Although chromosome fragments lacking a centromere would be expected to show severe defects in their segregation during anaphase, they do exhibit poleward movement by an unclear mechanism. Karg et al. now show how microtubules and the chromokinesin Klp3a can work together to successfully segregate chromosome fragments to daughter nuclei.
Abstract
Although poleward segregation of acentric chromosomes is well documented, the underlying mechanisms remain poorly understood. Here, we demonstrate that microtubules play a key role in poleward movement of acentric chromosome fragments generated in Drosophila melanogaster neuroblasts. Acentrics segregate with either telomeres leading or lagging in equal frequency and are preferentially associated with peripheral bundled microtubules. In addition, laser ablation studies demonstrate that segregating acentrics are mechanically associated with microtubules. Finally, we show that successful acentric segregation requires the chromokinesin Klp3a. Reduced Klp3a function results in disorganized interpolar microtubules and shortened spindles. Normally, acentric poleward segregation occurs at the periphery of the spindle in association with interpolar microtubules. In klp3a mutants, acentrics fail to localize and segregate along the peripheral interpolar microtubules and are abnormally positioned in the spindle interior. These studies demonstrate an unsuspected role for interpolar microtubules in driving acentric segregation.
Introduction
Eukaryotic cells possess robust mechanisms that safeguard against genomic damage. In the event of damage, cells activate checkpoint pathways that inhibit cell cycle progression, providing time for repair or elimination of the genetically compromised cells through apoptosis (Ermolaeva and Schumacher, 2014). Although we have learned a great deal regarding the function of checkpoint and DNA repair pathways during interphase (G1, S, and G2), little is known concerning the cellular response to genetic damage as cells progress through metaphase. Studies show that entry into metaphase with damaged DNA elicits either the spindle assembly or DNA damage checkpoint depending on the cell type (Mikhailov et al., 2002; Royou et al., 2005).
In spite of these safeguards, cells occasionally exit metaphase with unrepaired double-strand breaks (DSBs). The presence of DSBs at metaphase is particularly troublesome, because they can result in the formation of chromosome fragments lacking a centromere. Known as acentrics, these fragments are incapable of forming the microtubule–kinetochore attachments that drive poleward chromosome segregation. Consequently, acentrics are expected to lag on the cell equator and exhibit severe segregation defects. However, several recent studies demonstrate poleward migration of acentric chromosomes. In budding yeast and Drosophila melanogaster, acentrics are transmitted through many generations (Sandell and Zakian, 1993; Malkova et al., 1996; Ahmad and Golic, 1998; Galgoczy and Toczyski, 2001; Titen and Golic, 2008). Segregation of acentrics has been observed in Drosophila polyploid cells, Caenorhabditis elegans meiosis, Scadoxus multiflorus, and mammalian cells (Bajer, 1958; Liang et al., 1993; Khodjakov et al., 1996; Kanda et al., 1998; Kanda and Wahl, 2000; Muscat et al., 2015; Bretscher and Fox, 2016). Poleward directed movements of acentrics during mitosis have been previously reported in several different species (Bajer, 1958; Khodjakov et al., 1996; Kanda et al., 1998; Platero et al., 1999; Kanda and Wahl, 2000; Ishii et al., 2008; Titen and Golic, 2008). Proposed mechanisms of acentric segregation include neo-centromere formation and direct association of the acentrics with a kinetochore-bearing chromosome (Platero et al., 1999; Kanda et al., 2001; Ishii et al., 2008; Ohno et al., 2016). Despite these studies of poleward-directed movements of acentrics, the forces that power acentric segregation during mitosis remain poorly understood.
Recent studies have also demonstrated the presence of a DNA tether that connects acentrics to the main chromosome mass, suggesting that the tether is required for acentric segregation. The DNA tether consists primarily of DNA along with histones and is coated with Polo, Bub3, and BubR1 kinases and the chromosomal passenger proteins Aurora B and INCENP and Cdc20 (Royou et al., 2010; Derive et al., 2015). During metaphase, the acentric sisters are held together and are positioned toward the outer edge of the metaphase plate. At the onset of anaphase, through an unknown mechanism, acentric sisters remain held together on the spindle equator while the rest of the chromosome mass travels toward the pole. Eventually, the acentric sisters separate, migrate toward opposite spindle poles and rejoin the rest of the chromosome mass. The movement of acentrics toward the pole has been proposed to occur by the action of the DNA tether. Support for this hypothesis comes from experiments showing that disruptions in either BubR1 or Polo kinases, which decorate DNA tethers, result in defects in acentric segregation (Royou et al., 2010).
Several studies also suggest that interactions with microtubules can drive poleward segregation of acentrics. Acentric fragments generated in some plant cells (S. multiflorus) are transported from the spindle equator to the spindle poles during anaphase while associated with microtubules (Bajer, 1958; Khodjakov et al., 1996). Experiments in crane fly spermatocytes have shown that acentrics generated by laser microsurgery during metaphase can move poleward in association with microtubule flux, suggesting that the fluxing microtubule lattice can exert a force on acentrics (LaFountain et al., 2001). More recent work in C. elegans has demonstrated that chromosomes can move poleward during meiosis by a kinetochore-independent mechanism (Wignall and Villeneuve, 2009; Dumont et al., 2010; Muscat et al., 2015). In these studies, chromosomes lacking kinetochores were laterally associated with microtubules and relied on plus-end–directed kinesin motors to congress on the metaphase plate, whereas poleward-directed motion relied on dynein-based minus-end–directed forces (Wignall and Villeneuve, 2009; Muscat et al., 2015).
Here, we use a combination of genetic and laser ablation approaches to examine the role of microtubules and motor proteins in driving acentric segregation in Drosophila neuroblasts. Acentrics are efficiently produced in Drosophila neuroblasts by expressing the I-CreI endonuclease, which makes double-stranded DNA breaks in the ribosomal DNA repeats in the centric heterochromatin of the X chromosome (Rong et al., 2002; Royou et al., 2010). Our analysis reveals that acentric segregation relies on the chromokinesin Klp3a and microtubules. We refer to the population of microtubules that associate with acentrics as interpolar microtubules, because these microtubules extend from the centrosome toward the metaphase plate but do not bind to a kinetochore (Dumont and Mitchison, 2009). In these studies, we reveal an unsuspected role of interpolar microtubules and Klp3a in segregating acentric chromosome fragments during anaphase and telophase.
Results
Acentric sister chromosome separation and segregation is delayed relative to intact chromosomes
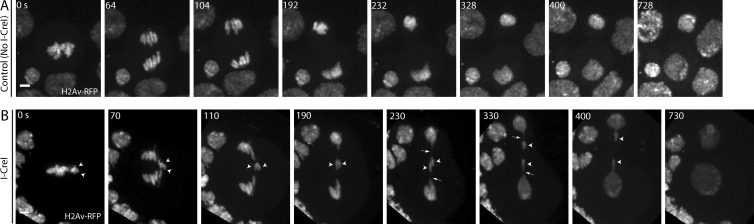
A dividing non–I-CreI–expressing neuroblast (control) with the chromosomes labeled with a red fluorescent protein (RFP)–tagged histone H2 variant (H2Av) is shown in Fig. 1 A. As previously reported by Royou et al. (2010) and shown again here in Fig. 1 B (Video 1), I-CreI expression produces sister acentrics positioned on the outer edge of the metaphase plate (Fig. 1 B, arrowheads, 0 s). These acentrics lag on the spindle equator well after the intact sister chromosomes have separated and moved poleward (70–230 s). Sister acentric separation occurs 230 s after the separation of intact chromosomes. In spite of the delayed segregation, the acentric chromosomes are successfully incorporated into the newly formed daughter nuclei (400–730 s). Although not visible in this image, a thin DNA tether (Fig. 1 B, arrow) connects the lagging acentrics to the main chromosome mass (Royou et al., 2010). The DNA tether has been proposed to facilitate the poleward movement of acentric chromosomes (Royou et al., 2010; Derive et al., 2015). By mid to late anaphase, the tether is thought to act like a rope facilitating the pulling of the acentric into daughter nuclei. An important prediction of this tether-based model for acentric segregation is that acentrics segregate poleward with their tether-associated broken end leading and their single telomere oriented toward the spindle equator.
Figure 1.
Acentric separation and segregation is delayed relative to intact chromosomes. (A) Still images from a time-lapse movie of a mitotic neuroblast without I-CreI. Chromosomes are labeled with H2Av-RFP. (B) Images from a time-lapse movie of a mitotic neuroblast with I-CreI–induced acentrics (Video 1). Acentrics (arrowheads) are pushed to edge of plate at 0 s. Acentrics lag on the spindle equator (0–190 s) but eventually move poleward (230–400 s). Bars, 2 µm. Time in seconds.
