Abstract
Oligonucleotides composed of 2′-O-methyl and locked nucleic acid residues complementary to HIV-1 trans-activation responsive element TAR block Tat-dependent trans-activation in a HeLa cell assay when delivered by cationic lipids. We describe an improved procedure for synthesis and purification under highly denaturing conditions of 5′-disulphide-linked conjugates of 3′-fluorescein labelled oligonucleotides with a range of cell-penetrating peptides and investigate their abilities to enter HeLa cells and block trans-activation. Free uptake of 12mer OMe/LNA oligonucleotide conjugates to Tat (48–58), Penetratin and R9F2 was observed in cytosolic compartments of HeLa cells. Uptake of the Tat conjugate was enhanced by N-terminal addition of four Lys or Arg residues or a second Tat peptide. None of the conjugates entered the nucleus or inhibited trans-activation when freely delivered, but inhibition was obtained in the presence of cationic lipids. Nuclear exclusion was seen for free delivery of Tat (48–58), Penetratin and R9 conjugates of 16mer phosphorothioate OMe oligonucleotide. Uptake into human fibroblast cytosolic compartments was seen for Tat, Penetratin, R9F2 and Transportan conjugates. Large enhancements of HeLa cell uptake into cytosolic compartments were seen when free Tat peptide was added to Tat conjugate of 12mer OMe/LNA oligonucleotide or Penetratin peptide to Penetratin conjugate of the same oligonucleotide.
INTRODUCTION
The HIV-1 trans-activation responsive element (TAR) RNA stem–loop interacts with the HIV trans-activator protein Tat and other cellular factors to stimulate transcriptional elongation from the viral long terminal repeat (LTR) (1,2). Inhibitors of these interactions would block full-length transcription and hence the ability of HIV to replicate. Much effort has gone into various approaches to design molecules capable of binding to TAR and inhibiting Tat-dependent trans-activation as potential candidates for anti-HIV therapies [reviewed in (3)], but so far no clinical candidates have emerged.
Oligonucleotides and their analogues have been much studied as potential trans-activation inhibitors because of their ability to form sequence-specific interactions with the TAR RNA target and hence block the ability of Tat and/or cellular factors to bind. One route involves the design of oligonucleotide aptamers that form kissing complexes with the apical TAR RNA stem–loop. Various nuclease-stabilized aptamer analogues have shown promise as trans-activation inhibition agents in vitro (4–6), but so far these have not led to significant cellular activity. In contrast, an antisense approach against TAR has yielded encouraging cellular inhibition results in the case of some oligonucleotide analogues (7,8).
Early studies showed that 18–26mer oligodeoxynucleotide phosphorothioates, in principle, would allow an RNase H type of antisense activity, when targeted to the apical stem–loop of TAR inhibited Tat-dependent trans-activation in a transient cell assay, but failed to show specific anti-viral activity (9). Based on previous studies of the ability of RNA and 2′-O-methyl (OMe) oligoribonucleotides to strand-invade the TAR target (10), we showed that short steric blocking OMe oligoribonucleotides of 12–16 residues bound TAR RNA strongly and blocked Tat binding very effectively (11,12). Such OMe oligoribonucleotides were able to inhibit sequence-specifically Tat-dependent in vitro transcription directed by HeLa cell nuclear extract on a DNA template containing the HIV-1 LTR (13). Similar levels of inhibition of in vitro transcription were observed for other steric block oligonucleotides such as a mixmer 16mer OMe containing five 5-methyl C locked nucleic acid (LNA) units and a 16mer peptide nucleic acid (PNA) (8).
We utilized a cellular assay involving inhibition of Tat-dependent HIV LTR trans-activation in a stably integrated plasmid system in HeLa cells and double luciferase reporters to show that, when delivered by a cationic lipid, a 12mer OMe/LNA mixmer oligonucleotide, but not its 12mer OMe counterpart, had cellular inhibitory activity (8). More recently, we carried out a detailed structure–function analysis of the requirements for cellular activity of LNA/OMe mixmer oligonucleotides with delivery by cationic gemini surfactant/lipid formulation GS11/DOPE (14). This showed oligomers of 12–16 residues were active, but that a 10mer was inactive under the assay conditions: 40–50% of LNA units scattered throughout the OMe oligonucleotide gave best results and the LNA residues could be placed at different locations without affecting activity.
Transfection of DNA and negatively charged oligonucleotides and their analogues is usually achieved for common laboratory cell lines in culture (such as HeLa cells) through complexation with cationic lipids (15). There is now a wide choice of such reagents, but there are many difficulties in their use. For example, the oligonucleotide must be formulated with the delivery reagent and ratios adjusted for each experimental case to optimize uptake whilst limiting lipid-associated cell toxicity. Different lipid-based delivery reagents can give rise to highly variable activity levels, and most formulations are not suitable for use with primary cells. Further, cationic lipid formulation has considerable disadvantages for potential therapeutic use. Accordingly, there has been considerable motivation to search for alternative methods of oligonucleotide delivery that do not involve lipid complex formation.
Much recent interest has been aroused by the suggestion that certain peptides [known as cell-penetrating peptides (CPP) or protein transduction domains (PTD)] can translocate into cells in culture and, when attached covalently, can carry in various cargoes [reviewed in (16,17)]. Amongst such cargoes are oligonucleotides and their analogues [reviewed in (18,19)]. Whereas there have been numerous reports showing enhanced cellular delivery and activity of PNA by covalently attached CPPs [reviewed in (18)], including a CPP–PNA oligomer targeted to the TAR RNA (20), the delivery of negatively charged phosphodiesters or phosphorothioates by CPPs has been more controversial.
Two early reports showed that a 16-residue cationic peptide from an Antennapedia homeodomain, called Penetratin, when covalently linked to phosphodiester oligonucleotides by disulphide bond, could enter neuronal cells and down-regulate different genes (21,22). More recently, disulphide-linked conjugates of a 20mer phosphorothioate oligodeoxyribonucleotide with Penetratin or with a second peptide, this time derived from the arginine-rich domain of the HIV-1 Tat protein, were found to be taken up by mouse 3T3 cells without additional carrier and to block expression of P-glycoprotein synthesis (23). More relevant to steric block applications, Tat or Penetratin peptide conjugates of an 18mer 2′-O-methyl phosphorothioate oligonucleotide were shown to be delivered to HeLa cell nuclei and to exhibit dose-dependent inhibition of splicing and up-regulate expression of a stably integrated luciferase gene (24).
In contrast, a number of disulphide-linked conjugates of phosphorothioate oligonucleotides with a hydrophobic signal sequence from K-FGF or the same domain extended at the C-terminus by a nuclear localization signal (NLS) from transcription factor κB failed to deliver to nuclei of monkey kidney fibroblast cells CVP-1 and failed to show antisense activity (25). However, all the conjugates were active when delivered to the nucleus by cationic lipid.
In preliminary studies, we found that fluorescently labelled, stable amide conjugates of a 12-long anti-TAR LNA/OMe mixmer with a 15mer Kaposi-fibroblast growth factor (K-FGF) peptide or with Transportan-TP10 [a shortened form of a hybrid of a section of the neuropeptide galanin and the wasp venom peptide mastoparan (26)] failed to enter HeLa cells, but were fully active in inhibition of Tat-dependent trans-activation when delivered by cationic gemini surfactant (14). It seemed clear therefore that the mere attachment of a CPP to an oligonucleotide is not itself sufficient to ensure free cell uptake and biological activity. Apart from the results of Antopolsky et al. (25), to our knowledge there have been few systematic attempts to establish the parameters important for efficient free cell uptake for CPP–oligonucleotide conjugates.
There are numerous methods reported for synthesis of peptide–oligonucleotide conjugates [comprehensively reviewed in (27)]. We have developed previously a number of methods for such conjugation, e.g. amide bond fragment condensation on solid support (28,29), native ligation of a cysteine-containing oligonucleotide with a thioester peptide (30), 2′-aldehyde oligonucleotide conjugations to form thiazolidine, oxime and hydrazine linkages (31) and total stepwise solid-phase synthesis on a single solid support (32). All of these methods give rise to linkages expected to be stable within cells. Since the few previously claimed successes for cell delivery and biological activity of CPP–oligonucleotide conjugates have all used disulphide linkages between oligonucleotide and peptide moieties (21–24), which are presumed to have greater lability within cells, it was appropriate therefore to prepare disulphide-linked conjugates and investigate cell activity and delivery.
In this study, we have improved significantly the chemical synthesis and purification of disulphide conjugates of CPPs with fluorescein-labelled oligonucleotides with particular emphasis on maintaining solubility through use of highly denaturing conditions. We have investigated HeLa and human fibroblast cell uptake properties of such conjugates and their ability to inhibit Tat-dependent trans-activation in our HeLa cell reporter assay. We show that in many cases attachment of a CPP does enhance free cell uptake of oligonucleotides, but that the uptake is confined to cytosolic compartments. The exclusion from the cell nucleus correlates with the lack of inhibition of Tat-dependent trans-activation for free delivery of oligonucleotide–CPP conjugates, suggesting that the barrier to nuclear activity is due to insufficient release from endosomal compartments. We also find that uptake is boosted dramatically by addition of extra free CPP to the conjugate, pointing to the need for extreme care in synthesis and purification of conjugates to avoid overestimation of cell delivery due to peptide complexation.
MATERIALS AND METHODS
HPLC purifications were carried out using a Waters HPLC 626 pump and 600S controller attached to a 996 photodiode array detector. MALDI-TOF mass spectra were recorded on a Voyager-DE workstation (PE Biosystems). Matrices used for preparing the MALDI-TOF samples were as follows: (a) α-cyano-4-hydroxycinnamic acid, 10 mg ml−1 in acetonitrile–3% aqueous trifluoroacetic acid (TFA) (1:1 v/v) for all peptides, (b) 2,6-dihydroxyacetophenone, 20 mg ml−1 and diammonium hydrogen citrate, 40 mg ml−1, in 50% aqueous methanol for phosphodiester oligonucleotide–peptide conjugates or (c) saturated solution of 2,4,6-trihydroxyacetophenone (Aldrich) in 50 mM diammonium hydrogen citrate/50% acetonitrile (33) for phosphorothioates and heavily salt-contaminated phosphodiester conjugates. The accuracy of the mass measurement is regarded as ±0.05%.
Peptide synthesis
Peptides were obtained either from Southampton Polypeptides Ltd or synthesized on an Advanced ChemTech APEX 396 robotic synthesizer (5–10 μmol scale) or a PerSeptive Biosystems Pioneer peptide synthesizer (100 μmol scale) using standard Fmoc/tert-butyl solid phase synthesis techniques. Arginine was Pbf protected. C-terminal amide peptides were prepared using NovaSyn TGR resin (Novabiochem), whereas C-terminal carboxylic acid peptides were synthesized with the C-terminal amino acid attached to a PEG-PS resin (PerSeptive Biosystems). Deprotection of all peptides and cleavage from solid support was achieved by treatment with TFA in the presence of triethylsilane and water (each 3%).
