Abstract
Using NMR methods, we have resolved the i-motif structures formed by d(AACCCC) and by d(CCCCAA), two versions of the DNA sequence repeated in the telomeric regions of the C-rich strand of tetrahymena chromosomes. Both oligonucleotides form fully symmetrical i-motif tetramers built by intercalation of two hemiprotonated duplexes containing four C•C+ pairs. The structures are extremely stable. In the tetramer of d(AACCCC), the outermost C•C+ pairs are formed by the cytidines of the 5′ ends of the cytidine tracts. A2 forms an A2•A2 (H6trans–N7) pair stacked to C3•C3+ and cross-strand stacked to A1. At 0°C, the lifetimes of the hemiprotonated pairs range from 1 ms for the outermost pair to ∼1 h for the innermost pairs. The tetramer of d(CCCCAA) adopts two distinct intercalation topologies in slow conformational exchange. One, whose outermost C•C+ pairs are built by the cytidines of the 5′ end and the other by those of the 3′ end. In both topologies, the adenosine bases are fairly well stacked to the adjacent C•C+ pairs. They are not paired but form symmetrical pseudo-pairs with their H6cis amino proton and N1 nitrogen pointing towards each other.
INTRODUCTION
Repetitive DNA sequences, dispersed into the eukaryote genome can form non-Watson–Crick structures providing potential specific recognition targets. In the chromosome ends, the telomeric DNA sequences include multiple repeats of short G-rich sequences on one strand and the complementary C-rich sequences on the opposite strand. These sequences, essential for chromosome maintenance, are highly conserved (1).
In the past 10 years, NMR and X-ray studies have established that the two complementary strands of telomeric regions can exist as four-stranded structures [for a review see (2,3)]. The G-rich strand can associate into a G-quadruplex built by stacked planar G-tetrads. The C-rich strand can form an i-motif, a structure composed of two parallel hemiprotonated duplexes intercalated into each other in a head-to-tail orientation (4). G-quadruplex and i-motif may be formed by association of four strands, by two hairpins containing two G- or C-rich stretches or by a folded single strand containing four G or C stretches as shown for the d(AGGGTTAGGGTTAGGGTTAGGG) (5) and (CCCTAACCCTAACCCTAACCCT) (6) oligonucleotides containing the sequences repeated at the ends of vertebrate chromosomes. The stability of monomeric i-motif structures at physiological pH and the discovery of specific proteic ligands (7,8) suggest a biological function.
The repeated sequences of the tetrahymena chromosome ends are d(GGGGTT) and d(AACCCC). Using NMR methods, we have investigated the i-motif structures of two versions of the C-rich repeat: d(AACCCC) and d(CCCCAA). The stoichiometry and the stability constant of the structures were determined by gel filtration chromatography. The i-motif of d(AACCCC) adopts a single intercalation topology. The quality of its NMR spectrum allowed the measure of the lifetime of the four C•C+ pairs intercalated in the i-motif core. The high definition structure, built on the base of distance restraints derived from NOESY experiments shows interesting differences with the crystal structure (9). d(CCCCAA) associates also into an i-motif but its NMR spectrum shows the formation of two distinct intercalation topologies of the C•C+ pairs. A possible interaction of [d(CCCCAA)]4 and [d(AACCCC)]4 with the G-quartet of d(TTGGGG) was examined in pH conditions selected to favor the i-motif stability.
Nomenclature
The nucleic acid bases have two faces. One, that will be designed as the black face, is oriented towards the 5′ direction and the white face that is oriented in the 3′ direction. In the i-motif, the intercalation in a head-to-tail orientation of two hemiprotonated duplexes brings in mutual contact the black faces of each other and the white faces of each other's bases, and results in alternating black and white steps.
MATERIALS AND METHODS
NMR samples
The oligonucleotides were synthesized on a 15 μM scale, purified by chromatography on an anionic DEAE column as described previously (10) and extensively dialyzed against 10 mM NaCl solutions and finally against water. The strand concentration of the NMR samples, 4–8 mM in the structural studies, and ∼1 mM for proton exchange measurements was determined from the absorbance measured at neutral pH using the A260 values of 52 400 for d(AACCCC), 52 900 for d(CCCCAA) and 54 100 for d(TTGGGG). The samples were dissolved either in H2O/2H2O (9:1, v/v) or in 99.98% 2H2O after repeated lyophilization and dissolution in 2H2O. The NMR samples contained 0.1 mM dimethyl silapentane sulphonate for chemical shift reference and 1 mM ethylene diamine tetra-acetic acid. Unless otherwise stated, the sample pH was adjusted close to the cytidine pK (pKN3 = 4.4).
Multimer stoichiometry
The multimer stoichiometry was determined by gel filtration chromatography in 0.3 M NaCl, pH 4.5 solutions on a Synchropack GPC-100 column (flow rate 2 ml/min). The eluted oligomers were detected by UV absorbance. The column was calibrated with 12 oligonucleotides containing from 3 to 76 residues. In order to reach the monomer–tetramer equilibrium, the samples were incubated during one week at room temperature before chromatography. The multimer stoichiometry was determined from the elution time and from the concentration dependence of the multimer–monomer ratio (11).
NMR methods
The NMR experiments were performed on a 500 MHz Varian Inova spectrometer equipped with a penta probe.
In H2O solution, the jump and return (JR) sequence with maximum sensitivity at 13.5 p.p.m. was used for water suppression (12). The spectral width and the acquisition time were typically 12 kHz and 0.17 s. The repetition rate was 1.5 and 2 s in the experiments performed at 0 and 15°C, respectively. In order to correct the frequency response to JR excitation, the NOESY spectra were multiplied by the ad hoc 1/sin function in the t2 dimension prior to cross-peaks measurements.
The 1H 2D experiments were performed with 4096 points in the first dimension and 256–512 t1 increments. In 2H2O the spectral width was 4.4 kHz, the acquisition time 0.46 s, and the repetition rate 3 s. The residual HDO signal was saturated by a low power 2 s pulse followed by a relaxation delay of 0.3 s allowing partial relaxation of the sugar protons at the water frequency. For NOESY experiments in 2H2O, the mixing times, tmix, was randomly varied between 0.95 tmix and 1.05 tmix in order to reduce the spurious contribution of zero-quantum coherence at short mixing time (13). The TOCSY experiments used 10 MLEV-17 repetitions (total time 15 ms) (14). In the 1H–31P hetero-TOCSY experiment (15) designed to connect together the sequentially adjacent residues of the i-motif of d(AACCCC), the proton spectral width was 1.6 kHz and the acquisition time 0.64 s. The spectral width in the 31P dimension was 1 kHz, corresponding to t1 incrementation up to 0.2 s (200 increments). The HMBC experiments in natural abundance (16) were performed at 15°C with spectral widths of 4.4 and 7 kHz in the 1H and 13C dimension, respectively. The t1 delay was incremented up to 0.11 s (128 increments). The acquisition time was 0.512 s and the repetition delay 3 s. The dephasing–rephasing τ delay was set to 50 ms. HMBC experiments performed with a strand concentration of 4 mM were typically 16–24 h long.
The 2D data were processed with the Felix 97.2 software. Apodization in both dimensions included a 1 Hz exponential broadening, a sine-bell with a phase shift of typically 60° and a Felix skew factor of 1. The remaining water signal was reduced using the time domain convolution function of the Felix software.
Structure determination
Proton identification and NMR determinants of the i-motif. The sugar protons of each residue were connected together by TOCSY cross-peaks and related to the base protons by the H1′–H6/H8 and H3′–H6/H8 NOESY intra-residue cross-peaks. The exchangeable protons of cytidines were identified by the H5-amino–imino NOESY cross-peaks. The H2 and H8 protons of each adenosine were matched together by HMBC experiments. The H2′ and H2″ protons were distinguished from each other by the stronger intensities of the H2″–H1′ and H2′–H3′ intra-residue cross-peaks on NOESY spectra at short mixing time.
