Abstract
Detecting the phosphorylation substrates of multiple kinases in a single experiment is a challenge, and new techniques are being developed to overcome this challenge. Here, we utilized a multiplexed assay for kinase specificity (MAKS) to identify the substrates directly and to map the phosphorylation site(s) of plant symbiotic receptor-like kinases. The symbiotic receptor-like kinases Nodulation Receptor-like Kinase (NORK) and Lysin motif domain-containing receptor-like kinase 3 (LYK3) are indispensable for the establishment of root nodule symbiosis. Although some interacting proteins have been identified for these symbiotic receptor-like kinases, very little is known about their phosphorylation substrates. Using this high-throughput approach, we identified several other potential phosphorylation targets for both these symbiotic receptor-like kinases. In particular, we also discovered the phosphorylation of LYK3 by NORK itself which was also confirmed by pair-wise kinase assays. Motif analysis of potential targets for these kinases revealed that the acidic motif xxxsDxxx was common to both of them. In summary, this high-throughput technique catalogs the potential phosphorylation substrates of multiple kinases in a single efficient experiment, the biological characterization of which should provide a better understanding of phosphorylation signaling cascade in symbiosis.
Keywords: Receptor-like Kinase, protein phosphorylation, kinase assay, isobaric tags, IMAC, tandem mass spectrometry, symbiosis, signaling, Medicago truncatula
INTRODUCTION
Proteins are vital cogs for normal cell functioning. In addition to providing structural support to the cells, proteins are involved in catalysis, transport, storage, or signal transmission (Duan and Walther, 2015). Even in this era of systems biology, studying proteomes is more challenging due to their complex and dynamic nature. A single gene can encode several proteins due to their multi-level regulation, such as alternative splicing, multiple reading frames, and post-translational modifications (PTMs)(Silva-Sanchez et al., 2014). For instance, the human genome, which consists of about 30,000 genes, can encode anywhere between 200,000 to 2 million proteins (Van Eyk and Dunn, 2006). Chief among the mechanisms that increase the functional diversity of the cellular proteome is the PTMs, which are modification(s) either during or after the synthesis of proteins by the ribosome (Silva-Sanchez et al., 2015; Pawson and Scott, 1997). At least 300 different PTMs are known so far, and it is widely believed that PTMs are essential for the biological functioning of synthesized proteins (Silva-Sanchez et al., 2014; Witze et al., 2007).
Protein phosphorylation is the most prominent reversible PTM, catalyzed by two classes of enzymes—protein kinases and phosphatases—that alter the localization, binding, configuration, protein interactions, and complex formations of proteins. Modified proteins, in turn, regulate intracellular biological functions (Bond et al., 2011; Khoury et al., 2011). Protein kinases are one of the largest gene families in eukaryotes (Manning, 2002) and due to recent whole-genome duplication events in flowering plants, this family of genes is significantly larger than in other eukaryotic lineages, suggesting their importance in plant species (Lehti-Shiu and Shiu, 2012). Receptor-like kinases (RLKs) are protein kinases which consist of a ligand-binding extracellular domain, a transmembrane domain, and an intracellular kinase domain. They are involved in plant growth and development, stress response, cell regulation, metabolism, and plant–microbe interactions (Shiu and Bleecker, 2001; Lehti-Shiu and Shiu, 2012; Stone and Walker, 1995). Despite the importance of protein kinases in plants, very few kinases have been characterized functionally. Only a small fraction of the identified plant RLKs have a known function (Shiu and Bleecker, 2001; Gish and Clark, 2011). The identification of potential phosphorylation targets/substrates of these kinases will immensely aid in the functional characterization of these kinases. However, this process is both challenging and limiting. Current approaches for target identification of protein kinases in plants consist of performing a protein–protein interaction assay, such as yeast two-hybrid (Y2H) or co-immunoprecipitation (CO-IP) assays, using the kinase(s) of interest. Protein interaction studies are often followed by pair-wise in vitro kinase assays using radiolabeled ATP to determine the phosphorylation status of interacting protein(s).
Although widely used, the major drawback of the classical Y2H screen is false positive or false negative results (Brückner et al., 2009; Phizicky and Fields, 1995). Several factors, such as steric hindrance of a reporter protein, choice of the protein used for fusion in bait or prey, or proteins for which posttranslational modifications is a prerequisite for interaction but that does not occur in yeast cells, may contribute to false negatives. Conversely, high expression of bait or prey proteins, mis-localization of these proteins in compartments not similar to their native cell environment, mis-folding of proteins, and interaction of prey proteins with reporter proteins may lead to false positives (Brückner et al., 2009). Similar to Y2H assays, CO-IPs have been used in thousands of protein–protein interaction studies, but this technique is not very sensitive and often fails to detect transient protein–protein interactions—especially those involving kinases and phosphatases (Phizicky and Fields, 1995; Daniel et al., 2013). Also, the interactors identified may be a part of the larger protein complex and therefore a direct interaction cannot be inferred (Phizicky and Fields, 1995; Daniel et al., 2013). Although not yet common in plant research, chip-based approaches have been used for the identification of phosphorylation substrates of kinases. These include a screening of random peptide libraries containing degenerate penta- or heptapeptides and identifying the putative substrate(s) using radioactivity. Kinome profiling, using chips spotted with consensus kinase substrates selected from the phosphorylated peptides database, Protein Kinase Research, can be used for identifying kinase substrates. This chip consists of substrates from different organisms including bacteria, plants, animals, and fungi, making the identification of substrates specific to plant kinases cumbersome (Ritsema et al., 2007; Ritsema and Peppelenbosch, 2009). Also, positional scanning peptide libraries, where a semi-degenerate 20-mer peptide library with the phospho-acceptor amino acid fixed in the center position, are also used for identifying the phosphorylation motif of plant kinases (Sugiyama and Ishihama, 2016; Sirichandra et al., 2010). However, the approaches mentioned above usually use radioactivity to identify putative targets, and individual phosphorylation sites cannot be identified. Kinase-client assay (KiC assay) is a further improvement in the chip–based technology. In this approach, kinase activity can be quantified by comparing the spectral counts of phosphorylated and non-phosphorylated peptides, and also individual phosphosites can be mapped. Using KiC several putative targets of 17 different kinases were identified, and phosphorylation of Arabidopsis thaliana protein phosphatase inhibitor-2 (AtPPI-2) by three different kinases was confirmed (Ahsan et al., 2013). Instead of peptide libraries, arrayed-protein chips, where individual proteins are expressed, purified and immobilized on chips and incubated with kinase of interest are used to identify potential substrate of kinases. This method was utilized for the identification of phosphorylation targets of Arabidopsis thaliana mitogen-activated protein kinases (MAPK) in plants (Feilner et al., 2005). Although not impossible, recombinant expression and purification of all the proteins are expensive and difficult. Furthermore, scaling-up of these chip-based libraries (either peptide or protein libraries) are expensive and limits the number of kinases being screened. Most peptide library approaches identify the phosphorylation motif likely to be phosphorylated by the kinases. Since plant genomes are large there may be hundreds if not thousands of proteins with those phosphorylation motifs, screening all those proteins to identify the actual phosphorylation substrate becomes a limiting factor for these approaches (Miller and Turk, 2016).
In the current study, we a used a multiplexed assay for kinase specificity (MAKS), previously used in yeast and human stem cells, to identify the phosphorylation substrates and sites of two symbiotic-receptor-like kinases: Lysin motif domain-containing receptor-like kinase 3 (LYK3) and Nodulation receptor-like kinase (NORK) sometimes referred to as Does not make infections2 (DMI2). These proteins are required for legume nodulation and arbuscular mycorrhizal associations in the model legume Medicago truncatula (Arrighi et al., 2006; Gabriella Endre et al., 2002; Patrick Smit et al., 2007). This approach provides information about putative targets of kinases which can be further confirmed by a targeted approach. Here we used crude protein extracts as the target instead of a peptide library. The main advantage of MAKS is that in a single experiment, multiple kinases can be monitored for their activity using isobaric tagging technology (Brumbaugh et al., 2014). Also, we can identify phosphorylation substrates without any of the methods used for detecting protein–protein interactions, thus avoiding their drawbacks. Furthermore, the putative targets obtained from our approach are much more relevant than those obtained using peptide library because protein extracts were used instead of peptides. MAKS identified several substrates for both symbiotic receptor-like kinases that we tested. Of particular interest is the phosphorylation of LYK3 by NORK. We confirmed this interaction using pair-wise in vitro kinase experiments, indicating that this method can be used to identify the substrates and interaction partners of other receptor-like kinases. Also, we identified the motifs that were most likely to be phosphorylated by these symbiotic receptor-like kinases. In summary, MAKS is a valid and efficient method broadly applicable to many kinases in plant sciences.
