Auckland et al. describe a previously overlooked step during chromosome congression that provides insight into how kinetochore maturation is linked to attachment load-bearing, microtubule occupancy, and spindle assembly checkpoint signaling.
Abstract
Kinetochores mediate chromosome congression by either sliding along the lattice of spindle microtubules or forming end-on attachments to their depolymerizing plus-ends. By following the fates of individual kinetochores as they congress in live cells, we reveal that the Ska complex is required for a distinct substep of the depolymerization-coupled pulling mechanism. Ska depletion increases the frequency of naturally occurring, force-dependent P kinetochore detachment events, while being dispensable for the initial biorientation and movement of chromosomes. In unperturbed cells, these release events are followed by reattachment and successful congression, whereas in Ska-depleted cells, detached kinetochores remain in a futile reattachment/detachment cycle that prevents congression. We further find that Ska is progressively loaded onto bioriented kinetochore pairs as they congress. We thus propose a model in which kinetochores mature through Ska complex recruitment and that this is required for improved load-bearing capacity and silencing of the spindle assembly checkpoint.
Introduction
Congression is the process by which the initially scattered chromosomes become aligned at the spindle equator, forming the metaphase plate (Auckland and McAinsh, 2015; Maiato et al., 2017). The force required for this process is generated by kinetochores, large protein machines that assemble on the centromeric DNA of each sister chromatid and form attachment sites for spindle microtubules (Westhorpe and Straight, 2013; Cheeseman, 2014). Two distinct congression mechanisms have been identified, which together ensure efficient chromosome alignment. Kinetochores located at the periphery of the spindle after nuclear envelope breakdown (NEB) engage the side wall of spindle microtubules, forming lateral attachments (Kapoor et al., 2006; Barisic et al., 2014; Auckland and McAinsh, 2015). Such kinetochores are enriched in the Kinesin-7 CENP-E, which steps toward the microtubule plus-end, pulling chromosomes to the equator. However, loss of CENP-E activity still allows the vast majority of chromosomes to congress (McEwen et al., 2001; Barisic et al., 2014; Bancroft et al., 2015) and only one-quarter of PtK1 cells contain laterally attached kinetochores (Kapoor et al., 2006). This is in part explained by the observation that sister kinetochore pairs can biorient in the first minutes after NEB (Magidson et al., 2011). Indeed, biorientation is an absolute requirement for the eventual accurate segregation of sister chromatids and is promoted by (a) the conversion of lateral to end-on attachments (Magidson et al., 2011, 2015; Shrestha and Draviam, 2013; Drpic et al., 2015) and (b) stabilization of the bioriented state via the dephosphorylation of outer-kinetochore Aurora B substrates (Lampson et al., 2004; Liu et al., 2009; Welburn et al., 2010; Lampson and Cheeseman, 2011).
Sister pairs that instantaneously biorient do not necessarily require congression, as they are preferentially located at the spindle equator (Magidson et al., 2011). However, those that biorient in a pole-proximal position must generate a directional force to align. This force is thought to be produced by microtubule plus-end depolymerization at the kinetochore, which allows the pulling of chromosomes to the equator via the maintenance of attachment to the shortening fiber (Cassimeris and Salmon, 1991; Skibbens et al., 1993, 1995; Khodjakov and Rieder, 1996; McEwen et al., 1997; Kapoor et al., 2006). Originally termed Pac-man (Gorbsky et al., 1987), this force-generating mechanism can be described as depolymerization-coupled pulling (DCP; Auckland and McAinsh, 2015). DCP demands that the leading (poleward-moving [P]) kinetochore can maintain its attachment to depolymerizing microtubules, whereas the trailing (away-from-the-pole–moving [AP]) kinetochore is attached to polymerizing microtubules.
In vitro reconstitution experiments have suggested that the heterotrimeric spindle and kinetochore associated (Ska) complex (Ska1-Ska2-Ska3/RAMA1) could mediate P kinetochore coupling to depolymerizing microtubules, because it can autonomously track depolymerizing plus-ends, bind curved protofilaments, and transduce the force generated by depolymerization to a polystyrene bead (Welburn et al., 2009; Schmidt et al., 2012). Moreover, siRNA-mediated depletion of the Ska complex in human cells has been shown to cause congression defects, consistent with its involvement in DCP (Hanisch et al., 2006; Daum et al., 2009; Gaitanos et al., 2009; Theis et al., 2009; Welburn et al., 2009; Jeyaprakash et al., 2012; Schmidt et al., 2012; Abad et al., 2014). Here, we use live-cell imaging of single kinetochores during congression to reveal how the Ska complex is required for a specific substep of DCP. We further show how bioriented kinetochores are maturing through progressive recruitment of the Ska complex and that this may reflect a mechanical self-check that is coupled to signaling from the spindle assembly checkpoint (SAC). These findings contribute to explaining how kinetochores ensure that anaphase initiates only when all sister-pairs have formed mature bioriented attachments and congressed to the spindle equator.
Results
The Ska complex is required for the maintenance of biorientation during congression
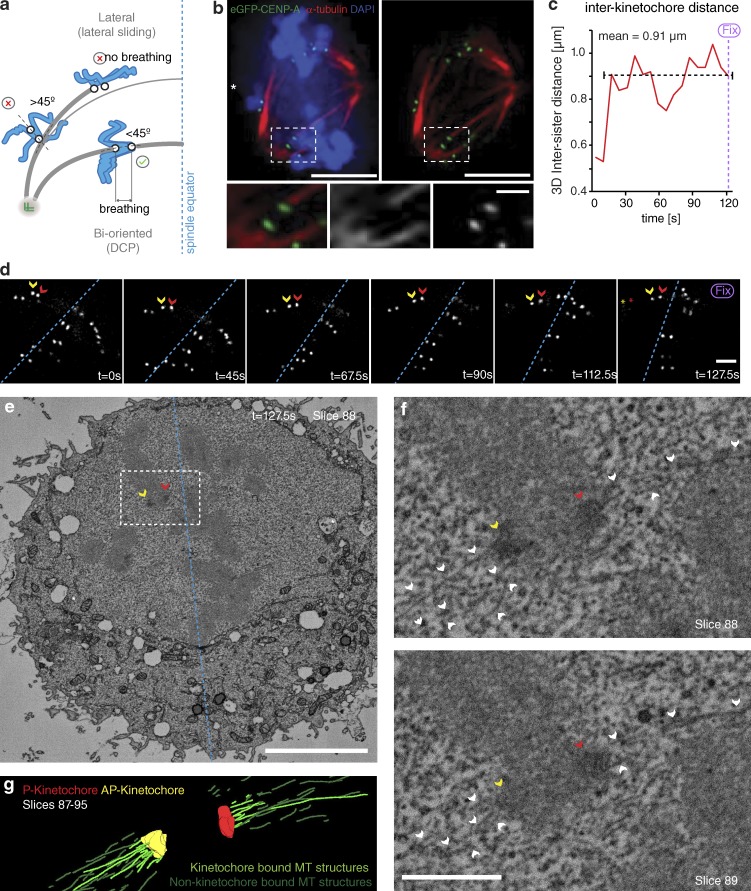
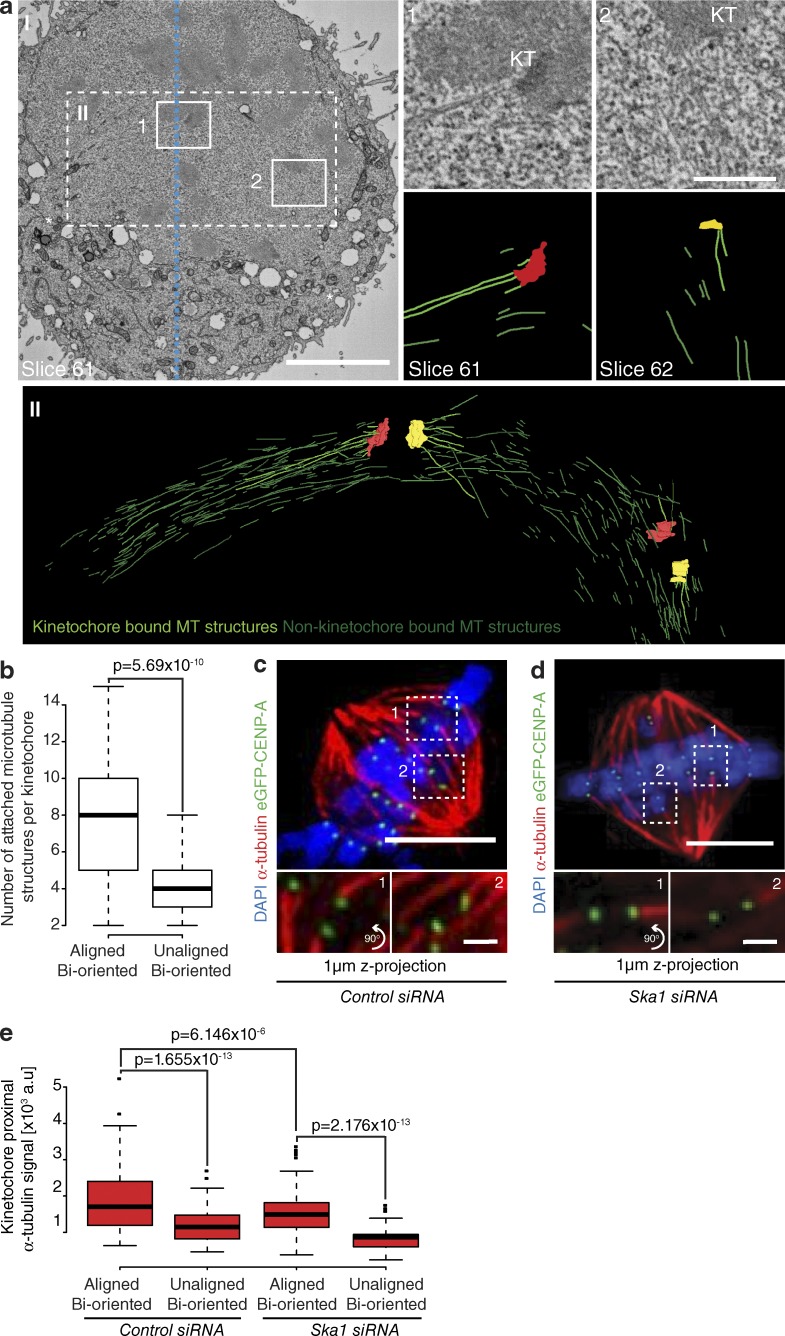
To assay the behavior of congressing chromosomes, we imaged mid-to-late prometaphase HeLa cells expressing eGFP-CENP-A at 7.5-s intervals for 5 min. Because kinetochores can congress by both lateral sliding and DCP (Fig. 1 a), it was important to identify the latter events in our videos. First, we focused on kinetochores located within the spindle (between pole and equator), because lateral sliding is largely restricted to the peripheral chromosomes (McEwen et al., 2001; Barisic et al., 2014). These kinetochores appeared bioriented based on glutaraldehyde fixation and α-tubulin staining (Fig. 1 b). Second, we constrained our analysis to sisters whose kinetochore–kinetochore (K–K) axis was <45° relative to local spindle microtubule path (Fig. 1 a), a geometry in which end-on attachment to opposite poles is possible. Third, we ensured that kinetochore pairs were undergoing interkinetochore breathing with a mean separation of >0.9 µm, an essential characteristic of biorientation (Cai et al., 2009; Jaqaman et al., 2010; Fig. 1 a). To ensure that kinetochores fulfilling these criteria can be bioriented, we filmed sister-pairs as they congressed (Fig. 1, c and d) and then fixed/processed the cell for serial block-face scanning EM (SBF-SEM). Kinetochores were clearly visible in the SBF-SEM images, enabling us to correlate the live cell and SEM (Fig. 1, d–f). Importantly, microtubule fibers terminated at both the AP and P kinetochores on the sister-pair that we had tracked (Fig. 1, c–g), confirming that such chromosomes are congressing by DCP.
