Abstract
Granulocyte-macrophage colony stimulating factor (GM-CSF) plays a key role in myeloid cell function and is rapidly and transiently expressed in T cells in response to immune or inflammatory stimuli. Induction of GM-CSF gene expression is accompanied by changes in chromatin structure across the proximal promoter region of the gene. We show that the promoter remodelling and subsequent gene transcription occurs with distinct signal and transcription factor requirements. Activation of the protein kinase C (PKC) signalling pathway is sufficient to induce changes in chromatin structure across the promoter, but both the PKC and calcium signalling pathways are required for efficient gene transcription. Although NFAT transcription factors contribute to GM-CSF gene transcription, they are not required for promoter remodelling. However, the presence of the nuclear factor-κB transcription factor, c-Rel, in the nucleus is strongly correlated with and required for the events of chromatin remodelling.
INTRODUCTION
Activation of T cells by antigen results in the expression of a range of cytokines, which are important for the orchestration of an immune response. Generally, the genes encoding these cytokines are maintained in a transcriptionally silent state, and are rapidly but transiently expressed following T cell receptor stimulation. Induction of cytokine gene expression is controlled largely at the level of transcription, by the assembly of specific transcription factor complexes at the promoter and enhancer regions of the genes (1,2). However, within the cell nucleus, genes are found assembled into chromatin and it is now clear that chromatin can provide an additional level of control, by regulating the accessibility of the promoter to the transcription machinery [reviewed in (3,4)].
A number of studies have clearly demonstrated that changes in chromatin structure accompany induction of cytokine gene transcription in response to immune stimulation (5–9). However, the exact sequence of events, which leads to chromatin remodelling and subsequent gene activation is understood in detail for only a small number of eukaryotic genes, most notably, the induction of the yeast Pho5 gene in response to low phosphate (10) and the IFNβ gene following viral infection (11). An emerging theme is that gene-specific changes in the chromatin structure can be generated following the recruitment of chromatin modifying activities to gene promoters by DNA bound transcription factors (12,13). In the case of IFNβ, gene expression in response to viral infection is initiated by the assembly of transcription factors into a highly organized enhanceosome complex on the nucleosome-free enhancer region of the gene (11). Chromatin modifying activities are then recruited by the enhanceosome to remodel an adjacent nucleosome, facilitating gene transcription (14). The Pho5 gene is activated in low-phosphate conditions by the binding of the Pho4 activator to a binding site in a nucleosome-free region. This facilitates the remodelling of adjacent nucleosomes, allowing the binding of additional transcription factors and gene activation (10,15). These remodelling events are a prerequisite for gene activation (16,17). However for many genes, such as those encoding the cytokines GM-CSF, IL-2 and IL-12, the transcription factor binding sites in the promoter regions of these genes are not found in nucleosome-free regions but appear to be assembled into nucleosomes (6,8,9). While chromatin remodelling has been found to accompany the activation of these cytokine genes following immune stimulation, the mechanisms involved remain to be elucidated.
GM-CSF plays an important role in the orchestration of an immune or inflammatory response (18). It is expressed in a variety of cell types, including T cells in response to immune or inflammatory stimuli. In T cells, GM-CSF expression is rapidly, but transiently induced following T cell receptor ligation and costimulation through the CD28 co-receptor (19). Induction of GM-CSF gene expression is regulated by a proximal promoter limited to 100 bp upstream of the transcription start site as well as an upstream enhancer [reviewed in (1)]. The proximal promoter consists of an array of transcription factor binding sites including sites for nuclear factor-κB (NF-κB) family members located within the CD28 response region (CD28RR) of the promoter and an adjacent region containing binding sites for a number of transcription factors, including NFAT and AP-1 (20,21).
Induction of GM-CSF gene expression upon T cell activation is accompanied by changes in chromatin structure across the GM-CSF promoter (7,9,22). A region of <200 bp encompassing the promoter becomes increasingly accessible upon T cell activation, suggesting the targeted remodelling of a single nucleosome (9). The chromatin remodelling events precede GM-CSF gene transcription and similar to gene transcription, are dependent on the presence of NF-κB proteins (9,21,22).
Here, we show that chromatin remodelling events across the GM-CSF promoter and GM-CSF gene transcription are distinct molecular events, which occur with distinct kinetics and have different signal and transcription factor requirements.
MATERIALS AND METHODS
Plasmids
The mouse GM-CSF construct, AOGM, was provided by Dr P. Cockerill and has been described previously (23). A mouse GAPDH–PCR plasmid, containing a 67 bp fragment of the mouse GAPDH cDNA was generated by cloning a PCR product amplified using the primers outlined below into pCR 2.1 (Invitrogen).
Cell culture
EL-4 T cells were cultured in RPMI as described previously (8). Cells were stimulated with 20 ng/ml of PMA (P) (Boehringer Mannheim) and 1 μM of calcium ionophore (I) (A23187; Sigma–Aldrich). Cells were pre-treated with inhibitors before stimulation as follows: 10 μg/ml of cyclohexamide (Calbiochem) for 30 min; 50 μg/ml of cyclosporin (Calbiochem) for 30 min; 10 μM of Ro-32-0432 (Roche) for 1 h; and 1–6 mg/ml of pentoxifylline (Sigma) for 30 min.
Primary T cell preparation
All mice were maintained in a pathogen-free environment in a barrier facility. Spleens were isolated from C57BL/6 mice and c-rel−/− mice (4–5 weeks old) and CD4+ T cells purified using MACS CD4+ beads, according to the manufacturer's instructions (Miltenyi Biotech). The cells were stained and analysed by flow cytometry with T cell populations demonstrated to be ∼90% CD4+. Cells were stimulated with P and I as described above and with an activating CD28 antibody (BD PharMingen) at 5 μg/ml. Anti-CD3 stimulation was performed by coating the plates with 10 μg/ml of anti-CD3 antibody (BD PharMingen) at 37°C for 4 h. The plates were washed with phosphate-buffered saline (PBS) before the addition of cells at a density of 1.25 × 106 cells/ml.
