Abstract
The tRNA 3′-processing endoribonuclease (tRNase Z or 3′ tRNase; EC 3.1.26.11) is an essential enzyme that removes the 3′ trailer from pre-tRNA. The long form (tRNase ZL) can cleave a target RNA in vitro at the site directed by an appropriate small-guide RNA (sgRNA). Here, we investigated whether this sgRNA/tRNase ZL strategy could be applied to gene therapy for AIDS. We tested the ability of four sgRNA-expression plasmids to inhibit HIV-1 gene expression in COS cells, using a transient-expression assay. The three sgRNAs guide inhibition of HIV-1 gene expression in cultured COS cells. Analysis of the HIV-1 mRNA levels suggested that sgRNA directed the tRNase ZL to mediate the degradation of target RNA. The observation that sgRNA was localized primarily in nuclei suggests that tRNase ZL cleaves the HIV-1 mRNA when complexed with sgRNA in this location. We also examined the ability of two retroviral vectors expressing sgRNA to suppress HIV-1 expression in HIV-1-infected Jurkat T cells. sgRNA-SL4 suppressed HIV-1 expression almost completely in infected cells for up to 18 days. These results suggest that the sgRNA/tRNase ZL approach is effective in downregulating HIV-1 gene expression.
INTRODUCTION
RNA-based gene-interference strategies for the treatment of HIV-1 infection have often used technology based on antisense oligonucleotides, ribozymes or double-stranded interference RNA (RNAi) (1–11). A more recent approach uses external guide sequences (EGSs) to induce cleavage of a target mRNA by endogenous RNase P. This strategy is unique in that cleavage of a specific target mRNA occurs after hybridization of the EGS to form a structure resembling a tRNA substrate (12,13). RNA-based EGSs have been expressed endogenously as transgenes in both bacteria and mammalian cells (12,14) and have been effective in inhibiting gene expression by HIV-1 (15,16). We previously designed a short RNase P-associated EGS to target HIV-1-U5 and evaluated its ability to inhibit HIV-1 replication (17). Mammalian cells contain the essential enzyme, tRNA 3′-processing endoribonuclease (tRNase Z or 3′-tRNase; EC 3.1.26.11), which removes 3′ trailers from pre-tRNAs (18). The human genome contains two tRNase Z genes, which encode a 362 amino acid short form (tRNase ZS) and an 829 amino acid long form (tRNase ZL) (19,20). Although the C-terminal half of tRNase ZL has a high level of similarity to the whole of tRNase ZS, they each require different reaction conditions for optimal activity (20). Interestingly, the human tRNase ZL gene was first identified as a candidate prostate cancer-susceptibility gene (21). Two types of mutation in the human tRNase ZL gene, an insertion/frameshift and a missense change, segregate with prostate cancer in two different pedigrees. Furthermore, two additional common missense mutations seem to be associated with prostate cancer. However, a causal association between the missense mutations and prostate cancer has not been proven, because these amino acid substitutions do not alter the enzymatic activities of tRNase ZL (22). The long-form enzyme is unique in that it can cleave any RNA at any site when directed by a small-guide RNA (sgRNA) in vitro (23–27). tRNase ZL functions in the same way as the 4 nt-recognizing RNA cutter RNase 65, by forming a complex with a 3′-truncated tRNA (23). Partial HIV-1 RNA targets can be cleaved site specifically by the enzyme, once the targets form pre-tRNA-like structures with the aid of appropriate 5′-half-tRNAs (24). RNA heptamers that form acceptor-stem-like duplexes with their targets through base pairing can also direct the specific cleavage of target RNAs by tRNase ZL in vitro with the same efficiency as the original 5′-half-tRNAs (25,26). However, in this case, the target sites are restricted to regions immediately downstream of stable hairpin structures resembling the T stem/loop. Together with such flexibility in substrate recognition, the ubiquity and constitutive expression of tRNase ZL suggests that this enzyme can be utilized for specific cleavage of cellular RNAs by introducing the appropriate sgRNAs into living cells (27).
Recently, we demonstrated the efficacy of this method in specifically targeting RNA in living cells by introducing sgRNAs encoded either by expression plasmids or by 2′-O-methyl RNAs (28). The expression of the exogenous reporter genes for Escherichia coli chloramphenicol acetyltransferase and firefly luciferase were downregulated for at least 48 h by appropriately designed sgRNAs in cultured human and dog cells. A 2′-O-methyl heptamer designed to target the endogenous Bcl-2 mRNA was also successful in Sarcoma 180 cells.
In the present study, we investigated whether the sgRNA/tRNase ZL strategy could also be an effective approach to gene therapy for AIDS. Several sgRNAs targeted against HIV-1 mRNA, and expressed by plasmid and retroviral vectors, were tested for their ability to repress its expression. Our findings confirmed that they were effective in both COS and Jurkat cells.
MATERIALS AND METHODS
Plasmid construction and retroviral vector production
Expression cassettes for the sgRNA were constructed under the control of the human methionine tRNA promoter. The expressed sgRNA was targeted against either the packaging signal or the gag portion of the HIV-1NL4-3 strain (Figure 1B). Enhanced green fluorescent protein (EGFP) is a red-shifted variant of wild-type GFP (29,30), which has been optimized for brighter fluorescence and higher expression in mammalian cells (excitation maximum = 488 nm; emission maximum = 507 nm). A DNA fragment containing EGFP was excised from the plasmid pLEGFP (Promega, Madison, WI) by digestion with BamHI–NaeI. The EGFP fragment was ligated to the EcoRI and XhoI sites of pSV2neo/sgR to generate pSV2neo/sgRG. The Moloney strain of the murine leukaemia virus (MoMLV)-based sgRNA pLsgRGSN (Figure 1B) was constructed by inserting the EcoRI and XhoI fragments from plasmid pSV2neo/sgRG, along with sgRNAs and EGFP genes, into the EcoRI and XhoI sites of the retroviral vector pLXSN (31).
