserved their antiproliferative ability in T cells (Fig. 4, I and J). In these experiments, primary human T cells from healthy donors expressing endogenous WT STING were used. Thus, it remained possible that STING V155M Δ354 heterodimerizes with endogenous WT STING to recruit TBK1. To exclude this possibility, we transduced WT and Tmem173-deficient mouse T cells with STING mutants. Expression of STING V155M, V155M S366A, and V155M Δ354 in mouse CD4+ T cells inhibited the proliferation of transduced cells in both WT and Tmem173-deficient cells, whereas STING V155M Δ342 had no effect (Fig. S3 E). Thus, the domain 343–354 in STING is required for the antiproliferative activity of STING V155M in T cells.
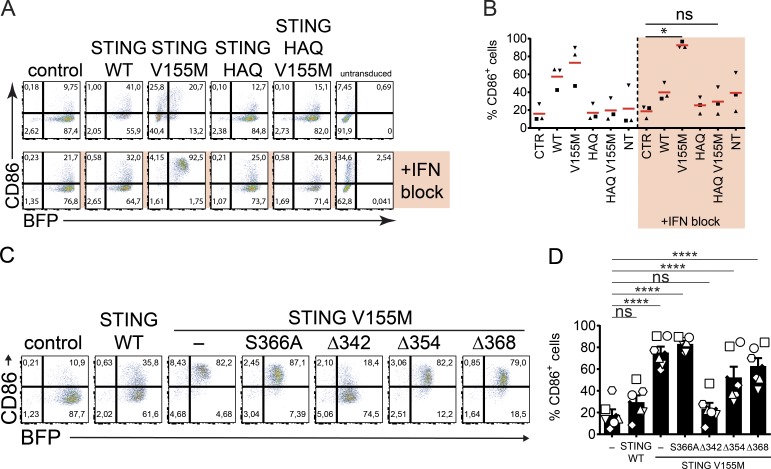
As this activity of STING occurred in the absence of TBK1 binding, we sought to determine its impact on innate immune activation. We tested the ability of the various STING mutants to induce expression of a co-stimulatory molecule such as CD86 in DCs, a hallmark of innate immune activation. We developed a lentiviral transduction assay in DCs. Transduction by STING V155M was low, and the ISG SIGLEC1 was induced, presumably because of the presence of type I IFN (Fig. S4, A and B). We combined STING lentivector transduction with type I IFN neutralization. Type I IFN neutralization abolished SIGLEC1 expression, and efficient transduction was restored. Unexpectedly, STING V155M induced expression of CD86 when type I IFN was neutralized, indicating DC activation through STING signaling, independently of type I IFN (Fig. S4, A and B). We examined the impact of the HAQ polymorphism on DC activation by STING. STING HAQ V155M lost the ability to induce expression of CD86, validating that the activated state of STING is required (Fig. 5, A and B). Next, we examined mutants of the CTT in STING V155M. STING V155M S366A induced CD86 expression, suggesting that IRF3 binding is not required for DC activation (Fig. 5, C and D). Strikingly, STING V155M Δ368 and Δ354 also induced CD86 expression despite the lack of TBK1 binding, whereas STING V155M Δ342 lost this ability (Fig. 5, C and D). Thus, the antiproliferative effect of STING in T cells corresponds to its ability to induce innate immune activation in DCs.
Figure 5.
miniCTT mediates activation of DCs. (A) BFP and CD86 expression in DCs 4 d after transduction with control, STING WT, STING HAQ, STING V155M, or STING HAQ V155M BFP lentivectors and neutralization of type I IFN. (B) CD86 expression as in A. n = 3 independent donors combined from two experiments. One-way ANOVA with Tukey’s posthoc test was used. *, P < 0.05. CTR, control; NT, not transduced. (C) CD86 and BFP expression in DCs transduced with control, STING WT, V155M, V155M S366A, V155M Δ342, V155M Δ354, and V155M Δ368 BFP lentivectors with type I IFN neutralization. (D) CD86 and BFP expression as in C. n = 6 independent donors combined from three independent experiments. One-Way ANOVA with Dunnett’s multiple comparisons test was used. Data are mean ± SEM. ****, P < 0.0001.
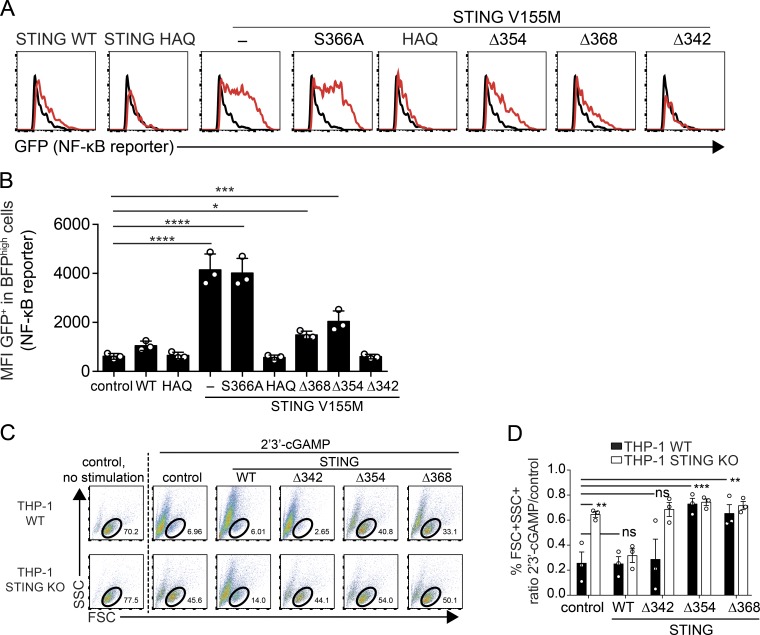
To gain mechanistic insights into this activity, we examined signaling by STING mutants. Given the ability of STING V155M Δ368 and Δ354 to activate DCs, we hypothesized that they retained the ability to activate NF-κB. Thus, we measured NF-κB reporter activation by the various STING mutants. After lentiviral transduction, STING V155M and V155M S366A strongly activated the reporter (Fig. 6, A and B). In contrast, STING WT exhibited a modest constitutive ability to activate the reporter, and HAQ mutations abrogated reporter activation (Fig. 6, A and B). Strikingly, STING V155M Δ368 and STING V155M Δ354 activated the reporter, whereas STING V155M Δ342 did not (Fig. 6, A and B). Thus, the CTT subdomain 343–354 is required for NF-κB activation by STING independently of TBK1 and IRF3 binding. We wished to establish that the identified CTT subdomain 343–354 is also a functional domain in WT STING. We noticed that cGAMP stimulation of THP-1 cells induces cell death (Fig. 6 C). cGAMP-induced IFN production and cell death were inhibited in STING KO THP-1 cells, and expression of STING WT in STING KO restored IFN expression and cell death induced by cGAMP (Fig. 6, C and D; and Fig. S4, C and D). STING Δ342, Δ354, and Δ368 in STING KO cells did not restore IFN expression, in agreement with the lack of TBK1 recruitment, or cGAMP-induced cell death (Fig. S4 D). Interestingly, expression of STING Δ354 and Δ368 protected WT THP-1 from cGAMP-induced cell death, whereas STING Δ342 did not have such an effect (Fig. 6, C and D). This indicates that the domain 343–354 is biologically active in STING WT. Overall, these findings define domain 343–354 as a minimal functional subdomain of the CTT (miniCTT) in STING V155M and STING WT, independent of TBK1 and IRF3 binding, that mediates NF-κB activation, DC activation, and inhibition of T cell proliferation.
Figure 6.
Signaling mediated by STING miniCTT. (A) GFP expression after NF-κB reporter activation in 293FT cells transduced with the indicated STING constructs. (B) GFP expression as in A. n = 3 independent experiments. One-Way ANOVA with Dunnett’s multiple comparison test was used. Data are mean ± SD. *, P < 0.05; ***, P < 0.0005; ****, P < 0.0001. MFI, mean fluorescence intensity. (C) Induction of cell death by 2′3′-cGAMP with Lipofectamine in THP-1 WT or THP-1 STING KO cells, transduced by the indicated BFP lentivector. FSC, forward scatter; SSC, side scatter. (D) Ratio of live cells (FSC+SSC+ live-cell gate) after 2′3′-cGAMP stimulation versus no stimulation is shown, as in C. n = 3 independent experiments. Data are mean ± SEM. One-Way ANOVA with Tukey's multiple comparisons test was used. **, P < 0.01; ***, P < 0.001.
