Abstract
Hybridization kinetics of DNA sequences with known secondary structures and random sequences designed with similar melting temperatures were studied in solution and when one strand was bound to 5 μm silica microspheres. The rates of hybridization followed second-order kinetics and were measured spectrophotometrically in solution and fluorometrically in the solid phase. In solution, the rate constants for the model sequences varied by almost two orders of magnitude, with a decrease in the rate constant with increasing amounts of secondary structure in the target sequence. The random sequences also showed over an order of magnitude difference in the rate constant. In contrast, the hybridization experiments in the solid phase with the same model sequences showed almost no change in the rate constant. Solid phase rate constants were approximately three orders of magnitude lower compared with the solution phase constants for sequences with little or no single-stranded structure. Sequences with a known secondary structure yielded solution phase rate constants as low as 3 × 103 M−1 s−1 with solid phase rate constants for the same sequences measured at 2.5 × 102 M−1 s−1. The results from these experiments indicate that (i) solid phase hybridization occurs three orders of magnitude slower than solution phase, (ii) trends observed in structure-dependent kinetics of solution phase hybridization may not be applicable to solid phase hybridization and (iii) model probes with known secondary structure decrease reaction rates; however, even random sequences with no known internal single-stranded structure can yield a broad range of reaction rates.
INTRODUCTION
Beginning in the early 1990s, gene microarrays that typically consisted of oligonucleotide probes of at most 100 bases in length bound to solid supports, had been used to provide genetic maps and understand gene functions. These arrays posed a stringent requirement on the kinetics of DNA hybridization. Quantitative measurements were needed for these assays that were typically run in 6–12 h. The assays were performed on two-dimensional surfaces as opposed to the three-dimensional solution phase environments of the past. It was assumed that short length DNA (<100 bases) of similar length would follow the same rates as those measured in solution and that all reactions would reach equilibrium within the time allotted. The evidence that emerged contradicted this assumption, i.e. the new solid support environment appeared to slow down the reaction rate (1–9). In many cases, equilibrium was not established in the allotted time for these experiments and, therefore, the rates of DNA hybridization became an important factor.
A large number of optimized probes were required for the assays performed both in solution and on microarrays for gene expression studies. It was essential that these probes behave similarly since the reactions were multiplexed and carried out in a single reaction container. Any variation in signal strengths due to variation in performance of the probes would yield false gene expression results and a major contributor to performance was the rate at which the probe hybridized to the target. Past conventions used calculated melting temperatures to ensure that the probes behaved similarly and algorithms were designed to help identify optimal probes. Despite this, the probes have often been rejected for gene expression studies based on their mediocre performance in assays. Understanding the relationships between sequence and hybridization rates as well as the extension of solution reactions to surfaces will greatly enhance this field.
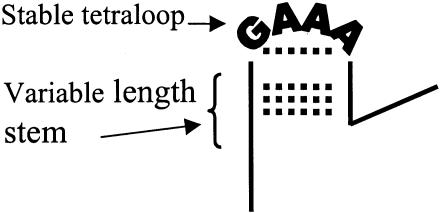
In this study, a number of different sequences were chosen to understand the effects of structure on hybridization reaction rates and to compare kinetics of these sequences in solution and on surfaces. Model sequences were chosen based on a series of stable single-stranded structures that form GNRA tetraloops where N can be A, G, C or T and R must be a purine (10–13). The hairpins that can form from the tetraloops are unique in that they do not consist of a series of repeating bases (14) or large multi-base loop regions (15). The stability of the four-base loop arises from a number of factors including base stacking between the third and fourth A, hydrogen bonding between an amino proton on the G and an oxygen from the phosphate between the second and third A, and base pairing between the G and the third A in the loop (10). In addition to the model tetraloop sequences, a series of random sequences, denoted as random coils, were used for comparison. These sequences have no known stable structure and have similar melting temperatures and identical lengths. To study the possible surface effects on the hybridization rates, kinetic data were obtained both in solution and when one strand was covalently bound to a silica microsphere.
MATERIALS AND METHODS
Solution phase
The DNA used for solution studies was synthesized by Keystone Biosource (Foster City, CA) and Integrated DNA Technologies (Coralville, IA). All hybridization reactions and melting curves were obtained in 5 mM Tris buffer with 25 mM KCl at pH 8.3. Melting curves were generated using 1 μM concentrations of duplexed DNA. For the hybridization reactions, 500 μl of a preheated solution of single-stranded DNA (1 μM) was added to a preheated complement (equimolar) in a thermostated quartz cell placed in either a CARY 3E spectrophotometer or an Ocean Optics UV spectrometer with a temperature regulated cell holder (final concentration of 0.5 μM). Solutions were mixed initially by pipetting the target into the probe solution. Since this took ∼5 s, this mixing time was added to the initial time for each experiment. For the data presented here, there was no additional mixing during data acquisition since the experiments with and without mixing were identical. The absorbance was monitored at 270 nm as a function of time. The hypochromic shift, resulting from the formation of the duplex, caused a decrease of ∼0.02 absorbance units for sequences with little or no stable single-stranded structure (for the fastest sequences, ∼10% of the overall absorbance change was lost during the initial mixing). For sequences containing partial hairpins, the observed hypochromicity decreased as the complementarity in the stem increased. This is expected if intramolecular base pairing is replaced by intermolecular base pairing since the intramolecular structure already has a substantial double-stranded character. All reactions were run at 20°C below the measured Tm of the duplex. In cases where the single-stranded structures were so stable that duplex melting temperatures could not be obtained under the experimental conditions, theoretical melting temperatures were used to calculate the hybridization temperature.
Solid phase
Reagents
Silica beads were obtained from Bangs Labs (Fishers, IN). Amino propyl triethoxy silane (APTES), 3,3′-Dithiobis (propionic acid N-hydroxysuccinimidyl ester) (DTPS), dimethyl formamide (DMF), HEPES buffer, ethanol, Tween-40 and triethyl amine were obtained from Sigma (St Louis, MO). DNA probes were synthesized with a 5′-amine and two penta (ethylene oxide) modifiers. Fluorescently labeled DNA was modified with a dye at either the 3′ or 5′ end. DNA with C6-amino penta (ethylene oxide) modifiers were synthesized using a 394 DNA synthesizer (Applied Biosystems, Inc., Foster City, CA).