Poleward segregating acentrics are randomly orientated
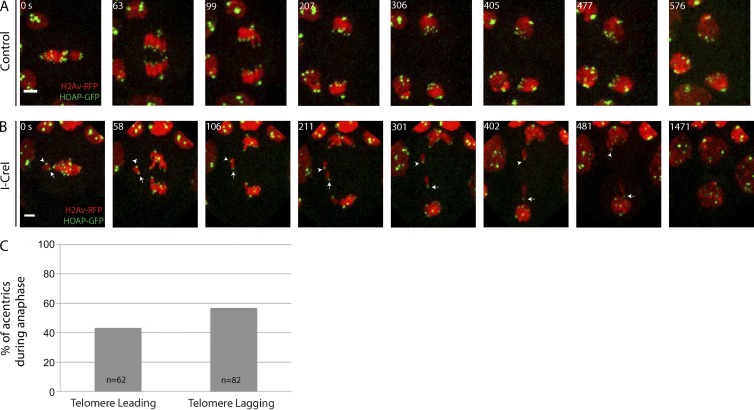
To investigate whether acentrics preferentially segregate poleward with their telomeres lagging, we performed live imaging taking advantage of GFP-tagged HOAP, a telomere-specific protein (Cenci et al., 2003). Time-lapse images of a control neuroblast expressing HOAP-GFP are shown in Fig. 2 A. No lagging chromosomes (red) are observed, and HOAP-GFP (green) marks the ends of chromosomes as expected. In I-CreI–expressing neuroblasts (Fig. 2 B and Video 2), acentrics travel poleward either with their telomeres lagging (facing the spindle equator as expected for intact chromosomes; arrowhead) or with their telomere leading (facing the spindle pole; arrow). Quantification of acentric telomere orientation reveals that acentric orientation is random, with approximately half the telomeres facing the pole (telomere leading, 43%, n = 62) and the other half of acentrics facing the spindle equator (telomere lagging, 57% n = 82; Fig. 2 C). χ2 analysis shows no statistical difference between the two telomere orientations (P = 0.096). The fact that nearly half of acentrics segregate poleward with their single telomere leading toward the pole suggests that forces other than or in addition to the tether propel acentrics poleward.
Figure 2.
Acentrics migrate toward the spindle pole during anaphase with their telomeres leading or lagging. Chromosomes labeled with H2Av-RFP (red) and the telomere protein HOAP labeled with GFP (green). (A) Images from a time-lapse movie of a control mitotic neuroblast. (B) Still images from a time-lapse movie of a mitotic neuroblast with I-CreI–induced acentrics (Video 2). Acentrics lag on the spindle equator (0–106 s) but eventually separate (211 s) and move poleward with their telomere either facing the spindle equator (arrowhead) or spindle pole (arrow). Bars, 2 µm. Time in seconds. (C) Percentages of acentrics that migrate with telomeres leading (n = 62) or lagging (n = 82).
Lagging acentrics associate with interpolar microtubules during anaphase
To determine whether microtubules provide a role in acentric segregation, we simultaneously imaged acentric chromosome segregation and microtubule dynamics in dividing neuroblasts. Microtubules were visualized by expressing a GFP-tagged microtubule-associated protein called Jupiter (Karpova et al., 2006). Time-lapse images from a control neuroblast (not expressing I-CreI) are shown in Fig. 3 A. In these control images, the intact sisters (red) are attached to the mitotic spindle (green) at metaphase (0 s). By anaphase (82–267 s), the central spindle forms while the separating sisters reach the poles.
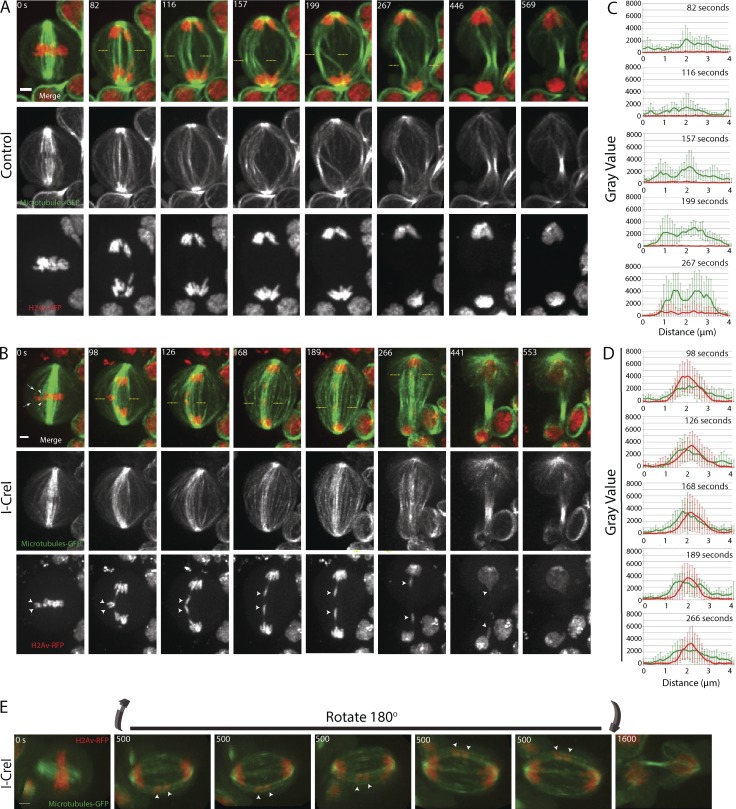
Figure 3.
Acentrics travel poleward in anaphase while highly associated with microtubules. Microtubules are in green and chromosomes in red. (A) Images from a time-lapse movie of a control neuroblast from metaphase (0 s) through telophase (569 s). (B) Still images from a time-lapse movie of a mitotic neuroblast with I-CreI–induced acentrics (Video 3). Acentrics are positioned to the edge of the metaphase plate while in contact with microtubules (arrows and arrowheads in merge panel at 0 s). Sister acentrics (arrowheads in H2Av-RFP panels) are held together on the cell equator after the intact sisters have separated (0 to 98 s). Sister acentrics eventually separate and move toward opposite poles while associated with microtubules. In all 47 cells imaged, the acentrics were strongly associated with microtubules. (C) Line graphs from a compilation of seven control videos showing the relative fluorescence intensities in arbitrary units (AU) of microtubules (green line) and chromosomes (red) calculated between the yellow dashed lines at the time points 82, 116, 157, 199, and 267 s after anaphase. (D) Line graphs from a compilation of seven videos of I-CreI–expressing neuroblasts showing the relative fluorescence intensities of microtubules (green line) and chromosomes (red) calculated between the yellow dashed lines at time points 98, 126, 168, 189, and 266 s after anaphase. Bars, 2 µm. Time in seconds. Error bars represent SDs of the fluorescent intensities at all points tested. (E) 3D rendering of a video of a neuroblast division with I-CreI induced acentrics (Video 4). Bar, 2 µm. The 3D rendering from a 180° rotation from multiple images taken at the same time point (500 s) show the association of acentrics with microtubules (arrowheads). Similar 3D rendering were generated from a total of seven videos.
The simultaneous live imaging of acentrics and microtubules (Fig. 3 B and Video 3) reveals several features suggesting that acentric movements are influenced by the spindle. First, during metaphase, sister acentrics (Fig. 3 B, arrowheads in H2Av-RFP–alone panels) are positioned at the outer edge of the metaphase plate away from the main mass of chromosomes (0 s). The positioning of acentrics on the edge of metaphase plate coincides with a gap between the acentrics and the main chromosome mass. As seen in the merged image in Fig. 3 B (0 s), this gap is associated with a high concentration of microtubules (arrowheads) that may play a role in positioning the acentrics. Similarly, an additional but less concentrated subset of microtubules associates with the outermost edge of the sister acentrics at metaphase (Fig. 3 B, dashed arrows in merge image at 0 s). The positioning of acentrics on the edge of the plate in Drosophila has been previously reported (Royou et al., 2010), as well as a similar positioning of chromosome fragments lacking kinetochores in human cell culture, which is purported to be dependent on astral microtubules (O’Connell et al., 2009). The acentrics are held together in a horizontal orientation (parallel to the cell equator), after the intact chromosomes migrate poleward (0–98 s). Concomitant with the separation and poleward segregation of these acentrics is their rotation from a perpendicular (Fig. 3 B, arrowheads in bottom panel at 98 s) to a parallel spindle orientation (Fig. 3 B, arrowheads in bottom panels at 98–168 s). Significantly, during the entire poleward migration of the acentrics, they are in close association and often appear embedded within microtubules (126–441 s). This analysis suggests that acentrics rely on a microtubule-based mechanism for their segregation and incorporation into daughter nuclei.