RESULTS AND DISCUSSION
Multiplexed assay to identify targets of protein kinases in plants
Phosphorylation/dephosphorylation is a dynamic process and various properties of the phosphorylated proteins, such as low stoichiometry, heterogeneity, along with the dynamic range of the technique used, can affect the coverage detection and quantification of phosphoproteins/peptides (Grimsrud et al., 2010; Jayaraman et al., 2012; Silva-Sanchez et al., 2014; Wenger et al., 2011). Hence there is a continuous need to develop and advance techniques including those related to sample preparation, enrichment, and fragmentation to achieve higher coverage and detection (Grimsrud et al., 2010; Silva-Sanchez et al., 2014). Although research in the last two decades using model legumes has led to the identification of several components of the symbiotic signaling cascade, the underlying mechanism of signal processing from symbiotic receptor-like kinases localized in the plasma membrane to the nucleus is not fully understood. Because of the involvement of several symbiotic receptor-like kinases, we suppose that a symbiotic signaling cascade, mediated by protein phosphorylation and dephosphorylation, might be established, thereby transducing the signals from the plasma membrane to the nucleus. Therefore, this study was conducted to study the phosphorylation specificity of these symbiotic receptor-like kinases with the aim of identifying novel components and substrates of symbiotic receptor-like kinases to dissect the pathway more precisely. We focused on identifying the autophosphorylation sites and substrates for LYK3 and NORK. One of the bottlenecks of studying phosphorylation specificity is the low-throughput nature of traditional in vitro kinase assays. Conventionally, kinase assays involve an in vitro reaction, after which the transfer of a radiolabeled phosphoryl group to a given substrate is detected, directly measuring phosphorylation but requiring the use of hazardous materials (Brumbaugh et al., 2014). Also, this method cannot multiplex kinases and substrates or directly localize phosphorylation to a single amino acid when more than one potential site is present. To multiplex kinase assays and identify potential phosphorylation substrates and the phosphorylation sites of the symbiotic receptor-like kinase(s), we used a multiplexed assay for kinase specificity. To our knowledge, this is the first time that this approach is used for plant proteins (Brumbaugh et al., 2014).
A heterologous Escherichia coli bacterial expression system was used for the expression of the kinase domains of both these proteins. For this, the kinase domains of LYK3 and NORK were tagged to glutathione s-transferase (GST) and maltose binding protein (MBP) tags, respectively. The expressed kinase domains of LYK3 and NORK were purified using Glutathione Superflow and amylose resin, respectively. In the initial step, crude protein extracts from one week old Medicago seedlings were mixed with these purified candidate kinases plus kinase buffer. As a mock control kinase buffer alone mixed with crude Medicago protein(s) was used (Figure 1). After a 30 minute incubation, each sample was enzymatically digested and differentially labeled with isobaric tags, rendering a unique chemical signature to each peptide and simultaneously linking it to the reaction from which it originated (Thompson et al., 2003). Following labeling, the six samples were combined and enriched for phosphopeptides using immobilized metal affinity chromatography (IMAC). Peptides were separated by reversed-phase (RP) chromatography and introduced to the mass spectrometer, where precursor charges were isolated and subjected to tandem mass spectrometry (MS/MS), providing sequence identity, phosphorylation site localization, and quantitative information in a single scan. During fragmentation, the isobaric tags are cleaved, creating reporter ions. The intensity of these reporter ions can serve as a proxy for phosphorylation abundance following the kinase reaction. The spectrum in Figure 2A shows an MS/MS scan mapped to AsNILLDK2+, a tryptic peptide of cysteine-rich receptor-kinase-like protein. Fragment ions allow the phosphorylation event to be confidently localized to Ser488. The inset depicting the reporter tag mass-to-charge (m/z) indicates strong up-regulation of this phosphorylation site in response to the LYK3 kinase. Using this multiplexed assay for kinase specificity approach, we identified multiple putative substrates of both these symbiotic receptor-like kinases, and they fall into various categories, such as receptor-like kinases, storage proteins, RNA binding proteins, transporters, catalytic enzymes, transcription factors, and ubiquitin ligases.
Figure 1. Overview of the analysis strategy.
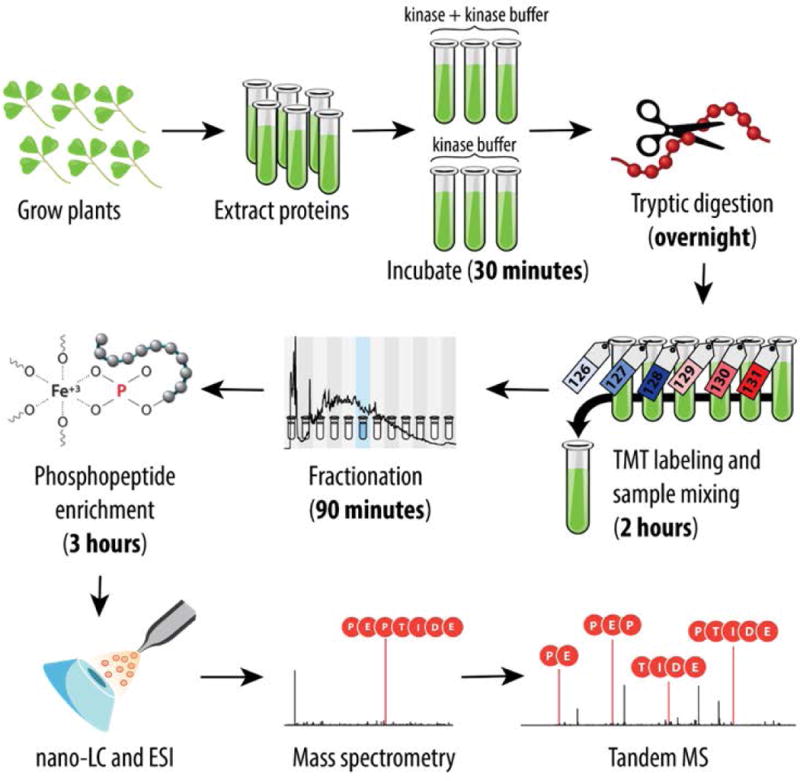
Six plant replicates were harvested, and proteins were extracted through a chloroform/methanol precipitation. Three of the replicates were incubated with a kinase reaction buffer (controls), while the other three replicates were incubated with a kinase reaction buffer and a purified kinase (either LYK3 or NORK). The resultant proteins were lysed, and trypsin digested, and each replicate was TMT-labeled and combined. Samples were fractionated, then analyzed via nanoLC-MS/MS. Estimated time period for some of the major steps is represented in brackets below each step.
Figure 2. Identification of phosphorylation sites for the substrates of symbiotic receptor-like kinases.
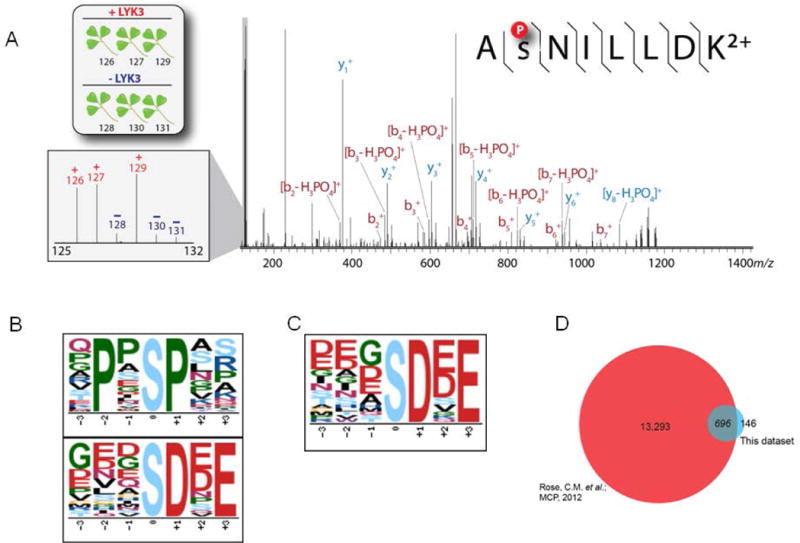
(A) Tandem mass spectrum of the phosphorylated peptide to AsNILLDK2+, a tryptic peptide of cysteine-rich receptor-kinase-like protein. Reporter tags are present in the low mass region (inset). The intensity of these reporter tags can be used to determine the amount of kinase-directed phosphorylation. Here, the phosphorylation intensity of the LYK3 treated channels (126, 127 and 129 m/z) show an increase in abundance compared to the control channels (128, 130, 131 m/z). Motif analysis of Medicago lysate treated with either (B) LYK3 or (C) NORK purified kinase. (D) Comparison of phosphopeptides from our previous work to the current work. Previously we identified 13293 phosphopeptides specific to symbiotic signaling in Medicago (Rose et al., 2012) and the current studies a total of 842 phosphopeptides were identified out of which 696 overlapped with our previous work and 146 were unique to this study.
Broad classification of phosphorylated proteins
Tables 1 and 2 present proteins displaying the greatest increase in phosphorylation after incubation with either LYK3 (Table 1) or NORK (Table 2). These proteins have never been characterized in Medicago symbiosis, suggesting that they may be the possible missing links in connecting plasma membrane signals to the nuclear events in the symbiotic signaling cascade. In addition to these proteins, other interesting proteins with significant fold changes (p-value < 0.10) are presented in Supplementary Tables S1 and S2. This compiled list offers some insight into the potential interactors of the tested symbiotic receptor-like kinases.
Table 1. A list of the top 15 putative phosphorylation targets of LYK3.