Derivatization of silica beads
Silica beads from Bangs Labs contained an excessive amount of ammonium-based ions, which changed the surface chemistry of the particles. Beads were suspended in 1 M sodium chloride solutions at 100°C for 2 h to exchange the cation from ammonium to sodium. The beads were washed with copious amounts of water followed by ethanol and dried. Derivatizations were carried out in microtubes and solutions were mixed in a Thermomixer (Eppendorf) at 1200 r.p.m.
For every 10 mg of silica beads (equivalent to 107 particles), 100 μl of 1 mM solution of APTES in 200-proof ethanol was added and reacted for 30 min. The beads were washed with ethanol and dried in a concentrator (Eppendorf) at 60°C overnight to remove non-specifically bound silanes. The number of reactive amines was determined using the ninhydrin assay (16).
A bifunctional linker, DTPS, was attached to the aminated surface of the bead. Aminated beads were reacted with 10 mM DTPS, ∼10-fold excess to the available amino groups, with a drop of triethyl amine in DMF. The NHS-terminated surface was washed with DMF and ethanol (200-proof). Unreacted amine groups were estimated with the ninhydrin assay.
The C6 amino linker on the 5′ end of the DNA was attached to the activated beads. DNA probes in 100 mM HEPES buffer (pH 7.5–8) were reacted with NHS-terminated beads for 12 h and washed with HEPES buffer several times and vacuum dried. The probes were in a 10-fold excess over the determined number of reactive functional groups on each bead. The unreacted functional groups on the beads were passivated using 0.1 M acetic anhydride in THF and subsequently washed with DMF and ethanol. The Cy5-labeled probes were attached to the beads in the same way as the unlabeled probes. The number of probes attached to the beads was determined by cleaving the disulfide bond using 0.1 M DTT in methanol at 55°C for 12 h. The supernatant containing Cy5 was measured fluorescently.
Hybridization on silica beads
Hybridization assays were carried out in 10 mM Tris buffer with 50 mM KCl at pH ∼8.0. Typically, 4–5 mg of DNA-derivatized silica beads was added to a 2 ml tube along with 600 μl of the buffer solution containing 0.1% Tween-40. The beads were thermally equilibrated at 35–40°C for 5 min. Known amounts of dye labeled target (5′-FAM labeled) were incubated in separate tubes at 35–40°C for 5 min. The target solutions were added to the bead solutions and placed on a mini-rotator (with a speed of 100–200 r.p.m.) in an oven. Each sequence was set up in triplicates so that statistical information could be obtained. Since most microarray assays are not set up to measure the Tm of the duplex bound to the surface, solution phase values are typically assumed and the hybridization assays are run from 15 to 25°C below the (calculated) solution phase Tm. In these experiments, hybridization was carried out at 35 ± 5°C for each of the sequences reported here. At various time intervals, tubes were removed from the oven and the beads were spun down. The supernatant liquid was removed and 600 μl of hybridization buffer was added. This procedure was repeated three times to remove the non-hybridized target. Beads were washed with water, followed by ethanol, and then dried and weighed. Hybridization buffer was added to each tube and fluorescent measurements were taken for the bead solution. For comparison measurements, the duplex DNA was denatured using 0.1 N NaOH for 2 min at 50°C and the labeled target was measured in solution. During the experiment, up to 50% bead loss was observed due to the multiple washing and drying steps. This resulted in fluctuations in the target FAM intensity for a given time point. These fluctuations were larger than the change in intensity from one time point to the next. In order to compensate for the bead loss, a 3′ Cy5-labeled probe of the same sequence was used. The labeled probe was spiked in to the unlabeled probe mixture during conjugation. The Cy5-labeled probe accounted for ∼8–10% of the total number of probes bound to the surface. The Cy5 measurement was made before bead washing for each time point and, therefore, the intensity remained constant throughout the kinetic experiment. In order to verify whether Cy5 quenched the FAM intensity following hybridization, the FAM intensity was measured on bead and after denaturation. The difference in the FAM signal strengths on bead and in solution were the same whether or not Cy5 was present. Each Cy5 measurement was used to correct for bead loss by calculating the ratio of the target FAM intensity to it.
Kinetic measurements
Two methods were used to measure fluorescence. In order to correct for bead loss, the bead solutions were measured in hybridization buffer at 520 and 665 nm (FAM and Cy5 emission, respectively), using front-face detection in a SPEX Fluorolog 3 fluorometer (Edison, NJ). The FAM-labeled target was also measured by denaturing it from the surface-bound probe in 0.1 N NaOH. The denatured FAM target intensity was used to calculate the second-order rate constant, k. To determine k, it was necessary to convert the number of probes bound to beads to a concentration. To convert to concentration, the total number of probe molecules within a given reaction was measured and used with the total volume of the bead suspension to calculate a pseudo-concentration for the surface-bound probe that had units of molarity.
RESULTS
Solution phase
Solution phase experiments were designed to study the effect of sequence conformation on rate constant. The results presented here on the partial hairpins are similar to those reported by Armitage and co-workers (17); however, only deoxyribonucleic acids were used here and the length of the sequences in this study were more closely matched to those typically used in microarray assays. The list of sequences is shown in Table 1 along with the corresponding measured rate constants and melting temperatures. The first set of sequences (RC A–G) was chosen because they had similar calculated and measured melting temperatures. They include seven 24-base sequences that appear to be random coils with no known secondary structure. Although all the melting temperatures are within 10°C of each other, the corresponding spread in rate constants spans an order of magnitude. The second set of sequences (PH series) includes three variations of a 20-base partial hairpin and a random sequence 20mer that was used for comparison (RC H). The partial hairpin sequences were designed based on the stability of the GNRA tetraloops. Figure 1 shows a schematic diagram of the theoretical structure of the stable GAAA tetraloop (10). The partial hairpin structures designed for these experiments have a varying number of complementary base pairs (0–4) along the stem. Both the sequence and its complement contain the same number of base pairs along the stem; however, only one sequence contains the tetraloop as well. The most stable set of single-stranded sequences corresponds to the set of CGAAAG sequences (PH A–E). Control sequences were designed by replacing the base pair adjacent to the loop region from CG to a lesser stable GC (PH B′E′) (11). In order to observe the stability effects of the tetraloop, the loop region was replaced with TGTT (PH B″–E″), keeping the other bases along the stem the same.
