Abstract
Aspartic acid residues comprising the -D-(aa)n-Y-L-D-D- DNA polymerase active site triad of reverse transcriptase from the Saccharomyces cerevisiae long terminal repeat-retrotransposon Ty3 (Asp151, Asp213 and Asp214) were evaluated via site-directed mutagenesis. An Asp151→Glu substitution showed a dramatic decrease in catalytic efficiency and a severe translocation defect following initiation of DNA synthesis. In contrast, enzymes harboring the equivalent alteration at Asp213 and Asp214 retained DNA polymerase activity. Asp151→Asn and Asp213→Asn substitutions eliminated both polymerase activities. However, while Asp214 of the triad could be replaced by either Asn or Glu, introducing Gln seriously affected processivity. Mutants of the carboxylate triad at positions 151 and 213 also failed to catalyze pyrophosphorolysis. Finally, alterations to the DNA polymerase active site affected RNase H activity, suggesting a close spatial relationship between these N- and C-terminal catalytic centers. Taken together, our data reveal a critical role for Asp151 and Asp213 in catalysis. In contrast, the second carboxylate of the Y-L-D-D motif (Asp214) is not essential for catalysis, and possibly fulfills a structural role. Although Asp214 was most insensitive to substitution with respect to activity of the recombinant enzyme, all alterations at this position were lethal for Ty3 transposition.
INTRODUCTION
Reverse transcriptase (RT) (1) belongs to the general class of polynucleotide polymerases, for which a two-metal ion-mediated mechanism of catalysis has been proposed (2,3). In brief, while one metal ion (conventionally designated Metal A) reduces the pKa of the 3′-OH of the primer strand to facilitate nucleophilic attack on the α-phosphate of the incoming deoxynucleoside triphosphate, a dual role of the second metal ion (Metal B) is envisaged. This includes stabilizing the negative charge building up on the leaving oxygen, and chelating the β- and γ-phosphates of the incoming deoxynucleoside triphosphate to facilitate their release as pyrophosphate. A central feature of the DNA polymerase active site of these enzymes is a triad of carboxylates in their palm subdomains, two of which are within a conserved -Y-X-D-D- motif, while a third is ∼60–100 residues N-terminal to this (4,5). Based on a combination of biochemical and structural studies, focusing primarily on the RTs of HIV-1 (6–13), Moloney murine leukemia virus (14) and Rous sarcoma virus (15,16), these residues have been generally considered as invariant. However, when the structures of several polynucleotidyl polymerases were compared, an absolute requirement for the second carboxylate of the -Y-X-D-D- motif has been challenged (17). In support of this notion are findings that (i) the second aspartate of this motif is rarely conserved in DNA-dependent RNA polymerases (5) and (ii) functional polymerases of minus-strand RNA viruses contain asparagine in place of the second aspartic acid (4).
In contrast to the wealth of data available for retroviral RT (8,15,16,18–24), there are only limited enzymatic studies on its counterpart from long terminal repeat (LTR)-containing retrotransposons, such as Ty1 and Ty3 from the budding yeast Saccharomyces cerevisiae. However, the recent availability of recombinant forms of each enzyme (25,26) has permitted analysis of both their DNA polymerase and ribonuclease H (RNase H) functions, as well as the structure of cis-acting signals governing the initiation of both (−) and (+) strand DNA synthesis (26–30). A study of Uzun and Gabriel (31) assessed the three conserved aspartic acid residues of the DNA polymerase catalytic center of Ty1 RT (Asp129, Asp210 and Asp211), indicating that, while the second aspartate of the Ty1 -Y-L-D-D- motif could be altered without affecting DNA polymerase activity, this mutation was lethal for transposition. More recently, Pandey et al. (32) have provided a detailed kinetic analysis of Ty1 RT mutant Asp211→Asn, but gave no details for other mutations at this position or at other positions of the active site triad. Furthermore, the consequence of mutating the DNA polymerase active site for RNase H activity was of particular interest to us, since a phylogenetic analysis of retroviral and retrotransposon RTs suggests that the latter lack the ‘tether’ or connection domain linking the DNA polymerase and RNase H catalytic centers (33). The proximity of the active sites suggests that mutations within one might have consequences for catalysis at the other.
In this communication, the contribution of Asp151, Asp213 and Asp214, constituting the triad of conserved carboxylates at the DNA polymerase active site Ty3 RT, was evaluated. Since analogous mutants within the active site of HIV-1 RT have a severe effect on DNA polymerase activity (34), it has been assumed that a precise geometry of the three carboxylates in the catalytic center is absolutely essential. Our results with Ty3 RT suggest more relaxed structural constrains for this catalytic triad. We demonstrate here that while substituting Asn for Asp151 and Asp213 completely disrupts Mg++-dependent activities, Asn and Glu can replace Asp214. In contrast, an Asp214→Gln mutation dramatically decreases catalytic activity. The Asp151→Glu mutation also has severe consequences for RNase H activity, suggesting close contact between the catalytic centers of Ty3 RT. In several instances, including mutant Asp151→Glu, metal rescue of DNA polymerase activity could be achieved by substitution of Mn++ for Mg++. Finally, although recombinant RTs containing Asn or Glu substitutions for Asp214 retain DNA polymerase function, these mutations are lethal for Ty3 transposition, which our data suggest may be due to reduced processivity during minus-strand, RNA-dependent DNA synthesis.
MATERIALS AND METHODS
Materials, strains and culture conditions
Oligodeoxynucleotides were obtained from Integrated DNA Technologies (Coralville, IA), and oligoribonucleotides from Dharmacon (Lafayette, CO). All other reagents were of the highest purity and purchased from Sigma. Escherichia coli and S.cerevisiae strains were cultured and transformed by standard methods. S.cerevisiae yTM443 (35) (MATa trp1-H3 ura3-52 his3 Δ200 ade2-101 lys2-1 leu1-12 can1-100 ΔTy3 ba r::hisG Gal3+), a derivative of yVB110 containing no endogenous copies of Ty3 (36), was used for transposition assays. Transposition analysis was performed as described previously (29). E.coli strain CJ236 (New England Biolabs) was used for the production of single-stranded DNA for site-directed mutagenesis (37). E.coli XL-10 Gold (Stratagene) was used to transform the mutated DNA plasmid according to the QuickChange XL Site-Directed Mutagenesis Kit (Stratagene).
Site-directed mutagenesis
Point mutations in the DNA polymerase domain of Ty3 RT expressed on plasmid p6HTy3RT (26) were introduced using the QuickChange XL Site-Directed Mutagenesis Kit (Stratagene). Colony screening was by DNA sequencing. The same mutations were introduced into plasmid pEGTy3-1 (38). Antisense primers homologous to the mutation site were used for site-directed mutagenesis as described previously (37).
Expression and purification of Ty3 RT mutants
Ty3 RT variants were purified by a modification of the procedure of Le Grice et al. (39). Enzymes were initially purified from logarithmically grown, isopropyl-β-d-thiogalactopyranoside-induced E.coli cultures by metal chelate chromatography (Ni++-NTA Sepharose; Qiagen) and subsequently by size exclusion chromatography (Superdex 200; Pharmacia). The latter purification step was conducted in a buffer containing 50 mM NaH2PO4/Na2HPO4 (pH 7.8), 0.7 M NaCl. Purified enzymes were stored at −20°C in a 50% glycerol-containing buffer (50 mM NaH2PO4/Na2HPO4 (pH 7.8), 0.7 M NaCl). Under these conditions, we observed minimal loss of activity over several months.
Enzymatic analyses
RNA-dependent DNA polymerase activity was evaluated on a 152 nt RNA template hybridized to a 5′ end-labeled 21 nt DNA primer. Template–primer was annealed by incubation at 95°C in 10 mM Tris/HCl (pH 7.8), 100 mM NaCl and slow cooling to room temperature. A reaction mixture containing 50 nM template–primer and 250 μM dNTPs was prepared in 10 mM Tris/HCl (pH 7.8), 80 mM NaCl, 5 mM DTT and 0.01% Triton X-100. An aliquot of 9 mM MgCl2 or 1 mM MnCl2 was used for Mg++- or Mn++-dependent polymerization, respectively Optimal Mg++ and Mn++ conditions for Ty3 RT were previously determined by Lener et al. (29). DNA synthesis was initiated at 30°C by adding varying amounts of wild-type or mutant RT as indicated in the text in a final reaction volume of 10 μl. Reactions were stopped after 30 min by mixing with an equal volume of 89 mM Tris-borate, pH 8.3, 2 mM EDTA (TBE), and 95% (v/v) formamide containing 0.1% (w/v) bromophenol blue and xylene cyanol. Polymerization products were resolved by high-voltage denaturing 10% PAGE and visualized by phosphorimaging. Processivity during RNA-dependent DNA polymerase activity was evaluated using the substrate indicated above. The reaction was modified as follows: a reaction mixture containing 50 nM template–primer was prepared in a buffer of 10 mM Tris/HCl (pH 7.8), 9 mM MgCl2 or 1 mM MnCl2, 80 mM NaCl, 5 mM DTT and 0.01% Triton X-100. Wild-type or mutant RT (final concentration of 200 nM in a final reaction volume of 10 μl) was added and the reaction incubated at room temperature for 5 min. DNA synthesis was initiated at 30°C by the addition of 250 μM dNTPs and 2 mg/ml heparin. Polymerization products were resolved as described above. DNA-dependent DNA polymerase activity and processivity during DNA-dependent DNA polymerase activity were evaluated on a 71 nt DNA template hybridized to a 5′-32P-end-labeled 36 nt DNA primer (40,41), using the same reaction conditions. Control reactions were performed adding heparin prior to Ty3 RT in order to verify the effectiveness of the trap. Under such conditions, DNA synthesis was eliminated (data not shown).
