Abstract
We have recently sequenced the genome of a novel thermophilic bacteriophage designated as TS2126 that infects the thermophilic eubacterium Thermus scotoductus. One of the annotated open reading frames (ORFs) shows homology to T4 RNA ligase 1, an enzyme of great importance in molecular biology, owing to its ability to ligate single-stranded nucleic acids. The ORF was cloned, and recombinant protein was expressed, purified and characterized. The recombinant enzyme ligates single-stranded nucleic acids in an ATP-dependent manner and is moderately thermostable. The recombinant enzyme exhibits extremely high activity and high ligation efficiency. It can be used for various molecular biology applications including RNA ligase-mediated rapid amplification of cDNA ends (RLM-RACE). The TS2126 RNA ligase catalyzed both inter- and intra-molecular single-stranded DNA ligation to >50% completion in a matter of hours at an elevated temperature, although favoring intra-molecular ligation on RNA and single-stranded DNA substrates. The properties of TS2126 RNA ligase 1 makes it very attractive for processes like adaptor ligation, and single-stranded solid phase gene synthesis.
INTRODUCTION
Thermophilic bacteriophages infecting bacteria from the Thermus genus have not been studied extensively to date. While some bacteriophages have been isolated from the extreme thermophilic eubacterium Thermus thermophilus, little is known of phages in other Thermus species (1,2). In the thermophilic bacteriophage discovery program at Prokaria Ltd, a number of bacteriophages infecting Thermus species have been identified and characterized. One of those is a thermophilic bacteriophage designated as TS2126, in accordance with the Thermus scotoductus strain it infects (3). The phage is chloroform-sensitive and infectious up to 85°C. The bacteriophage genome is double-stranded DNA with a size of ∼90 kb. The genome of the TS2126 phage has been sequenced and its genome annotated (S. Hjorleifsdottir, personal communication).
T.scotoductus is a gram negative, thermophilic heterotropic eubacteria first isolated from hot tap water in Iceland. T.scotoductus has a thick peptidoglycan layer, as well as a cytoplasmic membrane and an outer membrane, and produces dark brown melanin-like pigments. The bacteria are non-motile and do not form spores. Its optimum growth conditions are at 65°C and pH 7.5, and it has a GC-content of 64.5% (3).
The RNA ligase 1 (EC 6.5.1.3) family members can be found in a diverse group of viruses and fungi with the T4 RNA ligase 1 as the prototype and the most-studied family member (4–7). In addition to bacteriophage T4 RNA ligase 1, members of this family are found in the RM378 bacteriophage (6) and in the RNA ligase part of the pnk/pnl gene (ORF 86) from the baculovirus Autographa californica nucleopolyhedrovirus (ACNV) as well as some other baculoviruses (5,8).
These ligases belong to a superfamily of covalent nucleotidyl transferases that are both ATP and NAD+ dependent and include RNA ligase 2 family (Rnl2/REL), RNA capping enzymes and DNA ligases (9–11). These enzymes share a number of sequence motifs, which are largely confined to the central active site region bordering a presumed nucleic acid-binding cleft, as seen in the available structures of DNA ligases and mRNA capping enzyme (12,13). They presumably share a similar fold and analogous catalytic mechanism determined by their common origin and evolutionary history (11).
RNA ligases 1 have the ability to ligate single-stranded nucleic acids by catalyzing the ATP-dependent formation of phosphodiester bonds between 5′-phosphate and 3′-hydroxyl termini of single-stranded RNA or DNA (6,7,14). The biological role of bacteriophage RNA ligases has been primarily studied in T4 where the enzyme, in cooperation with polynucleotide kinase, repairs cleaved tRNA molecules. This is a counter-defense against a suicidal mechanism of the host, triggered by the anticodon nuclease (ACNase) system in Escherichia coli prr+ strains, which is induced after inactivation of the restriction-modification system EcoprrI by the T4 Stp polypeptide (14–17). The actual biological roles of RNA ligases in the RM378 bacteriophage and ACNV viruses have not been determined, although it is likely that these enzymes are a part of a repair machinery that responds to RNA degradation by the host (5,6,15).
The T4 RNA ligase has become a very important tool in molecular biology and is used in numerous protocols. Applications include RNA ligase mediated rapid amplification of cDNA ends (RLM-RACE) (18,19), ligation of oligonucleotide adaptors to cDNA or single-stranded primer extension products for PCR (20,21), oligonucleotide synthesis (22) and various 5′ nucleotide modifications of nucleic acids (23). Here, we describe the isolation and characterization of a thermostable RNA ligase 1 from bacteriophage TS2126, displaying efficient inter- and intra-molecular ligations of single-stranded DNA (ssDNA).
