Summary
Epstein–Barr virus (EBV) is a well‐documented aetiological factor for multiple sclerosis (MS). EBV encodes at least 44 microRNAs (miRNAs) that are readily detectable in the circulation of human. Previous studies have demonstrated that EBV‐encoded miRNAs regulate host immune response and may serve as biomarkers for EBV‐associated diseases. However, the roles of EBV miRNAs in MS are still unknown. To fill the gap, we conducted a comprehensive profiling of 44 mature EBV miRNAs in 30 relapsing–remitting MS (RRMS) patients at relapse and 30 matched healthy controls. Expression levels of ebv‐miR‐BHRF1‐2‐5p and ebv‐miR‐BHRF1‐3 were elevated significantly in the circulation and correlated positively with the expanded disability status scale (EDSS) scores of MS patients. Receiver operating characteristic (ROC) analyses confirmed that the expression of these two miRNAs distinguished MS patients clearly from healthy controls. Luciferase assays revealed that ebv‐miR‐BHRF1‐2‐5p may directly target MALT1 (mucosa‐associated lymphoid tissue lymphoma transport protein 1), a key regulator of immune homeostasis. In conclusion, we described the expression of EBV miRNAs in MS and preliminarily validated the potential target genes of significantly altered EBV miRNAs. The findings may pave the way for prospective study about the pathogenesis of MS.
Keywords: Epstein–Barr virus, MALT1, microRNA, multiple sclerosis
Introduction
Multiple sclerosis (MS) is a disease characterized by chronic inflammatory demyelination and axonal injury of white matter in the central nervous system (CNS) 1. MS is caused by intricate interactions between genetic predisposition and environmental triggers, including viral infection 2. Large‐scale studies have confirmed that Epstein–Barr virus (EBV) is the only viral agent that is associated positively with MS with no signs of bias 3, 4. The seropositivity rate of EBV in MS patients was 99·5% and both anti‐EBV capsid antigen (EBVCA) and anti‐EB nuclear antigen (EBNA) titres were increased in MS patients 5, 6, 7, 8, 9. EBV infection may be a prerequisite for the development of MS 10, 11, 12, 13, 14, 15. However, the underlying molecular mechanisms of this association remain unclear.
Interestingly, recent studies have demonstrated that EBV encodes 44 evolutionarily conserved microRNAs (miRNAs) which are classified into two clusters, the BamHI fragment H rightward open reading frame 1 (BHRF1) and BamHI A rightward transcripts (BART) clusters 16, 17. EBV miRNAs may serve as biomarkers for EBV‐related diseases, including nasopharyngeal carcinoma, leukaemia and chronic active EBV infection 18, 19, 20, 21. Additionally, EBV miRNAs are a novel class of pathogenic molecules of EBV that can specifically reflect the EBV proliferation and influence the immune response by regulating viral and host cellular genes 22, 23, 24, 25, 26. However, the expression of EBV miRNAs in MS patients and its connection with the pathogenesis of MS have not been investigated systematically. To fill this gap, we performed a study measuring the expression levels of EBV miRNAs in MS and preliminarily validated the potential target genes of the significantly altered EBV miRNAs.
Materials and methods
Study subjects
Conforming to the ethical guidelines of the Declaration of Helsinki, the study protocol was approved by the Institutional Review Board of Harbin Medical University, Harbin, China. A signed informed consent was obtained from each participant before collecting blood samples. Clinically diagnosed relapsing–remitting MS (RRMS) patients at relapse (n = 30) and age/gender‐matched healthy controls (n = 30) were recruited from the Neurology Department of the 2nd Affiliated Hospital of Harbin Medical University between 1 September 2014 and 15 June 2016. The mean time between the onset of symptoms of a relapse and the time of blood collection is 5·5 days. The baseline characteristics of the study subjects are listed in Table 1. Regarding inclusion criteria, (1) clinical diagnoses were confirmed using the 2010 revisions to the McDonald diagnostic criteria for multiple sclerosis; and (2) all participants were on periods of relapse and EBV‐immunoglobulin (Ig)G‐positive but EBV‐IgM‐negative. Regarding exclusion criteria, (1) primary progressive and paediatric MS cases were excluded; (2) patients who met the diagnostic criteria of other systemic autoimmune diseases involving central nervous system (CNS) demyelination were excluded; and (3) patients who received treatment within the previous 60 days were also excluded.
Table 1.
