Abstract
mRNA polyadenylation is a critical cellular process in eukaryotes. It involves 3′ end cleavage of nascent mRNAs and addition of the poly(A) tail, which plays important roles in many aspects of the cellular metabolism of mRNA. The process is controlled by various cis-acting elements surrounding the cleavage site, and their binding factors. In this study, we surveyed genome regions containing cleavage sites [herein called poly(A) sites], for 13 942 human and 11 155 mouse genes. We found that a great proportion of human and mouse genes have alternative polyadenylation (∼54 and 32%, respectively). The conservation of alternative polyadenylation type or polyadenylation configuration between human and mouse orthologs is statistically significant, indicating that alternative polyadenylation is widely employed by these two species to produce alternative gene transcripts. Genes belonging to several functional groups, indicated by their Gene Ontology annotations, are biased with respect to polyadenylation configuration. Many poly(A) sites harbor multiple cleavage sites (51.25% human and 46.97% mouse sites), leading to heterogeneous 3′ end formation for transcripts. This implies that the cleavage process of polyadenylation is largely imprecise. Different types of poly(A) sites, with regard to their relative locations in a gene, are found to have distinct nucleotide composition in surrounding genomic regions. This large-scale study provides important insights into the mechanism of polyadenylation in mammalian species and represents a genomic view of the regulation of gene expression by alternative polyadenylation.
INTRODUCTION
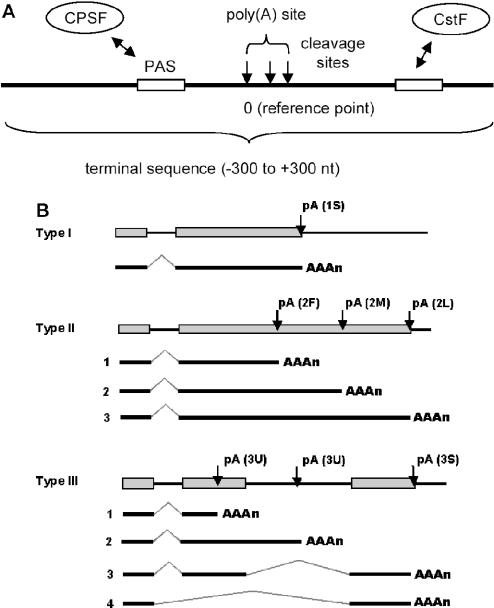
The 3′ ends of all fully processed eukaryotic mRNAs (except most histone genes) have a poly(A) tail. Poly(A) tails have been shown to influence mRNA stability, translation and transport (1–3). As has been understood more thoroughly in recent years, 3′ end formation is interconnected to other transcriptional and post-transcriptional processes, such as splicing and transcriptional termination (4–7). The cellular process of making poly(A) tails, called polyadenylation, is a two-step reaction (8–12), first involving specific endonucleolytic cleavage at a site determined by binding of polyadenylation factors. The second step involves polymerization of the adenosine tail to a length specific to the species, e.g. ∼150–250 in mammals and ∼55–90 in yeasts (13). Since the cleavage can be imprecise (14,15), resulting in mRNAs with variable ends, we refer to the cleavage site as a location where mRNA cleavage takes place, and poly(A) site as a region containing cleavage site(s) (Figure 1A). It should be pointed out that this variable 3′ end formation is different than alternative polyadenylation involving multiple polyadenylation sites, which is discussed below. When multiple cleavage sites exist in a single poly(A) site, the location of the first (or 5′-most) cleavage site is used to represent the location of the poly(A) site. In addition, terminal sequence is referred to as a genomic sequence (−300 to +300 nt) containing a poly(A) site (Figure 1A).
Figure 1.
(A) Schematic representation of a poly(A) site and polyadenylation configuration. In this study, a poly(A) site is a region containing cleavage site(s) (arrowed lines). The 5′-most cleavage site is the reference point (position 0) for the poly(A) site. Thus, the genomic location of a poly(A) site is represented by the location of the 5′-most cleavage site it contains. The sequence −300 to +300 is defined as a terminal sequence. The sites for CPSF and CstF are also depicted. (B) Three types of polyadenylation configuration. A type 1 gene has a single poly(A) site; a type II gene has alternative poly(A) sites all located in the 3′-most exon; and a type III gene has alternative poly(A) sites located in different exons. Types of poly(A) sites are also marked. 1S, a single poly(A) site; 2F, the 5′-most poly(A) site in a type II gene; 2L, the 3′-most poly(A) site in a type II gene; 2M, a middle poly(A) site between 2F and 2L in a type II gene; 3U, a poly(A) site located upstream of the 3′-most exon; and 3S, a single site in the 3′-most exon of a type III gene. Not shown in the graph are 3F, 3M and 3L, which are similar to 2F, 2M and 2L, respectively, except that the formers are located in the 3′-most exon of a type III gene. Exons are represented as boxes; pA, poly(A) site.
It has been reported that over 29% of human genes have more than one polyadenylation site (16). Alternative poly(A) sites can be located in the last or 3′-most exon, usually giving rise to mRNAs with variable 3′-untranslated regions (3′-UTRs), or in different exons, which can lead to mRNAs with variable 3′-UTRs as well as distinct protein products (17). Here, we use polyadenylation configuration to refer to the type of alternative polyadenylation (Figure 1B). As 3′-UTRs often include crucial sequence elements important for mRNA stability, mRNA localization and protein translation, the effect of alternative polyadenylation is multifold. The impact of alternative polyadenylation on protein variants has also been studied for a number of genes (17). It is generally believed that the choice of polyadenylation site is related to tissue types and development stages (12,14,17). A well-known example of alternative polyadenylation is the IgM heavy chain gene (18). During B lymphocyte activation, IgM heavy chain gene switches from using one poly(A) site to another, resulting in a shift in protein production from the membrane-bound form to the secreted form due to the deletion of a C-terminal hydrophobic region responsible for membrane interaction. This switch is an essential step in immune response.
The choice of a particular poly(A) site probably involves specific cis-acting elements and trans-acting factors. The terminal sequence of most mammalian genes contains the consensus AAUAAA hexamer (or a close variant) between 10 and 30 nucleotides upstream of the actual cleavage site, which serves as the binding site for the cleavage and polyadenylation specificity factor (CPSF). It has been called polyadenylation signal (PAS), upstream core element or AAUAAA motif. Here, we use PAS to denote this element. Several studies on PAS suggested that while most human polyadenylation sites contain the canonical AAUAAA (∼70%), a large portion of genes have single-nucleotide variants, with AUUAAA as the most common one (16,19,20). In addition, U-rich or U/G-rich elements that are located 20–40 nt downstream of the cleavage site are involved in directing polyadenylation by serving as the binding site for the cleavage stimulation factor (CstF). Together, the PAS and the CstF binding site are thought to determine the polyadenylation reaction. In addition, some auxiliary elements upstream of the PAS and downstream of the cleavage site have been characterized that can enhance polyadenylation efficiency in both viral and cellular systems (21–27). In general, the nucleotide composition surrounding human poly(A) site is U-rich (28), and can be used to computationally predict poly(A) sites, indicating the importance of the nucleotide composition of regions harboring poly(A) sites in polyadenylation (28,29).
In this study, we have surveyed a large number of human and mouse polyadenylation sites, delineated their configurations in the context of the genomic structure of genes, and studied the conservation of polyadenylation configuration between human and mouse orthologs. We have also investigated human and mouse PAS hexamers, heterogeneity of polyadenylation cleavage, the relationship between gene function and polyadenylation configuration, and nucleotide composition surrounding various types of poly(A) sites. This comprehensive study will shed light on the understanding of alternative polyadenylation in mammalian species, and provide insights into the mechanisms of mRNA polyadenylation.