RNase H activity was determined on a 5′ end-labeled 40 nt RNA template (5′-UCAUGCCCUGCUAGCUACUCGAUAUGGCAAUAAGACUCCA-3′) annealed to a 30 nt DNA primer (5′-TGGAGTCTTATTGCCATATCGAGTAGCTAG-3′). A reaction mixture containing 50 nM template–primer was prepared in a buffer containing 10 mM Tris/HCl (pH 7.8), 80 mM NaCl, 5 mM DTT, 0.01% Triton X-100 and 9 mM MgCl2. Hydrolysis was initiated by adding enzyme to a final concentration of 100 nM in a final volume of 10 μl. Reactions were stopped after 30 min by mixing with an equal volume of TBE and 95% (v/v) formamide containing 0.1% (w/v) bromophenol blue and xylene cyanol. Hydrolysis products were resolved by high-voltage denaturing PAGE.
Pyrophosphorolysis was evaluated in the buffer described above for DNA synthesis, using a 21 nt 5′-32P-end-labeled DNA primer (5′-GACAGGGATGGAAAGGATCAC-3′) hybridized to a 62 nt DNA template (5′-GGGACTAGGCCATTCTAATTCCCGTTTACGCCTCTCCTGGTGATCCTTTCCATCCCTGTCCC-3′) and a final concentration of 1 mM sodium pyrophosphate. An aliquot of 10 nM template/primer was incubated with 100 nM wild-type or mutant Ty3 RT for 60 min at 30°C in the presence of 9 mM MgCl2. Reaction products were stopped by the addition of EDTA, and resolved on 15% polyacrylamide/8 M urea gels by high-voltage electrophoresis.
Determination of dissociation constants
A 40 nt DNA template (5′-TCATGCCCTGCTAGCTACTCGATATGGCAATAAGACTCCA-3′) and 30 nt DNA primer (5′-TGGAGTCTTATTGCCATATCGAGTAGCTAG-3′) duplex (T/P, 0.3 nM) was incubated with varying concentrations of wild-type Ty3 RT and its mutant derivatives in a total volume of 30 μl containing 10 mM Tris/HCl (pH 7.8), 9 mM MgCl2, 80 mM NaCl, 5 mM DTT and 0.01% (v/v) Triton X-100. The reaction was incubated for 10 min at 30°C and the nucleoprotein complexes were resolved at 4°C through a 7% non-denaturing polyacrylamide gel using Tris-borate buffer (85 mM Tris/HCl, 85 mM boric acid, pH 8.0). The amount of T/P in the binary complex (E-T/P) and in free form with respect to the fraction of the bound DNA was plotted against enzyme concentration, and the dissociation constant (Kd) was determined using a hyperbolic binding curve.
Steady-state kinetics of polymerization
Steady-state kinetic parameters kcat and Km were determined using homopolymeric poly(rA)/oligo(dT)15, poly(rC)/(dG)18 and poly(dC)/(dG)18 as template–primer with the corresponding dNTP as the substrate. A modification of the procedure described by Kaushik et al. (42) was used. The reaction mixture contained 10 mM Tris/HCl, pH 7.8, 9 mM MgCl2, 80 mM NaCl, 5 mM DTT, 0.01% Triton X-100 and 9 μM oligo(dT)15 or oligo(dG)18 hybridized to complementary homopolymeric template [poly(rA) template at a 1:1 (w/w) ratio, poly(rC) and poly(dC) at 1:5 (w/w) ratio]. The concentration of Ty3 RT used in the assay was 9 nM for wild-type RT and most of the mutants, except Asp151→Glu and Asp214→Gln, where this increased to 150 nM. DNA synthesis was initiated at 30°C using variable concentrations of the corresponding α-[32P]dNTP. Reactions were stopped by mixing with an equal volume of 89 mM Tris-borate, pH 8.3, 2 mM EDTA, and 95% (v/v) formamide containing 0.1% (w/v) bromophenol blue and xylene cyanol. Polymerization products were resolved by high-voltage denaturing 10% PAGE and analyzed using phosphorimaging. Velocities for each substrate concentration were fit to the Michaelis–Menten equation and Km and Vmax values were determined. kcat was determined from the equation Vmax = kcat[E]total.
RESULTS
RNA-dependent DNA polymerase activity
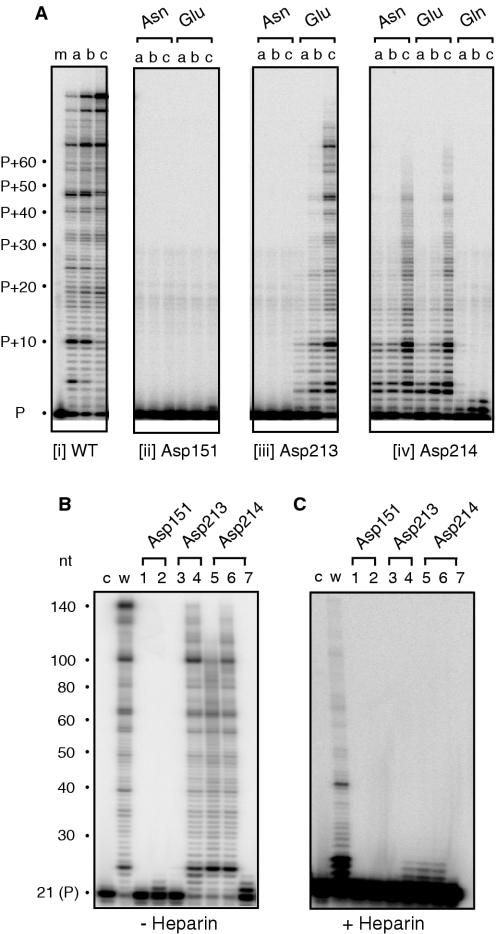
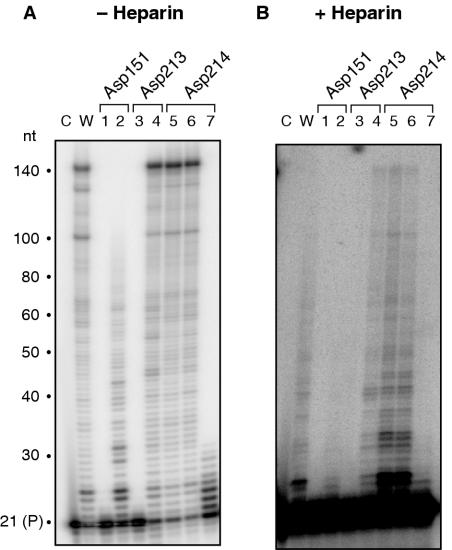
The RNA-dependent DNA polymerase activity of wild-type and mutant Ty3 enzymes was characterized under conditions allowing multiple rounds of enzyme binding (Figure 1A and B) or, by including the competitor heparin, restricting this to a single binding event (Figure 1C). In Figure 1A, the template–primer concentration was maintained at 50 nM and incubated with 0.8, 5 and 35 nM RT to assure steady-state conditions. Asp151→Asn and Asp 213→Asn mutations abolished catalytic activity, demonstrating the critical function for both of the aspartates (Figure 1A, panels ii and iii, respectively). However, introducing Glu at position 213 restores considerable RT activity (Figure 1A, panel iii). Primer extension products of ∼60–70 nt were generated with this mutant, and a further increase in enzyme concentration yielded full-length product (Figure 1B, lane 4). This result suggests that while a carboxylate function is required at position 213, the active site can accommodate different side-chain lengths. Under conditions of enzyme excess, mutant Asp151→Glu displays polymerase activity, but this is restricted to the addition of 1–2 nt to the primer (Figure 1B, lane 2).
Figure 1.
