Summary
We have functionally characterized the role of two putative mitochondrial enzymes in valine degradation using insertional mutants. Prior to this study, the relationship between branched-chain amino acid degradation (named for leucine, valine, and isoleucine) and seed development was limited to leucine catabolism. Using a reverse genetics approach we show that disruptions in the mitochondrial valine degradation pathway affect seed development and germination in Arabidopsis thaliana. A null mutant of 3-hydroxyisobutyryl-CoA hydrolase (CHY4, At4g31810) resulted in an embryo lethal phenotype, while a null mutant of methylmalonate semialdehyde dehydrogenase (MMSD, At2g14170) resulted in seeds with wrinkled coats, decreased storage reserves, elevated valine and leucine, and reduced germination rates. These data highlight the unique contributions CHY4 and MMSD make to the overall growth and viability of plants. It also increases our knowledge of the role branched-chain amino acid catabolism plays in seed development and amino acid homeostasis.
Keywords: valine catabolism, seed, development, embryo lethal, Arabidopsis
Introduction
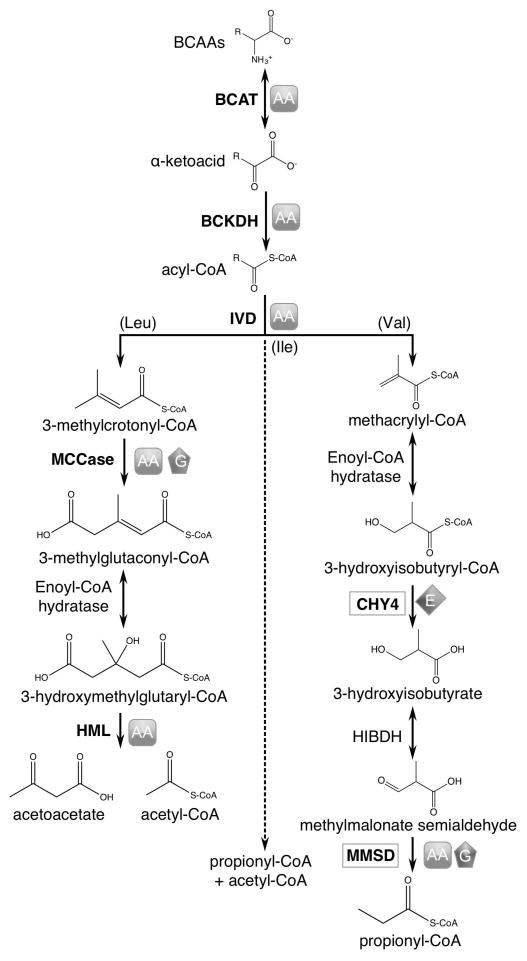
Branched-chain amino acids (leucine, valine, isoleucine) serve as alternative energy sources for mammals (Harper et al., 1984), plants (Binder, 2010), and bacteria (Massey et al., 1976). Their similar catabolic pathways produce energy-rich intermediates such as acetyl-CoA and propionyl-CoA. Specific interest in the plant metabolism of leucine, valine, and isoleucine has centered on finding mechanisms to either prevent the breakdown or increase the biosynthesis of these branched-chain amino acids (BCAAs), as they are essential in human diets and a requirement for animal feed (Ufaz and Galili, 2008). The biosynthetic pathways of BCAAs are well characterized, yet many questions remain unanswered regarding their degradation, particularly for valine and isoleucine (recently reviewed by Hildebrandt et al., 2015). BCAA degradation provides energy for plants during periods of extended darkness, early phases of germination, and late phases of senescence (Peng et al., 2015; Ding et al., 2012; Ishizaki et al., 2005; Araújo et al., 2010; Che et al., 2002; Dunford et al., 1990). However, research over the last decade has shown that BCAA degradation may also play a critical role maintaining amino acid homeostasis and ensuring normal seed development (Angelovici et al., 2013; Peng et al., 2015; Gu et al., 2010; Araújo et al., 2010; Lu et al., 2011; Ding et al., 2012). Our understanding of these additional roles is based on studies of the three shared reactions of BCAA degradation and those specific to leucine catabolism (Figure 1).
Figure 1.
Branched-chain amino acid degradation noting phenotypes of characterized genetic mutants. The symbols represent phenotypes observed in associated mutants. Those for CHY4 and MMSD are based on data presented in this work. AA: increased levels of leucine, valine, and/or isoleucine in seeds or seedlings; G: decreased germination rates; E: defects in embryo development. BCAT: branched-chain amino transferase; BCKDH: branched-chain α-ketoacid dehydrogenase; IVD: isovaleryl-CoA dehydrogenase; MCCase: methylcrotynyl-CoA carboxylase; HML: 3-hydroxymethylgutaryl-CoA lyase; CHY: 3-hydroxyisobutryl-CoA hydrolase; HIBDH: 3-hydroxyisobutyrate dehydrogenase; MMSD: methylmalonate semialdehyde dehydrogenase.
The initial reactions of BCAA degradation are catalyzed by branched-chain aminotransferase (BCAT) followed by branched-chain α-ketoacid dehydrogenase (BCKDH), and then isovaleryl-CoA dehydrogenase (IVD) before the pathway becomes amino acid-specific (Figure 1). Recent studies in Arabidopsis thaliana showed that genetic mutations affecting BCAT, BCKDH, and IVD all resulted in plants exhibiting early senescence under prolonged darkness and altered free amino acid levels (in seeds and plants under darkness) compared to wild-type plants (Angelovici et al., 2013; Peng et al., 2015; Gu et al., 2010; Araújo et al., 2010). In the mitochondrial leucine catabolism pathway, genetic mutations in methylcrotonyl-CoA carboxylase (MCCase) and 3-hydroxymethylgutaryl-CoA lyase (HML) also resulted in plants with altered free amino acid levels and early senescence (Peng et al., 2015; Lu et al., 2011). Notably, MCCase mutants also showed abnormal reproductive growth phenotypes and decreased germination rates in seeds (Ding et al., 2012).
Since BCAA degradation, and leucine in particular is involved in maintaining amino acid homeostasis and ensuring seed development, we hypothesized that there are enzymes in valine degradation that are also involved in these processes. In this paper, we present the growth phenotypes of mutants of two putative enzymes in the valine degradation pathway: 3-hydroxyisobutyryl-CoA hydrolase (CHY, EC: 3.1.2.4) and methylmalonate semialdehyde dehydrogenase (MMSD, EC: 1.2.1.27).
Earlier work predicted that eight 3-hydroxyisobutyryl-CoA hydrolases (CHY1 through CHY8) exist in Arabidopsis, which are thought to be localized to the mitochondrion, peroxisome, and cytosol (Zolman et al., 2001). These eight CoA hydrolases have long fueled a debate on the subcellular localization of valine degradation generally, with evidence pointing towards both peroxisomal and mitochondrial pathways (Gerbling and Gerhardt, 1988; Gerbling, 1993; Gerbling and Gerhardt, 1989; Zolman et al., 2001; Taylor et al., 2004; Angelovici et al., 2013; Daschner, 2001). The peroxisomal enzyme, CHY1, is the only confirmed CHY enzyme in Arabidopsis to date. Current reports suggest that CHY1 has a role in fatty acid β-oxidation (Zolman et al., 2001), cold stress signaling (Dong et al., 2009), and benzoic acid metabolism (Ibdah and Pichersky, 2009), but likely not seed development and germination. Conversely, we describe here an embryo lethal phenotype associated with CHY4. This putative, homologous CoA hydrolase is localized to the mitochondrion, as determined by proteomic experiments (Millar et al., 2001; Heazlewood et al., 2004; Taylor et al., 2011). Our findings suggest that CHY1 and CHY4 have distinct roles and that unlike the peroxisomal pathway, the mitochondrial valine pathway appears to have an important role in seed development.
