Caspase-2 appears to be activated by the PIDDosome, but the role of PIDD in this pathway remains controversial. Ando et al. demonstrate that caspase-2 can be activated by two distinct complexes: one in the nucleolus that comprises PIDD and the nucleolar phosphoprotein NPM1, and one in the cytoplasm that is PIDD independent. Therefore, the nucleolus is a novel site for caspase-2 activation that appears to be essential for caspase-2 function.
Abstract
The PIDDosome (PIDD–RAIDD–caspase-2 complex) is considered to be the primary signaling platform for caspase-2 activation in response to genotoxic stress. Yet studies of PIDD-deficient mice show that caspase-2 activation can proceed in the absence of PIDD. Here we show that DNA damage induces the assembly of at least two distinct activation platforms for caspase-2: a cytoplasmic platform that is RAIDD dependent but PIDD independent, and a nucleolar platform that requires both PIDD and RAIDD. Furthermore, the nucleolar phosphoprotein nucleophosmin (NPM1) acts as a scaffold for PIDD and is essential for PIDDosome assembly in the nucleolus after DNA damage. Inhibition of NPM1 impairs caspase-2 processing, apoptosis, and caspase-2–dependent inhibition of cell growth, demonstrating that the NPM1-dependent nucleolar PIDDosome is a key initiator of the caspase-2 activation cascade. Thus we have identified the nucleolus as a novel site for caspase-2 activation and function.
Introduction
Despite being the most evolutionarily conserved among the caspases, the role of caspase-2 in apoptosis remains unclear. However, the need to understand the mechanisms of regulation of this caspase has been underscored by growing evidence that caspase-2 can function as a tumor suppressor (Puccini et al., 2013a). Loss of caspase-2 has been associated with acceleration of tumorigenesis in murine models of Eµ-Myc lymphoma, ATM-deficient lymphoma, MMTV/c-neu mammary tumors, and K-ras–induced lung tumors (Ho et al., 2009; Pan et al., 2009; Parsons et al., 2013; Puccini et al., 2013b; Terry et al., 2015). Such phenotypes are often accompanied by increases in genomic instability and aneuploidy (Dorstyn et al., 2012; Parsons et al., 2013; Puccini et al., 2013b). This strongly suggests that caspase-2 is activated in response to DNA-damaging insults to remove damaged cells. Caspase-2 has been shown to be activated by DNA damage in both p53-dependent (Lassus et al., 2002; Robertson et al., 2002) and p53-independent manners (Sidi et al., 2008; Myers et al., 2009; Pan et al., 2009; Ando et al., 2012). However, the upstream regulators of caspase-2 in this context remain poorly defined.
Caspase-2 is activated by proximity-induced dimerization of inactive monomers after recruitment to specific high molecular weight protein complexes, which can be considered “activation platforms” (Muzio et al., 1998; Salvesen and Dixit, 1999; Boatright et al., 2003; Baliga et al., 2004). Although there is some evidence that caspase-2 can be recruited to the TNF receptor death-inducing signaling complex (Ahmad et al., 1997; Duan and Dixit, 1997), the most well-defined activation platform for caspase-2 is the PIDDosome. This complex includes the scaffold protein p53-induced protein with a death domain (PIDD), which binds and promotes the oligomerization of the adaptor protein RIP-associated ICH-1/CAD-3 homologous protein with a death domain (RAIDD) via a protein–protein interaction motif present in both proteins called death domain (DD; Tinel and Tschopp, 2004). RAIDD, in turn, binds caspase-2 via a related motif called caspase recruitment domain (CARD; Duan and Dixit, 1997; Jang and Park, 2013). The PIDDosome was first described as spontaneously forming after a temperature shift of cell extracts (heated to 37°C for 30 min; Tinel and Tschopp, 2004) and has a predicted molecular weight of 696.8 kD (Park et al., 2007).
The requirement of PIDD for caspase-2 activation has been subject to some controversy (Bouchier-Hayes and Green, 2012). PIDD can engage caspase-2 in response to genotoxic stress, and PIDD overexpression induces growth suppression that is entirely dependent on RAIDD and partially dependent on caspase-2 (Lin et al., 2000; Berube et al., 2005). However, studies of cells from PIDD-deficient mice show that caspase-2 processing can proceed in the absence of PIDD in response to etoposide and γ-irradiation (IR) and that the caspase-2–containing high molecular weight complex formed upon temperature shift still assembles in the absence of PIDD (Manzl et al., 2009, 2012). One likely explanation for these discrepancies resides in the recent identification of potent inhibitors of PIDDosome assembly that act to counter platform formation after IR. These negative regulators include the mitotic checkpoint factor BubR1, which competes with RAIDD for docking onto the PIDD DD, and the DNA-damage checkpoint kinase Chk1, which negates ATM-mediated phosphorylation of the PIDD DD through an unknown mechanism, thereby decreasing the affinity of PIDD for RAIDD (Ando et al., 2012; Thompson et al., 2015). Although the significance of Chk1- or BubR1-mediated PIDDosome control is not yet completely clear, genetic or pharmacologic reduction of either protein relieves PIDD from its endogenous inhibitors, unveiling a bona fide PIDDosome pathway that mediates apoptosis after IR (Ando et al., 2012; Thompson et al., 2015).
To directly assess the role of PIDD in caspase-2 activation in response to DNA damage, we used caspase-2 bimolecular fluorescence complementation (BiFC). BiFC uses nonfluorescent fragments of the yellow fluorescent protein Venus (“split Venus”) that can associate to reform the fluorescent complex when fused to interacting proteins (Shyu et al., 2006). When the prodomain of caspase-2 is fused to each half of split Venus, recruitment of caspase-2 to its activation platform and the subsequent induced proximity results in enforced association of the two Venus halves. Thus Venus fluorescence acts as a readout for caspase-2–induced proximity, the proximal step in its activation (Bouchier-Hayes et al., 2009). Using this approach, we reveal that in response to DNA damage, caspase-2 activation platforms assemble in the cytoplasm as well as, surprisingly, the nucleolus. This differential localization of activation platform assembly appears to be determined by PIDD, because PIDD is required only for caspase-2–induced proximity in the nucleolus. Furthermore, we identify a direct PIDD-interaction partner in the nucleolus, the phosphoprotein nucleophosmin (NPM1). NPM1 is the most frequently mutated gene in adult acute myeloid leukemia (AML) and has been previously implicated in apoptosis albeit through unknown mechanisms (Falini et al., 2005; Grisendi et al., 2005). Here, we show that NPM1 is directly and specifically responsible for PIDDosome assembly in the nucleolus, an event essential for PIDDosome-mediated apoptosis after DNA damage.
Results
DNA damage induces caspase-2 activation platform assembly in the cytoplasm and the nucleolus
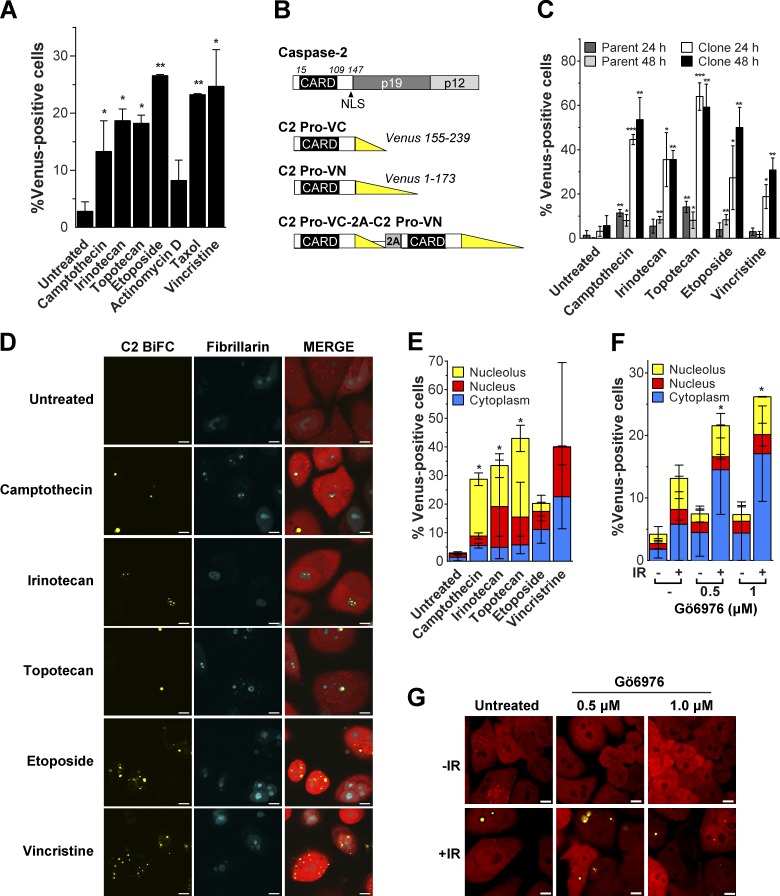
To investigate caspase-2 activation platform assembly induced by DNA damage, we used the caspase-2 BiFC assay (Bouchier-Hayes et al., 2009). We transiently expressed the C2-Pro BiFC pair (aa 1–147) in HeLa cells and observed induction of BiFC in response to a range of DNA-damaging agents, including the topoisomerase II inhibitor etoposide, as we have previously reported (Bouchier-Hayes et al., 2009; Fig. 1 A). We also observed an increase in caspase-2 BiFC in response to the previously untested topoisomerase I inhibitors camptothecin, irinotecan, and topotecan. As controls for the assay, we used the tubulin disruptors taxol and vincristine, known caspase-2 activators that induced BiFC as expected. Actinomycin D, a proapoptotic stimulus that does not engage caspase-2 (O’Reilly et al., 2002; Bouchier-Hayes et al., 2009), had little effect.
Figure 1.
DNA-damaging agents induce caspase-2 BiFC in the nucleolus. (A) HeLa cells transfected with C2-Pro VC (20 ng), C2-Pro VN (20 ng), and dsRed-mito as a transfection reporter (10 ng) were treated with camptothecin (100 µM), irinotecan (100 µM), topotecan (100 µM), etoposide (100 µM), actinomycin D (1 µM), taxol (10 µg/ml), or vincristine (10 µM) plus qVD-OPH (20 µM) to prevent cell detachment caused by apoptosis. Cells were assessed for the percentage of dsRed-positive transfected cells that were Venus+ at 24 h, determined from at least 30 microscopy images per well. Results are the mean of three independent experiments ± SD. *, P < 0.05; **, P < 0.005 compared with untreated sample. (B) Schematic representation of caspase-2 structure and BiFC constructs. Constructs containing the caspase-2 prodomain (C2 Pro) are shown, each fused to the C- or N-terminal fragment of Venus. The bicistronic construct consists of C2 Pro-VC and C2 Pro-VN linked by a 2A self-cleaving peptide. (C) HeLa cells stably expressing C2 Pro-VC-2A-C2 Pro-VN-2A-mCherry (HeLa.C2 Pro-BiFC; parent) were single-cell cloned (clone), and each was treated with the indicated drugs (100 µM) plus qVD-OPH (20 µM). After 24 and 48 h, the percentage of mCherry-positive cells that were Venus+ was determined from at least 30 microscopy images per well. Results are the mean of three independent experiments ± SD. *, P < 0.05; **, P < 0.005; ***, P < 0.0005 compared with the untreated sample in each group. (D) HeLa.C2 Pro-BiFC cells transfected with fibrillarin-CFP were treated with camptothecin (100 µM), irinotecan (100 µM), topotecan (100 µM), etoposide (100 µM), or vincristine (10 µM) plus qVD-OPH (20 µM) for 24 h. Representative images show cells (red) with caspase-2 BiFC (yellow) in the nucleolus (blue) or cytoplasm after treatment. Bars, 10 µm. (E) Percentage of cells treated as in D that were Venus+ in the nucleolus, nucleus, or cytoplasm was determined from at least 30 microscopy images per well. Results are the mean of three independent experiments ± SD. *, P < 0.05 determined from the percentage of nucleolar Venus-positive cells compared with untreated. (F) HeLa.C2 Pro-BiFC cells were treated with or without the Chk1 inhibitor, Gö6976 (0.5 µM, 1 µM), ± IR (10 Gy) plus qVD-OPH (20 µM). The percentage of cells that were Venus+ in the nucleolus, nucleus, or cytoplasm was determined at 24 h from at least 50 microscopy images per well. Results are the mean of three independent experiments ± SD. *, P < 0.05 determined from the percentage of nucleolar Venus-positive cells compared with untreated. (G) Representative confocal images show caspase-2 BiFC (yellow) and mCherry expression (red) in cells treated as in F. Bars, 10 µm.
To overcome the limitations of transient transfection and to validate the results shown in Fig. 1 A, we created stable caspase-2 BiFC reporter cell lines. We designed a bicistronic construct in which the C2 Pro-VC and C2 Pro-VN are expressed in a single vector separated by the viral 2A self-cleaving peptide (Fig. 1 B). This ensures that the caspase-2 BiFC components are expressed at equal levels as they are translated from a single mRNA transcript. The parental HeLa line showed very small yet specific responses to the panel of drugs tested, comprising camptothecin, irinotecan, topotecan, etoposide, and vincristine (Fig. 1 C). To enhance the sensitivity of the stable cell line, we made single-cell clones and identified a clone that showed robust and reproducible responses to each of the agents (Fig. 1 C). The cloned line showed negligible background BiFC fluorescence, indicating minimal spontaneous Venus reassociation. Thus the “cleavage” mediated by the 2A peptide was complete and produced two spatially separated C2 Pro-BiFC proteins in unstimulated cells. The topoisomerase I inhibitors camptothecin, irinotecan, and topotecan each induced high levels of caspase-2 BiFC, identifying a novel class of DNA-damaging agents that induce caspase-2 activation platform assembly. We analyzed the cells under the same conditions by microscopy (Figs. 1 C and S1 A) and flow cytometry (Fig. S1 B), and the results were nearly identical.