N-terminal thiol-activated cysteine peptides were synthesized using Boc-L-Cys(Npys)-OH obtained from Bachem. The only exceptions being CR9-OH and C-Transportan-NH2 which were activated (after synthesis and purification) with 2-Aldrithiol (Aldrich) as follows: to a solution of the thiol peptide (1 mM in 0.1 M NH4HCO3) was added 10 equivalents of 2-Aldrithiol in DMF (10 mg ml−1). The solution was mixed thoroughly and left to stand for 1 h. The peptide was analysed and purified using RP–HPLC on a Vydac C8 column (20 × 250 mm, 10 ml min−1, buffer A = 1% TFA (aqueous), buffer B = acetonitrile + 10% buffer A, gradient 5–50% B in 30 min). All C-terminal cysteine-containing peptides were activated (if required) and purified in the same manner.
Named peptide sequences (excluding terminal cysteines) are Tat (residues 48–58): GRKKRRQRRRP, Penetratin: RQIKIWFQNRRMKWKKGG, Transportan: GWTLNSAGYLLGKINLKALAALAKKIL.
Oligonucleotides
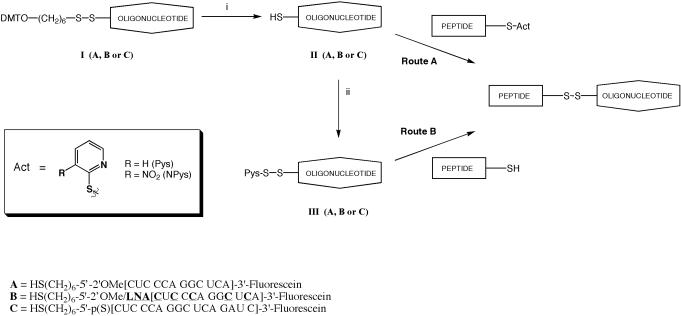
Oligonucleotides were synthesized on a 1 μmol scale according to previously reported protocols except that the activator was changed to 5-ethylthio-1H-tetrazole (a 0.25 M solution in acetonitrile, Link Technologies). LNA and 2′-OMe amidites were obtained from Link Technologies. The 5′-C6 thiol linker and Beaucage Reagent (to introduce phosphorothioate linkages) were purchased from Glen Research. 3′-Fluorescein support for the synthesis of OMe/LNA and OMe/PS oligonucleotides B and C was obtained from Glen Research [3′-(6-Fluorescein)CPG]. The 3′-(6-carboxyfluorescein) OMe oligonucleotide A was synthesized as described previously (34). All other reagents were obtained from Proligo. Oligonucleotides were simultaneously cleaved and deprotected from the CPG support using saturated aqueous ammonia solution and were subsequently purified by ion exchange HPLC using a DNAPac™ PA-100 (9 × 250) Dionex column [3 ml min−1, buffer A: 25% formamide, 1 mM NaClO4, 20 mM Tris–HCl (pH 6.8), buffer B: 1 mM NaClO4, 20 mM Tris–HCl (pH 6.8), 400 mM NaClO4], gradient 15–55% B in 20 min. Products were recovered by dialysis against water followed by lyophilization. The sequences of the oligonucleotides are shown in Figure 1.
Figure 1.
Scheme for formation of disulphide-linked conjugates of cysteine-containing peptides with 5′-thiol-substituted oligonucleotides and sequences of the oligonucleotides A, B and C. Reagents: (i) DTT, 0.1 M TEAA and (ii) dipyridyl disulphide, 0.1 M TEAA.
Preparation of 5′-thiol oligonucleotides
The thiol was liberated by reduction of the disulfide bond of the 5′-linker of the oligonucleotide [1 mM in a 0.05 M triethylammonium acetate (TEAA) solution containing 1% triethylamine (TEA)] using aqueous DTT (10 equivalents added from 1 M stock solution). After 1.5 h, the reaction mixture was desalted using size exclusion chromatography (NAP 10 or NAP 25 column, Amersham Biosciences) and lyophilized. The amount of residual DTT impurity was ascertained by comparing the A260 value for the oligonucleotide with the amount of thiol present as determined by the Ellman's test (measurement at A412, with adjustment to take into account the absorption at that wavelength of the fluorescein).
Preparation of 5′-Pys-activated thiol oligonucleotides
To the 5′-thiol oligonucleotide (1 mM in a 0.05 M TEAA solution) was added a solution of Aldrithiol-2 in DMF (10 equivalents, 10 mg ml−1). The reaction mixture was vortexed and left to stand for 1 h whereupon the crude product was purified by ion exchange HPLC as above.
Conjugations
Route A: Conjugation of activated thiol peptide to thiol oligonucleotide (conjugates 1–16, 18). General procedure. To a microfuge tube containing the oligonucleotide (20 nmol in 50 μl water) formamide (150 μl) and 2 M TEAA buffer (10 μl) were added. The solution was thoroughly mixed and the activated peptide was added (80 nmol in 8 μl). The solution was briefly mixed and allowed to stand for 30 min, whereupon the conjugate was purified by HPLC using a 1 ml Resource Q column (Amersham Biosciences) [1 ml min−1, buffer A: 20 mM Tris–HCl (pH 6.8), 50% formamide, buffer B: 20 mM Tris–HCl (pH 6.8), 50% formamide, 400 mM NaClO4], gradient 0–100% B buffer in 20 min. For highly cationic peptide conjugates 14, 15, 16 and 18, due to aggregation, the reaction mixture needed to be treated with extra TEAA and formamide (order of addition not important) in order to fully solubilize the product which had precipitated in the microfuge tube. The product was collected, dialysed and lyophilized. The lyophilized product was taken up in the solution as described in Table 2 for further analysis.
Table 2.
Properties of conjugates synthesized: retention time on RP–HPLC, calculated and observed masses by MALDI-TOF mass spectrometry, isolated yields and salts used for dissolution of conjugates
| Conjugate | Retention time (min) | Calculated molecular mass | Observed MALDI molecular mass | Isolated yield (%), salts |
|---|---|---|---|---|
| 1 | 10.84 | 6315.22 | 6315.52 | 55, 0 |
| 2 | 11.13 | 6315.22 | 6312.94 | 49, 0 |
| 3 | 12.99 | 6316.25 | 6315.34 | 69, 0 |
| 4 | 13.22 | 6316.25 | 6316.07 | 74, 0 |
| 5 | 13.28 | 7183.31 | 7181.99 | 68, 0 |
| 6 | 13.87 | 7183.31 | 7183.36 | 75, 0 |
| 7 | 12.66 | 6247.15 | 6244.28 | 50, 0 |
| 8 | 12.54 | 6247.15 | 6243.30 | 36, 0 |
| 9 | 15.24 | 7871.20 | 7869.28 | 70, 0 |
| 10 | 18.45 | 8738.26 | 8730.89 | 63, 0.01 M TEAA |
| 11 | 18.04 | 7802.1 | 7801.39 | 40, 0.01 M TEAA |
| 12 | 12.31 | 6540.52 | 6539.62 | 56, 0.1 M TEAA |
| 13 | 15.95 | 7663.92 | 7659.08 | 78, 0 |
| 14 | 9.80 | 6941.00 | 6936.68 | 24, 0.1 M TEAA |
| 15 | 9.94 | 6941.00 | 6944.49 | 55, 0.1 M TEAA |
| 16 | 8.10 | 7792.01 | 7799.79 | 15, 1 M TEAA, 1 M NH4Cl |
| 17 | 8.02 | 7792.01 | 7788.30 | 12, 1 M TEAA |
| 18 | 10.27 | 6828.94 | 6827.88 | 58, 0.1 M TEAA |
| 19 | 8.85 | 8120.43 | 8161.83 (Broad hump) | Not possible to completely dissolve in 2 M TEAA |
Route B: Conjugation of thiol peptide to activated thiol oligonucleotide (conjugates 4, 6, 15, 17–19). General procedure. To a microfuge tube containing the activated oligonucleotide (20 nmol in 50 μl water) formamide (150 μl) and 2 M TEAA buffer (10 μl) were added. The solution was thoroughly mixed and the thiol peptide was added (50 nmol in 5 μl water). The solution was briefly mixed and allowed to stand for 30 min, whereupon the conjugate was purified by HPLC using a 1 ml Resource Q column [1 ml min−1, buffer A: 20 mM Tris–HCl (pH 6.8), 50% formamide, buffer B: 20 mM Tris–HCl (pH 6.8), 50% formamide, 400 mM NaClO4], gradient 0–100% B buffer in 20 min. For highly cationic peptides 15, 17–19, due to aggregation, the reaction mixture needed to be treated with extra TEAA and formamide (order of addition not important) in order to fully solubilize the product which had precipitated in the microfuge tube. The product was collected, dialysed and lyophilized. The lyophilized product was taken up in the solution as described in Table 2 for further analysis. In the case of conjugate 19 (R6-Penetratin-oligo B), there was considerable precipitation seen during dialysis, and upon lyophilization, the resultant product could not be dissolved fully even in 2 M TEAA.
Binding of oligonucleotides and conjugates to TAR RNA
A polyacrylamide gel mobility shift assay was used to measure the apparent dissociation constant of oligos or their conjugates to a 32P-labelled TAR 39mer RNA as previously described (8,13).
Inhibition of Tat-dependent trans-activation in cells
Inhibition of HIV-1 Tat-mediated transcription trans-activation by oligonucleotide analogues in HeLa cells was carried out in a similar way to that described previously (8,14). Briefly, in each experiment two identical 96-well plates were prepared with 10 × 103 HeLa Tet-Off/Tat/luc-f/luc-R cells per well and incubated at 37 °C for 24 h. One of the plates was used for the luciferase assay and the other for the cytotoxicity assay. For delivery by cationic lipid mixture of 1 μM oligonucleotides or conjugates and 10 μl ml−1 of Lipofectamine2000 in Opti-MEM (both Invitrogen) serum-free medium were prepared according to manufacturer's manual. After 20 min at room temperature, subsequent dilutions were prepared from oligonucleotide/Lipofectamine2000 mixture. Cells were incubated with oligonucleotide/Lipofectamine2000 mixtures for 3 h, washed with phosphate-buffered saline (PBS) and left for additional incubation in DMEM/10% FBS for 18 h. For free conjugate delivery, conjugates were prepared at 2.5 μM concentration in Opti-MEM, subsequently diluted and added to the cells for 6 or 24 h incubation followed by 18 h incubation in DMEM/10% FBS.
Luciferase assay: Cell lysates were prepared and analysed using the Dual Luciferase Reporter Assay System (Promega) and relative light units for both firefly and Renilla luciferase read sequentially using a Berthold Detection Systems Orion Microplate luminometer. Each data point was averaged over two replicates of three separate experiments.
