The coat protein complex II (COPII) is essential for the secretion of large cargo, such as procollagen I (PC1), but evidence that COPII vesicles act as PC1 transport carriers from the ER was lacking. Using high-resolution microscopy and in vitro reconstituted vesicle budding assays, Gorur et al. show that COPII vesicles carry PC1.
Abstract
The coat protein complex II (COPII) is essential for the transport of large cargo, such as 300-nm procollagen I (PC1) molecules, from the endoplasmic reticulum (ER) to the Golgi. Previous work has shown that the CUL3-KLHL12 complex increases the size of COPII vesicles at ER exit sites to more than 300 nm in diameter and accelerates the secretion of PC1. However, the role of large COPII vesicles as PC1 transport carriers was not unambiguously demonstrated. In this study, using stochastic optical reconstruction microscopy, correlated light electron microscopy, and live-cell imaging, we demonstrate the existence of mobile COPII-coated vesicles that completely encapsulate the cargo PC1 and are physically separated from ER. We also developed a cell-free COPII vesicle budding reaction that reconstitutes the capture of PC1 into large COPII vesicles. This process requires COPII proteins and the GTPase activity of the COPII subunit SAR1. We conclude that large COPII vesicles are bona fide carriers of PC1.
Introduction
As an essential step in conventional protein secretion, coat protein complex II (COPII) mediates vesicular transport from the ER to the Golgi apparatus in eukaryotes. The GTPase SAR1, inner coat proteins SEC23/SEC24, and outer coat proteins SEC13/SEC31 are five cytosolic components of the COPII complex, and they are sufficient to generate COPII-coated vesicles from synthetic liposomes (Matsuoka et al., 1998; Kim et al., 2005). COPII vesicles were observed by EM to be ∼60–80 nm in diameter, which potentially limits the transport of large cargos such as the 300-nm-long procollagen I (PC1) rigid rod (Bächinger et al., 1982; Barlowe et al., 1994; Kim et al., 2005; Noble et al., 2013). However, human genetic evidence showed that COPII is required to secrete procollagens. Mutations in genes that code for the human COPII paralogs SEC23A and SEC24D were identified as causing the genetic diseases cranio-lenticulo-sutural dysplasia and osteogenesis imperfecta and their characteristic collagen deposition defects during development (Boyadjiev et al., 2006; Kim et al., 2012; Garbes et al., 2015).
The requirement for COPII to secrete PC has been independently demonstrated in multiple model systems. Mutation of the sec-23 gene in Caenorhabditis elegans disrupts collagen secretion and leads to aberrant cuticle, dissociated hypodermal cells, and late embryonic lethality (Roberts et al., 2003). In Drosophila melanogaster, tissue-specific knockdown of Sar1 or Sec23 in collagen-secreting fat body cells leads to intracellular accumulation of collagen and cell lethality (Pastor-Pareja and Xu, 2011). The zebrafish mutants crusher and bulldog result from mutations in sec23a and sec24d genes, respectively, and their chondrocytes retain procollagen II in the ER. These mutants also show defects during craniofacial development, with phenotypes reminiscent of human cranio-lenticulo-sutural dysplasia (Lang et al., 2006; Sarmah et al., 2010). Sec23A-null mice are embryonically lethal, and skin fibroblasts accumulate ER-localized collagen I and III (Zhu et al., 2015). Knockdown of SEC13 in primary human dermal fibroblasts also selectively blocks PC1 secretion (Townley et al., 2008). Hence, the requirement for COPII in the ER exit of PC is evolutionarily conserved in metazoans.
The necessary role of COPII in large-cargo secretion is further supported by the discovery of a large transmembrane protein, TANGO1 (MIA3), which has been shown to have a general role in the secretion of large cargos, including many members of the collagen family, laminin, and large lipoprotein complexes such as prechylomicrons (Saito et al., 2009; Wilson et al., 2011; Petley-Ragan et al., 2016; Santos et al., 2016). The luminal Src homology 3 domain of TANGO1 interacts with the PC-specific chaperone HSP47 to recognize a broad range of PC isoforms (Saito et al., 2009; Ishikawa et al., 2016). The cytosolic side of TANGO1 was shown to interact with multiple COPII components: its proline-rich domain binds to the inner COPII coat protein SEC23 directly, and its second coiled-coil domain recruits cTAGE5, a spliced variant of a TANGO1 isoform, which binds SEC12, an initiating factor of COPII assembly (Saito et al., 2009, 2011, 2014; Ma and Goldberg, 2016). Therefore, TANGO1 plays an important role in coordinating large-cargo sensing and COPII recruitment, which further supports the involvement of COPII in large-cargo secretion.
Although the requirement for COPII to export the large-cargo PC out of the ER is clear, the precise role that COPII plays in this process is poorly understood. A conventional model was proposed in which COPII concentrates large cargos at ER exit sites (ERESs) and orchestrates the packaging of large cargos into vesicles and the formation of vesicles with structured coats (Fromme and Schekman, 2005). An alternative model suggests that COPII functions only to concentrate large cargos and other factors required for the ER export at ERESs, and large cargos exit the ER in carriers not coated with COPII proteins (Mironov et al., 2003; Siddiqi et al., 2003, 2010).
The conventional model is paradoxical unless a cellular mechanism exists to increase the size of COPII-coated vesicles. The Rape and Schekman laboratories previously reported that the E3 ubiquitin ligase, CUL3, and one of its substrate adaptors, KLHL12, regulate the size of COPII-coated vesicles and collagen I and IV secretion (Jin et al., 2012). Overexpression of KLHL12 induces the formation of large COPII structures that are decorated with KLHL12. Most of these structures are more than 300 nm in diameter and large enough to accommodate cargo of the size of PC1. Unfortunately, the coincident localization of PC1 in these large carriers was not clearly elucidated in that work. Here, we reexamined these large COPII structures and showed that they are bona fide membranous carriers of PC1 in cells. Moreover, we reconstituted PC1 capture into vesicles formed in a cell-free reaction. These carriers were isolated and visualized as large COPII-coated vesicles.
Results
Large COPII vesicles colocalize with PC1
The ubiquitin ligase CUL3 and its substrate adaptor KLHL12 were shown to regulate collagen secretion and the size of COPII vesicles, possibly through the monoubiquitylation of the COPII protein SEC31A (Jin et al., 2012). To study whether the large COPII vesicles observed after overexpression of KLHL12 are collagen carriers, we engineered human fibrosarcoma cells (KI6) to stably overexpress the human pro-α1(I) collagen and inducible KLHL12 under doxycycline-controlled transcriptional activation. KI6 cells synthesized PC1 in contrast to the parent cell line as confirmed by an immunofluorescence (IF) signal for PC1 (Fig. S1 A) and immunogold-labeled PC1 in the ER and large vesicles (Fig. S1 B). KLHL12 overexpression was reported to accelerate the ER export of PC1: more cells had secreted PC1 after 30 min of ascorbate treatment when KLHL12 was overexpressed, and no significant difference was observed at a later time point (1 h) after ascorbate treatment (Jin et al., 2012). Consistent with our previous study, we also observed accelerated ER-to-Golgi trafficking of PC1 in KI6 cells 20 min after ascorbate treatment in cells that had induced KLHL12 overexpression for 7.5 h (Fig. S1 C; Jin et al., 2012). KI6 cells secreted PC1 through the conventional secretory pathway, shown by the inhibitory effect of brefeldin A (BFA) on the export of PC1 into the culture medium (Fig. S1 D). Because TANGO1-knockout mice are defective in collagen I secretion, we knocked down TANGO1 in KI6 cells and observed a decreased level of PC1 secretion, as detected by immunoblots of the culture medium, with a corresponding intracellular accumulation of PC1 detected in cell lysates (Fig. S1 E; Wilson et al., 2011).
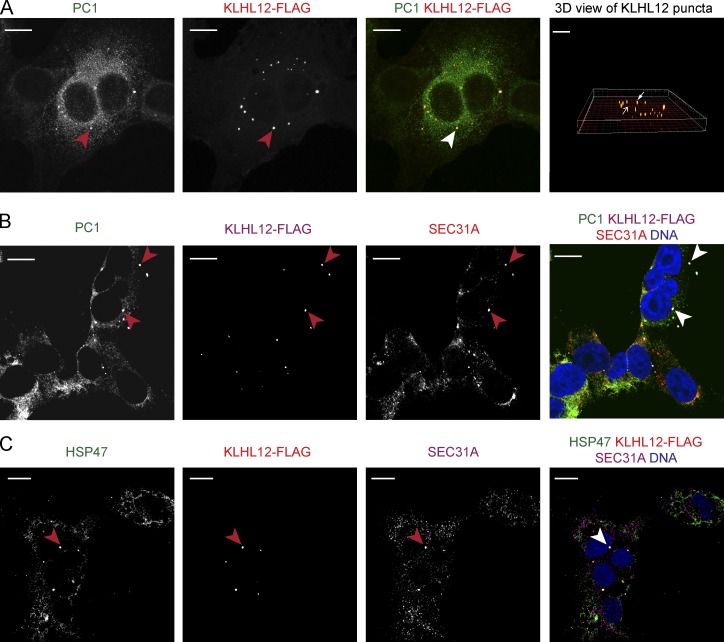
In our previous study (Jin et al., 2012) using a polyclonal antibody (LF-67) raised against synthetic human α1(I) collagen C-telopeptide (Bernstein et al., 1996), we observed partial colocalization between PC1 and KLHL12 but could not unambiguously document that the large COPII vesicles, generated by overexpression of KLHL12, carried PC1. In contrast, with the use of two monoclonal antibodies raised against N- or C-propeptides of PC1 (Fig. S2 A; Foellmer et al., 1983), we observed clear colocalization of PC1 and KLHL12 in KI6 cells after 7.5 h of induction (Figs. 1 A and S2 B). Puncta positive for both PC1 and KLHL12 also colocalized with SEC31A, a subunit of the outer coat of COPII vesicles (Fig. 1 B).
Figure 1.
Large COPII vesicles colocalize with PC1. (A) PC1 (green) and KLHL12 (red) colocalize in KI6 cells that were treated with doxycycline for 7.5 h to induce the expression of KLHL12. Images shown are mean intensity projected views of a z-stack shown for each and merged channel. Red arrowheads indicate KLHL12-positive puncta, whereas white arrowheads indicate colocalized puncta. Rightmost panel shows 3D view of KLHL12 puncta obtained from the z-stack using the Spot feature in Imaris 8.1.2. An example of colocalizing yellow puncta is indicated by a white arrowhead, and KLHL12-positive puncta not colocalizing with PC1 is indicated by a red arrowhead. Total number of KLHL12-positive puncta determined in the z-stack was 27, and 78% of these spots were positive for PC1 calculated using CoLoc feature in Imaris 8.1.2. (B) PC1 (green), KLHL12 (magenta), and SEC31A (red) colocalize in KI6 cells (white arrowhead). DAPI is shown in blue. Half of the total number of KLHL12-positive puncta (n = 14) were positive for PC1 and SEC31A. (C) HSP47 (green), KLHL12 (red), and SEC31A (magenta) colocalize in KI6 cells (white arrowhead). Of the total number of KLHL12-positive puncta (n = 24) counted in 11 cells, 79% were positive for both SEC31 and HSP47. Bars, 10 µm. n = 3.
We reasoned that if large COPII vesicles are functional secretory transporters of collagen, they should also contain HSP47, a collagen-specific chaperone. HSP47 binds to the triple helical region of PC in the ER and promotes the correct folding of trimerized PC (Nagai et al., 2000; Tasab et al., 2000, 2002; Ishida and Nagata, 2011; Ono et al., 2012; Widmer et al., 2012). HSP47 accompanies PC to the ER–Golgi intermediate compartment (ERGIC) or cis-Golgi membrane, where it dissociates because of a lower pH, after which it is recycled back to the ER via its C-terminal RDEL sequence (Satoh et al., 1996; Oecal et al., 2016). We found that HSP47 colocalized with the large COPII vesicles as visualized by triple IF labeling of HSP47, SEC31A, and KLHL12 in KI6 cells, consistent with a role for the large COPII vesicles as secretory carriers of PC1 (Fig. 1 C).
