Abstract
In mammals, interferon-inducible transmembrane proteins (IFITMs) prevent infections by various enveloped viruses. The expression of IFITMs in chicken was herein examined in the adult and embryonic organs using a quantitative reverse-transcription-polymerase chain reaction. The results obtained revealed that IFITM3 was expressed at a higher level than IFITM1, 2 and 5, in both embryonic and adult organs. However, the expression levels of IFITMs in embryonic organs were less than 5 % of those in adult lungs. Among the embryonic tissues examined, primordial germ cells (PGCs) at day 2.5 expressed relatively higher levels of IFITM3. IFITM3 expression levels were 1.5-fold higher in the chicken cell line DF-1 than in PGCs. The knockdown of IFITM3 in DF-1 cells by siRNA increased the infectivity of a vesicular stomatitis virus G protein-pseudotyped lentiviral vector, suggesting that lower levels of IFITM3 are still sufficient to restrict this viral vector.
Keywords: Interferon-inducible transmembrane protein, Chicken, Primordial germ cell, Transgenic chicken, Vaccine, Virus
Introduction
Interferon-inducible transmembrane protein (IFITM) family proteins restrict various pathogenic viruses including influenza A virus and Ebola virus. They block viral replication at early steps by preventing the fusion of viral and host cell membranes after endocytosis. Five IFITM species have been identified in humans and mice (IFITM1, 2, 3, 5, 10 and IFITM1, 2, 3, 5, 6 in humans and mice, respectively) (Bailey et al. 2014; Perreira et al. 2013). Among them, IFITM3 blocks viral infections through late endosome such as influenza viruses, West Nile virus and Yellow fever virus (Perreira et al. 2013). Chicken orthologs of IFITMs have recently been identified, and IFITM3 was shown to restrict infections by influenza viruses and lyssaviruses in vitro (Smith et al. 2013). It has been serious problem that influenza viruses prevail worldwide in chickens. Although the incidence of humans being infected with chicken viruses remains small, it still occurs. Therefore, reducing the prevalence of chicken influenza viruses may contribute to decreasing the probability of pandemics involving chicken influenza viruses. Furthermore, chicken eggs have been used to prepare vaccines for influenza viruses. Thus, studies on chicken IFITMs that may restrict infections by influenza viruses in chickens are becoming increasingly important.
IFITMs may also affect gene transfer using viral vectors such as those used in the establishment of transgenic chickens. Transgenic techniques have been rapidly developed using various livestock species as an alternate method to produce biologically active substances (Houdebine 2000; Kues and Niemann 2004; Rudolph 1999). Retroviral vectors, including the vesicular stomatitis virus G protein (VSV-G)-pseudotyped viral vector, have been widely used in the establishment of transgenic chickens (Nishijima and Iijima 2013). In our research, the viral vectors have been injected into the subgerminal cavity at the blastodermal stage or into the heart of developing embryos (at day 2.5) in order to generate transgenic chickens (Kamihira et al. 2005, 2009, Kodama et al. 2008; Kyogoku et al. 2008). In general, the frequency of obtaining G1 transgenic descendants using this method is very low. One possible reason for this may be the host restriction of viral vectors, because infections by VSV are restricted by IFITMs in mammals (Perreira et al. 2013).
Primordial germ cells (PGCs) are the progenitor cells of ova and spermatozoa. Chicken PGCs are found at the center of the area pellucida in stage X blastoderms (freshly laid eggs). After gastrulation, they exist in the germinal crescent. They then enter and circulate in the blood vessels of the embryo, and migrate to the germinal ridge, which develops into the mature gonads (D’Costa et al. 2001). Relatively low frequency in transgene transmission indicated the low efficiency to infect these stages of PGCs (blood circulating stages at day 2.5) by retroviral vectors used in transgenesis.
In the present study, we examined the expression of IFITMs in adult and embryonic chickens, which may hamper infections by viruses that require endosomes to successfully infect their hosts. We also determined whether lower levels of IFITM3 in PGCs inhibit infections by the VSV-G-pseudotyped virus in order to know the effects of this IFITM protein on gene delivery during the establishment of transgenic chickens.
Materials and methods
Reagents and cells
Chicken interferon (IFN)-α was obtained from Abcam (Cambridge, UK). An anti-stage-specific embryonic antigen (SSEA)-1 antibody was purchased from Santa Cruz Biotechnology (Santa Cruz, CA, USA). A phycoerythrin (PE)-labeled goat anti-mouse IgM antibody was obtained from Rockland (Gilbertsville, PA, USA). The chicken fibroblast cell line DF-1 (CRL-12203) was purchased from the American Type Culture Collection (ATCC, Manassas, VA, USA) and maintained in Dulbecco’s modified Eagle’s medium (Sigma-Aldrich, St. Louis, MO, USA) containing 10 % fetal bovine serum (FBS) (Biological Industries, Kibbutz Beit Haemek, Israel), 100 U/ml penicillin G, and 100 μg/ml streptomycin.
