Abstract
The thousands of serial images used for medical pedagogy cannot be included in a printed book; they also cannot be efficiently handled by ordinary image viewer software. The purpose of this study was to provide browsing software to grasp serial medical images efficiently. The primary function of the newly programmed software was to select images using 3 types of interfaces: buttons or a horizontal scroll bar, a vertical scroll bar, and a checkbox. The secondary function was to show the names of the structures that had been outlined on the images. To confirm the functions of the software, 3 different types of image data of cadavers (sectioned and outlined images, volume models of the stomach, and photos of the dissected knees) were inputted. The browsing software was downloadable for free from the homepage (anatomy.co.kr) and available off-line. The data sets provided could be replaced by any developers for their educational achievements. We anticipate that the software will contribute to medical education by allowing users to browse a variety of images.
Keywords: Visible Human Projects, Cadaver, Anatomy, Body Image, Computer Software
Graphical Abstract
INTRODUCTION
In the clinical setting, countless serial images are obtained and reviewed, including those from computed tomography (CT) and magnetic resonance imaging (MRI). To view these images on a personal computer, existing image browsers (e.g., MRIcro [Neuropsychology Lab, Columbia, SC, USA]) are not appropriate. For example, the browsers lack the ability to automatically label the outlined anatomical structures (1,2,3).
The Visible Korean research team has developed software to view serial sectioned images of a cadaver with convenience and efficiency. Specifically, the software has the ability to display the names of outlined structures, which would aid students in learning sectional anatomy (4,5,6). Additionally, the team has accumulated raw images and processed images of different subjects, anatomical regions, and organs. The image data include color-filled images, volume models, and photographs from cadavers (7,8,9). To view a variety of images, previous software having one scroll bar is insufficient.
The purpose of this study was to help medical trainees grasp numerous serial images by providing the advantageous browsing software. For that purpose, the new software was programmed to be equipped not only with buttons, scroll bars, and checkbox to select images, but also with other improved functions such as zoom capability. To confirm applicability of the software, 3 different types of image data were inputted into the software and then examined.
MATERIALS AND METHODS
Manufacture of the software
The new browsing software consisted of program files and data files. The program files in the main folder were composed by the C# language of Microsoft Visual Studio.NET 2013 (Microsoft Corporation, Redmond, WA, USA; www.microsoft.com). The accessory text files such as config.txt (Table 1), color.txt, and help.txt were also put in the main folder.
Table 1. Significant variants in config.txt of the 2 browsing software packages.
| Variables | Functions | Software of horizontal images | Software of stomach images |
|---|---|---|---|
| Software_title | Software title, shown on the window | Browsing software (Male whole body, male head, female pelvis, ver. 2.0) | Browsing software (Male - Peeled and piled stomach, ver. 2.0) |
| Margin_red_value | R value of the software margin | 0 | 255 |
| Margin_green_value | G value of the software margin | 0 | 255 |
| Margin_blue_value | B value of the software margin | 0 | 255 |
| X_axis_scroll_bar | Buttons (0) or scroll bar (1) on the x-axis | 0 | 1 |
| X_axis_button_number | No. of buttons (maximum 10) | 3 | - |
| X_axis_button_name_1 | Name of the 1st button | Male whole body | - |
| X_axis_button_name_2 | Name of the 2nd button | Male head | - |
| X_axis_button_name_3 | Name of the 3rd button | Female pelvis | - |
| X_axis_initial | Value on the x-axis in the initial view | 1 | 0 |
| Checkbox_off_folder_name | Folder name of the images that appear with checkbox off | Sectioned images | Volume models |
| Checkbox_on_folder_name | Folder name of the images that appear with checkbox on | Color-filled images | Reference sectioned images |
| Y_axis_initial | Value on the y-axis in the initial view | 851 | 0 |
R = red, G = green, B = blue.