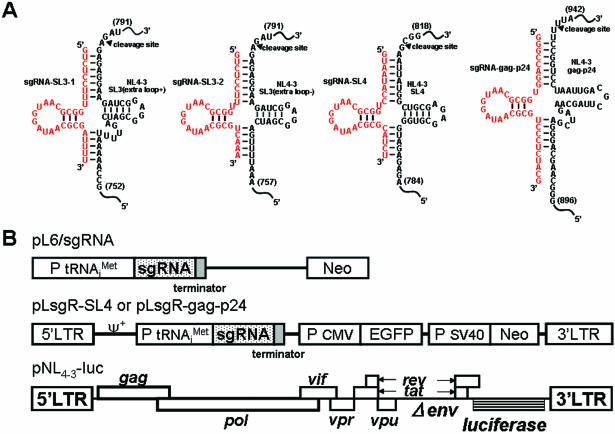
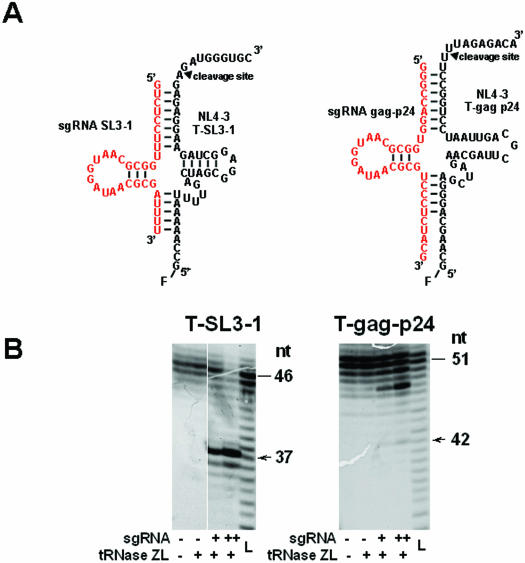
Figure 1.
(A) Plausible secondary structures of complexes of the three target sites within the HIV-1 genome [the ψ site (SL3-stem loop), the SL4-stem-loop site and gag-24 within the HIV-1 gene] with the modified 5′-half-tRNAArg (sgRNA-SL3-1, 2, SL-4 and gag-p24) containing 7 and 5 nt sequences complementary to the target HIV-1 ψ site and the gag gene. The arrow indicates the tRNase Z cleavage point. (B) Schematic diagrams of the sgRNA-expression plasmids, the retroviral vector and the HIV-1 luciferase reporter-vector constructs. Methods for the construction of the sgRNA-expression plasmids (pL6-sgRNA-SL3-1, pL6-sgRNA-SL3-2, pL6-sgRNA-SL4 and pL6-sgRNA-gag-p24) and the retroviral vectors (pLsgRGSN-SL4 and pLsgRGSN-gag-p24) are detailed in Materials and Methods. The envelope-defective HIV-1 NL4-3-based retroviral vector contained a luciferase-expression marker (pNL4-3-luc). This HIV-1-based vector was generated by substituting the nef gene sequences of the HIV-1NL4-3 genome with the firefly luciferase gene, and deleting the envelope gene sequences located between two Bgl II restriction endonuclease sites (32).
Vesicular stomatitis virus glycoprotein-pseudotyped retrovirus vector supernatants were generated by transient transfection of 293T cells, as described previously (32), using 20 μg of vector plasmid, 10 μg of pMLVΔψΔenv (33) and 10 μg of pVSVG envelope plasmid (32,34). Thereafter, supernatants were collected every 12 h for 3 days, filtered through a 0.45 μm pore-size filter (Nalgene, Rochester, NY) and concentrated 100- to 1000-fold by ultracentrifugation (35,36). Pellets were resuspended in serum-free DMEM and stored at −80°C until they were used.
Transduction of target cells
A total of 3 × 105 Jurkat cells per well were plated out in 6-well plates 1 day before transduction. After 24 h, virus supernatant was added together with polybrene (final concentration = 5–8 μg/ml) and the cells were incubated at 25°C overnight. The medium was then replaced with fresh medium containing G418 (500 μg/ml; GIBCO-BRL, Rockville, MD). After 10 days, cell pools that were resistant to G418 were established.
Cells and transfection
COS and Jurkat cells were grown in RPMI 1640 medium (Sigma, St. Louis, MO) supplemented with 10% (v/v) heat-inactivated fetal bovine serum (FBS) at 37°C in a 5% CO2 atmosphere. Transfection was carried out using the FuGENE™6 reagent (Roche Diagnostics, Tokyo, Japan) according to the manufacturer's protocol.
Luciferase assay
Luciferase activity was measured with the PicaGene kit (Toyo-inki, Tokyo, Japan) according to the manufacturer's protocol. The envelope-defective HIV-1 NL4-3-based retroviral vector containing a luciferase-expression marker (pNL4-3-luc) (32) was generated as follows. The nef gene sequences of the HIV-1 NL4-3 genome were substituted with the firefly luciferase gene, and the envelope gene sequences located between two Bgl II restriction endonuclease sites were deleted (Figure 1B). The transfection reagent, FuGENE™6, was used to transfect COS cells with the plasmids expressing sgRNA and pNL4-3-luc. The COS cells were lysed using 200 μl of PicaGene cell lysis buffers (Toyo-inki) for 15 min and detached from the plate by scraping. Cellular debris were then removed by centrifugation. The luminescent signal was quantitated by adding 10 μl clarified lysate to 100 μl luminous substrate, and the level of fluorescence was recorded immediately using a luminometer (Lumat LB 9507; Berthold, Bad Wildbad, Germany).
