Abstract
Single-molecule fluorescence spectroscopy can reveal mechanistic and kinetic details that may not be observed in static structural and bulk biochemical studies of protein synthesis. One approach requires site-specific and stable attachment of fluorophores to the components of translation machinery. Fluorescent tagging of the ribosome is a prerequisite for the observation of dynamic changes in ribosomal conformation during translation using fluorescence methods. Modifications of the ribosomal particle are difficult due to its complexity and high degree of sequence and structural conservation. We have developed a general method to label specifically the prokaryotic ribosome by hybridization of fluorescent oligonucleotides to mutated ribosomal RNA. Functional, modified ribosomes can be purified as a homogenous population, and fluorescence can be monitored from labeled ribosomal complexes immobilized on a derivatized quartz surface.
INTRODUCTION
Translation of genetic information into polypeptide by the ribosome is a fundamental and universal cellular process. The ribosome is a highly conserved and complex macromolecular assembly that has been extensively studied by bulk biochemical and structural techniques. Aided by protein elongation factors Tu and G (EF-Tu and EF-G), the ribosome reads trinucleotide codons on the mRNA and selects tRNAs carrying specific amino acids to synthesize the encoded polypeptide (1,2). The general scheme of translation has been outlined by classical biochemical and biophysical approaches, and the 3D structures of ribosomal particles have recently been solved. Despite this rich body of data, further dynamic and kinetic experiments are required to understand ribosome function and mechanism. Structural and biochemical evidence strongly support the functional importance of ribosome dynamics during protein synthesis (3–6).
Single-molecule fluorescence spectroscopy allows analysis of complex multistep and repetitive biological processes that are impossible to synchronize at the molecular level (7). Observation of dynamic behavior of individual particles yields spatial and kinetic information that is typically lost in bulk experiments due to ensemble averaging of signals from an asynchronous population of molecules. For single-molecule experiments, ribosomal complexes are tethered to derivatized quartz surfaces through a streptavidin–biotin linkage, such that fluorescence emission and fluorescence resonance energy transfer (FRET) can be monitored over time periods ranging from 25 ms to hours. Using fluorescently labeled tRNAs, we have applied single-molecule methods to observe dynamic equilibria between classical and hybrid tRNA conformations on the ribosome (8). Analysis of ribosome fidelity using the same approach led to the discovery of a novel transient state during tRNA accommodation (9). Although much can be learned about translation from analysis of ribosomal substrate conformations, more direct observation of the dynamic behavior of the ribosome itself is needed. Site-specific and stable attachment of fluorescent dyes to ribosomal domains is necessary to study their conformations using single-molecule fluorescence spectroscopy.
Several approaches have been explored previously to label the ribosome for biochemical and structural studies. Covalent attachment of chelated Fe2+ to unique cysteine residues of an isolated ribosomal protein was used to monitor local ribosome structure through the generation of hydroxyl radicals (10). Early bulk fluorescence studies used covalent modification of the rRNAs to label their 3′ ends with fluorescent dyes (11). Both methods involved labeling of a purified ribosomal protein or rRNA followed by reconstitution of the entire ribosomal particle from isolated components. This approach can result in an inhomogeneous population of particles whose activity might be compromised due to missing or misfolded constituents. Hybridization of oligonucleotides to the wild-type rRNA has been used as a non-covalent tagging method for crystallographic and electron microscopy experiments (12,13). Oligonucleotides were also used for structural and functional probing of rRNA regions (14–16). Similar methods using fluorescent oligonucleotides have been used for monitoring microbial populations with flow cytometry (17,18). The hybridization approach might have deleterious effects on the functionality of the ribosomal particles for a variety of reasons, such as steric hindrance or interference with proper fold of the wild-type rRNA.
Here, we present an efficient and flexible approach to label ribosomes for fluorescence studies by hybridization of fluorescent oligonucleotides to helical extensions engineered in the rRNA. Unlike ribosome reconstitution from isolated components, our approach allows the preparation of homogenous populations of modified ribosomal particles that retain functionality in vivo and in vitro. Engineering of hybridization sites in surface-accessible regions of rRNA ensures binding specificity and efficiency. Purified labeled ribosome complexes can be immobilized on derivatized quartz surfaces and assayed for function using single-molecule fluorescence spectroscopy.
MATERIALS AND METHODS
Ribosome mutagenesis
Mutations were introduced in selected helices of the Escherichia coli 16S rRNA by a two-step PCR using as template the pKK3535 plasmid (19) carrying the wild-type rrnB rRNA operon under the control of the native constitutive promoter P1P2. First, portions of the gene upstream and downstream of the mutation site were amplified with complementary extensions (Table 1), which were then used as a composite template for amplification of the entire mutant fragment. Mutant 16S rDNA fragments flanked by AflIII/BglII or BglII/XbaI restriction sites were ligated with the remaining portions of pKK3535 and transformed into the DH5α strain. Plasmids isolated from viable clones were sequenced and retransformed into the TA531 strain, which lacks all seven rrn operons and expresses its rRNA off the pHK-rrnC plasmid (20). Transformants were grown on carbenicillin media to select for pKK3535 derivatives, and loss of pHK-rrnC was confirmed by their sensitivity to kanamycin.
Table 1.