Figure 1.
Congressing chromosomes that are oriented and breathing are bioriented. (a) Schematic showing the selection criteria used to sample for kinetochore pairs that are congressing via DCP. (b) Image of a prometaphase HeLa cell expressing eGFP-CENP-A stained with DAPI and an antibody against α-tubulin. Zoom boxes depict an unaligned kinetochore pair that has formed a bioriented attachment. Bars: (top) 5 µm; (bottom) 1 µm. (c) Measurement of 3D intersister distance over time for the congressing kinetochore pair depicted in d, showing that it is undergoing interkinetochore breathing (mean 0.91 µm) after attachment (seen as rise in interkinetochore distance at t = 15 s). (d) Video stills of an eGFP-CENP-A–marked kinetochore-pair congressing to the metaphase plate. The cell was fixed at t = 127.5 s and processed for SBF-SEM. Red and yellow arrows, P and AP kinetochores; red and yellow stars, position of the kinetochore pair at t = 0 s; dotted blue line, metaphase plate periphery. Bar, 1 µm. (e) A single slice (84) from an SBF-SEM stack of the cell displayed in d after fixation at t = 127.5 s. Red and yellow arrows, P and AP kinetochores; dotted blue line, metaphase plate periphery; white asterisks, spindle poles. Bar, 5 µm. (f) Zoom of the white box in d. Here, the congressing kinetochore pair followed in c and d can clearly be seen with end-on microtubule attachment at both sisters, confirming that it is bioriented. White arrows, microtubules; red and yellow arrows, P and AP kinetochores. Bar, 1 µm. (g) Rendered image of slices 87–95 from the SBF-SEM stack of the cell depicted in d–f.
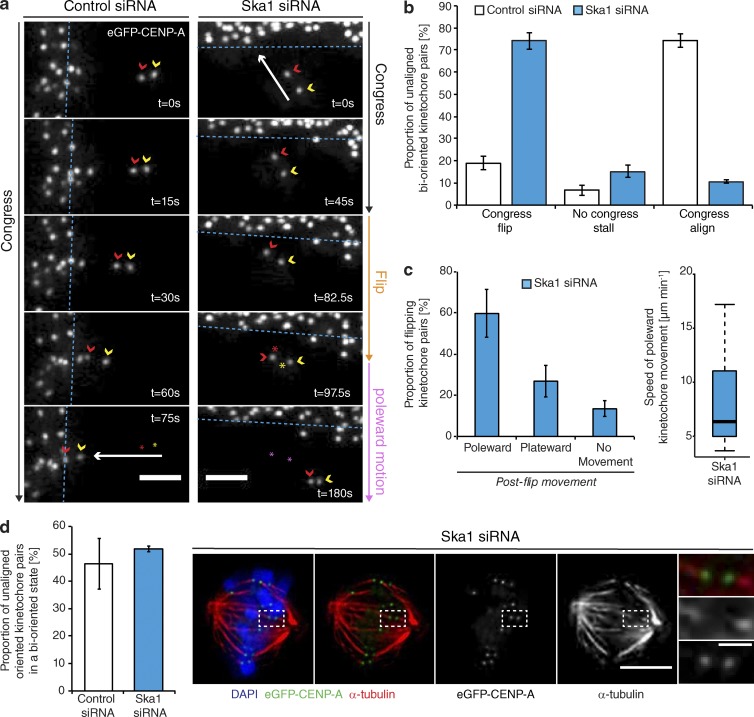
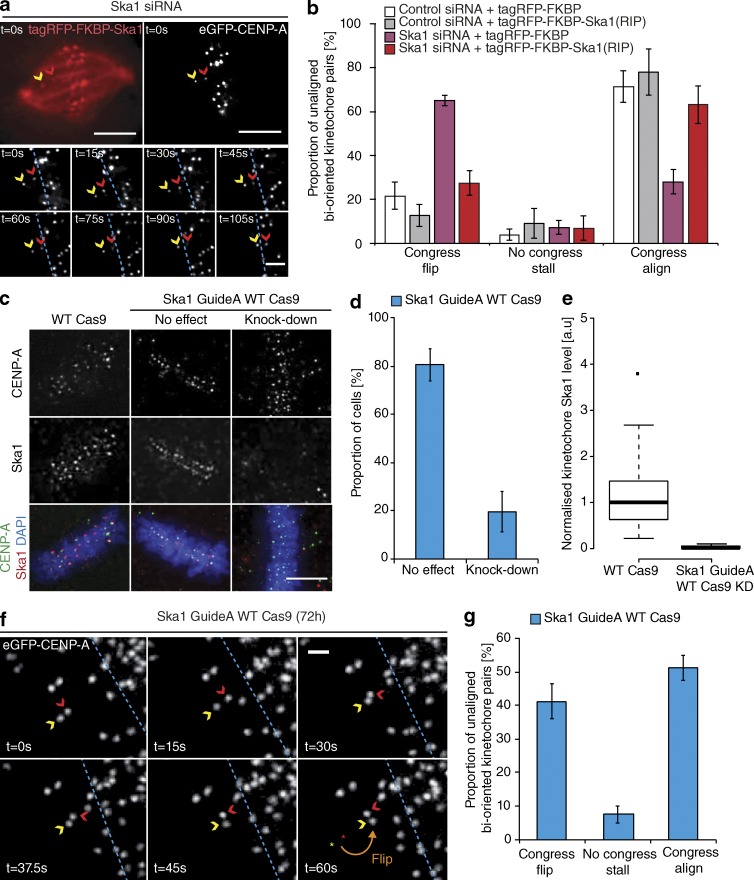
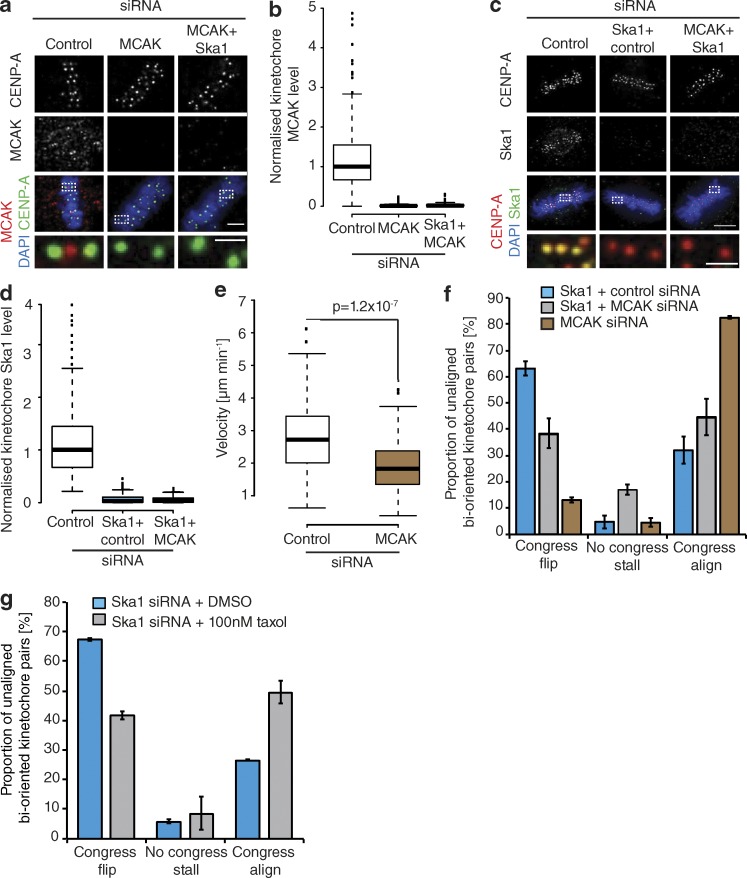
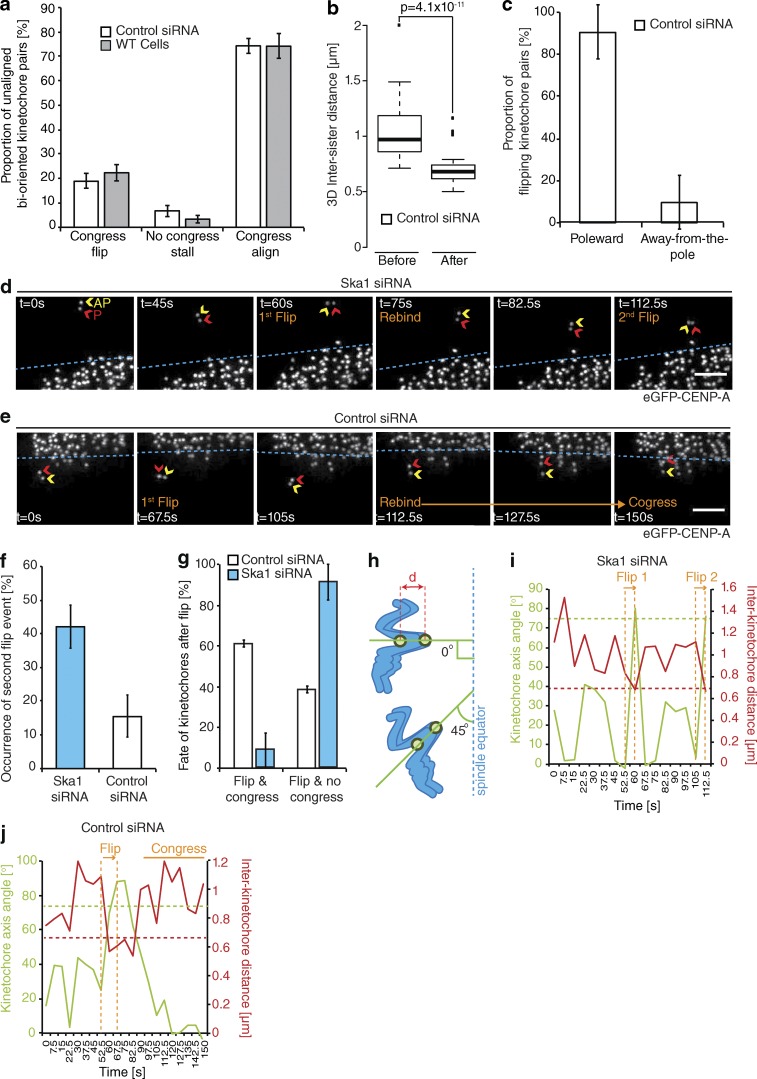
By tracking the behavior of individual bioriented kinetochore pairs, we documented three phenotypes associated with DCP: (a) successfully congressing to the spindle equator (74.3 ± 3.3%), (b) stalling, where the sister-pair would persist in an immobile state (6.8 ± 2.4%), and (c) “flipping” (18.8 ± 3%), where the sister-pair would initiate congression but then rotate through 90° relative to the spindle axis as it approached the metaphase plate (Fig. 2, a and b). (This phenotype is distinct from the previously reported “kinetochore wobbling” by Magidson et al. [2011], where laterally attached kinetochore pairs in early prometaphase changed their orientation by >45° in the absence of directed motion.) We then sought to investigate how these phenotypes differed in cells depleted of Ska1 (Fig. S1). We found a dramatic reduction in successful congression to the spindle equator (to 10.7 ± 1%), which corresponded with a large increase in kinetochore flipping (to 70 ± 3.7%; Fig. 2, a and b). The majority of these flipped sisters were then transported poleward with a median velocity of ∼7 µm/min (Fig. 2 c). Importantly, the flipping phenotype was not the consequence of a change in attachment state, because the fraction of unaligned bioriented kinetochores in control and Ska1-depleted cells was unchanged (Fig. 2 d). The metaphase plate was identified in these images as the region of highest kinetochore density in a region roughly equidistant from either pole. To control for siRNA off-target effects, we transfected eGFP-CENP-A–expressing cells depleted of Ska1 with an siRNA resistant tagRFP-FKBP-Ska1 transgene. This successfully rescued the flipping phenotype, with the majority of unaligned bioriented kinetochore pairs (63 ± 8.5%) now congressing (Fig. 3, a and b). To validate this phenotype independently of siRNA, we targeted the first exon of Ska1 using CRISPR/Cas9. Quantitative immunofluorescence revealed that ∼20% of cells demonstrated a >95% Ska1 knockdown upon Ska1-GuideA-WTCas9 transfection for 72 h (Fig. 3, c–e; and Fig. S1). In agreement with our siRNA data, transfection with the Ska1-targeted Cas9 augmented kinetochore flipping (to 41 ± 5.1%; Fig. 3, f and g). Together, these data demonstrate that the Ska complex is required for the maintenance of biorientation during congression.
