Retinal stem cells yield all retinal cell types, but their embryonic origin is elusive. Using a Zebrabow-based clonal analysis and in vivo lineage analyses, Tang et al. identify bipotent progenitors as the cell of origin of retinal stem cells.
Abstract
In lower vertebrates, retinal stem cells (RSCs) capable of producing all retinal cell types are a resource for retinal tissue growth throughout life. However, the embryonic origin of RSCs remains largely elusive. Using a Zebrabow-based clonal analysis, we characterized the RSC niche in the ciliary marginal zone of zebrafish retina and illustrate that blood vessels associated with RSCs are required for the maintenance of actively proliferating RSCs. Full lineage analysis of RSC progenitors reveals lineage patterns of RSC production. Moreover, in vivo lineage analysis demonstrates that these RSC progenitors are the direct descendants of a set of bipotent progenitors in the medial epithelial layer of developing optic vesicles, suggesting the involvement of the mixed-lineage states in the RSC lineage specification.
Introduction
In lower vertebrates, the ciliary marginal zone (CMZ), the most peripheral region of the retina, provides the retinal stem cell (RSC) niche, where cells proliferate and differentiate into new neurons and glial cells over a lifetime (Straznicky and Gaze, 1971; Johns, 1977; Fraser and Hunt, 1980; Negishi et al., 1982; Reh and Levine, 1998; Marcus et al., 1999; Perron and Harris, 2000; Reh and Fischer, 2001, 2006; Otteson and Hitchcock, 2003; Hitchcock et al., 2004; Moshiri et al., 2004; Fischer et al., 2014). Comparative studies of multiple vertebrate species have revealed a gradual reduction in the neurogenic capacity of CMZ cells over evolution (Kubota et al., 2002; Amato et al., 2004; Todd et al., 2016). In chicks, CMZ cells continue to add new retinal neurons of restricted types for a short period after hatching (Willbold and Layer, 1992; Fischer and Reh, 2000). In rodents, there is as yet no evidence of active RSCs at the adult retinal ciliary margin, analogous to the CMZ of lower vertebrates, even after injury (Kubota et al., 2002; Fischer et al., 2013), although the marginal cells contribute to retinogenesis before birth (Marcucci et al., 2016; Bélanger et al., 2017). However, increasing evidence suggests that cells at the retinal margin of homeothermic vertebrates, including birds and mammals, might hold the neurogenic potential beyond embryonic development. In the postnatal chick, cells at the retinal margin express the genes that are present in embryonic retinal progenitors and are capable of proliferating and producing new neurons under certain conditions (Willbold and Layer, 1992; Fischer and Reh, 2000, 2001, 2002; Fischer et al., 2002; Spence et al., 2004; Fischer, 2005; Moshiri et al., 2005). In ptc+/− mice, marginal progenitors are able to persist up to 3 mo (Moshiri and Reh, 2004). Neurogenic characteristics at the retinal margin of primate species, including humans, were also documented (Fischer et al., 2001; Martínez-Navarrete et al., 2008; Bhatia et al., 2009; Kiyama et al., 2012). Consistently, in culture assays, cells from the mouse pigmented ciliary margin are able to clonally proliferate and differentiate into retinal pigmented and nonpigmented cells (Tropepe et al., 2000). Similar results also were described in the rat and human retina (Ahmad et al., 2000; Coles et al., 2004; Bhatia et al., 2011). Interestingly, cells in self-organizing CMZ-like organoids derived from human embryonic stem cells behave similarly to the CMZ cells of lower vertebrates (Kuwahara et al., 2015). All these results support the idea that RSCs are silenced rather than lost from the ciliary margin of mammalian retinas during vertebrate evolution. Therefore, better understanding of CMZ cells of lower vertebrate retinas ultimately may guide the activation of dormant RSCs in mammals.
In lower vertebrates, CMZ cells are capable of generating both neural cell types and pigmented cell types in the retina, implying that RSCs and pigmented stem cells are in the CMZ (Wetts et al., 1989). A recent clonal study has demonstrated that RSCs and pigmented stem cells are actually two distinct cell populations that are maintained independently (Centanin et al., 2011). Intriguingly, many genetic mutants exhibit a phenotype where the reduction of the CMZ is accompanied by an expansion of the retinal pigment epithelium (RPE) and vice versa (Wehman et al., 2005; Cerveny et al., 2010; Miesfeld et al., 2015). It was thus proposed that there is the crosstalk during the production and maintenance of both of these stem cell populations within the CMZ.
Genetic-based lineage mapping has shown that RSCs comprise a subset of rx2-expressing cells that reside in the tip region of the CMZ (Reinhardt et al., 2015; Wan et al., 2016). The topology of RSC-derived clones indicates that RSCs proliferate asymmetrically to generate one self-renewing RSC and one differentiating retinal progenitor (Centanin et al., 2014). Individual RSCs are multipotent, capable of producing arched continuous strips composed of all retinal cell types over time (Wetts and Fraser, 1988; Centanin et al., 2011). The multipotency of RSCs is reminiscent of embryonic retinal progenitors (Turner and Cepko, 1987; Holt et al., 1988; Wetts and Fraser, 1988). In fact, extensive molecular analysis has uncovered dozens of signaling molecules distributed in characteristic patterns in the CMZ (Harris and Perron, 1998; Wehman et al., 2005; Raymond et al., 2006; Agathocleous and Harris, 2009; Kubo and Nakagawa, 2009; Cerveny et al., 2012; Parain et al., 2012; Cabochette and Vega-Lopez, 2015). The genes expressed in early embryonic retinal progenitors are expressed in the most peripheral region of the CMZ where RSCs are presumed to reside, whereas the genes expressed in late embryonic retinal progenitors are expressed in the CMZ closer to the central retina where the differentiating CMZ progenitors reside. These results indicate that the spatial unfolding of CMZ cells recapitulates the temporal progression of embryonic retinal progenitor lineages, suggesting that RSCs and their embryonic counterparts use similar developmental programs to produce cell lineages (Harris and Perron, 1998; Wan et al., 2016). This similarity between RSCs and their embryonic counterparts highlights a longstanding hypothesis: namely, that RSCs are simply embryonic retinal progenitors that persist beyond embryonic development, raising the intriguing question as to how embryonic retinal progenitors are specified into the RSC lineage.
In zebrafish, the eye develops from the optic vesicle and optic stalk by evagination of the anterior part of the neural tube ∼12 h post fertilization (hpf). By 16 hpf, the optic vesicle, detached from the neural tube, is composed of the lateral epithelial layer (LL) and the medial epithelial layer (ML). Cells of the ML migrate dorsally and ventrally into the LL such that the ML becomes reduced while the LL enlarges. At 24 hpf, the optic vesicle transforms into the optic cup, with the ML developing into the RPE and the LL enveloping to form the neural retina (NR; Schmitt and Dowling, 1994; Li et al., 2000; Kwan et al., 2012). Early studies have revealed that CMZ cells originate from migrating cells of the ML (El Yakoubi et al., 2012; Heermann et al., 2015). However, the precise lineage patterns of RSC generation, in particular at the single-cell resolution, is still missing.
In this study, we developed a Zebrabow-based method in zebrafish to provide the first characterization of cells within the CMZ tip region at an unprecedented spatial resolution. Clonal analysis identified RSC progenitors (SPs), and full lineage analysis of these progenitors uncovered a complete set of lineage patterns through which RSCs were produced. Moreover, we discovered a new set of bipotent progenitors in the developing optic vesicles as the embryonic origin of RSCs. Thus, this study provides the framework of the embryonic development of RSCs.
Results
Clonal analysis revealing the cell cluster of the CMZ tip as the RSCs niche
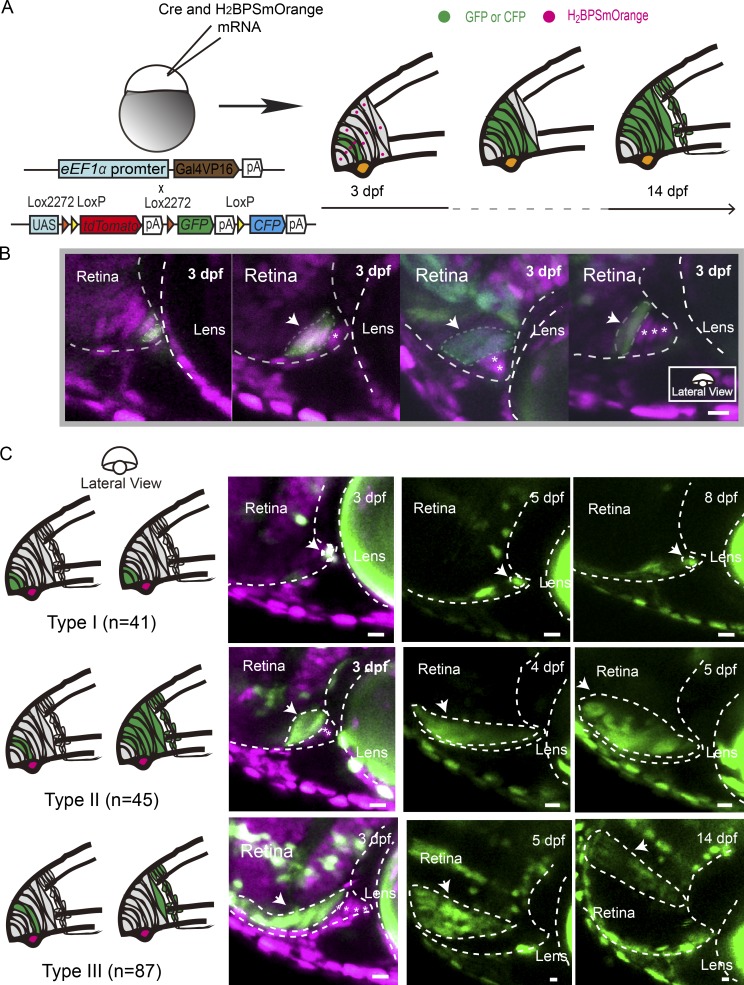
RSCs of lower vertebrates have been thought to reside at the very periphery of the CMZ (Straznicky and Gaze, 1971; Johns, 1977; Wetts and Fraser, 1988; Centanin et al., 2011). To characterize RSCs, we developed a Zebrabow-based strategy. Individual CMZ cells marked stochastically with the Cre-dependent combination of CFP, GFP, and tdTomato were followed over time (Figs. 1 A and S1 A). Simultaneously, all cell nuclei were labeled by H2BPSmOrange, a nucleus-localized fluorescent protein representing the original position of a given labeled cell relative to the CMZ tip boundary (Fig. 1 B). A systematic analysis of a total of 226 clones was performed from 72 hpf until 14 d post fertilization (dpf; Fig. 1 C). RSCs have been reported to undergo asymmetric cell divisions, thereby expanding clones while maintaining a constant physical origin at the edge of the CMZ (Centanin et al., 2014). We therefore categorized individual clones according to whether a clone stayed in contact with its physical origin over time or not (Fig. 1 C). We identified three types of clones. Progenitors giving rise to Type I clones (Fig. 1 C, Type I; n = 41) were the first- or second-position cells of the CMZ. To our surprise, these cells rarely divided and remained at their original locations over time. We termed these cells as dormant CMZ tip cells. Type I clones were also confirmed by the lineage tracing of individual CMZ tip cells using photoconverted Kaede (Fig. S1 C). In contrast, Type II clones, which initiated at the positions arranging from cells 2 to 4, proliferated while remaining attached to the same spatial location in the CMZ where they originated (Fig. 1 C, Type II; n = 45). These continuously growing clones have been recognized as characteristic of RSCs in fish retinas previously (Centanin et al., 2011, 2014). Meanwhile, Type III clones, which originated from cells located beyond the first four cells, rapidly detached from the CMZ and formed differentiated clones within 1–2 d, leaving no RSCs behind in the CMZ (Fig. 1 C, Type III; n = 87). Therefore, these cells were likely to represent retinal progenitors, which have a limited ability to proliferate before giving rise to differentiated progeny (Wan et al., 2016). As expected, if >4 CMZ tip cells were labeled, the clones entirely occupied the CMZ over time (Fig. S1 B; n = 53). Our clonal analysis clearly showed that actively proliferating RSCs were confined within the four-cell cluster of the CMZ tip, and at the very tip, one or two cells were dormant.