The table contains both localized and non-localized proteins calculated at 1% FDR with a p-value less than 0.10. Non-Localized phosphorylation sites are represented by (NL).
| Gene ID | Annotation | Phospho-isoform | Fold Change (linear) |
|---|---|---|---|
| Medtr1g021635 | Cysteine-rich receptor-kinase-like protein | S488 | 12.05 |
| Medtr8g089695 | Transmembrane protein; putative | T189 | 11.28 |
| Medtr3g086520 | E3 ubiquitin-protein ligase RGLG2-like protein | S395 | 4.72 |
| Medtr4g061610 | Nonphototropic hypocotyl protein | S152 | 4.07 |
| Medtr4g074930 | RNA recognition motif-RBD protein | T368 | 3.15 |
| Medtr3g098420 | 110 kDa 4SNc-tudor domain protein | (NL) | 3.05 |
| Medtr4g075410 | MAP kinase kinase kinase | S409 | 2.66 |
| Medtr4g125330 | Hypothetical protein | S167 | 2.63 |
| Medtr8g089785 | Plant-specific eukaryotic initiation factor | S464 | 2.58 |
| Medtr4g074930 | RNA recognition motif; a.k.a. RRM; RBD protein | (NL) | 2.48 |
| Medtr7g097000 | Legumin A2 | (NL) | 2.38 |
| Medtr4g052540 | Fission ELM1 protein | S11 | 2.32 |
| Medtr4g106980 | Serine/Threonine-kinase HT1-like protein | (NL) | 2.26 |
| Medtr5g065490 | Transport inhibitor response protein; putative | (NL) | 2.25 |
| Medtr4g005510 | Ubiquitin-specific protease family C19 protein | (NL) | 2.14 |
Table 2. A list of the top 15 putative phosphorylation targets of NORK.
The table contains both localized and non-localized proteins calculated at 1% FDR with a p-value less than 0.10. Non-Localized phosphorylation sites are represented by (NL).
| Gene ID | Annotation | Phospho-isoform | Fold Change (linear) |
|---|---|---|---|
| Medtr4g081675 | S-locus lectin kinase family protein | (NL) | 5.56 |
| Medtr2g028340 | Elongation factor Tu GTP-binding domain protein | (NL) | 5.06 |
| Medtr1g103550 | DUF1677 family protein | S100 | 2.74 |
| Medtr4g083230 | Transcription factor Pur-alpha-like protein | (NL) | 2.69 |
| Medtr6g077870 | OPT family oligopeptide transporter | S395 | 2.68 |
| Medtr5g086130 | LysM receptor kinase K1B (LYK3) | (NL) | 2.68 |
| Medtr5g012420 | VAMP-associated protein | (NL) | 2.41 |
| Medtr1g072610 | Glycinin G4 | S549 | 2.26 |
| Medtr0168s0110 | Plant/T23J7-180 protein; putative | S296 | 2.25 |
| Medtr5g095120 | Receptor-like protein | (NL) | 2.25 |
| Medtr7g058460 | Transcriptional corepressor leunig-like protein | S347 | 2.17 |
| Medtr7g096990 | Legumin A2 | S51 | 2.14 |
| Medtr3g084870 | Lipoprotein | S229 | 1.96 |
| Medtr4g080160 | Trehalose-6-phosphate synthase domain protein | S5 | 1.96 |
| Medtr4g030140 | dynamin-2B-like protein | (NL) | 1.95 |
A striking feature of proteins phosphorylated (directly or indirectly) by both these kinases is the prevalence of proteins directly or indirectly involved in ubiquitination and protein trafficking. Previous studies have shown that both LYK3 and NORK/SYMRK interact with E3 ubiquitin ligases (SYMRK is the ortholog of NORK in the other model legume Lotus Japonicus). LYK3 interacts with PUB1, while NORK/SYMRK interacts with PUB1 and the seven in absentia E3-ubiquitin ligase SINA4 and both these interactions negatively affect nodulation (Den Herder et al., 2012; Mbengue et al., 2010; Vernié et al., 2016). This study identifies other ubiquitin ligases that may interact with and act as potential substrates for LYK3 and NORK. It should be mentioned that in this study that PUB1, the characterized interactor and phosphorylation substrate of LYK3 and NORK, was not detected. Nonetheless, we detect several proteins/ligases that are involved in ubiquitination, such as an E3 ubiquitin-protein ligase RGLG2-like protein (Medtr3g086520), a ubiquitin-specific protease family C19 protein (Medtr4g005510), and an ARM repeat RING/U-box protein (Medtr3g466220), as putative targets of LYK3. Similarly, an ubiquitin-conjugating enzyme family protein (Medtr8g015590) and a regulatory component of the 26S proteasome (Medtr8g106120) were found as putative targets of NORK.
Besides these, several proteins that are involved in trafficking were identified as the potential targets for both of these tested symbiotic receptor-like kinases. For instance, two ARF GTPase activators Medtr6g027300 and Medtr6g086630 are putative targets of LYK3 and NORK, respectively. ARF family proteins are involved in the synthesis of transport vesicles on the donor membrane (Nielsen et al., 2008). Also, LYK3 potentially phosphorylates a clathrin heavy chain protein (Medtr5g082900), whereas a sorting nexin 2B (Medtr5g073280), a vacuolar protein sorting protein (Medtr4g056480), and an SH3 domain protein (Medtr8g100105) are putative targets of NORK. Recently a clathrin heavy chain protein CHC1 which interacts with ROP6, an interaction partner of NFR5 (an ortholog of LYK3 in another model legume Lotus japonicus) has been implicated in nodulation and Medtr5g082900 may play a similar role in Medicago (Wang et al., 2015). Taken together these symbiotic-receptor-like kinases phosphorylate proteins involved in ubiquitination or vesicle trafficking either directly or indirectly. There may be ubiquitination-based degradation or receptor endocytosis, which in turn regulates cell signaling by determining the abundance of receptors that are available at any given time (Haffani et al., 2004; Hartmann et al., 2008; Vanoosthuyse et al., 2003; Moling et al., 2014).
Potential phosphorylation targets of LYK3
Klaus-Heisen et al. (2011) demonstrated that phosphorylation of the activation loop is essential for the kinase activity of LYK3 which in turn is required for its biological functions. Phosphorylation sites such as Ser-471, Ser-523, Thr-268, Thr-319, Thr-472, Thr-475, and Thr-520, previously identified and characterized by Klaus-Heisen et al. (2011), were also identified in this study. Also, other potential sites such as Ser-307, Ser-323, Ser-584, Thr-311, and Tyr-559 were also identified, the characterization of which will doubtless provide further insights into the biological activity of LYK3.
Among the putative targets trans-phosphorylated by LYK3, Cysteine-rich receptor-kinase-like protein (Medtr1g021635) exhibited the highest fold change (12.05). Besides this, a MAP triple kinase (Medtr4g075410), a serine/threonine kinase HT1-like protein (Medtr4g106980), and a cyclin-dependent kinase (Medtr8g080190) are the other kinases putatively phosphorylated by LYK3. Also, Medtr5g032060, a kinase interacting protein (KIP1-like), is a potential target of LYK3 (Table S1). Although the role of some receptor-like kinases and protein kinases is extensively characterized in symbiotic signaling, the above-mentioned putative targets of LYK3 have not yet been characterized in symbiotic signaling. The use of multiplexed assay for kinase specificity has enabled us to identify these potential targets of LYK3, and the role of some of them in symbiotic signaling will be investigated.
MAP kinase cascades operate via a phosphorelay mechanism, and in symbiotic signaling, the role of a MAP double kinase (SIP2, which interacts with NORK/SYMRK) has been established before (Chen et al., 2012). However, to our knowledge, no MAP triple kinase has been implicated directly in early symbiotic signaling so far, and it would be interesting to investigate the relationships between SIP2 and our MAP triple kinase. The characterization of this newly identified MAP triple kinase should further our knowledge of this pathway.
At least eight different cyclin-dependent kinases are known in plants that have been implicated in cell cycle transition (Tank and Thaker, 2011). These kinases are activated by cyclins, with which they form a complex and phosphorylate target proteins, thereby facilitating cell cycle transition (Tank and Thaker, 2011). In Medicago, cyclins and cell cycle switch components have been implicated in both the early stages of infection and late stages of nodule development.
PIN genes are implicated in auxin transport and knocking down several PIN genes decreased nodulation in Medicago, implying a crucial role of auxin transport in nodulation (Huo et al., 2006). In our current study, an auxin efflux carrier, MtPIN2 (Medtr4g127100), was identified as a putative target of LYK3. A significant reduction in nodule numbers was observed in prior studies using the RNA interference lines of MtPIN2 possibly due to impaired auxin transport (Grunewald et al., 2009; van Noorden et al., 2007; Huo et al., 2006).
Potential phosphorylation targets of NORK
Similar to LYK3 the kinase activity of SYMRK/NORK is affected by its phosphorylation status (Yoshida and Parniske, 2005). At least 37 phosphorylation sites of NORK were identified in this study including three sites (Ser-754, Thr-760, and Thr-593) identified using Q-ToF/MS and characterized by Yoshida et al. (2005). The biological implications of these sites need to be ascertained.
In addition to the NORK phosphorylation sites, we also detected several putative trans-phosphorylation targets of NORK. A trehalose-6-phosphate synthase domain protein (Medtr4g080160) and a peptidylprolyl cis–trans isomerase (Medtr3g037570) are among the top putative targets exhibiting phosphorylation changes upon incubation with NORK. In addition to their role in metabolism, sugars are also implicated in signal transduction in particular in stress responses (Fernandez et al., 2010; Ponnu et al., 2011). Although the bulk of trehalose is synthesized by the microbial symbiont, plants also produce trace amounts of trehalose, and in Medicago, Phaseolus vulgaris, and Glycine max, trehalose accumulation has been detected in nodules (Brechenmacher et al., 2010).
In addition to these proteins, it is interesting to note that NORK also phosphorylates LYK3 which will be discussed later in detail.