Toxicity assay: The extent of toxicity was determined by measurement of the proportion of live cells colorimetrically using CellTiter 96 AQueous One Solution Assay (Promega). The absorbance at 490 nm was read using a Molecular Devices Emax microplate Reader. Each data point was averaged over two replicates of three separate experiments.
The luciferases' activities results were normalized to the absorbance data from the toxicity assay, which reflects the amount of live cells.
Confocal microscopy
HeLa cells or human fibroblasts (15 × 103 cells) were plated on 8-well Lab-Tek chambered coverglass (Fisher Scientific) in DMEM/10%FBS and cultured overnight. The medium was discarded and cells were washed with PBS followed by incubation with 300 μl of 0.5 μM oligonucleotide/Lipofectamine2000 complex in OptiMEM for 3 h, free 2.5 μM conjugates or conjugate/peptide mixtures in OptiMEM for 5 or 24 h. The conjugate/peptide mixtures were sonicated for 1 min prior to addition to the cells. For nuclear staining, cells were washed three times with PBS before addition of 300 μl OptiMEM containing Hydroethidine (20 μg/ml) and incubated for 1 h at 37 °C. After two washes, 300 μl of OptiMEM (without phenol red) (Invitrogen) medium containing HEPES buffer was added into the wells for observation of living cells.
The cells were observed with a Radiance 2100 confocal system on a Nikon Eclipse TE300 inverted microscope using a 60× Planapo objective N.A. 1.4. A 488 nm Argon-laserline was used to excite fluorescein and a HQ 515/30 emission filter was used for observation of the green emission. Hydroethidine was excited with a 543 nm (green) HeNe-laser and detected using a HQ 570LP (orange) emission filter. A dual fluorescence method was used with a Differential Interference Contrast (DIC) transmission channel. The images in the three channels were acquired sequentially at ∼1 frame per second with a scanning resolution of 512 × 512 pixels and a Kalman average of 6 frames was used. When comparing the uptake or activity of the oligonucleotides, the imaging conditions (such as photomultiplier gain/offset, laser intensities and confocal aperture size) were kept constant for the observation of the different oligos, so that the intensities represent the true differences in uptake/activity.
Quantitation of fluorescence in cells
A 96-well plate containing HeLa cells was prepared as described above and cells were transfected either with carboxyfluorescein-labelled oligonucleotide–peptide conjugate for 5 h prepared as for microscopy or for 3 h with 0.5 μM oligonucleotides complexed with transfection reagent as above. Cells were washed twice with PBS and lysed for 20 min with 20 μl of passive lysis buffer (Promega), which is the same as used in the Dual Luciferase Reporter Assay System. After 5 min centrifugation at 2500 g, the cell lysate was transferred to a 96-well assay plate. The cell debris were washed with 100 μl of water, and after centrifugation the supernatant was added to the cell lysate followed by fluorescence measurement. The fluorescence of conjugates or oligonucleotides was detected using a microplate fluorimeter (Fluoroskan, Labsystems) supplied with a 460/536 nm excitation/emission filter pair. The resulting fluorescence was normalized to the background amount of fluorescence of untreated cells and expressed as the percentage of added cellular fluorescence over background. Each data point was averaged over two replicates of three separate experiments.
RESULTS
Synthesis of disulphide conjugates of oligonucleotides with CPPs
The first published method of disulphide-bond formation involved reaction of a thiol-modified oligonucleotide with a 3-nitropyridinesulphenyl (Npys) activated N-terminal cysteine residue of an NLS peptide (35). Subsequently, the most reliable method seems to be pyridyl sulphide (Pys) activation of the peptide and reaction with the oligonucleotide (Figure 1, Route A) or activation of a thiol-modified oligonucleotide with 2,2′-dipyridyldisulphide and reaction with a cysteine-containing peptide (Route B) (23,25,36–38). Simple peptide conjugates of oligonucleotides can be prepared in high yield and purified by RP–HPLC (30). However, cationic peptides (including many well-known CPPs) associate with negatively charged phosphodiester or phosphorothioate oligonucleotides and cause precipitation, especially when excess peptide is used to drive the reaction to completion (25,38,39). To overcome this problem, Vivès and Lebleu reported that a Tat peptide could be coupled to a deoxyoligonucleotide in the presence of 0.4 M potassium chloride solution and 40% acetonitrile to maintain solubility (38). For hydrophobic peptides, Antopolosky et al. (25) suggested coupling in the presence of 90% formamide. We have investigated the latter denaturing agent for conjugation of CPPs to oligonucleotides.
In Route A, peptides with N-terminal cysteine were prepared on solid-phase either to carry a Npys group on the side-chain of the cysteine or synthesized as the free peptide and the cysteine residue activated post-assembly by reaction with 2,2′-dipyridyldisulphide to form the Pys derivative and then purified by HPLC. C-terminal cysteine-containing peptides were all activated to form the Pys derivative before conjugation. An advantage of Route A is that the component that is prepared on smaller scale (the oligonucleotide) requires less manipulation prior to conjugation. Conjugations were carried out in 70% formamide with a TEAA buffer and with 4-fold excess peptide component for 15–30 min. In some cases, Route B was used. Here a smaller excess of peptide was required (2.5-fold) over oligonucleotide, and there was generally a slightly better conversion into conjugate. This may be due in Route A to traces of reducing agent DTT remaining in the oligonucleotide component before conjugation, whereas in Route B the oligonucleotide component is activated with 2,2′-dipyridyldisulphide. The disadvantage is that the oligonucleotide component must be purified again by HPLC after activation, resulting in some losses. Thus, there is little to choose between the two routes and the product is identical in each case.
Our first targets were disulphide conjugates of several well-known CPPs [Tat (48–58) (40), Penetratin (21), the homopeptide R9 (41), R9F2 (42) and Transportan (26)]. Tat was coupled with a 12mer OMe (oligo A) to give conjugates 1 and 2 (Table 1). All 5 CPPs were conjugated with a 12mer OMe/LNA mixmer (oligo B) to give conjugates 3–8, 12 and 13. C-terminal Tat, Penetratin and R9 conjugates were made with a 16mer 2′-O-methylphosphorothioate (OMe/PS) oligonucleotide (oligo C) to give conjugates 9–11. All three oligonucleotides are complementary to the HIV-1 TAR RNA apical loop as previously described (8,14). Each oligonucleotide was synthesized with a 5′ C6-thiol linker and a 3′ 6-carboxyfluorescein attachment (Figure 1). Peptides contained additionally either an N- or C-terminal cysteine residue (for sequences, see Materials and Methods).
Table 1.
List of conjugates synthesized
| Peptide | Conjugate-Oligo A | Conjugate-Oligo B | Conjugate-Oligo C |
|---|---|---|---|
| C-Tat-NH2 | 1 | 3 | |
| Tat-C-NH2 | 2 | 4 | 9 |
| C-Penetratin-NH2 | 5 | ||
| Penetratin-C-NH2 | 6 | 10 | |
| CR9-OH | 7 | ||
| R9C-OH | 8 | 11 | |
| R9F2C-NH2 | 12 | ||
| C-Transportan-NH2 | 13 | ||
| C-R4-Tat-NH2 | 14 | ||
| R4-Tat-C-NH2 | 15 | ||
| C-Tat-Tat-NH2 | 16 | ||
| Tat-Tat-C-NH2 | 17 | ||
| K4-Tat-C-NH2 | 18 | ||
| R6-Penetratin-C-NH2 | 19 |
The sequences of the oligonucleotides are shown in Figure 1.
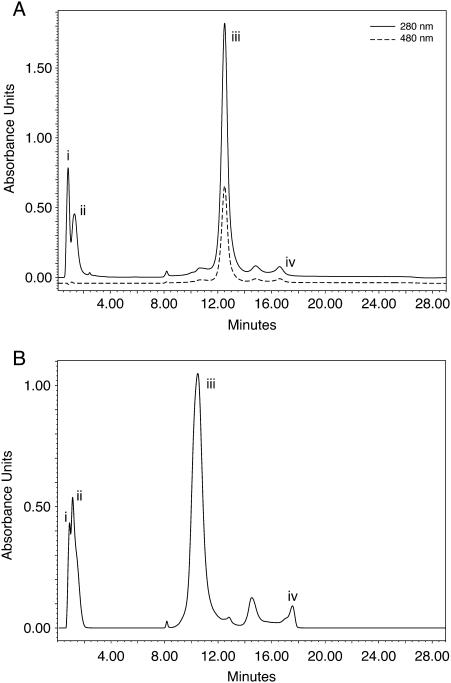
Products were purified by short path ion exchange chromatography (Resource Q) under highly denaturing conditions (50% formamide) to ensure that the desired conjugate was quickly separated from excess peptide and to prevent precipitation. A typical HPLC chromatogram is shown in Figure 2A, which shows that the conjugate is well resolved and always by far the major component (conversions >90%). The conjugate could then be recovered by dialysis and lyophilization and generally redissolved without difficulty: conjugates 1–9, 12 and 13 in water, conjugates 10 and 11 in low strength buffer (0.01 M TEAA). All products showed a single conjugate peak in the expected mass range by MALDI-TOF mass spectrometry (Table 2 and Supplementary Material). In no case using matrix 3, in which both peptides and oligonucleotide–peptide conjugates can be readily observed (see Materials and Methods), was more than a trace of peak seen in the expected mass region of free peptide for all conjugates prepared, which confirms the homogeneity of the products. Isolated yields of conjugates were 36–78%.
Figure 2.
HPLC chromatograms showing purification of conjugates. (A) Conjugation of Penetratin-Cys with OMe/LNA oligo B activated with Pys (Route B); peaks (i) salts and formamide, (ii) excess Penetratin-Cys (iii) conjugate product, (iv) unconjugated oligoB (Pys). The solid trace is at 280 nm and the dashed trace is at 480 nm, which identifies the fluorescein label on the oligonucleotide. (B) Conjugation of K4Tat-Cys(Pys) + OMe/LNA oligo B (free SH) (Route A); peaks (i) salts and formamide, (ii) excess K4Tat-Cys(Pys), (iii) conjugate product and (iv) unconjugated oligo B (free SH).
We have not had success in purification of conjugates from conjugation reactions by use of an alternative ion exchange column (e.g. DNAPac PA 100) or a reversed phase column commonly used for oligonucleotide or peptide purifications, due to precipitation and aggregation. These experiences are similar to those of Prater and Miller who recently failed to obtain a conjugate of Tat peptide with a fluorescently labelled 2′-O-methyl oligonucleotide after trying several options (39). In contrast, conjugations and Resource Q purifications in the presence of formamide as denaturing agent in all cases were successful and therefore can be recommended for a range of cationic or other CPPs to oligonucleotides containing phosphodiester or phosphorothioate backbones.