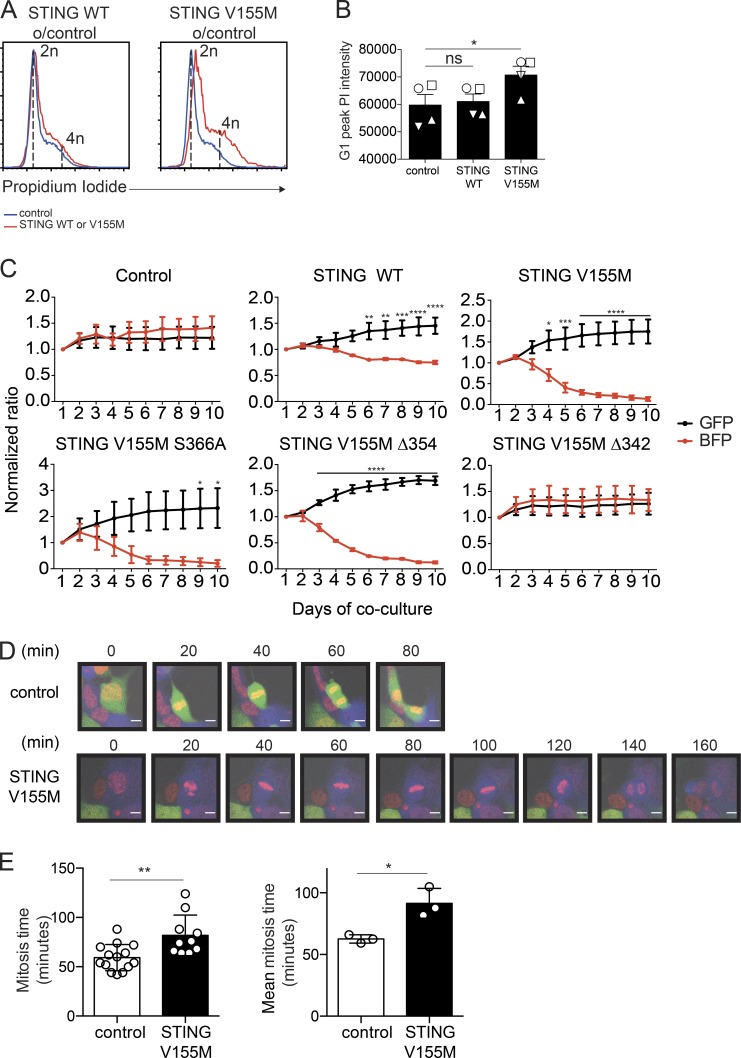
To examine how STING V155M could inhibit T cell proliferation, we assessed DNA content during cell cycle. T cells expressing STING V155M displayed a shift in their G1 peak and accumulated cells with >4n DNA (Fig. 7, A and B), indicating mitotic errors (Pillai et al., 2015). We performed live imaging of primary T cells to examine mitosis, but cells were too motile to track mitotic events. We developed an assay for the antiproliferative activity of STING in STING-deficient human 293FT cells (Fig. 4 B; Burdette et al., 2011; Gentili et al., 2015) based on growth competition between GFP control–transduced cells co-cultured and BFP-2A–STING construct–transduced cells. Cells expressing STING V155M, V155M S366A, and V155M Δ354 were rapidly diluted out, whereas cells expressing STING V155M Δ342 remained stable in culture (Fig. 7 C). Thus, this assay recapitulated the antiproliferative effect of STING V155M observed in T cells and confirmed that this effect is cell intrinsic. Interestingly, STING WT–expressing cells were also slowly diluted in this assay. We examined mitosis in this assay for the STING V155M condition by live microscopy. The duration of mitosis was increased in cells expressing STING V155M, with strikingly unstable and long metaphases, as compared with control cells (Fig. 7, D and E).
Figure 7.
STING V155M induces mitotic errors. (A) PI staining in naive CD4+ T cells 4 d after transduction with control, STING WT, or STING V155M BFP lentivectors. o/control, over control. (B) PI intensity of G1 peak (mode of linear PE) as in A. n = 4. Data are mean ± SEM. One-way ANOVA with Dunnett’s correction was used. (C) Competition between GFP control-transduced cells and the indicated BFP control or the indicated BFP-2A–STING–transduced cells. n = 3 independent experiments. Data are mean ± SEM. Two-way ANOVA with Sidak’s multiple comparison test was used. (D) Snapshots of a representative control cell expressing GFP (top) or STING V155M (bottom) and undergoing cell division. DNA (red), BFP-2A–V155M (blue), and control cells expressing GFP (green) are shown. Bars, 10 µM. (E, left) Mitosis time calculated in 293FT cells expressing STING V155M or control (GFP or untransduced). (Right) Mean mitosis time from three independent experiments. n = 3. Data are mean ± SD. Unpaired Student’s t test was used. *, P < 0.05; **, P < 0.005; ***, P < 0.0005; ****, P < 0.0001.
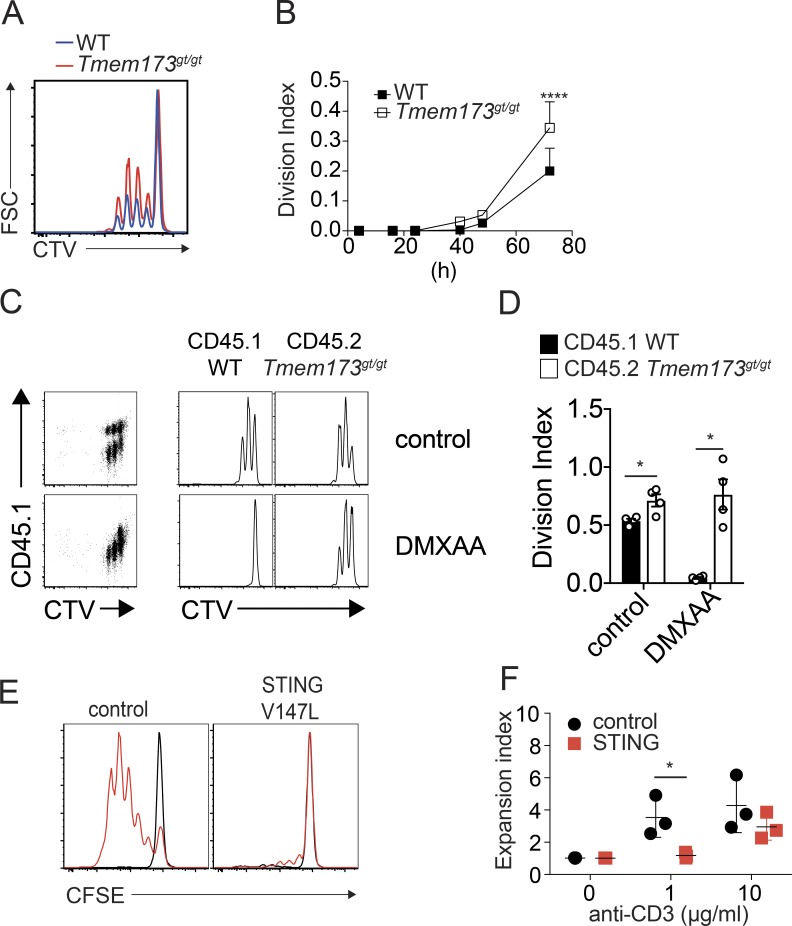
Finally, we sought to determine whether endogenous STING could impact proliferation of T cells. First, we examined Tmem173-deficient mice (Fig. S5 A). Spleen and lymph nodes of Tmem173-deficient mice did not show defects in the proportion of T cell subsets (Fig. S5, B–E). In contrast, stimulation of T cells revealed that Tmem173-deficient cells had an increased proliferation capacity (Fig. 8, A and B). Mixed cultures of WT and Tmem173-deficient cells confirmed that the antiproliferative activity of STING is cell intrinsic (Fig. 8, C and D). Next, we tested whether activation of endogenous STING could reduce T cell proliferation. We used the mouse STING agonist DMXAA (5,6-dimethylxanthenone 4-acetic acid). Importantly, because DMXAA can induce apoptosis (Ching et al., 2002), we specifically monitored live cells. We found that DMXAA treatment abrogated proliferation of WT live cells but had no impact on Tmem173-deficient live cells (Fig. 8, C and D). To confirm the T cell–intrinsic antiproliferative effect of STING in vivo, we generated mixed bone marrow chimeras in which only T cells were fully deficient for Tmem173 or WT and measured expression of the cell proliferation marker Ki67. We found a significant increase in the percentage of Ki67+ memory CD8+ T cells in peripheral and mesenteric lymph nodes in Tmem173-deficient conditions and no impact on the corresponding naive cells (Fig. S5 F). We did not observe a significant effect in splenic CD8+ T cells or CD4+ T cells at the time point tested in vivo, which could suggest that the antiproliferative activity of STING may be tissue specific. Finally, we examined the ability of T cells from patients with activating TMEM173 mutations to undergo cell division. Using a moderate TCR stimulation, patients’ T cells exhibited a profound proliferation defect compared with healthy controls (Fig. 8, E and F).