Figure 2.
The Ska complex is required for the maintenance of biorientation during congression. (a) Example image sequence of a sister kinetochore pair labeled with eGFP-CENP-A congressing to the metaphase plate in a cell treated with control siRNA (left) or initiating congression toward the metaphase plate in a Ska1-depleted cell, but then rotating through 90° relative to the spindle axis (flipping; right). Red and yellow arrows, P and AP kinetochores; red and yellow stars, position of the kinetochore pair at t = 0 (control) and preflip (Ska1 siRNA); pink stars, postflip position of the Ska1-depleted kinetochore pair; dotted blue line, metaphase plate periphery. Bar, 2 µm. (b) Quantification of unaligned bioriented kinetochore-pair behavior in eGFP-CENP-A–expressing cells treated with control or Ska1 siRNA. Error bars ± SD; n (control) = 157 KT from 90 cells; n (Ska1) = 114 KT from 67 cells. (c, left) Quantification of postflip kinetochore behavior in cells treated with Ska1 siRNA. Error bars ± SD, n = 114 KT from 67 cells. (right) Quantification of postflip kinetochore P-movement velocity in cells treated with Ska1 siRNA. (d, left) Proportion of unaligned sister-pairs oriented along their local microtubule path that are bioriented in cells treated with control or Ska1 siRNA. Error bars ± SD; n (control) = 93 KT from 59 cells; n (Ska1) = 138 KT from 82 cells. (right) Images of a prometaphase HeLa cell expressing eGFP-CENP-A depleted of Ska1 and stained with DAPI and an antibody against α-tubulin. Zoom boxes depict an unaligned kinetochore pair that is bioriented. Bars: (left) 5 µm; (right) 1 µm.
Figure 3.
Confirming the role of Ska in congression. (a) Example image sequence of a congressing bioriented kinetochore pair labeled with eGFP-CENP-A in a cell depleted of Ska1 and subsequently rescued with an siRNA-resistant tagRFP-FKBP-Ska1 transgene. Red and yellow arrows, P and AP kinetochores; dotted blue line, metaphase plate periphery. Bars: (top) 5 µm; (bottom) 1 µm. (b) Quantification of unaligned bioriented kinetochore behavior during the Ska1 siRNA rescue experiment. Error bars ± SD; n (control+tagRFP-FKBP) = 126 KT from 39 cells; n (control+tagRFP-FKBP-Ska1) = 132 KT from 27 cells; n (Ska1+tagRFP-FKBP) = 251 KT from 77 cells; n (Ska1+tagRFP-FKBP-Ska1) = 57 KT from 29 cells. (c) Images of HeLa cells transfected with either an untargeted (WT Cas9) or Ska1-targeted (Ska1 Guide A WT Cas9) CRISPR/Cas9 vector and stained with DAPI and antibodies against Ska1 and CENP-A. Bar, 5 µm. (d) Quantification of cells treated with the Ska1-targeted CRISPR/Cas9 that displayed a knockdown of Ska1 as judged by a reduction in kinetochore staining intensity. Error bars ± SD; n = 200 cells. (e) Quantification of Ska1 staining intensity in the cells transfected with either an untargeted (WT Cas9) or Ska1-targeted (Ska1 Guide A WT Cas9) CRISPR/Cas9 vector that display a knockdown phenotype. n = 200 KT from 20 cells per condition. (f) Example image sequence of a flipping kinetochore pair labeled with eGFP-CENP-A in a cell treated with Ska1 GuideA WT Cas9. Red and yellow arrows, P and AP kinetochores; dotted blue line, metaphase plate periphery. Bar, 1 µm. (g) Quantification of unaligned bioriented kinetochore behavior in cells treated with Ska1 GuideA WTCas9. Error bar ± SD; n = 191 KT from 60 cells.
Kinetochore flipping corresponds to lead sister detachment
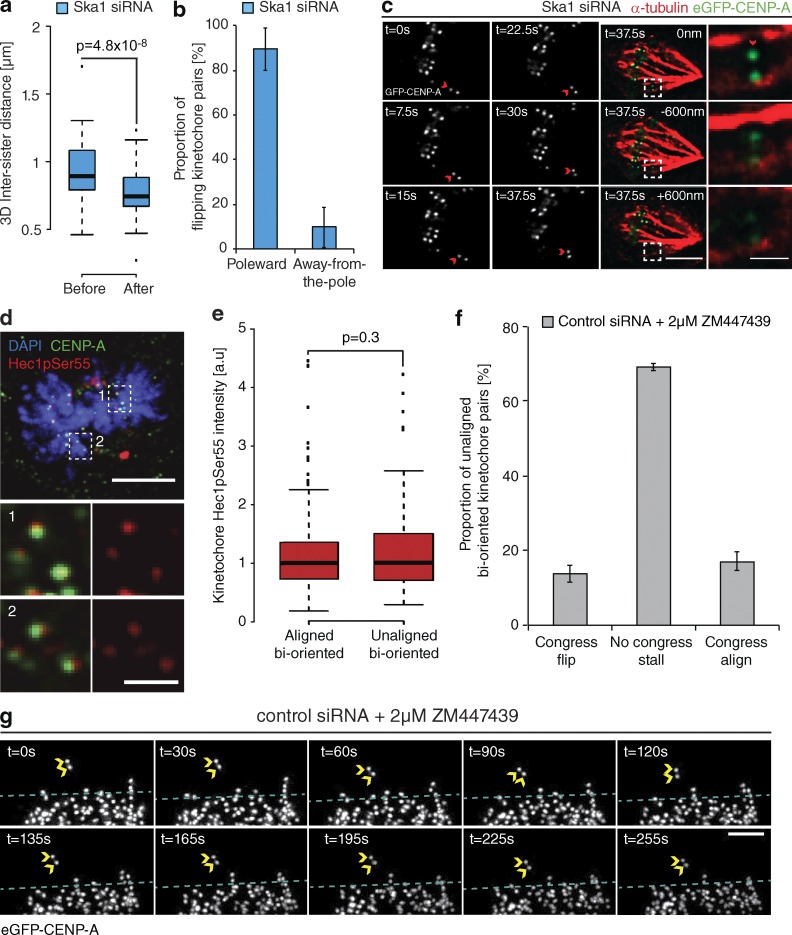
The rotation of a bioriented sister-pair through 90° relative to the spindle axis (Figs. 2 a and 3 f) suggests that a flip event corresponds to the loss of microtubule attachment at the sister-pair. To validate this, we measured the eGFP-CENP-A–based intersister distance of flipping kinetochore pairs at two time points before and after a flip event in cells depleted of Ska1. Consistent with loss of attachment, we observed a significant reduction of the intersister distance after a flip event (from 900 ± 195 nm to 740 ± 190 nm) to a level comparable to that measured for tensionless (nonattached) kinetochores in the presence of the microtubule-depolymerizing drug nocodazole (Fig. 4 a). Given the proposed function of the Ska complex in DCP, where force generation occurs at the P kinetochore, we sought to identify whether there was a P/AP bias in the flipping kinetochore. By determining which kinetochore rotated around its sister, we found that microtubule detachment occurred at the P kinetochore in ∼90% of cases (Fig. 4 b). To confirm this, we imaged a congressing kinetochore pair depleted of Ska1 and, when a flip initiated, we immediately fixed the cell and stained with α-tubulin antibodies. In Fig. 4 c, the P kinetochore can clearly be seen rotated through 90° and not bound to any microtubules, whereas the AP kinetochore can still be seen bound to a K-fiber.
Figure 4.
Kinetochore flipping corresponds to lead sister detachment. (a) Quantification of interkinetochore distance before and after the flip event in eGFP-CENP-A cells treated with Ska1 siRNA. Measurements were taken manually from the two time frames before the initiation of a flip and the two after the 90° rotation. n = 47 KT from 39 cells; P-value calculated using two-sample t test. (b) The proportion of flip events that occurred at either the P or AP kinetochore in cells treated with Ska1 siRNA. The flipping kinetochore was identified as the kinetochore that rotated ∼90° around its roughly stationary sister. Error bars ± SD; 39 KT from 37 cells. (c) The lead sister is no longer associated with microtubules after a flip event. Cells expressing eGFP-CENP-A were depleted of Ska1, and an unaligned bioriented kinetochore pair was followed during a flip. Immediately after the 90° rotation was observed, the cell was fixed with glutaraldehyde and microtubules were visualized using an anti–α-tubulin antibody. Insets show the flipped kinetochore in three z-sections. Red arrow indicates the P (flipped) kinetochore. Bars: (left) 3 µm; (right) 1 µm. (d) Image of a prometaphase cell stained with DAPI and antibodies against CENP-A and phosphorylated Hec1S55. Zoom boxes depict the staining intensities at aligned bioriented (1) and unaligned bioriented (2) kinetochore pairs. Bars: (top) 5 µm; (bottom) 1 µm. (e) Quantification of Hec1pS55 staining intensity relative to CENP-A at aligned and unaligned bioriented kinetochore pairs. n = 256 aligned KT and 135 unaligned KT from 26 cells; P-value calculated using two-sample t test. (f) Quantification of unaligned bioriented kinetochore behavior in eGFP-CENP-A cells treated with control siRNA and 2 µM ZM1 for ≥10 min. Error bars ± SD; 123 KT from 29 cells. (g) Video stills of a stalled kinetochore pair in an eGFP-CENP-A cell treated with control siRNA and 2 µM ZM1. Yellow arrows, kinetochore position; dotted blue line, metaphase plate periphery. Bar, 3 µm.
One possibility is that kinetochore flipping may represent Aurora B–mediated error correction. However, it has been reported that bioriented kinetochore pairs irreversibly antagonize error correction caused by recruitment of PP1 (DeLuca et al., 2011). Nevertheless, it is unknown whether this antagonism occurs once sister-pairs have congressed or immediately after biorientation in a position-independent manner. To investigate this, we quantified the phosphorylation of Ndc80 at serine 55 (a key error correction target; DeLuca et al., 2011) at bioriented sister-pairs that were in either an aligned or unaligned state. We found no difference in Ndc80pS55 levels between these two sister-pair subgroups (Fig. 4, d and e). Importantly, unlike their phospho-state, the rate of kinetochore flipping in control cells is not equal between these two groups. Instead, flipping is restricted to bioriented sister-pairs undergoing congression, suggesting that an alternate mechanism underpins this process. We did attempt to directly test whether Aurora B is required for flipping by treating cells with ZM1 (Ditchfield et al., 2003), but the treatment caused unaligned bioriented kinetochore pairs to stall (Fig. 4, f and g). We have observed flipping only at kinetochore pairs undergoing active congression, so this result is uninformative.