MATERIALS AND METHODS
Cloning of the RlnA gene from TS2126
The gene was amplified by the PCR from the TS2126 genomic DNA using Dynazyme™ DNA polymerase according to the manufacturer's instructions (Finnzymes Oy, Espoo, Finland), with primers TSligFwd-NdeI: d(CGGGCTCGAGCATATGAGCTCTTTGGCCCCGTG); and TsligRev-EcoRI: d(CCGGAATTCAATGATGATGATGATGGTGAAAAATAAGCTCCCCGC). The PCR product was then digested with NdeI and EcoRI and ligated into pET-23b expression vector (Novagen Inc., Madison, WI) containing a His-tag on the C-terminus. A few pET-Rlig122 clones were verified by DNA sequencing. The pET-Rlig122H#6 clone was selected and transformed into E.coli BL21 (DE3)-RIL strain (Stratagen Inc., La Jolla, CA). The strain was cultivated at 37°C in a BioFlow 3000 fermentor (10 liter) and induced with 1 mM isopropyl-β-d-thiogalactopyranoside (IPTG). The cells were harvested and disrupted by sonication. The crude cell extract was centrifuged in a SA-600 rotor (Sorvall Inc.) at 10 400 g for 1 h. The supernatant was collected and applied to XK 26/10 50 ml Column (Amersham Biosciences Corp., Piscataway, NJ), packed with chelating sepharose charged with nickel ions. The column was washed using washing buffer (10 mM sodium phosphate buffer, pH 7.5, 0.5 M NaCl and 25 mM imidazole) and the recombinant TS2126 RNA ligase protein eluted using the same buffer with 100, 200 and 300 mM imidazole in a stepwise manner. The eluted protein was then put through HiPrep sephacryl 26/60 S200 High Resolution Column (Amersham Biosciences Corp.) and eluted in 2× RNA ligase storage buffer (20 mM Tris-HCl, pH 8, 100 mM KCl, 2 mM DTT and 0.2 mM EDTA). The protein solution was finally mixed with 1:1 (v/v) 100% glycerol and stored at −20°C. Measurements of protein concentrations were performed as described by Bradford (24). The TS2126 RNA ligase was tested for the following contaminating activities.
Endonuclease assay: An aliquot of 0.25 μg of λ-DNA EcoRI/HindIII fragments was incubated with 1× RNA ligase buffer and 2 μg of TS2126 RNA ligase in 25 μl reactions at 37 and 64°C for 4 h and checked for changes in band pattern on 1% agarose gel.
Nicking assay: An aliquot of 0.25 μg of supercoiled pBR322 DNA was incubated with 1× RNA ligase buffer and 2 μg of TS2126 RNA ligase in 25 μl reactions at 37 and 64°C for 4 h, and checked for relaxation of the supercoiled structure of pBR322 DNA on 1% agarose gel.
Exonuclease assay: The incubation was done in 50 μl reaction containing 1× RNA ligase buffer, 1 μg of 3H-labeled E.coli double- or single-stranded DNA and 2 μg of TS2126 RNA ligase at 37 and 64°C for 4 h. The samples were spotted onto DE81 filters and washed in sodium phosphate buffer (pH 7), dried and measured for release of tritium from the chromosomal DNA in a liquid scintillation counter.
RNAse assay: RNaseAlert™ Lab Test Kit (catalog no. 1964) from Ambion Inc. was used to detect RNAse activity according to the manufacturer's protocol. An aliquot of 2 μg of TS2126 RNA ligase was incubated for 1 h at 37 or 64°C and the samples were checked for degradation of RNA.
Characterization of the RNA ligase
We used the standard RNA ligase assay developed by Silber et al. (7) to characterize the TS2126 RNA ligase. The conditions for the standard assay were 50 mM MOPS-sodium salt, pH 7.5, 10 mM MgCl2, 1 mM DTT, 25 μg/ml BSA, 0.5 mM ATP, 5–10 μM [5′-32P]r(A20) (EurogenTech Inc., Philadelphia, PA) and 0.001–0.01 mg/ml enzyme. Reaction times and protein concentration varied, based on the assay, and the substrate(s). The reactions were terminated by incubation at 90°C for 5 min, followed by cooling on ice, addition of 30 μl solution of shrimp alkaline phosphatase (0.2 U/μl) in its standard buffer (USB Corp., Cleveland, OH) and incubating for 4 h at 37°C. The alkaline phosphatase-resistant 5′- phosphoryl termini were captured on DE81 filters, which were washed twice in a 500 mM sodium phosphate buffer and dried. Radioactivity on the filters was counted in a scintillation counter (Packard Inc.) after addition of 5 ml of scintillation fluid. All assays were done in triplicates and the mean value was calculated. One unit of RNA ligase activity was determined as the amount of enzyme needed to convert 1 nmol of 5′-phosphoryl termini in 32P-r(A20) to a phosphatase-resistant form in 30 min at 65°C (7).
