Abstract
DNA-PKcs and Ku are essential components of the complex that catalyzes non-homologous end joining (NHEJ) of DNA double-strand breaks (DSBs). Ku, a heterodimeric protein, binds to DNA ends and facilitates recruitment of the catalytic subunit, DNA-PKcs. We have investigated the effect of DNA strand orientation and sequence bias on the activation of DNA-PK. In addition, we assessed the effect of the position and strand orientation of cisplatin adducts on kinase activation. A series of duplex DNA substrates with site-specific cisplatin–DNA adducts placed in three different orientations on the duplex DNA were prepared. Terminal biotin modification and streptavidin (SA) blocking was employed to direct DNA-PK binding to the unblocked termini with a specific DNA strand orientation and cisplatin–DNA adduct position. DNA-PK kinase activity was measured and the results reveal that DNA strand orientation and sequence bias dramatically influence kinase activation, only a portion of which could be attributed to Ku-DNA binding activity. In addition, cisplatin–DNA adduct position resulted in differing degrees of inhibition depending on distance from the terminus as well as strand orientation. These results highlight the importance of how local variations in DNA structure, chemistry and sequence influence DNA-PK activation and potentially NHEJ.
INTRODUCTION
DNA double-strand breaks (DSBs) are one of the most lethal types of DNA damage, and eukaryotic cells have evolved two major pathways to repair DSBs, homologous recombination (HR) and non-homologous end joining (NHEJ). The NHEJ pathway requires the heterodimeric protein Ku, which consists of a 70 kDa subunit and 80 kDa subunit (1). Ku binds to the ends of DSBs and recruits the 470 kDa DNA-PK catalytic subunit (DNA-PKcs). Together Ku and DNA-PKcs make up the complex termed DNA-PK holoenzyme. DNA-PK activation, an essential step in the repair process, occurs once the kinase assembles at the site of DSB. The activation of DNA-PK may signal to other downstream components involved in signal transduction of the damage event (2). This active DNA-PK complex may then recruit other factors including the MRN complex (3), Artemis (4) and the DNA ligase IV/XRCC4 complex (5), which are potentially involved in processing of the termini and completing the repair process.
The interaction of Ku with DNA has been the subject of extensive analysis. DNA double-stranded termini display the greatest affinity for Ku (6). A significant advance in our understanding of Ku structure and function was revealed when the co-crystal structure of Ku bound to a duplex DNA was determined (7). In this structure, each subunit of Ku encircles the duplex DNA in a ring-like structure, with a large base on which the DNA rests. Two pillars support a bridge-like structure through which the DNA strand can thread. Further analysis of this crystal structure reveals that while there are no direct interactions with the DNA bases, specific amino acids protrude into the major and minor grooves of the DNA so as to provide a helical guide through the ring structure of Ku (7). Additional studies have revealed that Ku binds and orients itself at a DNA terminus in a very specific fashion. This was highlighted in a series of photo-cross-linking experiments where short duplex DNA substrates were used to demonstrate that at a DNA terminus, Ku 80 is positioned more internally and Ku 70 is positioned closer to the terminus (8,9). In addition, Ku 70 was observed to make specific contacts within the major groove in the DNA helix, while Ku 80 does not appear to contact the major groove but is oriented in proximity of the minor groove (9). These results indicate that while Ku binding is not sequence specific, alterations in duplex structure could influence the DNA binding activity of Ku.
After Ku binds to a double-stranded DNA terminus, it recruits DNA-PKcs to the site of the break. Electron crystallography at 22 Å resolution revealed an open channel in the DNA-PKcs structure that could interact with double-stranded DNA (10). Studies show that on short DNA substrates, the association of DNA-PKcs causes Ku to move further along the length of the duplex DNA (8). DNA-PKcs was demonstrated to make direct contact with approximately the first 10 bp along a duplex DNA substrate (8). Electron micrographic studies also show that dramatic conformational changes accompany DNA-PKcs binding to DNA (11). These conformational changes were postulated to be involved in activation of the kinase, which in turn could facilitate recruitment of other proteins involved in the repair process (11). Considering that DNA-PKcs is maximally activated by double-stranded DNA containing un-annealed single-strand segments (10), how these substrates influence the DNA-PKcs conformational changes is of significant interest.
While much is known about the repair of single sites of DNA damage or strand breaks, considerably less is known concerning the repair of compound lesions on DNA. One study has assessed the effect of terminal nucleotide modification on the activation of DNA-PK. The results demonstrate that DNA-PK is largely unaffected by terminal base modification (12). The presence of a cisplatin adduct in the vicinity of a DNA DSB represents a compound lesion potentially encountered by NHEJ proteins in cells treated with concurrent cisplatin and IR. Cisplatin is a DNA damaging agent that forms DNA adducts preferentially at adjacent guanine bases in duplex DNA (13). The effect of cisplatin lesions on Ku binding and DNA-PKcs activation has been studied, and results show that Ku has a similar binding affinity for cisplatin-damaged DNA as compared to a control DNA of identical sequence without the cisplatin adducts (14). Cisplatin adducts on duplex DNA strands were also demonstrated to decrease Ku's ability to translocate along the DNA, away from the terminus to which it was originally bound (15). Interestingly, despite Ku and DNA-PKcs binding, activation of DNA-PKcs kinase activity was significantly reduced on cisplatin-damaged DNA (14,15). To further investigate the effect of compound lesions and the specificity of the DNA on the DNA/DNA-PK interaction, we prepared a series of modified DNA duplex substrates focusing on sequence bias and strand orientation. These results are discussed with respect to Ku and DNA-PK structure and function, and NHEJ-catalyzed repair of DNA DSBs.
MATERIALS AND METHODS
Preparation of DNA substrates
The 30mer single-strand oligonucleotides used in this study are presented in Table 1. Oligonucleotides were purchased from Integrated DNA Technology (Coralville, IA), gel purified by preparative denaturing polyacrylamide gel electrophoresis, and treated with cisplatin as described previously (14), with the following modification. The cisplatin was pre-aquated by incubation with AgNO3 (16) and the ratio of cisplatin to GG site was 10:1 for all cisplatin modification reactions. The degree of cisplatin modification was assessed by analytical DNA sequencing gel analysis or exonuclease digestion, and in some cases the platinated DNA strands were purified by preparative gel electrophoresis, to assure that 100% of the DNA substrates were platinated. The control and platinated DNA strands were eluted from the gel, ethanol precipitated, quantified and annealed to the appropriate complement to generate the duplex DNA substrates depicted in Figure 1. Duplex DNA substrates were purified by native polyacrylamide gel electrophoresis, eluted from the gel, precipitated and quantified.
Table 1.