The rules allowing the resolution of the intercalation topology of i-motif tetramers have already been described (10,17). The intercalation of two duplexes results in large distances between the covalently linked residues and in the quasi-absence of sequential NOE connectivities at short mixing time. The head-to-tail orientation of the duplexes generates two distinct steps between the stacked pairs (cf. Nomenclature in Introduction). Each step is characterized by specific short inter-residue 1H–1H distances corresponding to direct NOE cross-peaks providing redundant information to characterize the intercalation topology. At the white steps, the sugars of the residues on each side of the narrow grooves are connected by H1′–H2′/H2″ cross-peaks and the residues on each side of the wide grooves are related by amino-H2′/H2″ cross-peaks. At the black steps, the residues are connected across the narrow grooves by H1′–H4′ and H4′–H4′ cross-peaks. At both steps, the sugars on each side of the narrow groove are connected by H1′–H1′ cross-peaks and the stacked base pairs by imino–imino and inter-residues H5-amino proton cross-peaks.
Distance restraint and molecular dynamics. The structures were determined on the basis of the inter-proton distances obtained from the build-up of NOE cross-peaks measured at 0 and 15°C with mixing times of 30, 50, 70, 90 ms and 200 ms in 2H2O and of 50, 70, 90 and 150 ms in H2O.
The sugar glycosidic angles were determined by the H6/H8–H1′ and H6/H8–H3′ intra-residue distances and the sugar puckers by the H6/H8–H3′, H2″–H4′ and H1′–H4′ distances. The generation of distance restraints from the NOE cross-peak volumes (18) and the strategy used to overcome the ambiguities resulting from the symmetry of the structures have been described previously (10).
The structures were calculated on an INDIGO Silicon Graphics workstation using the simulated annealing method of the X-PLOR 3.851 program (19) with the standard harmonic potential for covalent geometry. Potential energy terms related to electrostatics were omitted. The force constants related to distance and to H-bonding restraints were set to 50 kcal mol−1 and 500 kcal mol−1 Å−2, respectively. The computation started with an initial model formed of four strands in an extended conformation. The energy of the initial structure was first minimized by five Powell cycles and the molecular dynamics computation was run for 1500 steps of 2 ps with an initial velocity corresponding to 2400 K. H-bonding energy was introduced during the cooling procedure to 300 K in step of 25 K, each step being followed by a 0.1 ps dynamics computation.
The inter-proton distances shorter than 4.7 Å on the computed conformers were systematically searched using the MOLMOL 2.4 software (20). Some conformers displayed short inter-proton distances between proton pairs that are not connected by NOE cross-peaks. The short inter-proton distances incompatible with the NOESY spectra were excluded in further calculations by repulsive constraints of 4.2 Å. This procedure was used only for well-resolved non-exchangeable protons.
Finally, the energy was minimized by 600 Powell cycles. The computed conformers were sorted according to the energy related to NOE violations. The 10 structures of lowest energy were selected for structural analysis. The pairwise r.m.s.d., the geometry parameters and SD values were computed using X-PLOR and home-made softwares. The structures were visualized using MOLMOL 2.4.
Imino proton exchange
The proton exchange formalism and methodology have been described extensively (21). Most of the imino proton exchange times were determined by magnetization transfer from water. Exchange times shorter than 10 ms were derived from the imino proton longitudinal relaxation rates and those longer than minutes were obtained from the deuteration rate of a 40 μl protonated sample diluted into 400 μl 2H2O.
RESULTS
Multimer stoichiometry
The elution profile of d(AACCCC) and d(CCCCAA) solutions on the gel-filtration chromatograms shows two components in proportions depending on pH and concentration. One corresponds to a monomer (six residues) and the other to a tetramer (24 residues). When the monomer–tetramer equilibrium is reached, the proportion of the tetrameric species increases with the power of four of the monomer concentration.
The tetramers are very stable: at room temperature, pH 4.5, one half of the oligonucleotides are in tetrameric structures when the strand concentration is 2.4 × 10−5 and 3.6 × 10−5 M3, respectively for d(CCCCAA) and d(AACCCC). The proportion of monomer is therefore negligible in the NMR experiments performed with strand concentrations in the millimolar range.
NMR spectra
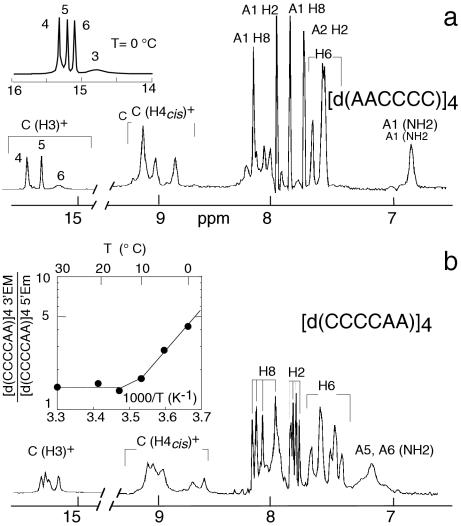
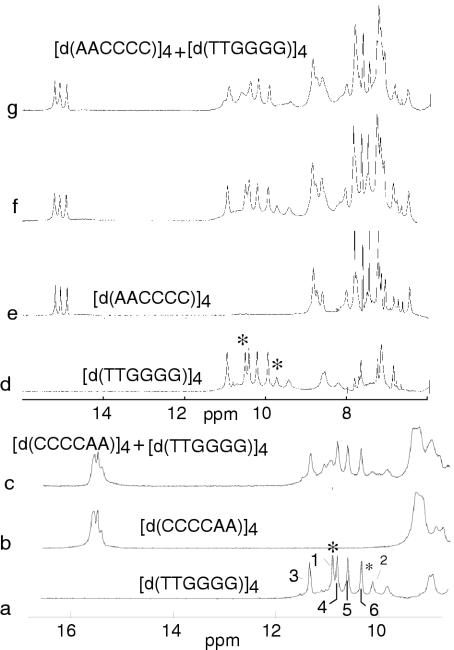
The proton spectra of the tetramers of d(AACCCC) and d(CCCCAA) are displayed in Figure 1. At −5°C, pH 4.5, the spectrum of [d(AACCCC)]4 exhibits three narrow C•C+ imino protons peaks (1515.5 p.p.m.) and a much broader one (14.8 p.p.m.) indicating the formation of four hemiprotonated pairs. In the aromatic region, between 7.7 and 8.2 p.p.m., four narrow NMR lines identified (cf. below) as those of the adenosine H8 and H2 protons show immediately that the tetramer adopts a single conformation fully symmetrical on the NMR time scale.
Figure 1.
Exchangeable and aromatic proton spectrum of d(AACCCC) and d(CCCCAA) at 25°C, pH 4.5. (a) The spectrum of [d(AACCCC)]4 shows a single peak for each proton indicating the formation of a single, fully symmetrical, structure. The imino protons of four C•C+ pairs are detected on the spectrum at −5°C (inset). (b) The spectrum of d(CCCCAA) shows four H8 and four H2 proton peaks and reveals an equilibrium between two i-motif species. The ratio of the two species (designed 5′EM and 3′Em) is displayed versus temperature in the inset.
In contrast, the spectrum of [d(CCCCAA)]4 exhibits two spin systems for each residues indicating two coexisting structures. The equilibrium between the two species is independent of pH between pH 3.8 and 5.5, and of the oligonucleotide concentration. The absence of exchange cross-peaks between the two spin systems establishes that the structures are in slow conformational exchange. The ratio of the two species measured versus temperature from the intensities of the H6–H5 cross-peaks of TOCSY spectra collected with a mixing time of 40 ms vary with temperature from 4.2 at 0°C to 1.5 at 15°C (Figure 1, inset). The minor (m) and the major (M) species of [d(CCCCAA)]4 will be henceforth designed as the 5′Em and 3′EM species (see below for justification of this designation) and the residues of the 5′Em species will be labeled by a star (Figure 2). Addition of NaCl shifts the equilibrium towards the 5′Em species with a time constant of several days.