Oligonucleotides used for 16S rRNA mutagenesis
| Name | Sequence | Clone |
|---|---|---|
| 16S.A-1a | AAAAAGCGAAGCGGCACTG | External primer |
| 16S.A-1d | AATCCTGTTTGCTCCCCACG | External primer |
| 16S.G-1a | GTGTAGCGGTGAAATGCGTAGAG | External primer |
| 16S.G-1d | TCACAAACCAGCAAGTGGCG | External primer |
| 16S.A-1b | GAAGCGAGGCGACAGGAGTGGCGACGCTTCTTCCTGTTACCGTTCGACTT | helix 6 hairpin 1 |
| 16S.A-1c | GAAGCGTCGCCACTCCTGTCGCCTCGCTTCTTTGCTGACGAGTGGCG | helix 6 hairpin 1 |
| 16S.B-1b | TCCCGAGGCGACAGGAGTGGCGACGGGCACATTCTCATCTCTGAAAAC | helix 33a hairpin 1 |
| 16S.B-1c | GCCCGTCGCCACTCCTGTCGCCTCGGGAACCGTGAGACAGGTGCTGC | helix 33a hairpin 1 |
| 16S.B-2b | TCCCGGGGAGACAGGACTGGCGACGGGCACATTCTCATCTCTGAAAAC | helix 33a hairpin 2 |
| 16S.B-2c | GCCCGTCGCCAGTCCTGTCTCCCCGGGAACCGTGAGACAGGTGCTGC | helix 33a hairpin 2 |
| 16S.B-3b | TCCCGGAGCGACAGGACTGGCGACGGGCACATTCTCATCTCTGAAAAC | helix 33a hairpin 3 |
| 16S.B-3c | GCCCGTCGCCAGTCCTGTCGCTCCGGGAACCGTGAGACAGGTGCTGC | helix 33a hairpin 3 |
| 16S.B-4b | TCCCGGAGCGATCAGGAGTGGCGACGGGCACATTCTCATCTCTGAAAAC | helix 33a hairpin 4 |
| 16S.B-4c | GCCCGTCGCCACTCCTGATCGCTCCGGGAACCGTGAGACAGGTGCTGC | helix 33a hairpin 4 |
| 16S.B-5b | TCCCGGGGAGATCAGGATAGGCGACGGGCACATTCTCATCTCTGAAAAC | helix 33a hairpin 5 |
| 16S.B-5c | GCCCGTCGCCTATCCTGATCTCCCCGGGAACCGTGAGACAGGTGCTGC | helix 33a hairpin 5 |
| 16S.C-1b | TCCCGAGGCGACAGGAGTGGCGACGGGTTAAGCTACCTACTTCTTTTGC | helix 44 hairpin 1 |
| 16S.C-1c | ACCCGTCGCCACTCCTGTCGCCTCGGGAGGGCGCTTACCACTTTGTG | helix 44 hairpin 1 |
| 16S.D-1b | CGGCGAGGCGACAGGAGTGGCGACGCCGCTGGCAACAAAGGATAAGGG | helix 39 hairpin 1 |
| 16S.D-1c | CGGCGTCGCCACTCCTGTCGCCTCGCCGGGAACTCAAAGGAGACTGCC | helix 39 hairpin 1 |
| 16S.E-1b | GCAAAGCGAGGCGACAGGAGTGGCGACGCTTTACTCCCTTCCTCCCCGC | helix 17 hairpin 1 |
| 16S.E-1c | GTAAAGCGTCGCCACTCCTGTCGCCTCGCTTTGCTCATTGACGTTACCCGC | helix 17 hairpin 1 |
| 16S.F-1b | GGCCCGAGGCGACAGGAGTGGCGACGGGTCCCCCTCTTTGGTCTTGCG | helix 10 hairpin 1 |
| 16S.F-1c | GACCCGTCGCCACTCCTGTCGCCTCGGGCCTCTTGCCATCGGATGTGCC | helix 10 hairpin 1 |
| 16S.G-1b | CGCCCGAGGCGACAGGAGTGGCGACGGGCACAACCTCCAAGTCGACATCG | helix 26 hairpin 1 |
| 16S.G-1c | TGCCGTCGCCACTCCTGTCGCCTCGGGCGTGGCTTCCGGAGCTAACGC | helix 26 hairpin 1 |
Primers with names ending in ‘a’ and ‘b’ were used to amplify the upstream portion of the gene, while primers ‘c’ and ‘d’ were used to amplify the downstream portion. The resultant PCR products with extensions (shown in bold) were then used to amplify the entire mutant fragment.
Preparation of ribosomes and cell extracts
Ribosomes were purified from TA531 cells expressing wild-type or mutant pKK3535 following published protocols with slight modifications (21,22). Cell extracts for translation assays were prepared from MRE600 cells following the same protocols to remove ribosomes from clarified lysates. Ion exchange chromatography on DEAE cellulose (Whatman) was used to prepare tRNA-free extract for tRNA aminoacylation.
Preparation of translation factors, mRNA, tRNAs and oligonucleotides
Initiation factors IF-1, IF-2 and IF-3 were purified from overexpressing E.coli strains following published protocols (23,24). Elongation factors EF-Tu, EF-Ts and EF-G were expressed and purified as described previously (8). Short 5′-biotinylated mRNA containing a spacer region, strong Shine–Dalgarno sequence and the upstream portion of the T4 gene 32 was chemically synthesized (Dharmacon Research). Aminoacylation and formylation of tRNAfMet (Sigma) followed standard methods (25) using tRNA-free cell extract and 10-formyltetrahydrofolate as the formyl donor (26). The acp3U47 position of tRNAPhe (Sigma) was labeled with Cy5-NHS ester (Amersham Biosciences) following published protocols (27). Labeled tRNAPhe was purified by high-performance liquid chromatography using a TSK-phenyl 5-PW column (Tosohaas) and aminoacylated as described previously (28). Short synthetic DNA oligonucleotides (IDT DNA) were 5′ labeled with 32P and purified by gel filtration. Synthetic DNA oligonucleotides with amino modification at the 5′ end (IDT DNA) were tagged with Cy3-NHS ester (Amersham Biosciences) according to the manufacturer's recommendations and purified by elution from polyacrylamide gels.