Large COPII vesicles are hollow membranous containers
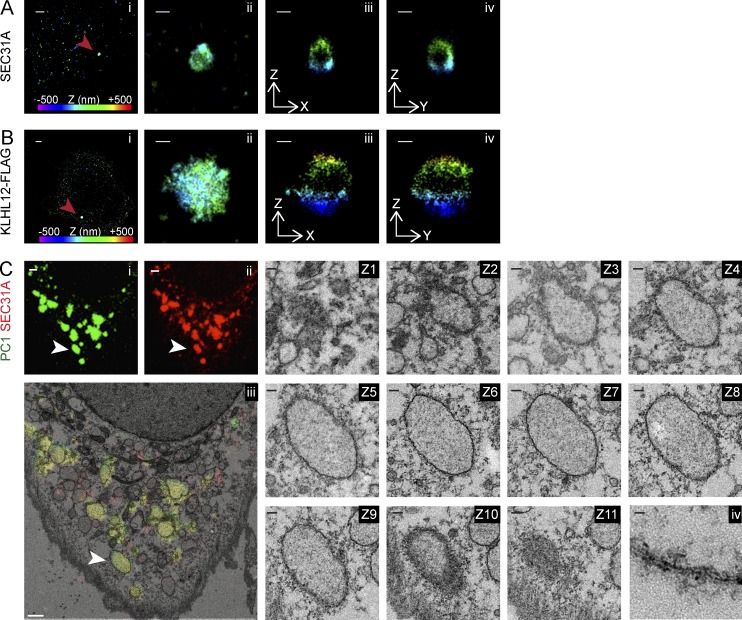
Given that conventional confocal microscopy images revealed colocalized diffraction-limited puncta with no discernible morphological details, we sought to resolve these structures using stochastic optical reconstruction microscopy (STORM; Rust et al., 2006; Huang et al., 2008), a type of superresolution microscopy, and correlative light and electron microscopy (CLEM). With 3D STORM analysis, we observed large, hollow, cage-like COPII structures that are 300–1,000 nm in diameter, using a SEC31A antibody and a secondary antibody conjugated with Alexa Fluor 647 fluorophore in KI6 cells induced for the overexpression of KLHL12 (Fig. 2 A). A virtual z-stack of a single structure confirmed the cage-like COPII protein localization completely surrounding a cavity in three dimensions. Similar analysis was conducted on vesicles immunolabeled with FLAG antibody, which targeted the overexpressed KLHL12-FLAG, and large hollow cages that were 300–1,000 nm in diameter were also observed (Fig. 2 B).
Figure 2.
Large COPII vesicles are hollow membranous containers. 3D STORM images of large SEC31A (A) or KLHL12-FLAG (B) puncta marked as red arrowheads in KI6 cells after 7.5-h induction. Position in depth (z axis) is represented by color according to the color bar in Ai and Bi. (i) Overview of an area of the cell imaged by STORM: SEC31A (A) or FLAG (B). (ii) Magnified maximum xy projection of a large SEC31A (A) or FLAG (B) structure. Virtual cross sections of this structure in XZ (iii) and YZ (iv) planes reveal a hollow compartment encapsulated by SEC31A (A) or KLHL12-FLAG (B). (C) CLEM-resolved overlapping immunofluorescent PC1 (green; i) and SEC31A (red; ii) puncta as single membrane-bound compartments in cultured Saos-2 cells expressing endogenous levels of PC1 and KLHL12. (iii) Representative cell showing colocalizing puncta (n = 36; an example was indicated by white arrowhead) was processed for CLEM. Image shows overlay of the merged fluorescent channels (green and red) on a thin-section (70-nm) TEM image (Z1-11). Serial sections of 70-nm thicknesses through structure of interest. (iv) Magnified view of lipid bilayer from an area of the single membrane enclosing the organelle. n = 2. Bars: (Ai) 1 µm; (Bi) 1 µm; (Aii–Aiv) 200 nm; (Bii–Biv) 200 nm; (Ci–Ciii) 1 µm; (Z1–Z11) 100 nm; (Civ) 10 nm.
We observed a similar morphological feature by CLEM on regions of colocalized endogenous PC1 and SEC31A without KLHL12 overexpression in human osteosarcoma Saos-2 cells, a cell line that secretes endogenous PC1 (Fig. 2 C; Pautke et al., 2004). Correlative serial thin-section EM revealed that fluorescent puncta containing colocalized PC1 and SEC31A were large, single membrane–bounded compartments and were not clusters of small vesicles. These correlated structures were spherical to ovoid in shape and 350 nm to 1.7 µm in diameter, decorated with what may be remnants of coat protein. Collectively, the diffraction-limited PC1 and COPII colocalized puncta were resolved by STORM and CLEM to be large protein-coated, single membrane–bounded containers.
PC1 is completely encapsulated in large COPII-coated membranes
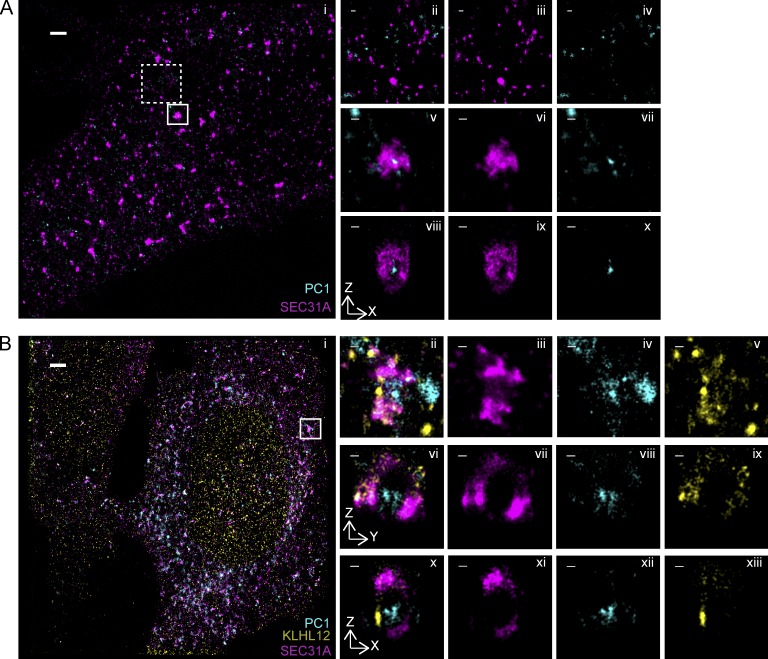
We next sought to dissect the precise localization of PC1 with respect to the COPII coat within puncta that showed colocalization of the two markers. To achieve this, we performed dual- and triple-color 3D STORM imaging on SEC31A/PC1 colocalizing puncta from cells with overexpressed and endogenous levels of KLHL12. Large COPII cages with hollow cavities were observed in KI6 cells (Fig. 3 Aix), consistent with our observation using single-color STORM (Fig. 2, Aiii and Aiv), and PC1 was resolved to be inside of the hollow cavities, entirely encapsulated by the COPII cage (Fig. 3, Av–Ax; and Video 1). This was also evident in KLHL12/SEC31A/PC1 colocalizing puncta in Saos-2 cells, where endogenous KLHL12 and SEC31A was found surrounding the endogenous PC1 (Fig. 3, Bvi–Bxiii). The appearance of confined labeling of the PC1 fiber in relation to the expansive COPII coat may reflect a restricted orientation of the fiber within the vesicle or steric hindrance of the PC1 monoclonal antibody by the polyclonal antibody used to label the coat. In contrast, we did not observe PC1 in canonical COPII cages ranging in size from 80 to 100 nm (Fig. 3, Aii–Aiv). Interestingly, we also observed puncta that appeared to have the COPII coat only partially enveloping PC1, possibly representing an intermediate in the shedding of COPII subunits or a nascent budding event at the ERES (Fig. 3, Bii–Bxiii; and Fig. S3, A–C).
Figure 3.
PC1 is completely encapsulated in large COPII-coated vesicles. (A) Two-color 3D STORM revealed PC1 (cyan) residing inside of large SEC31A (magenta) cages in KI6 cells after 7.5 h of induced KLHL12 overexpression. (i) Overview of an area of the cell imaged by STORM. Small (80- to 100-nm-diameter) canonical SEC31A puncta did not colocalize with PC1 (0%, n = 76 puncta, n = 8 experiments). Examples in dashed inset are magnified in ii as merged image, in iii as SEC31A, and in iv as PC1. Large SEC31A puncta of diameter larger than 300 nm colocalized with PC1 (76.3%, n = 36 puncta, n = 8 experiments). An example in solid inset is magnified in v as merged image, in vi as SEC31A, and in vii as PC1. A virtual cross section in the xz plane of the large COPII vesicle is shown in viii as merged image, in ix as SEC31A, and in x as PC1. (B) Triple-color 3D STORM imaging shows endogenous PC1 (cyan) encapsulated by endogenous KLHL12 (yellow) and SEC31A (magenta) in Saos-2 cells grown at steady state. xy maximum projection of the overview is shown in i and of a magnified vesicle in ii–v. Virtual cross sections in the xz plane are shown in x–xii and yz in vi–ix. Merged three-color channel is shown in i, ii, vi, and x; SEC31A in iii, vii, and xi; PC1 in iv, viii, and xii; and KLHL12 in v, ix, and xiii. Bars: (Ai) 1 µm; (Bi) 2 µm; (magnified views) 100 nm.
PC1-containing giant COPII vesicles exhibit movement
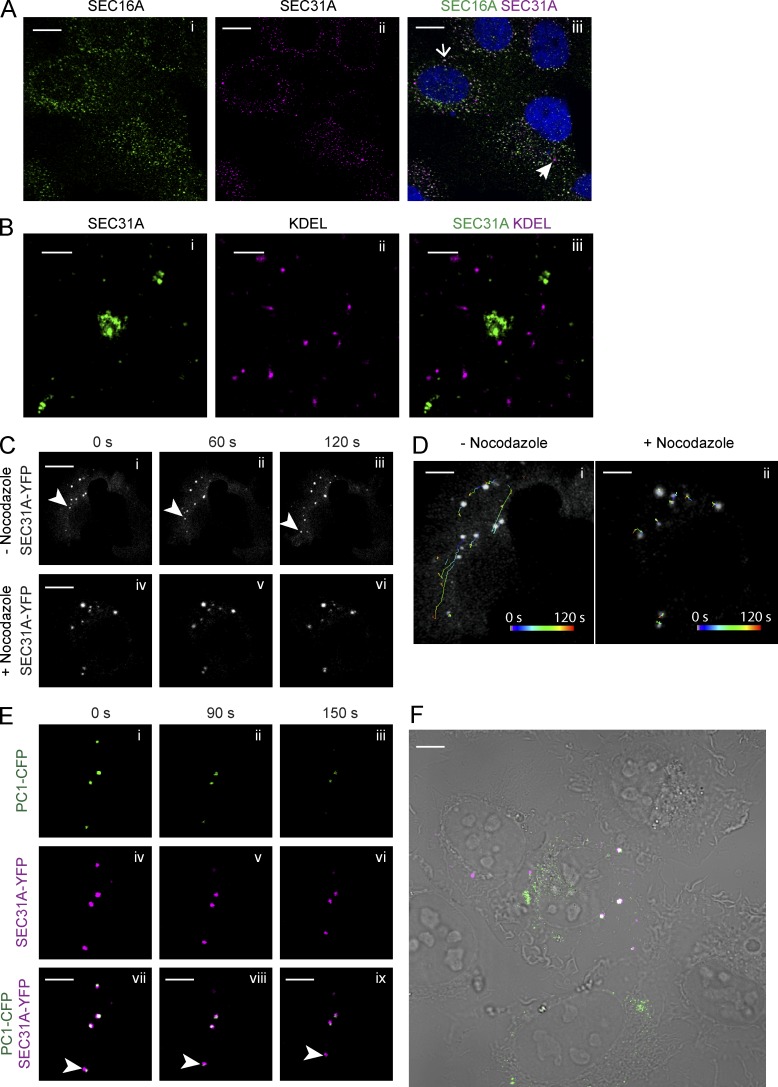
The dynamic nature and organization of ER-to-Golgi transport have been visualized previously with the use of fluorescent labeling and time-lapse microscopy (Presley et al., 1997; Scales et al., 1997; Shima et al., 1999). We sought to understand whether the large COPII vesicles were capable of functioning as mobile transport carriers independent of the ER. The localization of large COPII vesicles relative to the ERES and ER was examined in KI6 cells by dual-color confocal and STORM microscopy using antibodies against SEC31A, SEC16A, a scaffolding protein at the ERES, and KDEL, the ER retrieval signal at the C terminus of ER resident proteins. Images from both approaches showed clear separation of large COPII and ERES or ER marker proteins (Fig. 4, A and B). Large puncta positive for both SEC31A and SEC16A were also observed by confocal microscopy (Fig. 4 A). These coincidently labeled structures may represent prebudding complexes of COPII vesicles that remained attached to the ER.