Figure 8.
STING inhibits T cell proliferation in normal and pathological conditions. (A) Proliferation profile (CTV) of WT (blue) or Tmem173gt/gt (red) mouse CD4+ T cells 72 h after activation with 1 µg/ml αCD3/αCD28. FSC, forward scatter. (B) Division index as in A. n = 4 mice combined from two independent experiments. Data are mean + SEM. Two-way ANOVA with Sidak’s multiple comparison test was used. (C) Proliferation profile of mixed CD45.1+ WT or CD45.2+ Tmem173gt/gt CD4+ T cells cultured for 48 h in the presence of 10 µg/ml αCD3/αCD28 with or without 12.5 µM DMXAA. Live cells were gated. (D) Division index as in C. n = 4 mice combined from two independent experiments. Data are mean ± SEM. One-way ANOVA with Tukey correction was used. (E) Proliferation profile (CFSE) of CD3+ T cells from a patient carrying an activating TMEM173 mutation (V147M) or from a healthy control upon PBMC stimulation with 1 µg/ml anti-CD3 and 1 µg/ml anti-CD28 for 4 d. Black peaks indicate unstimulated cells, and red peaks correspond to the stimulated condition. (F) Expansion index as in E for two patients carrying the V155M mutation and one patient carrying the V147M mutation in TMEM173. n = 3 donors. Data are mean ± SEM. Two-Way ANOVA with Sidak’s multiple comparison test was used. *, P < 0.05; ****, P < 0.0001.
Discussion
Overall, our findings show that, in T cells, STING is a negative regulator of lymphocyte proliferation under normal and pathological conditions. Stimulation of STING was previously shown to induce apoptosis (Liu et al., 2014; Tang et al., 2016), but we could not find any evidence of apoptosis in STING V155M–expressing CD4+ T cells. Instead, our results are consistent with a loss of proliferative capacity. The antiproliferative activity of STING required STING localization to the Golgi and a subdomain in the C-terminal domain, termed miniCTT, distinct from TBK1 and IRF3 recruitment domains. The antiproliferative activity of STING in T cells mediated by the miniCTT corresponded to the ability of STING to activate DCs and an NF-κB reporter. Nevertheless, STING V155M Δ368 and V155M Δ354 activated the NF-κB reporter to a lesser extent than STING V155M and V155M S366A, indicating that TBK1 also contributes to NF-κB reporter activation in this assay. These results suggest that STING possesses at least two mechanisms of innate immune stimulation through NF-κB: One mode requires TBK1 and IRF3 binding, and another mode requires the miniCTT. NF-κB activation through the miniCTT may represent an ancestral function of STING, before the full CTT was acquired in vertebrates (Kranzusch et al., 2015). Further studies will be required to determine how NF-κB signaling differs between these two modes. Using a THP-1 assay of 2′3′-cGAMP–induced cell death and IFN production, we also found that STING comprising the miniCTT protected from 2′3′-cGAMP–induced cell death. This evokes the possibility that TBK1-IRF3 signaling, which induces IFN production on one hand, and miniCTT signaling, which mediates the antiproliferative activity on the other hand, are competing activities in STING.
Similar to our findings in patients with a TMEM173-activating mutation, patients with loss of function mutations in IKBKB, CARD11, and BCL10 and dominant gain-of-function mutations in IKBA show an imbalance in the peripheral T cell subset characterized by an increase in the naive and decrease in the memory subset (Courtois et al., 2003; Greil et al., 2013; Pannicke et al., 2013; Stepensky et al., 2013; Torres et al., 2014). Isolated T cells from these patients also show a defect in ex vivo T cell proliferation. Thus, T cell subset balance and proliferation is critically dependent on a functional NF-κB signaling pathway. We thus propose that STING V155M interferes with NF-κB–related signals required for T cell proliferation. DC activation also typically requires NF-κB signaling (Janeway and Medzhitov, 2002). Hence, the miniCTT of STING may converge on NF-κB signaling in T cells and DCs. Interestingly, NF-κB activation has been previously implicated in the control of senescence (Chien et al., 2011). In T cells, we envision that the NF-κB stimulatory activity of STING intersects with antiproliferative pathways associated with cell-cycle arrest and senescence, consistent with the induction of mitotic errors by STING V155M. Importantly, although the role of NF-κB activation during TCR stimulation is well described, knowledge on its role during ongoing T cell proliferation is limited.
The antiproliferative activity of STING required its localization to the Golgi. This was observed independently by introducing the HAQ polymorphism in STING or by deleting the CTT of STING. The Golgi is a critical organelle for cell mitosis entry and progression (Colanzi and Sütterlin, 2013), and it was also recently proposed to function as a signaling platform (Pourcelot et al., 2016). However, these properties are poorly characterized, and it will be important to study the interplay between STING and the Golgi in these processes. STING induced mitotic errors characterized by aberrant cellular DNA content, extended mitosis, and unstable metaphases. Interestingly, endogenous STING expression level was found to correlate with chromatin compaction in a selection of cell lines (Malik et al., 2014). Mitotic error and aberrant cellular DNA content have also been reported in cell types that are dependent on TBK1 for mitosis when TBK1 was inhibited or sequestered (Pillai et al., 2015; Onorati et al., 2016). We found that TBK1 binding to STING is not required for its antiproliferative activity. Thus, distinct proteins involved in innate sensing may converge to induce mitotic errors.
We found that STING-deficient mouse T cells had a constitutive increased cell proliferation rate in vitro, indicating that endogenous WT STING has an antiproliferative activity. In addition, exogenous stimulation of STING in mouse CD4+ T cells with DMXAA at the time of TCR activation abrogated their proliferation in a STING-dependent manner. Altogether, these data imply the existence of an endogenous STING-activating signal in T cells, the nature of which is currently unknown. In vivo, the proportion of T cell subsets was not affected by STING deficiency in spleen and lymph nodes. This may imply that the activating signal is missing or that the intrinsic antiproliferative activity of endogenous STING is cancelled by another pathway in this specific pathogen–free in vivo context. In human T cells, we found cGAS to be induced upon in vitro TCR activation, which could contribute to an endogenous STING-activating signal. We do not exclude that the STING antiproliferative activity may also be triggered by yet unknown cGAS-independent signals.