Surprisingly, imaging of the HeLa.C2 Pro-VC-2A-C2 Pro-VN-2A-mCherry clone (HeLa.C2 Pro-BiFC) revealed that caspase-2 BiFC induced in response to DNA-damaging agents often manifested as one or more large fluorescent puncta that appeared to be located in the nucleolus (Fig. S1 A). In contrast, caspase-2 BiFC induced by vincristine was primarily detected as a series of small fluorescent complexes localized in the cytoplasm (Fig. S1 A). To confirm that caspase-2 BiFC localized to the nucleolus in response to topoisomerase I inhibition, we expressed fibrillarin-CFP, a nucleolar protein, in the HeLa.C2 Pro-BiFC cells (Fig. 1 D). A substantial number of cells treated with camptothecin, irinotecan, or topotecan induced caspase-2 BiFC in the nucleolar compartment that was delineated by concentrated fibrillarin-CFP fluorescence. Thus a large proportion of cells treated with topoisomerase I inhibitors showed caspase-2 activation localized to the nucleolus, whereas other cells induced caspase-2 BiFC that localized to either the cytoplasm or nucleus (Fig. 1 E). In comparison, in cells exposed to etoposide, only a small fraction of the cells induced nucleolar-localized caspase-2 BiFC, whereas vincristine induced caspase-2 BiFC only in the cytoplasm or nucleus (Fig. 1 E). Similar results were observed in U2OS cells stably expressing the C2 Pro-BiFC components (Fig. S1, C and D). Cells treated with IR plus Chk1 inhibitor (Chk1i, Gö6976), a genotoxic stimulus known to trigger endogenous PIDDosome assembly (Ando et al., 2012), also engaged caspase-2 BiFC in both nucleolar and cytoplasmic compartments (Fig. 1, F and G). Lower doses of IR that are not expected to induce apoptosis had little effect in the absence of Chk1 inhibition (Fig. S1 E). Importantly, HeLa cells transiently transfected with the full-length caspase-2 BiFC components also formed caspase-2 BiFC puncta in the nucleolus in response to camptothecin, albeit with reduced efficiency, further underscoring the enhanced sensitivity of the stable reporter (Fig. S1 F). Together, these results suggest that DNA damage can induce the assembly of activation platforms that recruit caspase-2 in at least two distinct compartments in the cell: the nucleolus and the cytoplasm.
Caspase-2 activation in the nucleolus
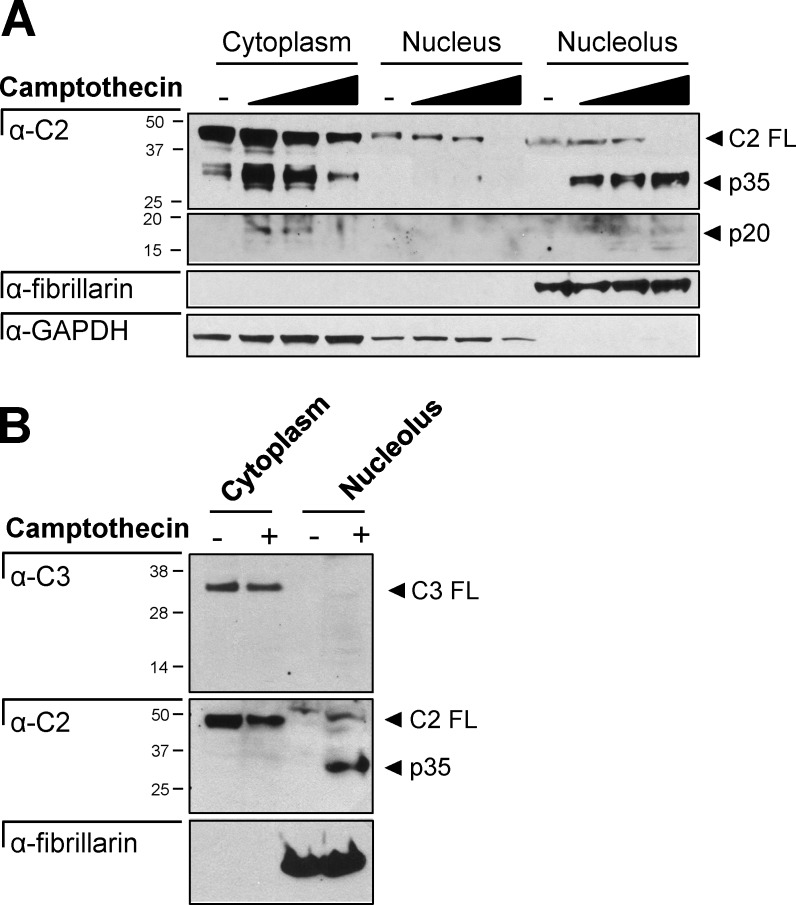
To verify that the localization of caspase-2 BiFC represented the activity of the endogenous caspase-2 protein, we fractionated cells into cytoplasmic, nuclear, and nucleolar fractions. Full-length caspase-2 was detected in all three fractions, and caspase-2 cleavage was detected in the cytosol after camptothecin treatment (Fig. 2 A). In the nucleolus, increased doses of camptothecin resulted in disappearance of full-length caspase-2 and the concomitant appearance of the p35 and p20 cleavage fragments. Caspase-3 was detected only in the cytoplasmic fraction (Fig. 2 B). Therefore, the caspase-2 cleavage detected is unlikely to be a result of caspase-3–mediated cleavage. These results are consistent with our findings that caspase-2 activation platforms assemble in the nucleolus in response to DNA damage, unveiling a novel site for caspase-2 activation.
Figure 2.
Caspase-2 is cleaved in the nucleolus. (A) HeLa cells treated with or without camptothecin (0, 50, 100, or 200 µM) for 16 h were fractionated into the cytosol, nucleoplasm, and nucleolus and immunoblotted for caspase-2, GAPDH (cytosol), or fibrillarin (nucleolus). (B) HeLa cells treated with or without camptothecin (200 µM) for 16 h were fractionated into the cytosol and nucleolus and immunoblotted for caspase-3, caspase-2, or fibrillarin (nucleolus).
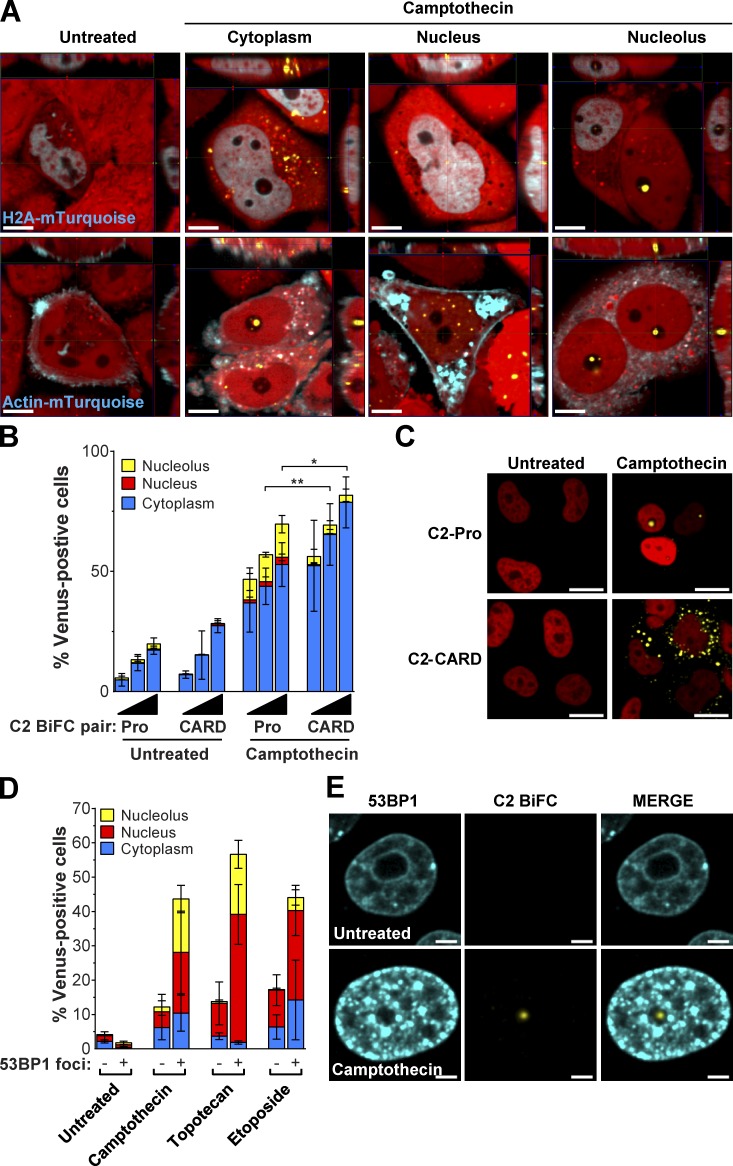
We used colocalization and 3D imaging to more clearly visualize where caspase-2 is activated within the cell. We expressed mTurquoise-tagged versions of Histone H2A and actin to identify the nuclear and cytoplasmic regions, respectively. Fig. 3 A shows representative images of camptothecin-treated cells with caspase-2 BiFC in the cytoplasm, nucleus, and nucleolus in relation to these markers. Caspase-2 contains a classic nuclear localization sequence (NLS) in its prodomain (aa 131–138; Baliga et al., 2003). To determine whether the NLS is required for nucleolar caspase-2 BiFC, we compared cells transfected with the C2-Pro BiFC pair that contains the NLS and a C2-CARD BiFC pair that lacks the NLS, constructs that behave identically in cells exposed to heat shock (Bouchier-Hayes et al., 2009). After treatment with camptothecin, there was a significant decrease in the amount of C2 CARD-BiFC in the nucleolus compared with the C2 Pro-BiFC reporter containing the NLS (Fig. 3, B and C). This indicates that recruitment of caspase-2 to the nucleolus is not just a CARD-specific event but is highly dependent on the NLS.
Figure 3.
Caspase-2 activation in the nucleolus requires the NLS and is a response to DNA breaks. (A) 3D reconstructions of HeLa.C2 Pro-BiFC cells transfected with Histone H2A-mTurquoise or Actin-mTurquoise and treated with camptothecin (100 µM) plus qVD-OPH (20 µM) were composed from 0.2-µm serial confocal images through the z-plane of the cell. Representative orthogonal-slice views show cells (red) with caspase-2 BiFC (yellow) and the indicated coexpressed proteins (blue). The middle, right, and top panels are the xy, yz, and xz planes, respectively. The yz and xz planes intersect according to the crosshairs. Bars, 10 µm. (B) HeLa cells transfected with the C2-Pro (1–147) BiFC or the C2-CARD (1–109) BiFC plasmid pair (25, 50, or 100 ng of each) with Histone H2B RFP (50 ng) were treated with or without camptothecin (100 µM). Cells were assessed 24 h later for the percentage of RFP-positive transfected cells that were Venus+ in the nucleolus, nucleus, or cytoplasm determined from at least 30 microscopy images per well. Results are the mean of three independent experiments ± SD. *, P < 0.05; **, P < 0.005 determined from the percentage of Venus-positive cells in the nucleolus. (C) Representative confocal images show caspase-2 BiFC (yellow) and nuclear Histone H2B expression (red) in cells treated as in B. Bars, 20 µm. (D) HeLa.C2 Pro-BiFC cells transfected with 53BP1-TFP (2 µg) were treated with the indicated drugs (100 µM) plus qVD-OPH (20 µM). TFP-positive cells were counted for absence or presence of 53BP1 foci and the percentage of cells that were Venus+ in the nucleolus, nucleus, or cytoplasm. Results are the mean of three independent experiments ± SD. The association of cells with caspase-2 BiFC in the nucleolus and 53BP1 foci was significant across experiments (camptothecin, P = 7.58 × 10−6; topotecan, 3.06 × 10−7; etoposide, 1.61 × 10−3) as calculated by the Cochran–Mantel–Haenszel test. (E) Representative confocal images of cells treated as in D show 53BP1 (blue) and caspase-2 BiFC (yellow). Bars, 5 µm.
Next, we determined where caspase-2 BiFC localized in cells with DNA double-strand breaks using 53BP1–teal fluorescent protein (TFP), a fusion protein of the p53 binding protein 1 (53BP1) that localizes to double-strand breaks and lacks the functional domains of 53BP1 (Dimitrova et al., 2008). We expressed 53BP1-TFP in Hela.C2 pro-BiFC cells and analyzed the subcellular localization of caspase-2 BiFC in cells with and without 53BP1 fluorescent DNA damage foci. There was a strong correlation between the appearance of caspase-2 BiFC in the nucleolus and the appearance of 53BP1 foci, even in cells treated with etoposide (P = 1.61 × 10−3), which induces minimal nucleolar caspase-2 BiFC (Fig. 3, D and E). This suggests that caspase-2 activation in the nucleolus is a direct response to the presence of DNA damage.