MATERIALS AND METHODS
Align cDNA/ESTs to genomic sequences
We used all sequences listed in human and mouse UniGene databases (NCBI, March, 2004 versions) that are associated with LocusLink IDs, and aligned them to genome sequences (human genome Build 34.2 and mouse genome Build 32, both from NCBI). RefSeq mRNA and cDNA sequences (NCBI GenBank March, 2004 release) and ESTs (NCBI dbEST March, 2004 release) were aligned to genomes using BLAST and MegaBLAST suites with default settings (J. Hu and B. Tian, unpublished data). Briefly, MegaBLAST was first used to find the genomic location of a sequence, and BLAST was used to fill gaps left behind by MegaBLAST. In both steps, high scoring pairs (HSPs) were assembled using the Longest Increasing Subsequence (LIS) algorithm (30) with a modification on the calculation of sequence length (J. Hu and B. Tian, unpublished data). Exon ends were located by using scoring matrices for canonical and non-canonical splicing sites (31) at the 5′ and 3′ ends of assembled HSPs. The results were comparable to those obtained from using BLAT (32) (data not shown). The transcriptional orientation of a sequence on the genome was first determined by its splicing sites, e.g. GT.AG (5′ and 3′ splice sites, respectively) indicates a sense orientation whereas CT.AC indicates an anti-sense orientation, and/or its poly(A) tail (see below) since polyadenylation only occurs at the 3′ end. If the orientation of a sequence indicated by splicing is in conflict with that by its poly(A) tail, the sequence is discarded. If neither piece of information can be obtained from a sequence, which automatically indicates that the sequence does not have a poly(A) tail (thus not of interest in this study), the sequence is also discarded.
In this study, genes are represented by LocusLink entries, which were obtained from NCBI (http://www.ncbi.nih.gov/LocusLink/). RefSeq mRNA sequences were used to represent transcripts of genes. If a gene has more than one RefSeq sequence, their corresponding genomic regions are required to overlap, and their transcriptional orientations are required to be the same. Genes whose RefSeq sequences do not meet these two criteria are discarded. Thus each gene's orientation and genomic location can be unequivocally determined using its RefSeq(s). cDNA/ESTs that are associated with a gene, as listed in the UniGene database, are required to meet the following criteria: (i) A sequence's transcriptional orientation is required to be in agreement with that of its associated gene. (ii) The genomic regions aligned with a cDNA/EST are required to overlap with those of RefSeq sequences of its corresponding gene by either a 32 nt sequence or an entire exon (either the cDNA/EST's or RefSeq's). This is to eliminate sequences incorrectly associated with LocusLink IDs in the UniGene database. Also, it discards sequences that reside in the intron region of other genes.
For each gene, the 3′-most intron/exon junction of its RefSeqs is arbitrarily defined as the gene's 3′-most intron/exon junction. Thus a gene's 3′-most exon is determined by its RefSeqs. The stop codon of a gene is located using the RefSeq GenBank annotation file. If a gene has more than one stop codon, the 3′-most is used.
Poly(A) site determination
cDNA/EST sequences aligned to genomic sequences were examined for poly(A) tails after the alignment. Unaligned sequences at both 5′ and 3′ termini of the cDNA/EST were checked for a stretch of T and A, respectively. For the 3′ end, a sequence is considered to have a poly(A) tail if after the unaligned position (i) the sequence contains 8 or more consecutive As, or (ii) if it has one other nucleotide, it has 8 or more consecutive As after the other nucleotide. The criteria are the same for the 5′ end except that consecutive Ts are searched.
For sequences that contain poly(A) tails, the poly(A) cleavage site on the genome is considered to be right after the 3′-most position of the alignment of cDNA/EST with the genome. To address the internal priming issue, the genomic sequence −10 to +10 nt surrounding the cleavage site was examined. If the sequence has six continuous As or more than 7 As in a 10 nt window, it is considered as internal priming candidate, similar to what was used by other groups (16,33). However, if an internal priming candidate site is supported by more than one cDNA/EST and has one of the 12 PAS hexamers in −40 to −1 nt region (16), it is believed to be a real site. Because of the heterogeneity of polyadenylation cleavage, we iteratively clustered poly(A) cleavage sites that are located next to each other within 24 nt, i.e. starting from the 5′-most one, the process of clustering cleavage sites is continued until no adjacent cleavages sites are located within 24 nt. We used the 5′-most cleavage site of a set of clustered sites as the reference site for a poly(A) site. The number of cDNA/ESTs associated with a poly(A) site is the sum of all cDNA/ESTs supporting its constituent cleavage sites. After clustering of cleavage sites, if a poly(A) site has at least two supporting cDNA/EST sequences, or has a PAS hexamer (AAUAAA or 11 variants) in the −40 to −1 region of the poly(A) site, it is considered to be a genuine poly(A) site.
Identification of PAS
We used a method developed by Beaudoing et al. (16) to identify PAS. Briefly, the most frequently occurring hexamer in the −40 to −1 region of all poly(A) sites is detected, and sequences containing the hexamer are removed. The process is repeated until less than 5% of sequences are remaining.
Conservation study of human and mouse orthologous genes
Human and mouse orthologous genes were obtained from HomoloGene database (ftp://ftp.ncbi.nih.gov/pub/HomoloGene/). Only reciprocal best BLAST hits of protein sequences were used. Pearson's χ2-test was used to determine the conservation of polyadenylation configuration between orthologs. Expected values were obtained by the χ2-test function in the R program (http://www.r-project.org/).
Gene Ontology analysis
Gene Ontology annotations of genes were obtained from the LocusLink database of NCBI. The full list of GO terms in three categories, namely Biological Process (BP), Cellular Component (CC) and Molecular Function (MF), was downloaded from Gene Ontology Consortium website (http://www.geneontology.org/). For each GO term, all associated GO terms were found by a recursive method which searches the whole gene ontology tree for related entries through either ‘is a’ or ‘part of’ relationship. A total number of 7315 GO terms (3607 BP, 690 CC and 3039 MF) were found to be associated with 9057 human and 7700 mouse genes. Fisher's exact test using 2 × 2 table was applied to assess the bias of polyadenylation configuration for each GO term. In the table, two columns are constitutive polyadenylation and alternative polyadenylation, and two rows are ‘having the GO term’ and ‘not having the GO term’. The test for type III gene was carried out in a similar manner except that one column is ‘is a type III gene’, and the other is ‘is not a type III gene’. The Benjamini and Hochberg method was applied to eliminate false positives generated as a result of multiple testing (34).
Heat map and two-way clustering of poly(A) site types and PAS hexamers
Heat map was generated by the image function in R (35). Hierarchical clustering using Euclidean distance was used to make dendrograms.
Sequence analysis of poly(A) sites
For each poly(A) site, terminal sequence −300 to +300 nt neighboring the cleavage site was obtained from the genome. If a cleavage site is located less than 300 nt from the end of a genome contig, the sequence between the cleavage site and the end of the contig is used.
RESULTS
Mapping poly(A) sites on human and mouse genomes
The availability of human and mouse nearly complete genomic sequences and a large number of their respective cDNA and EST sequences (herein collectively called cDNA/EST, the difference being that an EST is usually a partial sequence of a transcript and a cDNA is usually full-length) provide an unprecedented opportunity to study polyadenylation. Using a series of databases from NCBI, including UniGene, LocusLink, RefSeq, dbEST and GenBank databases (see Materials and Methods), we mapped a large number of human and mouse poly(A) sites on the genomes (Table 1). We took a gene-centered approach by using LocusLink entries to represent genes and their associated RefSeq mRNAs to represent transcripts. cDNA/ESTs are then mapped to RefSeq sequences.
Table 1.