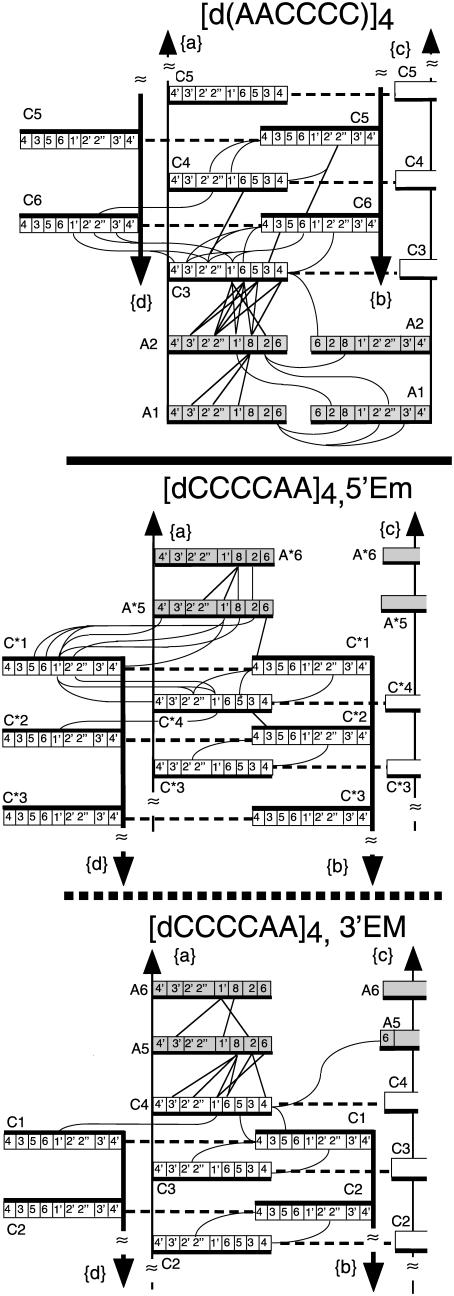
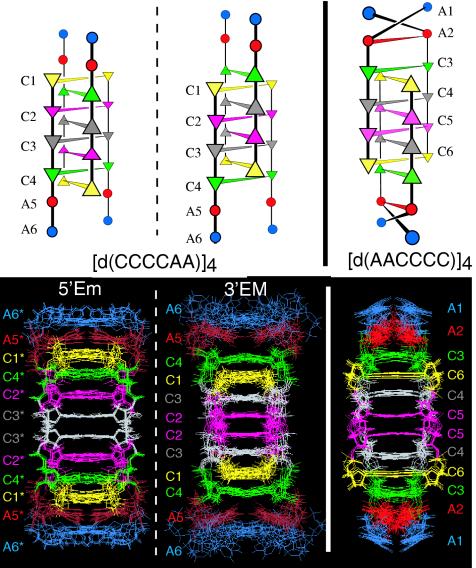
Figure 2.
Schematic representation of the inter-residue distances shorter than 4.7 Å derived from NOESY experiments in [d(AACCCC)]4 and in the 5′Em and 3′EM intercalation topologies of [d(CCCCAA)]4. One half of the symmetrical structures are shown and only the distances issuing from strand {a} are displayed. Owing to the structure symmetry, any strand is connected similarly by symmetry-related connectivities. The faces oriented in the 5′ direction are underlined by thick black lines. The dotted lines indicate the base-paired cytidines. The adenosines are shaded gray.
Proton identification and intercalation topology
[d(AACCCC)]4. The sugar, H6/H8 and H5 protons of the six spin systems of [d(AACCCC)]4 were identified as reported in Materials and Methods by the standard pattern of intra-residue connectivities of TOCSY spectra in 2H2O solution and of NOESY spectra in 2H2O and H2O solution. The H2 and the H8 proton of each adenosine were matched together by their connectivities to a common 13C4 detected on an HMBC spectrum in natural abundance. Starting from A1, the six residues were identified on an 1H–31P hetero-TOCSY experiment by the cross-peaks connecting the H3′ and the H4′ protons of sequentially adjacent residues to the same 31P. At last, the intercalation topology of [d(AACCCC)]4, A1A2C3C6C4C5C5C4C6C3A2A1, was characterized by the inter-residue amino-H2′/H2″ (Figure 4) and H1′/H1′ NOESY connectivities. At 0°C, the spectrum in H2O solution shows the NMR lines (8.21 and 7.64 p.p.m.) of two exchangeable protons connected by a strong cross-peak. They were assigned to the amino protons of A2 by their NOESY cross-peaks with C3, the adjacent cytidine. Both A2 amino protons are also connected to A2(H8) by NOESY cross-peaks whose intensities are inconsistent with the amino–H8 intra-residue distances (4.8 and 6.1 Å, respectively for the trans and cis amino protons). The largest cross-peak connects the 8.21 p.p.m. amino proton to A2(H8) and corresponds to a distance of 2.7 Å. Slow rotation of the A2 amino group and the downfield shift of the 8.21 p.p.m. proton are strong evidences for H-bonding. The short distance between the 8.21 p.p.m. amino proton and A2(H8) is characteristic of A2•A2 (H6trans–N7) base-pairing, and thus identify the 8.21 p.p.m. amino proton peak to A2(H6 trans). When the temperature is raised above 10°C, the A2 amino proton peaks broaden and disappear. Above 30°C, a new NMR line, around 7 p.p.m., indicates disruption of the inter-strand A2(H6 trans)–(N7)A2 bond and fast rotation of the A2(NH2) group. Another NMR line integrating for two exchangeable protons is observed between 0 and 35°C around 6.8 p.p.m. (Figure 1). It was assigned by its NOE connectivities with A1 and A2 to the amino protons of A1 in fast rotational exchange. The A2•A2 base pair was aligned on the adjacent C3•C3+ pair by 14 inter-residue NOE cross-peaks (Figure 2). A1 was aligned on pair A2•A2 by the distance restraints derived from five inter-residue NOE cross-peaks.
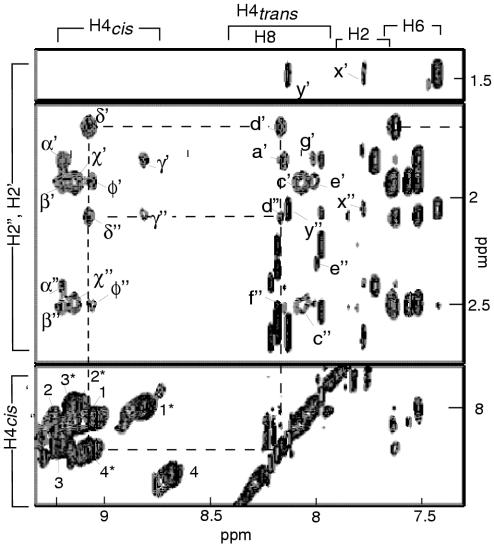
Figure 4.
Amino-H2′/H2″ inter-residue NOESY connectivities used to determine the intercalation of the C•C+ pairs in the 3′EM and 5′EM topologies of [d(CCCCAA)]4. The H4cis–H4trans intra-residue cross-peaks are labeled by the residue number. The residues of the 5′E topology are labeled with a star. α′: C3(H4cis)-C1(H2′); α′: C3(H4cis)-C1(H2″); a′ C1(H4trans)-C3(H2′); β′: inter-residue C2(H4cis)–C2(H2′); β″: inter-residue C2(H4cis)–C2(H2″); χ″ unresolved C3*(H4cis)–C2*(H2′) and C2*(H4cis)–C3*(H2′); χ″ unresolved C3*(H4cis)–C2*(H2″) and C2*(H4cis)–C3*(H2″); c′: unresolved C3*(H4trans)–C2*(H2′) and C2*(H4trans)–C3*(H2′); c″: unresolved C3*(H4trans)–C2*(H2″) and C2*(H4trans–C3*(H2″). δ′: C4*(H4cis)–C1*(H2′); δ″: C4*(H4cis)–C1*(H2″); d′ C4*(H4trans)–C1*(H2′); d″: C4*(H4trans)–C1*(H2″). φ′: C1(H4cis)–C3(H2′); φ″: C1(H4cis)–C3(H2″; γ″: C1(H4trans)-C3(H2″); e′ C3*(H4cis)-C2*(H2′), e′ C3*(H4cis)-C2*(H2″). y′ C4 (H2′)–A5(H8), y″ C4 (H2″)–A5(H8), x′ C4 (H2′)–A5(H2), y″ C4 (H2″)–A5(H2). The NOE connectivities between C4* amino protons and C1* H2′/H2″ protons are traced with dashed lines. T = 0°C, mixing time 150 ms.