The last reaction of valine degradation is catalyzed by the enzyme methylmalonate semialdehyde dehydrogenase (MMSD, also known as ALDH6B2), also found to be mitochondrial by several proteomic-based experiments (Taylor et al., 2004; Millar et al., 2001; Sweetlove et al., 2002; Klodmann et al., 2011). We show that mutations affecting MMSD result in phenotypes distinct from chy4-1 mutants, but still similar in that they impact seed development. Null mutants (mmsd-1) produced viable seeds, but exhibit wrinkled seed coats with reduced storage reserves and germination rates.
The reverse genetics approach revealed that disruptions in the mitochondrial valine degradation pathway affected seed development and germination. This suggests that the collective degradation of BCAAs is critical to the growth and viability of plants, but that CHY and MMSD have unique contributions compared to the other BCAA degradation enzymes.
Results
Reverse genetics provides the opportunity to critically analyze gene function and subsequently the function of proteins in specific reactions or the overall health and viability of an organism. Here we used insertional mutants to more fully characterize the role of CHY4 and MMSD in plant development. Heterozygous T-DNA insertion seeds were obtained for mitochondrial CHY4 (chy4-1: SALK_002356) and MMSD (mmsd-1: WiscDSLox242A12) from the Arabidopsis Biological Resource Center (ABRC) or Nottingham Arabidopsis Stock Centre (mmsd-2: GK-849G06). Seed lines were screened by PCR for viable homozygous mutants using primers that were gene or T-DNA specific. The location of the T-DNA insert for each gene was confirmed by PCR and DNA sequencing (Figure S1).
CHY4 is essential for embryo development
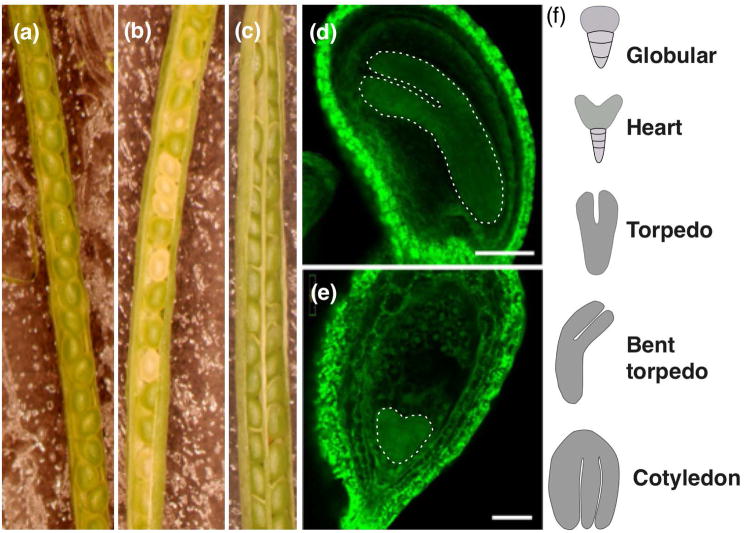
Genotyping chy4 heterozygous plants resulted in no plants homozygous for the T-DNA (approximately 500 screened). Failure to obtain homozygous mutant plants suggested homozygous lethality for this gene. Further investigation of the seeds within mature siliques of chy4-1/CHY4 heterozygotes showed that 24% (of 378) of the seeds were phenotypically different than wild-type (Figure 2a,b), which suggested a deficiency in endosperm or embryo development (Meinke, 1994a). Confocal microscopy revealed that 21% (of 518) of the embryos arrested at heart stage (Figure 2d–f), which confirmed our hypothesis that homozygous plants were embryo lethal. To ensure that the embryo lethality phenotype was indeed due to the null mutant, chy4-1/CHY4 heterozygous plants were complemented with the full-length CHY4 gene under a 35S promoter. The resulting genetically complemented plants produced fully developed and viable seeds similar to wild-type (Figure 2c).
Figure 2.
Microscopy images showing chy4-1 embryo lethality. Light microscopy images of (a) wild-type, (b) chy4-1/CHY4 heterozygous, and (c) chy4-1 35S::CHY4 complement siliques. All siliques were collected at the same time after flowering and captured at the same magnification. Confocal images of seeds from the same chy4-1/CHY4 heterozygous silique showed most embryos had progressed to torpedo or bent cotelydon stage (d), scale bar = 100 μm. Within a heterozygous silique, ~21% of the embryos had arrested at heart stage (e), scale bar = 50 μm. Shape of embryo is artificially highlighted to aid visualization. (f) Stages of embryo development (Baud et al., 2002).
MMSD is involved in seed development
We genotyped plants from heterozygous T-DNA insertion lines of MMSD and confirmed the presence of two homozygous lines (mmsd-1 and mmsd-2) by PCR and DNA sequencing (Figure S1). Reverse transcription-PCR (RT-PCR) studies of mmsd-1 plants confirmed complete loss of MMSD expression in Arabidopsis (Figure S2a). Plants homozygous for the mmsd-2 T-DNA insertion did not produce a null allele, but expression of the gene was reduced (Figure S2b).
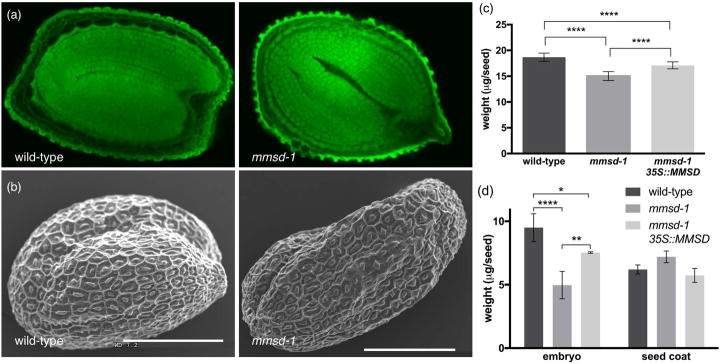
The homozygous mmsd-1 line produced embryos that reached the full cotyledon stage of development, as seen in wild-type seeds (Figure 3a). However desiccated seeds exhibited a wrinkled appearance and weighed 20% less than wild-type seeds (Figure 3b,c). Dissected mmsd-1 seeds produced less massive embryos (by approximately 48%) compared to wild-type, which could be the reason for the wrinkled seed coat. Null mmsd-1 plants complemented with a partial sequence of MMSD under a 35S promoter restored these phenotypes to near wild-type levels of seed weight, storage reserves, and germination rates (Figure 3c, 3d, 4, 5b). The homozygous mmsd-2 seeds were also observed to have a wrinkled seed coat (Figure S3a) and also weighed less than wild-type seeds (Figure S3b).
Figure 3.
Phenotypic analysis of mmsd-1 seeds. (a) Confocal images of mature seeds from wild-type and mmsd-1 null mutants. Images were acquired at the same magnification. (b) SEM images of desiccated mature wild-type and mmsd-1 null mutant seeds. Scale bars are 200 μm. (c) Average weight of desiccated mature seeds (per seed +/− SD) ***P<0.01, **P<0.05 as determined by one-way ANOVA for two independent trials of 100 seeds each. (d) Average weight per seed of embryo or seed coat +/− SD. ****P<0.001, **P<0.05, and *P<0.1 as determined by two-way ANOVA for n = 3 of 100 seed samples. Embryos were extracted from desiccated mature seeds after imbibition to soften the seed coat for dissection.