The software title shown on the window and the color (red [R], green [G], and blue [B] values) shown on the margin of the images were listed in config.txt (Fig. 1; Table 1). The essential function of the software was to select the desired images using 3 types of interfaces. The first interface on the x-axis was buttons or a scroll bar, of which one was selected in config.txt. In the case of buttons, the number and names of the buttons were fixed in config.txt. To decide the initial view, the code of x-axis button (value 1) or coordinate of the x-axis scroll bar (value 0) was designated in config.txt (Fig. 1; Tables 1 and 2).
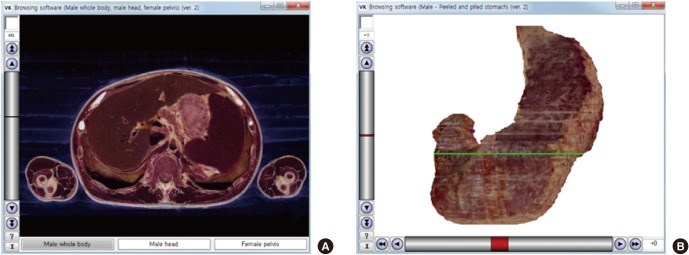
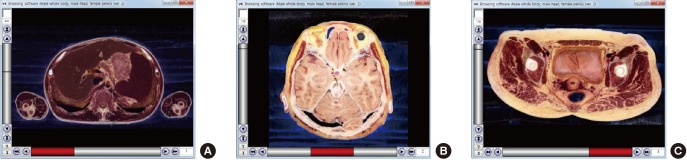
Fig. 1.
Initial views of the 2 new browsing software packages. (A) The software shows horizontal sectioned image with the x-axis represented by the 3 buttons. The image file shown in the initial view is named “1_851.png.” The “1” indicates the first button (male whole body), and “851” indicates the 851st sectioned image. (B) The software shows stomach volume model with the x-axis as the scroll bar. The image file shown in the initial view is named “0_0.png,” where the first “0” indicates 0 degrees of rotation and the second “0” indicates the volume model with no peeling or piling.
Table 2. Significant variants in config.txt of the 2 browsing software packages.
| Data sets | X-axis (horizontal direction on screen) | Y-axis (vertical direction on screen) | Checkbox |
|---|---|---|---|
| Horizontal images | To select one among male whole body, male head, female pelvis (buttons) | To select levels of images (scroll bar) | To show color-filled images |
| Stomach images | To rotate stomach (scroll bar) | To peel and pile stomach (scroll bar) | To show a reference sectioned image |
| Knee images | To select one among slightly dissected knee, moderately dissected knee, much dissected knee, and flexed knee of embalmed cadaver (buttons) | To rotate knee (scroll bar) | To show images of fresh cadaver |
The second interface on the y-axis was the scroll bar. The third interface located above the y-axis scroll bar was a checkbox. The folder names of the images appearing with checkbox off/on were listed in config.txt. To decide the initial view (checkbox off), the y-axis coordinate (value 851 or 0) was designated in config.txt (Fig. 1; Tables 1 and 2).
Apart from the program files, the data files refer to the image files to be shown on the software. Five major image formats were readable using this software: portable network graphics (PNG), bit map (BMP), graphics interchange format (GIF), joint photographic experts group (JPEG), and tagged image file format (TIFF). It was because most of viewer software used the 5 standardized formats; these formats were reliable for creating and using software because they had an authorized library.
The image files in 2 corresponding folders were named based on the x-axis interface. If the x-axis used buttons, the image file name (e.g., 1_851) consisted of the x-axis button code (value 1) and the y-axis coordinate (value 851). If the x-axis used the scroll bar, the image file name (e.g., 0_0) consisted of the x-axis coordinate (value 0) and the y-axis coordinate (value 0) (Fig. 1; Table 1).