Potential phosphorylation targets of LYK3 and NORK
At least 13 different proteins were found to be a common putative target of both LYK3 and NORK, indicating that both these symbiotic receptor-like-kinases act through some common signaling intermediates, as expected during legume–rhizobium symbiosis (Table 3). Among these 13 proteins, 3 of them, a putative ATP/GTP-binding family protein (Medtr5g042000), a drug resistance transporter-like ABC domain protein (Medtr8g014360), and a P-loop nucleoside triphosphate hydrolase superfamily protein (Medtr8g099065) were phosphorylated at the same residues, Ser-975, Ser-848, and Ser-200, respectively, by both LYK3 and NORK. This result is consistent with the observation by Ahsan et al., 2013 that AtPPI–2 Ser–140 can be phosphorylated by three different kinases.
Table 3. A list of putative phosphorylation targets of both NORK and LYK3.
The table contains both localized and non-localized proteins calculated at 1% FDR with a p-value less than 0.10. Non-Localized phosphorylation sites are represented by (NL). The proteins are listed in ascending order by gene ID.
| Gene ID | Annotation | Phospho-isoform | Fold Change (linear) | ||
|---|---|---|---|---|---|
| LYK3 | NORK | LYK3 | NORK | ||
| Medtr1g072600 | Legumin storage protein | (NL) | (NL) | 1.15 | 1.45 |
| Medtr1g103400 | Vicilin 47 kDa protein | Y639 | S391 | 1.15 | 1.41 |
| Medtr3g006760 | Serrate RNA effector molecule-like protein | S364 | S15 | 1.19 | 1.43 |
| Medtr3g098420 | 110 kDa 4SNc-tudor domain protein | (NL) | (NL) | 3.05 | 1.37 |
| Medtr4g030140 | Dynamin-2B-like protein | (NL) | (NL) | 1.53 | 1.95 |
| Medtr4g108140 | IQ calmodulin-binding motif protein | (NL) | (NL) | 1.79 | 1.23 |
| Medtr5g012420 | VAMP-associated protein | (NL) | (NL) | 1.12 | 2.41 |
| Medtr5g042000 | ATP/GTP-binding family protein; putative | S975 | S975 | 1.16 | 1.63 |
| Medtr6g477980 | Argonaute protein 1A | (NL) | (NL) | 1.33 | 1.37 |
| Medtr7g018610 | Nuclear matrix constituent-like protein | (NL) | (NL) | 1.34 | 1.67 |
| Medtr7g097000 | Legumin A2 | S77 | T436 | 1.31 | 1.23 |
| Medtr8g014360 | Drug resistance transporter-like ABC domain protein | S848 | S848 | 1.16 | 1.60 |
| Medtr8g099065 | P-loop nucleoside triphosphate hydrolase superfamily protein | S200 | S200 | 1.21 | 1.18 |
P-loop nucleoside triphosphate hydrolase is chiefly implicated in programmed cell death, response to biotic (disease), and abiotic stresses in plants (Leipe et al., 2004). As they are a key component of the 26S proteasome complex, they may play a role in protein degradation and turnover (JM et al., 2002).
In the model legume, Lotus at least 91 ABC proteins were detected, and an AtPDR12-like gene was upregulated upon inoculation with symbiotic rhizobial bacteria (Sugiyama et al., 2006). It is hypothesized that these ABC transporters, along with the PIN family of genes, play a role in polar auxin transport which affects nodulation (Sugiyama et al., 2006).
A dynamin-2B-like protein, Medtr4g030140, is a putative phosphorylation target of both these symbiotic-receptor-like kinases. Dynamins or dynamin-related proteins (DRP) are vesicles transporting proteins implicated in ligand-induced endocytosis in mammalian systems and plants (Smith et al. 2014). It is possible that the dynamin-2B-like protein (Medtr4g030140) potentially phosphorylated by both these symbiotic receptor-like-kinases may play a similar role in symbiosis which needs to be investigated (Smith et al., 2014).
VAMP-associated protein (Medtr5g012420) mostly implicated in vesicle trafficking is also a putative target of both of these symbiotic receptor-like kinases. (Murray et al., 2011). In Medicago, Vapyrin (VPY), has been implicated in both legume–rhizobia and arbuscular mycorrhizal symbioses (Pumplin et al., 2010; Murray et al., 2011). It is reasonable to speculate that Medtr5g012420, phosphorylated by both LYK3 and NORK, may be involved in late stages of symbiosis.
Overall the multiplexed assay for kinase specificity approach has led to the identification of several putative targets of LYK3 and NORK, respectively (both localized and non-localized) with more than 1-fold change in phosphorylation and a significant p-value. In total, we have identified about 842 phosphopeptides being phosphorylated by both these receptor-like kinases (Figure 2D and supplementary data file 1). While this approach is high throughput, it is also possible that we may have some false positives, similar to those frequently observed with the yeast-two-hybrid approach. The use of cell lysates as the experimental material enables two proteins that are otherwise in different compartments of the cell and with minimal opportunity to interact, to come into proximity with each other and interact, leading to false positives. Also, when protein kinases are removed from their native cell environment, they may lose their regulatory mechanisms, which can also result in non-specific phosphorylation of targets. Furthermore, it is also possible that the kinase of interest may activate other downstream kinases, so there is a possibility of misinterpreting the indirect targets of phosphorylation from downstream kinases as direct phosphorylation substrates for the kinases of interest. Also, background signals, add to the false positives as several active kinases are present in the cell lysates. To address the extent of false positives, we compared the phosphopeptides obtained in this study with our previous large-scale phosphoproteomic analysis of Medicago in which we identified 13293 phosphopeptides specific to symbiotic signaling (Rose et al., 2012, and supplementary data file 1). We found that 696 phosphopeptides overlapped significantly between these studies and 146 phosphopeptides were unique to this work (Figure 2D, and supplementary data file 1). This high level of overlap indicates that a majority of the phosphopeptides identified using our MAKS approach are indeed related to symbiotic signaling, and the degree of false positives may be low. Despite these limitations multiplexed assay for kinase specificity is a powerful tool for the large-scale identification of phosphorylation substrates of multiple kinases under multiple conditions, and as a proof of concept of this technology’s applicability to plants, we confirmed the phosphorylation of LYK3 by NORK using a pair-wise in vitro kinase assay.
Pair-wise In vitro kinase assay confirms LYK3 phosphorylation by NORK
Among the proteins phosphorylated by NORK, LYK3 showed the maximum number of hits and exhibited the 6th highest fold change. Furthermore, the role of LYK3 and NORK in the symbiotic signal transduction pathway has been extensively characterized, and both these genes are essential for the establishment of legume nodulation symbiosis with rhizobia (Arrighi et al., 2006; G Endre et al., 2002; P Smit et al., 2007). Also, NORK is essential for the formation of a symbiotic association with arbuscular mycorrhizal fungi (Delaux et al., 2013; G Endre et al., 2002). Because of their significance in symbiotic signaling and also to validate the results of multiplexed assay for kinase specificity approach in which we could not localize the site of LYK3 phosphorylation by NORK, we wanted to confirm the phosphorylation of LYK3 by NORK using a traditional in vitro kinase assay technique.
Both LYK3 and NORK exhibit auto-phosphorylation activity (Arrighi et al., 2006; Yoshida and Parniske, 2005). Therefore, wild-type versions of these kinases cannot be used to test the trans-phosphorylation of one by the other. A glycine to glutamic acid substitution at the 334th position of the LYK3 kinase domain abolished its kinase activity in vitro which in turn affected nodule formation in planta (Klaus-Heisen et al., 2011). In the case of NORK, a mutant allele (R38) with a non-synonymous substitution converting glycine to glutamic acid at the 794th position had no effect on mycorrhization of plants but impaired the ability of plants to form nodules (nod−, Myc+)(G Endre et al., 2002). It was hypothesized that the kinase activity was abolished in these mutants affecting the nodulation. Here we demonstrate that the kinase activity is indeed abolished in the R38 mutant’s version of NORK (Figure 3) and used the kinase-inactive versions of these symbiotic receptor-like kinases to test the trans-phosphorylation of NORK by LYK3 and vice versa. Because the LYK3 and NORK kinase domains are approximately the same sizes, the LYK3, and NORK kinase domains were tagged with GST and MBP, respectively, to differentiate them based on their size on an SDS-PAGE gel. As expected, the wild-type versions of LYK3 and NORK exhibited auto-phosphorylation (Figure 3A and 3B) and trans-phosphorylation (as indicated by the phosphorylation of casein) activities (Figure 3A and 3B). Furthermore, their corresponding mutants lacked auto-phosphorylation activity (Figure 3A and 3B). In a reaction mixture of wild-type LYK3 and NORK(G794E), a band corresponding to the autophosphorylation of LYK3 was observed. However, no band corresponding to the size of NORK was observed, indicating that LYK3 indeed failed to trans-phosphorylate the NORK mutant (Figure 3C). In the reaction mixture involving NORK-wild-type and LYK3(G334E), a band corresponding to the size of LYK3 was observed in addition to the expected NORK band (Figure 3C). Because LYK3(G334E) lacks autophosphorylation activity, the detection of the LYK3 band implies that LYK3 is indeed trans-phosphorylated by NORK. We identified that sites Ser-269, Ser-273, Ser-307, Ser-323 and Ser-471 of LYK3 (as a control only mLYK3 was used) were phosphorylated by NORK using Mass spectrometry, thereby validating the results obtained using the multiplexed assay for kinase specificity approach.
Figure 3. In vitro kinase assay demonstrating the phosphorylation of LYK3 by NORK.