Because T cells undergo clonal expansion during immune responses, the intrinsic antiproliferative activity of STING may be a primitive innate defense mechanism to limit pathogen replication in infected T cells. Conversely, the antiproliferative activity of STING could be implicated in T cell–related pathogenic processes such as HIV infection (Yan et al., 2010; Gao et al., 2013; Lahaye et al., 2013). It will be important to determine the relative contribution of STING activities in different cell types to antimicrobial defenses, antitumor responses, and autoimmunity. In the case of viruses, STING activation may have a direct impact on viral replication by directly contributing to the establishment of a cell-intrinsic antiviral state. STING activation also leads to cytokine production, such as type I IFN, and can thus impact viral replication in bystander cells. Finally, the antiproliferative activity of STING in T cells implies that STING may impact the development of the adaptive antiviral responses. In autoimmunity, the T cell–intrinsic antiproliferative activity of STING could contribute to the reported suppression of systemic autoimmunity in a mouse model of systemic lupus erythematosus (Sharma et al., 2015). The STING mutants that we describe will be instrumental to decipher these activities in vitro and in vivo. In summary, our results extend the paradigm of innate control of adaptive immunity (Iwasaki and Medzhitov, 2010) by establishing that inhibition of proliferation is a lymphocyte-intrinsic activity of the innate sensor STING.
Materials and methods
Cells
293FT and HL116 were cultured as previously described (Lahaye et al., 2013). Human peripheral blood mononuclear cells were isolated from buffy coats from normal human donors (approved by the Institut National de la Santé et de la Recherche Médicale ethics committee) using Ficoll-Paque PLUS (GE Healthcare). Naive CD4 T cell enrichment was performed with an EasySep Human Naive CD4 T cells Enrichment kit (no. 19155; STEMCELL Technologies) before sorting naive CD4 T cells (CD4+, CD27+, CD45RO−, CD25−, CD8−, CD14−, CD16−, CD19−, and CD123−) with MoFlo Astrios (Beckman Coulter). Sorted naive CD4+ T cell purity was superior to 98%. Then, cells were labeled with CFSE (no. 85-0850-84; eBioscience) or CPD670 (no. 65-0840-85; eBioscience), counted and resuspended at 106/ml in X-VIVO15 medium (Lonza) and penicillin-streptomycin (Invitrogen), and incubated at 37°C overnight. The next day, CD4+ T cells were cultured at one million per ml (200 µl/well in round-bottom 96-well plates) in X-VIVO15 medium and penicillin-streptomycin and activated with the T Cell Activation/Expansion kit (Miltenyi Biotec) at a ratio of two cells per bead. For immunoblots, total CD4+ T cells were preenriched by a positive selection with anti–human CD4 magnetic beads (Miltenyi Biotec) before sorting of naive (CD4+, CD27+, CD8−, CD45RO−, and CD25−) and central memory (CD4+, CD8−, CD45RO+, CD27+, and CD25−) cells. Sorted CD4+ T cells were directly lysed or activated with 5 µg/ml PHA-L (no. 61764; Sigma-Aldrich) and 1,000 U/ml IL-2 (Novartis) in RPMI Glutamax medium, 10% decomplemented FBS, 10 mM Hepes (Invitrogen), and 50 µg/ml penicillin-streptomycin and gentamicin (Invitrogen). 48 h after activation, fresh media was added with 1,000 U/ml IL-2, and cells were resuspended at 106/ml. Cells were collected and counted from day 0 to day 6 after activation for immunoblot or cytometry analysis.
Plasmids
Expression plasmids psPAX, CMV–VSV-G, and HXB2 envelopes were previously described (Lahaye et al., 2016). The NF-κB reporter lentivector was pTRH1_NF-κB_dscGFP (System Biosciences). pTRIP-SFFV–BFP-2A was generated from pTRIP-SFFV (Lahaye et al., 2016) by replacing GFP with mTagBFP (Evrogen) followed by the 2A peptide from porcine teschovirus-1 (Kim et al., 2011). The STING WT and STING V155M coding sequences were previously described (Jeremiah et al., 2014). The STING HAQ sequence was cloned by PCR from pUNO1-hSTING-HAQ (InvivoGen). The V155M mutation was introduced by site-directed mutagenesis. STING V155M S366A, STING V155M Δ342, STING V155M Δ354, STING V155M Δ368, STING Δ342, STING Δ354, and STING Δ368 were obtained by overlapping PCR mutagenesis and cloned in pTRIP-SFFV–tagBFP-2A. In immunoprecipitation experiments, pcDNA3.1-Hygro+ (Invitrogen) and pEF-BOS huTBK1 Flag-His (no. 27241; Addgene) were used. STING WT, STING V155M, STING HAQ, and STING HAQ V155M were cloned in pMSCV-Hygro, used in a type I IFN reporter assay in 293FT. In all final constructs, the DNA fragments originating from the PCR and encompassing the restriction sites used for cloning were fully verified by sequencing.
Lentiviral transductions in human CD4+ T cells
293FT cells were transfected for lentivector production as previously described (Lahaye et al., 2016). The ratio of plasmids for transfection was, per well of a 6-well plate: 0.2 µg HXB2 envelope expression plasmid, 0.2 µg CMV–VSV-G, 1 µg psPAX2, and 1.6 µg pTRIP lentivector. Sorted naive CD4+ T cells were activated for 24 h in 96-well plates as described in the Cells section. 100 µl of media was removed, and 100 µl 293FT supernatant containing the indicated lentivectors with 8 µg/ml of protamine was added to cells. Cells were spinoculated at 1,200 g for 2 h at 25°C. At day 2 after transduction (day 3 after activation), cells were seeded at 0.5 million per milliliter, and media was changed in presence of 1,000 U/ml IL-2 (Novartis). At day 4 after transduction (day 5 after activation), cells were harvested for analyses.
Human CD4+ T cell treatments
For type I IFN neutralization experiments, a cocktail of 3 µg/ml B18R (eBioscience), 2.5 µg/ml anti–IFN-β (eBioscience), 1.2 µg/ml anti–IFN-α (eBioscience), and 0.6 µg/ml anti-IFNAR (EMD Millipore) was added at the time of CD4+ T cell transduction. Reverse transcription inhibitors azidothymidine (25 µM) and nevirapine (10 µM) were used to inhibit lentiviral transduction as a negative control. To inhibit caspases, 50 µM Z-VAD-FMK (R&D Systems) was added at the time of transduction and maintained during the course of the experiment. 25 µM etoposide (Sigma-Aldrich) was added for 24 h at day 3 after transduction. For TNF neutralization experiments, a cocktail of 10 µg/ml anti–human TNFRI (R&D Systems), 10 µg/ml anti–human TNFRII (R&D Systems), and 10 µg/ml anti–human TNF (R&D Systems) was added at the time of CD4+ T cell transduction and maintained during the course of the experiment. Recombinant TNF (R&D Systems) was added to CD4+ T cells transduced with NF-κB reporter lentivector 24 h before analysis.
Flow cytometry analysis of human CD4+ T cells
Staining was performed in FACS buffer composed of PBS, 1% BSA (Euromedex), and 1 mM EDTA (Gibco). The antibodies used were anti–human CD25 and anti–human CD69. Cells were stained for 15 min at 4°C, washed twice, and fixed in 1% paraformaldehyde (PFA; Electron Microscopy Sciences). For intracellular MX1 expression analysis, cells were fixed with 100 µl/well intracellular fixation buffer (eBioscience) for 30 min at room temperature and then permeabilized with 200 µl of 1× permeabilization buffer (eBioscience) at room temperature. Then, cells were centrifuged at 1,700 g for 3 min. Cells were incubated for 30 min at room temperature in 50 µl of 1× permeabilization buffer. Anti-MX1 antibodies (Ab95926; Abcam) or normal rabbit IgG (Thermo Fisher Scientific) were diluted. Cells were washed in 200 µl of 1× permeabilization buffer and incubated for 30 min at room temperature with secondary antibody F(ab′)2 fragment donkey anti–rabbit IgG (H+L) phycoerythrin R-PE (Jackson ImmunoResearch Laboratories, Inc.). Cells were washed and resuspended in FACS buffer. For annexin V and propidium iodide (PI) staining, cells were washed twice with cold PBS and resuspended with 50 µl of 1× annexin V binding buffer (no. 51-66121E; BD) and 5 µl annexin V FITC (no. 130-093-060; Miltenyi Biotec). Cells were incubated at room temperature for 15 min. 150 µl of 1× binding buffer with 2 µg/ml PI (Thermo Fisher Scientific) was added before acquisition. Acquisition occurred within 15 min after PI addition to cells. Data were acquired on a FACSVerse flow cytometer (BD) and analyzed in FlowJo (Tree Star).