Force-dependent release of the leading kinetochore
An alternative model is that flipping may result from the mechanical failure of attachment, possibly because of phases of high pulling force on the lead sister. To test this idea, we depleted the microtubule depolymerase mitotic centromere-associated kinesin (MCAK; Hunter et al., 2003), which reduces the speed of kinetochore movement within the metaphase plate as a result of changing the balance of microtubule depolymerization at the P kinetochore (Wordeman et al., 2007; Jaqaman et al., 2010; Armond et al., 2015). Quantitative immunofluorescence revealed that both MCAK and Ska1 were codepleted efficiently (Fig. 5, a–d). In agreement with studies of aligned kinetochore-pairs (Jaqaman et al., 2010), MCAK depletion slowed the speed of congressing kinetochore-pairs from 2.93 ± 1.2 µm/min in control cells to 1.95 ± 0.81 µm/min in MCAK-depleted cells (Fig. 5 e). We then compared the number of flip events in Ska1 and Ska1/MCAK double-depleted cells and found that MCAK codepletion with Ska1 reduced the number of flip events from 63.1 ± 2.7% to 38.3 ± 5.6%, enabling a larger proportion of chromosomes to congress (Fig. 5 f). Interpretation of this result is potentially problematic, as MCAK has been proposed to function in Aurora B–mediated error correction (Bakhoum et al., 2009; Ems-McClung et al., 2013). Therefore, we treated Ska1-depleted cells with 100 nM taxol for 1 h, which reduces microtubule dynamicity and decreases intersister tension (DeLuca et al., 2011). We found that the treatment reduced the rate of flipping to 41.7 ± 1.55%, allowing more kinetochore pairs to successfully congress (Fig. 5 g). Thus, we favor a model in which flip events are caused by excess pulling forces being exerted on the kinetochore-depolymerizing microtubule attachment, which can lead to mechanical failure.
Figure 5.
Force-dependent release of the leading kinetochore. (a) Images of cells treated with control, MCAK, or Ska1 + MCAK siRNA and stained with antibodies against CENP-A and MCAK. Zoom boxes depict the staining at individual kinetochores. Bars: (top) 5 µm; (bottom) 1 µm. (b) Quantification of MCAK intensities in cells treated with control, MCAK, or Ska1 + MCAK siRNA. n = 200 KT from 20 cells per condition. (c) Left, Images of cells treated with control, Ska1 + control, or MCAK siRNA and stained with antibodies against CENP-A and Ska1. Insets show individual kinetochores. Bars: (top) 5 µm; (bottom) 1 µm. (d) Quantification of Ska1 staining intensities in cells treated with control, Ska1 + control, or Ska1 + MCAK siRNA. n = 200 KT from 20 cells per condition. (e) Quantification of kinetochore speed during congression in eGFP-CENP-A–expressing cells treated with control or MCAK siRNA. Measurements were taken from tracks of persistent movement that lasted at least three time frames. n (MCAK) = 76 KT from 42 cells; n (control) = 75 KT from 53 cells; P-value calculated using two-sample t test. (f) Quantification of unaligned bioriented kinetochore behavior in eGFP-CENP-A–expressing cells treated with Ska1 + control, MCAK, or Ska1 + MCAK siRNA. Error bars ± SD; n (Ska1 + control) = 218 KT from 46 cells; n (Ska1 + MCAK) = 215 KT from 72 cells; n (MCAK) = 323 KT from 99 cells. (g) Quantification of unaligned bioriented kinetochore behavior in cells depleted of Ska1 by siRNA and treated with DMSO or 100 nM taxol for 1 h. Error bars ± SD; n (DMSO) = 132 KT from 35 cells; n (taxol) = 127 KT from 38 cells.
Congression is coupled to an increase in microtubule occupancy at kinetochores
We suspected that the increase in load on the P kinetochore may be related to the observation that metaphase kinetochores are bound to more microtubules that those in prometaphase (McEwen et al., 1997). However, this experiment did not rule out that differences were caused by cell cycle effects; we needed a comparison of unaligned and congressed kinetochores in the same cell. To do this, we used our SBF-SEM images to quantify the number of microtubules terminating at bioriented kinetochore pairs. We note that the resolution limit of SEM means that the observed microtubule fibers may reflect both single microtubules and small bundles. Hence, this assay reads out microtubule density rather than absolute microtubule number. We found a ∼50% increase in microtubule density at aligned bioriented sister-pairs compared with those that had not yet congressed (Fig. 6, a and b; and Fig. S2). To substantiate this finding, we analyzed prometaphase HeLa cells fixed using glutaraldehyde and stained with an α-tubulin antibody. The tubulin signal increased by ∼27% at aligned bioriented sister-pairs (Fig. 6, c and e), a relationship that remained in Ska1-depleted cells (Fig. 6, d and e). We do note that depletion of Ska reduces the number of microtubules bound to the kinetochore, a finding that is consistent with previous work (Gaitanos et al., 2009; Raaijmakers et al., 2009; Chan et al., 2012). Together, these data demonstrate that kinetochores recruit additional microtubules in a Ska complex–independent manner during congression.
Figure 6.
Congression is coupled to an increase in microtubule occupancy at kinetochores. (a, I) Single slice from an SBF-SEM image of a prometaphase HeLa cell; boxes show aligned bioriented (1) and unaligned bioriented (2) kinetochore pairs. Zoom boxes display single slices and their associated segmentation. Kinetochore-attached and non–kinetochore-attached microtubule structures are in light green and dark green, respectively. Dotted blue line, metaphase plate center; white asterisks, spindle pole positions. Bars: (left) 5 µm; (right) 1 µm. (II, top) Box depicting the spindle region used to render microtubule attachment at the sister pairs indicated in 1 and 2. (bottom) Z-projection (slices 56–65) of the kinetochore and microtubule model generated from the cell in I. Kinetochore-attached and non–kinetochore-attached microtubule structures are indicated in light green and dark green, respectively. (b) Quantification of microtubule structures terminating at kinetochores in either aligned bioriented or unaligned bioriented state by SBF-SEM. n = 100 aligned KT and 20 unaligned KT from three cells; P-value calculated using two-sample t test. Image of a prometaphase eGFP-CENP-A cell treated with control (c) or Ska1 (d) siRNA and stained with DAPI and anti–α-tubulin. Zoom boxes depict the staining intensities at aligned bioriented (1) and unaligned bioriented (2) kinetochore pairs, respectively. Bars: (top) 5 µm; (bottom) 1 µm. (e) Quantification of kinetochore-proximal α-tubulin signal at aligned bioriented and unaligned bioriented kinetochore pairs in cells treated with either control or Ska1 siRNA. n (control) = 199 aligned KT and 75 unaligned KT from 20 cells; n (Ska1) = 152 aligned KT and 38 unaligned KT from 16 cells; P-value calculated using two-sample t test.
The fates of flipped kinetochore pairs
During our initial analysis of congression phenotypes associated with DCP, we found that siRNA control cells have a ∼20% baseline level of flipping (Fig. 2 b), which is also observed in untreated HeLa cells (flip rate of 22.3 ± 3.3%; Fig. 7 a). Additionally, flipping can be observed in RPE1 cells expressing eGFP-CENP-A (Fig. S3); however, because of the accelerated speed of congression and the instantaneous biorientation of kinetochore pairs in these cells (Magidson et al., 2011), such events are less frequent. In unperturbed cells, this flipping behavior is identical to that in Ska1-depleted cells, in which the P kinetochore detaches from its associated K-fiber during congression, resulting in a reduction in intersister distance (Fig. 7, b and c). Importantly, however, the fates of these flipping kinetochore pairs are distinct from those in Ska1-depleted cells. In Ska1-depleted cells, kinetochores flip, reattach to spindle microtubules (the K–K axis stabilizes at <45°), and resume congression. However, 42 ± 6.4% of these kinetochores undergo a second flip (Fig. 7, d and f), and 91 ± 8.7% of all flipped kinetochores failed to congress during the 5-min video (Fig. 7, d and g). This sequential flipping can be illustrated by tracking the intersister distance and K–K axis angle of the sister-pair (Fig. 7, h and i). We observed a decrease in sister separation (indicative of detachment), which corresponds with an increase in K–K axis angle after both flip events (Fig. 5 i). In both cases, these flip events were preceded by a period of low K–K axis angle and intersister breathing ∼1 µm, demonstrating that the sister-pair was bioriented before the first flip and reattached to microtubules before the second flip (Fig. 7 i). In contrast, only 15 ± 6.2% of kinetochores in control cells undergo a second flip (Fig. 7 f), and 61 ± 1.7% of all flipped kinetochores successfully congress (Fig. 7, e, g, and j). Given that we have only observed a 5-min snapshot of congression, it is likely that all flipped kinetochores in control cells reach the metaphase plate. This is supported by 12-h time-lapse videos of HeLa cells expressing Histone-2B-GFP, in which 100% of chromosomes in control cells congressed by 24 min, compared with 50% in Ska1-depleted cells (Fig. S4). The resolution of attachment and subsequent congression of flipped kinetochore pairs in control cells highlights two points: (a) flipping does not represent a subset of defective kinetochores, and (b) flipping is a normal feature of chromosome congression because flips can lead to reattachment and successful congression in unperturbed human cells.
Figure 7.
Fates of flipped kinetochore pairs. (a) Quantification of unaligned bioriented kinetochore behavior in untreated (WT) eGFP-CENP-A–expressing cells and those treated with control siRNA. Error bars ± SD; n (control) =157 KT from 90 cells; n (WT) = 134 KT from 43 cells. (b) Quantification of interkinetochore distance before and after the flip event in control siRNA-treated cells; measurements were taken as in Fig. 4 a; n = 52 KT from 38 cells; P-value calculated using two-sample t test. (c) Proportion of flip events that occurred at the either P or AP kinetochore in control siRNA-treated cells; measurements were taken as in Fig. 4 b. Error bars ± SD; n = 52 KT from 43 cells. (d) Example image sequence of a Ska1-depleted kinetochore pair undergoing two sequential flip events (t = 60 and 112.5 s) interspersed by a period of reorientation (t = 75–82.5 s). Red and yellow arrows, P and AP kinetochores; dotted blue line, metaphase plate periphery. Bar, 2 µm. (e) Example image sequence of a kinetochore pair undergoing a flip event followed by reorientation and congression. Red and yellow arrows, P and AP kinetochores; dotted blue line, metaphase plate periphery. Bar, 2 µm. (f) Proportion of flipped kinetochore pairs that undergo a second flip event in cells treated with control or Ska1 siRNA. Error bar ± SD; n (control) = 28 KT from 27 cells; n (Ska1) = 78 KT from 59 cells. (g) Quantification of kinetochore pair fate after a flip event in cells treated with control or Ska1 siRNA. Error bar ± SD; n (control) = 28 KT from 27 cells; n (Ska1) = 78 KT from 59 cells. (h) Schematic illustrating the measurement of interkinetochore distance (d) and K–K axis angle (the angle of sister kinetochores [green lines] relative to metaphase plate). K–K axis angle is normalized so that a 90° orientation to the metaphase plate is 0. (i and j) Plot of K–K axis angle and interkinetochore distance during two sequential flip events in the Ska1-depleted kinetochore pair shown in d (i) or during a flip event followed by successful congression in the control siRNA treated kinetochore pair shown in e (j). Dotted green line, kinetochore axis angle of 75°; dotted red line, intersister distance in absence of microtubules (i.e., rest length).