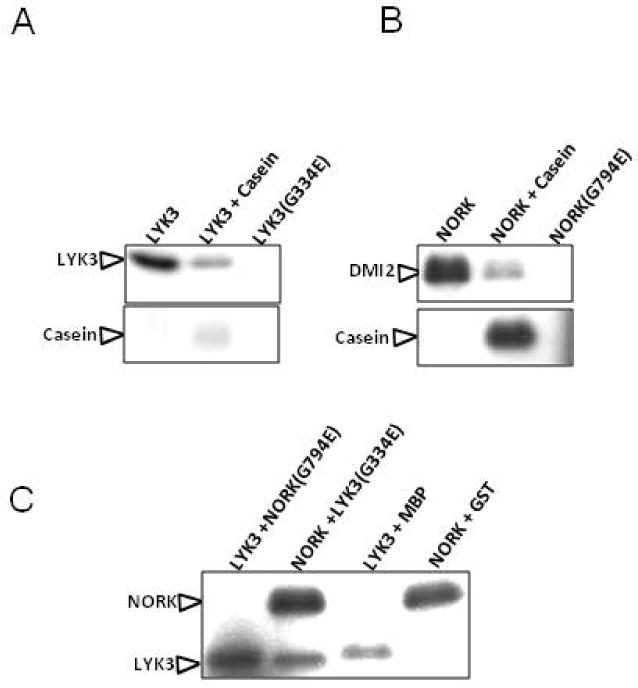
Auto phosphorylation and transphosphorylation activity of LYK3 tagged with GST(indicated by phosphorylation of casein) along with the lack of kinase activity for the mutant version (G334E) of LYK3 (A), autophosphorylation and transphosphorylation activity of NORK tagged with MBP (indicated by phosphorylation of casein) along with the lack of kinase activity for the mutant version (G794E) of NORK (B), lack of phosphorylation of NORK by LYK3 indicated by absence of band corresponding to the size of mutant NORK and a band corresponding to the size of LYK3 in a LYK3 + mutant NORK reaction mix, phosphorylation of LYK3 by NORK indicated by presence of band corresponding to the size of mutant LYK3 and a band corresponding to the size of NORK in a mutant LYK3 + NORK reaction mix, no non-specific phosphorylation indicated by a band corresponding to LYK3 alone in a LYK3 + purified MBP alone reaction mix or a band corresponding to NORK alone in a NORK + purified GST reaction mix (C).
Motif analysis
Phosphorylation motifs were identified from localized phosphorylation sites using the Motif-x algorithm (Schwartz and Gygi, 2005). Phosphorylated peptides were centered on the phosphorylated residue and aligned to the Arabidopsis thaliana database. A central character of S (serine), 20 occurrences, and a significance threshold of 0.000001 were specified. The human IPI database was used as the background dataset to normalize the score against a random distribution of amino acids. Using these settings, two phosphorylation motifs were identified in the LYK3 treated plants (Figure 2B), and one phosphorylation motif was identified in the NORK treated plants (Figure 2C). The acidic motif xxxsDxxx was common to both kinases studied (Figure 2B and 2C). However, as indicated before some of these phosphorylation substrates may not be the direct targets of these symbiotic RLKs and may be the targets of some downstream kinase(s) activated by these symbiotic RLKs.
Conclusion
Strategies targeting the activity of a specific kinase often require the use of phosphorylation-specific antibodies to determine the phosphorylation status of specific residues of protein kinases or their substrates, but the information gained from such studies is not necessarily quantitative. Traditionally, a biochemical approach to kinase activity has been the radioisotope filtration binding assay, where radiolabeled phosphate is incorporated into the kinase substrate. Kinase phosphoryl transfer activity, which is proportional to the amount of phosphorylated substrate, is detected. This method is not well-suited for high throughput screening of multiple phosphorylation targets, as numerous wash steps are required. Additionally, there are inherent dangers associated with the use of radioisotopes. Alternatively, MS-based phosphopeptide analysis provides a high-throughput alternative for identifying, localizing, and quantifying phosphorylation sites on a single amino acid, though it can be difficult to determine the kinase responsible for this activity. Recently, an MS-based assay to determine phosphorylation rates, utilizing synthetic peptides with known phosphorylation sites combined with cell lysate serving as a kinase source, was described (Yu et al. 2009). Although the phosphorylation rate of up to 90 different phosphorylation sites was determined with this method, it requires an extensive library of synthetic peptides, and cannot directly determine the activating kinase. A strategy employing stable isotope labeling allows the quantitative comparison of a control and kinase phosphorylated substrate, but is limited to the analysis of one or two samples (Singh et al., 2012). The MS-based in vitro kinase activity used here utilizes isobaric tag technology, allowing the analysis of up to 10 samples within a single experiment (Brumbaugh et al., 2014). This multiplexing capability allowed us to combine replicates of both kinase-treated and non-treated controls within a single experiment, allowing for statistical analysis of phosphorylation events and increasing the experimental throughput.
In the present study, proteins involved in ubiquitination or those associated with the ubiquitination pathway are among the major proteins phosphorylated by both these symbiotic receptor-like kinases. Vesicle trafficking, regulated by ubiquitination, directs the movement of receptor-like kinases back and forth from the plasma membrane (Furlan et al., 2012). Recent experimental evidence demonstrates that cell signaling and endocytic recycling of membrane proteins are closely associated events rather than two independent processes (Scita and Di Fiore, 2010). Phosphorylation of several proteins related to ubiquitination in our study suggests a possible depletion of signal transduction by protein degradation via the 26S proteasome pathway or receptor endocytosis culminating in the sorting of receptors to another destination. Previous studies have demonstrated the alteration in the localization of LYK3 upon treatment with rhizobia and the interaction of LYK3 and SYMRK with E3 ubiquitin ligases PUB1 and SINA4, respectively, but the underlying mechanism is not understood (Haney et al., 2011). Our current work, due to its inherent high throughput nature, has identified several potential phosphorylation candidates for these symbiotic receptor-like kinases which would otherwise be time-consuming and cumbersome to identify using the conventional methods. The characterization of these candidates should lead not only to the further understanding of the function of individual components but also in deciphering the mechanism of signal transduction from the plasma membrane to the nucleus in symbiotic signaling.
MATERIALS AND METHODS
In vitro protein purification and site-directed mutagenesis
The kinase domain of LYK3 was cloned into the pGEX-6P-1 vector (Amersham Biosciences, Piscataway, NJ, USA) as described in Arrighi et al., (2006). For the NORK kinase domain, a sequence-verified construct was recombined using the LR recombination reaction (Invitrogen, La Jolla, CA, USA). A maltose binding protein (MBP)::NORK (KD) construct was generated in the Gateway® vector pVP16 (Center for Eukaryotic Structural Genomics, Madison, WI, USA). The QuickChangeII® site-directed mutagenesis kit (Stratagene, Santa Clara, CA, USA.) was used to induce point mutations on pGEX-6p-1::LYK3 (kinase domain) and pVP16:: NORK (kinase domain). The following primers were used: LYK3G334EF (5ʹ-TCAAGGTGGATTTGAAGCTGTCTATT) and LYK3G334ER (5ʹ-CATAATAGACAGCTTCAAATCCACCTT) for LYK3 and NORKR38F (5ʹ-CTTGAAATTGTAAGCGAACGGGAACC) and NORKR38R (5ʹ-TTGAGAGGTTCCCGTTCGCTTACAAT) for NORK. The sequence-verified constructs were used for protein expression and purification.
In vitro kinase assays
An in vitro kinase assay was performed following the protocol that was described by Arrighi et al., (2006). Briefly, the aforementioned purified proteins were checked by SDS–PAGE and quantified by Bradford assay (Stoscheck,1990). Then, 1 μg of protein in 20 μl of kinase buffer (50 mM HEPES, pH 7.4, 10 mM MgCl2, 10 mM MnCl2, 1 mM dithiothreitol, 600 nM ATP, 110 nM [10 mCi] [γ-32P]ATP) was incubated at 30°C for 1.5 hours. The reaction was stopped by boiling with SDS-PAGE sample buffer at 95°C for 5 minutes and analyzed by SDS-PAGE. The gels were dried, and the phosphorylated proteins were analyzed by autoradiography (Fujifilm, New Berlin, WI, USA).
Protein extraction
Protein was obtained from 7-day old seedlings of Medicago through chloroform/methanol extraction following the protocol used in Rose et al., 2012. One volume of chloroform and three volumes of water were added to the plant extract. The solution was vortexed and then centrifuged for five minutes (4,696× g, 4°C). Following centrifugation, the top layer was removed and discarded using a serological pipette. Three volumes of methanol were subsequently added, and the solution was vortexed and centrifuged for five minutes (4,696× g, 4°C). The supernatant was removed and discarded using a serological pipette. The resulting protein pellet was washed three times by vortexing with ice-cold 80% acetone followed by five minutes of centrifugation (10,000× g). The pellet was dried on ice for 30 minutes and stored at −80°C.
Protein lysis and kinase reaction
The protein pellets were resuspended in ice-cold lysis buffer containing 8 M urea, 40 mM Tris (pH 8), 2 mM MgCl2, 1 mM CaCl2, 1 mM sodium orthovanadate, 6 mM sodium pyrophosphate, 1× mini ethylenediaminetetraacetic acid-free protease inhibitor (Roche Diagnostics) and 1× phosSTOP phosphatase inhibitor (Roche Diagnostics, Indianapolis, IN, USA). The lysate protein concentration was measured by bicinchoninic acid assay (Thermo Pierce, Rockford, IL, USA). A total of twelve 1 mg aliquots of protein lysate were prepared. These aliquots were divided for two separate experiments, with each experiment comprising three control samples and three samples that were treated with the kinase of interest. Each aliquot was resuspended in a kinase reaction buffer containing 5 mM ATP, 5 mM MgCl2, 50 mM Tris (pH 7.7) and 7.5 mM β-glycerophosphate. Purified NORK or LYK3 was added to half of the lysate aliquots at a 1:75 kinase:protein ratio. Kinase was omitted from the other lysate samples. The lysate was incubated with the kinase reaction buffer at room temperature for thirty minutes.