Nuclear extracts and western blotting
Nuclear extracts were prepared after a modification in the method of Schreiber et al. (24). Briefly, EL-4 T cells at a density of 5 × 105 cells/ml were centrifuged for 5 min at 500 g, washed with PBS, resuspended in 1 ml of ice-cold Buffer A (10 mM Tris, pH 7.4, 10 mM NaCl, 3 mM MgCl2, 0.1 mM EDTA and 0.5% NP-40, containing protease inhibitors) per 5 × 106 cells, and incubated on ice for 5 min. Nuclei were recovered by centrifugation at 900 g for 5 min, washed in Buffer A without NP-40, resuspended in 75 μl Buffer C (400 mM NaCl, 7.5 mM MgCl2, 0.2 mM EDTA, 0.1 mM EGTA, 1 mM DTT and protease inhibitors) per 5 × 107 cells and incubated on ice with shaking for 15 min. Nuclear debris was removed by centrifugation at 13 000 g for 5 min. Protein concentrations were determined by Bradford Assay (Bio-Rad).
Proteins were resolved by SDS–PAGE through 12% polyacrylamide, transferred onto nitrocellulose membrane and subjected to western-blot analysis using anti-RelA, anti-c-Rel, anti-NFATp, anti-NFATc and anti-Sp1 antibodies (Santa Cruz) and the corresponding peroxidase-conjugated secondary antibodies (DAKO). Proteins were visualized using the Supersignal West Pico Chemiluminescent kit (Pierce), according to the manufacturer's instructions.
RNA isolation and real-time PCR analysis
Total RNA was isolated using Tri-Reagent (Sigma) and reverse transcribed using Superscript II Reverse Transcriptase (Life Technologies), as described previously (9). SYBR Green PCR amplification was performed on the Rotor-Gene 2000 real-time cycler (Corbett Research) using the QuantiTect SYBR Green PCR kit (Qiagen), according to the manufacturer's instructions in a total volume of 25 μl, containing 50 ng of cDNA. Cycling conditions were as follows: 95°C for 15 min; 95°C for 15 s, 60°C for 1 min for 35 cycles, acquiring on channel 1, followed by melt analysis from 60 to 95°C.
The GM-CSF primers used were as follows:
GM-CSF for, 5′-AAGGGTCCTGAGGAGGATGTG-3′;
GM-CSF rev, 5′-GAGGTTCAGGGCTTCTTTGA-3′.
PCR reactions were conducted in parallel to normalize for differences in cDNA synthesis and RNA input, using the following primers:
GAPDH for, 5′-AAGTATGATGACATCAAGAAGGTGGT-3′;
GAPDH rev, 5′-AGCCCAGGATGCCCTTTAGT-3′.
To correlate the threshold (Ct) values from the amplification plots to copy number, a standard curve was generated using the mouse GM-CSF plasmid AOGM and pCR2.1–GAPDH plasmid. PCR product melt curves were analysed for a single peak and the products were visualized by agarose gel electrophoresis and ethidium bromide staining to ensure that a single product was generated in the PCR.
Chromatin accessibility by real-time PCR
Accessibility of DNA to digestion with restriction enzymes and micrococcal nuclease (MNase) was analysed using chromatin accessibility by real-time PCR (CHART–PCR) (8). EL-4 T cell nuclei (5 × 106 nuclei per 100 μl) were treated with 150 U of HinfI enzyme (New England BioLabs) at 37°C for 45 min or with 25 U of MNase (Boehringer) for 5 min at 20°C, as determined empirically. In each case, control samples without the enzyme were incubated in a similar manner to monitor for endonuclease activity. Genomic DNA was subsequently isolated using a QIAamp blood kit (Qiagen), according to the manufacturer's instructions. Genomic DNA (50 ng) was analysed by SYBR Green real-time PCR in a total volume of 25 μl, using the QuantiTect SYBR Green PCR kit (Qiagen), according to the manufacturer's instructions. The primer sets used were as follows:
set−I For, 5′-GCCTGACAACCTGGGGGAAG-3′;
set−I Rev, 5′-TGATTAATGGTGACCACAGAACTC-3′;
set+I For, 5′-GAGTTCTGTGGTCACCATTAATCA-3′; and
set+I Rev, 5′-CACATCCTCCTCAGGACCTT-3′,
as described previously (9). Accessibility was determined by correlating the Ct values from the amplification plots to a standard curve generated with the AOGM plasmid, and was expressed as a percentage of undigested genomic DNA for each primer set.
RESULTS
Distinct signals are required for chromatin remodelling and gene transcription at the GM-CSF promoter
Upon T cell activation, changes in the chromatin structure occur across the GM-CSF promoter and are limited to the region between −174 and +24, suggesting the remodelling of a single nucleosome encompassing the proximal promoter (9). To determine the relationship between the signal requirements for GM-CSF promoter remodelling and gene transcription, these processes were examined following the activation of different intracellular signalling pathways in T cells. T cell receptor ligation results in the activation of two major intracellular signal pathways involving mobilization of protein kinase C (PKC) and increases in free intracellular calcium levels, which can be mimicked by treatment of cells with either P or I, respectively. Therefore, murine EL-4 T cells were either left unstimulated or stimulated with P, I or both (P/I) for 4 h. Following the stimulation, the accessibility of the GM-CSF promoter to micrococcal nuclease (MNase) or restriction enzyme digestion was measured by a real-time PCR assay [CHART–PCR, (8)] using primer sets, which span the GM-CSF promoter [Figure 1A and (9)]. Accessibility of a HinfI site in the CD28RR of the GM-CSF promoter (at −97) was assessed using primer set −I (−155 to −40, Figure 1A), which spans the CD28RR. PCR amplification was monitored by SYBR green incorporation and the amount of PCR product generated from digested samples was plotted as a percentage of the amount generated from undigested genomic DNA samples. As described previously (9), in unstimulated cells, the HinfI site in the promoter displayed an inherent accessibility to restriction enzyme digestion (Figure 1B) and the accessibility increased significantly upon stimulation with P/I. While stimulation with I alone had no effect on the accessibility of the HinfI site, stimulation with P alone increased accessibility to the levels seen with P/I combined (Figure 1B). To determine whether the accessibility changes at the HinfI site were also seen across the entire promoter, the effect of the different stimuli on the accessibility of the promoter region to MNase digestion was assessed using primer set −I, which covers the CD28RR and primer set +I (−63 to +44), which covers the adjacent NFAT/AP-1 region (Figure 1A). In unstimulated cells, both the CD28RR and the NFAT/AP-1 region displayed an inherent accessibility to MNase, which increased following stimulation with P/I or with P alone (Figure 1C and D). However, no increase in accessibility was detected following treatment with I alone (Figure 1C and D).
Figure 1.