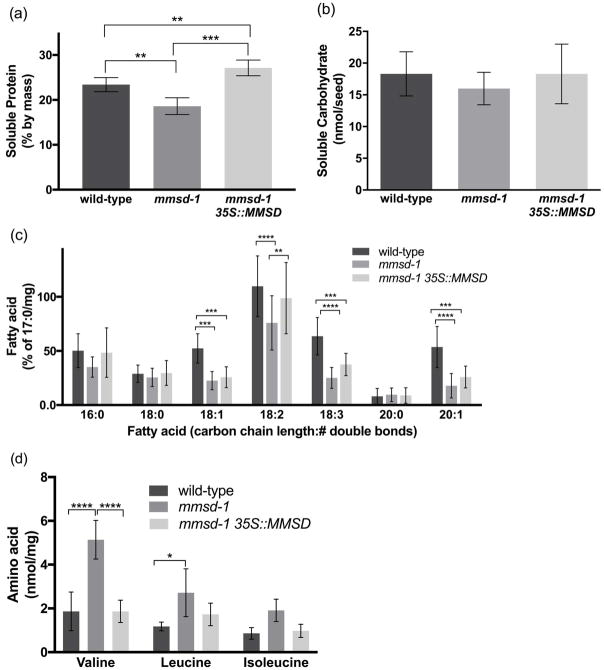
Figure 4.
Soluble storage reserves in mmsd-1 seeds. (a) Average total soluble protein per sample (% by mass), n = 4 of 15–20 mg sample of desiccated mature seeds. (b) Average total soluble carbohydrate (nmol/seed), n = 6 samples of 24 desiccated mature seeds each. (c) Relative amounts of fatty acids normalized to the internal standard, heptadecanoic acid (%/mg), n = 10 of 10 mg sample of desiccated mature seeds. (d) Amount of the free amino acids valine, leucine, and isoleucine (nmol/mg), n = 3 of 5–10 mg sample of desiccated mature seeds. (a) and (b) statistically analyzed by one-way ANOVA; (c) and (d) statistically analyzed by two-way ANOVA. In all experiments: ****P<0.001, ***P<0.01, **P< 0.05, and *P<0.1. Error bars represent SD.
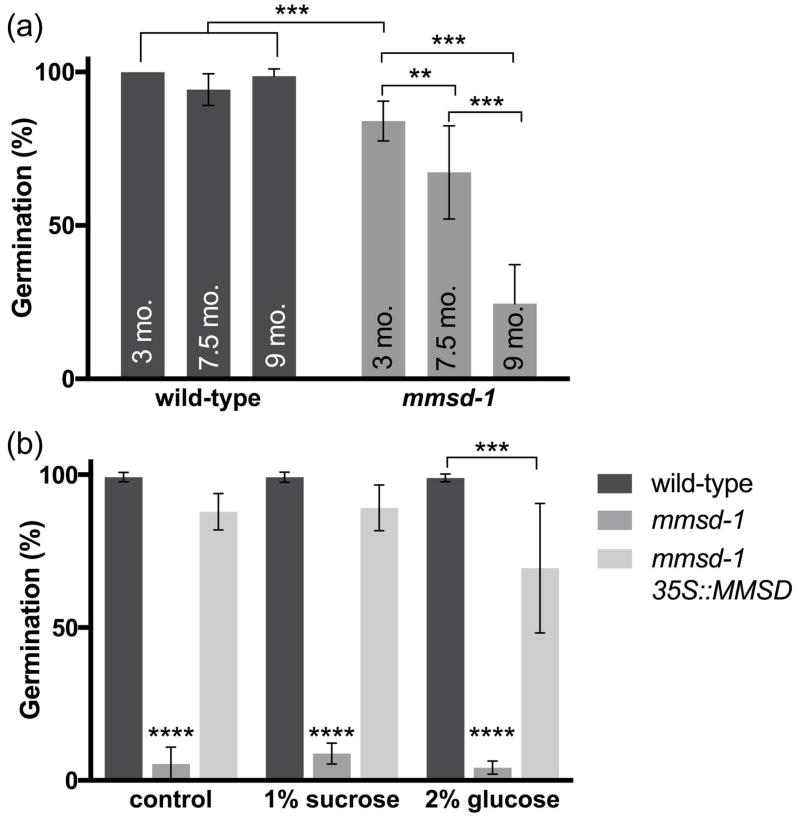
Figure 5.
Germination studies of mmsd-1 seeds. (a) Germination rates of different aged seeds at eight days following imbibition. Age of the seed was determined from time post harvest to planting and each biological replicate contained 14–25 seedlings. (b) Germination rates of mmsd-1 with wild-type and mmsd-1 35S::MMSD complement seeds in the absence (control) or presence of 1% sucrose or 2% glucose at least five days following imbibition. The seeds used in this experiment were less than three months old (from time of harvest) and were at least 5th generation. Each biological replicate contained approximately 60 seedlings. Germination rates for mmsd-1 were statistically different from the other seedlings. There was no statistical difference between treatments for mmsd-1 seedlings. For both experiments: ****P< 0.001, ***P< 0.01, and **P< 0.05, as determined by two-way ANOVA for n = 3 or 4. Error bars represent SD.
We hypothesized that the differences in embryo mass was the result of decreased storage reserves. Proteins, lipids, and carbohydrates constitute the majority of a seed’s mass and serve as the necessary fuel sources for pre-photosynthetic growth (Baud et al., 2002). Soluble proteins were decreased by 20% in mmsd-1 seeds (Figure 4a). Additionally, fatty acid levels were decreased, on average, by 50% in the more abundant types of fatty acids (18:1, 18:2, 18:3, 20:1, Figure 4b). However, in the three samples there were no statistically significant differences in less abundant fatty acids (16:0, 18:0, 20:0) or in soluble carbohydrate levels (Figure 4c).
Free BCAAs constitute a very small portion of a seed’s mass but still serve as additional sources of energy (Taylor et al., 2004; Engqvist et al., 2009; Ishizaki et al., 2005; Araújo et al., 2010). Given that other BCAA degradation mutants showed increased levels of valine, leucine, and isoleucine, we measured BCAA levels in mmsd-1 seeds (Figure 1). Valine content in mmsd-1 seeds was almost three-fold greater than wild-type (Figure 4d) and leucine content was also increased over wild-type, but to a lesser extent (two-fold greater). Average isoleucine content was higher in mmsd-1 seeds, but not statistically different from wild-type or mmsd-1 35S::MMSD complement seeds.
MMSD is involved in seedling establishment
Given the decreased levels of storage reserves and altered BCAA levels, we measured germination rates of mmsd-1 seeds. Compared to wild type seeds, the null mutants had low germination rates that continued declining with seed age. Figure 5a shows germination rates of seeds after various times of storage, ranging from 3 months to 9 months after harvest. Older generations of seeds exhibited very low germination rates and could not be rescued by the addition of sucrose or glucose, regardless of planting time post-harvest (Figure 5b).