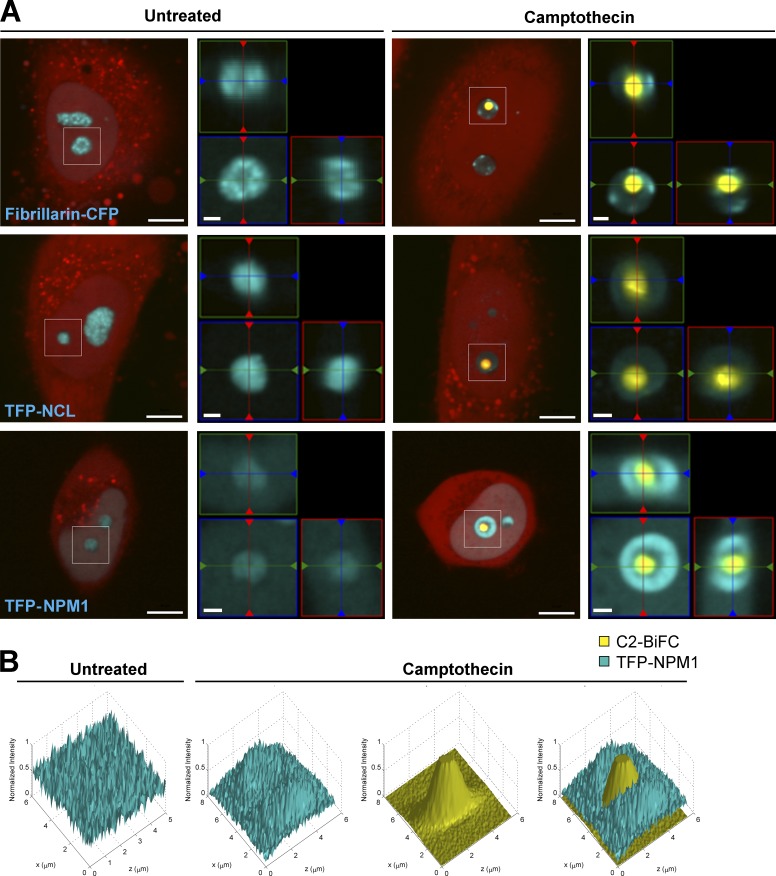
The nucleolus consists of three compartments: the fibrillar center (FC), the dense fibrillar center (DFC), and the granular component (GC). To more precisely determine the compartment where caspase-2 is predominantly activated, we expressed CFP-tagged versions of fibrillarin, which localizes to the DFC; nucleolin (NCL), which localizes to the GC and DFC; and NPM1, which localizes to the GC. Using 3D imaging, we found that the nucleolar caspase-2 BiFC signal, represented by one or more distinct puncta, was encapsulated by these various markers, with signals for NPM1 and NCL closely adjacent to the circumference of the BiFC signal (Fig. 4 A, Fig. S2 A, and Video 1). This suggests that caspase-2 activation platforms assemble predominantly in the FC and the internal boundary of the surrounding GC. Interestingly, the assembly of the caspase-2 activation platform appeared to induce changes in the organization of NPM1 and NCL, from being diffuse throughout the nucleolus in untreated cells to appearing as a ring around the caspase-2 BiFC complex in camptothecin-treated cells. Analysis of the localization of the NPM1 signal or the NCL signal relative to the C2 BiFC signal confirms this, showing that NPM1 and NCL redistributed with treatment to form a pocket where the caspase-2 activating complex resided, in contrast to fibrillarin, which did not change its distribution (Fig. 4 B and Fig. S2, B and C).
Figure 4.
Localization of caspase-2 activation platforms within the nucleolus. (A) HeLa.C2 Pro-BiFC cells transfected with Fibrillarin-CFP (2 µg), TFP-Nucleolin (NCL; 2 µg), or TFP-NPM1 (2 µg) were treated with or without camptothecin (10 µM) plus qVD-OPH (20 µM). Representative images show cells (red) with caspase-2 BiFC (yellow) and the indicated nucleolar proteins (blue). Bars, 10 µm. 3D reconstructions of the white boxed region composed from 0.2-µm serial confocal images through the z-plane of the cell are shown as orthogonal-slice views (right). The middle, right, and top panels are the xy, yz, and xz planes, respectively. The yz and xz planes intersect according to the crosshairs. Bars, 2 µm. (B) 3D graphs of pixel intensities of TFP-NPM1 and C2-BiFC signals in a representative untreated and camptothecin-treated cell.
Nucleolar caspase-2 activation is driven by the PIDDosome
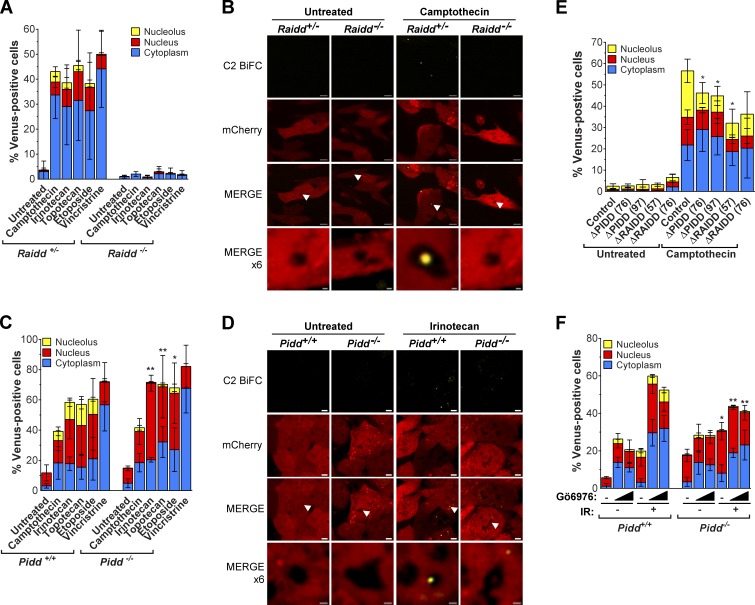
Next, we determined the components of the activation platform that assembled in the nucleolus by investigating the roles of the known PIDDosome proteins, PIDD and RAIDD. We stably expressed the bicistronic C2 Pro-BiFC vector in litter-matched Raidd+/− and Raidd−/−, as well as litter-matched Pidd+/+ and Pidd−/−, E1A/Ras transformed mouse embryonic fibroblasts (MEFs). For each pair of cell lines, we sorted on mCherry fluorescence to select for matched lines expressing equal levels of the linked caspase-2 BiFC reporter (Fig. S3, A and B). Raidd+/− and Pidd+/+ MEFs both engaged caspase-2 BiFC in the cytoplasm, nucleus, and nucleolus in response to the DNA-damaging agents, whereas nucleolar caspase-2 BiFC was not detected in response to vincristine. Deletion of Raidd completely blocked the ability of each of the genotoxic drugs to induce caspase-2 BiFC (Fig. 5, A and B; and Fig. S3 C). This demonstrates that RAIDD is required for caspase-2 BiFC and hence caspase-2 induced proximity in all three cellular compartments. Surprisingly, we found that PIDD was dispensable for cytosolic or nuclear caspase-2 activation in response to all drugs tested (Fig. 5 C). In striking contrast, PIDD was essential for the DNA damage–induced nucleolar BiFC component (Fig. 5, C and D; and Fig. S3 D). These MEF data were confirmed in HeLa.C2 Pro-BiFC cells bearing RAIDD- or PIDD-null alleles generated by CRISPR/Cas9 as well as siRNA-mediated silenced PIDD (Fig. 5 E and Fig. S3, E and F). An essential role for PIDD in nucleolar caspase-2 activation was also observed in response to IR plus Chk1i in MEFs, and in this context, the gene’s requirement extended from nucleolar BiFC to the cytoplasmic compartment (Fig. 3 F). These results indicate that the nucleolus is the predominant site for PIDDosome-mediated caspase-2 activation in response to DNA damage.
Figure 5.
RAIDD is required for caspase-2 BiFC, but PIDD is required for caspase-2 BiFC only in the nucleolus. (A) Litter-matched Raidd+/− or Raidd−/− MEFs stably expressing C2 Pro-BiFC were treated with camptothecin (250 nM), irinotecan (250 nM), topotecan (250 nM), etoposide (250 nM), or vincristine (25 nM) plus qVD-OPH (20 µM). The percentage of cells that were Venus+ in the nucleolus, nucleus, or cytoplasm was determined at 24 h from at least 30 microscopy images per well. Results are the mean of three independent experiments ± SD. The differences in total Venus-positive cells between the Raidd+/− and Raidd−/− groups is significant to P = 0.0034. (B) Representative confocal images show caspase-2 BiFC (yellow) and mCherry expression (red) in Raidd+/− and Raidd−/− C2 Pro-BiFC MEFs. Bars, 5 µm. Arrowheads indicate locations of nucleoli that are magnified in the bottom panel (bars, 1 µm). See Fig. S3 C for full dataset images. (C) Litter-matched Pidd+/+ or Pidd−/− MEFs stably expressing C2 Pro-BiFC were treated with camptothecin (250 nM), irinotecan (250 nM), topotecan (250 nM), etoposide (250 nM), or vincristine (25 nM) plus qVD-OPH (20 µM). The percentage of cells that were Venus+ in the nucleolus, nucleus, or cytoplasm was determined at 24 h from at least 30 microscopy images per well. Results are the mean of three independent experiments ± SD. *, P < 0.05; **, P < 0.005 determined from the percentage of nucleolar Venus-positive cells between Pidd+/+ and Pidd−/− cells for each treatment. (D) Representative confocal images show caspase-2 BiFC (yellow) and mCherry expression (red) in Pidd+/+ and Pidd−/− C2 Pro-BiFC MEFs. Bars, 5 µm. Arrowheads indicate locations of nucleoli that are magnified in the lower panel (bars, 1 µm). See Fig. S3D for full dataset images. (E) PIDD or RAIDD was deleted from HeLa.C2 Pro-BiFC cells with CRISPR/Cas9 using two independent sgRNAs per gene. The percentage of cells treated with or without camptothecin (100 µM) plus qVD-OPH (20 µM) that were Venus+ in the nucleolus, nucleus, or cytoplasm was determined after 24 h from at least 50 microscopy images per well. Results are the mean of three independent experiments ± SD. *, P < 0.05 determined from the percentage of nucleolar Venus-positive cells compared with the control treated group. The difference in total Venus-positive cells compared with camptothecin-treated control for ΔRAIDD cells is significant to P < 0.0001 (ΔRAIDD(57)) and P = 0.0187 (ΔRAIDD(76)). (F) Pidd+/+ and Pidd−/− C2-Pro BiFC MEFs were treated with or without Gö6976 (0.125, 0.25, or 0.5 µM) ± IR (10 Gy) plus qVD-OPH (20 µM). The percentage of cells that were Venus+ in the nucleolus, nucleus, or cytoplasm was determined 24 h after treatment from at least 50 microscopy images per well. Results represent triplicate counts ± SD. *, P < 0.05; **, P < 0.005 determined from the percentage of Venus-positive cells in the nucleolus between Pidd+/+ and Pidd−/− cells for each treatment.
PIDD interacts with the nucleolar resident protein NPM1
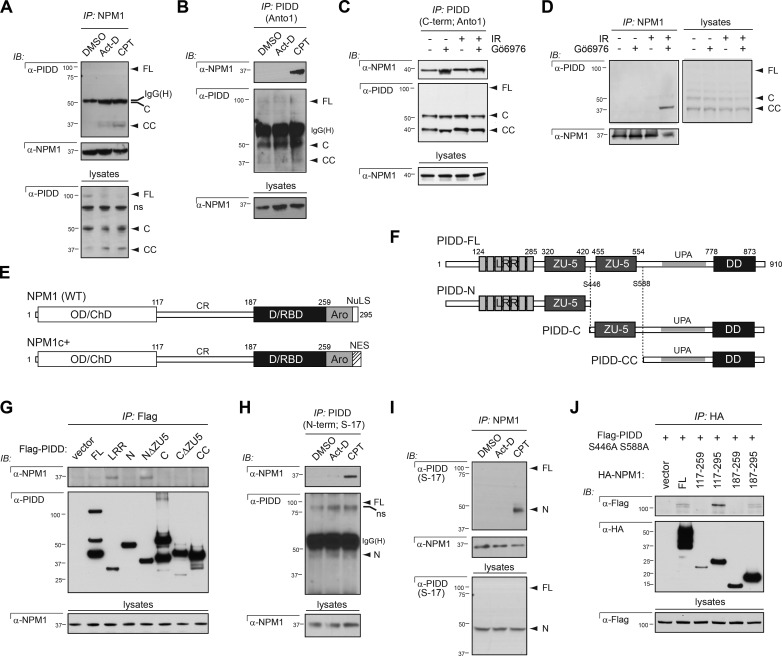
Having identified the nucleolus as a novel site for PIDDosome signaling, we set out to explore the mechanisms underlying platform recruitment to this organelle. Ongoing proteomic analyses of Flag-PIDD immunoprecipitates detected six unique peptides derived from NPM1 (Table S1). The spatial vicinity between NPM1 and caspase-2 BiFC within the nucleolus, including the close association of signals (Fig. 4, A and B), prompted us to ask whether endogenous PIDD and NPM1 physically interact in cells exposed to DNA damage. An interaction between the two proteins was readily detected in reciprocal NPM1 and PIDD pulldowns from cells treated with camptothecin, but not (or less so) in untreated cells or cells treated with actinomycin D (which failed to trigger caspase-2 BiFC; Fig. 6, A and B; and Fig. 1 A). An endogenous interaction between NPM1 and PIDD was also observed in response to IR plus Chk1i, as detected in PIDD or NPM1 immunoprecipitates obtained with multiple antibodies (Fig. 6, C and D; and Fig. S4 A). Thus a PIDDosome component, PIDD, and a nucleolar protein, NPM1, physically interact in response to DNA-damaging stimuli that trigger nucleolar and PIDDosome-dependent caspase-2 BiFC.
