Abstract
The basic leucine zipper (bZIP) transcription factor HY5 plays a multifaceted role in plant growth and development. Here the apple MdHY5 gene was cloned based on its homology with Arabidopsis HY5. Expression analysis demonstrated that MdHY5 transcription was induced by light and abscisic acid treatments. Electrophoretic mobility shift assays and transient expression assays subsequently showed that MdHY5 positively regulated both its own transcription and that of MdMYB10 by binding to E-box and G-box motifs, respectively. Furthermore, we obtained transgenic apple calli that overexpressed the MdHY5 gene, and apple calli coloration assays showed that MdHY5 promoted anthocyanin accumulation by regulating expression of the MdMYB10 gene and downstream anthocyanin biosynthesis genes. In addition, the transcript levels of a series of nitrate reductase genes and nitrate uptake genes in both wild-type and transgenic apple calli were detected. In association with increased nitrate reductase activities and nitrate contents, the results indicated that MdHY5 might be an important regulator in nutrient assimilation. Taken together, these results indicate that MdHY5 plays a vital role in anthocyanin accumulation and nitrate assimilation in apple.
Introduction
Apple (Malus×domestica) is an important fruit crop that is widely cultivated worldwide. Peel color is an important trait that determines apple market value; red fruits are more attractive to consumers. As a kind of secondary metabolite, anthocyanins are responsible for the red coloration in apple peel.1,2 In general, anthocyanins are synthesized via the phenylpropanoid pathway, and multiple enzymes are involved in this pathway, including phenylalanine ammonia lyase, chalcone isomerase, chalcone synthase, flavanone 3-hydroxylase (F3H), dihydroflavonol 4-reductase (DFR), UDP glucose: flavonoid 3-O-glucosyltransferase (UF3GT) and anthocyanidin synthase (ANS).3,4
Three main protein families—MYB, bHLH and WD40—are involved in the regulation of anthocyanin biosynthesis by forming the MYB-bHLH-WD40 protein complex.5,6 In apple, three MYB transcription factors (TFs) (MdMYB1, MdMYB10 and MdMYBA) have been functionally identified and confirmed to be responsible for anthocyanin accumulation, as these TFs directly regulate the expression of anthocyanin biosynthesis structural genes.7,8
In addition to genetic components, nutritional components such as nitrogen, phosphorus and potassium can affect fruit coloration as well as plant growth.9,10 Among these nutritional components, nitrogen is the most important; NO3− and NH4+ are the major sources of nitrogen in aerobic and anoxic soils, respectively.11 In recent years, the physiological process and genetic mechanism of nitrogen uptake and transport have been studied in depth, and protein families, such as NRT1/2, NAR2 and AMT1/2, participate in the absorption and transport of NO3− and NH4+.12,13,14,15 In addition, NO3− is widely regarded as a crucial signal molecule that modulates multiple aspects of plant growth and development, such as nitrate-mediated root development,16,17 stress tolerance18 and crop yield and quality.19,20 Therefore, identification of the functions of genes involved in nitrogen uptake and transport in apple is essential.
TFs are a group of key regulatory proteins that play important roles in controlling the expression of signal response genes. They can be divided into many families, such as the basic leucine zipper (bZIP), bHLH, MYB, zinc-finger and NAC families, according to their conserved domains. In addition, different types of TFs play diverse roles in plant growth and development. An important TF in plants is the bZIP protein HY5, which was first identified to positively regulate plant photomorphogenesis based on the light insensitivity of the hy5 mutant.21,22 This factor has also been subsequently implicated in abscisic acid (ABA) signaling.23
HY5 acts as a master regulator that integrates signals from multiple pathways to coordinate plant stress tolerance and development.24 HY5 binds the T/G-box (CACGTT), E-box (CAATTG), GATA-box (GATGATA), ACE-box (ACGT), Z-box (ATACGGT) and C-box (GTCANN) as well as the hybrid C/G- (G) and C/A-boxes in the promoters of many genes that are involved in various signaling pathways, such as light signaling,25 anthocyanin and chlorophyll biosynthesis,26,27 nutrient signaling28 and defense signaling.29 In addition, HY5 regulates the expression of microRNAs by directly binding to their promoters,30 which indicates that HY5 regulates gene expression at both the transcriptional and post-transcriptional levels. The HY5 protein is unstable and is degraded in darkness by COP1 via the 26S proteasome pathway,31,32 but HY5 stability is indirectly promoted in blue light by SPA1 by dissociating with COP1 and associating with CRY1.33 Moreover, recent studies have shown that low-temperature and short-heat shock treatments also stabilize the HY5 protein.26,34 Thus, HY5 is dynamic in plants and plays a central role in the hub of transcriptional networks.
In the present study, we cloned and functionally characterized the apple bZIP protein MdHY5. MdHY5 may be involved in anthocyanin biosynthesis by directly activating MdMYB10 and nitrogen signaling by promoting the expression of nitrate reductase (NR) genes and nitrate uptake genes. In brief, these results indicate that MdHY5 plays a vital role in anthocyanin accumulation and nitrate assimilation in apple.
Materials and methods
Plant materials and growth conditions
Calli of the ‘Orin’ apple (wild type) were subcultured on Murashige and Skoog (MS) medium containing 0.5 mg L−1 indole acetic acid and 1.5 mg L−1 6-butyric acid (BA) at room temperature (24 °C) in continuous darkness; calli were subcultured at 20 days intervals. For high-light and low-temperature treatments, transgenic and wild-type (WT) apple calli were transferred to a phytotron at 15 °C under constant high light (photon flux density of ~100 μmol s−1 m−2).
For light treatment, apomictic crabapple (Malus hupehensis) seedlings grown in darkness for 3 days at room temperature were transferred to white light conditions. For ABA treatment, apomictic crabapple seedlings were treated with 50 μM ABA. RT-qPCR and semi-quantitative RT-PCR were performed to monitor the expression level of MdHY5.
Gene cloning of MdHY5
Using the NCBI database with the Basic Local Alignment Search Tool (BLAST) program, we found two potential homologous genes (MDP0000586302 and MDP0000264514) that differed by only one amino acid. On the basis of these results, we speculate that these two genes might have similar functions and possibly be allelic. MDP0000586302 was selected for functional identification.
RT-qPCR and semi-quantitative RT-PCR
Apple seedlings and 4 g of apple calli collected from three plates were used for RNA extraction. RNAs were extracted from the apple seedlings and calli using RNA plant plus Reagent (Tiangen, Beijing, China) followed by reverse transcription using a PrimeScript first-strand cDNA synthesis kit (Takara, Dalian, China). RT-qPCR assays were conducted with the UltraSYBR mixture (SYBR Green I) (Takara) using an ABI7500 RT-qPCR system. The concentration of cDNA was diluted to 1–10 ng μL−1. One microliter of diluted cDNA was used for RT-qPCR, and the 2−ΔΔCt calculation method of RT-qPCR was used. The results were normalized to those of MdACTIN. A minimum of three biological replicates per sample were used for RT-qPCR.