Distinct signals are required for chromatin remodelling at the GM-CSF promoter and GM-CSF gene transcription. (A) Schematic representation of the GM-CSF promoter showing transcription factor binding sites and the CD28RR. DNA fragments amplified by PCR primer sets −I and +I and the HinfI restriction enzyme site are shown. (B) Nuclei from non-stimulated EL-4 T cells (NS) or cells stimulated for 4 h with P, I or P/I as indicated, were incubated with HinfI. Genomic DNA was analysed by real-time PCR using primer set −I. The mean and standard error of three replicate assays are shown. (C and D) Nuclei from cells treated as in (B) were incubated with MNase and genomic DNA was analysed using primer set −I (C) or primer set +I (D). (E) GM-CSF mRNA levels were determined by real-time PCR analysis of cDNA prepared from EL-4 T cells treated as in (B). Data were graphed as a fold change in the mRNA levels compared with NS.
To compare the changes in chromatin structure across the GM-CSF promoter with gene transcription, GM-CSF mRNA levels were analysed by quantitative real-time PCR analysis of RNA isolated from EL-4 T cells. While treatment with P alone induced GM-CSF gene transcription (20-fold) and a dramatic increase in mRNA (∼200-fold) was detected in cells stimulated with P/I for 4 h (Figure 1E), little increase in mRNA levels was detected in cells treated with I alone (∼4-fold). Therefore, while the signals generated from treatment of cells with P alone are sufficient to induce changes in chromatin structure at the GM-CSF promoter, signals generated by I cooperate with P signals to lead to high levels of GM-CSF gene transcription.
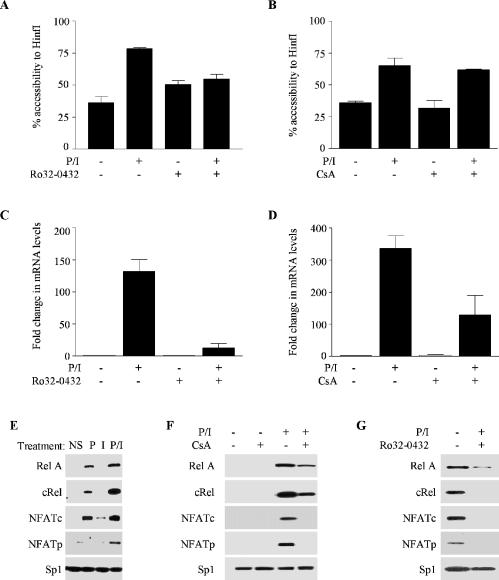
To confirm that the PKC signalling pathway is required for chromatin changes at the GM-CSF promoter, EL-4 T cells were pre-treated with the PKC inhibitor Ro32-0432 and then either left unstimulated or stimulated with P/I for 4 h and the accessibility of the HinfI restriction site was assessed by CHART–PCR. Pre-treatment with the PKC inhibitor, prevented chromatin remodelling in response to P/I stimulation (Figure 2A). In contrast, pre-treatment of cells with cyclosporin A (CsA), which inhibits the calcium signalling pathway (25), had no impact on chromatin remodelling at the Hinf1 site in the GM-CSF promoter in response to P/I stimulation (Figure 2B). In contrast to their differential effects on chromatin remodelling, both Ro32-0432 and CsA reduced but did not completely inhibit the accumulation of GM-CSF mRNA in response to P/I (Figure 2C and D), which agrees with the partial impact of each of these signals on transcription (Figure 1E).
Figure 2.
The PKC, but not the calcium signalling pathway is required for chromatin remodelling of the GM-CSF promoter. (A) EL-4 T cells were either left untreated or pre-treated with Ro32-0432, then incubated with or without P/I for 4 h, as indicated. Nuclei were incubated with HinfI and the genomic DNA analysed by real-time PCR using primer set −I. The mean and standard error of three replicate assays are shown. (B) EL-4 T cells were either untreated or pre-treated with cyclosporin A (CsA), then incubated with or without P/I for 4 h, as indicated. Nuclei were incubated with HinfI and the genomic DNA analysed as in (A). (C) GM-CSF mRNA levels were determined by real-time PCR analysis of cDNA prepared from EL-4 T cells treated as in (A). Data were graphed as a fold change in the mRNA levels compared with unstimulated, untreated cells. The mean and standard error of three replicate assays are shown. (D) GM-CSF mRNA levels were determined by real-time PCR analysis of cDNA prepared from EL-4 T cells treated as in (B). Data were graphed as a fold change in the mRNA levels compared with unstimulated, untreated cells. (E) Nuclear extracts from non-stimulated EL-4 T cells (NS) or cells stimulated with P, I or P/I for 4 h were subjected to SDS–PAGE and analysed by western blotting with the indicated antibodies. (F) Nuclear extracts were isolated from untreated cells or cells pre-treated with CsA, then incubated for 4 h with or without P/I. Extracts were analysed as in (E). (G) Nuclear extracts were isolated from untreated cells or cells pre-treated with Ro32-0432, then stimulated for 4 h with P/I. Extracts were analysed as in (E).
Therefore, the PKC pathway appears to be necessary and sufficient for chromatin remodelling across the GM-CSF promoter, while the calcium signalling pathway is not required for chromatin remodelling events but contributes to efficient GM-CSF gene transcription.
The presence of NF-κB proteins in the nucleus correlates with chromatin remodelling events
The GM-CSF proximal promoter contains an array of binding sites for the related transcription factor families NF-κB and NFAT, which have distinct but overlapping signal requirements. These transcription factors have been previously implicated in promoter function [Figure 1A and (20,21,26)], and we have shown that chromatin remodelling was dependent on NF-κB proteins (9). However, it was not clear if this was a unique role or if other transcription factor families, such as NFAT also contributes to chromatin remodelling. Therefore, western-blot analysis was used to correlate the nuclear activation of both NFAT and NF-κB factors with promoter accessibility and gene transcription. Nuclear extracts were prepared from unstimulated cells and cells treated with P, I and P/I for 4 h. As expected, RelA, c-Rel, NFATc and NFATp proteins were translocated to the nucleus following stimulation with P/I (Figure 2E). Treatment with I led to a low-level nuclear accumulation of NFATc alone, whereas treatment with P alone caused a nuclear accumulation of RelA, c-Rel and NFATc, but not NFATp (Figure 2E). Nuclear levels of the Sp1 protein, which is known to be constitutively present in the nucleus, were constant in all extracts, demonstrating equal protein loading in each lane (Figure 2E).