Figure 6.
NPM1 interacts with PIDD-CC and PIDD-N after DNA damage. (A) HeLa cells treated with DMSO, actinomycin D (Act-D; 500 nM) or camptothecin (CPT; 200 µM) were harvested 24 h after treatment, lysed, and immunoprecipitated (IP) with NPM1 antibody. Immunoprecipitates were analyzed by Western blot (IB). (B) HeLa cells treated with DMSO, Act-D (500 nM), or CPT (200 µM) were harvested 24 h after treatment, lysed, and immunoprecipitated with monoclonal Anto-1 antibody to PIDD C terminus. Immunoprecipitates were analyzed by Western blot. (C) HeLa cells treated with or without Gö6976 (1 µM) ± IR (10 Gy) were harvested 24 h after IR, lysed, and immunoprecipitated with monoclonal anti-PIDD antibody. Immunoprecipitates were analyzed by Western blot. (D) HeLa cells treated with or without Gö6976 (1 µM) ± IR (10 Gy) were harvested 24 h after IR, lysed, and immunoprecipitated with anti-NPM1 antibody. Immunoprecipitates were analyzed by Western blot. (E) Schematic representation of NPM1 (WT) and NPM1c+ (mutated in AML) protein domain structure. Indicated are amino acids defining the extremities of deletion constructs used in J. OD/ChD, oligomerization/chaperone domain; D/RBD, DNA/RNA binding domain; Aro, aromatic region; NuLS, nucleolar localization signal; NES, nuclear exclusion signal. (F) Schematic representation of PIDD-FL, PIDD autocleavage products. Indicated are amino acids defining the extremities of deletion constructs used in G and autocleavage sites in PIDD. LRR, LRR domain; ZU-5, ZO-1 and UNC5-like; UPA, uncharacterized protein domain in UNC5, PIDD, and Ankyrin family of proteins (designates the putative PIDD oligomerization domain; Janssens and Tinel, 2012). (G) HeLa cells transfected with the indicated Flag-PIDD constructs were harvested 24 h after transfection. Flag immunoprecipitates were analyzed by Western blot. (H) HeLa cells treated with DMSO, Act-D (500 nM), or CPT (200 µM) were harvested 24 h after treatment, lysed, and immunoprecipitated with S-17 antibody to PIDD N terminus. Immunoprecipitates were analyzed by Western blot. ns, nonspecific band. (I) HeLa cells treated with DMSO, Act-D (500 nM), or CPT (200 µM) were harvested 24 h after treatment, lysed, and immunoprecipitated with NPM1 antibody. Immunoprecipitates were analyzed by Western blot. (J) HeLa cells cotransfected with the indicated HA-NPM1 constructs and the noncleavable mutant form of PIDD were harvested 24 h after transfection. HA immunoprecipitates were analyzed by Western blot.
Full-length PIDD is constitutively autoprocessed into three fragments termed PIDD-N, PIDD-C, and PIDD-CC (Fig. 6 F). Of these, only PIDD-CC can bind RAIDD and mobilize the caspase-2–activating PIDDosome (Tinel and Tschopp, 2004; Tinel et al., 2007). PIDD-C anchors the pro-survival PIDD-RIP1-NEMO PIDDosome (Janssens et al., 2005), whereas PIDD-N serves an as-yet-uncharacterized regulatory role (Tinel et al., 2007). Transfections with Flag-PIDD deletion constructs identified the leucine-rich repeats (LRR) domain on PIDD-N as sufficient for NPM1 binding (Fig. 6 G). NPM1 docking onto PIDD-N was antagonized by the proximal ZO-1 and UNC5-like (ZU-5) domain, whose deletion restored full NPM1 binding to PIDD-N (Fig. 6 G, lane 5). Pulldowns with a specific N-terminal PIDD antibody (S-17; Fig. S4 C) confirmed the NPM1-PIDD-N interaction at the endogenous level (Fig. 6, H and I). Importantly, NPM1 also interacts with PIDD-CC at the endogenous level. This is evident from the analysis of NPM1 pulldowns, which contain both PIDD-N and PIDD-CC (Fig. 6, A, D, and I). An NPM1–PIDD-CC interaction is further supported by the presence of NPM1 in PIDD immunoprecipitates containing PIDD-C and PIDD-CC, but not PIDD-N or PIDD-FL (see PIDD IPs with C-terminal antibodies Anto1 and AL233 in Fig. 6, B and C, and Fig. S4 A). Overall, these experiments identify two NPM1 binding sites on PIDD, enabling interactions with both PIDD-N (via the LRR domain) and PIDD-CC.
Similar mapping experiments exploiting HA-NPM1 deletion constructs (Korgaonkar et al., 2005) identified the central region (CR; spanning aa 117–187) as the PIDD binding site on NPM1 (Fig. 6, E and J). The CR contains conserved acidic tracts and has been implicated in histone binding yet remains largely uncharacterized (Grisendi et al., 2006; Mitrea and Kriwacki, 2013). PIDD binding to the CR assigns a new functional role for this domain. These experiments identify NPM1 as a PIDD interaction partner in the nucleolus.
NPM1 is required for PIDDosome signaling in the nucleolus
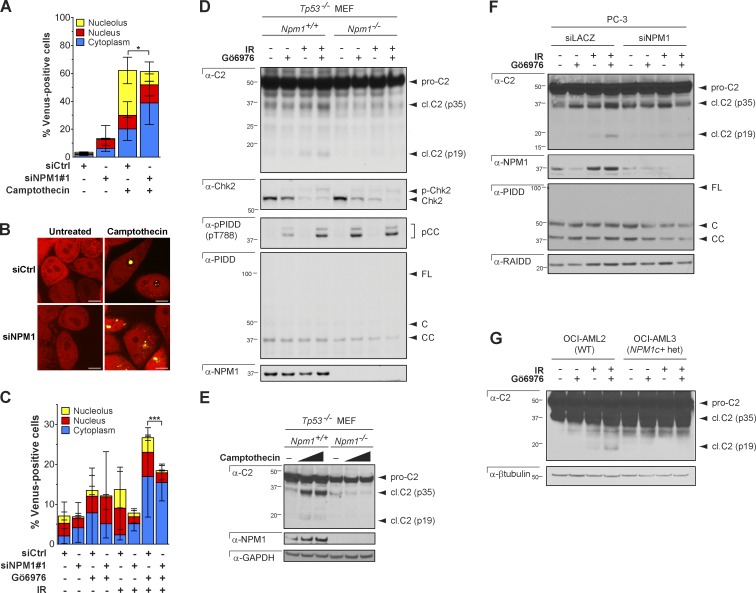
Next, we asked whether NPM1 is required for PIDDosome signaling. SiRNA-mediated silencing of NPM1 in HeLa.C2 Pro-BiFC cells substantially reduced the levels of caspase-2 BiFC in the nucleolus in response to camptothecin compared with control cells but had no effect on total caspase-2 BiFC or the size or morphology of the nucleoli (Fig. 7, A and B; and Fig. S5 A). This result was very similar to the effect of PIDD deficiency on nucleolar caspase-2 BiFC (Fig. 5, C and E). In response to IR plus Chk1i, we also observed a reduction in nucleolar caspase-2 BiFC (Fig. 7 C) in addition to a reduction in overall caspase-2 BiFC. These data identify a critical role for NPM1 in DNA damage–induced nucleolar PIDDosome signaling. In the context of IR plus Chk1i, the requirement for NPM1 appears to extend to all cellular PIDDosomes, a seemingly surprising observation for a predominantly nuclear protein. However, cells treated with IR plus Chk1i showed an unusual distribution of NPM1: in many cells, the nuclei were highly fragmented, with NPM1 accumulating in micronuclei in addition to the nucleus and nucleolus, compared with camptothecin-treated cells, in which NPM1 accumulated only in the nucleus and nucleolus (Fig. S5 B). Thus the cytoplasmic caspase-2 BiFC that is dependent on NPM1 is likely caused by the fragmentation of the nucleus and nucleolus dispersing the localization of NPM1.
Figure 7.
NPM1 is required for PIDDosome signaling. (A) HeLa.C2 Pro-BiFC cells were transfected with the indicated siRNAs in the presence of qVD-OPH (20 µM). 48 h later, cells were treated with or without camptothecin (100 µM). The percentage of cells that were Venus+ in the nucleolus, nucleus, or cytoplasm was determined at 24 h from at least 50 microscopy images per well. Results are the mean of three independent experiments ± SD. *, P < 0.05. (B) Representative confocal images show caspase-2 BiFC (yellow) and mCherry expression (red) from cells treated as in A. Bars, 10 µm. (C) HeLa.C2 Pro-BiFC cells transfected with the indicated siRNAs in the presence of qVD-OPH (20 µM) for 48 h were treated with or without IR (50 Gy) ± Gö6976 (1 µM). The percentage of cells that were Venus+ in the nucleolus, nucleus, or cytoplasm was determined at 24 h from at least 50 microscopy images per well. Results are the mean of four independent experiments ± SD. ***, P < 0.0005. (D) Tp53−/− MEFs of indicated Npm1 genotypes were treated with or without Gö6976 (1 µM) ± IR (10 Gy) and harvested 24 h later. Lysates were analyzed by Western blot. (E) Tp53−/− MEFs of indicated Npm1 genotypes were treated with or without camptothecin (100 or 150 µM) and harvested 24 h later. Lysates were analyzed by Western blot. (F) PC3 cells transfected with the indicated siRNAs were treated with or without Gö6976 (1 µM) ± IR (10 Gy) and harvested 24 h later. Lysates were analyzed by Western blot. (G) OCI-AML2 and OCI-AML3 cells, of indicated NPM1 genotypes, were treated with or without Gö6976 (1 µM) ± IR (10 Gy) and harvested 24 h later. Lysates were analyzed by Western blot.
To confirm the requirement of NPM1 for caspase-2 activation in response to IR plus Chk1i or camptothecin, we assessed caspase-2 processing in multiple mammalian cell types including NPM1-depleted PC3, HCT-116, and HeLa cell lines, as well as Npm1−/− MEFs (Fig. 7, D–F; and Fig. S5, C and D). In all cases, reduction or loss of NPM1 reduced or completely blocked caspase-2 cleavage. Importantly, knockdown or loss of NPM1 did not affect the expression levels of any PIDDosome component, nor did it affect ATM-mediated phosphorylation of PIDD on Thr788, the initiating step in PIDDosome assembly after IR plus Chk1i (Fig. 7, D and F; Ando et al., 2012).
We then tested the effects of the NPM1c+ mutation, found in 30–50% of patients with adult AML (Falini et al., 2005). This four-base insertion at the 3′ end of the NPM1 coding sequence converts a nucleolar localization signal into a nuclear export signal but does not otherwise affect the protein’s sequence or its ability to oligomerize (Fig. 6 E; Falini et al., 2005; Grisendi et al., 2005). Thus in NPM1c+ heterozygous AML cells, all NPM1 protein (wild-type [WT] and mutant) relocates from the nucleolus into the cytoplasm, providing a setting in which to investigate the unique consequences of loss of nucleolar NPM1. NPM1c+ cells failed to engage caspase-2 cleavage after IR plus Chk1i, similar to NPM1-depleted or -knockout cells (Fig. 7 G). Thus, not only does NPM1 specifically orchestrate nucleolar PIDDosome signaling (Fig. 7, A–C), but such nucleolar signaling is also critical to overall PIDDosome activity after DNA damage.