RT-PCR was conducted to examine the expression levels of MdHY5 in response to light and ABA. Each PCR mixture contained 200 ng of cDNA, 1× Taq buffer, 2.5 mM dNTPs, 0.5 μL of Taq DNA polymerase (Trans, Beijing, China) and each primer at 10 μm in a total volume of 25 μL. The reactions were performed as follows: denaturation at 95 °C for 10 min; 25–28 cycles at 95 °C for 30 s, 58 °C for 30 s and 72 °C for 30 s; and a final cycle at 72 °C for 5 min.
The transcription levels of MdHY5, anthocyanin biosynthesis genes and nitrogen signaling-related genes (MdNIA1: MDP0000585462; MdNIA2: MDP0000280001; MdNRT1.1: MDP0000163192; MdNRT1.5: MDP0000260604; MdNRT1.7: MDP0000681768; MdNRT2.1: MDP0000239537; MdNRT2.4: MDP0000897809; MdNRT2.5: MDP0000266497; and MdNRT2.7: MDP0000131368) were examined using specific primers.35 All of the primers used are shown in Supplementary Table 1.
Amino-acid sequence analysis and phylogenetic tree construction
The Protein BLAST program (http://www.ncbi.nlm.nih.gov/BLAST/) was used to obtain homologs of Arabidopsis HY5. The amino-acid secondary structure of MdHY5 was predicted using the Simple Modular Architecture Research Tool (SMART) software program (http://smart.embl-heidelberg.de/).
The phylogenetic tree was constructed using MEGA 5.0 software.
Plasmid construction and genetic transformation of apple calli
The overexpression vector MdHY5-pCAMBIA1300 was constructed by inserting the MdHY5 open reading frame into the transformed vector pCAMBIA1300.
To generate MdHY5 transgenic apple calli, the recombinant plasmid was introduced into Agrobacterium tumefaciens LBA4404 as described by An et al.36 Regarding the transformation of apple calli, 15-day-old ‘Orin’ calli (WT) were co-cultured with Agrobacterium carrying MdHY5-pCAMBIA1300. The calli were co-cultured on MS medium containing 1.5 mg L−1 2, 4-dichlorophenoxyacetic acid and 0.5 mg L−1 6-butyric acid for 2 days at room temperature. The calli were then washed three times with sterile water and transferred to selective media supplemented with 300 mg L−1 carbenicillin and 35 mg L−1 hygromycin for transgene selection. The transgenic apple calli were co-cultivated in selective media that contained appropriate concentrations of antibiotics.
Measurement of anthocyanins
Total anthocyanins were extracted as described by Lee and Wicker.37 Three grams of apple calli were incubated in an anthocyanin extraction solution for 12 h at 4 °C in darkness. The absorbance of each sample was measured at 530, 620 and 650 nm using a spectrophotometer (Shimadzu UV-2450, Kyoto, Japan). The relative anthocyanin contents were normalized according to the following formula: optical density (OD)=(A530−A620)−[0.1×(A650−A620)]. One unit of anthocyanin content was expressed as a change of 0.1×OD (units×103 g−1 of fresh weight (FW)). Measurements were performed in triplicate.
MdHY5-GST fusion protein expression and purification
The open reading frames of MdHY5 were cloned into the pGEX-4T-1 vector, which contained a glutathione (GST) tag sequence, and the recombinant vector was then transformed into Escherichia coli BL21 (DE3). The BL21 bacteria were subsequently treated with 3 mM isopropyl β-D-1-thiogalactopyranoside to induce the production of the MdHY5-GST fusion protein.
The fusion protein was then added to a cobalt chelate affinity resin containing the immobilized GST-tag and incubated at 4 °C for 2 h under rotation. After three washings, the proteins were collected and detected via western blot with GST antibodies (Abcam, Cambridge, UK).
Electrophoretic mobility shift assay (EMSA)
EMSA was conducted using a LightShift Chemiluminescent EMSA Kit (Thermo, Waltham, MA, USA). Briefly, biotin-labeled probes were incubated in a 1× binding buffer containing 2.5% glycerol, 10 mM EDTA, 5 mM MgCl2 and 50 mM KCl with or without proteins at 24 °C for 25 min. An unlabeled probe was added to the reactions for unlabeled probe competition.
Transient expression assay in Nicotiana benthamiana leaves
Transient expression assays in N. benthamiana leaves were performed as previously described.38,39 The MdHY5 and MdMYB10 promoters were amplified and cloned into pGreenII 0800-LUC vectors, which generated the reporter constructs MdHY5pro:Luc and MdMYB10pro:Luc, respectively. Site-directed mutagenesis was performed using the Takara MutanBEST kit. The effector (35Spro:MdHY5) was constructed by cloning the open reading frame of the MdHY5 into the pGreenII 62-SK vector. A charge-coupled device imaging apparatus (NightOWL II LB983 in conjunction with Indigo software) was used to collect the LUC images and quantify luminescence intensity. Transformed leaves were sprayed with and soaked in 100 mM luciferin, after which they were placed in darkness for 6 min before luminescence examination.
Measurement of NR activity
NR activity was measured as previously described.40 Five-hundred-milligram samples (apple calli) were transferred to test tubes after being washed with distilled water and weighed; 1 mL of trichloroacetic acid served as the control. Then, 9 mL of 0.1 M phosphate buffer (pH=7.5) mixed with 3% propanol and 0.1 M KNO3 was added to the tubes, and the samples were vacuum infiltrated until the samples sunk to the bottom of the tubes. The reactions were performed at 30 °C in the dark for 30 min, and 1 mL of trichloroacetic acid was added to stop the reaction. After standing for 2 min, 4 mL of sulfanilamide mixed with 3 M HCl and 4 mL of 0.2% N-1-naphthyl-ethylene-diamine were added to the 2 mL of supernatant, which was transferred into a new tube. Finally, after 30 min, the absorbance was measured at 540 nm. Next, 0–2 g of NaNO2 per reaction was used to generate a standard curve. A regression equation was calculated according to the standard curve. NR activity is presented as nanomoles of nitrite produced per hour per gram of FW (nmol nitrite h−1 g−1 FW). Measurements were performed in triplicate.
Measurement of nitrate content
Nitrate content was measured according to the salicylic acid method.41,42 First, samples of ~1 g (apple calli) were frozen in liquid nitrogen and milled into powder. Next, 10 mL of deionized water was added to the tubes. The samples were boiled at 100 °C for 20 min and centrifuged at 15 000 g for 10 min, after which 0.1 mL of the supernatant was transferred to a new tube. Then, 0.4 mL of a 5% salicylic acid–sulfuric acid solution was then added to the tubes. The reactions occurred at room temperature for 20 min, after which 9.5 mL of 8% NaOH was slowly added to the reactions. After the samples cooled to room temperature, the optical density at 410 nm (OD410 value) was measured; deionized water served as the control. The nitrate content was calculated according to the following equation: N=C×V/W (N, nitrate content; C, nitrate concentration calculated using OD410 and the regression equation; V, total volume of extracted sample; W, weight of sample). Concentrations of 10–120 mg L−1 KNO3 were used to generate a standard curve. The regression equation was calculated according to the standard curve. Measurements were performed in triplicate.