Poly(A) sites identified in human and mouse genes
| Homo sapiens | Mus musculus | |
|---|---|---|
| cDNA/EST useda | 3 619 860 | 2 676 296 |
| cDNA/EST with poly(A) tailb | 396 908 | 108 691 |
| Cleavage sites | 67 440 | 31 179 |
| Poly(A) sites | 29 283 | 16 282 |
| LocusLink entries | 13 942 | 11 155 |
| Poly(A) sites per gene | 2.10 | 1.46 |
aNumber of cDNA/EST sequences in the UniGene database.
bAfter sequence clean-up using approaches detailed in Materials and Methods. cDNA/EST sequences listed in the UniGene database were downloaded from GenBank and dbEST. Sequences were aligned to human and mouse genomes, and poly(A) cleavage sites were identified and clustered (see Materials and Methods for details). cDNA/ESTs were mapped to LocusLink IDs according to the UniGene database.
We identified 67 440 cleavage sites that are supported by cDNA/ESTs for 29 283 poly(A) sites in 13 942 human genes, and 31 179 cleavage sites for 16 282 poly(A) sites in 11 155 mouse genes. This mapping is by far the most comprehensive to date to the best of our knowledge. The average poly(A) sites per gene is 2.1 for human genes and ∼1.5 for mouse genes. The difference between human and mouse figures is mainly attributable to the fact that there are fewer cDNA/EST sequences for mouse genes and a smaller proportion of them have poly(A) tails (11% human cDNA/ESTs versus 4% mouse cDNA/ESTs), resulting in human genes having ∼3.7 times as many poly(A) tail-containing cDNA/ESTs than mouse genes (Table 1).
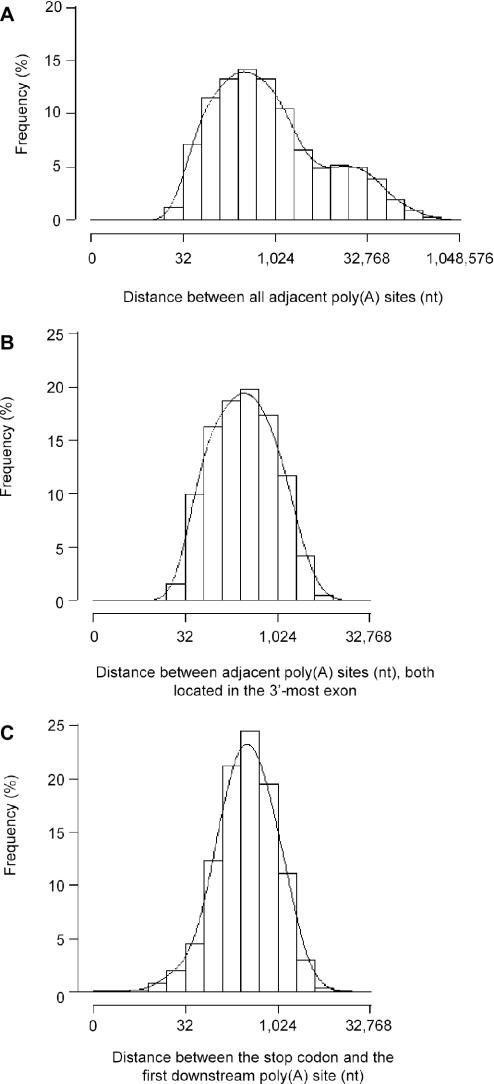
The distribution of the genomic distance between adjacent poly(A) sites in a gene, when multiple sites are present, shows two modes (Figure 2A), with one peak at ∼300 nt and the other at ∼14 kb for human genes (mouse genes show similar distribution, see Supplementary Materials). These two modes appear to correspond to alternative poly(A) sites located in the 3′-most exon and alternative poly(A) sites located in different exons, respectively. This is based on the observation that when we only used poly(A) sites located in the 3′-most exon defined by RefSeq sequences (see Materials and Methods), we could see a single-mode distribution similar to the left peak in Figure 2A (Figure 2B). The distance between adjacent poly(A) sites in the 3′-most exon has a median value of 288 nt for human genes and 345 nt for mouse genes. The distance between the stop codon of a gene to its closest downstream poly(A) site has a median value of 324 nt for human genes (Figure 2C), and 385 nt for mouse genes (Supplementary Figure 2C).
Figure 2.
Poly(A) sites of human genes. (A) Histogram of the genomic distance between adjacent poly(A) sites in a gene. (B) Histogram of the distance between adjacent poly(A) sites, both located in the 3′-most exon of a gene (median = 288 nt). (C) Histogram of the distance between the stop codon and its closest downstream poly(A) site (median = 324 nt). The x-axes in all graphs are in base-2 logarithmic scale. For each histogram, a Gaussian smoothing kernel method was used to generate a density line.
We studied PAS in the −40 to −1 region upstream of poly(A) sites using a method developed by Beaudoing et al. (16). As shown in Table 2, top occurring PAS hexamers for human and mouse poly(A) sites are very similar in their frequency, with the top five hexamers having identical ranks. AAUAAA and 11 single nucleotide variants are prominent hexamers detected. UUUAAA is the most frequent hexamer among other hexamers. The frequencies of top PAS hexamers of human poly(A) sites are similar to those reported by Beaudoing et al., with the exception of AAGAAA, whose occurrence is higher in our result. Thus in this study, we focused on AAUAAA and 11 single nucleotide variants, which are associated with ∼92% human and 93% mouse poly(A) sites.
Table 2.
Top detected PAS hexamers
| Frequency (%) | Rank | |||||
|---|---|---|---|---|---|---|
| Hs | Mm | Hs.B | Hs | Mm | Hs.B | |
| AAUAAA | 53.18 | 59.16 | 58.2 | 1 | 1 | 1 |
| AUUAAA | 16.78 | 16.11 | 14.9 | 2 | 2 | 2 |
| UAUAAA | 4.37 | 3.79 | 3.2 | 3 | 3 | 3 |
| AGUAAA | 3.72 | 3.28 | 2.7 | 4 | 4 | 4 |
| AAGAAA | 2.99 | 2.15 | 1.1 | 5 | 5 | 10 |
| AAUAUA | 2.13 | 1.71 | 1.7 | 6 | 7 | 5 |
| AAUACA | 2.03 | 1.65 | 1.2 | 7 | 8 | 8 |
| CAUAAA | 1.92 | 1.80 | 1.3 | 8 | 6 | 6 |
| GAUAAA | 1.75 | 1.16 | 1.3 | 9 | 9 | 7 |
| AAUGAA | 1.56 | 0.90 | 0.8 | 10 | 11 | 11 |
| UUUAAA | 1.20 | 1.08 | 1.2 | 11 | 10 | 9 |
| ACUAAA | 0.93 | 0.64 | 0.6 | 12 | 12 | 13 |
| AAUAGA | 0.60 | 0.36 | 0.7 | 13 | 15 | 12 |
Human and mouse genomic sequences located −40 to −1 nt upstream of poly(A) sites were used to detect hexamers that may function as polyadenylation signals. Hs, human sequences; Mm, mouse sequences; and Hs.B, human results reported by Beaudoing et al. (16).