RNA-dependent DNA polymerase activity of Ty3 DNA polymerase active site mutants. (A) An aliquot of 50 nM template/primer was incubated with 0.8 nM (lane a), 5 nM (lane b) and 35 nM Ty3 RT (lane c) for 5 min, respectively. The active site residues analyzed and nature of the mutation are indicated below and above each panel, respectively. P, unextended primer. (B) RNA-dependent DNA polymerase activity in the absence of competitor heparin. For this analysis, 50 nM template/primer was incubated with 200 nM RT for 5 min. Lane c, 32P-end-labeled primer; lane w, wild-type Ty3 RT; lane 1, Asp151→Asn; lane 2, Asp151→Glu; lane 3, Asp213→Asn; lane 4, Asp213→Glu; lane 5, Asp214→Asn; lane 6, Asp214→Glu; and lane 7, Asp214→Gln. (C) RNA-dependent DNA polymerase activity in the presence of competitor heparin. Lane notations are as in (B).
The requirement for the second aspartate of the -Y-L-D-D- motif (Asp214) appears less critical, since its substitution with Asn and Glu yielded mutants with significant RT activity (Figure 1A, panel iv). The results with mutant Asp214→Glu also suggest that, similar to position 213, the increased side-chain length (relative to Asp) could be tolerated. However, increasing the side-chain length but introducing an amide function (Asp214→Gln) restricts DNA synthesis to the addition of 1–3 nt to the primer (Figure 1A, panel iv). The pausing pattern of mutants Asp213→Glu, Asp214→Asn and Asp214→Glu are similar to wild-type RT. Including heparin during RNA-dependent DNA synthesis restricts DNA synthesis wild-type Ty3 RT primarily to the addition of ∼4 nt to the primer (Figure 1C, lane w) most likely indicating frequent dissociation when secondary structure in the template is encountered. RNA-dependent DNA polymerase activity of mutants Asp213→Glu, Asp214→Asn and Asp214→Glu was likewise restricted to the addition of ∼4 nt (Figure 1C, lanes 4–6, respectively), while no activity was observed for the remaining active site mutants.
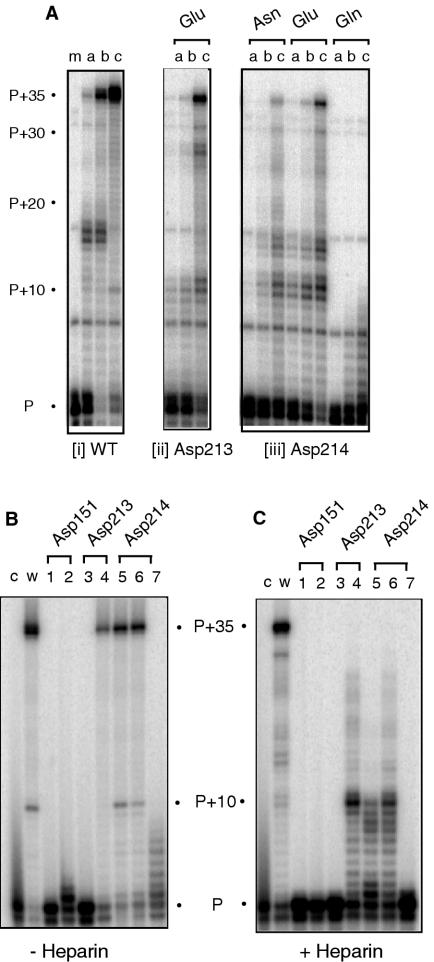
DNA-dependent DNA polymerase activity
DNA-dependent DNA polymerase activity was characterized in a similar manner to the experiments of Figure 1. The substrate for these studies was previously used for chemical and enzymatic footprinting experiments (40,41) and assumes a stem-loop structure through intramolecular base pairing 10 nt ahead of the template–primer duplex. This structure has proven useful in highlighting HIV-1 RT mutants defective or impaired in strand-displacement activity (43,44) through transient pausing or cessation of DNA synthesis, following the addition of ∼10 nt to the primer. Consistent with the RNA-dependent DNA polymerase activity assay of Figure 1, Asp151→Asn and Asp213→Asn substitutions eliminated activity on the DNA template (data not shown). Mutant Asp213→Glu was only slightly less active than wild-type RT, but showed a different pattern of pausing (Figure 2A, panel ii). Mutants where Asp214 was replaced with either Asn or Glu also displayed reduced DNA-dependent DNA polymerase activity, with significant pausing at position P+10, where the single-stranded template assumes an intramolecular base-paired structure (Figure 2A, panel iii). A similar alteration in the pausing pattern was observed by Pandey et al., (32) using a Ty1 RT mutant altered at the second aspartate of its of -Y-X-D-D- motif (Asp211). Finally, mutant Asp214→Gln catalyzed the addition of ∼8 nt to the primer (Figure 2A, panel iii).
Figure 2.
(A) DNA-dependent DNA polymerase activity of Ty3 DNA polymerase active site mutants Asp213→Glu, Asp214→Asn, Asp214→Glu and Asp214→Gln. Enzyme:substrate ratios and lane notations are as indicated in the legend to Figure 1A. (B and C) DNA-dependent DNA polymerase activity evaluated in the absence (B) and presence of heparin (C), respectively. Lane notations are as in the legend to Figure 1B.
In Figure 2B, the enzyme:substrate ratio was increased to 5:1 for all mutants to determine the severity of the DNA polymerase defect. As might be predicted, mutants Asp151→Asn and Asp213→Asn were still inactive (Figure 2B, lanes 1 and 3, respectively), while mutants Asp213→Glu, Asp214→Asn and Asp214→Glu were capable of fully extending the primer (Figure 2B, lanes 4–6, respectively). Under these conditions, however, mutant Asp151→Glu extended almost 50% of the primer by a single nucleotide (Figure 2B, lane 2), suggesting this alteration induced a translocation defect of the ternary complex. Surprisingly, while the substrate was almost exhausted, mutant Asp214→Gln still failed to extend the primer by more than 7–8 nt (Figure 2B, lane 7). The data of Figure 2A differ from recent findings of Uzun and Gabriel (31) who showed that an Asp211→Glu211 substitution of Ty1 RT (the equivalent of Asp214→Glu in Ty3) eliminated DNA polymerase function, but are in general agreement that the second Asp of the -Y-X-D-D- motif can tolerate substitution without loss of DNA polymerase activity. Finally, Figure 1C evaluates DNA synthesis in the presence of heparin. Although Asp213→Glu, Asp214→Asn and Asp214→Glu substitutions retained polymerase function, decreased processivity is evident, with the majority of primer extension products terminating on the template where intramolecular strand displacement activity is required (Figure 2C, lanes 4–6, respectively). Heparin also decreased processivity of mutant Asp214→Gln (Figure 2C, lane 7), although the addition of a single nucleotide was still possible. In contrast, mutant Asp151→Glu failed to add a single dNTP in the presence of heparin (Figure 2C, lane 2).
Affinity of mutant enzymes for duplex DNA
Complete loss of Mg++-dependent DNA polymerase activity for mutants Asp151→Asn and Asp213→Asn might indicate a severe catalytic defect following binding of Ty3 RT to template/primer. Alternatively, this could reflect reduced affinity of mutant enzyme for its substrate or a global conformational change incompatible with enzymatic function. Dissociation constants (Kds) for duplex DNA were therefore determined to distinguish between these possibilities (Table 1). Although such data have not been reported for retrotransposon RTs, our values of 3.6 ± 0.3 nM for wild-type Ty3 RT is consistent with Kds of 1–5 nM determined for retroviral RTs on duplex DNA or synthetic heteropolymers (45–47). The affinity of most active site mutants for duplex DNA shows little variation, suggesting that differences in enzymatic function are more related to a catalytic deficiency rather than reduced substrate binding. Mutants Asp213→Glu and Asp213→Asn showed a 2- to 3-fold increase in Kd (7.7 and 10.4 nM, respectively), indicating slightly reduced affinity for their substrate. However, these two mutants retain RNase H function, indicating that their global conformation is not compromised (see below).
Table 1.
Affinity of Ty3 mutants for duplex DNA
| Mutation | Kd (nM) |
|---|---|
| WT | 3.6 ± 0.3 |
| Asp151→Asn151 | 3.0 ± 1.2 |
| Asp151→Glu151 | 5.5 ± 0.3 |
| Asp213→Asn213 | 10.4 ± 1.3 |
| Asp213→Glu213 | 7.7 ± 2.3 |
| Asp214→Asn214 | 3.4 ± 1.2 |
| Asp214→Glu214 | 2.9 ± 1.7 |
| Asp214→Gln214 | 2.9 ± 1.3 |
Dissociation constants for each enzyme are the average of duplicate experiments. Values shown ± SD are averaged from two independent experiments.