Dynamic maturation of the Ska complex during congression
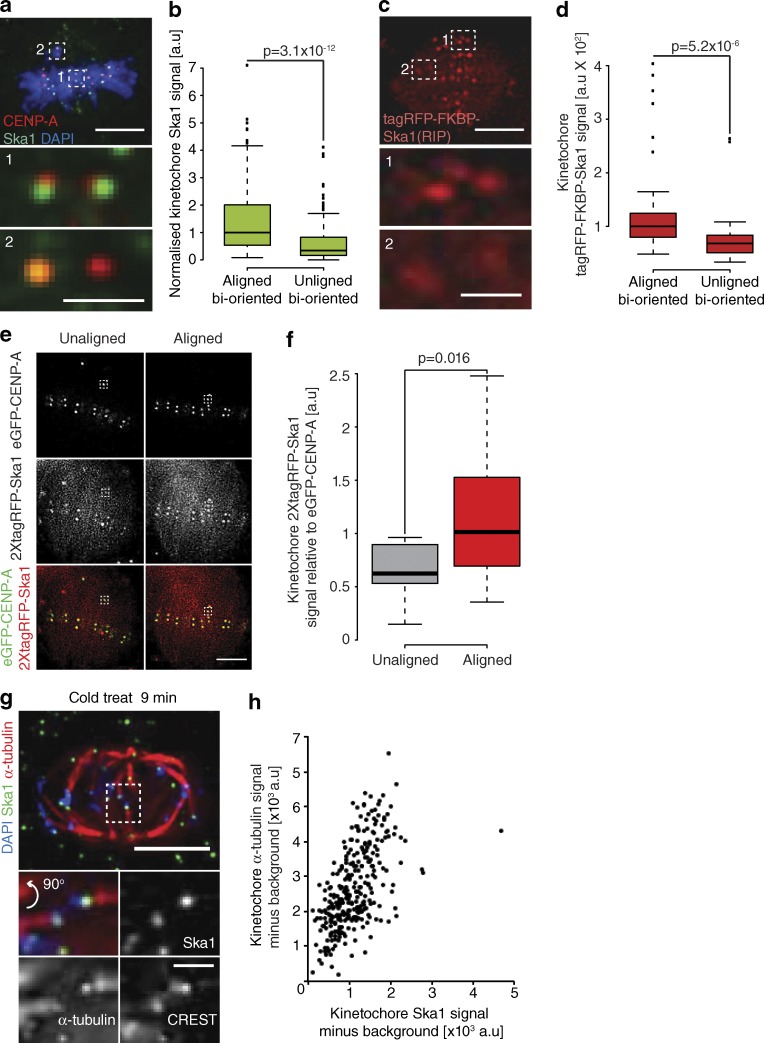
Our data show that the Ska complex is required to limit the number of lead kinetochore detachment events. Interestingly, we noticed in our immunostaining experiments that Ska1 levels appeared to be reduced on unaligned bioriented kinetochore pairs compared with those that had aligned. Indeed, quantification revealed that Ska1 staining intensity was reduced by 51% at unaligned bioriented kinetochore pairs, a finding we confirmed in live cells using a tagRFP-FKBP–tagged Ska1 transgene (Fig. 8, a–d). This finding suggested that bioriented kinetochores load the Ska complex during congression. To test this, we took a single image of an unaligned kinetochore pair expressing 2xtagRFP-Ska1, allowed it to congress, and imaged the same kinetochore pair once more. We found that the 2xtagRFP-Ska1 signal increased 178% during this alignment (Fig. 8, e and f; and Fig. S5). Thus, bioriented kinetochores progressively load Ska complexes as they congress.
Figure 8.
Dynamic maturation of the Ska complex during congression. (a) Image of a prometaphase cell stained with DAPI and antibodies against CENP-A and Ska1. Zoom boxes show aligned (1) and unaligned (2) bioriented kinetochore pairs. Bars: (top) 5 µm; (bottom) 1 µm. (b) Quantification of Ska1 staining intensity at aligned and unaligned bioriented kinetochore pairs. n ≥ 157 KT from 37 cells per condition; P-value calculated using two-sample t test. (c) Image of a prometaphase eGFP-CENP-A–expressing cell transfected with tagRFP-FKBP-Ska1 (tagRFP channel is shown). Zoom boxes depict the staining at aligned (1) and unaligned (2) bioriented kinetochore pairs. Bars: (top) 5 µm; (bottom) 1 µm. (d) Quantification of tagRFP-FKBP-Ska1 intensities at aligned and unaligned bioriented kinetochore pairs after background subtraction. n ≥ 53 KT from 15 cells per condition; P-value calculated using two-sample t test. (e) Left, image of a late-prometaphase HeLa cell with a single unaligned kinetochore pair expressing eGFP-CENP-A and transfected with 2×tagRFP-Ska1. Right, image of the same cell after the unaligned kinetochore pair has congressed. Bar, 5 µm. (f) Quantification of 2×tagRFP-Ska1 intensity at single kinetochores when in an unaligned state and after congression. n = 12 KT from 10 cells; P-value calculated using two-sample t test. (g) Image of a cold-treated metaphase cell fixed and stained with DAPI and antibodies against α-tubulin and Ska1. Zoom boxes depict the staining intensities at individual kinetochore pairs. Bars: (top) 5 µm; (bottom) 1 µm. (h) Scatterplot of Ska1 and kinetochore proximal α-tubulin signals at single kinetochores after background subtraction in cold-treated metaphase cells. n = 300 KT from 30 cells.
This result raised the possibility that Ska complex maturation is coupled to the recruitment of microtubules (Fig. 6, a–f). Ideally, we would have correlated Ska complex and α-tubulin intensities at single kinetochores in different spindle positions. However, our Ska1 antibody was ineffective with glutaraldehyde fixation, and the noise from non–K-fiber microtubules confounds quantification in cells fixed with paraformaldehyde. To overcome these obstacles, we used cold treatment to remove nonkinetochore microtubules, fixed the cells using paraformaldehyde, and then quantified the Ska1 and α-tubulin signals at single aligned kinetochores (Fig. 8, g and h). It is important to note that a limitation of this approach is that spindle length decreases in the cold, forming a broad metaphase plate that contains a mix of sister-pairs that were aligned and unaligned before treatment (Fig. 8 g). Nonetheless, our analysis revealed a strong linear correlation (R2 = 0.3444) between the levels of Ska1 and α-tubulin (Fig. 8, g and h). Together, these data demonstrate that bioriented kinetochores can exist in both mature and immature states, and this Ska1 maturation is correlated with both congression and the number of K-fiber microtubules.
Ska maturation correlates with loss of Bub1 from kinetochores
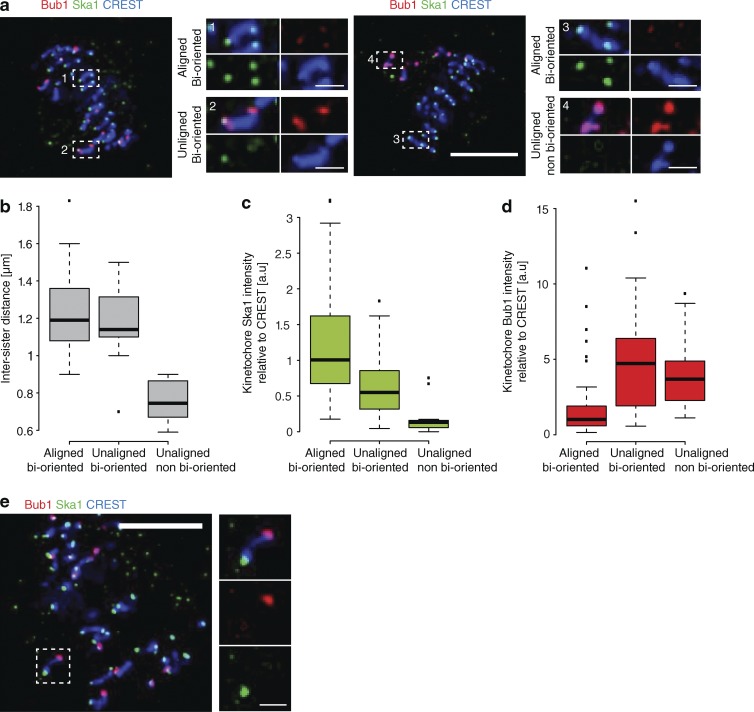
It is well established that unattached kinetochores generate a “wait-anaphase” signal through activation of the SAC. This involves the recruitment of Bub and Mad proteins to the kinetochore (Musacchio, 2015). Our data suggest that immature bioriented kinetochores may also generate a wait-anaphase signal. To investigate this, we stained cells with anti-Bub1 as a readout of checkpoint signaling and anti-Ska1 antibodies as a marker for maturation (Fig. 9 a). First, we measured the levels of both proteins on the same single kinetochores that were aligned, unaligned/bioriented, or unaligned/non-bioriented. These states were defined by the spindle position, orientation, and intersister distance of each sister kinetochore pair (Fig. 9 b). As expected, unaligned/non-bioriented kinetochores had high levels of Bub1 and low levels of Ska1 (Fig. 9, a, c, and d). In contrast, the aligned kinetochores had the opposite phenotype, with high levels of Ska1 and low levels of Bub1 (Fig. 9, a, c, and d). As we previously found, the unaligned but bioriented kinetochores (intersister distances equivalent to the aligned population) had Ska1 levels twofold lower than the aligned kinetochores but significantly higher than the non-bioriented population. However, unlike the aligned kinetochores, Bub1 was still loaded to a level similar to that measured on non-bioriented kinetochores (Fig. 9, a, c, and d). This behavior was also sister kinetochore autonomous, as we observed instances in which one sister was Bub1+/Ska– and the other Bub1–/Ska1+ (Fig. 9 e), thus providing evidence that immature bioriented kinetochores produce an independent SAC signal.
Figure 9.
Ska maturation correlates with loss of Bub1 from kinetochores. (a) Images of prometaphase HeLa cells stained with antibodies against Ska1 and Bub1 and CREST antisera. Zoom boxes depict the staining intensities at aligned bioriented (1 and 3), unaligned bioriented (2), and unaligned nonbioriented (4) kinetochore pairs. Bars: (zoom-out) 5 µm; (zoom-in) 1 µm. (b) Quantification of CREST-based intersister distance at the kinetochore pair subgroups shown in a. n = 159 KT from 16 cells. (c) Quantification of Ska1 staining intensity at the kinetochore pair subgroups defined in a. n = 159 KT from 16 cells. (d) Quantification of Bub1 staining intensity at the kinetochore pair subgroups defined in a. n = 159 KT from 16 cells. (e) Images of prometaphase cells stained with antibodies against Ska1 and Bub1 and CREST antisera. Zoom boxes depict an unaligned bioriented kinetochore pair with opposing Ska1/Bub1 loading. Bars: (left) 5 µm; (right) 1 µm.