Heterogeneity of cleavage sites
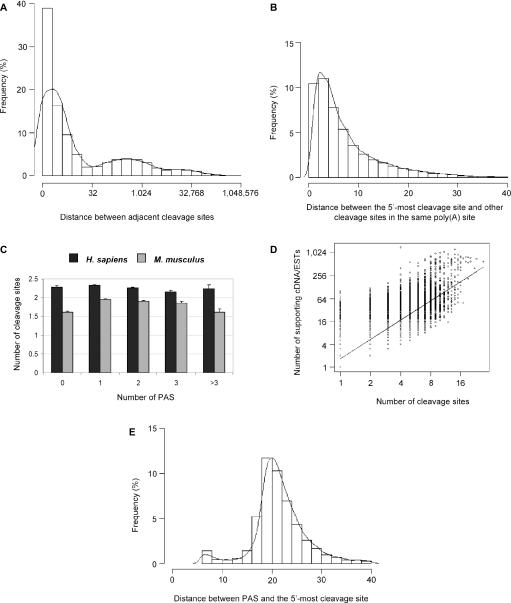
We found that a large number of poly(A) sites have more than one cleavage site, as also noted before by other groups (14,15). Specifically, 51.25% of human and 46.97% mouse poly(A) sites have more than one cleavage site; and human poly(A) sites have an average of 2.30 cleavage sites per poly(A) site with SD of 1.91, and mouse sites have an average of 1.92 with SD of 1.33. In either case, the distribution of the number of cleavage sites per poly(A) site is not normal (Gaussian). In fact, if we do not cluster adjacent cleavage sites, the histogram of the distance between adjacent cleavage sites will give rise to three modes (Figure 3A), with the first mode (the first peak in Figure 3A) corresponding to the distance between heterogeneous cleavage sites. The other two modes are similar to the two peaks derived from alternative poly(A) sites shown in Figure 2A. Since there is a valley at ∼24 nt between the first and second modes, derived from the fitted density line (Figure 3A), we therefore iteratively clustered all cleavage sites located within 24 nt next to each other. When there are multiple cleavage sites in a poly(A) site, the distance between the 5′-most cleavage site and other ones after clustering is shown in Figure 2B. Since the majority (>95%) of the distances are below 24 nt, we conclude that the heterogeneity of poly(A) cleavage usually occurs within 24 nt after the 5′-most cleavage site.
Figure 3.
Multiple cleavage sites in a poly(A) site. (A) Histogram of the genomic distance between adjacent cleavage sites in genes. (B) Histogram of the distance between the 5′-most cleavage site and other downstream cleavage sites when multiple cleavage sites are present in a poly(A) site (mean = 7.9 nt, median = 5 nt). (C) The relationship between the number of PAS hexamers (AAUAAA and other 11 variants) associated with a poly(A) site and the number of cleavage sites in the poly(A) site. Error bars are standard error of the mean (SEM). (D) Correlation between the number of cleavage sites and the number of supporting cDNA/EST sequences for poly(A) sites (Pearson correlation coefficient R = 0.83). (E) Histogram of the distance between a poly(A) site and the associated PAS when only one PAS is present. Only human poly(A) sites are used in (A–E).
To study the mechanism leading to this heterogeneity, we studied PAS and supporting cDNA/ESTs for cleavage sites. We found that the heterogeneity of mRNA cleavage does not appear to be a result of multiple PAS upstream of a poly(A) site (Figure 3C), but seems to correlate with the number of supporting cDNA/ESTs that a poly(A) site has (Figure 3D). The Pearson correlation R is 0.83 for human poly(A) sites and 0.75 for mouse poly(A) sites when both the number of supporting cDNA/ESTs and the number of cleavage sites are in logarithmic scales. Therefore, the heterogeneity of cleavage sites is in general due to stochastic selection of cleavage position. In other words, the more sequences investigated, the higher the chance of finding heterogeneous cleavage sites. Since our method to align cDNA/ESTs with the genomic sequence will always match the 5′-most adenosine of a poly(A) tail to the genome sequence if the −1 position is an adenosine, we do not know if there is any nucleotide preference for the cleavage site, such as the A > U > C ≫ G order reported previously (36).
For poly(A) sites that are associated with only one PAS, we measured the distance between the 5′-most cleavage site and the first nucleotide of the PAS (Figure 3E). The median value is 21 nt, which is in agreement with previous surveys (16,36). Interestingly, a small population of poly(A) sites are very close to the PAS. A careful inspection of all 12 PAS hexamers indicates that they are mainly poly(A) sites associated with AAGAAA (Supplementary Figure 3), and they are usually located in exons upstream of the 3′-most exon (see below). Possible biological implications of this finding are provided in the discussion section.
Widespread alternative polyadenylation in human and mouse genes
The two-mode distribution of genomic distance between poly(A) sites (Figure 2A) prompted us to classify genes according to their poly(A) site location. Edwalds-Gilbert et al. (17) proposed three types of alternative polyadenylation, i.e. (i) tandem poly(A) sites, (ii) coupled with composite in/terminal exons and (iii) coupled with skipped exons. In line with that, we have classified genes based on their polyadenylation configuration as delineated in Figure 1B. Genes with only one poly(A) site are classified as type I genes, genes with multiple poly(A) sites all in the 3′-most exon as type II genes, and genes with multiple poly(A) sites located in different exons as type III genes. For simplicity, we did not differentiate types B and C alternative polyadenylation proposed by Edwalds-Gilbert et al. (17), which result from two distinct mechanisms of alternative splicing. Instead, they are collectively called type III genes, where alternative polyadenylation is related to splicing. Thus type I genes have a single constitutive poly(A) site, whereas types II and III genes have alternative poly(A) sites.
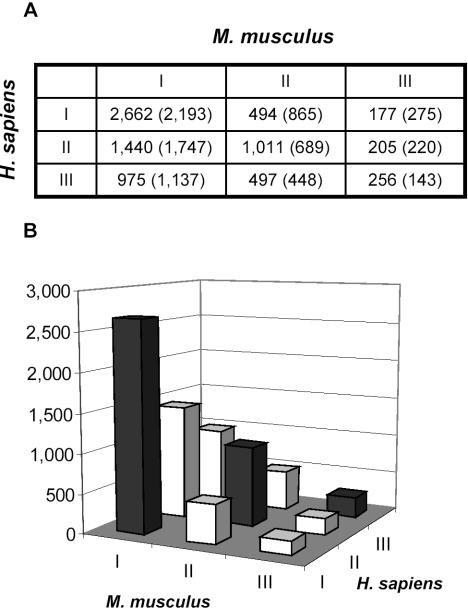
We found that ∼54% of human genes and ∼32% of mouse genes have multiple poly(A) sites (Table 3). These numbers are significantly higher than that previously thought about the occurrences of alternative polyadenylation. The difference between human and mouse figures is at least partially attributable to fewer mouse poly(A) tail-containing cDNA/ESTs than human ones (Table 1). Nevertheless, the conservation of polyadenylation configuration between human and mouse orthologs is statistically significant (P-value of 2.0 × 10−132 using a χ2-test; Figure 4). All values for corresponding human and mouse polyadenylation configurations are more than 1.2-fold above the expected values, indicating that alternative polyadenylation is an evolutionarily conserved cellular process between human and mouse orthologous genes.
Table 3.
Classification of genes according to the configuration of mRNA polyadenylation
| H.sapiens | M.musculus | |
|---|---|---|
| Number of genes | 13 942 | 11 155 |
| Type I genes | 6418 | 7576 |
| Type II genes | 4416 | 2681 |
| Type III genes | 3108 | 898 |
| Constitutive poly(A) sites | 46.03% | 67.92% |
| Alternative poly(A) sites | 53.97% | 32.08% |
Genes are classified according to the configuration of mRNA polyadenylation depicted in Figure 1B.
Figure 4.
Conservation of polyadenylation configuration between human and mouse orthologs. (A) Conservation of polyadenylation configuration between human (rows) and mouse (columns) orthologs (χ2-test, P-value = 2.0 × 10−132). Expected values, based on the null hypothesis that there is no correlation, are shown in parentheses. Observed values in (A) are plotted in (B), with the closed bars corresponding to conserved configurations, i.e. human type I versus mouse type I, etc.
To address the question of whether some functional groups of genes have biased polyadenylation configuration, we studied the relationship between gene functions, as indicated by Gene Ontology (GO) annotations of a gene, and the polyadenylation configuration. We used genes whose polyadenylation configuration is conserved between human and mouse orthologs (Figure 4). A total number of 7315 GO terms belonging to three categories were studied, namely biological process, cellular component and molecular function (Table 4). Genes whose functions are related to cell surface receptor-linked site transduction (biological process), extracellular genes (cellular component) and site transducer activity (molecular function) are found to be disproportionately associated with type I constitutive polyadenylation (Table 4). Genes that are found to be disproportionately associated with alternative polyadenylation include those encoding intracellular proteins (cellular component), proteins involved in intracellular protein transport (biological process) and proteins having protein transporter activity (molecular functions). Thus, it appears that genes with extracellular protein products tend to have constitutive polyadenylation sites, whereas genes with intracellular protein products are biased for alternative polyadenylation. In addition, genes whose protein products are located in the nucleus and have RNA-binding activities are found to be disproportionately associated with type III configuration, i.e. related to splicing.