Table 1.
A list of the sequences, second-order rate constants and experimental melting temperatures used in this study
| Sequence name | Sequence | Number of complements in stem | Second-order rate constant, k (M−1 s−1) | Melting temperature, Tm (°C) |
|---|---|---|---|---|
| RC A | AAAGCCATGCTTTCCCTCCCAAAG | 4.0 × 104 | 55 | |
| RC B | CGTTAGGATGCGTACAGTGCGTGC | 1.2 × 105 | 60 | |
| RC C | CTTGTGCGTGCGCCATTCTGCTGT | 5.3 × 104 | 60 | |
| RC D | GGTCTGAGGCACCGCAGTCCACGA | 1.2 × 105 | 64 | |
| RC E | CGTGCGTGACAGCAAGGCACGACA | 1.2 × 104 | 65 | |
| RC F | CGATGCACGTCCCACTCGTGCTTT | 1.6 × 104 | 62 | |
| RC G | AAGCCATCCTTTCGATTTGCCAAG | 1.1 × 105 | 55 | |
| RC H | CAGGACATGGGTTCTCTTCT | 7.4 × 104 | 53 | |
| PH A | CCAATAACGAAACCCGCCAC | 0 | 1.9 × 105 | 55 |
| PH B | CCAATAACGAAAGCCGCCAC | 1 | 1.6 × 105 | 55 |
| PH C | CCAATAGCGAAAGCCGCCAC | 2 | 2.4 × 104 | 57 |
| PH D | CCAATGGCGAAAGCCGCCAC | 3 | 3 × 103a | — |
| PH E | CCAACGGCGAAAGCCGCCAC | 4 | 3 × 103a | — |
| PH B′ | CCAATAAGGAAACCGGCCAC | 1 | 1.3 × 105 | 53 |
| PH C′ | CCAATAGGGAAACCGGCCAC | 2 | 1.3 × 105 | 55 |
| PH D′ | CCAATCGGGAAACCGGCCAC | 3 | 9.6 × 104 | 56 |
| PH E′ | CCAACCGGGAAACCGGCCAC | 4 | 1.3 × 104 | 62 |
| PH B″ | CCAATAAGTGTTCCGGCCAC | 1 | 1.3 × 105 | 55 |
| PH C″ | CCAATAGGTGTTCCGGCCAC | 2 | 9.7 × 104 | 56 |
| PH D″ | CCAATCGGTGTTCCGGCCAC | 3 | 1.5 × 105 | 60 |
| PH E″ | CCAACCGGTGTTCCGGCCAC | 4 | 2.8 × 104 | 62 |
RC A–G are random coil 24mer sequences that have similar measured Tms. RC H is a random coil 20mer, which was used for comparison with the partial hairpin (PH) sequences. For sequences PH B′–E′, the bases in italics were changed from the most stable CGAAAG to a lesser stable form: GGAAAC. For sequences PH A″–E″, the GAAA tetraloop in PH A′–E′ was replaced with TGTT. The number of complementary pairs along the stem for the PH sequences is indicated.
aThese values are upper limits for k.
Figure 1.
A schematic representation of the theoretical structure of the stable GAAA tetraloop within a single strand of DNA showing base pairing between the G and the last A in the loop and complementarity within the stem (hydrogen bonding is represented as black dots).
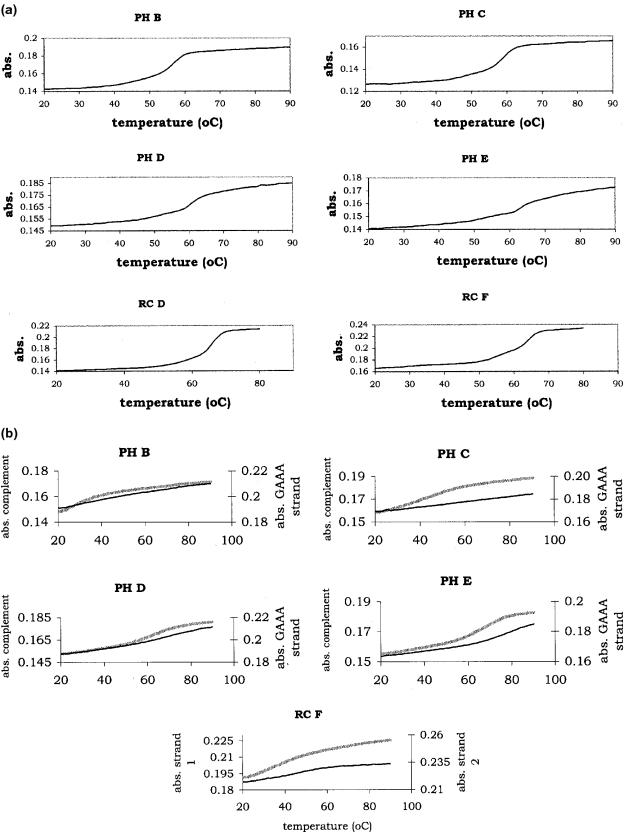
Table 1 shows the variation in k for solution phase PH series rate constants from as low as 3 × 103 to >1.5 × 105 M−1 s−1. For PH D and PH E, there is a greater uncertainty in the values of k since equilibrium values for the absorbances were approximated (reactions did not reach an equilibrium on the time-scale of the experiment). In addition, it was not possible to extract an accurate melting temperature for the duplex since the melting curves were complex (see Figure 2a). Instead of using the experimental melting temperature to calculate the temperature for hybridization, PH D and PH E hybridization reactions were run at 20°C below the calculated Tm. The reactions for PH D were run at 40°C and for PH E, the reaction was run at 42°C. Figure 2a shows the melting curves for the partial hairpin sequences, PH B–E, and for two of the random coil 24mers, RC D and F. PH B, PH C, RC D and RC F show fairly smooth transitions from double-stranded to single-stranded structures. The curves for PH D and E are more complex and may be indicative of biphasic transitions (17,18). Because of this, it was impossible to extract a true Tm for the PH D and PH E duplexes. For these sequences, the melting curves include phase transitions for the single strands and possibly more complex intermediates. It was not possible to simply subtract the single-stranded melts from the duplex melts since the single-stranded curves did not always yield clear transitions and for some cases (PH E) the melts were too high to measure in the buffer and salt concentrations used.
