Abstract
The melting of tetramolecular DNA or RNA quadruplexes is kinetically irreversible. However, rather than being a hindrance, this kinetic inertia allows us to study association and dissociation processes independently. From a kinetic point of view, the association reaction is fourth order in monomer and the dissociation first order in quadruplex. The association rate constant kon, expressed in M−3·s−1 decreases with increasing temperature, reflecting a negative activation energy (Eon) for the sequences presented here. Association is favored by an increase in monocation concentration. The first-order dissociation process is temperature dependent, with a very positive activation energy Eoff, but nearly ionic strength independent. General rules may be drawn up for various DNA and RNA sequence motifs, involving 3–6 consecutive guanines and 0–5 protruding bases. RNA quadruplexes are more stable than their DNA counterparts as a result of both faster association and slower dissociation. In most cases, no dissociation is found for G-tracts of 5 guanines or more in sodium, 4 guanines or more in potassium. The data collected here allow us to predict the amount of time required for 50% (or 90%) quadruplex formation as a function of strand sequence and concentration, temperature and ionic strength.
INTRODUCTION
The inclination of GMP or guanine-rich poly- and oligonucleotides to self-assemble into G-quadruplexes has been recognized for over 40 years (1–3). A G-quartet is a planar association of four guanines held together by eight hydrogen bonds (Figure 1A, left); G-quadruplexes result from the hydrophobic stacking of several quartets (Figure 1A, right) (4). A cation (typically Na+ or K+) is located between two quartets forms cation–dipole interactions with eight guanines, reducing the repulsion of the 2 × 4 central oxygen atoms, enhancing hydrogen bond strength and stabilizing quartet stacking.
Figure 1.
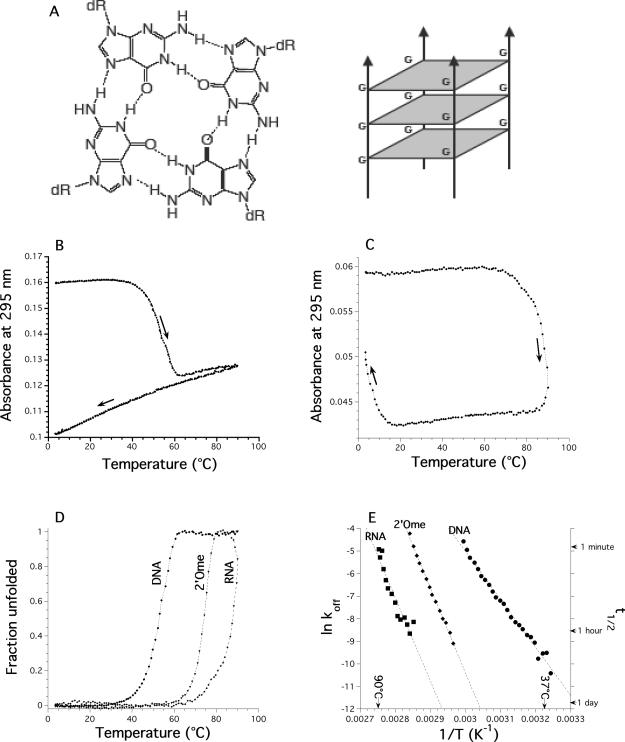
Quadruplex melting and cooling profiles. (A) Scheme of a G-quartet (left) and a tetramolecular quadruplex (right). (B) d-(TG4T)4 (20 μM) in 0.11 M Na+. Melting profile: at this strand concentration, once melted, the tetramolecular quadruplex does not refold. Absorbance at 295 nm is recorded every 6 min with a thermal gradient of ∼0.18°C/min in a 10 mM sodium cacodylate buffer (pH 7.0) supplemented with 0.1 M NaCl. Arrows indicate directions of temperature changes (heating or cooling). (C) r-(UG4U)4 (10 μM) in 0.11 M Na. Absorbance at 295 nm is recorded every 6 min with a thermal gradient of ∼0.18°C/min in a 10 mM sodium cacodylate buffer (pH 7.0) supplemented with 0.1 M NaCl. Arrows indicate directions of temperature changes (heating or cooling). (D) Fraction unfolded as a function of temperature for the DNA d-(TG4T)4, RNA and 2′-O-methyl r-(UG4U)4 samples, deduced from the heating profiles shown in Figure 1B (DNA) or 1C (RNA). (E) Arrhenius plot of ln(koff) versus 1/T for the DNA d-(TG4T)4, RNA and 2′-O-methyl r-(UG4U)4 samples. Calculated lifetimes of the complexes are shown on the right y-scale.
G-quartets may have applications in areas ranging from supramolecular chemistry to medicinal chemistry [for a recent review see (5)]. Several reports suggest that DNA may be used as a building block for novel nano-sized objects (6). Quadruplex DNA is an excellent module for the design of devices for nano-technology, because of its extreme rigidity and self-recognition properties. G-quadruplexes are also likely to form higher-order structures such as synapsable DNA (7,8) or G-wires (9,10). We and others have described DNA nano-devices based on quadruplex–duplex interconversion (11,12) or biosensors (13). The self-assembly of G-rich sequences can constitute liquid crystals (14) and may serve as the scaffold for artificial ion channels or receptors (15,16). Quadruplex-prone segments may also be found in biologically significant regions such as telomeres (17–21) or oncogene promoter regions (22), and a number of proteins or small molecules bind to G-quadruplexes (23–25).
In the tetramolecular quadruplex configuration (also called G4-DNA), all strands are parallel, and all guanines are in the anti conformation. These tetramolecular quadruplexes offer an interesting paradox: on the one hand, their conformation is very well known, and a number of high or very high [0.61 Å; (26)] resolution X-ray or NMR structures are available. This structural wealth might be explained in part by the extraordinary stiffness of this motif, as demonstrated by molecular dynamic simulations (27,28). On the other hand, one cannot but notice the paucity of thermodynamic and even more kinetic data [with one notable exception (29)] on these structures. This is rather unfortunate, as these quadruplexes may play important roles. First, several tetramolecular quadruplexes strongly interact with the HIV gp120 protein, and act as specific inhibitors of infection in vitro (30). Second, the conformation of the stem of these quadruplexes is very close to the central core of intramolecular parallel quadruplexes found for human telomeric repeats (20,31) or other motifs. These quadruplexes may be seen as simpler models of biologically relevant tetrads, and several groups have used such motifs to obtain high resolution data on drug–DNA interactions (32–34). All in vitro experiments would benefit from precise knowledge of the kinetics of these structures, in order to define acceptable incubation times, choice of temperature and buffer. Finally, the intracellular ionic environment, where potassium is present at high concentrations, favors tetramolecular parallel quadruplexes over foldback structures (35–37).
In this manuscript, we will demonstrate that the slow kinetics of association and dissociation of tetramolecular G-quadruplexes actually make equilibrium measurement impractical (29). We show here that, as for other nucleic acid structures, kon, but not koff, is strongly ionic strength dependent. General rules will be drawn up for various DNA and RNA sequence motifs, involving 3–6 consecutive guanines and a variable number of 5′ and/or 3′ protruding bases. This allows us to calculate the amount of time required for 50% (or 90%) quadruplex formation as a function of strand sequence and concentration (in the micromolar to millimolar range), temperature (between 2 and 37°C) and ionic strength.
MATERIALS AND METHODS
Oligonucleotides and nomenclature
The DNA and RNA oligonucleotides presented here were synthesized by Eurogentec (Seraing, Belgium) on the 200 or 1000 nmol scale and dissolved in 200–500 μl of double-distilled water. Once formed, some quadruplexes are difficult to disrupt and 30 min at 100°C did not lead to significant quadruplex unfolding. When undesired quadruplex formation is observed before a kinetic experiment, the addition of limited amounts of sodium hydroxide followed by HCl neutralization is a convenient and fast method to disrupt these complexes (38,39), but results in a net increase in salt concentration. Concentrations of all oligodeoxynucleotides were estimated by UV absorption using published sequence-dependent extinction coefficients (40) (Table 1).
Table 1.
Oligonucleotides chosen for this study
| Sequence (5′ → 3′) | G-tract length | εa | konb | T1/2c Na+ | T1/2d K+ | Reference(s) |
|---|---|---|---|---|---|---|
| DNA: d- | ||||||
| TGGGf | 3 | 39 200 | n.d. | n.d. | 51 | |
| TGGGT | 3 | 47 700 | 5.2 × 107 | 16 | 48 | (72,75,76) |
| TTAGGG | 3 | 61 300 | 1.8 × 107h | 17 | 50 | (77) |
| TTAGGGT | 3 | 69 800 | 2.1 × 106 | 24 | 55 | (77,78) |
| TGGGG | 4 | 49 300 | 2.8 × 109e | 47 | — | (46) |
| GGGGTf | 4 | 50 300 | e | 63 | — | |
| TTGGGG | 4 | 57 400 | n.d. | 48.5 | — | (77,79) |
| GGGGTTf | 4 | 58 400 | 1.5 × 109e | 64.5 | — | |
| TGGGGT | 4 | 57 800 | 3.8 × 108 | 54.5 | — | (49–56) |
| AGGGG | 4 | 55 300 | 3.9 × 1010e,f | 54 | — | |
| AGGGGT | 4 | 63 800 | 1.3 × 1010 | 59.5 | — | |
| TTGGGGT | 4 | 65 900 | 4.3 × 107 | n.d. | n.d. | (56,77,80) |
| TTGGGGTT | 4 | 74 000 | 1.5 × 106 | 71 | — | (29,56,64) |
| TTTGGGGT | 4 | 74 000 | 1.3 × 106 | 67 | n.d. | |
| TGGGGG | 5 | 59 400 | 3.1 × 1011 | — | — | |
| TTGGGGG | 5 | 67 500 | 2.2 × 1010 | — | — | |
| TGGGGGT | 5 | 67 900 | 6.1 × 109 | — | — | |
| TTGGGGGTT | 5 | 84 100 | 6.1 × 107h | — | — | |
| TGGGGGGTf | 6 | 78 000 | 1.4 × 1011 | — | — | |
| RNA: r- | ||||||
| UGGGU | 3 | 49 900 | n.d. | 50 | n.d. | |
| UGGGGU | 4 | 60 000 | 5.0 × 1012 | 89 | — | (81) |
| UGGGGUg | 4 | 60 000 | 2.0 × 1011 | 75 | — | |
| UUGGGGU | 4 | 69 700 | 5.0 × 1012 | >85 | n.d. | |
| UGGGGUUU | 4 | 79 400 | 2.1 × 1011 | 69 | n.d. | |
| UUUGGGGU | 4 | 79 400 | 6.3 × 1011 | >85 | n.d. | |
| UUGGGGUU | 4 | 79 400 | 2.0 × 1011 | 79 | n.d. | |
n.d.: not done. Reference oligomers (TG4T and UG4U) are shown in bold.