Figure 1.
Characterization of CMZ cells at single-cell resolution. (A) Schematic illustrating the clonal analysis of CMZ cells. Cre mRNA and H2BPSmOrange mRNA were coinjected into zebrafish embryos (eEF1α:Gal4VP16::UAS:Zebrabow) at one cell stage. Starting at 72 hpf, sparsely labeled cells originating at different CMZ loci were selected and followed for up to 14 dpf. (B) Micrographs illustrating clones derived from the cells at the different loci of the CMZ tip region at 72 hpf (the cells at positions 1–4 are indicated from the left to the right). Gray dashed lines indicate the time window between 3 dpf and 14 dpf. (C) Type I clones originated from the tip-most first or second cell in the CMZ tip and remained in their original loci without cell proliferation as observed until 14 dpf (n = 41). Tracing of one representative Type I clone is shown until 8 dpf. Type II clones consist of marked cells, which originated from cells that were one or two cells further away from the very CMZ tip. These clones remained attached to their original location while expanding continuously (n = 45). One representative type II clone, traced until 5 dpf, is shown. In contrast, Type III clones that originated from cells that were more than three cells away from the CMZ tip at 72 hpf (Type III; n = 87) detached rapidly from the CMZ tip and formed differentiated clones within 1 or 2 d. An example trace of such Type III clone is shown from 3 until 14 dpf. Arrows indicate traced cells. Asterisks indicate CMZ tip cells labeled by H2BPSmOrange but not GFP, GFP, and tdTomato. White dashed lines outline the boundaries of retinas and lenses. Bars, 10 µm.
Characterization of dormant CMZ tip cells
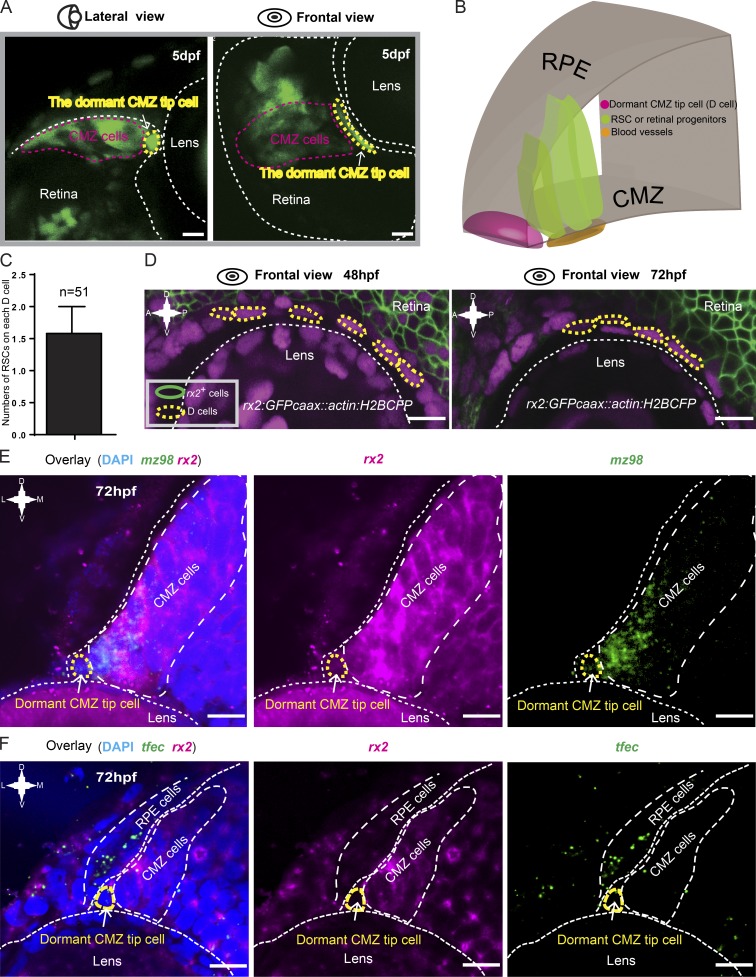
The discovery of dormant CMZ tip cells was unexpected. Interestingly, from a front view, these cells actually appeared elongated and fusiform perpendicular to the remaining CMZ cells (Fig. 2, A and B). Quantitative analysis showed that each dormant CMZ tip cell was able to directly contact approximately one and a half putative RSCs on average (Fig. 2 C). We then examined whether these cells expressed rx2, a retinal identity gene expressed in RSCs and retinal progenitors (Reinhardt et al., 2015). In the transgenic Tg(rx2:GFPcaax) line, in which GFPcaax expression is driven by the rx2 promoter (Heermann et al., 2015), dormant CMZ tip cells exhibited no expression of the GFP signal, suggesting the absence of rx2 expression (Fig. 2 D). To faithfully monitor rx2 dynamics, we generated a new transgenic line in which a destabilized version of GFP was driven by the rx2 promoter (Fig. S2 A). This transgenic line showed that rx2 expression was absent from dormant CMZ tip cells but was expressed in other CMZ cells (Fig. S2 B). FISH further confirmed the absence of rx2 from dormant CMZ tip cells, which, however, expressed mz98, a gene that has been reported to be expressed in the very tip region of the CMZ (Fig. 2 E; Gonzalez-Nunez et al., 2010). Moreover, these tip cells also had very little or no expression of tfec, a gene required for RPE fate specification (Fig. 2 F; Miesfeld et al., 2015). Therefore, the exact fate of dormant CMZ tip cells requires further exploration. Collectively, these results reveal a set of dormant CMZ tip cells that are morphologically and molecularly (mz98+rx2−tfec−) distinct from actively proliferating RSCs (mz98+rx2+tfec−) and pigmented cells (mz98−rx2−tfec+).
Figure 2.
Characterization of dormant CMZ tip cells. (A) Lateral and frontal views of a representative 72-hpf Type I clone (in green) labeled by the Zebrabow-based strategy showing the characteristic cell morphology of dormant CMZ tip cells. (B) 3D schematic of the CMZ tip region, which is composed of dormant CMZ tip cells (D cells; in magenta), RSCs, and retina progenitors (in dark green). (C) Imaging analysis showing that each dormant CMZ cell directly contacts with ∼1.5 RSCs on average. The error bar shows the mean ± SD. (D) In the retina of the transgenic fish Tg(rx2:GFPcaax::actin:H2BCFP) at 48 and 72 hpf, dormant CMZ tip cells (circled by yellow dashed lines) did not have the GFPcaax signal driven by the rx2 promoter. (E) Double FISH showing the dormant CMZ tip cells express mz98 but not rx2. (F) Double FISH results showing that the tfec gene is either absent or expressed at extremely low levels in dormant CMZ tip cells. A, anterior; D, dorsal; P, posterior; V, ventral. Bars, 10 µm.
Blood vessels are required for actively proliferating RSCs
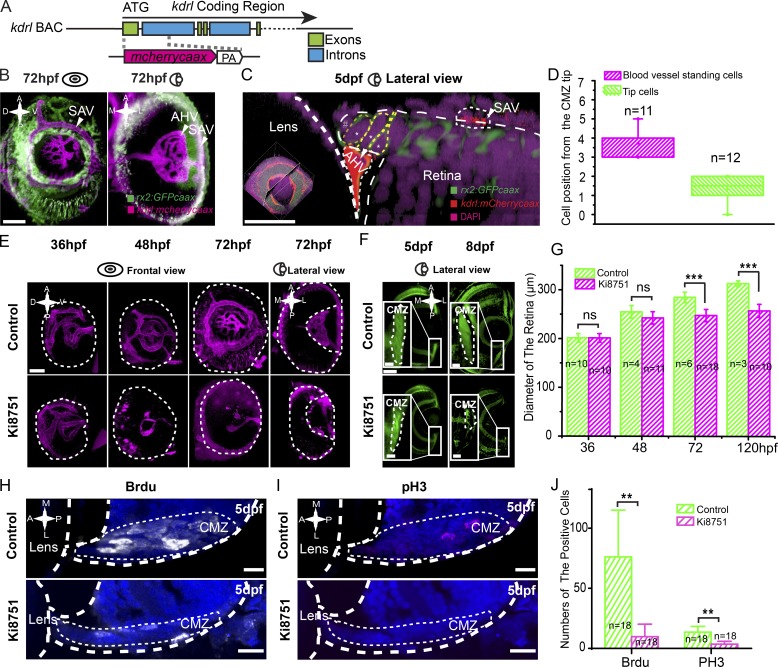
The gene kinase insert domain receptor like (kdrl) is expressed specifically in endothelial cells of blood vessels (Fouquet et al., 1997; Liao et al., 1997; Sumoy et al., 1997). To further characterize the RSC niche, we generated a kdrl BAC transgenic line to label vascular structures, and we identified two major blood vessels surrounding the CMZ: anterior hyaloid vessels (AHVs), residing at the basal surface of the CMZ tip region, and superficial annular vessels (SAVs), located at the side of the RPE (Fig. 3, A and B; and Video 1; Kitambi et al., 2009; Hartsock et al., 2014; Kaufman et al., 2015). Imaging analysis showed that the first one to two cells at the very peripheral tip of the CMZ (1.3 ± 0.7 cells; n = 12) had no direct contact with the AHV but that the next three to four cells directly apposed the AHV (3.7 ± 0.6 cells; n = 11; Fig. 3, C and D; and Video 2). This cell–vessel association again supports the distinction between dormant CMZ tip cells and RSCs.
Figure 3.
Blood vessels are required for actively proliferative RSCs. (A) Schematic of the design of BAC transgene kdrl:mCherrycaax. The initiation codon ATG was used. (B) 3D illustration (frontal and lateral views) of blood vessels (in magenta) in the eye of the transgenic fish line Tg(rx2:GFPcaax::kdrl:mCherrycaax). SAVs and AHVs surround the CMZ tip region. (C) A high-resolution image of a four-cell cluster of the CMZ tip region that is associated with the local blood vessels. (D) Quantification, measured by the physical cell positions of the CMZ tip region, showing that the second to fourth cells (n = 11) often directly associated with the local blood vessel (AHV), whereas dormant CMZ tip cells did not (n = 12). (E) 3D representation of the blood vessels in control eyes and eyes treated with 50 nM Ki8751 at 36, 48, and 72 hpf. (F) Representative images of the CMZ labeled in the Tg(rx2:GFPcaax) with the treatment of 50 nM Ki8751 at 5 and 8 dpf showing the reduction of the CMZ. Insets represent the enlarged CMZ. (G) Quantifications of eye diameters in embryos with or without Ki8751 treatment show a significant reduction of the eye size in the treated embryos at 72 hpf onwards. The p-values in groups of 48 hpf, 72 hpf, and 5 dpf were 0.124, 1.42 × 10−6, and 3.1 × 10−5, respectively. (H–J) BrdU and pH3 staining of the retina in embryos with or without Ki8751 treatment shows a significant reduction of cell proliferation in the CMZ of the retina in the treated embryos at 5 dpf. The p-values in groups of BrdU and pH3 were 0.002 and 0.001, respectively. Students’ t test: **, P < 0.01; ***, P < 0.001. Error bars indicate means ± SD. A, anterior; D, dorsal; P, posterior; V, ventral. Dashed lines outline the boundaries of retinas, lenses, and CMZs. Bars: (B, E, and F [main images]) 50 µm; (C) 20 µm; (F [insets], H, and I) 10 µm.