Km constants for ATP in the substrate adenylation were determined in the following manner: 0.01 mg/ml of enzyme was incubated with 20 μM dideoxy-blocked 32P-d(A10) or 32P-r(A10) (EurogenTech Inc.) at 60°C for 60 min with different amounts of ATP (1, 10, 25, 50, 100, 250, 500 and 1000 μM) followed by the protection assay outlined above.
The single-stranded intra-molecular DNA ligations were analyzed with the exonuclease protection assay (EPA) as described by Blondal et al. (6). We used 86 nt. oligomer-2 PO4-d(ATGCTGTGCTGGGTCACTTTCCTTATCCCGACCCTTCCCAGTGGACAGATGAGGAATTGGGTATCCCTCCGGATGATGAAGACTGA) as a substrate. The reactions were carried out at 60–70°C for 1–5 h. The results were quantified using the EPA process and OliGreen ssDNA quantification kit (Molecular Probes Inc.) and also by running the samples on 20% native PAGE.
For end-to-end ligations, we used two ssDNA oligomers: oligomer-1 donor, P-d(ATGCTGTGCTGGGTCACTTTCCTTATCCCGACCCTTCCCAGTGGACAGAT) with Oregon® Green internal label and dideoxy modification on 3′ end; and oligomer-3 acceptor, d(GGCGGCTGGAGCCCGTGGCGTCCGTCGGGCTGGCGGAGGTGCG-CACATTGAGCCCCGGTACAGACAGTTCCCGCAGCTGA). The reactions were carried out at 50–70°C for 1–10 h. After incubation, the samples were diluted to 1:100 and mixed with 100% formamide solution and size standard (Applied Biosystems Inc.). The samples were run on ABI 3730 genetic analyzer and the results analyzed using the ABI Prism Genetic Analyzer™ software (Applied Biosystems Inc.).
RLM-RACE assay
Human testis Poly(A) mRNA (Ambion Inc.) was used as a substrate for RLM-RACE protocol using the components of the First Choice RLM-RACE kit, according to the the manufacturer's instructions (Ambion Inc.). In the ligation step, 0.1 μg of TS2126 RNA ligase and the standard buffer was used for incubation at 65°C for 1 h. The cDNA was made using AMV First Strand Synthesis Kit (Invitrogen Inc.) with oligo(dT) primer, according to the manufacturer's instructions. The template was then amplified using standard PCR protocol with AmpliTaq Gold (Applied Biosystems Inc.) as recommended by the manufacturer. The products were run on 1.2% agarose gel, dyed with ethidium bromide and visualized under UV light. The products were cloned into TOPO TA vectors and sequenced for verification using BigDye 2.0 DNA sequencing kit and analyzed on ABI Prism 3730 genetic analyzer (Applied Biosystems Inc.).
RESULTS
Isolation, expression and purification of the RNA ligase
When annotating the putative open reading frames (ORFs) of the TS2126 phage genome, it became apparent that one of the ORFs showed homology to RNA ligase family 1. The putative gene was 1188 bp in size and coded for a 395 amino acids polypeptide. The nucleotide sequence was submitted to GenBank™ under the accession number CQ796353. Sequence comparisons to T4, ACNV and RM378 RNA ligases, revealed greater sequence similarity to T4 RNA ligase 1 (18% identity overall) than to the moderately thermostable RM378 RNA ligase 1 (15% identity overall). Sequence similarity to the RNA ligase part of the ACNV pnk/pnl gene (ORF 86) was low (9% identity overall). The sequence alignment shown in Figure 1 further shows that significant inserts found in RM378 RNA ligase 1 are absent in TS2126 RNA ligase 1, which further underlines its similarity to the mesophilic T4 enzyme rather than its thermophilic counterparts in bacteriophage RM378.
Figure 1.
Amino acid sequence alignment of bacteriophage TS2126, T4, RM378 and ACNV RNA ligase 1 are shown. The motifs I, Ia, IV and V are boxed. The black-boxed amino acids represent ≥3 identical residues. The gray-boxed amino acids represent ≥3 amino acids with similar properties. The alignment was made with Clustal X software (29).