Figure 4.
Procollagen-carrying vesicles exhibit movement. (A) Confocal microscopy imaging on KI6 cells treated with doxycycline for 7.5 h were labeled with SEC16A and SEC31A antibodies. Open arrow in merged image points to a colocalized spot; closed arrow points to a large SEC31A-labeled vesicle not colocalizing with SEC16A. (B) Two-color STORM imaging was performed on KI6 cells induced for KLHL12 overexpression for 7.5 h and then labeled with KDEL and SEC31A antibodies. Panels show magnified xy view of a SEC31A (green)-labeled large COPII cage (i); KDEL marker labeling ER (magenta; ii); and merged image of the two channels (iii). (C) KI6 cells were transfected with YFP-tagged SEC31A and induced for KLHL12 expression for 7.5 h. (i–iii) Time lapse imaging of SEC31A-YFP puncta in cells not treated with nocodazole. Arrowheads indicate a puncta exhibiting long-range transport (>10 µm distance). n = 3. (iv–vi) Time-lapse images of SEC31A-YFP puncta after the treatment of 5 µM nocodazole for 30 min. (D) Images shown are the first frame of the time-lapse movie overlaid with the trajectories of SEC31A-YFP puncta in the absence (i) and presence (ii) of nocodazole. Trajectories in time (0–120 s) represented by color according to the color scale in D. n = 3. (E) Time-lapse imaging of SEC31A-YFP– and PC1-CFP–positive puncta in KI6 cells. Arrowheads point to a vesicle that displays a spatial displacement in time. (F) Overview of the cell imaged in E; overlay of merged fluorescent image over DIC image shows PC1-CFP in green and SEC31A-YFP in magenta. Bars: (A) 5 µm; (B) 500 nm; (C)10 µm; (D–F) 5 µm.
We then visualized the spatiotemporal dynamics of these vesicles by imaging YFP-tagged, SEC31A-labeled structures with time-lapse microscopy. KLHL12 was induced in KI6 cells for 7.5 h, and the cells were treated with ascorbate for 10 min to promote the formation of hydroxyproline residues essential to the folding and ER exit of PC1 (Stephens and Pepperkok, 2002). We observed large COPII vesicles exhibiting both long-range (>2 µm displacement) and short-range (<2 µm displacement) transport, as previously observed for fluorescently tagged small COPII vesicles (Fig. 4, C and D; and Video 2 Stephens et al., 2000). Long-range transport was not evident in cells treated with nocodazole, a microtubule-depolymerizing agent, as determined by particle tracking of individual vesicles (Fig. 4, C and D; and Video 3). The long-range mobility of large COPII vesicles was observed to be independent of a CFP-tagged ER marker (Stephens et al., 2000), which is consistent with our observations in fixed samples (Video 4). To confirm that these large mobile COPII vesicles are PC1 carriers, we tracked vesicles positive for both YFP-tagged SEC31A and CFP-tagged PC1, which also exhibited directional movements (Fig. 4 E and Video 5).
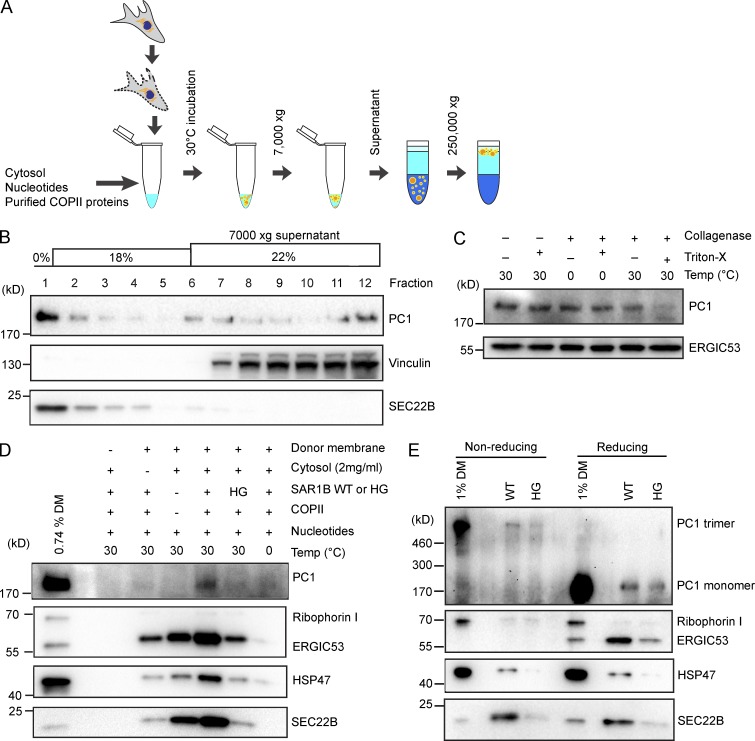
COPII is required to package PC1 into vesicles that bud from the ER in a cell-free reaction
To further investigate whether the large-cargo PC1 exits the ER within COPII-coated vesicles, we modified a cell-free reaction designed to detect the formation of transport vesicles that bud from ER membranes in a preparation of permeabilized cells (Fig. 5 A; Kim et al., 2005; Merte et al., 2010). Donor ER membrane was prepared from IMR-90 cultured human fibroblasts and incubated at 30°C with purified recombinant human COPII proteins, cytosol, and nucleotides for 1 h to allow the formation of transport vesicles. Budded vesicles were isolated by a modification of our previous procedures to allow the detection of large vesicular carriers. A low centrifugal speed of 7,000 g was used to sediment donor membrane from a complete incubation, and the supernatant fraction, which contained budded vesicles, was applied to the bottom of an Optiprep step flotation gradient. After a high-speed centrifugation step of 250,000 g, 12 fractions were collected, and their contents were analyzed by immunoblotting (Fig. 5 B). Because lipid vesicles are buoyant, they floated to the top of the gradient as shown by the enrichment of a standard COPII cargo SEC22B in fraction 1. In contrast, a soluble cytosolic protein marker, vinculin, was observed at the bottom of the gradient in fractions 7–12. Most PC1 signal floated to the top of the gradient and cofractionated with SEC22B, indicating that most PC1 in the reaction was associated with membrane.
Figure 5.
COPII is required to export PC1 in vesicles that bud from the ER in a cell-free reaction. (A) Scheme depicting the experimental procedure of the cell-free vesicle budding reactions. In brief, donor membranes prepared from IMR-90 cells were incubated at 30°C with cytosol (from HT1080 or HTPC1.1 cells), nucleotides (an ATP-regenerating system and GTP), and purified recombinant human COPII proteins. Vesicles in 7,000-g supernatant fractions from budding reactions were isolated by flotation. (B) Fractions (12) were taken from the top of an Optiprep gradient after flotation and analyzed by immunoblotting. SEC22B is found in conventional COPII vesicles and serves as a positive control. Vinculin serves as a control for cytosolic proteins that do not float with vesicles. n = 3. (C) Top floated fraction was treated with or without collagenase (0.1 U/µl) in the presence or absence of the detergent Triton X-100 (1%) at the indicated temperature for 10 min. ERGIC53 is found in conventional COPII vesicles and serves as loading control. n = 4. (D) Budding requirements of PC1 and HSP47 were assessed under different incubation conditions. The top fraction after flotation was taken from each sample and analyzed by immunoblotting. Ribophorin I is an ER resident protein that serves as a negative control. ERGIC53 and SEC22B are found in conventional COPII vesicles and serve as positive controls. n = 3. (E) To detect trimeric PC1, we treated donor membrane or floated fractions with nonreducing denaturing sample buffer before gel electrophoresis. Donor membrane or floated fractions treated with reducing denaturing sample buffers were used as controls. n = 2.
To test whether PC1 detected in the floated fractions was packaged inside of intact membrane vesicles, we used a collagenase protection assay. When the top floated fraction was treated with collagenase at 30°C, most of the PC1 was protected from collagenase digestion and became susceptible only when detergent (1% Triton X-100) was included to lyse the membrane (Fig. 5 C). Because collagenase digestion was specific to collagens, another standard COPII cargo, ERGIC53, was used as loading control.
We tested the requirement for COPII in PC1 packaging by supplementing the reaction with the inhibitory SAR1B H79G GTPase-defective mutant protein in place of wild-type SAR1B (Fig. 5 D). Significantly less PC1 was detected in the top floated fraction, consistent with the observation that microinjection of SAR1 H79G into live cells arrested PC in the ER (Stephens and Pepperkok, 2002). As further controls, the cell-free reaction was performed in the absence of donor membrane, cytosol, or purified recombinant COPII proteins, respectively. Immunoblotting of the top floated fractions showed that donor membrane, cytosol, purified COPII proteins, and a physiological temperature of 30°C were required to reconstitute export of PC1 (Fig. 5 D). PC1 packaging appears to be completely dependent on both cytosolic proteins and COPII for packaging; thus, soluble factors in addition to the COPII coat may be essential to sort PC1 for transport.
PC1 is known to trimerize in the ER, and only the correctly folded PC1 trimers are chaperoned by HSP47 to exit the ER en route to the Golgi membrane. Consistent with our observation in cells where HSP47 was found to be present on the large COPII vesicles, the budding of HSP47 was also observed in the cell-free reaction, which required similar conditions as the budding of PC1 (Figs. 5 D and 1 C). To further demonstrate the physiological relevance of our cell-free reaction, we analyzed the content of the floated fractions under a nonreducing, denaturing condition, which would preserve the interchain disulfide bonds and allow the detection of trimeric PC1 (Fig. S2 A). In cell-free reactions supplemented with wild-type SAR1B, lower-mobility ∼570-kD PC1 (the predicted size of PC1 trimer) was detected in the floated fraction under the nonreducing, denaturing condition, whereas monomeric ∼190-kD PC1 was detected under a reducing, denaturing condition (Fig. 5 E). Consistent with observations under the reducing, denaturing condition, the amount of trimeric PC1 observed in the floated fraction was significantly decreased upon incubation with SAR1B H79G (Fig. 5 E). Thus, we conclude that the COPII-dependent packaging of trimeric PC1 and its chaperone HSP47 is sustained in our cell-free reaction.
PC1 is exported from the ER in large COPII-coated vesicles
To visualize the morphology of vesicular PC1 carriers in the floated fraction, we modified the cell-free reaction so that both the COPII coat and the large-cargo PC1 were fluorescently labeled. For fluorescence visualization of isolated COPII-coated vesicles, we added purified Alexa Fluor 647–conjugated SEC23A/24D to the reaction mixture (Bacia et al., 2011). For fluorescence visualization of PC1, we prepared donor membrane from cells that were transiently transfected with a C-terminally GFP-tagged construct of the pro-α1(I) chain of human PC1. This construct was shown to produce GFP signal that exits the ER in cells incubated in ascorbate-containing medium (Stephens and Pepperkok, 2002). We confirmed that the GFP tag did not interfere with trimerization and ER-to-Golgi transport of PC1-GFP, as GFP signal was found at the Golgi apparatus after the addition of ascorbate (Fig. S4 A). Moreover, PC1-GFP was packaged in a COPII-dependent manner in the vesicle budding reaction, as detected by immunoblotting of GFP in the floated fraction (Fig. S4 B).
Vesicles observed by structured illumination microscopy (SIM) displayed PC1 colocalized with large COPII-coated structures of ∼400 nm in diameter (Fig. 6 A). With the resolution of SIM, the PC1-GFP signal appeared fully within the signal of SEC23A/24D (Fig. 6 A and Videos 6 and 7). Smaller COPII structures devoid of PC1-GFP, likely to be conventional COPII vesicles, were also observed. Not all large COPII structures observed contained the PC1-GFP signal (quantified in Fig. 6, Biii and Biv), possibly because only 10–30% of the donor membrane contained GFP signal as a result of low transfection efficiency. It is also possible that PC1 was not the only large cargo exiting the ER in large COPII-coated vesicles. Not all PC1-GFP observed colocalized with SEC23A/24D: the nonoverlapping GFP signal may represent PC1 not enclosed within membranes, possibly coinciding with the amount of PC1 that was sensitive to collagenase digestion (Fig. 5 C); alternatively, the signal may emanate from transport vesicles from which the COPII coat had been shed (Fig. 3 B; Fig. S3, A–C; and Fig. S5 A).
Figure 6.
