Abstract
We present evidence of remarkable genome-wide mobility and evolutionary expansion for a class of protein domains whose borders locate close to the borders of their encoding exons. These exon-bordering domains are more numerous and widely distributed in the human genome than other domains. They also co-occur with more diverse domains to form a larger variety of domain architectures in human proteins. A systematic comparison of nine animal genomes from nematodes to mammals revealed that exon-bordering domains expanded faster than other protein domains in both abundance and distribution, as well as the diversity of co-occurring domains and the domain architectures of harboring proteins. Furthermore, exon-bordering domains exhibited a particularly strong preference for class 1-1 intron phase. Our findings suggest that exon-bordering domains were amplified and interchanged within a genome more often and/or more successfully than other domains during evolution, probably the result of extensive exon shuffling and gene duplication events. The diverse biological functions of these domains underscore the important role they play in the expansion and diversification of animal proteomes.
INTRODUCTION
Expansion in the size and/or complexity of genomes and proteomes during evolution was accomplished in many ways. One important force for species evolution is gene duplication and the ensuing mutation and selection (1). A duplication event can occur on different scales from a gene fragment to the whole genome, and it can be produced by a broad range of biological mechanisms from slippage during recombination to horizontal transfer. While many duplicate copies of genes become pseudogenes and are eventually lost, some duplicates evolve to serve new functions for the organism and eventually form families of paralogues and orthologues.
The exon–intron structure of eukaryotic genes provides another way to generate new functional genes. When the coding exons correlate with protein functional modules, the duplication, permutation and rearrangement of these exons results in novel genes with diverse functions (2,3). Exon shuffling can occur through mechanisms such as retrotransposition (4) or illegitimate recombination (5). Since the inception of exon shuffling theory, it has been bolstered by studies on individual protein families as well as large-scale studies on the correlation between exons and protein structural/functional modules (6–9). In addition, exon shuffling has been shown by a number of intron phase matching studies (8,10–12) to result in the over-representation of domains bound by same phase introns, due to their preservation of open reading frame (13).
Most recently, we have demonstrated that correlation between the borders of protein domains and their encoding exons is a genome-wide phenomenon in multiple eukaryotic organisms (14). More importantly, in that work we have found that the statistically significant match between the borders of domain instances and the borders of exons improved from worms and insects to fish and mammals as the more complex organisms consistently displayed stronger exon–domain correlations. The overwhelming body of evidence for exon–domain correlation invites a number of interesting conjectures and questions, intensifying the interest in the exon shuffling theory from a genomics perspective. For example, would the exon–domain border correlation be beneficial for domain propagation by facilitating the mobility and spread of domains within a genome? Could this domain mobility be a driving force behind the formation of mosaic proteins? Could the exon-bordering domains contribute more to the expansion and diversification of proteomes than other domains as a result of duplications and exon shuffling?
We sought to address these questions by investigating a special group of protein domains whose borders are located close (defined statistically) to exon–intron junctions. Using the genomes of nine animal species, we demonstrate that these exon-bordering domains exhibited remarkable mobility in their genomic distributions. The exon-bordering domains are on average more abundant and present in more genes than other protein domains. They also co-occur with a larger number of different domains to form mosaic proteins with diverse domain architectures. Evolutionarily, exon-bordering domains appear to have an advantage as they proliferated faster in both numbers and distribution in animal genomes compared to other domains. Taken together, these results suggest that exon-bordering domains were subject to positive selection as a result of their success in creating functional diversity in genes after duplication and shuffling events and probably contributed significantly to the proteome expansion and diversification during evolution.
MATERIALS AND METHODS
Selection and scoring of exon-bordering domains
Gene structure and protein annotations came from EnsEMBL v.11 confirmed cDNA and protein sets (15) with domain annotations performed by us using HmmPfam 2.0 g (16) against Pfam (17). We defined ‘domain border box’ as a flexible range overlapping the borders of protein domains to allow for some variability in the correlation of exon and domain borders and possible domain definition inaccuracy (Figure 1a). A border box was not defined for the domain borders with no degree of freedom, e.g. domain borders that coincide with start or end of the protein. We collected statistics for only one multi-exon transcript per gene whose protein translation had at least one domain. Thus our dataset from nine animal genomes comprised nearly 100 000 proteins, 200 000 domains and 650 000 exon borders.
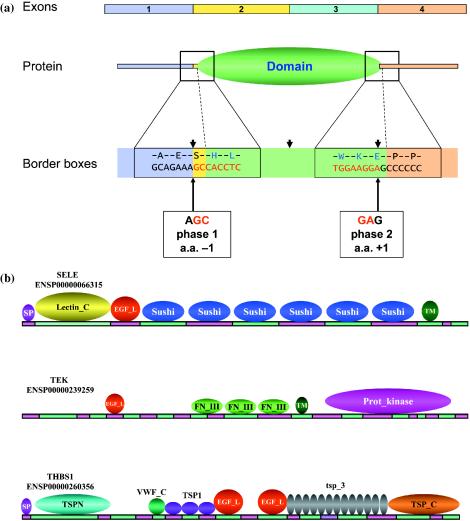
Figure 1.
Exon-bordering domains. (a) Nomenclature. The exon borders were examined for their positions in relation to domain borders as described previously (14). Nucleotide positions of exon borders were translated into amino acid positions with codon phase information preserved. The first amino acid outside domain was designated as position −1 and the first amino acid inside domain was position +1, and so on. Domain border boxes were defined as short ranges covering the proximity of the start or end position of domains, i.e. border box of [−10, +10] covers 10 amino acids outside and 10 amino acids inside domain. A hypothetical 4-exon transcript, its protein translation, a domain instance on the protein and the two border boxes for the domain are illustrated. Inside the two border boxes, partial nucleotide/amino acid sequences covering the junctions between exons 1 (purple) and 2 (yellow), domain (green) and exons 4 (light brown) are shown. Inside the border boxes, the exon junctions are indicated by the arrowheads and font colors in nucleotide sequences (black and red). Domain border positions in the border boxes are indicated by dotted lines as well as background and font colors. In the illustration, the particular domain instance correlates with exon 2 at N-terminus at amino acid position −1, and exon 3 at C-terminal position +1. Based on the position of the exon border in relation to triplet codon, the N-terminal exon border for the domain is of phase 1 (after the first nucleotide of the triplet codon for Serine at amino acid position −1), while the C-terminal exon border is of phase 2. A domain is considered an exon-bordering domain only when the observed number of exon borders inside all of its border boxes is significantly higher than expected (as detailed in the Methods). (b) EGF-like domain examples. The cDNA and protein domain structures of three EGF-like domain-containing proteins are illustrated proportionally. Each mosaic protein is annotated with gene name and EnsEMBL protein ID on top left. Exons are alternatively colored pink and green. In each case, EGF-like domain (abbreviated as EGF_L) is encoded by one exon. A few other exon-matching exon-bordering domain instances such as Sushi, Fibronectin type III (abbreviated as FN_III) domains were also illustrated. TM, transmembrane domain; SP, signal peptide; Lectin_C, Lectin C-type domain; TSPN, Laminin G domain; VWF_C, von Willebrand factor type C domain; TSP1, Thrombospondin type 1 domain; TSP3, Thrombospondin type 3 repeat; and TSP_C, Thrombospondin C-terminal region.