DNA oligonucleotides
| DNA | Sequencea | Function |
|---|---|---|
| 3P | 5′-CCTCTCTCCTTCTTTTCCTCTTCGGTCTCC-3′ | Platinum adduct at 3′ terminus |
| 5P | 5′-CCCCTGGCTTTCTTTTCCTCTTCCTTCCCC-3′ | Platinum adduct at 5′ terminus |
| MP | 5′-CCCCTCTCCTTCTTGGCTTCTTCCTTCCCC-3′ | Platinum adduct central |
| 3Pc3B | 5′-GGAGACCGAAGAGGAAAAGAAGGAGAGAGGbiotin-3′ | Complement to 3P |
| 5Pc5B | 5′-biotinGGGGAAGGAAGAGGAAAAGAAAGCCAGGGG-3′ | Complement to 5P |
| MPc3B | 5′-GGGGAAGGAAGAAGCCAAGAAGGAGAGGGGbiotin-3′ | Complement to MP |
| MPc5B | 5′-biotinGGGGAAGGAAGAAGCCAAGAAGGAGAGGGG-3′ | Complement to MP |
| 3P3B | 5′-CCTCTCTCCTTCTTTTCCTCTTCGGTCTCCbiotin-3′ | |
| 5B-mixed | 5′-biotin-CGATGCTCAGTAGAT-3′ | |
| T-5B-mixed | 5′-TTTTTTTTTTTTTTTCGATGCTCAGTAGAT-3′ | |
| A-5B-mixed | 5′-AAAAAAAAAAAAAAACGATGCTCAGTAGAT-3′ | |
| c-mixed-T | 5′-TTTTTTTTTTTTTTTTTTTTTTTTTTTTTTATCTACTGAGCATCG-3′ | |
| c-mixed-A | 5′-AAAAAAAAAAAAAAAAAAAAAAAAAAAAAAATCTACTGAGCATCG-3′ | |
| c-mixed-15 | 5′-TCGGTAGTCCATCTAATCTACTGAGCATCG-3′ | |
| c-mixed-15(T) | 5′-TTTTTTTTTTTTTTTATCTACTGAGCATCG-3′ | |
| c-mixed-15(A) | 5′-AAAAAAAAAAAAAAAATCTACTGAGCATCG-3′ | |
| c-mixed-T(GG) | 5′-TTTTTGGTTTTTTTTTTTTTTTTTTTTTTTATCTACTGAGCATCG-3′ | |
| c-mixed-A(CC) | 5′-AAAAACCAAAAAAAAAAAAAAAAAAAAAAAATCTACTGAGCATCG-3′ | |
| 2.1 | 5′-CCCCTATCCTTTCCGCGTCCTTACTTCCCC-3′ | EMSA substrate |
| 2.2 | 5′-GGGGAAGTAAGGACGCGGAAAGGATAGGGG-3′ | Complement to 2.1 |
aThe position of the cisplatin modification is indicated by the underlined base.
Figure 1.
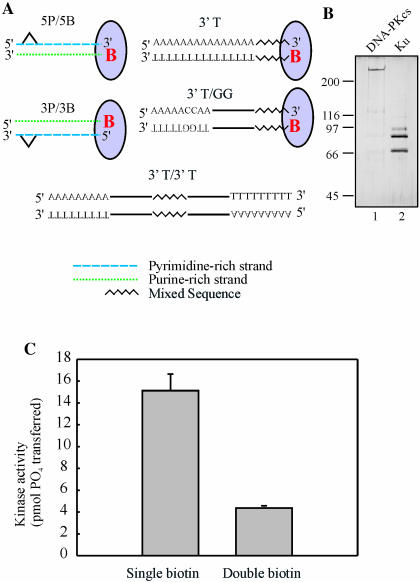
DNA substrates used in this study. (A) Duplex DNA substrates were prepared from the single-stranded oligonucleotides presented in Table 1 as described in Materials and Methods. The addition of SA, as indicated by the oval, results in blocking of the biotinylated terminus, indicated by the red letter ‘B’. The cisplatin adducts are positioned either 6 or 15 bp from the accessible terminus in the 5P and 3P series, and MP series of substrates, respectively. The substrates generated have either a 5′ pyrimidine-rich strand (5P series) or a 3′ pyrimidine-rich strand (3P series) accessible to Ku when SA blocking is employed. The remaining substrates represent duplexes that were designed to contain a mixed 15 base sequence, indicated by the black jagged line, and either 15 bases of dAs or dTs on the 3′ strand, 45 bases of dAs or dTs with two dCs or dGs placed 6 bp in, or substrates with the 15 base mixed sequence containing 15 bases of dTs or dAs on both sides. (B) Ku and DNA-PKcs were purified as described in Materials and Methods: 200 ng of DNA-PKcs and 400 ng of Ku were separated on an 8% SDS–PAGE gel and silver stained. The position and molecular weights (kDa) of the standards are indicated on the left-hand side. (C) In the presence of SA, the 3P/3B substrate and the same substrate with an additional biotin modification on the remaining 3′ end were assayed for DNA-PK kinase activation as described in Materials and Methods. Reactions contained 20 nM DNA-PKcs, 3.3 nM Ku and 5 nM SA–DNA, as indicated. Results are presented as the pmol of 32P transferred to the synthetic p53 peptide in a 30 min reaction. Reactions were performed in triplicate and the mean and standard error of the mean are presented.
Additional oligonucleotides were designed to generate DNA substrates containing homopolymeric termini. A 15 base 5′ biotinylated oligonucleotide was designed to be annealed to complementary 15 base oligonucleotides that have various homopolymeric 5′ extensions. Subsequent extension of the 5′ biotinylated primer results in the formation of a duplex DNA with one terminus capable of being blocked by streptavidin (SA) and the other terminus having a defined homopolymeric 3′ and 5′ accessible terminus. Extension of the biotinylated strand was performed with 100 μM dNTPs and 10 U/reaction of Sequenase polymerase, incubated for 30 min at 37°C, and the reaction stopped with 100 mM EDTA. The duplexes generated from the extension were purified using a G-50 mini-column and quantified. A similar procedure was followed to generate the duplexes containing 15 bases of a single nucleotide at both ends of a mixed 15 base sequence (Figure 1A).
Protein purification
Human Ku was purified from recombinant baculovirus-infected Sf-9 insect cells. Transfer vectors containing N-terminal [His]6 tagged Ku 70 and full-length Ku 80 were generously provided by Dr Dale Ramsden (University of North Carolina). Sf-9 cells were co-infected with Ku 70 and Ku 80 recombinant virus at a multiplicity of infection of 5 and 10, respectively. Cells were infected for 48 h, after which cell-free extracts were prepared and Ku was purified via sequential Ni-NTA agarose and Q-Sepharose column chromatography, as previously published (5). Fractions containing Ku were identified using SDS–PAGE and Coomassie staining, and peak fractions were pooled, dialyzed and stored at −80°C until use.
DNA-PKcs was purified from cell-free extracts prepared from 16 l of HeLa cells. Extracts were prepared as previously described (17), and fractionated on a 50 ml cisplatin-damaged DNA–Sepharose column and phosphocellulose column, as previously described (14). Further purification and separation of the DNA-PK catalytic subunit from Ku subunits was achieved by chromatography on a 5 ml hydroxylapatite column. Protein (0.3 mg) was loaded on a 5 ml column and washed with buffer containing 10 mM KPi. Proteins were eluted with a 50 ml linear gradient from 10 to 500 mM KPi. DNA-PKcs containing fractions were identified by Coomassie staining of SDS gels and Ku-DNA dependent protein kinase activity, as previously described (14). Active fractions were pooled, dialyzed and stored at −80°C until use.