Protein digestion and isobaric tagging
Each aliquot was reduced using 5 mM dithiothreitol and incubated for 30 minutes at room temperature. Free thiols were alkylated with 15 mM iodoacetamide (IAA) in the dark for 30 minutes. The alkylation reaction was quenched with 5 mM dithiothreitol. The urea concentration was diluted to 1.5 M with 50 mM Tris (pH 8.0). The proteins were digested with trypsin (Promega, Madison, WI, USA) at a 1:50 enzyme:protein ratio and were incubated at ambient temperature overnight. Following digestion, the peptides were acidified with trifluoroacetic acid and desalted over a tC18 Sep-Pak (Waters, Milford, MA, USA). TMT labeling was carried out according to the manufacturer’s instructions (Thermo Pierce). Following tagging, the proteins were mixed in a 1:1:1:1:1:1 ratio according to BCA results, with three channels serving as controls and three channels that were treated with either LYK3 or NORK kinase.
Strong cation exchange fractionation
Strong cation exchange (SCX) fractionation was performed at a flow rate of 3.0 ml/minute using a polysulfoethylaspartamide column (9.4 × 200 mM, PolyLC) on a Surveyor LC quaternary pump. The tagged samples were resuspended in buffer A and separated using the following gradient: 0–2 minutes, 100% buffer A; 2–5 minutes, 0–15% buffer B; and 5–35 minutes, 15–100% buffer B. Buffer B was then held at 100% for 10 minutes. The column was washed with buffer C and water and re-equilibrated after each use. The buffer compositions that were used were as follows: buffer A: 5 mM KH2PO4, 30% acetonitrile (pH 2.6); buffer B: 5 mM KH2PO4, 30% acetonitrile, 350 mM KCl (pH 2.6); and buffer C: 50 mM KH2PO4, 500 mM KCl (pH 7.5). All of the fractions were collected by hand, frozen, lyophilized and desalted over a tC18 Sep-Pak.
Phosphopeptide sample preparation
The peptide fractions were enriched for phosphopeptides using immobilized metal affinity chromatography (IMAC) with metal beads (Qiagen, Valencia, CA, USA). Following re-equilibration with water, the beads were incubated with 40 mM ethylenediaminetetraacetic acid (pH 8.0) for 30 minutes with shaking. Ethylenediaminetetraacetic acid was removed by three successive water washes, and the beads were then incubated with 100 mM FeCl3 for thirty minutes while shaking. The beads were then washed with four rounds of 80% acetonitrile and 0.1% trifluoroacetic acid. The peptides were resuspended in 80% acetonitrile and 0.1% trifluoroacetic acid and incubated with the beads for one hour with shaking. Non-phosphorylated peptides were removed by washing the beads with 80% acetonitrile and 0.1% trifluoroacetic acid. The phosphorylated peptides were eluted from the beads with 50% acetonitrile, 0.7% NH4OH and immediately acidified with 4% formic acid (FA).
Liquid chromatography-tandem mass spectrometry
Online reversed-phase chromatography was performed using a NanoACQUITY UPLC system (Waters, Milford, MA, USA). The peptides were loaded onto a 75 μm inner diameter, 360 μm outer diameter bare fused silica capillary that was packed with 10 cm of Magic C18 particles (Bruker-Michrom) for twelve minutes at a flow rate of 1 μl/minute. The peptides were then eluted onto a 5-μm inner diameter, 360-μm outer diameter analytical column that was packed with 17 cm of Magic C18 particles. The peptides were separated over a 90-minutes gradient by ramping from 2% to 35% acetonitrile, 0.2% FA at a flow rate of 0.3 μl/minute.
The eluting peptide cations were converted to gas-phase ions by electrospray ionization and analyzed on a Thermo Orbitrap Elite. Survey scans of peptide precursors from 350 to 1500 m/z were performed at 60K resolution (at 400 m/z) with a 1×106 ion count target. The dynamic exclusion duration was set to 45 s, with a maximum exclusion list of 500 and with an exclusion width of 0.5 Th below and 1.5 Th above the selected average mass. Tandem MS was performed with higher-energy C-trap dissociation fragmentation with a normalized collision energy of 35. The analysis was performed in the Orbitrap at a resolution 15K. The MS2 ion count target was set to 1×105 with a maximum injection time of 350 msec.
Database searching
All of the MS/MS data were analyzed using the Coon OMSSA Proteomics Software Suite (COMPASS)(Wenger et al., 2011). The spectra were searched using the Open Mass Spectrometry Search Algorithm (OMSSA)2 against a concatenated target-decoy database consisting of Medicago protein sequences that were downloaded from UniProt. Tryptic peptides were created in silico allowing up to three missed cleavages. The mass tolerance was set to 20 ppm for precursors and 0.01 Th for fragment ions. The carbamidomethylation of cysteines and the TMT 6-plex on lysine and N-terminus were searched as fixed modifications. The oxidation of methionine, the TMT 6-plex on tyrosine, the phosphorylation of tyrosine and the phosphorylation with a neutral loss on serine and threonine were searched as variable modifications. The results were filtered to 1% FDR at the unique peptide level. The TMT-labeled peptides were quantified using TagQuant according to previously published procedures (Wenger et al., 2011). The peptides were combined into protein groups and filtered to 1% FDR. The proteins were quantified by summing the intensity of the reporter ion tags of each channel for each protein. Phosphinator software was used to localize the phosphosites. If the site(s) of phosphorylation can be mapped to a specific amino acid residue with absolute certainty, it is termed ‘localized phosphorylation’, whereas if the site(s) of phosphorylation cannot be determined with absolute certainty often due to the presence of multiple potential phosphorylation residues in close proximity (even though we are confident that the peptide is phosphorylated), it is termed ‘non-localized phosphorylation’. Following phosphosite localization, all of the quantitative measurements were log2-transformed and mean-normalized. The fold changes between the conditions were determined by averaging the protein-normalized values for each condition and calculating the difference of averages. The p-values were then calculated using Microsoft Excel using Student’s t-test Complete lists of localized and non-localized phosphorylation sites identified in this study, using both these symbiotic-receptor-like kinases, along with fold-changes in comparison to the control are presented in Supplementary data file 1.
Supplementary Material
Table S1. A list of other top putative phosphorylation targets of LYK3.
Table S2. A list of other top putative phosphorylation targets of NORK.
Data S1. Lists of localized and non-localized phosphorylation sites detected in this study.
Significance Statement.
Multiplexed assay for kinase specificity, a recently developed radioisotope-independent high-throughput technique, was used to identify and map the phosphorylation substrates of up to 10 kinases in a single experiment. Using this technique for plants, we identified potential phosphorylation substrates of symbiotic receptor–like kinases, and these substrates might be the missing link connecting plasma membrane events to nuclear events during symbiotic signal transduction.
Acknowledgments
This research was supported by grants from the National Science Foundation (NSF No. 1237936 and 1546742) to JJC and JMA. ALR was supported by an NIH-funded Genomic Sciences Training Program (5T32HG002760).
Footnotes
Author contributions
DJ, ALR, MSW, JJC, JMA designed the experiments; DJ, ALR performed the experiments; JJC, JMA provided materials; and DJ, ALR, MWP, JJC, JMA wrote the paper.
Author Information
The authors declare no conflict of interests.