Figure 2.
(a) Melting curves for some of the sequences shown in Table 1 hybridized to their fully matched complementary strands. The first two sequences, corresponding to PH B and PH C show a smooth transition from ds to ss; PH D and PH E are more complex and Tms were not obtained from these curves. RC D and RC F correspond to random coils with Tms similar to each other. (b) Single-stranded melting curves for the PH series and one of the random coils shown in (a). The absorbance of the GAAA strand is shown on the right axis and the absorbance of the complement is shown on the left axis for the first four traces. The solid, thin lines in PH B–E correspond to strands containing the GAAA tetraloop. The heavier, lighter lines in PH B–E correspond to the complements of the tetraloop strand. The dashed and solid line for RC F corresponds to two complementary strands.
Figure 2b shows a series of single-stranded melts for PH B–E and a typical set of melting curves from single strands of the random coil sequences (RC F is shown). The single-stranded melts were measured under the same conditions as the duplexed structures. Examining the melts from the tetraloop strand (shown as a solid line), PH B and PH C show a steady increase in absorbance with an increase in temperature but none of the sequences show a clear transition temperature that might be correlated with the melting temperature of the single strand. The curve for the tetraloop PH D shows the beginning of a melting transition that becomes clearer for the tetraloop PH E. However, neither PH D nor PH E reaches a plateau. The complementary strands (shown as dashed lines in the figure) show transitions that shift to a higher temperature with increasing numbers of complements along the stem. The complementary strands for PH D and PH E appear to have lower melting temperatures, compared with the tetraloop strands since they both reach a plateau. Both strands from the random coil sequence look similar to the complementary strand for PH C. Although the melts from both strands show evidence of a transition, the transitions appear to take place at or below the hybridization temperature for the duplex implying that the single-stranded conformation for RC F appears only at temperatures below ∼42°C.
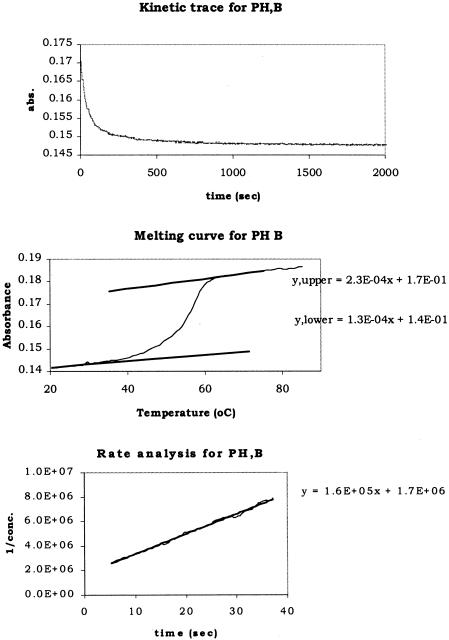
The upper panel in Figure 3 shows a typical kinetic trace from the hybridization of PH B with its complement. The second-order rate constants were extracted from curves like these by converting absorbance data into concentration using absorbances of measured single-stranded (ss) and double-stranded (ds) DNA obtained from the melting curve, shown in the middle panel of Figure 3. The absorbance of dsDNA was obtained by fitting the lower portion of the melting curve to a straight line. The fit was then used to calculate the absorbance of dsDNA at the hybridization temperature. The absorbance of the two ssDNAs were calculated in the same way, fitting the upper portion of the melting curve to a straight line. The concentration of ssDNA at time t, Ct, was then calculated using the following equation:
| 1 |
where At is the absorbance at time t, A∞ is the absorbance at infinite time (at equilibrium), C0 is the initial concentration of ssDNA, and Ass and Ads are the absorbances of ssDNA and dsDNA, respectively. Alternatively, the values of absorbance of ssDNA and dsDNA from the kinetic curve could have been used, providing the reaction had reached equilibrium. The lower panel of Figure 3 shows the analysis used to determine the second-order rate constant, k. Absorbance data up to three times the half-life was converted into concentration and the inverse was plotted as a function of time. The slope of the line gave the second-order rate constant. For PH B, k = 1.6 × 105 M−1 s−1 (±25%). This is approximately twice that of the control sequence, RC H, k = 7.4 × 104 M−1 s−1. Although PH B contains the stable tetraloop, a single base complement along the stem was not enough to stabilize intramolecular structure. The measured half-life, t1/2, for the PH B hybridization was 12 s.
Figure 3.
Upper panel: typical kinetic trace of a second-order solution phase hybridization of a partial hairpin (PH B). Middle panel: melting curve for PH B showing a linear fit to the upper and lower arms. lower panel: graph of the data above, where the units for the y-axis are M−1, fit to a straight line using a linear regression. The slope of the line corresponds to the second-order rate constant.