One powerful function of the software was to automatically label the names of the outlined structures (Fig. 2). This was possible by referring to both the color-filled images and color.txt. In color.txt, every color-filled structure’s name and its red, green, and blue values were listed. Two software packages of the stomach and knee images did not include the color-filled images, so this function was not feasible, and color.txt was meaningless (Figs. 3 and 4; Table 2).
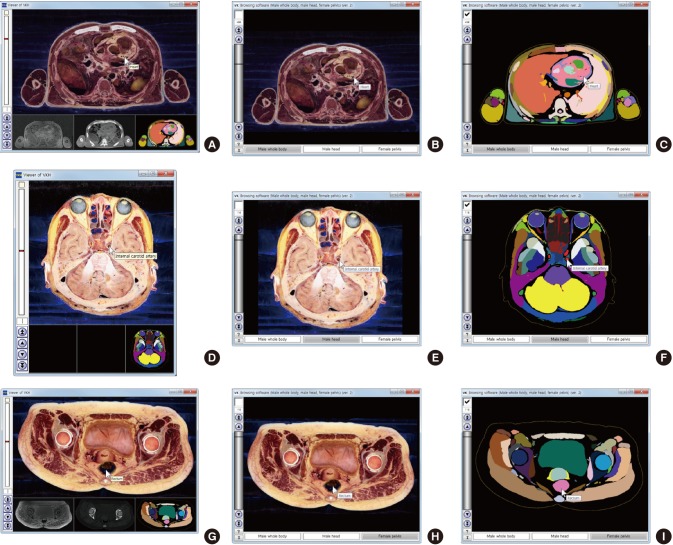
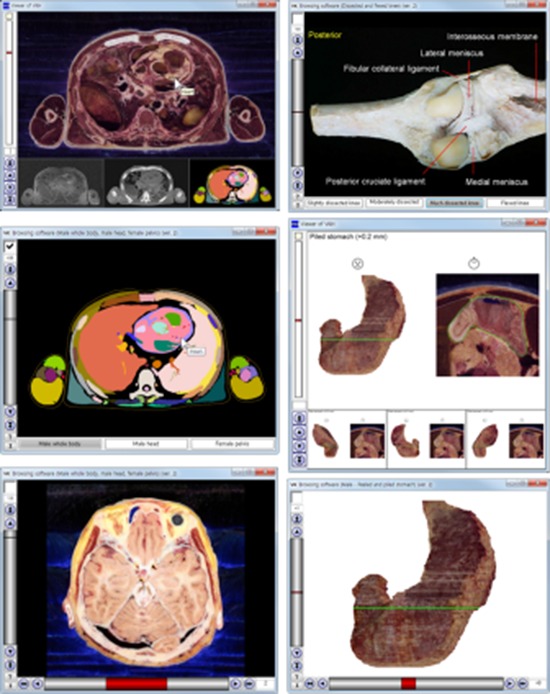
Fig. 2.
Comparison of previous and new software that automatically label the names of the outlined structures of 3 cadavers. (A, B, C) Male whole body. (D, E, F) Male head. (G, H, I) Female pelvis. (A, D, G) Previous software to simultaneously show the sectioned and color-filled images. (B, E, H) New software to show the sectioned images when the checkbox is off. (C, F, I) New software to show the color-filled images when the checkbox is on.