Characteristics of the study subjects
| Parameter | HC (n = 30) | MS patients (n = 30) | P‐value |
|---|---|---|---|
| Age (mean ± s.d.) | 34·43 ± 9·35 | 31·13 ± 13·44 | 0·921 |
| Sex | 12 | ||
| Female | 20 | 20 | |
| Male | 10 | 10 | |
| Smoking status | 0·762 | ||
| Ever/current | 8 | 6 | |
| Never | 22 | 24 | |
| Alcohol consumption | 0·732 | ||
| Ever/current | 4 | 6 | |
| Never | 26 | 24 | |
| Expanded disability status scale (EDSS) (mean ± s.d.) | n.a. | 2·80 ± 1·61 | |
HC = healthy controls; MS = multiple sclerosis; n.a. = not applicable; s.d. = standard deviation.
Serum collection and RNA isolation
Venous blood (10 ml) was drawn into a sterile tube with no anti‐coagulant prior to therapy. The tubes were placed upright for 20 min, and samples were centrifuged at 1500 g for 10 min at 20°C. Supernatant sera were harvested immediately and stored at −80°C for analysis.
For quantitative real‐time polymerase chain reaction (qRT–PCR) assay, a well‐vortexed mixture of 100 μl of serum, 200 μl of acid phenol, 200 μl of chloroform and 300 μl of RNase‐free water was made. Then the mixture was centrifuged at 16 000 g at 20°C for 15 min for phase separation. The aqueous layer was harvested and mixed with 1·5 volumes of isopropyl alcohol and 0·1 volume of sodium acetate (3 mol/l, pH = 5·3). The solution was conserved at −20°C for 2 h. The RNA pellet was collected by centrifugation at 16 000 g for 15 min at 4°C. The resulting pellet was washed twice with 750 ml/l ethanol and dried for 20 min at 20°C. The pellet was dissolved in 20 μl of RNase‐free water and conserved at −80°C for analysis.
Individual quantitative reverse‐transcription polymerase chain reaction assay
According to the manufacturer's instructions (7500 Sequence Detection System; Applied Biosystems, Foster City, CA, USA) with a minor modification, TaqMan probe‐based qRT–PCR assays were performed to determine the miRNAs threshold cycle (Ct) value of each serum sample. Briefly, the reverse transcription reaction was performed in 10 μl solution containing RNA (2 μl), 10 mmol/l deoxynucleotides (dNTPs) (1 μl; TaKaRa, Shiga, Japan), avian myeloblastosis virus (AMV) reverse transcriptase (0·5 μl; TaKaRa), a stem‐loop RT primer (1 μl; Applied Biosystems), 5× reverse transcription buffer (2 μl) and diethylpyrocarbonate (DEPC) water (3·5 μl). For cDNA synthesis, the mixtures were incubated at 16°C for 30 min, 42°C for 30 min, 85°C for 5 min and then held at 4°C. Real‐time PCR was performed using an Applied Biosystems 7500 Sequence Detection System as follows: 5 min initial hold at 95°C, followed by 40 cycles of 95°C for 15 s and 60°C for 1 min. Reactions were performed in a final volume of 20 μl containing cDNA (1 μl), TaqMan hydrolysis probe (0·33 μl; Applied Biosystems), rTaq (0·3 μl; TaKaRa), 10 mmol/l dNTPs (0·4 μl; TaKaRa), 25 mmol/l MgCl2(1.2 μl), 10× PCR buffer (2 μl) and DEPC water (14·77 μl). All qPCR reactions were performed in triplicate. After qPCR reactions, the Ct values were acquired by using a fixed threshold value. The expression levels of EBV miRNAs were normalized to the levels of U6 snRNA, which was demonstrated to be expressed stably in the serum according to previous studies. The 2–ΔΔCt equation [ΔΔCt = (Ct miRNA – Ct U6)target – (Ct miRNA – Ct U6)control] was used as a calculation method to test the amount of miRNA relative to the internal control U6 snRNA. In controls, the Ct value of U6 snRNA was 27·90 ± 0·10; in MS patients, the Ct value of U6 snRNA was 27·86 ± 0·20 (P = 0·86). To quantify further and evaluate serum EBV miRNAs as disease markers, absolute miRNAs concentrations were determined using a calibration curve method. Calibration curves for each assay were prepared using 10‐fold serial dilutions of synthetic single‐strand miRNAs for the target miRNAs (synthesized by GenePharma, Shanghai, China). Using the calibration curves, the concentrations of miRNAs were calculated and then normalized to the sample volume.
Target prediction of miRNAs
TargetScan Human Custom (release 5.2) 27 and RNAhybrid software 28 were used to predict the target mRNAs containing the binding site of the seed region (positions 2–7 of miRNA) of up‐regulated circulating EBV miRNAs.