Table 4.
Gene Ontology terms disproportionately associated with different types of polyadenylation configuration
| GO termsa | Typeb | H.sapiens | M.musculus |
|---|---|---|---|
| Biological process | |||
| GO:0007166 (cell surface receptor linked site transduction) | I | 1.15E−04 (171, 37, 3) | 9.38E−06 (154, 30, 1) |
| GO:0046907 (intracellular transport) | II and III | 1.29E−07 (60, 59, 7) | 1.57E−08 (64, 68, 5) |
| GO:0015031 (protein transport) | 1.41E−07 (49, 53, 5) | 1.53E−07 (54, 59, 3) | |
| GO:0006886 (intracellular protein transport)c | 7.25E−08 (46, 52, 5) | 5.95E−07 (51, 55, 3) | |
| Cellular component | |||
| GO:0005576 (extracellular) | I | 9.98E−05 (168, 32, 8) | 2.54E−10 (518, 115, 24) |
| GO:0005622 (intracellular) | II and III | 6.32E−08 (819, 337, 109) | 1.62E−04 (826, 335, 92) |
| GO:0005524 (nucleus) | III | 7.84E−05 (393, 162, 66) | 2.50E−04 (376, 143, 53) |
| Molecular function | |||
| GO:0004871 (site transducer activity) | I | 4.94E−06 (344, 77, 18) | 3.81E−06 (315, 71, 13) |
| GO:0008565 (protein transporter activity) | II and III | 4.52E−07 (30, 42, 1) | 3.41E−07 (25, 40, 0) |
| GO:0003723 (RNA binding) | III | 1.17E−05 (46, 32, 19) | 1.12E−04 (40, 22, 14) |
aFor each GO term, its GO ID and annotation (in parentheses) are given.
bType is the type of polyadenylation configuration (shown in Figure 1B).
cGO:0006886 (intracellular protein transport) is associated with both GO:0046907 (intracellular transport) and GO:0015031 (protein transport) through an ‘is a’ relationship (for details see Materials and Methods). Three categories of GO (Biological Process, Cellular Component and Molecular Function) were studied to find correlation with polyadenylation configuration. For each GO term in one species, a P-value from Fisher's exact test is provided, which indicates the significance of the association between this GO term and polyadenylation configuration, i.e. the lower the P-value the more significant the association is. Multiple testing adjustment using the Benjamanini and Hochberg method was applied to the selection of significant GO terms. The numbers of genes in three polyadenylation configurations, i.e. type I, II and III, are listed in parentheses. For example, (171, 37, 3) means that 171 type I genes, 37 type II genes and 3 type III genes.
Characteristics of poly(A) sites of different types
To further characterize poly(A) sites in genes with different poly(A) configuration, we classified poly(A) sites according to their location in a gene (Figure 1B). A poly(A) site can be one of the following nine types: 1S, the single poly(A) site in a type I gene; 2F, the 5′-most poly(A) site in a type II gene; 2L, the 3′-most poly(A) site in a type II gene; 2M, a middle poly(A) site between 2F and 2L in a type II gene, if it has more than two sites; 3U, a poly(A) site located upstream of the 3′-most exon in a type III gene, either in the intronic region or exonic region; 3F, the first site in the 3′-most exon of a type III gene; 3L, the 3′-most site in a type III gene; 3M, a middle poly(A) site between 3F and 3L, if there are more than two sites in the 3′-most exon; and 3S, the single poly(A) site in the 3′-most exon of a type III gene.
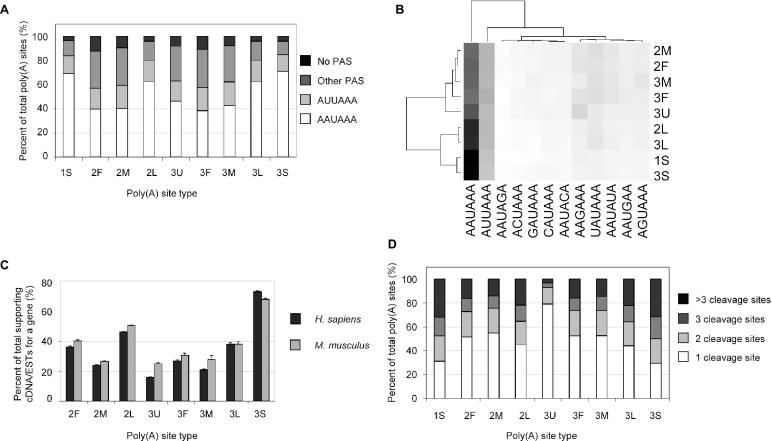
We studied the −40 to −1 region of all types of poly(A) sites for the usage of 12 types of PAS (16). As shown in Figure 5A, poly(A) sites of different types use different PAS hexamers to various extent. For constitutive poly(A) sites (1S), ∼70% of them have AAUAAA, 15% have AUUAAA, 12% have other 10 types of PAS hexamers and ∼4% of them do not have any known PAS hexamers. Almost an identical pattern is observed for 3S. Similar usage of PAS is also observed between 2F and 3F, between 2M and 3M, and between 2L and 3L. Overall, nine types of poly(A) sites appear to fall into two groups according to the usage of PAS hexamers. One group includes 1S, 2L, 3L and 3S. They tend to use AAUAAA (>60%) more than other types of PAS hexamer. The other group includes 2F, 2M, 3F, 3M and 3U, and <50% of the sites of each type contain AAUAAA, and >35% use PAS hexamers other than AAUAAA and AUUAAA. Interestingly, the percentage of sites containing AUUAAA does not vary much (14–19%) between all types of poly(A) sites, compared to AAUAAA (38–70%). This grouping is also supported by cluster analysis using the distribution of PAS hexamers in different types of poly(A) sites (Figure 5B). PAS hexamers can also be grouped by a similar manner. Interestingly, AAGAAA seems to be present in the 3U type more than other non-AUUAAA variants. Indeed, if we set the total number of poly(A) site for each PAS hexamer to 100%, we can see a more conspicuous presence of AAGAAA in 3U (data not shown).
Figure 5.
Characteristics of different types of poly(A) sites. (A) Association of various PAS hexamers with different types of poly(A) sites [for detailed definition of nine types of poly(A) sites see Figure 1B and Results]. (B) Cluster analysis of PAS hexamers and poly(A) types. The grayscale heat map represents the percentages of usage of PAS hexamers in different poly(A) types, with the sum of all values for each poly(A) type set to 100%. The shade of a cell indicates its value, with darker ones corresponding to higher values. Two-way hierarchical clustering was conducted using Euclidean distance as the metric. (C) Percentage of the number of supporting cDNA/EST sequences for different types of poly(A) sites. The total number of supporting cDNA/EST sequences for a gene is set to 100%. (D) Distribution of the number of cleavage sites per poly(A) site for different types of poly(A) site. Different shades are used to represent the number of cleavage sites per poly(A) site.
This grouping is in agreement with the notion that the 3′-most poly(A) sites usually are the ‘strongest’ site among all sites in a gene (28). Further supporting this is the number of cDNA/EST sequences (Figure 5C). The 3′-most poly(A) site of a gene, such as 2L, 3L and 3S, has a greater number of supporting cDNA/EST sequences than other types of poly(A) sites. The difference is expected to be even larger if only non-normalized cDNA libraries are used. Owing to the correlation between the number of supporting cDNA/ESTs and the number of cleavage sites, a similar pattern is observed, as expected, when we investigated the number of cleavage sites per poly(A) site for different types of poly(A) sites (Figure 5D). Approximately 70% of 1S and 3S poly(A) sites have more than one cleavage site; ∼55% for 2L and 3L; 45–49% for 2F, 2M, 3F and 3M; and only ∼20% for 3U.