Kinetic parameters of wild-type and mutant enzymes
The catalytic efficiency (kcat/Km) of each enzyme was evaluated using steady-state kinetic analysis with the homopolymeric duplexes poly(rA)/(dT)15, poly(rC)/(dG)18 and poly(dC)/(dG)18 (48). Catalytic parameters were not determined for Ty3 RT mutants Asp151→Asn and Asp213→Asn, which showed severe polymerization defects (Figures 1 and 2). Parameters determined for wild-type Ty3 RT were in the range previously reported for MMLV RT (49). Wild-type Ty3 RT exhibited the highest catalytic rate (kcat) on poly(dC)/(dG)18. This rate, 24 s−1, was 4- to 10-fold greater than that observed on the RNA homopolymeric templates (Table 2). Substituting Asp151 with Glu caused a decrease in kcat of approximately five orders of magnitude, indicating a strict requirement for aspartic acid at this position. Consistent with data of Figures 1 and 2, the substitutions Asp213→Glu, Asp214→Asn and Asp214→Glu were less deleterious for DNA polymerase function, decreasing the catalytic rate 9- to 122-fold on a homopolymeric RNA template and 28- to 550-fold on a homopolymeric DNA template. However, the difference between the wild type and the mutants may be underestimated because of the nature of the steady-state assay (i.e. RT dissociation contributes to the catalytic rate). However, the relatively high catalytic rates for wild-type RT (ranging from 2.44–24.3 s−1) suggest that enzyme dissociation does not contribute significantly to kcat. The decrease is less dramatic compared with similar steady-state report with analogous substitutions at the HIV-1 RT catalytic center (Asp185 and Asp186) (34), where the decrease in kcat was approximately three orders of magnitude. In contrast, the Asp213→Glu mutant had a 37-fold increase in Km on poly(rA)/(dT)15. In this case, the change in the Km is, however, offset by a relatively high kcat resulting in a catalytic efficiency in the same range as those determined for mutants Asp214→Asn and Asp214→Glu.
Table 2.
Kinetic parameters for wild-type and mutant Ty3 RT variants
| Enzyme | KmdTTP (μM) | Kcat (s−1) | Kcat/Km (μM−1 S−1) | |
|---|---|---|---|---|
| poly(rA):oligo(dT)15 | WT | 14 | 2.4 | 0.18 |
| Asp151→Glu | 690 | 2.1 × 10−5 | 0.3 × 10−7 | |
| Asp213→Glu | 510 | 0.15 | 0.29 × 10−3 | |
| Asp214→Asn | 34 | 0.02 | 0.59 × 10−3 | |
| Asp214→Glu | 78 | 0.031 | 0.41 × 10−3 | |
| Asp214→Gln | 26 | 0.0016 | 0.62 × 10−4 | |
| Enzyme | KmdGTP (μM) | Kcat (s−1) | Kcat/Km (μM−1 S−1) | |
| poly(rC):oligo(dG)18 | WT | 42 | 6.2 | 0.15 |
| Asp151→Glu | ND | ND | ND | |
| Asp213→Glu | 260 | 0.67 | 0.26 × 10−2 | |
| Asp214→Asn | 42 | 0.12 | 0.28 × 10−2 | |
| Asp214→Glu | 64 | 0.19 | 0.3 × 10−2 | |
| Asp214→Gln | ND | ND | ND | |
| poly(dC):oligo(dG)18 | ||||
| WT | 19 | 24 | 1.3 | |
| Asp151→Glu | ND | ND | ND | |
| Asp213→Glu | 140 | 0.88 | 0.64 × 10−2 | |
| Asp214→Asn | 4 | 0.044 | 0.012 | |
| Asp214→Glu | 14 | 0.12 | 0.86 × 10−2 | |
| Asp214→Gln | ND | ND | ND |
The steady-state parameters were derived using the template/primer indicated and corresponding dNTP substrate as described in Materials and Methods. ND, not determined.
A similar trend was observed for all homopolymeric templates. However, the lower Km for the Asp214→Asn and Asp214→Glu substitutions compared with the Asp213→Glu mutant and wild-type RT with poly(dC)/(dG)18 is interesting. In general, Km reflects both the apparent affinity of the enzyme for substrate as well as the catalytic rate. However, for processive polymerases, Kd is the major component of Km (50). Comparing the effect of substitution Asp213→Glu and Asp214→Asn on the Km with the changes in the catalytic rates for the respective mutants suggests that substitution at position 214 does not significantly affect the affinity of the enzyme for dNTP. A carboxylate group contains two virtually equivalent negatively charged oxygens that have the potential to interact with two independent cations, while an amide can only interact with a single cation. Since Asn and Glu potentially interact with 1 or 2 cations, respectively, yet induce similar kinetic effects when substituted for Asp in the protein, it can be reasoned that 214 only coordinates one Mg ion. In contrast, a different phenotype was observed for Asp213, where enzyme substituted with Glu retained significant polymerase activity, but introducing Asn was inhibitory.
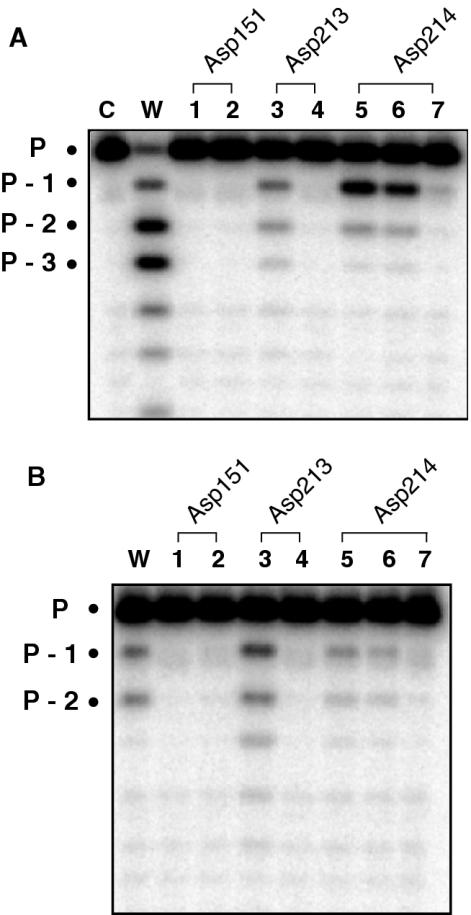
Pyrophosphorolysis activity
Active site residues implicated in phosphodiester bond formation should also be those that catalyze the reverse reaction, namely pyrophosphorolysis, or pyrophosphate-catalyzed reduction in primer length and release of dNTP. Figure 3A illustrates the pyrophosphorolysis activity of each Ty3 active site mutant. In Figure 3B, wild-type and mutant enzymes were incubated with substrate, but in the absence of pyrophosphate in order to determine the level of contaminating 3′→5′ exonuclease activity which might be incorrectly interpreted as pyrophosphorolysis. Taking into account the low level of contaminating exonuclease activity indicates that mutating Asp151 and Asp213 to either Asn or Glu eliminates pyrophosphorolysis (Figure 3A and B, lanes 1–4). In contrast, the equivalent mutations at position 214 reduce, but do not eliminate pyrophosphorolysis (Figure 3A and B, lanes 5 and 6), while activity is lost when Gln is introduced at position 214. Thus, in conjunction with alterations to DNA polymerase activity, the data of Figure 3 argue that Ty3 RT residues Asp151 and Asp213 coordinate divalent metal ions A and B required for catalysis. Assuming that the interactions of catalytic carboxylates with pyrophosphate are not direct but mediated by metal ion B, tolerance of the pyrophosphorolysis reaction to substitution of Asp214 suggests that an interaction of this residue with metal ion B is unlikely. This would agree with the model of HIV RT proposed by Huang et al. (12), which localizes the second aspartic acid of the -Y-X-D-D- motif in the vicinity of the metal A but distant to metal B. However, our data do not provide a definitive conclusion regarding the involvement of Asp214 in metal A coordination. Although direct evidence has not been provided, a structural contribution of Asp214 in the form of maintaining active site architecture might be envisaged. The notion that only one aspartate of the Ty3 -Y-L-D-D- motif is required for metal ion coordination is also supported by crystallographic studies on DNA polymerase from the bacteriophages T7 (51) and RB69 (52).
Figure 3.
Pyrophosphorolysis activity of Ty3 RT active site mutants. (A and B) Incubation of Ty3 RT with substrate in the presence and absence of pyrophosphate, respectively. Lane C, no RT; lane w, wild-type Ty3 RT; lane 1, mutant Asp151→Asn; lane 2, mutant Asp151→Glu; lane 3, mutant Asp213→Asn; lane 4, mutant Asp213→Glu; lane 5, mutant Asp214→Asn; lane 6, mutant Asp214→Glu; and lane 7, mutant Asp214→Gln. Migration position of the intact primer is designated P.