To examine the influence of blood vessels on RSCs, we found that transient treatment with 50 nM Ki8751, a potent and selective inhibitor of mammalian vascular endothelial growth factor receptor 2 (VEGFR2), for the specific time window between 38 and 44 hpf could selectively block the generation of the vessels surrounding the CMZ (Fig. 3 E and Video 3) with the minor influence on the development of the retinas and entire embryos (Fig. S3, A and B). In retinas lacking blood vessels, the CMZ shrank, and at 8 dpf the entire zone was reduced into a narrow band (Fig. 3 F). Meanwhile, eye size became significantly smaller in the Ki8751-treated group compared with that of wild type at 72 hpf onwards (n = 10; P < 0.001; Fig. 3, F and G). To quantify this CMZ reduction, we used a short pulse of BrdU, a nucleotide analogue incorporated into newly synthesized DNA of proliferating cells in the S phase, marked 10 ± 10 cells in the CMZ of Ki8751-treated animals compared with 76 ± 39 cells in the control (n = 18 slices from six eyes; P < 0.01; Fig. 3, H and J), suggesting the absence of mitotic cells. Consistent with BrdU results, there were significantly less phosphorylated histone H3 (pH3)-positive proliferating cells in Ki8751-treated group (4 ± 2 cells) compared with 14 ± 5 cells in the control (n = 18 slices from six eyes; P < 0.01; Fig. 3, I and J). No cell death was observed in the CMZ as a result of the vessel removal (n = 18 slices from six eyes; Fig. S3 C). Our results thus reveal a novel dependence of actively proliferating RSCs on local blood vessels.
Identification of SPs
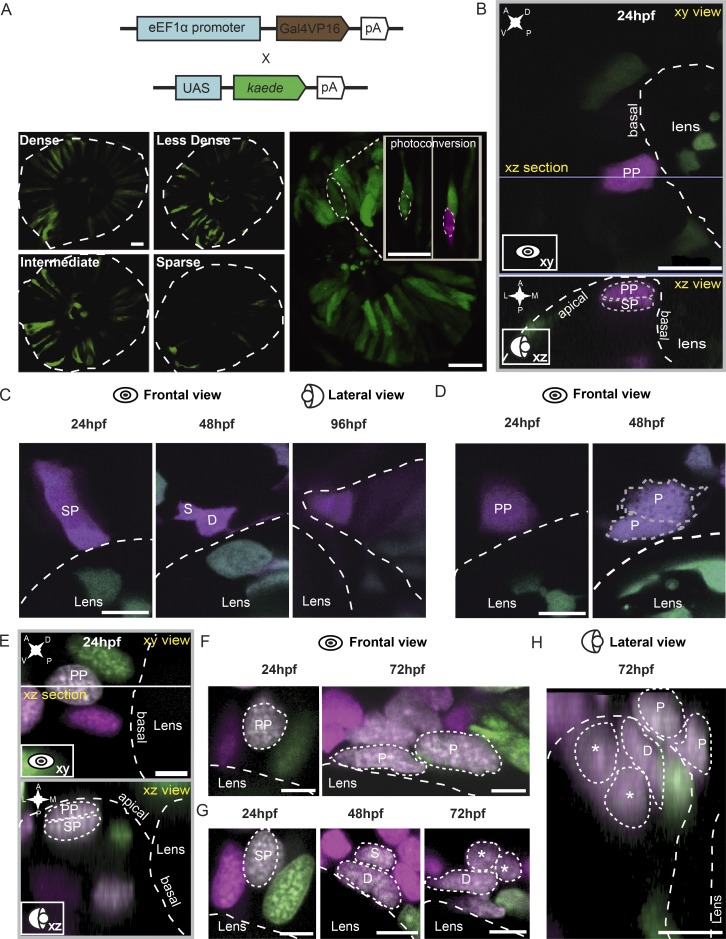
We next asked about the cell origin of RSCs. Mapping the early progenitors generating RSCs is very difficult because such progenitors are likely to constitute a very small portion within the progenitor pool. To overcome this, we used a mosaic strategy. The Tg(eEF1α:Gal4VP16::UAS:kaede) transgenic line produced mosaic labeling of early retinal progenitors with great variability (Fig. 4 A). Within densely labeled retinas, we were therefore able to mark a large number of individual progenitors using photoconverted Kaede protein at 24 hpf and follow them at 48 and 72 hpf, then retrospectively identifying ones giving rise to cell clusters of the CMZ (Fig. 4, B and C). A total of 231 progenitors were analyzed. Retrospective analysis revealed that the second layer of peripheral cells in the 24-hpf optic cups were SPs capable of giving rise to cell clusters of the CMZ tip, including dormant CMZ tip cells, RSCs, and retinal progenitors (n = 38; Fig. 4, B and C; and Video 4). In contrast, the first layer of peripheral cells produced RPE progenitors (PPs), eventually giving rise to pigmented cells that had the characteristics of dark pigmentation as well as typical pigmented cell morphology (Figs. 4 D and S4 A; Burgoyne et al., 2015).
Figure 4.
Identification of SPs. (A) Individual retinal progenitors were mosaically labeled to different levels in the transgenic fish Tg(eEF1α:Gal4VP16::UAS:kaede). At 24 hpf, individual cells were labeled by Kaede photoconversion. (B) Representative images of peripheral cells located in the first and second layers labeled by photoconverted Kaede in the 24 hpf optic cup from the frontal (xy) and lateral view (xz). (C) Representative images of a peripheral cell of the second layer (SP) dividing into one dormant CMZ tip cell (D) and one RSC (S) at 48 hpf and further generating the cell cluster of the CMZ tip at 96 hpf. (D) Representative images of a peripheral cell of the first layer (PP) dividing into two pigmented cells (P) at 48 hpf. (E) Representative images of peripheral cells of the first and second layers labeled by photoconverted Kaede in the 24-hpf optic cup from the frontal (xy) and lateral view (xz). (F) Representative images of a peripheral cell of the first layer (the same cell as in E) producing two pigmented cells at 72 hpf. (G) Representative images of a peripheral cell of the second layer dividing into one dormant CMZ tip cell and an RSC. The dormant cell did not divide further, whereas the RSC divided into two cells (marked by asterisks) at 72 hpf. (H) The dorsal view of the cells in F and G. Asterisks indicate two daughter cells derived from RSCs. White and gray dashed lines outline the boundaries of retinas, lenses, and cells. A, anterior; D, dorsal; L, lateral; M, medial; P, posterior; V, ventral. Bars: (A and B) 20 µm; (C–H) 5 µm.
To confirm the identity of SPs, we also introduced nlsKaede, a nucleus-localized photoconverted protein that conferred better imaging resolution to distinguish cells in 3D space. To create mosaic expression, nlsKaede mRNA was injected into the embryos at 4- to 32-cell stages. Using this new method, we analyzed 165 progenitors labeled at 24 hpf. Of these, 44 were SPs, which eventually gave rise to cell clusters of the CMZ at 72 hpf (Fig. 4, E, G, and H), whereas 17 were PPs capable of generating pigmented cells (Fig. 4, F and H; and Fig. S4, A and B). This analysis confirmed that cells of the first and second layer of the 24-hpf optic cup were PPs and SPs, respectively. Of note, SPs’ nuclei had an oval shape, in contrast to PPs, which had more rounded nuclei. This facilitated the identification of SPs from the peripheral retinas at 24 hpf (Fig. 4, F and G).
Lineage patterns of RSC generation
Next, we set out to derive the complete lineages of SPs. We therefore collected 82 complete lineages of individual SPs and found that the majority of them divided approximately every 24 h to give rise to cell clusters of the CMZ by 72 hpf (n = 76; Fig. 5, A–C and E). Detailed analysis showed that the cell cycle length of SPs ranged from 12 to 28 h (Fig. 6 I). In contrast, retinal progenitors of the central optic cup during the same developmental stage had a much shorter cell cycle length of ∼8 h (He et al., 2012). Therefore, SPs residing at the periphery of optic cups were a set of slowly cycling cells.
Figure 5.
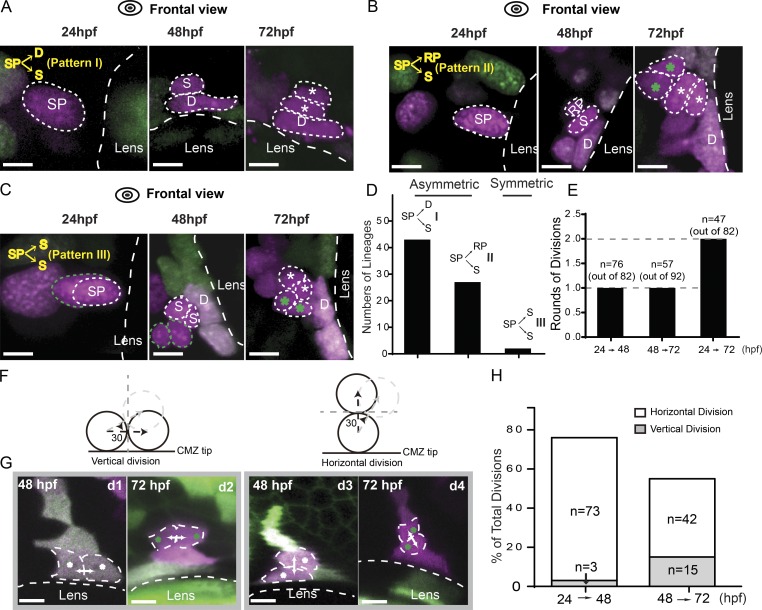
Lineage patterns of RSC generation. (A) Representative images of the lineage pattern I showing one SP dividing into one dormant CMZ tip cell (D) and one RSC (S) between 24 and 48 hpf. From 48 to 72 hpf, the dormant CMZ tip cell did not divide any more, whereas the RSC continued to proliferate into more cells (indicated by asterisks). (B) Representative images of the lineage pattern II showing that one SP divided into one RSC and one retinal progenitor (RP) from 24 to 48 hpf. Between 48 and 72 hpf, the RSC produced two more cells closer to one dormant CMZ tip cell, whereas the retinal progenitor gave rise to two cells (indicated by green asterisks) far away from the dormant cell. (C) One SP cell divided into two RSCs between 24 and 48 hpf, and both of them divided once during 48 to 72 hpf to form the CMZ tip region. (D) Plot for the numbers of different lineage patterns (n = 43, 27, and 2 from I–III). Patterns I and II were asymmetric, whereas Pattern III was symmetric. (E) Plot showing the numbers of cell divisions that SPs completed within various time windows, including 24–48 hpf, 48–72 hpf, and 24–72 hpf. On average, each cell finished one round of cell divisions every 24 h. (F) Schematics of vertical and horizontal divisions. (G) Representative images of vertical (d 1 and d2) and horizontal divisions (d3 and d4) at 48 and 72 hpf. The white arrows represent cell migration directions. (H) Plot showing the frequency of the vertical and horizontal divisions within 24–48 hpf and 48–72 hpf. Dashed lines outline the boundaries of the lens and cells. Bars, 5 µm.
Figure 6.