The amount of firefly luciferase activity was normalized with reference to the protein concentration in the lysate. The protein was quantitated using the BCA Protein Assay Reagent kit (Pierce, IL), which is based on bicinchoninic acid.
Localization of sgRNA
The cytoplasmic fraction was prepared from collected cells after washing twice with phosphate-buffered saline (PBS). The cells were resuspended in digitonin lysis buffer (50 mM HEPES-KOH, pH 7.5, 50 mM potassium acetate, 8 mM MgCl2, 2 mM EGTA and 50 μg/ml digitonin) and incubated on ice for 10 min. The lysate was centrifuged at 1000 g for 5 min and the resultant supernatant was collected and used as the cytoplasmic fraction. The pellets were resuspended in NP-40 lysis buffer (20 mM Tris–HCl, pH 7.5, 50 mM KCl, 10 mM NaCl, 1 mM EDTA and 0.5% NP-40) and incubated on ice for 10 min. The resultant lysate was used as the nuclear fraction. Cytoplasmic and nuclear RNA were extracted and purified from their respective fractions using ISOGEN reagent (Wako, Osaka, Japan). RNA samples were treated with DNase I (Takara Shuzo, Shiga, Japan) according to the manufacturer's instructions. RT–PCR assays were performed using an RT–PCR High-Plus kit (Toyobo, Osaka, Japan) according to the manufacturer's protocol. The nucleotide sequences of the sgRNA-SL4 primers were 5′-TACTGTGAGACCGTGTGCTT-3′ (F-primer) and 5′-TACTGTGAGACCGTGTGCTT-3′ (R-primer). The nucleotide sequences of the U6 primers were 5′-CAGACATGATAAGATACATTGATGAGTTTG-3′ (F-primer) and 5′-CGGGATCCCGGCAATAGCATCACAAATTTC-3′ (R-primer).
Cleavage activities of sgRNA in COS cells
COS cells were grown to ∼80% confluence (3 × 105 cells) and transfected with 3 μg each of the sgRNA vector and the pNL4-3-luc plasmid. The cells were incubated for 2 days before total cellular RNA was isolated. RNA samples were treated with DNase I according to the manufacturer's specifications, and RT–PCR assays were carried out as described above. The levels of cleaved and uncleaved HIV-1 mRNAs were quantified by RT–PCR with an endogenous internal standard, GAPDH. A sample of 1 μg of total RNA was used as the template with the SL3, SL4, p24 and GAPDH primers (20 pmol each). Reverse transcription in a final reaction volume of 50 μl was carried out at 60°C for 30 min. The cDNA products were then amplified by PCR using amplification conditions comprising 40 cycles of 94°C for 60 s and 60°C for 90 s. The SL3-F1 (5′-TTGCTGAAGCGCGCACGGCA-3′) and p24-F1 (5′-TAAGGCCAGGGGGAAAGAAACAATATAAAC-3′) primer pair, and the SL3-R and SL4-R (5′-GTTCTTCTGATCCTGTCTGAAGGGATGGTT-3′) and p24-R (5′-GCCCCTGGAGGTTCTGCACTATAGGGTAAT-3′) primer pair, only generated a cDNA product from the uncleaved HIV-1 mRNA (RT–PCR products I and III, SL3 = 310 bp; RT–PCR product V, gag-p24 = 360 bp). The SL3-F2 (5′-TAAATGGGAAAAAATTCGGT-3′), SL4-F2 (5′-ATTCGGTTAAGGCCAGGGGG-3′) and p24-F2 (5′-AGACAAATACTGGGACAGCTACAACCATCC-3′) primer pairs were used to generate cDNA products corresponding to the cleaved and uncleaved sgRNA sequences, respectively, thereby identifying both sets of products. The SL3, SL4 and p24 yielded the RT–PCR products II (SL3 = 182 bp), IV (SL4 = 168 bp) and VI (p24 = 250 bp), whereas the GAPDH-F and R-primers amplified a fragment of the GAPDH gene (0.45 kb) as an internal control.
In vitro RNA synthesis
The partial HIV-1 RNA targets (T-SL3-1 and T-gag-p24) and the sgRNAs (sgRNA-SL3-1 and sgRNA-gag-p24) were synthesized from the corresponding synthetic DNA templates with an additional unencoded ‘G’ at the 5′ end corresponding to the −1 in a tRNA to enhance its transcriptional efficiency, using T7 RNA polymerase (Takara Shuzo). The sequences of the target RNAs and sgRNAs were as follows: 5′-GCCAAAAAUUUUGACUAGCGGAGGCUAGAAGGAGAGAGAUGGGUGC-3′ (T-SL3-1); 5′-GCAAGCAGGGAGCUAGAACGAUUCGCAGUUAAUCCUGGCCUUUUAGAGACA-3′ (T-gag-p24); 5′-GUCUCCUUUGGCGCAAUGGAUAACGCGAUUUU-3′ (sgRNA-SL3-1); and 5′-GGGCCAGGUGGCGCAAUGGAUAACGCGUCCCUCUACG-3′ (sgRNA-gag-p24). The transcription reactions were carried out under the conditions recommended by the manufacturer (Takara Shuzo), and after synthesis the RNAs were purified by denaturing gel electrophoresis. The RNA transcripts for T-SL3-1 and T-gag-p24 were subsequently labelled with fluorescein (F) according to the manufacturer's protocol (Amersham Pharmacia Biotech, NJ). Briefly, after the removal of the 5′-phosphates of the transcribed RNAs using bacterial alkaline phosphatase (Takara Shuzo), the RNAs were phosphorylated with T4 polynucleotide kinase (Takara Shuzo) and ATPγS. Then, a single fluorescein moiety was appended to the 5′-phosphorothioate site. The resulting fluorescein-labelled RNAs were gel-purified before being used in the assays.