Mn++-catalyzed DNA polymerase activity
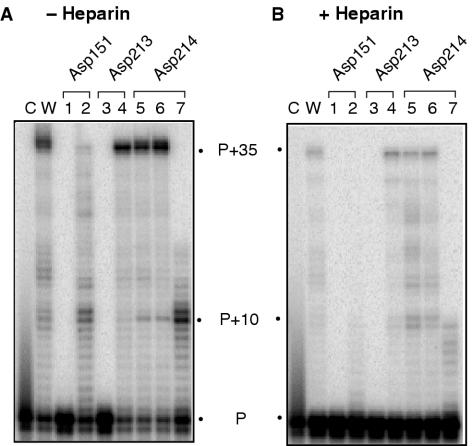
Our previous studies of the RNase H domain of HIV-1 (53) and Ty3 RT (29) demonstrated that substituting Mn++ for Mg++ would restore activity to catalytic site mutants. Mn++ has also been shown to partially restore catalytic function to an active site mutant of φ29 DNA polymerase (54), proposed by these authors to reflect distortions of the active site induced by the larger divalent metal ion. We therefore determined whether DNA polymerase activity of Ty3 active site mutants could be rescued by altering the divalent metal ion requirement. The results of this analysis are presented in Figures 4 and 5.
Figure 4.
Mn++-supported DNA-dependent DNA polymerase activity of wild-type (lane w) and mutant Ty3 derivatives (lanes 1–7). Lane c, no enzyme. Enzymes were analyzed in the absence (A) and presence of heparin (B). Substrate and lane notations are according to the legend of Figure 1B.
Figure 5.
Mn++-supported RNA-dependent DNA polymerase activity of wild-type (lane w) and mutant Ty3 derivatives (lanes 1–7). Lane c, no enzyme. Enzymes were analyzed in the absence (A) and presence of heparin (B). Substrate and lane notations are according to the legend of Figure 1B.
Under conditions allowing multiple turnover, Mn++ failed to rescue DNA-dependent DNA polymerase activity of mutants Asp151→Asn and Asp213→Asn (Figure 4A, lanes 1 and 3), while synthesis only slightly beyond the P+10 pause site was observed for mutant Asp214→Gln (Figure 4A, lane 7). Asp213→Glu, Asp214→Asn and Asp214→Glu substitutions supported Mn++-dependent DNA synthesis as efficiently as wild-type Ty3 RT (Figure 4A, lanes 4–6, respectively). Surprisingly, a substantial recovery in activity was evident for mutant Asp151→Glu in the presence of Mn++. In contrast to only extending the primer by 1–2 nt in the presence of Mg++ (Figure 2B, lane 2), fully extended primer is evident in the presence of Mn++ (Figure 4A, lane 2).
The ability of mutant Asp214→Gln to extend the primer to the P+10 pause site in the presence of Mn++, albeit at low levels, suggests that Mn++ can stimulate recovery of processivity in the presence of heparin (cf. lane 7 of Figures 4B and 2C. In support of this notion, wild-type Ty3 RT and mutants Asp213→Glu, Asp214→Asn and Asp214→Glu were equally processive in the presence of heparin while these mutants had reduced processivity in the presence of Mg++ (lane w versus lanes 4–6 of Figures 4B, 2B, 2C). However, inclusion of heparin virtually eliminated Mn++- stimulated DNA-dependent DNA polymerase activity of mutant Asp151→Glu (Figure 4B, lane 2), suggesting significantly reduced catalytic efficiency.
Similar results were obtained when RNA-dependent DNA polymerase activity was examined (Figure 5), with the exception of mutant Asp214→Gln. In the presence of Mn++, elevated levels of DNA synthesis were evident for this mutant, but restricted predominantly to P+1–P+7 products, which are only slightly larger than those obtained in the presence of Mg++ (cf. lane 7 of Figures 1B and 5A) This contrasts with mutant Asp151→Glu, where substantially increased processivity (i.e. product length) was obtained in the presence of Mn++ (cf. lane 2 of Figures 1B and 5A).
RNase H activity
A recent phylogenetic study, comparing the RNase H domains of retroviruses and LTR-retrotransposons (33), suggests that the latter lack the ‘tether’ or connection subdomain between the DNA polymerase and RNase H active centers characteristic of retroviral enzymes. The proximity of the Ty3 RT catalytic centers raised the possibility that altering one active site might have consequences for the other, potentially through altering metal ion coordination. Using heteropolymeric RNA/DNA hybrids (26,29), we previously showed that Ty3 RT hydrolyzes the RNA strand at positions corresponding to 21 and 13 bp behind its DNA polymerase active center (defined as −21 and −13, respectively), which indirectly illustrates an altered spatial relationship of the two active sites relative to the more extensively characterized retroviral enzymes (15,55,56). Our RNase H analysis of the Ty3 DNA polymerase active site mutants is presented qualitatively in Figure 6A and quantified in Figure 6B.
Figure 6.
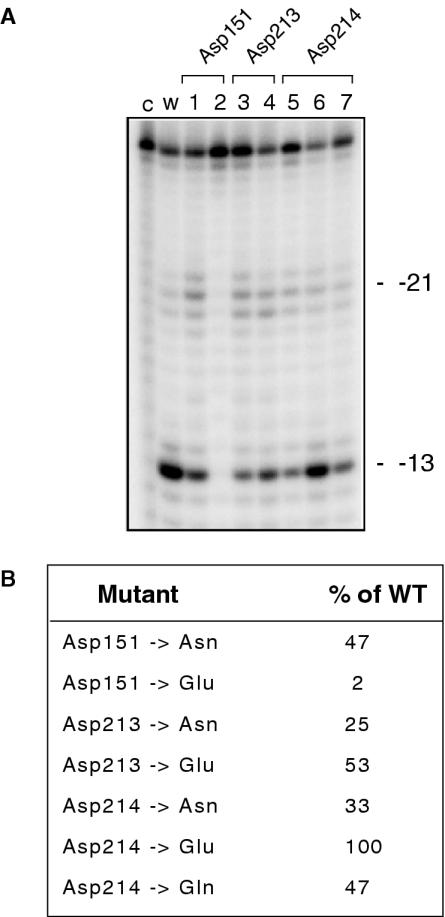
(A) RNase H activity of Ty3 RT DNA polymerase mutants. Lane notations are as described in the legend to Figure 1B. Migration positions of the −21 and −13 RNase H cleavage products have been indicated. (B) Quantification of RNase H hydrolysis data. Total hydrolysis products are expressed as a percentage of the wild-type value.
Although data of Figure 1A indicate that Asp151 is essential for DNA polymerase activity, its replacement differentially affects RNase H function, i.e. mutant Asp151→Glu is virtually inactive (Figure 6A, lane 2), while RNase H activity of mutant Asp151→Asn is ∼50% of wild-type RT (Figure 6A, lane 1). Asp213 is more tolerant to substitution with respect to RNase H activity, i.e. replacement with Asn and Glu reducing this to ∼25 and 50%, respectively, of wild-type levels (Figure 6A, lanes 3 and 4, respectively). Finally, although replacing Asp214 with Glu does not affect RNase H function (Figure 6A, lane 6), introducing the amide-containing side chain of Asn or Gln is deleterious, reducing activity ∼2- (Asp214→Asn) to 3-fold (Asp214→Gln). Thus, although we cannot distinguish between mutations that eliminate metal binding and those which might change the coordination geometry of the bound cation (57), the data of Figure 6 strongly suggest a greater degree of communication between the N- and C-terminal catalytic centers of Ty3 RT relative to the retroviral enzymes.
Asp214 substitutions are lethal for Ty3 transposition
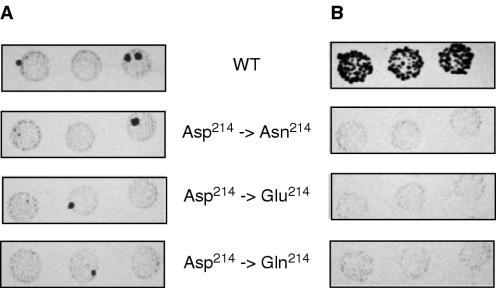
Taken together, the data of Figures 1–6 indicate that the second carboxylate (Asp214) of the Ty3 -Y-L-D-D- active site motif is more tolerant to substitution than Asp151 or Asp213 with respect to DNA polymerase and RNase H activity, suggesting that it may not participate in metal ion coordination. At the same time, DNA polymerase activity of mutants Asp214→Glu and Asp214→Asn was almost unaffected on a DNA template (Figure 2) and only slightly on an RNA template (Figure 1) under conditions allowing multiple rounds of synthesis. In order to determine the biological consequences of mutations that appear to minimally influence polymerase activity, all mutations at position 214 were introduced into the RT domain of pEGTy3-1, a plasmid harboring a replication-competent Ty3 element (38). This system follows integration of Ty3 into the HIS3-marked target plasmid pCH2bo19V between divergent tRNA genes sup2bio and tRNAVal(AAC), activating of the former and suppressing the ade2-101 lys2-1 ochre nonsense mutations in the yeast host yTM443. Transposition is then scored as papillations on a synthetic complete medium lacking adenine and lysine (29). Figure 7 indicates that, while transposition was achieved with the wild-type clone, all mutations at position Asp214 were inhibitory. These strains could be rescued by co-transfection of a plasmid, containing a copy of the wild-type RT gene (data not shown), indicating that loss of transposition activity was directly related to alterations in the Ty3 RT DNA polymerase active site and not to artifacts arising from cloning.