aExtinction coefficient, in M−1·cm−1, according to (40).
bAssociation rate constant at 4°C, pH 7, with 0.11 M Na+, in M−3·s−1. kon is given ±30%. Note that, as in Wyatt et al., we defined kon as d[A]/dt = −4·d[A4]/dt = −kon[A]4.
c(Non-equilibrium) melting temperature of the preformed quadruplex, in °C, in 0.11 M Na+, determined with a temperature gradient of 0.18°C/min. —: no melting of the quadruplex, even at the highest temperature recorded (94°C).
d(Non-equilibrium) melting temperature of the preformed quadruplex, in °C, in 0.11 M K+ determined with a temperature gradient of 0.18°C/min. —: no melting of the quadruplex, even at the highest temperature recorded (94°C). T1/2 is provided with a 0.5°C accuracy.
ePoor fit.
fAnomalous migration/several bands (for a simple tetramolecular quadruplex) on a non-denaturing gel; see Supplementary Figure S4 for details.
g2′-O-methyl.
hSingle point determination.
All oligonucleotides studied here contain only one block of guanines and form tetramolecular species. In this manuscript, the tetramolecular quadruplex resulting from the association of four d-TGGGGT strands will be called d-(TG4T)4.
Thermal denaturation and renaturation
Formation and dissociation of the different duplexes and quadruplexes were estimated by heating/cooling experiments, recording the UV absorbance at several wavelengths as a function of temperature on Kontron Uvikon 940 spectrophotometers thermostated with an external ThermoNeslab RTE111 or a ThermoHaake Phoenix C25P1 waterbath. The temperature of the bath was increased or decreased at a rate of 0.02–3.4°C/min. All temperatures were measured in a buffer-containing cuvette as significant differences between water bath and sample temperatures may be observed. Hence, the temperature values reported here reflect the real temperature of the sample, even when fast heating/cooling experiments are performed. All experiments were carried out in 10 mM sodium cacodylate or lithium cacodylate buffer (pH 7.0 or 7.2) containing 50–400 mM NaCl or KCl. The thermal denaturation of the G-quadruplex gives rise to hyperchromism at 240–245 nm and to a large hypochromism at 295 nm (41). Following absorbance at 240 or 295 nm is therefore a convenient way to monitor quadruplex dissociation or formation. Depending on strand concentration, 0.2 or 1 cm pathlength cuvettes were used. In order to convert absorbance values into unfolded fraction, linear baselines were manually chosen, as previously described (42). For tetramolecular quadruplexes, this conversion was simple, especially for the low temperature baseline, as the absorbance of the folded species was nearly temperature independent (see Figure 1B and C).
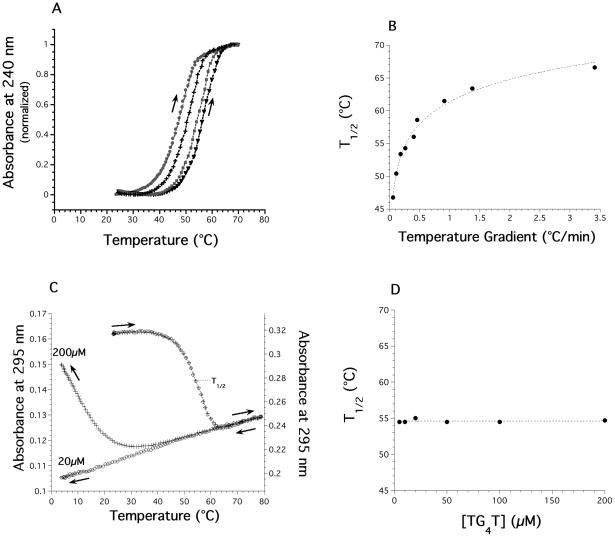
A prerequisite for the recovery of thermodynamic, that is equilibrium, parameters from these curves is that they are true equilibrium curves. A simple and useful criterion for this is the coincidence of the heating and cooling curves at the chosen rate of heating and cooling. This is generally the case for double-stranded helix–coil transitions of oligodeoxynucleotides. We have previously observed that a rapid increase in temperature may lead to experimental curves that do not correspond to the equilibrium curves (43,44). Most melting curves recorded by heating a preformed quadruplex do not correspond to equilibrium melting curves; the ‘Tm’ deduced from these experiments (which depends on the heating rate) is therefore inaccurate. It is still informative, though, as it reflects the temperature dependency of the dissociation process. In order to distinguish it from the true thermodynamic Tm, we will call this value T1/2. For the tetramolecular quadruplex structures studied here, the hysteresis phenomenon is extreme: no renaturation is observed for the DNA sample at low strand concentration (Figure 1B) whereas a 60°C difference may be found between the ‘Tm’ obtained during heating and cooling of a RNA quadruplex (Figure 1C). The apparent melting temperature (T1/2) was found to be strongly dependent on the temperature gradient chosen for the experiment (i.e. the rate of heating; see Figure 2A and B for an example).
Figure 2.
T1/2 is dependent on the temperature gradient, not on strand concentration. (A) Normalized melting profiles recorded at 240 nm of the preformed d-(TG4T)4 quadruplex (20 μM strand concentration) in 10 mM lithium cacodylate buffer (pH 7.2) with 0.11 M NaCl at four different heating rates (circles: 0.054; crosses: 0.107; squares: 0.26 and triangles 0.4°C/min). (B) T1/2 of the d-(TG4T)4 quadruplex as a function of the average temperature gradient. Note that the ‘true’ equilibrium Tm, which would be obtained with an infinitely slow gradient is calculated to be below 30°C. T1/2 is provided with a 0.5°C accuracy. (C) Melting and cooling profiles recorded at 295 nm of the preformed d-(TG4T)4 quadruplex in 10 mM lithium cacodylate buffer (pH 7.2) with 0.11 M NaCl at two different strand concentrations (20 μM: circles and 200 μM: squares. y-scale values shown on the left and right axis, respectively. 1 cm pathway cuvette for 20 μM, 0.2 cm for 200 μM). Thermal gradient: 0.24°C/min. (D) Dependence of the T1/2 value (obtained upon heating at a rate of 0.24°C/min) of the preformed d(TG4T)4 quadruplex (5–200 μM strand concentration) in 10 mM lithium cacodylate buffer (pH 7.2) with 0.11 M NaCl.
Isothermal kinetic analysis for association
Isothermal experiments were performed on a Kontron Uvikon 940 UV/Vis spectrophotometer. In general, association experiments were carried out at various strand concentrations (up to 500 μM), depending on the kinetics. In order to fit the experimental data (using Kaleidagraph 3.5) with a proper mathematical model, we had to make the following assumptions or determinations:
A two-state analysis is required. Our data were analyzed within the framework of a concerted ‘two-state’ or ‘all-or-none’ model of dissociation. In this model, it is assumed that species with incomplete base pairing are not significantly populated, so that the only absorbing species are the separated strands and the full quadruplex. Note that this model does not necessarily imply a simple kinetic process. A useful criterion was recently proposed by Wallimann et al. (45): a dual wavelength parametric test allowed us to verify the two state model for various experiments (Supplementary Figure S3).
The order (n) of the reaction in separate strands was initially floating (the kinetic order, the stoichiometry and the molecularity of the reaction may be different). For all further analysis, n was fixed equal to 4.
Denaturation may be neglected: once formed, the quadruplex lifetime is long compared to the duration of the experiment (1–3 days).
For each set of experimental conditions (temperature, buffer) experimental curves were obtained at several strand concentrations. However, because of the extreme dependence of reaction rate on oligonucleotide concentration, the useful concentration range for kinetic analysis is often limited. Assuming that unfolding is negligible, the association of four identical A strands leads to the formation of the tetramer A4: 4A → A4. At each wavelength, the absorbance Abs of the sample is linked to the fraction unfolded α by:
| 1 |
where εss and εq are the extinction coefficients of the unstructured single-strand and of the oligonucleotide involved in a quadruplex, respectively. α is the fraction of unfolded strand [α = [A](t)/C0]. The order of the reaction n may be experimentally estimated by analyzing the concentration dependency of the association process d[A]/dt = −kon [A]n. Assuming that at t = 0; α = 1, one can demonstrate that (29)
| 2 |
where C0 is the initial strand concentration and kon the association rate constant. Experimental curves were fitted with this formula using the Kaleidagraph 3.5 software.
In most cases, this equation gave excellent fits, with parameters independent of the wavelength chosen for analysis (usually 240 or 295 nm, sometimes 230, 260 and/or 273 nm; Supplementary Figure S2a). Therefore, the observed reactions seem to obey a simple kinetic pathway and there is no evidence that significant concentrations of intermediate species are present. Nevertheless, one should note that oligomers ending with a terminal 3′ guanine are more likely to form higher-order molecular species. The time-dependent association of a few oligonucleotides, such as d-G4T was much more complicated (Figure S2c). In our hands, profiles obtained with some guanine-rich sequences such as TG3–5 or G3–5T at various wavelengths do not always agree, as pointed out with CD by Lieberman and Hardin (46). This does not necessarily imply a measurable buildup of intermediate species, but could also be the result of the presence of several distinct tetra- or higher molecular end products having different kinetics (47,48). It should be noted that there is no need and no theoretical reason to fit these profiles with bi-exponential forms. In any case, results obtained with oligomers that do not involve guanine as a terminal 5′ or 3′ base (such as d-TG4T for example) are usually simpler.