Binding assays, in vitro translation and toeprinting
Binding assays were carried out with 10 nM 32P-labeled oligonucleotides and 100 nM purified ribosomes preheated at 42°C and incubated for 2 h at 37°C (unless indicated otherwise) in Tris-polymix buffer (8). Reactions were resolved on native composite gels containing 0.5% agarose, 2.75% acrylamide, 25 mM Tris OAc, pH 7.5, 6 mM KCl, 2 mM MgCl2, 1 mM DTT, 1% sucrose and 0.45% DMAPN (29), and quantified on PhosphorImager (Molecular Dynamics).
Translation assays were carried out according to the published protocols (30) using 1 μM purified ribosomes prehybridized with excess unlabeled oligonucleotide in Tris-polymix buffer. Reaction products were resolved on 12% SDS–PAGE and visualized on PhosphorImager (Molecular Dynamics). Ribosomes prepared in the same manner were used for toeprinting analysis as described previously (8).
Preparation and immobilization of initiation complexes and tRNA delivery
Purified ribosomes prehybridized with excess Cy3-labeled oligonucleotide were assembled onto 5′-biotinylated synthetic mRNA in the presence of fMet-tRNAfMet and initiation factors in Tris-polymix buffer following published protocols with slight modifications (31). Initiation complexes were purified using 10–40% (w/v) sucrose gradients in Tris-polymix buffer with 20 mM Mg(OAc)2 and immobilized on streptavidin-derivatized quartz surfaces as described previously (8). The preparation and delivery of EF-Tu/GTP/Phe-tRNAPhe (Cy5-acp3U47) ternary complex was according to the earlier protocols (8). Single-molecule fluorescence data collection using total internal reflection microscopy was carried out as described previously (8).
RESULTS
Choice of labeling strategy
The ribosome is a multicomponent assembly of rRNA and proteins, both of which are potential targets for attachment of fluorescent dyes. To ensure homogeneity and functionality of our ribosomal particles, we devised a method that involves the expression of pure mutant ribosomes that can be labeled by hybridization of fluorescent oligonucleotides to extensions in rRNA hairpins. The oligonucleotide hybridization approach also ensures maximum labeling specificity and stability, while allowing a wide distribution range of target sites throughout the ribosome structure.
Design and construction of labeling sites
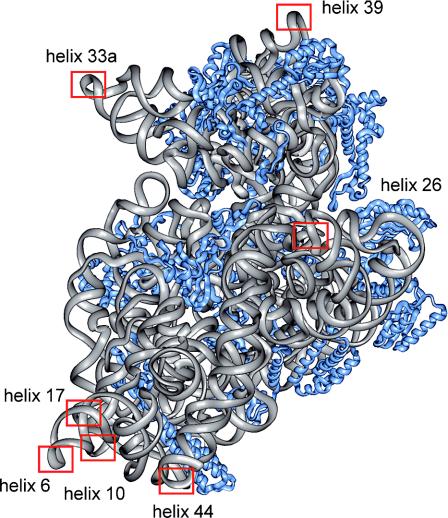
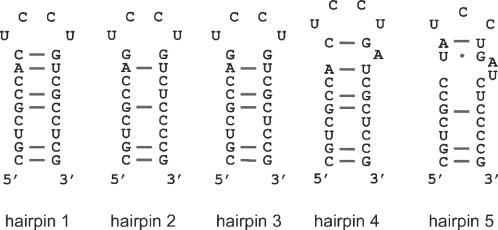
Sites for oligonucleotide hybridization were introduced into phylogenetically variable regions of the E.coli 16S rRNA by cloning 22–23 nt extensions that replaced the loops of surface-exposed rRNA helices. Using atomic resolution models of the prokaryotic ribosome as a physical map (32–34) and phylogenetic conservation analysis (based on the comparative RNA website http://www.rna.icmb.utexas.edu/), seven sites were selected for mutagenesis in the initial approach: helices 6, 10, 17 and 44 in the lower body of the 30S subunit, helices 33a and 39 in the head region, and helix 26 on the platform (Figure 1). The sequence of the rRNA insertion was designed to form a stable, partially mismatched helical extension that would not interfere with the native rRNA fold but would readily unfold and hybridize with an oligonucleotide to achieve efficient rRNA labeling (hairpin 1 shown in Figure 2). The insertions were cloned into rrnB rRNA operon-carrying plasmid pKK3535 (19) and transformed into the TA531 strain. In this strain, all seven chromosomal rrn operons have been deleted and wild-type rRNA is expressed from the pHK-rrnC plasmid (20). All mutant rRNA plasmids, except for those with helix 17 and helix 26 extensions, encoded viable functional rRNAs as demonstrated by the efficient replacement of pHK-rrnC with mutant pKK3535 derivative. The doubling times of E.coli expressing mutant ribosomes were comparable to wild-type strains (data not shown).
Figure 1.
The atomic resolution model of Thermus thermophilus 30S subunit (32) viewed from the subunit interface. Blue ribbons represent proteins and gray ribbons represent 16S rRNA. Sites of helix extensions introduced in corresponding regions of Escherichia coli 16S rRNA are highlighted.
Figure 2.
Hairpin extensions introduced into E.coli 16S rRNA. The insertion sequence was designed to form a partially mismatched stem loop that could unfold and base pair with a fluorescently labeled oligonucleotide.