[d(CCCCAA)]4. The six spin systems of the minor and major species of d(CCCCAA)]4 were assigned to the 3′EM or 5′EM species according to the intensity of their NMR lines on the spectrum at 0°C. Proton identification of the six spin systems of each species proceeded as described above. The H2 and H8 protons of each adenosine were connected together by an HMBC experiment (Figure 3). The terminal A6 residue of each species was identified as the adenosine that shows no connectivity to cytidine and A5 by its sequential NOE connectivities with A6 and with a single cytidine (Figure 2).
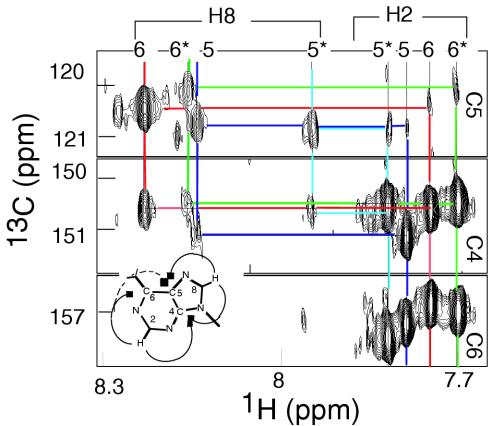
Figure 3.
Identification of the adenosine H2 and H8 protons in the two topologies of [d(CCCCAA)]4 by an HMBC experiment in natural abundance. The spectrum shows the cross-peaks of 13C5 with H8 (strong) and H2 protons (weak), of 13C4 with H8 (medium to weak) and H2 (strong) and of 13C6 with H2. The adenosine numbers are indicated. The residues of the 5′EM species are labeled with a star. T = 15°C, pH 4.5.
The reciprocal cross-peaks connecting the amino and the H2′/H2″ protons of the C•C+ pairs stacked by the faces oriented in the 3′ direction (white faces, Figure 2) provide a strategic information to resolve the intercalation topology of i-motif structures.
In the 3′EM species of [d(CCCCAA)]4, the C•C+ pair connected by NOE to A5 shows no amino-H2′/H2″ connectivity with the other cytidines (Figure 4). This indicates that its black face is oriented in the direction of the adjacent cytidine and therefore that its white face is oriented towards A5. This establishes that this pair is C4•C4+, the pair sequentially adjacent to A5. Starting from C4•C4+, the stacking order of 3′EM, A6A5C4C1C3C2C2C3C1C4A5A6, was determined by the inter-residue amino-H2′/H2″ cross-peaks connecting C1•C1+ to C3•C3+ and the two C2•C2+ symmetry related pairs (Figure 4).
In the 5′Em species, the four C•C+ pairs are connected two by two by amino-H2′/H2″ cross-peaks (Figures 2 and 4). Using the same line of arguments as above, this shows that all the stacked C•C+ pairs have contacting white faces. The black face of the outermost pair is therefore oriented towards A5* and this establishes that it is intercalated between A5* and C4*. The stacking order of the C•C+ pairs of 5′Em, A6*A5*C1*C4*C2*C3*C3*C2*C4*C1*A5*A6*, was read from the amino-H2′/H2″cross-peaks connecting C1*•C1*+ to C4*•C4*+ and C2*•C2*+ to C3*•C3*+ (Figure 4).
As shown in Figure 2, the topology of each species, was confirmed by redundant inter-residue NOE connecting the sugar protons of cytidines across the narrow groove (cross-peaks H1′–H1′, H1′–H2′/H2″) and the base protons of stacked C•C+ pairs.
The two tetramers of d(CCCCAA) differ by the intercalation topology of the C•C+ pairs. The outermost C•C+ pairs of structure 3′EM are from the 3′ end cytidines whereas those of the 5′Em species are from the 5′ end.
Proton exchange and base-pair opening kinetics
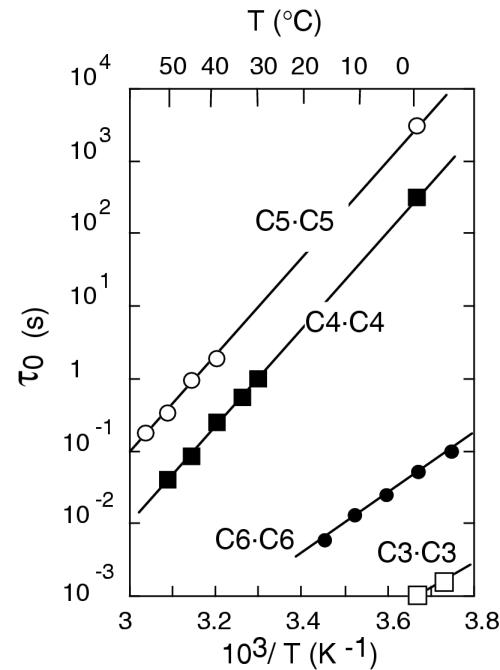
The excellent resolution of the imino proton spectrum of [d(AACCCC)]4 allowed the determination of the lifetime of the four intercalated C•C+ pairs. As it is always observed, the exchange rate with water of the imino proton of C•C+ pairs is limited by the base-pair opening rate. Hence, the imino proton exchange times may be identified with the base-pair lifetimes (22). At 0°C, the lifetime of the C•C+ pairs increases from 1 ms for the outermost C3•C3+ pair to 3400 s for the central C5•C5+ pair (Figure 5). The Arrhenius plot of the lifetimes yields activation energies of 130 kJ/mol for C4•C4+ and C5•C5+ and of 79 kJ/mol for C6•C6+.
Figure 5.
Base-pair lifetimes versus temperature of the four C•C+ pairs of [d(AACCCC)]4.
Real time proton exchange experiments performed at 0°C, pH 4.5 show that all the exposed cytidine amino protons are deuterated 3 min after dilution of a concentrated protonated sample into 2H2O. The exchange times of the H-bonded amino protons are much longer: 2000 s for that of the outer C3•C3+ pair and about one day (8 × 104 s) for those of the three other pairs. It is noteworthy that the rotation time around the C–N4 bond of the amino group of protonated and neutral cytidine monomers is 3.8 and 0.07 s, respectively at 0°C (23). The large exchange time difference of the exposed and H-bonded amino protons reveals hindered rotation of the amino group in the tightly packed i-motif core (22). The remarkable difference of the exchange times of the exposed and H-bonded amino protons of the outermost C3•C3+ pair discloses hindered rotation of C3(NH2) and provides indirect indication for a short stacking interval with the adjacent A2•A2 pair.
Magnetization transfer from water and real time exchange experiments were performed on [d(CCCCAA)]4. They show that the innermost pairs of conformations 5′Em and 3′EM, i.e. C3*•C3*+ and C2•C2+, have comparable lifetimes: τ0 = 3100 ± 400 s. As observed for [d(AACCCC)]4, the exchange times of the external amino protons of the four C•C+ pairs of 5′Em and 3′EM are shorter than 3 min, and those of the poorly resolved H-bonded amino protons of the three inner C•C+ pairs are extremely long: 6 × 103 to 3.5 × 104 s. But, in contrast with [d(AACCCC)]4, the exchange times of the H-bonded amino protons of the outermost C1*.C1*+ and C4•C4+ pairs are close to 0.3 s. This indicates that the rotation of the amino group of the outermost C•C+ pairs is not hindered in the 5′Em and 3′EM conformations of [d(CCCCAA)]4.
Structure of the i-motif tetramers of d(AACCCC) and d(CCCCAA)
Figure 6 shows the structures computed using the distance restraints displayed in Figure 2 and Table 1. Ten conformers selected for their low energy related to NOE violations were used for structural analysis (Table 2). The deviations from ideal geometry and the distance restraint violations are listed in Table 2.
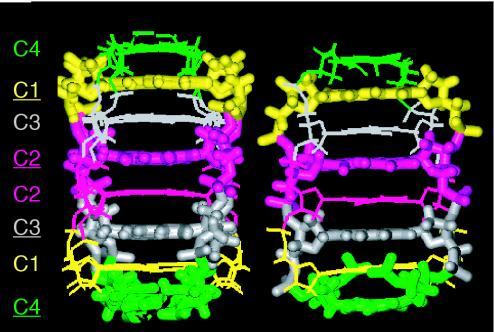
Figure 6.