Kinetic analysis for dissociation
The analysis of non-equilibrium melting profiles is based on the methods we developed for triplexes (43) and i-DNA (44). In our case, the calculations are even simpler as re-association was not measurable in the temperature range where melting occurs (kon ≈ 0). As a result, rather than solving at each temperature a system of two equations with two unknowns (koff and kon), one has to deal with a single equation and one unknown parameter (koff). As reformation of the complex is negligible, d[A]/dt = koff·[A4], hence
| 3 |
where α is the fraction of unfolded strand and koff the rate constant for appearance of single strands. koff may simply be deduced from Equation 3 at each temperature T where d[α(T)]/dt (the time-dependent variation of the unfolded fraction at a given temperature) and α may be determined with some confidence (generally 0.05 < α < 0.95).
RESULTS
All quadruplex-forming oligonucleotides studied here are presented in Table 1. For most sequences, quadruplex formation has been demonstrated previously, and relevant references are provided. Our goal was not to re-demonstrate quadruplex formation for these oligomers; nevertheless, one should note that the thermal difference spectra of all these structures (Supplementary Figure S1) are in agreement with the formation of quadruplexes (41). Furthermore, non-denaturing gel electrophoresis confirmed that, at least for some sequences, a single major retarded band was observed when the oligonucleotides were preincubated at high concentration in the presence of NaCl or KCl (Supplementary Figure S4). Long G-runs such as TG6T sometimes led to more complicated migration patterns. We aimed to obtain reliable thermodynamic and kinetic data on these structures. We initially concentrated our efforts on well-known tetramolecular quadruplexes d-(TG4T)4 (49–56) and r-(UG4U)4 formed by the tetramerization of 6-base long oligonucleotides before analyzing other DNA and RNA guanine-rich oligomers.
Dissociation of the preformed d-(TG4T)4 and r-(UG4U)4 quadruplexes
Hybridization to a complementary oligonucleotide is a convenient method to study the dissociation process of a quadruplex (31,57–59). Unfortunately, in this work, most sequences were too short to allow the formation of a stable DNA duplex, making this method inapplicable. The other method commonly used relies on concentration jumps (31,60), but the thermal inertia of preformed tetramolecular quadruplexes is so high that lifetimes of the complexes are very long (30) (see below). We therefore had to use melting experiments to study the dissociation process.
Starting from preformed quadruplexes (several days at 0°C and high strand concentration: 200–500 μM), one can simply follow the denaturation of this structure by recording the absorbance at 295 nm (41) in a 0.11 M Na+ buffer. This leads to a ‘nice’ inverted and ‘cooperative’ curve (Figure 1B), which resembles data collected by CD experiments. As shown in Figure 1B, this profile does not reflect an equilibrium denaturation curve. Upon cooling, no renaturation of the DNA quadruplex is obtained, and further heating/cooling cycles led to a similar monotonous variation of absorbance with no evidence for quadruplex reformation or denaturation. On the other hand, one should note that partial renaturation of the RNA quadruplex is observed below 10°C (Figure 1C). Furthermore, this apparent melting temperature strongly depends on the rate of heating (Figure 2A and B) again indicating that this profile does not correspond to an equilibrium curve. It should be noted that if sodium is replaced by potassium, no melting of this quadruplex is observed, in agreement with a previous report (55).
This apparently annoying behavior has an interesting consequence: the melting profile actually solely reflects the dissociation of the quadruplex and it is possible to extract koff values at each temperature. In this temperature range, what we see in practice is a simple one-way reaction from a folded quadruplex to a dissociated state. As the reverse reaction (refolding) is not considered here, the only species for which concentration matters is the folded tetramer. This requires several assumptions or verifications. (i) Renaturation at this temperature has to be minimal. This was checked by using different strand concentrations (from 5 to 200 μM): no variation at all of the T1/2 was observed, in agreement with the complete absence of renaturation in that temperature range (Figure 2C and D). Another indication of negligible reassociation is provided by following the second heating profiles: starting from completely unfolded species, one does not observe any trace of reassociation (data not shown). (ii) koff values extracted from a melting curve had to be confirmed with another technique: we and others performed T-jump experiments in which a preformed quadruplex (at low temperature) is suddenly transferred to high temperature, and the time-course of isothermal quadruplex dissociation is recorded (29,60). Values obtained through this method were, within experimental error, in agreement with the koff values deduced from melting experiments (data not shown). (iii) The melting temperature depends on the temperature gradient and knowing Eoff (see below), it is even possible to predict the dependence of T1/2 on the heating rate (29,60).
In the temperature range in which the dissociation occurs (35–60°C for the DNA quadruplex; 75–90°C for the RNA quadruplex) one can determine α, the fraction of unfolded oligonucleotide (Figure 1D) and d(α)/dt, the time-dependent variation of the unfolded fraction. From Equation 3 (see Materials and Methods) one can write:
| 4 |
The temperature range for which d(α(T))/dt may be accurately determined is relatively narrow (the melting transition is spread over a limited temperature range). Hence, the Arrhenius plots of ln[koff(T)] as a function of temperature (presented in Figure 1E) involve a relatively limited number of points, especially for RNA. Note that this temperature range may be extended by using different temperature gradients which lead to different apparent melting temperatures T1/2 (Figure 2A and B). For d-(TG4T)4, this apparent melting temperature variation (between 46 and 66°C, Figure 2B) extends the practical range for d(α(T))/dt determination to ≈36–76°C (data not shown). This ‘trick’ is not applicable to the RNA sample: faster scan rates lead to partial denaturation only (T1/2 too high), whereas slower scans lead to unacceptable evaporation and degradation of the sample. Therefore, Eoff values deduced from the slope of Arrhenius plots of ln[koff(T)] as a function of temperature (+43 and +74 kcal/mol for DNA and RNA, respectively) are determined with a much greater confidence for DNA than for RNA.
Association of the isolated strands at low temperature: effects of concentration
Isothermal renaturation experiments were used to study the formation of the quadruplexes. We first determined that the low temperature renaturation was minimal for the DNA sample at concentrations <20 μM (Figure 2C). Starting with a concentration of 30 μM or higher, a time-dependent increase of absorbance at 295 nm was observed (Figure 3A). An opposite trend was seen at 240 nm, with a time-dependent decrease of absorbance. These variations reflect the spectral differences between the initial single-strand and the quadruplex (Supplementary Figure S1).
Figure 3.
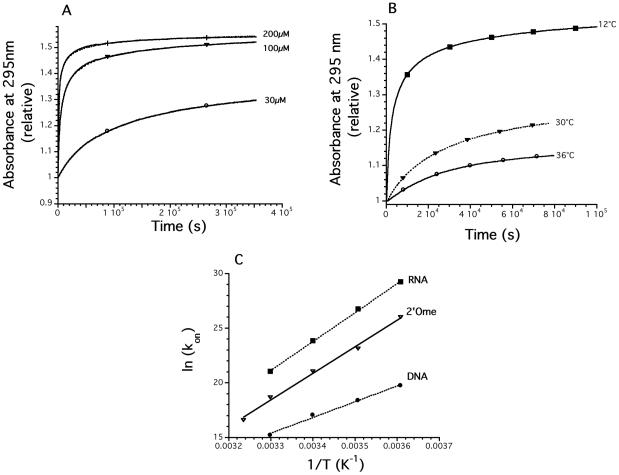
Kinetics of association. (A) Effect of strand concentration (30–200 μM) for d-TG4T at 3°C in 0.11 M Na+ on the relative absorbance at 295 nm. Quadruplex formation leads to an increase of absorbance at this wavelength (see Figure S1). Mathematical fits (Equation 2) are shown as dotted lines; data points are displayed as solid lines (each curve results from the recording of 900 experimental points; only a few are shown). (B) Effect of temperature for d-TG4T (178 μM) on the relative absorbance at 295 nm. Circles: 36°C, triangles: 30°C, squares: 12°C (each curve results from the recording of 300 experimental points; only a few are shown). (C) Arrhenius plots [ln(kon) versus 1/T] for the association of the DNA d-TG4T (circles), RNA (squares) and 2′-O-methyl (triangles) r-UG4U oligonucleotides. All experiments were performed in a 10 mM sodium cacodylate buffer (pH 7.0) supplemented with 0.1 M NaCl.
Concentration played a dramatic role in the kinetics of association. Formation of the quadruplex at 3°C was nearly complete in 2 h at 200 μM whereas much longer times are required at 30 μM (Figure 3A). It was then possible to fit the experimental profiles with the mathematical function defined in the experimental section (Equation 2) for the determination of the order of the reaction. Data could be fitted with 3.4 ≤ n ≤ 4.1; we defined n = 4 for all further studies. As illustrated in Figure 3A, these fits (dotted lines) are in nearly perfect agreement with the experimental curve (solid line; the overlap between the two precludes seeing the mathematical fit over most of the curve. This agreement is also reflected by R > 0.995 and χ2 as low as 10−4 in most cases). Moreover, the kon values determined from the curves at three different concentrations or at two different wavelengths (240 and 295 nm; see Supplementary Figure S2A for an example) were in excellent agreement [see Supplementary Figure S3 for a dual wavelength parametric test (45)]. The association rate constant at 4°C was 3.75 × 108 and 5.0 × 1012 M−3·s−1 for the DNA and RNA samples, respectively (see Table 1). The value found for RNA (>1012) may look extremely high when compared with bimolecular rate constants (in the 105 to 106 M−1·s−1 range for duplexes); one should note, however, that these quadruplex rate constants reflect fourth-order reactions and are expressed in different units (M−3·s−1) which prevents a direct comparison of the numerical values. These values allowed us to calculate the amount of time required for half formation of the DNA, 2′Ome and RNA quadruplex at various strand concentrations (Table 2).