Nucleolar PIDDosome signaling contributes to PIDDosome-mediated apoptosis
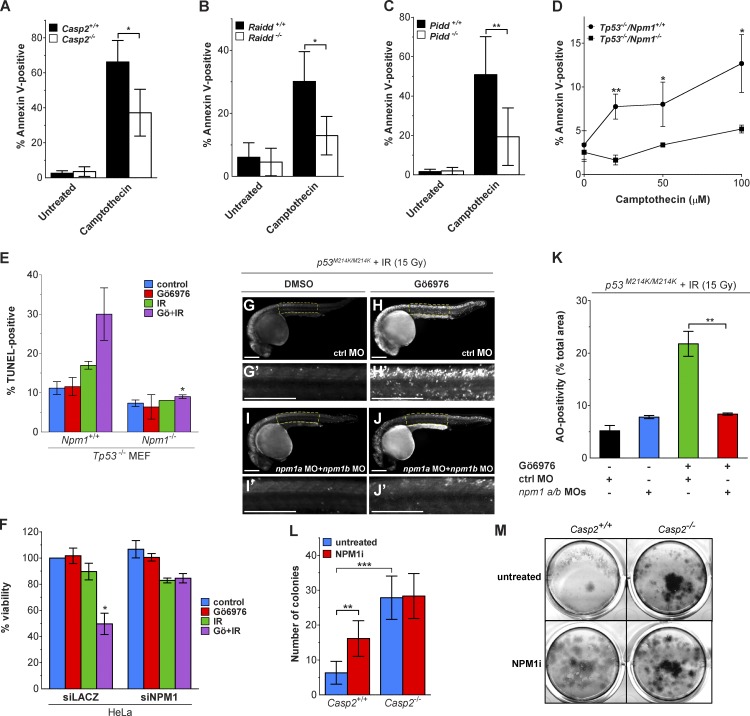
To investigate the significance of nucleolar PIDDosome signaling, we analyzed the effects of caspase-2, PIDD, RAIDD, or NPM1 loss on the apoptotic response to DNA damage. We noted a significant reduction in cell death in cells exposed to camptothecin in caspase-2–, RAIDD-, and PIDD-deficient MEFs (Fig. 8, A–C). Npm1−/− MEFs were also highly resistant to apoptosis induced by camptothecin, irinotecan, and IR plus Chk1i (Fig. 8, D and E; and Fig. S5 E). Npm1-depleted HeLa cells also showed significant resistance to IR plus Chk1i (Fig. 8 F). The latter form of cell death depends on the PIDDosome in multiple cell types and species (Ando et al., 2012; Manzl et al., 2013; Thompson et al., 2015), and this result phenocopies Casp2−/− MEFs (Ho et al., 2009; Ando et al., 2012). Therefore, the requirement of NPM1 for caspase-2 cleavage in response to IR plus Chk1i (Fig. 7, D–F; and Fig. S5, C and D), appears to be an essential component of the resulting apoptosis. To further test this hypothesis, we used zebrafish embryos that are devoid of WT p53 (similar to the Npm1-deficient MEFs) and thus lack alternative apoptotic responses to IR (Sidi et al., 2008; Ando et al., 2012; Thompson et al., 2015). Zebrafish embryos depleted of all endogenous NPM1 via combined injections of specific npm1a and npm1b morpholinos (Bolli et al., 2010) failed to engage apoptosis in response to IR plus Chk1i (compare Fig. 8 H/H′ and Fig. 8 J/J′; acridine orange signal quantifications are shown in Fig. 8 K), again phenocopying loss of caspase-2 (Sidi et al., 2008).
Figure 8.
Inhibition of NPM1 blocks apoptosis and caspase-2–dependent suppression of cell growth. (A–C) Litter-matched Casp2+/+ and Casp2−/− MEFs (A), Raidd+/+ and Raidd−/− MEFs (B), or Pidd+/+ and Pidd−/− MEFs (C) were treated with or without camptothecin (250 nM). Apoptosis was assessed by flow cytometry for Annexin V binding 16 h later. Results are the mean of three to four independent experiments ± SD. *, P < 0.05. (D) Tp53−/− MEFs of indicated Npm1 genotypes were treated with or without camptothecin. Apoptosis was assessed by flow cytometry for Annexin V binding 16 h after treatment. Results are the mean of three independent experiments ± SD. *, P < 0.05; **, P < 0.005. (E) Tp53−/− MEFs of indicated Npm1 genotypes were treated with or without Gö6976 (1 µM) ± IR (10 Gy), harvested 24 h after IR, stained for TUNEL, and analyzed by flow cytometry. Results are the mean of three independent experiments ± SD. *, P < 0.05. (F) HeLa cells transfected with the indicated siRNAs were treated with or without Gö6976 (1 µM) ± IR (10 Gy) and stained with alamarBlue 72 h after IR. Results are the mean of three independent experiments ± SD.*, P < 0.05 (two-tailed Student’s t test). (G–J′) p53M214K/M214K zebrafish embryos were injected at the one-cell stage with standard control (ctrl) or npm1a+npm1b morpholinos, incubated 17 h later with or without Gö6976 at indicated concentrations (μM), treated with or without 15 Gy IR, and stained with the cell death marker acridine orange (AO) after 7 h. All embryos were imaged live at 24 h postfertilization. Panels G′, H′, I′, and J′ are blowups of indicated spinal cord areas in corresponding whole-embryo panels. Bars, 250 µm. (K) Quantification of AO stains shown in G–J′. Results are the mean of three independent experiments (≥10 embryos per condition) ± SEM. **, P < 0.005. (L) Litter-matched Casp2+/+ and Casp2−/− MEFs, plated at low density, were treated with or without a chemical inhibitor of NPM1 (3 µM) for 7 h. Colonies were stained with methylene blue 5 d after treatment. The number of colonies under each treatment condition is shown. Results are the mean of six individual wells across two independent experiments ± SD. **, P < 0.005; ***, P < 0.0005. (M) Representative images of methylene blue–stained colonies from cells treated as in L.
Numerous studies have shown that deficiency of caspase-2 increases cell proliferation (Berube et al., 2005; Ho et al., 2009; Parsons et al., 2013; Puccini et al., 2013b), suggesting a role in regulating cell cycle in addition to inducing apoptosis. To explore the role of NPM1 in this phenomenon, we examined clonogenic outgrowth in Casp2+/+ and Casp2−/− MEFs treated with or without NSC348884, a chemical inhibitor of NPM1 that disrupts a defined hydrophobic pocket of NPM1 required for oligomerization (Qi et al., 2008). As expected, we observed increased cell growth of Casp2−/− cells compared with the WT cells. Inhibition of NPM1 resulted in increased growth of the Casp2+/+ cells but had no effect when caspase-2 was absent (Fig. 8, L and M). Thus we conclude that NPM1-driven nucleolar PIDDosome signaling is critical to PIDDosome-mediated cell cycle regulation and apoptosis in response to DNA damage.
Discussion
The PIDDosome was characterized more than a decade ago as an activation platform that assembles in response to genotoxic stress to recruit and activate caspase-2 (Tinel and Tschopp, 2004). Since then, there have been no robust descriptions of alternative activation platforms for caspase-2. Nonetheless, there has been some controversy over the requirement of PIDD for caspase-2 activation (Bock et al., 2012; Bouchier-Hayes and Green, 2012). The data presented here indicate that the PIDD requirement for caspase-2 activation depends on not only the type of caspase-2–activating stimulus, but also where it is activated in the cell. Moreover, our results support a model in which NPM1 forms a scaffold for PIDDosome assembly in the nucleolus in response to DNA damage (Fig. 9).
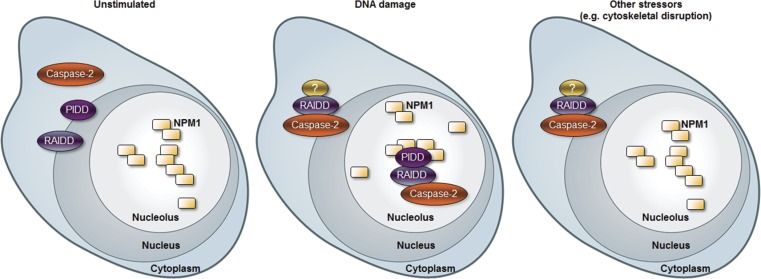
Figure 9.
Model for DNA damage induces caspase-2 activation. Schematic representation of model for two distinct caspase-2 activation platforms that assemble in the nucleolus or cytoplasm after DNA damage. See text for details.
It has been reported that caspase-2 activation can proceed normally in cells from PIDD-deficient mice (Manzl et al., 2009). PIDD-knockout mice also fail to phenocopy the tumor acceleration effects of caspase-2 deficiency in the Eµ-Myc model (Manzl et al., 2012). These observations have challenged the importance of PIDD for caspase-2 activation and function. In MEFs or human cells with reduced Chk1 or BubR1 dosage, however, caspase-2 activation in response to IR is completely dependent on PIDD (Ando et al., 2012; Thompson et al., 2015). Thus these different observations may be caused by varying levels of the PIDDosome’s endogenous inhibitors. However, the possibility that distinct activation platforms for caspase-2 exist had not been thoroughly explored. Using a highly sensitive and specific method to visualize caspase-2 activation platform assembly, we show that PIDD is required only for the proportion of cells that induce caspase-2 activation platform assembly in the nucleolus in response to many DNA-damaging agents. Therefore, we propose a model in which at least two distinct activation platforms for caspase-2 exist: one in the nucleolus that requires PIDD, and one in the cytoplasm or nucleus that is PIDD independent (Fig. 9).
Although this is the first evidence that caspase-2 is activated in the nucleolus, the presence of caspase-2 in the nucleus has been reported by several groups. Because of the NLS in the prodomain of caspase-2, GFP-tagged caspase-2 prodomain localizes to the nucleus upon overexpression (Baliga et al., 2003), and immunostaining for endogenous caspase-2 shows accumulation in nucleus, cytosol, and Golgi apparatus (Mancini et al., 2000). However, our previous results using the caspase-2 BiFC reporter showed no caspase-2 activation in the nucleus in response to heat shock (Bouchier-Hayes et al., 2009). In addition, a specific role for caspase-2 activation in the nucleus has not been described. In contrast to our previous results, our results here show some caspase-2–induced proximity in the nucleus, distinguishable from nucleolar and cytoplasmic BiFC signals. This is likely because of the enhanced sensitivity of the stably expressed caspase-2 BiFC reporter, which ensures equal expression of both BiFC components, hence reducing false-negative results caused by the preferential recruitment of one of the components to the platform. However, the levels of nuclear caspase-2 BiFC varied greatly between experiments. It is therefore unclear whether the nuclear caspase-2 BiFC represents an independent caspase-2 activation platform or is contiguous with the cytosolic platform. Given that both are RAIDD dependent and PIDD independent, we suggest that this represents the dynamic nature of the cytosolic platform. The nucleolar complex, on the other hand, appears distinct, as it is dependent on PIDD and NPM1 for its assembly.
The existence of a unique activation platform that assembles in the nucleolus to recruit caspase-2 suggests that this nucleolar complex may also have distinct functions. Traditionally known as the cell’s ribosome factory, the nucleolus has recently emerged as a complex organelle with multiple nonribosomal functions. This long list of functions includes apoptosis, cell proliferation, regulation of protein stability, tumor surveillance, telomere metabolism, and DNA damage repair (Antoniali et al., 2014). Nucleolar structure is often compromised in cancer cells, resulting in increased nucleolar size and number (Pianese, 1896). A clear difference in the caspase-2 requirement for apoptosis between transformed cells and primary cells has been reported: transformed caspase-2–deficient embryonic fibroblasts were robustly resistant to IR-induced apoptosis, whereas the primary cells were not (Ho et al., 2009). Thus, it is possible that the increased size and number of nucleoli provides more ample sites for caspase-2 activation in cancer cells. The primary role of NPM1 is thought to be in ribosome biogenesis, functioning as a nuclear chaperone that shuttles between the cytoplasm and nucleus (Lindström, 2011). However, it is also a strong tumor suppressor and has important roles in regulating cell cycle and cell survival. Knockout of Npm1 in mice is embryonic lethal, and Npm1 haploinsufficiency leads to uncontrolled centrosome duplication, resulting in aneuploidy, genomic instability, and acceleration of Eµ-Myc–driven lymphomagenesis (Grisendi et al., 2005).
Our results suggest that NPM1-driven activation of caspase-2 is a key initiator of the apoptotic pathway. However, the specific role of NPM1 in apoptosis is unclear. Inhibition of NPM1 has been shown to induce apoptosis or sensitize cells to apoptosis (Colombo et al., 2005; Grisendi et al., 2005; Cullen et al., 2009; Balusu et al., 2011). This is thought to be caused by an inhibitory effect on p53 activity by NPM1, since knockout of p53 rescues apoptosis as a result of NPM1 deficiency (Colombo et al., 2005). We have shown that when p53 is absent, loss of NPM1 profoundly protects against apoptosis induced by topoisomerase I inhibition or IR plus Chk1i (Fig. 8, D–J). However, using different experimental systems, NPM1 has also been shown to enhance p53 stability and activity (Colombo et al., 2002). Therefore it remains possible that NPM1-induced caspase-2 activation can engage p53 to induce apoptosis.
In addition to apoptosis, caspase-2 has been implicated in the regulation of cell proliferation. Loss of caspase-2 has been associated with higher proliferation rates, and caspase-2 has been observed to induce cell cycle arrest after DNA damage (Ho et al., 2009). Our data indicate that the caspase-2–associated inhibition of cell proliferation is dependent on NPM1, further underscoring a biological role for the caspase-2 nucleolar complex. Loss of caspase-2 has also been associated with telomere shortening (Dorstyn et al., 2012) and increased genomic instability (Dorstyn et al., 2012; Parsons et al., 2013). Thus, there is considerable overlap between the nonribosomal functions of the nucleolus and NPM1 with some of the putative functions of caspase-2. Therefore, the nucleolus may provide a site for PIDDosome assembly to initiate the caspase-2 activation cascade, resulting in regulation of both its proapoptotic and nonapoptotic functions.