In vitro RNA-cleavage assays
The in vitro RNA-cleavage assays for the fluorescein-labelled target RNA, T-SL3-1 or T-gag-p24 (0.1 pmol), were performed with pig liver tRNase ZL (20 ng) in the presence of unlabelled sgRNA-SL3-1 or sgRNA-gag-p24 (5 or 10 pmol) in a mixture (6 μl) containing 10 mM Tris–HCl (pH 7.5), 1.5 mM DTT and 10 mM MgCl2 at 50°C for 10 min. After resolution of the reaction products by electrophoresis through a 10% polyacrylamide/8 M urea sequencing gel, the gel was analysed using a Typhoon 9210 (Amersham Pharmacia Biotech).
HIV-1-challenge assay
G418-resitant cell pools were incubated for 4 h with HIV-1NL4-3 at a multiplicity of infection of 0.01. After two washes with PBS, the cells were cultured in RPMI 1640 medium (Sigma) supplemented with 10% (v/v) heat-inactivated FBS. The supernatant was collected on days 1, 3, 6, 9, 12, 15 and 18 after viral infection, and the culture medium was assayed for HIV-1 gag-p24 antigen using CLEIA (Lumipulse, Fujirebio, Tokyo, Japan) according to the manufacturer's protocol (37).
RESULTS AND DISCUSSION
sgRNAs can inhibit HIV-1 gene expression
We demonstrated in a previous study that antisense phosphorothioate oligonucleotides (S-ODNs) complementary to the gag mRNA (SL4-stem loop), containing the HIV-1 gag AUG initiation codon, have potent anti-HIV activity in infected cultured cells. This activity was strong compared with oligonucleotides targeted to the splice acceptor of the tat gene and the AUG initiation codon of the rev gene (38). We have also shown in vitro that mammalian tRNase ZL with the aid of a 5′-half-tRNA-like sgRNA can cleave a partial HIV-1 mRNA substrate containing the p24 site of the HIV-1 gag gene (24). Therefore, it would be reasonable to select the gag AUG site (designated SL4) and the gag-p24 site to examine the efficacy of the sgRNA method in cultured cells. The HIV-1 packaging signal (ψ) that efficiently targets genomic RNA into nascent virions (39,40) and the ψ site (designated SL3) was chosen as an additional target site for tRNase ZL.
The 5′-half-tRNA-type sgRNAs, designated sgRNA-SL3-1 and sgRNA-SL3-2 (extra loop-), were designed to form pre-tRNA-like complexes with SL3. They contained or lacked an extra loop, respectively (Figure 1A). sgRNA-SL4 without an extra loop was intended to make a pre-tRNA-like complex with SL4, and sgRNA-gag-p24 with an extra loop was designed to form a pre-tRNA-like complex with the gag-p24 site (Figure 1A).
The sgRNA-expression plasmids were constructed in the mammalian expression plasmid pL6 (41,42) and (Figure 1B) by inserting synthetic DNA sequences between the human promoter sequence and the RNA polymerase III termination-signal sequence. RT–PCR analysis was used to examine the expression of the sgRNA by these plasmids in transfected COS cells. A high level of sgRNA was expressed in the transfected cells, driven by a promoter (data not shown).
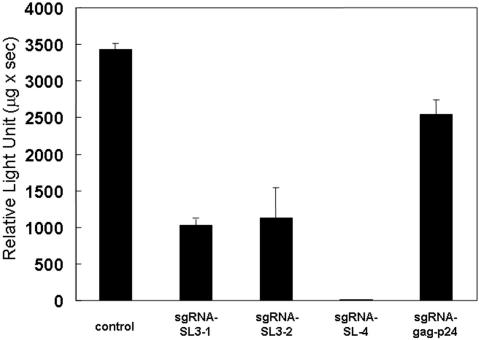
A transient-expression assay was used to test the ability of pL6-expressed sgRNA-SL3-1 and 2, sgRNA-SL4 and sgRNA-gag-p24 to inhibit HIV-1 expression. COS cells were co-transfected with the sgRNA plasmids and an HIV-1NL4-3-based vector containing a luciferase-expression maker (pNL4-3-luc) (31), then suppression of HIV-1 was assessed in a single-cycle infectivity assay (Figure 2). HIV-1NL4-3-based luciferase activity was recorded using the control plasmid vector L6-ter with only the promoter and terminator sequences, rather than the sgRNA-expression plasmid. Both sgRNA-SL3-1 and sgRNA-SL3-2 showed good inhibition of HIV-1 expression in the cultured cells, suggesting that the extra loop in the pre-tRNA-like complex is not important for tRNase ZL recognition (Figure 2). Amazingly, HIV-1 gene expression was almost completely inhibited by sgRNA-SL4, but only moderately suppressed by sgRNA-gag-p24. This might be because endogenous tRNase ZL has difficulty in recognizing the sgRNA-gag-p24/target complex, possibly due to the lack of a stable ‘T-stem-loop’ structure in the target, as indicated by the in vitro cleavage assays described below. These results imply that the sgRNA/tRNase ZL strategy is effective in reducing the level of the HIV-1 gene expression, although the efficiency of inhibition differs according to the sgRNAs used.