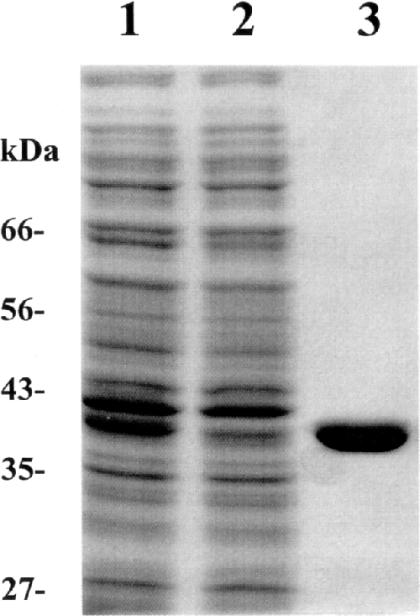
The RNA ligase gene was amplified by PCR from the viral genome and inserted into a pET23b expression vector with a C-terminal His6-tag. The vector was transformed into E.coli strain BL21 and few colonies were selected for plasmid harvesting and purification. The clones were sequenced to verify correct sequence and insertion site. The recombinant protein was expressed using IPTG induction and purified to near homogeneity (>95%) as estimated on SDS–PAGE (Figure 2) (25). Protein concentrations measurements were done as described by Bradford (24). Yields of the recombinant TS2126 RNA ligase 1 were estimated to be 1.15 mg/g cells (wet weight). The TS2126 RNA ligase was tested for a number of contaminating nuclease activities and found to be free of RNAse, exonuclease, endonuclease and nicking activities at both 37 and 64°C (data not shown).
Figure 2.
Purification of the His-tagged recombinant TS2126 RNA ligase is shown. Lane 1, crude extract; lane 2, XK 26/10 His-Column flow through; and lane 3, purified TS2126 RNA ligase elute in 300 mM imidazole. The purification was estimated over 95% by SDS–PAGE analysis.
Characterization of the RNA ligase
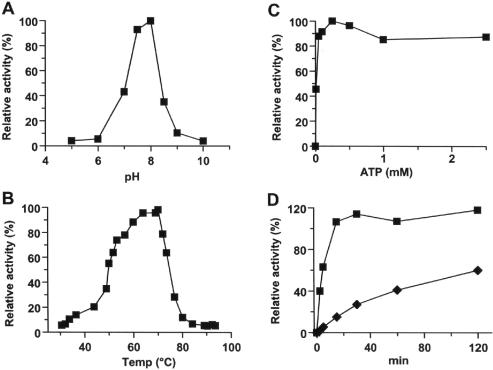
The basic characterization of the recombinant TS2126 RNA ligase 1 was performed using the phosphatase resistance assay developed by Silber et al. (7). First, the optimum pH was determined to be 7.5–8 in MOPS buffer (Figure 3A). The apparent optimum temperature was determined by measuring the activity at different reaction temperatures for 1 h under standard conditions. The enzyme had a rather broad temperature profile with apparent optimum temperature between 65 and 70°C (Figure 3B). The enzyme showed linear accumulation of product over a 2 h reaction time at 65°C, but started to lose activity at higher temperatures. The enzyme lost all activity at temperatures >80°C (data not shown). The enzyme should be considered as moderately thermostable since it was relatively stable at 65°C but not at higher temperatures. The protein showed optimum activity at 5–10 mM for both Mg2+ and Mn2+ ions, with Mn2+ showing slightly higher activity than Mg2+ (data not shown). The enzyme showed relatively good activity (>75%) at ATP concentrations of 0.025–2.5 mM ATP (Figure 3C); however, we observed inhibition of the overall ligation at higher ATP concentrations, resulting in complete inhibition at 10 mM ATP. This inhibition could be partially rescued by adding more Mg2+ to the reaction (data not shown) and indicates that the inhibition is, at least in part, due to chelation of the divalent cation by ATP.
Figure 3.
Characterization of the TS2126 RNA ligase using the phosphatase resistant assay is shown. (A) The pH profile using MOPS buffer. The pH optimum was between 7.5 and 8.0. (B) Temperature optimum. The apparent optimum temperature of the enzyme activity was between 60 and 70°C. (C) The effect of ATP concentration on the TS2126 RNA ligase activity is shown. Optimum activity was obtained in the range of 0.025–2.5 mM ATP. (D) The comparison of specific activity for TS2126 RNA ligase (squares) and T4 RNA ligase (diamonds) in the phosphatase resistant assay. TS2126 RNA ligase showed ∼10-fold specific activity when compared with commercial T4 RNA ligase.
To compare the activity of the TS2126 RNA ligase 1 with the commercially available T4 RNA ligase 1 activity, a circularization assay using 10 μM 32P-r(A20) substrate, was performed under standard phosphatase resistance conditions (0.1 μg protein per 10 μl reaction volume). A time curve of phosphatase protection of the substrate is shown in Figure 3D. The specific activity of the TS2126 RNA ligase under saturated substrate conditions was 20 μmol/mg h, or 10.000 U/mg under the definition of Silber et al. (7). However under identical conditions at 37°C, T4 RNA ligase activity was only ∼1000 U/mg. Note that the T4 RNA ligase controls used from three different commercial sources, only showed ∼1/10 of its reported activity in our phosphatase protection assay. The reason for this is unknown, but probably owing to the different RNA ligase assays. We utilized synthesized, deprotected, phosphorylated r(A20) oligonucleotides but not digested RNA transcripts, which was commonly used in this assay.