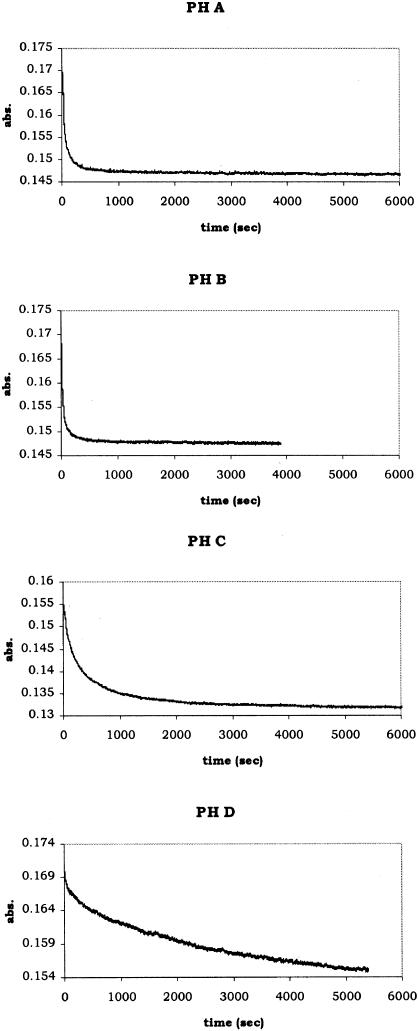
A comparison of the kinetic traces from PH A, B, C and D is shown in Figure 4. These sequences are from the partial hairpins that include CGAAAG. The top trace corresponds to the sequence with no complements along the stem, the second trace contains 1 complement, the sequence from the third trace contains 2 complementary pairs and the sequence from the fourth trace contains 3 complementary pairs. As the number of complementary pairs increases along the stem, the rate constant decreases, as can be seen by the slower decay. Another important feature is the overall change in absorbance from two single strands to the duplex. The greatest change occurs for PH A, with ΔA = 0.026 and the least amount of change occurs for PH D, where ΔA = 0.014. Even though the reaction for PH D has not reached equilibrium, the overall change at the point the reaction was terminated is significantly less than that for PH A at a similar point along the curve. With only five bases on either end of PH D available for hybridization with the complement initially, it seems unlikely that the small change in the kinetic curve is a result of partial hybridization of these five bases since this would yield a hybrid with a melting temperature well below the temperature of the reaction (40°C for PH D). The smaller change in absorbance upon hybridization is more likely because of the initial intramolecular structure. The single strands have an increasing number of base pairs from PH B to PH D and, therefore, the net number of base pairs formed upon hybridization (or the overall absorbance change) decreases from PH B to PH D.
Figure 4.
A series of solution phase kinetic traces from the hybridization of four partial hairpin sequences. Each reaction was carried out at 20°C below the Tm (the calculated Tm was used for PH D) at single strand concentrations of 0.5 μM in 5 mM Tris buffer and 25 mM KCl, pH 8.3.
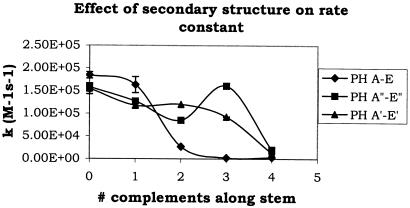
Figure 5 shows the trend in rate constants as a function of the number of complements along the stem for all three PH series. The most dramatic change occurs when 2 bp are added to the stem (from PH B to PH C). The second-order rate constant decreases by almost seven times. In contrast, the rate constants for the sequences containing GGAAAC with 0–3 complements in the stem vary from 1.6 × 105 to 9.6 × 104 M−1 s−1, respectively. These sequences only show a large change in k when 4 or more complements exist in the stem region. Then, the rate constant decreases by approximately seven times, as it did for the sequences containing CGAAAG. For the control sequences containing GTGTTC and 0–3 complements in the stem, the rate constants vary from 1.6 × 105 to 9.7 × 104 M−1 s−1; however, in this case the values fluctuate. The rate constants from 0 to 2 complements in the stem show a steady decrease but the sequence with 3 complements within the stem increases and are close to the value for the 0 complement. These fluctuations in the measured rate constants are small compared to the jumps described above (changes of ∼1.5 times as opposed to almost 7 times). However, they can be used as another example of how a small change in sequence composition can change the rate constant of the reaction. In this case, increasing the number of complements within the stem region from 2 to 3 did not decrease the rate. This sequence change corresponded to the replacement of the third A from the 5′ end to a C. This change may have given the nucleation site better exposure thereby increasing the rate constant. When a fourth complement was present in the stem region for the GTGTTC sequence, the rate decreased by a much larger amount (over five times). It is likely that the decrease in rates of 5–7 times for both GTGTTC and GGAAAC sequences is solely due to the larger number of complements within the stem and not from stability within the loop region.
Figure 5.
A plot of the second-order rate constant as a function of the number of complements along the stem for the PH series. Diamonds correspond to PH A–E; squares correspond to PH A″–E″; and triangles correspond to PH A′–E′. The values for k from PH C–E, PH E′ and PH E″are approximate since the reactions were stopped before equilibrium was reached. Error bars correspond to the standard deviation calculated from multiple runs.
Solid phase
Solid phase experiments were performed using some of the same sequences that were used in the solution experiments to study the effects of the surface on the rates of hybridization. Non-porous silica beads (5 μm) were used as supports and the PH and PH′ series sequences were attached. Each milligram of silica beads contained 7.8 × 106 beads with a surface area of ∼79 m2 g−1. The silanol groups on the bead were functionalized with APTES in order to obtain reactive amine groups. Beads were further derivatized to obtain reactive NHS groups so that the free amine at the 5′ end of the DNA could react and form a covalent bond. A ninhydrin assay showed 2–4 amine groups per 100 Å2 after silanization with APTES. Dye-labeled probes that were coupled to the reactive NHS-terminated beads were used to quantitate the number of DNA molecules per bead. There were 5–9 × 106 molecules attached to each 5 μm bead. At this density, there is ∼10 nm between covalently bound probes and, therefore, we did not expect probe–probe interactions to occur. The intensity from fluorometric measurements of bead solutions and of cleaved dye-labeled DNA solutions showed that the fluorophores were quenched by ∼10–20% when measured on-bead. This may be a result of the surface interaction with the dye. The DNA probe density varied from sequence to sequence by ∼50% possibly because of conformational changes within the single strand, which inhibited the reaction between the 5′ amine and the surface.