Figure 7.
Transposition analysis of Ty3 mutants altered at codon 214 of the DNA polymerase active site. Transformants were patched onto selective (−His, −URA) SD to repress transposition, and grown for 2 days. Cells were then replica plated onto SD medium lacking lysine and adenine (A) or onto selective SG medium to induce transposition. Cells, grown on selective SG medium, were then replica plated onto SD medium lacking lysine and adenine. (B) Papillations on SD medium lacking lysine and adenine indicate transposition.
DISCUSSION
Despite sharing the general enzymatic properties of their retroviral counterparts, significant differences in (i) primer binding site structure (27), (ii) the position within the tRNA primer from which (−) strand synthesis is initiated (58), (iii) size and sequence of their (+) strand polypurine tract primers (28) and (iv) residues comprising the RNase H active center (29) have been noted for LTR-retrotransposon RTs. A more comprehensive understanding of reverse transcription would therefore benefit from comparing and contrasting these enzymes. We have focused here on the triad of carboxylate residues, Asp151, Asp213 and Asp214, of the Ty3 RT DNA polymerase catalytic center. While a strict requirement for aspartic acid at position 151 is clear, we found that those of the -Y-L-D-D- motif were surprisingly tolerant to substitution. The simplest phenotype to explain is loss of function, evidenced by Asp151→Asn and Asp213→Asn mutants, most likely indicating their involvement in metal ion coordination. In contrast, cessation of synthesis following the addition of the first dNTP (mutant Asp151→Glu) represents more complicated phenotypes. In the subsequent sections, the properties of each residue and its requirement for catalysis will be discussed individually.
Asp151. Collectively, our data indicate that Asp151 is the most critical residue of the carboxylate triad, since substitution with either Asn or Glu affects Mg++-dependent DNA polymerase, pyrophosphorolysis and RNase H activity. At the same time, the affinity of mutant enzymes for duplex DNA is only marginally affected, indicating that their structure is not globally compromised. Data of Figure 2B indicate that at an elevated enzyme:substrate ratio, mutant asp151→Glu extends ∼50% of the primer by a single nucleotide. Such a result would suggest a translocation defect, possibly caused by the inability to release pyrophosphate, thereby trapping the primer terminus at the pre-translocation stage, (N) site defined by Sarafianos et al. (59). Surprisingly, the deleterious effects of increasing side-chain length can be partially overcome by replacing Mg++ with Mn++, indicating that active site architecture can be re-modeled in the presence of a larger divalent cation.
Asp213. Although loss of activity by introducing the amide-containing side chain of Asn at position 213 might be expected, retention of DNA polymerase activity with mutant Asp213→Glu was unexpected, especially in light of a report that the equivalent mutation in HIV-1 RT (Asp185→Glu185) reduces kcat by three orders of magnitude (34). The unexpectedly small loss in catalytic rate (ranging between 10- and 28-fold for different templates) is partially compensated by a more significant increase in Km (ranging between 7- and 37-fold) compared with wild-type Ty3 RT (Table 2). The relatively high Km determined for the Asp213→Glu mutant most likely accounts for the severe defect in dNTP binding observed for this enzyme. While mutant Asp213→Glu retains some polymerase activity compared with Asp214→Asn and Asp214→Glu substitutions, this mutant lacks pyrophosphorolysis activity. Despite slightly reduced affinity for duplex DNA, mutant Asp213→Glu retains DNA-dependent DNA polymerase activity under conditions permitting a single binding event, pausing predominantly at a position where the single-stranded template assumes an intramolecular base-paired structure (60). Although RNA-dependent DNA synthesis is more severely affected under single-round conditions (Figure 1C), pausing is likewise affected after the addition of ∼3 deoxynucleotides, where structural predictions have indicated the potential for the RNA template to assume an intramolecular duplex (data not shown). Taken together with the loss of pyrophosphorolysis activity, we propose that Asp213, together with Asp151, represents the carboxylate functions critical for Mg++ coordination. However, the relatively high catalytic efficiency of mutant Asp213→Glu indicates that the flexibility in side-chain length at this position can be tolerated.
Asp214. Catalytic rate constants for mutants Asp214→Asn and Asp214→Glu were decreased 32- to 545-fold, depending on the template used (Table 2). The ability to substitute both Asn and Glu for Asp214 without compromising DNA polymerase activity in combination with the retention of pyrophosphorolysis activity indicates that the carboxylate function of the second aspartate of the Ty3 RT -Y-X-D-D- motif is less essential than the others of the catalytic triad, although we cannot rule out its involvement in catalysis. Interestingly, Asn and Glu substitutions at position 214 do not significantly affect the Km, which is in sharp contrast to mutant Asp213→Glu. It is therefore unlikely that Asp214 is involved in dNTP binding, which agrees with previous data of Pandey et al. (32). A role in active site architecture might be considered for Asp214, which would be consistent with a proposal for the third conserved carboxylate of φ29 DNA polymerase (54). Alternatively, Asp214 may contribute to the overall negative charge within the catalytic site to facilitate translocation, as suggested by Sarafianos et al. (59) for HIV-1 RT.
Our data suggest the importance of Asp151 in translocation. However, altering the divalent cation requirement from Mg++ to Mn++ alleviates this defect, enhancing processive DNA synthesis on both RNA and DNA templates (lane 2 of Figures 4A and 5A). Although indirect, such a result supports a role for Asp151 in metal ion coordination, presumably through alterations in active site architecture induced by the larger divalent cation. The proximity of the polymerase and RNase H domains in RTs of LTR-retrotransposons (33) also suggests that compromising the architecture of the former might induce a conformational change at the RNaseH active site, and possibly incorrect coordination of the divalent cation required for hydrolysis. However, it is also possible that even slight alteration in the mode of binary complex interaction may not manifest themselves as a significant decrease in the dissociation constant but still affect RNase H activity. Finally, while our biochemical analyses have indicated that substitutions at Asp214 do not eliminate activity of recombinant RT, such mutations are lethal in the context of transposition. Although our data cannot assess directly which step in reverse transcription is affected, we believe that reduced processivity and RNase H activity will combine to prevent synthesis of an integration-competent double-stranded DNA. Nonetheless, retention of biochemical activity in selected mutants, coupled with modulation of this activity by altering the divalent cation requirement, will allow in vitro analysis of the multiple steps of DNA synthesis catalyzed by an LTR-retrotransposon RT to be examined for the first time in detail.
Acknowledgments
Funding to pay the Open Access publication charges for this article was provided by the National Cancer Institute, National Institutes of Health, Department of Health and Human Services.