Statistical analysis
Statistical analysis was conducted as previously described using appropriate methods and R (3.0.2) software with the R Commander package.43 Differences were considered statistically significant when P<0.05 and P<0.01. The results were analyzed in triplicate.
Results
Identification of the HY5 gene in apple
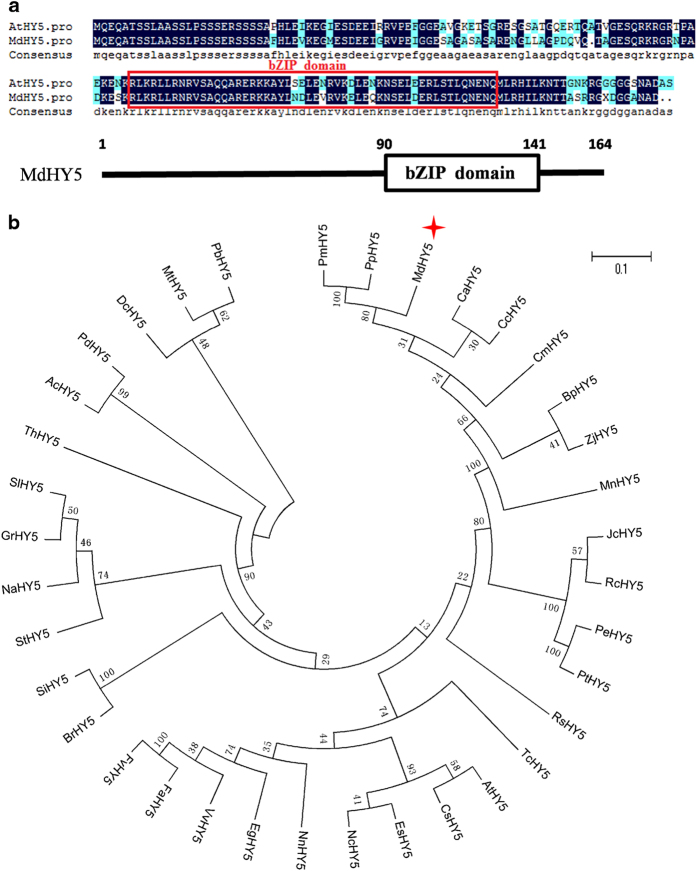
Regarding the identification of the HY5 gene in the apple genome, the Arabidopsis HY5 gene was used as a query to search similar sequences in apple by mining the NCBI database with the BLAST program. Two potential homologous genes were found: GenBank accession numbers MDP0000586302 and MDP0000264514, which differed by only one amino acid (Supplementary Figure S1). The gene MDP0000586302 was selected for functional identification. This gene contained a 495-bp open reading frame that encoded a protein containing 164 amino acids and was named MdHY5. In addition, the protein secondary structure of MdHY5 showed that it contained a bZIP domain on its C-terminal side (amino acids 90–141) (Figure 1a), which indicated the correlation between structure and function as observed in other bZIP proteins.
Figure 1.
Sequence alignment and phylogenetic analysis of MdHY5. (a) Protein alignment of MdHY5 and its homologs in Arabidopsis. The zinc-finger domain is boxed. (b) Phylogenetic analysis of MdHY5 and 35 other plants HY5 protein sequences obtained from the NCBI database. MdHY5 is denoted by the red asterisk, and the scale bar indicates the branch length. RsHY5: Raphanus sativus, XP_018445811.1; AtHY5: Arabidopsis thaliana, AT5G11260.1; BrHY5: Brassica rapa, XP_009121971.1; ThHY5: Tarenaya hassleriana, XP_010541629.1; PeHY5: Populus euphratica, XP_011039711.1; MdHY5: Malus domestica, MDP0000586302; PbHY5: Pyrus bretschneideri, XP_009355719.1; FvHY5: Fragaria vesca, XP_004291469.1; ZjHY5: Ziziphus jujuba, XP_015885857.1; PmHY5: Prunus mume, XP_008219477.1; FaHY5: Fragaria ananassa, AKG58815.1; PpHY5: Prunus persica, ONI34365.1; CcHY5: Citrus clementina, XP_006450470.1; BpHY5: Betula platyphylla, AHY20043.1; EgHY5: Eucalyptus grandis, XP_010048982.1; JcHY5: Jatropha curcas, XP_012076602.1; CaHY5: Camptotheca acuminate, APD29065.1; PtHY5: Populus trichocarpa, XP_002308656.1; CmHY5: Cucumis melo, NP_001284656.1; TcHY5: Theobroma cacao, XP_007013841.2; RcHY5: Ricinus communis, XP_002515537.1; VvHY5: Vitis vinifera, XP_010648648.1; MnHY5: Morus notabilis, XP_010110356.1; SiHY5: Sesamum indicum, XP_011081579.1; NnHY5: Nelumbo nucifera, XP_010250037.1; NcHY5: Noccaea caerulescens, JAU18721.1; EsHY5: Eutrema salsugineum, XP_006399627.1; StHY5: Solanum tuberosum, XP_006361723.1; MtHY5: Medicago truncatula, XP_013459310.1; SlHY5: Solanum lycopersicum, NP_001234820.1; PdHY5: Phoenix dactylifera, XP_008785002.1; AcHY5: Ananas comosus, XP_020097860.1; NaHY5: Nicotiana attenuate, XP_019265660.1; DcHY5: Daucus carota, XP_017229054.1; GrHY5: Gentiana rigescens, AIC64080.1; CsHY5: Camelina sativa, XP_010419684.1.
To analyze the phylogenetic relationship between MdHY5 and HY5 proteins from other plant species, a phylogenetic tree of 36 plant HY5 proteins was constructed using MEGA 5.0. Figure 1b shows that MdHY5 was most closely related to and exhibited the highest homology with PpHY5 and PmHY5 from Prunus persica and Prunus mume, respectively.