Solution structure and schematic representation of the i-motif of the 5′E (left-hand side) and 3′E topologies (center) of d(CCCCAA) and of d(AACCCC) (right-hand side). The schematic representation of [d(AACCCC)]4 emphasis cross-strand stacking at step A1–A2. The superposition of 10 computed conformer is displayed for each structure. The views are normal to the wide groove at the center of the structures.
Table 1.
Distance restraints and quality of the computed structures of d(AACCCC)]4 and of the 5′Em and 3′EM intercalation topologies of d(CCCCAA)]4
| [d(AACCCC)]4 | [d(CCCCAA)]4 | ||
|---|---|---|---|
| 5′E | 3′E | ||
| Number of distance restraints | |||
| Intra-residue | 27 | 31 | 16 |
| Inter-residue | 53 | 40 | 32 |
| Repulsive | 19 | 16 | 18 |
| Base-pairing restraints | 12 | 12 | 12 |
| Violations and deviations from ideal geometrya | |||
| Number of NOE violations >0.2 Å | 2.6 ± 0.7 | 3 ± 2 | 0.8 ± 0.5 |
| Largest NOEviolation (Å) | 0.27 | 0.36 | 0.23 |
| R.m.s.d. on NOE violations (Å) | 0.076 ± 0.002 | 0.072 ± 0.002 | 0.04 ± 0.004 |
| H-bond length violations >0.05 Å | 0 | 0 | 0 |
| R.m.s.d. on bond length violations (Å) | 7 × 10−3 ± 5 × 10−4 | 6 × 10−3 ± 7 × 10−4 | 7 × 10−3 ± 7 × 10−4 |
| Number of angle violations >6° | 1.3 ± 0.8 | 1.1 ± 0.9 | 1.2 ± 0.8 |
| R.m.s.d. on angle violations (°) | 1.3 ± 0.03 | 1.4 ± 0.03 | 1.3 ±0.02 |
| Number of improper violations >6° | 0 | 0 | 0 |
| R.m.s.d. on improper violations (°) | 0.43 ± 0.1 | 0.44 ± 0.1 | 0.43 ± 0.04 |
aComputed for 10 conformers of each structure, selected for their low energy related to NOE violations.
Table 2.
Geometrical parameters of the 5′E and 3′E intercalation topologies of d(CCCCAA)]4 and of d(AACCCC)]4
| Residue | χ (°) | P (°) | Buckle (°) | Propeller twist (°) | Helical twist (°) | Helical rise (Å) |
|---|---|---|---|---|---|---|
| C4A2 5′Em | ||||||
| C1* | 233 ± 4 | 13 ± 6 | −2 ± 5 | 5 ± 1 | ||
| C2* | 240 ± 3 | 36 ± 14 | 4 ± 3 | 3 ± 3 | 19 ± 6 | 6.6 ± 0.2 |
| C3* | 241 ± 3 | 70 ± 35 | −2 ± 3 | 3 ± 1 | 11 ± 7 | 6.8 ± 0.5 |
| C4* | 257 ± 3 | 18 ± 5 | 11 ± 3 | −2 ± 2 | 13 ± 6 | 7.0 ± 0.3 |
| A5* | 254 ± 16 | 125 ± 14 | 9 ± 10 | 12 ± 11 | 18 ± 5 | 6.0 ± 0.2 |
| A6* | 223 ± 5 | 72 ± 20 | 23 ± 25 | 5 ± 8 | 46 ± 67 | 3.2 ± 0.2 |
| C4A2 3′EM | ||||||
| C1 | 214 ± 4 | 2 ± 6 | 2 ± 9 | −4 ± 3 | ||
| C2 | 238 ± 4 | 48 ± 8 | 0 ± 5 | 1 ± 3 | 10 ± 8 | 6.9 ± 0.5 |
| C3 | 224 ± 5 | 72 ± 37 | −14 ± 6 | −1 ± 3 | 19 ± 10 | 6.4 ± 0.2 |
| C4 | 238 ± 8 | 27 ± 16 | −12 ± 5 | −7 ± 2 | 13 ± 11 | 6.9 ± 0.2 |
| A5 | 139 ± 6 | 237 ± 9 | 2 ± 7 | −16 ± 3 | 80 ± 6 | 4.4 ± 0.2 |
| A6 | 196 ± 19 | 253 ± 16 | 24 ± 25 | 0 ± 27 | 4 ± 6 | 3.4 ± 1.5 |
| A2C4 | ||||||
| A1 | 3 ± 19 | 102 ± 51 | 26 ± 9 | −14 ± 6 | ||
| A2 | 219 ± 1 | 119 ± 18 | 20 ± 1 | −21 ± 1 | 26 ± 9 | 4.4 ± 0.3 |
| C3 | 226 ± 3 | 31 ± 3 | 6 ± 2 | −11 ± 1 | 20 ± 1 | 3.0 ± 0.05 |
| C4 | 214 ± 5 | 9 ± 8 | 3 ± 3 | −9 ± 2 | 6 ± 2 | 6.2 ± 0.3 |
| C5 | 224 ± 3 | 16 ± 12 | 7 ± 2 | −10 ± 3 | 3 ± 3 | 6.7 ± 0.3 |
| C6 | 217 ± 2 | 9 ± 8 | 7 ± 1 | 5 ± 1 | 7 ± 2 | 6.7 ± 0.3 |
Glycosidic angle (χ), pseudo-rotation angles (P), Buckle, Propeller twist, helical twist and helical rise. The parameters and SD values were measured from 10 selected conformers. The helical rise is computed as the average vertical distance between the adjacent base planes of each duplex along the direction perpendicular to the base planes.
Mixing the G- and C-rich four-stranded structures
The competition between the G- and C-rich tetramers of complementary sequences and the Watson–Crick duplex has been already examined for several oligonucleotides (24–26). It is generally observed that, in the pH range optimal for the i-motif stability, i.e. between pH 4 and 5, the equilibrium is shifted towards the tetrameric species. The stability of the tetramers of d(CCCCAA), d(AACCCC) and d(TTGGGG) (27) together with the good resolution of the NMR spectra of these fully symmetrical structures provided the examination of a potential interaction between i-motif and G-tetramer.
The proton spectra of the parallel tetramer of d(TTGGGG), of [d(AACCCC)]4 and [d(CCCCAA)]4 are compared in Figure 7 with the spectra of the 1/1 mixtures of the G-rich and C-rich tetramers. All the spectra were obtained at pH 4.6 with the same total strand concentration: [d(TTGGGG) + C-rich strand] = 0.7 mM.
Figure 7.
Comparison of the NMR spectra of the tetramer of d(TTGGGG), d(CCCCAA) and d(AACCCC) with that of 1/1 mixtures of the G- and C-rich tetramers. (a) Imino and amino proton region of [d(TTGGGG)]4. (b) Spectrum of the i-motif of the complementary d(CCCCAA) strand. (c) Spectrum at equilibrium of the 1/1 mixture of [d(TTGGGG)]4 and [d(CCCCAA)]4. (d) Reference spectrum of the exchangeable and aromatic region of [d(TTGGGG)]4 (line broadening 3 Hz). (e) Reference spectrum of [d(AACCCC)]4 (line broadening 3 Hz). (f) Spectrum built by addition of spectra (c) and (d) processed with a line broadening of 18 Hz. (g) Spectrum of a solution containing the 1/1 mixture of [d(TTGGGG)]4 and [d(AACCCC)]4 (line broadening 3 Hz). The comparison of spectra (f) and (g) shows that most of the NMR lines are broadened by ∼15 Hz in the 1/1 mixture of the tetramers. Some resolved peaks, indicated by stars, exhibits a larger line broadening on the spectra of the 1/1 mixtures. The spectra were obtained with the same total strand concentration of 0.7 mM. T = 0°C, KCl = 0.3 M, pH 4.6. The identification of the G imino protons of [d(TTGGGG)]4 was obtained from ref. (27).