Binding of CPP–oligonucleotide conjugates to the TAR target
In a previous study, we found that stably linked K-FGF and Transportan-TP10 conjugates of OMe/LNA 12mer oligo B were able to bind the TAR RNA target without loss of binding strength (14). Since disulphide conjugates are potentially unstable within cells, we measured the binding strength in transcription buffer of oligo B containing a 5′-C6-thiol linker (which would be obtained on reduction of the disulphide linkage) to a synthetic 39mer TAR RNA. The apparent Kd was 2.2 ± 0.2 nM, very similar to that of unfunctionalized oligo B (8,14). We also measured the apparent Kd of binding to TAR of typical cationic CPP conjugates of oligo B Tat conjugate 4: 1.7 ± 0.4 nM and oligo B Penetratin conjugate 6: 6.2 ± 1.1 nM. Thus, whether or not a CPP peptide is removed from the oligo during passage through the cell, binding strength to the target TAR site is unlikely to be significantly affected by the conjugation chemistry. Note that in the case of the Tat peptide conjugate, the peptide part is unlikely to compete with the oligo B part significantly for binding to TAR, since it was previously reported by us that a slightly longer peptide that includes the basic domain and the C-terminal Glu-rich region of Tat (residues 48–72) has a much higher Kd of about 300 nM, as judged by mobility shift analysis (43). The Kd in transcription buffer of PS/OMe 16mer oligo C containing 5′-C6-thiol linker was found to be 8.9 ± 0.8 nM, slightly weaker than that of OMe/LNA 12mer oligo B.
Trans-activation inhibition of CPP–oligonucleotide conjugates
We have published previously the use of a stably transformed HeLa cell line that contains three plasmids (8,14). Tat is produced from the first plasmid and trans-activates the HIV-1 LTR on a second plasmid in order to produce firefly luciferase. The third plasmid produces the control Renilla luciferase under constitutive CMV promoter regulation. Specific knockdown of firefly luciferase expression, but not Renilla luciferase, can be obtained only if the oligonucleotide is taken up by a large majority of cells and is able to inhibit the HIV LTR by sterically blocking the TAR RNA target over the assay period. Thus, this assay system measures both the cellular activity of the oligonucleotide or its conjugate as well as its cell delivery ability. We showed previously specific inhibition of Tat-dependent trans-activation by a 12mer OMe/LNA mixmer (oligo B), when delivered by cationic lipids, but not mismatched or scrambled oligomers (8) and carried out a range of structure–activity studies (14). In contrast, OMe 12mer (oligo A) was entirely inactive in this cell assay.
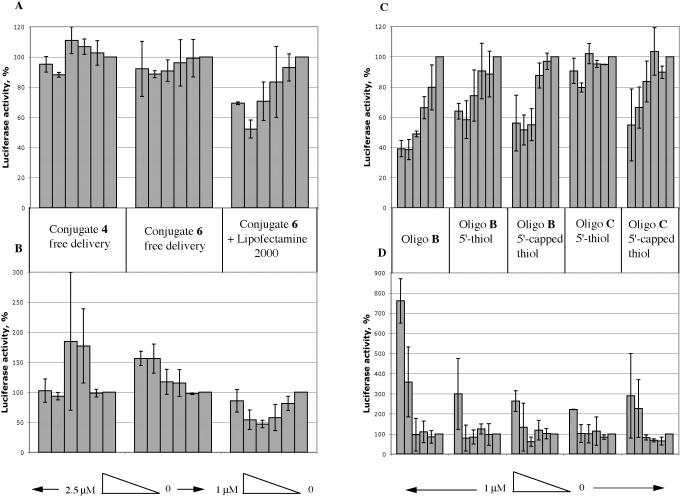
Activity of conjugates by free uptake. We tested all the CPP conjugates of the OMe/LNA mixmer oligo B (conjugates 3–8, 12 and 13) in the cell assay by free uptake. None of these conjugates showed inhibition of firefly luciferase (Tat-dependent trans-activation) or control Renilla Luciferase up to 2.5 μM concentration tested (Figure 3A and B, left and centre panels, which shows two representative examples of Tat and Penetratin conjugates 4 and 6). In order to simulate as closely as possible the conjugates of Astriab-Fisher et al. (24), we also tested in our assay system Tat, Penetratin and R9 conjugates of a 16mer OMe/PS oligonucleotide targeted to TAR (oligo C, conjugates 9–11). No activity was observed up to 2.5 μM for all three conjugates (data not shown).
Figure 3.
Inhibitory effects of oligonucleotides and conjugates in the HeLa cell reporter assay. (A and C) Firefly luciferase activity. (B and D): Control Renilla luciferase activity. A and B panels, left sections: free delivery of conjugate 4; centre sections: free delivery of conjugate 6; right sections: Lipofectamine 2000 delivery of conjugate 6. (C and D) Lipofectamine 2000 delivery from left to right: oligo B without linker, oligo B with 5′-thiol, oligo B with 5′-capped thiol, oligo C with 5′-thiol and oligo C with 5′-capped thiol.
Activity of oligos B and C and their conjugates by cationic lipid delivery. OMe/LNA 12mer (oligo B), either without or with the 5′-thiol linker, when delivered by cationic lipid Lipofectamine2000 showed dose-dependent inhibition of firefly luciferase expression but not Renilla luciferase expression (Figure 3C and D). This is in line with our previous findings for delivery of the same oligonucleotide with another cationic lipid as well as a cationic surfactant (8,14). A 16mer OMe/PS oligo C that contains a 5′-thiol linker did not show dose-dependent inhibition of firefly luciferase or Renilla luciferase, whereas the same oligo B containing the 5′-DMT-C6-S-S-linker before treatment with reducing agent (5′-thiol capped) with delivery by Lipofectamine2000 showed selective inhibition of firefly luciferase in the HeLa cell assay (Figure 3C and D). Thus oligo C in principle does have target-specific activity when delivered by cationic lipid, but we have no explanation for the lack of activity of the 5′-thiol oligo C.
In general, we found that CPP–oligo B conjugates showed specific firefly luciferase inhibition activity in the HeLa cell assay in the presence of cationic lipid, but at varying levels. For example, conjugate 6 (OMe/LNA oligo B conjugated to Penetratin) showed a moderately good dose-dependent reduction in firefly luciferase activity without concomitant reduction in Renilla luciferase activity (Figure 3A and B, right panels). Similar results were seen for the Tat conjugate 4 but the Transportan conjugate 13 showed both firefly and Renilla luciferase inhibition (data not shown). The varying levels of activity may reflect varying abilities of the conjugates to be packaged by cationic lipid, depending on the particular attached peptide.
Cell penetration of CPP–oligonucleotide conjugates
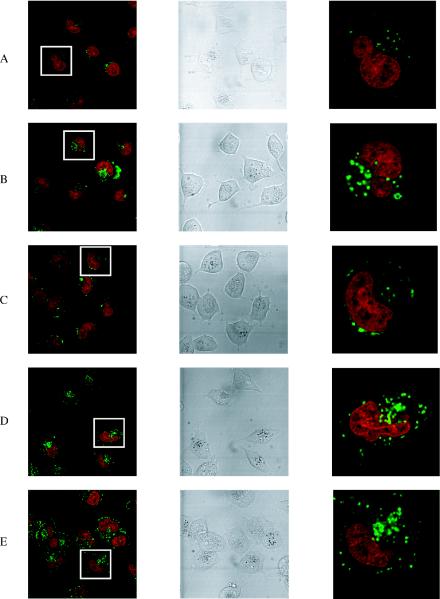
Since the CPP–conjugates failed to show significant cellular activity by free delivery, we asked whether 3′-carboxyfluorescein labelled CPP–oligonucleotide conjugates could penetrate HeLa cells and where they might be localized. We therefore incubated them at 2.5 μM concentration with HeLa cells for 5 h, and observed the uptake by confocal microscopy (Figure 4). The 3′-fluorescent label allows any oligonucleotide component that has been taken up by cells to be seen as a green fluorescence. In addition, a hydroethidine dye was used to stain the cell nucleus. Not only does this make it easier to observe nuclear uptake (as a green or yellow colour), but also only the nuclei of live cells are stained red, ensuring that only healthy cells are included in observations.
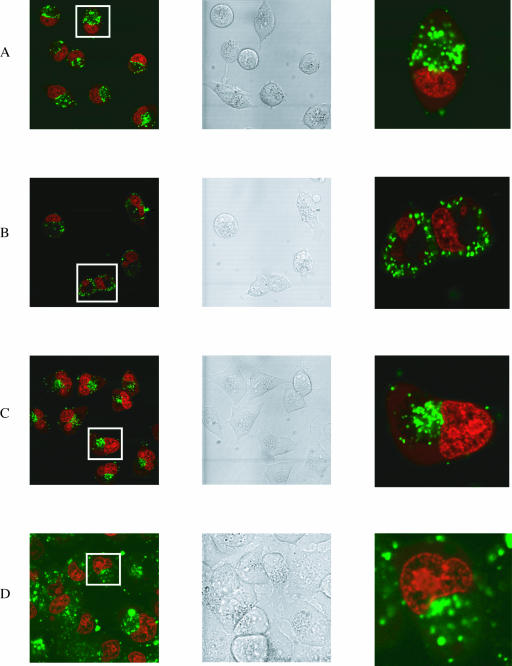
Figure 4.
Confocal microscope images of the free uptake of fluorescein-labelled CPP conjugates (green) into HeLa cells. Left panels show a range of cells (nucleus stained red). Central panels are the same cells in DIC. Right panels show a magnification of a single cell boxed in the left panels. Horizontally, (A) conjugate 4 (OMe/LNA oligo conjugated with the C-terminus of Tat peptide), (B) conjugate 6 (OMe/LNA oligo B conjugated with the C-terminus of Penetratin), (C) conjugate 5 (OMe/LNA oligo B conjugated with the N-terminus of Penetratin), (D) conjugate 12 (OMe/LNA oligo B conjugated with R9F2) and (E) conjugate 10 (PS/OMe oligo C conjugated with the C-terminus of Penetratin).
Free uptake of CPP–oligonucleotide conjugates. In the absence of cationic lipid, unconjugated oligos A, B and C do not enter HeLa cells [(14) and data not shown]. Conjugates 1 and 2 (OMe oligo conjugates) and conjugate 3 (OMe/LNA oligo conjugated with the N-terminus of Tat peptide) showed only a very weak fluorescence (data not shown). In contrast, conjugate 4 (OMe/LNA oligo-conjugated with the C-terminus of Tat peptide) showed a little more noticeable internalization for most cells (Figure 4A). Fluorescence was not seen in the nucleus, but in vesicular compartments within the cytosol. Very similar uptake patterns were observed for conjugate 6 (OMe/LNA oligo conjugated with the C-terminus of Penetratin peptide) (Figure 4B) but only very weakly for conjugate 5 (OMe/LNA oligo conjugated with the N-terminus of Penetratin peptide) (Figure 4C). Neither conjugates 7 or 8 (containing R9 in different orientations) showed significant cell uptake (data not shown). However, the conjugate of OMe/LNA oligo B with R9F2 (conjugate 12) showed substantial cellular fluorescence, once again confined to vesicular compartments (Figure 4D). Thus, unlike R9, the CPPs Tat, Penetratin and R9F2 do enhance free HeLa cell uptake of oligo B somewhat when conjugated at the C-terminus, but no evidence of release was seen from endosomal compartments. No significant improvement in uptake for any of these conjugates was seen after 24 h incubation nor was any nuclear fluorescence seen (data not shown).