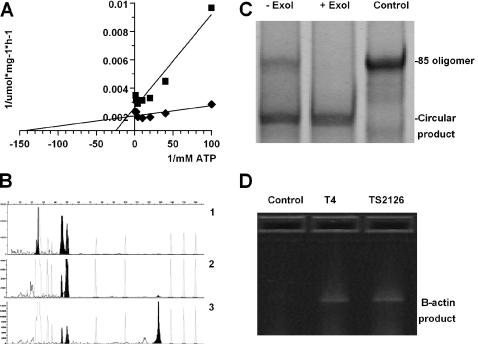
Formation of donor-adenylate intermediates by TS2126 RNA ligase was studied by using substrates 32P-r(A10)- and 32P-d(A10)-dideoxy blocked oligomers and different amounts of ATP. The adenylation reaction reached optimum velocity of 0.39 and 0.50 μmol/mg h for the 32P-d(A10) and 32P-r(A10) substrates, respectively, under saturated conditions. As seen in Figure 4A, the Km constants for ATP in the adenylation reaction were calculated to be 7 and 40 μM on the RNA and DNA substrates, respectively. Comparison of the velocity of the adenylation reaction and the overall reaction revealed that the velocity of the adenylation reaction was more than one order of magnitude less than the overall ligation.
Figure 4.
Characterization of the TS2126 RNA ligase is shown. (A) The Km constants of ATP in adenylation of the saturated donor substrate and Vmax of the adenylation reaction using both 32P-r(A10) (diamonds) and 32P-d(A10) (squares) dideoxy blocked oligomers as substrates. The Km constants for ATP in the adenylation reaction were calculated to be 7 and 40 μM for the r(A10) and d(A10), respectively. (B) End-to-end ligation of oligomers-1 and -3 in the reaction buffer with 25 μM ATP and 7.5% PEG6000, incubated for 2.5 h at 60°C. The samples were run on ABI 3730 DNA analyzer. (i) Control without ligase, (ii) ligation with 10 mM Mg2+ and (iii) ligation in the presence of 10 mM Mg2+ and 2.5 mM Mn2+. The 130 nt reaction product was estimated to be 5 and 60%, respectively. (C) Native 15% PAGE of oligomer-2 after ligation reaction as described in Materials and Methods. Lane 1, ligation reaction without exonuclease I (−ExoI) digestion. Lane 2, ligation reaction products after exonuclease I (+ExoI) digestion. Exonuclease I resistant product was present and estimated to be 65% of the total oligomer concentration. Lane 3, unligated control oligomer-2. (D) RLM-RACE PCR products of the 5′ end of human beta-actin gene, run on 1% agarose gel electrophoresis stained with ethidium bromide. Bands of 850 bp sizes appear both when the 5′ adaptor was ligated using T4 RNA ligase and TS2126 RNA ligase.
To investigate potential applications of the enzyme, for example, in gene synthesis, two ssDNA oligomers were ligated in an end-to-end manner. Ligations using 50 pmol of oligomer-1 donor and 100 pmol of oligomer-3 acceptor, and 0.1 mg/ml of TS2126 RNA ligase 1 only yielded 5–10% of the ligated product under conditions that gave >50% circularization of oligomer-2. However, by lowering the ATP concentration to 25 μM, using 5–15% PEG 6000 and increasing the incubation time (2.5–5 h at 60–65°C) slightly increased the yields. Further improvement was observed when Mn2+ was used instead of Mg2+ and even better yields were obtained, by using 2–3 mM MnCl2 in the Mg2+ containing reaction buffer (Figure 4B).
Ligation of ssDNA was also studied extensively using the EPA (6). Circularization experiments, using oligomer-2 (86 nt oligonucleotide) as a substrate, in the standard buffer with 2.5 mM MnCl2 and 50 μM ATP for 1 h at 65°C, resulted in the ssDNA activity of 100 nmol/mg h. Figure 4C shows that the product of ligation is resistant to exonuclease I digestion. The ligation yield was determined to be ∼65% by Oligreen™ ssDNA quantification (6). Overall, we have assayed a number of ssDNA oligonucleotides ranging in size from 18 to 85 nt, and routinely got >50% circularization in 1 h using 0.05–0.1 mg/ml protein per reaction, containing 100 pmol of oligomers under the standard assay conditions (data not shown).