For each domain, we collected counts of total amino acids and exon borders for proteins containing the domain, which were used to estimate the probability p of exon border falling at any amino acid position. Total number of amino acids and observed number of exon borders at each position inside domain border boxes and probability p were used to derive one-tailed Binomial P-values. For the set of top 112 human exon-bordering domains, we collected only domains that have at least one position with P < 10−7 or three positions with P < 10−5 inside domain border boxes.
Wilcoxon test on exon-bordering domains
For each domain in the human exon-bordering domain group defined above, we collected in all the nine species, the total number of domain instances, total number of genes containing the domain, total number and type of co-occurring domains that co-exist on the same gene as the given domain as well as the total number of domain architectures existing in proteins containing the domain. The domain architecture of a protein is defined as a sequence of different domains it contains, omitting tandem repetitive instances of one domain (e.g. if a protein contains a total of six instances of domains A, B and C arranged in itself as AABCCB, the architecture for the protein will be accounted as ABCB). Hence, we take into account only the lower bound estimate and the actual number of architectures of exon-bordering domains is even higher. For each domain, the architecture of any protein is counted only once however many instances of the domain are present on the protein.
One-tailed Wilcoxon test was performed with statistical package R (http://www.r-project.org/) using alternative hypothesis that exon-bordering domains have a greater mean than the background domains, domains that do not correlate with exons. In each of the above-mentioned five data categories, we compared the 112 human exon-bordering domains with 2266 background domains. To generate the amplification ratio between the total number of a domain in human and the total number of the same domain in another evolutionary group, we only included domains that were present in both human and the other evolutionary group. For tests on the top 112 exon-bordering domains, 86 exon-bordering domains were compared with 1802 background domains for worm versus human; for insect versus human, 88 exon-bordering domains were compared with 1907 other domains; for fish, 107 exon-bordering domains were compared with 2076 other domains; for rodent, 112 exon-bordering domains were compared with 2201 other domains.
Intron phase study and chi-square test
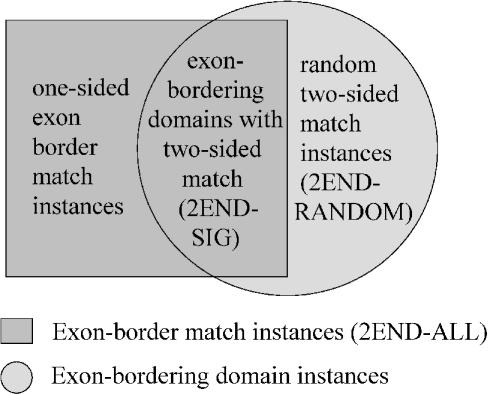
We collected a group of domain instances in human genome where we found exon border inside the border boxes ([−10, +10]) at both ends of the domain instance (2END-ALL, Figure 2). We separately recorded the intersection of this group with the exon-bordering domain group to produce a set 2END-SIGNIFICANT, as well as the remainder of this group, named 2END-RANDOM. The phases of the exon borders at both ends of the domain instances were recorded separately for both of the subsets (2END-SIGNIFICANT and 2END-RANDOM).
Figure 2.
Naming convention for domain instances. The exon-border match instances (2END-ALL) include all the domain instances in genome whose border boxes on both ends contain exon borders. Exon-bordering domain instances also contain the instances of one-sided exon-border match. 2END-SIGNIFICANT (2END-SIG in illustration) instances contain the intersection of exon-bordering domain instances and 2END-ALL, while 2END-RANDOM denotes the resulting set by subtracting 2END-SIGNIFICANT from 2END-ALL.
In chi-square test, we calculated the expected number of domain instances for each phase class by first obtaining the probability PN and PC for each phase of 0, 1, 2 at N- or C-terminus of exon-coinciding domain instances, then deriving the expected number with PN × PC × Total number of domain instances in the group being tested. The chi-square P-values for both individual phase class and the symmetrical phase classes are generated using two categories where first category is the given phase class(es), the second category is all the remaining phase classes.
RESULTS
Exon-bordering domains are more abundant and widespread than other domains
Using predefined domain border boxes spanning amino acid positions [−10, +10] (Figure 1a), we examined the positional relationship between borders of the protein domain instances and borders of their encoding exons. In our method, a substantially higher than expected number of exon borders recorded at some amino acid position(s) inside the domain border boxes for a domain would indicate a strong correlation between the borders of exons and the domain. Our set of exon-bordering domains includes domains that match with exon borders on only one end or both ends, as well as the ‘one exon–many domains’ and ‘one domain–many exons’ cases. This allows us to take into account the possible effects of intron gain and loss in the vicinity of a domain.
We selected the top 112 exon-bordering domains in the human genome using the statistical criteria described in Materials and Methods. We list the top 10 exon-bordering domains based on the P-values of overall exon-correlation in Table 1 (for full list refer to Supplementary Material). All of these domains have highly significant P-values varying from around 10−72 to practically zero. Some of the domains were identified by previous studies as individual examples for domains involved in exon shuffling (see Figure 1b for examples), such as EGF-like domain and Sushi domain (18). However, the Krueppel-associated box (KRAB) domain, which has a particularly good correlation with exons (P-value practically 0), was not strongly associated with exon shuffling previously.
Table 1.
