Abstract
DNA triplexes are formed by both isomorphic (structurally alike) and non-isomorphic (structurally dissimilar) base triplets. It is espoused here that (i) the base triplet non-isomorphism may be articulated in structural terms by a residual twist (Δt°), the angle formed by line joining the C1′…C1′ atoms of the adjacent Hoogsteen or reverse Hoogsteen (RH) base pairs and the difference in base triplet radius (Δr Å), and (ii) their influence on DNA triplex is largely mechanistic, leading to the prediction of a high (t + Δt)° and low (t − Δt)° twist at the successive steps of Hoogsteen or RH duplex of a parallel or antiparallel triplex. Efficacy of this concept is corroborated by molecular dynamics (MD) simulation of an antiparallel DNA triplex comprising alternating non-isomorphic G*GC and T*AT triplets. Conformational changes necessitated by base triplet non-isomorphism are found to induce an alternating (i) high anti and anti glycosyl and (ii) BII and an unusual BIII conformation resulting in a zigzag backbone for the RH strand. Thus, base triplet non-isomorphism causes DNA triplexes into exhibiting sequence-dependent non-uniform conformation. Such structural variations may be relevant in deciphering the specificity of interaction with DNA triplex binding proteins. Seemingly then, residual twist (Δt°) and radial difference (Δr Å) suffice as indices to define and monitor the effect of base triplet non-isomorphism in nucleic acid triplexes.
INTRODUCTION
An exquisite property of nucleic acids is their ability to exhibit a repertoire of secondary structures. Triple helix is one of them, and is formed when a third oligonucleotide strand, referred to as the Triplex Forming Oligonucleotide (TFO), wraps along the major groove of a DNA duplex, forming sequence-specific Hoogsteen or reverse Hoogsteen (RH) hydrogen bonds. A number of physicochemical studies have demonstrated that such triplexes are thermodynamically stable at physiological or near physiological conditions. The high level of sequence-specific recognition interaction between the TFOs and the target DNA duplex, along with the ability of the former to compete with regulatory proteins, endow triplex forming strategy with the potential to down regulate gene expression. Efficacy of this has been demonstrated both in vitro and in vivo. A number of reviews have appeared addressing the various facets of DNA triplexes (1–6). Triplexes are also found to interfere with the binding of enzymes such as DNA polymerase (7), endonuclease (8) and methylase (9). They are shown to create permanent heritable changes in the genome by inducing mutagenesis (5) and, by enhancing recombination (5). Even intra-molecular triplexes are believed to cause transcription (10,11) as well as replication inhibition (12).
Base triplet non-isomorphism and the proviso of residual Hoogsteen/reverse Hoogsteen twist
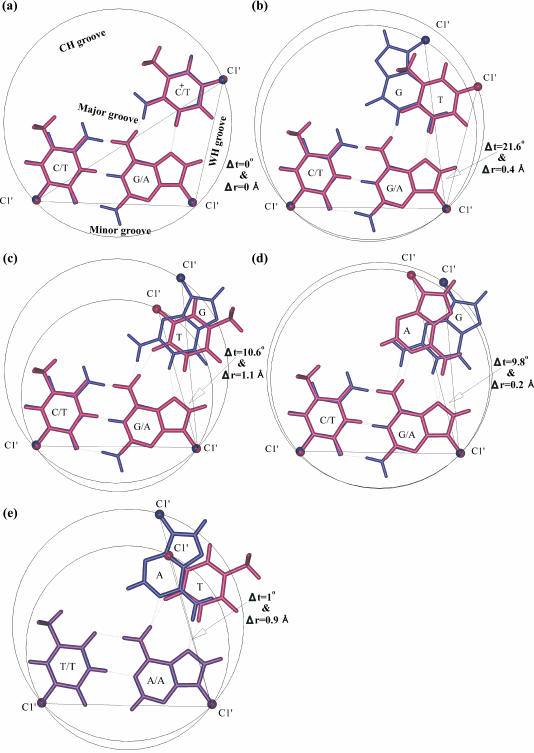
Both parallel and antiparallel DNA triplexes can be formed, and a number of base triplets, such as T*AT, C+*GC, G*GC and A*AT, figure in them depending on the base composition of the target DNA duplex. Among these, the T*AT and C+*GC base triplets are distinctive (Figure 1a) in that the three C1′ atoms of the former coincide with the corresponding C1′ atoms of the latter, resulting in the overlap of the lines joining the C1′…C1′ atoms of the Watson and Crick pairs, and those of the Hoogsteen pairs. These also render the diameters of the T*AT and C+*GC base triplets to be identical. Such features leave them to be isosteric/isomorphic/isostructural. On the other hand, superposition of other base triplet combinations, such as T*AT and G*GC (parallel orientation) (Figure 1b), T*AT and G*GC (antiparallel) (Figure 1c), A*AT and G*GC (antiparallel) (Figure 1d) and A*AT and T*AT (antiparallel) (Figure 1e), reveals that such a structural overlap of all the three C1′ atoms of the triplets does not occur. Instead, an angle (Δt°) is made, as is apparent from Figure 1b–d. This characterizes them to be non-isomorphic. Since angle Δt° persists even in the absence of a given triple helical twist ‘t’, it is tantamount to a pre-existing twist between the base pairs of Hoogsteen or the reverse Hoogsteen duplex of a triplex. As such, it represents an intrinsic feature of a pair of non-isomorphic base triplets, and is therefore referred to as the residual Hoogsteen twist or residual reverse Hoogsteen twist (Δt°). Its magnitude reflects the extent of base triplet non-isomorphism. Table 1 lists the values of Δt° for a few combinations of non-isomorphic base triplet pairs.
Figure 1.
Definition of intrinsic residual Hoogsteen twist and residual reverse Hoogsteen (Δt°) twist. Superposition of (a) isomorphic T*AT (maroon) and C+*GC (blue) (parallel orientation) and non-isomorphic base triplets: (b) G*GC (blue) and T*AT (maroon) (parallel orientation) (c) G*GC (blue) and T*AT (maroon) (antiparallel orientation) (d) G*GC (blue) and A*AT (maroon) (antiparallel orientation) and (e) A*AT (blue) and T*AT (maroon) (antiparallel orientation). Dotted lines represent the hydrogen bonds. Filled circles represent the C1′ atoms. Coincidence of C1′ atoms and the lines joining C1′…C1′ atoms of WC and Hoogsteen base pairs is seen only in (a). Non-coincidence of all the C1′ atoms leads to the formation of an angle between the lines joining C1′…C1′ atoms of the Hoogsteen and reverse Hoogsteen base pairs (b–d). This is tantamount to a pre-existing helical twist and is referred to as residual Hoogsteen twist or residual reverse Hoogsteen twist, and is denoted by ‘Δt°’. Difference in triplet radii (Δr Å) reflects yet another factor contributing to base triplet non-isomorphism (c and e), besides ‘Δt°’.
Table 1.