DNA-PK kinase assays
Assays were performed at 37°C in a final volume of 20 μl containing 20 mM HEPES, pH 7.5, 8 mM MgCl2, 1 mM DTT, 5% glycerol, [γ-32P]ATP (0.5 μCi, 125 μM), 2.1 nM DNA-PKcs, 3.3 nM Ku, 5 nM DNA and 500 μM p53 synthetic peptide. In reactions using SA–DNA complexes, the DNA was mixed with 1 ng SA/fmol DNA and incubated on ice for 5 min. Ku and DNA-PKcs were added to reaction buffer containing the peptide and incubated on ice for 5 min. The DNA substrate was then added and incubation continued for an additional 5 min on ice. Reactions were then initiated by the addition of ATP and incubated for 15 min at 37°C. Reactions were terminated by the addition of an equal volume of 30% acetic acid and were spotted onto P81 phosphocellulose filter paper. The filters were washed five times for 5 min each in 15% acetic acid, once in 100% methanol, and allowed to dry. Samples were quantified by PhosphorImager analysis using ImageQuant software (Molecular Dynamics).
Competition binding analyses
Electrophoretic mobility shift assays (EMSAs) were performed in 20 μl reactions containing 50 mM Tris–HCl, 10 mM MgCl2 and 50 mM NaCl. In reactions containing SA–DNA complexes, SA was pre-incubated with competitor DNA substrates (1 ng SA/fmol DNA) for 5 min. Increasing amounts of the unlabeled competitor SA–DNA complex were then mixed with 2.5 nM 32P-labeled double-stranded 2.1/2.2 DNA (18), followed by 5 nM Ku. Reactions without SA were carried out in the same manner, except that the competitor DNA was added to the labeled DNA directly. Reactions were mixed and loaded onto a 6% native polyacrylamide gel. Gels were dried and exposed to a PhosphorImager and quantified using ImageQuant software.
RESULTS
DNA substrates to assess the effect of DNA strand polarity, sequence bias and platinum adduct position on DNA-PK activation
Ku, a major player in the DNA DSB repair process, arrives at the site of a break and binds to the terminus created by the break. This facilitates the recruitment of DNA-PKcs, which in turn initiates the repair process. While it is known that Ku has a high binding affinity for double-stranded DNA, the effect of different duplex structures or sequences on Ku binding and DNA-PKcs activation have not been determined. To assess the relative binding affinity of Ku for different DNA substrates and the subsequent activation of DNA-PK, substrates were designed that would limit binding of the protein complex to the specific DNA terminus in question. Biotin modification with SA blocking of the specific ends of the substrates was employed to create the desired substrates (Figure 1A). When SA was bound to the biotin-modified ends of the 30mer substrates, that end of the DNA duplex was sufficiently blocked so that Ku and DNA-PKcs could not bind. This results in the accessible terminus containing 3′ pyrimidine- and 5′ purine-rich strands and 3′ purine- and 5′ pyrimidine-rich strands (Figure 1A). Individual substrates 3P/3B, 5P/5B, MP/3B and MP/5B were designed to also allow the incorporation of site-specific cisplatin adducts at two positions, 6 bp or 15 bp from the accessible terminus. These sites were positioned on the 3′ pyrimidine (3P/3B and MP/3B) or 5′ pyrimidine (5P/5B and MP/5B) rich strands to allow the effect of strand polarity on adduct position to be determined. Additional oligonucleotides consisting of a mixed complement sequence with an additional either 5′-T15 or 5′-A15 were synthesized and annealed to the biotin-modified oligonucleotide. The modified oligonucleotide was then extended by polymerase-catalyzed extension to create a blunt-ended, fully duplex DNA substrate, and after blocking with SA, the resulting substrate contains either a 3′-T15·5′-A15 termini (3′-T, Figure 1A) or a 3′-A15· 5′-T15 termini. To assess the influence of purine bases within the homopolymeric pyrimidine sequence, an oligonucleotide was designed to contain a 5′-A5G2A23, such that when annealed and extended, two guanines were positioned six bases from the 3′ termini of a pyrimidine-rich sequence (3′-T/GG, Figure 1A). The corresponding 3′-A/CC was also prepared using a similar methodology. Biotin modification was eliminated completely on another set of substrates (3′-T/3′-T, Figure 1A) that had a 15 base mixed sequence flanked by 3′ dTs or dAs. Using this combination of substrates, differences in DNA strand polarity, sequence bias and position of the cisplatin adduct on DNA-PKcs kinase activation and Ku binding could be determined.
Ku and DNA-PKcs were purified separately and the DNA-PK holoenzyme was reconstituted to enable the analysis of DNA-PK activation by the different DNA substrates. This approach also allowed the determination of Ku binding to the DNA substrates, independent of DNA-PKcs. Human Ku was prepared from insect cells infected with recombinant baculovirus (5). DNA-PKcs was purified from HeLa cell extracts (14,19). The final pools of protein were subjected to analysis by SDS–PAGE and stained with silver (Figure 1B). As expected, the Ku subunits migrated consistent with molecular masses of 70 and 80 kDa, and DNA-PKcs migrated slower than the 200 kDa marker and is consistent with the predicted molecular mass of 460 kDa. The DNA PKcs preparation does contain a low level of Ku judged to be <5% of the DNA-PKcs. This was confirmed in kinase assays where the activity of DNA-PKcs was significantly stimulated by the addition of the recombinant Ku (data not shown). A series of preliminary experiments were performed to determine the optimal concentrations for the kinase assays of Ku, DNA-PKcs and DNA for in vitro kinase activation (data not shown). In other experiments, increasing concentrations of SA were titrated with the biotinylated DNA to determine the minimal amount necessary to quantitatively bind the DNA substrate (data not shown). The efficiency of SA binding and blocking of the biotinylated terminus was assessed on a substrate prepared with both termini bound by SA (Figure 1C). The results demonstrate that with both ends bound by SA, an 80% decrease in activity was observed compared to a substrate with one terminus bound by SA.
Effect of cisplatin–DNA adduct position and DNA strand polarity on activation of DNA-PK
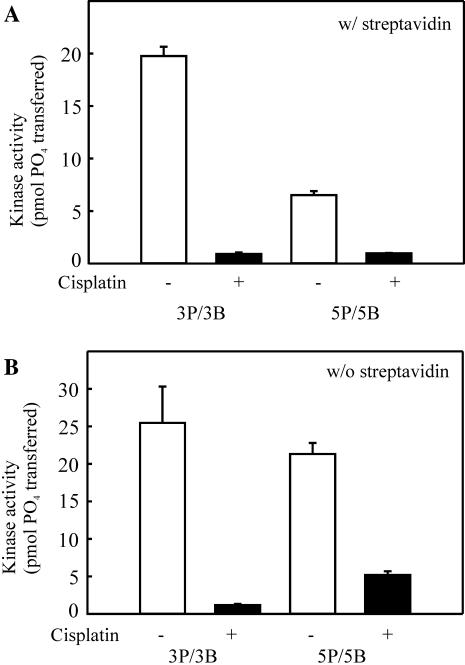
We have previously shown that Ku is able to bind to cisplatin-damaged double-stranded DNA with a similar affinity as to undamaged duplex DNA, but that cisplatin-damaged DNA is a less effective activator of DNA-PK (19). To date, however, the effect of cisplatin adduct position in relation to strand polarity has not been determined. Using the substrates presented in Figure 1A, we have determined that adduct position influences DNA-PK kinase activation. The DNA-PK kinase activity supported by the 3P/3B and 5P/5B substrates was assessed in reactions performed with SA (Figure 2A). SA–biotin blocking of the 3P/3B results in a 3′ pyrimidine-rich accessible terminus, and SA–biotin blocking of the 5P/5B substrate generates a 5′ pyrimidine-rich accessible terminus. These results demonstrate that the activity of DNA-PK was reduced to near background levels when either the SA-3P/3B and SA-5P/5B substrates were modified with cisplatin such that the cisplatin–DNA adduct was 6 bp from termini (Figure 2A, closed bars). Therefore, inhibition of DNA-PKcs activity by cisplatin is not influenced by the strand, 5′ or 3′, on which the cisplatin–DNA adduct resides. These data suggests that cisplatin-dependent inhibition of DNA-PK is a function of the structural alteration in the duplex induced by the cisplatin–DNA adduct.