Exon-bordering domains are abundant and widely distributed
| Domain name | Score | Peak score | P-value rank | Domain # rank | Gene # rank | Co-occur # rank | Co-occur type rank | Architecture # rank |
|---|---|---|---|---|---|---|---|---|
| KRAB box | 0 | 0 | 1 | 14 | 5 | 1 | 70 | 28 |
| EGF-like domain | 0 | 1.5E−251 | 2 | 5 | 13 | 2 | 2 | 1 |
| Sushi domain (SCR repeat) | 2.0E−308 | 5.2E−197 | 3 | 17 | 56 | 77 | 53 | 34 |
| Fibronectin type III domain | 2.4E−213 | 2.3E−119 | 4 | 11 | 18 | 8 | 13 | 10 |
| Low-density lipoprotein receptor domain class A | 2.4E−105 | 3.1E−77 | 5 | 26 | 81 | 17 | 24 | 20 |
| Ankyrin repeat | 2.9E−104 | 1.4E−43 | 6 | 3 | 10 | 21 | 8 | 9 |
| Leucine Rich Repeat | 5.1E−93 | 5.6E−49 | 7 | 10 | 16 | 16 | 14 | 13 |
| Immunoglobulin domain | 9.1E−80 | 5.7E−11 | 8 | 2 | 3 | 5 | 6 | 8 |
| B-box zinc finger | 5.4E−74 | 5.4E−74 | 9 | 52 | 37 | 43 | 82 | 37 |
| Laminin EGF-like (Domains III and V) | 5.8E−73 | 2.1E−38 | 10 | 24 | 101 | 22 | 22 | 27 |
We listed the top 10 exon-bordering domains selected based on the statistical significance of their correlation to exons (Materials and Methods). Domain score is the overall P-value calculated for the correlation of given domain with exons as described in Methods. Peak score is the lowest P-value for all positions inside the [−10, +10] domain border boxes. P-value column ranks all exon-bordering domains based on P-values of exon-domain correlation; Domain # column ranks all human domains based on the total number of domain instances in genome; Gene # ranks domains based on the total number of human genes containing the given domain; Co-occur # ranks domains based on the total number of co-occurring domains for a domain; Co-occur type ranks domains based on the total number of different types of co-occurring domains for the domain; Architecture # ranks domains based on the total number of different domain architectures for the genes containing the domain. All of these data categories measure the abundance and distribution of a given domain.
The strong correlation between the borders of exon-bordering domains and exons could confer a selective advantage in creating new functional domain instances in proteins following exon shuffling events. If exon shuffling was a prevalent evolutionary mechanism, one would predict that exon-bordering domains would be preferentially amplified during evolution and, as a result, become more abundant and widely distributed than other domains. To test this hypothesis, we created five criteria to rank all protein domains in the human genome based on their abundance and distribution. The first criterion measures the abundance of a domain by the total number of its instances in the genome. The second criterion ranks a domain by the total number of genes containing it; this gives indication about both the abundance and distribution of the domain. The rest of the criteria gauge the tendency of a domain to be present within multi-functional proteins. The third criterion is based on the total number of domain instances co-occurring (in the same protein) with the instances of a given domain and the fourth criterion is based on the total number of different co-occurring domain types for a domain. The last criterion utilizes a concept of domain architecture, which represents protein domain organization as a sequence of the domain types found in a protein (see Materials and Methods). A high domain architecture ranking would indicate that a domain frequently co-occurs with diverse domains and suggest that the given domain is heavily ‘reused’ to constitute novel multi-functional mosaic proteins.
Consistent with our expectations, the top 10 exon-bordering domains based on P-values of correlations ranked high for all of these five criteria (Table 1). Indeed, out of a total of 2378 human domains in the rankings, the top exon-bordering domain KRAB box is the 14th in abundance, 5th in total genes containing KRAB box, co-occurs with the largest number of domain instances, at 70th in the ranking for most types of co-occurring domains, and ranks 28th in terms of diverse domain architectures. In all, the 10 most significant exon-bordering domains are among the top 4.2% of all human domains in any of the 5 rankings for abundance and distribution.
To further evaluate whether exon-bordering domains tend to be more abundant and widely distributed than other domains, we performed one-tailed Wilcoxon test on the group of 112 exon-bordering domains and a background group that contains all the other protein domains in the human genome (Table 2, (a), column ‘Human’). Measured by the total number of domain instances in the genome, the exon-bordering domain (EBD) group had an unequivocally higher mean than the background group (EBD: 128.3 versus Non-EBD: 8.9, P < 2.2 × 10−16), indicating that on average exon-bordering domains are more numerous than other domains in the human genome. When we examined the number of genes containing the domains in the two groups, again the exon-bordering domain group displayed a clear advantage over the background group (EBD: 48.5 versus Non-EBD: 7.4, P < 2.2 × 10−16), demonstrating higher abundance and wider distribution. Exon-bordering domains also tend to co-occur with more domain instances (EBD: 119.9 versus Non-EBD: 15.9, P < 2.2 × 10−16) of more diverse types (EBD: 12.0 versus Non-EBD: 4.0, P < 6 × 10−13) than domains found in the background group, suggesting that exon-bordering domains have a stronger tendency to form mosaic proteins whose modular structures are comprised of multiple diverse domains. Additionally, there are usually multiple ways of using exon-bordering domains as building blocks for mosaic proteins, as demonstrated by the finding that genes containing exon-bordering domains have more diversified domain architectures than those with the background domains (EBD: 12.8 versus Non-EBD: 2.4, P < 2.2 × 10−16).
Table 2.
Exon-bordering domains were preferentially amplified during evolution
| Category | Human | Worm | Insect | Fish | Rodent |
|---|---|---|---|---|---|
| (a) | |||||
| Domain # | <2.2E−16 | 4.9E−10 | 2.3E−12 | 0.52 | 4.7E−05 |
| Gene # | <2.2E−16 | 1.6E−09 | 1.2E−12 | 0.83 | 5.9E−04 |
| Coocur # | <2.2E−16 | 5.4E−12 | 2.3E−11 | 0.047 | 0.11 |
| Coocur type | 5.2E−13 | 3.9E−08 | 3.9E−07 | 0.61 | 0.67 |
| Architecture # | <2.2E−16 | <2.2E−16 | <2.2E−16 | 0.069 | 5.5E−03 |
| (b) | |||||
| Domain # | 42 | 19 | 15 | 31 | 35 |
| Gene # | 24 | 15 | 6 | 18 | 20 |
| Coocur # | 39 | 15 | 11 | 26 | 33 |
| Coocur type | 20 | 7 | 5 | 16 | 18 |
| Architecture # | 26 | 7 | 5 | 18 | 20 |
(a) P values in Wilcoxon tests. We collected data in five categories (see Table 1 legend) that gauge the abundance and distribution of domains in genome. We compared the exon-bordering domain group to the background group (that contained all the remaining human domains) in Wilcoxon test with alternative hypothesis that exon-bordering domains had a higher mean (column ‘Human’). In the other data columns, a ratio was first produced using numbers obtained in human against the total numbers from the labeled evolutionary group. This ratio indicates the amplification fold for a domain in the data category. We used only domains present in both the evolutionary group and human in generating ratios for these data categories. A better mean amplification ratio in Wilcoxon test suggests a preferential amplification of the exon-bordering domain group in these categories. (b) Percentage of exon-bordering domains in different species. For each of the five data categories, we collected the number contributed by exon-bordering domains as a percentage of the total number. For example, in worm, 19% of all domain instances come from exon-bordering domain instances. In contrast, 42% of all domain instances in human are exon-bordering domain instances.