To quantify the association of acentrics with microtubules, we calculated the fluorescence intensities of the microtubules (green) and acentrics (red) from a collection of images from at least seven movies like the images shown in Fig. 3 (A and B). A compilation of these line-scan analyses of both control and I-CreI–expressing neuroblasts (between yellow dashed lines) is shown in Fig. 3 (C and D, respectively). The absence of a lagging acentric in control (non–I-CreI–expressing) neuroblasts shows a high fluorescence intensity of midzone microtubules (green line) without any fluorescence signal from chromosomes (red line) at all time points tested (Fig. 3 C, 82, 116, 157, 199, and 267 s). Line-scan analysis of images from neuroblasts expressing I-CreI shows a striking overlap of microtubule fluorescence intensity (green line) with the fluorescence intensity from acentrics (red line) at all time points tested (Fig. 3 D; 98, 126, 168, 189, and 266 s). Collectively, these data demonstrate that acentrics are intimately associated with microtubules during their poleward segregation.
To determine whether the association of microtubules and acentrics is maintained for a significant period of time, as would be expected if they are physically associated, we imaged live over multiple Z-planes to generate 3D renderings of the metaphase-to-telophase transition in acentric bearing neuroblasts. These 3D reconstructions of microtubules (green) and chromosomes (red) show lagging acentrics (Fig. 3 E, arrowheads; and Video 4) positioned at the peripheral of the spindle in association with the interpolar microtubules. The path of the acentric poleward segregation follows this arc of the interpolar microtubules. 3D renderings generated from seven different videos show similar acentric and microtubule associations as seen in Fig. 3 E. These 3D movies reveal that the segregating acentrics are embedded in a distinct pool of bundled microtubules. The images in the 3D reconstructions in Fig. 3 E are not from the same videos used to make still frames in panels A and B.
Acentrics are mechanically linked to microtubules
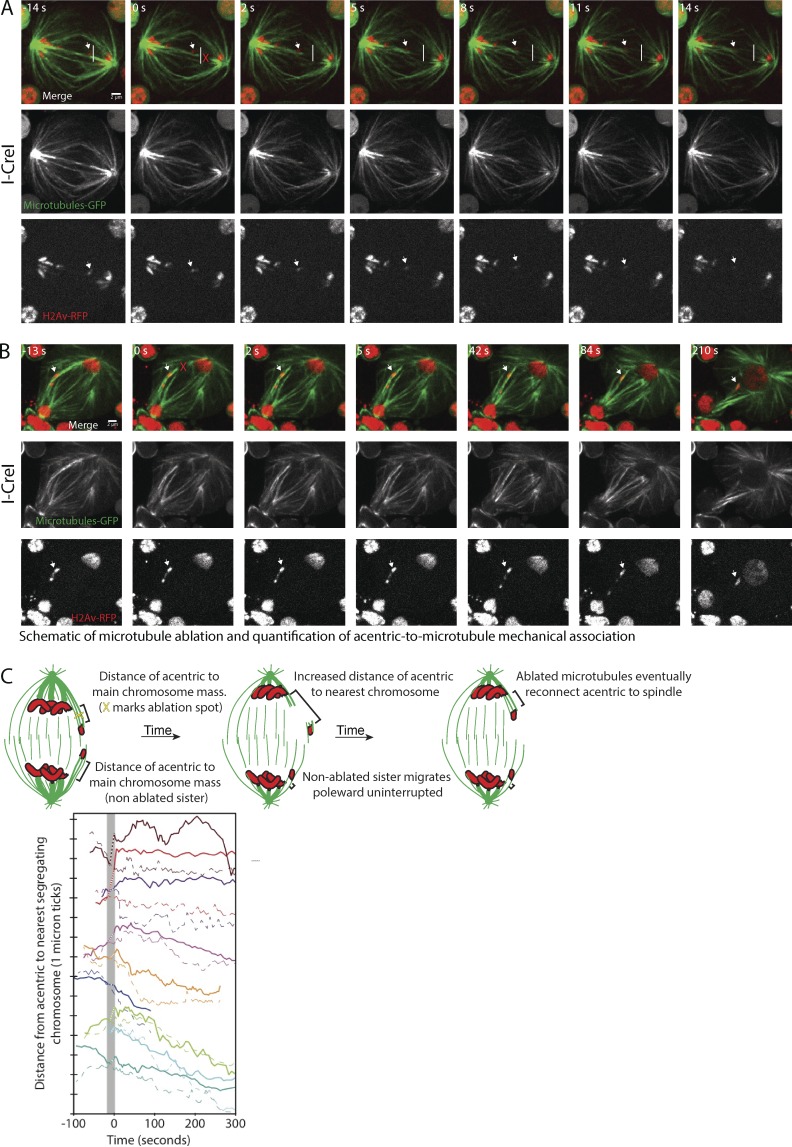
The prominent association of microtubules with segregating acentrics suggests that they rely on microtubules for their poleward transport. To directly test the role of microtubules in acentric poleward movement, we used laser ablation to sever the bundled microtubules connecting acentrics delayed on the spindle equator to the spindle pole. As shown in Fig. 4 A (Video 5), acentrics (arrow) segregate toward a spindle pole. The white line in Fig. 4 A marks the furthest distance the acentric has segregated toward a pole. As shown in Fig. 4 A, laser ablation of the microtubules positioned between the lagging acentric and the pole (red X, 0 s) results in retraction toward the spindle equator of acentrics and associated microtubules and, on the other side, depolymerization of putative microtubule plus ends (see Materials and methods). Importantly, we find that the acentric (Fig. 4 A, arrow) retracts with and to the same extent as the microtubules retracting (or possibly depolymerizing) toward the spindle equator, suggesting that the acentric is physically linked to microtubules. In five out of nine ablated dividing neuroblasts (Fig. 4 C, top five traces), we observed such a retraction toward the spindle equator of the acentric after ablation (solid traces), whereas sister acentrics continued their normal segregation (dotted traces). In each of these cases, this acentric retraction was accompanied by a corresponding movement of acentric-associated microtubules, as in Fig. 4 A. In some of these cases (Fig. 4 C, top three traces: dark red, red, and purple), acentric segregation stopped or stalled after retraction, whereas in other cases (magenta and orange), the acentrics seemed to reconnect to the pole and resumed segregation. The concurrent retractions of the microtubules and acentrics suggest that acentrics are mechanically linked to microtubules as acentrics travel from the spindle equator to the pole. Retraction of the acentrics and microtubules toward the spindle equator suggests that before ablation, either the associated microtubule bundles or the acentrics are under tension, which is released once they are detached from the pole. Consistent with acentric tension, previous studies show that separating acentrics remain connected by thin stretches of DNA, potentially from DNA catenations (Royou et al., 2010; Derive et al., 2015). According to this model, the persistent DNA connections between acentrics may contain elastic properties that may contribute to acentric retraction toward the spindle equator and the opposing acentric sister after ablation. Although this model may partly account for acentric retraction, the fact that acentrics and microtubule retract in association with each other suggest a physical linkage between the acentric and microtubules.
Figure 4.
Acentrics are mechanically linked to microtubules during anaphase. (A) Still images from a time-lapse movie (Video 5) of a mitotic neuroblast with I-CreI–induced acentrics. Acentrics (arrows) are associated with microtubules in midzone that extend to spindle pole just before ablation (−14 s). The white line indicates the furthest edge the acentric has traveled toward right hand spindle pole just before ablation (−14 s). The ablated (red X at 0 s) microtubules show an immediate retraction or depolymerization at both ends of microtubules. Acentric (arrow) retracts in association with ablated microtubules, suggesting the two structures are physically linked. (B) Still images from a time-lapse movie of a mitotic neuroblast with I-CreI–induced acentrics. Acentrics (arrow) are associated with microtubules in midzone that extend to spindle pole just before ablation (−13 s). The ablated microtubules (red X at 0 s) show an immediate retraction, however, the ablated microtubules are reassociated with neighboring microtubules by 42 s after ablation, providing tracks for acentrics to continue poleward segregation. Chromosomes are labeled in red (H2Av-RFP) and microtubules are labeled in green (Jupiter-GFP). Bars, 2 µm. (C, top) Schematic showing ablation site (yellow X) of microtubules that extend from pole to acentrics during anaphase and distances from the acentrics to the nearest chromosome mass. (bottom) The distance of example acentrics from the nearest normal (undamaged) chromosome after ablation is plotted over time. Acentrics with ablated microtubules are shown as solid lines, and their corresponding sisters are shown as dotted lines, with each color indicating an individual acentric pair. The first frame after ablation is set to 0 s, and dotted sections of the solid traces (highlighted with a gray bar) connect the last frame before ablation and first frame after ablation. Because ablation has already occurred by the first frame after ablation, sometimes a mechanical response starts before that first frame.