We reported previously that 3′ fluorescently labelled OMe/LNA oligo B conjugated stably with Transportan-TP10 showed only weak cytosolic uptake in HeLa cells (14). Similarly, the uptake of oligo B disulphide linked to full-length Transportan (conjugate 13) was extremely weak (data not shown).
The C-terminal Penetratin conjugate of PS/OMe oligo C (conjugate 10) showed a very similar level of uptake into vesicular compartments (Figure 4E) as for the same conjugate of oligo B (Figure 4B). Once again no nuclear uptake was seen, in contrast to the recent report of Astriab-Fisher et al. (24) for a very similar disulphide Penetratin-PS/OMe oligonucleotide. Conjugates of Tat (conjugate 9) and R9 (conjugate 11) showed only very weak uptake into the cytosol (data not shown).
Synthesis and activity of conjugates of OMe/LNA oligo B with CPPs containing additional cationic charge
Since the formal charge balance of conjugates 1–12 remains negative overall, we wondered whether addition of further cationic charge to the CPP Tat or Penetratin could improve the level of uptake into HeLa cells and/or aid release from endosomes. Hence, we synthesized a number of Tat and Penetratin peptides having additional Arg or Lys residue extensions as well as containing N- or C-terminal Cys residues and conjugated these to OMe/LNA Oligo B (Table 1). Conjugates 14 and 15 contained Tat with 4 additional Arg residues in different orientations, 16 and 17 contained double Tat peptides in different orientations, 18 contained Tat and 4 additional Lys residues on the N-terminus and a C-terminal Cys, and 19 contained Penetratin with 6 additional Arg residues on the N-terminus and a C-terminal Cys. These conjugation reactions were in general more difficult to carry out due to precipitation which almost always occurred, requiring the addition of extra TEAA buffer and formamide to maintain solubility. However, purifications of the conjugates by HPLC were readily achieved in all cases on the Resource Q column (Table 2 and Figure 2B). Lower isolation yields were obtained (12–55%) due to tendencies of the conjugates to precipitate during dialysis or to be partially insoluble after lyophilization. In all cases, a high salt buffer was necessary for dissolution (1 M TEAA ± 1 M ammonium chloride solution). Conjugate 19 could not be fully dissolved in any aqueous buffer. Products were characterized by MALDI-TOF mass spectrometry and in all cases showed within experimental error the expected mass values (Table 2).
None of these conjugates showed significant inhibition of trans-activation (firefly luciferase expression) in the HeLa cell assay under free delivery for 5 h (data not shown). When viewed by confocal microscopy, OMe/LNA oligo B conjugates 15 (R4-Tat), 17 (Tat-Tat) and 18 (K4-Tat) all showed substantial cytosolic uptake into vesicular structures (Figure 5A–C). The uptake levels appeared to be generally stronger for these more cationic conjugates than for those with a CPP alone, but still less than that for cationic lipid delivery (cf. Figure 7C). Further, there was no evidence of nuclear uptake in any case. After 24 h incubations, some further cytosolic uptake was seen for the K4-Tat conjugate 18 (Figure 5D) and for the R4-Tat conjugate 15 but no nuclear uptake nor trans-activation inhibition activity was seen in the HeLa cell assay (data not shown).
Figure 5.
Confocal microscope images of the free uptake of fluorescein-labelled CPP conjugates containing additional cationic charge (green) into HeLa cells (nuclei stained red). Vertical Panels as in Figure 4 legend. Horizontally, (A) Conjugate 15 (OMe/LNA oligo B conjugated with R4-Tat), (B) Conjugate 17 (OMe/LNA oligo B conjugated with Tat-Tat), (C) Conjugate 18 (OMe/LNA oligo B conjugated with K4-Tat) and (D) Conjugate 18 (OMe/LNA oligo B conjugated with K4-Tat), 24 h experiment.
Figure 7.
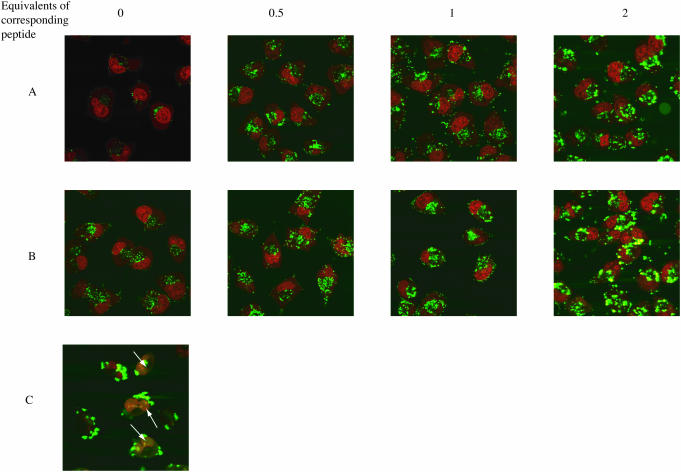
Confocal microscope images of the free uptake of fluorescein-labelled CPP conjugates (green) into HeLa cells (nuclei stained red). (A) Effect of additional equivalents of Tat peptide on uptake of 2.5 μM Tat-Cys-Oligo B, 5 h. (B) Effect of additional equivalents of Penetratin peptide on uptake of 2.5 μM Penetratin-Cys-Oligo B, 5 h. (C) Lipofectamine 2000 delivery of 0.5 μM oligo B, 3 h. Note here the nuclear localization seen (white arrows) in addition to cytosolic uptake.
Uptake of conjugates in human fibroblasts
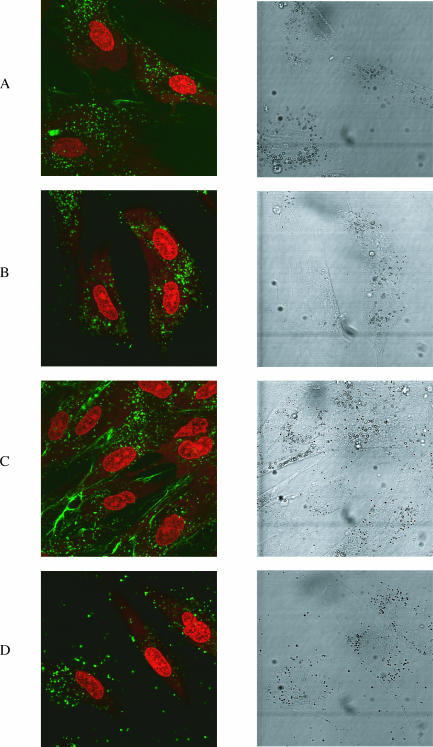
We asked whether the restriction of uptake of CPP conjugates of oligonucleotides into cytosolic vesicles is a feature only of HeLa cells or whether this also extends to other cell lines. We therefore incubated cultured human foreskin fibroblasts (44) with certain fluorescein-labelled conjugates that showed good uptake into HeLa cells. For example, the Penetratin–C-terminal conjugate 6 showed a very similar cytosolic uptake to that seen in HeLa cells except that there was a more defined punctate nature of the fluorescence and a more even distribution through the cytosol (Figure 6A). No nuclear uptake was observed. Interestingly, the difference in uptake levels for the C-terminally linked conjugates of Tat and Penetratin conjugates 4 and 6, and N-terminally linked conjugates 3 and 5 were less pronounced than for HeLa cells (data not shown). A strong cytosolic pattern of uptake was also observed for the R9F2 conjugate 12 (Figure 6B) and for the R4-Tat conjugate 15 (Figure 6D), but uptake of R9 conjugates 7 and 8 was very weak (data not shown). The uptake level of Transportan conjugate 13 into fibroblasts was much better than that into HeLa cells (Figure 6C). In addition to the punctate cytosolic distribution, there was more membrane association seen, which may reflect the greater hydrophobicity of the Transportan peptide. Nevertheless, there was still no nuclear uptake observed in any case. No further improvements in uptake were seen for 24 h incubation (data not shown). Thus in general, uptake of conjugates into human fibroblast cells was not dissimilar to that of HeLa cells.
Figure 6.
Confocal microscope images of the free uptake of fluorescein-labelled CPP conjugates (green) into human fibroblasts (nucleus stained red). Left panels show a range of cells (nucleus stained red). Right panels are the same cells in DIC. Horizontally, (A) conjugate 6 (OMe/LNA oligo B conjugated with the C-terminus of Penetratin), (B) conjugate 12 (OMe/LNA oligo B conjugated with R9F2), (C) conjugate 13 (OMe/LNA oligo B conjugated with Transportan) and (D) conjugate 15 (OMe/LNA oligo B conjugated with R4-Tat).
Uptake of CPP–oligo conjugates into HeLa cells in the presence of additional free CPP
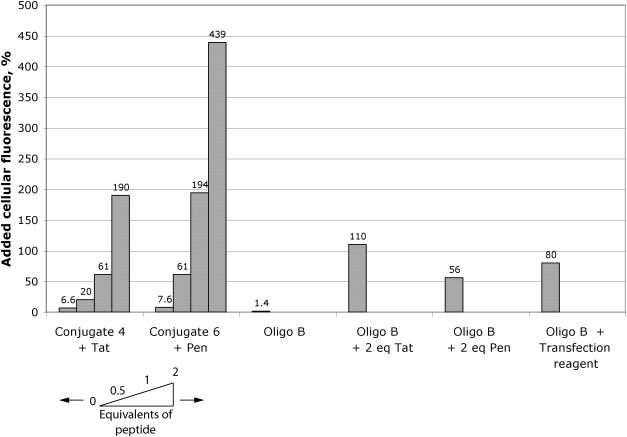
The above activity and cellular uptake results contrast with some previous reports of antisense or steric block activity and nuclear delivery of Penetratin and Tat conjugates of oligonucleotides (21–24). Noting the tendency of CPP conjugates to aggregate or precipitate in conjugation reactions, we took considerable care in isolation and purification of conjugates to remove excess CPP peptide. Therefore, we questioned whether uptake of conjugates into HeLa cells could be influenced by addition of further equivalents of unconjugated CPP. Addition of half, one or two molar equivalents of Tat peptide to 2.5 μM conjugate 4 (Tat C-terminally conjugated to OMe/LNA oligo B) resulted in each case in substantially increased uptake as observed by confocal microscopy (Figure 7 panel A). Extraction and measurement of the fluorescence showed that the fluorescence was enhanced ∼190% in the case of 2 equivalents of Tat (Figure 8). The effect was even more dramatic with Penetratin addition to conjugate 6 (Penetratin C-terminally conjugated to OMe/LNA oligo B) with a 440% enhancement of uptake (Figure 7 panel B and Figure 8). The amount of fluorescence internalized in the latter case was substantially more as compared to that delivered by 0.5 μM oligo B using Effectin 12 cationic lipid for 3 h which was enhanced 80% (Figure 8). When 2 equivalents of Tat or Penetratin were added to unconjugated oligo B alone, the improvement in uptake, 110% and 56%, respectively, was less than that for the conjugates (Figure 8). However, in no case did we observe significant nuclear fluorescence using the HeLa cell line. The results show that free CPPs Tat and Penetratin act to help substantially the conjugates to enhance entry into the cell, presumably through complexation, but that they do not help release from the endosomal compartments.