Since the high statistical significance displayed by exon-bordering domains to some extent relates to the relative abundance of these domains, to further confirm the results we collected the average number of co-occurring domain instances per gene for every domain in the human genome, which more accurately reflects the tendency of a domain to exist in multi-domain proteins. We also collected the average number of co-occurring domain types for each domain per gene, which better gauges the functional diversity of the proteins containing each domain. Again, we obtained exceedingly significant P-values (For average instance, EBD: 4.3 versus Non-EBD: 2.2, P < 2.2 × 10−16; For average type number, EBD: 2.2 versus Non-EBD: 1.7, P < 9 × 10−15) in one-tailed Wilcoxon test, indicating a higher mean of exon-bordering domain group in both data categories. These results are consistent with the expectation that the ability to preserve functionality after exon shuffling events makes exon-bordering domains the ideal building blocks for multi-function proteins. If such events are positively selected, this may explain why exon-bordering domains are more prevalent in mosaic proteins and diverse genes than the domains that do not strongly correlate with exons.
Exon-bordering domains are preferentially amplified during evolution
We were particularly interested to learn whether exon-bordering domains were always so numerous and widely distributed or if they had acquired or improved such features during evolution. While it has always been predicted that such domains would possess advantage through exon shuffling events, it was not demonstrated that this advantage actually benefited the expansion of exon-bordering domains during evolution. We reasoned that through comparing the expansion rates of all exon-bordering domains over an evolutionary distance with those of the other background domains, we could determine if exon-bordering domains as a group expanded faster through evolution.
To cover the spectrum of animal genomes, we took eight species other than human and separated them into four different evolutionary groups: worm (Caenorhabditis elegans and Caenorhabditis briggsae), insect (fruitfly and mosquito), fish (fugu and zebrafish) and rodent (mouse and rat). To represent the proliferation of a domain over the evolutionary distance between the group and human, we used a ratio between the total number of the domain instances in the human genome and the total number of the domain instances in both of the genomes of each evolutionary group. For example, the domain number ratio for KRAB domain between human (h) and the worm group (w) would be
where Nh, Nce, Ncb are the total numbers of KRAB instances in human, C.elegans and C.briggsae, respectively. When we compared the mean proliferation ratios, for the exon-bordering domain (REhw) and background (RBhw) groups in Wilcoxon test, we found strong evidence that on average the total number of instances of an exon-bordering domain increased faster than that of a background domain from both worm (REhw = 2.7 versus RBhw = 1.2, P < 5 × 10−10) and insect (REhi = 2.0 versus RBhi = 1.1, P < 3 × 10−12) to human. This result argues that compared to the evolutionarily ancient worm and insect, exon-bordering domains in human became preferentially amplified over the domains not correlating with exons. When we compared the rodent group to human, there is also a significant statistical difference (REhr = 0.68 versus RBhr = 0.60, P < 5 × 10−5) between the amplification ratios of exon-bordering domains and other domains, albeit the difference is much lower than that observed for the invertebrates. However, we found no statistically significant difference (REhm = 1.1 versus RBhm = 0.9, P = 0.52) between the amplification ratios of exon-bordering domains and other domains when fish group was compared to human.
As a result of faster expansion rate in the total domain number in the genome, it is expected that exon-bordering domains could spread out faster into more genes, as well as into more diversified mosaic proteins. Indeed, data obtained by comparing worm and insect groups to human lent strong support to this notion as we investigated the amplification ratios of total gene numbers, total co-occurring domain number, total co-occurring domain types and total number of domain architectures for both mobile and background domain groups. In each of these categories, we observed a P-value lower than 4 × 10−7, suggesting a faster expansion rate of exon-bordering domain in terms of both abundance and distribution in genome [Table 2, (a)]. Most notably, exon-bordering domains compared especially favorably with other domains in the evolutionary expansion ratio for the total number of domain architectures, suggesting that genes containing exon-bordering domains have a much stronger tendency to diversify domain organizations, a likely result of exon shuffling events.
Our statistics are so far based on the evolutionary properties of individual domains. However, it is interesting to evaluate the collective prevalence of all exon-bordering domain instances in each species, which gives a broad assessment for the expansion of exon-bordering domains during animal evolution. Therefore, we compared the relative abundance of the exon-bordering domains to all domains in the five evolutionary groups for the nine species [Table 2, (b)]. We found that for each of the four data categories, exon-bordering domains accounted for an increasingly large proportion of all domains in the more complex organisms. When the overall prevalence of exon-bordering domains were measured by the total number of domains, 15% and 19% of all domains were exon-bordering in insect and worm groups, respectively. However, exon-bordering domains in vertebrates ranged from 31% in fish to 42% in human. In the other three data categories, vertebrates again commanded a substantially higher percentage of exon-bordering domains when compared to invertebrates. It is notable that worm group contains a considerably higher percentage of exon-bordering domains than the fly group. We found that the increase in worm group was almost entirely due to a surprisingly high number of exon-bordering domains in C.elegans. Specifically, we found various exon-bordering domains that were either specific to C.elegans or largely expanded in C.elegans, most notably the 7tm chemoreceptor domains (PF01461 and PF01604) and a number of other domains of unknown functions. These domains contributed greatly to the total number of exon-bordering domains in C.elegans, potentially skewing the data towards higher exon-bordering domain percentage. Because Pfam was initially developed for C.elegans genome project (19), it is possible that the other invertebrate genomes were somewhat under-represented in Pfam database compared to C.elegans. Notwithstanding, these results collectively suggest that exon-bordering domains became increasingly dominant during evolution from invertebrates to vertebrates including human. Exon-bordering domains likely expanded into more genes and constructed more mosaic proteins through co-occurrence with domains of diverse types, all at a faster evolutionary pace than non-bordering domains. Such a trend eventually led to the presence of a larger number of exon-bordering domains in more diversified genes in vertebrate genomes.
Exon-bordering domains tend to be bounded by same phase introns
The splice frame rule (13) for exon shuffling stipulates that shuffling domains are likely bounded by same phase introns, meaning that intron/exon borders at both sides of the domain fall onto the same nucleotide position of triplet codons—phase 0 intron falls right between borders of triplet codons in coding sequence, phase 1 intron falls after the first nucleotide of a triplet while phase 2 intron falls after the second nucleotide of a triplet.
For testing that rule, we collected statistics on the phase of introns for all domain instances with exon-border matches on both ends and called that set 2END-ALL (Figure 2). It includes only a portion of our exon-bordering domain instances because we defined exon-bordering domains by their statistically significant correlations with exons on at least one end. We call this portion 2END-SIGNIFICANT, in contrast to 2END-RANDOM (the remainder of 2END-ALL) which contains the non-exon-bordering domains. Because overall the domains in 2END-RANDOM do not display statistically significant correlation with exons, it is likely that the match with exon borders by the instances in that set is coincidental.