Residual twist (Δt°) and radial difference (Δr Å) between adjacent base triplets
| Antiparallel triplexes | Parallel triplexes | ||||||
|---|---|---|---|---|---|---|---|
| Triplets | T*AT | A*AT | T*CGa | Triplets | T*AT | G*TAa | T*CGa |
| G*GC | G*GC | ||||||
| Δt° | 10.6 | 9.8 | 6.5 | Δt° | 21.6 | 5.0 | 9.4 |
| Δr Å | 1.1 | 0.2 | 0.1 | Δr Å | 0.4 | 0.5 | 0.6 |
| A*AT | T*AT | ||||||
| Δt° | 1.0 | 0.0 | 3.2 | Δt° | 0.0 | 16.8 | 12.4 |
| Δr Å | 0.9 | 0.0 | 0.3 | Δr Å | 0.0 | 0.9 | 1.0 |
| T*AT | C+*GC | ||||||
| Δt° | 0.0 | 1.0 | 4.0 | Δt° | 0.0 | 16.8 | 12.4 |
| Δr Å | 0.0 | 0.9 | 1.2 | Δr Å | 0.0 | 0.9 | 1.0 |
aSingle hydrogen bonded mismatch triads that occasionally juxtapose the familiar two hydrogen bonded Hoogsteen or reverse Hoogsteen pairs.
Radial difference as yet another indicator of base triplet non-isomorphism
It is also clear from Figure 1 that the diameters of some of the overlapping base triplets (Figure 1c and e) differ to a noticeable extent, contributing to the structural dissimilarity of base triplets. Hence, the radial difference (Δr Å) is yet another indicator that reflects non-isomorphic nature of the base triplets. Presence of either this alone (Figure 1e) or residual twist alone (Figure 1b and d), or both (Figure 1c), can contribute to the base triplet non-isomorphism. Table 1 lists the values of Δr Å for a few combinations of non-isomorphic base triplets.
Thus, Δt° and Δr Å together provide a holistic description of base triplet non-isomorphism in structural terms. Their magnitudes reflect the nature and degree of non-isomorphism, which are distinctive for a given pair of base triplets and, are governed by the scheme of Hoogsteen or reverse Hoogsteen hydrogen bonding.
Existence of base triplet non-isomorphism, although realized quite early (13,14), there has been no attempt either to characterize (and quantify) it or to systematically investigate its influence on the structure of DNA triplexes. Thus, it is shown here for the first time, that non-isomorphism between a pair of base triplets may be adequately and conveniently articulated by the pre-existing or residual twist (Δt°), between the adjacent Hoogsteen base pairs in parallel DNA triplexes, and reverse Hoogsteen (RH) base pairs in antiparallel DNA triplexes, and by their radial difference (Δr Å). Occurrence of the residual twist, at once, predicts the need for the prevalence of an alternating high and low twist angles at successive steps of the Hoogsteen or reverse Hoogsteen duplex of the triplex; the nature and magnitude of which being predetermined by the magnitude of the residual twist (Δt°). Likewise, it is likely that the difference in radius of the triplets (Δr Å) would also affect the triplex conformation. In order to examine their influence, molecular dynamics (MD) simulations on the antiparallel DNA triplex comprising alternating non-isomorphic G*GC and T*AT triplets (Figure 1c) have been carried. Triplexes formed by them are known to be stable, and also effective in down regulating gene expression (15–18). Identification of proteins interacting with these triplexes has indicated possible other biological roles for them (19–21). The structural distortions due to base triplet non-isomorphism might therefore be relevant in understanding triplex…protein recognition interactions.
MATERIALS AND METHODS
A 14mer antiparallel DNA triplex comprising alternating G*GC and T*AT triplets (Scheme 1) is generated conforming to a 12-fold helix (22,23).
Scheme 1.
A 14mer antiparallel DNA triplex with alternating non-isomorphic G*GC and T*AT triplets. Base sequence is numbered to facilitate discussion. RH hydrogen bond pairs are represented by ‘*’. Here onwards, the pyrimidine and purine strands of the WC duplex is referred to as Crick and Watson strand, respectively, and the third strand is referred to as the reverse Hoogsteen (RH) strand.
Stereochemistry of the third strand is regularized by constrained–restrained molecular geometry optimization and by van der Waal's energy minimization using X-PLOR (24). The model is subjected to a steepest descent energy minimization using the Sander module of AMBER 4.1 suit (25). This is used as the starting model for the MD simulation. Using the LEaP module of AMBER 6 (26), 39 net neutralizing Na+ counter ions and, periodic box of TIP3P waters measuring 9 Å in all the directions from the triplex, are added, resulting in a periodic box of ∼52 Å × ∼51 Å × ∼75 Å. Equilibration is effected in several steps. Initially, minimization for 2500 cycles using steepest descent algorithm is pursued followed by a dynamics for 40 ps with positional restraints of 500 kcal/(mol Å2) on the triplex. Later, particle mesh Ewald (PME) (27) is introduced using cubic B-spline interpolation order, and a tolerance of 0.00001 (10−5) for direct space sum cut-off. Then, dynamics is continued for an additional period of 40 ps. Positional restraints on the solute are reduced to 25 kcal/(mol Å2) in the next step and, a minimization for 2500 cycles is carried out followed by a dynamics for 10 ps. During the last five rounds of minimization, each lasting 1500 cycles, positional restraints on the solute are slowly reduced in steps of 5 kcal/(mol Å2). Finally, minimization is carried out for 800 cycles without any positional restraints, but with hydrogen bond restraints for the base pairs. Subsequently, a completely free minimization for 1000 cycles is carried out. The entire system is heated from 100 to 300K over 4 ps, and the production run is continued up to 4 ns using Sander module of AMBER6. Isobaric and isothermal conditions are used during the production run and nonbonded pair list is updated for every 10 steps. Simulation is performed with SHAKE (tolerance = 0.0005 Å) on the hydrogens (28), a 2 fs integration time and a cut-off distance of 9 Å for Lennard–Jones interaction. The centre of mass translation is periodically removed (every 10 ps) during the production run. The protocol used here is similar to that described by Cheatham and Kollman (29).
Analysis of the trajectory
Average structure and root mean square (r.m.s.) deviation are calculated using Ptraj Ver 6.4 (http://www.chpc.utah.edu/~cheatham/software.html). Insight II program from Biosym Technologies (30) is used for the visualization of the molecules and moil-view Ver 9.0 program is used for trajectory analysis (31).
Helical parameters are calculated separately for the WC and reverse Hoogsteen duplexes. Twist angles of the reverse Hoogsteen duplex are calculated with respect to C1′…C1′ vector. This is adopted in view of the fact that, the line joining C6 and C8 atoms (or C8 and C8 atoms in the case of Purine…Purine pairing) of the RH base pairs does not pass through the centre of gravity of the RH pairs, unlike in WC pairs. This is logical and also appropriate in keeping with the definition of the intrinsic residual twist (Figure 1). For the sake of uniformity, helical parameters of the WC duplex are also calculated using this scheme. These, along with the other parameters of the helix, stacking diagrams and torsion angles are extracted from the output of 3DNA Ver 1.5 (32) using in-house programs.
Water…DNA interaction
Water distribution around DNA over the last 2.5 ns of the trajectories is calculated using Ptraj Ver 6.4. Water oxygen positions obtained by r.m.s. fit carried out over all DNA atoms, at 1 ps interval, are binned into 0.5 Å3 grids, which are then contoured using the program O (33). Subsequently, water oxygens are fitted into the density.