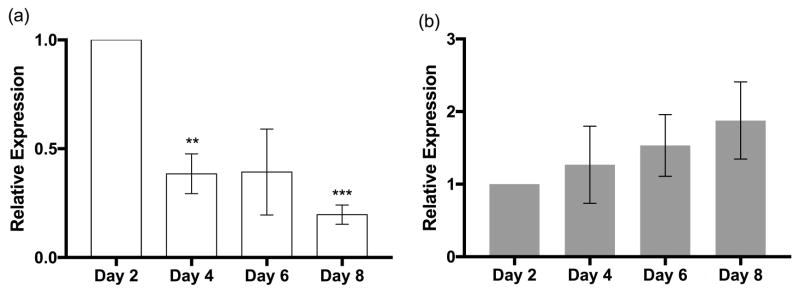
While defects in seed development could explain the germination phenotype, it is important to keep in mind that BCAA degradation continues through post-germination growth (Anderson et al., 1998; Lange et al., 2004; Peng et al., 2015; Czarna et al., 2016). To determine if CHY4 and MMSD had elevated expression during early stages of seedling establishment, we isolated total RNA and measured gene expression from seedlings grown over 8 days following imbibition. CHY4 showed higher expression levels on day 2 compared to days 4–8 (Figure 6a), similar to other genes in BCAA catabolism (Lange et al., 2004; Anderson et al., 1998; Peng et al., 2015). In contrast, MMSD showed a trend towards increased expression over time, although not statistically different across each day (Figure 6b). Therefore, the reduced germination rates of mmsd-1 could be due to a combination of lower seed storage reserves and the seedling’s inability to catabolize valine, which supplies the plant with necessary metabolites during this period of high metabolic activity.
Figure 6.
Gene expression of (a) CHY4 and (b) MMSD during the first eight days of development. Total RNA was collected from whole seedlings at the same time each day and gene expression levels were measured from 15 ng RNA by RT-qPCR. Data was normalized to methionine aminopeptidase 2B (MAP2B) and then compared to expression on day 2. ***P< 0.01, and **P< 0.05, as determined by one-way ANOVA for n = 3 biological replicates, each containing three technical replicates. Error bars represent SD.
mmsd-1 35S::MMSD complement
We attempted to complement mmsd-1 null mutants with the full-length MMSD gene. However, we were unable to express the full-length gene using primers at the annotated start and stop codons of At2g14170.1 (www.arabidopsis.org). Upon further investigation, sequence alignment with other MMSD homologs revealed high sequence identity with Brassica (90%, XP_013661420), rice (75%, AAC03055), human (60%, NP_005580) and rat (59%, NP_112319) proteins. It also revealed that MMSD from Arabidopsis best aligned with the other sequences beginning at Met74 and not the annotated start codon (Figure S4). Primers located at the annotated start codon and 64 bases downstream from the annotated start codon would not amplify cDNA in multiple reactions. Furthermore, when a primer was placed at the downstream ATG (corresponding to Met74), PCR resulted in amplification of a cDNA for MMSD (see XhoI and XmaI sites in Figure S1). Using this partial cDNA, we complemented mmsd-1 plants, which produced larger embryos and seeds with greater storage reserves and better germination rates compared to the null mutant line (Figure 3–5).
Discussion
Seed development and germination can be characterized by seed shape and size, quantification of storage reserves and free amino acid levels, and germination rates. Changes in these parameters have been linked to numerous metabolic pathways, including BCAA degradation. The first three enzymes of BCAA degradation and those specific to leucine play a critical role during this developmental phase. These enzymes function in maintaining amino acid homeostasis and ensure growth during periods of high energy demand, likely through the more significant mitochondrial pathway. Our research focused on two proteins associated with mitochondrial valine degradation, a relatively unstudied pathway. Here, we demonstrated their importance in seed development and germination.
3-Hydroxyisobutyryl-CoA hydrolase
In one of the first committed reactions of valine degradation, 3-hydroxyisobutyryl-CoA hydrolase converts 3-hydroxyisobutyryl-CoA to 3-hydroxyisobutyrate. This key metabolic reaction prevents the accumulation of the upstream metabolite, methacrylyl-CoA, which can serve as a Michael acceptor and react readily and irreversibly with free sulfhydryl groups such as coenzyme A, glutathione, and cysteine (Brown et al., 1982). In humans, the congenital metabolic disease associated with HIBCH (human 3-hydroxyisobutyryl-CoA hydrolase) deficiency is characterized by neurodegeneration during the early stages of life, and is attributed to the accumulation of methacrylyl-CoA (Stiles et al., 2015). Whereas the human genome codes for one CoA hydrolase, Arabidopsis codes for eight distinct hydrolases. This redundancy is presumed to prevent the accumulation of methacrylyl-CoA during high-energy stages of growth. Contrary to this apparent redundancy, null mutants of mitochondrial CHY4 are embryo lethal; growth is arrested at the heart stage of embryogenesis. The question then arises as to the function of the other CHY hydrolases as they could be sources for rescuing the chy4-1 lethality. Therefore, it is likely that they each have distinct roles, either in other metabolic reactions (as is the case for CHY1) or during other phases of development and growth.
However, it is unclear as to the fate of the products of the CHY reactions in the peroxisomes (Zolman et al., 2001), as the final reaction in valine degradation, which is catalyzed by MMSD, is only localized to the mitochondrion. The complete degradation to propionyl-CoA would require a specific shuttling system between the organelles and such a system has yet to be identified. The lack of an apparent mitochondrial-peroxisomal shuttle and lethality of null mitochondrial CHY4 supports the hypothesis that the mitochondrial pathways of BCAA degradation play a critical role than during these periods of development.
Methylmalonate semialdehyde dehydrogenase
The last reaction in valine degradation includes the conversion of methylmalonate semialdehyde to propionyl-CoA by MMSD. When MMSD activity is compromised, seeds go through all the stages of embryo development but result in embryos with less mass. One possibility is that these embryos are actually smaller, and that during the desiccation phase of seed development the seed coat contracts and folds around the smaller embryo, giving the appearance of a wrinkled seed coat. The mmsd-1 seeds also exhibited reduced storage reserves, particularly fatty acids and soluble proteins, which are necessary for early germination events. This phenotype has similarities to that of wri-1 (wrinkled-1) mutants, which exhibits wrinkled seed coats and a significant reduction in seed oils due to compromised carbohydrate metabolism during seed filling (Focks and Benning, 1998).
Additionally, mmsd-1 seeds have increased levels of valine and leucine compared to wild-type. This supports previous evidence that defects in BCAA degradation affect amino acid homeostasis during seed development (Angelovici et al., 2013; Peng et al., 2015). Furthermore, the increased BCAA levels, particularly valine, could be the result of the metabolic switch that occurs when the seed enters desiccation (Fait et al., 2006).
Phenotypes of a complement seed line and an additional mmsd mutant further support the observed characteristics in mmsd-1 seeds. Phenotypes were restored to near wild-type levels when mmsd-1 null mutants were complemented with a partial sequence of the annotated MMSD gene under a 35S promoter. Complements could not be generated with the full-length sequence due to lack of clarity regarding the location of the start codon and mitochondrial leader sequence. It is also unclear as to where the stop codon is located, since the splice variants show two possible sites (Figure S1). Therefore, more work will need to be completed to determine the correct genetic sequence for MMSD.
A second homozygous mmsd allele also displayed a wrinkled seed coat and increased valine in seeds. Despite expression of MMSD transcript in this mutant, it is still possible that MMSD protein expression is compromised during development (Monte, 2003). Immunochemical tests of MMSD will confirm this hypothesis. While mmsd-2 is not a null mutant, it displayed phenotypes similar to mmsd-1 and supports the significance of MMSD and valine degradation in seed development and germination.