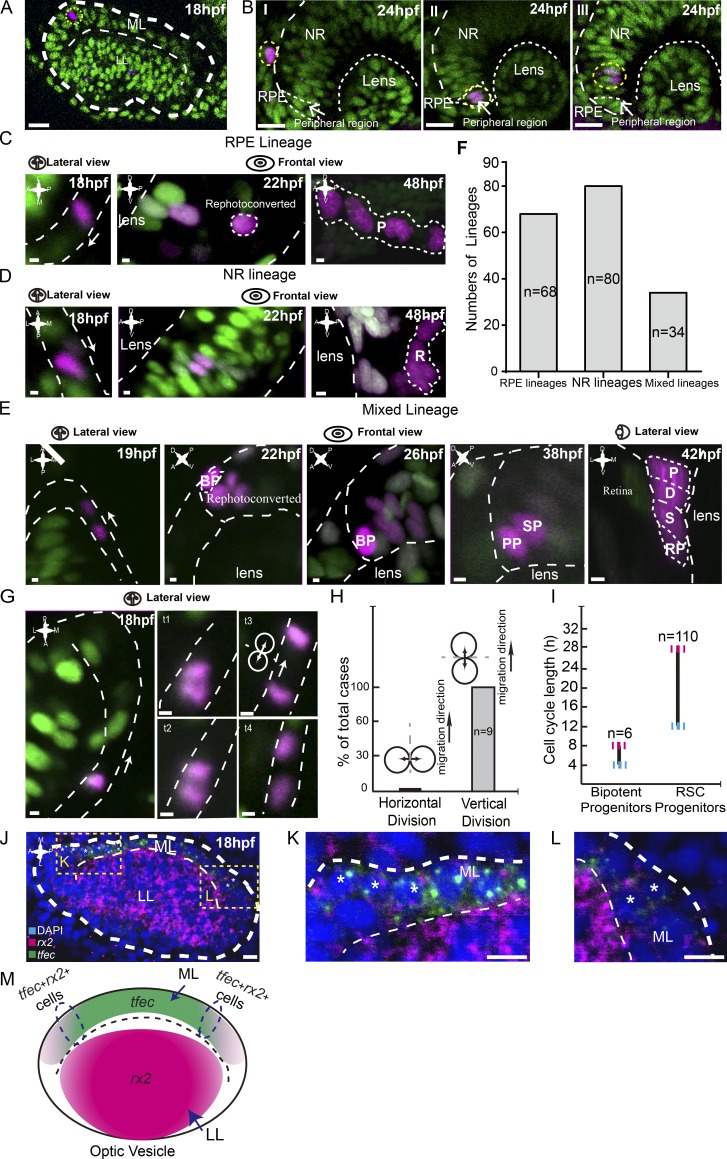
Bipotent progenitors as the embryonic origin of SPs. (A) An example of a single photoconverted cell (in magenta) in the ML of the optic vesicle at 18 hpf. Yellow dashed lines indicate traced clones. (B) Representative clones derived from single cells labeled at 18 hpf reached distinct final destinations of the optic cup at 24 hpf, including the RPE, peripheral region, and NR. (C–E) Three types of lineages originated from individual cells in the ML. RPE lineages (in C) contained RPE cells only (n = 68). NR lineages (in D) were composed of NR cells only (n = 80). Mixed lineages (in E) were composed of pigmented cells (P), dormant CMZ tip cells (D), RSCs (S), and retinal progenitors (RPs; n = 34). Mixed lineages were derived from bipotent cells (BPs). To distinguish from neighboring cells, bipotent cells were rephotoconverted in brighter red by exposure to the UV laser once more. (F) Plot of numbers of different lineages derived from individual 18-hpf cells in the ML, including RPE lineages, NR lineages, and mixed lineages. (G) Time lapse of the representative vertical division by the migrating cell in the ML. White arrows represent cell migration direction. (H) Plot showing all migrating cells in the ML divided vertically. (I) Plot showing that the cell cycle length of bipotent progenitors ranges from 4–8 h (n = 6), whereas SPs, the bipotent progenitors’ daughters, had a cell cycle length of between 12 and 28 h (n = 110). (J) Double FISH showing a set of cells in the ML expressing both rx2 and tfec, suggesting the mixed lineage state of RPE and RSCs. Yellow dashed lines indicate zoomed areas. (K and L) Zoomed images of the regions in J marked with yellow dashed squares. Asterisks indicate cells expressing both tfec and rx2 signals. (M) A schematic of the expression patterns of tfec and rx2 in the optic vesicle at 18 hpf. Some cells in the region (circled by dashed lines) express both genes. Green cells, nlsKaede; magenta cells, photoconverted nlsKaede; A, anterior; D, dorsal; L, lateral; M, medial; P, posterior; V, ventral. White dashed lines outline the boundaries of retinas, lenses, LLs, MLs, cells, and clones. The white arrows in C, D, E, and G indicate the migration directions of cells in the ML. Bars: (A and B) 20 µm; (C–E and G) 2.5 µm; (J–L) 10 µm.
We then asked how RSCs are produced within lineages. In the clonal dataset, we characterized three lineage patterns that were capable of producing RSCs (Fig. 5, A–C). The most frequently observed pattern involved one progenitor asymmetrically giving rise to one dormant CMZ tip cell and an RSC, which constituted ∼60% of all analyzed SP lineages (n = 43 out of 72 lineages; Fig. 5, A and D). A total of 38% of SP lineages involved a progenitor asymmetrically giving rise to one RSC and one retinal progenitor, which eventually differentiated into retinal cells (n = 27 out of 72 lineages; Fig. 5, B and D). Lineages in which an SP symmetrically gave rise to two RSCs were also observed at a very low frequency (n = 2 out of 72 lineages; Fig. 5, C and D). Additionally, 50 PP lineages were analyzed. These RPE produced two reproducible types of lineages. One included RPE giving rise to two pigmented cells (∼64%; n = 32; Figs. 7 and S4), and the other lineages involved a PP generating one pigmented cell and one dormant CMZ tip cell (∼36%; n = 18; Figs. 7 and S4 B).
Figure 7.
Working model for RSC lineage specification. Fast-cycling bipotent cells (BPs; in blue) of the ML in developing optic vesicles migrate and proliferate into one PP (in black) and one SP (in red) between 18 and 26 hpf. Subsequently, slow-cycling SPs proliferate and produce three types of lineages, including two asymmetric lineage types and one symmetric lineage type. Two asymmetric lineages were composed of one RSC (S; in red) and one dormant CMZ tip cell (D; in black/red; 60% of total RSC-generating lineages) or one RSC and a retinal progenitor (RP; in red; 38%). One symmetric lineage contained two RSCs (in red; 2%). Meanwhile, PPs proliferated and produced two types of lineages, including the lineages of two pigmented cells (Ps; in black; 64%) and the lineages of one pigmented cell (in black) and one CMZ dormant tip cell (36%). Note that dormant CMZ tip cells are produced by both PPs and SPs.
Because the fates of daughter cells have previously correlated with the axis of cell division (Agathocleous and Harris, 2009), we next asked whether cell division orientation affected lineage outcomes of SPs. RSCs progenitors were able to divide horizontally or vertically (Fig. 5 F). In horizontal divisions, the division axis was orthogonal to the circumference of the CMZ tip such that one daughter stayed at the original tip position while the other daughter cell moved away from the tip. However, vertical cell divisions gave rise to two daughter cells remaining side by side at the original tip position. According to the topology of sister cells’ position (Fig. 5, A–C), we examined whether horizontal divisions contributed to two asymmetric lineage patterns, with one RSC being born together with one dormant CMZ tip cell or one retinal progenitor (Fig. 5, A and B), whereas the symmetric lineages of two RSCs (Fig. 5 C) resulted from the vertical divisions. If this is the case, one would expect that the ratio of vertical divisions to horizontal divisions would predict the ratio of symmetric and asymmetric lineages derived from SPs. Our analysis showed that horizontal divisions constituted ∼96% of all divisions of SPs between 24 and 48 hpf (n = 73 out of 76; Fig. 5, G [d3] and H; and Fig. S5), whereas vertical divisions accounted for <4% (Fig. 5 G, d1). This agrees with our results showing that the majority of SPs produced asymmetric lineages composed of one RSC and one dormant CMZ tip cell or one retinal progenitor (∼97%; n = 70 out of 72), whereas only 3% lineages were symmetric with the production of two RSCs (n = 2 out of 72). This result indicates that vertical divisions and horizontal divisions resulted in symmetric and asymmetric lineages, respectively, and that SPs preferentially underwent horizontal cell divisions (∼97% of total cell divisions). Interestingly, the daughter cells of SPs subsequently underwent a shift toward more vertical cell divisions between 48 and 72 hpf (∼26% of total divisions; Fig. 5, G [d2 and d4] and H). This result suggests that RSCs might symmetrically expand the RSC pool through the vertical divisions shortly after their birth. Collectively, SPs were capable of producing RSCs through two types of cell divisions. Horizontal divisions gave rise to the asymmetric lineages wherein one progenitor produced one RSC together with a dormant CMZ tip cell or retinal progenitor, whereas vertical divisions resulted in symmetric lineages in which two RSCs were produced.
Bipotent progenitors as the embryonic origin of SPs
Finally, we asked how SPs were specified. The coincidence of labeled SPs and PPs in our analysis (Fig. 4, B and E) led us to examine whether the SPs shared a direct lineage origin with PPs.
During the formation of optic cups, embryonic retinal progenitors are distributed into two distinct zones, the LL and the ML, which finally develop into the NR and RPE, respectively (Heermann et al., 2015). Until 24 hpf, cells of the ML continue to migrate into the LL, thereby becoming CMZ cells (Kwan et al., 2012). To address the embryonic origin of SPs, we performed clonal analysis of individual cells in the ML marked with nlsKaede or H2BPSmOrange, the nucleus-localized photoconvertable proteins, starting at 18 hpf (n = 182; Fig. 6 A). Cells of the ML proliferated, migrated, and formed clones in different regions of the optic cup at 24 hpf, which included the RPE, NR, and, in particular, the peripheral region of optic cups where future RSCs reside (Fig. 6 B and Video 5). At 48 hpf, these cells produced three types of clones, including the lineages composed of pigmented cells only (RPE lineages; n = 68; Fig. 6, C and F), retinal cells only (NR lineages; n = 80; Fig. 6, D and F), and intriguingly, mixed lineages, which contained pigmented cells, dormant CMZ tip cells, RSCs, and their derivatives (mixed lineages; n = 34; Fig. 6, E and F). Of note, all RSCs were generated by such mixed lineages. The presence of mixed lineages indicates that a set of cells in the ML were bipotent, i.e., capable of producing two sister progenitors, one being an SP and the other being a PP (Fig. 6 E). Lineage tracing experiments also showed that the cell cycle length of these bipotent cells ranged from 4–8 h, which is much faster than that of their daughter cells, SPs (Figs. 5 E and 6 I). Time lapse analysis revealed that while migrating, all bipotent cells underwent a horizontal division, and as a result, the leading daughter progenitors became RSCs, whereas the following ones became PPs (Fig. 6, E, G, and H). To look into bipotent cells, we examined the expression of rx2 and tfec, genes specifically expressed in SPs and PPs, respectively, in the developing optic vesicles at 18 hpf. The rx2 gene was abundant in the LL and became weak and eventually disappeared in the ML, whereas the expression of tfec was the opposite (Fig. 6, J–M). Notably, there was a small subset of cells in the LL that expressed both genes (Fig. 6, J–M), suggesting that these bipotent cells were in mixed lineage states and retained the intrinsic potential to produce both RSC and RPC lineages.