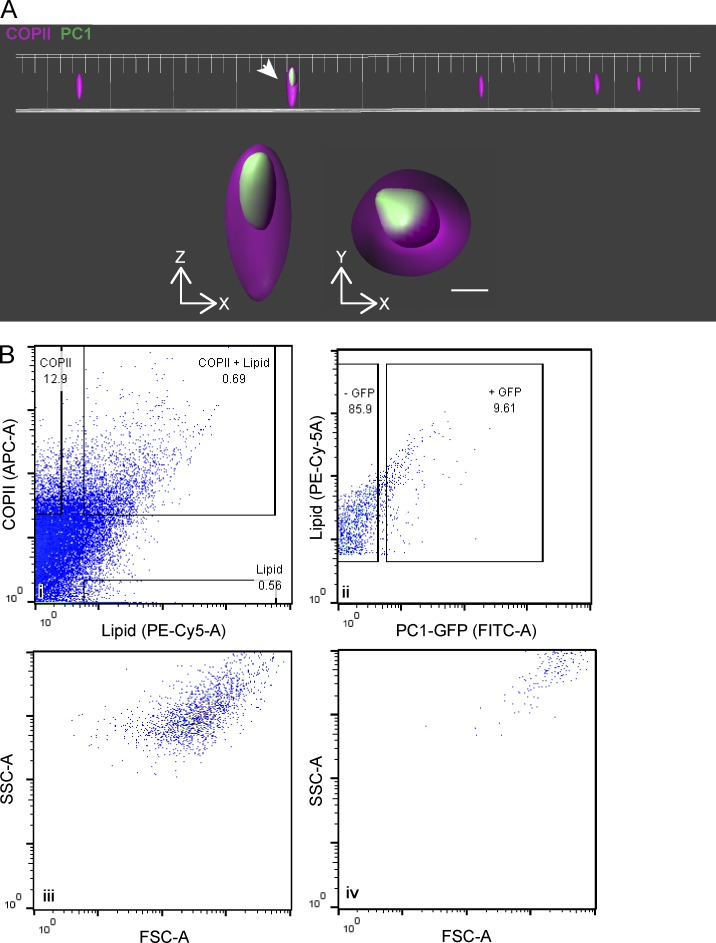
PC1 is exported out of the ER in large COPII-coated vesicles. (A) A representative field of COPII vesicles isolated from cell-free vesicle budding reaction visualized by SIM. One (indicated by the arrow) contained PC1-GFP (green) that was entirely encapsulated by the COPII inner coat proteins SEC23A/24D-647 (magenta). The magnified xz and yz views are displayed underneath the overview. Approximately 20 images were collected from each experiment with fluorescence controls. n = 3. Bar, 200 nm. (B) Relative size of COPII carriers of PC1 compared with regular COPII vesicles quantified by flow cytometry; numbers in graphs represent the percentage of particles in the respective subpopulation. (i) Singlets were plotted for the fluorescence intensities of the COPII coat proteins SEC23A/24D-647 (APC-A) and the lipid dye FM4-64 (PE-Cy5-A). (ii) Particles positive for both COPII and lipid are COPII vesicles and were plotted for the fluorescence intensity of PC1-GFP (FITC-A). (iii and iv) The values of side scatter (SSC-A) versus forward scatter (FSC-A) of COPII vesicles that are PC1-GFP negative (iii) or GFP positive (iv) are plotted. n = 3.
We developed an unbiased flow cytometry approach to quantify the relative fluorescence signal associated with both conventional and large COPII vesicles. Single particles (singlets) were gated for their content of fluorescently labeled COPII proteins SEC23A/24D and the fluorescent lipid dye FM4-64. Particles that satisfied such criteria accounted for ∼0.7% of all singlets detected in the 7,000-g supernatant fraction (Fig. 6 Bi). Of these COPII-coated vesicles, 9.6% were GFP+, indicating that they carried the large cargo PC1-GFP (Fig. 6 Bii). This population of collagen-carrying COPII vesicles exhibited higher side and forward light scatter, both of which positively correlate with larger particle sizes (Fig. 6, Biii and Biv). The sizes of reconstituted COPII-coated vesicles were estimated using nanoparticle tracking analysis (NTA), which tracks and analyzes the Brownian motion of each particle (Fig. S5 A; Dragovic et al., 2011). COPII-coated vesicles in the range of 50–150 nm were ∼17.6× more prevalent than larger COPII-coated vesicles in the range of 300–1,000 nm. A medium-sized category of 150–300 nm was also observed, and it was ∼4.3× less prevalent than the 50- to 150-nm category and 4.1× more numerous than the larger vesicles. We used thin-section EM to independently confirm the existence of medium to large coated membrane vesicles, with a coat of 10–15 nm (Fig. S5 B).
Recruitment of functional KLHL12 to COPII-coated vesicles
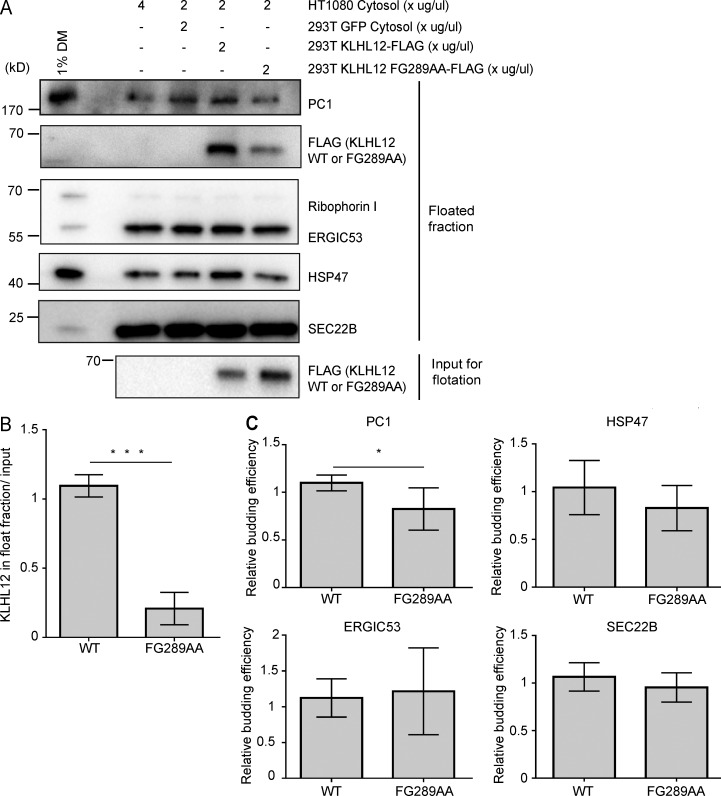
To test whether our cell-free COPII vesicle formation reaction reconstitutes the cellular behavior of KLHL12, we supplemented the reaction with cytosol collected from cells transfected with either FLAG-tagged wild-type KLHL12 or a mutant, FG289AA, that fails to promote the formation of large COPII vesicles (Jin et al., 2012; Fig. 7). Densitometry analyses of immunoblots showed 6.8× more wild-type KLHL12 in the vesicle floated fraction compared with the FG289AA mutant protein (P < 0.0001, paired t test, n = 7; Fig. 7, A and B). This result coincided with our previous observation that KLHL12 FG289AA mutant protein rarely colocalized with COPII markers in cells (Jin et al., 2012). The relative budding efficiency of PC1 and control COPII cargos was also calculated from densitometry analyses of immunoblots, and PC1 budding was only marginally stimulated by KLHL12 wild-type cytosol compared with KLHL12 FG289AA cytosol (P = 0.0412, paired t test, n = 7; Fig. 7, A and C). Thus, although we have reconstituted the association of functional KLHL12 with budded vesicles, we were unable to assess the role of this protein in the capture of PC1 in our cell-free reaction. There may be other rate-limiting components, such as the calcium-binding coadaptors PEF1 and ALG2, which may fail to be recruited to the site of vesicle budding under the conditions of this incubation (McGourty et al., 2016).
Figure 7.
The cellular behavior of KLHL12 is recapitulated in the cell-free reaction. (A) Budding reactions were supplemented with 2 µg/µl 293T cytosol containing FLAG-tagged KLHL12 wild-type or FG289AA mutant in addition to 2 µg/µl HT1080 cytosol. Cytosol (293T cells) that contained GFP and 4 µg/µl HT1080 cytosol were used as controls with endogenous levels of wild-type KLHL12. Top fractions were collected after density gradient flotation for immunoblotting analyses. A representative experiment of seven independent repeats. (B) Relative enrichment of KLHL12 detected in the floated fraction. Densitometry quantification of FLAG intensities in the floated fraction was divided by the corresponding FLAG intensity in the 7,000-g supernatant (input for flotation). Error bars = SD. Paired t test. ***, P < 0.0001. n = 7. Mean of wild-type/mean of FG289AA = 6.80. (C) Relative budding efficiency of PC1, HSP47, ERGIC53, and SEC22B in reactions that were supplemented with 293T cytosol that contained wild-type KLHL12 or KLHL12 FG289AA. Vesicle budding efficiencies were calculated from densitometry quantification relative to the control lane, where 293T cytosol that contained GFP was supplemented. Paired t test was used to analyze the difference between wild-type and FG289AA samples of each cargo; p-values were 0.0412 for PC1 (*, P < 0.05), 0.1215 for HSP47, 0.5307 for ERGIC53, and 0.2993 for SEC22B. Error bars = SD. n = 7.
Discussion
The function of canonical (60- to 80-nm) COPII vesicles in conventional cargo transport is well established: COPII subunits concentrate cargo proteins into vesicles that bud from the ER and form a structure that dictates the size and contour of the bud and completed vesicle (Zanetti et al., 2011). Although a wide range of studies have shown that PC secretion is COPII dependent, it has remained unclear whether COPII coats the membranes surrounding the large cargo or instead plays an indirect role in cargo packaging and transport carrier biogenesis. A primary requirement for the transport of large cargos would be to overcome the structural constraints of a small cage (Noble et al., 2013). KLHL12 was previously shown to fulfill this requirement, as overexpressing KLHL12 induced the formation of large COPII structures (Jin et al., 2012). Here, we confirmed that these large COPII-coated structures appeared hollow, presumably with a space to contain large cargos (Fig. 2, A and B). Similar observations were also made in PC1-secreting osteosarcoma cells (Saos-2) without any overexpression of KLHL12 (Figs. 2 C and 3 B). Notably, CLEM analysis of these structures in Saos-2 showed a COPII-coated single-membrane bilayer envelope surrounding a lumenal space filled with PC1 (Fig. 2 C).
Evidence for the contrary model that PC is transported out of the ER in non-COPII carriers came from studies of fibroblasts released from a PC1 hydroxylation block (Mironov et al., 2003). That study reported that PC1 exits from saccular extensions of the ER that appeared devoid of the COPII coat based on IF microscopy and immunogold EM studies. Those conclusions were based on the use of a polyclonal antibody, LF-68, that was generated against synthetic peptides of a unique sequence selected from the C-telopeptide (nontriple-helical segment exposed on the C terminus of mature collagen after a C-terminal propeptide is cleaved) of human α1(I) collagen (Fig. S2 A; Fisher et al., 1995; Bernstein et al., 1996). Here, using two monoclonal antibodies, QED-42024 and sp1.d8, directed against C- and N- terminal propeptides cleaved from triple-helical PC1, respectively (Fig. S2 A; Foellmer et al., 1983), we observed colocalization with large COPII structures in KI6 cells where both PC1 and KLHL12 were overexpressed (Fig. 1, A–C; and Fig. 3 A), natural collagen I–secreting svIMR-90 cells that overexpressed KLHL12 (Fig. S2 C), and natural collagen I–secreting Saos-2 and IMR-90 cells where PC1 and KLHL12 are expressed endogenously (Figs. 2 C, 3 B, and S2 D). We have independently confirmed the results of Mironov et al. (2003) using the same polyclonal antibody (LF-68) for IF localization (Fig. S2 E). However, we suspect that the C-telopeptides are not fully accessible in nondenatured PC1 trimers, possibly hindered by the presence of C-terminal propeptides, which are cleaved in post-Golgi vesicles or after secretion to the extracellular space (Fig. S2 A). Thus this polyclonal antibody may have limited utility in IF detection of folded PC1 species in early secretory transport vesicles.
Independent of PC1 antibody labeling, the colocalization of PC1-CFP and the COPII coat protein SEC31A-YFP was also observed in live cells (Fig. 4 E). Moreover, the PC-specific chaperone HSP47 also colocalized with KLHL12 and COPII (Fig. 1 C). HSP47 binds to trimerized PC in the ER and assists its correct folding by preventing lateral aggregation (Nagai et al., 2000; Tasab et al., 2000; Ono et al., 2012; Widmer et al., 2012). This chaperone traverses the early secretory pathway with PC to ERGIC or cis-Golgi membranes, where it is released because of lower pH, before being recycled back to the ER via its C-terminal RDEL sequence (Satoh et al., 1996; Oecal et al., 2016). In HSP47-knockout mouse embryonic fibroblasts, misfolded PC1 trimers were observed to accumulate in the ER and were degraded by autophagy, independent of the ERAD-mediated clearance of monomeric PC1 aggregates (Ishida et al., 2006). Hence, the observation of HSP47 colocalized with large COPII confirms that these large COPII structures contain correctly folded triple helical PC1 and are physiologically relevant.