RESULTS
Mechanistic influence of residual twist and radial difference on DNA triplex
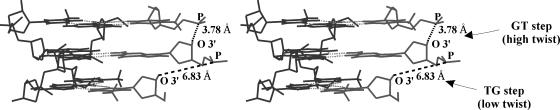
Nature of influence of residual twist (Δt°) on DNA triplex conformation can be readily figured out when it is recognized that the helical twist between adjacent Watson and Crick pairs as well as of the add-on Hoogsteen pairs remains the same (and is equal to the given triple helical twist ‘t’) in triplexes formed with either isomorphic T*AT and C+*GC triplets (Figure 1a) or homo triplets, where Δt is 0°. On the contrary, when the residual twist (Δt°) has a distinct value, say 10.6° (Table 1), as found between the adjacent T…A and G…G reverse Hoogsteen base pairs (Figure 1c) of an antiparallel DNA triplex, effective helical twist between them is rendered t + (Δt) 10.6° and t − (Δt) 10.6°. When t = 30° as in a 12-fold DNA triplex, a higher twist angle (t = 40.6°) is expected at the GT (5′GT/5′AG) step and a lower twist angle (t = 19.4°) at the TG (5′TG/5′GA) step of the RH duplex, while the twist angle (t) between the adjacent Watson and Crick pairs remains at 30° (due to their isomorphic character). This naturally leads to an over winding and an under winding at alternate steps of the RH duplex resulting in non-uniform twist angles in the triplex. It can be seen (Figure 2) that this causes the RH strand to be disjointed, with a gap of over a nucleotide length especially, at the low twist TG (6.8 Å) step. In addition, this brings about steric overlap between the methyl group (T) and C2′ atom of the sugar (G) at the high twist GT step. The steric overlap is even more severe between the sugar (T) and base (G) of the adjacent residues, at the low twist TG step. The large radial difference between the T*AT and G*GC triplets (Δr = 1.1 Å) also contributes to this (Figure 1c). Triplex formation under these circumstances therefore, warrants considerable adjustments in the conformations of the triplex. Clearly then, distortions arising from the base triplet non-isomorphism are due to mechanistic effects rather than electronic causes. The latter, in addition, may have their characteristic influence. It is anticipated that similar effects of mechanistic nature are likely in DNA triplexes formed out of other non-isomorphic base triplet combinations.
Figure 2.
Stereo diagram showing the disjointed nature of the reverse Hoogsteen (RH) strand (grey) caused by Δt = 10.6° and Δr Å in an antiparallel DNA triplex (t = 30° and h = 3.26 Å) comprising alternating G*GC and T*AT triplets. Gaps of nearly one nucleotide length at TG step may be seen. WC duplex is coloured black.
DNA triplex comprising alternating non-isomorphic T*AT and G*GC base triplets
It is clear from above that the residual twist and the radial difference entails conformational modulations in a DNA triplex formed out of non-isomorphic base triplets. In order to examine this, MD simulation has been carried out on an antiparallel DNA triplex comprising alternating non-isomorphic T*AT and G*GC triplets (Scheme 1). Alternating G*GC and T*AT triplets are deliberately chosen to accentuate any distinguishing effects that may arise due to base triplet non-isomorphism. Prior to MD simulation, the RH strand of the triplex is regularized by establishing backbone connectivity at both the steps by energy minimization (see Materials and Methods).
Non-uniform Helical twist in the RH duplex
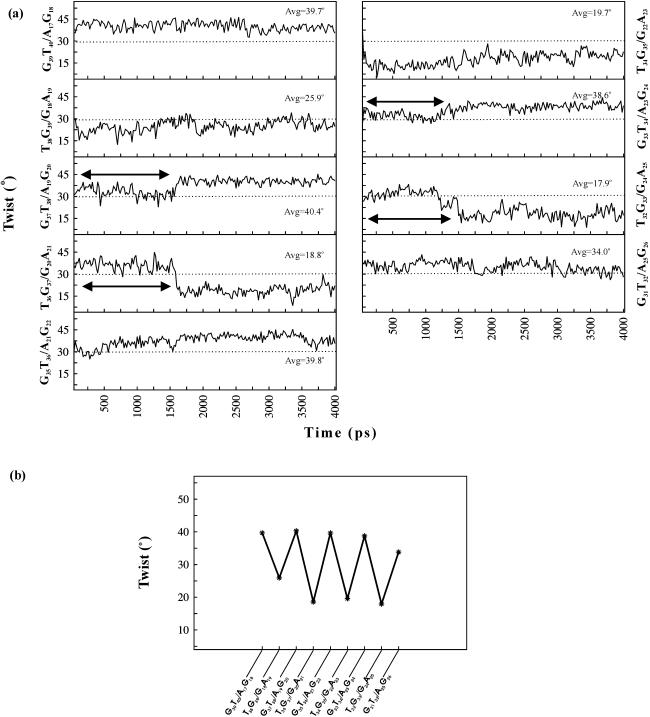
Since the anticipated primary influence of base triplet non-isomorphism concerns non-uniform twist angle variations in the RH duplex, helical twist angles at the alternating TG and GT steps are monitored over the entire simulation. In view of possible end-effects, only the central decamer of the 14mer RH duplex is considered, and the nature of twist angle variation is shown Figure 3a. Mean value of the helical twists at various GT and TG steps, calculated over the final 2.4 ns of dynamics varies between 33.9–40.4° and 17.9–25.9°, respectively. Average values of these correspond to 38.5° and 20.6° at the GT and TG steps, respectively. These, incidentally, also match with the average values of the twist angles obtained from the average structure, corresponding to the last 2.4 ns (Figure 3b). Interestingly, they are also very close to the twist angles of 40.6° (t + Δt) and 19.4° (t − Δt) anticipated from the residual twist angle (Δt = 10.6°) between the non-isomorphic G*GC and T*AT triplets (Figure 1c). This indicates that the relative orientation of the base triplets has remained essentially unaffected. Apparently then, the effects of residual twist such as the unfavourable steric interactions, and disjointed nature of the RH strand mentioned above, are somehow taken care of by the torsion angle flexibility of the polynucleotide chains (see below). In any case, the results are clearly indicative of the non-uniform helical twist along the RH strand of the DNA triple helix.
Figure 3.
(a) Variation of helical twist angles at the alternating steps of the central 10mer of the RH d(GT)7*d(AG)7 duplex over 4 ns dynamics. Triplex structure at every 20 ps interval is considered for calculation of twists. Note the alternating high and low twist at alternating steps. Dotted lines correspond to the assigned twist angle of 30°. Double-headed arrows indicate unusual variations seen at different steps. Average values of twist calculated over the last 2.4 ns are also given. (b) Variations of helical twist angles at different steps of the average structure of the central 10mer RH duplex. Alternating high and low twist angles at GT/AG and TG/GA steps result in a saw–tooth-like pattern. Base sequences corresponding to different steps of the RH duplex are shown for clarity. However, base sequence along the RH strand alone is referred in the text.