RLM-RACE
The RLM-RACE protocol was performed using the First Choice RACE kit (Ambion Inc.) as described by the manufacturer and also with TS2126 RNA ligase as a substitute for the T4 RNA ligase. The reaction temperature was 65°C and incubation time was 1 h. Analysis of the products showed 850 bp bands of similar intensity in both the T4 and TS2126 RNA ligase-ligated cDNA pools as seen in Figure 4D. The products were cloned and verified by DNA sequencing. The results clearly showed that TS2126 RNA ligase could substitute for T4 RNA ligase in RLM-RACE protocols.
DISCUSSION
Isolation, expression and purification of RNA ligase
The DNA genome sequencing of TS2126 bacteriophage revealed the RNA ligase gene reported here but no evidence was found for a polynucleotide kinase (pnk) gene, although such a gene might be expected by analogy with bacteriophages T4 and ACNV (5,7,26). However, the presence of a potential PNK gene in the genome of TS2126 may have escaped discovery by conventional sequence comparisons. For example, we have recently isolated a novel PNK from the RM378 bacteriophage, which did not show much similarity to T4 PNK (unpublished results).
The overall identity of the aligned RNA ligase 1 sequences was relatively low. The amino acid sequence of RNA ligase 1 from the TS2126 bacteriophage showed more similarity to T4 (18%) than RM378 RNA ligase 1 (15%), indicating that the two proteins of thermophilic origin have evolved and adapted independently to the elevated thermal conditions.
Sequence comparison of the four RNA ligases shows clearly, three of the six previously identified motifs in the superfamily, namely the motifs I, IV and V, which comprise the NMP binding site and affect the nucleotidyl transfer (adenylation) of the proteins (step 1) (11,27,28). The roles of the motifs I, IV and V in the RNA ligase family 1 are supported by mutational analysis and indirect structural evidence from other members of the superfamily. For T4 RNA ligase, motif I (K99 and G102) and motif IV (E227 and G228) are essential for the adenylation of the protein (step 1) as well as the phosphodiester bond formation (step 3) (27). On the other hand, motif V (K240 and K242) is essential for the step 1 but not step 3 for T4 RNA ligase 1 (27). The motifs II, III and IIIa are not well conserved within the group of RNA ligase family 1 and the importance of the corresponding residues within the group seems unclear. However, we note that a new conserved motif, upstream of motif I, is emerging as more RNA ligase 1 sequences become available. This motif corresponds to amino acid residues 76–89 in TS2126 RNA ligase 1 and amino acid residues 68–81 in the T4 RNA ligase 1 (Figure 1). Mutational data from T4 suggest that residues in this motif (Lys75 and Phe77) are functionally important in the step 1 of the ligation reaction mechanism (the adenylation of the protein) (27). The TS2126 RNA ligase sequence also confirms the newly suggested motif Ia (S/T/D)-(R/K) for ATP-dependent RNA and DNA ligase and GTP-dependent capping enzymes, although the RM378 RNA ligase 1 contains aliphatic L instead of the basic R/K amino acid. However, the structure–function relationship between residues in the different motifs will be difficult to fully resolve without high-resolution structure(s) of RNA ligase(s) from family 1. Such structures may also give insights into the evolution of the covalent nucleotidyl-transferase superfamily.
Characterization of the TS2126 RNA ligase 1
The characterization of the TS2126 RNA ligase 1 reveals that its thermostability is moderate. This is not unexpected since the phage and its host, live at similar temperature (3). The TS2126 RNA ligase 1 is stable at 60–65°C for extended time but loses activity at higher temperatures, which is similar to the properties of RM378 RNA ligase 1 (6). However, when comparing the activity of TS2126 and RM378 and a commercial T4 RNA ligase 1, the TS2126 showed ∼30 and 10 times higher specific activity, respectively, as studied using the 32P-r(A20) phosphatase protection assay (6,7). This striking difference in ligation efficiency was also observed in the ssDNA ligation experiments where TS2126 RNA ligase 1 was much more effective than RNA ligases from RM378 and T4. The TS2126 RNA ligase 1 has a clear preference for circularization when the experiments are performed in the presence of Mg2+.
Comparison of the velocity of the adenylation of donor template experiments (step 2) versus the circularization reaction experiments (overall ligation) suggests that the completion of phosphodiester bond formation is essential for the dissociation of the enzyme–donor complex, allowing a new donor to bind to the enzyme. In other words, the adenylated-donor–enzyme complex is tightly bound, waiting for the appropriate acceptor. Therefore, it is not surprising that the bound adenylated donor is much more efficiently ligated to itself (intra-molecular ligation), than to a second nucleic acid due to a higher effective concentration of the already bound donor molecule. The 3′ end of the donor could also have an inhibitory effect on acceptor binding.