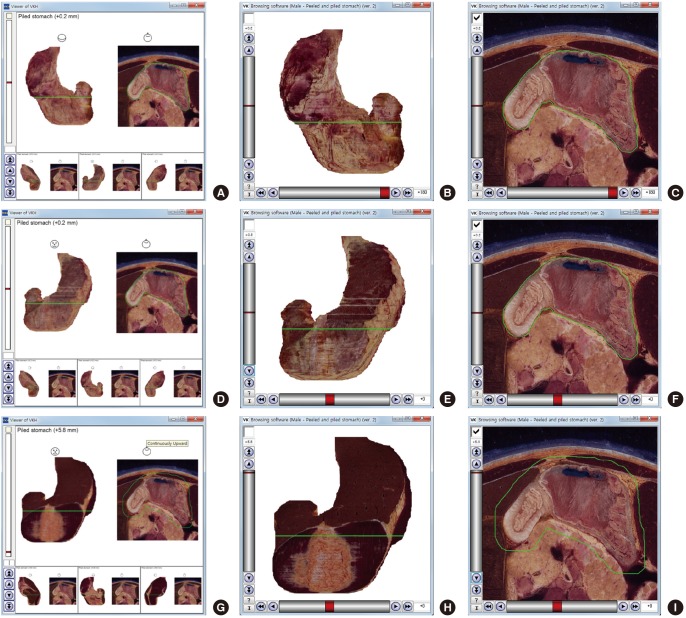
Fig. 3.
Comparison of previous and new software that show the volume models and sectioned images of stomach. (A, B, C) Posterior view of stomach. (D, E, F) Anterior view of stomach. (G, H, I) Anterior view of piled stomach. (A, D, G) Previous software to simultaneously show the volume models and the sectioned image of the stomach. (B, E, H) New software to alternatively show the volume models when the checkbox is off. (C, F, I) New software to alternatively show the sectioned image when the checkbox is on.
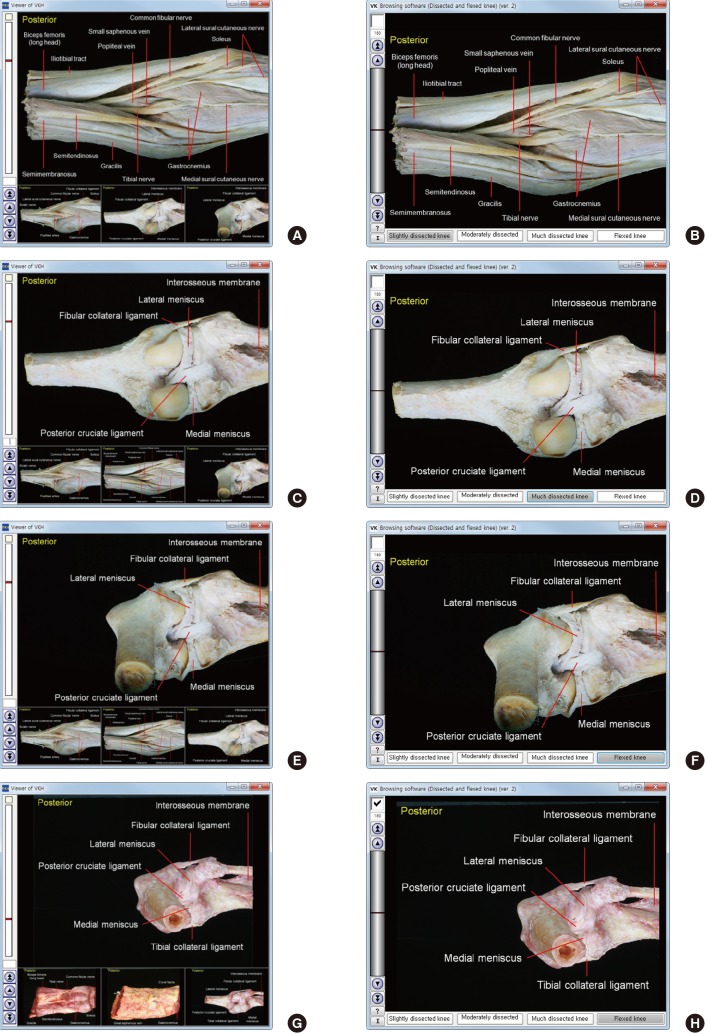
Fig. 4.
Comparison of previous and new software that show cadaveric knees. (A, B) Slightly dissected knee of embalmed cadaver. (C, D) Much dissected knee of embalmed cadaver. (E, F) Flexed knee of embalmed cadaver. (G, H) Flexed knee of fresh cadaver. (A, C, E, G) Previous software. (B, D, F, H) New software.