Chickens and eggs
White Leghorn fertilized eggs were purchased from Nisseiken (Yamanashi, Japan) or Takeuchi Farm (Nara, Japan), or were obtained in our laboratory from the descendants of those from Nisseiken. After the eggs had been incubated at 38 °C for 10 days with rocking, chicken embryonic fibroblast cells (CEFs) were obtained using a standard method with trypsin. Cells were cultured in Dulbecco’s modified Eagle’s medium supplemented with 10 % FBS and antibiotics, and were used for experiments after 2–3 passages. Chickens were housed in our laboratory. Organs were isolated from adult chickens and 5.5-day embryos by dissection, cut into small pieces, washed with phosphate-buffered saline (PBS), and then used for further analyses. Total RNA was isolated using ISOGEN II (Nippon Gene, Tokyo, Japan). In some experiments, the egg shell and choriomembrane were carefully removed from 9-day embryonated eggs, and the chorioallantoic membrane and amnion were collected. Tissues from a chicken housed under specific pathogen-free conditions were kindly provided by Dr. Inayoshi (Nisseiken). All animal experiments were performed according to the ethical guidelines for animal experimentation of Nagoya University.
Isolation of PGCs
PGCs were isolated using a fluorescence-activated cell sorter (EPICS ALTRA, Beckman-Coulter, Fullerton, CA, USA or FACSJazz, BD Biosciences, San Jose, CA, USA) based on the expression of SSEA-1, as described previously (Motono et al. 2008, 2010). Briefly, fertilized eggs were incubated at 38 °C and 65 % humidity for 55 h [Hamburger–Hamilton stages 13–16 (Hamburger and Hamilton 1951)] and 5.5 days (stages 27–28) to isolate circulating PGCs (cPGCs) and gonadal PGCs (gPGCs), respectively. In order to obtain cPGCs, blood was drawn with a glass micropipette from embryos, and the cells were washed with PBS containing 0.5 % FBS. To obtain gPGCs, embryonic gonads were isolated with forceps and were dispersed in 0.1 % trypsin and 0.02 % EDTA. Dispersed blood and gonadal cells were incubated with the anti-SSEA-1 antibody for 30 min and then with the PE-labeled goat anti-mouse IgM antibody for 30 min on ice. Cells expressing SSEA-1 were sorted as the PGC fraction. In order to obtain PGCs in the germinal ridge (grPGCs), the germinal ridges of 3.5-day embryos were dissected, and SSEA-1+ cells were purified using a similar method to that for gPGCs.
Quantitative real-time reverse transcription-polymerase chain reaction (qRT-PCR)
Real-time PCR was performed using LightCycler (Roche Diagnostics, Mannheim, Germany) or LightCycler 96 (Roche Diagnostics). RNA was extracted using ISOGEN II, reverse-transcribed by ReverTra Ace (Toyobo, Osaka, Japan), and subjected to real-time PCR using SYBR Green I dye (Thunderbird qPCR Mix, Toyobo). Due to the small number of PGCs available, the ReliaPrep RNA Cell Miniprep System (Promega, Madison, WI, USA) and ReverTra Ace qPCR RT Master Mix with gDNA Remover (Toyobo) were used to extract total RNA and perform the reverse-transcription of PGCs, respectively. LightCycler amplification involved denaturation at 95 °C for 60 s, followed by amplification of the target DNA for 40 cycles (95 °C for 3 s, 60 °C for 10 s, and 72 °C for 30 s). The amount of each gene was first determined with LightCycler Software (Roche Diagnostics) based on the standard curve using a corresponding plasmid after linearization, and expression levels were then normalized by calculating the ratio of the mRNA of interest to the amount of glyceraldehyde-3-phosphate dehydrogenase (GAPDH) mRNA. cDNA was amplified by PCR with the following primers: IFITM1, forward: cacaccagcatcaacatgcc and reverse: cctacgaagtccttggcgat; IFITM3, forward: tcacggcccatctgatcaac and reverse: gggtccaatgaattcggggt; IFITM5, forward: gactcatctcccaccactgc and reverse: ttgacagagaaggcgagagc; GAPDH, forward: gggcacgccatcactatc and reverse: gtgaagacaccagtggactcc.
cDNAs for chicken IFITM1, 3 and 5 were amplified by PCR from the cDNA of PGCs (White Leghorn Line-M, Nisseiken). The following primers were used for amplification. IFITM1, forward: catgaattcgccaccatgcagagctaccctcagcacacc and reverse: catggatccgggccgcacagtgtacaacggga; IFITM3, forward: catgcggccgcaccatggagcgggtacgcgcttc and reverse: catagatcttcagtgggtccaatgaattcgggg; IFITM5, forward: catgaattcaccatggatacatcctacccacgg and reverse: catagatcttccttgtcctcgtcatcgctgg. Underlined nucleotides indicate the restriction enzyme sites used for cloning. Amplified DNA fragments were cloned into the p3xFLAGCMV14 vector (Sigma-Aldrich). The recombinant plasmids were propagated in Escherichia coli. The purity of the plasmid was confirmed by the ratio of OD260/OD280 to be 1.60–1.75. After being digested with appropriate restriction enzymes and purified, these plasmid DNAs were used for the control of qRT-PCR. Regarding IFITM2, we were unable to clone cDNA since the relevant data were not available in the database. Thus, primers for qRT-PCR were synthesized, as previously reported (Smith et al. 2013), and data were normalized with GAPDH.