Binding assays and optimization of hybridization sequence
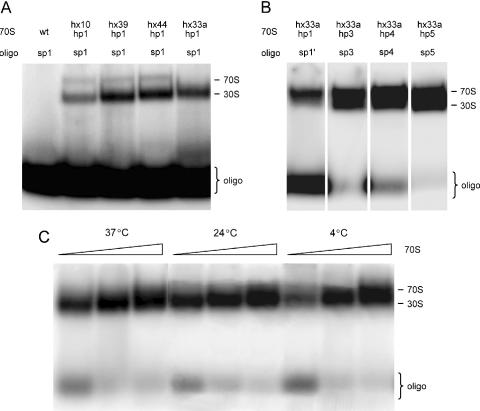
Mutant ribosomes expressed from plasmid pKK3535 containing hairpin 1 extensions in helices 10, 33a, 39 and 44 were isolated as pure populations from the TA531 strain; wild-type ribosomes were also expressed and isolated from this plasmid system. To test for binding of the oligonucleotide to the designed ribosomal extension and to optimize annealing conditions, we performed hybridization with 32P-labeled DNA oligonucleotides targeting various portions of hairpin 1 extension (Table 2). Relative binding affinities were measured using electrophoretic mobility shift assays on native composite gels. The results demonstrate that all sites are accessible for oligonucleotide hybridization, and that ribosome-oligonucleotide interaction is specific, as wild-type ribosomes do not bind oligonucleotides at the concentrations tested (Figure 3A). However, oligonucleotide binding is not very efficient since only small portion of oligonucleotide binds to excess ribosomes. We estimate a KD of ∼1 μM, which is orders of magnitude greater than that predicted by simple base-pairing thermodynamics of single strands (35). These results indicate that under our conditions the thermodynamics of the target hairpin favors its native helical conformation rather than the oligonucleotide-hybridized form. Increasing the hybridization temperature and time, and testing various salt conditions, only slightly improved binding affinity and was not optimal for ribosome activity (data not shown). Binding affinity was somewhat higher for oligonucleotides targeting the 3′ region of the target hairpin and leaving its 5′ portion single stranded and flexible. Coaxial stacking of the native rRNA helix and the rRNA–oligonucleotide duplex might increase the stability of the complex (data not shown).
Table 2.
Selected oligonucleotides used for hybridization to the 16S rRNA insertions
| Name | Sequence | Target insertion |
|---|---|---|
| sp1 | AGGCGACAGGAGTGGCGA | hairpin 1 |
| sp1′ | AGGCGACAGGAGTG | hairpin 1 |
| sp3 | GAGCGACAGGACTG | hairpin 3 |
| sp4 | GAGCGATCAGGAGT | hairpin 4 |
| sp5 | GGGAGATCAGGATA | hairpin 5 |
Figure 3.
Gel shift assays showing the binding of 32P-labeled oligonucleotides to 70S ribosomes isolated from various mutant strains. Oligonucleotides were hybridized to ribosomes preheated at 42°C and slowly cooled to 37°C for 2 h. Free oligonucleotide runs at the bottom of the gel. Oligonucleotide bound to mutant ribosomes forms a slower migrating complex on top of the gel. The two bands observed on top of some lanes are due to the oligonucleotide interaction with both intact 70S ribosomes and 30S subunits resulting from partial 70S dissociation in this low Mg2+ gel system. (A) Hybridization of oligonucleotide sp1 to hairpin 1 (hp1) insertions in different regions of the 16S rRNA: wild-type ribosomes (wt), insertion in helix 10 (hx10), helix 39 (hx39), helix 44 (hx44) and helix 33a (hx33a). (B) Binding of oligonucleotides to different hairpin constructs cloned into helix 33a: hairpin 1 (hp1), hairpin 3 (hp3), hairpin 4 (hp4) and hairpin 5 (hp5). (C) Lower temperature hybridization of oligonucleotide sp5 to increasing amounts of ribosomes with hairpin 5 extension in helix 33a.
To improve binding affinity, a series of target hairpins were designed, in which more mismatches were introduced into the hairpin stem (selected constructs, hairpin 2 through 5, are shown in Figure 2). We expected that destabilization of the rRNA hairpin structure should shift the ribosome–oligonucleotide binding equilibrium towards the bound form. The insertions were cloned into helix 33a and functionality of mutant ribosomes was again tested in vivo in the TA531 strain. Mutants containing insertions with too many mismatches (e.g. hairpin 2) were lethal, possibly due to unfolding, misassembly or degradation of rRNA. Functional ribosomes purified from viable mutants (hairpins 3, 4 and 5) showed very efficient hybridization of oligonucleotide with KD of ∼15 nM (Figure 3B) even under gentle non-inactivating conditions (Figure 3C).
Activity assays of mutant ribosomes
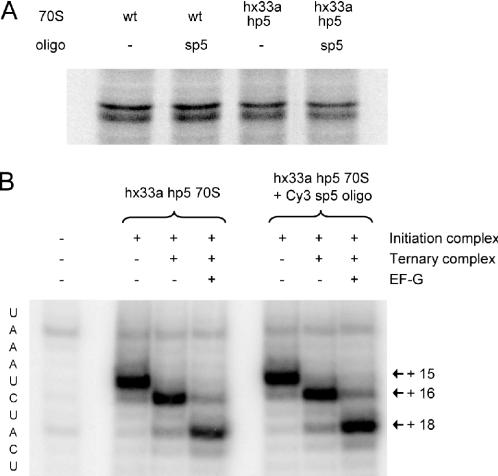
The functionality of purified mutant ribosomes containing hairpin 5 extension in helix 33a prehybridized with the sp5 oligonucleotide was demonstrated using in vitro translation assays. Comparison of the radioactive protein product yields in translation reactions with and without the oligonucleotide confirms that its presence on the ribosomes does not affect translational activity. Mutant ribosomes translate at more than 90% activity of wild-type ribosomes both in the presence and absence of the oligonucleotide (Figure 4A). In addition, mutant ribosomes prehybridized with the oligonucleotide and assembled into initiation complexes with mRNA and initiator tRNA were shown to bind aminoacyl-tRNA and translocate using standard toeprinting analysis (Figure 4B). In toeprinting assays, reverse transcription of the mRNA is blocked when the polymerase encounters the ribosome. Thus, the length of the reaction product is indicative of the position of the ribosome on the mRNA and can be used to monitor ternary complex binding and translocation. These results demonstrate that our hybridization approach does not interfere with the proper function of the ribosomal particle.