Table 2.
| Oligonucleotide/concentration | 1 μM | 10 μM | 100 μM | 1 mM |
|---|---|---|---|---|
| d-TG4T (DNA) | >100 years | 110 days | 2.6 h | 6 s |
| o-UG4U (2′Ome) | 120 days | 2.9 h | 11 s | 10−2·s |
| r-UG4U (RNA) | 3 days | 4.4 min | 0.25 s | <10−3·s |
aCalculated from Equation 2. To determine the time required for 90% quadruplex formation, multiply by 143. For 99% quadruplex formation, multiply by 143 000!
bCalculated at 4°C in a 0.1 M NaCl, 10 mM sodium cacodylate pH 7.0 buffer (total Na+ concentration 0.11 M). At 21°C, the reaction will take 15 times longer (for DNA) and 150–200 times longer for RNA and 2′Ome. In 0.11 M KCl (instead of NaCl) divide these durations by ≈50. For sequences other than dTGGGGT, multiply these numbers by the ratio between kon (d-TG4T) and kon of the chosen sequence (values found in Table 1) (higher kon means faster association!).
Next, we performed the same renaturation experiment at various temperatures. As shown in Figure 3B, an increase in temperature has a deleterious impact on the kinetics of association: at a given strand concentration, folding was much slower at 36°C (circles) than at 12°C (squares). Similar experiments were done for the RNA and 2′Ome samples at various temperatures in the 2–37°C range. The association rates are shown in Figure 3C (Arrhenius plot: natural logarithm of kon as a function of 1/T). At low temperature (3°C), the RNA sample folded 104 times faster than DNA (note the logarithmic scale on the y-axis). At 30°C, this difference was less pronounced: for the RNA UG4U sample, the association process was even more temperature-dependent: a 10°C increase led to a >20 times lower kon.
Thermodynamics of the d-(TG4T)4 and r-(UG4U)4 quadruplexes
From the Arrhenius representations of association (Figure 3C) and dissociation (Figure 1E) processes, it is possible, at least in theory, to recalculate the equilibrium constant at every temperature. However, one should note that the experimental points are not experimentally determined in the same temperature range (2–37°C for association, 37–90°C for dissociation): an extrapolation of these data is therefore required. Fortunately, a reasonable linear fit may be obtained for these points: their slopes allow the determination of the association and dissociation activation energies (Eon and Eoff). The ΔH° of the reaction may be deduced from the relation ΔH° = Eon − Eoff (Table 3). The values found for the DNA and RNA quadruplexes were −72 and −127 kcal/mol, suggesting that the RNA quadruplex is significantly more enthalpy driven. The value for DNA is in close agreement with calorimetric data for the same quadruplex under slightly different conditions (0.2 M NaCl) (54). Note, however, that the values found for RNA are less precise, as Eoff is difficult to measure accurately.
Table 3.
Kinetic parameters for the d(TG4T)4 and r(UG4U)4 quadruplexes
| Oligonucleotide | Eona | Eoffa | ΔH°a,b | T1/2c | t1/2d |
|---|---|---|---|---|---|
| d-TG4T (DNA) | −29 ± 2 | 43 ± 1 | −72 ± 3 | 54.5 | 6 h |
| o-UG4U (2′Ome) | −49 ± 2 | 75 ± 2 | −124 ± 4 | 75 | 3 years |
| r-UG4U (RNA) | −53 ± 4 | 74 ± 7 | −127 ± 11 | 89 | >100 years |
All values determined in a 0.1 M NaCl, 10 mM sodium cacodylate pH 7.0 buffer (total Na+ concentration 0.11 M).
aIn kcal/mol.
bDeduced from ΔH° = Eon − Eoff.
cIn °C, determined with a temperature gradient of 0.2°C/min. T1/2 is provided with a 0.5°C accuracy. The true Tm value (in °C), calculated at 100 μM strand concentration is at least 20°C lower (data not shown).
dLifetime of the tetramolecular quadruplex at 37°C.
Buffer effects on the association and dissociation of the d-(TG4T)4 quadruplex
All profiles shown before were obtained in a buffer containing Na+ ions at 110 mM. Next, we investigated the effects of varying sodium concentration, replacing sodium by potassium, changing the pH or adding divalent or multivalent cations.
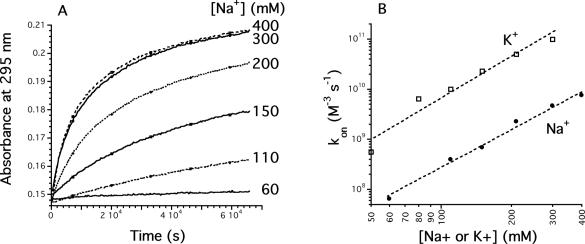
First, sodium was replaced by potassium. As previously described, K+ stabilizes quadruplexes, as demonstrated by an increase in melting temperature, which reflects a slower dissociation at a given temperature. However, potassium also increases the association rate constant by a factor of 20–50 (data not shown). In other words, potassium affects kon and koff. This difference in kon between sodium and potassium was more or less conserved over a 2–37°C temperature range, suggesting that the activation energies of association were close. The 50-fold difference in kon between Na+ and K+ was more or less constant in the 50–300 mM NaCl or KCl concentration range (Figure 4B).
Figure 4.
Effect of NaCl concentration. (A) Examples of kinetics of association at 3.5°C at various NaCl concentrations (shown on the right in a 10 mM cacodylate buffer). Absorbance recorded at 295 nm as a function of time, starting from an unfolded d-TG4T oligodeoxynucleotide (25 μM). Quadruplex formation leads to a time-dependent increase of absorbance at this wavelength. (B) kon values determined as a function of NaCl (circles) or KCl (squares) concentration.
Second, various concentrations of sodium and potassium were tested in the 50–400 mM range. Increasing NaCl concentration played little, if any role in the thermal dissociation of the d-(TG4T)4 quadruplex (ΔT1/2 < 2°C; data not shown). However, varying the sodium concentration had a dramatic effect on the association process, as shown in Figure 4A. This illustrates the fact that the stability of parallel quadruplexes is indeed dependent on the cation concentration, and that this effect is mainly reflected in kon, as for many other nucleic acid structures such as duplexes or triplexes. A 10-fold increase in Na+ concentration leads to a 1 × 103 to 2 × 103 increase in the association rate constant, in agreement with the involvement of several (≈3) Na+ or K+ ions in the association process.
Third, pH was varied between 5.0 and 7.8. No variation of association and of thermal stability was found between pH 5.5 and 7.8 (Supplementary figures S5A and S5B). However, the association rate was significantly faster at pH 5.0. This stabilizing effect (on association) is compensated by a destabilizing effect on the denaturation process (evidenced by a 4°C decrease in T1/2). These mildly acidic conditions have a paradoxical role on the stability of the quadruplexes, as a low pH both accelerates association and dissociation.
Fourth, magnesium chloride was added to a sodium- or potassium-containing buffer. The results are shown in Supplementary Figure S6A for the association rate at 4°C. Magnesium ions accelerate the association process in a concentration-dependent manner. This effect was observed both in 0.11 M Na+ and 0.11 M K+. At the highest tested Mg2+ concentration (15 mM), the association rate constant was six times larger than in the absence of magnesium. We next investigated whether magnesium affected the melting temperature (T1/2) of the preformed quadruplex. As shown in Supplementary Figures S6B–S6C, increasing magnesium concentration led to a decrease of the apparent melting temperature, hence, led to an increase of the dissociation process.
Finally, multivalent cations (spermine and spermidine, in the 0.04–0.2 mM concentration range) were added to a 10 mM sodium cacodylate buffer at pH 7.2 containing 0.1 M NaCl (data not shown). Results were in qualitative agreement with the effects of magnesium (i.e. a net acceleration of association and dissociation at 0.2 mM spermine or spermidine).
Sequence effects
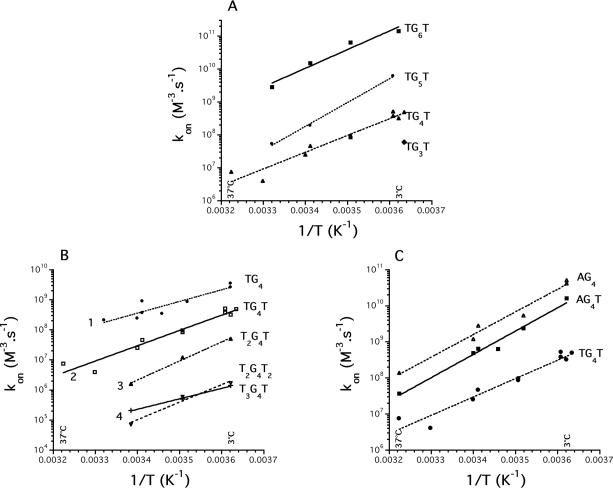
So far, all results were obtained with the d-(TG4T)4 quadruplex or its RNA equivalent. We next investigated if the results obtained for this sequence could be extended to other motifs. The sequences used for this analysis are shown in Table 1, and some of the results are summarized in Figure 5. For most oligomers, the association process could well be fitted with the same mathematical models.
Figure 5.
Sequence effects on kon. (A) Impact of G-tract length on association rate constant. Squares: d-TG6T; circles: d-TG5T; triangles: d-TG4T; diamonds: d-TG3T (single determination at 3°C). (B) Impact of non-guanine bases on association. Total number of 5′ and 3′ thymines shown on the left. Circles d-TG4; squares: d-TG4T; triangles: d-T2G4T; crosses: d-T3G4T; inverted triangles: d-T2G4T2. (C) Influence of a 5′ terminal adenine on association. Sequence shown on the right. All experiments performed in a pH 7.0 or 7.2 10 mM sodium cacodylate buffer containing 100 mM NaCl between 2 and 37°C (total Na+ concentration: 110 mM).