Induction of IFITMs by IFN-α
CEFs were seeded at 1.0 × 105 cells per well of a 24-well plate. After 24 h, IFN-α was added at 200 ng/ml, and cells were incubated for an additional 18 h. RNAs were then isolated using ISOGEN II, and IFITM3 RNA was quantified by qRT-PCR.
VSV-G-pseudotyped lentiviral vector production
Lentiviral vectors were produced as described previously (Motono et al. 2010) with some modifications. Briefly, 293FT cells seeded on 60-mm collagen-coated dishes were transfected with 3.8 μg of a lentiviral vector [pLSi/ΔAeGFP, expressing eGFP under the control of a chicken actin promoter (Motono et al. 2010)], 2 μg of a gag/pol plasmid (pLP1), 1.1 μg of a rev plasmid (pLP2), and 1.1 μg of a VSV-G plasmid (pLP VSV-G) (Invitrogen, Carlsbad, CA, USA) using Lipofectamine 2000 (Invitrogen). The culture supernatant was collected 72 h post-transfection. The viral titer was determined with HeLa cells in terms of the percentage of cells expressing the eGFP gene.
Knockdown of IFITM3 and estimation of infectivity
In order to knockdown IFITM3, siRNA (Sigma-Aldrich) was synthesized as previously reported (Smith et al. 2013). DF-1 cells (1.25 × 104) were seeded on 24-well plates and transfected with specific or control (siTrio; B-Bridge International Inc., Santa Clara, CA, USA) siRNAs using Lipofectamine 3000 (Invitrogen) 16 h after seeding according to the supplier’s recommendation. The viral vector was infected to cells 48 h after siRNA transfection at a MOI (multiplicity of infection) = 0.1. Infectivity was measured 9 h post-infection. In order to extract viral cDNA, trypsinized cells were collected by centrifugation (3500 rpm, 5 min), and DNA was extracted with the QIAamp DNA Mini Kit (QIAGEN, Tokyo, Japan) and then subjected to qPCR in order to quantify the vector cDNA sequence (eGFP region). By this, viral cDNA in cytosol that had been reverse-transcribed by viral reverse-transcriptase was quantified as an indication of the magnitude of infection. The amounts of viral cDNA were normalized by the amount of chicken genomic GAPDH corresponding to the cell number. The same primers for GAPDH mRNA quantification were used, which discriminate mRNA (cDNA) and genomic DNA by the sizes of amplified DNA fragments (98 and 182 bp for mRNA and genomic DNA, respectively). The primers used were eGFP, forward: cggcaactacaagacccgc and reverse: gaagttcaccttgatgccgttc. The efficiency for knockdown was also confirmed at the same time. Total RNA was extracted using the ReliaPrep RNA Cell Miniprep System (Promega, Madison, WI, USA) and subjected to qRT-PCR. The significance of differences was analyzed by Student’s t-test.
Results
Chicken IFITM3 was expressed at high levels in adult chicken lungs
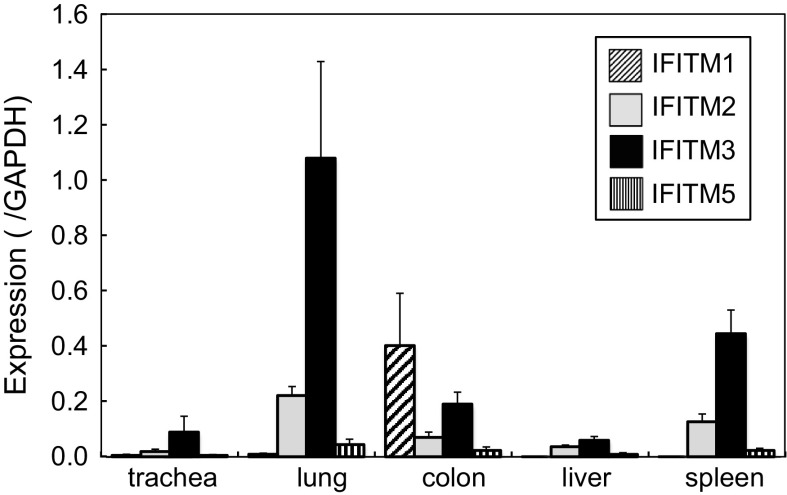
The expression of IFITMs in adult chicken organs was examined using qRT-PCR (Fig. 1). To date, the expression levels of IFITMs have only been investigated in a semi-quantitative manner (Smith et al. 2013). Therefore, we herein analyzed expression levels by qRT-PCR and normalized the values obtained using the expression level of GAPDH in order to compare the basal expression levels of IFITMs under physiological conditions. IFITM3 was expressed at a high level in the lungs, and at lower level in the spleen. Certain levels of IFITM3 were expressed in the colon and trachea. Our results also showed that the copy numbers of IFITM3 in these organs were comparable to those of GAPDH: almost equal to (lung) or 0.4-fold (spleen) those of GAPDH. The expression of IFITM2 was approximately one-fifth that of IFITM3 in the lung, and was also expressed in several other organs. The expression levels of IFITM1 and 5 were relatively low in all organs tested, except for IFITM1 in the colon (0.4-fold those of GAPDH). These results suggest that IFITM3, which exhibits strong antiviral activity (Smith et al. 2013), was strongly expressed in the lungs, which is a major target organ for infections by avian influenza viruses. On the other hand, lower levels of IFITM1 and 3 were major IFITMs in colon, another target for influenza viruses. Since IFNs caused by viral infections possibly induced IFITMs, a chicken housed under specific pathogen-free conditions was analyzed. Similar results were observed, indicating that expression levels of IFITMs in Fig. 1 were those not induced by infection but represent the basal levels of healthy adult chickens.