STORM and CLEM analyses revealed the ultrastructural details of these large COPII structures: collagen became completely encapsulated in a large COPII coat during vesicle formation, suggesting that COPII proteins acted directly by forming a complete cage around a single membrane bilayer vesicle that carried PC1 (Fig. 2). Furthermore, fluorescently labeled large COPII vesicles showed microtubule-dependent vectorial movement in live cells (Fig. 4, C–E). To sum up, bona fide large COPII-coated single-membrane carriers of PC1 were observed in fixed as well as live cells.
In an independent effort to test the role of COPII-coated vesicles in the sorting of PC1, we used a cell-free transport vesicle budding assay that was previously developed in our laboratory to probe the requirements for sorting of conventional COPII cargo proteins (Kim et al., 2005; Merte et al., 2010). Here, we report an alternative fractionation method to detect the ER export of the large-cargo PC1 and its chaperone HSP47 (Fig. 5). Using the combination of a buoyant density flotation protocol, a collagenase protection assay, and SIM, we showed that PC1 exits the ER inside of large membrane vesicles that are coated with COPII proteins (Fig. 5, B and C; and Fig. 6 A). Furthermore, the packaging of PC1 requires the addition of recombinant COPII proteins, as well as other unspecified cytosolic proteins, and depends on the GTPase activity of SAR1, the GTP-binding protein that initiates the assembly of the COPII coat (Fig. 5 D).
Although we were unable to identify a function for KLHL12 in our cell-free reaction, we observed the wild-type but not a ubiquitylation-defective mutant form of the protein recruited to budded vesicles (Fig. 7). The requirement for cytosolic proteins in addition to COPII is consistent with reports in the literature of other factors such as Sedlin, Sly1, TFG, ALG2, and PEF1 in the export of PC from the ER (Venditti et al., 2012; Nogueira et al., 2014; McCaughey et al., 2016; McGourty et al., 2016). Further resolution of the cytosolic proteins required in the cell-free reaction may illuminate those that cooperate with KLHL12 to stimulate PC1 packaging.
In summary, we examined the role of COPII during the ER export of large-cargo PC1 using a combination of in vivo morphological analyses and in vitro reconstitution studies. Our results support the conventional model in which COPII participates directly by forming a large COPII-coated membrane vesicle to transport bulky cargo out of the ER. Although the exact mechanism by which the size of COPII vesicles is regulated awaits further investigation, the cell-free PC1 budding reaction described here should aid in such studies.
Materials and methods
Antibodies and plasmids
Commercially available antibodies used for IF and immunoblotting (IB) were as follows: mouse anti-PC1 (clone 42024; QED Biosciences; 1:200 [1 mg/ml stock concentration] for IF); rabbit anti-SEC31A (Bethyl Laboratories; for IF, at 1:200 for confocal and 1:2,000 for STORM); mouse anti-FLAG (Thermo Fisher Scientific; 1:5,000 for IB); goat anti-FLAG (Novus Biologicals; for IF at 1:1,000 for confocal and 1:5,000 for STORM); chicken anti-KLHL12 (Novus Biologicals; for IF at 1:200 for confocal and 1:2,000 for STORM); mouse anti-KLHL12 (clone 2G2; Cell Signaling Technology; at 1:1,000 for IB); rabbit anti-calnexin (Abcam; at 1:200 for IF); mouse anti-HSP47 (Enzo Life Sciences; at 1:200 for IF and 1:5,000 for IB); sheep anti-TGN46 (AbD Serotec; at 1:200 for IF); mouse anti-KDEL (clone 10C3; Enzo Life Sciences; at 1:200 for IF); mouse anti-vinculin (Abcam; 1:5,000 for IB); and rabbit anti-GFP (Torrey Pines Biolabs; at 1:1,000 for IB). Rabbit anti–ribophorin I, ERGIC53, and SEC22B were made in-house and used at 1:5,000 for IB. Rabbit anti–PC1 LF-41, -67, and -68 antibodies were a gift from L. Fisher (National Institute of Dental and Craniofacial Research, Bethesda, MD; Fisher et al., 1989, 1995; Bernstein et al., 1996). LF-67 and LF-68 were raised against the same synthetic peptide of the human α1(I) collagen C-telopeptide (aa 1,192–1,218; Bernstein et al., 1996), and LF-68 was used at 1:1,000 for IF only to repeat previous studies (Mironov et al., 2003; Jin et al., 2012). LF-41 was raised against a synthetic peptide of the C terminus of the human α1(I) collagen (aa 1,443–1,464; Fisher et al., 1989) and was used for all anti-PC1 IB at 1:5,000, but not for IF in this study. The mouse anti-PC1 antibody sp1.d8 was purified in-house from culture medium of mouse hybridoma cells obtained from Developmental Studies Hybridoma Bank at the University of Iowa (Iowa City, IA) using standard procedures and used at 3.75 ng/µl for IF. Expression constructs for SEC31A-YFP, PC-CFP, PC-GFP, and pCFP-ER (encodes a fusion protein consisting of enhanced CFP flanked by an N-terminal signal peptide of calreticulin and a C-terminal ER retrieval sequence, KDEL) were provided by D. Stephens (University of Bristol, Bristol, England, UK).
Cell culture, transfection, and drug treatments
Human lung fibroblasts IMR-90 and svIMR-90 (IMR-90 immortalized with SV-40) were obtained from Coriell Cell Repositories at the National Institute on Aging, Coriell Institute for Medical Research. Human osteosarcoma Saos-2 and U-2OS and human fibrosarcoma HT-1080 were obtained from ATCC. IMR-90, svIMR-90, Saos-2, U-2OS, and HT-1080 were maintained in DMEM plus 10% FBS (GE Healthcare). The HT-PC1.1 cell line was generated from HT-1080 as previously described (Jin et al., 2012) by stable expression of COL1A1 in a pRMc/CMV plasmid (gift from N. Bulleid, University of Glasgow, Glasgow, Scotland, UK) and maintained in 0.4 mg/ml G418. The doxycycline-inducible KLHL12-3xFLAG stable cell line (KI6) was generated through sequential clonal selection of HT-PC1.1 cells that stably integrated pcDNA6/TR and KLHL12-3xFLAG in a pcDNA5/FRT/TO vector (Flp-In T-REx Core kit; Thermo Fisher Scientific) and were selected in the presence of 6 µg/ml blasticidin and 0.2 mg/ml hygromycin, respectively. Cells were kept in a 37°C incubator with 5% CO2. Transfection of DNA constructs into svIMR-90 and KI6 cells was performed using Lipofectamine 2000 as detailed in the manual provided by Invitrogen. Transfection of DNA constructs to Saos-2, svIMR-90, and U-2OS cells for donor membrane preparation and HEK293T cells for cytosol preparation was performed using polyethylenimine (PEI) at a DNA/PEI ratio of 1:3. Ascorbate treatment used 0.25 mM ascorbic acid (Sigma-Aldrich) and 1 mM ascorbic-2-phosphate (Sigma-Aldrich) were used for ascorbic treatment. This concentration was in addition to the amount of ascorbic acid present in FBS (0.08 mM) as supplied by the manufacturer. Doxycycline (Sigma-Aldrich) was used at 1 µg/ml. BFA (Sigma-Aldrich) was used at 10 µg/ml. Nocodazole (Sigma-Aldrich) was used at 5 µM.
Immunofluorescence
Cells growing on poly-l-lysine–coated glass coverslips were fixed in 4% PFA for 20 min at RT or 30 min at 4°C, washed five times with PBS, and incubated with permeabilization buffer (PBS containing 0.1% Triton X-100 and 0.2 M glycine) at RT for 15 min. Cells were incubated with blocking buffer (0.5% BSA in PBS) for 30 min at RT followed by incubation for 1 h each at RT with primary antibody and then secondary antibody. Antibody incubations were followed by five washes with PBS. Coverslips were mounted in ProLong-Gold antifade mountant with DAPI (Thermo Fisher Scientific) overnight, before imaging. Images were acquired using Zen 2010 Software on an LSM 710 confocal microscope system (ZEISS). The objectives used were Plan-Apochromat 100×, 1.4 NA, and Plan-Apochromat 100×, 1.4 NA. The excitation lines and laser power used were 488 (4%), 543 (6%), 405 (2%), and 633 (6%). Colocalization analysis was performed using the spots detection algorithm in the image-processing module Imaris 8.1.2. In brief, a total number of KLHL12 labeled puncta in the z-stack was obtained, and the percentage of puncta positive for both KLHL12 and PC1 was calculated.
Immunoblotting
Standard immunoblotting procedures were followed. In brief, samples were heated at 65°C for 10 min, resolved on 4–20% polyacrylamide gels (15-well, Invitrogen; 26-well, Bio-Rad Laboratories), and transferred to PVDF (EMD Millipore) at constant 0.15A for 16 h. The PVDF membrane was incubated with antibodies (primary for 2.5 h and secondary for 1 h at RT), and bound antibodies were visualized by the enhanced chemiluminescence method (Thermo Fisher Scientific) on a ChemiDoc Imaging System (Bio-Rad Laboratories) with ImageLab software v4.0 (Bio-Rad Laboratories).
Immunoelectron microscopy
KI6 cells were grown on 35-mm glass-bottom dishes (MatTek Corp.) and fixed with 4% PFA and 0.05% glutaraldehyde for 30 min. Cells were washed with PBS and incubated with blocking buffer (0.5% BSA and 0.02% saponin in PBS) at RT for 20 min. Primary antibody labeling was done at RT for 1 h followed by overnight incubation at 4°C in the presence of saponin. Secondary antibody labeling (1:50 dilution of 1.4 nm Nanogold-conjugated goat anti-mouse Fab′ fragment; Nanoprobes) was done at RT for 2 h. The Nanogold particles were gold enhanced in the dark for 5–7 min per the manufacturer’s instructions using GoldEnhance (Nanoprobes). Cells were postfixed with 1% osmium tetroxide and processed for thin-section EM.
Correlative light and electron microscopy
Saos-2 cells were grown on gridded 35-mm glass-bottom dishes (MatTek Corp.) and fixed with PFA (4% in culture medium; Electron Microscopy Sciences). The cells were processed for IF as described earlier with the exception that 0.02% saponin was used as the permeabilizing agent, a concentration well below the established protocol, which typically uses 0.1% saponin (Mironov et al., 2005). The cells (in PBS) displaying colocalized PC1/SEC31A spots were imaged at RT on a Zeiss 710 confocal microscope using a C-Plan Apochromat 63×/1.2 W objective followed by marking the alphanumeric location using a 5×/0.12-NA objective. A z-stack series of the cell of interest was also collected using Zen 2010 software. The cells were then fixed for 30 min in 0.1 M cacodylate buffer, pH 7.2, containing 2% glutaraldehyde and washed with 0.1 M sodium cacodylate buffer before postfixation with 1% osmium tetroxide for 30 min on ice. This was followed by staining with 1% aqueous uranyl acetate for 30 min at RT. For dehydration with progressive lowering of temperature, each incubation period was 10 min, with exposure to 35% ethanol at 4°C, 50% ethanol and 70% ethanol at −20°C, and 95% and 100% ethanol at −35°C. Cells were restored to RT in 100% ethanol before embedding in an epon resin. The cell of interest was identified by the grid location on the resin and thin (70- to 100-nm) serial sections were collected on Formvar-coated 200-mesh copper grids and poststained with 2% aqueous uranyl acetate and 2% tannic acid. The sections were imaged at 120 kV using a Tecnai 12 Transmission Electron Microscope (FEI). Regions of interest were identified manually by correlating the z-slices of the confocal stack with the serial sections and laying the fluorescent image over the TEM image by marking down prominent cell landmarks such as the nuclear boundary, vacuoles, and mitochondria. The overlay was confirmed using the Icy software package (de Chaumont et al., 2012).