Expression analysis of the MdHY5 gene
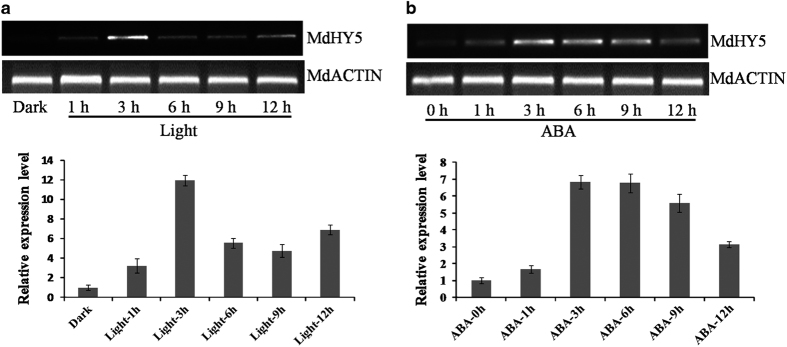
Arabidopsis HY5 plays a vital role in light signaling and ABA responses.23,25 To investigate the functions of MdHY5 in planta, the expression patterns of MdHY5 were determined by RT-qPCR and semi-quantitative RT-PCR using cDNA isolated from apomictic crabapple seedlings treated with light or subjected to ABA. These results demonstrated that the expression levels of MdHY5 increased in response to light and ABA treatments. The expression level increased 1 h after light treatment and peaked at 3 h (Figure 2a). In addition, treatment of apple seedlings with ABA led to a peak in the expression of MdHY5 at 3–6 h (Figure 2b). Therefore, MdHY5 plays an important role in light signaling and ABA responses.
Figure 2.
Effects of light and ABA on the transcript level of MdHY5. (a) Apomictic crabapple (Malus hupehensis) seedlings grown in darkness for 3 days at room temperature were treated with continuous white light for 1, 3, 6, 9 and 12 h. The transcript levels of MdHY5 were determined using RT-PCR (above) and RT-qPCR (below); the value for dark-treated plants was set to 1. (b) Apomictic crabapple (M. hupehensis) seedlings were treated with 50 μM ABA at the indicated time intervals. The expression levels of MdHY5 were monitored by RT-qPCR (above) and RT-PCR (below); the value for untreated plants was set to 1.
MdHY5 positively regulates its own transcription
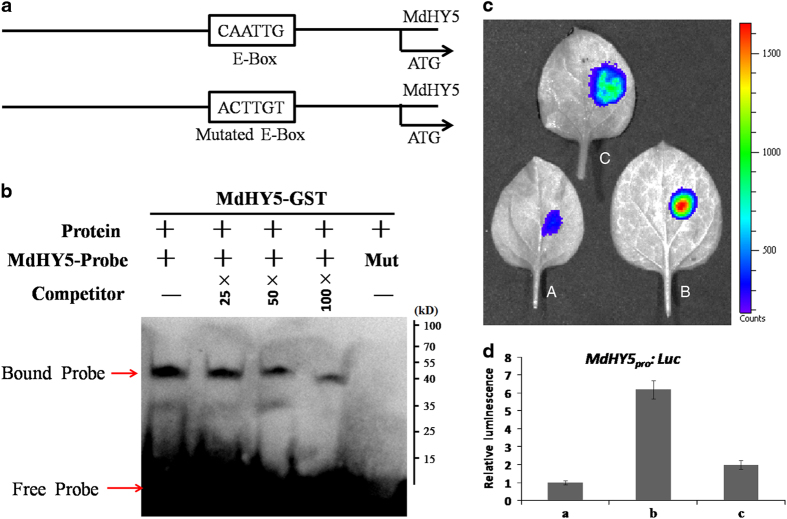
HY5 and its homolog HYH can regulate HY5 gene induction in Arabidopsis.44 To test whether MdHY5 could also directly interact with the MdHY5 promoter in vitro, we expressed and purified recombinant MdHY5-GST fusion protein and prepared MdHY5 promoter fragments containing versions of the E-box motif. As predicted, EMSAs demonstrated that MdHY5 bound to the oligonucleotide sequence of the CAATTG-box of MdHY5 (Figures 3a and b).
Figure 3.
MdHY5 directly and positively regulates its own expression. (a) Schematic diagram showing both the predicted E-box and mutated E-box sites in black boxes. (b) EMSA results showing that the MdHY5-GST fusion protein directly binds to its own promoter at the E-box in vitro. Biotin-labeled probes were incubated with MdHY5-GST protein, and the free and bound DNAs (arrows) were separated in an acrylamide gel. Unlabeled probes served as competitors. The 5′-CAATTG-3′ motif in mutated probes (Mut) was replaced with 5′-ACTTGT-3′. (c) Transient expression assays showing that MdHY5 promotes its own expression. A: MdHY5pro:Luc; B: MdHY5pro:Luc-35Spro:MdHY5; C: MdHY5pro(Mut):Luc-35Spro:MdHY5. Representative images of Nicotiana benthamiana leaves 72 h after infiltration are shown. (d) Quantitative analysis of luminescence intensity in c. The value for MdHY5pro:Luc was set to 1.
To investigate whether MdHY5 could activate its own transcription, we carried out a transient assay to compare the activation effect of MdHY5 on the expression of MdHY5pro:Luc and MdHY5pro(Mut):Luc reporters containing the MdHY5 promoter fragments or mutated MdHY5 promoter fragments fused with LUC genes. As shown in Figures 3c and d, co-expression of MdHY5pro:Luc with 35Spro:MdHY5 markedly increased the luminescence intensity. In contrast, 35Spro:MdHY5 failed to induce the expression of MdHY5pro(Mut):Luc. This result suggests that MdHY5 is also capable of positively regulating its own expression.
MdHY5 promotes anthocyanin accumulation by directly binding to the MdMYB10 promoter
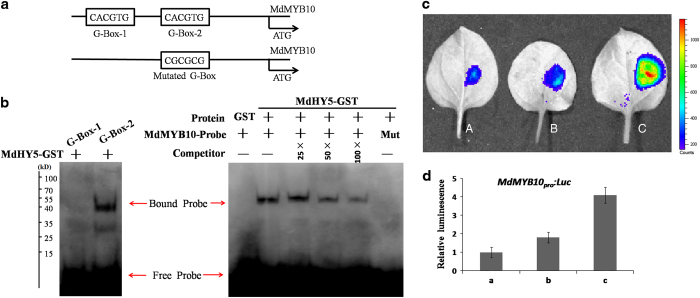
Arabidopsis HY5 promotes anthocyanin biosynthesis by inducing the expression of PAP1.27 To investigate this in apple, we investigated the DNA–protein interaction between MdHY5 and MdMYB10 promoter fragments containing G-boxes. As shown in Figure 4a, the upstream region of the MdMYB10 gene contained two MdHY5-binding sites (G-box-1 and G-box-2). Interestingly, the EMSA assays showed that MdHY5 interacted with an MdMYB10 promoter fragment (G-box-2) (Figure 4b). In addition, the transient assay suggested that MdHY5 could induce the transcriptional activation of MdMYB10 in apple.
Figure 4.
MdHY5 promotes the expression of MdMYB10 by directly interacting with its promoter. (a) Schematic diagram of the MdMYB10 promoter showing the potential MdHY5-binding sites. The predicated two G-box (G-box-1 and G-box-2) and mutated G-box sites and sequences are indicated with black boxes. (b) EMSA results showing that MdHY5-GST fusion protein directly binds to the MdMYB10 promoter at G-box-2 in vitro. Biotin-labeled probes were incubated with MdHY5-GST protein, and the free and bound DNAs (arrows) were separated in an acrylamide gel. Unlabeled probes served as competitors. The 5′-CACGTG-3′ motif in mutated probes (Mut) was replaced with 5′-CGTGTG-3′. (c) Transient expression assays showing that MdHY5 promotes the expression of MdMYB10. A: MdMYB10pro:Luc; B: MdMYB10pro(Mut):Luc-35Spro:MdHY5; C: MdMYB10pro:Luc-35Spro:MdHY5. Representative images of Nicotiana benthamiana leaves 72 h after infiltration are shown. (d) Quantitative analysis of luminescence intensity in c. The value for a MdMYB10pro:Luc was set to 1.