Luciferase reporter assays
Luciferase reporter experiments were conducted to confirm the direct targets of up‐regulated circulating EBV miRNAs. The fragment containing the full‐length 3' untranslated region (UTR) of the target genes was cloned into the pEZX‐MT05 plasmid. HEK 293 cells were seeded at a density of 1 × 106 per well in six‐well culture plates, cultured in Dulbecco's modified Eagle's medium (DMEM) containing 10% fetal bovine serum. Luciferase assays were performed in triplicate using the Luc‐Pair™ miR luciferase assay kit, according to the manufacturer's protocol.
Statistical analysis
The results of miRNAs levels are expressed as the mean ± standard error of the mean (s.e.m.). Statistical differences are considered significant at P < 0·05. We used Student's t‐test to compare the concentrations of serum EBV miRNAs between MS patients and healthy controls. Spearman's rank correlation coefficient was used to examine the correlation between miRNAs expression levels and the expanded disability status scale (EDSS) scores. Receiver operating characteristic (ROC) analysis was used to evaluate the identified miRNAs as diagnostic biomarkers. Area under the ROC curve (AUC) was estimated and reported with its correspondent 95% confidence interval (CI). Statistical analysis was performed using spss version 22.0 software.
Results
Ebv‐miR‐BHRF1‐2‐5p and ebv‐miR‐BHRF1‐3 are up‐regulated in the sera of MS patients
To explore the expression of EBV miRNAs in patients with MS, we conducted a comprehensive profiling of 44 mature EBV miRNAs using commercially available hydrolysis stem‐loop TaqMan probes (Supporting information, Table S1). The expression levels were examined by qRT–PCR using a set of serum samples from 30 RRMS patients at relapse and 30 matched healthy controls. Twenty‐four EBV miRNAs were undetectable or barely detectable in more than 90% of the samples (average Ct value > 35 or the approximate background Ct value). Twenty EBV miRNAs were expressed robustly and detectable in all tested samples. Ebv‐miR‐BHRF1‐2‐5p and ebv‐miR‐BHRF1‐3 showed significantly increased expression in MS patients compared with healthy controls (fold changes = 1·48 and 1·33, P = 0·0005 and 0·0004, respectively) (Fig. 1a,b), while the remaining 18 miRNAs showed no significant difference (Supporting information, Table S2).
Figure 1.
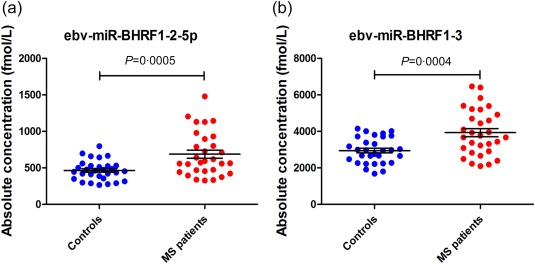
Scattergram of ebv‐miR‐BHRF1‐2‐5p and ebv‐miR‐BHRF1‐3 serum concentrations. (a) The concentrations of ebv‐miR‐BHRF1‐2‐5p in controls and multiple sclerosis (MS) patients were 464·72 ± 24·55 and 687·26 ± 55·08 fmol/l, respectively (P = 0·0005, fold change = 1·48). (b) The concentrations of ebv‐miR‐BHRF1‐3 in controls and MS patients were 2946·14 ± 129·68 and 3929·94 ± 229·82 fmol/l, respectively (P = 0·0004, fold change = 1·33). [Colour figure can be viewed at wileyonlinelibrary.com]
Expression of ebv‐miR‐BHRF1‐2‐5p and ebv‐miR‐BHRF1‐3 in MS patients is correlated positively with EDSS score and allows for discrimination of MS patients from healthy controls
We further analysed the association between the expression levels of ebv‐miR‐BHRF1‐2‐5p and ebv‐miR‐BHRF1‐3 with the EDSS scores of MS patients using Spearman's rank correlation coefficient. Here we showed that the elevated expression of ebv‐miR‐BHRF1‐2‐5p (r s = 0·62, P = 0·0002) and ebv‐miR‐BHRF1‐3 (r s = 0·43, P = 0·017) were associated positively with EDSS scores of MS patients (Fig. 2a,b). ROC analyses further revealed the value of ebv‐miR‐BHRF1‐2‐5p and ebv‐miR‐BHRF1‐3 as biomarkers for MS. ROC curves were drawn using the absolute expression levels of individual miRNAs and their combinations. When independently using ebv‐miR‐BHRF1‐2‐5p and ebv‐miR‐BHRF1‐3, the analyses showed that the AUCs were 0·74 (95% CI = 0·61–0·86) (Fig. 2c) and 0·72 (95% CI = 0·60–0·85) (Fig. 2d), respectively. Using a combination of the two miRNAs, the AUC was 0·76 (95% CI = 0·64–0·88) (Fig. 2e).