Figure 2.
Effect of strand bias, sequence specificity and cisplatin–DNA adduct position on kinase activation. (A) The cisplatin-damaged or control double-stranded SA-3P/3B and SA-5P/5B 30mers were assayed for DNA-PK kinase activation as described in Materials and Methods. Reactions contained 20 nM DNA-PKcs, 3.3 nM Ku and 5 nM SA–DNA, as indicated. Results are presented as the pmol of 32P transferred to the synthetic p53 peptide in a 30 min reaction. Reactions were performed in triplicate and the mean and standard error of the mean are presented. (B) The cisplatin damaged or control double-stranded 3P/3B and 5P/5B 30mers were assayed for DNA-PK kinase activation as described above, except SA was not included in the reaction. Results are presented as the pmol of 32P transferred to the synthetic p53 peptide in a 30 min reaction. Reactions were performed in triplicate and the mean and standard error of the mean are presented.
Interestingly, in control reactions performed using DNA substrates prepared without cisplatin, a significant difference in the degree of kinase activation was observed with the two different substrates (Figure 2A, open bars). Kinase assays performed using the 5P/5B substrate resulted in significantly lower DNA-PK activity compared to that obtained with the 3P/3B substrate. The only major difference between the 3P/3B and 5P/5B substrates is the base composition in reference to the DNA strand polarity of the accessible termini. Therefore, these data are consistent with DNA-PK being differentially activated dependent on sequence bias and strand polarity, with a preference being displayed for a duplex terminus consisting of 3′ pyrimidine- and 5′ purine-rich strands.
In a series of control experiments, we assessed the activation of DNA-PK by each substrate without the use of SA (Figure 2B). The results obtained are consistent with either the 3′ biotin modification or 3′ purine-rich terminus resulting in significantly less DNA-PK activity. The 3′ biotin modification inhibition was ruled out, as biotin modification of both 3′ termini resulted in kinase activation at levels similar to reactions using substrates of identical sequence without the biotin modification (data not shown).
Ku binding
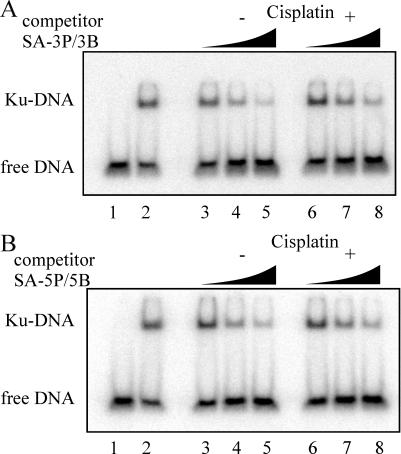
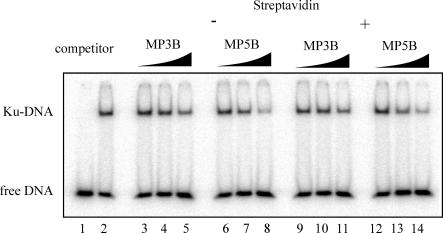
To determine if differential kinase activity was attributable to the affinity of Ku for the DNA substrates, a series of competition DNA binding assays were performed. For competition assays, Ku was added to reactions containing a 32P-labeled duplex DNA and increasing concentrations of the unlabeled competitor DNA (3P/3B, 5P/5B). These assays allowed for the indirect determination of the relative affinity of Ku for the different DNA substrates without having to 32P-label each DNA to be tested. The results of a typical competition assay are presented in Figure 3A and B. Increasing concentrations of SA-3P/3B DNA (lanes 3–5) were titrated into reactions containing fixed concentrations of Ku and 32P-labeled duplex DNA, and the results show a decrease in the level of Ku bound to 32P-labeled DNA. This decrease in Ku binding to the labeled DNA corresponds to the increased amount of Ku bound to the competitor DNA. The analysis of the identical sequence, with the addition of the site-specific cisplatin lesion (SA-3P/3B + Pt), is presented in lanes 6–8. Again, a decrease in Ku bound to 32P-labeled DNA was observed, although it was not as dramatic as the decrease observed for 3P/3B − Pt. Quantification of the results by PhosphorImager analysis revealed a 2-fold difference in Ku binding to the labeled DNA in the presence of the highest concentration of competitor DNA. In the presence of the undamaged competitor DNA, Ku binding was 10% of the control value observed in the absence of competitor (compare Figure 3A, lanes 2 and 5). In the presence of cisplatin-damaged competitor, the level of Ku binding the labeled substrate was 20% of control (lanes 2 and 8). This 2-fold difference in binding the labeled DNA calculates to a 10% difference in Ku affinity for the competitor DNAs. Similar quantification of the intermediate concentration of competitor was performed and the difference was similar, at ∼22%. These results indicate that the cisplatin adduct on the substrate only partially hinders Ku's ability to bind. The same effect of cisplatin is observed in reactions comparing Ku binding to the SA blocked 5P/5B series (Figure 3B). Again, only slightly less binding was observed with the cisplatin-damaged DNA compared to the undamaged control DNA (lanes 5 and 8). Quantification of these data revealed the differences in binding were <5% and not significant. Therefore, the dramatic decrease in kinase activation observed on cisplatin-damaged DNA substrates (Figure 2A) is not accompanied by a dramatic decrease in Ku binding. This is apparent in the comparison of Ku binding activity with the cisplatin-damaged and control undamaged DNA substrates, where only a slight decrease in Ku binding is observed, while DNA-PK kinase activity is decreased by ∼95%.
Figure 3.
Effect of strand bias, sequence specificity and cisplatin–DNA adduct position on Ku binding. (A) Competition binding assays were performed in 20 μl reactions and contained 2.5 nM 32P-labeled double-stranded 2.1/2.2 DNA and 5 nM Ku. Increasing concentrations of the SA-bound competitor DNA (3P/3B), 5, 50 and 200 nM is indicated by the triangles. Controls for the assay include 32P-labeled double-stranded 2.1/2.2 DNA alone (lane 1) and 32P-labeled double-stranded 2.1/2.2 DNA with 5 nM Ku in the absence of any competitor (lane 2). (B) Competition assays were carried out identically to those in (A) except reactions included 5P/5B competitor DNA.