Discussion
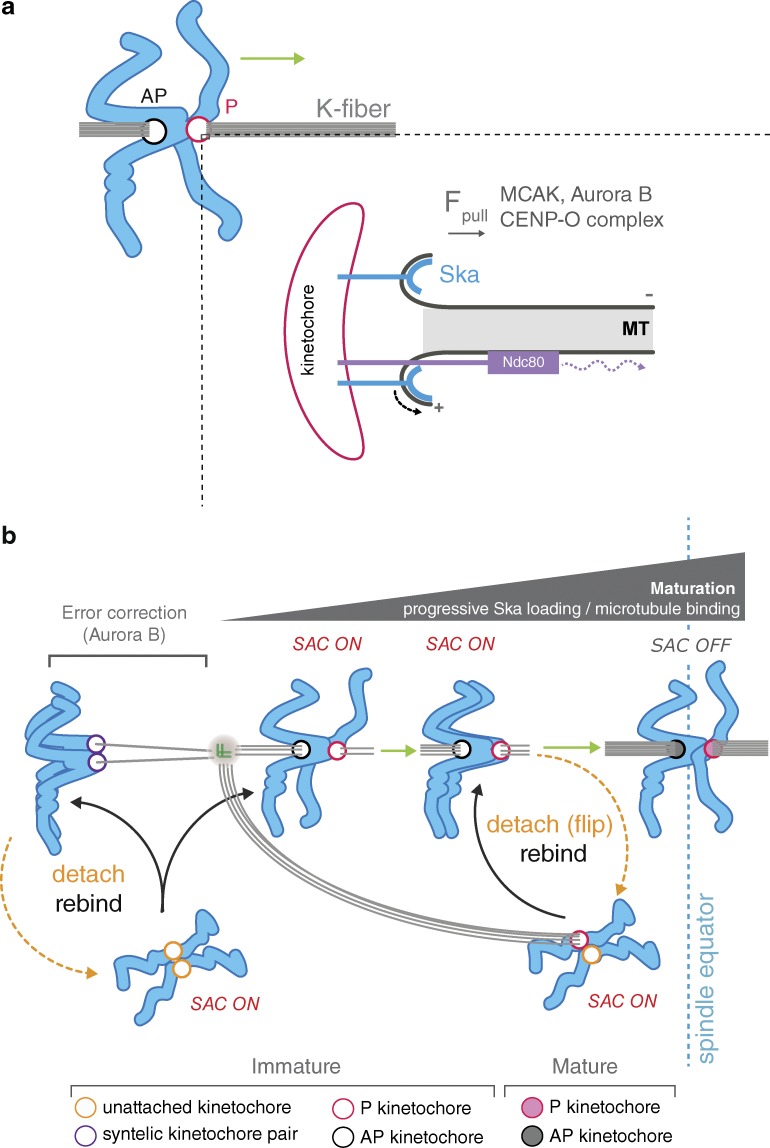
Our data show that the Ska complex is required for the P kinetochore to withstand microtubule pulling forces—a key feature of DCP (Fig. 10 a). This finding is consistent with structural and biochemical studies that show how the Ska complex mediates coupling to depolymerizing microtubule plus-end by binding and remaining attached to curved protofilaments that are extruded during disassembly (Welburn et al., 2009; Jeyaprakash et al., 2012; Schmidt et al., 2012; Abad et al., 2014). However, the Ska complex is not essential for congression per se, because the majority of kinetochores can align in its absence. Presumably, these sisters were not subject to excess load or congressed by lateral sliding (Kapoor et al., 2006), by instantaneous biorientation (Magidson et al., 2011), or through contributions from other redundant factors, with CENP-F being a prime candidate (Volkov et al., 2015).
Figure 10.
Model for congression via DCP and integration with checkpoint signaling. (a) Schematic depicting the contribution of kinetochore factors to movement (green arrow) of bioriented chromosomes by DCP. Inset shows single microtubule-kinetochore attachment site. The Ska complex attaches to, and tracks with, the curving protofilaments as they peel away from the lattice, thus generating a pulling force. This coupling is essential for the maintenance of P kinetochore attachment when under load. Attachments absolutely require the Ndc80 complex, which may also contribute to force generation by biased diffusion (dotted curved arrow). In addition to these coupling factors, MCAK, Aurora B, and CENP-Q—a subunit of CENP-O complex (Bancroft et al., 2015)—contribute to force generation (Fpull). (b) Model for bioriented kinetochore maturation and attachment regulation during prometaphase. The canonical Aurora B error correction pathway destabilizes erroneous attachments (i.e., syntelic), promoting the formation of bioriented attachments. These unaligned bioriented kinetochore pairs then congress by DCP. During congression, each kinetochore progressively (and autonomously) loads Ska complexes. This increases the load-bearing capacity of the structure, enabling kinetochores to efficiently track the depolymerizing K-fiber microtubules, which are increasing in number and stability. Unaligned bioriented, but immature, kinetochore pairs do not silence the SAC. We propose that maturation is part of a mechanical microtubule attachment self-check: if the kinetochore does not recruit sufficient Ska complex, or microtubule recruitment is too rapid, the kinetochore will detach from its associated K-fiber. As a result, only mature bioriented sister kinetochores reach the metaphase plate and silence the SAC.
Here, we have documented a previously overlooked event during mitosis: the detachment and flipping of the P kinetochore during congression (Fig. 10 b). Our data support a model that involves the failure of the kinetochore-microtubule attachment, which would activate the SAC (Fig. 10 b). It could be suggested that we are observing error-correction events mediated by Aurora B. However, several lines of evidence argue against this idea: (a) our own observations and those from DeLuca et al. (2011) suggest that error correction is antagonized once a bioriented attachment has formed; (b) we observed flip events at high intersister distance, thus ruling out correction caused by a loss of tension; (c) it is hard to reconcile how a symmetric cue, the loss of tension across the sister-pair, leads to a highly asymmetric response with only the P sister detaching; and (d) the Ska complex has recently been reported to promote Aurora B activity (Redli et al., 2016). Therefore, Ska1-depleted kinetochores display a high rate of microtubule detachment despite having compromised error correction. Nevertheless, the cause of P kinetochore detachment remains obscure. We favor the idea that kinetochores come under increasing load, perhaps because of the increase in kinetochore-bound microtubules. Moreover, we also found that the poleward-facing sister had more bound microtubules than the sister facing the metaphase plate (unpublished data). This may also explain why the P kinetochore needs to recruit microtubules during congression and why it is more sensitive to pulling forces.
We have directly observed that the Ska complex is progressively recruited to bioriented kinetochore pairs as they congress. This shows that once kinetochores biorient, they load Ska complexes in a maturation step dependent on active congression. We propose that this is to handle the escalating pulling forces that would occur as microtubules are recruited to the kinetochore (Fig. 10 b). Consistently, the level of kinetochore-bound Ska complex is positively correlated with the number of stably attached microtubules. Importantly, Ska1-depleted cells still display the increase in kinetochore-bound microtubules at aligned bioriented sister-pairs, which suggests that the Ska complex is not required for microtubule recruitment to the kinetochore per se, but functions immediately after binding to maintain the attachment during congression. Given that loss of Ska increases the probability of flipping, we suggest that it is the slow maturation of Ska that triggers detachment in normal mitosis. Interestingly, the Ska complex has been reported to recruit PP1, and so it is possible this increase in kinetochore-associated phosphatase activity contributes to maturation (Sivakumar et al., 2016).
Our working model is that these mechanics would allow only bioriented kinetochores that fully mature (in terms of Ska complexes and microtubules) to persist. Indeed, in wild-type cells, flip events are typically followed by reattachment and successful congression, whereas in Ska1-depleted cells, kinetochores are trapped in a futile cycle of detachment-reattachment-detachment. Thus, episodes of lead kinetochore detachment-reattachment in wild-type cells may represent some kind of mechanical self-check on the attachment (Fig. 10 b).
This work provides evidence that unaligned, but bioriented, kinetochores generate a SAC signal that is only extinguished after the kinetochore matures, an event that coincides with congression. This does not, however, mean that the SAC is monitoring congression. In fact, our data show that the maturation state is independent of position; i.e., we can observe kinetochores that are Bub1+/Ska– within the metaphase plate. Our data are consistent with the previous observation that KNL1 and the Bub1:Bub3 complex (KBB pathway) is required for producing a SAC signal when kinetochores are bioriented but not aligned (Silió et al., 2015). The alternative RZZ pathway can activate the SAC only when the kinetochore is unattached. Our data thus provide further evidence that the KBB pathway is required for immature bioriented kinetochores to generate a SAC signal. The nature of the molecular signals that silence the KBB pathway and drive accumulation of Ska complexes at the metaphase plate remains a crucial open question.
Materials and methods
Cell culture, siRNA transfection, and drug treatments
HeLa-Kyoto (K) cells were grown in a humidified incubator at 37°C and 5% CO2 in DMEM (Gibco) containing 10% FCS, 100 U/ml penicillin, and 100 µg/ml streptomycin supplemented with 0.1 µg/ml puromycin (Invitrogen) for maintenance of the eGFP-CENP-A cell line (Jaqaman et al., 2010). The HeLa H2B-GFP cell line was maintained in nonselective medium. The hTERT-RPE1 eGFP-CENP-A cell line was maintained in DMEM/F-12 medium containing 10% FCS, 2.3 g/l sodium bicarbonate, 100 U/ml penicillin, and 100 µg/ml streptomycin. siRNA oligonucleotides (53 nM) were transfected using oligofectamine (Invitrogen) according to the manufacturer’s guidelines and analyzed at 48 h. The following sequences were used: control, 5′-GGACCUGGAGGUCUGCUGU-3′; Ska1, 5′-CCGCUUAACCUAUAAUCAA-3′; and MCAK, 5′-GAUCCAACGCAGUAAUGGU-3′. For drug treatments, cells were treated with 2 µM ZM447439 (Tocris Bioscience) for 30 min or 100 nM taxol (Sigma-Aldrich) for 1 h before live-cell imaging.
Plasmid construction and siRNA rescue experiments
To generate tagRFP-FKBP-Ska1, eGFP was first replaced with tagRFP in pEGFP-C1 (Takara Bio Inc.) using NheI and XhoI, creating pMC387. FKBP12 was then inserted in the XhoI and HindIII restriction sites creating pMC390. Next, full-length siRNA-resistant Ska1 was ligated into pMC390 using PstI/MfeI to create tagRFP-FKBP-Ska1 (RIP; pMC393). Ska1 was rendered resistant to the Ska1 siRNA oligonucleotide (Hanisch et al., 2006) using site-directed mutagenesis with the following primer pairs: 5′-CTTCGTACATGAAATCCCGGTTAACCTATAATCAAATTAA-3′/5′-TTAATTTGATTATAGGTTAACCGGGATTTCATGTACGAAG-3′, 5′-ATGAAATCCCGGTTAACCTACAATCAAATTAATGATGTTA-3′/5′-TAACATCATTAATTTGATTGTAGGTTAACCGGGATTTCAT-3′, and 5′-AATCCCGGTTAACCTACAACCAAATTAATGATGTTATTAA-3′/5′-TTAATAACATCATTAATTTGGTTGTAGGTTAACCGGGATT-3′. For siRNA rescue experiments, eGFP-CENP-A cells were transfected with either Ska1 or control siRNA and grown for 12 h in MEM. The medium was then changed to DMEM containing 0.1 µg/ml puromycin, and the cells were transfected with 1 µg of tagRFP-FKBP-Ska1 (pMC393) or tagRFP-FKBP (pMC390) transgene using FuGene6 (Roche) according to the manufacturer’s instructions and incubated for a further 48 h. For Ska1 loading analysis, FKBP was excised from pMC393 and replaced with PCR-amplified tagRFP fragment using XhoI and HindIII sites, creating pMC463. For expression in cells, 1.5 µg of DNA was transfected using FuGene6 (Roche) and incubated for 48 h.
CRISPR/Cas9
To target Ska1 exon 1, the guide 5′-TAATTGTTCCAGATCTGACG-3′ (Ska1 GuideA top) was cloned into the human codon optimized SpCas9 and chimeric guide expression plasmid (pX330; Addgene) using BbsI. For BbsI compatibility, the sequence CACCG was added to the 5′ end (creating 5′-CACCG TAATTGTTCCAGATCTGACG-3′). Because insertion requires double-stranded DNA, a complimentary oligo 5′-CGTCAGATCTGGAACAATTA-3′ (Ska1 GuideA bottom) was ordered with 5′-AAAC and 3′-C additions for BbsI compatibility (creating 5′-AAACCGTCAGATCTGGAACAATTAC-3′). Together, these modifications allow for scarless cloning into the pX330 vector. The oligos were phosphorylated and annealed by incubation with T4 PNK (New England Biolabs, Inc.) and T4 ligation buffer (New England Biolabs, Inc.; replacing the supplied PNK buffer). The product was ligated into the pX330 vector using a single-step digestion-ligation with FastDigest BbsI (Thermo Fisher Scientific) and T4 ligase (New England Biolabs, Inc.). Any residual linearized DNA was digested using the PlasmidSafe exonuclease (EpiBio). Cells were transfected with 1.5 µg plasmid using Fugene6 (Roche) in 1.5 ml DMEM according to the manufacturer’s instructions. After 24 h, 500 µl fresh DMEM was added, and the cells were grown for a further 24 h, at which point the medium was replaced with 2 ml DMEM containing selective antibiotics if necessary and grown for 24 h. Cells were imaged, harvested, and fixed after 72 h of transfection. For Surveyor analysis of guide-induced indel mutation, HeLa-K cells were harvested from a single 35-mm dish after guide transfection for 72 h, and genomic DNA was extracted using a QIAamp DNA blood mini kit (QIAGEN). A ∼1-kb fragment containing the target site was amplified using PCR with the primer pair 5′-TTAGACCCTCCCCTTCTCTCTC-3′ and 5′-CGCTTTTGTCAGAACACATCTC-3′. 200 ng of PCR product was denatured and reannealed in NEB buffer 2 (New England Biolabs, Inc.; final volume 19 µl) using a thermocycler and the following conditions: 95°C for 5 min; 95°C to 85°C at –2°C/s; and 85°C to 25°C at –0.1°C/s. After cycling, the reaction mix was incubated with 1 µl T7 endonuclease (New England Biolabs, Inc.) for 15 min at 37°C.