A primary target of CsA is the calcium/calmodulin-dependent phosphatase calcineurin, which regulates NFAT translocation to the nucleus and as expected, accumulation of NFATp and NFATc in the nucleus, following stimulation with P/I for 4 h is inhibited in cells pre-treated with CsA (Figure 2F). In contrast, CsA pre-treatment reduces, but does not completely inhibit nuclear translocation of the NF-κB proteins, RelA and c-Rel (Figure 2F). On the other hand, the inhibitor of the PKC pathway, Ro32-0432 prevented nuclear accumulation of all of these proteins, although there was a residual amount of RelA seen in the nucleus (Figure 2G).
Taken together, there is a clear correlation between chromatin remodelling at the GM-CSF promoter, activation of the PKC signalling pathway and the presence of NF-κB proteins, but not NFAT proteins, in the nucleus. However, NFAT proteins appear to contribute to GM-CSF transcription and mRNA accumulation.
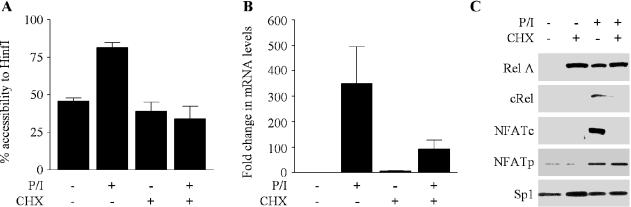
Protein synthesis is required for chromatin remodelling at the GM-CSF promoter
We have shown previously that chromatin remodelling at the GM-CSF promoter required at least 1 h to occur and was stable over a 6 h period (9). These delayed kinetics raised the possibility that new protein synthesis might be required for this event. The various NF-κB and NFAT proteins can also be distinguished by their requirement for new protein synthesis (27,28). Pre-treatment of EL-4 T cells with the translation inhibitor cyclohexamide inhibited chromatin remodelling at the promoter in response to P/I (Figure 3A), demonstrating that new protein synthesis is required for chromatin changes at the promoter. However, GM-CSF gene transcription still occurred in the presence of cyclohexamide and in the absence of chromatin remodelling, although at reduced levels (Figure 3B). In addition, c-Rel but not RelA nuclear accumulation was dependent on new protein synthesis (Figure 3C). Similarly, NFATp, which pre-exists in the cytoplasm of unstimulated cells (28), but not NFATc, which is synthesized de novo, accumulated in the nucleus of cells pre-treated with cyclohexamide following stimulation.
Figure 3.
Protein synthesis is required for chromatin remodelling but not transcription at the GM-CSF promoter. (A) EL-4 T cells were either untreated or pre-treated with cyclohexamide (CHX), then incubated with or without P/I for 4 h, as indicated. Nuclei were incubated with HinfI and the genomic DNA analysed by real-time PCR using primer set −I. The mean and standard error of three replicate assays are shown. (B) GM-CSF mRNA levels were determined by real-time PCR analysis of cDNA prepared from EL-4 T cells treated as in (A). Data were graphed as a fold change in the mRNA levels compared with NS. (C) Nuclear extracts from cells treated as in (A), were subjected to SDS–PAGE and analysed by western blotting with the indicated antibodies.
These data show that chromatin remodelling requires new protein synthesis and implicates proteins, such as c-Rel and NFATc, in this process. It should also be noted that gene transcription can proceed even in the absence of any detectable chromatin remodelling.
The stability of GM-CSF promoter remodelling
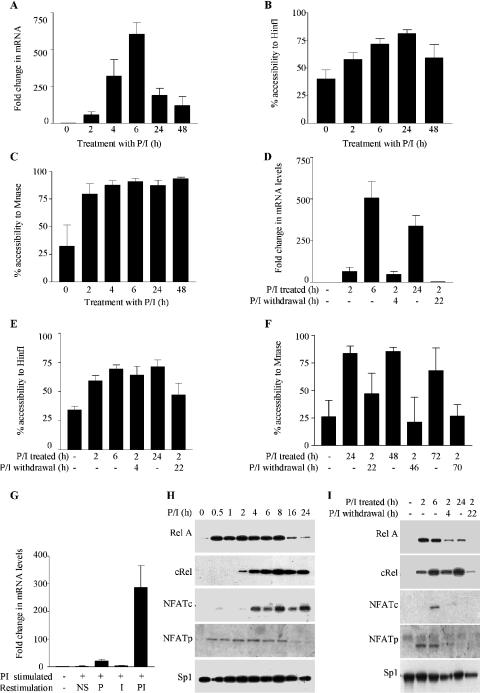
We have shown previously that GM-CSF mRNA accumulation followed the event of chromatin remodelling at the promoter (7) but do not know if transcription levels and chromatin accessibility follow similar kinetics over longer time frames. Following P/I stimulation of EL-4 T cells, the GM-CSF mRNA levels increased to ∼600-fold, 6 h after stimulation (Figure 4A), but had decreased significantly by 24 and 48 h, post-stimulation. This decrease is probably a combination of reduced gene transcription as well as increased mRNA instability [reviewed in (18)]. The GM-CSF promoter displayed an increased accessibility to HinfI and MNase digestion within 2 h of stimulation with P/I, and was maintained at maximal levels for up to 24 h (Figure 4B and C). At 48 h post-stimulation HinfI accessibility had decreased to some extent, but the promoter still displayed maximal accessibility to MNase (Figure 4B and C).
Figure 4.
GM-CSF promoter remodelling and gene transcription display distinct kinetics. (A) GM-CSF mRNA levels were determined by real-time PCR analysis of cDNA prepared from EL-4 T cells stimulated with P/I for the indicated time periods. The mean and standard error of three replicate assays are shown. (B and C) Nuclei from cells stimulated with P/I for the indicated time periods, were incubated with HinfI (B) or MNase (C). Genomic DNA was analysed by real-time PCR using primer set −I. The mean and standard error of three replicate assays are shown. (D) EL-4 T cells were either left unstimulated or stimulated with P/I for 2 h, then the stimulus was withdrawn and the cells incubated for the indicated time periods. GM-CSF mRNA levels were determined by real-time PCR analysis of cDNA prepared from cells harvested at the indicated time points. The mean and standard error of three replicate assays are shown. (E and F) Nuclei were isolated from the cells treated as in (D) for the time periods indicated, then incubated with HinfI (E) and MNase (F). Genomic DNA was analysed by real-time PCR using primer set −I. The mean and standard error of three replicate assays are shown. (G) EL-4 T cells were either left unstimulated or stimulated with P/I for 2 h as indicated, then the stimulus withdrawn for 22 h. Cells were then either left unstimulated (NS) or restimulated for 4 h with P, I or PI. GM-CSF mRNA levels were determined by real-time PCR analysis of cDNA prepared from the cells. The mean and standard error of three replicate assays are shown. (H) Nuclear extracts were prepared from either unstimulated EL-4 T cells or cells stimulated with P/I for the indicated time periods and analysed by western blotting with the indicated antibodies. (I) EL-4 T cells were either left unstimulated or stimulated with P/I for 2 h, then the stimulus was withdrawn and the cells were incubated for the indicated time periods. Nuclear extracts were prepared and analysed by western blotting with the indicated antibodies.