Figure 8.
Quantitation of internalized fluorescence. Percentage increase in cellular fluorescence over background levels for (left to right) addition of Tat peptide to 2.5 μM conjugate 4, addition of Penetratin peptide to 2.5 μM conjugate 6, compared to 2.5 μM Oligo B alone, 2.5 μM Oligo B with 2 equivalents of Tat peptide or 2 equivalents of Penetratin peptide and 0.5 μM oligo B with transfection agent Lipofectamine 2000.
DISCUSSION
Synthesis and cell uptake of peptide–oligonucleotide conjugates
Hitherto, the synthesis of conjugates of oligonucleotides with highly cationic cell-penetrating peptides has been extremely hard. A key area of concern has been the homogeneity of the conjugate product. This is particularly the case for cationic CPPs when conjugation is carried out by fragment coupling and where typically excess peptide is added to the oligonucleotide. We and others (38,39) have observed how aggregation and precipitation can occur during conjugation reactions and purifications under non-denaturing conditions, presumably as a result of tight binding of excess cationic peptide to the conjugate and starting oligonucleotide. We have now developed rapid and reliable methods of disulphide conjugation of oligonucleotides to cationic and other CPP peptides, and short path anion exchange chromatography purification under highly denaturing conditions to ensure that significant excess cationic peptide does not remain bound to the conjugate. The principles should also be applicable to other chemistries of conjugation that tolerate denaturing agents. These methods allow for the first time a more systematic investigation of cell uptake of CPP–oligonucleotide conjugates.
We have shown here that in many cases, CPP conjugation does indeed enhance free uptake of OMe/LNA oligonucleotides into HeLa cells and human fibroblasts in culture, but that the level of uptake does not approach the level that can be achieved with cationic lipid delivery. The addition of extra cationic charge to Tat peptide, or a double Tat peptide, enhances uptake further beyond that of Tat alone, but this is at the cost of increased difficulties in purification, isolation and solubility of the conjugate. In contrast, a dramatic effect in enhancement of uptake into HeLa cells was observed when extra equivalents of cationic CPPs Tat or Penetratin were added to purified oligonucleotide–CPP conjugates (Figures 7 and 8). This points to a significant delivery role that Tat and Penetratin have on the conjugates, presumably by complexing the conjugate to shield better the negative charges. The delivery was greater for conjugates than that seen for complexation of free oligonucleotide. These results highlight concerns about cell uptake results from a number of previous cationic peptide conjugate studies of oligonucleotides or siRNA where a strong denaturing agent has not been used during synthesis and purification (21,22,45–48).
The mechanism of CPP-directed cell delivery
The mechanism of CPP-directed cell uptake has been controversial for some time. Initially, it was thought that such peptides translocate across cell membranes in an energy-independent way that avoids the endosomal uptake pathways (49). More recently, cellular uptake of CPPs has been re-evaluated following the realization that the use of cell fixation agents, such as methanol or formaldehyde, can give rise to artefacts during confocal microscopy and that only data from unfixed cells may be reliable. Thus, fluorescein-labelled Tat peptide was found to enter HeLa cells by an endosomal pathway and was found in cytosolic vesicles (50). Similarly studies of TAMRA-labelled R9 into live CHO-K1 cells have confirmed that this peptide also enters via endocytosis, and is seen only in cytosolic vesicles and not the nucleus (51). Our own data for Tat (48–58), Penetratin and R9F2 peptides fluorescein-labelled on a C-terminal cysteine residue have shown in all cases delivery entirely into cytosolic compartments in HeLa cells (Arzumanov, A., Turner, J. and Gait, M.J., unpublished data). By contrast, a recent report showed that TAMRA-labelled Tat peptide was taken up by HeLa cells and located predominantly in nuclear bodies thought to be the nucleolus (48) and nuclear localization was also reported recently for live HeLa cell delivery of C-terminally FITC-labelled Tat peptide (52). Recent re-evaluation of fluorescently labelled Penetratin uptake showed endosomal uptake, but some polyarginine analogues (such as R7W) have shown evidence of energy-independent uptake (53). Thus for CPPs alone, there remains conflicting data about uptake and localization.
Antopolsky et al. reported that disulphide conjugates of hydrophobic peptides and phosphorothioate oligonucleotides were taken up and localized in endosomal compartments, and they also reported that the disulphide bond appeared to be intact after isolation of such conjugates from cells after several hours (25). Ours is the first detailed study aimed at correlation of nuclear steric block activity with cell uptake for a range of conjugates of cationic and other well-known CPPs to OMe/LNA phosphodiester and phosphorothioate oligonucleotides. In no case of free uptake of conjugates into HeLa cells did we see nuclear localization. Instead all the fluorescence was found within cytosolic compartments. This explains why with free conjugate delivery, we did not observe significant reduction in firefly luciferase expression resultant from inhibition of Tat-dependent trans-activation. In contrast, our main test OMe/LNA 12mer oligonucleotide B is active when delivered by cationic lipids, with or without a 5′-thiol linker. A free 5′-thiol would be obtained if the disulphide bond is cleaved during cell uptake, but this seems unlikely given the results of Antopolsky et al. (25). CPP–oligo B conjugates were also found to be active when delivered by cationic lipids, but with variable efficiency, presumably due to lipid packaging variations.
Our results showing only cytosolic localization in both HeLa cells and human fibroblasts for our complete range of conjugates is consistent with recently reported studies of a C-terminal linked Tat conjugate of a molecular beacon phosphodiester oligonucleotide where the linkage (thiol–maleimide or disulphide) is through a base residue. Uptake into human dermal fibroblasts was apparent and a strong fluorescent signal, presumably, due to opening of the beacon upon hybridization with GAPDH mRNA, was seen in cytosolic compartments within 30–60 min (46). No fluorescent signal was seen in the cell nucleus. Interestingly, a sequence-specific molecular beacon signal was only seen with peptide delivery and not with cationic lipid delivery, suggesting that localization may not be to the same endosomal compartments in the two cases.
It is believed that there are several endosomal portals in mammalian cells, for example clathrin-dependent and cavaeloae-dependent endocytosis, as well as macropinocytosis [reviewed in (54)]. Although the particular portal or portals used by CPPs or CPP–protein conjugates are now being studied [e.g. (51,55,56)], this has yet to be established for oligonucleotide–peptide conjugates. Further, there is only limited knowledge of factors affecting release from endosomal compartments (or leakage) into the cytosol. Once in the cytosol, short oligonucleotides are known to track readily into the nucleus by passive diffusion across the nuclear membrane (57). It is clear that endosomal release of peptide conjugates of oligonucleotides is inefficient in both HeLa cells and human fibroblasts. Therefore, at least with the current list of well-known CPPs, peptide conjugation is unlikely to be a panacea for the nuclear delivery of anionic oligonucleotides unless we can find better sequences that can cause sufficient endosomal membrane disruption. However, some cell lines of different lineage may have less demanding endosomal release characteristics, e.g. Jurkat T cells (58).
We are unable to reconcile our results fully with the study of Astriab-Fisher et al. who prepared disulphide conjugates of a 3′-TAMRA-2′-O-methylphosphorothioate oligonucleotides to Tat or Penetratin (24). In this study, denaturing agents were used both in the conjugations and in purifications. These conjugates were reported to localize both in endosomal compartments and strongly in the nucleus of HeLa 705 cells. There was no indication of the use of cell fixation during confocal microscopy. Up-regulation of luciferase expression was seen due to altered splicing, but the authors conceded that the assay used was extremely sensitive and that ‘maximal effects attained with the conjugates were several-fold less’ than for cationic lipid delivery of similar amounts of the same oligonucleotide. In contrast, in our trans-activation assay, the Tat-dependent HIV-1 promoter driving production of firefly luciferase is very strong, and inhibition of expression will only be seen if there is significant nuclear delivery into most cells. Thus, weak nuclear delivery would be insufficient to see activity in our robust assay.
Role of charge and hydrophobicity in cell uptake
It is interesting to note that the cationic charge of the peptide is not the only factor mediating cell uptake, since neither of the R9-conjugates of oligo B showed significant uptake into HeLa cells whereas R9F2, Tat and Penetratin did. This result is in contrast to a recent report for good cytosolic internalization of a disulphide-linked, FITC-labelled R9-conjugate of an unmodified phosphodiester oligonucleotide into HepG-2 cells (45). Our results point instead to the importance beyond cationic charge for additional residues within the peptide that act either to space the cationic charge more amphipathically or perhaps to add some hydrophobic character to enhance membrane crossing. Combinations of hydrophobic and cationic residues are well known for peptides used as complexing agents for the delivery of nucleic acids (59,60). Note also in this context that a stearylated Tat peptide has been shown to significantly enhance transfection of DNA into COS-7 cells (61), and that myristoylation of polyarginine peptides resulted in enhanced uptake of the peptide into HeLa cells (52). For our conjugates, the interplay of cationic charge and hydrophobic components of the peptide together with the anionic charge and hydrophobic characteristics (e.g. LNA) of the oligonucleotide cargo in cell uptake is clearly complex and will require further investigation. Although 3′-fluorescein oligo B is not taken up by HeLa cells (data not shown), we cannot rule out the possibility that the fluorescein label itself influences the free cell uptake of the CPP conjugates.
This study has focussed entirely on anionic oligonucleotides and their cell delivery and activity. Peptide conjugates of electrically neutral peptide nucleic acids and morpholino oligonucleotides have been studied recently for cellular uptake and biological activity (62–65). A disulphide-linked Transportan conjugate of a 16mer PNA targeted to the TAR apical stem–loop was found to inhibit Tat-dependent activity in the 1–5 μM range in a transient luciferase reporter assay in CEM and Jurkat cells, as well as inhibition of HIV-1 production in chronically infected H9 cells in the same concentration range (20). Our own results for PNA–peptide conjugates against the TAR target will be reported shortly, but our preliminary findings [as well as that of others (64)] indicate that difficulties in endosomal release in HeLa cells extends also to these conjugates (Arzumanov, A., Turner, J., Verbeure, B., Williams, D. and Gait, M.J., unpublished data).