Discussion
In this study, we describe the lineage development of RSCs in the zebrafish CMZ in its entirety. The results (Fig. 7) show that RSCs originate from a subset of migrating cells in the ML of developing optic vesicles at 18 hpf. These migrating cells are bipotent, capable of producing two types of progenitors, PPs and SPs. PPs are able to produce pigmented cells predominantly (∼82%) as well as some dormant CMZ tip cells (∼18%). However, SPs proliferate and differentiate into RSCs via three terminal lineage patterns, including one dormant CMZ tip cell and one RSC (∼60%), one retinal progenitor and one RSC (∼38%), and two RSCs (∼2%). Our analysis lays out a novel framework regarding the lineage progression of RSC generation during embryonic development.
Characterization of dormant CMZ tip cells
Using Zebrabow-based clonal analysis, we characterized the cell cluster at the CMZ tip region where RSCs reside. Within the cell cluster, our analysis revealed RSCs to be the second to third cells (Fig. 1) that grew continuous retinal clones that remained attached to the original positions. Previously, this clonal topology has been described as the gold standard to identify genuine RSCs in frogs and fish (Wetts and Fraser, 1988; Centanin et al., 2011, 2014; Wan et al., 2016).
Surprisingly, we also discovered dormant CMZ cells located as the very first cells within the CMZ tip region (Fig. 2 B). To our knowledge, these cells have not been analyzed previously, but our results suggest they might function as a critical part of the RSC niche. These cells shared a characteristic cell shape polarity, forming a cell layer at the most peripheral circumference of the retina proximal to the lens. In the sagittal view, they look prolonged, fusiform, and perpendicular to other CMZ cells in 3D space (Fig. 2 B), whereas they appeared rounded from a front view. Distinct from RSCs, these tip-most cells were not in direct contact with local blood vessels, which were apposed to the tip CMZ region.
An overarching characteristic of these tip cells was dormancy, as they were cycling extremely slowly. Indeed, we rarely observed any cell division during our experimental time window from 3–14 dpf (Fig. 1 C). Considering that the retina grows significantly over this time (Centanin et al., 2011), the dormant CMZ tip cells were certainly not an active contributor to retinal growth before 14 dpf. However, we cannot exclude an important role for these tip cells in maintaining RSCs, as they are direct neighbors in the CMZ. Also, it will be particularly interesting to assess whether the dormant CMZ tip cells observed here are the pigmented stem cells that have been described previously (Wetts and Fraser, 1988; Centanin et al., 2011).
The role of local blood vessels
In our analysis, another highlighted characteristic of the niche is its association with local blood vessels. Two rings of vessels surround the most peripheral circumference of the CMZ: the AHVs residing at the basal side of the CMZ tip region and the SAVs located at the side of the RPE (Kaufman et al., 2015). This spatial organization raises an interesting question as to whether the cells located between the two blood vessel rings provide niches for RSCs and pigmented stem cells. Interestingly, the vessels directly contact putative RSCs but not the dormant CMZ tip cells, suggesting a potential interaction specifically with the stem cells. In fact, the association of blood vessels and neural stem cells is a highly conserved phenotype in other central nervous system (CNS) niches, including the subventricular zone and the subgranular zone (Palmer et al., 2000; Bovetti et al., 2007; Mirzadeh et al., 2008; Shen et al., 2008; Tavazoie et al., 2008; Fuentealba et al., 2012; Gómez-Gaviro et al., 2012; Licht and Keshet, 2015; Otsuki and Brand, 2017). In culture assays, vessel endothelial cells facilitate the proliferation and differentiation of neural stem cells (Shen et al., 2004; Sun et al., 2010). In this study, we demonstrated that removal of local vessels using the drug Ki8751 dramatically reduces cell proliferation of CMZ cells as well as the CMZ size and results in smaller eyes. However, how the vascular cells interact with RSCs remains elusive. Single cell profiling of RSCs might reveal molecular cues underlying the cell–cell interaction.
Lineage specification of RSCs
How is the RSC lineage specified? In the current study, we first identified SPs as the cells located in the second most peripheral layer of cells in developing optic cups at 24 hpf. These RSCs gave rise to cell clusters of the CMZ tip region, including the dormant tip cells and RSCs. Inferring from lineage patterns of SPs (Fig. 7), the ratio of the number of RSCs to that of dormant CMZ tip cells was estimated to be ∼1:5 upon birth, which is in perfect agreement with the factor of ∼1:5 concluded from our topology analysis of dormant CMZ tip cells and RSCs (Fig. 2 C). This suggests that the balance in numbers of both cell types is achieved as early as their birth. However, we cannot exclude the possibility that the additional expansion of newborn RSCs through vertical divisions within a short period after RSC occurs, because an increase in vertical divisions was observed between 48–72 hpf (Fig. 5 H).
Intriguingly, our lineage analysis identifies a novel population of bipotent embryonic cells capable of generating both RSC lineages and RPE lineages as the origin of the SPs. The existence of such bipotent cells indicates a role for the mixed-lineage state having an ability to produce multiple cell lineages. In fact, the loss or significant reduction of the CMZ in many mutants often accompanies a significant increase in the pigment cells (Wehman et al., 2005). We speculate that a disturbance of lineage choices of these bipotent cells between RSCs and RPE might account for the phenotype of thickening the RPE in CMZ-deficit mutants. Recently, similar mixed-lineage states manifested in hematopoietic stem cells by single-cell RNA sequencing have been shown to be essential in cell lineage divergence and specification (Olsson et al., 2016). In addition, our findings highlight a role for the cell cycle in RSC lineage specification. The SPs are slowly cycling cells, which generated a four-cell clone from 24–72 hpf, whereas their embryonic counterparts in the developing retina divide every ∼6–10 h and form clones of 11–12 cells over the same development period (He et al., 2012). Moreover, we observed a dramatic slowdown of the cell cycle length from bipotent cells (∼4–8 h) to the SPs (∼12–28 h). In the mammalian CNS, slow cycling also has been shown to be a property of neural stem cell–producing precursors (Furutachi et al., 2015). Our findings suggest mixed-lineage states of bipotent cells as well as lengthening of the cell cycle as characteristics of lineage specification of RSCs in lower vertebrate retinas. Collectively, our anatomical and clonal data provide an insightful demonstration of embryonic development of RSC lineages and thereby lays out an integrated framework of the lineage basis of the production of a vertebrate CNS stem cell niche.
Materials and methods
Zebrafish maintenance and embryo manipulations
Zebrafish were maintained and bred at 27.5°C. Embryos were harvested and kept in the embryo medium (0.294 g/liter NaCl, 0.0127 g/liter KCl, 0.0485 g/liter CaCl2 · 2 H2O, 0.0813 g/liter MgSO4 · 7 H2O, 0.3 g/liter sea salt, and 2 × 10−4 g/liter methylene blue) at 28.5°C and staged in hpf. Embryos were treated with 0.003% phenylthiourea (Sigma-Aldrich) from 10 hpf to delay pigmentation and anaesthetized by 0.04% MS-222 (Sigma-Aldrich) before live imaging.
Transgenic lines and constructs
Tg(UAS:Zebrabow) was a gift from A. Pan (Georgia Regents University, Augusta, GA); Tg(eEF1α:GAL4VP16) was a gift from A. Fleming (University of Cambridge, Cambridge, England, UK); TG(rx2:GFPcaax) was a gift from J. Wittbrodt and L. Centamin (University of Heidelberg, Heidelberg, Germany); Tg(UAS:kaede) was a gift from K. O’Hare (University of Cambridge, Cambridge, England, UK); and Tg(actin:H2BGFP) and the multicolor transgenic line (Almeida et al., 2014) were gifts from W.A. Harris (University of Cambridge, Cambridge, England, UK). All other transgenic lines used in this study were generated by DNA injection at the one-cell stage.
To create a kdrl BAC construct, the BAC clone CH211-276G21 was obtained from the BACPAC Resources Center, and the BAC plasmid kdrl:mCherrycaax was constructed as described previously (Suster et al., 2011). In brief, the iTol2 cassette was amplified from the piTol2-Amp plasmid and introduced into the BAC DNA by the recombination reaction. The insertion plasmid of PIGCN21-mCherrycaax-PA-frt-neo-frt was constructed, and the 50-bp recombination arms were added by the PCR amplification. After DNA recombination in the SW105 bacterial strain, the neo selection maker was removed by 0.1% arabinose (Sigma-Aldrich) at 32°C for 1 h. All used primers are listed as followed: pTARBAC_iTol2 forward, 5′-GCGTAAGCGGGGCACATTTCATTACCTCTTTCTCCGCACCCGACATAGATCCCTGCTCGAGCCGGGCCCAAGTG-3′; pTARBAC_iTol2 reverse, 5′-GCGGGGCATGACTATTGGCGCGCCGGATCGATCCTTAATTAAGTCTACTAATTATGATCCTCTAGATCAGATCT-3′; left arm mCherry forward, 5′-TTATCATACATCAAGAGAAATTAAGTTCTGGACTACTGCAGCCGCATCCGCCACCATGGTGAGCAAGGGC-3′; and right arm PIGCN21R reverse, 5′-GTGCTTCTTGAGTGGTTAATTTACTATCAGAGTGGAGAACACTCACTCCTGAGGAGGCTTTTTTGGAGGCT-3′.
To create Tg(rx2-dsGFP), an rx2:dsGFP plasmid was constructed by replacing the GFPcaax DNA fragment with a destabilized GFP (dsGFP) DNA fragment in the plasmid rx2:GFPcaax. The dsGFP fragment was amplified from the plasmid pCAG-GFPd2 (Addgene) using the following primers: dsGFP forward (EcoRV), 5′-GCAGATATCGCCACCATGGTGAGCAAGGG-3′; and dsGFP reverse (NotI), 5′-ATAAGAATGCGGCCGCGAGTGCGGCCGCGCATCT-3′.
To create Tg(actin:H2BCFP), the actin:H2BCFP plasmid was constructed by using the gateway method. For mRNA synthesis, the plasmids of pCS2-H2BPSmOrange, pCS2-Cre, and pCS2-nlskaede were constructed.
mRNA injection
For synthesis mRNA, H2BPSmOrange, Cre, or nlskaede, the plasmids pCS2-H2BPSmOrange, pCS2-Cre, or pCS2-nlskaede were digested by the NotI enzyme, and the linearized DNA was used as the template for mRNA synthesis by using an mMESSAGE mMACHINE SP6 Transcription kit (AM1340M; Ambion).
Drug treatment
To remove the blood vessels surrounding the CMZ, embryos of Tg(rx2:GFPcaax::kdrl:mCherrycaax) were treated with 50 nM Ki8751 in 0.05% DMSO (SelleckChem) at 28.5°C from 38–44 hpf. The control group was treated with 0.05% DMSO from 38–44 hpf. After treatment, the embryos were washed with the embryo medium at least three times and raised in the medium at 28.5°C until desired time points for imaging analysis.
Single cell labeling and lineage analysis
To label CMZ cells, 150 pg H2BPSmOrange mRNA and 5 pg Cre mRNA were injected into Tg(eEF1α:Gal4VP16::UAS:Zebrabow) embryos at the single-cell stage. To label SPs and bipotent cells, 8 pg nlskaede mRNA was injected into AB embryos at 4- to 32-cell stages, and individual cells were labeled by the photoconversion of nlskaede at 18 or 24 hpf. Developing clones were examined at various time points, such as at 22, 26, and 48 hpf for the clones labeled at 18 hpf and at 24, 48, and 72 hpf for the ones labeled at 24 hpf. To label SPs using Tg(eEF1α:Gal4VP16::UAS:kaede), individual cells were labeled by the photoconversion of kaede at 24 hpf and followed at 48 and 72 hpf.