The spectra observed right after tetramer mixing and after incubation at room temperature during one month are the same. The absence of NMR lines in the Watson–Crick imino proton region (14.5–12 p.p.m.) on the spectrum of the 1/1 mixture of [d(TTGGGG)]4 and [d(CCCCAA)]4 shows that the formation of the [d(TTGGGG)•d(CCCCAA)] duplex is negligible. The comparison of the reference spectra of the isolated G- and C-rich tetramers with that of the 1/1 mixtures of [d(TTGGGG)]4 + [d(AACCCC)]4 or of [d(TTGGGG)]4 + [d(CCCCAA)]4 shows that all the resolved proton peaks are broadened by at least 15 Hz in the 1/1 mixtures. Both thymidine imino protons of the G-tetramer exhibit a larger line broadening. Magnetization transfer experiments shows that the exchange times of G-quartet thymidine imino protons are much faster in the presence of the i-motif structures.
The spectrum of the 1/1 mixture of [d(TTGGGG)]4 + [d(AACCCC)]4 shows also a larger line broadening for the A2 (H8). The line broadening observed in the 1/1 tetramer mixtures at the strand concentration of 0.7 and 0.17 mM is the same.
These observations first establish that in pH conditions of optimal stability for the i-motif, the tetramers are much more stable than the Watson–Crick duplex. The comparison of the spectra of the isolated tetramers and of the 1/1 mixtures gives no indication for structural change. Nevertheless, the line broadening observed in the 1/1 mixtures suggests interaction of the G-rich and C-rich tetramers. The line broadening of the adenosine H8 proton and the faster thymidine imino proton exchange rates in 1/1 mixtures are indicative of end-to-end stacking or of transient base-pairing interactions between the terminal adenosines and thymidines. Such interactions result in larger effective hydrodynamic volume and longer correlation time. It is likely that exchange with water of the thymidine imino protons of the G-quartet is catalyzed in the 1/1 mixtures by the N3 nitrogen of the A residues of the i-motif structures. It must be clear that the conclusions derived from the study of the interaction between the i-motif tetamers of d(CCCCAA) or d(AACCCC) and [d(TTGGGG)]4 cannot be extended to the folded structures of the four repeats of d[(AACCCCAACCCCAACCCCAACCCC] and of the complementary G-rich stand that may by extended in vivo since their A and T bases are located in the loops connecting the paired G and C segments (28,29).
At pH 6, 0°C, in condition of marginal stability for the i-motif structure of d(CCCCAA), the NMR spectrum at equilibrium of the 1/1 mixture of [d(CCCCAA)]4 + [d(TTGGGG)]4 shows that only 1/3 of the strand concentration is in tetrameric structures and that the remaining 2/3 is in the duplex form (data not shown).
DISCUSSION
The i-motif core structure
The tetramers of [d(CCCCAA)]4 and of [d(AACCCC)]4 are fully symmetrical. They are built by two anti-parallel duplexes held together by four intercalated C•C+ pairs forming a core of eight stacked hemiprotonated pairs. The i-motif core of the tree structures do not show significant differences. The cytidine glycosidic angles are anti and the cytidine sugar puckers are in the C3′-endo or C4′-endo conformational ranges. This allows sugar-phosphate stretching up to helical rises in the range of 6.2–7 Å (Table 2), large enough for duplex intercalation. The stacking interval between the C•C+ pairs ranges from 3 ± 0.2 to 3.7 ± 0.4 Å (Table 2). The propeller twist and buckle angles displayed in Table 2 show that the C•C+ pairs are flat with deviations from planarity smaller than 20°. The structures exhibit two narrow and two wide grooves. The groove widths were computed as the average distance between the sugar C1′, C2′, C3′, C4′ and O4′ atoms of the stacked cytidines. They range between 10.5 and 11.5 Å for the wide groove, and between 5 and 7 Å for the narrow grooves. As previously observed (18), the narrow groove width is systematically broader by ∼0.7 Å at the steps between C•C+ pairs stacked by the faces oriented in the 3′ direction (white step). The intercalated duplexes are right handed. The helical twist between the covalently linked C•C+ pairs varies between 10 ± 8° and 19 ± 10° (Table 2).
Adenine pairing and stacking interactions with the i-motif core
The adenine bases of each structure are symmetrically stacked at the ends of the i-motif core. The chemical shifts of the H2 and H8 protons are independent of pH between pH 6 and 4.3 and give no indication for adenosine protonation. The adenosines at the 5′ end of [d(AACCCC)]4 and of those at the 3′ end of the two topologies of [d(CCCCAA)]4 are oriented differently.
In both topologies of [d(CCCCAA)]4, the adenosines form symmetrical arrangements of A•A pseudo-pairs with the Hcis amino proton oriented towards the N1 nitrogen of the same adenine on the complementary strand (Figure 8). The small number of NOE between A5 and A6 (Figure 2) shows that A6 is not closely stacked. The lack of restraints results in poor definition of the A6 residues whose r.m.s.d. values are respectively 3.2 and 5.5 Å in the 5′E and 3′E topologies.
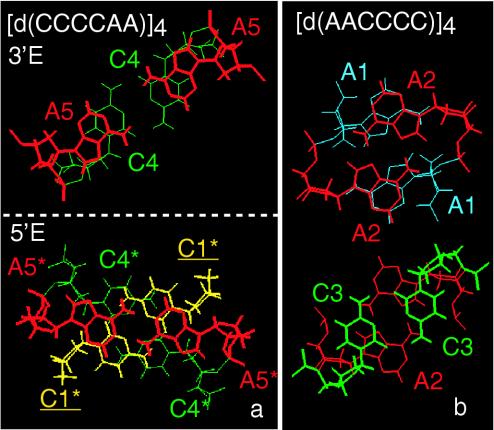
Figure 8.
Adenine base-pairing and stacking in [d(AACCCC)]4 and [d(CCCCAA)]4. On each structure, the adenosine (red) and the covalently linked green cytidine belong to the same duplex. The yellow cytidine belongs to the other duplex. The structures are shown looking down the helix axis from the 5′ end. The bond widths decrease with the base elevation. (a) [d(CCCCAA)]4: stacking at the C4–A5 step in the 3′E intercalation topology and inter-duplex stacking of C1* between C4* and A5* in the 5′E topology. (b) [d(AACCCC)]4: cross-strand stacking of A1 and A2 and intra-strand stacking at the A2–C3 step.
In the computed structure of 5′Em, the A5*(H6cis)–A5*(N1) cross-strand distance, 2.7 ± 0.9 Å, is compatible with H-bonding, nevertheless, the fast rotation of the amino group and the chemical shift of the A5* amino protons, 7.04 p.p.m., argue against A5* pairing. The NOE cross-peaks connecting the sugar protons of C1* and A5* (cross-peaks H1′–H1′ and H1′–H4′, Figure 2) are characteristic of those observed in i-motif structures between the sugar protons of bases stacked by the faces oriented in the 5′ direction (black step). This suggests a similarity of the groove geometry at the C1*–A5* step and at the i-motif black steps. The short H1′–O4′ distance across the narrow groove at the black steps that allows the formation of CH1′–O4′ bonds (30,31), which could contribute to the free energy of the 5′Em topology.
In the 3′EM species, the cross-strand A5(H6 cis)–A5(N1) distance, 4.9 ± 0.12 Å, is too large for H-bonding.
In [d(AACCCC)]4, the H trans amino proton of each adenine is oriented towards the N7 nitrogen of the same adenine of the opposite strand (Figure 8). The short A2(H6 trans)–A2(N7) cross-strand distance, 1.75 ± 0.15 Å, together with the slow rotation rate of A2(NH2) and the low field shift of A2(H6 trans) indicates A2•A2 (N7–H6 trans) pairing. The short stacking intervals between pairs A2•A2 and C3•C3+, 3 ± 0.05 Å, probably account for the hindered rotation of the amino group of C3. The fast rotation rate of A1(NH2) and the A1(H6 trans)–A1(N7) cross-strand distance, 4.5 ± 0.6 Å, show that A1 is not base-paired. The strong A1 H8–H1′ intra-residue cross-peak reveals a syn glycosidic angle (Table 2). Syn orientation of A1 results in cross-strand stacking to A2 with nearly orthogonal orientation of the stacked bases and overlapping of their six membered rings (Figure 8). Cross-strand stacking has been observed previously in another parallel duplex, at the G3–A4 step of [d(TCGA)]2 (32).