In conclusion, we have made significant improvements to the synthesis of peptide–oligonucleotide conjugates that are disulphide-linked to ensure high purity and shown that CPP conjugation enhances uptake into HeLa and human fibroblast cells, but that further complexation with additional equivalents of peptide results in substantial uptake enhancement. However, such conjugates are not released from endosomal compartments in HeLa cells and do not allow the oligonucleotide to inhibit Tat-dependent trans-activation that must occur in the cell nucleus. These results provide important information towards a better understanding of oligonucleotide cell delivery potential.
SUPPLEMENTARY MATERIAL
Supplementary Material is available at NAR Online.
Supplementary Material
Acknowledgments
We thank Leann Tait and Duncan Graham (Department of Chemistry, University of Strathclyde) for information regarding the use of Resource Q and Meredith Ross (MRC Dunn Nutrition Unit, Cambridge) for the human foreskin fibroblast cells. We are very grateful for measurements of binding constants carried out by Gabriele Ivanova, for synthesis of peptides by David Owen, for synthesis of fluorescein-labelled oligonucleotides by Donna Williams and Matthew Watson and helpful advice on the manuscript from Marsha Rosner. This work is funded in part by a grant from EC Framework 5 (contract QLK3-CT-2002-01989). Funding to pay the Open Access publication charges for this article was provided by the Medical Research Council.
REFERENCES
- 1.Karn J. Tackling Tat. J. Mol. Biol. 1999;293:235–254. doi: 10.1006/jmbi.1999.3060. [DOI] [PubMed] [Google Scholar]
- 2.Rana T.M., Jeang K.-T. Biochemical and functional interactions between HIV-1 Tat protein and TAR RNA. Arch. Biochem. Biophys. 1999;365:175–185. doi: 10.1006/abbi.1999.1206. [DOI] [PubMed] [Google Scholar]
- 3.Krebs A., Ludwig V., Boden O., Göbel M.W. Targeting the HIV trans-activation responsive region-approaches towards RNA-binding drugs. Chembiochem. 2003;4:972–978. doi: 10.1002/cbic.200300652. [DOI] [PubMed] [Google Scholar]
- 4.Darfeuille F., Arzumanov A., Gait M.J., Di Primo C., Toulmé J.J. 2′-O-methyl RNA hairpins generate loop–loop complexes and selectively inhibit HIV-1 Tat-mediated transcription. Biochemistry. 2002;41:12186–12192. doi: 10.1021/bi025974d. [DOI] [PubMed] [Google Scholar]
- 5.Darfeuille F., Arzumanov A., Gryaznov S., Gait M.J., Di Primo C., Toulmé J.J. Loop–loop interaction of HIV-1 TAR RNA with N3′-5′ deoxyphosphoramidate aptamers inhibits in vitro Tat-mediated transcription. Proc. Natl Acad. Sci. USA. 2002;99:9709–9714. doi: 10.1073/pnas.122247199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Darfeuille F., Hansen J.B., Orum H., Di Primo C., Toulmé J.J. LNA/DNA chimeric oligomers mimic RNA aptamers targeted to the TAR RNA element of HIV-1. Nucleic Acids Res. 2004;32:3101–3107. doi: 10.1093/nar/gkh636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mayhood T., Kaushik N., Pandey P.K., Kashanchi F., Deng L., Pandey V.N. Inhibition of Tat-mediated transactivation of HIV-1 LTR transcription by polyamide nucleic acid targeted to the TAR hairpin element. Biochemistry. 2000;39:11532–11539. doi: 10.1021/bi000708q. [DOI] [PubMed] [Google Scholar]
- 8.Arzumanov A., Walsh A.P., Rajwanshi V.K., Kumar R., Wengel J., Gait M.J. Inhibition of HIV-1 Tat-dependent trans-activation by steric block chimeric 2′-O-methyl/LNA oligoribonucleotides. Biochemistry. 2001;40:14645–14654. doi: 10.1021/bi011279e. [DOI] [PubMed] [Google Scholar]
- 9.Vickers T.A., Baker B.F., Cook P.D., Zounes M., Buckheit R.W., Germany J., Ecker D.J. Inhibition of HIV-LTR gene expression by oligonucleotides targeted to the TAR element. Nucleic Acids Res. 1991;19:3359–3368. doi: 10.1093/nar/19.12.3359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ecker D.J., Vickers T.A., Bruice T.W., Freier S.M., Jenison R.D., Manoharan M., Zounes M. Pseudo-half knot formation with RNA. Science. 1992;257:958–961. doi: 10.1126/science.1502560. [DOI] [PubMed] [Google Scholar]
- 11.Mestre B., Arzumanov A., Singh M., Boulmé F., Litvak S., Gait M.J. Oligonucleotide inhibition of the interaction of HIV-1 Tat protein with the trans-activation responsive region (TAR) of HIV RNA. Biochim. Biophys. Acta. 1999;1445:86–98. doi: 10.1016/s0167-4781(99)00019-6. [DOI] [PubMed] [Google Scholar]
- 12.Arzumanov A., Gait M.J. Inhibition of the HIV-1 tat protein–TAR RNA interaction by 2′-O-methyl-oligoribonucleotides. In: Holy A., Hocek M., editors. Collection Symposium Series. Vol. 2. pp. 168–174. Academy of Sciences of the Czech Republic. [Google Scholar]
- 13.Arzumanov A., Walsh A.P., Liu X., Rajwanshi V.K., Wengel J., Gait M.J. Oligonucleotide analogue interference with the HIV-1 Tat protein-TAR RNA interaction. Nucleosides, Nucleotides and Nucleic Acids. 2001;20:471–480. doi: 10.1081/NCN-100002321. [DOI] [PubMed] [Google Scholar]
- 14.Arzumanov A., Stetsenko D.A., Malakhov A.D., Reichelt S., Sørensen M.D., Babu B.R., Wengel J., Gait M.J. A structure–activity study of the inhibition of HIV-1 Tat-dependent trans-activation by mixmer 2′-O-methyl oligoribonucleotides containing locked nucleic acid (LNA), α-LNA or 2′-thio-LNA residues. Oligonucleotides. 2003;13:435–453. doi: 10.1089/154545703322860762. [DOI] [PubMed] [Google Scholar]
- 15.Bennett C.F., Chiang M.-Y., Chan H., Shoemaker J.E.E., Mirabelli C.K. Cationic lipids enhance cellular uptake and activity of phosphorothioate antisense oligonucleotides. Mol. Pharmacol. 1992;41:1023–1033. [PubMed] [Google Scholar]
- 16.Lindsay M.A. Peptide-mediated cell delivery: application in protein target validation. Curr. Opin. Pharmacol. 2002;2:587–594. doi: 10.1016/s1471-4892(02)00199-6. [DOI] [PubMed] [Google Scholar]
- 17.Wadia J.S., Dowdy S.F. Protein transduction technology. Curr. Opin. Biotechnol. 2002;13:52–56. doi: 10.1016/s0958-1669(02)00284-7. [DOI] [PubMed] [Google Scholar]
- 18.Gait M.J. Peptide-mediated cellular delivery of antisense oligonucleotides and their analogues. Cell. Mol. Life Sci. 2003;60:1–10. doi: 10.1007/s00018-003-3044-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Thierry A.R., Vivès E., Richard J.-P., Prevot P., Martinand-Mari C., Robbins I., Lebleu B. Cellular uptake and intracellular fate of antisense oligonucleotides. Curr. Opin. Mol. Ther. 2003;5:133–138. [PubMed] [Google Scholar]
- 20.Kaushik N., Basu A., Palumbo P., Nyers R.L., Pandey V.N. Anti-TAR polyamide nucleotide analog conjugated with a membrane-permeating peptide inhibits Human Immunodeficiency Virus Type I production. J. Virol. 2002;76:3881–3891. doi: 10.1128/JVI.76.8.3881-3891.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Allinquant B., Hantraye P., Mailleux P., Moya K., Bouillot C., Prochiantz A. Downregulation of amyloid precursor protein inhibits neurite outgrowth in vitro. J. Cell Biol. 1995;128:919–927. doi: 10.1083/jcb.128.5.919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Troy C.M., Derossi D., Prochiantz A., Greene L.A., Shelanski M.L. Downregulation of Cu/Zn superoxide dismutase lead to cell death via the nitric oxide-peroxynitrite pathway. J. Neurosci. 1996;16:253–261. doi: 10.1523/JNEUROSCI.16-01-00253.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Astriab-Fisher A., Sergueev D.S., Fisher M., Ramsay Shaw B., Juliano R.L. Antisense inhibition of P-glycoprotein expression using peptide-oligonucleotide conjugates. Biochem. Pharmacol. 2000;60:83–90. doi: 10.1016/s0006-2952(00)00310-5. [DOI] [PubMed] [Google Scholar]
- 24.Astriab-Fisher A., Sergueev D., Fisher M., Ramsay Shaw B., Juliano R.L. Conjugates of antisense oligonucleotides with the Tat and Antennapedia cell-penetrating peptides: effect on cellular uptake, binding to target sequences, and biologic actions. Pharm. Res. 2002;19:744–754. doi: 10.1023/a:1016136328329. [DOI] [PubMed] [Google Scholar]
- 25.Antopolsky M., Azhayeva E., Tengvall U., Auriola S., Jääskeläinen I., Rönkkö S., Honkakoski P., Urtti A., Lönnberg H., Azhayev A. Peptide-oligonucleotide phosphorothioate conjugates with membrane translocation and nuclear localization properties. Bioconjug. Chem. 1999;10:598–606. doi: 10.1021/bc980133y. [DOI] [PubMed] [Google Scholar]
- 26.Pooga M., Soomets U., Hällbrink M., Valkna A., Saar K., Rezaei K., Kahl U., Hao J.-X., Xu X.-J., Wiesenfeld-Hallin Z., et al. Cell penetrating PNA constructs regulate galanin receptor levels and modify pain transmission in vivo. Nat. Biotechnol. 1998;16:857–861. doi: 10.1038/nbt0998-857. [DOI] [PubMed] [Google Scholar]
- 27.Zubin E.M., Romanova E.A., Oretskaya T.S. Modern methods for the synthesis of peptide-oligonucleotide conjugates. Russ. Chem. Rev. 2002;71:239–264. [Google Scholar]
- 28.Peyrottes S., Mestre B., Burlina F., Gait M.J. The synthesis of peptide-oligonucleotide conjugates by a fragment coupling approach. Tetrahedron. 1998;54:12513–12522. [Google Scholar]
- 29.Kachalova A.V., Stetsenko D.A., Romanova E.A., Tashlitsky V.N., Gait M.J., Oretskaya T.S. A new and efficient method for synthesis of 5′-conjugates of oligonucleotides through amide-bond formation on solid phase. Helv. Chim. Acta. 2002;85:2409–2416. [Google Scholar]