The question then arises, why do disruptions of two different reactions in valine degradation result in different phenotypes? The answer is likely related to the associated metabolites of valine catabolism (Figure 1). 3-Hydroxyisobutyryl-CoA hydrolase (e.g. CHY4) helps to ensure that methacrylyl-CoA does not accumulate in the cell. In chy4-1 mutants, methacrylyl-CoA likely accumulates and reacts with cysteine-containing proteins, glutathione, or coenzyme A, preventing critical metabolic processes from occurring during early stages of seed development and subsequently resulting in embryo lethality. On the other hand, methylacrylyl-CoA is not likely to accumulate in mmsd mutants since functioning CHY proteins will keep the equilibrium shifted towards the production of methylmalonate semialdehyde (the substrate for MMSD). This is further supported by evidence in humans of the accumulation of other non-toxic BCAA metabolites such as 3-hydroxyisobutyrate in patients with mutations in ALDH6A1, which codes for the human MMSD (Marcadier et al., 2013). The toxicity of methylmalonate semialdehyde is unknown; however there is some evidence in E. coli that malonate semialdehyde (another proposed substrate for MMSD) forms adducts to free amino groups (Kim et al., 2010). Based on the relative reactivities of methacrylyl-CoA and malonate semialdehyde, it is not surprising that chy4-1 mutants result in embryo lethality whereas mmsd mutants are somewhat viable.
The observed phenotypes of the Arabidopsis mmsd mutants could be due to the absence of downstream products like propionyl-CoA or acetyl-CoA (Hildebrandt et al., 2015). Propionyl-CoA likely gets converted to acetyl-CoA by the very same enzymes in valine catabolism (Lucas et al., 2007). Acetyl-CoA then has the potential to be converted to citrate synthase and further utilized by the citric acid cycle in the mitochondria. Reduced citrate synthase activity would inhibit acetyl-CoA entry into the cycle, resulting in phenotypes similar to those seen with mmsd mutants. However, Sienkiewicz-Porzucek et al. showed that reduced mitochondrial citrate synthase activity had “no effect on plant growth” (Sienkiewicz-Porzucek et al., 2008). Conversely, the major source of acetyl-CoA comes from pyruvate via pyruvate dehydrogenase. Yu et al. did find that a mutation in the E2 subunit of pyruvate dehydrogenase does affect plant growth and development and free amino acid levels (Yu et al., 2012). Therefore, since BCAA degradation is not the major source of acetyl-CoA for the citric acid cycle, it is likely that the phenotypes observed for mmsd mutants are not from the absence of downstream products, but instead from the accumulation of upstream metabolites.
Ultimately, our work contributes to the growing body of knowledge on the variety of approaches for increasing storage reserves and essential amino acids found in seeds and plants, an issue that is likely to only intensify as the world population increases and the farmable acreage remains inadequate (Plant Science Research Summit, 2013). One could speculate that engineering BCAA catabolism could produce amino acid-rich seeds and positively impact human nutrition (Ufaz and Galili, 2008), but several key issues remain. In particular we demonstrated that disruptions in the valine pathway lead to embryo lethality or poorly developed seeds with reduced germination rates. This issue would need to be rectified before a greater impact could be accurately estimated. Until then, there is still much to learn regarding the involvement of BCAA degradation on plant growth and development. This is especially true for enzymes specific for valine and isoleucine degradation, as most have yet to be functionally or kinetically characterized.
Experimental Procedures
All chemicals were purchased from Sigma-Aldrich (http://www.sigmaaldrich.com) or Fisher Scientific (http://www.fishersci.com) unless otherwise noted.
Plant materials and growth conditions
All seed lines were obtained from The Arabidopsis Biological Resource Center (Columbus, OH) or Nottingham Arabidopsis Stock Centre (University of Nottingham, UK). Wild type Arabidopsis ecotype Columbia (Col-0) was used in all studies (obtained from ABRC). Unless noted, seedlings were surface sterilized and planted on soil (Sun Gro Metro-Mix 360), placed in 4 °C for two to three days, then transferred to growth chambers. Plants were grown under 16-h-light/8-h-dark photoperiods at 21–23 °C. Seeds were also surface sterilized and grown on plates using ½-strength Murashige and Skoog (MS) medium (Murashige and Skoog, 1962) and 1% (w/v) sucrose solidified with 0.8% (w/v) agar (Teknova Inc.) under the same growth conditions described above.
The heterozygous T-DNA mutant lines, SALK_002356 (chy4-1), WiscDsLox24A12 (mmsd-1), and GK-849G06 (mmsd-2) were screened using the primers listed in Table S1. T-DNA insertion sites were confirmed by DNA sequencing. A homozygous chy4 mutant line was not isolated (see Results). Seedling establishment of homozygous mmsd mutants must be initiated by first growing seeds on 1% sucrose plates as described above. After 6–10 days, seedlings can be transferred to well-hydrated soil and covered during root establishment. The cover was removed once plant growth was established.
Both chy4-1 and mmsd-1 mutant plant lines (heterozygous and homozygous, respectively) were complemented with their respective genes. For CHY4 the full-length cDNA (U10305, including mitochondrial leader sequence, obtained from ABRC) was PCR-amplified using primers containing restriction sites for XhoI and XbaI. For MMSD, a 1605bp sequence fragment containing XhoI and XmaI sites was PCR-amplified from expressed cDNA (Figure S1). Following restriction digestion, the products were gel purified (Qiagen, http://www.qiagen.com) and ligated directly into pFGC5941 (GenBank Accession no. AY310901, obtained from Dr. Chris Makaroff, Miami University) at 16°, 16 hrs. Transformants were selected with kanamycin (50 μg/mL) and positive colonies were confirmed by PCR and restriction digests of purified plasmid. Constructs were transformed into GV3103/PMP90 Agrobacterium cells and transferred into heterozygous chy4-1/CHY4 and homozygous mmsd-1 plants (Chung et al., 2000). BASTA resistant transformants were selected and confirmed by PCR screening, restriction digests, and evidence for rescue of phenotype.
Measuring gene expression levels
Total RNA was isolated using PureLink kit (Life Technologies, http://www.lifetechnologies.com) according to the manufacturer’s instructions except for the following modifications: frozen, powdered plant material was resuspended in lysis buffer, vortexed, lysed with 21G syringe and then centrifuged at top speed in a microcentrifuge for three minutes at room temperature. The supernatant was then used according to the instructions. On-column DNase treatment was performed as recommended by the manufacturer. cDNA was synthesized using the Verso cDNA synthesis kit with random hexamers. For semi-quantitative PCR experiments, cDNA was amplified using a touchdown thermocycle (Korbie and Mattick, 2008). For real-time qPCR experiments, 15 ng RNA was amplified using SYBR green master mix (ThermoFisher Scientific, https://www.thermofisher.com) with associated no-template and no-RT controls using a 7500 Real-time PCR system (ThermoFisher Scientific, https://www.thermofisher.com). Methionine aminopeptidase 2B (MAP2B) was used as the housekeeping gene (Dekkers et al., 2012). See Table S1 for associated primers.
Microscopy
Siliques from heterozygous chy4-1/CHY4 plants, chy4-1 35S::CHY4 complements, and wild-type plants were dissected (Meinke, 1994b) and examined using an Olympus SZX-12 Stereoscope and pictures taken with a Nikon D200 (Miami University). Confocal microscopy was performed on an Olympus FV500 Laser Scanning Confocal System using Fluoview software (Miami University). Seeds were fixed and stained (Braselton et al., 1996), followed by clearing and mounting with methyl salicylate. SEM images were acquired on a Hitachi, S-3000N microscope (Doane University).