Immunofluorescence microscopy
For anti–CENP-A (mouse; ab13939; Abcam), anti-Ska1 (rabbit; ab46826; Abcam), anti-Hec1PSer55 (rabbit; no longer available; Thermo Fisher Scientific), and anti-Bub1 (mouse; ab54893; Abcam) primary antibodies, cells were fixed for 10 min at RT with 20 mM Pipes, pH 6.8, 10 mM EGTA, 1 mM MgCl2, 0.2% Triton X-100, and 4% formaldehyde. For the cold stable assay, cells were placed on ice for 9 min before fixation. For anti-MCAK antibodies (rabbit; AKIN05; Cytoskeleton, Inc.), cells were fixed for 10 min at −20°C in methanol. In both cases, the cells were then washed three times in PBS for 5 min before blocking in 3% BSA in PBS for 30 min. Cells were incubated for 1 h with primary antibodies: anti–CENP-A (1:500), anti-Ska1 (1:400), anti-Hec1pSer55 (1:300), anti-MCAK (1:500), anti-Bub1 (1:500), and CREST antisera (1:250) and washed three times in PBS for 5 min. Secondary antibodies were Alexa Fluor–conjugated secondary antibodies (Invitrogen) used at 1:500 for 30 min. For anti–α-tubulin (mouse; T6074-200UL; Sigma-Aldrich) antibodies, cells were preextracted for 30 s in 80 mM Pipes, pH 6.8, 1 mM MgCl2, 4 mM EGTA, and 0.5% Triton X-100 and fixed by adding glutaraldehyde at 0.5% for 10 min. Glutaraldehyde was quenched using a 7-min treatment with 0.1% NaBH4. Cells were washed three times in PBS for 5 min, followed by blocking in TBS containing 0.1% Triton X-100 and 2% BSA for 10 min. Cells were then incubated with anti–α-tubulin (1:1,000) primary antibody for 30 min, followed by four 5-min washes in TBS containing 0.1% Triton X-100 and incubation with 647 nm Alexa Fluor–conjugated secondary antibodies (Invitrogen) in TBS containing 0.1% Triton X-100 and 2% BSA for 30 min. Cells were mounted and imaged in Vectashield (Vector Laboratories). 3D image stacks were acquired (75 × 0.2-µm z-sections) using a 100× oil NA 1.4 objective on an Olympus Deltavision Elite microscope (Applied Precision Ltd.) equipped with a DAPI, FITC, Rhodamine, or Texas Red and Cy5 filter set, solid-state light source, and a CoolSNAP HQ2 camera (Roper Technologies) at 37°C. Images were acquired, the stacks were deconvolved using SoftWorx, and fluorescence intensity measurements were made manually after background subtraction and normalization to the CENP-A or CREST signal. Figures were constructed in Illustrator CS5 (Adobe Systems).
Statistical methods
Staining intensity distributions were compared using a two-sample t test in R. Data distributions were assumed to be normal, but this was not formally tested.
Live-cell imaging
To film kinetochore fates, HeLa-K eGFP-CENP-A–expressing cells were seeded in FluoroDishes (World Precision) and imaged in DMEM supplemented with 10% FCS, 100 U/ml penicillin, 100 µg/ml streptomycin, and 0.1 µg/ml puromycin. 3D image stacks (25 × 0.5-µm z-sections) were acquired every 7.5 s using a 100× oil NA 1.4 objective on an Olympus Deltavision Elite microscope equipped with a eGFP and mCherry filter set, Quad-mCherry dichroic mirror, solid-state light source, and CoolSNAP HQ2 camera. Environment was tightly controlled at 37°C and 5% CO2 using a stage-top incubator (Tokai Hit) and a weather station (Precision Control). Image stacks were deconvolved using SoftWorx (Applied Precision Ltd.), and kinetochore fates were determined manually. Measurements of kinetochore velocity were taken manually from tracks of persistent movement that lasted at least three time frames. Images were deconvolved using SoftWorx, and fluorescence intensity measurements were made manually after background subtraction. To film kinetochore fates in the hTERT-RPE1 eGFP-CENP-A cell line, cells were seeded in FluoroDishes and imaged in Leibovitz L-15 supplemented with 10% FCS. 3D image stacks (25 z-sections, 0.5 µm) were acquired every 2 s for 5 min using a 100× 1.4 NA oil objective on a confocal spinning-disk microscope (VOX Ultraview; PerkinElmer) equipped with a Hamamatsu Photonics ORCA-R2 camera, controlled by Volocity 6.0 (PerkinElmer) running on a Windows 7 64-bit (Microsoft) PC (IBM) at 37°C. Images were deconvolved with Huygens 4.1 (SVI) using a point spread function measured from microbead images (using the Huygens 4.1 PSF distiller), and visualized in Fiji.
EM
For correlative light EM experiments, HeLa-K eGFP-CENP-A–expressing cells were imaged on gridded glass dishes (P35G-1.5-14-CGRD; MatTek Corporation) using a Deltavision microscope as described earlier. The photo-etched grid coordinate containing the cell of interest was recorded using bright-field illumination at 20×. The same cell was then relocated, and live-cell imaging was acquired at 100×. Kinetochore pairs of interest where tracked as described earlier, and once the congression event had occurred, cells were immediately chemically fixed in 3% glutaraldehyde, 0.5% paraformaldehyde, and 0.1% tannic acid in 0.05 M phosphate buffer, pH 7.4, for 1 h and processed for SBF-SEM. Samples were processed and images segmented as previously described (Nixon et al., 2016 Preprint). In brief, cells were postfixed in reduced osmium (2% osmium tetroxide, 1.5% potassium ferricyanide) for 1 h, incubated in 0.1% thiocarbohydrazide in dH2O for 20 min, and incubated in 2% osmium tetroxide in dH2O for 30 min before being incubated overnight in 1% uranyl acetate dH2O at 4°C. Cells were incubated in Walton’s lead aspartate at 60°C for 30 min and then dehydrated in graded series ethanol before being infiltrated and embedded in EPON 812 hard premixed resin (TAAB). Once the resin had fully polymerized, the coverslip was removed and the cell of interest was located using previously acquired grid coordinates. The block of resin containing the cell of interest was then excised and mounted on to a cryo pin using Silver Dag (Agar Scientific). Excess resin was trimmed away with an Ultramicrotome (Leica Biosystems), leaving a 200 × 200-µm block face. To reduce charging, the block was first painted with silver conductive paste (TAAB) before 10 nm gold/palladium was evaporated onto it (Quorum Technologies). Imaging was done using Gatan 3View System and Digital Micrograph software (Gatan) and FEI Quanta 250 (FEI). To image the whole cell including all chromosomes and visualize both microtubules and kinetochores at a resolution of ∼5 nm in x and y and 60 nm in z, the following imaging parameters were used: 4,000 × 4,000 frame size, 12-µs dwell time, 2.3 KeV, 50 Pa, and magnification 9500×. Kinetochores were identified in the images as electron-dense regions on chromatin that span two to three z-slices (120–180 nm).
Online supplemental material
Fig. S1 shows effectiveness of siRNA and CRISPR/Cas9 targeting Ska1. Fig. S2 shows slice by slice representation of the EM and its associated render for the kinetochore pairs shown in Fig. 6. Fig. S3 shows an example flip event from an untreated RPE1 cell. Fig. S4 shows changes in eGFP-CENP-A signal do not contribute to the observed increase in Ska1 at congressed kinetochore pairs. Fig. S5 shows quantification of eGFP-CENP-A and tagRFP intensity.
Supplementary Material
Acknowledgments
We thank H. Drechsler, E. Vladimirou, J. Armond, E. Roscioli, P. Meraldi, and J. Millar for discussion and comments on the manuscript. We thank C. Smith for generation of the tagRFP-C1 vector and imaging of kinetochores in RPE1 cells and P. Jallepalli for the Ska1 cDNA. We acknowledge technical assistance from A. Beckett (Liverpool Biomedical EM Unit).
P. Auckland is funded by the Medical Research Council Doctoral Training Partnership grant MR/J003964/1, N.I. Clarke by a Cancer Research UK studentship (C25425/A16141), and S.J. Royle by a Senior Fellowship from Cancer Research UK (C25425/A15182). A.D. McAinsh was supported by a Wellcome Trust Senior Investigator Award (grant 106151/Z/14/Z) and a Royal Society Wolfson Research Merit Award (grant WM150020).
The authors declare no competing financial interests.
Author contributions: Project conception, planning, data interpretation, and manuscript preparation were carried out by A.D. McAinsh and P. Auckland. All experiments were carried out by P. Auckland, except for the SBF-SEM, which was completed and analyzed by N.I. Clarke and S.J. Royle.