Western blotting analysis of human CD4+ T cells
1.5 million sorted naive and central memory or activated CD4+ T cells were lysed in sample buffer (4% SDS, 20% glycerol, 0.1 M Tris-HCl, pH 6.8, 0.05% bromophenol blue, and 0.1 M dithiothreitol). Cellular protein lysates were resolved on 4–20% precast SDS-PAGE gels (Bio-Rad Laboratories) and transferred on a nitrocellulose membrane. The membrane was blocked in 1× Tris-buffered saline, 0.1% Tween 20 (AMRESCO), and 5% BSA (Euromedex). Proteins were blotted with antibodies as follow: IFI16 (1G7; Santa Cruz Biotechnology, Inc.), cGAS (HPA031700; Sigma-Aldrich), STING (D2P2F; Cell Signaling Technology), and actin (C4; EMD Millipore) in 1× Tris-buffered saline, 0.1% Tween 20 (AMRESCO), and 5% BSA (Euromedex). For immunoprecipitation experiments, proteins were blotted with antibodies as follow: TBK-1 (D1B4; Cell Signaling Technology), STING (D2P2F; Cell Signaling Technology), or STING (clone 723505; R&D Systems). Membranes were washed three times in 1× PBS and 0.1% Tween 20 (AMRESCO). Enhanced chemiluminescence signal was recorded on the ChemiDoc XRS Imager (Bio-Rad Laboratories). Data were analyzed and quantified with Image Lab software (Bio-Rad Laboratories).
Type I IFN and TNF quantification
TNF concentration was measured with a human TNF cytometric assay (BD) according to the manufacturer’s protocol. Data were acquired on a FACSVerse flow cytometer and analyzed in FCAP Array (BD). Type I IFN measurement was performed using the HL116 as previously described (Lahaye et al., 2013).
Immunoprecipitations
0.8 million 293FT cells were plated in 6-well plates and transfected as previously described (Lahaye et al., 2013) with 1.5 µg pEF-BOS huTBK1 Flag-His or pcDNA3.1-Hygro+ combined with 1.5 µg of empty pTRIP-SFFV–tagBFP-2A or pTRIP-SFFV–tagBFP-2A coding for STING. 24 h later, supernatant was removed, and 1 ml of cold PBS was used to collect three wells per condition and transferred to an Eppendorf tube. Then, cells were centrifuged at 240 g for 5 min. The cell pellet was resuspended with cold PBS and centrifuged again. Cells were lysed in 500 µl of cold lysis buffer (20 mM Tris-HCl, pH 7.5, 150 mM NaCl, and 0.5% NP-40 as previously described [Liu et al., 2015] complemented with 1× EDTA-free protease inhibitor cocktail [Roche], 50 mM NaF, and 1 mM sodium orthovanadate). Lysis was performed on ice for 30 min, followed by centrifugation at 7,000 g for 7 min at 4°C. Supernatant was transferred into a new Eppendorf tube, and 10% of the total volume was removed and collected as input cell lysate before adding anti-FLAG M2 Affinity gel (no. A-2220; Sigma-Aldrich). 40 µl per sample of anti-FLAG M2 Affinity gel was washed once with 1 ml of complemented lysis buffer and resuspended in 100 µl of lysis buffer. 100 µl of anti-FLAG M2 Affinity gel was added to 400 µl of lysates and incubated at 18 rpm constant wheel rotation at 4°C overnight. Immunoprecipitates were washed five times with 1 ml of lysis buffer. The resulting 30 µl of immunoprecipitates were complemented with 30 µl of 2× Laemmli buffer and incubated at 95°C for 20 min. 15 µl of input cell lysate and of immunoprecipitates were loaded and analyzed by Western blotting as described in the Western blotting analysis of human CD4+ T cells section.
Lentiviral transductions in DCs
Lentiviral particles and simian immunodeficiency virus of macaques–like particles were produced as previously described from 293FT cells (Lahaye et al., 2013). Monocytes were freshly isolated from blood of healthy donors and cultured as previously described (Gentili et al., 2015). 50,000 freshly isolated monocytes were seeded in 96-well U-bottom plates and infected in a final volume of 250 µl (100 µl of cells at 0.5 × 106cells/ml, 100 µl of freshly produced lentiviral particles, and 50 µl of freshly produced simian immunodeficiency virus–like particles) with Protamine (Sigma-Aldrich) at 8 µg/ml in presence of 10 ng/ml human recombinant GM-CSF (Miltenyi Biotec) and 50 ng/ml IL-4 (Miltenyi Biotec).
Type I IFN was inhibited using a combination of reagents consisting of 3 µg/ml B18R carrier-free (eBioscience), 1.2 µg/ml anti–human IFN-α antibody (clone EBI-1; eBioscience), 2.5 µg/ml anti–human IFN-β (functional grade purified; clone A1; eBioscience), and 1.5 µg/ml anti–IFN-α/β receptor chain 2 antibody (clone MMHAR-2; EMD Millipore). The type I IFN–blocking reagents were added to cells 1–2 h before infection.
Confocal microscopy
293FT cells were plated on fibronectin (10 µg/ml; Sigma-Aldrich)-coated coverslips before transduction with STING or control BFP lentivectors. 2 d after transduction, cells were fixed with 4% PFA, quenched with PBS-glycine, and permeabilized with 0.1% Triton X-100. Then, cells were blocked with PBS–2% BSA–10% goat serum (Sigma-Aldrich), incubated with the primary antibodies STING (MAB7169; R&D Systems) and GM130 (610823; BD) in PBS–2% BSA, followed by five washes and incubation with fluorescence-labeled secondary antibodies (A21242 and A21121; Thermo Fisher Scientific) in PBS–2% BSA followed by four washes. Fluoromount-G (0100-01; Clinisciences) was used as the mounting solution, and slides were dried in a 37°C chamber for 10 min. Images were acquired on a confocal microscope (SP8; Leica Biosystems) equipped with a 60× oil immersion objective (NA = 1.4). For analysis of mitosis, 293FT cells transduced with pTRIP-SFFV (coding for GFP) were co-cultured with cells transduced with pTRIP-SFFV–BFP-2A–STING-V155M on fibronectin-coated fluordishes (World Precision Instruments). Nuclear DNA was stained using a SiR-DNA kit (Spirochrome AG) 1 h before acquisition. Live imaging was performed in a temperature-controlled CO2 chamber (Stage Top Chamber; Okolab) using an SP8 microscope equipped with a 20× objective (NA = 0.75). Image analysis was done on Fiji software. Mitosis time was calculated from nucleus envelope breakdown to the appearance of daughter cells.
IFN reporter assay in 293FT cells
The IFN reporter assay in 293FT cells was performed as previously described (Jeremiah et al., 2014; Gentili et al., 2015). In brief, 45,000 293FT cells were plated in a 24-well plate. The next day, cells were transfected with 500 ng of total DNA comprising 200 ng IFN-β–pGL3 and 300 ng of the empty vector pMSCV-Hygro or pMSCV-Hygro encoding the STING variants with TransIT-293 (Mirus). The next day, the medium was removed, and 2′3′-cGAMP (InvivoGen) was delivered with Lipofectamine 2000 (Invitrogen) transfection (1 µg 2′3′-cGAMP/1 µl Lipofectamine 2000) in a final volume of 500 µl (final concentrations: 4, 1.3, and 0.4 µg/ml). Fresh medium was added in the case of nonstimulated cells. After 24 h, cells were washed with PBS and lysed with Passive Lysis Buffer (Promega), and 10 µl of the lysate was used to perform the luciferase assay. Luciferase activity was measured using Luciferase Assay Reagent (Promega). Luminescence was acquired on a FLUOstar OPTIMA microplate reader (BMG Labtech).
NF-κB reporter assay in 293FT
293FT cells were transduced with pTRH1-NFkB-dscGFP, split, and transfected with BFP-2A–STING or control BFP lentiviral vector plasmids. Cells were detached and fixed with 1% PFA 48 h later. Data were acquired on a FACSVerse flow cytometer and analyzed in FlowJo.