Fig. 1.
Expression of chicken IFITMs in various adult chicken organs. Adult chicken organs were subjected to qRT-PCR. Expression levels are shown as a ratio to that of GAPDH. Data are the mean ± standard error of three different chickens
Chicken IFITMs were enhanced by IFN-α
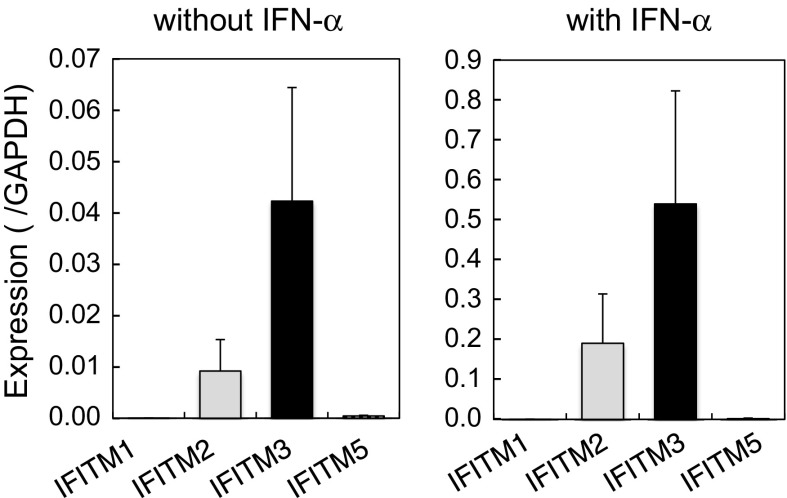
Unstimulated CEFs from 10-day embryos expressed IFITM3 and 2, but not IFITM1 or 5 (Fig. 2). IFITM3 expression levels were approximately 4 % those of GAPDH. Lower levels of IFITM2 (approximately one quarter of IFITM3) were also expressed. These levels were markedly lower than those in adult organs and similar to those in embryonic organs at the earlier stage (see below). The antiviral effects of IFN-α were partly mediated by its inducible genes including IFITMs. The stimulation of CEFs with IFN-α significantly enhanced the expression of IFITMs: an approximately 13-fold induction of IFITM3 was observed. IFN-α also induced IFITM2: approximately 21-fold. The induction of IFITM1 and 5 was also observed, but at markedly lower levels than that of IFITM3. These results are consistent with the notion that chicken IFITMs participate in the antiviral activity of IFN-α in chickens.
Fig. 2.