Although we observe a clear mechanical association with acentrics in five out of nine ablated acentric associated microtubules, in the other four cases (Fig. 4, B and C, bottom four traces in dark blue, green, light blue, and cyan), we did not detect a retraction of either microtubules or acentrics after ablation. In these cases, microtubules connecting acentrics to the pole may not be completely severed, or their connection to the pole may be restored more quickly than we can detect a retraction. Such reconnection is also likely to occur in cases where we observed a retraction that was not accompanied by an extended pause in acentric segregation after ablation. For example, in Fig. 4 B, by 42 s after ablation, microtubule ends have clearly established indirect connections (through other microtubules) to poles, and microtubules in other planes may do so even more quickly. Indeed, previous studies have found that minus ends of ablated microtubules undergo dynein-powered poleward movement via an indirect connection to the pole through neighboring microtubules within ∼15 s of ablation (Elting et al., 2014; Sikirzhytski et al., 2014), quickly enough to potentially prevent our detection of a pause.
Poleward movement of the acentric after ablation of BubR1-coated tethers
As previously reported, acentric fragments remain connected to the main mass of chromosomes by a thin stretch of DNA that is coated with BubR1, Bub3, Polo, Aurora B, INCENP, and Cdc20 (Royou et al., 2010; Derive et al., 2015). This thin stretch of DNA, which is referred to as a “DNA tether,” is required for successful acentric segregation (Royou et al., 2010; Derive et al., 2015). To determine whether or not the force for acentric segregation was exclusively generated by the tether or from microtubules, we used laser ablation to severe the DNA tether. The DNA tether was identified by a stretch of BubR1 signal that extended from the acentric to the main chromosome mass (Fig. S1, BubR1-GFP, green arrow). We ablated the tether at the location indicated by the arrowheads in Fig. S1 (12 s). Although difficult to fully verify complete severance, a mechanical response of the late segregating acentric after ablation and/or a loss of BubR1 signal between the lagging acentric and the main chromosome mass (Fig. S1, 12 s) under the same ablation conditions were consistent with successful tether ablation. Although some BubR1-GFP signal persisted on the lagging acentric, we did not see evidence of reformation of the BubR1-GFP signal that extended from the acentric back to the main chromosome mass, suggesting that the tether may not fully repair during mitosis. In spite of ablating the tether, the acentric migrated poleward and incorporated into the daughter telophase nucleus (Fig. S1, 32–475 s). We saw similarly correct acentric segregation after BubR1-GFP ablation in a total of four movies. We conclude that forces other than or in addition to the DNA tether provide the force driving acentric segregation.
The motor protein Klp3a is required for acentric segregation
Given the aforementioned studies that provide strong evidence for a role of microtubules in acentric segregation, we hypothesized that chromokinesins may be involved as well. Chromokinesins are motor proteins that interact with both microtubules and chromatin (Mazumdar and Misteli, 2005; Vanneste et al., 2011). Two well-characterized chromokinesin genes in Drosophila are the kinesin-10–related motor Nod (Theurkauf and Hawley, 1992) and the kinesin-4–related motor Klp3a (Williams et al., 1995). To genetically test this idea, we took advantage of the fact that in a wild-type background, I-CreI–induced generation of acentrics in the late larval stage does not induce lethality. However, in a genetically compromised background such as under conditions of reduced levels of the BubR1 or Polo kinases associated with the DNA tether, I-CreI induction results in significant lethality. Presumably, this is a result of increased aneuploidy caused by incorrect acentric segregation (Royou et al., 2010). To determine if a reduction in Klp3a levels also show an increase in synthetic lethality upon I-CreI induction, we expressed I-CreI in klp3a1124 mutant male third-instar larvae. klp3a1124 is an X-linked gene containing missense mutation that results in a glutamic acid to lysine exchange at residue 829 (Page and Hawley, 2005). I-CreI–induced synthetic lethality was also tested in male third instars bearing a loss-of-function mutation in the X-linked gene nod4 (Theurkauf and Hawley, 1992). As shown in Table 1, klp3a1124 males produce a synthetic lethal interaction upon I-CreI induction. klp3a males in which I-CreI is expressed survive only 36% as well as klp3a males in which I-CreI is not expressed. The same experiment performed with nod males revealed that they survive 45% as well upon I-CreI expression (Table 1).
Table 1. Percentage of survival of third-instar to adulthood.
| Genotype | I-CreI expression | Number of experiments | Total number of larvae | Percentage of survival into adulthood (mean ± SD) |
|---|---|---|---|---|
| Wild type | No | 4 | 76 | 92 ± 9 |
| Yes | 4 | 85 | 78 ± 30 | |
| klp3a1124/Y | No | 9 | 139 | 44 ± 17 |
| Yes | 8 | 89 | 16 ± 15 | |
| nod4/Y | No | 3 | 33 | 51 ± 42 |
| Yes | 3 | 16 | 23 ± 20 |
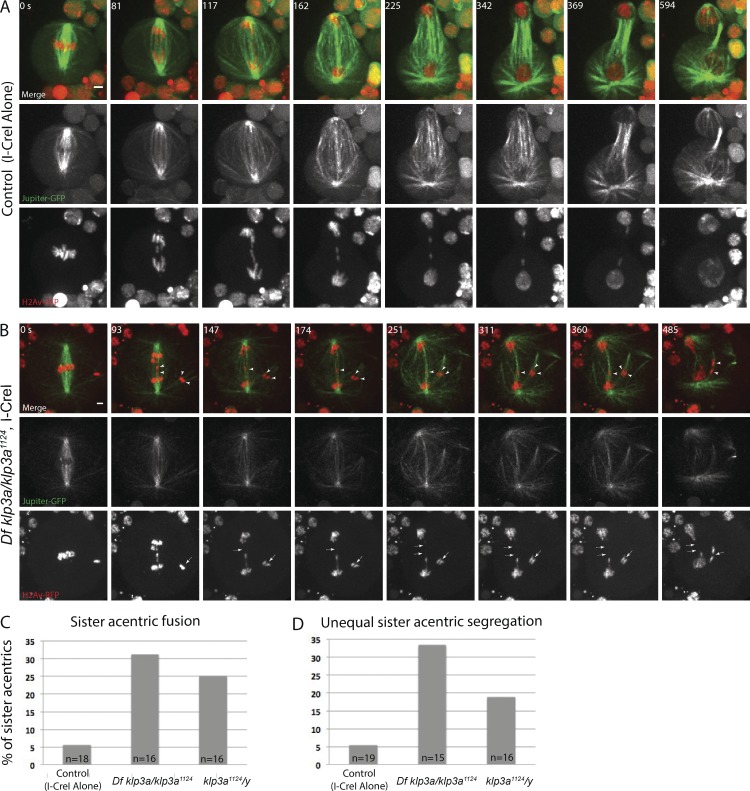
Because of the more pronounced synthetic lethality, we focused our efforts on cytologically examining acentric segregation in klp3a1124 mutant neuroblasts. In klp3a1124 neuroblasts, we find several defects in acentric segregation compared with controls (Fig. 5, A and B). First, the acentrics are often observed much further off the metaphase plate in the klp3a mutants. We also find an increase in the number of sister acentrics that fail to separate from each other during anaphase (Fig. 5 B, arrowheads in merged images; and Video 7). Instead, these sister acentrics appear to remain fused with each other or with the opposite daughter nucleus (Fig. 5 B, dashed arrows in H2Av-RFP–alone panels). Compared with acentrics generated in a wild-type background, acentrics generated in klp3a mutant males results in a statistically significant (P < 0.05; χ2) increase in sister acentric fusion rates (Fig. 5 C). We also observed an increased percentage in sister acentrics that unequally segregate into daughter cells compared with controls (Fig. 5 B, arrowheads). Compared with acentrics generated in a wild-type background, acentrics generated in klp3a mutant males results in a statistically significant (P < 0.05; χ2) increase in unequal segregation rates (Fig. 5 D). Collectively, these data reveal that Klp3a is required for normal acentric segregation and loss of Klp3a function results in an increased rate of aneuploidy.