Figure 4.
Translational activity assays. (A) Purified wild-type ribosomes (wt) and mutant ribosomes with hairpin 5 insertion in helix 33a (hx33a hp5) were incubated without or with excess sp5 oligonucleotide. The ribosomes were then used for in vitro translation in cell extracts. Activity was monitored by 35S-methionine incorporation into the T4 gene 32 protein product visualized on 12% SDS–PAGE. (B) Toeprinting assays using mutant ribosomes preincubated with or without excess oligonucleotide and assembled into initiation complexes on the T4 gene 32 mRNA. Extension of a primer complementary to the mRNA with reverse transcriptase gives a toeprint at position +15. Delivery of tRNA by ternary complex and subsequent translocation with EF-G cause toeprint shift to position +16 and +18, respectively. The lane on the left shows reverse transcription of ribosome-free mRNA. Reaction products were resolved on denaturing 8% acrylamide gels.
Surface attachment and visualization of single mutant ribosomes
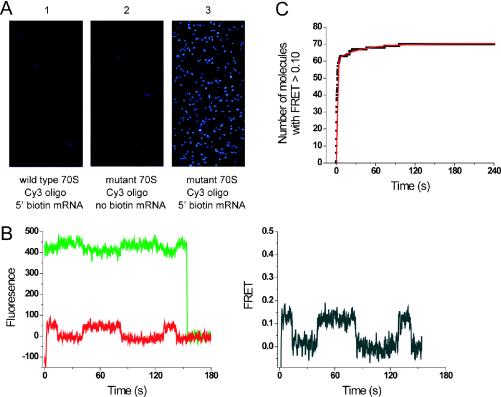
To examine the applicability of the oligonucleotide hybridization method for tagging ribosomal particles for use in single-molecule fluorescence experiments, mutant ribosomes with the hairpin 5 extension in helix 33a were hybridized with the Cy3-labeled sp5 oligonucleotide. Labeled ribosomes were then assembled into initiation complexes with 5′-biotinylated or non-biotinylated mRNA and initiator tRNA, and immobilized on streptavidin-derivatized quartz surfaces, as described previously (8). Laser illumination of quartz surfaces resulted in significant biotin-dependent Cy3 fluorescence from spatially separated mutant ribosomes (Figure 5A, panel 3, on average 275 particles per 60 μm × 120 μm field of view) but only background fluorescence from wild-type ribosomes (Figure 5A, panel 1, on average six spots per field of view). This is consistent with over 98% specificity of our labeling method.
Figure 5.
Single-molecule fluorescence observation of surface-immobilized, fluorescently labeled ribosomes. (A) Wild-type (panel 1) and mutant ribosomes with hairpin 5 extension in helix 33a (panels 2 and 3) were incubated with excess Cy3-labeled sp5 oligonucleotide, assembled into initiation complexes on non-biotinylated (panel 2) or biotinylated mRNA (panels 1 and 3) and incubated on streptavidin-derivatized quartz surfaces. Excitation of Cy3 with 532 nm laser resulted in emission from single, spatially dispersed mutant ribosomes. (B) Delivery of EF-Tu/GTP/Phe-tRNAPhe (Cy5-acp3U47) ternary complex to surface-immobilized Cy3-labeled ribosomal complexes. Interaction of Phe-tRNAPhe with the ribosome positions Cy3 and Cy5 within close distance (∼70 Å) and results in FRET upon 532 nm illumination. Fluorescence emission from Cy3 (green trace) and Cy5 (red trace) co-localized on the single ribosome can be monitored simultaneously (left panel). FRET efficiency (gray trace, right panel) is calculated from Cy3 and Cy5 intensities according to the equation FRET = ICy5/(ICy3+ICy5). (C) Time plot of delivery of EF-Tu/GTP/Phe-tRNAPhe (Cy5-acp3U47) to Cy3-labeled ribosomal complexes as detected by an increase in the number of particles with FRET signal ≥ 0.10. The plot can be fitted with a double exponential equation: number of molecules = A − B exp (−t/τ1) − C exp (−t/τ2), where A = 70, B = 64, τ1 = 1.12 ± 0.01 s, C = 9 and τ2 = 37.40 ± 0.42 s.
Observation of single-molecule FRET signal with labeled ribosomes
To observe dynamic distance changes using FRET, two fluorophores must be introduced on the particle of interest. Excitation of the donor fluorophore (Cy3 in this study) with laser light will result in non-radioactive transfer of energy and fluorescence emission from the acceptor fluorophore (Cy5 in this study) if the two dyes are within certain distance determined by their spectral properties (7). The activity of our mutant ribosomes was verified using single-molecule FRET. Mutant ribosomes carrying a Cy3-labeled sp5 oligonucleotide prehybridized to the hairpin 5 extension in helix 33a were immobilized on the surface as described above, and a cognate Cy5-labeled tRNA in ternary complex with EF-Tu/GTP was added using a stopped-flow delivery apparatus. Based on structural models of ribosomal complexes, we predicted a 70–80 Å distance between Cy5 on the accommodated tRNA and Cy3 on the 30S subunit. Upon excitation with 532 nm laser light, anti-correlated changes in fluorescence intensity of Cy3 and Cy5 were observed in a population of particles that were indicative of a FRET signal defined as FRET = ICy5/(ICy3 + ICy5), where ICy3 and ICy5 are fluorescence intensities of both dyes (Figure 5B). The average FRET value was found to be 0.12 ± 0.04. This corresponds to a distance of ∼70 Å between the two fluorophores [assuming R0 = 50 Å and random dye orientation as determined previously (8)]. The observed transitions to zero FRET result from photophysical event known as blinking, which involves temporary loss of Cy5 fluorescence.