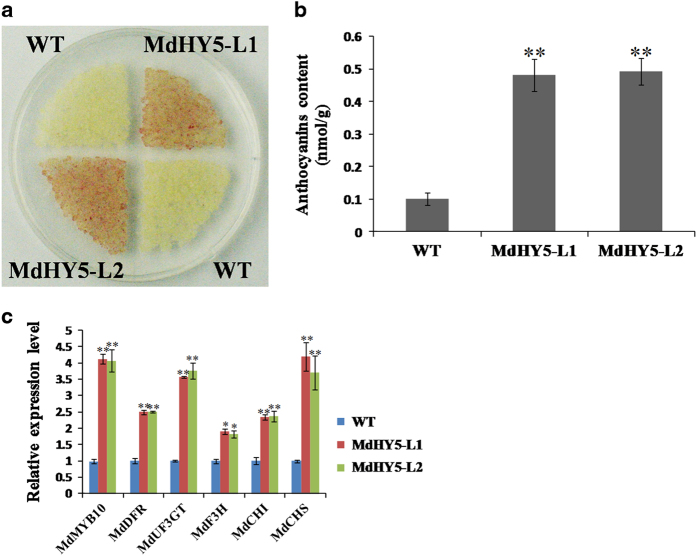
To examine whether MdHY5 regulates anthocyanin accumulation, we transformed the overexpression construct MdHY5-pCAMBIA1300 into ‘Orin’ apple calli through Agrobacterium-mediated genetic transformation (Supplementary Figure S2). The observation of the appearance of the apple calli revealed that the MdHY5 overexpression calli (MdHY5-L1 and MdHY5-L2) appeared redder in color under low-temperature and high-light conditions than did the WT control (Figure 5a). Spectrophotometric analysis demonstrated that the apple calli overexpressing MdHY5 produced much higher contents of anthocyanins (Figure 5b). In addition, the expression levels of flavonoid structural genes in the WT and transgenic calli were analyzed by RT-qPCR. The overexpression of MdHY5 significantly upregulated the expression of primary genes involved in the anthocyanin biosynthesis pathway, including MdMYB10, MdDFR, MdUF3GT, MdF3H, MdCHS and MdCHI (Figure 5c). This result suggests that MdHY5 affects the accumulation of anthocyanins. Taken together, these results demonstrate that MdHY5 binds to the G-box-2 site of the MdMYB10 promoter to induce anthocyanin biosynthesis in apple.
Figure 5.
MdHY5 increases anthocyanin accumulation in transgenic apple calli. (a, b) Anthocyanin contents in transgenic calli (MdHY5-L1 and MdHY5-L2) and wild-type (WT) control grown on medium under low-temperature and high-light conditions. (c) Relative expression levels of MdMYB10, MdDFR, MdUF3GT, MdF3H, MdCHI and MdCHS in transgenic calli and WT control. The value for WT was set to 1.
MdHY5 is involved in nitrate assimilation
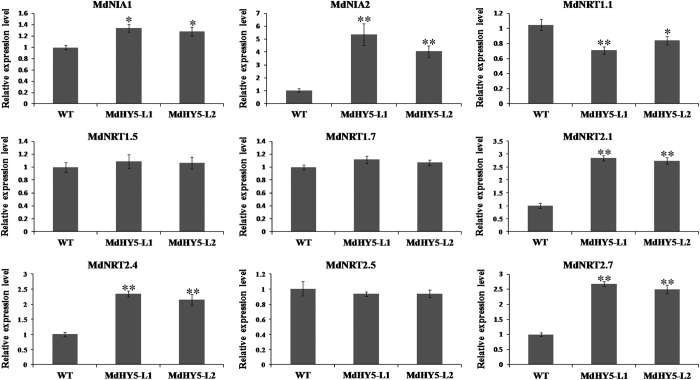
Arabidopsis HY5 and HYH upregulate NR genes but negatively regulate nitrate uptake genes.45 To examine whether MdHY5 influenced nitrogen signaling in apple, we attempted to identify the possible target genes of MdHY5; we therefore investigated the cis elements in the NR genes and nitrate uptake genes in apple. Supplementary Figure S3 shows that four genes related to nitrogen signaling (MdNIA1, MdNRT1.1, MdNRT2.4 and MdNRT2.7) contained potential MdHY5-binding sites. The expression levels of a series of NR genes and nitrate uptake genes were subsequently detected. As shown in Figure 6, MdHY5 positively regulated the expression of the NR gene MdNIA2, which is in agreement with the observation in Arabidopsis. In contrast to the results for MdNIA2, MdHY5 negatively regulated the expression of the nitrate uptake gene MdNRT1.1 in apple. In addition, overexpression of MdHY5 promoted the expression of MdNRT2.1, MdNRT2.4 and MdNRT2.7, which might help coordinate the acquisition of plant carbon and nitrogen.28
Figure 6.
MdHY5 influences the expression of nitrogen signaling-related genes. RT-qPCR analysis of genes related to nitrogen signaling in WT and MdHY5 transgenic calli (MdHY5-L1 and MdHY5-L2) under low-temperature and high-light conditions. The value for WT was set to 1.
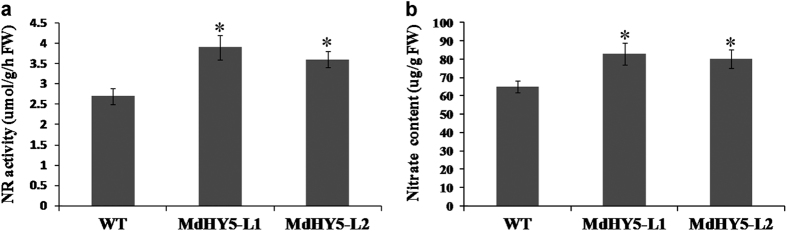
We also examined NR activity and nitrate content. As shown in Figure 7a, NR activities increased in MdHY5-overexpressing apple calli that were grown under low-temperature and high-light conditions, which indicates that MdHY5 might be a positive regulator of NR in apple. In addition, the nitrate contents of transgenic apple calli markedly increased (Figure 7b), which might be associated with increased demand for metabolites during plant growth.
Figure 7.
MdHY5 influences NR activity and nitrate acquisition. (a) NR activities and (b) nitrate contents of WT and MdHY5 transgenic calli (MdHY5-L1 and MdHY5-L2) under low-temperature and high-light conditions are shown.