The relative affinity of Ku for the 3′ pyrimidine-rich DNA (3P/3B) versus 3′ purine-rich DNA (5P/5B) can also be determined from these binding data. Comparison of the results obtained with the undamaged control DNA substrates (Figure 3A and B, lanes 3–5) suggests that Ku has a slightly higher affinity for the 3P/3B substrate. When comparing the results obtained in the Ku binding assays to kinase activation with identical substrates, it is clear that differences in kinase activation cannot be attributed to differences in Ku binding. This conclusion is supported by a series of experiments where increasing concentrations of DNA were titrated into kinase reactions verifying that saturating DNA concentrations were employed (data not shown).
Distance of cisplatin from terminus differentially activates DNA-PK
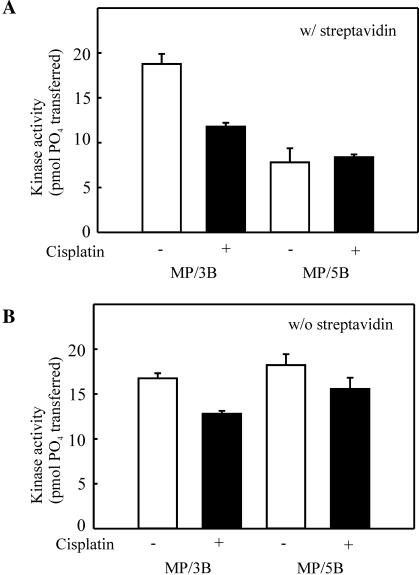
Previous work from our laboratory has demonstrated that the distance of the cisplatin adduct from the terminus to which Ku and DNA-PKcs bind influences kinase activation (15). Considering the dramatic reduction in kinase activity on the cisplatin-damaged substrates with the cisplatin adduct positioned 6 bp from the terminus, it was difficult to assess the significance of cisplatin adduct position in terms of strand polarity. Therefore, substrates were modeled after the 3P/3B and 5P/5B substrates, with the cisplatin adduct moved to 15 bp from the terminus (MP/3B and MP/5B). In reactions performed with SA, cisplatin modification results in a decrease in kinase activity compared to undamaged control DNA substrates on the 3P/3B series of substrates (Figure 2A). The degree of cisplatin-dependent inhibition of kinase activity in the presence of SA observed with the MP series of substrates (Figure 4A) compared to that observed on the 3P series of substrates (Figure 2A) is significantly less, 60% compared to the 95% inhibition. These results are consistent with previous results from our lab which demonstrated that positioning a cisplatin-modification further from a terminus reduces the degree of inhibition (15). Analysis of substrates with cisplatin-modification of the 5′ strand yielded negligible inhibition when compared to control undamaged DNA. These data are consistent with the interpretation that positioning the cisplatin–DNA adduct further from the terminus limits the degree of inhibition by the cisplatin lesion. Importantly, the kinase activity supported by the MP/3B and MP/5B cisplatin-damaged substrates was nearly equivalent, again demonstrating that the strand polarity has a limited influence on the degree of cisplatin inhibition of DNA-PKcs. Importantly, in the absence of cisplatin modification, the 3′ pyrimidine-rich strand again was a more effective activator of the kinase than the 5′ pyrimidine-rich strand (Figure 4A, open bars). These results confirm the sequence bias and strand polarity preference presented in Figure 2. In the absence of SA and cisplatin, both MP/3B and MP/5B have an accessible 3′ pyrimidine-rich end, resulting in similar activity levels with the two different substrates (Figure 4B). In the presence of cisplatin and the continued absence of SA, these substrates result in a somewhat lower activity of DNA-PKcs, as expected based on the position of the cisplatin adduct to the DNA terminus.
Figure 4.
Effect of cisplatin–DNA adduct position, strand bias and sequence specificity on kinase activation. (A) The cisplatin-damaged or control double-stranded SA-MP/3B and SA-MP/5B 30mers were assayed for DNA-PK kinase activation as described in the legend for Figure 2. (B) Reactions were performed as described in Figure 2, except SA was not included in the reactions. Results are presented as the pmol of 32P transferred to the synthetic p53 peptide in a 30 min reaction. Reactions were performed in triplicate and the mean and standard error of the mean are presented.
The affinity of Ku for the MP series of substrates was also assessed in competition binding studies. The results obtained with the cisplatin-damaged substrates are presented in Figure 5. If the affinity of Ku for a DNA substrate was responsible for the differences observed in kinase activation, we would expect Ku to display a higher affinity for the MP/3B substrate, as higher kinase activity was observed with this substrate. Interestingly, the Ku binding analyses revealed a somewhat higher affinity for the MP/5B substrate compared to the MP/3B substrate. This effect was observed for both the unbound and SA-bound competitor DNA (Figure 5, lanes 3–8 and 9–14). Quantification of the results revealed a 10–20% difference in binding the competitor DNA (data not shown). A similar result was also obtained in competition analyses performed with the control series of undamaged MP substrates (data not shown). Therefore, differences in the activation of DNA-PK obtained with different DNA substrates are not dictated by the affinity of Ku for that specific substrate.
Figure 5.
Effect of cisplatin–DNA adduct position on Ku binding. Competition binding assays were performed in 20 μl reactions and contained 2.5 nM 32P-labeled double-stranded 2.1/2.2 DNA and 5 nM Ku. Increasing concentrations of the cisplatin-damaged competitor DNA 5, 50 and 200 nM, is indicated by the triangles. The MP/3B and MP/5B substrates were pre-incubated with SA as indicated. Controls for the assay include 32P-labeled double-stranded 2.1/2.2 DNA alone (lane 1) and 32P-labeled double-stranded 2.1/2.2 DNA with 5 nM Ku in the absence of any competitor (lane 2).
DNA-PK is preferentially activated by 3′ pyrimidine and 5′ purine strands
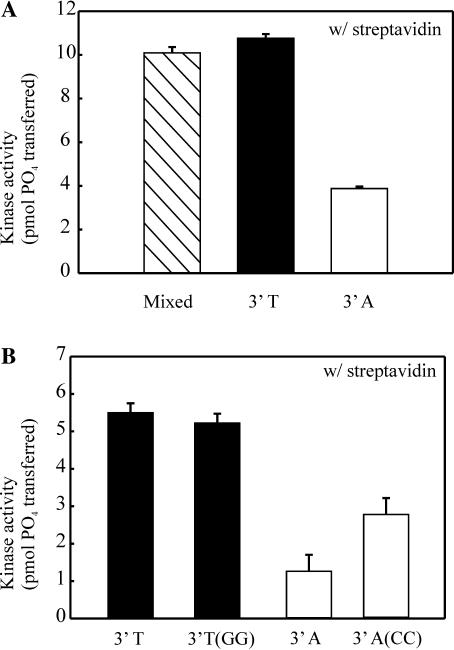
After having preliminarily established that DNA-PK is preferentially activated based on strand bias and polarity, duplex substrates were designed to allow a more rigorous determination of the sequence and polarity bias for DNA-PK activation. Three 30mer substrates were designed to assess sequence bias and polarity. The control substrate is a duplex 30 base mixed sequence with equal base composition and a 5′ biotin. The two test substrates contain a biotinylated 15 base mixed sequence followed by either 15 homopolymeric dTs to generate a 3′ poly T15 (3′-T) or the substrate was prepared to contain a 3′ poly dA (3′-A). Streptavidin blocking was employed to ensure that specific ends of the duplex were made accessible for DNA-PK binding, and kinase activity was assessed based on these substrates. Results show that while DNA-PK is highly activated by the random sequence (Figure 6A, hatched bar), it is also highly activated by a duplex that has a 3′-T15·A15 (Figure 6A, closed bar). In contrast, a duplex made up of a 3′-A15·T15 results in significantly reduced DNA-PK activity (Figure 6A, open bar). This data supports our previous data that DNA-PK is preferentially activated by duplex ends containing a 3′ pyrimidine-rich strand and a 5′ purine-rich strand. A substrate prepared with a 15 base 3′-G15·C15 gave results similar to those observed with the 3′-A terminus (data not shown). Difficulty in chemical synthesis of a poly dG template prohibited analysis of 3′-C15 terminus.