STORM imaging
Immunofluorescently labeled cell samples were mounted on glass slides with a standard STORM imaging buffer consisting of 5% (wt/vol) glucose, 100 mM cysteamine, 0.8 mg/ml glucose oxidase, and 40 µg/ml catalase in Tris-HCL, pH 7.5 (Rust et al., 2006; Huang et al., 2008), and sealed using Cytoseal 60 (Thermo Fisher Scientific). STORM imaging was performed on a home-built setup based on a modified Eclipse Ti-U inverted fluorescence microscope (Nikon) using a Nikon CFI Plan Apo λ 100× oil-immersion objective (NA 1.45). Dye molecules were photo switched to the dark state and imaged using either 647- or 560-nm lasers (MPB Communications); the lasers were passed through an acousto-optic tunable filter and introduced through an optical fiber into the back focal plane of the microscope and onto the sample at intensities of ∼2 kW/cm. A translation stage was used to shift the laser beams toward the edge of the objective so that light reached the sample at incident angles slightly smaller than the critical angle of the glass-water interface. A 405-nm laser was used concurrently with either the 647- or 560-nm lasers to reactivate fluorophores into the emitting state. The power of the 405-nm laser (typical range 0–1 W/cm) was adjusted during image acquisition so that at any given instant, only a small, optically resolvable fraction of fluorophore in the sample was in the emitting state. For 3D STORM imaging, a cylindrical lens was inserted into the imaging path so that images of single molecules were elongated in opposite directions for molecules on the proximal and distal sides of the focal plane (Huang et al., 2008). The raw STORM data were analyzed according to previously described methods (Rust et al., 2006; Huang et al., 2008). Data were collected at a frame rate of 110 Hz using an iXon Ultra 897 EM-CCD camera (Andor), for a total of ∼80,000 frames per image. Three-color imaging was performed on targets labeled by Alexa Fluor 647, CF680, and CF568 via sequential imaging with 647- and 560-nm excitation. With 647-nm excitation, a ratiometric detection scheme (Bossi et al., 2008; Testa et al., 2010) was first used to concurrently collect the emission of single Alexa Fluor 647 and CF680 molecules. Emission of single molecules was split into two light paths (channels) using a long-pass dichroic mirror (T685lpxr; Chroma), each of which was projected onto one-half of an iXon Ultra 897 EM-CCD camera. We performed fluorophore assignment by localizing and recording the intensity of each single molecule in the two channels. Excitation at 560 nm was subsequently used to image CF568 through the reflected light path of the dichroic mirror.
Live-cell imaging, particle tracking, and image analysis
KI6 cells were transfected with plasmids encoding SEC31A-YFP and PC1-CFP for a period of 9 h and induced for 7.5 h with doxycycline within that time period to induce the overexpression of KLHL12. The cells were imaged using a temperature-controlled LSM 710 Axio Observer microscope (ZEISS; Plan-Apochromat 63×, 1.4 NA oil DIC M27; standard ZEISS PMT detectors) at 37°C in prewarmed phenol red–free culture medium. Main beam splitters were MBS-405 and MBS-458/514. Emission wavelengths collected were at 454–516 nm and 519–621 nm. Cells were imaged every 5 s for 2 min starting at the end of 10-min treatment with ascorbate and recorded using Zen 2010 software. For experiments requiring nocodazole treatment, the drug was added for 30 min before imaging. Particle detection and tracking were performed using Imaris v8.1 (Bitplane USA). Images were first subjected to background subtraction, and the Spots module was used to automatically detect point-like particles with spot diameter of 200 nm. Appropriate threshold values were confirmed. Object tracing through sequential time frames was done using an autoregressive motion particle-tracking algorithm. A maximum search distance of 5 µm was defined. A gap-closing algorithm was also implemented to link track segment ends to track segment starts. Track outputs were then visually inspected.
Vesicle budding reaction
Vesicle budding reactions were performed as previously described in Kim et al. (2005) with the following modifications. Donor ER membrane was prepared fresh for each reaction by permeabilizing young (under PDL 37.5) IMR-90 cells (95% confluent in 3 × 10-cm dishes) treated with 20 µg/ml digitonin (5 min on ice) in B88 (20 mM Hepes, pH 7.2, 250 mM sorbitol, 150 mM potassium acetate, and 5 mM magnesium acetate) and washed with 0.5 M LiCl in B88 then B88 and resuspended in 200 µl B88-0 (20 mM Hepes, pH 7.2, 250 mM sorbitol, and 150 mM potassium acetate) to a final concentration of OD600 2–6. Each 100-µl reaction contained ATP regeneration system (1 mM ATP, 40 mM creatine phosphate, and 0.2 mg/ml creatine phosphokinase), 3 mM GTP, purified human COPII proteins (2 µg SAR1B, 1 µg SEC23A/24D, and 1 µg SEC13/31A), cytosol (2 µg/µl unless otherwise stated, see “Cytosol preparation” for details), and a final concentration of 5 OD600/ml donor ER membrane in B88-0. Vesicles generated in vitro were isolated from the reaction mixture in two steps. First, donor membranes were sedimented by centrifuging twice at 7,000 g at 4°C for 5 min each in a swinging bucket rotor (S-24-11-AT; Eppendorf). Then, 85 µl of the supernatant was mixed with 50 µl of 60% OptiPrep (Sigma-Aldrich) gently until homogeneous, placed at the bottom of a 7 × 20-mm tube (Beckman Coulter), and overlaid with 100 µl of 18% and 10 µl of 0% OptiPrep in B88. The OptiPrep gradient was centrifuged at 250,000 g for 90 min at 4°C (Beckman TLS-55 with adaptors for 7 × 20-mm tubes) with slow acceleration and deceleration, after which 20-µl fractions were collected from the top and mixed with sample buffer for immunoblotting analysis. When a vesicle budding reaction was analyzed by SIM or flow cytometry, the donor ER membrane was prepared from cells (2 × 10-cm, 80% confluent Saos-2, 40% confluent U-2OS, or 60% confluent svIMR-90 at the time of transfection) transiently transfected with 7.5 µg COL1A1-GFP per 10-cm plate using PEI (Sigma-Aldrich) for 72 h when cells were 100% confluent. The reaction was supplemented with fluorescently labeled SEC23A/24D. Fluorescent SEC23A/24D was produced as previously described in Bacia et al. (2011) using Alexa Fluor 647 C2 maleimide (Thermo Fisher Scientific). An extended version of the protocol for the cell-free reconstitution of COPII-coated procollagen I carriers can be found in Bio-protocol (https://doi.org/10.21769/BioProtoc.2450).
Protein purification
Human SAR1 proteins were overexpressed in Escherichia coli and purified as cleaved GST-fusions, as described for hamster Sar1 purifications in Kim et al. (2005). In brief, a bacterial lysate was first centrifuged at 43,000 g for 15 min, then the supernatant fraction was further centrifuged at 185,000 g for 1 h. The supernatant was incubated with prewashed glutathione agarose (1 ml slurry/l bacteria; Thermo Fisher Scientific) for 1 h at 4°C. Agarose was washed with wash buffer (50 mM Tris, pH 7.4, 150 mM NaCl, 0.1% Tween, 5 mM MgCl2, and 100 µM GDP), and SAR1 was eluted by cleaving with 20 U/ml thrombin (Roche) in TCB (50 mM Tris, pH 8, 250 mM KoAc, 5 mM CaCl2, 5 mM MgCl2, and 100 µM GDP). Human SEC13/31A and SEC23A/24D were purified from lysates of baculovirus-infected insect cells, as described previously (Kim et al., 2005). In brief, insect cell lysates were centrifuged at 185,000 g for 1 h, and 30% ammonium sulfate was added to the supernatant fraction at 4°C. The precipitant was collected by centrifugation at 30,000 g for 30 min and solubilized in no-salt buffer (20 mM Hepes, pH 8, 10% glycerol, 250 mM sorbitol, 0.1 mM EGTA, 5 mM β-mercaptoethanol, and 10 mM imidazole). The solubilized 30% ammonium sulfate precipitant was cleared at 30,000 g for 20 min, and the supernatant was incubated with prewashed Ni-NTA resin (1.25 ml slurry/l insect cells; Thermo Fisher Scientific) for 1 h at 4°C. Ni-NTA was washed with 20 mM Hepes, pH 8, 10% glycerol, 250 mm sorbitol, 500 mM KoAc, 0.1 mM EGTA, 5 mM β-mercaptoethanol, and 50 mM imidazole and eluted with 250 mM imidazole. Ni-NTA–eluted protein was further purified using an anion exchange column (MonoQ) on an AKTA FPLC system (GE Healthcare).
Cytosol preparation
Plates (20 × 15 cm) of HT-1080 or HT-PC1.1 cells were cultured to confluence, washed with PBS, and gently scraped in B88 with protease inhibitors (Roche). Plates (5 × 15 cm) of HEK293T cells were transiently transfected with 18 µg of empty pEGFP_n1 plasmid, wild-type, or mutant KLHL12-3xFLAG in a pcDNA5/FRT/TO vector per 15-cm plate, and cells were collected 24 h after transfection by pipetting with PBS after gently washing with PBS. The collected cells were permeabilized with gentle rocking for 30 min in 80 µg/ml digitonin in 5 ml B88 per five 15-cm plates with protease inhibitors at the concentration suggested by manufacturer (Roche) at 4°C. The cell lysate was collected as a supernatant fraction after centrifugation at 300 g for 5 min. Digitonin was removed from the cell lysate by incubating with washed Bio-Beads SM2 (dry weight of 1 g beads per five 15-cm dishes; Bio-Rad Laboratories) at 4°C overnight. Bio-Beads were removed by centrifugation at 300 g for 5 min. The supernatant fraction was further centrifuged at 160,000 g for 30 min, and the clarified supernatant was collected and concentrated with an Amicon Ultra-3k (EMD Millipore) filter to 40–80 mg/ml. Small aliquots were frozen in liquid nitrogen and stored at −80°C for future use in vesicle budding reactions.
Collagenase protection assay
Top fractions collected after Optiprep gradient flotation were pooled and redistributed to each reaction to ensure that all samples had the equal starting material. Samples were mixed with or without 0.1 U/µl collagenase (Sigma-Aldrich) in the presence or absence of 1% Triton X-100 (Sigma-Aldrich) and incubated at 0°C or 30°C for 10 min.
Structured illumination microscopy Imaging
Donor ER membrane isolated from cells that expressed PC-GFP was incubated with purified Alexa Fluor 647–conjugated SEC23A/24D and other components, as specified, and budded vesicles were isolated from the top of the flotation gradient as described in “Vesicle budding reaction.” Budded vesicles were mounted in Prolong Diamond (Invitrogen) under no. 1.5H coverslips (ZEISS) and set for at least 5 d before imaging to eliminate drift. z-stacks of 1–2 µm were collected in three rotations and five phases using the 100×/1.46 objective on Elyra PS.1 superresolution microscope (ZEISS) with less than 2% laser power and less than 400-ms exposure in all channels, which was determined using no fluorescence and single-channel fluorescence controls. Images were reconstructed and channels were aligned using Zen Black software (ZEISS). Images and videos of 3D iso-surface renderings of each channel were done using Imaris v8.1 (Bitplane USA). Image acquisition, processing, and 3D rendering were done using equipment and software in the CNR Biological Imaging Facility at the University of California, Berkeley.
Flow cytometry
Vesicle budding reactions were scaled up 6× for flow cytometry analyses. After a 7,000-g, 10-min centrifugation of a 600-µl cell-free reaction, 540 µl supernatant was collected from the top. The lipid dye FM4-64 (Thermo Fisher Scientific) was added to the supernatant at 5 µg/ml immediately before the flow cytometry analyses. 100,000 particles were collected for each sample. FSC-A and FSC-H were used to gate for single particles (singlets), which were used for further analysis. Gating of each fluorescent channel was determined by comparing a control sample without any fluorescent labeling and a control that was labeled in a single channel. Data were collected on a BD LSR Fortessa (BD) and analyzed by FlowJo software. Instrument and software were provided by the LKS flow core facility at the University of California, Berkeley.