Fluorescent labeling of surface-immobilized ribosomes had no effect on their function. The efficiency of tRNA binding was 50% as visualized by FRET in 143 out of 287 ribosome complexes observed in four separate experiments. The rate of tRNA delivery was monitored as the increase in the number of ribosomes with FRET ≥ 0.10 over time (Figure 5C). The labeled mutant ribosomes bind 10 nM ternary complex with a rate constant of 9 × 107 M−1 s−1. This value is consistent with the tRNA binding rate reported previously for wild-type ribosomes (8), and provides further evidence that our labeling method does not interfere with ribosome activity. These preliminary results demonstrate the applicability of our oligonucleotide hybridization approach to label ribosomal particles for single-molecule FRET studies of translation.
DISCUSSION
The high degree of conservation and complexity of the ribosome does not make it very amenable to genetic or chemical modification. Our approach to hybridize fluorescent oligonucleotides to target sites engineered into the 16S rRNA allows stable and specific labeling of the prokaryotic ribosome for single-molecule fluorescence studies. To ensure that fluorescence labeling does not disrupt biological function, the activity of engineered ribosomal particles was tested both genetically and biochemically. Plasmids encoding the E.coli rRNA modified at specific sites were introduced into an rRNA knockout strain and grown under selective conditions to demonstrate activity of mutant ribosomes in vivo. Ribosomes purified from viable mutant strains and hybridized with oligonucleotides were equally active as wild-type ribosomes in protein synthesis using in vitro translation and toeprinting assays.
Ribosomes labeled with fluorescent oligonucleotides were used in single-molecule fluorescence experiments. Cy3-labeled ribosomes assembled efficiently into initiation complexes and were immobilized onto derivatized quartz surfaces through an mRNA–biotin–streptavidin linkage. Fluorescence corresponding to single Cy3 dyes was observed over long time periods, indicating stable association of the dye-labeled DNA with the ribosomal particle. This is consistent with the slow dissociation rates of nucleic acid helices. Binding of fluorescently labeled tRNA to surface-immobilized labeled ribosomes was observed using single-molecule FRET. The efficiency and rates of aminoacyl-tRNA delivery to the labeled ribosomes were comparable with that of unlabeled ribosomes, as previously measured. The fidelity of mutant ribosomes was also determined by testing their selectivity in binding cognate tRNA species on the surface (M. Dorywalska and J. D. Puglisi, unpublished data).
The advantages of our approach include high degree of labeling specificity and efficiency, versatility in the selection of tagging sites and ability to assemble functional particles in vivo without the limitations of ribosome reconstitution in vitro as encountered when using other methods for covalent tagging of ribosomal components. Since the hybridization sites are surface exposed, oligonucleotide tags could be used for in vivo analysis of ribosome function. The high efficiency of hybridization at low temperatures under low salt conditions makes this approach especially useful for potential in vivo applications. The drawback in our method arises from the high level of conservation of the most functionally interesting sites of the prokaryotic rRNA. Introduction of hybridization extensions within such sites often results in nonviable ribosomes. We are currently exploring alternate labeling approaches that involve smaller changes in rRNA. The eukaryotic ribosome with a high degree of sequence variability should also be a good target to label with our current methodology, especially within the multiple extension regions.
Insertions of short sequences into rRNA have been previously used as means for determining the location of the extended helices on the E.coli ribosome using electron microscopy (36,37). Insertions that were designed to bind an affinity tag can also be used to purify selectively ribosomes that contain mutation of interest in another region of rRNA (38,39). The high stability and specificity of nucleic acid base pairing makes our oligonucleotide hybridization approach potentially useful for affinity purification or surface tethering of ribosomal particles. The method widens the spectrum of tools available to modify and manipulate ribosomes for both in vivo and in vitro studies of translation mechanism.
Acknowledgments
M.D. is supported by the Howard Hughes Predoctoral Fellowship and Stanford Graduate Fellowship, S.C.B. was supported by the Giannini Family Foundation and R.L.G. is supported by the American Cancer Society. Funded by NIH grant GM51266 (J.D.P.), grants from NSF and NASA (S.C.) and Packard Foundation grant 2000-01671 (J.D.P. and S.C.). The authors thank Dr Claudio Gualerzi for initiation factor overexpression strains, Dr Cathy Squires for the TA531 strain, Dr Tae-Hee Lee for help with FRET data analysis, Eric Lau for the preparation of initiation factors, and Drs Eric Jan and Brian Gibbons for comments on the manuscript. Funding to pay the Open Access publication charges for this article was provided by NIH grant GM51266.