Footnotes
Abbreviations used:
- AP
- away from the pole
- DCP
- depolymerization-coupled pulling
- K–K axis
- kinetochore–kinetochore axis
- MCAK
- mitotic centromere-associated kinesin
- NEB
- nuclear envelope breakdown
- P
- poleward
- SAC
- spindle assembly checkpoint
- SBF-SEM
- serial block-face scanning EM
References
- Abad M.A., Medina B., Santamaria A., Zou J., Plasberg-Hill C., Madhumalar A., Jayachandran U., Redli P.M., Rappsilber J., Nigg E.A., and Jeyaprakash A.A.. 2014. Structural basis for microtubule recognition by the human kinetochore Ska complex. Nat. Commun. 5:2964 10.1038/ncomms3964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armond J.W., Vladimirou E., Erent M., McAinsh A.D., and Burroughs N.J.. 2015. Probing microtubule polymerisation state at single kinetochores during metaphase chromosome motion. J. Cell Sci. 128:1991–2001. 10.1242/jcs.168682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Auckland P., and McAinsh A.D.. 2015. Building an integrated model of chromosome congression. J. Cell Sci. 128:3363–3374. 10.1242/jcs.169367 [DOI] [PubMed] [Google Scholar]
- Bakhoum S.F., Thompson S.L., Manning A.L., and Compton D.A.. 2009. Genome stability is ensured by temporal control of kinetochore-microtubule dynamics. Nat. Cell Biol. 11:27–35. 10.1038/ncb1809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bancroft J., Auckland P., Samora C.P., and McAinsh A.D.. 2015. Chromosome congression is promoted by CENP-Q- and CENP-E-dependent pathways. J. Cell Sci. 128:171–184. 10.1242/jcs.163659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barisic M., Aguiar P., Geley S., and Maiato H.. 2014. Kinetochore motors drive congression of peripheral polar chromosomes by overcoming random arm-ejection forces. Nat. Cell Biol. 16:1249–1256. 10.1038/ncb3060 [DOI] [PubMed] [Google Scholar]
- Cai S., O’Connell C.B., Khodjakov A., and Walczak C.E.. 2009. Chromosome congression in the absence of kinetochore fibres. Nat. Cell Biol. 11:832–838. 10.1038/ncb1890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cassimeris L., and Salmon E.D.. 1991. Kinetochore microtubules shorten by loss of subunits at the kinetochores of prometaphase chromosomes. J. Cell Sci. 98:151–158. [DOI] [PubMed] [Google Scholar]
- Chan Y.W., Jeyaprakash A.A., Nigg E.A., and Santamaria A.. 2012. Aurora B controls kinetochore–microtubule attachments by inhibiting Ska complex–KMN network interaction. J. Cell Biol. 196:563–571. 10.1083/jcb.201109001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheeseman I.M. 2014. The kinetochore. Cold Spring Harb. Perspect. Biol. 6:a015826 10.1101/cshperspect.a015826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daum J.R., Wren J.D., Daniel J.J., Sivakumar S., McAvoy J.N., Potapova T.A., and Gorbsky G.J.. 2009. Ska3 is required for spindle checkpoint silencing and the maintenance of chromosome cohesion in mitosis. Curr. Biol. 19:1467–1472. 10.1016/j.cub.2009.07.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeLuca K.F., Lens S.M., and DeLuca J.G.. 2011. Temporal changes in Hec1 phosphorylation control kinetochore-microtubule attachment stability during mitosis. J. Cell Sci. 124:622–634. 10.1242/jcs.072629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ditchfield C., Johnson V.L., Tighe A., Ellston R., Haworth C., Johnson T., Mortlock A., Keen N., and Taylor S.S.. 2003. Aurora B couples chromosome alignment with anaphase by targeting BubR1, Mad2, and Cenp-E to kinetochores. J. Cell Biol. 161:267–280. 10.1083/jcb.200208091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drpic D., Pereira A.J., Barisic M., Maresca T.J., and Maiato H.. 2015. Polar ejection forces promote the conversion from lateral to end-on kinetochore-microtubule attachments on mono-oriented chromosomes. Cell Reports. 13:460–469. 10.1016/j.celrep.2015.08.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ems-McClung S.C., Hainline S.G., Devare J., Zong H., Cai S., Carnes S.K., Shaw S.L., and Walczak C.E.. 2013. Aurora B inhibits MCAK activity through a phosphoconformational switch that reduces microtubule association. Curr. Biol. 23:2491–2499. 10.1016/j.cub.2013.10.054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaitanos T.N., Santamaria A., Jeyaprakash A.A., Wang B., Conti E., and Nigg E.A.. 2009. Stable kinetochore-microtubule interactions depend on the Ska complex and its new component Ska3/C13Orf3. EMBO J. 28:1442–1452. 10.1038/emboj.2009.96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorbsky G.J., Sammak P.J., and Borisy G.G.. 1987. Chromosomes move poleward in anaphase along stationary microtubules that coordinately disassemble from their kinetochore ends. J. Cell Biol. 104:9–18. 10.1083/jcb.104.1.9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanisch A., Silljé H.H., and Nigg E.A.. 2006. Timely anaphase onset requires a novel spindle and kinetochore complex comprising Ska1 and Ska2. EMBO J. 25:5504–5515. 10.1038/sj.emboj.7601426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter A.W., Caplow M., Coy D.L., Hancock W.O., Diez S., Wordeman L., and Howard J.. 2003. The kinesin-related protein MCAK is a microtubule depolymerase that forms an ATP-hydrolyzing complex at microtubule ends. Mol. Cell. 11:445–457. 10.1016/S1097-2765(03)00049-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaqaman K., King E.M., Amaro A.C., Winter J.R., Dorn J.F., Elliott H.L., McHedlishvili N., McClelland S.E., Porter I.M., Posch M., et al. 2010. Kinetochore alignment within the metaphase plate is regulated by centromere stiffness and microtubule depolymerases. J. Cell Biol. 188:665–679. 10.1083/jcb.200909005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeyaprakash A.A., Santamaria A., Jayachandran U., Chan Y.W., Benda C., Nigg E.A., and Conti E.. 2012. Structural and functional organization of the Ska complex, a key component of the kinetochore-microtubule interface. Mol. Cell. 46:274–286. 10.1016/j.molcel.2012.03.005 [DOI] [PubMed] [Google Scholar]
- Kapoor T.M., Lampson M.A., Hergert P., Cameron L., Cimini D., Salmon E.D., McEwen B.F., and Khodjakov A.. 2006. Chromosomes can congress to the metaphase plate before biorientation. Science. 311:388–391. 10.1126/science.1122142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khodjakov A., and Rieder C.L.. 1996. Kinetochores moving away from their associated pole do not exert a significant pushing force on the chromosome. J. Cell Biol. 135:315–327. 10.1083/jcb.135.2.315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lampson M.A., and Cheeseman I.M.. 2011. Sensing centromere tension: Aurora B and the regulation of kinetochore function. Trends Cell Biol. 21:133–140. 10.1016/j.tcb.2010.10.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lampson M.A., Renduchitala K., Khodjakov A., and Kapoor T.M.. 2004. Correcting improper chromosome-spindle attachments during cell division. Nat. Cell Biol. 6:232–237. 10.1038/ncb1102 [DOI] [PubMed] [Google Scholar]
- Liu D., Vader G., Vromans M.J.M., Lampson M.A., and Lens S.M.. 2009. Sensing chromosome bi-orientation by spatial separation of aurora B kinase from kinetochore substrates. Science. 323:1350–1353. 10.1126/science.1167000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magidson V., O’Connell C.B., Lončarek J., Paul R., Mogilner A., and Khodjakov A.. 2011. The spatial arrangement of chromosomes during prometaphase facilitates spindle assembly. Cell. 146:555–567. 10.1016/j.cell.2011.07.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magidson V., O’Connell C.B., Lončarek J., Paul R., Mogilner A., and Khodjakov A.. 2011. The spatial arrangement of chromosomes during prometaphase facilitates spindle assembly. Cell. 146:555–567. 10.1016/j.cell.2011.07.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magidson V., Paul R., Yang N., Ault J.G., O’Connell C.B., Tikhonenko I., McEwen B.F., Mogilner A., and Khodjakov A.. 2015. Adaptive changes in the kinetochore architecture facilitate proper spindle assembly. Nat. Cell Biol. 17:1134–1144. 10.1038/ncb3223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maiato H., Gomes A.M., Sousa F., and Barisic M.. 2017. Mechanisms of chromosome congression during mitosis. Biology (Basel). 6:6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McEwen B.F., Heagle A.B., Cassels G.O., Buttle K.F., and Rieder C.L.. 1997. Kinetochore fiber maturation in PtK1 cells and its implications for the mechanisms of chromosome congression and anaphase onset. J. Cell Biol. 137:1567–1580. 10.1083/jcb.137.7.1567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McEwen B.F., Chan G.K.T., Zubrowski B., Savoian M.S., Sauer M.T., and Yen T.J.. 2001. CENP-E is essential for reliable bioriented spindle attachment, but chromosome alignment can be achieved via redundant mechanisms in mammalian cells. Mol. Biol. Cell. 12:2776–2789. 10.1091/mbc.12.9.2776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Musacchio A. 2015. The molecular biology of spindle assembly checkpoint signaling dynamics. Curr. Biol. 25:R1002–R1018. 10.1016/j.cub.2015.08.051 [DOI] [PubMed] [Google Scholar]
- Nixon F.M., Honnor T.R., Starling G.P., Beckett A.J., Johansen A.M., Brettschneider J.A., Prior I.A., and Royle S.J.. 2016. Microtubule organization within mitotic spindles revealed by serial block face scanning EM and image analysis. bioRxiv. doi:10.1101/087866 (Preprint posted November 15, 2016) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raaijmakers J.A., Tanenbaum M.E., Maia A.F., and Medema R.H.. 2009. RAMA1 is a novel kinetochore protein involved in kinetochore-microtubule attachment. J. Cell Sci. 122:2436–2445. 10.1242/jcs.051912 [DOI] [PubMed] [Google Scholar]
- Redli P.M., Gasic I., Meraldi P., Nigg E.A., and Santamaria A.. 2016. The Ska complex promotes Aurora B activity to ensure chromosome biorientation. J. Cell Biol. 215:77–93. 10.1083/jcb.201603019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt J.C., Arthanari H., Boeszoermenyi A., Dashkevich N.M., Wilson-Kubalek E.M., Monnier N., Markus M., Oberer M., Milligan R.A., Bathe M., et al. 2012. The kinetochore-bound Ska1 complex tracks depolymerizing microtubules and binds to curved protofilaments. Dev. Cell. 23:968–980. 10.1016/j.devcel.2012.09.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shrestha R.L., and Draviam V.M.. 2013. Lateral to end-on conversion of chromosome-microtubule attachment requires kinesins CENP-E and MCAK. Curr. Biol. 23:1514–1526. 10.1016/j.cub.2013.06.040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silió V., McAinsh A.D., and Millar J.B.. 2015. KNL1-bubs and RZZ provide two separable pathways for checkpoint activation at human kinetochores. Dev. Cell. 35:600–613. 10.1016/j.devcel.2015.11.012 [DOI] [PubMed] [Google Scholar]
- Sivakumar S., Janczyk P.L., Qu Q., Brautigam C.A., Stukenberg P.T., Yu H., and Gorbsky G.J.. 2016. The human SKA complex drives the metaphase-anaphase cell cycle transition by recruiting protein phosphatase 1 to kinetochores. eLife. 5:pii:e12902 10.7554/eLife.12902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skibbens R.V., Skeen V.P., and Salmon E.D.. 1993. Directional instability of kinetochore motility during chromosome congression and segregation in mitotic newt lung cells: A push-pull mechanism. J. Cell Biol. 122:859–875. 10.1083/jcb.122.4.859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skibbens R.V., Rieder C.L., and Salmon E.D.. 1995. Kinetochore motility after severing between sister centromeres using laser microsurgery: Evidence that kinetochore directional instability and position is regulated by tension. J. Cell Sci. 108:2537–2548. [DOI] [PubMed] [Google Scholar]
- Theis M., Slabicki M., Junqueira M., Paszkowski-Rogacz M., Sontheimer J., Kittler R., Heninger A.K., Glatter T., Kruusmaa K., Poser I., et al. 2009. Comparative profiling identifies C13orf3 as a component of the Ska complex required for mammalian cell division. EMBO J. 28:1453–1465. 10.1038/emboj.2009.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volkov V.A., Grissom P.M., Arzhanik V.K., Zaytsev A.V., Renganathan K., McClure-Begley T., Old W.M., Ahn N., and McIntosh J.R.. 2015. Centromere protein F includes two sites that couple efficiently to depolymerizing microtubules. J. Cell Biol. 209:813–828. 10.1083/jcb.201408083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welburn J.P., Grishchuk E.L., Backer C.B., Wilson-Kubalek E.M., Yates J.R. III, and Cheeseman I.M.. 2009. The human kinetochore Ska1 complex facilitates microtubule depolymerization-coupled motility. Dev. Cell. 16:374–385. 10.1016/j.devcel.2009.01.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welburn J.P., Vleugel M., Liu D., Yates J.R. III, Lampson M.A., Fukagawa T., and Cheeseman I.M.. 2010. Aurora B phosphorylates spatially distinct targets to differentially regulate the kinetochore-microtubule interface. Mol. Cell. 38:383–392. 10.1016/j.molcel.2010.02.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westhorpe F.G., and Straight A.F.. 2013. Functions of the centromere and kinetochore in chromosome segregation. Curr. Opin. Cell Biol. 25:334–340. 10.1016/j.ceb.2013.02.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wordeman L., Wagenbach M., and von Dassow G.. 2007. MCAK facilitates chromosome movement by promoting kinetochore microtubule turnover. J. Cell Biol. 179:869–879. 10.1083/jcb.200707120 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.