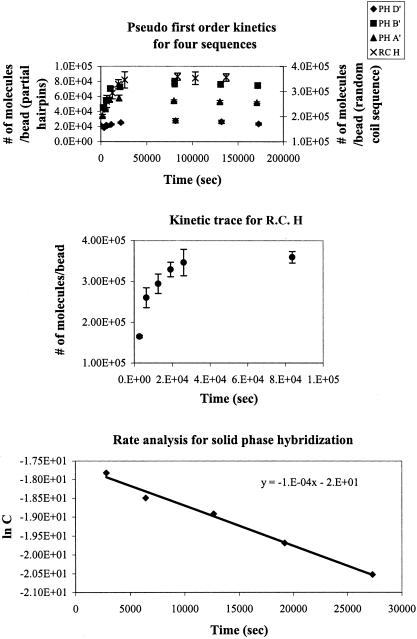
The number of hybridizable probes was determined by reacting the beads with an excess amount of dye-labeled target (10-fold excess). RC H showed the highest number of hybridizable oligos, compared to all of the other sequences. However, we did not expect the high-hybridization efficiency for this sequence to affect the kinetics since the spacing between the probes was still ∼10 nm. The upper panel in Figure 6 shows kinetic traces for four sequences, PH A′, B′, D′ and RC H. The solid phase experiments were set up as pseudo first-order reactions where the number of surface-bound probes was in 10-fold excess over the labeled target (which was typically ∼50 nM). All four sequences reached equilibrium in ∼11 h. At equilibrium, the total number of bound targets varied because different amount of beads were used for each kinetics experiment. In contrast to the solution phase results, the solid phase rate constants showed no significant sequence dependence for the sequences studied. The middle panel in Figure 6 shows an expanded view of the RC H kinetic trace where the error bars associated with each point correspond to the standard deviation calculated from the triplicate runs. The lower panel shows the analysis used in calculating the pseudo first-order rate constant for the RC H sequence. For each sequence, the number of surface-bound probes was converted to a concentration by calculating the number of moles and dividing by the volume of the bead suspension. The pseudo first-order rate constant was obtained by plotting the natural log of the concentration of target as a function of time. The slope of the line gave kpseudo, the pseudo first-order rate constant. Second-order rate constants were calculated by dividing the pseudo first-order values by the initial pseudo-concentrations of the probe.
Figure 6.
Upper panel: Kinetic traces for four different pseudo first-order hybridization reactions on 5 μm silica beads. Both a random coil (RC H) and the partial hairpin sequences (PH A′, B′ and D′) are shown. Middle panel: an expanded view of the RC H kinetic trace from above. lower panel: rate analysis for the RC H sequence. The data were fit to a straight line using a linear regression. The slope of the line gives the pseudo first-order rate constant.
Table 2 summarizes the second-order rate constants obtained in solution and in the solid phase for the PH series. The rate constant for PH B, the sequence expected to have almost no cooperative intramolecular structure, is almost three orders of magnitude lower when measured on beads compared to that measured in solution. Although the solution phase rate constants showed a strong dependence on sequence, this dependence was not seen in the bead-based studies. The half-life for the PH B–E sequences measured on beads is in the order of 104 s, two orders of magnitude longer than that measured in solution.
Table 2.
A table showing the comparisons between second-order rate constants obtained from solution and from beads along with the corresponding half times
| Sequence name | Number of complements in stem | k, solid phase (M−1 s−1) | k, solution phase (M−1 s−1) | t1/2, solid (s) | t1/2, solution (s) |
|---|---|---|---|---|---|
| PH B | 1 | 2.5 × 102 | 1.60 × 105 | 3.45 × 104 | 12 |
| PH D | 3 | 5.1 × 102 | <3 × 103a | 1.73 × 104 | >670 |
| PH E | 4 | 2.5 × 102 | <3 × 103a | 3.45 × 104 | >670 |
aThe rate constants for PH D and PH E are upper limits since the melting curves were complex and single strand absorbances could not be extracted from them.
DISCUSSION
The solution phase studies show that the rate constants are strongly dependent on sequence. Previous experimental results have shown temperature (19), ionic strength, denaturing solvents and solvent viscosity dependence on the rates (20–23); however, base composition has only just begun to be used to help in explaining rate constant variations (7,17). The model sequences that were chosen for this study, the PH series, show how increasing single-stranded structure can slow down the rates of hybridization by several orders of magnitude. This is owing to the stability of the hairpin at the hybridization temperature. However, the model sequences also show complex melting curves. What is more surprising is that the random sequences studied, RC A–G, which were designed to have similar melting temperatures, have rate constants that vary by over an order of magnitude. There are no known stable single-stranded conformations for these sequences and their melting curves show smooth transitions, as shown in Figure 2. The broad range of reaction rates found for probe hybridization can have significant effects. For example, in gene expression assays, target sequences are used to quantify expression levels. Unless equilibrium has been reached or the rates for the sequences used as probes are known and taken into consideration, it would be impossible to quantitate and compare the amount of target present.
The model partial hairpin sequences, PH A–E, that were generated to test the effects of stable single-stranded conformations, show an even greater spread in rate constants although their calculated melting temperatures are all within 10°C of each other. The incorrect prediction of the melting temperature is due to the common algorithms used to calculate these values. In this case, the algorithms used did not predict the stable single-stranded conformation (24–26). In addition, it is unlikely that most software used in designing optimal probes for hybridization applications would include these types of conformations since most of the algorithms are based on thermodynamic properties obtained from calculated melting temperatures. Even if the model sequences were to be included, our data show that there are other sequences that have similar experimental melting temperatures but have vastly different reaction rates.