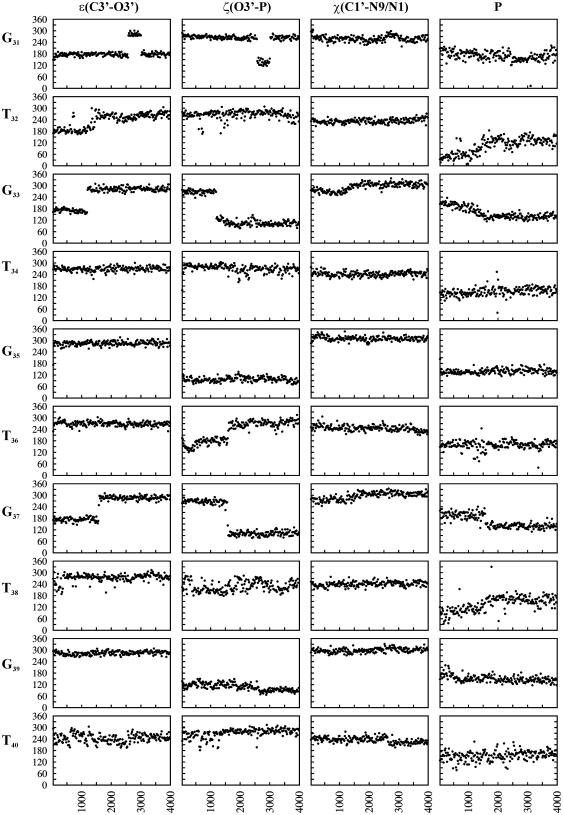
One of the windfalls of MD simulations is the feasibility of identifying the behaviour over and above the average trend. Some of which could have arisen due to interactions with water and/or ions that might prevail under experimental conditions. Prominent among such observations are the unusual twist angles found at the T32G33, G33T34, T36G37 and G37T38 steps and, these are indicated by double-headed arrows in Figure 3a. In contrast to the average behaviour of high twist at the GT step and a low twist at the TG step, G33T34 and G37T38 steps exhibit a low twist (∼25–35°) accompanied by a high twist (∼30–45°) at the preceding T32G33 and T36G37 steps, respectively, during the initial 1600 ps. This trend, however, is reversed beyond 1600 ps, conforming to the average behaviour. These can be traced to the effects of deviations from the preferred nucleotide conformation involving the C3′–O3′(ε) and O3′–P(ζ) bond torsions (Figure 4), and to interactions involving water (Figures 5 and 6). It is found that the BI conformation (34,35) characterized by (ε,ζ,α) = (t,g−,g−), predominates at these two GT steps (G33T34 and G37T38) during the initial 1600 ps in contrast to the BII conformation characterized by (ɛ,ζ,α) = (g−,t,g−), that prevails at all the other GT steps (Figure 4). On the other hand, at the T32G33 step, BI conformation reigns during the initial 1600 ps instead of an all gauche− conformation, (ɛ,ζ,α) = (g−,g−,g−), (hereafter referred to as BIII conformation) favoured at all the other TG steps. Likewise, occurrence of BII conformation (34,35) at the T36G37 step, instead of the BIII conformation, is responsible for the high twist during the initial 1600 ps dynamics.
Figure 4.
Variation of conformational angles ɛ(C3′–O3′), ζ(O3′–P) and χ(C1′–N9/N1) and P (phase angle of pseudo rotation) in the central 10mer of the RH strand over 4 ns dynamics. Alternating high anti (G) and anti (T) glycosyl conformation along with alternating BII (GT step) and BIII (TG step) conformation may be seen.
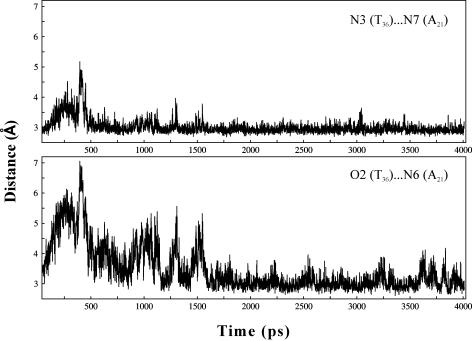
Figure 5.
Variation of N3(T36)…N7(A21) and O2(T36)…N6(A21) RH hydrogen bond distances over 4 ns simulation. Note the large fluctuation in N3(T36)…N7(A21) hydrogen bond distance during the first 400 ps, and in O2(T36)…N6(A21) hydrogen bond distance during the first 1600 ps. These are correlated with water-mediated interactions shown in Figure 6 (see also text). Hydrogen bond distances at every 1 ps interval are calculated.
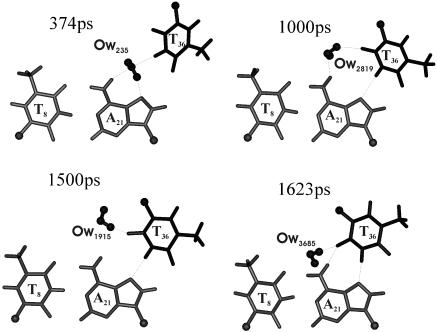
Figure 6.
Interaction of water with RH T36…A21 pair during the dynamics. The spikes in Figure 5 correspond to a situation when water comes exactly between A21 and T36 around 400 ps. Water molecules are represented by ball and stick.
In addition, it is found that marked deviations in twist angles at the T32G33, G33T34, T36G37 and G37T38 steps are also influenced by the guanines, G33 and G37, assuming the lower range of high anti glycosyl conformations during the initial ∼1600 ps, instead of the characteristic high anti conformation (Figure 4). Sugar pucker transition from C3′exo (P ∼ 200°) to C2′endo (P ∼ 150°) is also observed in these guanine residues (Figure 4). In addition, sugar puckers of the neighbouring T32 and T38 residues undergo transition from the C3′endo (T32, P ∼ 30–70°)/O4′endo (T38, P ∼ 70–120°) to the C2′endo (P ∼ 120–170°) range of conformations.
Interestingly, the above conformation variations at the T36G37 and G37T38 steps result in a water-mediated T36…A21 RH pair during the initial 1600 ps, as indicated in Figures 5 and 6. During the first 500 ps of simulations, O2(T36)…N6(A21) and N3(T36)…N7(A21) hydrogen bonds of the T36…A21 RH base pair are lost (Figure 5). In fact, around 400 ps, both O2(T36)…N6(A21) and N3(T36)…N7(A21) distances increase to ∼7 and ∼5 Å, respectively. At this juncture, a water molecule engages A21 and T36 in hydrogen bond interactions (374 ps in Figure 6). Soon after this, N3(T36)…N7(A21) distance reverts to ∼3 Å. A variety of water-mediated hydrogen bonding, as illustrated in Figure 6, is found till ∼1600 ps before the canonical RH O2(T36)…N6(A21) hydrogen bond is restored.
An alternating anti and high anti glycosyl conformation in the RH strand
MD simulations reveal substantial conformation modulations in the RH strand of the DNA triplex. One of the more prominent observations has been the transition from the anti to high anti (χ ∼ 270–340°) glycosyl conformation of all the guanines (Figure 4), while thymines retain the preferred anti (χ ∼ 210–270°) conformation. This leads to an alternating anti and high anti glycosyl conformation for the RH strand. This is yet another significant influence of the base triplet non-isomorphism entrenched between G*GC and T*AT triplets. Fraying tendencies of the terminal residues (G29 and T30) afford sufficient flexibility for G31 to retain the anti glycosyl conformation.
An alternating phosphodiester and zig-zag backbone conformation for the RH strand
Backbone of the RH strand also exhibits unusual conformational features. It is found that ɛ(C3′–O3′) favour predominantly the gauche− range of conformations at all the GT and TG steps (Figure 4). This conformation is normally associated with a near trans conformation around the ζ(O3′–P) bond leading to a BII conformation (ɛ,ζ,α) = (g−,t,g−) (34,35). This, in fact, prevails at four of the five GT steps, with the O3′–P torsions favouring values in the range 90–150°. These result in a (t/g+,g−) phosphodiester conformation at the high twist GT steps of the RH strand. On the other hand, (g−,g−) phosphodiester prevails at all the TG steps, notwithstanding the gauche− conformation around ɛ(C3′–O3′), resulting in an all gauche− (ɛ,ζ,α) = (g−,g−,g−) conformation. We refer to this as the BIII conformation to distinguish from the other two frequently observed conformations. Thus, an alternating (g−,g−) and (t/g+,g−) phosphodiester conformations occur along the TG and GT steps of the RH strand. This, together with the alternating anti and high anti glycosyl conformation described above, leaves the RH strand to exhibit an alternating conformation with a zig-zag backbone. MD simulation (2 ns) carried out with a different starting model, wherein all the three strands are unlinked (Figure S1 in Supplementary Material), essentially retains all the features detailed above indicating that the results are independent of the starting models.