This result indicates that the predominant form of an mRNA sequence is usually the longest one, generated by the 3′-most poly(A) site, and alternative polyadenylation is employed to shorten the mRNA. The exact implications of this mode of regulation may differ from gene to gene as sequences in the regulated region may contain cis elements involved in various aspects of RNA metabolism, such as RNA localization, translation and RNA stability.
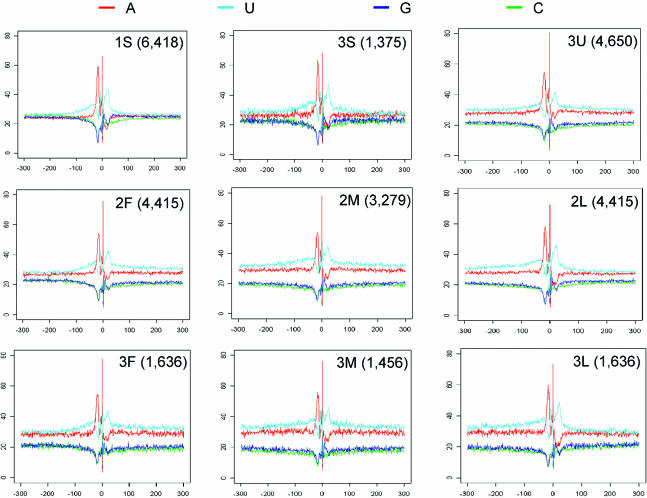
We next examined the nucleotide composition of the genomic sequence of poly(A) sites. For each poly(A) site, we selected terminal sequences spanning −300 to +300 nt surrounding the cleavage site (Figure 6). For poly(A) sites of all types, the −100 to +100 region has different nucleotide composition than regions upstream (<−100) or downstream (>+100) of the cleavage site. Invariably, within this window, it is U-rich, as reported previously (28), with a peak at around +20, which is generally known as the CstF-binding sites. The region −40 to −10 is A-rich, which causes a drop in U content. This region coincides with the region containing PAS hexamers.
Figure 6.
Nucleotide composition of human terminal sequences. Human terminal sequences containing nine types of poly(A) sites are plotted. The poly(A) site type is marked in each graph, and the number of sequences used for each graph is shown in parentheses. The y-axis for each graph is the percentage of a nucleotide (%) and the x-axis is the genomic location (nt) relative to the poly(A) site. See Figure 1B and Results for detailed definitions of nine poly(A) site types.
In spite of similarities in −100 to +100 region, there are conspicuous differences between poly(A) sites of different types. Poly(A) sites of 1S and 3S types have similar contents of GC and AU in regions upstream of −100 and downstream of +100, whereas other types of poly(A) sites have higher content of AU than GC in these regions. However, for the first poly(A) sites in the 3′-most exon (2F and 3F), the difference between AU and GC contents in the upstream region (−300 to −100) is less than that in the downstream region (+100 to +300). Interestingly, for the 3′-most poly(A) sites (2L and 3L), the downstream region appear to have smaller difference between AU and GC contents than that of upstream regions. In other words, the upstream region of the 5′-most alternative poly(A) site and the downstream region of the 3′-most alternative poly(A) site resemble the upstream and downstream regions of constitutive sites. In contrast, 3U, 2M and 3M poly(A) sites have similar nucleotide compositions in the sequence upstream of −100 and the sequence downstream of +100, and both of them are AU-rich. This AU richness is also discernible in mouse poly(A) sites, but to a lesser degree (Supplementary Figure 5).
DISCUSSION
We surveyed a large number of human and mouse genes for poly(A) sites. Strikingly, we found ∼54% human genes and 32% of mouse genes have multiple alternative poly(A) sites. The conservation of polyadenylation configuration between human and mouse orthologs was found to be statistically significant. These two numbers are higher than the 29% obtained in a previous study for human genes (16). We think this is mainly attributable to two factors (1). In this study, we used far more cDNA/ESTs. In Beaudoing and colleagues' study, they used 157 775 poly(A) tail containing ESTs for 8775 UTR sequences. After sequence clean-up, which eliminated >23% sequences, on average a UTR sequence was supported by <14 ESTs. In this study, we used 396 908 poly(A) tail containing cDNA/ESTs (using criteria designed to discard problematic cDNA/ESTs both at the genome alignment step and the poly(A) tail identification step; see Materials and Methods) for 13 942 human genes, giving rise to about 28 supporting cDNA/ESTs per gene. Given the difference between human and mouse numbers (54% versus 32%), which is also due to the disparity in the number of supporting cDNA/EST per gene, the difference between the numbers of identified poly(A) sites in these two studies is not surprising. However, we think that most additional poly(A) sites we identified are probably weak sites that are elusive with small numbers of cDNA/ESTs (2). Beaudoing and colleagues took a UTR-centered approach, where ESTs were mapped to UTR sequences. UTRs were used to represent mRNAs in order to find the real 3′ end of an mRNA sequence. Therefore, Beaudoing and colleagues' study in effect focused on alternative polyadenylation happening in the 3′-most exon, since the majority of 3′-UTRs are located in the 3′-most exon (data not shown). Here, we used a gene-centered approach, where cDNA/ESTs were mapped to genomes, which allowed us to study alternative polyadenylation coupled with splicing. In fact, if we do not consider poly(A) sites located upstream of the 3′-most exon, the percentage of genes having alternative polyadenylation will drop by 10% to about 44%. Nevertheless, both studies found similar percentages of usage of PAS hexamers AAUAAA (58.2% in Beaudoing et al. study and 53.2% in this study) and AUUAAA (14.9% in their study and 17% in this study). But the percentages of poly(A) sites containing other types of PAS hexamers and no PAS hexamer differ between two studies. In Beaudoing et al.'s work, about 15% poly(A) sites contain one of the 10 PAS hexamer variants, and ∼22% poly(A) sites do not have any PAS hexamer. These two numbers are ∼22% and ∼8% respectively in our study. The difference is because that in our study, we required more than one cDNA/EST sequence if there are no PAS hexamers found.
Our study indicates that alternative polyadenylation is widespread in both humans and mice. In addition, a large number of human and mouse orthologs have conserved polyadenylation configuration highlighting its importance in producing variable gene products, i.e. mRNAs and/or proteins. Some groups of genes are found disproportionately associated with certain types of poly(A) configuration. While genes encoding extracellular proteins often have single poly(A) sites, and genes encoding intracellular proteins tend to have alternative poly(A) sites, the biological implications have yet to be elucidated. Interestingly, genes encoding RNA binding proteins tend to have alternative poly(A) sites and are found disproportionately associated with the type III polyadenylation configuration, i.e. alternative polyadenylation coupled with splicing.
Our observation that there are a large number of poly(A) sites located upstream of the 3′-most exon, i.e. 3U type poly(A) sites, raises an intriguing question as to the role of this type of alternative polyadenylation in gene regulation. The biological consequences of this include change of protein sequence, exemplified by IgM genes, or non-stop mRNA decay if the resulting transcript lacks a stop codon (37). An extensive study is needed to delineate the significance of mRNA polyadenylation upstream of the 3′-most exon. Interestingly, the AAGAAA PAS hexamer was found disproportionately associated with 3U type poly(A) sites. By an exhaustive hexamer search, AAGAAA as a PAS was found to be statistically significant (16). However, previous biochemical works have shown that mutating AAUAAA to AAGAAA could abrogate polyadenylation (22,38). AAGAAA was also shown to be an exonic splicing enhancer (39). It is possible that this element can function in both processes. Based on current knowledge of the relationship between splicing and polyadenylation, an attractive model is that there is competition between splicing factors and polyadenylation factors for binding to AAGAAA. If splicing factors can compete favorably with polyadenylation factors, a splicing will occur which also prevents adjacent polyadenylation. Likewise, if polyadenylation factors are successful in binding, polyadenylation will take place instead of splicing. Since the distance between AAGAAA to the cleavage site is shorter than that from other types of PAS hexamers (Supplementary Material), we think the polyadenylation machinery involved may not be identical to the one involved in the polyadenylation process in the 3′-most exon. However, this hypothesis will require considerable experimental evidence to assess its validity.