References
- Ahsan N, Huang Y, Tovar-Mendez A, Swatek KN, Zhang J, Miernyk JA, Xu D, Thelen JJ. A versatile mass spectrometry-based method to both identify kinase client-relationships and characterize signaling network topology. J Proteome Res. 2013;12:937–948. doi: 10.1021/pr3009995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ansari MW, Trivedi DK, Sahoo RK, Gill SS, Tuteja N. A critical review on fungi mediated plant responses with special emphasis to Piriformospora indica on improved production and protection of crops. Plant Physiol Biochem. 2013;70:403–410. doi: 10.1016/j.plaphy.2013.06.005. [DOI] [PubMed] [Google Scholar]
- Arrighi JFFF, Barre A, Amor B Ben, et al. The Medicago truncatula lysin [corrected] motif-receptor-like kinase gene family includes NFP and new nodule-expressed genes. Plant Physiol. 2006;142:265–79. doi: 10.1104/pp.106.084657. Available at: http://www.ncbi.nlm.nih.gov/pubmed/16844829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bond AE, Row PE, Dudley E. Post-translation modification of proteins; methodologies and applications in plant sciences. Phytochemistry. 2011;72:975–996. doi: 10.1016/j.phytochem.2011.01.029. Available at: <Go. [DOI] [PubMed] [Google Scholar]
- Brechenmacher L, Lei ZT, Libault M, Findley S, Sugawara M, Sadowsky MJ, Sumner LW, Stacey G. Soybean metabolites regulated in root hairs in response to the symbiotic bacterium Bradyrhizobium japonicum. Plant Physiol. 2010;153:1808–1822. doi: 10.1104/pp.110.157800. Available at: <Go. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brückner A, Polge C, Lentze N, Auerbach D, Schlattner U. Yeast two-hybrid, a powerful tool for systems biology. Int J Mol Sci. 2009;10:2763–2788. doi: 10.3390/ijms10062763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brumbaugh J, Russell JD, Yu P, Westphall MS, Coon JJ, Thomson JA. NANOG is multiply phosphorylated and directly modified by ERK2 and CDK1 in vitro. Stem Cell Reports. 2014;2:18–25. doi: 10.1016/j.stemcr.2013.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen T, Zhu H, Ke DX, Cai K, Wang C, Gou HL, Hong ZL, Zhang ZM. A MAP kinase kinase interacts with SymRK and regulates nodule organogenesis in Lotus japonicus. Plant Cell. 2012;24:823–838. doi: 10.1105/tpc.112.095984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniel CJ, Zhang X, Sears RC. Detection of c-Myc protein-protein interactions and phosphorylation status by immunoprecipitation. Methods Mol Biol. 2013;1012:65–76. doi: 10.1007/978-1-62703-429-6_5. [DOI] [PubMed] [Google Scholar]
- Delaux PM, Sejalon-Delmas N, Becard G, Ane JM. Evolution of the plant-microbe symbiotic “toolkit”. Trends Plant Sci. 2013;18:298–304. doi: 10.1016/j.tplants.2013.01.008. [DOI] [PubMed] [Google Scholar]
- Duan G, Walther D. The roles of post-translational modifications in the context of protein interaction networks. PLoS Comput Biol. 2015;11 doi: 10.1371/journal.pcbi.1004049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endre G, Kereszt A, Kevei Z, Mihacea S, Kalo P, Kiss GB. A receptor kinase gene regulating symbiotic nodule development. Nature. 2002;417:962–966. doi: 10.1038/nature00842. [DOI] [PubMed] [Google Scholar]
- Endre G, Kereszt A, Kevei Z, Mihacea S, Kaló P, Kiss GBB, Kalo P, Kiss GBB. A receptor kinase gene regulating symbiotic nodule development. Nature. 2002;417:962–966. doi: 10.1038/nature00842. [DOI] [PubMed] [Google Scholar]
- Van Eyk JE, Dunn MJ. Proteomic and Genomic Analysis of Cardiovascular Disease 2006 [Google Scholar]
- Feilner T, Hultschig C, Lee J, et al. High throughput identification of potential Arabidopsis mitogen-activated protein kinases substrates. Mol Cell Proteomics. 2005;4:1558–1568. doi: 10.1074/mcp.M500007-MCP200. Available at: http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=16009969%5Cnhttp://www.mcponline.org/content/4/10/1558.full.pdf. [DOI] [PubMed] [Google Scholar]
- Fernandez O, Bethencourt L, Quero A, Sangwan RS, Clement C. Trehalose and plant stress responses: friend or foe? Trends Plant Sci. 2010;15:409–417. doi: 10.1016/j.tplants.2010.04.004. Available at: <Go. [DOI] [PubMed] [Google Scholar]
- Furlan G, Klinkenberg J, Trujillo M. Regulation of plant immune receptors by ubiquitination. Front Plant Sci. 2012;3 doi: 10.3389/fpls.2012.00238. Available at: <Go. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gish LA, Clark SE. The RLK/Pelle family of kinases. Plant J. 2011;66:117–127. doi: 10.1111/j.1365-313X.2011.04518.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimsrud PA, Os D, den Wenger CD, Swaney DL, Schwartz D, Sussman MR, Ane JM, Coon JJ. Large-scale phosphoprotein analysis in Medicago truncatula roots provides insight into in vivo kinase activity in legumes. Plant Physiol. 2010;152:19–28. doi: 10.1104/pp.109.149625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grunewald W, Noorden G, van Isterdael G, Van Beeckman T, Gheysen G, Mathesius U. Manipulation of auxin transport in plant roots during rhizobium symbiosis and nematode parasitism. Plant Cell. 2009;21:2553–2562. doi: 10.1105/tpc.109.069617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haffani YZ, Silva NF, Goring DR. Receptor kinase signalling in plants. Can J Bot. 2004;82:1–15. Available at: http://dx.doi.org/10.1139/b03-126. [Google Scholar]
- Haney CH, Riely BK, Tricoli DM, Cook DR, Ehrhardt DW, Long SR. Symbiotic rhizobia bacteria trigger a change in localization and dynamics of the Medicago truncatula receptor kinase LYK3. Plant Cell. 2011;23:2774–2787. doi: 10.1105/tpc.111.086389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartmann C, Chami M, Zachariae U, Groot BL, de Engel A, Grutter MG. Vacuolar protein sorting: two different functional states of the AAA-ATPase Vps4p. J Mol Biol. 2008;377:352–363. doi: 10.1016/j.jmb.2008.01.010. [DOI] [PubMed] [Google Scholar]
- Herder G, Den Yoshida S, Antolin-Llovera M, Ried MK, Parniske M. Lotus japonicus E3 ligase Seven In Absentia4 destabilizes the symbiosis receptor-like kinase SYMRK and negatively regulates rhizobial infection. Plant Cell. 2012;24:1691–1707. doi: 10.1105/tpc.110.082248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huo XY, Schnabel E, Hughes K, Frugoli J. RNAi phenotypes and the localization of a protein :: GUS fusion imply a role for Medicago truncatula PIN genes in nodulation. J Plant Growth Regul. 2006;25:156–165. doi: 10.1007/s00344-005-0106-y. Available at: <Go. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jayaraman D, Forshey KL, Grimsrud PA, Ané JM. Leveraging proteomics to understand plant-microbe interactions. Front Plant Sci. 2012;3:44. doi: 10.3389/fpls.2012.00044. Available at: http://www.ncbi.nlm.nih.gov/pubmed/22645586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- JM B, JL T, L S. Biochemistry 5th edition Chapter 23, Protein turnover and amino acid catabolism. New York: W H Freeman; 2002. [Google Scholar]
- Kang J, Park J, Choi H, Burla B, Kretzschmar T, Lee Y, Martinoia E. Plant ABC transporters. Arab B. 2011;9:e0153. doi: 10.1199/tab.0153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khoury GA, Baliban RC, Floudas CA. Proteome-wide post-translational modification statistics: frequency analysis and curation of the swiss-prot database. Sci Rep. 2011;1:90. doi: 10.1038/srep00090. Available at: http://www.nature.com/srep/2011/110913/srep00090/full/srep00090.html. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klaus-Heisen D, Nurisso A, Pietraszewska-Bogiel A, et al. Structure-function similarities between a plant receptor-like kinase and the human Interleukin-1 receptor-associated kinase-4. J Biol Chem. 2011;286:11202–11210. doi: 10.1074/jbc.M110.186171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehti-Shiu MD, Shiu SH. Diversity, classification and function of the plant protein kinase superfamily. Phil Trans R Soc B. 2012;367:2619–2639. doi: 10.1098/rstb.2012.0003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leipe DD, Koonin EV, Aravind L. STAND, a class of P-loop NTPases including animal and plant regulators of programmed cell death: multiple, complex domain architectures, unusual phyletic patterns, and evolution by horizontal gene transfer. J Mol Biol. 2004;343:1–28. doi: 10.1016/j.jmb.2004.08.023. [DOI] [PubMed] [Google Scholar]
- Lin W, Li B, Lu D, Chen S, Zhu N, He P, Shan L. Tyrosine phosphorylation of protein kinase complex BAK1/BIK1 mediates Arabidopsis innate immunity. Proc Natl Acad Sci U S A. 2014;111:3632–3637. doi: 10.1073/pnas.1318817111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manning G. The protein kinase complement of the human genome. Science (80-) 2002;298:1912–1934. doi: 10.1126/science.1075762. Available at: http://www.sciencemag.org/content/298/5600/1912.abstract\nhttp://www.ncbi.nlm.nih.gov/pubmed/12471243\nhttp://www.sciencemag.org/cgi/doi/10.1126/science.1075762. [DOI] [PubMed] [Google Scholar]