Induction of chicken IFITMs by IFN-α. CEFs were stimulated with IFN-α and the expression levels of IFITMs were analyzed by qRT-PCR. Left without IFN-α; right with IFN-α. Data are the mean ± standard error of three independent experiments
IFITM3 was expressed in several embryonic organs and PGCs
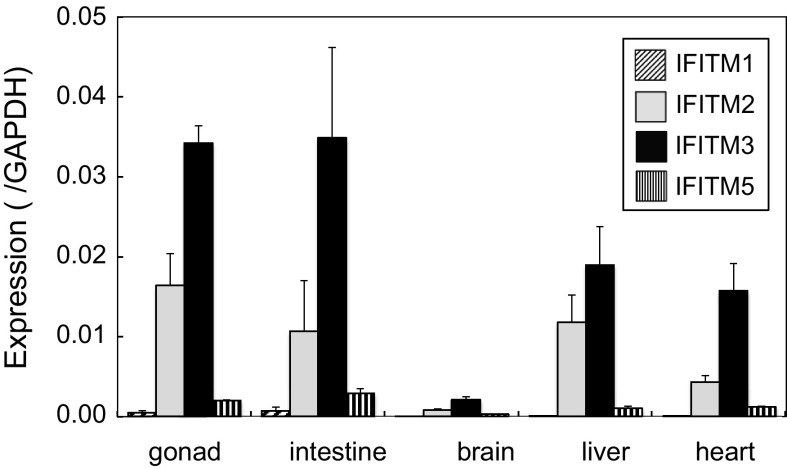
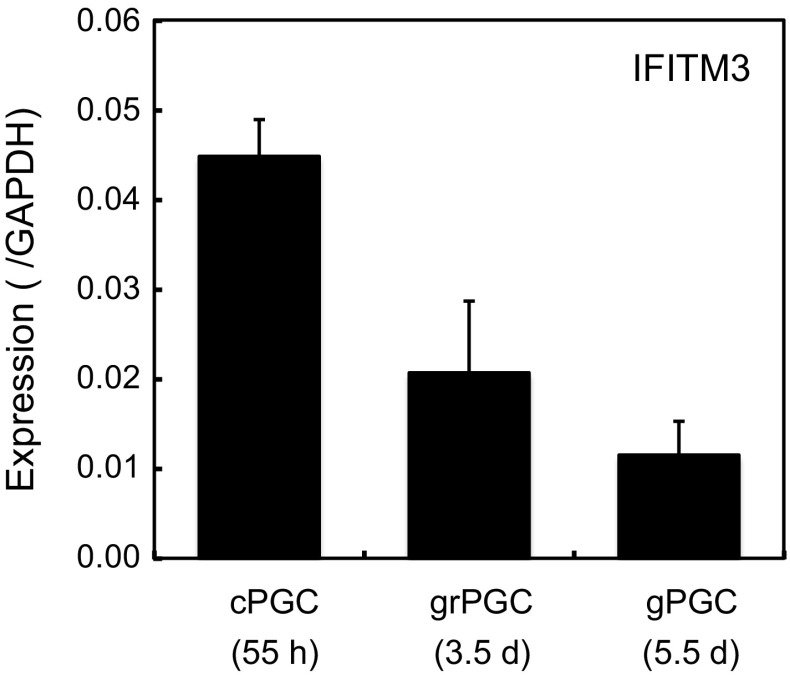
The expression of IFITMs in chicken embryos has not yet been reported. Several organs were isolated from 5.5-day embryos and the expression of IFITM1, 2, 3 and 5 was analyzed by qRT-PCR (Fig. 3). IFITM3 was expressed at high levels in the gonads and intestines, and at lower levels in the liver and heart. The brain did not strongly express IFITM3. IFITM2 was substantially expressed in the gonads (approximately 50 % those of IFITM3), livers and intestines, whereas those of IFITM1 and 5 were low. Overall, the expression pattern of IFITMs differed in each embryonic organ, with expression levels being markedly lower than those in adult organs: 3 % at most of those in the adult lungs for IFITM3. Among the embryonic organs tested, the gonads showed the highest expression levels of IFITM3. Therefore, we examined the expression of IFITM3 in PGCs (Fig. 4). The mRNA level of IFITM3 in cPGCs was 4.5 % that of GAPDH, then gradually decreased, and its expression in gPGCs was 1.2 % that of GAPDH.
Fig. 3.
Expression of chicken IFITMs in various embryonic organs. Organs from two 5.5-day embryos were pooled and subjected to qRT-PCR in order to quantify the expression of IFITMs. Note that several organs including the lungs and spleen did not develop well at this stage, thus, were excluded from the analysis. Data are the mean ± standard error of three independent experiments
Fig. 4.