Figure 2.
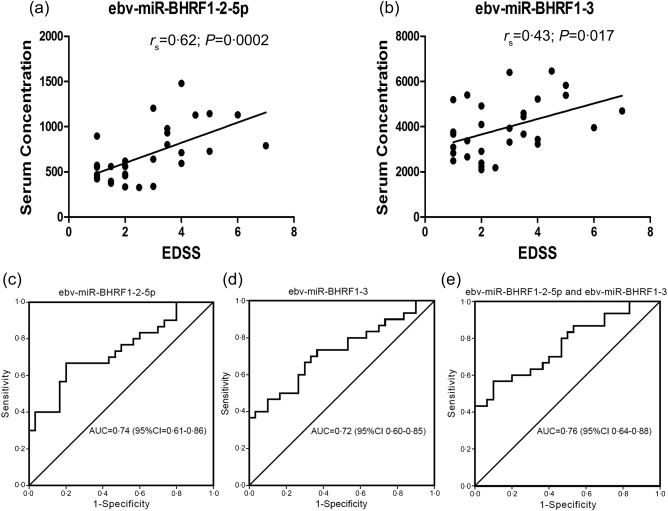
The correlation between serum Epstein–Barr virus (EBV) miRNAs concentration and expanded disability status scale (EDSS) scores and receiver operating characteristic (ROC) analyses. (a,b) Ebv‐miR‐BHRF1‐2‐5p (r s = 0·62, P = 0·0002) and ebv‐miR‐BHRF1‐3 (r s = 0·43, P = 0·017) were correlated positively with EDSS scores of MS patients. (c,d) When independently using ebv‐miR‐BHRF1‐2‐5p and ebv‐miR‐BHRF1‐3, the areas under the curve (AUCs) were 0·74 [95% confidence interval (CI) = 0·61–0·86] and 0·72 (95% CI = 0·60–0·85), respectively. (e) ROC curve of ebv‐miR‐BHRF1‐2‐5p combined with ebv‐miR‐BHRF1‐3; the AUC was 0·76 (95% CI = 0·64–0·88).
Prediction and validation of mucosa‐associated lymphoid tissue lymphoma transport protein 1 (MALT1) as a direct target of ebv‐miR‐BHRF1‐2‐5p
Two computational algorithms (TargetScan Human Custom 27, RNAhybrid 28) and literature reviews were used in combination to identify the target genes of ebv‐miR‐BHRF1‐2‐5p and ebv‐miR‐BHRF1‐3. Among the hundreds of candidate genes, MALT1 was predicted to have two potential binding sites for ebv‐miR‐BHRF1‐2‐5p on the 3' untranslated region (3' UTR). The predicted conjugation between the seed region of ebv‐miR‐BHRF1‐2‐5p and the binding sites (on positions 3519 and 4319) within the MALT1 mRNA is illustrated in Fig. 3a. Furthermore, literature reviews showed that MALT1 played considerable roles in immune homeostasis 29, 30. Therefore, we presumed that MALT1 was a potential target of ebv‐miR‐BHRF1‐2‐5p. Subsequently, we co‐transfected HEK293 cells with luciferase report vectors containing MALT1 3' UTR binding sites and ebv‐miR‐BHRF1‐2‐5p or scrambled control RNA. Luciferase reporter assays confirmed that ebv‐miR‐BHRF1‐2‐5p suppressed the luciferase activity significantly in contrast to the control RNA (Fig. 3b). Additionally, we discovered a binding site for ebv‐miR‐BHRF1‐3 within the mRNA of PTEN (phosphatase and tensin homologue deleted on chromosome 10), which has been confirmed previously as a target of ebv‐miR‐BHRF1‐3 31.
Figure 3.