The assembly of the PIDDosome in the nucleolus, as mediated by NPM1, is likely to provide activated caspase-2 access to a set of substrates distinct from those available in the cytosol. Very few verified substrates for caspase-2 exist, the most well-characterized being the BH3-only Bcl-2 protein, Bid. Although cleavage of Bid is considered the primary route to apoptosis by caspase-2 (Guo et al., 2002; Bonzon et al., 2006), Bid-deficient cells do not phenocopy caspase-2–deficient cells with respect to DNA damage–induced cell cycle arrest (Kaufmann et al., 2007) and tumor suppression (Manzl et al., 2012). Therefore, additional caspase-2 substrates are likely to exist. Many of the proteomic screens to identify caspase substrates have focused primarily on proteins in the cytosol (Wejda et al., 2012) and may not have been able to identify substrates that reside in the nucleolus or substrates that are cleaved in the nucleolus. Our results suggest that interrogating the nucleolar proteome may identify novel caspase-2 substrates. For example, MDM2 is a caspase-2 substrate that is sequestered in the nucleolus by p19ARF to help promote p53 stabilization in response to IR (Weber et al., 1999). Cleavage by caspase-2 is proposed to produce a fragment of MDM2 that can bind to p53 but does not degrade it (Oliver et al., 2011). NPM1 acts to tether ARF to the nucleolus, which is required to maintain ARF stability (Bertwistle et al., 2004; Colombo et al., 2005). Thus the proximity of the caspase-2 activation platform to its substrate may facilitate this function of MDM2, promoting the ability of nucleolar ARF to induce apoptosis.
In conclusion, we have found that PIDDosome assembly in response to DNA damage is highly regulated by the presence and localization of NPM1. This complex is primarily assembled in the nucleolus, providing a novel localization for PIDDosome assembly. In addition, our data show that caspase-2 is also recruited to a distinct cytosolic activation platform that is PIDD independent while still dependent on RAIDD. These data, which provide a model of context-specific regulation of caspase-2 by PIDD, may alleviate the controversy surrounding the role of PIDD—and by extension the PIDDosome—in caspase-2 activation. Importantly, our observations indicate that a cytosolic sensor exists that initiates assembly of the RAIDD–caspase-2 signaling platform in the cytoplasm. Further exploration of this complex is required to both identify this sensor and fully understand the functional consequences of this PIDD-independent complex.
Materials and methods
Chemicals and antibodies
The following antibodies were used: anti–caspase-2 (clone 11B4; EMD Millipore); anti-fibrillarin (C13C3; Cell Signaling Technology); anti-GAPDH (Santa Cruz Biotechnology, Inc.); anti–caspase-3 and anti–cleaved caspase-3 (9662 and 9664; Cell Signaling Technology); anti-NPM1 (clone FC8229, Abcam; clone FC-61991, Thermo Fisher Scientific; rabbit anti-NPM1 3542, Cell Signaling Technology); anti-RAIDD (clone 4B12; MBL); anti-PIDD mAbs (AL233 and Anto-1; Alexis Biochemicals); anti-PIDD pAbs (clones H-300 and s-17; Santa Cruz Biotechnology, Inc.); anti-pPIDD (pT788, as previously described [Ando et al., 2012]); anti-Chk2 mAb (clone 7; EMD Millipore), anti–Flag M2 (Sigma-Aldrich); anti–β-tubulin (2144; Cell Signaling Technology), and anti-actin (C4; MP Biomedicals). All cell culture media reagents were purchased from Invitrogen. Gö6976 was purchased from Cell Signaling Technology. Unless otherwise indicated, all other reagents were purchased from Sigma-Aldrich.
Plasmids
The pBiFC.VC155 and pBiFC.VN173 plasmids encoding C2 Pro, C2 CARD, or C2 FL (C320S) were described previously (Bouchier-Hayes et al., 2009) and are available from Addgene. The bicistronic vector consists of C2 Pro-VC and C2 Pro-VN linked with a 2A peptide. Silent mutations were introduced into the second C2 Pro nucleotide sequence to prevent the sequence recombining out during cloning because of the presence of two identical C2 Pro sequences. The sequence was generated by Bionexus and cloned into pRRL-MND-MCS-2A-mCherry-2A-Puro. The nucleolar and 53BP1 protein sequences were cloned by standard PCR strategies, with primers designed to incorporate the appropriate restriction enzyme sites, from cDNA for NCL (GFP-Nucleolin, a gift from M. Kastan [Duke University, Durham, NC; plasmid 28176; Addgene]); NPM1 (B23(NPM), a gift from M. Dasso [National Institutes of Health, Bethesda, MD; plasmid 34553; Addgene]); and 53BP1 (mCherry-BP1-2-pLPC-puro [plasmid 19835; Addgene; Dimitrova et al., 2008]) into mTFP-C1 (a gift from R. Campbell [University of Alberta, Edmonton, Alberta, Canada] and M. Davidson [Florida State University, Tallahassee, FL]). Fibrillarin-CFP was a gift from L. Parent (Pennsylvania State University, State College, PA). HA-NPM1 deletion constructs were provided by D.E. Quelle (University of Iowa Carver College of Medicine, Iowa City, IA). Histone H2B was purchased from Addgene (plasmid #20972; Addgene; Nam and Benezra, 2009). PIDD-deletion constructs were provided by E. Logette and J. Tschopp (University of Lausanne, Lausanne, Switzerland). mTurquoise-H2A-10 and mTurquoise-Actin-C-18 were gifts from M. Davidson (plasmids # 55556 and #55520, respectively; Addgene). Each construct was verified by sequencing.
Cell culture and generation of cell lines
Human embryonic kidney (HEK) 293T, U2OS, and HeLa cells were grown in DMEM containing FCS (10% [vol/vol]), l-glutamine (2 mM), and penicillin/streptomycin (50 IU/50 µg/ml). MEFs were grown in the same medium supplemented with sodium pyruvate (1 mM), 1× nonessential amino acids, and β-mercaptoethanol (55 µM). Litter-matched SV40-transformed Npm1+/+/Tp53−/− and Npm1+/+/Tp53−/− MEFs were a gift from P.P. Pandolfi (Beth Israel Deaconess Medical Center, Boston, MA). OCI-AML2 (NPM1 WT) and OCI-AML3 (NPM1c+) cells were provided by A. Kentsis (Sloan-Kettering Institute, New York, NY). Litter-matched Pidd+/+ and Pidd−/− MEFs were a gift from A. Villunger (Innsbruck Medical University, Innsbruck, Austria). Early-passage MEFs were simultaneously transduced with frozen supernatants of the retroviral expression vectors pBabePuro.H-ras (G12V) and pWZLH.E1A (provided by S.W. Lowe [Memorial Sloan Kettering Cancer Center, New York, NY] and G. Hannon [Cold Spring Harbor Laboratory, Cold Spring Harbor, NY]. After 48 h, the cells were harvested by mild trypsinization, seeded at 105 cells/well in six-well plates, and cultured for 10 d in media containing 0.5 µg/ml puromycin and 40 µg/ml hygromycin for selection of the transduced viruses. Litter-matched Raidd+/− and Raidd−/− MEFs were generated and transduced with E1A and Ras as previously described (Bouchier-Hayes et al., 2009). Stable cell lines were generated by lentiviral transduction. 293T cells were transiently transfected with pRRL.C2 Pro-VC-2A-C2 Pro-VN-2A-mCherry along with the packaging vector, pSPAX-2, and the envelope vector, pVSV-G, using Lipofectamine 2000 transfection reagent (Invitrogen) according to manufacturer’s instructions. After 48 h, virus-containing supernatants were cleared by centrifugation and incubated with HeLa, U2OS, or MEF lines. HeLa and U2OS cells were selected in 1 µg/ml puromycin followed by FACS for high mCherry expression. The HeLa.C2 Pro BiFC clone was generated by single-cell plating of the parental cell line, and single-cell clones were screened for induction of Venus fluorescence in response to DNA-damaging agents. Raidd+/− and Raidd−/− C2 Pro-BiFC MEF and Pidd+/+ and Pidd−/− C2 Pro-BiFC MEFs were selected by FACS for equal mCherry expression across the matching cell lines.
CRISPR/Cas9 gene editing
PIDD and RAIDD were deleted from HeLa.C2 Pro-BiFC cells using an adaptation of the protocol described in Gundry et al. (2016). Protospacer sequences for each target gene were identified using the CRISPRscan scoring algorithm (Moreno-Mateos et al., 2015). DNA templates for single-guide RNAs (sgRNAs) were made by PCR using pX459 plasmid containing the sgRNA scaffold sequence and using the following primers: ΔPIDD(76) sequence: 5′-TTAATACGACTCACTATAGGCGGTTGTGTGTCACTGTGGTTTTAGAGCTAGAAATAGC-3′; ΔPIDD(97) sequence: 5′-TTAATACGACTCACTATAGGCTTGGACCTGTACCCCGGGTTTTAGAGCTAGAAATAGC-3′; ΔRAIDD(57) sequence: 5′-TTAATACGACTCACTATAGGCCCAGGGAAACTCCTGTAGTTTTAGAGCTAGAAATAGC-3′; ΔRAIDD(76) sequence: 5′-TTAATACGACTCACTATAGGAGGAGCATTGTTTTCCGGGTTTTAGAGCTAGAAATAGC-3′; and universal reverse primer: 5′-AGCACCGACTCGGTGCCACT-3′. sgRNAs were generated by in vitro transcription using the Hiscribe T7 high-yield RNA synthesis kit (New England Biolabs, Inc.). Purified sgRNA (0.5 µg) was incubated with Cas9 protein (1 µg, PNA Bio) for 10 min at room temperature. HeLa.C2 Pro-BiFC cells were electroporated with the sgRNA/Cas9 complex using the Neon transfection system (Thermo Fisher Scientific) at 1,005 V, 35 ms, and two pulses.
Transient transfection and siRNA assays
105 HeLa cells were transfected with appropriate plasmids using Lipofectamine 2000 transfection reagent (Invitrogen) according to manufacturer’s instructions. Cells were transfected with amounts of the relevant expression plasmids as described in the figure legends. SiRNA transfections were performed using Hiperfect (QIAGEN) or Dharmafect (GE Healthcare) according to the manufacturer’s instructions. Cells were exposed to drugs at 48 h posttransfection. Control siRNAs were siLACZ or siCyclophilin B. NPM1 siRNAs were HP-validated siRNAs from QIAGEN (NPM1-1 sequence, 5′-AAGAATTGCTTCCGGATGACT-3′; NPM1-8 sequence 5′-AATGTCTGTACAGCCAACGGT-3′). Stable shRNA-expressing HeLa cell lines were described previously (Ando et al., 2012). PIDD siRNAs were ON-TARGETplus PIDD1 SMARTpool (target sequences: 5′-GAGCAGAUCCGUCACAUGC-3′; 5′-GGACGUGGCUGAAGAGGUG-3′; 5′-CCAGAAAUGCCCAGACUGU-3′; and 5′-CCUCAGAUUUGGACAGCUU-3′; GE Healthcare).
Microscopy
Cells were imaged using a spinning disk confocal microscope (ZEISS) equipped with a CSU-X1A 5000 spinning disk unit (Yokogawa Electric Corp.), multilaser module with wavelengths of 458, 488, 514, and 561 nm, and an Axio Observer Z1 motorized inverted microscope equipped with a precision motorized XY stage (ZEISS). Images were acquired with a Zeiss Plan-Neofluar 40× 1.3-NA or 64× 1.4-NA objective on an Orca R2 CCD camera using Zen 2012 software (ZEISS). Cells were plated on dishes containing coverslips coated with fibronectin (MatTek Corporation) 24 h before treatment or transfection.
Image analysis
At least 30 individual images were acquired for each treatment. Cells expressing the BiFC components were identified by fluorescence of the linked mCherry protein in stable cell lines or fluorescence of a transfection reporter in transient transfection experiments. Images were analyzed using Zen 2012 by counting the total number of cells per image and those with fluorescence in the cytosol, nucleus, or nucleolus. For 3D imaging, Venus and Cerulean channel image data were analyzed to determine the relative localization of caspase-2 BiFC and nuclear (Histone H2A), cytoskeletal (actin), or nucleolar proteins (NPM1, nucleolin, and fibrillarin), respectively. To analyze z-stacks, a line profile was drawn along the x-direction at the same y-position in each image. For each image, the yellow and cyan intensities along the line profile were linearly normalized to have a minimum of 0 and maximum of 1. Surface plots of normalized intensity in the x–z plane were generated in Matlab using the surfl function and custom color maps.
Immunofluorescence
Cells were washed in 3× 2 ml PBS and fixed in 3% (wt/vol) PFA in PBS, pH 7.2, for 10 min. Cells were washed for 3× 5 min in PBS followed by permeabilization in PBS and 0.15% (vol/vol) Triton S-100 for 10 min. Cells were blocked in FX image enhancer (Invitrogen) for 30 min and then stained with the anti-NPM1 antibody at a 1:100 dilution in PBS and 2% (wt/vol) BSA for 1 h. After washing in PBS and 2% (wt/vol) BSA, the cells were incubated with anti-mouse Alexa Fluor 488–conjugated secondary antibody (Invitrogen) at a 1:500 dilution in 2% (wt/vol) BSA for 45 min. Cells were washed in PBS and imaged as described earlier.