The software also had a help function so that clicking the “?” button at the left bottom (Fig. 1) revealed help.txt. The txt file was written not only about how the software could be used by clients but also about how the image data and config.txt could be changed (Table 1) by other investigators.
After the software programming was finished, 3 data sets of the images, prepared in the Visible Korean laboratory, were inputted (Table 2).
Input of the horizontal images
The sectioned images and color-filled images of the male whole body (10), the male head (11), and the female pelvis (12) were inputted into the software. The software was configured such that the x-axis buttons were used to select one of the 3 subjects (male whole body, male head, and female pelvis). The serial sectioned images (intervals of 1 mm) were selected using the y-axis scroll bar. The sectioned images and corresponding color-filled images were selected using the checkbox (Fig. 2; Tables 1 and 2).
Input of the stomach images
The peeled and piled volume models of the stomach, accompanied by a reference sectioned image (13), were inputted into the software. The serial rotated volume models (intervals of 30 degrees) were selected using the x-axis scroll bar. The piled and peeled volume models (intervals of 0.4 mm) were selected using the y-axis scroll bar. The piling and peeling thickness was designated with plus (+) and minus (−) signs. The checkbox was used to display the reference sectioned image with a green line to indicate the extent of piling and peeling (Fig. 3; Tables 1 and 2).
Input of the knee images
Images of dissected and then flexed knees from 2 subjects (embalmed and fresh cadavers) (7) were inputted into the software. The x-axis buttons were used to select views of the slightly dissected knee, moderately dissected knee, much dissected knee, and flexed knee. The y-axis scroll bar selected serial rotated views of the knee (intervals of 5 degrees). The checkbox was used to select either the knee from the embalmed cadaver (initial view) or the knee from the fresh cadaver, which did not necessarily correspond to one another (Fig. 4; Table 2).
RESULTS
Three new browsing software packages (compressed as ZIP files) could be freely downloaded from our homepage (anatomy.co.kr) without registration: male whole body, male head, and female pelvis (ver. 2); male peeled and piled stomach (ver. 2); and dissected and flexed knee (ver. 2). Once the compressed ZIP files were extracted, the programs could be used on the Microsoft Windows environment without installation. Each software package consisted of the program files, config.txt (Table 1), color.txt, help.txt and the 2 folders containing image files.
Images could be selected by clicking the buttons or using the scroll bar. The chosen button was highlighted; the x- and y-coordinates of the selected image were shown on the 2 axes. The scroll bar function could be substituted by typing an image number. The neighboring images could be browsed stepwise or continuously by clicking the arrow buttons. If the checkbox was clicked, the image switched to the corresponding one. At any time, the initial image was brought back by clicking the “I” button on the left bottom corner of the window (Figs. 1,2,3,4; Table 2).
In the case of the stomach, serial rotated images were browsed with the x-axis scroll bar because the rotation axis was vertical (Fig. 3). However, in the case of the knee, serial rotated images were browsed with the y-axis scroll bar because the rotation axis was horizontal (Fig. 4; Table 2).
The selected image could be zoomed in or zoomed out by rolling the mouse wheel. The zoomed in image was shifted by moving the mouse with its left button pressed.
If the user located the mouse pointer on a structure in the color-filled image, the structure’s name was shown as the tool tip text beside the mouse pointer. The structure’s name could also be seen in the sectioned image because the 2 images were equivalent with each other (Fig. 2).
All the software testing was performed in real time because the software was operated off-line rather than on-line and because the image files were saved as PNG files with a small file size and a reduced image resolution. Nevertheless, in the case of horizontal images, the reduced resolution (1,920 × 1,080) was as good as that of a high-definition television (Fig. 1A).