Figure 5.
The chromokinesin Klp3a is required for segregating acentrics. (A) Still images from a time-lapse movie of a control neuroblast (I-CreI alone) showing sister acentrics (red) separating and moving to opposite poles while in close association with microtubules (green). (B) Still images from a time-lapse movie of a neuroblast from a female Df klp3a/klp3a1124 with I-CreI–induced acentrics (Video 7). In neuroblasts depleted of Klp3a, acentric sisters remain fused by a thin stretch of chromatin (dashed arrow) with each other or with the opposite daughter nucleus and segregate to the same pole. Bars, 2 µm. Time in seconds. (C and D) Bar graphs showing the percentage of control and klp3a-defective neuroblasts with sister acentric fusions and the percentage of control and klp3a-defective neuroblasts with unequal acentric segregation.
Acentric segregation along peripheral interpolar microtubules is disrupted in klp3a mutants
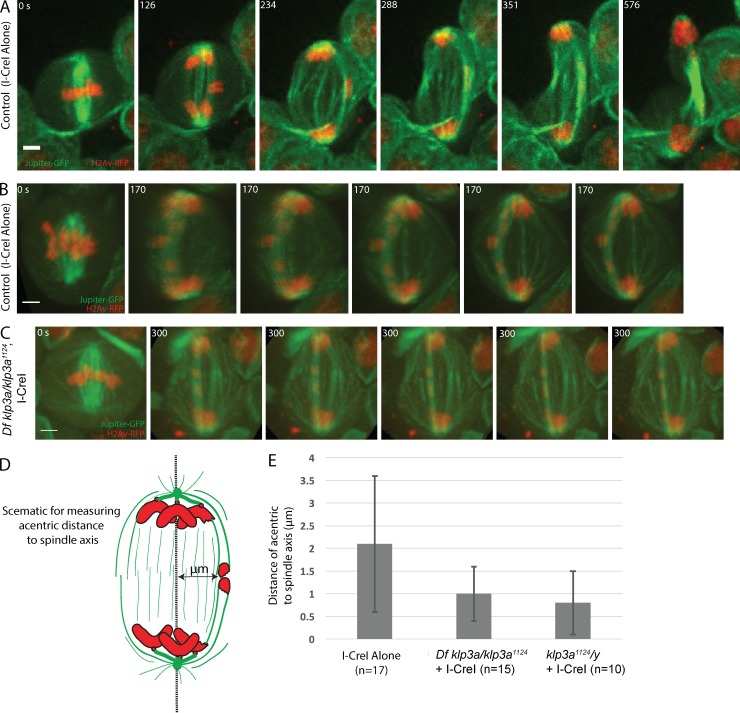
Live analysis revealed acentrics preferentially segregate along a peripheral arc of the mitotic spindle suggesting transport relies on interpolar microtubules (Fig. 6 A). To quantify this, we generated 3D movies of acentrics segregating in a wild-type and a klp3a mutant background (Fig. 6, B and C; and Video 8). For each movie, we measured the maximal distance of newly separated acentrics from a medial line drawn between centrosomes (Fig. 6 D). As shown in the graph in Fig. 6 E, the distance is significantly reduced from 2.1 ± 1.5 µm in I-CreI–alone controls to 0.8 ± 0.7 µm in a klp3a mutant background (P < 0.05, Bonferroni multiple comparison test). As previously reported (Kwon et al., 2004) and shown here (Fig. S2), the organization of the interpolar microtubules is disrupted in klp3a mutants. Given acentrics normally segregate along the arc of interpolar spindle microtubules and this class of microtubules is disrupted in klp3a mutants, these findings suggest a model in which acentric chromosome segregation preferentially relies on interpolar microtubules. That is, Klp3a is required for normal acentric segregation primarily because of its role in organizing interpolar microtubules.
Figure 6.
Acentric segregation along peripheral interpolar microtubules is disrupted in klp3a mutants. (A) Still images from a time-lapse movie of a mitotic neuroblast with I-CreI induced acentrics migrating poleward on an arch of microtubules on the periphery of spindle. (B) Still frames from a 3D rendering of a control I-CreI expressing neuroblast showing acentrics (red) positioned to the outer edge of spindle midzone during anaphase, while associated with an arch of pole-pole microtubules (green). (C) Still frames from a 3D rendering (Video 8) from a female Df klp3a/ klp3a1124 neuroblast showing that acentrics are within the middle of spindle midzone during anaphase. Bars, 2 µm. Time in seconds. (D) Schematic showing measurements of the maximal distance of newly separated acentrics from a medial line drawn between centrosomes. (E) Bar graphs showing the maximal distance (μm) of acentric from a medial line between centrosomes to the outer edge of the anaphase spindle midzone in control and Klp3a-defective neuroblast. Error bars represent one SD from the mean.
Discussion
Studies in a variety of organisms demonstrate that acentric chromosomes are capable of efficient poleward segregation during mitosis (Sandell and Zakian, 1993; Malkova et al., 1996; Ahmad and Golic, 1998; Galgoczy and Toczyski, 2001; Titen and Golic, 2008; Bretscher and Fox, 2016). Proposed mechanisms include neocentromere formation and direct association of acentric chromosomes with a kinetochore-bearing chromosome (Platero et al., 1999; Kanda et al., 2001; Ishii et al., 2008; Ohno et al., 2016). In addition, X-chromosome acentrics generated through I-CreI endonuclease induction in Drosophila neuroblasts are connected to the main chromosome mass by DNA tethers that are coated with Bub3, Cdc20, BubR1, Polo, INCENP, and Aurora B (Royou et al., 2010; Derive et al., 2015). Disruption of these components resulted in failed acentric segregation, suggesting that the DNA tethers may provide an elastic pulling force that drives acentric segregation (Royou et al., 2010; Derive et al., 2015). Here, we tested this idea using GFP-labeled telomeres (Cenci et al., 2003) and discovered that acentrics segregate poleward with their telomeres leading and lagging in approximately equal frequencies (Fig. 2). Because acentrics are connected to their centric partner via a DNA tether, if the tether were the primary force driving acentric segregation, one would expect segregation with telomeres lagging would be the predominant form of segregation. These results imply that forces in addition to the DNA tether drive acentric segregation.
Insight into these kinetochore-independent forces comes from studies examining nonkinetochore forces imposed on chromosome arms. For example, plus end–directed polar ejection forces can push on chromosome arms to assist in aligning chromosomes on the metaphase plate (Mazumdar and Misteli, 2005; Barisic et al., 2014). Additional studies demonstrate that kinetochore-independent lateral interactions can drive chromosome movements (Wignall and Villeneuve, 2009; Dumont et al., 2010; Muscat et al., 2015). In accord with these studies, we find I-CreI–generated acentrics maintain a close lateral association with microtubules during their entire journey from the spindle equator to the spindle pole (Figs. 3 and 4). These images suggest that microtubules are directly involved in acentric segregation. In accord with this interpretation, our laser ablation studies reveal a mechanical association between microtubules and acentrics. In addition, live 3D imaging reveals a robust bundle of microtubules encompassing the segregating acentrics. These images are strikingly similar to images of chromosome segregation in C. elegans meiosis, which involve lateral associations between microtubules and the segregating chromosomes (Muscat et al., 2015). Poleward transport of chromosome fragments via lateral microtubule interactions has also been observed in plant cells (Bajer, 1958; Bajer and Vantard, 1988; Khodjakov et al., 1996). Thus, our study adds to a growing body of literature indicating that lateral chromosome–microtubule associations may be a common alterative mechanism for transporting chromosomes poleward during anaphase. As end-on interactions between the kinetochore and k-fiber microtubules are typically the dominant force in driving poleward chromosome segregation, we suspect that these lateral interactions are more readily observed on chromatin lacking a kinetochore. Lateral interactions between chromosomes and microtubules are well documented during chromosome congression (Barisic et al., 2014; Drpic et al., 2015). It may be that the factors promoting these lateral interactions during congression at prometaphase may also promote the observed lateral interactions between microtubules and acentrics during anaphase.