Nanoparticle tracking analysis
Size of vesicles budded in vitro were estimated using the NanoSight NS300 instrument equipped with a 405-nm laser (Malvern Instruments). Qdot525-positive particles were analyzed with a 500-nm filter (fluorescent mode); nonfluorescent particles were analyzed in the scatter mode without a filter. Silica 100-nm microspheres (Polysciences) were analyzed to check instrument performance and determine the viscosity coefficient of B88. Vesicles were collected from the top 20 µl of the flotation gradient as described in the Vesicle budding reaction section and diluted 40× with 780 µl filtered B88 (0.02 µm; Whatman). To label COPII with the fluorescent Qdot525, the 7,000-g supernatant fraction was incubated with mouse anti-SEC31A primary antibody (1:100; BD) and donkey anti–mouse IgG secondary antibody Qdot525 conjugate (1:100; Thermo Fisher Scientific) for 1 h at RT. As a labeling control, primary antibody was omitted. The labeled supernatants were used as inputs for flotation (see details in Vesicle budding reaction), and 2× top 20 µl of floated fractions was collected from duplicates, to double the labeling material, and diluted 20× with 760 µl filtered B88 (0.02 µm; Whatman). The samples were automatically introduced into the sample chamber at a constant flow rate of 50 (arbitrary manufacturer unit, ∼10 µl/min) during five repeats of 60-s captures at camera level 13 in scatter mode and level 15 in fluorescent mode with Nanosight NTA 3.1 software (Malvern Instruments). Each sample was measured at two different positions of the syringe introduction to minimize the heterogeneous flow of vesicles. The particle size was estimated with detection threshold 3 for fluorescent mode and 5 for scatter mode using the Nanosight NTA 3.1 software, after which “experiment summary” and “particle data” were exported. Particle numbers in each size category was calculated from the particle data, in which “true” particles with track length >3 were pooled, binned, and counted with Excel (Microsoft). GraphPad Prism 7 was used for graphing and statistical analyses.
Electron microscopy of budded vesicles
For morphological analysis of the budded vesicles, the top 80 µl of a supernatant fraction after the 7,000-g, 10-min spin of a 100-µl reaction was fixed with 2% PFA and 0.2% glutaraldehyde in B88 buffer for 15 min at 4°C and centrifuged at 25,000 g (Beckman TLS-55 with adaptors for 11 × 34-mm tubes) for 30 min with slow acceleration and deceleration on to a 0.5-ml agarose bed (2% low melting point; Sigma-Aldrich). This centrifugation speed was optimized to collect large COPII-coated vesicles. After three washes in B88 buffer, the pellet was embedded in 12% gelatin, cut in small blocks, and infiltrated with 2.3 M sucrose in 0.1 M phosphate buffer at pH 7.4 overnight at 4°C. The blocks were mounted on pins and stored frozen in liquid nitrogen. Ultrathin cryosections were collected on Formvar and carbon-coated nickel grids, poststained with 2% uranyl acetate, and imaged at 120 kV using a Tecnai 12 Transmission Electron Microscope.
Online supplemental material
Fig. S1 shows the validity of using the KI6 cell line stably transfected with PC1 and doxycycline-inducible KLHL12 for functional assays. Fig. S2 shows IF data comparing two monoclonal antibodies (sp1.d8 and QED-42024) with polyclonal antibody LF-68. Fig. S3 shows triple IF (PC1/SEC31A/KLHL12) labeling of partially coated large COPII vesicles by STORM microscopy. Fig. S4 shows that overexpressed PC1-GFP acts similarly to endogenous PC1: PC1-GFP was observed in the Golgi apparatus after an ascorbate chase in cells and in the floated vesicle fractions generated by the in vitro budding assay in a COPII-dependent manner. Fig. S5 shows size distribution of the vesicles generated in the budding assay using NTA and EM. Video 1 shows z-sections through a PC1/SEC31A-immunolabeled vesicle by STORM analysis. Video 2 shows movement of YFP-tagged SEC31A vesicles by live-cell imaging. Video 3 shows live-cell imaging of the YFP-tagged SEC31A vesicles in the presence of 5 µM nocodazole. Video 4 shows movement of YFP-tagged SEC31A vesicles separated from ER marked by a CFP-tagged ER marker. Video 5 shows movement of SEC31A-YFP and PC1-CFP tagged vesicles. Videos 6 and 7 show more examples of the reconstituted COPII-coated PC-1 carriers and regular COPII-coated vesicles visualized by SIM.
Supplementary Material
Acknowledgments
We thank the staff at the University of California Berkeley shared facilities including Ann Fisher and Alison Kililea (Tissue Culture facility), Steven Ruzin and Denise Schichnes (CNR Biological Imaging facility), Kartoosh Heydari (Flow Cytometry facility), and Reena Zalpuri (Electron Microscopy laboratory). We are grateful for advice and guidance by Manfred Auer (Lawrence Berkeley National Laboratory) and Kent McDonald (Electron Microscopy Laboratory). We also thank past and present members of the Schekman Lab, in particular Kanika Bajaj, Yusong Guo, Liang Ge, and David Melville.
Research reported in this publication was performed in part at CRL Molecular Imaging Center and the CNR Biological Imaging facility, supported in part by the Gordon and Betty Moore Foundation and the National Institutes of Health S10 program under award numbers 1S10RR026866-01 and 1S10OD018136-01. R. Schekman is supported as an Investigator of the Howard Hughes Medical Institute and the University of California Berkeley Miller Institute for Basic Research in Science. L. Yuan was supported in part by the Tang Family Fellowship. A. Gorur was supported in part by the Employee Development Program at Lawrence Berkeley National Laboratory. S.J. Kenny and K. Xu acknowledge support from National Science Foundation under CHE-1554717, the Pew Biomedical Scholars Award, and the Sloan Research Fellowship.
The authors declare no competing financial interests.
Author contributions: A. Gorur, L. Yuan, K. Xu, and R. Schekman conceptualized the experiments. A. Gorur and L. Yuan identified the monoclonal antibodies for collagen 1 and performed the immunofluorescence assays. A. Gorur performed electron microscopy and live-cell imaging. S.J. Kenny performed and analyzed STORM microscopy data. L. Yuan established the in vitro budding protocol. L. Yuan performed and analyzed data from in vitro budding experiments. S. Baba carried out the initial optimization of the in vitro budding experiments. A. Gorur, L. Yuan, and R. Schekman prepared the original draft and revised the manuscript.
Footnotes
Abbreviations used:
- BFA
- brefeldin A
- CLEM
- correlative light electron microscopy
- COPII
- coat protein complex II
- ERES
- ER exit site
- ERGIC
- ER–Golgi intermediate compartment
- IF
- immunofluorescence
- PC
- procollagen
- SIM
- structured illumination microscopy
- STORM
- stochastic optical reconstruction microscopy
References
- Bächinger H.P., Doege K.J., Petschek J.P., Fessler L.I., and Fessler J.H.. 1982. Structural implications from an electronmicroscopic comparison of procollagen V with procollagen I, pC-collagen I, procollagen IV, and a Drosophila procollagen. J. Biol. Chem. 257:14590–14592. [PubMed] [Google Scholar]
- Bacia K., Futai E., Prinz S., Meister A., Daum S., Glatte D., Briggs J.A., and Schekman R.. 2011. Multibudded tubules formed by COPII on artificial liposomes. Sci. Rep. 1:17 10.1038/srep00017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barlowe C., Orci L., Yeung T., Hosobuchi M., Hamamoto S., Salama N., Rexach M.F., Ravazzola M., Amherdt M., and Schekman R.. 1994. COPII: A membrane coat formed by Sec proteins that drive vesicle budding from the endoplasmic reticulum. Cell. 77:895–907. 10.1016/0092-8674(94)90138-4 [DOI] [PubMed] [Google Scholar]
- Bernstein E.F., Chen Y.Q., Kopp J.B., Fisher L., Brown D.B., Hahn P.J., Robey F.A., Lakkakorpi J., and Uitto J.. 1996. Long-term sun exposure alters the collagen of the papillary dermis. Comparison of sun-protected and photoaged skin by northern analysis, immunohistochemical staining, and confocal laser scanning microscopy. J. Am. Acad. Dermatol. 34:209–218. 10.1016/S0190-9622(96)80114-9 [DOI] [PubMed] [Google Scholar]
- Bossi M., Fölling J., Belov V.N., Boyarskiy V.P., Medda R., Egner A., Eggeling C., Schönle A., and Hell S.W.. 2008. Multicolor far-field fluorescence nanoscopy through isolated detection of distinct molecular species. Nano Lett. 8:2463–2468. 10.1021/nl801471d [DOI] [PubMed] [Google Scholar]
- Boyadjiev S.A., Fromme J.C., Ben J., Chong S.S., Nauta C., Hur D.J., Zhang G., Hamamoto S., Schekman R., Ravazzola M., et al. . 2006. Cranio-lenticulo-sutural dysplasia is caused by a SEC23A mutation leading to abnormal endoplasmic-reticulum-to-Golgi trafficking. Nat. Genet. 38:1192–1197. 10.1038/ng1876 [DOI] [PubMed] [Google Scholar]
- de Chaumont F., Dallongeville S., Chenouard N., Hervé N., Pop S., Provoost T., Meas-Yedid V., Pankajakshan P., Lecomte T., Le Montagner Y., et al. . 2012. Icy: An open bioimage informatics platform for extended reproducible research. Nat. Methods. 9:690–696. 10.1038/nmeth.2075 [DOI] [PubMed] [Google Scholar]
- Dragovic R.A., Gardiner C., Brooks A.S., Tannetta D.S., Ferguson D.J., Hole P., Carr B., Redman C.W., Harris A.L., Dobson P.J., et al. . 2011. Sizing and phenotyping of cellular vesicles using Nanoparticle Tracking Analysis. Nanomedicine (Lond.). 7:780–788. 10.1016/j.nano.2011.04.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher L.W., Lindner W., Young M.F., and Termine J.D.. 1989. Synthetic peptide antisera: Their production and use in the cloning of matrix proteins. Connect. Tissue Res. 21:43–48. 10.3109/03008208909049994 [DOI] [PubMed] [Google Scholar]
- Fisher L.W., Stubbs J.T. III, and Young M.F.. 1995. Antisera and cDNA probes to human and certain animal model bone matrix noncollagenous proteins. Acta Orthop. Scand. Suppl. 266:61–65. [PubMed] [Google Scholar]
- Foellmer H.G., Kawahara K., Madri J.A., Furthmayr H., Timpl R., and Tuderman L.. 1983. A monoclonal antibody specific for the amino terminal cleavage site of procollagen type I. Eur. J. Biochem. 134:183–189. 10.1111/j.1432-1033.1983.tb07549.x [DOI] [PubMed] [Google Scholar]
- Fromme J.C., and Schekman R.. 2005. COPII-coated vesicles: Flexible enough for large cargo? Curr. Opin. Cell Biol. 17:345–352. 10.1016/j.ceb.2005.06.004 [DOI] [PubMed] [Google Scholar]
- Garbes L., Kim K., Rieß A., Hoyer-Kuhn H., Beleggia F., Bevot A., Kim M.J., Huh Y.H., Kweon H.-S.S., Savarirayan R., et al. . 2015. Mutations in SEC24D, encoding a component of the COPII machinery, cause a syndromic form of osteogenesis imperfecta. Am. J. Hum. Genet. 96:432–439. 10.1016/j.ajhg.2015.01.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang B., Wang W., Bates M., and Zhuang X.. 2008. Three-dimensional super-resolution imaging by stochastic optical reconstruction microscopy. Science. 319:810–813. 10.1126/science.1153529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishida Y., and Nagata K.. 2011. Hsp47 as a collagen-specific molecular chaperone. Methods Enzymol. 499:167–182. 10.1016/B978-0-12-386471-0.00009-2 [DOI] [PubMed] [Google Scholar]
- Ishida Y., Kubota H., Yamamoto A., Kitamura A., Bächinger H.P., and Nagata K.. 2006. Type I collagen in Hsp47-null cells is aggregated in endoplasmic reticulum and deficient in N-propeptide processing and fibrillogenesis. Mol. Biol. Cell. 17:2346–2355. 10.1091/mbc.E05-11-1065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishikawa Y., Ito S., Nagata K., Sakai L.Y., and Bächinger H.P.. 2016. Intracellular mechanisms of molecular recognition and sorting for transport of large extracellular matrix molecules. Proc. Natl. Acad. Sci. USA. 113:E6036–E6044. 10.1073/pnas.1609571113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin L., Pahuja K.B., Wickliffe K.E., Gorur A., Baumgärtel C., Schekman R., and Rape M.. 2012. Ubiquitin-dependent regulation of COPII coat size and function. Nature. 482:495–500. 10.1038/nature10822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J., Hamamoto S., Ravazzola M., Orci L., and Schekman R.. 2005. Uncoupled packaging of amyloid precursor protein and presenilin 1 into coat protein complex II vesicles. J. Biol. Chem. 280:7758–7768. 10.1074/jbc.M411091200 [DOI] [PubMed] [Google Scholar]