Figure 2.
Luciferase activity of the pNL4-3-luciferase (pNL4-3-luc) fusion gene in COS cells. The cells were co-transfected with the target-expressing plasmid pNL4-3-luc and either the pL6 plasmid encoding the sgRNAs or pL6-ter serving as a negative control for inhibition using the FuGENE™6 transfection reagent. The plotted data were averaged from three independent experiments, and the bars represent ±SD.
Intracellular localization of the –sgRNA
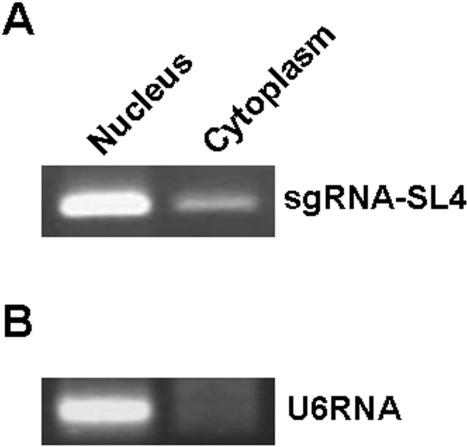
Co-localization of sgRNAs, their RNA targets and tRNase ZL is important for efficient downregulation; therefore, we examined the intracellular location of sgRNA-SL4 (41,42). Transformed cells expressing sgRNA-SL4 were separated into nuclear and cytoplasmic fractions, and the total RNA was extracted from each. sgRNA-SL4 was detected using RT–PCR analysis with a primer specific for the sgRNA. This revealed that they were located almost exclusively in the nucleus (80%) (Figure 3).
Figure 3.
Intracellular localization of the –sgRNA. To analyse the degree of intracellular localization of –sgRNA, nuclear and cytoplasmic fractions were prepared from transformants that expressed sgRNA-SL4 and the total RNA was extracted from each. The transcribed sgRNA-SL4 was detected using RT–PCR analysis with a primer specific for the sgRNA. (A) RT–PCR analyses revealed that –sgRNA-SL4 was located almost exclusively in the nucleus, as predicted. (B) Nuclear and cytoplasmic fractions were examined with a probe specific for the transcript of the U6 gene (control).
The inhibitory effect of sgRNA occurs through target RNA degradation by tRNase ZL
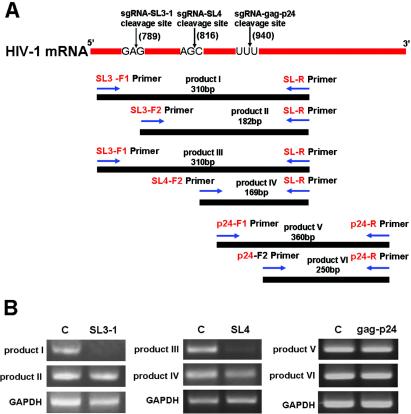
The contribution of HIV-1 mRNA cleavage to the sgRNA-mediated anti-HIV-1 effect was examined by measuring HIV-1 mRNA levels (43,44). Two sets of RT–PCR reactions were used to establish the level of HIV-1 mRNA uncleaved at the target site by tRNase ZL [products I (310 bp), III (310 bp) and V (360 bp)], and the total amount of HIV-1 mRNA at the target site [products II (182 bp), IV (169 bp) and VI (250 bp)], i.e. both cleaved and uncleaved. The uncleaved HIV-1 mRNA was amplified by primers SL3-F1 and gag-p24-F1, and the SL3-R, SL4-R and gag-p24-R primers (Figure 4A). The levels of products I, III and V were expected to decrease after cleavage of the HIV-1 mRNA. The levels of products II, IV and VI reflected the total amount of HIV-1 mRNA (cleaved and uncleaved SL3, SL4 and gag-p24), as the 3′ fragment of cleaved HIV-1 mRNA remained a viable template for amplification in this PCR. The results, which are shown in Figure 4B, indicate that the sgRNA-SL3-1- and SL4-dependent expression system reduced the amount of HIV-1 mRNA [products I (92%) and IV (97%)], whereas transfection with pNL4-3-luc/sgRNA-gag-p24 did not significantly alter uncleaved HIV-1 mRNA expression in COS cells (Figure 4B). These data are consistent with the results of the luciferase assays and suggest that the inhibitory effect of sgRNA is achieved through target RNA degradation by tRNase ZL.
Figure 4.