Discussion
The bZIP TF HY5 is a central regulator of plant growth and development and has been functionally characterized to be involved in multifaceted developmental processes such as cell elongation and proliferation,46,47 chloroplast development,48,49 photomorphogenesis, pigment accumulation,27,50,51, 52 nutrient assimilation and carbon/nitrogen balance.28,53 Other HY5-mediated response pathways, such as hormone signaling,23,54 reactive oxygen species and defense signaling,29,55 as well as temperature responses26 have recently been uncovered. Among these responses, pigment accumulation and nutrient assimilation are economically important characteristics that influence apple quality and yield.
Here we cloned and isolated a gene from the apple genome that contains a highly conserved bZIP motif in its C terminus and is highly homologous to HY5 proteins from other species; this finding indicated that MdHY5 might have similar functions as other HY5 proteins. As we predicted, our molecular and genetic analyses implied that MdHY5 was highly similar in structure and function to Arabidopsis HY5 in terms of the conserved bZIP domain (Figure 1) and response to light and ABA signaling (Figure 2). In addition, EMSA assays showed that MdHY5 could bind to the E-box motif of its promoter (Figure 3), which suggests a potential autoregulatory loop for MdHY5 transcription. These results indicate that the functions of MdHY5 are conserved across different species.
To further investigate the functions of MdHY5 in planta, we used Agrobacterium-mediated transformation to obtain transgenic apple calli overexpressing MdHY5 under the control of the 35S promoter (Supplementary Figure S2). HY5 is a positive regulator of flavonoid biosynthesis by modulating the expression of MYB75/PAP1 in Arabidopsis.27,30 We also investigated the role of MdHY5 in the regulation of flavonoid biosynthesis on the basis of EMSA-binding assays and apple calli coloration detection. As hypothesized, the EMSA results showed that MdHY5 was able to bind the G-box site of the MdMYB10 promoter (Figure 4). Furthermore, the MdHY5-overexpressing apple calli produced much higher amounts of anthocyanins than did the WT control, and the RT-qPCR analysis indicated that anthocyanin biosynthesis genes, including MdDFR, MdUF3GT, MdF3H, MdCHI and MdCHS, were clearly upregulated in the MdHY5 transgenic lines (Figure 5). These results suggest that MdHY5 promotes anthocyanin accumulation by directly binding to the MdMYB10 promoter.
In addition, HY5 positively regulates NR activity and coordinates plant carbon and nitrogen acquisition by affecting the expression of both NR genes and nitrate uptake genes in Arabidopsis.28,45,53 To determine whether MdHY5 is also required for nitrogen assimilation in apple, we measured the transcripts of a series of NR genes and nitrate uptake genes in transgenic and WT apple calli. MdHY5 positively regulated the expression of MdNIA2, MdNRT2.1, MdNRT2.4 and MdNRT2.7 but negatively regulated the expression of MdNRT1.1, which was in agreement with observations in Arabidopsis (Figure 6).28,53 In addition, the upstream region of the MdNIA1 gene may contain a putative MdHY5-binding site (G-box) (Supplementary Figure S3), and overexpression of MdHY5 slightly induced the transcription of MdNIA1, despite AtHY5 having no impact on the expression of MdNIA1.28 The function of MdHY5 might be distinct from that of Arabidopsis HY5, or the function of MdHY5 might be a secondary metabolic effect caused by the upregulation of MdNIA2. In addition, the increased NR activities and nitrate contents in MdHY5-overexpressing apple calli might be associated with the increased demand for metabolites during plant growth (Figure 7). Therefore, MdHY5 may play a vital role in nitrogen assimilation in apple. However, these results are not sufficient and require further investigation.
On the basis of our results and previous studies in model plant species, we propose a hypothetical working model for MdHY5-regulated anthocyanin accumulation and nitrate assimilation (Figure 8). MdHY5 is released from MdCOP1-mediated degradation after detection of light signaling. MdHY5 then directly binds to the G-box site of the MdMYB10 promoter to induce its expression to regulate anthocyanin biosynthesis. In addition, MdHY5 may also be involved in nitrate assimilation by regulating the transcription of both NR genes and nitrate uptake genes. At the same time, MdHY5 induces its own expression in all light conditions by directly binding to the E-box site of its promoter. Improved comprehension of MdHY5 function and signaling in apple could be highly useful for improving fruit traits, such as pigment accumulation and nitrogen use efficiency, for producing high-quality fruits.
Figure 8.
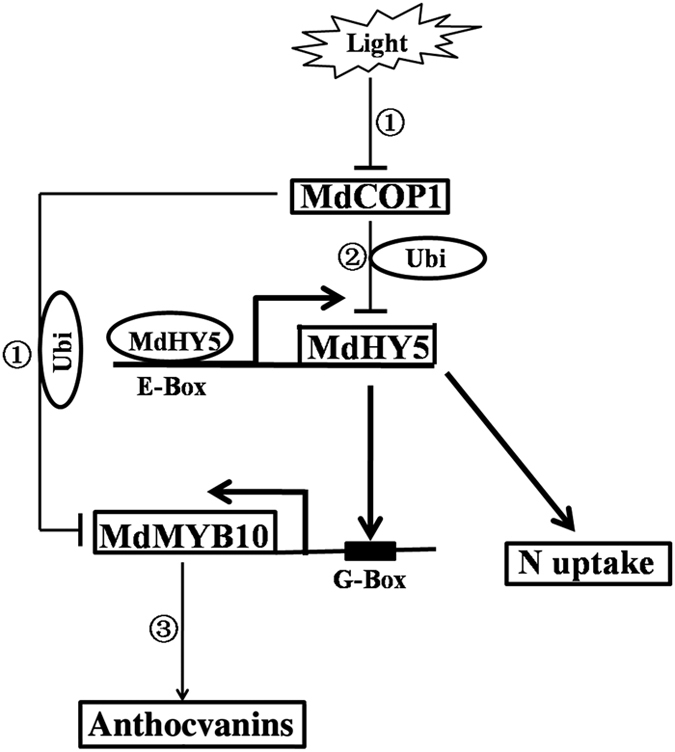
Model of MdHY5-mediated anthocyanin accumulation and nitrate assimilation. The bold arrows indicate the pathway verified in apple in the present work. The arrows indicate pathways that have been previously reported in apple or Arabidopsis. MdCOP1: apple ubiquitin E3 ligase CONSTITUTIVE PHOTOMORPHOGENIC 1; Ubi: ubiquitination. (1) Li et al.56 and Maier et al.;57 (2) Ang et al.;22 (3) Takos et al.58 and Ban et al.59
Acknowledgments
This work was supported by grants from the Natural Science Foundation of China (31325024 and 31601742), the Ministry of Education of China (IRT15R42) and Shandong Province Government (SDAIT-06-03).
Footnotes
Supplementary Information for this article can be found on the Horticulture Research website (http://www.nature.com/hortres)
The authors declare no conflict of interest.