- Mbengue M, Camut S, de Carvalho-Niebel F, et al. The Medicago truncatula E3 ubiquitin ligase PUB1 interacts with the LYK3 symbiotic receptor and negatively regulates infection and nodulation. Plant Cell. 2010;22:3474–3488. doi: 10.1105/tpc.110.075861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller CJ, Turk BE. Rapid identification of protein kinase phosphorylation site motifs using combinatorial peptide libraries. Methods Mol Biol. 2016;1360:203–16. doi: 10.1007/978-1-4939-3073-9_15. Available at: http://www.scopus.com/inward/record.url?eid=2-s2.0-84946058087&partnerID=tZOtx3y1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moling S, Pietraszewska-Bogiel A, Postma M, Fedorova E, Hink Ma, Limpens E, Gadella TWJ, Bisseling T. Nod factor receptors form heteromeric complexes and are essential for intracellular infection in Medicago nodules. Plant Cell. 2014;26:4188–4199. doi: 10.1105/tpc.114.129502. Available at: http://www.ncbi.nlm.nih.gov/pubmed/25351493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray JD, Muni RRD, Torres-Jerez I, et al. Vapyrin, a gene essential for intracellular progression of arbuscular mycorrhizal symbiosis, is also essential for infection by rhizobia in the nodule symbiosis of Medicago truncatula. Plant J. 2011;65:244–252. doi: 10.1111/j.1365-313X.2010.04415.x. [DOI] [PubMed] [Google Scholar]
- Nakagami H, Sugiyama N, Mochida K, Daudi A, Yoshida Y, Toyoda T, Tomita M, Ishihama Y, Shirasu K. Large-scale comparative phosphoproteomics identifies conserved phosphorylation sites in plants. Plant Physiol. 2010;153:1161–1174. doi: 10.1104/pp.110.157347. Available at: http://www.ncbi.nlm.nih.gov/pubmed/20466843\nhttp://www.plantphysiol.org/content/153/3/1161.full.pdf. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen E, Cheung AY, Ueda T. The regulatory RAB and ARF GTPases for vesicular trafficking. Plant Physiol. 2008;147:1516–1526. doi: 10.1104/pp.108.121798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noorden GE, van Kerim T, Goffard N, Wiblin R, Pellerone FI, Rolfe BG, Mathesius U. Overlap of proteome changes in Medicago truncatula in response to auxin and Sinorhizobium meliloti. Plant Physiol. 2007;144:1115–1131. doi: 10.1104/pp.107.099978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pawson T, Scott JD. Signaling through scaffold, anchoring, and adaptor proteins. Science (80-) 1997;278:2075–2080. doi: 10.1126/science.278.5346.2075. Available at: http://www.biomednet.com/db/medline/98071002\npapers2://publication/uuid/B63D0B80-280F-40D4-83D9-2CE7EF11CD64. [DOI] [PubMed] [Google Scholar]
- Phizicky EM, Fields S. Protein-protein interactions: methods for detection and analysis. Microbiol Rev. 1995;59:94–123. doi: 10.1128/mr.59.1.94-123.1995. Available at: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=239356&tool=pmcentrez&rendertype=abstract. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pitzschke A, Schikora A, Hirt H. MAPK cascade signalling networks in plant defence. Curr Opin Plant Biol. 2009;12:421–426. doi: 10.1016/j.pbi.2009.06.008. [DOI] [PubMed] [Google Scholar]
- Ponnu J, Wahl V, Schmid M. Trehalose-6-phosphate: connecting plant metabolism and development. Front Plant Sci. 2011;2 doi: 10.3389/fpls.2011.00070. Available at: <Go. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pumplin N, Mondo SJ, Topp S, Starker CG, Gantt JS, Harrison MJ. Medicago truncatula Vapyrin is a novel protein required for arbuscular mycorrhizal symbiosis. Plant J. 2010;61:482–494. doi: 10.1111/j.1365-313X.2009.04072.x. [DOI] [PubMed] [Google Scholar]
- Ritsema T, Joore J, Workum W, van Pieterse CMJ. Kinome profiling of Arabidopsis using arrays of kinase consensus substrates. Plant Methods. 2007;3:3. doi: 10.1186/1746-4811-3-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritsema T, Peppelenbosch MP. Kinome profiling of sugar signaling in plants using multiple platforms. Plant Signal Behav. 2009;4:1169–73. doi: 10.4161/psb.4.12.10022. Available at: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=2819448&tool=pmcentrez&rendertype=abstract. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz D, Gygi SP. An iterative statistical approach to the identification of protein phosphorylation motifs from large-scale data sets. Nat Biotechnol. 2005;23:1391–1398. doi: 10.1038/nbt1146. Available at: <Go. [DOI] [PubMed] [Google Scholar]
- Scita G, Di Fiore PP. The endocytic matrix. Nature. 2010;463:464–473. doi: 10.1038/nature08910. [DOI] [PubMed] [Google Scholar]
- Shiu SH, Bleecker AB. Expansion of the receptor-like kinase/Pelle gene family and receptor-like proteins in Arabidopsis. Plant Physiol. 2003;132:530–543. doi: 10.1104/pp.103.021964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiu SH, Bleecker AB. Plant receptor-like kinase gene family: diversity, function, and signaling. Sci STKE. 2001;2001:re22. doi: 10.1126/stke.2001.113.re22. Available at: http://www.ncbi.nlm.nih.gov/pubmed/11752632. [DOI] [PubMed] [Google Scholar]
- Silva-Sanchez C, Li H, Chen S. Recent advances and challenges in plant phosphoproteomics. Proteomics. 2014 doi: 10.1002/pmic.201400410. [DOI] [PubMed] [Google Scholar]
- Silva-Sanchez C, Li H, Chen S. Recent advances and challenges in plant phosphoproteomics. Proteomics. 2015;15:1127–1141. doi: 10.1002/pmic.201400410. [DOI] [PubMed] [Google Scholar]
- Singh SA, Winter D, Bilimoria PM, Bonni A, Steen H, Steen JA. FLEXIQinase, a mass spectrometry-based assay, to unveil multikinase mechanisms. Nat Methods. 2012;9:504–508. doi: 10.1038/nmeth.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sirichandra C, Davanture M, Turk BE, Zivy M, Valot B, Leung J, Merlot S. The arabidopsis ABA-activated kinase OST1 phosphorylates the bZIP transcription factor ABF3 and creates a 14-3-3 binding site involved in its turnover. PLoS One. 2010;5 doi: 10.1371/journal.pone.0013935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smit P, Limpens E, Geurts R, Fedorova E, Dolgikh E, Gough C, Bisseling T. Medicago LYK3, an entry receptor in rhizobial nodulation factor signaling. Plant Physiol. 2007;145:183–191. doi: 10.1104/pp.107.100495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith JM, Leslie ME, Robinson SJ, et al. Loss of Arabidopsis thaliana Dynamin-related protein 2B reveals separation of innate immune signaling pathways. PLoS Pathog. 2014;10:e1004578. doi: 10.1371/journal.ppat.1004578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stone JM, Walker JC. Plant protein kinase families and signal transduction. Plant Physiol. 1995;108:451–457. doi: 10.1104/pp.108.2.451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoscheck C. Guide to protein purification. Methods in enzymology. 1990:1–818. [PubMed] [Google Scholar]
- Sugiyama A, Shitan N, Sato S, Nakamura Y, Tabata S, Yazaki K. Genome-wide analysis of ATP-binding cassette (ABC) proteins in a model legume plant, Lotus japonicus: comparison with Arabidopsis ABC protein family. DNA Res. 2006;13:205–228. doi: 10.1093/dnares/dsl013. Available at: <Go. [DOI] [PubMed] [Google Scholar]
- Sugiyama N, Ishihama Y. Large-scale profiling of protein kinases for cellular signaling studies by mass spectrometry and other techniques. J Pharm Biomed Anal. 2016;130:264–272. doi: 10.1016/j.jpba.2016.05.046. Available at: http://linkinghub.elsevier.com/retrieve/pii/S0731708516302941. [DOI] [PubMed] [Google Scholar]
- Tank JG, Thaker VS. Cyclin dependent kinases and their role in regulation of plant cell cycle. Biol Plant. 2011;55:201–212. Available at: <Go. [Google Scholar]
- Thompson A, Schafer J, Kuhn K, Kienle S, Schwarz J, Schmidt G, Neumann T, Hamon C. Tandem mass tags: a novel quantification strategy for comparative analysis of complex protein mixtures by MS/MS. Anal Chem. 2003;75:1895–1904. doi: 10.1021/ac0262560. [DOI] [PubMed] [Google Scholar]
- Vanoosthuyse V, Tichtinsky G, Dumas C, Gaude T, Cock JM. Interaction of calmodulin, a sorting nexin and kinase-associated protein phosphatase with the Brassica oleracea S locus receptor kinase. Plant Physiol. 2003;133:919–929. doi: 10.1104/pp.103.023846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vernié T, Camut S, Camps C, et al. PUB1 interacts with the receptor kinase DMI2 and negatively regulates rhizobial and arbuscular mycorrhizal symbioses through its ubiquitination activity in Medicago truncatula. Plant Physiol. 2016;170:2312–2324. doi: 10.1104/pp.15.01694. Available at: http://www.plantphysiol.org.ezproxy.library.wisc.edu/content/170/4/2312.full. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C, Zhu M, Duan L, et al. Lotus japonicus Clathrin Heavy Chain 1 is associated with ROP6 GTPase and involved in nodule formation. Plant Physiol. 2015;167:114.256107. doi: 10.1104/pp.114.256107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang P, Heitman J. The cyclophilins. Genome Biol. 2005;6:226. doi: 10.1186/gb-2005-6-7-226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wenger CD, Phanstiel DH, Lee MV, Bailey DJ, Coon JJ. COMPASS: a suite of pre- and post-search proteomics software tools for OMSSA. Proteomics. 2011;11:1064–1074. doi: 10.1002/pmic.201000616. Available at: <Go. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witze ES, Old WM, Resing KA, Ahn NG. Mapping protein post-translational modifications with mass spectrometry. Nat Methods. 2007;4:798–806. doi: 10.1038/nmeth1100. Available at: http://www.ncbi.nlm.nih.gov/pubmed/17901869. [DOI] [PubMed] [Google Scholar]
- Yap Y, Kodama Y, Waller F, et al. Activation of a novel transcription factor through phosphorylation by WIPK, a wound-induced mitogen activated protein kinase in tobacco plants. Plant Physiol. 2005;139:127–137. doi: 10.1104/pp.105.065656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida S, Parniske M. Regulation of plant symbiosis receptor kinase through serine and threonine phosphorylation. J Biol Chem. 2005;280:9203–9209. doi: 10.1074/jbc.M411665200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. A list of other top putative phosphorylation targets of LYK3.
Table S2. A list of other top putative phosphorylation targets of NORK.
Data S1. Lists of localized and non-localized phosphorylation sites detected in this study.