Weighing seeds
Batches of 100 seeds were weighed on an AD 6000 Ultra Microbalance (Perkin Elmer, http://www.perkinelmer.com).
Protein extraction and analysis
Proteins were extracted with the following modifications (Cocuron et al., 2014). Biological replicates of wild-type, mmsd-1, and mmsd-1 35S::MMSD complement seeds (15–20 mg) were finely ground and re-suspended in hexane:isopropanol (2:1). Lipids were removed by centrifugation at 2,000 xg for 5 min. The remaining pellet was placed in a heating block at 60 °C and dried under a gentle stream of N2. Dried pellet was vortexed (on setting 8) for 15 min at 42 °C in prewarmed extraction buffer (0.5 mL, 20 mM Tris HCl, pH 7.5, 150 mM NaCl, 1% SDS). Soluble material was separated by centrifugation at 17,000 xg for 10 min and subsequently transferred to a clean tube. A second extraction from the same pellet was performed as outlined except the sample was mixed for 10 min. Soluble protein was quantified using the DC Protein Assay (Bio-Rad, http://www.bio-rad.com) with BSA as the standard.
Carbohydrate extraction and analysis
Soluble carbohydrates were extracted from wild-type, mmsd-1, and mmsd-1 35S::MMSD complement seeds according to (Masuko et al., 2005): 500 μL of 80% ethanol was added to microfuge tubes containing 24 seeds and homogenized using a glass Dounce homogenizer. Homogenization was repeated once more and then samples were centrifuged at 15,000 xg at 4 °C for 10 min. The supernatant was placed in vacufuge for 3–4 hours to dry. Samples were stored in freezer until ready for analysis. Soluble carbohydrates were resuspended in water and analyzed using glucose as the standard and water as the diluent. Samples were quantified by adding 150 μL of conc. H2SO4 and 30 μL of 5% phenol solution with 50 μL of sample or standard in a 96-well plate. The plate was heated in a 90 °C water bath for 5 min, cooled and analyzed by reading the absorbance at 493 nm.
Fatty acid extraction and analysis
Seeds (~10 mg) were processed for GC-MS analysis by direct methylation (Larson and Graham, 2008). Seeds were heated at 85 °C for 1.5 h in 500 μL methanolic 1 N HCl and 450 μL hexane along with 0.206 μmol heptadecanoic (C17:0) as an internal standard. After heating, the solution was cooled to room temperature and mixed with 250 μL 0.9% NaCl (w/v). The organic phase (~200 μL) containing fatty acid methyl esters (FAMEs) was removed to a GC vial, evaporated under N2, and resuspended in 400 μL hexane. Extracts were analyzed using the Agilent 7890A GC System equipped with a Zebron ZB-AAA 10 m × 0.25 mm column (Phenomenex, http://www.phenomenex.com) with helium as a carrier gas and 5975C VL MSD (Agilent Technologies, http://www.agilent.com). Injection volume was 1 μL with splitless injection; oven was heated to 110 °C for 1 min, then ramped to 180 °C at 20 °C/min then to 221 °C at 2.5 °C/min. Mass spectrometer limits were a low mass of 20 and a high mass of 550.
Amino Acid Extraction and Analysis
Amino acids were extracted from 5–10 mg of seeds (Cocuron et al., 2014). Samples were homogenized in 1 mL of approximately 100 °C dH2O using a glass Dounce homogenizer. The homogenizer was rinsed with 500 μL 100 °C dH2O and added to sample. The homogenate was heated for 10 min at 100 °C, then chilled on ice before centrifuging at 14,000 xg at 4 °C. The supernatant was then lyophilized to concentrate the sample. Each sample was resuspended in 300 μL of dH2O and centrifuged at 14,000 xg at 4 °C to pellet any insoluble material. 100 μL of the supernatant was then derivatized using the EZ:FAAST Physiological Amino Acid Kit (Phenomenex, http://www.phenomenex.com) and analyzed by GC-MS as described above except 2 μL of sample was injected at 250 °C with split 15:1 injection. Oven was heated from 110 °C to 300 °C at 10°C/min with MS limits between 45 and 450 m/z.
Supplementary Material
Gene maps for CHY4 and MMSD showing T-DNA insertion sites, relevant primers, and cloning sites for complementation lines.
mRNA expression of MMSD in mutant plant lines.
Phenotypes of mmsd-2 seeds.
Sequence alignment of MMSD from Arabidopsis, Brassica, rice, human, and rat.
Primers used for genotyping heterozygous T-DNA plants and measuring gene expression levels.
Significance Statement.
In plants, there is little research on the enzymes that catalyze the mitochondrial pathway of valine degradation. Here, we show the functional characterization of two putative proteins and find them to play a significant role in seed development and germination.
Acknowledgments
We thank Mithzy Peña for early experimental work on this project and Scott Freeburg and Emma Mairson for images of mmsd-2 seeds. We are grateful to Karen Hicks and John Hofferberth for critical comments on the paper. This work was supported at Kenyon College by a Cottrell College Science Award from the Research Corporation and the Jean Dreyfus Boissevain Lectureship in the Chemical Sciences for Undergraduate Institutions. This work was also supported at Doane University by a grant from the National Institute for General Medical Science (NIGMS) (5P20GM103427), a component of the National Institutes of Health (NIH). The contents of this paper are the sole responsibility of the authors and do not necessarily represent the official views of NIGMS or NIH. The authors declare that there are no conflicts of interest.