- Kim S.-D.D., Pahuja K.B., Ravazzola M., Yoon J., Boyadjiev S.A., Hammamoto S., Schekman R., Orci L., and Kim J.. 2012. SEC23-SEC31 interface plays critical role for export of procollagen from the endoplasmic reticulum. J. Biol. Chem. 287:10134–10144. 10.1074/jbc.M111.283382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lang M.R., Lapierre L.A., Frotscher M., Goldenring J.R., and Knapik E.W.. 2006. Secretory COPII coat component Sec23a is essential for craniofacial chondrocyte maturation. Nat. Genet. 38:1198–1203. 10.1038/ng1880 [DOI] [PubMed] [Google Scholar]
- Ma W., and Goldberg J.. 2016. TANGO1/cTAGE5 receptor as a polyvalent template for assembly of large COPII coats. Proc. Natl. Acad. Sci. USA. 113:10061–10066. 10.1073/pnas.1605916113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuoka K., Orci L., Amherdt M., Bednarek S.Y., Hamamoto S., Schekman R., and Yeung T.. 1998. COPII-coated vesicle formation reconstituted with purified coat proteins and chemically defined liposomes. Cell. 93:263–275. 10.1016/S0092-8674(00)81577-9 [DOI] [PubMed] [Google Scholar]
- McCaughey J., Miller V.J., Stevenson N.L., Brown A.K., Budnik A., Heesom K.J., Alibhai D., and Stephens D.J.. 2016. TFG promotes organization of transitional ER and efficient collagen secretion. Cell Reports. 15:1648–1659. 10.1016/j.celrep.2016.04.062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGourty C.A., Akopian D., Walsh C., Gorur A., Werner A., Schekman R., Bautista D., and Rape M.. 2016. Regulation of the CUL3 ubiquitin ligase by a calcium-dependent co-adaptor. Cell. 167:525–538.e14. 10.1016/j.cell.2016.09.026 [DOI] [PubMed] [Google Scholar]
- Merte J., Jensen D., Wright K., Sarsfield S., Wang Y., Schekman R., and Ginty D.D.. 2010. Sec24b selectively sorts Vangl2 to regulate planar cell polarity during neural tube closure. Nat. Cell Biol. 12:41–46: 1–8. 10.1038/ncb2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mironov A.A., Mironov A.A. Jr., Beznoussenko G.V., Trucco A., Lupetti P., Smith J.D., Geerts W.J., Koster A.J., Burger K.N., Martone M.E., et al. . 2003. ER-to-Golgi carriers arise through direct en bloc protrusion and multistage maturation of specialized ER exit domains. Dev. Cell. 5:583–594. 10.1016/S1534-5807(03)00294-6 [DOI] [PubMed] [Google Scholar]
- Mironov A.A., Beznoussenko G.V., Luini A., and Polishchuk R.S.. 2005. Visualizing intracellular events in vivo by combined video fluorescence and 3-D electron microscopy. Methods Enzymol. 404:43–57. 10.1016/S0076-6879(05)04005-X [DOI] [PubMed] [Google Scholar]
- Nagai N., Hosokawa M., Itohara S., Adachi E., Matsushita T., Hosokawa N., and Nagata K.. 2000. Embryonic lethality of molecular chaperone hsp47 knockout mice is associated with defects in collagen biosynthesis. J. Cell Biol. 150:1499–1506. 10.1083/jcb.150.6.1499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noble A.J., Zhang Q., O’Donnell J., Hariri H., Bhattacharya N., Marshall A.G., and Stagg S.M.. 2013. A pseudoatomic model of the COPII cage obtained from cryo-electron microscopy and mass spectrometry. Nat. Struct. Mol. Biol. 20:167–173. 10.1038/nsmb.2467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nogueira C., Erlmann P., Villeneuve J., Santos A.J.J., Martínez-Alonso E., Martínez-Menárguez J.Á.Á., and Malhotra V.. 2014. SLY1 and Syntaxin 18 specify a distinct pathway for procollagen VII export from the endoplasmic reticulum. eLife. 3:e02784 10.7554/eLife.02784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oecal S., Socher E., Uthoff M., Ernst C., Zaucke F., Sticht H., Baumann U., and Gebauer J.M.. 2016. The pH-dependent client release from the collagen-specific chaperone HSP47 is triggered by a tandem histidine pair. J. Biol. Chem. 291:12612–12626. 10.1074/jbc.M115.706069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ono T., Miyazaki T., Ishida Y., Uehata M., and Nagata K.. 2012. Direct in vitro and in vivo evidence for interaction between Hsp47 protein and collagen triple helix. J. Biol. Chem. 287:6810–6818. 10.1074/jbc.M111.280248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pastor-Pareja J.C., and Xu T.. 2011. Shaping cells and organs in Drosophila by opposing roles of fat body-secreted Collagen IV and perlecan. Dev. Cell. 21:245–256. 10.1016/j.devcel.2011.06.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pautke C., Schieker M., Tischer T., Kolk A., Neth P., Mutschler W., and Milz S.. 2004. Characterization of osteosarcoma cell lines MG-63, Saos-2 and U-2 OS in comparison to human osteoblasts. Anticancer Res. 24:3743–3748. [PubMed] [Google Scholar]
- Petley-Ragan L.M., Ardiel E.L., Rankin C.H., and Auld V.J.. 2016. Accumulation of laminin monomers in Drosophila glia leads to glial endoplasmic reticulum stress and disrupted larval locomotion. J. Neurosci. 36:1151–1164. 10.1523/JNEUROSCI.1797-15.2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Presley J.F., Cole N.B., Schroer T.A., Hirschberg K., Zaal K.J., and Lippincott-Schwartz J.. 1997. ER-to-Golgi transport visualized in living cells. Nature. 389:81–85. 10.1038/38891 [DOI] [PubMed] [Google Scholar]
- Roberts B., Clucas C., and Johnstone I.L.. 2003. Loss of SEC-23 in Caenorhabditis elegans causes defects in oogenesis, morphogenesis, and extracellular matrix secretion. Mol. Biol. Cell. 14:4414–4426. 10.1091/mbc.E03-03-0162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rust M.J., Bates M., and Zhuang X.. 2006. Sub-diffraction-limit imaging by stochastic optical reconstruction microscopy (STORM). Nat. Methods. 3:793–795. 10.1038/nmeth929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito K., Chen M., Bard F., Chen S., Zhou H., Woodley D., Polischuk R., Schekman R., and Malhotra V.. 2009. TANGO1 facilitates cargo loading at endoplasmic reticulum exit sites. Cell. 136:891–902. 10.1016/j.cell.2008.12.025 [DOI] [PubMed] [Google Scholar]
- Saito K., Yamashiro K., Ichikawa Y., Erlmann P., Kontani K., Malhotra V., and Katada T.. 2011. cTAGE5 mediates collagen secretion through interaction with TANGO1 at endoplasmic reticulum exit sites. Mol. Biol. Cell. 22:2301–2308. 10.1091/mbc.E11-02-0143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito K., Yamashiro K., Shimazu N., Tanabe T., Kontani K., and Katada T.. 2014. Concentration of Sec12 at ER exit sites via interaction with cTAGE5 is required for collagen export. J. Cell Biol. 206:751–762. 10.1083/jcb.201312062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos A.J.J., Nogueira C., Ortega-Bellido M., and Malhotra V.. 2016. TANGO1 and Mia2/cTAGE5 (TALI) cooperate to export bulky pre-chylomicrons/VLDLs from the endoplasmic reticulum. J. Cell Biol. 213:343–354. 10.1083/jcb.201603072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarmah S., Barrallo-Gimeno A., Melville D.B., Topczewski J., Solnica-Krezel L., and Knapik E.W.. 2010. Sec24D-dependent transport of extracellular matrix proteins is required for zebrafish skeletal morphogenesis. PLoS One. 5:e10367 10.1371/journal.pone.0010367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satoh M., Hirayoshi K., Yokota S., Hosokawa N., and Nagata K.. 1996. Intracellular interaction of collagen-specific stress protein HSP47 with newly synthesized procollagen. J. Cell Biol. 133:469–483. 10.1083/jcb.133.2.469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scales S.J., Pepperkok R., and Kreis T.E.. 1997. Visualization of ER-to-Golgi transport in living cells reveals a sequential mode of action for COPII and COPI. Cell. 90:1137–1148. 10.1016/S0092-8674(00)80379-7 [DOI] [PubMed] [Google Scholar]
- Shima D.T., Scales S.J., Kreis T.E., and Pepperkok R.. 1999. Segregation of COPI-rich and anterograde-cargo-rich domains in endoplasmic-reticulum-to-Golgi transport complexes. Curr. Biol. 9:821–824. 10.1016/S0960-9822(99)80365-0 [DOI] [PubMed] [Google Scholar]
- Siddiqi S.A., Gorelick F.S., Mahan J.T., and Mansbach C.M. II. 2003. COPII proteins are required for Golgi fusion but not for endoplasmic reticulum budding of the pre-chylomicron transport vesicle. J. Cell Sci. 116:415–427. 10.1242/jcs.00215 [DOI] [PubMed] [Google Scholar]
- Siddiqi S., Saleem U., Abumrad N.A., Davidson N.O., Storch J., Siddiqi S.A., and Mansbach C.M. II. 2010. A novel multiprotein complex is required to generate the prechylomicron transport vesicle from intestinal ER. J. Lipid Res. 51:1918–1928. 10.1194/jlr.M005611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephens D.J., and Pepperkok R.. 2002. Imaging of procollagen transport reveals COPI-dependent cargo sorting during ER-to-Golgi transport in mammalian cells. J. Cell Sci. 115:1149–1160. [DOI] [PubMed] [Google Scholar]
- Stephens D.J., Lin-Marq N., Pagano A., Pepperkok R., and Paccaud J.P.. 2000. COPI-coated ER-to-Golgi transport complexes segregate from COPII in close proximity to ER exit sites. J. Cell Sci. 113:2177–2185. [DOI] [PubMed] [Google Scholar]
- Tasab M., Batten M.R., and Bulleid N.J.. 2000. Hsp47: A molecular chaperone that interacts with and stabilizes correctly-folded procollagen. EMBO J. 19:2204–2211. 10.1093/emboj/19.10.2204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tasab M., Jenkinson L., and Bulleid N.J.. 2002. Sequence-specific recognition of collagen triple helices by the collagen-specific molecular chaperone HSP47. J. Biol. Chem. 277:35007–35012. 10.1074/jbc.M202782200 [DOI] [PubMed] [Google Scholar]
- Testa I., Wurm C.A., Medda R., Rothermel E., von Middendorf C., Fölling J., Jakobs S., Schönle A., Hell S.W., and Eggeling C.. 2010. Multicolor fluorescence nanoscopy in fixed and living cells by exciting conventional fluorophores with a single wavelength. Biophys. J. 99:2686–2694. 10.1016/j.bpj.2010.08.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Townley A.K., Feng Y., Schmidt K., Carter D.A., Porter R., Verkade P., and Stephens D.J.. 2008. Efficient coupling of Sec23-Sec24 to Sec13-Sec31 drives COPII-dependent collagen secretion and is essential for normal craniofacial development. J. Cell Sci. 121:3025–3034. 10.1242/jcs.031070 [DOI] [PubMed] [Google Scholar]
- Venditti R., Scanu T., Santoro M., Di Tullio G., Spaar A., Gaibisso R., Beznoussenko G.V., Mironov A.A., Mironov A. Jr., Zelante L., et al. . 2012. Sedlin controls the ER export of procollagen by regulating the Sar1 cycle. Science. 337:1668–1672. 10.1126/science.1224947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Widmer C., Gebauer J.M., Brunstein E., Rosenbaum S., Zaucke F., Drögemüller C., Leeb T., and Baumann U.. 2012. Molecular basis for the action of the collagen-specific chaperone Hsp47/SERPINH1 and its structure-specific client recognition. Proc. Natl. Acad. Sci. USA. 109:13243–13247. 10.1073/pnas.1208072109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson D.G., Phamluong K., Li L., Sun M., Cao T.C., Liu P.S., Modrusan Z., Sandoval W.N., Rangell L., Carano R.A., et al. . 2011. Global defects in collagen secretion in a Mia3/TANGO1 knockout mouse. J. Cell Biol. 193:935–951. 10.1083/jcb.201007162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zanetti G., Pahuja K.B., Studer S., Shim S., and Schekman R.. 2011. COPII and the regulation of protein sorting in mammals. Nat. Cell Biol. 14:20–28. 10.1038/ncb2390 [DOI] [PubMed] [Google Scholar]
- Zhu M., Tao J., Vasievich M.P., Wei W., Zhu G., Khoriaty R.N., and Zhang B.. 2015. Neural tube opening and abnormal extraembryonic membrane development in SEC23A deficient mice. Sci. Rep. 5:15471 10.1038/srep15471 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.