Therefore, while GM-CSF gene transcription occurs transiently following T cell activation, GM-CSF promoter remodelling is more stable, with the promoter maintained in an accessible state for >48 h post-stimulation.
To determine whether the maintenance of a remodelled state at the GM-CSF promoter requires persistent stimulation, the cells were stimulated with P/I for 2 h, which gives sufficient time to induce changes in chromatin accessibility (Figure 4B and C), and the stimulus then removed for various times. After stimulus removal GM-CSF mRNA levels declined by 4 h and returned to basal levels within 22 h (Figure 4D). In contrast, following stimulus withdrawal, the accessibility of the HinfI site in the promoter was maintained at near maximal levels for at least 4 h, but by 22 h it was decreased by ∼50% (Figure 4E). Accessibility to MNase was also examined over an extended time course. Cells were stimulated for 2 h with P/I to facilitate complete chromatin remodelling (Figure 4C) and then incubated for up to 72 h either in the presence of P/I or following stimulus withdrawal. In the continual presence of P/I stimulation, promoter accessibility was maintained for 48 h, and although decreasing by 72 h, had still not returned to basal levels (Figure 4F). In contrast, if the stimulus was withdrawn at 2 h, accessibility decreased within 22 h, and had returned to basal levels 46 h after stimulus withdrawal. Therefore, the remodelled GM-CSF promoter returns to its basal state following stimulus withdrawal but with significantly delayed kinetics compared to mRNA levels.
Chromatin remodelling across the promoter is induced by stimulation with P, but additional signals generated by the treatment of cells with I are required to induce significant transcription (Figure 1). To determine whether these stimuli are both required for transcriptional activation once chromatin changes are induced, cells were stimulated for 2 h, then the stimulus withdrawn for 22 h to allow the mRNA levels, but not the chromatin accessibility to return to basal levels. Cells were then either left unstimulated or restimulated with P, I or P/I (Figure 4G). While a small increase in the GM-CSF mRNA levels was detected upon stimulus with P alone, both P and I were required to induce a significant increase in the mRNA levels. Therefore, the signals generated by treatment of cells with P are required to induce chromatin remodelling events across the promoter, but are also required to induce GM-CSF transcriptional activation.
The kinetics of transcription factor activation in the nucleus was then compared with those of chromatin remodelling. As expected, RelA and NFATp were translocated to the nucleus rapidly following stimulation and, mirroring the GM-CSF mRNA levels (Figure 4A), had returned to near basal levels by 24 h post-stimulation (Figure 4H). In contrast, c-Rel and NFATc accumulation in the nucleus occurred more slowly and were still present in the nucleus for up to 24 h (Figure 4H). If the presence of c-Rel and/or NFATc in the nucleus is required for the maintenance of the remodelled state, then these proteins may persist following stimulus withdrawal. If the stimulus is removed after 2 h, RelA levels decrease dramatically within 4 h and are completely depleted within 22 h (Figure 4I), mirroring GM-CSF transcription events (Figure 4D). Similarly, both NFATp and NFATc are present in the nucleus 6 h post-stimulation but are depleted from the nucleus rapidly following stimulus withdrawal (Figure 4I). However, the c-Rel levels in the nucleus are maintained 4 h after stimulus removal, and although at somewhat reduced levels, are still evident in the nucleus, 22 h after stimulus withdrawal (Figure 4I). Therefore, the nuclear accumulation of c-Rel is the most stable of these proteins.
Thus, the presence of c-Rel correlates with the more stable kinetics of chromatin remodelling and with the ability to maintain chromatin accessibility following stimulus withdrawal.
c-Rel is required for chromatin remodeling events at the GM-CSF promoter
Chromatin remodelling events at the GM-CSF promoter correlate with the presence of NF-κB proteins, and c-Rel in particular, in the cell nucleus. To investigate the requirement for c-Rel, cells were pre-treated with pentoxifylline before stimulation with P/I for 4 h. Treatment of cells with 6 mg/ml of pentoxifylline inhibited c-Rel but not RelA nuclear accumulation (29) (Figure 5A). This concentration of pentoxifylline also inhibited nuclear accumulation of NFATc but not NFATp (Figure 5B). Pre-treatment with pentoxifylline inhibited chromatin remodelling at the GM-CSF promoter in response to P/I stimulation (Figure 5C), implicating c-Rel in GM-CSF promoter remodelling. While the nuclear activation of NFATc was also inhibited under these conditions, the NFAT proteins were previously found not to contribute to promoter remodelling events (Figure 2). GM-CSF transcription still occurred in pentoxifylline-treated cells although at reduced levels (Figure 5D). This parallels the observations made in the presence of cyclohexamide.
Figure 5.
c-Rel is required for GM-CSF promoter remodeling. (A and B) Nuclear extracts were prepared from EL-4 T cells pre-treated with the indicated concentrations of pentoxifylline (PTX) and then stimulated with P/I. Extracts were analysed by western blotting with the indicated antibodies. (C) EL-4 T cells were either untreated or pre-treated with 6 mg/ml of pentoxifylline, then incubated with or without P/I for 4 h, as indicated. Nuclei were incubated with HinfI and the genomic DNA was analysed by real-time PCR using primer set −I. The mean and standard error of three replicate assays are shown. (D) GM-CSF mRNA levels were determined by real-time PCR analysis of cDNA prepared from EL-4 T cells treated as in (C). Data were graphed as a fold change in the mRNA levels compared with unstimulated, untreated cells. The mean and standard error of three replicate assays are shown. (E) CD4+ T cells isolated from wild-type or c-Rel−/− mice were incubated with P/I and anti-CD28 antibodies for 4 h, as indicated. Nuclei were incubated with HinfI and the genomic DNA was analysed by real-time PCR using primer set −I. Data were graphed as a relative accessibility compared to unstimulated wild-type T cells. The data from one representative experiment are shown. (F) CD4+ T cells isolated from wild-type or c-Rel−/− mice were incubated with anti-CD3 and anti-CD28 antibodies for 4 h, as indicated. Nuclei were incubated with MNase and analysed as in (E).