References
- Anderson M, Che P, Song J, Nikolau B, Wurtele E. 3-Methylcrotonyl-coenzyme A carboxylase is a component of the mitochondrial leucine catabolic pathway in plants. [Accessed June 4, 2016];Plant Physiol. 1998 118:1127–38. doi: 10.1104/pp.118.4.1127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angelovici R, Lipka AE, Deason N, Gonzalez-Jorge S, Lin H, Cepela J, Buell R, Gore MA, Dellapenna D. Genome-wide analysis of branched-chain amino acid levels in Arabidopsis seeds. [Accessed June 4, 2016];Plant Cell. 2013 25:4827–43. doi: 10.1105/tpc.113.119370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Araújo WL, Ishizaki K, Nunes-Nesi A, et al. Identification of the 2-hydroxyglutarate and isovaleryl-CoA dehydrogenases as alternative electron donors linking lysine catabolism to the electron transport chain of Arabidopsis mitochondria. [Accessed June 4, 2016];Plant Cell. 2010 22:1549–63. doi: 10.1105/tpc.110.075630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baud S, Boutin JP, Miquel M, Lepiniec L, Rochat C. An integrated overview of seed development in Arabidopsis thaliana ecotype WS. [Accessed July 8, 2016];Plant Physiol Biochem. 2002 40:151–60. [Google Scholar]
- Binder S. Branched-chain amino acid metabolism in Arabidopsis thaliana. [Accessed October 11, 2016];Arabidopsis Book. 2010 8:e0137. doi: 10.1199/tab.0137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braselton JP, Wilkinson MJ, Clulow SA. Feulgen staining of intact plant tissues for confocal microscopy. [Accessed June 4, 2016];Biotech Histochem. 1996 71:84–7. doi: 10.3109/10520299609117139. [DOI] [PubMed] [Google Scholar]
- Brown GK, Hunt SM, Scholem R, et al. Beta-hydroxyisobutyryl coenzyme A deacylase deficiency: a defect in valine metabolism associated with physical malformations. [Accessed July 8, 2016];Pediatrics. 1982 70:532–8. [PubMed] [Google Scholar]
- Che P, Wurtele ES, Nikolau BJ. Metabolic and environmental regulation of 3-methylcrotonyl-coenzyme A carboxylase expression in Arabidopsis. [Accessed June 30, 2016];Plant Physiol. 2002 129:625–37. doi: 10.1104/pp.001842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung MH, Chen MK, Pan SM. Floral spray transformation can efficiently generate Arabidopsis. [Accessed June 4, 2016];Transgenic Res. 2000 9:471–86. doi: 10.1023/a:1026522104478. [DOI] [PubMed] [Google Scholar]
- Cocuron JC, Anderson B, Boyd A, Alonso AP. Targeted metabolomics of Physaria fendleri, an industrial crop producing hydroxy fatty acids. [Accessed June 30, 2016];Plant Cell Physiol. 2014 55:620–33. doi: 10.1093/pcp/pcu011. [DOI] [PubMed] [Google Scholar]
- Czarna M, Kolodziejczak M, Janska H. Mitochondrial proteome studies in seeds during germination. [Accessed July 12, 2016];Proteomes. 2016 4:19. doi: 10.3390/proteomes4020019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daschner K. The mitochondrial isovaleryl-coenzyme A dehydrogenase of Arabidopsis oxidizes intermediates of leucine and valine Catabolism. [Accessed June 30, 2016];Plant Physiol. 2001 126:601–12. doi: 10.1104/pp.126.2.601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dekkers BJW, Willems L, Bassel GW, van Bolderen-Veldkamp RPM, Ligterink W, Hilhorst HWM, Bentsink L. Identification of reference genes for RT-qPCR expression analysis in Arabidopsis and tomato seeds. [Accessed October 13, 2016];Plant Cell Physiol. 2012 53:28–37. doi: 10.1093/pcp/pcr113. [DOI] [PubMed] [Google Scholar]
- Ding G, Che P, Ilarslan H, Wurtele ES, Nikolau BJ. Genetic dissection of methylcrotonyl CoA carboxylase indicates a complex role for mitochondrial leucine catabolism during seed development and germination. [Accessed May 27, 2016];Plant J. 2012 70:562–77. doi: 10.1111/j.1365-313X.2011.04893.x. [DOI] [PubMed] [Google Scholar]
- Dong CH, Zolman BK, Bartel B, Lee B, Stevenson B, Agarwal M, Zhu JK. Disruption of Arabidopsis CHY1 reveals an important role of metabolic status in plant cold stress signaling. [Accessed January 12, 2017];Mol Plant. 2009 2:59–72. doi: 10.1093/mp/ssn063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunford R, Kirk D, ap Rees T. Respiration of valine by higher plants. [Accessed July 8, 2016];Phytochemistry. 1990 29:41–43. [Google Scholar]
- Engqvist M, Drincovich MF, Flügge UI, Maurino VG. Two D-2-hydroxy-acid dehydrogenases in Arabidopsis thaliana with catalytic capacities to participate in the last reactions of the methylglyoxal and beta-oxidation pathways. [Accessed July 8, 2016];J Biol Chem. 2009 284:25026–37. doi: 10.1074/jbc.M109.021253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fait A, Angelovici R, Less H, Ohad I, Urbanczyk-Wochniak E, Fernie AR, Galili G. Arabidopsis seed development and germination is associated with temporally distinct metabolic switches. [Accessed July 8, 2016];Plant Physiol. 2006 142:839–54. doi: 10.1104/pp.106.086694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Focks N, Benning C. wrinkled1: A novel, low-seed-oil mutant of Arabidopsis with a deficiency in the seed-specific regulation of carbohydrate metabolism. [Accessed June 30, 2016];Plant Physiol. 1998 118:91–101. doi: 10.1104/pp.118.1.91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerbling H. Peroxisomal degradation of 2-oxoisocaproate. Evidence for free acid intermediates. [Accessed June 30, 2016];Bot Acta. 1993 106:380–7. Available at: http://doi.wiley.com/10.1111/j.1438-8677.1993.tb00764.x. [Google Scholar]
- Gerbling H, Gerhardt B. Oxidative decarboxylation of branched-chain 2-oxo fatty acids by higher plant peroxisomes. [Accessed June 30, 2016];Plant Physiol. 1988 88:13–15. doi: 10.1104/pp.88.1.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerbling H, Gerhardt B. Peroxisomal degradation of branched-chain 2-oxo acids. [Accessed June 30, 2016];Plant Physiol. 1989 91:1387–92. doi: 10.1104/pp.91.4.1387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu L, Jones AD, Last RL. Broad connections in the Arabidopsis seed metabolic network revealed by metabolite profiling of an amino acid catabolism mutant. [Accessed May 27, 2016];Plant J. 2010 61:579–90. doi: 10.1111/j.1365-313X.2009.04083.x. [DOI] [PubMed] [Google Scholar]
- Harper AE, Miller RH, Block KP. Branched-Chain Amino Acid Metabolism. [Accessed October 11, 2016];Annu Rev Nutr. 1984 4:409–54. doi: 10.1146/annurev.nu.04.070184.002205. [DOI] [PubMed] [Google Scholar]
- Heazlewood JL, Tonti-Filippini JS, Gout AM, Day DA, Whelan J, Millar AH. Experimental analysis of the Arabidopsis mitochondrial proteome highlights signaling and regulatory components, provides assessment of targeting prediction programs, and indicates plant-specific mitochondrial proteins. [Accessed October 13, 2016];Plant Cell. 2004 16:241–56. doi: 10.1105/tpc.016055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hildebrandt TM, Nunes Nesi A, Araújo WL, Braun HP. Amino acid catabolism in plants. [Accessed May 27, 2016];Mol Plant. 2015 8:1563–79. doi: 10.1016/j.molp.2015.09.005. [DOI] [PubMed] [Google Scholar]
- Ibdah M, Pichersky E. Arabidopsis Chy1 null mutants are deficient in benzoic acid-containing glucosinolates in the seeds. Plant Biol. 2009;11:574–81. doi: 10.1111/j.1438-8677.2008.00160.x. Available at: http://onlinelibrary.wiley.com/doi/10.1111/j.1438-8677.2008.00160.x/full. [DOI] [PubMed] [Google Scholar]