First, concerning DNA oligomers, one can observe that the longer the G-tract, the faster the association (Figure 5A). This effect is surprisingly large: each extra guanine leads to ∼10-fold larger association rate constant. As expected, a stabilizing effect could also be seen on dissociation: the longer the G-tract, the higher the T1/2. Unfortunately, this effect was harder to quantify as many quadruplexes did not dissociate at 90°C (Table 1).
Second, one can also notice that extra non G-bases at the 5′ or 3′ end play a detrimental role in the association of DNA and RNA (Figure 5B and Table 1). The addition of one thymine/uracil to a DNA or RNA oligonucleotide decreased the association rate constant by a factor of roughly 10. This effect was cumulative; the slowest forming quadruplexes involved strands containing four thymines. The effect on koff was more variable. As shown in Table 1, the presence of extra 5′ or 3′ thymines was often stabilizing (compare TTAGGG and TTAGGGT, ΔT1/2 = +5°C in K+, +7°C in Na+ or TGGGGT and TTTGGGGT, ΔT1/2 = +12.5°C in Na+). A counter-example is provided by TGGG and TGGGT, ΔT1/2 = −3°C in K+. In other words, the presence of extra thymines was always detrimental to the association rate constant but often beneficial to the thermal stability of the quadruplex.
Third, the replacement of a 5′ thymine by adenine had a strong beneficial effect on the kinetics of association (Figure 5C). One may observe that d-AG4T has a 5–40 times higher association rate constant than d-TG4T: the presence of a terminal 5′ adenine is much more favorable than a 5′ thymine, especially at 4°C. This is the first direct evidence of the impact of the nature of a 5′ terminal base on the kinetics of quadruplex formation.
Generally, association processes for all these sequences were temperature dependent, with a large negative energy of activation of association (Eon). Plots of kon versus temperature for a variety of sequences are shown in Figure 5 and as Supplementary Material (Supplementary Figures S7A–H). Association was studied in 0.11 M Na+ or K+. For most sequences, and whatever the temperature studied, faster association was obtained in potassium. The ratio varied between 1 (for d-TTAGGG, Supplementary Figure S7H) and 400 (d-TGGGGGT at 4°C, Supplementary Figure S7B). The energies of activation of association (Eon) were negative in sodium and potassium (and in some cases very close, see Figure S7A for d-TTGGGGG).
DISCUSSION
In the present study, absorbance spectroscopy was used to measure kinetics of association and dissociation. In agreement with the seminal paper by Wyatt et al. (29), who used size exclusion chromatography to study this process, we found that dissociation of quadruplexes appeared first order and association fourth order with respect to strand concentration. We actually took advantage of the unusual concentration dependency of the association process to measure association rate constants that differed by 105 or more: a 100-fold increase in concentration (from 5 to 500 μM in this manuscript) allowed us to analyze processes with a 106 slower association rate constant. Most experiments were performed on the d-(TG4T)4 DNA quadruplex and its corresponding RNA tetraplex r-(UG4U)4. These oligomers adopt relatively similar folded conformations: the four strands are parallel, all nucleotides are in the anti conformation and both of these structures exhibit right-handed helicity. Differences may be found in the organization of the thymine/uracil and in sugar pucker: in the DNA and RNA motifs, the sugar conformations are mostly of the S-type and mixed type, respectively (61).
Temperature effects
We took advantage of the large hysteresis or quasi-irreversible melting of these complexes to study the association and dissociation processes independently. This difference between heating and cooling (and between the first and second heating) should lead to great caution concerning conclusions reached solely by the likening of the heating curves to thermodynamic equilibrium curves. For most oligonucleotides, it was possible to find the following:
A temperature range for which association could be studied with negligible dissociation (once formed, the structure was so inert that no dissociation occurred in the time range of the experiment, making the association process a quasi-irreversible reaction).
A (higher) temperature range where association could be neglected, and stability of the complexes was sufficiently lowered to observe their dissociation. Again, denaturation of the complex at these high temperatures could be considered as an irreversible process.
This uncoupling between the association and dissociation processes allowed easy determination of kon and koff. Unfortunately, this uncoupling necessarily means that kon and koff values were not obtained at the same temperature. The calculation of the equilibrium constant required the determination of kon and koff at several temperatures and an extrapolation (which in turn required a precise measurement of the association and dissociation activation energies Eon and Eoff) to the desired temperature range. In the temperature range delimited in Figures 1E and 3C where these values are obtained with some confidence, straight lines are obtained with a positive slope for kon and a negative one for koff. In other words, the activation energy Eon for the association is negative and the activation energy Eoff for the dissociation is positive.
Once formed, the structure is extremely inert, as the ring protons of the guanines involved in central quartets require days to exchange even at 60°C (62). This is consistent with the exceptional thermal stability of this structure, which is still partially formed at 95–100°C in the presence of potassium (36,63). In our hands, it was difficult or impossible to observe the melting of a pre-formed DNA quadruplex that involves four or more guanines (in 0.11 M K+) or five or more guanines (in 0.11 M Na+). Hence, data on koff could only be collected for a subset of the sequences shown in Table 1, corresponding to oligomers containing relatively short runs of guanines. For these oligomers, it was clear that (i) stability was strongly temperature dependent; the activation energy for dissociation Eoff was strongly positive, (ii) at a given temperature koff was smaller in potassium than in sodium, (iii) at physiological temperature or lower, the time for half dissociation of these complexes was generally long (days or more), (iv) a decrease in NaCl concentration had little effect on G4 dissociation but strongly decreased the rate constant of association hence, the equilibrium constant.
Concentration effects
For the T2G4T2 sequence, Shida et al. noted that the transformation of the single-stranded form into the quadruple-stranded form was undetectable at 43 μM strand concentration at room temperature. However, at a 50-fold higher strand concentration, the single-stranded oligomer was gradually converted into the quadruplex (64). The rates of association of the DNA, RNA and 2′Ome oligomers depend strongly on oligonucleotide strand concentration (see Figures 2C and 3A): fourth-order reactions are not common in biochemistry, and the practical consequences of this reaction order are important. At some concentrations (below 30 μM for d-TG4T) no detectable association may be measured over a 3-day period. In contrast, at slightly higher concentrations, the association is so rapid that a majority of the oligonucleotide is already folded when the first measurement is performed. This is illustrated in Table 2, where the times required for 50% quadruplex formation is indicated for the DNA, RNA and 2′Ome oligomers.
Mechanism for quadruplex association
Concerning association, two important results were found: (i) the association process is fourth order in monomer; (ii) the activation energy of association is negative. These observations were valid for a majority of different DNA and RNA sequences under a variety of experimental conditions.
A fourth-order reaction does not imply that an elementary kinetic step involves a four-body collision. Such mechanism is extremely unlikely and other processes could lead to this fourth order. For example, our data are in full agreement with the pathway proposed by Wyatt et al. (29), where single strands and dimers are in a rapid pre-equilibrium that favors single strands, and tetramer formation from dimers is rate limiting. The structure of this elusive dimer intermediate remains unknown: Stefl et al. have recently demonstrated that a Hoogsteen G–G duplex is an improbable intermediate (28). Its identification will be experimentally difficult, as numerical simulations indicate that it may not be present at detectable levels (29).
Concerning the activation energy of association, Pörschke and Eigen (65) obtained negative ‘apparent’ activation energies for duplex formation and interpreted their results within the now so-called nucleation-zipping model. Our data is consistent with the observed fourth-order dependence of the association rate and with the negative energy of activation for association, which implies a rapid pre-equilibrium step (29). The negative values we have obtained for Eon are in favor of the nucleation-zipping model. Our finding that kon strongly decreases upon reducing ionic strength whereas koff is independent of NaCl concentration is also in good agreement with what is observed in the duplex case. Other nucleic acid structures such as duplexes (65,66) triplexes (43) and i-DNA (44) also exhibit a negative Eon. The observation that ionic strength and nature of the monocation plays an important role in the value of kon indicates that several ions are involved in this rate-limiting step and participate in the early stages of the quadruplex stem assembly (28): kon strongly decreases upon reducing the NaCl concentration but koff is independent of ionic strength.
In most (but not all cases, see d-TGn and d-GnT), there is no clear evidence for complicated folding pathways proceeding through stable intermediate states, as observed by Hardin et al. (46,67). Nevertheless, the order of the reaction n, when left unknown in Equation 2 is often experimentally found between 3.4 and 4 (i.e. lower than 4; data not shown); this could be the signature of small amounts of stable intermediates. Finally, one may note that low pH, magnesium, spermine and spermidine seem to act as catalysts for quadruplex association and dissociation. These cations lead to an increased rate of both association and dissociation; it is difficult to determine whether these effects cancel each other out, in other words if the equilibrium constant is unaffected.
DNA versus RNA and other nucleic acid analogs
It is important to emphasize the extreme stability of parallel RNA quadruplexes. r-UG4U forms a much more stable quadruplex than its DNA counterpart (d-TG4T). This increased stability results from a faster association (350 times faster at 30°C) and a slower dissociation (at least 1000 times slower at 60°C). Besides a larger number of sugars in the C3′-endo conformation in the RNA quadruplex, the main structural difference between the two structures is the organization of the uracyl/thymine groups (61). This is in contrast with phosphorothioate (PS) oligomers, which exhibited a slower association and faster dissociation as compared to DNA (29). Data collected from this study, Wyatt et al. (29) and unpublished observations from our group (B. Saccà and J.L. Mergny, in preparation) allow oligomers to be ranked in the following order of thermodynamic stability:
Besides methylphosphonate oligonucleotides (MP) which are, in our hands, totally unable to form tetramolecular quadruplexes, it should be noted that all other modifications (RNA, DNA, 2′O-methyl and phosphorothioates) exhibit a fourth-order dependence of the association rate, a negative energy of activation for association and a strongly positive energy of activation for dissociation. The extremely high stability of RNA parallel quadruplexes was unexpected: bi- and intra-molecular RNA quadruplexes are less stable than their DNA counterparts in sodium (B. Saccà and J.L. Mergny, in preparation). This difference might be explained by the greater difficulty in adopting a syn conformation for the ribonucleoside residues, making the formation of quadruplexes involving an antiparallel strand alignment unfavorable (68).