RT–PCR analyses of HIV-1 mRNA expression. RT–PCR analyses of uncleaved (product I), and total cleaved and uncleaved (product II), HIV-1 mRNA were performed using HIV-1 gag-specific primers with concurrent amplification of GAPDH mRNA. (A) Schematic representation of HIV-1-specific primer sites with respect to HIV-1 mRNA: F1 primers, SL3 and p24; F2 primers, SL3, SL4 and p24; R-primers, SL3, SL4 and p24. (B) RT–PCR amplification products analysed by 2% agarose gel electrophoresis with ethidium bromide staining.
sgRNA-gag-p24 barely directs in vitro HIV-1 RNA cleavage by tRNase ZL
sgRNA-gag-p24 was much less effective than the other sgRNAs in suppressing HIV-1 gene expression in cells. We examined the reason for this by assessing its ability to guide HIV-1 RNA cleavage by tRNase ZL in vitro. The partial HIV-1 RNAs T-SL3-1 and T-gag-p24 containing SL3 and the gag p24 site, respectively, were synthesized in vitro with T7 RNA polymerase and 5′ end-labelled with fluorescein. sgRNA-SL3-1 and sgRNA-gag-p24 were also transcribed by T7 RNA polymerase in vitro. The ability of each sgRNA to induce cleavage of the corresponding target RNA by tRNase ZL was tested in vitro. The target T-SL3-1 was cleaved efficiently under the direction of sgRNA-SL3-1, whereas cleavage of the target T-gag-p24 by sgRNA-gag-p24 was highly inefficient (Figure 5), this was in agreement with the in vivo observations. Cleavage of each target occurred primarily 1 nt downstream of the nucleotide corresponding to the discriminator (Figure 5). These results indicate that sgRNA bound to its target HIV-1 mRNA, and cleavage of the pre-tRNA complexes with tRNase ZL occurred. It was therefore important to determine the localization of both the sgRNA and its target HIV-1 in the cells. The reduction in functional HIV-1 mRNA was consistent with tRNase ZL cleavage occurring at the post-transcriptional level.
Figure 5.
sgRNA guided specific HIV-1 mRNA cleavage by in vitro tRNase ZL assays. (A) Secondary structures of the substrate-HIV-1 SL3-1 (extra loop) and gag-24 complexes with sgRNA-SL3-1 and sgRNA-gag-p24. (B) The assays for the fluorescein (F)-labelled target RNA T-SL3-1 or T-gag-p24 (0.1 pmol) were performed with pig liver tRNase ZL (20 ng) in the presence of the unlabelled sgRNA-SL3-1 or sgRNA-gag-p24 (5 or 10 pmol) at 50°C for 10 min. The cleavage reactions were analysed using a 10% polyacrylamide/8 M urea sequencing gel. The target RNA and the primary 5′-cleavage product are indicated by a bar and arrow, respectively, together with their size in nucleotides. L denotes the alkaline ladder of each fluorescein-labelled target RNA.
Inhibition of HIV-1 gene expression by retroviral vector-mediated sgRNAs in human T cells
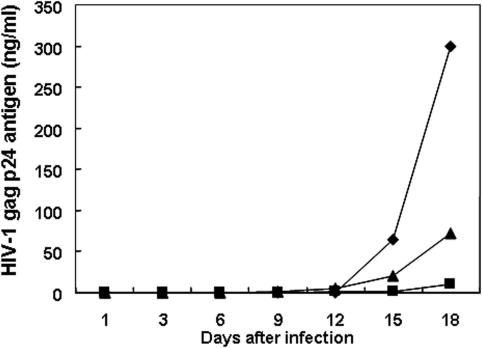
The inhibitory effect of HIV-1 expression by tRNase ZL-mediated sgRNAs was investigated in human T cells by constructing a MoMLV-based sgRNA retroviral vector. Most retroviral vectors used in experimental and clinical gene therapy are derived from the MoMLV (45). Retroviruses integrate into the chromosomal DNA, so their genome is stable in the host cells and is inherited by their progeny. Accordingly, long-term expression of a transduced gene can be achieved through retrovirus-mediated gene transfer. Other advantages of this vector include its broad host range and the availability of packaging cell lines for the large-scale production of high-titre vectors. It has previously been shown that an amphotropic MoMLV-based retrovirus vector can transduce a human T-cell line (46). We therefore expressed the sgRNA under the control of the promoter of a human gene via a retroviral vector (Figure 1B). The plasmid pLsgRGSN (Figure 1B) was constructed by inserting the following elements into the EcoRI and XhoI sites of the retroviral vector pLXSN: an EcoRI and XhoI fragment from the plasmid pSV2neo/sgRG, sgRNAs and EGFP genes. We then obtained transduced Jurkat T cells stably expressing the sgRNAs. These Jurkat T cells were infected with wild-type HIV-1NL4-3, and HIV-1gag-p24 antigen levels in the cell-free supernatant were measured at weekly intervals over 18 days. By day 18, the HIV-1 gag-p24 product was suppressed almost completely (∼97%) in the cell cultures expressing sgRNA-SL4 (Figure 6). In contrast, sgRNA-gag-p24 and sgRNA-SL4 failed to inhibit viral expression under these experimental conditions. The difference between the effects of sgRNA-SL4 and sgRNA-gag-p24 in the HIV-1-challenged assay was due to the lack of base pairing in the hairpin structure resembling the T-stem-loop region.
Figure 6.
Inhibition of HIV-1 gag-p24 product in Jurkat cells expressing stably retrovirus vector-mediated sgRNA. Cells were cultured for 18 days after infection with HIV-1NL4-3. Small aliquots of supernatant were prepared from each culture on days 1, 3, 6, 9, 12, 15 and 18. HIV-1 gag p24 antigen was determined using CLEIA (Lumipulse) according to the manufacturer's protocol (37): closed diamonds, pLGSN-ΔsgR; closed squares, pLsgRGSN-SL4; and closed triangles, pLsgRGSN-gag-p24.