Expression of chicken IFITM3 in PGCs under different developmental stages. PGCs were sorted from blood (cPGC), germinal ridges (grPGC) and gonads (gPGC), and subjected to a qRT-PCR analysis. Data are the mean ± standard error of three independent experiments
Knockdown of chicken IFITM3 enhanced infections by the VSV-G-pseudotyped lentiviral vector in DF-1 cells
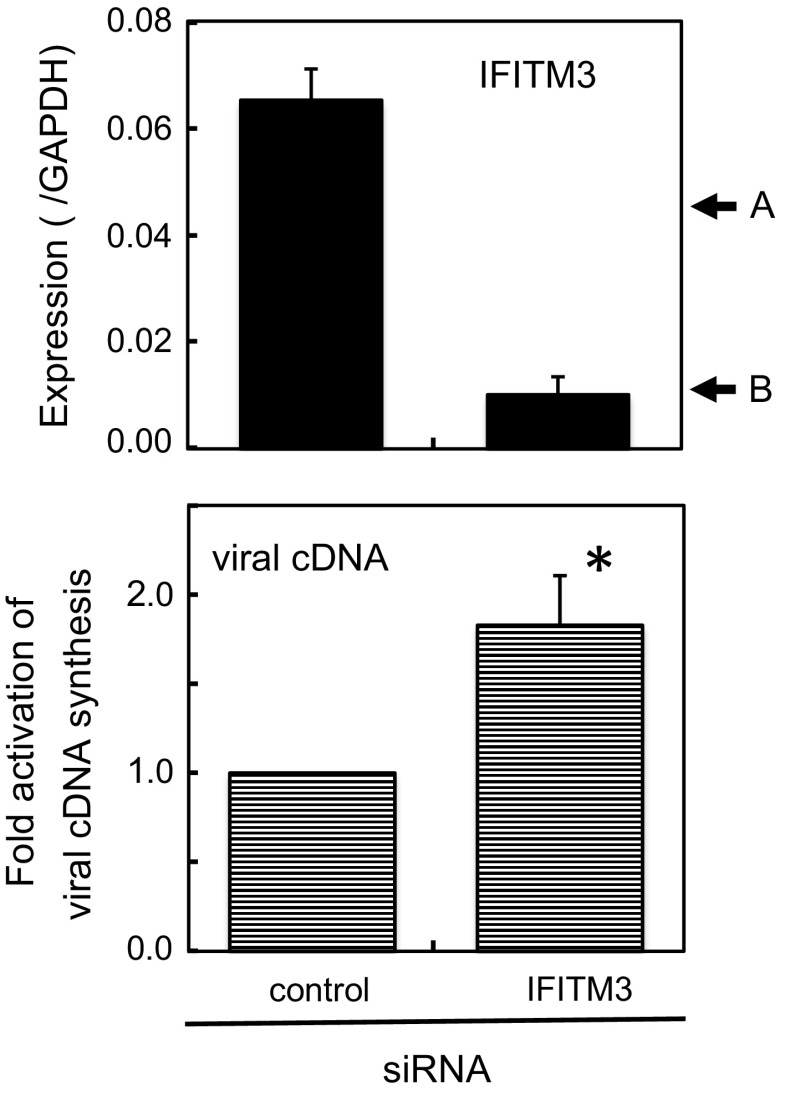
Mammalian IFITM3 strongly restricts infections by increasing numbers of viruses that infect through late endosomes, and IFITM1 blocks infections through early endosomes (Perrerira et al. 2013). In chickens, a previous study reported that IFITM3 inhibited infections by influenza A virus and lyssavirus in vitro (Smith et al. 2013). These findings indicated the potential of chicken IFITM3 for virus prevention; however, it currently remains unclear whether basal levels of IFITMs under physiological conditions are sufficient to block infections. Thus, we used DF-1 cells that expressed IFITM3 at a 1.5-fold higher level than that in cPGCs (see below). By knockdown with siRNA, the expression level of IFITM3 decreased to 15 % of the original level, roughly corresponding to that in gPGCs (Fig. 5). By the knockdown, infectivity of the VSV-G-pseudotyped virus increased by 1.8-fold, suggesting that this low level of IFITM3 was sufficient to prevent infections by the VSV-G-pseudotyped viral vector.
Fig. 5.
Knockdown of IFITM3 in DF-1 cells increased the infectivity of the VSV-G-pseudotyped lentiviral vector. Top Confirmation of the knockdown efficiency of IFITM3. IFITM3 levels were determined by qRT-PCR at the end of culture. Arrows A and B, IFITM3 expression levels in cPGCs and gPGCs, respectively. Bottom Enhancement of infectivity. Cells were infected with the VSV-G-pseudotyped lentiviral vector, and the amount of viral cDNA was quantified as an indication of the magnitude of infection. Viral cDNA levels with control siRNA were regarded as 1. Data are the mean ± standard error of three independent experiments. *p < 0.05 relative to control by Student’s t test
Chicken IFITM3 was expressed at low levels in embryonic tissues used for viral propagation in vaccine production
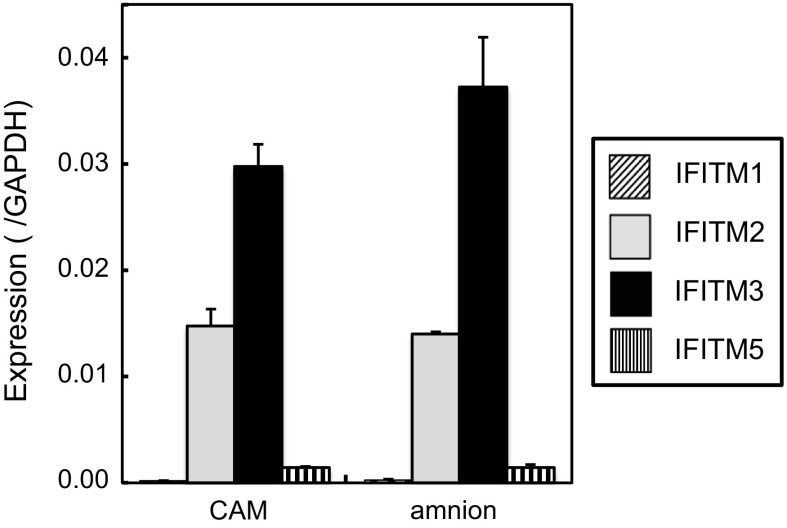
Since chicken eggs are used in the production of vaccines against various viruses, we analyzed the expression of IFITMs in the chorioallantoic membrane and amnion, which support the propagation of influenza viruses. The expression levels of IFITM3 in these organs were 3–4 % that of GAPDH and slightly lower than those in cPGCs (Fig. 6). IFITM2 expression levels were 40–50 % those of IFITM3. IFITM1 and 5 expression levels were very low. These results suggest that extraembryonic membranes express IFITM3 and 2 to some extent.