Induction of GM-CSF gene expression is impaired in the T cells from c-Rel−/− mice (30). To determine whether chromatin remodelling events at the GM-CSF promoter are also impaired in c-Rel−/− mice, HinfI accessibility of the GM-CSF promoter was examined in the CD4+ T cells isolated from wild-type or c-Rel−/− mice. While an increase in accessibility was detected in wild-type T cells following stimulation with P/I and anti-CD28 antibodies, no change in accessibility was detected in the T cells from c-Rel−/− mice (Figure 5E). Similarly, the stimulation of wild-type T cells with anti-CD3/anti-CD28 antibodies resulted in an increased promoter accessibility to MNase (Figure 5F), and while the basal accessibility to MNase was higher in the unstimulated T cells from c-Rel−/− mice, no increased accessibility was observed following stimulation (Figure 5F). Thus, these data clearly demonstrate that c-Rel is required for GM-CSF promoter remodelling in response to T cell stimulation.
DISCUSSION
Upon T cell activation, induction of GM-CSF gene transcription is accompanied by changes in chromatin accessibility across the proximal promoter (7,9,22). These changes are limited to a region of <200 bp, suggesting the remodelling of a single nucleosome (9). We show here that the changes in chromatin structure and GM-CSF gene transcription are distinct molecular events, having different transcription factor and signal requirements. Activation of the PKC signalling pathway is both necessary and sufficient to induce changes in chromatin accessibility at the GM-CSF promoter. The PKC pathway may signal chromatin remodelling events at the GM-CSF promoter in a number of ways. Transcription factors activated via this pathway may bind to the GM-CSF promoter and recruit chromatin remodelling activities, and there is evidence to support such a mechanism. PKC signalling induces nuclear expression of the NF-κB transcription factors, which have been implicated in the induction of chromatin remodelling events at the GM-CSF promoter, both here and in previous studies (9,22). In addition, the signals generated from PKC mobilization may activate coactivators or remodelling complexes required for GM-CSF promoter remodelling, or alternatively may signal to histone proteins directly. In support of this mechanism, the transcriptional activation of the LDL receptor via PKC signalling was recently found to correlate with histone H3-Ser10 phosphorylation at the LDL receptor gene (31). PKC was also found to phosphorylate H3-Ser10 in vitro. Further studies are required to determine whether chromatin remodelling events at the GM-CSF promoter involve similar hyperphosphorylation of histone proteins.
While the activation of the PKC signalling pathway is sufficient to induce changes in the chromatin structure at the GM-CSF promoter, additional signals generated by the activation of intracellular calcium are required to induce high levels of GM-CSF gene transcription. However, just as changes in chromatin structure can be induced across the GM-CSF promoter without significant gene transcription occurring, transcriptional activation of the GM-CSF gene can also be induced without any detectable changes in accessibility at the promoter. This suggests that the transcription machinery can be assembled at the GM-CSF promoter and drive transcription, at least at a low level, without reorganization of the chromatin structure across the promoter. This indicates that the mechanics of GM-CSF promoter activation are different to a gene such as IFNβ, where remodelling of a nucleosome positioned over the TATA box is a prerequisite for gene transcription (14,16) and may reflect the different chromatin environment of the IFNβ enhancer compared to the GM-CSF promoter. Whereas the IFNβ enhancer is found in a nucleosome-free linker region and an adjacent nucleosome must be remodelled to expose the TATA box and enable transcription to occur (11,14), the GM-CSF proximal promoter, and TATA box, appears to be assembled into a nucleosome structure (9). However, even in resting T cells, the GM-CSF promoter displays some inherent accessibility and this may enable the transcription machinery to be recruited and assembled at the promoter without significant changes to the chromatin structure. Remodelling of the proximal promoter region may be required for the binding of additional activators to facilitate an efficient recruitment of the transcription machinery and high-level gene transcription.
Chromatin remodelling of the GM-CSF promoter and gene transcription also occurs with different kinetics following T cell activation. While transcriptional activation occurs transiently following stimulation, with the mRNA levels maximal at 6–8 h, the remodelled chromatin structure is relatively stable in comparison and is maintained for several days. How the remodelled chromatin structure is retained and whether it is actively maintained or reset following cell division is unclear. At least in EL-4 T cells, which undergo rapid cell division, the maintenance of the remodelled state for up to 48 h following stimulation would suggest that the remodelling is not automatically reset during DNA replication, but is actively maintained through cell division. The stability of the remodelled chromatin state is mirrored by the stable expression of the NF-κB protein, c-Rel, in the nucleus, following stimulation, and it is therefore possible that the remodelled state is actively maintained as long as the c-Rel protein is present in the nucleus.
The NF-κB proteins have been previously implicated in chromatin remodelling events at the GM-CSF promoter (9,22), and here, we have demonstrated a role for the c-Rel protein in particular. From our data, it is clear that the presence of c-Rel in the nucleus mirrors the chromatin remodelling events at the promoter. The stability of the chromatin remodelling correlates with the stable nuclear accumulation of c-Rel, while the prevention of nuclear accumulation of c-Rel with a PKC inhibitor, pentoxifylline or cyclohexamide correlated with the inhibition of chromatin remodelling. Furthermore, the delayed depletion of c-Rel from the nucleus following stimulus withdrawal correlated with the delayed return of the promoter to a less accessible state. This correlation between c-Rel and the chromatin remodelling events was verified by data from c-Rel−/− mice. Stimulation of T cells from c-Rel−/− mice failed to induce changes in chromatin remodelling at the GM-CSF promoter. This is similar to chromatin remodelling events at the IL-2 promoter, which have also been found to be dependent on the c-Rel protein (32), but in contrast to remodelling events at the IL-12 promoter for which c-Rel is dispensable (33). It is not yet clear whether other NF-κB family members also contribute to remodelling events. The NF-κB family consists of five members; RelA, c-Rel, p50/p105, p52/p100 and RelB. While the role of RelB in T cell differentiation and activation is not well characterized, defects in lymphocyte activation have been observed in knockout mice of all the other family members [reviewed in (34)]. Since the other family members, particularly RelA and p50 have well-established roles in T cell activation, they may contribute to the GM-CSF gene activation. Chromatin remodelling at the GM-CSF promoter was inhibited in cyclohexamide-treated cells in which RelA but not c-Rel proteins were present in the cell nucleus. This suggests that the RelA protein alone is not sufficient to drive remodelling events. However, this does not rule out the possibility that it may act in conjunction with c-Rel to facilitate the chromatin remodelling, as both these proteins bind to the CK-1 element in the GM-CSF promoter at least in vitro (35). It is clear that the RelA family member is required at least for subsequent transcriptional events as microarray analysis of T cells from RelA−/− mice revealed a deficiency in GM-CSF gene activation (M. F. Shannon, unpublished data). In contrast to the NF-κB proteins, NFAT proteins are dispensable for the chromatin remodelling events at the GM-CSF promoter, but appear to cooperate with NF-κB proteins to direct efficient GM-CSF gene transcription.