We examined the phases of introns bounding 2END-ALL instances in the human genome [Table 3, (a)]. Among the nine phase classes, phase 1-1 is the predominant class—nearly a third of these instances have phase 1-1 compared to a fifth for phase 0-0, with other phase classes ranging from around 5% to 12%. We calculated the expected number of bounding phase classes and compared them with the observed numbers. There is a general bias towards same-phase introns: observed numbers for phase 1-1, 0-0, 2-2 are all well above expectations. Together, same-phase introns are much more prevalent than expected (P < 10−158).
Table 3.
Strong preference for the 1-1 phase class
| Phase class | Observed | % | Expected | P-value |
|---|---|---|---|---|
| (a) Set 2END-ALL | ||||
| 1-1 | 1535 | 32 | 1063 | 3.7E−60 |
| 0-0 | 1018 | 21 | 685 | 6.0E−43 |
| 0-1 | 573 | 12 | 901 | 9.1E−34 |
| 1-0 | 439 | 9 | 808 | 8.8E−46 |
| 0-2 | 296 | 6 | 301 | 0.77 |
| 2-0 | 313 | 6 | 278 | 0.029 |
| 1-2 | 252 | 5 | 355 | 1.4E−08 |
| 2-2 | 230 | 5 | 122 | 4.1E−23 |
| 2-1 | 222 | 5 | 365 | 6.2E−15 |
| Symmetrical | 2783 | 57 | 1870 | 3.1E−159 |
| (b) Set 2END-SIGNIFICANT | ||||
| 1-1 | 1253 | 45 | 603 | 3.8E−197 |
| 0-0 | 513 | 19 | 388 | 8.1E−12 |
| 0-1 | 300 | 11 | 511 | 4.9E−25 |
| 1-0 | 154 | 6 | 458 | 1.7E−54 |
| 0-2 | 106 | 4 | 171 | 3.3E−07 |
| 2-0 | 123 | 4 | 157 | 0.0048 |
| 1-2 | 96 | 4 | 201 | 1.3E−14 |
| 2-2 | 109 | 4 | 69 | 1.2E−06 |
| 2-1 | 111 | 4 | 207 | 3.8E−12 |
| Symmetrical | 1875 | 68 | 1060 | 5.1E−223 |
| (c) Set 2END-RANDOM | ||||
| 1-1 | 282 | 13 | 461 | 5.0E−21 |
| 0-0 | 505 | 24 | 297 | 6.2E−39 |
| 0-1 | 273 | 13 | 390 | 4.6E−11 |
| 1-0 | 285 | 13 | 350 | 1.5E−04 |
| 0-2 | 190 | 9 | 130 | 7.0E−08 |
| 2-0 | 190 | 9 | 120 | 5.7E−11 |
| 1-2 | 156 | 7 | 154 | 0.85 |
| 2-2 | 121 | 6 | 53 | 2.2E−21 |
| 2-1 | 111 | 5 | 158 | 9.3E−05 |
| Symmetrical | 908 | 43 | 810 | 1.2E−05 |
For domain instances that correlated with exons on both ends (sets 2END-ALL, 2END-SIGNIFICANT and 2END-RANDOM in Figure 2), we recorded the phase classes as x–y where x is the phase of exon border at N-terminus of domain while y is the phase at C-terminus. The observed numbers for each phase class were thus tallied in human. The percentage of each phase class in all cases is displayed in column marked ‘%’. The expected numbers and P-values for the significance of each phase class were calculated as described in Methods. The ‘symmetrical’ class comes from the total of phase classes 0–0, 1–1 and 2–2.
For the set 2END-SIGNIFICANT, we observed an even stronger preference for phase 1-1 class (close to half of all such instances in human), compared to other phase classes [Table 3, (b)]. Interestingly, the observed numbers for phase 2-0 and 0-2 classes were relatively close to expectation while phase 1-0 and 0-1 were much lower than expectation, similar to trends observed earlier [Table 3, (a)]. However, while still quite significant, the observed number of phase 0-0 cases was much closer to expectation compared to 2END-ALL instances, indicating a weaker bias towards phase 0-0 among exon-bordering domains. Nonetheless, same-phase introns were heavily favored by 2END-SIGNIFICANT instances with an even stronger statistical significance than 2END-ALL instances. The difference between these two groups is probably because 2END-ALL instances included random border match cases (2END-RANDOM) with little preference for any phase class, thereby somewhat diminishing the phase bias in 2END-ALL. Following this thought, we checked the phase preferences for 2END-RANDOM instances only (Figure 2) by subtracting 2END-SIGNIFICANT from 2END-ALL and found only a moderate preference for same phase introns for 2END-RANDOM [Table 3, (c)]. Interestingly, among the symmetrical classes, the phase 0-0 instances were considerably higher than expected, while phase 1-1 was substantially lower.
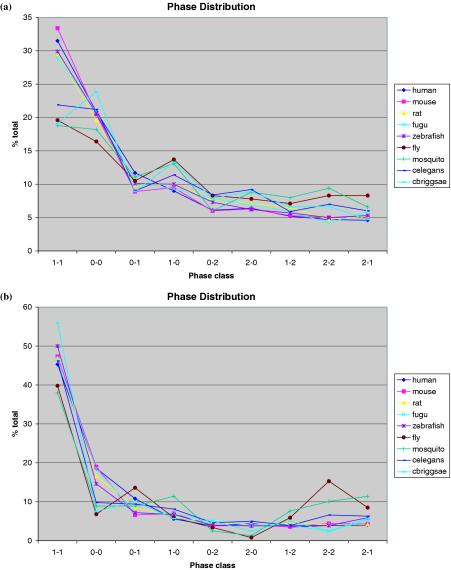
We expanded our intron phase studies to all nine animal species (Figure 3). Consistent with our observations in human, we found that phase 1-1 was one of the primary phase classes for 2END-ALL instances in these species. We noticed that from human to fugu, phase 1-1 remained the predominant phase class despite a slight but noticeable decrease in numbers (Figure 3a). However, there was a steep drop in the percentage of domains bounded by phase 1 introns in the four invertebrate species. Notably, phase 0-0 suffered little decline from vertebrates to invertebrates and became comparable to phase 1-1 in worms and insects. This result is consistent with our previous observation that the significance of genome-wide exon–domain correlation increases consistently from worm to human, with vertebrates exhibiting sharply higher correlation than invertebrates (14). It also agrees with our observation above that the expansion rate of exon-bordering domains and other domains among vertebrates do not seem to differ significantly [Table 2, (a)], yet exon-bordering domains expanded comparatively faster than other domains evolving from invertebrates to vertebrates.