Given these data demonstrating an intimate association between acentrics and microtubules, we suspected chromokinesins may be required for their poleward segregation. Chromokinesins are plus end motor proteins that bind to both chromatin and microtubules (Mazumdar and Misteli, 2005; Vanneste et al., 2011), and exert polar ejection forces on chromosome arms to promote metaphase congression (Vernos et al., 1995; Antonio et al., 2000; Funabiki and Murray, 2000; Barisic et al., 2014; Drpic et al., 2015). To determine if chromokinesins played a role in acentric segregation in Drosophila, we assayed for synthetic lethal interactions between chromokinesins and acentric induction. In a wild-type background, the segregation of sister acentrics to daughter nuclei is delayed but ultimately successful and, surprisingly, no lethality is associated with acentric induction. Previous studies identified proteins required for acentric segregation by screening for mutations that were synthetically lethal with the induction of I-CreI (Royou et al., 2010). Consequently, we tested klp3a and nod for synthetic lethality upon I-CreI induction. These are mutants in two well-characterized Drosophila chromokinesins that belong to the kinesin-4 family (klp3a) and the kinesin-10 family (nod; Theurkauf and Hawley, 1992; Williams et al., 1995). We found reduced rates of survival by expressing I-CreI in male larvae with hypomorphic mutations in klp3a and a loss-of-function mutation in nod. Live analysis revealed several defects in acentric segregation in neuroblast divisions with reduced Klp3a function but no evidence of acentric segregation defects in neuroblasts with defective Nod function (Fig. S3 and Video 9). Consequently, we focused on the role of Klp3a in acentric segregation.
Live analysis revealed that initial defects in klp3a1124 mutants include an increased rate of failed sister acentric separation and unequal poleward segregation of acentrics. How the microtubule plus end–directed activity of Klp3a could be involved with poleward segregation of acentrics was initially mysterious. Insight into how Klp3a, a plus end–directed motor protein, could impart acentric movement from the spindle equator toward the minus end of microtubules at the poles came from studies showing that loss of Klp3a results in defective mitotic spindle organization (Kwon et al., 2004). Because Klp3a is thought to be a plus end–directed motor and thus would oppose poleward transport of the acentric, we focused on the role of Klp3a in organizing the anaphase spindle. Reduced Klp3a activity leads to reduced interpolar microtubules of the spindle in metaphase, as well as decreased spindle elongation during anaphase B (Kwon et al., 2004). In accord with these findings, we also observe disruptions in the organization of interpolar microtubules in klp3a mutants (Figs. 5 and S2). In wild-type neuroblasts, we found that the acentrics segregate along an arc of microtubules following the outer edge of midzone microtubules. In contrast, in klp3a mutants, the acentrics tend to remain much closer to interior midzone microtubules (Fig. 6). Quantification clearly shows that acentrics are not appropriately positioned at the edge of the spindle arc in klp3a mutants suggesting that acentric segregation during anaphase and telophase preferentially relies on interpolar microtubules. Thus, it is likely the disruption in acentric segregation observed in the klp3a mutants is a direct result of a failure to properly organize interpolar microtubules. Additionally, we analyzed for tether formation in neuroblast with reduced Klp3a function. We found that BubR1 ectopically localized to I-CreI–induced acentrics and the tether in neuroblast from klp3a mutant larvae, indicating that reduced Klp3a activity does not disrupt tether formation (Fig. S4 and Video 10).
These studies provide strong evidence for a role of microtubules and chromokinesins for driving acentric segregation, suggesting that the associated DNA tether may have functions other than or in addition to force production. This idea is supported by the recent finding that the tether is required to delay completion of nuclear envelope assembly facilitating inclusion of the late segregating acentric into daughter telophase nuclei (Karg et al., 2015). Based on the studies presented here demonstrating that microtubules are required for acentric segregation, it is possible that a key function of the tether components Polo and BubR1 is to promote microtubule organization around the acentrics. Support of this idea comes from the well-established role Polo kinase in mitotic spindle organization (Das et al., 2016). Polo kinase localizes to the mitotic spindle where Polo functions to stimulate microtubule nucleation (Johmura et al., 2011). Similarly, reduced BubR1 activity has been shown to lead to abnormal spindle morphology in Drosophila third-instar neuroblasts, including reduced spindle length during metaphase (Rahmani et al., 2009). Alternatively, a key function of the tether may be to prevent drift of the acentric fragment far from the metaphase plate, facilitating its capture by spindle microtubules. Support for this idea comes from live imaging acentrics in which the tether has been compromised through reduced BubR1 function. In these instances, the acentric is often observed far from the metaphase plate (Royou et al., 2010; Derive et al., 2015).
Another contributing factor to failed acentric segregation may be diminished anaphase A and B spindle elongation in klp3a mutants (Kwon et al., 2004). Both lagging intact chromosome arms and lagging acentrics result in cell and spindle elongation during anaphase (Kotadia et al., 2012). This was viewed as an adaptive response facilitating complete separation of the lagging chromatin. Thus, the reduced length of the anaphase spindle may further inhibit proper and complete segregation of the acentric chromosome fragments. Accordingly, we find a reduction in spindle size in both metaphase and anaphase in klp3a mutant neuroblasts with I-CreI–induced acentrics compared with I-CreI–expressing control neuroblasts (Fig. S5). These results suggest that the short klp3a mutant spindles may also contribute to the observed increase in acentric segregation failure.
Previous studies, and our own observations, demonstrate that acentric segregation is greatly delayed compared with the segregation of the main mass of undamaged chromosomes (Royou et al., 2010; Derive et al., 2015; Karg et al., 2015). Here, we find that the acentrics rely on Klp3a to establish a segregation path distinct from the path traveled by the main chromosome mass. We suspect that this temporal and spatial separation between the potentially recombination prone acentric fragment and the normal chromosomes decreases the likelihood of deleterious recombination events.
Materials and methods
Fly stocks
All stocks were raised on standard Drosophila media at room temperature. The transgenic line bearing the I-CreI endonuclease with a heat shock 70 promoter was provided by K. Golic (University of Utah, Salt Lake City, UT). The GFP-tagged protein, Jupiter, was used to image microtubules. klp3a1124 was provided by M. Goldberg (Cornell University, Ithaca, NY), and nod4 was provided by the Bloomington Stock Center (stock number 3340). HOAP-GFP lines were provided by J. Tamkun (University of California, Santa Cruz, Santa Cruz, CA).
Live analysis of acentric behavior in Drosophila third-instar neuroblasts
Acentric chromosome fragments were induced by I-CreI expression (under heat shock 70 promoter) in third-instar larvae by a 1-h 37°C heat shock followed by a 1 h recovery period at room temperature. The larval brains from third-instar larvae were dissected in PBS and then transferred to a slide with 20 µl PBS. A coverslip was dropped on PBS solution with brain, and the excess PBS solution was wicked out from edge of coverslip to induce squashing of brain between slide and coverslip. For live analysis, the edge of coverslip was sealed with halocarbon and was imaged as described in the Microscopy and image acquisition section. Neuroblasts divisions in Fig. 5 were from males, and all other images were from female third instars.
Microscopy and image acquisition
Images in Figs. 1, 2, 3, 5, S1, and S2 were acquired with an inverted Eclipse TE2000-E spinning disc (CSLI-X1) confocal microscope (Nikon) with a 100× 1.4 NA oil-immersion objective. Images were captured with a EM-CCD camera (ImageE MX2; Hamamatsu Photonics). Images were acquired with MicroManager 1.4 software. Time-lapse fluorescent images of neuroblasts divisions were done with 120 and 100 ms exposures for GFP (508 nm) and RFP (585 nm), respectively, with 0.5 µm Z-steps. Time-lapse videos with both GFP and RFP were done every 5 to 9 s, and time-lapse movies with RFP alone were done every 5 s. Figures were assembled in Adobe Illustrator. Selected stills (both experimental and control) were processed with ImageJ (http://rsb.info.nih.gov/ij/).
Measurements
Relative fluorescent intensities of acentrics (H2Av-RFP) overlapped with microtubules (Jupiter-GFP) were done using the plot profile function in ImageJ with a line width of one pixel between yellow dashed lines or yellow arrows in grayscale images in Fig. 3 (A and B) and Fig. S3 B. 3D reconstructions in Figs. 3 and 6 were done with Imaris software. Statistical analysis (Bonferroni) in Fig. S5 and Fig. 6, χ2 analysis in Figs. 2 and 5, and Grubbs outlier test for Fig. 6, were done with Prism Version 5 (GraphPad Software).