In conclusion, we demonstrated the inhibition of HIV-1 gene products in cultured cells by inducing HIV-1 mRNA cleavage using a modified 5′-half-tRNAArg (sgRNA) and mammalian tRNase ZL. The sgRNA/target HIV-1RNA complex formed a pre-tRNA-like structure with 5′-half-tRNA and a stable hairpin (3′-half-tRNA) structure resembling the T-stem-loop region. The –sgRNA transcript was expressed at high levels and localized in the nucleus. The greatest inhibitory effect on HIV-1 expression was achieved using sgRNA-SL4 targeting the HIV-1 gag gene. These results suggest that both sgRNA and its target HIV-1 mRNA were located in the nucleus, allowing specific cleavage by tRNase ZL. Furthermore, MoMLV-based sgRNA-SL4 could suppress sgRNA-dependent HIV-1 expression in human T cells. We believe that the use of mammalian tRNase ZL in conjunction with guide sequences represents a promising tool for the inactivation of genes in mammalian cells. Furthermore, the inhibition of HIV-1 using this approach demonstrates its potential as a therapeutic agent for AIDS.
Acknowledgments
We thank Prof. Y. Koyanagi for providing some of the plasmids, and Dr H. Takeuchi for technical assistance. This work was supported by a Grant-in-Aid for High Technology Research (HTR) from the Ministry of Education, Science, Sports and Culture, Japan, by research grants from the Human Science Foundation (HIV-K-14719) and the Science Research Promotion Fund, and by an Academic Frontier Research Project Grant from the Promotion and Mutual Aid Corporation for the Private Schools of Japan. Y.H. is a Research Fellow of the HTR. Funding to pay the Open Access publication charges for this article was provided by HTR.
REFERENCES
- 1.Kelin A.S., Klebba C., Hoelzer D. Gene therapy of HIV-1 infection. In: Unger R.E., Kreuter J., Rubsaman-Waigmann H., editors. Antivirals Agent AIDS. NY: Marcel Dekker Inc.; 2000. pp. 219–248. [Google Scholar]
- 2.Rigden J.R., Ely J.A., Macpherson J.L., Gerlach W.L., Sun L.-Q., Symonds G.P. The use of ribozyme gene therapy for the inhibition of HIV replication and its pathogenic sequelae. In: Rossi J.J., Couture L.A., editors. Intracellular Ribozyme Applications, Principles and Protocols. England: Horizon Scientific Press; 1999. pp. 93–102. [Google Scholar]
- 3.Dornburg R., Pomerantz R.Z. Gene therapy and HIV-1 infection. In: Templeton N.S., Lasic D.D., editors. Gene Therapy, Therapeutic Mechanisms and Strategies. NY: Marcel Dekker Inc.; 2000. pp. 519–534. [Google Scholar]
- 4.Kurreck J. Antisense technologies, improvement through novel chemical modifications. Eur. J. Biochem. 2003;270:1628–1644. doi: 10.1046/j.1432-1033.2003.03555.x. [DOI] [PubMed] [Google Scholar]
- 5.Jen K.-Y, Gewirtz A.M. Suppression gene expression by targeted disruption of messenger RNA available options and current strategies. Stem Cells. 2000;18:307–319. doi: 10.1634/stemcells.18-5-307. [DOI] [PubMed] [Google Scholar]
- 6.Bunnell B.A., Morgan R.H. Gene therapy for infectious diseases. Clin. Microbiol. Rev. 1998;11:42–56. doi: 10.1128/cmr.11.1.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Martinez M.A., Clotet B., Este J. RNA interference of HIV replication. Trends Immunol. 2002;23:559–561. doi: 10.1016/s1471-4906(02)02328-1. [DOI] [PubMed] [Google Scholar]
- 8.Lori F., Guallini P., Gulluzzi L., Lisziewicz J. Gene therapy approaches to HIV infections. Am. J. Pharmacogenomics. 2002;2:245–252. doi: 10.2165/00129785-200202040-00004. [DOI] [PubMed] [Google Scholar]
- 9.Stevenson M. Dissecting HIV-1 through RNA interference. Nature Rev. Immunol. 2003;3:851–858. doi: 10.1038/nri1227. [DOI] [PubMed] [Google Scholar]
- 10.Scherer L.J., Rossi J.J. Approaches for the sequence-specific knockdown of mRNA. Nat. Biotechnol. 2003;21:1457–1465. doi: 10.1038/nbt915. [DOI] [PubMed] [Google Scholar]
- 11.Takaku H. Gene silencing of HIV-1 by RNA interference. Antivir. Chem. Chemother. 2004;15:57–65. doi: 10.1177/095632020401500201. [DOI] [PubMed] [Google Scholar]
- 12.Yuan Y., Hwang E.S., Altman S. Targeted cleavage of mRNA by human RNase P. Proc. Natl Acad. Sci. USA. 1992;89:8006–8010. doi: 10.1073/pnas.89.17.8006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yuan Y., Altman S. Selection of guide sequences that direct efficient cleavage of mRNA by human ribonuclease. Science. 1994;263:1269–1273. doi: 10.1126/science.8122108. [DOI] [PubMed] [Google Scholar]
- 14.Guerrier-Takada C., Li Y., Altman S. Artificial regulation of gene expression in Escherichia coli by RNase P. Proc. Natl Acad. Sci. USA. 1995;92:11115–11119. doi: 10.1073/pnas.92.24.11115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hnatyszyn H., Spruillm G., Youngm A., Seivrightm R., Kraus G. Long-term RNase P-mediated inhibition of HIV-1 replication and pathogenesis. Gene Ther. 2001;24:1863–1871. doi: 10.1038/sj.gt.3301606. [DOI] [PubMed] [Google Scholar]