The photoconversion of kaede and nlskaede was conducted by the exposure to 405-nm lasers with 2–3 pulses (3 s; 5% laser power/pulse) on a confocal microscope (Olympus). To distinguish bipotent cells, a rephotoconversion procedure was performed to mark one sister cell more in red by the exposure to 405-nm lasers with 2–3 pulses (2 s; 5% laser power/pulse) once more.
FISH
The open reading frame fragments of rx2, mz98, and tfec were amplified and inserted into T vectors using the following primers: tfec forward, 5′-GAGAACTCCAAGTACCACCTCAAC-3′; tfec reverse, 5′-GAGATCGTCTGTTTCCTCTGAGC-3′; rx2 forward, 5′-ATGCATCTGTCACCAGATACC-3′; rx2 reverse, 5′-TTACATGGGTTGCCAGGTTT-3′; mz98 forward, 5′-TTTGTGGCGAATGCGGG-3′; and mz98 reverse, 5′-GAAGTTCTGTATCAATGTCAGCATC-3′.
The FITC-labeled rx2 antisense probe was prepared by using the T7 RNA Polymerase kit (Promega) and FITC RNA Labeling kit (Roche). The digoxigenin (DIG)-labeled mz98 and tfec antisense probes were prepared by using the T7 RNA Polymerase kit and DIG RNA Labeling kit (Roche). Zebrafish embryos (18 hpf or 5 dpf) were fixed in 4% PFA (Electron Microscopy Services) at 4°C overnight followed by the dehydration in 30% sucrose and then were sectioned for 20-µm-thick slices transversely. The slices were dried, fixed in 4% PFA at RT for 15 min and washed with PBS at RT for 3 min. To block the activity of the endogenous peroxidase, all slices were treated with 0.1% H2O2 at RT for 30 min. After being washed twice with PBS at RT for 3 min, slices were treated with 10 µg/ml proteinase K (Sigma-Aldrich) in TE (10 mM Tris, pH 8.0, and 1 mM EDTA, pH 8.0) at 37°C for 5–12 min followed by the treatment with 4% PFA at RT for 10 min. Subsequently, all slices were washed with PBS at RT for 3 min followed by the incubation in 0.2 M HCl at RT for 10 min. After being washed with PBS for 3 min, all slices were sequentially incubated with 0.1 M triethanol amine–HCl (662.5 µl triethanol amine and 1.35 ml 1 M HCl; adding water to the final volume of 50 ml, pH 8.0) at RT for 1 min and in 0.1 M triethanol amine–HCl containing 0.25% acetic anhydrate at RT for 10 min. After being washed in PBS at RT for 3 min, samples were dehydrated in a series of 60%, 80%, 95%, and twice in 100% ethanol at RT for 90 s. Slices were then incubated in the hybridization buffer (50% formamide [Sigma-Aldrich], 10 mM Tris-HCl, pH 8.0, 200 µg/ml yeast tRNA [Invitrogen], and 10% dextran sulfate [Ambion]) containing 1 µg/ml probes at 60°C overnight. On the next day, slices were washed sequentially with 5×SSC at 65°C for 30 min, 2×SSC with 50% formamide at 65°C for 30 min, TNE buffer (100 ml TNE consisting of 1 ml 1 M Tris-Cl, pH 7.5, 10 ml 5 M NaCl, and 0.2 ml 0.5 M EDTA) at 37°C for 10 min and then with TNE buffer with 20 µg/ml RNaseA at 37°C for 30 min. Subsequently, samples were washed with 2×SSC at 60°C for 20 min, 0.2×SSC at 60°C for 20 min, and 0.1×SSC at RT for 20 min. Next, samples were blocked by TN buffer at RT for 5 min (200 ml TN buffer consisting of 20 ml 1 M Tris, pH 7.5, 6 ml 5 M NaCl, and 174 ml water) followed by TNB buffer (TN buffer + 0.5% blocking reagent; Roche) at RT for 5 min. Finally, slices were incubated in TNB buffer with sheep anti–DIG-AP (1:1,000; Roche) and sheep anti–FITC-HRP (1:1,000; Roche) at 4°C overnight. On the third day, the signal was detected by the Tyramide Signal Amplification kit (Invitrogen) followed by the 2-hydroxy-3-naphtoic acid-2’-phenylamilide phosphate kit (Roche).
BrdU, pH3, and Tunel staining
For BrdU and pH3 labeling, the control and ki8751-treated Tg(rx2:GFPcaax::kdrl:mCherrycaax) embryos at 5 dpf were treated with 10 mM BrdU (5-bromo-20-deoxyuridine; B5002; Sigma-Aldrich) at 28.5°C for 30 min. After being rinsed with the embryo medium three times, embryos were fixed with 10% formalin (Sigma-Aldrich) at 4°C overnight. After dehydration, embedding and sectioning were performed according to the standard procedure. 20-µm-thick slices were sectioned. After being washed with 1×PBS for 10 min three times, the sections were treated with 2 M HCl at RT for 1 h followed by neutralization with 0.1 M sodium borate solution at RT for 10 min. The sections were washed with 1×PBS for 10 min for three times and permeabilized in 1×PBS with 0.5% Triton X-100 for 10 min. After the incubation with 5% BSA solution (Sigma-Aldrich) at RT for 1 h, samples were incubated with the rat anti-BrdU antibody (1:600; Abcam) and mouse anti-pH3 antibody (1:500; Abcam) at 4°C overnight. Samples were then washed with 1×PBS and then incubated with Alexa Fluor 647–conjugated goat anti–rat IgG antibody (1:1,000; Invitrogen) and Alexa Fluor 594–conjugated donkey anti–mouse IgG antibody (1:1,000; Jackson ImmunoResearch Laboratories, Inc.) at RT for 2 h. DAPI staining was performed according to the standard protocol. Slices were finally mounted using the fluorescent mounting medium (Sigma-Aldrich). The Tunel assay was performed according to the protocol of Tunel Apoptosis Detection kit (FITC; 40306ES50; Yeason).
BrdU-, pH3-, and Tunel-positive cells from three slices of one retina were counted, and six retinas were analyzed for each group. The second, fourth, and sixth sections from a given retina were selected for the analysis to avoid the sampling bias.
Wholemount immunohistology
Tg(rx2:dsGFP) zebrafish larva at 5 dpf treated with 0.003% phenylthiourea were fixed with 10% formalin at RT for 1 h followed by washing with 1×PTw (PBS + 0.1% Tween-20) three times for 5 min. The embryos were then soaked in the scale solution at 37°C overnight and washed once with 1×PTw for 5 min. After the following incubation in blocking buffer (10% sheep serum, 1% BSA, and 0.8% Triton X-100 in 1×PTw) at 4°C overnight, the embryos were incubated in the GFP antibody (1:200; Proteintech) at 4°C for 2 d. After being washed with 1×PTw three times, these embryos were incubated with Alexa Fluor 488–conjugated goat anti–rabbit secondary antibody (1:1,000; Invitrogen) at 4°C for 2 d. The embryos were then washed and embedded in 1% low melting point agarose (Sigma-Aldrich) for imaging.
Scale method
To make brain tissues transparent for imaging, the embryos were fixed with 10% formalin for 1 h at RT and soaked in the scale solution (20% glycerol, 0.1% Triton X-100, and 1 M urea; Hama et al., 2011) at 37°C overnight. Treated tissues were kept in the scale solution at 4°C until imaging.
Imaging acquisition and analysis
Image acquisition was performed on an inverted confocal microscope system (FV1200) or an upright two-photon microscope (Olympus). For fixed samples, embryos were embedded in 1% agarose. For live embryos, they were anaesthetized by 0.04% MS222 and subsequently embedded in 1% agarose with 0.04% MS222. Images were taken by using a 30× 1.05 NA or 60× 1.3 NA silicon oil objective (Olympus) with 488-nm and/or 559-nm lasers.
For time lapse from 18–24 hpf, Tg(actin:H2BGFP) embryos injected with H2BPSmOrange mRNA were embedded in 1% agarose with 0.04% MS222. The image dish was filled with embryonic medium, and z stack images were taken every 135 s at 28.5°C. For time lapse from 24–48 hpf, Tg(eEF1α:Gal4VP16::UAS:kaede) embryos at 24 hpf were anesthetized with 0.04% MS222 and mounted in 0.6% agarose. Z stack images were taken every 1 h until the cell division of individual photoconverted cells at 28.5°C.
Fluoview software (Olympus) and ImageJ (1.49k; National Institutes of Health) were used to process acquired 2D images. 3D image stacks were analyzed using Imaris software (Bitplane). To achieve standard CMZ structure, a slicer was created along the diameter of the lens but perpendicular to blood vessels surrounding the CMZ using the orthogonal camera in Imaris. The detailed 3D structure of CMZ cells was then inspected by rotating the slicer. Time-lapse videos and all animations were created using the Imaris Animation module.
Statistical analysis
Excel (Microsoft) was used for data analysis. All values are given as means ± SD. Statistical analysis was performed by Student’s t test (**, P < 0.01; ***, P < 0.001).
Online supplemental material
Fig. S1 shows the characteristics of RSCs. Fig. S2 shows the characteristics of dormant CMZ tip cells. Fig. S3 shows the effects of Ki8751 on the development of retinas. Fig. S4 shows lineage analysis of PPs. Fig. S5 shows the cell division of an SP. Video 1 shows 3D animation of an eye of the Tg(rx2:GFPcaax::kdrl:mCherrycaax). Video 2 shows 3D animation of the optical sections of the CMZ in the Tg(rx2:GFPcaax::kdrl:mCherrycaax). Video 3 shows 3D animation of an eye of the Tg(kdrl:mCherrycaax) treated with Ki8751. Video 4 shows 3D animation of a PP and an SP at the peripheral retina. Video 5 shows a time-lapse video of a migrating cell in the ML.
Supplementary Material
Acknowledgments
We thank Dr. Patricia Jusuf for careful editing of the manuscript. We also thank Professor William A. Harris and Dr. Owen Randlett for critical reading of the manuscript. We are grateful to Professor Joachim Wittbrodt and Dr. Lazaro Centanin for generously providing the rx2:GFPcaax plasmid. Thanks to Dr. Albert Pan for the transgenic line of UAS:Zebrabow and the advice on the use of this line. Thanks to Dr. Angeleen Fleming for the transgenic line Tg(eEF1α:Gal4VP16) and to Dr. Kathy O’Hare for the transgenic line Tg(UAS:kaede).
This project is supported by the National Natural Science Foundation of China (grant 31471042) and the China Thousand Talents Program.
The authors declare no competing financial interests.
Author contributions: X. Tang and Y. Zhang did the lineage analysis of SPs; X. Tang and J. Gao did the lineage analysis of bipotent cells; X. Tang with the assistance of W. Zhao and W. Pan examined the role of the blood vessels; J. He did Zebrabow-based clonal analysis; X. Jia and X. Tang did in situ experiments; and J. He did experimental designs and wrote the manuscript together with X. Tang.