The software supported dynamic windows of various resolutions. It also supported window dragging mode in the corner or either side of monitor. If the window was dragged from either side; the buttons, scroll bars, and check box were dynamically changed.
DISCUSSION
The software we designed is an improvement over tools traditionally used for displaying medical images. In this study, the software contains 4,376 (= [1,702 + 235 + 251] × 2) horizontal images, 8,784 (= 72 × 61 × 2) stomach images, or 576 (= 4 × 72 × 2) knee images. The numerous serial medical images cannot be displayed easily in a printed book or in existing viewer software, which shows pictures one by one.
The new software features 2 axes (Table 1) compared to a single y-axis found in previous software (Fig. 2). The x-axis buttons function to choose the subjects (male whole body, male head, and female pelvis). These buttons enable us to unite 3 individual software packages (4,5,6) into one (Fig. 2).
On the other hand, the x-axis scroll bar has the merit to browse numerous images conveniently. The x-axis scroll bar allows a view of the stomach for every 30 degrees of rotation relative to every 90 degrees in the previous software (Fig. 3) (13). It is also possible to use the x-axis scroll bar with the 3 subjects, in which case the scroll bar handle elongates to occupy one-third of the length (Fig. 5).
Fig. 5.
Experimental software to show the sectioned images of 3 cadavers with the x-axis scroll bar. (A) Male whole body. (B) Male head. (C) Female pelvis.
Installation of the setup file results in production of the image data, which can then be copied and utilized freely (e.g., lecture slide or examination). The authors have kept the images available free of charge for the benefit of medical education.
Any investigators can change the software by replacing the included image data with their own images. For example, CT and MRI data of normal and diseased persons could be inputted into the software. Such flexibility might be useful for various needs in medical education (14, 15).
In order to replace data, it is necessary to follow the procedure described. First, image file names should be labeled with an x-axis value and a y-axis value (Fig. 1). Second, config.txt should be modified using Table 1 as a guide. Third, for the color-filled images, color.txt should be filled with structure names and their color values. Finally, help.txt can be modified to meet the needs of the users.
The previous software (ver. 1) has the ability to simultaneously show 4 corresponding images, which can be easily compared with one another (4,5,6); this is not possible with the new software (version 2), which alternatively shows 2 corresponding images using the checkbox (Figs. 2,3,4). Other developers can select either software package, considering the characteristics of their own images to be inputted. Therefore, we still offer the previous software on the same website.
Totally different way is for other researchers to develop new software with enhanced features. In the case of non-commercial products, it would be useful to allow data replacement as we have done with our software.
The present study describes the development of new software to browse medical images with convenient functions and describes in detail the methods of using and modifying the software. We expect that the software will contribute to medical education and possibly to other kinds of pedagogy, for example, to teach people about the human body as part of an exhibit in a science museum or natural history museum.
ACKNOWLEDGMENT
Raw data of the Visible Korean were acquired by the assistance from the Korea Institute of Science and Technology Information (KISTI).
Footnotes
Funding: This work was supported by the National Research Foundation of Korea (NRF) grant funded by the Korea government (Ministry of Science, ICT and Future Planning [MSIP]) (No. 2015R1A2A2A01008248). This research was financially supported by the Ministry of Trade, Industry and Energy (MOTIE) and Korea Institute for Advancement of Technology (KIAT) through the International Cooperative R & D program (Grant No. N0002249).
DISCLOSURE: The authors have no potential conflicts of interest to disclose.
AUTHOR CONTRIBUTION: Conceptualization: Kwon K, Chung MS, Park JS, Shin BS, Chung BS. Data curation: Kwon K, Chung BS. Formal analysis: Kwon K, Chung BS. Investigation: Kwon K, Park JS. Writing - original draft: Kwon K, Chung MS, Park JS, Shin BS, Chung BS.