References
- Ubi BE, Honda C, Bessho H et al. Expression analysis of anthocyanin biosynthetic genes in apple skin: effect of UV-B and temperature. Plant Sci 2006; 170: 571–578. [Google Scholar]
- Ban Y, Kondo S, Ubi BE, Honda C, Bessho H, Moriguchi T. UDP-sugar biosynthetic pathway: contribution to cyanidin 3-galactoside biosynthesis in apple skin. Planta 2009; 230: 871–881. [DOI] [PubMed] [Google Scholar]
- Honda C, Kotoda N, Wada M et al. Anthocyanin biosynthetic genes are coordinately expressed during red coloration in apple skin. Plant Physiol Bioch 2002; 40: 955–962. [Google Scholar]
- Telias A, Wang KL, Stevenson DE et al. Apple skin patterning is associated with differential expression of MYB10. BMC Plant Biol 2011; 11: 93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang F, Gonzalez A, Zhao M, Payne CT, Lloyd A. A network of redundant bHLH proteins functions in all TTG1-dependent pathways of Arabidopsis. Development 2003; 130: 4859–4869. [DOI] [PubMed] [Google Scholar]
- Koes R, Verweij W, Quattrocchio F. Flavonoids: a colorful model for the regulation and evolution of biochemical pathways. Trends Plant Sci 2005; 10: 236–242. [DOI] [PubMed] [Google Scholar]
- Espley RV, Hellens RP, Putterill J, Stevenson DE, Kutty-Amma S, Allan AC. Red coloration in apple fruit is due to the activity of the MYB transcription factor, MdMYB10. Plant J 2007; 49: 414–427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Zhang X, Liu Y et al. The effect of fruit bagging on the color, phenolic compounds and expression of the anthocyanin biosynthetic and regulatory genes on the ‘Granny Smith’ apples. Eur Food Res Technol 2013; 237: 875–885. [Google Scholar]
- Neilsen D, Neilsen G. Nutritional effects on fruit quality for apple trees. New York Fruit Quarterly 2009; 17: 21–24. [Google Scholar]
- Li X, Uddin MR, Park WT et al. Accumulation of anthocyanin and related genes expression during the development of cabbage seedlings. Process Biochem 2014; 49: 1084–1091. [Google Scholar]
- Robertson GP, Vitousek PM. Nitrogen in agriculture: balancing the cost of an essential resource. Annu Rev Environ Resour 2009; 34: 97–125. [Google Scholar]
- Tong Y, Zhou JJ, Li Z, Miller AJ. A two-component high-affinity nitrate uptake system in barley. Plant J 2005; 41: 442–450. [DOI] [PubMed] [Google Scholar]
- Ludewig U. Ion transport versus gas conduction: function of AMT/Rh-type proteins. Transfus Clin Biol 2006; 13: 111–116. [DOI] [PubMed] [Google Scholar]
- Orsel M, Chopin F, Leleu O et al. Characterization of a two-component high-affinity nitrate uptake system in Arabidopsis. Physiology and protein-protein interaction. Plant Physiol 2006; 142: 1304–1317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsay YF, Chiu CC, Tsai CB, Ho CH, Hsu PK. Nitrate transporters and peptide transporters. FEBS Lett 2007; 581: 2290–2300. [DOI] [PubMed] [Google Scholar]
- Mounier E, Pervent M, Ljung K, Gojon A, Nacry P. Auxin-mediated nitrate signalling by NRT1.1 participates in the adaptive response of Arabidopsis root architecture to the spatial heterogeneity of nitrate availability. Plant Cell Environ 2014; 37: 162–174. [DOI] [PubMed] [Google Scholar]
- Bouguyon E, Perrine-Walker F, Pervent M et al. Nitrate controls root development through posttranscriptional regulation of the NRT1.1/NPF6.3 Transporter/Sensor. Plant Physiol 2016; 172: 1237–1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu H, Zhao X, Guo C, Chen L, Li K. Spinach 14-3-3 protein interacts with the plasma membrane H+-ATPase and nitrate reductase in response to excess nitrate stress. Plant Physiol Biochem 2016; 106: 187–197. [DOI] [PubMed] [Google Scholar]
- Elia A, Santamaria P, Serio F. Nitrogen nutrition, yield and quality of spinach. J Sci Food Agr 1998; 76: 341–346. [Google Scholar]
- Hilbert G, Soyer JP, Molot C, Giraudon J, Milin M, Gaudillere JP. Effects of nitrogen supply on must quality and anthocyanin accumulation in berries of cv. Merlot. VITIS J Grapevine Res 2003; 42: 69–72. [Google Scholar]
- Koornneef M, Rolff E, Spruit CJP. Genetic control of light-inhibited hypocotyl elongation in Arabidopsis thaliana. Z Pflanzenphysiol 1980; 100: 147–160. [Google Scholar]
- Ang LH, Deng XW. Regulatory hierarchy of photomorphogenic loci: allele-specific and light-dependent interaction between the HY5 and COP1 loci. Plant Cell 1994; 6: 613–628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H, Zhang J, Neff MM et al. Integration of light and abscisic acid signaling during seed germination and early seedling development. Proc Natl Acad Sci USA 2008; 105: 4495–4500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gangappa SN, Botto JF. The multifaceted roles of HY5 in plant growth and development. Mol Plant 2016; 9: 1353–1365. [DOI] [PubMed] [Google Scholar]
- Abbas N, Maurya JP, Senapati D, Gangappa SN, Chattopadhyay S. Arabidopsis CAM7 and HY5 physically interact and directly bind to hy5 promoter to regulate its expression to promote photomorphogenesis. Plant Cell 2014; 26: 1036–1052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catala R, Medina J, Salinas J. Integration of low temperature and light signaling during cold acclimation response in Arabidopsis. Proc Natl Acad Sci USA 2011; 108: 16475–16480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin DH, Choi M, Kim K et al. HY5 regulates anthocyanin biosynthesis by inducing the transcriptional activation of the MYB75/PAP transcription factor in Arabidopsis. FEBS Lett 2013; 587: 1543–1547. [DOI] [PubMed] [Google Scholar]
- Chen X, Yao Q, Gao X, Jiang C, Harberd NP, Fu X. Shoot-to-root mobile transcription factor HY5 coordinates plant carbon and nitrogen acquisition. Curr Biol 2016; 26: 640–646. [DOI] [PubMed] [Google Scholar]
- Chai T, Zhou J, Liu J, Xing D. LSD1 and HY5 antagonistically regulate red light induced-programmed cell death in Arabidopsis. Front Plant Sci 2015; 6: 292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, He H, Wang X et al. Genome-wide mapping of the HY5-mediated gene networks in Arabidopsis that involve both transcriptional and posttranscriptional regulation. Plant J 2011; 65: 346–358. [DOI] [PubMed] [Google Scholar]
- Ang LH, Chattopadhyay S, Wei N et al. Molecular interaction between COP1 and HY5 defines a regulatory switch for light control of Arabidopsis development. Mol Cell 1998; 1: 213–222. [DOI] [PubMed] [Google Scholar]
- Osterlund MT, Hardtke CS, Wei N, Deng XW. Targeted destabilization of HY5 during light-regulated development of Arabidopsis. Nature 2000; 405: 462–466. [DOI] [PubMed] [Google Scholar]
- Liu B, Zuo Z, Liu H, Liu X, Lin C. Arabidopsis cryptochrome 1 interacts with SPA1 to suppress COP1 activity in response to blue light. Genes Dev 2011; 25: 029–1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karayekov E, Sellaro R, Legris M, Yanovsky MJ, Casal JJ. Heat shock-induced fluctuations in clock and light signaling enhance phytochrome B-mediated Arabidopsis deetiolation. Plant Cell 2013; 25: 2892–2906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li HH, Liu X, An JP, Hao YJ, Wang XF, You CX. Cloning and elucidation of the functional role of apple MdLBD13 in anthocyanin biosynthesis and nitrate assimilation. Plant Cell Tiss Org 2017, 1–13.