Other torsions, α(P–O5′), β(O5′–C5′) and γ(C5′–C4′) adopt their preferred gauche−, trans and gauche+ conformations, respectively. The deoxysugars of the RH strand favour P ∼ 120–170° covering C1′exo to C2′endo range of conformations.
Average structure of the triplex
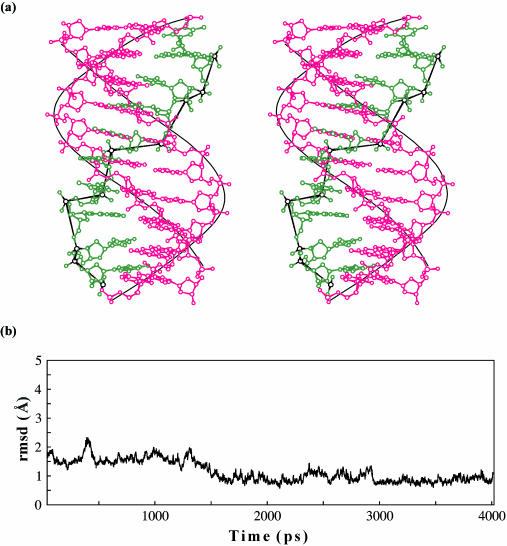
Average structure of the triplex corresponding to the last 2.4 ns of MD simulation is shown in Figure 7a. Root mean square deviation (r.m.s.d.) of the central decamer (taken at 1 ps interval over the entire simulation) varies between 0.6 and 2.3 Å from the average structure (Figure 7b). The fluctuations are rather large initially, but stabilize to r.m.s.d. of 1 Å after 3000 ps.
Figure 7.
(a) Stereo plot of the antiparallel DNA triplex comprising alternating G*GC and T*AT base triplets corresponding to the last 2.4 ns of the central 10mer of the average structure. Smooth curve linking the phosphates of the WC duplex and the zig-zag nature of the RH strand (green) may be readily seen. (b) RMS deviation (r.m.s.d.) of the central 10mer triplex structures from the average structure. Triplex structures at every 1 ps interval over 4 ns dynamics are taken for the calculation. Note the stabilization of the RMS deviation after 3 ns.
Helical rise of the RH duplex
Average value of the helical rise of the RH duplex varies between 2.6 and 3.7 Å at the GT steps, and from 3.6 to 4.0 Å at the TG steps. Overall average of rise at the GT step (3.1 Å) is slightly lower (0.8 Å) than that at the TG step (3.9 Å). Comparison of these with twist angle variations indicates that a high rise at the TG step is accompanied by a low twist and vice versa. A similar trend with a slightly larger difference in the helical rise at the GT and TG steps is found in the NMR structure of an antiparallel intra-molecular DNA triplex (36).
Base pair and base step parameters of the WC duplex
Twist angles at the CT (CT/AG) and TC (TC/GA) steps of the WC duplex vary from 22° to 35° with an overall average of 30°. Similarly, helical rise lies between 3.5 and 3.9 Å at the CT step and, between 3.5 and 3.7 Å at the TC step. Overall average of rise, x-displacement, slide and propeller twist correspond to 3.6 Å, −3.1 Å, −1.6 Å and −4.7°, respectively.
Conformation of the Watson and Crick strands
It is observed that the backbone torsions α(P–O5′), β(O5′–C5′), γ(C5′–C4′), ɛ(C3′–O3′) and ζ(O3′–P) and the side chain glycosyl C1′–N9/N1 (χ) torsion angles of the WC duplex strands essentially retain their preferred gauche−, trans, gauche+, trans, gauche− and anti conformations, respectively. Sugar puckers in both the strands of the WC duplex exhibit large fluctuations during the dynamics, covering O4′endo, C1′exo and C2′endo conformations (data not shown). But, they largely prefer C1′exo conformation in the average structure with P varying between 102° and 142° for pyrimidine sugars, and between 132° and 164° for the purine sugars. In any case, the WC duplex strands possess a uniform backbone conformation, unlike the RH strand (Figure 7a).
Groove widths
The average values of the major and minor groove widths correspond to 24 (1) Å and 11 (1) Å, respectively. A third strand along the major groove of DNA duplex, divides it into WH groove (formed by Watson and RH strands, see Figure 1) and CH groove (formed by Crick and RH strands, see Figure 1). Their widths vary at alternate phosphates, because of the alternating and zig-zag nature of the RH strand discussed above. The CH groove width alternates between 11.9 (3) and 15 (1) Å, while the width of the WH groove alternates between 10.3 (9) and 12.4 (9) Å. Low width of WH groove is matched with a higher width of the CH groove and vice versa, such that their sum corresponds to the major groove width.
Base stacking
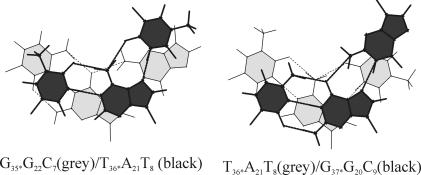
Base stacking at the representative GT and TG steps of the average structure of the WC duplex is shown in Figure 8. Intra-strand stacking is characterized by the overlap of the methyl group of thymine with the adjacent cytosine at the CT step of the pyrimidine strand, while stacking is negligible at the TC step. On the other hand, adjacent purines stack well both at the AG and GA steps. In general, stacking at GA step is marginally better than at the AG step. Intra-strand base stacking in the RH strand is only marginal both at the GT and TG steps. At the GT step, O4 (T) is involved in stacking with the imidazole ring of guanine.
Figure 8.
Stacking interactions at the G35T36 step (G35*G22C7/T36*A21T8) and T36G37 step (T36*A21T8/G37*G20C9) of the average structure. Hydrogen bonds are indicated by dotted lines. Figures are drawn using 3DNA (32).
Hydrogen bonds in the triplex
Except for the two RH hydrogen bonds (G29…G28, T30…A27) at the 5′ terminus of the TFO, all the others are retained. Incidentally, this correlates well with the observation that 5′ end of a (GT)n TFO interacts rather weakly with the purine strand of the duplex (37,38). Hydrogen bonds in the WC duplex of the triplex are retained.
Hydration around the triplex
Water is found to preferentially interact with the anionic oxygen atoms of the phosphates of all the three strands. Possibility of its interaction with other oxygen atoms of the sugar residue, O3′, O4′ and O5 is also noticed (Figure S2 in Supplementary Material).
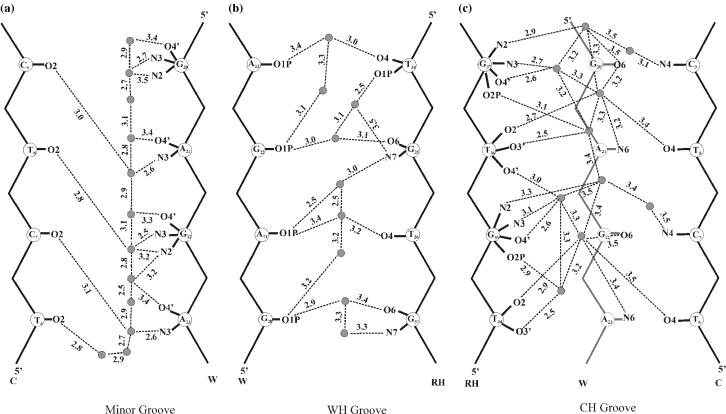
Water molecule is found to bridge N3 (purines) and O2 (pyrimidines) of all along the minor groove (Figure 9a). At the same time, N2(G) also interacts with this water. These, along with the water molecules that are hydrogen bonded to O4′(purines) of the sugar leads a continuous network of water along the minor groove.