In comparing the results from the solid and solution phase experiments, the rate constants measured when probes were attached to beads were as much as three orders of magnitude lower than those measured in solution (see PH B, Table 2). The constant temperature used for all sequences during the incubation times for the solid phase (35 ± 5°C) and the higher ionic strength of the buffer (50 mM KCl versus 25 mM for the solution phase experiments) was not enough to explain this difference. For example, running the solution phase experiments for PH D at 35°C decreased k by only ∼2.5 times. Similarly, there was no appreciable change in k for PH A or PH E hybridizing in the bead buffer. Although k should increase with increasing ionic strength, the detergent may be slowing down the rate so that the two changes end up canceling each other. The solid phase experiments were set up assuming that the melting curves were similar to those measured in solution [based on previous studies of oligonucleotide melting when one strand was surface-bound (27)].
The differences in solid versus solution phase rates are less when comparing PH D and PH E because the single-stranded structure shows a drastic effect on k in solution and virtually no effect on k in the solid phase reactions. The solid phase rate constants remain similar, regardless of sequence, implying that sequences, in the cases studied, do not impact reaction rates in the same way on surfaces as it does in solution. For the model sequences, attaching the strand with the stable partial hairpin to the surface may have disrupted the structure, rendering the strand closer to a random coil than a partial hairpin. This may have been caused by electrostatic interactions at the surface. For example, the bead surface has a negative charge due to unpassivated silanol groups and acid groups form the passivation steps that have not been coupled to linkers and probes. An electrostatic repulsion between the surface and the probe may be enough to change the original conformation of the strand. In addition, the single-stranded hairpin may have denatured due to residual DMF left on the surface following the coupling chemistry step. These factors may have also been responsible for the decrease in fluorescence intensity that was observed when the hybridized DNA was measured on the bead. Currently, additional experiments are being performed to test these hypotheses that include a series of surface hybridizations at increasing ionic strengths.
Although there are a number of reports in the literature on the kinetics of DNA–DNA hybridization (a sample have been referenced here) (1–9), only a few of them are quantitative (5–8). The second-order rate constants from these studies range from 4 × 104 to 1 × 105 M−1 s−1 for targets of similar length hybridizing to the probes bound to flat surfaces (6) and beads (8). These rate constants are at least two orders of magnitude higher than those quoted here. The differences in ionic strength of the hybridization buffers (4,6) and in the experiments described here may account for some of the difference in rates. Studies found that the solution phase rate constants increase with increasing ionic strength (19). An increase in salt concentration of 3 times can increase the rate constant ∼10 times; however, this is not enough of an increase to explain the differences. We encountered a number of factors that influenced the solid phase reaction rates by orders of magnitude. It was only through careful characterization at each step of the experiment that we were able to achieve the rates reported here. For instance, probe densities varied if the beads were not washed sufficiently before conjugation. We suspect this was owing to non-specifically bound probes that may have released from the bead surface at some point during incubation with the target. Improper mixing of the beads during incubation also led to variations in the rate constant. Since the silica beads settled to the bottom of the reaction tube quickly (in ∼10 min), it was difficult to keep the beads suspended in solution. If some of the beads settled during hybridization, the kinetic traces would not show smooth transitions to equilibrium and therefore the rate constants that were extracted from these curves would show large standard deviations. Finally, we needed to prevent the evaporation of the target solution over a period of days (accumulation of data from the bead-based studies was collected over 2 days). We found that the small amount of detergent used helps to prevent losses to the walls of the container. With these factors under control, we were able to obtain consistent results on solid phase hybridization.
In comparing the results presented here to microarray assays, it should be noted that the ionic strengths of the hybridization buffers typically used in microarray assays are much higher than those used here (sometimes these are as high as 1 M for the microarray assays compared to millimolar quantities used here). The higher ionic strength in the buffer should give rise to much higher rate constants compared to those measured here; however, they will also promote mismatchced hybridization since the high-salt concentration tends to stabilize base pairing. To compensate for these kinds of problems, many different protocols have been developed but in many cases, these include additions of several components to the buffer, including denaturants and detergents. Because of these additions, it is not possible to quantitatively predict the effects of the higher salt concentrations on the rates. If typical target concentrations were 50 nM, as was the case in these experiments, the microarray rates might be higher using a buffer with a higher ionic strength and a denaturant; however, in general, the microarray assays are run at much lower concentrations compared to those used here. In some cases, the microarrays are capable of detecting picomolar levels of the target. In these cases, the differences in concentrations, assuming the target is the limiting reagent, might counteract the effects of higher ionic strengths.
Acknowledgments
This work was supported in part by a cost-sharing grant from Ocean Optics, Inc. (Dunedin, FL). Funding to pay the Open Access publication charges for this article was provided by The School of Science and Engineering, SUNY New Paltz.