- An XH, Tian Y, Chen KQ et al. MdMYB9 and MdMYB11 are involved in the regulation of the JA-induced biosynthesis of anthocyanin and proanthocyanidin in apples. Plant Cell Physiol 2015; 56: 650–662. [DOI] [PubMed] [Google Scholar]
- Lee HS, Wicker L. Anthocyanin pigments in the skin of lychee fruit. J Food Sci 1991; 56: 466–468. [Google Scholar]
- Shang Y, Yan L, Liu ZQ et al. The Mg-chelatase H subunit of Arabidopsis antagonizes a group of WRKY transcription repressors to relieve ABA-responsive genes of inhibition. Plant Cell 2010; 22: 1909–1935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Q, Sun J, Zhai Q, Zhou W, Xu L, Li X. The basic helix-loop-helix transcription factor MYC2 directly represses PLETHORA expression during jasmonate-mediated modulation of the root stem cell niche in Arabidopsis. Plant Cell 2011; 23: 3335–3352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freschi L, Rodrigues MA, Domingues DS et al. Nitric oxide mediates the hormonal control of Crassulacean acid metabolism expression in young pineapple plants. Plant Physiol 2010; 152: 1971–1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cataldo DA, Maroon M, Schrader LE, Youngs VL. Rapid colorimetric determination of nitrate in plant tissue by nitration of salicylic acid. Commun Soil Sci Plant Anal 1975; 6: 71–80. [Google Scholar]
- Vendrell PF, Zupancic J. Determination of soil nitrate by transnitration of salicylic acid. Commun Soil Sci Plant Anal 1990; 21: 1705–1713. [Google Scholar]
- An JP, Li R, Qu FJ, You CX, Wang XF, Hao YJ. Apple F-box protein MdMAX2 regulates plant photomorphogenesis and stress response. Front Plant Sci 2016; 7: 1685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Binkert M, Kozma-Bognár L, Terecskei K, De Veylder L, Nagy F, Ulm R. UV-B-responsive association of the Arabidopsis bZIP transcription factor ELONGATED HYPOCOTYL5 with target genes, including its own promoter. Plant Cell 2014; 26: 4200–4213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jonassen EM, Lea US, Lillo C. HY5 and HYH are positive regulators of nitrate reductase in seedlings and rosette stage plants. Planta 2008; 227: 559–564. [DOI] [PubMed] [Google Scholar]
- Oyama T, Shimura Y, Okada K. The Arabidopsis HY5 gene encodes a bZIP protein that regulates stimulus-induced development of root and hypocotyl. Gene Dev 1997; 11: 2983–2995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jing Y, Zhang D, Wang X et al. Arabidopsis chromatin remodeling factor PICKLE interacts with transcription factor HY5 to regulate hypocotyl cell elongation. Plant Cell 2013; 25: 242–256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andronis C, Barak S, Knowles SM, Sugano S, Tobin EM. The clock protein CCA1 and the bZIP transcription factor HY5 physically interact to regulate gene expression in Arabidopsis. Mol Plant 2008; 1: 58–67. [DOI] [PubMed] [Google Scholar]
- Toledo-Ortiz G, Johansson H, Lee KP et al. The HY5-PIF regulatory module coordinates light and temperature control of photosynthetic gene transcription. PLoS Genet 2014; 10: e1004416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang CS, Li YH, Chen LT et al. LZF1, a HY5-regulated transcriptional factor, functions in Arabidopsis de-etiolation. Plant J 2008; 54: 205–219. [DOI] [PubMed] [Google Scholar]
- Huang X, Ouyang X, Yang P et al. Arabidopsis FHY3 and HY5 positively mediate induction of COP1 transcription in response to photomorphogenic UV-B light. Plant Cell 2012; 24: 4590–4606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen NH, Jeong CY, Kang GH, Yoo SD, Hong SW, Lee H. MYBD employed by HY5 increases anthocyanin accumulation via repression of MYBL2 in Arabidopsis. Plant J 2015; 84: 1192–1205. [DOI] [PubMed] [Google Scholar]
- Jonassen EM, Sévin DC, Lillo C. The bZIP transcription factors HY5 and HYH are positive regulators of the main nitrate reductase gene in Arabidopsis leaves, NIA2, but negative regulators of the nitrate uptake gene NRT1. 1. J Plant Physiol 2009; 166: 2071–2076. [DOI] [PubMed] [Google Scholar]
- Li Z, Zhang L, Yu Y, Quan R, Zhang Z, Huang R. The ethylene response factor AtERF11 that is transcriptionally modulated by the bZIP transcription factor HY5 is a crucial repressor for ethylene biosynthesis in Arabidopsis. Plant J 2011; 68: 88–99. [DOI] [PubMed] [Google Scholar]
- Chen D, Xu G, Tang W et al. Antagonistic basic helix-loop-helix/bZIP transcription factors form transcriptional modules that integrate light and reactive oxygen species signaling in Arabidopsis. Plant Cell 2013; 25: 1657–1673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li YY, Mao K, Zhao C et al. MdCOP1 ubiquitin E3 ligases interact with MdMYB1 to regulate light-induced anthocyanin biosynthesis and red fruit coloration in apple. Plant Physiol 2012; 160: 1011–1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maier A, Schrader A, Kokkelink L et al. Light and the E3 ubiquitin ligase COP1/SPA control the protein stability of the MYB transcription factors PAP1 and PAP2 involved in anthocyanin accumulation in Arabidopsis. Plant J 2013; 74: 638–651. [DOI] [PubMed] [Google Scholar]
- Takos AM, Jaffé FW, Jacob SR, Bogs J, Robinson SP, Walker AR. Light-induced expression of a MYB gene regulates anthocyanin biosynthesis in red apples. Plant Physiol 2006; 142: 1216–1232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ban Y, Honda C, Hatsuyama Y, Igarashi M, Bessho H, Moriguchi T. Isolation and functional analysis of a MYB transcription factor gene that is a key regulator for the development of red coloration in apple skin. Plant Cell Physiol 2007; 48: 958–970. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.