REFERENCES
- 1.Green R., Noller H.F. Ribosomes and translation. Annu. Rev. Biochem. 1997;66:679–716. doi: 10.1146/annurev.biochem.66.1.679. [DOI] [PubMed] [Google Scholar]
- 2.Rodnina M.V., Stark H., Savelsbergh A., Wieden H.J., Mohr D., Matassova N.B., Peske F., Daviter T., Gualerzi C.O., Wintermeyer W. GTPase mechanisms and functions of translation factors on the ribosome. Biol. Chem. 2000;381:377–387. doi: 10.1515/BC.2000.050. [DOI] [PubMed] [Google Scholar]
- 3.Ogle J.M., Murphy F.V., IV, Tarry M.J., Ramakrishnan V. Selection of tRNA by the ribosome requires a transition from an open to a closed form. Cell. 2002;111:721–732. doi: 10.1016/s0092-8674(02)01086-3. [DOI] [PubMed] [Google Scholar]
- 4.Agrawal R.K., Heagle A.B., Penczek P., Grassucci R.A., Frank J. EF-G-dependent GTP hydrolysis induces translocation accompanied by large conformational changes in the 70S ribosome. Nature Struct. Biol. 1999;6:643–647. doi: 10.1038/10695. [DOI] [PubMed] [Google Scholar]
- 5.Stark H., Rodnina M.V., Wieden H.J., van Heel M., Wintermeyer W. Large-scale movement of elongation factor G and extensive conformational change of the ribosome during translocation. Cell. 2000;100:301–309. doi: 10.1016/s0092-8674(00)80666-2. [DOI] [PubMed] [Google Scholar]
- 6.Frank J., Agrawal R.K. A ratchet-like intersubunit reorganization of the ribosome during translocation. Nature. 2000;406:318–322. doi: 10.1038/35018597. [DOI] [PubMed] [Google Scholar]
- 7.Weiss S. Fluorescence spectroscopy of single biomolecules. Science. 1999;283:1676–1683. doi: 10.1126/science.283.5408.1676. [DOI] [PubMed] [Google Scholar]
- 8.Blanchard S.C., Kim H.D., Gonzalez R.L., Jr, Puglisi J.D., Chu S. tRNA dynamics on the ribosome during translation. Proc. Natl Acad. Sci. USA. 2004;101:12893–12898. doi: 10.1073/pnas.0403884101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Blanchard S.C., Gonzalez R.L., Jr, Kim H.D., Chu S., Puglisi J.D. tRNA selection and kinetic proofreading in translation. Nature Struct. Mol. Biol. 2004;11:1008–1014. doi: 10.1038/nsmb831. [DOI] [PubMed] [Google Scholar]
- 10.Heilek G.M., Marusak R., Meares C.F., Noller H.F. Directed hydroxyl radical probing of 16S rRNA using Fe(II) tethered to ribosomal protein S4. Proc. Natl Acad. Sci. USA. 1995;92:1113–1116. doi: 10.1073/pnas.92.4.1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Odom O.W., Jr, Robbins D.J., Lynch J., Dottavio-Martin D., Kramer G., Hardesty B. Distances between 3′ ends of ribosomal ribonucleic acids reassembled into Escherichia coli ribosomes. Biochemistry. 1980;19:5947–5954. doi: 10.1021/bi00567a001. [DOI] [PubMed] [Google Scholar]
- 12.Auerbach T., Pioletti M., Avila H., Anagnostopoulos K., Weinstein S., Franceschi F., Yonath A. Genetic and biochemical manipulations of the small ribosomal subunit from Thermus thermophilus HB8. J. Biomol. Struct. Dyn. 2000;17:617–628. doi: 10.1080/07391102.2000.10506553. [DOI] [PubMed] [Google Scholar]
- 13.Oakes M.I., Clark M.W., Henderson E., Lake J.A. DNA hybridization electron microscopy: ribosomal RNA nucleotides 1392–1407 are exposed in the cleft of the small subunit. Proc. Natl Acad. Sci. USA. 1986;83:275–279. doi: 10.1073/pnas.83.2.275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tapprich W.E., Hill W.E. Involvement of bases 787–795 of Escherichia coli 16S ribosomal RNA in ribosomal subunit association. Proc. Natl Acad. Sci. USA. 1986;83:556–560. doi: 10.1073/pnas.83.3.556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Weller J.W., Hill W.E. Probing dynamic changes in rRNA conformation in the 30S subunit of the Escherichia coli ribosome. Biochemistry. 1992;31:2748–2757. doi: 10.1021/bi00125a015. [DOI] [PubMed] [Google Scholar]
- 16.Alexander R.W., Muralikrishna P., Cooperman B.S. Ribosomal components neighboring the conserved 518-533 loop of 16S rRNA in 30S subunits. Biochemistry. 1994;33:12109–12118. doi: 10.1021/bi00206a014. [DOI] [PubMed] [Google Scholar]
- 17.Fuchs B.M., Wallner G., Beisker W., Schwippl I., Ludwig W., Amann R. Flow cytometric analysis of the in situ accessibility of Escherichia coli 16S rRNA for fluorescently labeled oligonucleotide probes. Appl. Environ. Microbiol. 1998;64:4973–4982. doi: 10.1128/aem.64.12.4973-4982.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fuchs B.M., Syutsubo K., Ludwig W., Amann R. In situ accessibility of Escherichia coli 23S rRNA to fluorescently labeled oligonucleotide probes. Appl. Environ. Microbiol. 2001;67:961–968. doi: 10.1128/AEM.67.2.961-968.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Brosius J., Ullrich A., Raker M.A., Gray A., Dull T.J., Gutell R.R., Noller H.F. Construction and fine mapping of recombinant plasmids containing the rrnB ribosomal RNA operon of E.coli. Plasmid. 1981;6:112–118. doi: 10.1016/0147-619x(81)90058-5. [DOI] [PubMed] [Google Scholar]