STING-expressing cell competition assay
293FT cells were transduced with BFP-2A–STING, control BFP, or pTRIP-SFFV lentiviral vectors. 2 d after transduction, equal numbers of control GFP-transduced cells were co-cultured with each of the BFP-transduced cells. Cells were harvested every day up to 10 d, and the fraction of BFP-positive and GFP-positive cells was determined by flow cytometry.
cGAMP stimulation assay in THP-1 cells
Lentiviral vectors were produced as described in the Lentiviral transductions in human CD4+ T cells section. THP-1 WT and THP-1 STING KO were THP-1 cells, transduced with pLentiCRISPRv2 and pLentiCRISPRv2-STING_gRNA3 (gRNA sequence: 5′-AGGTACCGGAGAGTGTGCTC-3′), respectively, and selected with puromycin. KO efficiency was confirmed by Western blotting, and cells were maintained as bulk populations. 250,000 THP-1 WT or THP-1 STING KO cells were transduced with 1 ml of lentivector with 8 µg/ml protamine and 1 ml of media in a 6-well plate. Transduction rates were analyzed on a FACSVerse cytometer and were >95% in all cases. For stimulation, 100,000 THP-1 WT or THP-1 STING KO cells were transfected with 2′3′-cGAMP (InvivoGen) with Lipofectamine 2000 (1 µl Lipofectamine 2000/µg cGAMP). cGAMP final concentration in culture was 4 µg/ml. 2 d after transfection, cells and supernatants were harvested. Cells were fixed in 4% PFA in FACS buffer. Live cells were quantified on a FACSVerse cytometer using a forward scatter/side scatter live-cell gate. Supernatants were assayed for type I IFN activity with the HL116 cell line as described in the IFN reporter assay in 293FT cells section.
Flow cytometry for DCs
Cell surface staining was performed in PBS, 1% BSA (Euromedex), 1 mM EDTA (Gibco), and 0.01% NaN3 (AMRESCO). The antibodies used were anti–human CD86 PE (clone IT2.2; eBioscience) and anti–human CD169 (Siglec1) APC (clone 7-239; Miltenyi Biotec). Cells were stained for 15 min at 4°C, washed two times, and fixed in 1% PFA (Electron Microscopy Sciences). Data were acquired on a FACSVerse flow cytometer and analyzed in FlowJo.
Mouse ex vivo phenotyping and cell sorting
All animal procedures were in accordance with the guidelines and regulations of the French Veterinary Department in an accredited animal facility. The animal protocol was approved by the Animal Ethical Committee of Paris Centre (C2EA-59). C57BL/6J-Tmem173gt/J and C57BL/6J strains were obtained from The Jackson Laboratory. C57BL/6 controls or Tmem173gt/gt mutant mice were bred in house at Institut Curie but were not littermates. Rag2−/− (Shinkai et al., 1992) and Cd3e−/− mutant mice (Malissen et al., 1995) were obtained from the Centre National de la Recherche Scientifique Central Animal Facility and bred in house. Mice were sacrificed, and spleens and lymph nodes were collected. Tissues were homogenized and passed through 100-µm cell strainers. Homogenates were counted using a MACSQuant analyzer (Miltenyi Biotec) and phenotyped using antibodies against CD4, CD8a, CD62L, CD44, and CD25 (all BD). T cell subpopulations were defined as follows: naive CD4 T cells (CD4+, CD25−, CD62Lhi, and CD44lo), regulatory T cells (CD4+ and CD25+), and effector memory CD4 T cells (CD4+, CD25–, CD62Llo, and CD44hi).
For sorting, splenocytes and lymphocytes were pooled, and T cells were enriched using a negative selection kit for mouse T cells (Thermo Fisher Scientific) according to the manufacturer’s instructions. The enriched T cell fraction was stained with antibodies against CD4, CD8a, CD62L, CD44, CD25, and CD69 (all BD), and naive CD4 T cells (CD4+, CD25–, CD69–, CD62Lhi, and CD44lo) were sorted using a FACSAria II flow cytometer (BD).
In vitro stimulation of mouse CD4+ T cells
Sorted naive CD4+ T cells were stained with Cell Trace Violet (Thermo Fisher Scientific) according to the manufacturer’s instructions and plated onto flat-bottom 96-well plates coated with anti-CD3 (1 or 10 µg/ml and precoated at 4°C overnight) and stimulated with 0.1 or 1 µg/ml anti-CD28 (BD) in complete medium (RPMI, 100 mM Hepes, 2 mM Glutamax, 50 µM 2-mercaptoethanol, 100 U/100 µg/ml penicillin/streptomycin, 1× nonessential amino acids, 1 mM sodium pyruvate, [all Thermo Fisher Scientific], and 10% FCS [eurobio]) at 37°C. For mixed cultures, naive CD4+ T cells from CD45.2 Tmem173gt/gt were cultured with naive CD4+ T cells from CD45.1 C57BL/6 WT mice in a 1:1 ratio, in the presence or absence of the STING agonist DMXAA (Selleckchem). At the indicated time points, activated CD4+ T cells were stained with fixable viability dye (eBioscience) before being fixed using Cytofix/Cytoperm (BD). Cells were acquired using a MACSQuant flow cytometer, and data were analyzed using FlowJo software (Tree Star).
Lentiviral transductions in mouse CD4+ T cells
293FT cells were transfected for lentivector production as described in the Human CD4+ T cell treatments section. The ratio of plasmids for transfection was, per well of a 6-well plate: 0.4 µg CMV–VSV-G envelope expression plasmid, 1 µg psPAX2, and 1.6 µg pTRIP lentivector. Sorted naive CD4+ T cells were stained with CPD670 (no. 65-0840-85; eBioscience), and 0.2 million cells were activated for 24 h in 96-well plates as described in the previous paragraph. 100 µl of media was removed, and 100 µl of 293FT supernatant containing the indicated lentivectors with 8 µg/ml protamine was added to cells. Cells were spinoculated at 1,200 g for 2 h at 25°C. At day 2 after transduction (day 3 after activation), cells were seeded at 0.5 million per ml, and the media was changed in presence of 1,000 U/ml IL-2 (Novartis). At day 4 after transduction (day 5 after activation), cells were harvested for analyses.
Mixed bone marrow chimeras and Ki67 staining
Rag2−/− mice were irradiated with 10 Gy using an x-ray generator (Phillips) and reconstituted with an i.v. injection of 4 × 106 bone marrow cells from Cd3e−/− mice and 106 bone marrow cells from either Tmem173gt/gt or WT mice. After 12 wk, mice were sacrificed, and the spleen, peripheral lymph nodes, and mesenteric lymph nodes were removed. The organs were homogenized into single-cell suspensions using 100-µm cut-off cell strainers and 2.5-ml syringe plungers. Single-cell suspensions were counted on a MACSQuant flow cytometer and stained with antibodies against CD4, CD8a, CD44, CD25, and CD62L (all BD) for 30 min at 4°C. Cells were washed and fixed using the Foxp3 kit according to the manufacturer’s instructions (eBioscience). Intracellular staining was performed in permeabilization buffer using antibodies directed against Foxp3 and Ki67 (both eBioscience) for 30 min at room temperature. After a final wash, cells were acquired on a FACSVerse flow cytometer (BD).
Ethics
Written informed consent (parental consent for minors) was obtained from all participants. The study and protocols conformed to the 1975 Declaration of Helsinki and were approved by the Comité de protection des personnes Ile de France II and the French advisory committee on data processing in medical research.
Immunophenotyping of PBMCs
Leukocytes were counted in an automated hematological analyzer (ABXMicrosES60; Horiba Medical). Monoclonal antibodies (against CD3 [HIT3a], CD4 [OKT4], CD8 [RPA-T8], CD11a [2D7], CD14 [HCD14], CD15 [W6D3], CD19 [HIB19], CD27 [MT-271], CD31 [AC128], CD34 [8G12], CR45RA [HI100], CD45 [HI30], CD49d [9F10], CD56 [B159], CD57 [HNK-1], CD95 [DX2], CD178 [MFL3], HLADR [L243], IgM [G20-127], and IgD [IA6-2]); mouse IgM, IgG1k, IgG2a, and IgG2b isotype controls; and 7-aminoactinomycin D were obtained from BD. Monoclonal CD183 (CXCR4; clone 12G5), CD197 (CCR7; clone 4B12), and CD304 (BDCA4; clone 12C2) antibodies were purchased from BioLegend.