Figure 9.
Schematic diagram showing the water network around the tetrameric triplex T34*A23T6–G35*G22C7–T36*A21T8–G37*G20C9. Donor…acceptor distances within 3.5 Å are shown by dotted lines. Filled circles (grey) indicate water molecules.
Hydration sites around O6(G), N7(G) and O4(T) of the RH strand are observed along the WH groove (Figure 9b). This is in accord with the NMR data that suggested specific ‘pockets’ for water and counter ions along the WH groove involving these atoms (39). Water-mediated interaction involving N7(G) and O4(T) of the adjacent bases are seen at the GT step (Figure 9). Likewise, water-mediated interaction involving O4(T)/O6(G) with O1P of the opposing strand is found all along the WH groove.
CH groove is characterized by well-organized water network involving N3(G), N2(G) and O2(T) of the RH strand, N4(C) and O4(T) of the Crick strand and O6(G) and N6(A) of the Watson strand (Figure 9c). The network is further extended to include the backbone atoms, O3′, O4′ and O2P of the RH strand. A single water molecule is found to interact with O2(T), N6(A) and O4(T) of the same T*AT triplet. Interestingly, high anti glycosyl conformation of G leads to a water-mediated interaction between O4′ and N3 atoms of same G. Participation of O4′ of 5′T in this network is also observed. Water-mediated interaction involving O3′(T) and O2P(5′G) is also seen, wherein these atoms orient towards the CH groove due to the preference of BIII conformation at the TG step.
Triplex…ion interaction
Although Na+ ions show strong tendency for the coordination with anionic oxygens of the phosphates of all the three strands, it is prominent with one of the anionic oxygens (O1P) of the Watson strand that lined up in the WH groove (Figure S3 in Supporting Material). Other oxygen atoms like O3′, O5′ and O4′ of the sugar residue also interact with the counter ions.
In general, ion interacts rather weakly with the minor groove atoms. In the WH groove, ions directly coordinate with N7 of the RH base. On the other hand, interaction of ion with the triplex through first hydration shell is prominent in the CH groove compared to the WH groove (Figures S3 and S4 in Supplementary Material), resulting in good ion density in the CH groove (Figure S5 in Supplementary Material).
DISCUSSION
Although it is well documented that DNA duplex interacts with an additional strand along its major groove in a sequence-specific manner, to form a variety of triple helices, details concerning the effects of base sequential context are not completely understood. In this context, it may be noted that in WC paired DNA duplexes, sequence effects arise solely from electronic interactions in view of the isomorphic or isosteric nature of the WC base pairs. Indeed, this argument is expected to be true even in DNA triplexes, only when the triplets are either isomorphic, as in T*AT and C+*GC triplets, or when formed with homo triplets. But, such a simple and straightforward rationalization may not suffice, when DNA triplexes are formed with non-isomorphic base triplets. Here, sequence effects need to be decomposed, as arising from geometric or mechanistic effects caused by non-isomorphism of the base triplets and, those arising purely from electronic effects. While it may be difficult to isolate these from the experimental data, we have shown here for the first time that base triplet non-isomorphism, may be characterized in structural terms and they are amenable not only for quantitative description, but also their effects assessable. It is shown that non-isomorphism between a pair of base triplets may be conveniently and adequately expressed as a residual twist (Δt°) and/or radial difference (Δr Å) between them. Their influences on the DNA triplex structure are by and large mechanistic in nature, and are largely responsible for sequence-dependent structural variations in DNA triplexes.
A residual twist of 10.6° occurring between the non-isomorphic G*GC and T*AT triplets readily indicated that a DNA triplex formed out of them should exhibit a helical twist of 30 + 10.6° (t + Δt)° and 30 − 10.6° (t − Δt)° at the successive GT and TG steps of the RH duplex. In fact, MD simulation of a DNA triplex comprising alternating G*GC and T*AT triplets reveals that the major influence of the residual twist is to bring about non-uniform helical twist angles in the RH duplex, while the WC duplex remains unaffected. Alternating low (20.6°) and high (38.5°) helical twist angles are found at the alternating TG and GT steps, and interestingly these are very similar to the twist angles of ∼20° and ∼40° that are foreseen from the knowledge of residual RH twist (Figure 1c). This indicates that the relative orientations of the base triplets remain nearly unaltered from the initial position, despite the flexibility afforded by MD simulation. Further, it is clear that the helical twist variation is caused by mechanistic effect of base triplet non-isomorphism, and not by the electronic influences of adjacent bases. Good agreement between the predicted twist angles and those seen in the NMR data (36) establishes that the origin of such twist angle variations resides in the residual twist between the non-isomorphic triplets.
Analysis shows that, by and large, RH strand alone undergoes conformational changes, which are manifested in the backbone and glycosyl conformations. One such significant change is the transition from anti to high anti conformation of all the guanines in the RH strand. This facilitates in rendering the bases in proper orientation to maintain RH hydrogen bond, while at the same time brings to proximity the O3′ and P of the adjacent nucleotides to bridge proper stereochemistry, especially at the TG step. It is indeed noteworthy that this feature is observed in an NMR structure of an intra-molecular triplex with an adjacent non-isomorphic G*GC and T*AT base triplets (36).
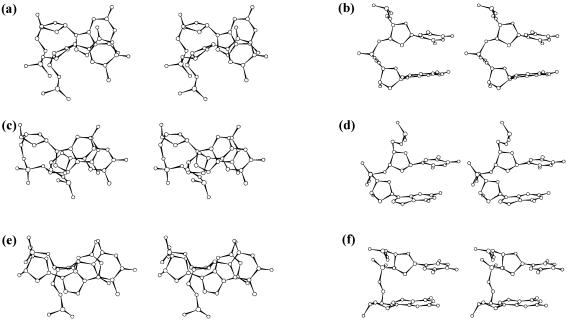
Another noticeable change is the occurrence of gauche− conformation around the C3′–O3′ torsion (ɛ) throughout the RH strand. Normally, this is accompanied by a change in the conformation around the P–O3′ bond (ζ) from a gauche− to an extended trans conformation leading to a (t, g−) phosphodiester conformation, commonly referred to as the BII conformation (34,35). A similar conformation is found at all the high twist GT steps, although it is noticed that the P–O3′ bond (ζ) assumes values in the range 90–150°. Interestingly, the low twist TG steps favour a (g−,g−) phosphodiester conformation, notwithstanding the gauche− conformation around the C3′–O3′ bond (ɛ), named as the BIII conformation to differentiate from the other conformational sub-states. This unusual BIII conformation has become necessary to bridge the large O3′…P separation at the low twist TG step, created by the cumulative effect of the residual twist (Δt°), and radial difference (Δr Å), of the T*AT and G*GC base triplets. Interestingly, BII conformation is associated with the higher twist (GT step) as in BDNA duplexes (40), while the low twist (TG step) is associated with the BIII conformation. Comparison of BI, BII and BIII conformation is shown in Figure 10. Thus, in addition to alternating anti and high anti glycosyl conformation, alternating BII and BIII conformation prevails, resulting in a zig-zag shape for the RH strand. RH duplex is, thus, heteronomous, as the conformations of the two strands of the duplex are distinct. The large conformational changes, mandatory for the formation of the RH DNA triplex with non-isomorphic G*GC and T*AT triplets, are expected to warrant much higher activation energy compared to that required for the formation of isomorphic DNA triplex. This gains support from the kinetic study which suggests that such triplex formation, in fact, needs higher activation energy of 88 ± 21 kJ m−1 compared to nearly zero value required for the formation of isomorphic T*AT and C+*GC containing triplexes (41).