- 20.Asai T., Zaporojets D., Squires C., Squires C.L. An Escherichia coli strain with all chromosomal rRNA operons inactivated: complete exchange of rRNA genes between bacteria. Proc. Natl Acad. Sci. USA. 1999;96:1971–1976. doi: 10.1073/pnas.96.5.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Powers T., Noller H.F. A functional pseudoknot in 16S ribosomal RNA. EMBO J. 1991;10:2203–2214. doi: 10.1002/j.1460-2075.1991.tb07756.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Robertson J.M., Wintermeyer W. Effect of translocation on topology and conformation of anticodon and D loops of tRNAPhe. J. Mol. Biol. 1981;151:57–79. doi: 10.1016/0022-2836(81)90221-7. [DOI] [PubMed] [Google Scholar]
- 23.Dahlquist K.D., Puglisi J.D. Interaction of translation initiation factor IF1 with the E.coli ribosomal A site. J. Mol. Biol. 2000;299:1–15. doi: 10.1006/jmbi.2000.3672. [DOI] [PubMed] [Google Scholar]
- 24.Soffientini A., Lorenzetti R., Gastaldo L., Parlett J.H., Spurio R., La Teana A., Islam K. Purification procedure for bacterial translational initiation factors IF2 and IF3. Protein Expr. Purif. 1994;5:118–124. doi: 10.1006/prep.1994.1018. [DOI] [PubMed] [Google Scholar]
- 25.Schmitt E., Blanquet S., Mechulam Y. Crystallization and preliminary X-ray analysis of Escherichia coli methionyl-tRNAfMet formyltransferase complexed with formyl-methionyl-tRNAfMet. Acta Crystallogr. D Biol. Crystallogr. 1999;55:332–334. doi: 10.1107/S0907444998011780. [DOI] [PubMed] [Google Scholar]
- 26.Dubnoff J.S., Maitra U. Isolation and properties of protein factors involved in polypeptide chain initiation in Escherichia coli. Meth. Enzymol. 1971;20:248–261. [Google Scholar]
- 27.Plumbridge J.A., Baumert H.G., Ehrenberg M., Rigler R. Characterisation of a new, fully active fluorescent derivative of E.coli tRNAPhe. Nucleic Acids Res. 1980;8:827–843. [PMC free article] [PubMed] [Google Scholar]
- 28.Carbon J., David H. Studies on the thionucleotides in transfer ribonucleic acid. Addition of N-ethylmaleimide and formation of mixed disulfides with thiol compounds. Biochemistry. 1968;7:3851–3858. doi: 10.1021/bi00851a010. [DOI] [PubMed] [Google Scholar]
- 29.Tokimatsu H., Strycharz W.A., Dahlberg A.E. Gel electrophoretic studies on ribosomal protein L7/L12 and the Escherichia coli 50S subunit. J. Mol. Biol. 1981;152:397–412. doi: 10.1016/0022-2836(81)90250-3. [DOI] [PubMed] [Google Scholar]
- 30.Chambliss G.H., Henkin T.M., Leventhal J.M. Bacterial in vitro protein-synthesizing systems. Meth. Enzymol. 1983;101:598–605. doi: 10.1016/0076-6879(83)01040-x. [DOI] [PubMed] [Google Scholar]
- 31.Pavlov M.Y., Ehrenberg M. Rate of translation of natural mRNAs in an optimized in vitro system. Arch. Biochem. Biophys. 1996;328:9–16. doi: 10.1006/abbi.1996.0136. [DOI] [PubMed] [Google Scholar]
- 32.Wimberly B.T., Brodersen D.E., Clemons W.M., Jr, Morgan-Warren R.J., Carter A.P., Vonrhein C., Hartsch T., Ramakrishnan V. Structure of the 30S ribosomal subunit. Nature. 2000;407:327–339. doi: 10.1038/35030006. [DOI] [PubMed] [Google Scholar]
- 33.Schluenzen F., Tocilj A., Zarivach R., Harms J., Gluehmann M., Janell D., Bashan A., Bartels H., Agmon I., Franceschi F., Yonath A. Structure of functionally activated small ribosomal subunit at 3.3 Å resolution. Cell. 2000;102:615–623. doi: 10.1016/s0092-8674(00)00084-2. [DOI] [PubMed] [Google Scholar]
- 34.Yusupov M.M., Yusupova G.Zh., Baucom A., Lieberman K., Earnest T.N., Cate J.H.D., Noller H.F. Crystal structure of the ribosome at 5.5 Å resolution. Science. 2001;292:883–896. doi: 10.1126/science.1060089. [DOI] [PubMed] [Google Scholar]
- 35.SantaLucia J., Jr, Turner D.H. Measuring the thermodynamics of RNA secondary structure formation. Biopolymers. 1997;44:309–319. doi: 10.1002/(SICI)1097-0282(1997)44:3<309::AID-BIP8>3.0.CO;2-Z. [DOI] [PubMed] [Google Scholar]
- 36.Spahn C.M.T., Grassucci R.A., Penczek P., Frank J. Direct three-dimensional localization and positive identification of RNA helices within the ribosome by means of genetic tagging and cryo-electron microscopy. Structure. 1999;7:1567–1573. doi: 10.1016/s0969-2126(00)88347-1. [DOI] [PubMed] [Google Scholar]
- 37.Matadeen R., Sergiev P., Leonov A., Pape T., van del Sluis E., Mueller F., Osswald M., von Knoblauch K., Brimacombe R., Bogdanov A., van Heel M., Dontsova O. Direct localization by cryo-electron microscopy of secondary structural elements in Escherichia coli 23S rRNA which differ from the corresponding regions in Haloarcula marismortui. J. Mol. Biol. 2001;307:1341–1349. doi: 10.1006/jmbi.2001.4547. [DOI] [PubMed] [Google Scholar]
- 38.Leonov A.A., Sergiev P.V., Bogdanov A.A., Brimacombe R., Dontsova O.A. Affinity purification of ribosomes with a lethal G2655C mutation in 23S rRNA that affects the translocation. J. Biol. Chem. 2003;278:25664–25670. doi: 10.1074/jbc.M302873200. [DOI] [PubMed] [Google Scholar]
- 39.Youngman E.M., Brunelle J.L., Kochaniak A.B., Green R. The active site of the ribosome is composed of two layers of conserved nucleotides with distinct roles in peptide bond formation and peptide release. Cell. 2004;117:589–599. doi: 10.1016/s0092-8674(04)00411-8. [DOI] [PubMed] [Google Scholar]