Even with a 3′ blocked donor and acceptor in 1:1–5 ratio we only got 5–10% end-to-end ligation products under normal ligation conditions. This was overcome by introducing Mn2+ to the Mg2+-containing reaction mixture, which dramatically increased the end-to-end ligation efficiency (from 5–10% to >50%). The reasons for the effect of Mn2+ on the end-to-end ligations are still not clear. The activity of the circularization reaction will increase with Mn2+ compared with Mg2+ but not in the manner that explains the rise in end-to-end product yields. The addition of Mn2+ enabled end-to-end ligation of ssDNA with >50% ligation efficiency, which is superior activity when compared with T4 RNA ligase 1. In addition to this, by introducing one ribonucleotide at the 3′ end of an acceptor oligomer, we were able to increase the yields of an Mg2+ reaction buffer up to 50% but even then, the reaction yielded more products when Mn2+ was also present.
TS2126 RNA ligase 1 can be used in the RLM-RACE assay at 65°C and can thereby substitute the use of T4 RNA ligase in this protocol. We suggest that performing RLM-RACE at elevated temperatures may produce better results if the 5′ donor end of mRNA molecules has a secondary structure that inhibits efficient ligations with T4 RNA ligase at 37°C. We also envisage that a number of other applications may benefit from the use of a thermostable RNA ligase with ssDNA activity. This includes primer extension/adaptor ligation-based PCR methods, for example on cDNA (RACE), and genomic DNA (for genome walking), ssDNA or RNA protection by circularization and solid phase single-stranded gene synthesis.
Acknowledgments
Funding to pay the Open Access publication charges for this article was provided by Prokaria Ltd.
REFERENCES
- 1.Pederson D.M., Welsh L.C., Marvin D.A., Sampson M., Perham R.N., Yu M., Slater M.R. The protein capsid of filamentous bacteriophage PH75 from Thermus thermophilus. J. Mol. Biol. 2001;309:401–421. doi: 10.1006/jmbi.2001.4685. [DOI] [PubMed] [Google Scholar]
- 2.Sakaki Y., Oshima T. Isolation and characterization of a bacteriophage infectious to an extreme thermophile, Thermus thermophilus HB8. J. Virol. 1975;15:1449–1453. doi: 10.1128/jvi.15.6.1449-1453.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kristjansson J.K., Hjorleifsdottir S., Marteinsson V.T., Alfredsson G.A. Thermus scotoductus, sp. nov., a pigment-producing thermophilic, bacterium from hot tap water in Iceland and including Thermus sp. X-1. System. Appl. Microbiol. 1994;17:44–50. [Google Scholar]
- 4.Phizicky E.M., Schwartz R.C., Abelson J. Saccharomyces cerevisiae tRNA ligase. Purification of the protein and isolation of the structural gene. J. Biol. Chem. 1986;261:2978–2986. [PubMed] [Google Scholar]
- 5.Martins A., Shuman S. An RNA ligase from Deinococcus radiodurans. J. Biol. Chem. 2004;279:50654–50661. doi: 10.1074/jbc.M407657200. [DOI] [PubMed] [Google Scholar]
- 6.Blondal T., Hjorleifsdottir S.H., Fridjonsson O.F., Aevarsson A., Skirnisdottir S., Hermannsdottir A.G., Hreggvidsson G.O., Smith A.V., Kristjansson J.K. Discovery and characterization of a thermostable bacteriophage RNA ligase homologous to T4 RNA ligase 1. Nucleic Acids Res. 2003;31:7247–7254. doi: 10.1093/nar/gkg914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Silber R., Malathi V.G., Hurwitz J. Purification and properties of bacteriophage T4-induced RNA ligase. Proc. Natl Acad. Sci. USA. 1972;69:3009–3013. doi: 10.1073/pnas.69.10.3009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Durantel D., Croizier L., Ayres M.D., Croizier G., Possee R.D., Lopez-Ferber M. The pnk/pnl gene (ORF 86) of Autographa californica nucleopolyhedrovirus is a non-essential, immediate early gene. J. Gen. Virol. 1998;79:629–637. doi: 10.1099/0022-1317-79-3-629. [DOI] [PubMed] [Google Scholar]
- 9.Altschul S.F., Gish W., Miller W., Myers E.W., Lipman D.J. Basic local alignment search tool. J. Mol. Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 10.Sugino A., Snoper T.J., Cozzarelli N.R. Bacteriophage T4 RNA ligase. Reaction intermediates and interaction of substrates. J. Biol. Chem. 1977;252:1732–1738. [PubMed] [Google Scholar]