It is likely then that the activation of the GM-CSF gene in response to T cell stimulation is a 2-step process in which the activation of the PKC signalling pathway results in the accumulation of NF-κB proteins in the cell nucleus. The inherent accessibility of nucleosomes across the promoter region may allow the binding of NF-κB proteins without prior remodelling events, and we have shown previously that this can occur, at least in vitro (9). The NF-κB proteins may then recruit chromatin remodelling activities such as the SWI/SNF complex to increase the accessibility of the promoter to other transcription factors. In support of this, we have demonstrated that the brg1 protein is recruited to the GM-CSF promoter in vitro, in an NF-κB dependent manner (9). Increased accessibility of the promoter may then facilitate binding of other transcription factors, such as NFAT, to enable an efficient recruitment of the transcription machinery.
Acknowledgments
We thank Dr Steve Gerondakis for providing the c-Rel−/− mouse line. This work was funded by a project grant from the National Health and Medical Research Council, Australia to M.F.S. and A.F.H. Funding to pay the Open Access publication charges for this article was provided by the NH&MRC, Australia.
REFERENCES
- 1.Shannon M.F., Coles L.S., Vadas M.A., Cockerill P.N. Signals for activation of the GM-CSF promoter and enhancer in T cells. Crit. Rev. Immunol. 1997;17:301–323. doi: 10.1615/critrevimmunol.v17.i3-4.30. [DOI] [PubMed] [Google Scholar]
- 2.Avots A., Escher C., Muller-Deubert S., Neumann M., Serfling E. The interplay between lymphoid-specific and ubiquitous transcription factors controls the expression of interleukin 2 gene in T lymphocytes. Immunobiology. 1995;193:254–258. doi: 10.1016/s0171-2985(11)80551-6. [DOI] [PubMed] [Google Scholar]
- 3.Workman J.L., Kingston R.E. Alteration of nucleosome structure as a mechanism of transcriptional regulation. Annu. Rev. Biochem. 1998;67:545–579. doi: 10.1146/annurev.biochem.67.1.545. [DOI] [PubMed] [Google Scholar]
- 4.Holloway A.F., Rao S., Shannon M.F. Regulation of cytokine gene transcription in the immune system. Mol. Immunol. 2002;38:567–580. doi: 10.1016/s0161-5890(01)00094-3. [DOI] [PubMed] [Google Scholar]
- 5.Ward S.B., Hernandez-Hoyos G., Chen F., Waterman M., Reeves R., Rothenberg E.V. Chromatin remodeling of the interleukin-2 gene: distinct alterations in the proximal versus distal enhancer regions. Nucleic Acids Res. 1998;26:2923–2934. doi: 10.1093/nar/26.12.2923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Weinmann A.S., Plevy S.E., Smale S.T. Rapid and selective remodeling of a positioned nucleosome during the induction of IL-12 p40 transcription. Immunity. 1999;11:665–675. doi: 10.1016/s1074-7613(00)80141-7. [DOI] [PubMed] [Google Scholar]
- 7.Cockerill P.N., Shannon M.F., Bert A.G., Ryan G.R., Vadas M.A. The granulocyte-macrophage colony-stimulating factor/interleukin 3 locus is regulated by an inducible cyclosporin A-sensitive enhancer. Proc. Natl Acad. Sci. USA. 1993;90:2466–2470. doi: 10.1073/pnas.90.6.2466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rao S., Procko E., Shannon M.F. Chromatin remodeling, measured by a novel real-time polymerase chain reaction assay, across the proximal promoter region of the IL-2 gene. J. Immunol. 2001;167:4494–4503. doi: 10.4049/jimmunol.167.8.4494. [DOI] [PubMed] [Google Scholar]
- 9.Holloway A.F., Rao S., Chen X., Shannon M.F. Changes in chromatin accessibility across the GM-CSF promoter upon T cell activation are dependent on nuclear factor kappaB proteins. J. Exp. Med. 2003;197:413–423. doi: 10.1084/jem.20021039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Almer A., Rudolph H., Hinnen A., Horz W. Removal of positioned nucleosomes from the yeast PHO5 promoter upon PHO5 induction releases additional upstream activating DNA elements. EMBO J. 1986;5:2689–2696. doi: 10.1002/j.1460-2075.1986.tb04552.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Agalioti T., Lomvardas S., Parekh B., Yie J., Maniatis T., Thanos D. Ordered recruitment of chromatin modifying and general transcription factors to the IFN-beta promoter. Cell. 2000;103:667–678. doi: 10.1016/s0092-8674(00)00169-0. [DOI] [PubMed] [Google Scholar]
- 12.Peterson C.L., Workman J.L. Promoter targeting and chromatin remodeling by the SWI/SNF complex. Curr. Opin. Genet. Dev. 2000;10:187–192. doi: 10.1016/s0959-437x(00)00068-x. [DOI] [PubMed] [Google Scholar]
- 13.Becker P.B., Horz W. ATP-dependent nucleosome remodeling. Annu. Rev. Biochem. 2002;71:247–273. doi: 10.1146/annurev.biochem.71.110601.135400. [DOI] [PubMed] [Google Scholar]
- 14.Lomvardas S., Thanos D. Nucleosome sliding via TBP DNA binding in vivo. Cell. 2001;106:685–696. doi: 10.1016/s0092-8674(01)00490-1. [DOI] [PubMed] [Google Scholar]