Proliferation assay on patient cells
PBMCs were isolated from whole blood on Ficoll gradient. Then, cells were stained with CFSE and immediately stimulated with coated anti-CD3 (eBioscience) and 1 µg soluble anti-CD28 (Sigma-Aldrich). Cells were acquired on an LSR Fortessa flow cytometer (BD) 4 d after stimulation, and data were analyzed using FlowJo.
Statistics
Statistical analyses were performed in Prism 6 (GraphPad Software) as indicated in the figure legends.
Online supplemental material
Fig. S1 shows the proliferation of BFP-negative T cells. Fig. S2 shows controls for STING V155M activation. Fig. S3 shows activities mediated by STING miniCTT in human and mouse T cells. Fig. S4 shows activities mediated by STING miniCTT in human DCs and STING KO THP-1 cells. Fig. S5 shows phenotypes of T cells from Tmem173-deficient mice. Table S1 shows the clinical immunophenotype of patients carrying an activating STING mutation.
Supplementary Material
Acknowledgments
We are grateful to M. Maurin and B. Jneid for technical assistance and O. Lantz, R. Basto, R. Weil, D. Arnoult, and L. Pace for scientific discussions. We acknowledge the flow cytometry facility at Institute Curie.
S. Cerboni was successively supported by Institut Curie and by Sidaction. N. Jeremiah was successively supported by Fondation pour la Recherche Médicale grant FDT20140930816, by Institut Curie, and by Agence Nationale de Recherches sur le Sida et les Hepatites Virales (ANRS). This work was supported by the ATIP-Avenir program, ANRS (France Recherche Nord & Sud Sida-hiv Hépatites), Ville de Paris Emergence program, European Seventh Framework Programme Marie Curie Actions (grant 268311), Labex Vaccine Research Institute (ANR-10-LABX-77), Labex DCBIOL (ANR-10-IDEX-0001-02 PSL* and ANR-11-LABX-0043), ACTERIA Foundation, Fondation Schlumberger pour l'Education et la Recherche, National Research Agency (ANR-14-CE14-0004-02), DIM Biothérapies (2015), and the European Research Council (grant 309848 HIVINNAC57BLTE) for N. Manel and by the National Research Agency (ANR-14-CE14-0026 Lumugène) and the Cancer national institute (INCa convention 2014-1-PL BIO-10-INSERM 5-1) for F. Rieux-Laucat.
N. Manel declares to be cofounder of the Stimunity company, which develops STING-targeting drugs. U. Gehrmann is currently employed by AstraZeneca Gothenburg but discloses no potential conflict of interest. The authors declare no further competing financial interests.
Footnotes
Abbreviations used:
- cGAMP
- cyclic guanosine monophosphate–adenosine monophosphate
- cGAS
- cGAMP synthase
- CTT
- C-terminal tail
- ISG
- IFN-stimulated gene
- PFA
- paraformaldehyde
- PI
- propidium iodide
- STING
- stimulator of IFN genes
References
- Abe T., and Barber G.N.. 2014. Cytosolic-DNA-mediated, STING-dependent proinflammatory gene induction necessitates canonical NF-κB activation through TBK1. J. Virol. 88:5328–5341. 10.1128/JVI.00037-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruchard M., Rebé C., Derangère V., Togbé D., Ryffel B., Boidot R., Humblin E., Hamman A., Chalmin F., Berger H., et al. 2015. The receptor NLRP3 is a transcriptional regulator of TH2 differentiation. Nat. Immunol. 16:859–870. 10.1038/ni.3202 [DOI] [PubMed] [Google Scholar]
- Burdette D.L., Monroe K.M., Sotelo-Troha K., Iwig J.S., Eckert B., Hyodo M., Hayakawa Y., and Vance R.E.. 2011. STING is a direct innate immune sensor of cyclic di-GMP. Nature. 478:515–518. 10.1038/nature10429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chien Y., Scuoppo C., Wang X., Fang X., Balgley B., Bolden J.E., Premsrirut P., Luo W., Chicas A., Lee C.S., et al. 2011. Control of the senescence-associated secretory phenotype by NF-κB promotes senescence and enhances chemosensitivity. Genes Dev. 25:2125–2136. 10.1101/gad.17276711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ching L.M., Cao Z., Kieda C., Zwain S., Jameson M.B., and Baguley B.C.. 2002. Induction of endothelial cell apoptosis by the antivascular agent 5,6-dimethylxanthenone-4-acetic acid. Br. J. Cancer. 86:1937–1942. 10.1038/sj.bjc.6600368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colanzi A., and Sütterlin C.. 2013. Signaling at the Golgi during mitosis. Methods Cell Biol. 118:383–400. 10.1016/B978-0-12-417164-0.00023-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Courtois G., Smahi A., Reichenbach J., Döffinger R., Cancrini C., Bonnet M., Puel A., Chable-Bessia C., Yamaoka S., Feinberg J., et al. 2003. A hypermorphic IκBα mutation is associated with autosomal dominant anhidrotic ectodermal dysplasia and T cell immunodeficiency. J. Clin. Invest. 112:1108–1115. 10.1172/JCI18714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crow Y.J., and Manel N.. 2015. Aicardi-Goutières syndrome and the type I interferonopathies. Nat. Rev. Immunol. 15:429–440. 10.1038/nri3850 [DOI] [PubMed] [Google Scholar]
- Gao D., Wu J., Wu Y.T., Du F., Aroh C., Yan N., Sun L., and Chen Z.J.. 2013. Cyclic GMP-AMP synthase is an innate immune sensor of HIV and other retroviruses. Science. 341:903–906. 10.1126/science.1240933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gentili M., Kowal J., Tkach M., Satoh T., Lahaye X., Conrad C., Boyron M., Lombard B., Durand S., Kroemer G., et al. 2015. Transmission of innate immune signaling by packaging of cGAMP in viral particles. Science. 349:1232–1236. 10.1126/science.aab3628 [DOI] [PubMed] [Google Scholar]
- Greil J., Rausch T., Giese T., Bandapalli O.R., Daniel V., Bekeredjian-Ding I., Stütz A.M., Drees C., Roth S., Ruland J., et al. 2013. Whole-exome sequencing links caspase recruitment domain 11 (CARD11) inactivation to severe combined immunodeficiency. J. Allergy Clin. Immunol. 131:1376–83.e3. 10.1016/j.jaci.2013.02.012 [DOI] [PubMed] [Google Scholar]
- Ishikawa H., and Barber G.N.. 2008. STING is an endoplasmic reticulum adaptor that facilitates innate immune signalling. Nature. 455:674–678. 10.1038/nature07317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishikawa H., Ma Z., and Barber G.N.. 2009. STING regulates intracellular DNA-mediated, type I interferon-dependent innate immunity. Nature. 461:788–792. 10.1038/nature08476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwasaki A., and Medzhitov R.. 2010. Regulation of adaptive immunity by the innate immune system. Science. 327:291–295. 10.1126/science.1183021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwasaki A., and Medzhitov R.. 2015. Control of adaptive immunity by the innate immune system. Nat. Immunol. 16:343–353. 10.1038/ni.3123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janeway C.A. Jr., and Medzhitov R.. 2002. Innate immune recognition. Annu. Rev. Immunol. 20:197–216. 10.1146/annurev.immunol.20.083001.084359 [DOI] [PubMed] [Google Scholar]