REFERENCES
- 1.Skalka A.M., Goff S.P. Reverse Transcriptase. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1993. [PubMed] [Google Scholar]
- 2.Beese L.S., Steitz T.A. Structural basis for the 3′–5′ exonuclease activity of Escherichia coli DNA polymerase I: a two metal ion mechanism. EMBO J. 1991;10:25–33. doi: 10.1002/j.1460-2075.1991.tb07917.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Steitz T.A. DNA- and RNA-dependent DNA polymerases. Curr. Opin. Struct. Biol. 1993;3:31–38. [Google Scholar]
- 4.Poch O., Sauvaget I., Delarue M., Tordo N. Identification of four conserved motifs among the RNA-dependent polymerase encoding elements. EMBO J. 1989;8:3867–3874. doi: 10.1002/j.1460-2075.1989.tb08565.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Delarue M., Poch O., Tordo N., Moras D., Argos P. An attempt to unify the structure of polymerases. Protein Eng. 1990;3:461–467. doi: 10.1093/protein/3.6.461. [DOI] [PubMed] [Google Scholar]
- 6.Larder B.A., Purifoy D.J., Powell K.L., Darby G. Site-specific mutagenesis of AIDS virus reverse transcriptase. Nature. 1987;327:716–717. doi: 10.1038/327716a0. [DOI] [PubMed] [Google Scholar]
- 7.Hizi A., McGill C., Hughes S.H. Expression of soluble, enzymatically active, human immunodeficiency virus reverse transcriptase in Escherichia coli and analysis of mutants. Proc. Natl Acad. Sci. USA. 1988;85:1218–1222. doi: 10.1073/pnas.85.4.1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Le Grice S.F., Naas T., Wohlgensinger B., Schatz O. Subunit-selective mutagenesis indicates minimal polymerase activity in heterodimer-associated p51 HIV-1 reverse transcriptase. EMBO J. 1991;10:3905–3911. doi: 10.1002/j.1460-2075.1991.tb04960.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kohlstaedt L.A., Steitz T.A. Reverse transcriptase of human immunodeficiency virus can use either human tRNA(3Lys) or Escherichia coli tRNA(2Gln) as a primer in an in vitro primer-utilization assay. Proc. Natl Acad. Sci. USA. 1992;89:9652–9656. doi: 10.1073/pnas.89.20.9652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jacobo-Molina A., Ding J., Nanni R.G., Clark A.D., Jr, Lu X., Tantillo C., Williams R.L., Kamer G., Ferris A.L., Clark P., et al. Crystal structure of human immunodeficiency virus type 1 reverse transcriptase complexed with double-stranded DNA at 3.0 A resolution shows bent DNA. Proc. Natl Acad. Sci. USA. 1993;90:6320–6324. doi: 10.1073/pnas.90.13.6320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rodgers D.W., Gamblin S.J., Harris B.A., Ray S., Culp J.S., Hellmig B., Woolf D.J., Debouck C., Harrison S.C. The structure of unliganded reverse transcriptase from the human immunodeficiency virus type 1. Proc. Natl Acad. Sci. USA. 1995;92:1222–1226. doi: 10.1073/pnas.92.4.1222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Huang H., Harrison S.C., Verdine G.L. Trapping of a catalytic HIV reverse transcriptase*template:primer complex through a disulfide bond. Chem. Biol. 2000;7:355–364. doi: 10.1016/s1074-5521(00)00113-7. [DOI] [PubMed] [Google Scholar]
- 13.Sarafianos S.G., Das K., Tantillo C., Clark A.D., Jr, Ding J., Whitcomb J.M., Boyer P.L., Hughes S.H., Arnold E. Crystal structure of HIV-1 reverse transcriptase in complex with a polypurine tract RNA:DNA. EMBO J. 2001;20:1449–1461. doi: 10.1093/emboj/20.6.1449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Georgiadis M.M., Jessen S.M., Ogata C.M., Telesnitsky A., Goff S.P., Hendrickson W.A. Mechanistic implications from the structure of a catalytic fragment of Moloney murine leukemia virus reverse transcriptase. Structure. 1995;3:879–892. doi: 10.1016/S0969-2126(01)00223-4. [DOI] [PubMed] [Google Scholar]
- 15.Werner S., Wohrl B.M. Homodimeric reverse transcriptases from rous sarcoma virus mutated within the polymerase or RNase H active site of one subunit are active. Eur. J. Biochem. 2000;267:4740–4744. doi: 10.1046/j.1432-1327.2000.01530.x. [DOI] [PubMed] [Google Scholar]
- 16.Werner S., Wohrl B.M. Asymmetric subunit organization of heterodimeric Rous sarcoma virus reverse transcriptase alphabeta: localization of the polymerase and RNase H active sites in the alpha subunit. J. Virol. 2000;74:3245–3252. doi: 10.1128/jvi.74.7.3245-3252.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Steitz T.A. DNA polymerases: structural diversity and common mechanisms. J. Biol. Chem. 1999;274:17395–17398. doi: 10.1074/jbc.274.25.17395. [DOI] [PubMed] [Google Scholar]
- 18.Boyer P.L., Ferris A.L., Hughes S.H. Mutational analysis of the fingers domain of human immunodeficiency virus type 1 reverse transcriptase. J. Virol. 1992;66:7533–7537. doi: 10.1128/jvi.66.12.7533-7537.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Boyer P.L., Ferris A.L., Clark P., Whitmer J., Frank P., Tantillo C., Arnold E., Hughes S.H. Mutational analysis of the fingers and palm subdomains of human immunodeficiency virus type-1 (HIV-1) reverse transcriptase. J. Mol. Biol. 1994;243:472–483. doi: 10.1006/jmbi.1994.1673. [DOI] [PubMed] [Google Scholar]
- 20.Sarafianos S.G., Pandey V.N., Kaushik N., Modak M.J. Glutamine 151 participates in the substrate dNTP binding function of HIV-1 reverse transcriptase. Biochemistry. 1995;34:7207–7216. doi: 10.1021/bi00021a036. [DOI] [PubMed] [Google Scholar]
- 21.Palaniappan C., Wisniewski M., Jacques P.S., Le Grice S.F., Fay P.J., Bambara R.A. Mutations within the primer grip region of HIV-1 reverse transcriptase result in loss of RNase H function. J. Biol. Chem. 1997;272:11157–11164. doi: 10.1074/jbc.272.17.11157. [DOI] [PubMed] [Google Scholar]
- 22.Beard W.A., Bebenek K., Darden T.A., Li L., Prasad R., Kunkel T.A., Wilson S.H. Vertical-scanning mutagenesis of a critical tryptophan in the minor groove binding track of HIV-1 reverse transcriptase. Molecular nature of polymerase–nucleic acid interactions. J. Biol. Chem. 1998;273:30435–30442. doi: 10.1074/jbc.273.46.30435. [DOI] [PubMed] [Google Scholar]
- 23.Jonckheere H., De Clercq E., Anne J. Fidelity analysis of HIV-1 reverse transcriptase mutants with an altered amino-acid sequence at residues Leu74, Glu89, Tyr115, Tyr183 and Met184. Eur. J. Biochem. 2000;267:2658–2665. doi: 10.1046/j.1432-1327.2000.01272.x. [DOI] [PubMed] [Google Scholar]
- 24.Shi Q., Singh K., Srivastava A., Kaushik N., Modak M.J. Lysine 152 of MuLV reverse transcriptase is required for the integrity of the active site. Biochemistry. 2002;41:14831–14842. doi: 10.1021/bi0258389. [DOI] [PubMed] [Google Scholar]
- 25.Wilhelm M., Boutabout M., Wilhelm F.X. Expression of an active form of recombinant Ty1 reverse transcriptase in Escherichia coli: a fusion protein containing the C-terminal region of the Ty1 integrase linked to the reverse transcriptase-RNase H domain exhibits polymerase and RNase H activities. Biochem. J. 2000;348:337–342. [PMC free article] [PubMed] [Google Scholar]
- 26.Rausch J.W., Grice M.K., Henrietta M., Nymark M., Miller J.T., Le Grice S.F. Interaction of p55 reverse transcriptase from the Saccharomyces cerevisiae retrotransposon Ty3 with conformationally distinct nucleic acid duplexes. J. Biol. Chem. 2000;275:13879–13887. doi: 10.1074/jbc.275.18.13879. [DOI] [PubMed] [Google Scholar]
- 27.Gabus C., Ficheux D., Rau M., Keith G., Sandmeyer S., Darlix J.L. The yeast Ty3 retrotransposon contains a 5′–3′ bipartite primer-binding site and encodes nucleocapsid protein NCp9 functionally homologous to HIV-1 NCp7. EMBO J. 1998;17:4873–4880. doi: 10.1093/emboj/17.16.4873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wilhelm M., Uzun O., Mules E.H., Gabriel A., Wilhelm F.X. Polypurine tract formation by Ty1 RNase H. J. Biol. Chem. 2001;276:47695–47701. doi: 10.1074/jbc.M106067200. [DOI] [PubMed] [Google Scholar]
- 29.Lener D., Budihas S.R., Le Grice S.F. Mutating conserved residues in the ribonuclease H domain of Ty3 reverse transcriptase affects specialized cleavage events. J. Biol. Chem. 2002;277:26486–26495. doi: 10.1074/jbc.M200496200. [DOI] [PubMed] [Google Scholar]