Footnotes
Abbreviations used:
- AHV
- anterior hyaloid vessel
- CMZ
- ciliary marginal zone
- CNS
- central nervous system
- DIG
- digoxigenin
- dpf
- days post fertilization
- dsGFP
- destabilized GFP
- hpf
- hours post fertilization
- kdrl
- kinase insert domain receptor like
- LL
- lateral epithelial layer
- ML
- medial epithelial layer
- NR
- neural retina
- PP
- RPE progenitor
- RPE
- retinal pigment epithelium
- RSC
- retinal stem cell
- SAV
- superficial annular vessel
- SP
- RSC progenitor
References
- Agathocleous M., and Harris W.A.. 2009. From progenitors to differentiated cells in the vertebrate retina. Annu. Rev. Cell Dev. Biol. 25:45–69. 10.1146/annurev.cellbio.042308.113259 [DOI] [PubMed] [Google Scholar]
- Ahmad I., Tang L., and Pham H.. 2000. Identification of neural progenitors in the adult mammalian eye. Biochem. Biophys. Res. Commun. 270:517–521. 10.1006/bbrc.2000.2473 [DOI] [PubMed] [Google Scholar]
- Almeida A.D., Boije H., Chow R.W., He J., Tham J., Suzuki S.C., and Harris W.A.. 2014. Spectrum of Fates: a new approach to the study of the developing zebrafish retina. Development. 141:1971–1980. 10.1242/dev.104760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amato M.A., Arnault E., and Perron M.. 2004. Retinal stem cells in vertebrates: parallels and divergences. Int. J. Dev. Biol. 48:993–1001. 10.1387/ijdb.041879ma [DOI] [PubMed] [Google Scholar]
- Bélanger M.C., Robert B., and Cayouette M.. 2017. Msx1-positive progenitors in the retinal ciliary margin give rise to both neural and non-neural progenies in mammals. Dev. Cell. 40:137–150. 10.1016/j.devcel.2016.11.020 [DOI] [PubMed] [Google Scholar]
- Bhatia B., Singhal S., Lawrence J.M., Khaw P.T., and Limb G.A.. 2009. Distribution of Müller stem cells within the neural retina: Evidence for the existence of a ciliary margin-like zone in the adult human eye. Exp. Eye Res. 89:373–382. 10.1016/j.exer.2009.04.005 [DOI] [PubMed] [Google Scholar]
- Bhatia B., Jayaram H., Singhal S., Jones M.F., and Limb G.A.. 2011. Differences between the neurogenic and proliferative abilities of Müller glia with stem cell characteristics and the ciliary epithelium from the adult human eye. Exp. Eye Res. 93:852–861. 10.1016/j.exer.2011.09.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bovetti S., Hsieh Y.C., Bovolin P., Perroteau I., Kazunori T., and Puche A.C.. 2007. Blood vessels form a scaffold for neuroblast migration in the adult olfactory bulb. J. Neurosci. 27:5976–5980. 10.1523/JNEUROSCI.0678-07.2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgoyne T., O’Connor M.N., Seabra M.C., Cutler D.F., and Futter C.E.. 2015. Regulation of melanosome number, shape and movement in the zebrafish retinal pigment epithelium by OA1 and PMEL. J. Cell Sci. 128:1400–1407. 10.1242/jcs.164400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabochette P., and Vega-Lopez G.. 2015. YAP controls retinal stem cell DNA replication timing and genomic stability. eLife. 4:e08488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Centanin L., Hoeckendorf B., and Wittbrodt J.. 2011. Fate restriction and multipotency in retinal stem cells. Cell Stem Cell. 9:553–562. 10.1016/j.stem.2011.11.004 [DOI] [PubMed] [Google Scholar]
- Centanin L., Ander J.J., Hoeckendorf B., Lust K., Kellner T., Kraemer I., Urbany C., Hasel E., Harris W.A., Simons B.D., and Wittbrodt J.. 2014. Exclusive multipotency and preferential asymmetric divisions in post-embryonic neural stem cells of the fish retina. Development. 141:3472–3482. 10.1242/dev.109892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerveny K.L., Cavodeassi F., Turner K.J., de Jong-Curtain T.A., Heath J.K., and Wilson S.W.. 2010. The zebrafish flotte lotte mutant reveals that the local retinal environment promotes the differentiation of proliferating precursors emerging from their stem cell niche. Development. 137:2107–2115. 10.1242/dev.047753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerveny K.L., Varga M., and Wilson S.W.. 2012. Continued growth and circuit building in the anamniote visual system. Dev. Neurobiol. 72:328–345. 10.1002/dneu.20917 [DOI] [PubMed] [Google Scholar]
- Coles B.L., Angénieux B., Inoue T., Del Rio-Tsonis K., Spence J.R., McInnes R.R., Arsenijevic Y., and van der Kooy D.. 2004. Facile isolation and the characterization of human retinal stem cells. Proc. Natl. Acad. Sci. USA. 101:15772–15777. 10.1073/pnas.0401596101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- El Yakoubi W., Borday C., Hamdache J., Parain K., Tran H.T., Vleminckx K., Perron M., and Locker M.. 2012. Hes4 controls proliferative properties of neural stem cells during retinal ontogenesis. Stem Cells. 30:2784–2795. 10.1002/stem.1231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer A.J. 2005. Neural regeneration in the chick retina. Prog. Retin. Eye Res. 24:161–182. 10.1016/j.preteyeres.2004.07.003 [DOI] [PubMed] [Google Scholar]
- Fischer A.J., and Reh T.A.. 2000. Identification of a proliferating marginal zone of retinal progenitors in postnatal chickens. Dev. Biol. 220:197–210. 10.1006/dbio.2000.9640 [DOI] [PubMed] [Google Scholar]
- Fischer A.J., and Reh T.A.. 2001. Transdifferentiation of pigmented epithelial cells: A source of retinal stem cells? Dev. Neurosci. 23:268–276. 10.1159/000048710 [DOI] [PubMed] [Google Scholar]
- Fischer A.J., and Reh T.A.. 2002. Exogenous growth factors stimulate the regeneration of ganglion cells in the chicken retina. Dev. Biol. 251:367–379. 10.1006/dbio.2002.0813 [DOI] [PubMed] [Google Scholar]
- Fischer A.J., Hendrickson A., and Reh T.A.. 2001. Immunocytochemical characterization of cysts in the peripheral retina and pars plana of the adult primate. Invest. Ophthalmol. Vis. Sci. 42:3256–3263. [PubMed] [Google Scholar]
- Fischer A.J., Dierks B.D., and Reh T.A.. 2002. Exogenous growth factors induce the production of ganglion cells at the retinal margin. Development. 129:2283–2291. [DOI] [PubMed] [Google Scholar]
- Fischer A.J., Bosse J.L., and El-Hodiri H.M.. 2013. The ciliary marginal zone (CMZ) in development and regeneration of the vertebrate eye. Exp. Eye Res. 116:199–204. 10.1016/j.exer.2013.08.018 [DOI] [PubMed] [Google Scholar]
- Fischer A.J., Bosse J.L., and El-Hodiri H.M.. 2014. Reprint of: The ciliary marginal zone (CMZ) in development and regeneration of the vertebrate eye. Exp. Eye Res. 123:115–120. 10.1016/j.exer.2014.04.019 [DOI] [PubMed] [Google Scholar]
- Fouquet B., Weinstein B.M., Serluca F.C., and Fishman M.C.. 1997. Vessel patterning in the embryo of the zebrafish: Guidance by notochord. Dev. Biol. 183:37–48. 10.1006/dbio.1996.8495 [DOI] [PubMed] [Google Scholar]
- Fraser S.E., and Hunt R.K.. 1980. Retinotectal specificity: Models and experiments in search of a mapping function. Annu. Rev. Neurosci. 3:319–352. 10.1146/annurev.ne.03.030180.001535 [DOI] [PubMed] [Google Scholar]
- Fuentealba L.C., Obernier K., and Alvarez-Buylla A.. 2012. Adult neural stem cells bridge their niche. Cell Stem Cell. 10:698–708. 10.1016/j.stem.2012.05.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furutachi S., Miya H., Watanabe T., Kawai H., Yamasaki N., Harada Y., Imayoshi I., Nelson M., Nakayama K.I., Hirabayashi Y., and Gotoh Y.. 2015. Slowly dividing neural progenitors are an embryonic origin of adult neural stem cells. Nat. Neurosci. 18:657–665. 10.1038/nn.3989 [DOI] [PubMed] [Google Scholar]
- Gómez-Gaviro M.V., Lovell-Badge R., Fernández-Avilés F., and Lara-Pezzi E.. 2012. The vascular stem cell niche. J. Cardiovasc. Transl. Res. 5:618–630. 10.1007/s12265-012-9371-x [DOI] [PubMed] [Google Scholar]
- Gonzalez-Nunez V., Nocco V., and Budd A.. 2010. Characterization of drCol 15a1b: A novel component of the stem cell niche in the zebrafish retina. Stem Cells. 28:1399–1411. 10.1002/stem.461 [DOI] [PubMed] [Google Scholar]
- Hama H., Kurokawa H., Kawano H., Ando R., Shimogori T., Noda H., Fukami K., Sakaue-Sawano A., and Miyawaki A.. 2011. Scale: a chemical approach for fluorescence imaging and reconstruction of transparent mouse brain. Nat. Neurosci. 14:1481–1488. 10.1038/nn.2928 [DOI] [PubMed] [Google Scholar]
- Harris W.A., and Perron M.. 1998. Molecular recapitulation: the growth of the vertebrate retina. Int. J. Dev. Biol. 42:299–304. [PubMed] [Google Scholar]
- Hartsock A., Lee C., Arnold V., and Gross J.M.. 2014. In vivo analysis of hyaloid vasculature morphogenesis in zebrafish: A role for the lens in maturation and maintenance of the hyaloid. Dev. Biol. 394:327–339. 10.1016/j.ydbio.2014.07.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He J., Zhang G., Almeida A.D., Cayouette M., Simons B.D., and Harris W.A.. 2012. How variable clones build an invariant retina. Neuron. 75:786–798. 10.1016/j.neuron.2012.06.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heermann S., Schütz L., Lemke S., Krieglstein K., and Wittbrodt J.. 2015. Eye morphogenesis driven by epithelial flow into the optic cup facilitated by modulation of bone morphogenetic protein. eLife. 4:e05216 10.7554/eLife.05216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hitchcock P., Ochocinska M., Sieh A., and Otteson D.. 2004. Persistent and injury-induced neurogenesis in the vertebrate retina. Prog. Retin. Eye Res. 23:183–194. 10.1016/j.preteyeres.2004.01.001 [DOI] [PubMed] [Google Scholar]
- Holt C.E., Bertsch T.W., Ellis H.M., and Harris W.A.. 1988. Cellular determination in the Xenopus retina is independent of lineage and birth date. Neuron. 1:15–26. 10.1016/0896-6273(88)90205-X [DOI] [PubMed] [Google Scholar]
- Johns P.R. 1977. Growth of the adult goldfish eye. III. Source of the new retinal cells. J. Comp. Neurol. 176:343–357. 10.1002/cne.901760304 [DOI] [PubMed] [Google Scholar]
- Kaufman R., Weiss O., Sebbagh M., Ravid R., Gibbs-Bar L., Yaniv K., and Inbal A.. 2015. Development and origins of zebrafish ocular vasculature. BMC Dev. Biol. 15:18 10.1186/s12861-015-0066-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitambi S.S., McCulloch K.J., Peterson R.T., and Malicki J.J.. 2009. Small molecule screen for compounds that affect vascular development in the zebrafish retina. Mech. Dev. 126:464–477. 10.1016/j.mod.2009.01.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiyama T., Li H., Gupta M., Lin Y.P., Chuang A.Z., Otteson D.C., and Wang S.W.. 2012. Distinct neurogenic potential in the retinal margin and the pars plana of mammalian eye. J. Neurosci. 32:12797–12807. 10.1523/JNEUROSCI.0118-12.2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubo F., and Nakagawa S.. 2009. Hairy1 acts as a node downstream of Wnt signaling to maintain retinal stem cell-like progenitor cells in the chick ciliary marginal zone. Development. 136:1823–1833. 10.1242/dev.029272 [DOI] [PubMed] [Google Scholar]