- Jeremiah N., Neven B., Gentili M., Callebaut I., Maschalidi S., Stolzenberg M.C., Goudin N., Frémond M.L., Nitschke P., Molina T.J., et al. 2014. Inherited STING-activating mutation underlies a familial inflammatory syndrome with lupus-like manifestations. J. Clin. Invest. 124:5516–5520. 10.1172/JCI79100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin L., Xu L.G., Yang I.V., Davidson E.J., Schwartz D.A., Wurfel M.M., and Cambier J.C.. 2011. Identification and characterization of a loss-of-function human MPYS variant. Genes Immun. 12:263–269. 10.1038/gene.2010.75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J.H., Lee S.R., Li L.H., Park H.J., Park J.H., Lee K.Y., Kim M.K., Shin B.A., and Choi S.Y.. 2011. High cleavage efficiency of a 2A peptide derived from porcine teschovirus-1 in human cell lines, zebrafish and mice. PLoS One. 6:e18556 10.1371/journal.pone.0018556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kranzusch P.J., Wilson S.C., Lee A.S., Berger J.M., Doudna J.A., and Vance R.E.. 2015. Ancient origin of cGAS-STING reveals mechanism of universal 2′,3′ cGAMP signaling. Mol. Cell. 59:891–903. 10.1016/j.molcel.2015.07.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lahaye X., Satoh T., Gentili M., Cerboni S., Conrad C., Hurbain I., El Marjou A., Lacabaratz C., Lelièvre J.D., and Manel N.. 2013. The capsids of HIV-1 and HIV-2 determine immune detection of the viral cDNA by the innate sensor cGAS in dendritic cells. Immunity. 39:1132–1142. 10.1016/j.immuni.2013.11.002 [DOI] [PubMed] [Google Scholar]
- Lahaye X., Satoh T., Gentili M., Cerboni S., Silvin A., Conrad C., Ahmed-Belkacem A., Rodriguez E.C., Guichou J.F., Bosquet N., et al. 2016. Nuclear envelope protein SUN2 promotes cyclophilin-A-dependent steps of HIV replication. Cell Reports. 15:879–892. 10.1016/j.celrep.2016.03.074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X.D., Wu J., Gao D., Wang H., Sun L., and Chen Z.J.. 2013. Pivotal roles of cGAS-cGAMP signaling in antiviral defense and immune adjuvant effects. Science. 341:1390–1394. 10.1126/science.1244040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S., Cai X., Wu J., Cong Q., Chen X., Li T., Du F., Ren J., Wu Y.T., Grishin N.V., and Chen Z.J.. 2015. Phosphorylation of innate immune adaptor proteins MAVS, STING, and TRIF induces IRF3 activation. Science. 347:aaa2630 10.1126/science.aaa2630 [DOI] [PubMed] [Google Scholar]
- Liu Y., Jesus A.A., Marrero B., Yang D., Ramsey S.E., Montealegre Sanchez G.A., Tenbrock K., Wittkowski H., Jones O.Y., Kuehn H.S., et al. 2014. Activated STING in a vascular and pulmonary syndrome. N. Engl. J. Med. 371:507–518. 10.1056/NEJMoa1312625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malik P., Zuleger N., de las Heras J.I., Saiz-Ros N., Makarov A.A., Lazou V., Meinke P., Waterfall M., Kelly D.A., and Schirmer E.C.. 2014. NET23/STING promotes chromatin compaction from the nuclear envelope. PLoS One. 9:e111851 10.1371/journal.pone.0111851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malissen M., Gillet A., Ardouin L., Bouvier G., Trucy J., Ferrier P., Vivier E., and Malissen B.. 1995. Altered T cell development in mice with a targeted mutation of the CD3-epsilon gene. EMBO J. 14:4641–4653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyts I., and Casanova J.L.. 2016. A human inborn error connects the α’s. Nat. Immunol. 17:472–474. 10.1038/ni.3420 [DOI] [PubMed] [Google Scholar]
- Onorati M., Li Z., Liu F., Sousa A.M., Nakagawa N., Li M., Dell’Anno M.T., Gulden F.O., Pochareddy S., Tebbenkamp A.T., et al. 2016. Zika virus disrupts phospho-TBK1 localization and mitosis in human neuroepithelial stem cells and radial glia. Cell Reports. 16:2576–2592. 10.1016/j.celrep.2016.08.038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pannicke U., Baumann B., Fuchs S., Henneke P., Rensing-Ehl A., Rizzi M., Janda A., Hese K., Schlesier M., Holzmann K., et al. 2013. Deficiency of innate and acquired immunity caused by an IKBKB mutation. N. Engl. J. Med. 369:2504–2514. 10.1056/NEJMoa1309199 [DOI] [PubMed] [Google Scholar]
- Pillai S., Nguyen J., Johnson J., Haura E., Coppola D., and Chellappan S.. 2015. Tank binding kinase 1 is a centrosome-associated kinase necessary for microtubule dynamics and mitosis. Nat. Commun. 6:10072 10.1038/ncomms10072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pourcelot M., Zemirli N., Silva Da Costa L., Loyant R., Garcin D., Vitour D., Munitic I., Vazquez A., and Arnoult D.. 2016. The Golgi apparatus acts as a platform for TBK1 activation after viral RNA sensing. BMC Biol. 14:69 10.1186/s12915-016-0292-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma S., Campbell A.M., Chan J., Schattgen S.A., Orlowski G.M., Nayar R., Huyler A.H., Nündel K., Mohan C., Berg L.J., et al. 2015. Suppression of systemic autoimmunity by the innate immune adaptor STING. Proc. Natl. Acad. Sci. USA. 112:E710–E717. 10.1073/pnas.1420217112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinkai Y., Rathbun G., Lam K.P., Oltz E.M., Stewart V., Mendelsohn M., Charron J., Datta M., Young F., Stall A.M., and Alt F.W.. 1992. RAG-2-deficient mice lack mature lymphocytes owing to inability to initiate V(D)J rearrangement. Cell. 68:855–867. 10.1016/0092-8674(92)90029-C [DOI] [PubMed] [Google Scholar]
- Stepensky P., Keller B., Buchta M., Kienzler A.K., Elpeleg O., Somech R., Cohen S., Shachar I., Miosge L.A., Schlesier M., et al. 2013. Deficiency of caspase recruitment domain family, member 11 (CARD11), causes profound combined immunodeficiency in human subjects. J. Allergy Clin. Immunol. 131:477–85.e1. 10.1016/j.jaci.2012.11.050 [DOI] [PubMed] [Google Scholar]
- Sun L., Wu J., Du F., Chen X., and Chen Z.J.. 2013. Cyclic GMP-AMP synthase is a cytosolic DNA sensor that activates the type I interferon pathway. Science. 339:786–791. 10.1126/science.1232458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka Y., and Chen Z.J.. 2012. STING specifies IRF3 phosphorylation by TBK1 in the cytosolic DNA signaling pathway. Sci. Signal. 5:ra20 10.1126/scisignal.2002521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang C.H., Zundell J.A., Ranatunga S., Lin C., Nefedova Y., Del Valle J.R., and Hu C.C.. 2016. Agonist-mediated activation of STING induces apoptosis in malignant B cells. Cancer Res. 76:2137–2152. 10.1158/0008-5472.CAN-15-1885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres J.M., Martinez-Barricarte R., García-Gómez S., Mazariegos M.S., Itan Y., Boisson B., Rholvarez R., Jiménez-Reinoso A., del Pino L., Rodríguez-Pena R., et al. 2014. Inherited BCL10 deficiency impairs hematopoietic and nonhematopoietic immunity. J. Clin. Invest. 124:5239–5248. 10.1172/JCI77493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J., Sun L., Chen X., Du F., Shi H., Chen C., and Chen Z.J.. 2013. Cyclic GMP-AMP is an endogenous second messenger in innate immune signaling by cytosolic DNA. Science. 339:826–830. 10.1126/science.1229963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan N., Regalado-Magdos A.D., Stiggelbout B., Lee-Kirsch M.A., and Lieberman J.. 2010. The cytosolic exonuclease TREX1 inhibits the innate immune response to human immunodeficiency virus type 1. Nat. Immunol. 11:1005–1013. 10.1038/ni.1941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi G., Brendel V.P., Shu C., Li P., Palanathan S., and Cheng Kao C.. 2013. Single nucleotide polymorphisms of human STING can affect innate immune response to cyclic dinucleotides. PLoS One. 8:e77846 10.1371/journal.pone.0077846 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.