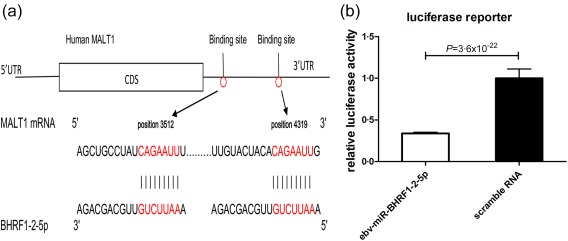
Prediction and validation of human MALT1 as a direct ebv‐miR‐BHRF1‐2‐5p target. (a) Schematic depicting the hypothetical duplexes formed by MALT1 3' untranslated region (UTR) and ebv‐miR‐BHRF1‐2‐5p interactions. The seed regions of ebv‐miR‐BHRF1‐2‐5p and the seed‐recognition sites in the MALT1 3' UTR are marked in red. Paired bases are marked with a black line. (b) The results were calculated as the ratio of luciferase activity in the ebv‐miR‐BHRF1‐2‐5p transfected cells normalized to the negative control RNA‐transfected cells. Over‐expression of ebv‐miR‐BHRF1‐2‐5p resulted in an approximately 66% reduction in luciferase reporter activity compared with the control. The results are presented as the mean ± standard deviation from three independent experiments (P = 3·6 × 10−22, ebv‐miR‐BHRF1‐2‐5p versus scramble RNA). [Colour figure can be viewed at wileyonlinelibrary.com]
Discussion
Previous studies have revealed that miRNAs encoded by EBV are associated with the aetiopathogenesis of neoplastic diseases, including nasopharyngeal carcinoma 19, leukaemia and Burkitt's lymphoma 20, and potentially exert the anti‐apoptotic function by suppressing the expression of PUMA (p53 up‐regulated modulator of apoptosis) and pro‐apoptotic protein Bid (the BH3‐interacting domain death agonist) 32, 33. Moreover, accumulating studies have highlighted the roles of EBV miRNAs in the regulation of the host's immune response. For example, ebv‐miR‐BHRF1‐3 could inhibit CXCL‐11 (C‐X‐C motif chemokine ligand 11), an interferon (IFN)‐inducible T cell attracting chemokine 34. Ebv‐miR‐BART15 potentially target NLRP3 (nucleotide‐binding oligomerization domain‐like receptor protein 3) and reduce endogenous NLRP3 protein levels and IL‐1β production 35. MICB (MHC class I‐related genes B) and IPO7 (Importin 7), which are involved in immune recognition of infected cells and the innate immunity, are the targets of ebv‐miR‐BART2‐5p and ebv‐miR‐BART3, respectively 26, 36. These findings showed that EBV miRNAs may lead to immune imbalance and autoimmune diseases. However, the expression of EBV miRNAs in MS patients and the connection with the pathogenesis of MS have not been studied systemically.
In this study, we investigated the expression profile of 44 EBV miRNAs in the sera of 30 RRMS patients at relapse and 30 matched controls using TaqMan probe‐based qRT–PCR assays. The expression levels of two BHRF1 miRNAs, including ebv‐miR‐BHRF1‐2‐5p and ebv‐miR‐BHRF1‐3, were elevated noticeably and correlated positively with EDSS scores of MS patients. In ROC analyses, the AUC was 0·76 when using a combination of the two miRNAs, confirming that ebv‐miR‐BHRF1‐2‐5p and ebv‐miR‐BHRF1‐3 allow discrimination of MS patients from healthy control subjects. Previous studies have shown that ebv‐miR‐BHRF1‐2 and ebv‐miR‐BHRF1‐3 were increased rapidly followed by the entry of EBV into the lytic cycle 37, which was related to the disease activity in MS patients 38. The specific elevated expressions of ebv‐miR‐BHRF1‐2‐5p and ebv‐miR‐BHRF1‐3 are related to the activation of EBV, which may contribute to the relapse of MS and suggest a potential role in the pathogenesis of MS. Recent studies have discovered that mature EBV miRNAs are transported by exosomes, which protect them from degradation by RNases 39. This implies that EBV‐encoded miRNAs are stable in the serum so that they can be used as a diagnostic marker and monitor of EBV‐associated disorders, including MS 40. EBV miRNAs may also be transferred into non‐infected recipient cells via exosomes, suggesting a role of intercellular communication in EBV biology 41.
Furthermore, we preliminarily validated that MALT1 may be a target of ebv‐miR‐BHRF1‐2‐5p. The latest researches demonstrated that MALT1 is indispensable for the development of regulatory T cells (Tregs) and plays a considerable role in immune homeostasis 29, 30. Mice with inactivation of the MALT1 protease activity showed absence of Tregs, increased T helper type 1 (Th1) and Th2 cells and elevated production of IFN‐γ and interleukin (IL)‐4, which induced imbalance of immunity, causing lymphocyte infiltration and multi‐organ inflammation 29, 30, 42. Two recent studies reported that MALT1 deficiency was associated with multi‐organ autoimmunity 43, 44. In addition, the MALT1 expression level was down‐regulated in peripheral blood mononuclear cells of patients with rheumatoid arthritis, which was also a systemic autoimmune disease that sometimes overlaps MS 45. Therefore, we postulated that the up‐regulation of ebv‐miR‐BHRF1‐2‐5p possibly affects the immune system through inhibiting MALT1. Additionally, ebv‐miR‐BHRF1‐3 was confirmed to target PTEN, a regulator of the phosphatidylinositol 3‐kinase (PIK3)/Akt (protein kinase B) survival pathway 31, 46. PTEN plays multiple regulatory roles in the pathogenesis of autoimmunity 47, including maintaining the expression of the transcription factor forkhead box protein 3 (FoxP3) and restraining T helper 1 and T follicular helper cell responses 47, 48. Deficiency of PTEN leads to systemic autoimmune disease because of decreased Tregs and increased IFN‐γ 49. Collectively, the up‐regulation of ebv‐miR‐BHRF1‐2‐5p and ebv‐miR‐BHRF1‐3 may induce immune imbalance by targeting host genes, suggesting a potential role of EBV miRNAs in the aetiopathogenesis of MS.