Adenosine ring currents
The chemical shifts induced by adenosine are much larger than those due to cytidine. The comparison of the chemical shifts of the cytidine sugar protons in the three investigated structures shows that several protons belonging to the cytidines stacked to adenosine exhibit large up-field chemical shifts. This is the case in particular for C1*(H1′) in the 5′Em topology of [d(CCCCAA)]4, for C4 (H1′, H2′ and H2″) in the 3′EM topology and for C3(H1′) in [d(AACCCC)]4 (Table S1). The ring current effects induced by the adenine bases on the chemical shift of these protons were computed according to a graphical approach (33). They are in excellent agreement with the observed shifts and provide an independent confirmation of the computed structures.
[d(AACCCC)]4: crystal and solution structures
The comparison of the solution and crystal structures of the i-motif of d(AACCCC) shows many similarities and some significant differences.
The solution structure of [d(AACCCC)]4 adopts the 5E′ topology with adenosine bases fairly well stacked to the i-motif core. In contrast, the crystal structure is composed by the assembly of the 3′E and 5′E topologies and the adenosine bases are roughly orthogonal to the C•C+ plan. The adenosines of tetramers adjacent in the crystal lattice form two kinds of adenosine clusters connecting together the tetramers along two orthogonal directions (9).
The comparison of the 5′E topology of the crystal structure with the solution structure shows the remarkable similarity of the organization of the C•C+ pairs (Figure 9). In the solution structure, the stacking intervals between the C•C+ pairs, 3.2 Å on the average, is comparable to that of the crystal structure: 3.16 Å. The average helical twist in the solution structure (13 ± 5)° is close to that of the crystal structure: (19 ± 8)°. These similarities results in r.m.s.d. between the C•C+ pairs of the crystal and solution structure of 0.65 Å, a value comparable to the r.m.s.d. of 0.58 Å measured from the pair-wise comparison of the 10 selected conformers of the solution structure. The cytidine glycosidic angles of the solution and crystal structures differ by less than 15%.
Figure 9.
Solution structure (left-hand side) and crystal structure (9) (right-hand side) of the 5′E topology of the i-motif core of [d(AACCCC)]4. For clarity, the adenosine residues, that have a different organization in the solution and crystal structures, are not shown. The r.m.s.d. between the base and sugar heavy atoms of each structure are 0.65 and 1.3 Å, respectively.
In contrast, the sugar-phosphate backbones of the solution and crystal structures show significant differences. The r.m.s.d. between the sugar heavy atoms of the solution and crystal structures, 1.3 Å, is almost twice larger than that measured between the computed conformers of the solution structure. The comparison of both structures shows in particular significant differences between the cytidine sugar puckers. The four cytidine sugars of the solution structure are in the C3′-endo conformation with pseudo-rotation angles ranging from 9° to 31° (Table 2). In the crystal structure, the cytidine pseudo-rotation angles range from 16° to 205°. In particular, 5 out of the 16 cytidine riboses of the crystal structure are in the C2′-endo or C3′-exo conformational range. It may be noted that these sugar conformations correspond to H6–H3′ intra-residue distances that should result in NOESY cross-peaks about five times weaker than those measured in NOESY spectra. Another difference between the solution and crystal structures is the narrow groove width which is ∼1 Å wider in the solution structure. In conclusion, the comparison of the crystal and solution structures of the 5′E topology of [d(AACCCC)]4 shows that the cytidine bases of both structures are nearly superimposable whereas the sugar-phosphate backbone presents significant differences.
3′E and 5′E intercalation topologies in i-motif structures
At 0°C, the proportion of the 3′EM and 5′Em species of [d(CCCCAA)]4 corresponds to a free energy difference of only 3.2 kJ/mol.
The intercalation in the 3′E topology of two hemiprotonated duplexes containing n cytidine pairs generates (n − 1) white steps (cf. Nomenclature in Introduction) and n black steps while intercalation in the 5′E topology generates n white step and (n − 1) black step. The factors influencing the balance between the two topologies have been previously discussed (31). Briefly, it has been proposed that the short H1′–O4′ distance across the narrow groove at the black steps allows the formation of CH1′–O4′ bonds that may contribute to stability of the 3′E topology.
Multiple intercalation topologies have been already reported for the solution structure of i-motif tetramers. The NMR spectra i-motif containing only a cytidine tract as in the case of the d(Cn) series (n = 3–6) (31) or a C-tract with anon-cytidine residues at the 3′ end such as d(C3T) and d(C4T) (34), d(CCCTAA) (35) and d(CCCCAA) show an equilibrium between the 3′E and 5′E intercalation topologies. The 3′E topology is always the major species. The proportion of each species is independent of the number of C•C+ pairs and correspond to free energy differences in the range of kJ/mol.
The presence of a thymidine at the 5′ end of the cytidine tract as in the case of the d(TCn) series [n = 2 (10), 3, 4 (unpublished data) and 5 (4)] increases the free energy difference between the two topologies in such way that the 5′E topology of these tetramers is not observed. In contrast, the adenosine at the 5′ end of [d(AACCCC)]4 favors the formation of the 5′E topology. This shows that the free energy difference between the 5′E and 3′E topologies may be balanced by stacking interactions of the outer non-cytidine residues. In [d(AACCCC)]4, the helical rise between the outermost C3•C3+ pair and A2•A2, 3 ± 0.05 Å, is indicative of stacking interaction favorable to the formation of this topology.
As shown for the d(Cn) oligonucleotides (31), the distribution of positive and negative charges is different in the 3′E and 5′E topologies and may also influence the proportion of each species. This may explain the equilibrium shift observed towards the 5′Em topology of [d(CCCCAA)]4 upon addition of NaCl. The comparison of the i-motifs structures of d(TCC) and d(T5mCC) shows that cytidine methylation in position C5 also influence the intercalation topology. In i-motif crystal structures, the packing interactions have probably a dominant contribution on the intercalation topology. Indeed, the crystal structure of [d(CCCC)]4 (36) adopts the 5′E intercalation topology whereas the solution structure shows the predominance of the 3′E topology (31) and in contrast with the solution structure, the crystal structure of [d(AACCCC]4 shows the association of both topologies.
Acknowledgments
We thank Renaud Charley for assistance in the determination of the intercalation topology of [d(CCCCAA)] and S. Nonin-Lecomte. Coordinates deposition: the coordinates of the lowest NOESY-related energy conformers of [d(AACCCC)]4 and of the 3′E and 5′E topology of [d(CCCCAA)]4 have been submitted with the distance restraints used in the molecular dynamics computations to the Protein Data Bank, (Chemistry Department, Brookhaven National Laboratory, Upton, NY) (acquisition numbers: 1YBL, 1YBN and 1YBR). Funding to pay the Open Access publication charges for this article was provided by CNRS.