Flow cytometry and TUNEL staining
Venus- and mCherry-positive cells were quantified by flow cytometry and analyzed with FloJo software. TUNEL assays were performed using the APO-BRDU kit (BD) according to the manufacturer’s recommendation. In brief, the cells were fixed with 1% PFA (Sigma-Aldrich) in PBS for 1 h on ice, washed in PBS, and incubated with 70% ethanol at −20°C overnight. The cells were then washed and incubated with DNA labeling solution containing TdT and BrdU for 4 h at 37°C. The cells were washed and incubated in the staining buffer containing FITC-labeled anti-BrdU antibody for 30 min at RT, and a mixture of PI/RNase was added. After 30-min incubation at RT, the cells were analyzed by flow cytometry. The threshold for TUNEL positivity was determined as the maximal TUNEL signal observed in nontreated cells. For annexin V binding, cells were resuspended in 200 µl annexin V binding buffer (10 mM Hepes, 150 mM NaCl, 5 mM KCl, 1 mM MgCl2, and 1.8 mM CaCl2) supplemented with 2 µl annexin V-FITC (Invitrogen). Annexin V–positive cells were quantitated by flow cytometry. AlamarBlue-based cell viability assays were performed as previously described (Thompson et al., 2015).
Nucleolar isolation
Nucleoli were isolated after a protocol adapted from Mitrea et al. (2016). 4 × 107 HeLa cells were treated overnight as indicated in the figure legends and collected by trypsinization. Cells were washed in 10 ml PBS and centrifuged at 400 g for 5 min. Cell pellets were resuspended in five pellet volumes of nucleolar isolation buffer (NIB; 10 mM Tris, 2 mM MgCl2, and 0.5 mM EDTA) with complete protease inhibitor (Roche) and incubated at RT for 2 min and then on ice for 10 min. Plasma membranes were disrupted by the addition of 10% (vol/vol) IGEPAL to a final concentration of 1% and mixed by gentle pipetting. Nuclei were isolated by centrifugation at 500 g for 3 min. The supernatant was removed and stored as the cytoplasmic fraction, and the pellets were washed in 10–15 pellet volumes of NIB and 1% IGEPAL. Nuclear pellets were then resuspended in 10 pellet volumes of NIB and sonicated on ice at 20% power for 12 cycles of 1 s on followed by 5 s off on an XL 2020 sonicator fitted with a microtip probe (Misonix). The samples were centrifuged, and the supernatant was stored as the nucleoplasmic fraction. The pellet was washed once more with 10–15 pellet volumes of NIB and finally resuspended in 50–100 µl NIB.
Immunoblotting and coimmunoprecipitation
Cell lysates were resolved by SDS-PAGE. The proteins were transferred to nitrocellulose (Bio-Rad Laboratories) and immunodetected using appropriate primary and peroxidase-coupled secondary antibodies (GE Healthcare). Proteins were visualized by West Pico and West Dura chemiluminescence Substrate (Thermo Fisher Scientific). Lysates for immunoprecipitation were prepared in 1% or 0.1% NP-40 buffer (1% or 0.1% NP-40, 50 mM Tris-HCl, pH 8.0, 150–250 mM NaCl, 5 mM EDTA, 1 mM PMSF, protease inhibitors cocktail [Complete Mini; Roche], and phosphatase inhibitor cocktail [PhosSTOP; Roche]). For endogenous IPs, whole-cell lysates or nuclear extracts (10–20 mg) were mixed with Protein-G magnetic beads (Invitrogen; 30 µl of a 50% slurry) and anti-NPM1 (FC-61991), C-terminal anti-PIDD (Anto1 or AL233), or N-terminal anti-PIDD (S-17) antibody (3–5 µg, 1 ml final volume) overnight at 4°C on a rotating wheel. Beads were then washed three times with PBS, resolved by SDS-PAGE, and probed with anti-PIDD (H-300 [1:200 dilution]), anti-PIDD (Anto-1), anti-NPM1 (#3542), or anti-PIDD (S-17) detected with the corresponding secondary antibodies or mouse TrueBlot HRP-conjugated secondary antibodies [eBioscience]). For anti-Flag IPs, whole-cell lysates (0.15–6.00 mg) were mixed with 20 µl beads (50% slurry) and mouse anti-Flag M2 antibody (3 µg) in 0.1% NP-40 buffer (500 µl final volume) overnight at 4°C on a rotating shaker. Beads were then washed three times with PBS, resolved by SDS-PAGE, and analyzed by Western blot.
mRNA injections and acridine orange labeling in zebrafish embryos
Previously validated antisense morpholino oligonucleotides (MOs) targeted to npm1a and npm1b (5′-TAATGTTATCCTCCATTTTTGCGCG-3′ and 5′-GACCCATCTGTTCGAGATCCATGTC-3′, respectively; Bolli et al., 2010) were synthesized by Gene Tools, LLC, and injected into one-cell-stage embryos as previously described (Sidi et al., 2008; Bolli et al., 2010). 24 h embryos were stained with acridine orange (10 µg/ml final) as previously described (Sidi et al., 2008) and imaged with an SMZ 1500 fluorescence microscope (Nikon).
Statistical analysis
Statistical comparisons were performed using two-tailed Student’s t test calculated using Prism 6.0 (GraphPad Software).
Online supplemental material
Fig. S1 shows that caspase-2 BiFC localizes to the nucleus in response to several DNA damage inducers. Fig. S2 shows detailed imaging analysis of localization of the nucleolar caspase-2 complex in relation to nucleolar proteins. Fig. S3 accompanies Fig. 5 and shows the full analysis of the PIDD and RAIDD knockout and knockdown cells. Fig. S4 shows that NPM1 binds to PIDD and demonstrates specificity of the PIDD s17 antibody. Fig. S5 shows the localization of NPM1 in response to DNA damage and that NPM1 is required for caspase-2 cleavage and apoptosis in response to DNA-damaging agents. Video 1 shows the localization of camptothecin-induced caspase-2 BiFC in the nucleolus. Table S1 lists the NPM1 peptide sequences pulled down with Flag-PIDD immunoprecipitation.
Supplementary Material
Acknowledgments
We thank J. Flanagan for statistical help, M. Mamonkin for technical assistance, and H. Steen for assistance with proteomic analysis.
Funding for this project includes Texas Children’s Hospital Pediatrics Pilot Award (L. Bouchier-Hayes); National Institutes of Health/National Cancer Institute R01CA178162; Searle Scholars Program Scholar Award 11-SSP-196; Pershing Square Sohn Prize, New York Community Trust P16-000373; and a JJR Foundation award (S. Sidi); National Institutes of Health/National Institute of Diabetes and Digestive and Kidney Diseases T32DK060445 (B.A. Rohrman); and National Institutes of Health/National Cancer Institute F30CA186448 (P.H. Liu). This project was supported by the Cytometry and Cell Sorting Core at Baylor College of Medicine with funding from the National Institutes of Health (P30 AI036211, P30 CA125123, and S10 RR024574) and the expert assistance of J.M. Sederstrom.
The authors declare no competing financial interests.
Author contributions: S. Sidi and L. Bouchier-Hayes conceived the project, designed experiments, analyzed data, wrote, and edited the paper. K. Ando, M.J. Parsons, R.B. Shah, C.I. Charendoff, S.L. Paris, P.H. Liu, and R. Thompson performed experiments and analyzed data. S.R. Fassio and A. Oberst designed and generated reagents. B.A. Rohrman analyzed data.
Footnotes
Abbreviations used:
- AML
- acute myeloid leukemia
- BiFC
- bimolecular fluorescence complementation
- CARD
- caspase recruitment domain
- CR
- central region
- DD
- death domain
- DFC
- dense fibrillar center
- FC
- fibrillar center
- GC
- granular component
- IR
- irradiation
- NLS
- nuclear localization sequence
- sgRNA
- single-guide RNA
- WT
- wild-type
References
- Ahmad M., Srinivasula S.M., Wang L., Talanian R.V., Litwack G., Fernandes-Alnemri T., and Alnemri E.S.. 1997. CRADD, a novel human apoptotic adaptor molecule for caspase-2, and FasL/tumor necrosis factor receptor-interacting protein RIP. Cancer Res. 57:615–619. [PubMed] [Google Scholar]
- Ando K., Kernan J.L., Liu P.H., Sanda T., Logette E., Tschopp J., Look A.T., Wang J., Bouchier-Hayes L., and Sidi S.. 2012. PIDD death-domain phosphorylation by ATM controls prodeath versus prosurvival PIDDosome signaling. Mol. Cell. 47:681–693. 10.1016/j.molcel.2012.06.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antoniali G., Lirussi L., Poletto M., and Tell G.. 2014. Emerging roles of the nucleolus in regulating the DNA damage response: The noncanonical DNA repair enzyme APE1/Ref-1 as a paradigmatical example. Antioxid. Redox Signal. 20:621–639. 10.1089/ars.2013.5491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baliga B.C., Colussi P.A., Read S.H., Dias M.M., Jans D.A., and Kumar S.. 2003. Role of prodomain in importin-mediated nuclear localization and activation of caspase-2. J. Biol. Chem. 278:4899–4905. 10.1074/jbc.M211512200 [DOI] [PubMed] [Google Scholar]
- Baliga B.C., Read S.H., and Kumar S.. 2004. The biochemical mechanism of caspase-2 activation. Cell Death Differ. 11:1234–1241. 10.1038/sj.cdd.4401492 [DOI] [PubMed] [Google Scholar]
- Balusu R., Fiskus W., Rao R., Chong D.G., Nalluri S., Mudunuru U., Ma H., Chen L., Venkannagari S., Ha K., et al. . 2011. Targeting levels or oligomerization of nucleophosmin 1 induces differentiation and loss of survival of human AML cells with mutant NPM1. Blood. 118:3096–3106. 10.1182/blood-2010-09-309674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertwistle D., Sugimoto M., and Sherr C.J.. 2004. Physical and functional interactions of the Arf tumor suppressor protein with nucleophosmin/B23. Mol. Cell. Biol. 24:985–996. 10.1128/MCB.24.3.985-996.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berube C., Boucher L.M., Ma W., Wakeham A., Salmena L., Hakem R., Yeh W.C., Mak T.W., and Benchimol S.. 2005. Apoptosis caused by p53-induced protein with death domain (PIDD) depends on the death adapter protein RAIDD. Proc. Natl. Acad. Sci. USA. 102:14314–14320. 10.1073/pnas.0506475102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boatright K.M., Renatus M., Scott F.L., Sperandio S., Shin H., Pedersen I.M., Ricci J.E., Edris W.A., Sutherlin D.P., Green D.R., and Salvesen G.S.. 2003. A unified model for apical caspase activation. Mol. Cell. 11:529–541. 10.1016/S1097-2765(03)00051-0 [DOI] [PubMed] [Google Scholar]
- Bock F.J., Peintner L., Tanzer M., Manzl C., and Villunger A.. 2012. P53-induced protein with a death domain (PIDD): Master of puppets? Oncogene. 31:4733–4739. 10.1038/onc.2011.639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolli N., Payne E.M., Grabher C., Lee J.S., Johnston A.B., Falini B., Kanki J.P., and Look A.T.. 2010. Expression of the cytoplasmic NPM1 mutant (NPMc+) causes the expansion of hematopoietic cells in zebrafish. Blood. 115:3329–3340. 10.1182/blood-2009-02-207225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonzon C., Bouchier-Hayes L., Pagliari L.J., Green D.R., and Newmeyer D.D.. 2006. Caspase-2-induced apoptosis requires bid cleavage: A physiological role for bid in heat shock-induced death. Mol. Biol. Cell. 17:2150–2157. 10.1091/mbc.E05-12-1107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouchier-Hayes L., and Green D.R.. 2012. Caspase-2: The orphan caspase. Cell Death Differ. 19:51–57. 10.1038/cdd.2011.157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouchier-Hayes L., Oberst A., McStay G.P., Connell S., Tait S.W., Dillon C.P., Flanagan J.M., Beere H.M., and Green D.R.. 2009. Characterization of cytoplasmic caspase-2 activation by induced proximity. Mol. Cell. 35:830–840. 10.1016/j.molcel.2009.07.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colombo E., Marine J.C., Danovi D., Falini B., and Pelicci P.G.. 2002. Nucleophosmin regulates the stability and transcriptional activity of p53. Nat. Cell Biol. 4:529–533. 10.1038/ncb814 [DOI] [PubMed] [Google Scholar]
- Colombo E., Bonetti P., Lazzerini Denchi E., Martinelli P., Zamponi R., Marine J.C., Helin K., Falini B., and Pelicci P.G.. 2005. Nucleophosmin is required for DNA integrity and p19Arf protein stability. Mol. Cell. Biol. 25:8874–8886. 10.1128/MCB.25.20.8874-8886.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cullen S.P., Afonina I.S., Donadini R., Lüthi A.U., Medema J.P., Bird P.I., and Martin S.J.. 2009. Nucleophosmin is cleaved and inactivated by the cytotoxic granule protease granzyme M during natural killer cell-mediated killing. J. Biol. Chem. 284:5137–5147. 10.1074/jbc.M807913200 [DOI] [PubMed] [Google Scholar]