Figure 3.
Intron phase distribution in all nine species. Phase classes were collected in all nine species for the sets (a) 2END-ALL and (b) 2END-SIGNIFICANT (see Figure 2 legend). Each of the nine possible phase classes was plotted as a percentage of the total phase classes in all species. Exon-bordering domain subgroup strongly favors phase 1-1.
As expected, the set 2END-SIGNIFICANT displayed an even stronger preference in all nine species for phase 1-1 class (Figure 3b). In all species, phase 1-1 was the predominant class with up to half of its instances bounded by phase 1 introns, compared with 7–19% by phase 0 introns and 1–15% by other phase classes. Interestingly, the phase 1-1 bias in the eight species other than human was not significantly different from what was observed in human. Instead, nearly all species had a similar level of bias for phase 1-1 class to that of human, although the insect group and C.briggsae deviated from human somewhat more than the others. This is in sharp contrast to the results obtained using 2END-ALL instances where vertebrates and invertebrates showed much difference in their preference for phase 1-1. It seems that 2END-ALL, which includes 2END-RANDOM instances has a rather different phase class preference favoring phases 0-0 and 2-2 but biased against 1-1. This suggests that the inclusion of random instances reversed the phase bias–against the phase 1-1. However, due to the statistical selection of exon-bordering domains, the exclusion of random instances from 2END-SIGNIFICANT instances allowed us to observe a strong phase bias even in the four invertebrates due to the strong preference for phase 1-1 of the exon-bordering domains.
DISCUSSION
Evolutionary expansion of exon-bordering domains and its implications
In the present study, we selected a group of protein domains based on the statistically significant correlation between their borders and exon borders. We systematically investigated the properties of these domains in nine animal species and found that these domains are highly mobile in the genomes. These exon-bordering domains are on average more abundant in the human genome and are more widely distributed as evidenced by larger numbers of genes containing them. More importantly, our results indicate a preferential expansion of the exon-bordering domain group during evolution.
We believe that the expansion of exon-bordering domains is driven by phenomena such as duplications and exon shuffling. Duplication and exon shuffling processes are not mutually exclusive—they share certain biological mechanisms, e.g. retrotransposition, and most likely run concurrently in the genomes. Duplication occurs at a relatively high frequency (20), while the frequency of shuffling events is harder to measure. In the case of exon-bordering domains, these two processes are probably complementary to each other. Gene duplications alone could lead to an increase in the gene number and domain instance number for domains, yet it could not explain the three most important observations for exon-bordering domain group—significant correlation of domain borders with exon borders, a strong preference for matching intron phases (phase 1-1 in particular), and a preferential increase in domain co-occurrence and the diversification of domain architectures during evolution. Exon shuffling could lead to diversified protein domain organizations and explain the interesting properties of exon-bordering domains, yet shuffling without duplication cannot increase the number of domain instances and generate a net gain of function in the genome of a species. Consequently, one can imagine a number of scenarios involving combinations of common elementary steps (shuffling events followed by duplication events, duplication followed by recombination/shuffling, loss of gene fragments, etc.). The outcome is likely to be gene- and domain-specific and to be selected based on the fitness gain associated with combining the domains into a new architecture or multiplying the architecture as a whole (or part). In either scenario, one should observe a net effect of amplified exon-bordering domains and an increase in proteome diversity.
Consistently, our exon-bordering domain collection contains prime examples of both gene duplication and exon shuffling processes while the combination of the two seems to best explain our observations. For example, gene duplication has been shown to play a role in expansion in the mammalian genomes of the top exon-bordering domain on our list, KRAB box domain (21,22). In contrast, the number two domain on our list, EGF-like domain (illustrated in Figure 1b), is a well-documented shuffling domain (18). Yet both domains have been significantly amplified in animal genomes over evolution, both strongly correlate with encoding exons whose borders preserve the integrity and functionality of these domains, both have an exceedingly large variety of domain architectures existing on the genes containing them, and both have a very strong and specific preference for certain intron phase classes. It is apparent that while KRAB box domain largely expanded through gene duplication events (with phase class 0-1), the increasingly larger number of domain architectures in its genes during mammalian evolution are not fully explained by gene duplication events but could arise from various mechanisms including exon shuffling.
How to estimate the extent of exon shuffling in a genome? In a simplified model, where a shuffling event would lead to new domain architecture, the number of successful shuffling events involving a domain could be approximated by the number of its architectures. In human, such estimate indicates some 1400 shuffling events involving exon-bordering domains. Of course, this rough calculation does not take into account many factors. For example, there could be shuffling events, leading to the same architecture independently, certain genes (and corresponding architectures) may have been lost, some architectural changes may not involve exon shuffling events, exons not containing detectable domains could be shuffled as well, etc. However, our lower-bound estimate still indicates that shuffling may have introduced significant modifications in the human genome.
According to exon shuffling theory, exon-bordering domains as we defined them could benefit from the correlation with exons and remain intact at new genomic location after exon shuffling events. Consequently, exon-bordering domains should have a higher success rate at forming novel functional proteins following exon shuffling events, making them valuable building blocks for proteome expansion and diversification. Positive selection on exon-bordering domains during evolution probably enhanced the preferential amplification of these domains relative to other domains, and contributed to the prevalence of exon-bordering domains in genomes as we described here. Our observations provide the strongest evidence yet that exon-bordering domains were indeed preferentially amplified over other domains in both abundance and distribution during evolution.
These findings are particularly important because they strongly support two points: (i) exon shuffling is a widespread evolutionary mechanism; (ii) exon-bordering domains face positive selection pressure as functional units with specific border and phase class properties. While the expansion of domains could be through various biological mechanisms such as duplications, retrotransposition, or even horizontal transfer, most of these mechanisms indiscriminately increase the protein domains encoded by the expanded DNA regions. In contrast, the ensuing exon shuffling events reuse exons to create new genes. Because of the correlation between the borders of exon-bordering domains and exons, the exon-bordering domains are likely preserved intact and are functional while non-bordering domains are lost following exon shuffling events. Accordingly, it is evident from our results that exon-bordering domains not only co-occur with more domains of more diversified types, their evolutionary expansion into these multi-domain, multi-architecture proteins also outpaced that of other domains. The dynamic interactions between exon shuffling events and exon-bordering domains thus could help shape the evolution of proteomes by creating novel functional proteins such as mosaic proteins, which are considered particularly important for the processes of development and signal transduction in multi-cellular organisms (23).