- 30.Lener D., Kvaratskhelia M., Le Grice S.F. Nonpolar thymine isosteres in the Ty3 polypurine tract DNA template modulate processing and provide a model for its recognition by Ty3 reverse transcriptase. J. Biol. Chem. 2003;278:26526–26532. doi: 10.1074/jbc.M302374200. [DOI] [PubMed] [Google Scholar]
- 31.Uzun O., Gabriel A. A Ty1 reverse transcriptase active-site aspartate mutation blocks transposition but not polymerization. J. Virol. 2001;75:6337–6347. doi: 10.1128/JVI.75.14.6337-6347.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pandey M., Patel S., Gabriel A. Insights into the role of an active site aspartate in Ty1 reverse transcriptase polymerization. J. Biol. Chem. 2004;279:47840–47848. doi: 10.1074/jbc.M406019200. [DOI] [PubMed] [Google Scholar]
- 33.Malik H.S., Eickbush T.H. Phylogenetic analysis of ribonuclease H domains suggests a late, chimeric origin of LTR retrotransposable elements and retroviruses. Genome Res. 2001;11:1187–1197. doi: 10.1101/gr.185101. [DOI] [PubMed] [Google Scholar]
- 34.Kaushik N., Rege N., Yadav P.N., Sarafianos S.G., Modak M.J., Pandey V.N. Biochemical analysis of catalytically crucial aspartate mutants of human immunodeficiency virus type 1 reverse transcriptase. Biochemistry. 1996;35:11536–11546. doi: 10.1021/bi960364x. [DOI] [PubMed] [Google Scholar]
- 35.Menees T.M., Sandmeyer S.B. Transposition of the yeast retroviruslike element Ty3 is dependent on the cell cycle. Mol. Cell. Biol. 1994;14:8229–8240. doi: 10.1128/mcb.14.12.8229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hansen L.J., Chalker D.L., Sandmeyer S.B. Ty3, a yeast retrotransposon associated with tRNA genes, has homology to animal retroviruses. Mol. Cell. Biol. 1988;8:5245–5256. doi: 10.1128/mcb.8.12.5245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kunkel T.A. Rapid and efficient site-specific mutagenesis without phenotypic selection. Proc. Natl Acad. Sci. USA. 1985;82:488–492. doi: 10.1073/pnas.82.2.488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hansen L.J., Sandmeyer S.B. Characterization of a transpositionally active Ty3 element and identification of the Ty3 integrase protein. J. Virol. 1990;64:2599–2607. doi: 10.1128/jvi.64.6.2599-2607.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Le Grice S.F., Cameron C.E., Benkovic S.J. Purification and characterization of human immunodeficiency virus type 1 reverse transcriptase. Methods Enzymol. 1995;262:130–144. doi: 10.1016/0076-6879(95)62015-x. [DOI] [PubMed] [Google Scholar]
- 40.Metzger W., Hermann T., Schatz O., Le Grice S.F., Heumann H. Hydroxyl radical footprint analysis of human immunodeficiency virus reverse transcriptase-template.primer complexes. Proc. Natl Acad. Sci. USA. 1993;90:5909–5913. doi: 10.1073/pnas.90.13.5909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wohrl B.M., Georgiadis M.M., Telesnitsky A., Hendrickson W.A., Le Grice S.F. Footprint analysis of replicating murine leukemia virus reverse transcriptase. Science. 1995;267:96–99. doi: 10.1126/science.7528942. [DOI] [PubMed] [Google Scholar]
- 42.Kaushik N., Talele T.T., Pandey P.K., Harris D., Yadav P.N., Pandey V.N. Role of glutamine 151 of human immunodeficiency virus type-1 reverse transcriptase in substrate selection as assessed by site-directed mutagenesis. Biochemistry. 2000;39:2912–2920. doi: 10.1021/bi991376w. [DOI] [PubMed] [Google Scholar]
- 43.Jacques P.S., Wohrl B.M., Ottmann M., Darlix J.L., Le Grice S.F. Mutating the “primer grip” of p66 HIV-1 reverse transcriptase implicates tryptophan-229 in template–primer utilization. J. Biol. Chem. 1994;269:26472–26478. [PubMed] [Google Scholar]
- 44.Ghosh M., Jacques P.S., Rodgers D.W., Ottman M., Darlix J.L., Le Grice S.F. Alterations to the primer grip of p66 HIV-1 reverse transcriptase and their consequences for template–primer utilization. Biochemistry. 1996;35:8553–8562. doi: 10.1021/bi952773j. [DOI] [PubMed] [Google Scholar]
- 45.Huber H.E., McCoy J.M., Seehra J.S., Richardson C.C. Human immunodeficiency virus 1 reverse transcriptase. Template binding, processivity, strand displacement synthesis, and template switching. J. Biol. Chem. 1989;264:4669–4678. [PubMed] [Google Scholar]
- 46.Reardon J.E., Furfine E.S., Cheng N. Human immunodeficiency virus reverse transcriptase. Effect of primer length on template–primer binding. J. Biol. Chem. 1991;266:14128–14134. [PubMed] [Google Scholar]
- 47.Kati W.M., Johnson K.A., Jerva L.F., Anderson K.S. Mechanism and fidelity of HIV reverse transcriptase. J. Biol. Chem. 1992;267:25988–25997. [PubMed] [Google Scholar]
- 48.Majumdar C., Abbotts J., Broder S., Wilson S.H. Studies on the mechanism of human immunodeficiency virus reverse transcriptase. Steady-state kinetics, processivity, and polynucleotide inhibition. J. Biol. Chem. 1988;263:15657–15665. [PubMed] [Google Scholar]
- 49.Kaushik N., Singh K., Alluru I., Modak M.J. Tyrosine 222, a member of the YXDD motif of MuLV RT, is catalytically essential and is a major component of the fidelity center. Biochemistry. 1999;38:2617–2627. doi: 10.1021/bi9824285. [DOI] [PubMed] [Google Scholar]
- 50.Wilson J.E., Porter D.J., Reardon J.E. Inhibition of viral polymerases by chain-terminating substrates: a kinetic analysis. Methods Enzymol. 1996;275:398–424. doi: 10.1016/s0076-6879(96)75024-3. [DOI] [PubMed] [Google Scholar]
- 51.Doublie S., Tabor S., Long A.M., Richardson C.C., Ellenberger T. Crystal structure of a bacteriophage T7 DNA replication complex at 2.2 A resolution. Nature. 1998;391:251–258. doi: 10.1038/34593. [DOI] [PubMed] [Google Scholar]
- 52.Wang J., Sattar A.K., Wang C.C., Karam J.D., Konigsberg W.H., Steitz T.A. Crystal structure of a pol alpha family replication DNA polymerase from bacteriophage RB69. Cell. 1997;89:1087–1099. doi: 10.1016/s0092-8674(00)80296-2. [DOI] [PubMed] [Google Scholar]
- 53.Cirino N.M., Cameron C.E., Smith J.S., Rausch J.W., Roth M.J., Benkovic S.J., Le Grice S.F. Divalent cation modulation of the ribonuclease functions of human immunodeficiency virus reverse transcriptase. Biochemistry. 1995;34:9936–9943. doi: 10.1021/bi00031a016. [DOI] [PubMed] [Google Scholar]
- 54.Saturno J., Lazaro J.M., Blanco L., Salas M. Role of the first aspartate residue of the “YxDTDS” motif of phi29 DNA polymerase as a metal ligand during both TP-primed and DNA-primed DNA synthesis. J. Mol. Biol. 1998;283:633–642. doi: 10.1006/jmbi.1998.2121. [DOI] [PubMed] [Google Scholar]
- 55.Miller J.T., Rausch J.W., Le Grice S.F. Evaluation of retroviral ribonuclease H activity. Methods Mol. Biol. 2001;160:335–354. doi: 10.1385/1-59259-233-3:335. [DOI] [PubMed] [Google Scholar]
- 56.Boyer P.L., Gao H.Q., Frank P., Clark P.K., Hughes S.H. The basic loop of the RNase H domain of MLV RT is important both for RNase H and for polymerase activity. Virology. 2001;282:206–213. doi: 10.1006/viro.2000.0827. [DOI] [PubMed] [Google Scholar]
- 57.Cowan J.A., Ohyama T., Howard K., Rausch J.W., Cowan S.M., Le Grice S.F. Metal-ion stoichiometry of the HIV-1 RT ribonuclease H domain: evidence for two mutually exclusive sites leads to new mechanistic insights on metal-mediated hydrolysis in nucleic acid biochemistry. J. Biol. Inorg. Chem. 2000;5:67–74. doi: 10.1007/s007750050009. [DOI] [PubMed] [Google Scholar]
- 58.Ke N., Gao X., Keeney J.B., Boeke J.D., Voytas D.F. The yeast retrotransposon Ty5 uses the anticodon stem-loop of the initiator methionine tRNA as a primer for reverse transcription. RNA. 1999;5:929–938. doi: 10.1017/s1355838299990015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sarafianos S.G., Clark A.D., Jr, Das K., Tuske S., Birktoft J.J., Ilankumaran P., Ramesha A.R., Sayer J.M., Jerina D.M., Boyer P.L., et al. Structures of HIV-1 reverse transcriptase with pre- and post-translocation AZTMP-terminated DNA. EMBO J. 2002;21:6614–6624. doi: 10.1093/emboj/cdf637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wohrl B.M., Tantillo C., Arnold E., Le Grice S.F. An expanded model of replicating human immunodeficiency virus reverse transcriptase. Biochemistry. 1995;34:5343–5356. doi: 10.1021/bi00016a005. [DOI] [PubMed] [Google Scholar]