- 15.McAndrew P.C., Svaren J., Martin S.R., Horz W., Goding C.R. Requirements for chromatin modulation and transcription activation by the Pho4 acidic activation domain. Mol. Cell. Biol. 1998;18:5818–5827. doi: 10.1128/mcb.18.10.5818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lomvardas S., Thanos D. Modifying gene expression programs by altering core promoter chromatin architecture. Cell. 2002;110:261–271. doi: 10.1016/s0092-8674(02)00822-x. [DOI] [PubMed] [Google Scholar]
- 17.Svaren J., Horz W. Transcription factors vs nucleosomes: regulation of the PHO5 promoter in yeast. Trends Biochem. Sci. 1997;22:93–97. doi: 10.1016/s0968-0004(97)01001-3. [DOI] [PubMed] [Google Scholar]
- 18.Gasson J.C. Molecular physiology of granulocyte-macrophage colony-stimulating factor. Blood. 1991;77:1131–1145. [PubMed] [Google Scholar]
- 19.Fraser J.D., Weiss A. Regulation of T-cell lymphokine gene transcription by the accessory molecule CD28. Mol. Cell. Biol. 1992;12:4357–4363. doi: 10.1128/mcb.12.10.4357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Thomas R.S., Tymms M.J., McKinlay L.H., Shannon M.F., Seth A., Kola I. ETS1, NFkappaB and AP1 synergistically transactivate the human GM-CSF promoter. Oncogene. 1997;14:2845–2855. doi: 10.1038/sj.onc.1201125. [DOI] [PubMed] [Google Scholar]
- 21.Jenkins F., Cockerill P.N., Bohmann D., Shannon M.F. Multiple signals are required for function of the human granulocyte-macrophage colony-stimulating factor gene promoter in T cells. J. Immunol. 1995;155:1240–1251. [PubMed] [Google Scholar]
- 22.Cakouros D., Cockerill P.N., Bert A.G., Mital R., Roberts D.C., Shannon M.F. A NF-kappa B/Sp1 region is essential for chromatin remodeling and correct transcription of a human granulocyte-macrophage colony-stimulating factor transgene. J. Immunol. 2001;167:302–310. doi: 10.4049/jimmunol.167.1.302. [DOI] [PubMed] [Google Scholar]
- 23.Osborne C.S., Vadas M.A., Cockerill P.N. Transcriptional regulation of mouse granulocyte-macrophage colony-stimulating factor/IL-3 locus. J. Immunol. 1995;155:226–235. [PubMed] [Google Scholar]
- 24.Schreiber E., Matthias P., Muller M.M., Schaffner W. Rapid detection of octamer binding proteins with ‘mini-extracts’, prepared from a small number of cells. Nucleic Acids Res. 1989;17:6419. doi: 10.1093/nar/17.15.6419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liu J., Farmer J.D., Jr, Lane W.S., Friedman J., Weissman I., Schreiber S.L. Calcineurin is a common target of cyclophilin–cyclosporin A and FKBP-FK506 complexes. Cell. 1991;66:807–815. doi: 10.1016/0092-8674(91)90124-h. [DOI] [PubMed] [Google Scholar]
- 26.Shannon M.F., Himes S.R., Coles L.S. GM-CSF and IL-2 share common control mechanisms in response to costimulatory signals in T cells. J. Leukoc. Biol. 1995;57:767–773. doi: 10.1002/jlb.57.5.767. [DOI] [PubMed] [Google Scholar]
- 27.Baeuerle P.A., Henkel T. Function and activation of NF-kappa B in the immune system. Annu. Rev. Immunol. 1994;12:141–179. doi: 10.1146/annurev.iy.12.040194.001041. [DOI] [PubMed] [Google Scholar]
- 28.Rao A., Luo C., Hogan P.G. Transcription factors of the NFAT family: regulation and function. Annu. Rev. Immunol. 1997;15:707–747. doi: 10.1146/annurev.immunol.15.1.707. [DOI] [PubMed] [Google Scholar]
- 29.Wang W., Tam W.F., Hughes C.C., Rath S., Sen R. c-Rel is a target of pentoxifylline-mediated inhibition of T lymphocyte activation. Immunity. 1997;6:165–174. doi: 10.1016/s1074-7613(00)80423-9. [DOI] [PubMed] [Google Scholar]
- 30.Gerondakis S., Strasser A., Metcalf D., Grigoriadis G., Scheerlinck J.Y., Grumont R.J. Rel-deficient T cells exhibit defects in production of interleukin 3 and granulocyte-macrophage colony-stimulating factor. Proc. Natl Acad. Sci. USA. 1996;93:3405–3409. doi: 10.1073/pnas.93.8.3405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Huang W., Mishra V., Batra S., Dillon I., Mehta K.D. Phorbolester promotes histone H3-Ser10 phosphorylation at low density lipoprotein receptor promoter in a protein kinase C-dependent manner in human hepatoma HepG2 cells. J. Lipid Res. 2004;45:1519–1527. doi: 10.1194/jlr.M400088-JLR200. [DOI] [PubMed] [Google Scholar]
- 32.Rao S., Gerondakis S., Woltring D., Shannon M.F. c-Rel is required for chromatin remodeling across the IL-2 gene promoter. J. Immunol. 2003;170:3724–3731. doi: 10.4049/jimmunol.170.7.3724. [DOI] [PubMed] [Google Scholar]
- 33.Weinmann A.S., Mitchell D.M., Sanjabi S., Bradley M.N., Hoffmann A., Liou H.C., Smale S.T. Nucleosome remodeling at the IL-12 p40 promoter is a TLR-dependent, Rel-independent event. Nature Immunol. 2001;2:51–57. doi: 10.1038/83168. [DOI] [PubMed] [Google Scholar]
- 34.Li Q., Verma I.M. NF-kappaB regulation in the immune system. Nature Rev. Immunol. 2002;2:725–734. doi: 10.1038/nri910. [DOI] [PubMed] [Google Scholar]
- 35.Himes S.R., Coles L.S., Reeves R., Shannon M.F. High mobility group protein I(Y) is required for function and for c-Rel binding to CD28 response elements within the GM-CSF and IL-2 promoters. Immunity. 1996;5:479–489. doi: 10.1016/s1074-7613(00)80503-8. [DOI] [PubMed] [Google Scholar]