Exon-bordering domains strongly prefer phase 1-1 bounding introns
Exon-bordering domains generally follow the splice frame rule (13) and tend to be bounded by same phase introns, particularly by the phase class 1-1. Our results strongly support this notion. Phase 1-1 dominated the other phase classes for exon-bordering domains while same phase introns overall were highly preferred over mix-phase introns by the exon-bordering domain group. The same pattern was observed in all nine animal species. The fact that exon-bordering domains prefer same phase introns was expected because such phase pairings would lead to less disruption of an existing open reading frame in a destination gene following exon shuffling events.
Interestingly, when we subtracted the exon-bordering domain instances from all exon-matching instances (2END-ALL in Figure 2), the subtract group (2END-RANDOM) displayed a strong preference to phase 0-0 class while biasing against phase 1-1 class based on the expected values calculated using 2END-ALL instances in chi-square test. As we reasoned, the removal of statistically significant exon-bordering domain instances (2END-SIGNIFICANT) would leave the subtract group with mostly random cases of exon–domain border match. In such random cases, the main phase class should be produced by the intron phase that dominates in numbers. Indeed, it is well documented that phase 0 introns are the most abundant among the three phases in eukaryotes (24,25). Therefore, in cases of random matches of exon–domain borders, phase class 0-0 should theoretically be the most prevalent one, consistent with our observation for the subtract group. This demonstrated that our selection of statistically significant exon-bordering domains is a superior method than simply selecting domain instances whose borders are close to the borders of exons due to our exclusion of random cases. This also suggests that exon-bordering domains truly have a strong preference for phase 1-1 class despite the fact that random instances of exon–domain border match preferred phase 0-0. We found that the phase distribution of 2END-ALL instances showed increasing bias for phase 1-1 from C.briggsae to human, yet when random border match instances were taken out, the remaining exon-bordering domain instances from all the nine species displayed similar phase bias.
Exon-bordering domains in vertebrates versus invertebrates
In the current study, we found that although there is a general expansion in the abundance and distribution for exon-bordering domains in human compared to other organisms, the increase seemed much more conspicuous from invertebrates to human than from other vertebrates to human. Further, the phase biases of vertebrates including human were quite similar while invertebrates exhibited much lower preference for phase 1-1 when we investigated the phase preference for 2END-ALL that includes all domain instances matching exons on both ends (Figure 3a). Combined with our previous observation that invertebrates exhibited much weaker genome-wide exon–domain correlation than vertebrates (14), these results consistently demonstrate a substantial difference between vertebrates and invertebrates in terms of exon–domain correlation.
Various explanations could be offered for the apparent difference between invertebrates and vertebrates in their exon–domain correlation. One possibility is a potential bias in the datasets. While our procedures do not introduce bias and the production of the databases we used were relatively unbiased in data inclusion, it is possible that the large number of vertebrate domain instances recorded in Pfam biased domain definitions or that the relatively incomplete sequence and annotation of some invertebrates species led to bias against them. However, because the extensive studies, sequencing and inclusion of domain data from C.elegans and fruitfly were comparatively stronger than for species like rat and fugu, a bias cannot fully explain the observed difference. Most importantly, this explanation cannot account for many other features we observed such as the stronger phase bias and preferential expansion of exon-bordering domains.
A more plausible explanation could be derived from observations that introns have apparently been lost in some invertebrate genes while the vertebrate homologues still retained those introns (12). As a result, the evidence for exon shuffling events in these invertebrate genes such as match to exon borders and phase bias might also have been partially lost. If such cases were representative of the evolution of invertebrate genomes, eventually vertebrates would display a much stronger exon–domain correlation simply due to their better preservation of ancient introns. Indeed, this possibility is supported by the observation that some invertebrate lineages including worms and insects seemed to have lost much of the ancestral introns (26) and by a recent study on modular proteins that demonstrated the loss of many phase 1 introns in fly and worm (27). Additionally, such a hypothesis could also readily explain an observation that a smaller percentage of exon-bordering domain instances correlate with exons in invertebrates versus vertebrates (M. Liu and A. Grigoriev, unpublished data). However, this hypothesis can explain neither the preferential increase in the number of exon-bordering domains over other domains, nor their wider distribution in mosaic proteins. Moreover, the observation that phase 0-0 for 2END-ALL domain instances (Figure 2) held constant while phase 1-1 class increased from invertebrates to vertebrates (Table 3) is not in agreement with this hypothesis, as intron loss should not have an obvious phase or position bias (27).
In contrast, a model that combines duplication and shuffling-mediated positive selection on exon-bordering domains could readily explain all the phenomena we observed. Duplication and exon shuffling events occur frequently in vertebrates and could increase the number of domain instances as well as the distribution of domains. The evolutionary influence of exon shuffling was perhaps most palpable at the earlier stages of evolution via combining singular domains into simple architectures. Later, the duplication of complex (optimized) architectures, e.g. KRAB and Zinc finger (21,22), may have provided a greater contribution to fitness gain, and our observations likely reflect the balance of these processes.
The strong phase bias of exon-bordering domains and the positive selection on exon-bordering domain instances following exon shuffling events could lead to the preferential proliferation of exon-bordering domains, the continuous expansion of domain architectures containing exon-bordering domains, the rise in the phase bias for phase 1-1 in vertebrates, as well as the overall increase in the exon–domain correlation in vertebrates. Additionally, this model is consistent with the vast amount of evidence from research on exon shuffling. While the intron-loss scenario might be relevant, our model seems the most likely explanation for not only the differential exon–domain correlation among invertebrates and vertebrates, but also for the overall significant exon–domain correlation in animal species.
We noticed that in addition to constituting the majority of extracellular matrix and receptor proteins, many exon-bordering domains are intimately involved in protein–protein interaction, and are often found in combination with DNA binding domains in intracellular mosaic proteins involved in transcription regulation. Examples for such domains include KRAB box, SCAN and LIM domains. Some exon-bordering domains are repeats that are involved in protein–protein interactions and form proteins of diverse functions such as transcription initiation, cell adhesion, signal transduction (e.g. Leucine-rich repeats and Ankyrin repeats). Yet some other exon-bordering domains such as Myosin head and Intermediate filament domains are involved in intracellular matrix construction. The most prominent enzymatic domain in our list is Protein Kinase domain, involved in a large number of diverse architectures (with nearly 100 architectures, it is ranked second after EGF-like domain). These examples demonstrate the diverse functionalities exon-bordering domains could provide to multicellular organisms in addition to their previously observed extracellular roles. Nonetheless, it seems that exon-bordering domains are generally involved in the intracellular or extracellular signal transduction to participate in activities crucial to the development and maintenance of multicellular organisms. Our results strongly suggest that exon-bordering domains in modern organisms were positively selected during evolution because of the critical role they played in the advanced biological activities of these species and the evolutionary advantage their proliferation conferred to the organisms.
SUPPLEMENTARY MATERIAL
Supplementary Material is available at NAR Online.