Fig. 6.
Expression of chicken IFITMs in the chorioallantoic membrane and amnion. The chorioallantoic membrane (CAM) and amnion were subjected to qRT-PCR. Data are the mean ± standard error of three independent experiments
Discussion
In the present study, we analyzed the expression patterns of IFITMs, antiviral cell-intrinsic restriction factors, in adult and embryonic chickens using qRT-PCR in order to directly compare the basal expression levels of various IFITMs. In previous studies, the distribution pattern of IFITMs in each organ was not clear; however, our results clearly showed that the main IFITM detected under physiological conditions in all organs tested (without an IFN stimulation) was IFITM3, except for the colon (Fig. 1). The copy number of IFITM3 mRNA in the lung was the highest, and was almost equal to that of GAPDH, even in the absence of infection. The expression levels of IFITM1, 2 and 5 were lower than those of IFITM3. Distribution patterns differed among organs: IFITM3 was the main IFITM in the lung, while lower levels of IFITM1 and 3 constituted IFITM expression in the colon.
In embryonic organs, the expression levels of IFITM3 were the highest among IFITMs, but were at most 3 % of those in the adult chicken lung (Fig. 3). Among embryonic tissues, PGC expressed certain levels of IFITM3, which decreased with development (Fig. 4). Transfection of specific siRNA considerably decreased mRNA level of IFITM3 (Fig. 5). We could not confirm the protein level of IFITM3 due to the lack of specific antibody. Precise analysis remains to be performed in future studies, since several modifications such as palmitoylation, ubiquitination and phosphorylation, which may affect protein level and/or localization, have been reported for mammalian IFITMs (Yount et al. 2012). Although the expression level of IFITM3 was very low in PGCs, the results of knockdown experiments suggested that this level of IFITM3 still prevented infections by the VSV-G-pseudotyped retroviral vector (Fig. 5). The underlying mechanism might include the inhibition of virus cell fusion in endosome/lysosome (Bailey et al. 2014; Perreira et al. 2013). Our results suggest that one of the mechanisms responsible for the difficulties associated with the establishment of transgenic chicken, limited germ-line transmission efficiency, may be due to the presence of IFITM3 in cPGCs. The low growth potential of germ cells may also prevent infections by viral vectors.
Embryonated eggs and CEFs are used to propagate several viruses for the production of vaccines. The low expression of IFITMs suggests that embryonic cells or tissues may be suitable for this purpose (Figs. 2, 6). In the production of influenza vaccines, a viral stock is injected into the allantois of 9-day embryonated eggs, which are further incubated for several days before harvesting virus-containing allantoic fluid. During this period, the chorioallantoic membrane supports influenza viral propagation. As shown in Figs. 5 and 6, the expression level of IFITM3 in DF-1 cells was twice that in the chorioallantoic membrane. Previously, it was reported that the knockdown of IFITM3 in DF-1 cells increased infections by influenza A virus (Smith et al. 2013). Thus, infections by influenza viruses may be partly blocked during the production of vaccines with chicken eggs. The amnion, which expresses different sugar chains from those of the chorioallantoic membrane (Ito et al. 1997) and is experimentally used in the propagation of human-isolated influenza viruses, also expressed similar levels of IFITM3. In addition to eggs, vaccines for rabies (lyssavirus), measles and mumps are prepared with cultured CEFs. A previous study reported that a retroviral infection pseudotyped with the envelope protein of the rabies virus was prevented by IFITM3 (Smith et al. 2013), although the effective level of IFITM3 needed to prevent viral infection was not determined. Thus, the preparation of vaccines for rabies may be partly affected by IFITM3 in CEFs. On the other hand, the measles and mumps viruses belong to Paramyxoviridae, in which membrane fusion occurs at neutral pH; these viruses are generally considered to enter cells at the plasma membrane (Smith et al. 2009). The restriction of these viruses by IFITMs has not yet been demonstrated. Taken together, the results of the present study suggest that further reductions in the expression of IFITMs in chicken embryos may enhance vaccine production for some viruses to a certain extent.
Acknowledgments
This work was supported by JSPS KAKENHI Grant Number 26289312.