This suggests that the kinetic model initially proposed for DNA and phosphorothioates (29) could apply to a variety of nucleic acid modifications. In particular, close inspection of the melting profile of the ‘Locked Nucleic Acid’ (LNA) parallel quadruplex (69) suggests that these locked nucleic acids could also obey to the same kinetic rules. LNA could actually induce a change from an antiparallel to parallel structure (70). On the other hand, the question remains open for ‘Peptide Nucleic Acids’ (PNA) quadruplexes (71,72): the PNA TG3 sequence exhibited a surprising independence of the presence/nature of the monocation (72). It should be noted that, contrary to most nucleic acids discussed above, interbase linkers in PNA are not charged, and that the global charge of these PNA at neutral pH is often positive because of an N- or C-terminal lysine group.
Sequence effects
Oligomers that have a single multi-guanine motif at their 3′ or 5′ end, with a guanine as the terminal base, also form higher-order products (47,48,63,73), which may sometimes be evidenced by an anomalous migration on a non-denaturing gel (Supplementary Figure S4). This is in agreement with the observation that mathematical fits were often of lower quality for GnTm sequences than for TGnTm (e.g. in Supplementary Figure S2C). Previous work indicated that the additional T at the 5′ or 3′ end not only inhibited formation of higher-order structures, but also stabilized the complex (63). As demonstrated here, this stabilization is reflected by a lower dissociation rate constant; on the other hand, this extra base has a strong detrimental effect on the association rate of the quadruplex. The net effect on the thermodynamic constant Kd = koff/kon may be detrimental. On the other hand, extra 5′ thymines are always detrimental, not only because the T1/2 is decreased (74), but also because the association rate constant is negatively affected. In that case, the effects on kon and koff are in the same direction. We would like to point out that an increase in T1/2 does not always imply a higher thermodynamic stability: it rather indicates a slower dissociation process. One should compare the association and dissociation processes of these modified quadruplexes to conclude that such modifications provide a net gain in free energy.
Conclusion and perspectives
Our data enable us to propose a quantification of the kinetic inertia of tetramolecular quadruplexes. We have summarized the impact of a number of parameters on the kinetics of association and dissociation of these complexes (Table 4). This might be helpful for a variety of applications. One may for instance study the interaction of small ligands or proteins with quadruplexes: are these molecules able to accelerate the kinetics of association of monomers giving the tetrameric form, or do they only bind to the structured quadruplex, lowering its dissociation rate constant? What happens with helicases of the RecQ family such as Werner or Bloom's? Knowledge of the kinetics of folding should also be useful for structural studies (X-ray or NMR) where high concentrations are often chosen: as shown in Table 2, millimolar strand concentrations lead to fast processes: tetrameric quadruplex formation is not necessarily a long timescale event with complicated kinetics. Finally, one should emphasize the extraordinary stability of RNA quadruplexes, coupled with their relatively fast kinetics of association. Runs of guanines are frequent in biological RNAs: or pose a specific problem, as cellular or viral machineries (such as reverse transcriptases) cannot displace these structures. Our results suggest that the presence of a few extra bases delays association: what are the effects of a much longer overhang, e.g. in the context of a natural RNA? This will be a crucial question for mRNA or genomic DNA, but also for antisense/triplex-forming ≈20-base-long oligonucleotides involving short stretches of guanines. It would be interesting to investigate whether some helicases are able to disrupt these RNA quadruplexes.
Table 4.
Summary of the effects of various parameters on G-quadruplexes
| Parameter | Association | Dissociation | Equilibrium Ka = kon/koff |
|---|---|---|---|
| Increased temperature | − | ++ | −− |
| Increased concentration | ++ | 0 | n/a |
| Increased ionic strength | + | 0 | + |
| Mg2+ addition | + | + | ≈0 |
| Spermine/spermidine add. | + | + | ≈0 |
| Lower pHa | 0 | − | − |
| Na+ → K+ | ++ | −− | ++ |
| DNA → RNA | ++ | −− | ++ |
| DNA → PSc | − | + | − |
| Longer G-stretch | ++ | −− | ++ |
| Longer non-G overhang | − | +/−b | variesb |
0: no effect. n/a: not applicable.
+/++: increase or strong increase (>30-fold), respectively of the association or dissociation process or of the equilibrium constant.
−/−−: decrease (strong decrease) of the parameter.
aBelow pH 5.5. No effect of pH in the 6.0–7.8 range.
bGenerally, dissociation is slower, but sequence-dependent effects may be observed, depending on length, base composition, side (5′ or 3′) of the extra thymines.
cFrom Wyatt et al. and B. Saccà et al., unpublished data.
SUPPLEMENTARY MATERIAL
Supplementary Material is available at NAR Online.
Supplementary Material
Acknowledgments
This manuscript is dedicated to the memory of Professor Claude Hélène (1938–2003). We thank M. Rougée (MNHN, Paris, France) and C. Gosse (CNRS/LPN Marcoussis, France) for helpful discussions and both referees for helpful suggestions. This work was supported by an ARC grant (#3365 to J.L.M.). Funding to pay the Open Access publication charges for this article was provided by the INSERM.
REFERENCES
- 1.Gellert M., Lipsett M.N., Davies D.R. Helix formation by guanylic acid. Proc. Natl Acad. Sci. USA. 1962;48:2013–2018. doi: 10.1073/pnas.48.12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fresco J.R., Massoulié J. Polynucleotides. V. Helix–coil transition of polyriboguanylic acid. J. Am. Chem. Soc. 1963;85:1352–1353. [Google Scholar]
- 3.Guschlbauer W., Chantot J.F., Thiele D. Four-stranded nucleic acid structures 25 years later: from guanosine gels to telomer DNA. J. Biomol. Struct. Dyn. 1990;8:491–511. doi: 10.1080/07391102.1990.10507825. [DOI] [PubMed] [Google Scholar]
- 4.Williamson J.R. G-quartet structures in telomeric DNA. Annu. Rev. Biophys. Biomol. Struct. 1994;23:703–730. doi: 10.1146/annurev.bb.23.060194.003415. [DOI] [PubMed] [Google Scholar]
- 5.Davis J.T. G-quartets 40 years later: from 5′-GMP to molecular biology and supramolecular chemistry. Angew. Chem. Int. Ed. 2004;43:668–698. doi: 10.1002/anie.200300589. [DOI] [PubMed] [Google Scholar]
- 6.Seeman N.C., Belcher A.M. Emulating biology: building nanostructures from the bottom up. Proc. Natl Acad. Sci. USA. 2002;99:6451–6455. doi: 10.1073/pnas.221458298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Venczel E.A., Sen D. Synapsable DNA. J. Mol. Biol. 1996;257:219–224. doi: 10.1006/jmbi.1996.0157. [DOI] [PubMed] [Google Scholar]
- 8.Fahlman R.P., Sen D. Cation-regulated self-association of ‘Synapsable’ DNA duplexes. J. Mol. Biol. 1998;280:237–244. doi: 10.1006/jmbi.1998.1875. [DOI] [PubMed] [Google Scholar]
- 9.Marsh T.C., Henderson E. G-wires: self-assembly of a telomeric oligonucleotide, d(GGGGTTGGGG), into large superstructures. Biochemistry. 1994;33:10718–10724. doi: 10.1021/bi00201a020. [DOI] [PubMed] [Google Scholar]
- 10.Poon K., Macgregor R.B. Formation and structural determinants of multi-stranded guanine-rich DNA complexes. Biophys. Chem. 2000;84:205–216. doi: 10.1016/s0301-4622(00)00106-x. [DOI] [PubMed] [Google Scholar]
- 11.Alberti P., Mergny J.L. DNA duplex-quadruplex exchange as the basis for a nanomolecular machine. Proc. Natl Acad. Sci. USA. 2003;100:1569–1573. doi: 10.1073/pnas.0335459100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li J.J., Tan W. A single DNA molecule nanomotor. Nano Letters. 2002;2:315–318. doi: 10.1021/nl9011694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ueyama Y., Takagi M., Takenaka A.J. A novel potassium sensing in aqueous media with a synthetic oligonucleotide derivative. Fluorescence resonance energy transfer associated with guanine. J. Am. Chem. Soc. 2002;124:14286–14287. doi: 10.1021/ja026892f. [DOI] [PubMed] [Google Scholar]