Figure 10.
Stereo diagram showing the BI (a and b), BII (c and d) and BIII (e and f) conformation that occurs at the low twist TG step of the RH strand during the dynamics.
It is also recognized that such drastic conformational changes in the RH strand are not observed when the constraints of RH hydrogen bonds, either in G*GC or T*AT triplet are absent, as in the case of terminal RH pairs or water-mediated RH pairs. Hence, when a G*GC and T*AT non-isomorphic triplet pair occurs in an isolated situation, say, flanked by isomorphic T*AT and C+*GC base triplets, either one of the two things may happen, (i) a local fluctuation in the triple helical conformation including the twist angle variation as discussed above with intact RH pair or (ii) water-mediated distortion of the RH pair so as to reduce the effect of residual twist on the conformation of the RH strand.
Although possibility of structural perturbation as a consequence of base triplet non-isomorphism has been indicated in earlier studies (36,42–44), none of these take into cognizance, the prospect of residual reverse Hoogsteen twist and the radial difference, the structural markers that characterize base triplet non-isomorphism, as the causative agents or the origin for the observed changes in the DNA triple helical structure. Instead, the observed changes are merely attributed to as due to the effect of non-isomorphism of G*GC and T*AT base triplets. On the contrary, the present study clearly points out that the structural perturbations predominantly stem from the effects of Δt° and Δr Å, which induce sequence-dependent non-uniform DNA triplex structure. Further, MD simulations carried out on DNA triplexes comprising alternating non-isomorphic G*GC and A*AT (Figure 1d, Δt = 9.8° and Δr = ∼0.2 Å) and A*AT and T*AT (Figure 1e, Δt = ∼0° and Δr = ∼0.9 Å) base triplets, in order to isolate the individual effects of Δt° and Δr Å, indicate that radial difference (Δr Å) brings forth conformational changes in third strand, while residual twist (Δt°) influences twist angle variations in the RH duplex besides contributing to conformational effects (R. Thenmalarchelvi and N. Yathindra, unpublished data). Therefore, it is important to take cognizance of the residual twist and radial difference of the base triplets and their consequence in the effective design of TFOs for antigene strategy of gene regulation. Structural details provided here about the DNA triplex comprising G*GC and T*AT base triplets may aid in the understanding of its interaction with proteins that are identified to interact with this triplex.
SUPPLEMENTARY MATERIAL
Supplementary Material is available at NAR Online.
Supplementary Material
Acknowledgments
R.T. acknowledges Mr Gold Smith for many useful discussions. R.T. thanks University Grants Commission (UGC) and Council of Scientific and Industrial Research for fellowships. Financial assistance to the Department under DSA and FIST programs of UGC and Department of Science and Technology is gratefully acknowledged. N.Y. thanks UGC for a grant. The Open Access publication charges for this article were waived by Oxford University Press.
REFERENCES
- 1.Soyfer V.N., Potaman V.N. Triple-Helical Nucleic Acids. New York: Springer-Verlag; 1996. [Google Scholar]
- 2.Gowers D.M., Fox K.R. Towards mixed sequence recognition by triple helix formation. Nucleic Acids Res. 1999;27:1569–1577. doi: 10.1093/nar/27.7.1569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Praseuth D., Guieysse A.L., Helene C. Triple helix formation and the antigene strategy for sequence-specific control of gene expression. Biochim. Biophys. Acta. 1999;1489:181–206. doi: 10.1016/s0167-4781(99)00149-9. [DOI] [PubMed] [Google Scholar]
- 4.Wang E., Feigon J. Structures of nucleic acid triplexes. In: Neidle S., editor. Oxford Handbook of Nucleic Acid Structure. New York: Oxford University Press; 1999. pp. 355–388. [Google Scholar]
- 5.Vasquez K.M., Glazer P.M. Triplex-forming oligonucleotides: principles and applications. Q. Rev. Biophys. 2002;35:89–107. doi: 10.1017/s0033583502003773. [DOI] [PubMed] [Google Scholar]
- 6.Zain R., Sun J.S. Do natural DNA triple-helical structures occur and function in vivo? Cell Mol. Life Sci. 2003;60:862–870. doi: 10.1007/s00018-003-3046-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Samadashwily G.M., Mirkin S.M. Trapping DNA polymerases using triplex-forming oligodeoxyribonucleotides. Gene. 1994;149:127–136. doi: 10.1016/0378-1119(94)90421-9. [DOI] [PubMed] [Google Scholar]
- 8.Francois J.C., Saison-Behmoaras T., Thoung N.T., Helene C. Inhibition of restriction endonuclease cleavage via triple helix formation by homopyrimidine oligonucleotides. Biochemistry. 1989;28:9617–9619. doi: 10.1021/bi00451a011. [DOI] [PubMed] [Google Scholar]
- 9.Maher L.J., III, Wold B., Dervan P.B. Inhibition of DNA binding proteins by oligonucleotide-directed triple helix formation. Science. 1989;245:725–730. doi: 10.1126/science.2549631. [DOI] [PubMed] [Google Scholar]
- 10.Sarkar P.S., Brahmachari S.K. Intramolecular triplex potential sequence within a gene down regulates its expression in vivo. Nucleic Acids Res. 1992;20:5713–5718. doi: 10.1093/nar/20.21.5713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sakamoto N., Ohshima K., Montermini L., Pandolfo M., Wells R.D. Sticky DNA, a self-associated complex formed at long GAA·TTC repeats in intron 1 of the frataxin gene, inhibits transcription. J. Biol. Chem. 2001;276:27171–27177. doi: 10.1074/jbc.M101879200. [DOI] [PubMed] [Google Scholar]
- 12.Krasilnikova M.M., Mirkin S.M. Replication stalling at Friedreich's ataxia (GAA)n repeats in vivo. Mol. Cell. Biol. 2004;24:2286–2295. doi: 10.1128/MCB.24.6.2286-2295.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Giovannangeli C., Rougee M., Garestier T., Thoung N.T., Helene C. Triple-helix formation by oligonucleotides containing the three bases thymine, cytosine, and guanine. Proc. Natl Acad. Sci. USA. 1992;89:8631–8635. doi: 10.1073/pnas.89.18.8631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sun J.S., de Bizemont T., Duval-Valentin G., Montenay-Garestier T., Helene C. Extension of the range of recognition sequences for triple helix formation by oligonucleotides containing guanines and thymines. C.R. Acad. Sci. Ser. III. 1991;313:585–590. [PubMed] [Google Scholar]