References
- 1.Plaisant C, Carr D, Shneiderman B. Image-browser taxonomy and guidelines for designers. IEEE Softw. 1995;12:21–32. [Google Scholar]
- 2.Rorden C, Brett M. Stereotaxic display of brain lesions. Behav Neurol. 2000;12:191–200. doi: 10.1155/2000/421719. [DOI] [PubMed] [Google Scholar]
- 3.Mojsilovic A, Gomes J. Semantic based categorization, browsing and retrieval in medical image databases. Proc Int Conf Image Proc. 2002;3:III145–8. [Google Scholar]
- 4.Shin DS, Chung MS, Park HS, Park JS, Hwang SB. Browsing software of the visible Korean data used for teaching sectional anatomy. Anat Sci Educ. 2011;4:327–332. doi: 10.1002/ase.249. [DOI] [PubMed] [Google Scholar]
- 5.Shin DS, Jang HG, Park JS, Park HS, Lee S, Chung MS. Accessible and informative sectioned images and surface models of a cadaver head. J Craniofac Surg. 2012;23:1176–1180. doi: 10.1097/SCS.0b013e31825657d8. [DOI] [PubMed] [Google Scholar]
- 6.Shin DS, Jang HG, Hwang SB, Har DH, Moon YL, Chung MS. Two-dimensional sectioned images and three-dimensional surface models for learning the anatomy of the female pelvis. Anat Sci Educ. 2013;6:316–323. doi: 10.1002/ase.1342. [DOI] [PubMed] [Google Scholar]
- 7.Chung BS, Park JS, Jang HG, Chung MS. Software to browse the pictures of two knees in diverse states of dissection, flexion and rotation. Int J Morphol. 2015;33:1009–1015. [Google Scholar]
- 8.Kwon K, Shin DS, Shin BS, Park HS, Lee S, Jang HG, Park JS, Chung MS. Virtual endoscopic and laparoscopic exploration of stomach wall based on a cadaver’s sectioned images. J Korean Med Sci. 2015;30:658–661. doi: 10.3346/jkms.2015.30.5.658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chung BS, Chung MS, Shin BS, Kwon K. Peeled and piled volume models of the kidney that show actual morphology. J Korean Med Sci. 2016;31:1514–1515. doi: 10.3346/jkms.2016.31.10.1514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Park JS, Chung MS, Hwang SB, Lee YS, Har DH, Park HS. Visible Korean human: improved serially sectioned images of the entire body. IEEE Trans Med Imaging. 2005;24:352–360. doi: 10.1109/tmi.2004.842454. [DOI] [PubMed] [Google Scholar]
- 11.Park JS, Chung MS, Shin DS, Har DH, Cho ZH, Kim YB, Han JY, Chi JG. Sectioned images of the cadaver head including the brain and correspondences with ultrahigh field 7.0 T MRIs. Proc IEEE Inst Electr Electron Eng. 2009;97:1988–1996. [Google Scholar]
- 12.Hwang SB, Chung MS, Hwang YI, Park HS, Har DH, Shin DS, Shin BS, Park JS. Improved sectioned images of the female pelvis showing detailed urogenital and neighboring structures. Korean J Phys Anthropol. 2010;23:187–198. [Google Scholar]
- 13.Chung BS, Kwon K, Shin BS, Chung MS. Peeled and piled volume models of the stomach made from a cadaver ‘s sectioned images. Int J Morphol. 2016;34:939–944. [Google Scholar]
- 14.Alvarez A, Gold GE, Tobin B, Desser TS. Software tools for interactive instruction in radiologic anatomy. Acad Radiol. 2006;13:512–517. doi: 10.1016/j.acra.2005.10.005. [DOI] [PubMed] [Google Scholar]
- 15.Biasutto SN, Caussa LI, Criado del Río LE. Teaching anatomy: cadavers vs. computers? Ann Anat. 2006;188:187–190. doi: 10.1016/j.aanat.2005.07.007. [DOI] [PubMed] [Google Scholar]