REFERENCES
- 1.Blackburn E.H. Structure and function of telomeres. Nature. 1991;350:569–573. doi: 10.1038/350569a0. [DOI] [PubMed] [Google Scholar]
- 2.Neidle S., Parkinson G.N. The structure of telomeric DNA. Curr. Opin. Struct. Biol. 2003;13:275–283. doi: 10.1016/s0959-440x(03)00072-1. [DOI] [PubMed] [Google Scholar]
- 3.Mills M., Lacroix L., Arimondo P.B., Leroy J.-L., Francois J.C., Klump H., Mergny J.-L. Unusual DNA conformations: implications for telomere. Curr. Med. Chem. Anti-Canc. Agents. 2002;2:627–644. doi: 10.2174/1568011023353877. [DOI] [PubMed] [Google Scholar]
- 4.Gehring K., Leroy J.-L., Guéron M. A tetrameric DNA structure with protonated cytidine–cytidine base pairs. Nature. 1993;363:561–565. doi: 10.1038/363561a0. [DOI] [PubMed] [Google Scholar]
- 5.Wang Y., Patel D.J. Solution structure of the human telomeric repeat d[AG3(T2AG3)3] G-tetraplex. Structure. 1993;1:263–282. doi: 10.1016/0969-2126(93)90015-9. [DOI] [PubMed] [Google Scholar]
- 6.Phan A.T., Guéron M., Leroy J.-L. The solution structure and internal motions of a fragment of the cytidine-rich strand of the human telomere. J. Mol. Biol. 2000;299:123–144. doi: 10.1006/jmbi.2000.3613. [DOI] [PubMed] [Google Scholar]
- 7.Lacroix L., Lienard H., Labourier E., Djavaheri-Mergny M., Lacoste J., Leffers H., Tazi J., Helene C., Mergny J.-L. Identification of two human nuclear proteins that recognise the cytosine-rich strand of human telomeres in vitro. Nucleic Acids Res. 2000;28:1564–1575. doi: 10.1093/nar/28.7.1564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cornuel J.-F., Moraillon A., Guéron M. Participation of yeast inosine 5′-monophosphate dehydrogenase in an in vitro complex with a fragment of the C-rich telomeric strand. Biochimie. 2002;84:279–289. doi: 10.1016/s0300-9084(02)01400-1. [DOI] [PubMed] [Google Scholar]
- 9.Cai L., Chen L., Raghavan S., Ratliff R., Moyzis R., Rich A. Intercalated cytosine motif and novel adenine clusters in the crystal structure of the Tetrahymena telomere. Nucleic Acids Res. 1998;26:4696–4705. doi: 10.1093/nar/26.20.4696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Leroy J.-L., Guéron M. Solution structure of the i-motif tetramers of d(TCC), d(5methylCCT) and d(T5methylCC): novel NOE connection between amino protons and sugar protons. Structure. 1995;3:101–120. doi: 10.1016/s0969-2126(01)00138-1. [DOI] [PubMed] [Google Scholar]
- 11.Phan A.T., Guéron M., Leroy J.-L. Investigation of unusual DNA motifs. Methods Enzymol. 1995;338:341–371. doi: 10.1016/s0076-6879(02)38228-4. [DOI] [PubMed] [Google Scholar]
- 12.Plateau P., Guéron M. Solvent signal suppression in NMR. J. Am. Chem. Soc. 1982;104:7310–7311. [Google Scholar]
- 13.Macura S., Wüthrich K., Ernst R.R. The relevance of J cross-peaks in two-dimentional NOE experiments of macromolecules. J. Magn. Reson. 1992;47:351–357. [Google Scholar]
- 14.Bax A., Davis D.G. MLEV-17 based two-dimensional homonuclear magnetization transfer spectroscopy. J. Magn. Reson. 1985;65:355–360. [Google Scholar]
- 15.Kellogg G.W. Proton-detected hetero-TOCSY experiments with application to nucleic acids. J. Magn. Reson. 1992;98:176–182. [Google Scholar]
- 16.van Dongen M.J.P., Wijmenga S.S., Eritja R., Azorin F., Hilbers C.W. Through-bond correlation of adenine H2 and H8 protons in unlabeled DNA fragments by HMBC spectroscopy. J. Biomol. NMR. 1996;8:207–212. doi: 10.1007/BF00211166. [DOI] [PubMed] [Google Scholar]
- 17.Malliavin T., Snoussi K., Leroy J.-L. The molecular structure of [d(XC2)]4 investigated by molecular dynamics simulations. Magn. Reson. Chem. 2002;43:18–25. [Google Scholar]
- 18.Snoussi K., Nonin-Lecomte S., Leroy J.-L. The RNA I-motif. J. Mol. Biol. 2001;309:139–153. doi: 10.1006/jmbi.2001.4618. [DOI] [PubMed] [Google Scholar]
- 19.Brünger A.T. New Haven, CT: Yale University; 1990. X-PLOR Version 3, a system for X-ray crystallography and NMR. [Google Scholar]
- 20.Koradi R., Billeter M., Wütrich K. MOLMOL: a program for display and analysis of macromolecular structures. J. Mol. Graphics. 1996;14:51–55. doi: 10.1016/0263-7855(96)00009-4. [DOI] [PubMed] [Google Scholar]
- 21.Guéron M., Leroy J.-L. Studies of base-pair kinetics by NMR measurement of proton exchange. Methods Enzymol. 1995;261:383–413. doi: 10.1016/s0076-6879(95)61018-9. [DOI] [PubMed] [Google Scholar]
- 22.Leroy J.-L., Gehring K., Kettani A., Guéron M. Acid multimers of oligo-cytidine strands: stoichiometry, base-pair characterization and proton exchange properties. Biochemistry. 1993;32:6019–6031. doi: 10.1021/bi00074a013. [DOI] [PubMed] [Google Scholar]
- 23.Kettani A., Guéron M., Leroy J.-L. Amino proton exchange process in mononucleotides. J. Am. Chem. Soc. 1997;119:1108–1115. [Google Scholar]
- 24.Phan A.T., Mergny J.-L. Human telomeric DNA: G-quadruplex, i-motif and Watson–Crick double helix. Nucleic Acids Res. 2002;30:4618–4625. doi: 10.1093/nar/gkf597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Miyoshi D., Matsumura S., Li W., Sugimoto N. Structural polymorphism of telomeric DNA regulated by pH and divalent cation. Nucleosides Nucleotides Nucleic Acids. 2003;22:203–221. doi: 10.1081/NCN-120019528. [DOI] [PubMed] [Google Scholar]
- 26.Li W., Miyoshi D., Nakano S., Sugimoto N. Structural competition involving G-quadruplex DNA and its complement. Biochemistry. 2003;42:11736–11744. doi: 10.1021/bi034168j. [DOI] [PubMed] [Google Scholar]
- 27.Wang Y., Patel D.J. Guanine residues in d(T2AG3) and d(T2G4) form parallel-stranded potassium cation stabilized G-quadruplexes with anti glycosidic torsion angles in solution. Biochemistry. 1992;31:8112–8119. doi: 10.1021/bi00150a002. [DOI] [PubMed] [Google Scholar]
- 28.Leroy J.-L., Guéron M., Mergny J.-L., Helene C. Intramolecular folding of a fragment of the cytosine-rich strand of telomeric DNA into an i-motif. Nucleic Acids Res. 1994;22:1600–1606. doi: 10.1093/nar/22.9.1600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang Y., Patel D.J. Solution structure of the Tetrahymena telomeric repeat d(T2G4)4 G-tetraplex. Structure. 1994;2:1141–1156. doi: 10.1016/s0969-2126(94)00117-0. [DOI] [PubMed] [Google Scholar]
- 30.Berger I., Egli M., Rich A. Inter-strand C-H…O hydrogen bonds stabilizing four-stranded intercalated molecules: stereoelectronic effects of O4′ in cytosine-rich DNA. Proc. Natl Acad. Sci. USA. 1996;93:12116–12121. doi: 10.1073/pnas.93.22.12116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Leroy J.-L., Snoussi K., Guéron M. Investigation of the CH…O hydrogen bonds in the DNA i-motif via the equilibrium between alternative intercalation topologies. Magn. Reson. Chem. 2001;39:S171–S176. [Google Scholar]
- 32.Wang Y., Patel D.J. Solution structure of the d(T-C-G-A) duplex at acidic pH. A parallel-stranded helix containing C•CC+, G•G and A•A pairs. J. Mol. Biol. 1994;242:508–526. [PubMed] [Google Scholar]
- 33.Giessner-Prettre C., Pullman B., Borer P.N., Kan L.-S., Ts'o P.O.P. Ring-current effects in the NMR of nucleic acids. Biopolymers. 1976;15:2277–2286. doi: 10.1002/bip.1976.360151114. [DOI] [PubMed] [Google Scholar]
- 34.Kanaori K., Maeda A., Kanehara H., Tajima K., Makino K. 1H nuclear magnetic resonance study on equilibrium between two four-stranded solution conformations of short d(CnT) Biochemistry. 1998;37:12979–12986. doi: 10.1021/bi980492g. [DOI] [PubMed] [Google Scholar]
- 35.Kanaori K., Shibayama N., Gohda K., Tajima K., Makino K. Multiple four-stranded conformations of human telomeresequence d(CCCTAA) in solution. Nucleic Acids Res. 2001;29:831–840. doi: 10.1093/nar/29.3.831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chen L., Cai L., Zhang X., Rich A. Crystal structure of a four-stranded intercalated DNA: d(C4) Biochemistry. 1994;33:13540–13546. doi: 10.1021/bi00250a005. [DOI] [PubMed] [Google Scholar]