In summary, we have demonstrated that ebv‐miR‐BHRF1‐2‐5p and ebv‐miR‐BHRF1‐3 are elevated significantly in the sera of RRMS patients at relapse and ebv‐miR‐BHRF1‐2‐5p may directly target MALT1, which is a key regulator of immune homeostasis. Although great advances have been made in understanding the protease activity of MALT1 and autoimmune diseases, further investigations of the potential role of EBV‐miRNA targeting MALT1 are still required. The specific aberrantly expressed EBV miRNAs in MS possibly regulate the immune response and contribute to the pathogenesis of MS.
Disclosure
The authors declare no conflicts of interest.
Author contributions
Y. W., H. L., X. C., J. F. and D. H. designed the experiments; Y. W. and D. H. performed experiments; Y. W., D. H., X. C., J. F. and H. L. analysed data and wrote the manuscript; D. Y., H. Y., X. Z., R. W., B. L., H. Y., Y. L., Y. C., Y. D. and C. Z. participated in part experiments and contributed materials. All authors approved the final manuscript.
Supporting information
Additional Supporting information may be found in the online version of this article at the publisher's web‐site:
Table S1. The mature miRNA sequence and product/catalogue numbers of all tested miRNAs
Table S2. Average Ct values (with standard error of the mean) of 20 expressed (average Ct < 35 and distinguishable from water background) Epstein–Barr virus (EBV) miRNAs in the sera of healthy controls and multiple sclerosis (MS) patients
Acknowledgements
This work was supported by grants from the National Basic Research Program of China (973 Program, 2012CB517603 and 2011CB504803), the National Natural Science Foundation of China (No. 81171120, 30988003, 30225037, 30471991 and 30570731).
Contributor Information
X. Chen, Email: xichen@nju.edu.cn
J. Fu, Email: fujin0612@126.com.
References
- 1. Milo R, Kahana E. Multiple sclerosis: geoepidemiology, genetics and the environment. Autoimmun Rev 2010; 9:A387–94. [DOI] [PubMed] [Google Scholar]
- 2. Frohman EM, Racke MK, Raine CS. Multiple sclerosis – the plaque and its pathogenesis. N Engl J Med 2006; 354:942–55. [DOI] [PubMed] [Google Scholar]
- 3. Pender MP, Burrows SR. Epstein–Barr virus and multiple sclerosis: potential opportunities for immunotherapy. Clin Transl Immunology 2014; 3:e27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Belbasis L, Bellou V, Evangelou E, Ioannidis JP, Tzoulaki I. Environmental risk factors and multiple sclerosis: an umbrella review of systematic reviews and meta‐analyses. Lancet Neurol 2015; 14:263–73. [DOI] [PubMed] [Google Scholar]
- 5. Larsen PD, Bloomer LC, Bray PF. Epstein–Barr nuclear antigen and viral capsid antigen antibody titers in multiple sclerosis. Neurology 1985; 35:435–8. [DOI] [PubMed] [Google Scholar]
- 6. Ascherio A, Munger KL, Lennette ET et al Epstein–Barr virus antibodies and risk of multiple sclerosis: a prospective study. JAMA 2001; 286:3083–8. [DOI] [PubMed] [Google Scholar]
- 7. Alotaibi S, Kennedy J, Tellier R, Stephens D, Banwell B. Epstein–Barr virus in pediatric multiple sclerosis. JAMA 2004; 291:1875–9. [DOI] [PubMed] [Google Scholar]
- 8. Kvistad S, Myhr KM, Holmoy T et al Antibodies to Epstein–Barr virus and MRI disease activity in multiple sclerosis. Mult Scler 2014; 20:1833–40. [DOI] [PubMed] [Google Scholar]
- 9. Mameli G, Cossu D, Cocco E et al EBNA‐1 IgG titers in Sardinian multiple sclerosis patients and controls. J Neuroimmunol 2013; 264:120–2. [DOI] [PubMed] [Google Scholar]
- 10. Fernandez‐Menendez S, Fernandez‐Moran M, Fernandez‐Vega I, Perez‐Alvarez A, Villafani E J. Epstein–Barr virus and multiple sclerosis. From evidence to therapeutic strategies. J Neurol Sci 2016; 361:213–9. [DOI] [PubMed] [Google Scholar]
- 11. Lossius A, Johansen JN, Vartdal F et al High‐throughput sequencing of TCR repertoires in multiple sclerosis reveals intrathecal enrichment of EBV‐reactive CD8+ T cells. Eur J Immunol 2014; 44:3439–52. [DOI] [PubMed] [Google Scholar]
- 12. Torring C, Andreasen C, Gehr N, Bjerg L, Petersen T, Hollsberg P. Higher incidence of Epstein–Barr virus‐induced lymphocyte transformation in multiple sclerosis. Acta Neurol Scand 2014; 130:90–6. [DOI] [PubMed] [Google Scholar]