- Dimitrova N., Chen Y.C., Spector D.L., and de Lange T.. 2008. 53BP1 promotes non-homologous end joining of telomeres by increasing chromatin mobility. Nature. 456:524–528. 10.1038/nature07433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorstyn L., Puccini J., Wilson C.H., Shalini S., Nicola M., Moore S., and Kumar S.. 2012. Caspase-2 deficiency promotes aberrant DNA-damage response and genetic instability. Cell Death Differ. 19:1288–1298. 10.1038/cdd.2012.36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan H., and Dixit V.M.. 1997. RAIDD is a new ‘death’ adaptor molecule. Nature. 385:86–89. 10.1038/385086a0 [DOI] [PubMed] [Google Scholar]
- Falini B., Mecucci C., Tiacci E., Alcalay M., Rosati R., Pasqualucci L., La Starza R., Diverio D., Colombo E., Santucci A., et al. GIMEMA Acute Leukemia Working Party . 2005. Cytoplasmic nucleophosmin in acute myelogenous leukemia with a normal karyotype. N. Engl. J. Med. 352:254–266. 10.1056/NEJMoa041974 [DOI] [PubMed] [Google Scholar]
- Grisendi S., Bernardi R., Rossi M., Cheng K., Khandker L., Manova K., and Pandolfi P.P.. 2005. Role of nucleophosmin in embryonic development and tumorigenesis. Nature. 437:147–153. 10.1038/nature03915 [DOI] [PubMed] [Google Scholar]
- Grisendi S., Mecucci C., Falini B., and Pandolfi P.P.. 2006. Nucleophosmin and cancer. Nat. Rev. Cancer. 6:493–505. 10.1038/nrc1885 [DOI] [PubMed] [Google Scholar]
- Gundry M.C., Brunetti L., Lin A., Mayle A.E., Kitano A., Wagner D., Hsu J.I., Hoegenauer K.A., Rooney C.M., Goodell M.A., and Nakada D.. 2016. Highly efficient genome editing of murine and human hematopoietic progenitor cells by CRISPR/Cas9. Cell Reports. 17:1453–1461. 10.1016/j.celrep.2016.09.092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Y., Srinivasula S.M., Druilhe A., Fernandes-Alnemri T., and Alnemri E.S.. 2002. Caspase-2 induces apoptosis by releasing proapoptotic proteins from mitochondria. J. Biol. Chem. 277:13430–13437. 10.1074/jbc.M108029200 [DOI] [PubMed] [Google Scholar]
- Ho L.H., Taylor R., Dorstyn L., Cakouros D., Bouillet P., and Kumar S.. 2009. A tumor suppressor function for caspase-2. Proc. Natl. Acad. Sci. USA. 106:5336–5341. 10.1073/pnas.0811928106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang T.H., and Park H.H.. 2013. PIDD mediates and stabilizes the interaction between RAIDD and caspase-2 for the PIDDosome assembly. BMB Rep. 46:471–476. 10.5483/BMBRep.2013.46.9.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janssens S., and Tinel A.. 2012. The PIDDosome, DNA-damage-induced apoptosis and beyond. Cell Death Differ. 19:13–20. 10.1038/cdd.2011.162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janssens S., Tinel A., Lippens S., and Tschopp J.. 2005. PIDD mediates NF-κB activation in response to DNA damage. Cell. 123:1079–1092. 10.1016/j.cell.2005.09.036 [DOI] [PubMed] [Google Scholar]
- Kaufmann T., Tai L., Ekert P.G., Huang D.C., Norris F., Lindemann R.K., Johnstone R.W., Dixit V.M., and Strasser A.. 2007. The BH3-only protein bid is dispensable for DNA damage- and replicative stress-induced apoptosis or cell-cycle arrest. Cell. 129:423–433. 10.1016/j.cell.2007.03.017 [DOI] [PubMed] [Google Scholar]
- Korgaonkar C., Hagen J., Tompkins V., Frazier A.A., Allamargot C., Quelle F.W., and Quelle D.E.. 2005. Nucleophosmin (B23) targets ARF to nucleoli and inhibits its function. Mol. Cell. Biol. 25:1258–1271. 10.1128/MCB.25.4.1258-1271.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lassus P., Opitz-Araya X., and Lazebnik Y.. 2002. Requirement for caspase-2 in stress-induced apoptosis before mitochondrial permeabilization. Science. 297:1352–1354. 10.1126/science.1074721 [DOI] [PubMed] [Google Scholar]
- Lin Y., Ma W., and Benchimol S.. 2000. Pidd, a new death-domain-containing protein, is induced by p53 and promotes apoptosis. Nat. Genet. 26:122–127. 10.1038/79102 [DOI] [PubMed] [Google Scholar]
- Lindström M.S. 2011. NPM1/B23: A multifunctional chaperone in ribosome biogenesis and chromatin remodeling. Biochem. Res. Int. 2011:195209 10.1155/2011/195209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mancini M., Machamer C.E., Roy S., Nicholson D.W., Thornberry N.A., Casciola-Rosen L.A., and Rosen A.. 2000. Caspase-2 is localized at the Golgi complex and cleaves golgin-160 during apoptosis. J. Cell Biol. 149:603–612. 10.1083/jcb.149.3.603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manzl C., Krumschnabel G., Bock F., Sohm B., Labi V., Baumgartner F., Logette E., Tschopp J., and Villunger A.. 2009. Caspase-2 activation in the absence of PIDDosome formation. J. Cell Biol. 185:291–303. 10.1083/jcb.200811105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manzl C., Peintner L., Krumschnabel G., Bock F., Labi V., Drach M., Newbold A., Johnstone R., and Villunger A.. 2012. PIDDosome-independent tumor suppression by Caspase-2. Cell Death Differ. 19:1722–1732. 10.1038/cdd.2012.54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manzl C., Fava L.L., Krumschnabel G., Peintner L., Tanzer M.C., Soratroi C., Bock F.J., Schuler F., Luef B., Geley S., and Villunger A.. 2013. Death of p53-defective cells triggered by forced mitotic entry in the presence of DNA damage is not uniquely dependent on Caspase-2 or the PIDDosome. Cell Death Dis. 4:e942 10.1038/cddis.2013.470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitrea D.M., and Kriwacki R.W.. 2013. Regulated unfolding of proteins in signaling. FEBS Lett. 587:1081–1088. 10.1016/j.febslet.2013.02.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitrea D.M., Cika J.A., Guy C.S., Ban D., Banerjee P.R., Stanley C.B., Nourse A., Deniz A.A., and Kriwacki R.W.. 2016. Nucleophosmin integrates within the nucleolus via multi-modal interactions with proteins displaying R-rich linear motifs and rRNA. eLife. 5:e13571 10.7554/eLife.13571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno-Mateos M.A., Vejnar C.E., Beaudoin J.D., Fernandez J.P., Mis E.K., Khokha M.K., and Giraldez A.J.. 2015. CRISPRscan: Designing highly efficient sgRNAs for CRISPR-Cas9 targeting in vivo. Nat. Methods. 12:982–988. 10.1038/nmeth.3543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muzio M., Stockwell B.R., Stennicke H.R., Salvesen G.S., and Dixit V.M.. 1998. An induced proximity model for caspase-8 activation. J. Biol. Chem. 273:2926–2930. 10.1074/jbc.273.5.2926 [DOI] [PubMed] [Google Scholar]
- Myers K., Gagou M.E., Zuazua-Villar P., Rodriguez R., and Meuth M.. 2009. ATR and Chk1 suppress a caspase-3-dependent apoptotic response following DNA replication stress. PLoS Genet. 5:e1000324 10.1371/journal.pgen.1000324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nam H.S., and Benezra R.. 2009. High levels of Id1 expression define B1 type adult neural stem cells. Cell Stem Cell. 5:515–526. 10.1016/j.stem.2009.08.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Reilly L.A., Ekert P., Harvey N., Marsden V., Cullen L., Vaux D.L., Hacker G., Magnusson C., Pakusch M., Cecconi F., et al. . 2002. Caspase-2 is not required for thymocyte or neuronal apoptosis even though cleavage of caspase-2 is dependent on both Apaf-1 and caspase-9. Cell Death Differ. 9:832–841. 10.1038/sj.cdd.4401033 [DOI] [PubMed] [Google Scholar]
- Oliver T.G., Meylan E., Chang G.P., Xue W., Burke J.R., Humpton T.J., Hubbard D., Bhutkar A., and Jacks T.. 2011. Caspase-2-mediated cleavage of Mdm2 creates a p53-induced positive feedback loop. Mol. Cell. 43:57–71. 10.1016/j.molcel.2011.06.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan Y., Ren K.H., He H.W., and Shao R.G.. 2009. Knockdown of Chk1 sensitizes human colon carcinoma HCT116 cells in a p53-dependent manner to lidamycin through abrogation of a G2/M checkpoint and induction of apoptosis. Cancer Biol. Ther. 8:1559–1566. 10.4161/cbt.8.16.8955 [DOI] [PubMed] [Google Scholar]
- Park H.H., Logette E., Raunser S., Cuenin S., Walz T., Tschopp J., and Wu H.. 2007. Death domain assembly mechanism revealed by crystal structure of the oligomeric PIDDosome core complex. Cell. 128:533–546. 10.1016/j.cell.2007.01.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parsons M.J., McCormick L., Janke L., Howard A., Bouchier-Hayes L., and Green D.R.. 2013. Genetic deletion of caspase-2 accelerates MMTV/c-neu-driven mammary carcinogenesis in mice. Cell Death Differ. 20:1174–1182. 10.1038/cdd.2013.38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pianese G. 1896. Beitrag zur Histologie und Aetiologie der Carcinoma. Histologische und experimentelle Untersuchungen. Beitr. Pathol. Anat. Allg. Pathol. 142:1–193. [Google Scholar]
- Puccini J., Dorstyn L., and Kumar S.. 2013a Caspase-2 as a tumour suppressor. Cell Death Differ. 20:1133–1139. 10.1038/cdd.2013.87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puccini J., Shalini S., Voss A.K., Gatei M., Wilson C.H., Hiwase D.K., Lavin M.F., Dorstyn L., and Kumar S.. 2013b Loss of caspase-2 augments lymphomagenesis and enhances genomic instability in Atm-deficient mice. Proc. Natl. Acad. Sci. USA. 110:19920–19925. 10.1073/pnas.1311947110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi W., Shakalya K., Stejskal A., Goldman A., Beeck S., Cooke L., and Mahadevan D.. 2008. NSC348884, a nucleophosmin inhibitor disrupts oligomer formation and induces apoptosis in human cancer cells. Oncogene. 27:4210–4220. 10.1038/onc.2008.54 [DOI] [PubMed] [Google Scholar]
- Robertson J.D., Enoksson M., Suomela M., Zhivotovsky B., and Orrenius S.. 2002. Caspase-2 acts upstream of mitochondria to promote cytochrome c release during etoposide-induced apoptosis. J. Biol. Chem. 277:29803–29809. 10.1074/jbc.M204185200 [DOI] [PubMed] [Google Scholar]
- Salvesen G.S., and Dixit V.M.. 1999. Caspase activation: The induced-proximity model. Proc. Natl. Acad. Sci. USA. 96:10964–10967. 10.1073/pnas.96.20.10964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shyu Y.J., Liu H., Deng X., and Hu C.D.. 2006. Identification of new fluorescent protein fragments for bimolecular fluorescence complementation analysis under physiological conditions. Biotechniques. 40:61–66. 10.2144/000112036 [DOI] [PubMed] [Google Scholar]
- Sidi S., Sanda T., Kennedy R.D., Hagen A.T., Jette C.A., Hoffmans R., Pascual J., Imamura S., Kishi S., Amatruda J.F., et al. . 2008. Chk1 suppresses a caspase-2 apoptotic response to DNA damage that bypasses p53, Bcl-2, and caspase-3. Cell. 133:864–877. 10.1016/j.cell.2008.03.037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terry M.R., Arya R., Mukhopadhyay A., Berrett K.C., Clair P.M., Witt B., Salama M.E., Bhutkar A., and Oliver T.G.. 2015. Caspase-2 impacts lung tumorigenesis and chemotherapy response in vivo. Cell Death Differ. 22:719–730. 10.1038/cdd.2014.159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson R., Shah R.B., Liu P.H., Gupta Y.K., Ando K., Aggarwal A.K., and Sidi S.. 2015. An inhibitor of PIDDosome formation. Mol. Cell. 58:767–779. 10.1016/j.molcel.2015.03.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tinel A., and Tschopp J.. 2004. The PIDDosome, a protein complex implicated in activation of caspase-2 in response to genotoxic stress. Science. 304:843–846. 10.1126/science.1095432 [DOI] [PubMed] [Google Scholar]
- Tinel A., Janssens S., Lippens S., Cuenin S., Logette E., Jaccard B., Quadroni M., and Tschopp J.. 2007. Autoproteolysis of PIDD marks the bifurcation between pro-death caspase-2 and pro-survival NF-κB pathway. EMBO J. 26:197–208. 10.1038/sj.emboj.7601473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber J.D., Taylor L.J., Roussel M.F., Sherr C.J., and Bar-Sagi D.. 1999. Nucleolar Arf sequesters Mdm2 and activates p53. Nat. Cell Biol. 1:20–26. 10.1038/8991 [DOI] [PubMed] [Google Scholar]
- Wejda M., Impens F., Takahashi N., Van Damme P., Gevaert K., and Vandenabeele P.. 2012. Degradomics reveals that cleavage specificity profiles of caspase-2 and effector caspases are alike. J. Biol. Chem. 287:33983–33995. 10.1074/jbc.M112.384552 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.