- 14.Bonazzi S., Capobianco M., De Morais M.M., Garbesi A., Gottarelli G., Mariani P., Ponzi B., Maria G., Spada G.P., Tondelli L. Four stranded aggregates of oligodeoxy-guanilates forming lyotropic liquid crystals: a study by circular dichroism, optical microscopy and X-ray diffraction. J. Am. Chem. Soc. 1991;113:5809–5816. [Google Scholar]
- 15.Forman S.L., Fettinger J.C., Pieraccini S., Gottareli G., Davis J.T. Toward artificial ion channels: a lipophilic G-quadruplex. J. Am. Chem. Soc. 2000;122:4060–4067. [Google Scholar]
- 16.Shi X.D., Mullaugh K.M., Fettinger J.C., Jiang Y., Hofstadler S.A., Davis J.T. Lipophilic G-quadruplexes are self-assembled ion pair receptors, and the bound anion modulates the kinetic stability of these complexes. J. Am. Chem. Soc. 2003;125:10830–10841. doi: 10.1021/ja035267n. [DOI] [PubMed] [Google Scholar]
- 17.Sen D., Gilbert W. Formation of parallel four-stranded complexes by guanine-rich motifs in DNA and its applications for meiosis. Nature. 1988;334:364–366. doi: 10.1038/334364a0. [DOI] [PubMed] [Google Scholar]
- 18.Williamson J.R., Raghuraman M.K., Cech T.R. Monovalent cation induced structure of telomeric DNA: the G-quartet model. Cell. 1989;59:871–880. doi: 10.1016/0092-8674(89)90610-7. [DOI] [PubMed] [Google Scholar]
- 19.Wang Y., Patel D.J. Solution structure of the human telomeric repeat d[AG3(T2AG3)3] G-tetraplex. Structure. 1993;1:263–282. doi: 10.1016/0969-2126(93)90015-9. [DOI] [PubMed] [Google Scholar]
- 20.Parkinson G.N., Lee M.P.H., Neidle S. Crystal structure of parallel quadruplexes from human telomeric DNA. Nature. 2002;417:876–880. doi: 10.1038/nature755. [DOI] [PubMed] [Google Scholar]
- 21.Neidle S., Parkinson G.N. The structure of telomeric DNA. Curr. Opin. Struct. Biol. 2003;13:275–283. doi: 10.1016/s0959-440x(03)00072-1. [DOI] [PubMed] [Google Scholar]
- 22.Rangan A., Fedoroff O.Y., Hurley L.H. Induction of duplex to G-quadruplex transition in the c-myc promoter region by a small molecule. J. Biol. Chem. 2001;276:4640–4646. doi: 10.1074/jbc.M005962200. [DOI] [PubMed] [Google Scholar]
- 23.Shafer R.H., Smirnov I. Biological aspects of DNA/RNA quadruplexes. Biopolymers. 2000;56:209–227. doi: 10.1002/1097-0282(2000/2001)56:3<209::AID-BIP10018>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- 24.Sun D., Thompson B., Cathers B.E., Salazar M., Kerwin S.M., Trent J.O., Jenkins T.C., Neidle S., Hurley L.H. Inhibition of human telomerase by a G-quadruplex-interactive compound. J. Med. Chem. 1997;40:2113–2116. doi: 10.1021/jm970199z. [DOI] [PubMed] [Google Scholar]
- 25.Neidle S., Parkinson G. Telomere maintenance as a target for anticancer drug discovery. Nature Rev. Drug. Des. 2002;1:383–393. doi: 10.1038/nrd793. [DOI] [PubMed] [Google Scholar]
- 26.Deng J.P., Xiong Y., Sundaralingam M. X-ray analysis of an RNA tetraplex (UGGGGU)(4) with divalent Sr2+ ions at subatomic resolution (0.61 angstrom) Proc. Natl Acad. Sci. USA. 2001;98:13665–13670. doi: 10.1073/pnas.241374798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Spackova N., Berger I., Sponer J. Nanoseconds molecular dynamics simulations of parallel and antiparallel guanine quadruplex DNA molecules. J. Am. Chem. Soc. 1999;121:5519–5534. [Google Scholar]
- 28.Stefl R., Cheatham T.E., Spackova N., Fadrna E., Berger I., Koca J., Sponer J. Formation pathways of a guanine-quadruplex DNA revealed by molecular dynamics and thermodynamic analysis of the substates. Biophys. J. 2003;85:1787–1804. doi: 10.1016/S0006-3495(03)74608-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wyatt J.R., Davis P.W., Freier S.M. Kinetics of G-quartet-mediated tetramer formation. Biochemistry. 1996;35:8002–8008. doi: 10.1021/bi960124h. [DOI] [PubMed] [Google Scholar]
- 30.Wyatt J.R., Vickers T.A., Roberson J.L., Buckheit R.W., Klimkait T., Debaets E., Davis P.W., Rayner B., Imbach J.L., Ecker D.J. Combinatorially selected guanosine-quartet structure is a potent inhibitor of human immunodeficiency virus envelope-mediated cell fusion. Proc. Natl Acad. Sci. USA. 1994;91:1356–1360. doi: 10.1073/pnas.91.4.1356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Phan A.T., Patel D.J. Two-repeat human telomeric d(TAGGGTTAGGGT) sequence forms interconverting parallel and antiparallel G-quadruplexes in solution: distinct topologies, thermodynamic properties, and folding/unfolding kinetics. J. Am. Chem. Soc. 2003;125:15021–15027. doi: 10.1021/ja037616j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fedoroff O.Y., Salazar M., Han H., Chemeris V.V., Kerwin S.M., Hurley L.H. NMR-based model of a telomerase inhibiting compound bound to G-quadruplex DNA. Biochemistry. 1998;37:12367–12374. doi: 10.1021/bi981330n. [DOI] [PubMed] [Google Scholar]
- 33.Read M., Neidle S. Structural characterization of a guanine-quadruplex ligand complex. Biochemistry. 2000;39:13422–13432. doi: 10.1021/bi001584k. [DOI] [PubMed] [Google Scholar]
- 34.Clark G.R., Pytel P.D., Squire C.J., Neidle S. Structure of the first parallel DNA quadruplex-drug complex. J. Am. Chem. Soc. 2003;125:4066–4067. doi: 10.1021/ja0297988. [DOI] [PubMed] [Google Scholar]
- 35.Miura T., Benevides J.M., Thomas G.J. A phase diagram for sodium and potassium ion control of polymorphism in telomeric DNA. J. Mol. Biol. 1995;248:233–238. doi: 10.1016/s0022-2836(95)80046-8. [DOI] [PubMed] [Google Scholar]
- 36.Sen D., Gilbert W. A sodium–potassium switch in the formation of four-stranded G4-DNA. Nature. 1990;344:410–414. doi: 10.1038/344410a0. [DOI] [PubMed] [Google Scholar]
- 37.Crnugelj M., Hud N.V., Plavec J. The solution structure of d(G(4)T(4)G(3))2: a bimolecular G-quadruplex with a novel fold. J. Mol. Biol. 2002;320:911–924. doi: 10.1016/s0022-2836(02)00569-7. [DOI] [PubMed] [Google Scholar]
- 38.Arimondo P., Riou J.F., Mergny J.L., Tazi J., Sun J.S., Garestier T., Hélène C. Interaction of human DNA topoisomerase I with intermolecular G-quartet structures. Nucleic Acids Res. 2000;28:4832–4838. doi: 10.1093/nar/28.24.4832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lyonnais S., Hounsou C., Teulade-Fichou M.P., Jeusset J., LeCam E., Mirambeau G. G-quartets assembly within a G-rich DNA flap. A possible event at the center of the HIV-1 genome. Nucleic Acids Res. 2002;30:5276–5283. doi: 10.1093/nar/gkf644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cantor C.R., Warshaw M.M., Shapiro H. Oligonucleotide interactions. 3. Circular dichroism studies of the conformation of deoxyoligonucleotides. Biopolymers. 1970;9:1059–1077. doi: 10.1002/bip.1970.360090909. [DOI] [PubMed] [Google Scholar]
- 41.Mergny J.L., Phan A.T., Lacroix L. Following G-quartet formation by UV-spectroscopy. FEBS Lett. 1998;435:74–78. doi: 10.1016/s0014-5793(98)01043-6. [DOI] [PubMed] [Google Scholar]
- 42.Mergny J.L., Lacroix L. Analysis of thermal melting curves. Oligonucleotides. 2003;13:515–537. doi: 10.1089/154545703322860825. [DOI] [PubMed] [Google Scholar]
- 43.Rougée M., Faucon B., Mergny J.L., Barcelo F., Giovannangeli C., Garestier T., Hélène C. Kinetics and thermodynamics of triple-helix formation: effects of ionic strength and mismatches. Biochemistry. 1992;31:9269–9278. doi: 10.1021/bi00153a021. [DOI] [PubMed] [Google Scholar]
- 44.Mergny J.L., Lacroix L. Kinetics and thermodynamics of i-DNA formation: phosphodiester vs. modified oligodeoxynucleotides. Nucleic Acids Res. 1998;26:4797–4803. doi: 10.1093/nar/26.21.4797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wallimann P., Kennedy R.J., Miller J.S., Shalango W., Kenp D.S. Dual wavelength parametric test of two-state models for circular dichroism spectra of helical polypeptides: anomalous dichroic properties of alanine-rich peptides. J. Am. Chem. Soc. 2003;125:1203–1220. doi: 10.1021/ja0275360. [DOI] [PubMed] [Google Scholar]
- 46.Lieberman D.V., Hardin C.C. Extraction of information on the buildup and consumption of reactive intermediates from quadruplex DNA assembly time courses. Biochim. Biophys. Acta. 2004;1679:59–64. doi: 10.1016/j.bbaexp.2004.03.006. [DOI] [PubMed] [Google Scholar]
- 47.Uddin M.K., Kato Y., Takagi Y., Mikuma T., Taira K. Phosphorylation at 5′ end of guanosine stretches inhibits dimerization of G-quadruplexes and formation of a G-quadruplex interferes with the enzymatic activities of DNA enzymes. Nucleic Acids Res. 2004;32:4618–4629. doi: 10.1093/nar/gkh766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Krishnan-Ghosh Y., Liu D., Balasubramanian S. Formation of an interlocked quadruplex dimer by d(GGGT) J. Am. Chem. Soc. 2004;126:11009–11016. doi: 10.1021/ja049259y. [DOI] [PubMed] [Google Scholar]