Laser ablation and imaging
Live imaging for Fig. 4 was done on an inverted microscope (Eclipse Ti-E; Nikon) equipped with a spinning disc confocal (CSU-X1; Yokogawa Electric Corporation), head dichroic Semrock Di01-T404/488/561 GFP, 488-nm (120 mW) and 561-nm (150 mW) diode lasers, emission filters ET525/36M (Chroma Technology Corp.) for GFP or ET630/75M for RFP, and an iXon3 camera (Andor Technology). Fluorescent imaging of neuroblasts was performed through Metamorph 7.7.8.0 (Molecular Devices) at 75–500 ms exposures every 3 to 14 s with a 100× 1.45 Ph3 oil objective through a 1.5× lens yielding 105 nm/pixel at bin = 1. Targeted laser ablation (several 3-ns pulses at 20 Hz) using 514-nm light was performed using a galvo-controlled MicroPoint Laser System (Photonic Instruments) operated through Metamorph. Depolymerization toward the spindle pole, as expected for uncapped (and unstable) microtubule plus ends, verified successful ablation of microtubules. Chromosome position data in Fig. 5 C were generated by manual tracking of acentrics and intact chromosomes (H2Av-RFP) in time-lapse videos using a custom MATLAB (R2012a version 7.4) program.
Online supplemental material
Fig. S1 shows that ablation of BubR1-coated tether does not prevent poleward migration of acentrics (Video 6). Fig. S2 shows that reduced Klp3a activity disrupts spindle morphology. Fig. S3 shows normal segregation of acentrics in neuroblasts from nod4 third instars (Video 9). Fig. S4 shows that reduced Klp3a function does not disrupt BubR1 localization to DNA tether (Video 10). Fig. S5 shows that reduced metaphase spindle size and diminished anaphase elongation may contribute to acentric segregation defects in klp3a mutant neuroblast. Video 1 shows third-instar neuroblast mitotic division with I-CreI–induced acentrics alone. Video 2 shows third-instar neuroblast mitotic division with I-CreI–induced acentrics and GFP-tagged telomeres. Video 3 shows I-CreI–expressing third-instar neuroblast mitotic division with I-CreI–induced acentrics and GFP-tagged microtubules. Video 4 is a 3D rendering of acentrics from an I-CreI–expressing third-instar neuroblast mitotic division with GFP-tagged microtubules. Video 5 shows third-instar neuroblast mitotic division with ablated (at time 00:00:45) GFP-tagged microtubules associated with I-CreI–induced acentrics. Video 6 shows third-instar neuroblast mitotic division with ablation (arrowhead at 00:33) of BubR1-coated tethers. Video 7 shows third-instar neuroblast mitotic division from klp3a1124 with poleward acentrics. Video 8 is a 3D rendering of a third-instar neuroblast mitotic division from klp3a1124 with poleward acentrics. Video 9 shows third-instar neuroblast mitotic division from nod4 with I-CreI–induced acentrics. Video 10 shows that BubR1-coated tethers form in I-CreI–expressing third-instar neuroblast from klp3a1124 mutants.
Supplementary Material
Acknowledgments
We would like to thank the Tamkun laboratory at University of California, Santa Cruz, for providing the HOAP-GFP fly line. We thank William Saxton and Susan Strome for use of their spinning disc microscope. We also thank Ben Abrams for microscope assistance and Jose Pietri for help with statistical analysis.
This work was supported by National Institutes of Health grant 1R01GM120321-01A1 to W. Sullivan and in part by National Institutes of Health grants R00GM09433 and DP2GM119177 to S. Dumont. M.W. Elting is a Damon Runyon Fellow supported by the Damon Runyon Cancer Research Foundation (DRG-2170-13).
The authors declare no competing financial interests.
Author contributions: T. Karg contributed to all aspects of planning, execution of the experiments, and manuscript preparation. M.W. Elting contributed to the planning, execution, and analysis of the laser ablation studies and manuscript preparation. S. Dumont contributed to the planning and analysis of the laser ablation studies and manuscript preparation. H. Vicars contributed to planning and execution of 3D live imaging. W. Sullivan contributed to all aspects of experimental planning and manuscript preparation.
Footnotes
Abbreviations used:
- DSB
- double-strand break
- HOAP
- Hp1/ORC–associated protein
- RFP
- red fluorescent protein
References
- Ahmad K., and Golic K.G.. 1998. The transmission of fragmented chromosomes in Drosophila melanogaster. Genetics. 148:775–792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antonio C., Ferby I., Wilhelm H., Jones M., Karsenti E., Nebreda A.R., and Vernos I.. 2000. Xkid, a chromokinesin required for chromosome alignment on the metaphase plate. Cell. 102:425–435. 10.1016/S0092-8674(00)00048-9 [DOI] [PubMed] [Google Scholar]
- Bajer A. 1958. Cine-micrographic studies on chromosome movements in β-irradiated cell. Chromosoma. 9:319–331. 10.1007/BF02568084 [DOI] [PubMed] [Google Scholar]
- Bajer A.S., and Vantard M.. 1988. Microtubule dynamics determine chromosome lagging and transport of acentric fragments. Mutat. Res. 201:271–281. 10.1016/0027-5107(88)90016-4 [DOI] [PubMed] [Google Scholar]
- Barisic M., Aguiar P., Geley S., and Maiato H.. 2014. Kinetochore motors drive congression of peripheral polar chromosomes by overcoming random arm-ejection forces. Nat. Cell Biol. 16:1249–1256. 10.1038/ncb3060 [DOI] [PubMed] [Google Scholar]
- Bretscher H.S., and Fox D.T.. 2016. Proliferation of double-strand break-resistant polyploid cells requires Drosophila FANCD2. Dev. Cell. 37:444–457. 10.1016/j.devcel.2016.05.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cenci G., Siriaco G., Raffa G.D., Kellum R., and Gatti M.. 2003. The Drosophila HOAP protein is required for telomere capping. Nat. Cell Biol. 5:82–84. 10.1038/ncb902 [DOI] [PubMed] [Google Scholar]
- Das A., Shah S.J., Fan B., Paik D., DiSanto D.J., Hinman A.M., Cesario J.M., Battaglia R.A., Demos N., and McKim K.S.. 2016. Spindle assembly and chromosome segregation requires central spindle proteins in Drosophila oocytes. Genetics. 202:61–75. 10.1534/genetics.115.181081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derive N., Landmann C., Montembault E., Claverie M.C., Pierre-Elies P., Goutte-Gattat D., Founounou N., McCusker D., and Royou A.. 2015. Bub3-BubR1-dependent sequestration of Cdc20Fizzy at DNA breaks facilitates the correct segregation of broken chromosomes. J. Cell Biol. 211:517–532. 10.1083/jcb.201504059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drpic D., Pereira A.J., Barisic M., Maresca T.J., and Maiato H.. 2015. Polar ejection forces promote the conversion from lateral to end-on kinetochore-microtubule attachments on mono-oriented chromosomes. Cell Reports. 13:460–469. 10.1016/j.celrep.2015.08.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dumont S., and Mitchison T.J.. 2009. Force and length in the mitotic spindle. Curr. Biol. 19:R749–R761. 10.1016/j.cub.2009.07.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dumont J., Oegema K., and Desai A.. 2010. A kinetochore-independent mechanism drives anaphase chromosome separation during acentrosomal meiosis. Nat. Cell Biol. 12:894–901. 10.1038/ncb2093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elting M.W.C.L., Hueschen C.L., Udy D.B., and Dumont S.. 2014. Force on spindle microtubule minus ends moves chromosomes. J. Cell Biol. 206:245–256. 10.1083/jcb.201401091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ermolaeva M.A., and Schumacher B.. 2014. Systemic DNA damage responses: Organismal adaptations to genome instability. Trends Genet. 30:95–102. 10.1016/j.tig.2013.12.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Funabiki H., and Murray A.W.. 2000. The Xenopus chromokinesin Xkid is essential for metaphase chromosome alignment and must be degraded to allow anaphase chromosome movement. Cell. 102:411–424. 10.1016/S0092-8674(00)00047-7 [DOI] [PubMed] [Google Scholar]
- Galgoczy D.J., and Toczyski D.P.. 2001. Checkpoint adaptation precedes spontaneous and damage-induced genomic instability in yeast. Mol. Cell. Biol. 21:1710–1718. 10.1128/MCB.21.5.1710-1718.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishii K., Ogiyama Y., Chikashige Y., Soejima S., Masuda F., Kakuma T., Hiraoka Y., and Takahashi K.. 2008. Heterochromatin integrity affects chromosome reorganization after centromere dysfunction. Science. 321:1088–1091. 10.1126/science.1158699 [DOI] [PubMed] [Google Scholar]