- 13. Magliozzi R, Serafini B, Rosicarelli B et al B‐cell enrichment and Epstein–Barr virus infection in inflammatory cortical lesions in secondary progressive multiple sclerosis. J Neuropathol Exp Neurol 2013; 72:29–41. [DOI] [PubMed] [Google Scholar]
- 14. Mameli G, Cossu D, Cocco E et al Epstein–Barr virus and Mycobacterium avium subsp. paratuberculosis peptides are cross recognized by anti‐myelin basic protein antibodies in multiple sclerosis patients. J Neuroimmunol 2014; 270:51–5. [DOI] [PubMed] [Google Scholar]
- 15. Pender MP. The essential role of Epstein–Barr virus in the pathogenesis of multiple sclerosis. Neuroscientist 2011; 17:351–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Barth S, Meister G, Grasser FA. EBV‐encoded miRNAs. Biochim Biophys Acta 2011; 1809:631–40. [DOI] [PubMed] [Google Scholar]
- 17. Pfeffer S, Zavolan M, Grasser FA et al Identification of virus‐encoded microRNAs. Science 2004; 304:734–6. [DOI] [PubMed] [Google Scholar]
- 18. Lung RW, Tong JH, To KF. Emerging roles of small Epstein–Barr virus derived non‐coding RNAs in epithelial malignancy. Int J Mol Sci 2013; 14:17378–409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Wong AM, Kong KL, Tsang JW, Kwong DL, Guan XY. Profiling of Epstein–Barr virus‐encoded microRNAs in nasopharyngeal carcinoma reveals potential biomarkers and oncomirs. Cancer 2012; 118:698–710. [DOI] [PubMed] [Google Scholar]
- 20. De Falco G, Antonicelli G, Onnis A, Lazzi S, Bellan C, Leoncini L. Role of EBV in microRNA dysregulation in Burkitt lymphoma. Semin Cancer Biol 2009; 19:401–6. [DOI] [PubMed] [Google Scholar]
- 21. Marquitz AR, Mathur A, Chugh PE, Dittmer DP, Raab‐Traub N. Expression profile of microRNAs in Epstein–Barr virus‐infected AGS gastric carcinoma cells. J Virol 2014; 88:1389–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kincaid RP, Sullivan CS. Virus‐encoded microRNAs: an overview and a look to the future. PLOS Pathog 2012; 8:e1003018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Kuzembayeva M, Hayes M, Sugden B. Multiple functions are mediated by the miRNAs of Epstein–Barr virus. Curr Opin Virol 2014; 7:61–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Cai X, Schafer A, Lu S et al Epstein–Barr virus microRNAs are evolutionarily conserved and differentially expressed. PLoS Pathog 2006; 2:e23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Barth S, Pfuhl T, Mamiani A et al Epstein–Barr virus‐encoded microRNA miR‐BART2 down‐regulates the viral DNA polymerase BALF5. Nucleic Acids Res 2008; 36:666–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Dolken L, Malterer G, Erhard F et al Systematic analysis of viral and cellular microRNA targets in cells latently infected with human gamma‐herpesviruses by RISC immunoprecipitation assay. Cell Host Microbe 2010; 7:324–34. [DOI] [PubMed] [Google Scholar]
- 27. Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell 2005; 120:15–20. [DOI] [PubMed] [Google Scholar]
- 28. Rehmsmeier M, Steffen P, Hochsmann M, Giegerich R. Fast and effective prediction of microRNA/target duplexes. RNA 2004; 10:1507–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Jaworski M, Marsland BJ, Gehrig J et al Malt1 protease inactivation efficiently dampens immune responses but causes spontaneous autoimmunity. EMBO J 2014; 33:2765–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Gewies A, Gorka O, Bergmann H et al Uncoupling Malt1 threshold function from paracaspase activity results in destructive autoimmune inflammation. Cell Rep 2014; 9:1292–305. [DOI] [PubMed] [Google Scholar]