Figure 6.
Effect of homopolymeric sequences in regard to strand polarity on kinase activiation. (A) 30mers containing 15 homopolymeric bases of 3′-dT or 3′-dA and a control 30mer of mixed sequence were constructed by annealing the 5B-mixed oligonucleotide to either c-mixed-15(A), c-mixed-15(T) or c-mixed-15, respectively. Full duplex substrates were generated by polymerase extension and each duplex was assayed for DNA-PK activation as described in Figure 2. (B) A control 45mer containing 30 homopolymeric bases of 3′-dT was compared to a 45mer containing 30 3′-dT homopolymeric bases with two dG six bases from the termini. These substrates were prepared by annealing 5B-mixed to a c-mixed-A or to c-mixed-A(CC), respectively, and extended. A similar 45mer control substrate containing 30 homopolymeric bases of 3′-dA was compared to a 45mer containing 30 3′-dA homopolymeric bases with two dC six bases from the termini. These substrates were prepared by annealing 5B-mixed to a c-mixed-T or to c-mixed-T(GG), respectively, and extended. Reactions were performed as described in Figure 2.
The original substrates used when we first discovered the sequence bias in reference to kinase activation (3P/3B and 5P/5B) were designed to contain two purines in the pyrimidine-rich strand (and subsequently two pyrimidines in the purine-rich strand) in order to form cisplatin–DNA adducts. After having established the preference of a 3′ pyrimidine-rich strand for the activation of DNA-PK, we were interested in determining what effect these two nucleotides in a purine-rich or pyrimidine-rich strand might have on activity of DNA-PK. Therefore, a substrate was designed to contain two Gs in the stretch of Ts on the 3′ strand, and kinase activity was assayed to determine if the two unfavored purines in the pyrimidine strand would decrease activity. Results show that having two purines (3′-T/GG) positioned six bases from the terminus does not adversely affect kinase activity otherwise observed with this substrate (Figure 6B, closed bars). DNA-PK exhibits very little activity with substrates designed with an accessible 3′ purine-rich end and 5′ pyrimidine-rich end. Interestingly, the addition of two Cs within the homopolymeric 3′-A results in a nearly 2-fold increase in activity compared to the control substrate containing only As (Figure 6B, open bars). This significant increase in activity further reinforces the hypothesis that DNA-PK is activated more by 3′ pyrimidines than by 3′ purines.
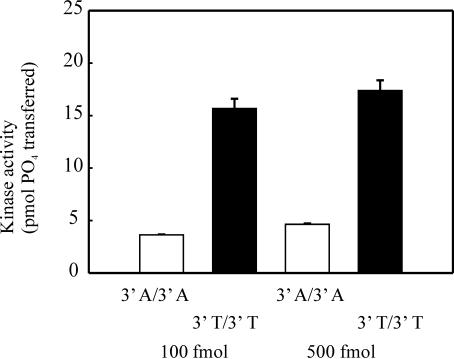
In order to finalize our findings that DNA-PK is preferentially activated by 3′ pyrimidine and 5′ purine ends, we designed a series of control substrates that eliminated the necessity of biotin and SA. 3′-T/3′-T and 3′-A/3′-A have a 15 base stretch of 3′ Ts or 3′ As, respectively, on both sides of the mixed sequence. We have shown that with a substrate that has been blocked with SA and presents only one accessible termini for DNA-PK binding that is made up of 3′ purine and 5′ pyrimidine strands, kinase activity is extremely low (Figures 2, 4 and 6). We have also shown that when SA is bound to biotin that is on opposite sides of a substrate (i.e. both 3′ ends), activity is reduced to negligible amounts (Figure 1C). Based on these findings, having 3′ As on both ends of a duplex substrate should be as ineffective in activating the kinase as a SA-bound terminus is. The results in Figure 7 show that a substrate with two 3′-dA ends (Figure 7, open bars) causes an 80% decrease in activity compared to a substrate with two 3′-dT ends.
Figure 7.
Effect of strand bias and sequence specificity in the absence of SA. The 45mer substrates containing either 15 homopolymeric 3′-dA on both sides of a 15 base sequence of mixed base composition (3′-A/3′-A) or 3′-dT on both ends (3′-T/3′-T) were constructed by annealing T-5B-mixed with c-mixed-15(T) or A-5B-mixed with c-mixed-15(A). Full duplex substrates were generated by polymerase extension and each duplex was assayed for DNA-PK activation at concentrations of 100 or 500 fmol of DNA as described in Figure 2, except SA was not included.
DISCUSSION
Ku and DNA-PKcs each have unique characteristics that contribute to their affinity for specific DNA substrates. The co-crystal structure of Ku bound to a duplex DNA reveals that the DNA is completely encircled by the Ku subunits, while not making any specific base contacts (7). Therefore, the specificity for different DNA substrates that we observe in kinase activation is largely dictated by the DNA-PKcs and how the DNA interacts with this subunit of the complex. Our data suggest that the sequence or chemical make-up of the specific DNA strands can modulate the allosteric activation of DNA-PK. Despite the structure-specific manner in which Ku binds DNA, with the Ku 70 subunit being positioned closer to the DNA end and the 80 subunit being positioned more internally (9), the sequence bias, polarity and position of cisplatin lesions have only a minimal effect on DNA binding. Experiments show that DNA-PKcs possess a channel that can accommodate double-stranded DNA, and upon binding DNA, undergoes a fairly dramatic conformational change that folds around the DNA, almost completely encircling the duplex (11). It is possible that specific structures either induce this change or accommodate the change better than others. By using specific substrates with varying strand polarity and sequence bias, as well as biotin and cisplatin modifications, we have demonstrated that in reference to strand polarity, different DNA substrates with specific sequences can in fact result in differential activation of DNA-PK.