- 30.Stetsenko D.A., Gait M.J. Efficient conjugation of peptides to oligonucleotides by ‘native ligation’. J. Org. Chem. 2000;65:4900–4908. doi: 10.1021/jo000214z. [DOI] [PubMed] [Google Scholar]
- 31.Zatsepin T.S., Stetsenko D.A., Arzumanov A., Romanova E.A., Gait M.J., Oretskaya T.S. Synthesis of peptide-oligonucleotide conjugates with single and multiple peptides attached to 2′-aldehydes through thiazolidine, oxime and hydrazine linkages. Bioconjug. Chem. 2002;13:822–830. doi: 10.1021/bc020016+. [DOI] [PubMed] [Google Scholar]
- 32.Stetsenko D.A., Malakhov A.D., Gait M.J. Total stepwise solid-phase synthesis of oligonucleotide (3′-N)-peptide conjugates. Org. Lett. 2002;4:3259–3262. doi: 10.1021/ol026502u. [DOI] [PubMed] [Google Scholar]
- 33.Jensen O.N., Kulkarni S., Aldrich J.V., Barofsky D.F. Characterization of peptide-oligonucleotide heteroconjugates by mass spectrometry. Nucleic Acids Res. 1996;24:3866–3872. doi: 10.1093/nar/24.19.3866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Stetsenko D.A., Gait M.J. A convenient solid-phase method for synthesis of 3′-conjugates of oligonucleotides. Bioconjug. Chem. 2001;12:576–586. doi: 10.1021/bc000157g. [DOI] [PubMed] [Google Scholar]
- 35.Eritja R., Pons A., Escarceller M., Giralt E., Albericio F. Synthesis of defined peptide-oligonucleotide hybrids containing a nuclear transport signal sequence. Tetrahedron. 1991;47:4113–4120. [Google Scholar]
- 36.Bongartz J.P., Aubertin A.M., Milhaud P.G., Lebleu B. Improved biological activity of antisense oligonucleotides conjugated to a fusogenic peptide. Nucleic Acids Res. 1994;22:4681–4688. doi: 10.1093/nar/22.22.4681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Corey C.R. 48000-fold acceleration of hybridisation by chemically modified oligonucleotides. J. Am. Chem. Soc. 1995;117:9373–9374. [Google Scholar]
- 38.Vivès E., Lebleu B. Selective coupling of a highly basic peptide to an oligonucleotide. Tetrahedron Lett. 1997;38:1183–1186. [Google Scholar]
- 39.Prater C.E., Miller P. 3′-Methylphosphonate-modified oligo-2′-O-methylribonucleotides and their Tat peptide conjugates: uptake and stability in mouse fibroblasts in culture. Bioconjug. Chem. 2004;15:498–507. doi: 10.1021/bc049977+. [DOI] [PubMed] [Google Scholar]
- 40.Vivès E., Brodin P., Lebleu B. A truncated HIV-1 Tat protein basic domain rapidly translocates through the plasma membrane and accumulates in the cell nucleus. J. Biol. Chem. 1997;272:16010–16017. doi: 10.1074/jbc.272.25.16010. [DOI] [PubMed] [Google Scholar]
- 41.Mitchell D.J., Kim D.T., Steinman L., Fathman C.G., Rothbard J.B. Polyarginine enters cells more efficiently than other polycationic homopolymers. J. Peptide Res. 2000;56:318–325. doi: 10.1034/j.1399-3011.2000.00723.x. [DOI] [PubMed] [Google Scholar]
- 42.Neuman B.W., Stein D.A., Kroeker A.D., Paulino A.D., Moulton H.M., Iversen P.L., Buchmeier M.J. Antisense morpholino-oligomers directed against the 5′-end of the genome inhibit coronovirus proliferation and growth. J. Virol. 2004;78:5891–5899. doi: 10.1128/JVI.78.11.5891-5899.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Churcher M., Lamont C., Dingwall C., Green S.M., Lowe A.D., Butler P.J.G., Gait M.J., Karn J. High affinity binding of TAR RNA by the human immunodeficiency virus tat protein requires amino acid residues flanking the basic domain and base pairs in the RNA stem. J. Mol. Biol. 1993;230:90–110. doi: 10.1006/jmbi.1993.1128. [DOI] [PubMed] [Google Scholar]
- 44.Ross M.F., Filipovska A., Smith R.A.J., Gait M.J., Murphy M.P. Cell-penetrating peptides do not cross mitochondrial membranes even when conjugated to a lipophilic cation: evidence against direct passage through phospholipid bilayers. Biochem. J. 2004;383:457–468. doi: 10.1042/BJ20041095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chen C.-P., Zhang L.-R., Peng Y.-F., Wang X.-B., Wang S.-Q., Zhang L.-H. A concise method for the preparation of peptide and arginine-rich peptide-conjugated antisense oligonucleotides. Bioconjug. Chem. 2003;14:532–538. doi: 10.1021/bc034004f. [DOI] [PubMed] [Google Scholar]
- 46.Nitin N., Santangelo P.J., Kim G., Nie S., Bao G. Peptide-linked molecular beacons for efficient delivery and rapid mRNA detection in living cells. Nucleic Acids Res. 2004;32:e58. doi: 10.1093/nar/gnh063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Muratovska A., Eccles M.R. Conjugate for efficient delivery of short interfering RNA (siRNA) into mamalian cells. FEBS Lett. 2004;558:63–68. doi: 10.1016/S0014-5793(03)01505-9. [DOI] [PubMed] [Google Scholar]
- 48.Chiu Y.-L., Ali A., Chu C., Cao H., Rana T.M. Visualizing a correlation between siRNA, localization, cellular uptake and RNAi in living cells. Chem. Biol. 2004;11:1165–1175. doi: 10.1016/j.chembiol.2004.06.006. [DOI] [PubMed] [Google Scholar]
- 49.Prochiantz A. Getting hydrophilic compounds into cells: lessons from homeopeptides. Curr. Opin. Neurobiol. 1996;6:629–634. doi: 10.1016/s0959-4388(96)80095-x. [DOI] [PubMed] [Google Scholar]
- 50.Richard J.-P., Melikov K., Vivès E., Ramos C., Verbeure B., Gait M.J., Chernomordik L.V., Lebleu B. Cell-penetrating peptides. A re-evaluation of the mechanism of cellular uptake. J. Biol. Chem. 2003;278:585–590. doi: 10.1074/jbc.M209548200. [DOI] [PubMed] [Google Scholar]
- 51.Fuchs S.M., Raines R.T. Pathway for polyarginine entry into mamallian cells. Biochemistry. 2004;43:2438–2444. doi: 10.1021/bi035933x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pham W., Kircher M.F., Weissleder R., Tung C.-H. Enhancing membrane permeability by fatty acylation of oligoarginine peptides. Chembiochem. 2004;5:1148–1151. doi: 10.1002/cbic.200400063. [DOI] [PubMed] [Google Scholar]
- 53.Thorén P.E.G., Persson D., Isakson P., Goksör M., Onfelt A., Nordén B. Uptake of analogs of penetratin, Tat(48-60) and oligoarginine in live cells. Biochem. Biophys. Res. Commun. 2003;307:100–107. doi: 10.1016/s0006-291x(03)01135-5. [DOI] [PubMed] [Google Scholar]
- 54.Connor S.D., Schmidt S.L. Regulated portals of entry into the cell. Nature. 2003;422:37–44. doi: 10.1038/nature01451. [DOI] [PubMed] [Google Scholar]
- 55.Fischer R., Köhler K., Fotin-Mleczek M., Brock R. A stepwise dissection of the intracellular fate of cationic cell-penetrating peptides. J. Biol. Chem. 2004;279:12625–12635. doi: 10.1074/jbc.M311461200. [DOI] [PubMed] [Google Scholar]
- 56.Wadia J.S., Stan R.V., Dowdy S.F. Transducible TAT-HA fusogenic peptide enhances escape of TAT-fusion proteins after lipid raft macropinocytosis. Nature Med. 2004;10:310–315. doi: 10.1038/nm996. [DOI] [PubMed] [Google Scholar]
- 57.Leonetti J.P., Mechti N., Degols G., Gagnor C., Lebleu B. Intracellular distribution of microinjected antisense oligonucleotides. Proc. Natl Acad. Sci. USA. 1991;88:2702–2706. doi: 10.1073/pnas.88.7.2702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kubo T., Kanno K., Ohba H., Rumiana B., Fujii M. Control of intracellular delivery of oligonucleotides by signal peptides and genetic expression in human cells. Nucleic Acids Symposium Series. 2004;48:303–304. doi: 10.1093/nass/48.1.303. [DOI] [PubMed] [Google Scholar]
- 59.Dokka S., Toledo-Velasquez D., Shi X., Wang L., Rojanasukul Y. Cellular delivery of oligonucleotides by synthetic import peptide carrier. Pharm. Res. 1997;14:1759–1764. doi: 10.1023/a:1012188014919. [DOI] [PubMed] [Google Scholar]
- 60.Morris M.C., Vidal P., Chaloin L., Heitz F., Divita G. A new peptide vector for efficient delivery of oligonucleotides into mammalian cells. Nucleic Acids Res. 1997;25:2730–2736. doi: 10.1093/nar/25.14.2730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Futaki S., Ohashi W., Suzuki T., Niwa M., Tanaka S., Ueda K., Harashima H., Sugiura Y. Stearylated arginine-rich peptides: a new class of transfection systems. Bioconjug. Chem. 2001;12:1005–1011. doi: 10.1021/bc015508l. [DOI] [PubMed] [Google Scholar]
- 62.Brandén L.J., Mohamed A.J., Smith C.I. A peptide nucleic acids-nuclear localization signal fusion that mediates nuclear transport of DNA. Nat. Biotechnol. 1999;17:784–787. doi: 10.1038/11726. [DOI] [PubMed] [Google Scholar]
- 63.Cutrona G., Carpaneto E.M., Ulivi M., Roncella S., Landt O., Ferrarini M., Boffa L.C. Effects in live cells of a c-myc anti-gene PNA linked to a nuclear localization signal. Nat. Biotechnol. 2000;18:300–303. doi: 10.1038/73745. [DOI] [PubMed] [Google Scholar]
- 64.Koppelhus U., Awasthi S.K., Zachar V., Holst H.U., Ebbeson P., Nielsen P.E. Cell-dependent differential cellular uptake of PNA, peptides and PNA-peptide conjugates. Antisense Nucleic Acid Drug Dev. 2002;12:51–63. doi: 10.1089/108729002760070795. [DOI] [PubMed] [Google Scholar]
- 65.Moulton H.M., Nelson M.H., Hatlevig S.A., Reddy M.T., P.L. I. Cellular uptake of antisense morpholino oligomers conjugated to arginine-rich peptides. Bioconjug. Chem. 2004;15:290–299. doi: 10.1021/bc034221g. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.