- Ishizaki K, Larson TR, Schauer N, Fernie AR, Graham IA, Leaver CJ. The critical role of Arabidopsis electron-transfer flavoprotein:ubiquinone oxidoreductase during dark-induced starvation. [Accessed June 30, 2016];Plant Cell. 2005 17:2587–600. doi: 10.1105/tpc.105.035162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim KS, Pelton JG, Inwood WB, Andersen U, Kustu S, Wemmer DE. The Rut pathway for pyrimidine degradation: novel chemistry and toxicity problems. [Accessed January 5, 2017];J Bacteriol. 2010 192:4089–102. doi: 10.1128/JB.00201-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klodmann J, Senkler M, Rode C, Braun HP. Defining the protein complex proteome of plant mitochondria. [Accessed October 13, 2016];Plant Physiol. 2011 157:587–98. doi: 10.1104/pp.111.182352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korbie DJ, Mattick JS. Touchdown PCR for increased specificity and sensitivity in PCR amplification. [Accessed July 9, 2014];Nat Protoc. 2008 3:1452–6. doi: 10.1038/nprot.2008.133. [DOI] [PubMed] [Google Scholar]
- Lange PR, Eastmond PJ, Madagan K, Graham IA. An Arabidopsis mutant disrupted in valine catabolism is also compromised in peroxisomal fatty acid beta-oxidation. [Accessed June 4, 2016];FEBS Lett. 2004 571:147–53. doi: 10.1016/j.febslet.2004.06.071. [DOI] [PubMed] [Google Scholar]
- Larson TR, Graham IA. Technical Advance: A novel technique for the sensitive quantification of acyl CoA esters from plant tissues. [Accessed July 8, 2016];Plant J. 2008 25:115–25. doi: 10.1046/j.1365-313x.2001.00929.x. Available at: http://doi.wiley.com/10.1111/j.1365-313X.2001.00929.x. [DOI] [PubMed] [Google Scholar]
- Lu Y, Savage LJ, Larson MD, Wilkerson CG, Last RL. Chloroplast 2010: a database for large-scale phenotypic screening of Arabidopsis mutants. [Accessed June 6, 2016];Plant Physiol. 2011 155:1589–600. doi: 10.1104/pp.110.170118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucas KA, Filley JR, Erb JM, Graybill ER, Hawes JW. Peroxisomal metabolism of propionic acid and isobutyric acid in plants. [Accessed January 5, 2017];J Biol Chem. 2007 282:24980–9. doi: 10.1074/jbc.M701028200. [DOI] [PubMed] [Google Scholar]
- Marcadier JL, Smith AM, Pohl D, et al. Mutations in ALDH6A1 encoding methylmalonate semialdehyde dehydrogenase are associated with dysmyelination and transient methylmalonic aciduria. [Accessed January 5, 2017];Orphanet J Rare Dis. 2013 8:98. doi: 10.1186/1750-1172-8-98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massey LK, Sokatch JR, Conrad RS. Branched-chain amino acid catabolism in bacteria. [Accessed October 11, 2016];Bacteriol Rev. 1976 40:42–54. doi: 10.1128/br.40.1.42-54.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masuko T, Minami A, Iwasaki N, Majima T, Nishimura SI, Lee YC. Carbohydrate analysis by a phenol-sulfuric acid method in microplate format. [Accessed March 29, 2016];Anal Biochem. 2005 339:69–72. doi: 10.1016/j.ab.2004.12.001. [DOI] [PubMed] [Google Scholar]
- Meinke DW. 10 Seed Development in Arabidopsis thaliana. [Accessed January 5, 2017];Cold Spring Harb Monogr Arch Vol 27 Arab. 1994a Available at: https://cshmonographs.org/index.php/monographs/article/view/3103.
- Meinke DW. Seed development in Arabidopsis. In: Meyerowitz E, Somerville C, editors. Arabidopsis thaliana. Cold Spring Harbor, New York: Cold Spring Harbor Laboratory Press; 1994b. pp. 253–95. [Google Scholar]
- Millar AH, Sweetlove LJ, Giegé P, Leaver CJ. Analysis of the Arabidopsis mitochondrial proteome. [Accessed July 8, 2016];Plant Physiol. 2001 127:1711–27. [PMC free article] [PubMed] [Google Scholar]
- Monte E. Isolation and characterization of phyC mutants in Arabidopsis reveals complex crosstalk between phytochrome signaling pathways. [Accessed July 12, 2016];Plant Cell. 2003 15:1962–80. doi: 10.1105/tpc.012971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murashige T, Skoog F. A revised medium for rapid growth and bio assays with tobacco tissue cultures. [Accessed November 15, 2014];Physiol Plant. 1962 15:473–97. Available at: http://doi.wiley.com/10.1111/j.1399-3054.1962.tb08052.x. [Google Scholar]
- Peng C, Uygun S, Shiu SH, Last RL. The impact of the branched-chain ketoacid dehydrogenase complex on amino acid homeostasis in Arabidopsis. [Accessed May 27, 2016];Plant Physiol. 2015 169:1807–20. doi: 10.1104/pp.15.00461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plant Science Research Summit. [Accessed January 5, 2017];Unleashing a decade of innovation in plant science: A vision for 2015–2025. 2013 Available at: https://plantsummit.wordpress.com/
- Sienkiewicz-Porzucek A, Nunes-Nesi A, Sulpice R, Lisec J, Centeno DC, Carillo P, Leisse A, Urbanczyk-Wochniak E, Fernie AR. Mild reductions in mitochondrial citrate synthase activity result in a compromised nitrate assimilation and reduced leaf pigmentation but have no effect on photosynthetic performance or growth. [Accessed January 5, 2017];Plant Physiol. 2008 147:115–27. doi: 10.1104/pp.108.117978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stiles AR, Ferdinandusse S, Besse A, Appadurai V, Leydiker KB, Cambray-Forker EJ, Bonnen PE, Abdenur JE. Successful diagnosis of HIBCH deficiency from exome sequencing and positive retrospective analysis of newborn screening cards in two siblings presenting with Leigh’s disease. [Accessed October 13, 2016];Mol Genet Metab. 2015 115:161–67. doi: 10.1016/j.ymgme.2015.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sweetlove LJ, Heazlewood JL, Herald V, Holtzapffel R, Day DA, Leaver CJ, Millar AH. The impact of oxidative stress on Arabidopsis mitochondria. [Accessed October 13, 2016];Plant J. 2002 32:891–904. doi: 10.1046/j.1365-313x.2002.01474.x. Available at: http://doi.wiley.com/10.1046/j.1365-313X.2002.01474.x. [DOI] [PubMed] [Google Scholar]
- Taylor NL, Heazlewood JL, Day DA, Millar AH. Lipoic acid-dependent oxidative catabolism of alpha-keto acids in mitochondria provides evidence for branched-chain amino acid catabolism in Arabidopsis. [Accessed June 30, 2016];Plant Physiol. 2004 134:838–48. doi: 10.1104/pp.103.035675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor NL, Heazlewood JL, Millar AH. The Arabidopsis thaliana 2-D gel mitochondrial proteome: Refining the value of reference maps for assessing protein abundance, contaminants and post-translational modifications. [Accessed October 13, 2016];Proteomics. 2011 11:1720–33. doi: 10.1002/pmic.201000620. [DOI] [PubMed] [Google Scholar]
- Ufaz S, Galili G. Improving the content of essential amino acids in crop plants: goals and opportunities. [Accessed July 8, 2016];Plant Physiol. 2008 147:954–61. doi: 10.1104/pp.108.118091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu H, Du X, Zhang F, Zhang F, Hu Y, Liu S, Jiang X, Wang G, Liu D. A mutation in the E2 subunit of the mitochondrial pyruvate dehydrogenase complex in Arabidopsis reduces plant organ size and enhances the accumulation of amino acids and intermediate products of the TCA Cycle. [Accessed January 5, 2017];Planta. 2012 236:387–99. doi: 10.1007/s00425-012-1620-3. [DOI] [PubMed] [Google Scholar]
- Zolman BK, Monroe-Augustus M, Thompson B, Hawes JW, Krukenberg KA, Matsuda SP, Bartel B. chy1, an Arabidopsis mutant with impaired beta-oxidation, is defective in a peroxisomal beta-hydroxyisobutyryl-CoA hydrolase. [Accessed May 27, 2016];J Biol Chem. 2001 276:31037–46. doi: 10.1074/jbc.M104679200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Gene maps for CHY4 and MMSD showing T-DNA insertion sites, relevant primers, and cloning sites for complementation lines.
mRNA expression of MMSD in mutant plant lines.
Phenotypes of mmsd-2 seeds.
Sequence alignment of MMSD from Arabidopsis, Brassica, rice, human, and rat.
Primers used for genotyping heterozygous T-DNA plants and measuring gene expression levels.