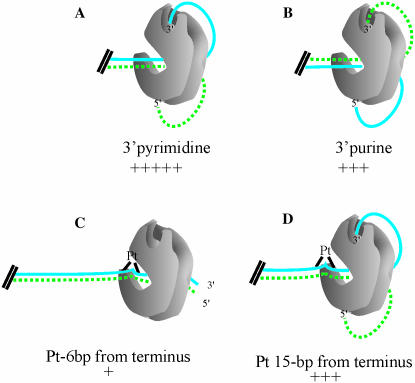
A model for DNA-PK activation and synapse activity in NHEJ was proposed based on in vitro analysis of kinase activation and electron microscopy of DNA–DNA-PKcs complexes (20,21). Once DNA-PKcs has been recruited to the ends of DNA with a DSB, it binds to the terminus of both pieces of the broken DNA. The double-stranded DNA protrudes through the kinase, and each frayed, single-stranded DNA end deviates further from the other (10). The single-stranded ends were then postulated to insert into potentially two separate cavities or single-stranded DNA binding sites. More recently, it has been shown that kinase activity is preferentially activated by three bases of single-stranded dT ends (12), but the preferred polarity of this nucleotide remains undefined. Our data demonstrates that in terms of activating DNA-PKcs, the cavities prefer a 3′ pyrimidine-rich strand and a 5′ purine-rich strand (Figure 8A). A 3′ purine-rich strand and 5′ pyrimidine-rich strand can reside in the cavities, but results in reduced activity of DNA-PK (Figure 8B), demonstrating that a 3′ pyrimidine-rich strand is optimal for kinase activation. Together, these results clearly demonstrate that the discrimination for the single-stranded termini lies on the 3′ strand where a pyrimidine-rich sequence is preferred. Our results, as well as those of Martensson et al. (12), are inconsistent with the discrimination being dictated by the 5′ strand. Our results are also consistent with in vitro analyses measuring the ability of 3′ and 5′ single-strand DNA overhangs to activate DNA-PKcs in the absence of Ku, with a preference being shown for 3′ single-strand overhangs (20). Considering the chemical structure of a pyrimidine is smaller than that of a purine base, it will be interesting to determine the degree to which chemical make-up and size of bases modulate DNA-PK activity.
Figure 8.
Model for effect of strand polarity, sequence bias and platinum position on the activation of DNA-PK. DNA-PKcs, denoted in gray, is bound to different duplex substrates. The duplex DNA separates once it is threaded through the kinase, and a single-strand can enter the 3′ cleft, depicted at the top of DNA-PKcs. The pyrimidine-rich strands of the substrates are represented by a solid blue line and purine-rich strands represented by a dotted green line. Cisplatin adducts are denoted by black triangles on the duplex DNA, and are representative of the structural distortions that the cisplatin–DNA adducts form in duplex DNA. The relative kinase activity supported by each DNA substrate is indicated by the ‘+’ symbols.
The experiments we conducted revealed several differences in DNA that can result in an inactive DNA-PK complex. Interestingly, the presence of a cisplatin–DNA adduct 6 bp from the terminus of a substrate results in significant inhibition of DNA-PKcs kinase activity. The cisplatin–DNA adduct results in a structural distortion in the DNA strand. Specifically, cisplatin modification of a duplex DNA results in a structural distortion of the strand, with a 78° bend in the DNA and a 25° untwisting of the structure (22). When the cisplatin adduct is in close proximity to an accessible end of DNA, over 80% inhibition of kinase activity compared to an undamaged control is observed. The degree of inhibition caused by the cisplatin adduct, however, is strand independent in that cisplatin modification of both the 3′ or 5′ strand resulted in dramatic reduction in kinase activity. Although the specific strand on which the cisplatin adduct resides does not affect kinase activity, the position of the cisplatin adduct with respect to the accessible terminus has a significant influence on the ability to activate DNA-PK. These results suggest that the mechanism of inhibition may be a result of larger structural differences in the duplex that influence either binding, conformational changes or correct positioning of the termini in the 3′ cleft. We propose that a cisplatin adduct positioned 6 bp from the terminus prevents the DNA from being fully threaded through the kinase, thus not allowing the single-stranded ends to wrap around and be placed in the activating cavities on DNA-PKcs (Figure 8C). Assuming the cisplatin modification 15 bp from the terminus also results in an inability to thread through DNA-PKcs, there is still sufficient DNA length from the modification to the end to result in the single-stranded termini to gain access to the cavities (Figure 8D). This model is consistent with other in vitro results demonstrating the requirement of 18 bp duplex DNA for successful binding of DNA-PKcs (11), and the ability of single-stranded DNA tails to activate the kinase (10).
Analysis of our results indicates that the degree of DNA-PK kinase activation is influenced by strand polarity, sequence bias and position of a cisplatin adduct. These conclusions can aid in forming a more detailed outline of how Ku and DNA-PKcs play a role in NHEJ. For example, in NHEJ-catalyzed repair of DNA double-strand breaks, DNA-PKcs binds to each DNA terminus to be joined, and once activated is thought to signal to other complexes involved in the repair process. Electron microscopic and additional in vitro analyses suggests that the DNA-PK complexes can be in close proximity to each other and DNA-PKcs can serve to promote the association of two DNA termini (21). Interestingly, this synaptic activity has also been attributed to Ku independent of DNA-PKcs (23). Independent of the mechanism, following synapsis, alignment must occur with the correct polarity of each broken strand, so that the 5′ and 3′ ends of the strands will be ligated. The presence of damage, like that associated with IR-induced DNA double-strand breaks, or having to search for regions of microhomology, may necessitate processing of the 5′ and 3′ termini (24). Processing of the termini in NHEJ is potentially catalyzed by two nuclease activities with opposing directionalities. The MRN complex is capable of 3′–5′ exonuclease activity and also contains an endonuclease activity capable of digesting a 3′ single-strand overhang (25,26). Artemis contains an exonuclease activity capable of degrading DNA in the 5′–3′ direction (27). The specific roles and substrate specificity of the MRN complex and Artemis in processing DNA termini have not been conclusively demonstrated. Our data, demonstrating the importance of the 3′ strand of a duplex DNA, and published data, demonstrating the preference for a 3′ single-strand to activate DNA-PKcs (20) suggests that the 5′–3′ exonuclease activity of Artemis may be responsible for generating a DNA structure capable of activating DNA-PK. Our data demonstrate a clear preference for a pyrimidine 3′ terminus for DNA-PK activation. This data suggests that either the 3′–5′ exonuclease activity of MRN or the endonuclease activity of MRN could also generate a substrate capable of activating DNA-PK and hence support NHEJ-catalyzed repair of a DNA DSB. Clearly, further investigation of how the chemical structure of the 3′ ends of a substrate influence the processing by MRN and Artemis, and activation of DNA-PK, is of great interest.
Acknowledgments
This work was supported by National Institutes of Health grant CA82741 awarded to J.J.T. Funding to pay the Open Access publication charges for this article was provided by WSU Research Incentive Funds.