REFERENCES
- 1.Wood S.J. DNA–DNA hybridization in real time using Biacore. Microchem. J. 1993;47:330–337. [Google Scholar]
- 2.Chan V., Graves D.J., McKenzie S.E. The biophysics of DNA hybridization with immobilized oligonucleotide probes. Biophys. J. 1995;69:2243–2255. doi: 10.1016/S0006-3495(95)80095-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Georgiadis R., Peterlinz K.P., Peterson A.W. Quantitative measurements and modeling of kinetics in nucleic acid monolayer films using SPR spectroscopy. J. Am. Chem. Soc. 2000;122:3166–3173. [Google Scholar]
- 4.Thiel A.J., Frutos A.G., Jordan C.E., Corn R.M., Smith L.M. In situ surface plasmon resonance imaging detection of DNA hybridization to oligonucleotide arrays on gold surfaces. Anal. Chem. 1997;69:4948–4956. [Google Scholar]
- 5.Jensen K.K., Ørum H., Nielsen P.E., Nordén B. Kinetics for hybridization of peptide nucleic acids (PNA) with DNA and RNA studied with the BIAcore technique. Biochemistry. 1997;36:5072–5077. doi: 10.1021/bi9627525. [DOI] [PubMed] [Google Scholar]
- 6.Liebermann T., Knoll W., Sluka P., Herrmann R. Complement hybridization from solution to surface-attached probe-oligonucleotides observed by surface-plasmon-field-enhanced fluorescence spectroscopy. Colloids Surf. A. 2000;169:337–350. [Google Scholar]
- 7.Riccelli P.V., Merante F., Leung K.T., Bortolin S., Zastawny R.L., Janeczko R., Benight A.S. Hybridization of single-stranded DNA targets to immobilized complementary DNA probes: comparison of hairpin versus linear capture probes. Nucleic Acids Res. 2001;29:996–1004. doi: 10.1093/nar/29.4.996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Henry M.R., Stevens P.W., Sun J., Kelso D.M. Real-time measurements of DNA hybridization on microparticles with fluorescence resonance energy transfer. Anal. Biochem. 1999;276:204–214. doi: 10.1006/abio.1999.4344. [DOI] [PubMed] [Google Scholar]
- 9.Ferguson J.A., Steemers F.J., Walt D.R. High density fiber optic DNA random microsphere array. Anal. Chem. 2000;72:5618–5624. doi: 10.1021/ac0008284. [DOI] [PubMed] [Google Scholar]
- 10.Heus H.A., Pardi A. Structural features that give rise to the unusual stability of RNA hairpins containing GNRA loops. Science. 1991;253:191–194. doi: 10.1126/science.1712983. [DOI] [PubMed] [Google Scholar]
- 11.Hirao I., Nishimura Y., Tagawa Y., Watanabe K., Miura K. Extraordinary stable mini-hairpins: electrophoretical and thermal properties of the various sequence variants of d(GCGAAAGC) and their effect on DNA sequencing. Nucleic Acids Res. 1992;20:3891–3896. doi: 10.1093/nar/20.15.3891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Antao V.P., Lai S.Y., Tinoco I., Jr A thermodynamic study of unusually stable RNA and DNA hairpins. Nucleic Acids Res. 1992;21:5901–5905. doi: 10.1093/nar/19.21.5901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yoshizawa S., Kawai G., Watanabe K., Miura K., Hirao I. GNA trinucleotide loop sequences producing extraordinarily stable DNA minihairpins. Biochemistry. 1997;36:4761–4767. doi: 10.1021/bi961738p. [DOI] [PubMed] [Google Scholar]
- 14.Senior M.M., Jone R.A., Breslauer K.H. Influence of loop residues on the relative stabilities of DNA hairpin structures. Proc. Natl Acad. Sci. USA. 1988;85:6242–6246. doi: 10.1073/pnas.85.17.6242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bonnet G., Krichevsky O., Kibchaber A. Kinetics of conformational fluctuations in DNA hairpin-loops. Proc. Natl Acad. Sci. USA. 1998;95:8602–8606. doi: 10.1073/pnas.95.15.8602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Moore S., Stein W.H. Photometric ninhydrin method for use in the chromatography of amino acids. J. Biol. Chem. 1948;176:367–388. [PubMed] [Google Scholar]
- 17.Kushon S.A., Jordan J.P., Seifert J.L., Nielsen H., Nielsen P.E., Armitage B.A. Effect of secondary structure on the thermodynamics and kinetics of PNA hybridization to DNA hairpins. J. Am. Chem. Soc. 2001;123:10805–10813. doi: 10.1021/ja016310e. [DOI] [PubMed] [Google Scholar]
- 18.Xodo L.E., Manzini G., Quadrifoglio F., van der Marel G.A., van Boom J.H. Oligodeoxynucleotide folding in solution: loop size and stability of B-hairpins. Biochemistry. 1988;27:6321–6326. doi: 10.1021/bi00417a018. [DOI] [PubMed] [Google Scholar]
- 19.Studier F.W. Effects of the conformation of single-stranded DNA on renaturation and aggregation. J. Mol. Biol. 1969;41:199–209. doi: 10.1016/0022-2836(69)90385-4. [DOI] [PubMed] [Google Scholar]
- 20.Wetmur J.G., Davidson N. Kinetics of renaturation of DNA. J. Mol. Biol. 1968;31:349–370. doi: 10.1016/0022-2836(68)90414-2. [DOI] [PubMed] [Google Scholar]
- 21.Wetmur J.G. Excluded volume effects on the rate of renaturation of DNA. Biopolymers. 1971;10:601–613. doi: 10.1002/bip.360100402. [DOI] [PubMed] [Google Scholar]
- 22.Chang C., Hain T.C., Hutton J.R., Wetmur J.G. Effects of microscopic and macroscopic viscosity on the rate of renaturation of DNA. Biopolymers. 1974;13:1847–1858. doi: 10.1002/bip.1974.360130915. [DOI] [PubMed] [Google Scholar]
- 23.Wetmur J.G. Acceleration of DNA renaturation rates. Biopolymers. 1975;14:2517–2524. doi: 10.1002/bip.1981.360200711. [DOI] [PubMed] [Google Scholar]
- 24.Breslauer K.J., Frank R., Blöcker H., Marky L.A. Predicting DNA duplex stability from the base sequence. Proc. Natl Acad. Sci. USA. 1986;83:3746–3750. doi: 10.1073/pnas.83.11.3746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Freier S.M., Kierzek R., Jaeger J.A., Sugimoto N., Caruthers M.H., Neilson T., Turner D.H. Improved free-energy parameters for predictions of RNA duplex stability. Proc. Natl Acad. Sci. USA. 1986;83:9373–9377. doi: 10.1073/pnas.83.24.9373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.SantaLucia J., Jr, Allawi H.T., Seneviratne P.A. Improved nearest-neighbor parameters for predicting DNA duplex stability. Biochemistry. 1996;35:3555–3562. doi: 10.1021/bi951907q. [DOI] [PubMed] [Google Scholar]
- 27.Jobs M., Fredriksson S., Brookes A.J., Landegren U. Effect of oligonucleotide truncation on single-nuclceotide distinction by solid-phase hybridization. Anal. Chem. 2002;74:199–202. doi: 10.1021/ac010555s. [DOI] [PubMed] [Google Scholar]