Supplementary Material
Acknowledgments
We are indebted to Li Na for his comments on statistical methods. We thank David Bancroft and Erica Evans for their suggestions on the manuscript and Jonathon Blake for early discussions. Funding to pay the Open Access publication charges for this article was provided by GPC Biotech.
REFERENCES
- 1.Ohno S. Evolution by Gene Duplication. Berlin: Springer-Verlag; 1970. [Google Scholar]
- 2.Gilbert W. Why genes in pieces? Nature. 1978;271:501. doi: 10.1038/271501a0. [DOI] [PubMed] [Google Scholar]
- 3.Blake C.C. Do genes-in-pieces imply proteins-in-pieces? Nature. 1978;273:267. [Google Scholar]
- 4.Eickbush T. Exon shuffling in retrospect. Science. 1999;283:1465–1467. doi: 10.1126/science.283.5407.1465. [DOI] [PubMed] [Google Scholar]
- 5.van Rijk A., Bloemendal H. Molecular mechanisms of exon shuffling: illegitimate recombination. Genetica. 2003;118:245–249. [PubMed] [Google Scholar]
- 6.Holmes N., Parham P. Exon shuffling in vivo can generate novel HLA class I molecules. EMBO J. 1985;4:2849–2854. doi: 10.1002/j.1460-2075.1985.tb04013.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Maki R., Traunecker A., Sakano H., Roeder W., Tonegawa S. Exon shuffling generates an immunoglobulin heavy chain gene. Proc. Natl Acad. Sci. USA. 1980;77:2138–2142. doi: 10.1073/pnas.77.4.2138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.de Souza S.J., Long M., Klein R.J., Roy S., Lin S., Gilbert W. Toward a resolution of the introns early/late debate: only phase zero introns are correlated with the structure of ancient proteins. Proc. Natl Acad. Sci. USA. 1998;95:5094–5099. doi: 10.1073/pnas.95.9.5094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.de Souza S.J., Long M., Schoenbach L., Roy S.W., Gilbert W. Intron positions correlate with module boundaries in ancient proteins. Proc. Natl Acad. Sci. USA. 1996;93:14632–14636. doi: 10.1073/pnas.93.25.14632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kaessmann H., Zollner S., Nekrutenko A., Li W.H. Signatures of domain shuffling in the human genome. Genome Res. 2002;12:1642–1650. doi: 10.1101/gr.520702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Long M., Rosenberg C., Gilbert W. Intron phase correlations and the evolution of the intron/exon structure of genes. Proc. Natl Acad. Sci. USA. 1995;92:12495–12499. doi: 10.1073/pnas.92.26.12495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Patthy L. Genome evolution and the evolution of exon-shuffling—a review. Gene. 1999;238:103–114. doi: 10.1016/s0378-1119(99)00228-0. [DOI] [PubMed] [Google Scholar]
- 13.Patthy L. Intron-dependent evolution: preferred types of exons and introns. FEBS Lett. 1987;214:1–7. doi: 10.1016/0014-5793(87)80002-9. [DOI] [PubMed] [Google Scholar]
- 14.Liu M., Grigoriev A. Protein domains correlate strongly with exons in multiple eukaryotic genomes—evidence of exon shuffling? Trends Genet. 2004;20:399–403. doi: 10.1016/j.tig.2004.06.013. [DOI] [PubMed] [Google Scholar]
- 15.Hubbard T., Barker D., Birney E., Cameron G., Chen Y., Clark L., Cox T., Cuff J., Curwen V., Down T., et al. The Ensembl genome database project. Nucleic Acids Res. 2002;30:38–41. doi: 10.1093/nar/30.1.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Eddy S.R. Profile hidden Markov models. Bioinformatics. 1998;14:755–763. doi: 10.1093/bioinformatics/14.9.755. [DOI] [PubMed] [Google Scholar]
- 17.Bateman A., Birney E., Cerruti L., Durbin R., Etwiller L., Eddy S.R., Griffiths-Jones S., Howe K.L., Marshall M., Sonnhammer E.L. The Pfam protein families database. Nucleic Acids Res. 2002;30:276–280. doi: 10.1093/nar/30.1.276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kolkman J.A., Stemmer W.P. Directed evolution of proteins by exon shuffling. Nat. Biotechnol. 2001;19:423–428. doi: 10.1038/88084. [DOI] [PubMed] [Google Scholar]
- 19.Sonnhammer E.L., Eddy S.R., Birney E., Bateman A., Durbin R. Pfam: multiple sequence alignments and HMM-profiles of protein domains. Nucleic Acids Res. 1998;26:320–322. doi: 10.1093/nar/26.1.320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lynch M., Conery J.S. The evolutionary fate and consequences of duplicate genes. Science. 2000;290:1151–1155. doi: 10.1126/science.290.5494.1151. [DOI] [PubMed] [Google Scholar]
- 21.Looman C., Abrink M., Mark C., Hellman L. KRAB zinc finger proteins: an analysis of the molecular mechanisms governing their increase in numbers and complexity during evolution. Mol. Biol. Evol. 2002;19:2118–2130. doi: 10.1093/oxfordjournals.molbev.a004037. [DOI] [PubMed] [Google Scholar]
- 22.Shannon M., Hamilton A.T., Gordon L., Branscomb E., Stubbs L. Differential expansion of zinc-finger transcription factor loci in homologous human and mouse gene clusters. Genome Res. 2003;13:1097–1110. doi: 10.1101/gr.963903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Patthy L. Modular assembly of genes and the evolution of new functions. Genetica. 2003;118:217–231. [PubMed] [Google Scholar]
- 24.Sverdlov A.V., Rogozin I.B., Babenko V.N., Koonin E.V. Evidence of splice signal migration from exon to intron during intron evolution. Curr. Biol. 2003;13:2170–2174. doi: 10.1016/j.cub.2003.12.003. [DOI] [PubMed] [Google Scholar]
- 25.Fedorov A., Merican A.F., Gilbert W. Large-scale comparison of intron positions among animal, plant, and fungal genes. Proc. Natl Acad. Sci. USA. 2002;99:16128–16133. doi: 10.1073/pnas.242624899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rogozin I.B., Wolf Y.I., Sorokin A.V., Mirkin B.G., Koonin E.V. Remarkable interkingdom conservation of intron positions and massive, lineage-specific intron loss and gain in eukaryotic evolution. Curr. Biol. 2003;13:1512–1517. doi: 10.1016/s0960-9822(03)00558-x. [DOI] [PubMed] [Google Scholar]
- 27.Banyai L., Patthy L. Evidence that human genes of modular proteins have retained significantly more ancestral introns than their fly or worm orthologues. FEBS Lett. 2004;565:127–132. doi: 10.1016/j.febslet.2004.03.088. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.