REFERENCES
- 1.Lees-Miller S.P., Anderson C.W. The DNA-activated protein kinase, DNA-PK: a potential coordinator of nuclear events. Cancer Cells. 1991;3:341–346. [PubMed] [Google Scholar]
- 2.Yang J., Yu Y.N., Hamrick H.E., Duerksen-Hughes P.J. ATM, ATR and DNA-PK: initiators of the cellular genotoxic stress responses. Carcinogenesis. 2003;24:1571–1580. doi: 10.1093/carcin/bgg137. [DOI] [PubMed] [Google Scholar]
- 3.Carney J.P., Maser R.S., Olivares H., Davis E.M., Le Beau M., Yates J.R., Hays L., Morgan W.F., Petrini J.H.J. The hMre11/hRad50 protein complex and Nijmegen breakage syndrome: linkage of double-strand break repair to the cellular DNA damage response. Cell. 1998;93:477–486. doi: 10.1016/s0092-8674(00)81175-7. [DOI] [PubMed] [Google Scholar]
- 4.Moshous D., Callebaut I., de Chasseval R., Corneo B., Cavazzana-Calvo M., Le Deist F., Tezcan I., Sanal O., Bertrand Y., Philippe N., et al. Artemis, a novel DNA double-strand break repair/V(D)J recombination protein, is mutated in human severe combined immune deficiency. Cell. 2001;105:177–186. doi: 10.1016/s0092-8674(01)00309-9. [DOI] [PubMed] [Google Scholar]
- 5.McElhinny S.A.N., Snowden C.M., McCarville J., Ramsden D.A. Ku recruits the XRCC4-ligase IV complex to DNA ends. Mol. Cell. Biol. 2000;20:2996–3003. doi: 10.1128/mcb.20.9.2996-3003.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Blier P.R., Griffith A.J., Craft J., Hardin J.A. Binding of Ku protein to DNA. Measurement of affinity for ends and demonstration of binding to nicks. J. Biol. Chem. 1993;268:7594–7601. [PubMed] [Google Scholar]
- 7.Walker J.R., Corpina R.A., Goldberg J. Structure of the Ku heterodimer bound to DNA and its implications for double-strand break repair. Nature. 2001;412:607–614. doi: 10.1038/35088000. [DOI] [PubMed] [Google Scholar]
- 8.Yoo S., Dynan W.S. Geometry of a complex formed by double strand break repair proteins at a single DNA end: recruitment of DNA-PKcs induces inward translocation of Ku protein. Nucleic Acids Res. 1999;27:4679–4686. doi: 10.1093/nar/27.24.4679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yoo S., Kimzey A., Dynan W.S. Photocross-linking of an oriented DNA repair complex–Ku bound at a single DNA end. J. Biol. Chem. 1999;274:20034–20039. doi: 10.1074/jbc.274.28.20034. [DOI] [PubMed] [Google Scholar]
- 10.Leuther K.K., Hammarsten O., Kornberg R.D., Chu G. Structure of DNA-dependent protein kinase: implications for its regulation by DNA. EMBO J. 1999;18:1114–1123. doi: 10.1093/emboj/18.5.1114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Boskovic J., Rivera-Calzada A., Maman J.D., Chacon P., Willison K.R., Pearl L.H., Llorca O. Visualization of DNA-induced conformational changes in the DNA repair kinase DNA-PKcs. EMBO J. 2003;22:5875–5882. doi: 10.1093/emboj/cdg555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Martensson S., Hammarsten O. DNA-dependent protein kinase catalytic subunit. Structural requirements for kinase activation by dna ends. J. Biol. Chem. 2002;277:3020–3029. doi: 10.1074/jbc.M106711200. [DOI] [PubMed] [Google Scholar]
- 13.Leng M., Brabec V. DNA adducts of cisplatin, transplatin and platinum-intercalating drugs. IARC Sci. Publ. 1994:339–348. [PubMed] [Google Scholar]
- 14.Turchi J.J., Henkels K.M. Human Ku autoantigen binds cisplatin-damaged DNA but fails to stimulate human DNA-activated protein kinase. J. Biol. Chem. 1996;271:13861–13867. doi: 10.1074/jbc.271.23.13861. [DOI] [PubMed] [Google Scholar]
- 15.Turchi J.J., Henkels K.M., Zhou Y. Cisplatin-DNA adducts inhibit translocation of the Ku subunits of DNA-PK. Nucleic Acids Res. 2000;28:4634–4641. doi: 10.1093/nar/28.23.4634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vaisman A., Lim S.E., Patrick S.M., Copeland W.C., Hinkle D.C., Turchi J.J., Chaney S.G. Effect of DNA polymerases and high mobility group protein 1 on the carrier ligand specificity for translesion synthesis past platinum-DNA adducts. Biochemistry. 1999;38:11026–11039. doi: 10.1021/bi9909187. [DOI] [PubMed] [Google Scholar]
- 17.Jayaraman L., Moorthy N., Murthy K., Manley J., Bustin M., Prives C. High mobility group protein-1 (HMG-1) is a unique activator of p53. Genes Dev. 1998;12:462–472. doi: 10.1101/gad.12.4.462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Andrews B.J., Turchi J.J. Development of a high-throughput screen for inhibitors of replication protein A and its role in nucleotide excision repair. Mol. Cancer Ther. 2004;3:385–391. [PubMed] [Google Scholar]
- 19.Turchi J.J., Patrick S.M., Henkels K.M. Mechanism of DNA-dependent protein kinase inhibition by cis- diamminedichloroplatinum(II)-damaged DNA. Biochemistry. 1997;36:7586–7593. doi: 10.1021/bi963124q. [DOI] [PubMed] [Google Scholar]
- 20.Hammarsten O., DeFazio L.G., Chu G. Activation of DNA-dependent protein kinase by single-stranded DNA ends. J. Biol. Chem. 2000;275:1541–1550. doi: 10.1074/jbc.275.3.1541. [DOI] [PubMed] [Google Scholar]
- 21.DeFazio L.G., Stansel R.M., Griffith J.D., Chu G. Synapsis of DNA ends by DNA-dependent protein kinase. EMBO J. 2002;21:3192–3200. doi: 10.1093/emboj/cdf299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gelasco A., Lippard S.J. NMR solution structure of a DNA dodecamer duplex containing a cis-diammineplatinum(II) d(GpG) intrastrand cross-link, the major adduct of the anticancer drug cisplatin. Biochemistry. 1998;37:9230–9239. doi: 10.1021/bi973176v. [DOI] [PubMed] [Google Scholar]
- 23.Bliss T.M., Lane D.P. Ku selectively transfers between DNA molecules with homologous ends. J. Biol. Chem. 1997;272:5765–5773. doi: 10.1074/jbc.272.9.5765. [DOI] [PubMed] [Google Scholar]
- 24.Delacote F., Guirouilh-Barbat J., Lambert S., Lopez B.S. Homologous recombination, non-homologous end-joining and cell cycle: genome's angels. Curr. Genomics. 2004;5:49–58. [Google Scholar]
- 25.Paull T.T., Gellert M. The 3′ to 5′ exonuclease activity of Mre11 facilitates repair of DNA double-strand breaks. Mol. Cell. 1998;1:969–979. doi: 10.1016/s1097-2765(00)80097-0. [DOI] [PubMed] [Google Scholar]
- 26.Trujillo K.M., Yuan S.S.F., Lee E.Y.H.P., Sung P. Nuclease activities in a complex of human recombination and DNA repair factors Rad50, Mre11, and p95. J. Biol. Chem. 1998;273:21447–21450. doi: 10.1074/jbc.273.34.21447. [DOI] [PubMed] [Google Scholar]
- 27.Ma Y.M., Pannicke U., Schwarz K., Lieber M.R. Hairpin opening and overhang processing by an Artemis/DNA-dependent protein kinase complex in nonhomologous end joining and V(D)J recombination. Cell. 2002;108:781–794. doi: 10.1016/s0092-8674(02)00671-2. [DOI] [PubMed] [Google Scholar]