References
- Bailey CC, Zhong G, Huang IC, Farzan M. IFITM-family proteins: the cell’s first line of antiviral defense. Annu Rev Virol. 2014;1:261–283. doi: 10.1146/annurev-virology-031413-085537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Costa S, Pardue SL, Petitte JN. Comparative development of avian primordial germ cells and production of germ line chimeras. Avian Poult Biol Rev. 2001;12:151–168. doi: 10.3184/147020601783698477. [DOI] [Google Scholar]
- Hamburger V, Hamilton HL. A series of normal stages in the development of the chick embryo. J Morphol. 1951;88:49–92. doi: 10.1002/jmor.1050880104. [DOI] [PubMed] [Google Scholar]
- Houdebine LM. Transgenic animal bioreactors. Transgenic Res. 2000;9:305–320. doi: 10.1023/A:1008934912555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito T, Suzuki Y, Takada A, Kawamoto A, Otsuki K, Masuda H, Yamada M, Suzuki T, Kida H, Kawaoka Y. Differences in sialic acid-galactose linkages in the chicken egg amnion and allantois influence human influenza virus receptor specificity and variant selection. J Virol. 1997;71:3357–3362. doi: 10.1128/jvi.71.4.3357-3362.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamihira M, Ono K, Esaka K, Nishijima K, Kigaku R, Komatsu H, Yamashita T, Kyogoku K, Iijima S. High-level expression of single-chain Fv-Fc fusion protein in serum and egg white of genetically manipulated chickens by using a retroviral vector. J Virol. 2005;79:10864–10874. doi: 10.1128/JVI.79.17.10864-10874.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamihira M, Kawabe Y, Shindo T, Ono K, Esaka K, Yamashita T, Nishijima K, Iijima S. Production of chimeric monoclonal antibodies by genetically manipulated chickens. J Biotechnol. 2009;141:18–25. doi: 10.1016/j.jbiotec.2009.02.022. [DOI] [PubMed] [Google Scholar]
- Kodama D, Nishimiya D, Iwata K, Yamaguchi K, Yoshida K, Kawabe Y, Motono M, Watanabe H, Yamashita T, Nishijima K, Kamihira M, Iijima S. Production of human erythropoietin by chimeric chickens. Biochem Biophys Res Commun. 2008;367:834–839. doi: 10.1016/j.bbrc.2008.01.020. [DOI] [PubMed] [Google Scholar]
- Kues WA, Niemann H. The contribution of farm animals to human health. Trends Biotechnol. 2004;22:286–294. doi: 10.1016/j.tibtech.2004.04.003. [DOI] [PubMed] [Google Scholar]
- Kyogoku K, Yoshida K, Watanabe H, Yamashita T, Kawabe Y, Motono M, Nishijima K, Kamihira M, Iijima S. Production of recombinant tumor necrosis factor receptor/Fc fusion protein by genetically manipulated chickens. J Biosci Bioeng. 2008;105:454–459. doi: 10.1263/jbb.105.454. [DOI] [PubMed] [Google Scholar]
- Motono M, Ohashi T, Nishijima K, Iijima S. Analysis of chicken primordial germ cells. Cytotechnology. 2008;57:199–205. doi: 10.1007/s10616-008-9156-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motono M, Yamada Y, Hattori Y, Nakagawa R, Nishijima K, Iijima S. Production of transgenic chickens from purified primordial germ cells infected with a lentiviral vector. J Biosci Bioeng. 2010;109:315–321. doi: 10.1016/j.jbiosc.2009.10.007. [DOI] [PubMed] [Google Scholar]
- Nishijima K, Iijima S. Transgenic chickens. Dev Growth Differ. 2013;55:207–216. doi: 10.1111/dgd.12032. [DOI] [PubMed] [Google Scholar]
- Perreira JM, Chin CR, Feeley EM, Brass AL. IFITMs restrict the replication of multiple pathogenic viruses. J Mol Biol. 2013;425:4937–4955. doi: 10.1016/j.jmb.2013.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudolph NS. Biopharmaceutical production in transgenic livestock. Trends Biotechnol. 1999;17:367–374. doi: 10.1016/S0167-7799(99)01341-4. [DOI] [PubMed] [Google Scholar]
- Smith EC, Popa A, Chang A, Masante C, Dutch RE. Viral entry mechanisms: the increasing diversity of paramyxovirus entry. FEBS J. 2009;276:7217–7227. doi: 10.1111/j.1742-4658.2009.07401.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith SE, Gibson MS, Wash RS, Ferrara F, Wright E, Temperton N, Kellam P, Fife M. Chicken interferon-inducible transmembrane protein 3 restricts influenza viruses and lyssaviruses in vitro. J Virol. 2013;87:12957–12966. doi: 10.1128/JVI.01443-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yount JS, Karssemeijer RA, Hang HC. S-palmitoylation and ubiquitination differentially regulate interferon-induced transmembrane protein 3 (IFITM3)-mediated resistance to influenza virus. J Biol Chem. 2012;287:19631–19641. doi: 10.1074/jbc.M112.362095. [DOI] [PMC free article] [PubMed] [Google Scholar]