- 31. Bernhardt K, Haar J, Tsai MH, Poirey R, Feederle R, Delecluse HJ. A viral microRNA cluster regulates the expression of PTEN, p27 and of a bcl‐2 homolog. PLoS Pathog 2016; 12:e1005405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Choy EY, Siu KL, Kok KH et al An Epstein–Barr virus‐encoded microRNA targets PUMA to promote host cell survival. J Exp Med 2008; 205:2551–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Shinozaki‐Ushiku A, Kunita A, Isogai M et al Profiling of virus‐encoded microRNAs in Epstein–Barr virus‐associated gastric carcinoma and their roles in gastric carcinogenesis. J Virol 2015; 89:5581–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Xia T, O'Hara A, Araujo I et al EBV microRNAs in primary lymphomas and targeting of CXCL‐11 by ebv‐mir‐BHRF1‐3. Cancer Res 2008; 68:1436–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Haneklaus M, Gerlic M, Kurowska‐Stolarska M et al Cutting edge: miR‐223 and EBV miR‐BART15 regulate the NLRP3 inflammasome and IL‐1beta production. J Immunol 2012; 189:3795–9. [DOI] [PubMed] [Google Scholar]
- 36. Nachmani D, Stern‐Ginossar N, Sarid R, Mandelboim O. Diverse herpesvirus microRNAs target the stress‐induced immune ligand MICB to escape recognition by natural killer cells. Cell Host Microbe 2009; 5:376–85. [DOI] [PubMed] [Google Scholar]
- 37. Amoroso R, Fitzsimmons L, Thomas WA, Kelly GL, Rowe M, Bell AI. Quantitative studies of Epstein–Barr virus‐encoded microRNAs provide novel insights into their regulation. J Virol 2011; 85:996–1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Angelini DF, Serafini B, Piras E et al Increased CD8+ T cell response to Epstein–Barr virus lytic antigens in the active phase of multiple sclerosis. PLOS Pathog 2013; 9:e1003220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Pegtel DM, Cosmopoulos K, Thorley‐Lawson DA et al Functional delivery of viral miRNAs via exosomes. Proc Natl Acad Sci USA 2010; 107:6328–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Gallo A, Tandon M, Alevizos I, Illei GG. The majority of microRNAs detectable in serum and saliva is concentrated in exosomes. PLOS ONE 2012; 7:e30679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Nanbo A, Kawanishi E, Yoshida R, Yoshiyama H. Exosomes derived from Epstein–Barr virus‐infected cells are internalized via caveola‐dependent endocytosis and promote phenotypic modulation in target cells. J Virol 2013; 87:10334–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Bornancin F, Renner F, Touil R et al Deficiency of MALT1 paracaspase activity results in unbalanced regulatory and effector T and B cell responses leading to multiorgan inflammation. J Immunol 2015; 194:3723–34. [DOI] [PubMed] [Google Scholar]
- 43. Charbit‐Henrion F, Jeverica AK, Begue B et al Deficiency in mucosa associated lymphoid tissue lymphoma translocation 1 (MALT1): a novel cause of IPEX‐like syndrome. J Pediatr Gastroenterol Nutr 2017; 64:378–84. [DOI] [PubMed] [Google Scholar]
- 44. Punwani D, Wang H, Chan AY et al Combined immunodeficiency due to MALT1 mutations, treated by hematopoietic cell transplantation. J Clin Immunol 2015; 35:135–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Wang X, Zhu L, Liao Z et al Alternative expression pattern of MALT1‐A20‐NF‐kappaB in patients with rheumatoid arthritis. J Immunol Res 2014; 2014:492872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Huynh A, DuPage M, Priyadharshini B et al Control of PI(3) kinase in Treg cells maintains homeostasis and lineage stability. Nat Immunol 2015; 16:188–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Liu X, Karnell JL, Yin B et al Distinct roles for PTEN in prevention of T cell lymphoma and autoimmunity in mice. J Clin Invest 2010; 120:2497–507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Shrestha S, Yang K, Guy C, Vogel P, Neale G, Chi H. Treg cells require the phosphatase PTEN to restrain TH1 and TFH cell responses. Nat Immunol 2015; 16:178–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Ray JP, Craft J. PTENtiating autoimmunity through Treg cell deregulation. Nat Immunol 2015; 16:139–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional Supporting information may be found in the online version of this article at the publisher's web‐site:
Table S1. The mature miRNA sequence and product/catalogue numbers of all tested miRNAs
Table S2. Average Ct values (with standard error of the mean) of 20 expressed (average Ct < 35 and distinguishable from water background) Epstein–Barr virus (EBV) miRNAs in the sera of healthy controls and multiple sclerosis (MS) patients