- 15.Postel E.H., Flint S.J., Kessler D.J., Hogan M.E. Evidence that a triplex-forming oligodeoxyribonucleotide binds to the c-myc promoter in HeLa cells, thereby reducing c-myc mRNA levels. Proc. Natl Acad. Sci. USA. 1991;88:8227–8231. doi: 10.1073/pnas.88.18.8227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ing N.H., Beekman J.M., Kessler D.J., Murphy M., Jayaraman K., Zendegui J.G., Hogan M.E., O'Malley B.W., Tsai M.J. In vivo transcription of a progesterone-responsive gene is specifically inhibited by a triplex-forming oligonucleotide. Nucleic Acids Res. 1993;21:2789–2796. doi: 10.1093/nar/21.12.2789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Musso M., Wang J.C., Van Dyke M.W. In vivo persistence of DNA triple helices containing psoralen-conjugated oligodeoxyribonucleotides. Nucleic Acids Res. 1996;24:4924–4932. doi: 10.1093/nar/24.24.4924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Carbone G.M., McGuffie E.M., Collier A., Catapano C.V. Selective inhibition of transcription of the Ets2 gene in prostate cancer cells by a triplex-forming oligonucleotide. Nucleic Acids Res. 2003;31:833–843. doi: 10.1093/nar/gkg198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Musso M., Bianchi-Scarra G., Van Dyke M.W. The yeast CDP1 gene encodes a triple-helical DNA-binding protein. Nucleic Acids Res. 2000;28:4090–4096. doi: 10.1093/nar/28.21.4090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nelson L.D., Musso M., Van Dyke M.W. The yeast STM1 gene encodes a purine motif triple helical DNA-binding protein. J. Biol. Chem. 2000;275:5573–5581. doi: 10.1074/jbc.275.8.5573. [DOI] [PubMed] [Google Scholar]
- 21.Ciotti P., Van Dyke M.W., Bianchi-Scarra G., Musso M. Characterization of a triplex DNA-binding protein encoded by an alternative reading frame of loricrin. Eur. J. Biochem. 2001;268:225–234. doi: 10.1046/j.1432-1033.2001.01859.x. [DOI] [PubMed] [Google Scholar]
- 22.Arnott S., Selsing E. Structures for the polynucleotide complexes poly(dA).poly(dT) and poly(dT).poly(dA).poly(dT) J. Mol. Biol. 1974;88:509–521. doi: 10.1016/0022-2836(74)90498-7. [DOI] [PubMed] [Google Scholar]
- 23.Chandrasekaran R., Giacometti A., Arnott S. Structure of poly (dT).poly (dA).poly (dT) J. Biomol. Struct. Dyn. 2000;17:1011–1022. doi: 10.1080/07391102.2000.10506589. [DOI] [PubMed] [Google Scholar]
- 24.Brunger A.T. Yale University; 1996. X-PLOR Ver 3.851. [Google Scholar]
- 25.Pearlman D.A., Case D.A., Caldwell J.W., Ross W.S., Cheatham T.E., III, DeBolt S., Ferguson D., Seibel G., Kollman P. AMBER, a package of computer programs for applying molecular mechanics, normal mode analysis, molecular dynamics and free energy calculations to simulate the structural and energetic properties of molecules. Comput. Phys. Commun. 1995;91:1–41. [Google Scholar]
- 26.Case D.A., Pearlman D.A., Caldwell J.W., Cheatham T.E., III, Ross W.S., Simmerling C.L., Darden T.A., Merz K.M., Stanton R.V., Cheng A.L., Vincent J.J., Crowley M., Tsui V., Radmer R.J., Daun Y., Pitera J., Massova I., Seibel G.L., Singh U.C., Weiner P.K., Kollman P.A. AMBER 6. San Francisco: University of California; 1999. [Google Scholar]
- 27.Essmann U., Perera L., Berkowitz M.L., Darden T., Lee H., Pedersen L.G. A smooth particle Mesh Ewald method. J. Chem. Phys. 1995;103:8577–8593. [Google Scholar]
- 28.Ryckaert J.P., Ciccotti G., Berendsen H.J.C. Numerical integration of the Cartesian equations of motion of a system with constraints: molecular dynamics of n-alkanes. J. Comp. Phys. 1977;23:327–341. [Google Scholar]
- 29.Cheatham T.E., III, Kollman P.A. Molecular dynamics simulations highlight the structural differences among DNA:DNA, RNA:RNA and DNA:RNA hybrid duplexes. J. Am. Chem. Soc. 1997;119:4805–4825. [Google Scholar]
- 30.Insight II User guide. San Diego: Biosym Technologies; 1992. Version 2.1.0. [Google Scholar]
- 31.Simmerling C., Elber R., Zhang J. MOIL-View—a program for visualization of structure and dynamics of biomolecules and STO. A program for computing stochastic paths. In: Pullman A., et al., editors. Modeling of Biomolecular Structure and Mechanisms. Netherlands: Kluwer; 1995. pp. 241–265. [Google Scholar]
- 32.Lu X.J., Olson W.K. 3DNA: a software package for the analysis, rebuilding and visualization of three-dimensional nucleic acid structures. Nucleic Acids Res. 2003;31:5108–5121. doi: 10.1093/nar/gkg680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jones T.A., Zou J.Y., Cowan S.W., Kjeldgard M. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Cryst. A. 1991;47:110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- 34.Gupta G., Bansal M., Sasisekharan V. Conformational flexibility of DNA: polymorphism and handedness. Proc. Natl Acad. Sci. USA. 1980;77:6486–6490. doi: 10.1073/pnas.77.11.6486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Prive G.G., Heinemann U., Chandrasegaran S., Kan L.S., Kopka M.L., Dickerson R.E. Helix geometry, hydration, and G.A mismatch in a B-DNA decamer. Science. 1987;238:498–504. doi: 10.1126/science.3310237. [DOI] [PubMed] [Google Scholar]
- 36.Radhakrishnan I., Patel D.J. Solution structure of a purine.purine.pyrimidine DNA triplex containing G.GC and T.AT triples. Strucure. 1993;1:135–152. doi: 10.1016/0969-2126(93)90028-f. [DOI] [PubMed] [Google Scholar]
- 37.Cheng A.J., Van Dyke M.W. Oligodeoxyribonucleotide length and sequence effects on intermolecular purine–purine–pyrimidine triple-helix formation. Nucleic Acids Res. 1994;22:4742–4747. doi: 10.1093/nar/22.22.4742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hardenbol P., Van Dyke M.W. Sequence specificity of triplex DNA formation: analysis by a combinatorial approach, restriction endonuclease protection selection and amplification. Proc. Natl Acad. Sci. USA. 1996;93:2811–2816. doi: 10.1073/pnas.93.7.2811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Radhakrishnan I., Patel D.J. Hydration sites in purine.purine.pyrimidine and pyrimidine.purine.pyrimidine DNA triplexes in aqueous solution. Structure. 1994;2:395–405. doi: 10.1016/s0969-2126(00)00041-1. [DOI] [PubMed] [Google Scholar]
- 40.Hartmann B., Piazzola D., Lavery R. BI–BII transitions in B-DNA. Nucleic Acids Res. 1993;21:561–568. doi: 10.1093/nar/21.3.561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Xodo L.E., Pirulli D., Quadrifoglio F. A kinetic study of triple-helix formation at a critical R•Y sequence of the murine c-Ki-ras promoter by (A,G)- and (G,T) oligonucleotides. Eur. J. Biochem. 1997;248:424–432. doi: 10.1111/j.1432-1033.1997.00424.x. [DOI] [PubMed] [Google Scholar]
- 42.Sun J.S., Helene C. Oligonucleotide-directed triple-helix formation. Curr. Opin. Struct. Biol. 1993;3:345–356. doi: 10.1016/s0959-440x(96)80051-0. [DOI] [PubMed] [Google Scholar]
- 43.Kiran M.R., Bansal M. Molecular mechanics studies of homopolymeric and mixed sequence Py.Pu*Pu triple helices. Indian J. Biochem. Biophys. 1995;32:391–403. [PubMed] [Google Scholar]
- 44.Avino A., Cubero E., Gonzalez C., Eritja R., Orozco M. Antiparallel triple helices. Structural characteristics and stabilization by 8-amino derivatives. J. Am. Chem. Soc. 2003;125:16127–16138. doi: 10.1021/ja035039t. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.