Heterogeneity of mRNA cleavage was noted experimentally in previous studies for individual genes (15,22,36,40). Here, using a large-scale survey, we have demonstrated that ∼51% human and 47% mouse poly(A) sites have more than one cleavage site. While the heterogeneity is probably not caused by the presence of multiple PAS hexamers, there appears to be a correlation between the number of supporting cDNA/ESTs and the number of cleavage sites. This indicates that cleavage site determination could be stochastic, albeit there might be preferences as to the exact cleavage position to choose, as shown before in biochemical assays (36). However, the involvement of other unidentified cis elements in the poly(A) site in determining cleavage sites cannot be ruled out. Nevertheless, this heterogeneity indicates the imprecise nature of RNA cleavage carried out by polyadenylation enzyme complex. In fact, it was suggested that the interaction between the basal polyadenylation machinery and the mRNA may provide some ‘opportunities’ for flexibility, regulation and heterogeneity (15,22). On the other hand, the quality of cDNA/EST sequence also complicates the situation, as errors occur more often at the ends of sequences. However, we do not expect this to be a major factor since an erroneous sequence would have to perfectly match the genome sequence to be considered in our study.
Our data showed that different types of poly(A) sites are surrounded by sequences with distinct nucleotide compositions. In general, the −100 to +100 sequence shows nucleotide bias for A and U. However, alternative poly(A) sites located between other poly(A) sites show this high AU content beyond the −100 to +100 region. This is unlikely to be caused by adjacent poly(A) sites since the median distance between alternative poly(A) sites in the 3′-most exon is about 288 nt. Also, when we used terminal sequences containing only one poly(A) site (no other poly(A) sites located within 300 nt upstream and downstream), we still saw this pattern (data not shown). On the other hand, protein coding sequence, while contributing to the nucleotide composition of terminal sequences, does not seem to change the overall trend (Supplementary Figure 6). This is partly because the median value of the distance between the stop codon and the first downstream poly(A) site is 324 nt. The unique AU-rich environment offers an opportunity for harboring cis elements rich in A and U. A notable example is the specific AU-rich elements known as AREs, which are responsible for targeting many mammalian mRNAs for rapid decay (41,42). Numerous proteins have been identified which bind to these elements. In some cases the protein–RNA interactions appear to stabilize the mRNA (43–45), while in other cases the interactions serve to destabilize the mRNA (46). The fact that an AU-rich region is associated with alternative poly(A) sites indicate that alternative polyadenylation can adeptly regulate mRNA stability. However, other cis elements involved in other aspects of mRNA metabolism can also be located in this AU-rich region, and thus are subject to alternative regulation by polyadenylation.
SUPPLEMENTARY MATERIAL
Supplementary Material is available at NAR Online.
Supplementary Material
Acknowledgments
We thank Melissa Rogers, Jeffrey Wilusz, and members of B.T. and C.S.L. labs for inspirational discussions and helpful suggestions, and three anonymous reviewers for many constructive comments. C.S.L. was supported by grants from the American Cancer Society RPG-00-265-01-GMC, Arthritis Investigator Award 2AI-LUT-A-5, NIH 5R03-HD042464-02 and NSF award MCB-0426195. Funding to pay the open access publication charges for this article was provided by UMDNJ.
REFERENCES
- 1.Lewis J.D., Gunderson S.I., Mattaj I.W. The influence of 5′ and 3′ end structures on pre-mRNA metabolism. J. Cell. Sci. Suppl. 1995;19:13–19. doi: 10.1242/jcs.1995.supplement_19.2. [DOI] [PubMed] [Google Scholar]
- 2.Jacobson A., Peltz S.W. Interrelationships of the pathways of mRNA decay and translation in eukaryotic cells. Annu. Rev. Biochem. 1996;65:693–739. doi: 10.1146/annurev.bi.65.070196.003401. [DOI] [PubMed] [Google Scholar]
- 3.Wickens M., Anderson P., Jackson R.J. Life and death in the cytoplasm: messages from the 3′ end. Curr. Opin. Genet. Dev. 1997;7:220–232. doi: 10.1016/s0959-437x(97)80132-3. [DOI] [PubMed] [Google Scholar]
- 4.Maniatis T., Reed R. An extensive network of coupling among gene expression machines. Nature. 2002;416:499–506. doi: 10.1038/416499a. [DOI] [PubMed] [Google Scholar]
- 5.Proudfoot N. New perspectives on connecting messenger RNA 3′ end formation to transcription. Curr. Opin. Cell. Biol. 2004;16:272–278. doi: 10.1016/j.ceb.2004.03.007. [DOI] [PubMed] [Google Scholar]
- 6.Neugebauer K.M. On the importance of being co-transcriptional. J. Cell. Sci. 2002;115:3865–3871. doi: 10.1242/jcs.00073. [DOI] [PubMed] [Google Scholar]
- 7.Calvo O., Manley J.L. Strange bedfellows: polyadenylation factors at the promoter. Genes Dev. 2003;17:1321–1327. doi: 10.1101/gad.1093603. [DOI] [PubMed] [Google Scholar]
- 8.Wahle E., Kuhn U. The mechanism of 3′ cleavage and polyadenylation of eukaryotic pre-mRNA. Prog. Nucleic Acid Res. Mol. Biol. 1997;57:41–71. doi: 10.1016/s0079-6603(08)60277-9. [DOI] [PubMed] [Google Scholar]
- 9.Keller W., Minvielle-Sebastia L. A comparison of mammalian and yeast pre-mRNA 3′-end processing. Curr. Opin. Cell. Biol. 1997;9:329–336. doi: 10.1016/s0955-0674(97)80004-x. [DOI] [PubMed] [Google Scholar]
- 10.Colgan D.F., Manley J.L. Mechanism and regulation of mRNA polyadenylation. Genes Dev. 1997;11:2755–2766. doi: 10.1101/gad.11.21.2755. [DOI] [PubMed] [Google Scholar]
- 11.Edmonds M. A history of poly A sequences: from formation to factors to function. Prog. Nucleic Acid Res. Mol. Biol. 2002;71:285–389. doi: 10.1016/s0079-6603(02)71046-5. [DOI] [PubMed] [Google Scholar]
- 12.Zhao J., Hyman L., Moore C. Formation of mRNA 3′ ends in eukaryotes: mechanism, regulation, and interrelationships with other steps in mRNA synthesis. Microbiol. Mol. Biol. Rev. 1999;63:405–445. doi: 10.1128/mmbr.63.2.405-445.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Brown C.E., Sachs A.B. Poly(A) tail length control in Saccharomyces cerevisiae occurs by message-specific deadenylation. Mol. Cell. Biol. 1998;18:6548–6559. doi: 10.1128/mcb.18.11.6548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Beaudoing E., Gautheret D. Identification of alternate polyadenylation sites and analysis of their tissue distribution using EST data. Genome Res. 2001;11:1520–1526. doi: 10.1101/gr.190501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pauws E., van Kampen A.H., van de Graaf S.A., de Vijlder J.J., Ris-Stalpers C. Heterogeneity in polyadenylation cleavage sites in mammalian mRNA sequences: implications for SAGE analysis. Nucleic Acids Res. 2001;29:1690–1694. doi: 10.1093/nar/29.8.1690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Beaudoing E., Freier S., Wyatt J.R., Claverie J.M., Gautheret D. Patterns of variant polyadenylation signal usage in human genes. Genome Res. 2000;10:1001–1010. doi: 10.1101/gr.10.7.1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Edwalds-Gilbert G., Veraldi K.L., Milcarek C. Alternative poly(A) site selection in complex transcription units: means to an end? Nucleic Acids Res. 1997;25:2547–2561. doi: 10.1093/nar/25.13.2547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Takagaki Y., Seipelt R.L., Peterson M.L., Manley J.L. The polyadenylation factor CstF-64 regulates alternative processing of IgM heavy chain pre-mRNA during B cell differentiation. Cell. 1996;87:941–952. doi: 10.1016/s0092-8674(00)82000-0. [DOI] [PubMed] [Google Scholar]