- 49.Aboul-ela F., Murchie A.I.H., Lilley D.M.J. NMR study of parallel-stranded tetraplex formation by the hexadeoxynucleotide d(TG4T) Nature. 1992;360:280–282. doi: 10.1038/360280a0. [DOI] [PubMed] [Google Scholar]
- 50.Aboul-ela F., Murchie A.I.H., Norman D.G., Lilley D.M.J. Solution structure of a parallel-stranded tetraplex formed by d(TG(4)T) in the presence of sodium ions by nuclear magnetic resonance spectroscopy. J. Mol. Biol. 1994;243:458–471. doi: 10.1006/jmbi.1994.1672. [DOI] [PubMed] [Google Scholar]
- 51.Laughlan G., Murchie A.I.H., Norman D.G., Moore M.H., Moody P.C.E., Lilley D.M.J., Luisi B. The high-resolution crystal structure of a parallel-stranded guanine tetraplex. Science. 1994;265:520–524. doi: 10.1126/science.8036494. [DOI] [PubMed] [Google Scholar]
- 52.Phillips K., Dauter Z., Murchie A.I.H., Lilley D.M.J., Luisi B. The crystal structure of a parallel-stranded guanine tetraplex at 0.95 angstrom resolution. J. Mol. Biol. 1997;273:171–182. doi: 10.1006/jmbi.1997.1292. [DOI] [PubMed] [Google Scholar]
- 53.Caceres C., Wright G., Gouyette C., Parkinson G., Subirana J.A. A thymine tetrad in d(TGGGGT) quadruplexes stabilized with Tl+/Na+ ions. Nucleic Acids Res. 2004;32:1097–1102. doi: 10.1093/nar/gkh269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Petraccone L., Erra E., Esposito V., Randazzo A., Mayol L., Nasti L., Barone G., Giancola C. Stability and structure of telomeric DNA sequences forming quadruplexes containing four G-tetrads with different topological arrangements. Biochemistry. 2004;43:4877–4884. doi: 10.1021/bi0300985. [DOI] [PubMed] [Google Scholar]
- 55.Dapic V., Abdomerovic V., Marrington R., Peberdy J., Rodger A., Trent J.O., Bates P.J. Biophysical and biological properties of quadruplex oligodeoxyribonucleotides. Nucleic Acids Res. 2003;31:2097–2107. doi: 10.1093/nar/gkg316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Schaffitzel C., Berger I., Postberg J., Hanes J., Lipps H.J., Plückthun A. In vitro generated antibodies specific for telomeric guanine quadruplex DNA react with Stylonychia lemnae macronuclei. Proc. Natl Acad. Sci. USA. 2001;98:8572–8577. doi: 10.1073/pnas.141229498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Li W., Miyoshi D., Nakano S., Sugimoto N. Structural competition involving G-quadruplex DNA and its complement. Biochemistry. 2003;42:11736–11744. doi: 10.1021/bi034168j. [DOI] [PubMed] [Google Scholar]
- 58.Green J.J., Ying L.M., Klenerman D., Balasubramanian S. Kinetics of unfolding the human telomeric DNA quadruplex using a PNA trap. J. Am. Chem. Soc. 2003;125:3763–3767. doi: 10.1021/ja029149w. [DOI] [PubMed] [Google Scholar]
- 59.Rosu F., Gabelica V., Shin-ya K., DePauw E. Telomestatin induced stabilization of the human telomeric DNA quadruplex monitored by electrospray mass spectrometry. Chem. Commun. 2003;34:2702–2703. doi: 10.1039/b309394h. [DOI] [PubMed] [Google Scholar]
- 60.Hardin C.C., Corregan M., Brown B.A., Frederick L.N. Cytosine-cytosine + base pairing stabilizes DNA quadruplexes and cytosine methylation greatly enhances the effect. Biochemistry. 1993;32:5870–5880. doi: 10.1021/bi00073a021. [DOI] [PubMed] [Google Scholar]
- 61.Cheong C.J., Moore P.B. Solution structure of an unusually stable RNA tetraplex containing G-Quartet and U-Quartet structures. Biochemistry. 1992;31:8406–8414. doi: 10.1021/bi00151a003. [DOI] [PubMed] [Google Scholar]
- 62.Gupta G., Garcia A.E., Guo Q., Lu M., Kallenbach N.R. Structure of a parallel-stranded tetramer of the Oxytricha telomeric DNA sequence dT4G4. Biochemistry. 1993;32:7098–7103. doi: 10.1021/bi00079a005. [DOI] [PubMed] [Google Scholar]
- 63.Lu M., Guo Q., Kallenbach N.R. Structure and stability of sodium and potassium complexes of dT4G4 and dT4G4T. Biochemistry. 1992;31:2455–2459. doi: 10.1021/bi00124a003. [DOI] [PubMed] [Google Scholar]
- 64.Shida T., Yokoyama K., Tamai S., Sekiguchi J. Self-association of telomeric short oligodeoxyribonucleotides containing a dG cluster. Chem. Pharm. Bull. (Tokyo) 1991;39:2207–2211. doi: 10.1248/cpb.39.2207. [DOI] [PubMed] [Google Scholar]
- 65.Pörschke D., Eigen M. Co-operative non-enzymic base recognition. 3. Kinetics of the helix–coil transition of the oligoribouridylic–oligoriboadenylic acid system and of oligoriboadenylic acid alone at acidic pH. J. Mol. Biol. 1971;62:361–381. doi: 10.1016/0022-2836(71)90433-5. [DOI] [PubMed] [Google Scholar]
- 66.Craig M.E., Crothers D.M., Doty P. Relaxation kinetics of dimer formation by self complementary oligonucleotides. J. Mol. Biol. 1971;62:383–401. doi: 10.1016/0022-2836(71)90434-7. [DOI] [PubMed] [Google Scholar]
- 67.Hardin C.C., Corregan M.J., Lieberman D.V., Brown II B.A. Allosteric interactions between DNA strands and monovalent cations in DNA quadruplex assembly: thermodynamic evidence for three linked association pathways. Biochemistry. 1997;36:15428–15450. doi: 10.1021/bi970488p. [DOI] [PubMed] [Google Scholar]
- 68.Liu H., Kugimiya A., Sakurai T., Katahira M., Uesugi S. A comparison of the properties and the solution structure for RNA and DNA quadruplexes which contain two GGAGG sequences joined with a tetranucleotide linker. Nucleosides Nucleotides Nucleic Acids. 2002;21:785–801. doi: 10.1081/NCN-120016481. [DOI] [PubMed] [Google Scholar]
- 69.Randazzo A., Esposito V., Ohlenschlager O., Ramachandran R., Mayol L. NMR solution structure of a parallel LNA quadruplex. Nucleic Acids Res. 2004;32:3083–3092. doi: 10.1093/nar/gkh629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Dominick P.K., Jarstfer M.B. A conformationally constrained nucleotide analogue controls the folding topology of a DNA G-quadruplex. J. Am. Chem. Soc. 2004;126:5050–5051. doi: 10.1021/ja039192z. [DOI] [PubMed] [Google Scholar]
- 71.Datta B., Schmitt C., Armitage B.A. Formation of a PNA(2)–DNA(2) hybrid quadruplex. J. Am. Chem. Soc. 2003;125:4111–4118. doi: 10.1021/ja028323d. [DOI] [PubMed] [Google Scholar]
- 72.Krishnan-Ghosh Y., Stephens E., Balasubramanian S. A PNA(4) quadruplex. J. Am. Chem. Soc. 2004;126:5944–5945. doi: 10.1021/ja031508f. [DOI] [PubMed] [Google Scholar]
- 73.Sen D., Gilbert W. Novel DNA superstructures formed by telomere-like oligomers. Biochemistry. 1992;31:65–70. doi: 10.1021/bi00116a011. [DOI] [PubMed] [Google Scholar]
- 74.Guo Q., Lu M., Kallenbach N.R. Effect of thymine tract length on the structure and stability of model telomeric sequences. Biochemistry. 1993;32:3596–3603. doi: 10.1021/bi00065a010. [DOI] [PubMed] [Google Scholar]
- 75.Jin R.Z., Gaffney B.L., Wang C., Jones R.A., Breslauer K.J. Thermodynamics and structure of a DNA tetraplex—a spectroscopic and calorimetric study of the tetramolecular complexes of d(TG3T) and d(TG3T2G3T) Proc. Natl Acad. Sci. USA. 1992;89:8832–8836. doi: 10.1073/pnas.89.18.8832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Esposito V., Randazzo A., Piccialli G., Petraccone L., Giancola C., Mayol L. Effects of an 8-bromodeoxyguanosine incorporation on the parallel quadruplex structure [d(TGGGT)](4) Org. Biomol. Chem. 2004;2:313–318. doi: 10.1039/b314672c. [DOI] [PubMed] [Google Scholar]
- 77.Wang Y., Patel D.J. Guanine residues in d(T2AG3) and d(T2G4) form parallel-stranded potassium cation stabilized G-quadruplexes with antiglycosidic torsion angles in solution. Biochemistry. 1992;31:8112–8119. doi: 10.1021/bi00150a002. [DOI] [PubMed] [Google Scholar]
- 78.Gavathiotis E., Searle M.S. Structure of the parallel-stranded DNA quadruplex d(TTAGGGT)(4) containing the human telomeric repeat: evidence for A-tetrad formation from NMR and molecular dynamics simulations. Org. Biomol. Chem. 2003;1:1650–1656. doi: 10.1039/b300845m. [DOI] [PubMed] [Google Scholar]
- 79.Sugimoto N., Ohmichi T., Sasaki M. The stability of DNA and RNA G-quartets. Nucleosides Nucleotides. 1996;15:559–567. [Google Scholar]
- 80.Wang Y., Patel D.J. Solution structure of a parallel-stranded G-quadruplex DNA. J. Mol. Biol. 1993;234:1171–1183. doi: 10.1006/jmbi.1993.1668. [DOI] [PubMed] [Google Scholar]
- 81.Kim J., Cheong C., Moore P.B. Tetramerization of an RNA oligonucleotide containing a GGGG sequence. Nature. 1991;351:331–332. doi: 10.1038/351331a0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.