- 11.Ho C.K., Wang L.K., Lima C.D., Shuman S. Structure and mechanism of RNA ligase. Structure. 2004;12:327–339. doi: 10.1016/j.str.2004.01.011. [DOI] [PubMed] [Google Scholar]
- 12.Doherty A.J., Suh S.W. Structural and mechanistic conservation in DNA ligases. Nucleic Acids Res. 2000;28:4051–4058. doi: 10.1093/nar/28.21.4051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Timson D.J., Singleton M.R., Wigley D.B. DNA ligases in the repair and replication of DNA. Mutat. Res. 2000;460:301–318. doi: 10.1016/s0921-8777(00)00033-1. [DOI] [PubMed] [Google Scholar]
- 14.Kaufmann G., David M., Borasio G.D., Teichmann A., Paz A., Amitsur M. Phage and host genetic determinants of the specific anticodon loop cleavages in bacteriophage T4-infected Escherichia coli CTr5X. J. Mol. Biol. 1986;188:15–22. doi: 10.1016/0022-2836(86)90476-6. [DOI] [PubMed] [Google Scholar]
- 15.Amitsur M., Levitz R., Kaufmann G. Bacteriophage T4 anticodon nuclease, polynucleotide kinase and RNA ligase reprocess the host lysine tRNA. EMBO J. 1987;6:2499–2503. doi: 10.1002/j.1460-2075.1987.tb02532.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tyndall C., Meister J., Bickle T.A. The Escherichia coli prr region encodes a functional type IC DNA restriction system closely integrated with an anticodon nuclease gene. J. Mol. Biol. 1994;237:266–274. doi: 10.1006/jmbi.1994.1230. [DOI] [PubMed] [Google Scholar]
- 17.Tomita K., Ogawa T., Uozumi T., Watanabe K., Masaki H. A cytotoxic ribonuclease which specifically cleaves four isoaccepting arginine tRNAs at their anticodon loops. Proc. Natl Acad. Sci. USA. 2000;97:8278–8283. doi: 10.1073/pnas.140213797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liu X., Gorovsky M.A. Mapping the 5′ and 3′ ends of Tetrahymena thermophila mRNAs using RNA ligase mediated amplification of cDNA ends (RLM-RACE) Nucleic Acids Res. 1993;21:4954–4960. doi: 10.1093/nar/21.21.4954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Maruyama K., Sugano S. Oligo-capping: a simple method to replace the cap structure of eukaryotic mRNAs with oligoribonucleotides. Gene. 1994;138:171–174. doi: 10.1016/0378-1119(94)90802-8. [DOI] [PubMed] [Google Scholar]
- 20.Zhang X.H., Chiang V.L. Single-stranded DNA ligation by T4 RNA ligase for PCR cloning of 5′-noncoding fragments and coding sequence of a specific gene. Nucleic Acids Res. 1996;24:990–991. doi: 10.1093/nar/24.5.990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tessier D.C., Brousseau R., Vernet T. Ligation of single-stranded oligodeoxyribonucleotides by T4 RNA ligase. Anal. Biochem. 1986;158:171–178. doi: 10.1016/0003-2697(86)90606-8. [DOI] [PubMed] [Google Scholar]
- 22.Kaluz S., Kaluzova M., Flint A.P. Enzymatically produced composite primers: an application of T4 RNA ligase-coupled primers to PCR. Biotechniques. 1995;19:182–184. [PubMed] [Google Scholar]
- 23.Kinoshita Y., Nishigaki K., Husimi Y. Fluorescence-, isotope- or biotin-labeling of the 5′-end of single-stranded DNA/RNA using T4 RNA ligase. Nucleic Acids Res. 1997;25:3747–3748. doi: 10.1093/nar/25.18.3747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bradford M.M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 25.Sambrook J., Fritsch E.F., Maniatis T. Molecular Cloning: A Laboratory Manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 26.Richardson C.C. Phosphorylation of nucleic acid by an enzyme from T4 bacteriophage-infected Escherichia coli. Proc. Natl Acad. Sci. USA. 1965;54:158–165. doi: 10.1073/pnas.54.1.158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang L.K., Ho C.K., Pei Y., Shuman S. Mutational analysis of bacteriophage T4 RNA ligase 1. Different functional groups are required for the nucleotidyl transfer and phosphodiester bond formation steps of the ligation reaction. J. Biol. Chem. 2003;278:29454–29462. doi: 10.1074/jbc.M304320200. [DOI] [PubMed] [Google Scholar]
- 28.Shuman S., Schwer B. RNA capping enzyme and DNA ligase: a superfamily of covalent nucleotidyl transferases. Mol. Microbiol. 1995;17:405–410. doi: 10.1111/j.1365-2958.1995.mmi_17030405.x. [DOI] [PubMed] [Google Scholar]
- 29.Higgins D.G., Sharp P.M. CLUSTAL: a package for performing multiple sequence alignment on a microcomputer. Gene. 1988;73:237–244. doi: 10.1016/0378-1119(88)90330-7. [DOI] [PubMed] [Google Scholar]