- Kubota R., Hokoc J.N., Moshiri A., McGuire C., and Reh T.A.. 2002. A comparative study of neurogenesis in the retinal ciliary marginal zone of homeothermic vertebrates. Brain Res. Dev. Brain Res. 134:31–41. 10.1016/S0165-3806(01)00287-5 [DOI] [PubMed] [Google Scholar]
- Kuwahara A., Ozone C., Nakano T., Saito K., Eiraku M., and Sasai Y.. 2015. Generation of a ciliary margin-like stem cell niche from self-organizing human retinal tissue. Nat. Commun. 6:6286 10.1038/ncomms7286 [DOI] [PubMed] [Google Scholar]
- Kwan K.M., Otsuna H., Kidokoro H., Carney K.R., Saijoh Y., and Chien C.B.. 2012. A complex choreography of cell movements shapes the vertebrate eye. Development. 139:359–372. 10.1242/dev.071407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z., Joseph N.M., and Easter S.S. Jr. 2000. The morphogenesis of the zebrafish eye, including a fate map of the optic vesicle. Dev. Dyn. 218:175–188. [DOI] [PubMed] [Google Scholar]
- Liao W., Bisgrove B.W., Sawyer H., Hug B., Bell B., Peters K., Grunwald D.J., and Stainier D.Y.. 1997. The zebrafish gene cloche acts upstream of a flk-1 homologue to regulate endothelial cell differentiation. Development. 124:381–389. [DOI] [PubMed] [Google Scholar]
- Licht T., and Keshet E.. 2015. The vascular niche in adult neurogenesis. Mech. Dev. 138:56–62. 10.1016/j.mod.2015.06.001 [DOI] [PubMed] [Google Scholar]
- Marcucci F., Murcia-Belmonte V., Wang Q., Coca Y., Ferreiro-Galve S., Kuwajima T., Khalid S., Ross M.E., Mason C., and Herrera E.. 2016. The ciliary margin zone of the mammalian retina generates retinal ganglion cells. Cell Reports. 17:3153–3164. 10.1016/j.celrep.2016.11.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcus R.C., Delaney C.L., and Easter S.S. Jr. 1999. Neurogenesis in the visual system of embryonic and adult zebrafish (Danio rerio). Vis. Neurosci. 16:417–424. 10.1017/S095252389916303X [DOI] [PubMed] [Google Scholar]
- Martínez-Navarrete G.C., Angulo A., Martín-Nieto J., and Cuenca N.. 2008. Gradual morphogenesis of retinal neurons in the peripheral retinal margin of adult monkeys and humans. J. Comp. Neurol. 511:557–580. 10.1002/cne.21860 [DOI] [PubMed] [Google Scholar]
- Miesfeld J.B., Gestri G., Clark B.S., Flinn M.A., Poole R.J., Bader J.R., Besharse J.C., Wilson S.W., and Link B.A.. 2015. Yap and Taz regulate retinal pigment epithelial cell fate. Development. 142:3021–3032. 10.1242/dev.119008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirzadeh Z., Merkle F.T., Soriano-Navarro M., Garcia-Verdugo J.M., and Alvarez-Buylla A.. 2008. Neural stem cells confer unique pinwheel architecture to the ventricular surface in neurogenic regions of the adult brain. Cell Stem Cell. 3:265–278. 10.1016/j.stem.2008.07.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moshiri A., and Reh T.A.. 2004. Persistent progenitors at the retinal margin of ptc+/- mice. J. Neurosci. 24:229–237. 10.1523/JNEUROSCI.2980-03.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moshiri A., Close J., and Reh T.A.. 2004. Retinal stem cells and regeneration. Int. J. Dev. Biol. 48:1003–1014. 10.1387/ijdb.041870am [DOI] [PubMed] [Google Scholar]
- Moshiri A., McGuire C.R., and Reh T.A.. 2005. Sonic hedgehog regulates proliferation of the retinal ciliary marginal zone in posthatch chicks. Dev. Dyn. 233:66–75. 10.1002/dvdy.20299 [DOI] [PubMed] [Google Scholar]
- Negishi K., Teranishi T., and Kato S.. 1982. New dopaminergic and indoleamine-accumulating cells in the growth zone of goldfish retinas after neurotoxic destruction. Science. 216:747–749. 10.1126/science.7079736 [DOI] [PubMed] [Google Scholar]
- Olsson A., Venkatasubramanian M., Chaudhri V.K., Aronow B.J., Salomonis N., Singh H., and Grimes H.L.. 2016. Single-cell analysis of mixed-lineage states leading to a binary cell fate choice. Nature. 537:698–702. 10.1038/nature19348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otsuki L., and Brand A.H.. 2017. The vasculature as a neural stem cell niche. Neurobiol. Dis. In press. [DOI] [PubMed] [Google Scholar]
- Otteson D.C., and Hitchcock P.F.. 2003. Stem cells in the teleost retina: persistent neurogenesis and injury-induced regeneration. Vision Res. 43:927–936. 10.1016/S0042-6989(02)00400-5 [DOI] [PubMed] [Google Scholar]
- Palmer T.D., Willhoite A.R., and Gage F.H.. 2000. Vascular niche for adult hippocampal neurogenesis. J. Comp. Neurol. 425:479–494. [DOI] [PubMed] [Google Scholar]
- Parain K., Mazurier N., Bronchain O., Borday C., Cabochette P., Chesneau A., Colozza G., El Yakoubi W., Hamdache J., Locker M., et al. . 2012. A large scale screen for neural stem cell markers in Xenopus retina. Dev. Neurobiol. 72:491–506. 10.1002/dneu.20973 [DOI] [PubMed] [Google Scholar]
- Perron M., and Harris W.A.. 2000. Retinal stem cells in vertebrates. BioEssays. 22:685–688. [DOI] [PubMed] [Google Scholar]
- Raymond P.A., Barthel L.K., Bernardos R.L., and Perkowski J.J.. 2006. Molecular characterization of retinal stem cells and their niches in adult zebrafish. BMC Dev. Biol. 6:36 10.1186/1471-213X-6-36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reh T.A., and Fischer A.J.. 2001. Stem cells in the vertebrate retina. Brain Behav. Evol. 58:296–305. 10.1159/000057571 [DOI] [PubMed] [Google Scholar]
- Reh T.A., and Fischer A.J.. 2006. Retinal stem cells. Methods Enzymol. 419:52–73. 10.1016/S0076-6879(06)19003-5 [DOI] [PubMed] [Google Scholar]
- Reh T.A., and Levine E.M.. 1998. Multipotential stem cells and progenitors in the vertebrate retina. J. Neurobiol. 36:206–220. [DOI] [PubMed] [Google Scholar]
- Reinhardt R., Centanin L., Tavhelidse T., Inoue D., Wittbrodt B., Concordet J.P., Martinez-Morales J.R., and Wittbrodt J.. 2015. Sox2, Tlx, Gli3, and Her9 converge on Rx2 to define retinal stem cells in vivo. EMBO J. 34:1572–1588. 10.15252/embj.201490706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitt E.A., and Dowling J.E.. 1994. Early eye morphogenesis in the zebrafish, Brachydanio rerio. J. Comp. Neurol. 344:532–542. 10.1002/cne.903440404 [DOI] [PubMed] [Google Scholar]
- Shen Q., Goderie S.K., Jin L., Karanth N., Sun Y., Abramova N., Vincent P., Pumiglia K., and Temple S.. 2004. Endothelial cells stimulate self-renewal and expand neurogenesis of neural stem cells. Science. 304:1338–1340. 10.1126/science.1095505 [DOI] [PubMed] [Google Scholar]
- Shen Q., Wang Y., Kokovay E., Lin G., Chuang S.M., Goderie S.K., Roysam B., and Temple S.. 2008. Adult SVZ stem cells lie in a vascular niche: A quantitative analysis of niche cell-cell interactions. Cell Stem Cell. 3:289–300. 10.1016/j.stem.2008.07.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spence J.R., Madhavan M., Ewing J.D., Jones D.K., Lehman B.M., and Del Rio-Tsonis K.. 2004. The hedgehog pathway is a modulator of retina regeneration. Development. 131:4607–4621. 10.1242/dev.01298 [DOI] [PubMed] [Google Scholar]
- Straznicky K., and Gaze R.M.. 1971. The growth of the retina in Xenopus laevis: an autoradiographic study. J. Embryol. Exp. Morphol. 26:67–79. [PubMed] [Google Scholar]
- Sumoy L., Keasey J.B., Dittman T.D., and Kimelman D.. 1997. A role for notochord in axial vascular development revealed by analysis of phenotype and the expression of VEGR-2 in zebrafish flh and ntl mutant embryos. Mech. Dev. 63:15–27. 10.1016/S0925-4773(97)00671-0 [DOI] [PubMed] [Google Scholar]
- Sun J., Zhou W., Ma D., and Yang Y.. 2010. Endothelial cells promote neural stem cell proliferation and differentiation associated with VEGF activated Notch and Pten signaling. Dev. Dyn. 239:2345–2353. 10.1002/dvdy.22377 [DOI] [PubMed] [Google Scholar]
- Suster M.L., Abe G., Schouw A., and Kawakami K.. 2011. Transposon-mediated BAC transgenesis in zebrafish. Nat. Protoc. 6:1998–2021. 10.1038/nprot.2011.416 [DOI] [PubMed] [Google Scholar]
- Tavazoie M., Van der Veken L., Silva-Vargas V., Louissaint M., Colonna L., Zaidi B., Garcia-Verdugo J.M., and Doetsch F.. 2008. A specialized vascular niche for adult neural stem cells. Cell Stem Cell. 3:279–288. 10.1016/j.stem.2008.07.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Todd L., Suarez L., Squires N., Zelinka C.P., Gribbins K., and Fischer A.J.. 2016. Comparative analysis of glucagonergic cells, glia, and the circumferential marginal zone in the reptilian retina. J. Comp. Neurol. 524:74–89. 10.1002/cne.23823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tropepe V., Coles B.L., Chiasson B.J., Horsford D.J., Elia A.J., McInnes R.R., and van der Kooy D.. 2000. Retinal stem cells in the adult mammalian eye. Science. 287:2032–2036. 10.1126/science.287.5460.2032 [DOI] [PubMed] [Google Scholar]
- Turner D.L., and Cepko C.L.. 1987. A common progenitor for neurons and glia persists in rat retina late in development. Nature. 328:131–136. 10.1038/328131a0 [DOI] [PubMed] [Google Scholar]
- Wan Y., Almeida A.D., Rulands S., Chalour N., Muresan L., Wu Y., Simons B.D., He J., and Harris W.A.. 2016. The ciliary marginal zone of the zebrafish retina: clonal and time-lapse analysis of a continuously growing tissue. Development. 143:1099–1107. 10.1242/dev.133314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wehman A.M., Staub W., Meyers J.R., Raymond P.A., and Baier H.. 2005. Genetic dissection of the zebrafish retinal stem-cell compartment. Dev. Biol. 281:53–65. 10.1016/j.ydbio.2005.02.010 [DOI] [PubMed] [Google Scholar]
- Wetts R., and Fraser S.E.. 1988. Multipotent precursors can give rise to all major cell types of the frog retina. Science. 239:1142–1145. 10.1126/science.2449732 [DOI] [PubMed] [Google Scholar]
- Wetts R., Serbedzija G.N., and Fraser S.E.. 1989. Cell lineage analysis reveals multipotent precursors in the ciliary margin of the frog retina. Dev. Biol. 136:254–263. 10.1016/0012-1606(89)90146-2 [DOI] [PubMed] [Google Scholar]
- Willbold E., and Layer P.G.. 1992. A hidden retinal regenerative capacity from the chick ciliary margin is reactivated in vitro, that is accompanied by down-regulation of butyrylcholinesterase. Eur. J. Neurosci. 4:210–220. 10.1111/j.1460-9568.1992.tb00869.x [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.