- 19.Graber J.H., Cantor C.R., Mohr S.C., Smith T.F. In silico detection of control signals: mRNA 3′-end-processing sequences in diverse species. Proc. Natl Acad. Sci. USA. 1999;96:14055–14060. doi: 10.1073/pnas.96.24.14055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.MacDonald C.C., Redondo J.L. Reexamining the polyadenylation signal: were we wrong about AAUAAA? Mol. Cell. Endocrinol. 2002;190:1–8. doi: 10.1016/s0303-7207(02)00044-8. [DOI] [PubMed] [Google Scholar]
- 21.Zarudnaya M.I., Kolomiets I.M., Potyahaylo A.L., Hovorun D.M. Downstream elements of mammalian pre-mRNA polyadenylation signals: primary, secondary and higher-order structures. Nucleic Acids Res. 2003;31:1375–1386. doi: 10.1093/nar/gkg241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Natalizio B.J., Muniz L.C., Arhin G.K., Wilusz J., Lutz C.S. Upstream elements present in the 3′-untranslated region of collagen genes influence the processing efficiency of overlapping polyadenylation signals. J. Biol. Chem. 2002;277:42733–42740. doi: 10.1074/jbc.M208070200. [DOI] [PubMed] [Google Scholar]
- 23.Chen F., Wilusz J. Auxiliary downstream elements are required for efficient polyadenylation of mammalian pre-mRNAs. Nucleic Acids Res. 1998;26:2891–2898. doi: 10.1093/nar/26.12.2891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Arhin G.K., Boots M., Bagga P.S., Milcarek C., Wilusz J. Downstream sequence elements with different affinities for the hnRNP H/H′ protein influence the processing efficiency of mammalian polyadenylation signals. Nucleic Acids Res. 2002;30:1842–1850. doi: 10.1093/nar/30.8.1842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schek N., Cooke C., Alwine J.C. Definition of the upstream efficiency element of the simian virus 40 late polyadenylation signal by using in vitro analyses. Mol. Cell. Biol. 1992;12:5386–5393. doi: 10.1128/mcb.12.12.5386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gilmartin G.M., Fleming E.S., Oetjen J., Graveley B.R. CPSF recognition of an HIV-1 mRNA 3′-processing enhancer: multiple sequence contacts involved in poly(A) site definition. Genes Dev. 1995;9:72–83. doi: 10.1101/gad.9.1.72. [DOI] [PubMed] [Google Scholar]
- 27.Moreira A., Takagaki Y., Brackenridge S., Wollerton M., Manley J.L., Proudfoot N.J. The upstream sequence element of the C2 complement poly(A) signal activates mRNA 3′ end formation by two distinct mechanisms. Genes Dev. 1998;12:2522–2534. doi: 10.1101/gad.12.16.2522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Legendre M., Gautheret D. Sequence determinants in human polyadenylation site selection. BMC Genomics. 2003;4:7. doi: 10.1186/1471-2164-4-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tabaska J.E., Zhang M.Q. Detection of polyadenylation signals in human DNA sequences. Gene. 1999;231:77–86. doi: 10.1016/s0378-1119(99)00104-3. [DOI] [PubMed] [Google Scholar]
- 30.Zhang H. Alignment of BLAST high-scoring segment pairs based on the longest increasing subsequence algorithm. Bioinformatics. 2003;19:1391–1396. doi: 10.1093/bioinformatics/btg168. [DOI] [PubMed] [Google Scholar]
- 31.Burset M., Seledtsov I.A., Solovyev V.V. SpliceDB: database of canonical and non-canonical mammalian splice sites. Nucleic Acids Res. 2001;29:255–259. doi: 10.1093/nar/29.1.255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kent W.J. BLAT—the BLAST-like alignment tool. Genome Res. 2002;12:656–664. doi: 10.1101/gr.229202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kan Z., Gish W., Rouchka E., Glasscock J., States D. UTR reconstruction and analysis using genomically aligned EST sequences. Proc. Int. Conf. Intell. Syst. Mol. Biol. 2000;8:218–227. [PubMed] [Google Scholar]
- 34.Benjamini Y., Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc. Ser. B. 1995;57:289–300. [Google Scholar]
- 35.Venables W.N., Ripley B.D. Modern Applied Statistics with S. New York, NY: Springer; 2002. [Google Scholar]
- 36.Chen F., MacDonald C.C., Wilusz J. Cleavage site determinants in the mammalian polyadenylation signal. Nucleic Acids Res. 1995;23:2614–2620. doi: 10.1093/nar/23.14.2614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Frischmeyer P.A., van Hoof A., O'Donnell K., Guerrerio A.L., Parker R., Dietz H.C. An mRNA surveillance mechanism that eliminates transcripts lacking termination codons. Science. 2002;295:2258–2261. doi: 10.1126/science.1067338. [DOI] [PubMed] [Google Scholar]
- 38.Wilusz J., Shenk T. A 64 kD nuclear protein binds to RNA segments that include the AAUAAA polyadenylation motif. Cell. 1988;52:221–228. doi: 10.1016/0092-8674(88)90510-7. [DOI] [PubMed] [Google Scholar]
- 39.Fairbrother W.G., Yeh R.F., Sharp P.A., Burge C.B. Predictive identification of exonic splicing enhancers in human genes. Science. 2002;297:1007–1013. doi: 10.1126/science.1073774. [DOI] [PubMed] [Google Scholar]
- 40.Sheets M.D., Ogg S.C., Wickens M.P. Point mutations in AAUAAA and the poly(A) addition site: effects on the accuracy and efficiency of cleavage and polyadenylation in vitro. Nucleic Acids Res. 1990;18:5799–5805. doi: 10.1093/nar/18.19.5799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shaw G., Kamen R. A conserved AU sequence from the 3′ untranslated region of GM-CSF mRNA mediates selective mRNA degradation. Cell. 1986;46:659–667. doi: 10.1016/0092-8674(86)90341-7. [DOI] [PubMed] [Google Scholar]
- 42.Chen C.Y., Shyu A.B. AU-rich elements: characterization and importance in mRNA degradation. Trends Biochem. Sci. 1995;20:465–470. doi: 10.1016/s0968-0004(00)89102-1. [DOI] [PubMed] [Google Scholar]
- 43.Ford L.P., Watson J., Keene J.D., Wilusz J. ELAV proteins stabilize deadenylated intermediates in a novel in vitro mRNA deadenylation/degradation system. Genes Dev. 1999;13:188–201. doi: 10.1101/gad.13.2.188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Fan X.C., Steitz J.A. Overexpression of HuR, a nuclear-cytoplasmic shuttling protein, increases the in vivo stability of ARE-containing mRNAs. EMBO J. 1998;17:3448–3460. doi: 10.1093/emboj/17.12.3448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Peng S.S., Chen C.Y., Xu N., Shyu A.B. RNA stabilization by the AU-rich element binding protein, HuR, an ELAV protein. EMBO J. 1998;17:3461–3470. doi: 10.1093/emboj/17.12.3461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Loflin P., Chen C.Y., Shyu A.B. Unraveling a cytoplasmic role for hnRNP D in the in vivo mRNA destabilization directed by the AU-rich element. Genes Dev. 1999;13:1884–1897. doi: 10.1101/gad.13.14.1884. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.