- Johmura Y., Soung N.K., Park J.E., Yu L.R., Zhou M., Bang J.K., Kim B.Y., Veenstra T.D., Erikson R.L., and Lee K.S.. 2011. Regulation of microtubule-based microtubule nucleation by mammalian polo-like kinase 1. Proc. Natl. Acad. Sci. USA. 108:11446–11451. 10.1073/pnas.1106223108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanda T., and Wahl G.M.. 2000. The dynamics of acentric chromosomes in cancer cells revealed by GFP-based chromosome labeling strategies. J. Cell. Biochem. Suppl. 35(S35, Suppl 35):107–114. [DOI] [PubMed] [Google Scholar]
- Kanda T., Sullivan K.F., and Wahl G.M.. 1998. Histone-GFP fusion protein enables sensitive analysis of chromosome dynamics in living mammalian cells. Curr. Biol. 8:377–385. 10.1016/S0960-9822(98)70156-3 [DOI] [PubMed] [Google Scholar]
- Kanda T., Otter M., and Wahl G.M.. 2001. Mitotic segregation of viral and cellular acentric extrachromosomal molecules by chromosome tethering. J. Cell Sci. 114:49–58. [DOI] [PubMed] [Google Scholar]
- Karg T., Warecki B., and Sullivan W.. 2015. Aurora B-mediated localized delays in nuclear envelope formation facilitate inclusion of late-segregating chromosome fragments. Mol. Biol. Cell. 26:2227–2241. 10.1091/mbc.E15-01-0026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karpova N., Bobinnec Y., Fouix S., Huitorel P., and Debec A.. 2006. Jupiter, a new Drosophila protein associated with microtubules. Cell Motil. Cytoskeleton. 63:301–312. 10.1002/cm.20124 [DOI] [PubMed] [Google Scholar]
- Khodjakov A., Cole R.W., Bajer A.S., and Rieder C.L.. 1996. The force for poleward chromosome motion in Haemanthus cells acts along the length of the chromosome during metaphase but only at the kinetochore during anaphase. J. Cell Biol. 132:1093–1104. 10.1083/jcb.132.6.1093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotadia S., Montembault E., Sullivan W., and Royou A.. 2012. Cell elongation is an adaptive response for clearing long chromatid arms from the cleavage plane. J. Cell Biol. 199:745–753. 10.1083/jcb.201208041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon M., Morales-Mulia S., Brust-Mascher I., Rogers G.C., Sharp D.J., and Scholey J.M.. 2004. The chromokinesin, KLP3A, dives mitotic spindle pole separation during prometaphase and anaphase and facilitates chromatid motility. Mol. Biol. Cell. 15:219–233. 10.1091/mbc.E03-07-0489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaFountain J.R. Jr., Oldenbourg R., Cole R.W., and Rieder C.L.. 2001. Microtubule flux mediates poleward motion of acentric chromosome fragments during meiosis in insect spermatocytes. Mol. Biol. Cell. 12:4054–4065. 10.1091/mbc.12.12.4054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang H., Wright W.H., Cheng S., He W., and Berns M.W.. 1993. Micromanipulation of chromosomes in PTK2 cells using laser microsurgery (optical scalpel) in combination with laser-induced optical force (optical tweezers). Exp. Cell Res. 204:110–120. 10.1006/excr.1993.1015 [DOI] [PubMed] [Google Scholar]
- Malkova A., Ross L., Dawson D., Hoekstra M.F., and Haber J.E.. 1996. Meiotic recombination initiated by a double-strand break in rad50 delta yeast cells otherwise unable to initiate meiotic recombination. Genetics. 143:741–754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazumdar M., and Misteli T.. 2005. Chromokinesins: Multitalented players in mitosis. Trends Cell Biol. 15:349–355. 10.1016/j.tcb.2005.05.006 [DOI] [PubMed] [Google Scholar]
- Mikhailov A., Cole R.W., and Rieder C.L.. 2002. DNA damage during mitosis in human cells delays the metaphase/anaphase transition via the spindle-assembly checkpoint. Curr. Biol. 12:1797–1806. 10.1016/S0960-9822(02)01226-5 [DOI] [PubMed] [Google Scholar]
- Muscat C.C., Torre-Santiago K.M., Tran M.V., Powers J.A., and Wignall S.M.. 2015. Kinetochore-independent chromosome segregation driven by lateral microtubule bundles. eLife. 4:e06462 10.7554/eLife.06462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Connell C.B., Lončarek J., Kaláb P., and Khodjakov A.. 2009. Relative contributions of chromatin and kinetochores to mitotic spindle assembly. J. Cell Biol. 187:43–51. 10.1083/jcb.200903076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohno Y., Ogiyama Y., Kubota Y., Kubo T., and Ishii K.. 2016. Acentric chromosome ends are prone to fusion with functional chromosome ends through a homology-directed rearrangement. Nucleic Acids Res. 44:232–244. 10.1093/nar/gkv997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Page S.L., and Hawley R.S.. 2005. The Drosophila meiotic mutant mei-352 is an allele of klp3A and reveals a role for a kinesin-like protein in crossover distribution. Genetics. 170:1797–1807. 10.1534/genetics.105.041194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Platero J.S., Ahmad K., and Henikoff S.. 1999. A distal heterochromatic block displays centromeric activity when detached from a natural centromere. Mol. Cell. 4:995–1004. 10.1016/S1097-2765(00)80228-2 [DOI] [PubMed] [Google Scholar]
- Rahmani Z., Gagou M.E., Lefebvre C., Emre D., and Karess R.E.. 2009. Separating the spindle, checkpoint, and timer functions of BubR1. J. Cell Biol. 187:597–605. 10.1083/jcb.200905026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rong Y.S., Titen S.W., Xie H.B., Golic M.M., Bastiani M., Bandyopadhyay P., Olivera B.M., Brodsky M., Rubin G.M., and Golic K.G.. 2002. Targeted mutagenesis by homologous recombination in D. melanogaster. Genes Dev. 16:1568–1581. 10.1101/gad.986602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Royou A., Macias H., and Sullivan W.. 2005. The Drosophila Grp/Chk1 DNA damage checkpoint controls entry into anaphase. Curr. Biol. 15:334–339. 10.1016/j.cub.2005.02.026 [DOI] [PubMed] [Google Scholar]
- Royou A., Gagou M.E., Karess R., and Sullivan W.. 2010. BubR1- and Polo-coated DNA tethers facilitate poleward segregation of acentric chromatids. Cell. 140:235–245. 10.1016/j.cell.2009.12.043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandell L.L., and Zakian V.A.. 1993. Loss of a yeast telomere: Arrest, recovery, and chromosome loss. Cell. 75:729–739. 10.1016/0092-8674(93)90493-A [DOI] [PubMed] [Google Scholar]
- Sikirzhytski V., Magidson V., Steinman J.B., He J., Le Berre M., Tikhonenko I., Ault J.G., McEwen B.F., Chen J.K., Sui H., et al. . 2014. Direct kinetochore-spindle pole connections are not required for chromosome segregation. J. Cell Biol. 206:231–243. 10.1083/jcb.201401090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theurkauf W.E., and Hawley R.S.. 1992. Meiotic spindle assembly in Drosophila females: Behavior of nonexchange chromosomes and the effects of mutations in the nod kinesin-like protein. J. Cell Biol. 116:1167–1180. 10.1083/jcb.116.5.1167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Titen S.W., and Golic K.G.. 2008. Telomere loss provokes multiple pathways to apoptosis and produces genomic instability in Drosophila melanogaster. Genetics. 180:1821–1832. 10.1534/genetics.108.093625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanneste D., Ferreira V., and Vernos I.. 2011. Chromokinesins: Localization-dependent functions and regulation during cell division. Biochem. Soc. Trans. 39:1154–1160. 10.1042/BST0391154 [DOI] [PubMed] [Google Scholar]
- Vernos I., Raats J., Hirano T., Heasman J., Karsenti E., and Wylie C.. 1995. Xklp1, a chromosomal Xenopus kinesin-like protein essential for spindle organization and chromosome positioning. Cell. 81:117–127. 10.1016/0092-8674(95)90376-3 [DOI] [PubMed] [Google Scholar]
- Wignall S.M., and Villeneuve A.M.. 2009. Lateral microtubule bundles promote chromosome alignment during acentrosomal oocyte meiosis. Nat. Cell Biol. 11:839–844. 10.1038/ncb1891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams B.C., Riedy M.F., Williams E.V., Gatti M., and Goldberg M.L.. 1995. The Drosophila kinesin-like protein KLP3A is a midbody component required for central spindle assembly and initiation of cytokinesis. J. Cell Biol. 129:709–723. 10.1083/jcb.129.3.709 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.