- 16.Kraus G., Geffin R., Spruill G., Young A.K., Seivright R., Cardona D., Burzawa J., Hnatyszyn H.J. Cross-clade inhibition of HIV-1 replication and cytopathology by using RNase P-associated external guide sequences. Proc. Natl Acad. Sci. USA. 2002;99:3406–3411. doi: 10.1073/pnas.052651199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Barnor J.S., Endo Y., Habu Y., Miyano-Kurosaki N., Kitano M., Yamamoto H., Takaku H. Effective inhibition of HIV-1 replication in cultured cells by external guide sequences and ribonuclease P. Bioorg. Med. Chem. Lett. 2004;14:4941–4944. doi: 10.1016/j.bmcl.2004.07.021. [DOI] [PubMed] [Google Scholar]
- 18.Nashimoto M. Distribution of both lengths and 5′ terminal nucleotides of mammalian pre-tRNA 3′ trailers reflects properties of 3′ processing endoribonuclease. Nucleic Acids Res. 1997;25:1148–1154. doi: 10.1093/nar/25.6.1148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Takaku H., Minagawa A., Takagi M., Nashimoto M. A candidate prostate cancer susceptibility gene encodes tRNA 3′ processing endoribonuclease. Nucleic Acids Res. 2003;31:2272–2278. doi: 10.1093/nar/gkg337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Takaku H., Minagawa A., Takagi M., Nashimoto M. The N-terminal half domain of the long form of tRNase Z is required for the RNase 65 activity. Nucleic Acids Res. 2004;32:4429–4438. doi: 10.1093/nar/gkh774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tavtigian S.V., Simard J., Teng D.H., Abtin V., Baumgard M., Beck A., Camp N.J., Carillo A.R., Chen Y., Dayananth P., et al. A candidate prostate cancer susceptibility gene at chromosome 17p. Nature Genet. 2001;27:172–180. doi: 10.1038/84808. [DOI] [PubMed] [Google Scholar]
- 22.Minagawa A., Takaku H., Takagi M., Nashimoto M. The missense mutations in the candidate prostate cancer gene ELAC2 do not alter enzymatic properties of its product. Cancer Lett. 2004 doi: 10.1016/j.canlet.2004.09.013. in press. [DOI] [PubMed] [Google Scholar]
- 23.Nashimoto M. Conversion of mammalian tRNA 3′ processing endoribonuclease to four-base-recognizing RNA cutters. Nucleic Acids Res. 1995;23:3642–3647. doi: 10.1093/nar/23.18.3642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nashimoto M. Specific cleavage of target RNAs from HIV-1 with 5′ half tRNA by mammalian tRNA 3′ processing endoribonuclease. RNA. 1996;2:523–534. [PMC free article] [PubMed] [Google Scholar]
- 25.Nashimoto M., Geary S., Tamura M., Kaspar R. RNA heptamers that direct RNA cleavage by mammalian tRNA 3′ processing endoribonuclease. Nucleic Acids Res. 1998;26:2565–2571. doi: 10.1093/nar/26.11.2565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nashimoto M. Anomalous RNA substrates for mammalian tRNA 3′ processing endoribonuclease. FEBS Lett. 2000;472:179–186. doi: 10.1016/s0014-5793(00)01462-9. [DOI] [PubMed] [Google Scholar]
- 27.Takaku H., Minagawa A., Takagi M., Nashimoto M. A novel four-base-recognizing RNA cutter that can remove the single 3′ terminal nucleotides from RNA molecules. Nucleic Acids Res. 2004;32:e91. doi: 10.1093/nar/gnh092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tamura M., Nashimoto C., Miyake N., Daikuhara Y., Ochi K., Nashimoto M. Intracellular mRNA cleavage by 3′ tRNase under the direction of 2′-O-methyl RNA heptamers. Nucleic Acids Res. 2003;31:4354–4360. doi: 10.1093/nar/gkg641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cormack B.P., Valdivia R.H., Falkow S. FACS-optimized mutants of the green fluorescent protein (GFP) Gene. 1996;173:33–38. doi: 10.1016/0378-1119(95)00685-0. [DOI] [PubMed] [Google Scholar]
- 30.Yang T.T., Cheng L., Kain S.R. Optimized codon usage and chromophore mutations provide enhanced sensitivity with the green fluorescent protein. Nucleic Acids Res. 1996;24:4592–4593. doi: 10.1093/nar/24.22.4592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Miller A.D., Rosman G.J. Improved retroviral vectors for gene transfer. BioTechniques. 1989;7:980–987. [PMC free article] [PubMed] [Google Scholar]
- 32.Akkina R.K., Walton R.M., Chen M.L., Li Q.-X., Planelles V., Chen I.Y. High-efficiency gene transfer into CD34+ cells with a human immunodeficiency virus type 1-based retroviral vector pseudotyped with vesicular stomatitis virus envelope glycoprotein G. J. Virol. 1996;70:2581–2585. doi: 10.1128/jvi.70.4.2581-2585.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Landau N.R., Littman D.R. Packaging system for rapid production of murine leukemia virus vectors with variable tropism. J. Virol. 1992;66:5110–5113. doi: 10.1128/jvi.66.8.5110-5113.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li X., Mukai T., Young D., Frankel S., Law P., Wong-Staal F. Transduction of CD34+ cells by a vesicular stomach virus protein G (VSV-G) pseudotyped HIV-1 vector. Stable gene expression in progeny cells, including dendritic cells. J. Hum. Virol. 1998;1:346–352. [PubMed] [Google Scholar]
- 35.Burns J.C., Friedmann T., Driever W., Burrascano M., Yee J.-K. Vesicular stomatitis virus G glycoprotein pseudotyped retroviral vectors: concentration to very high titer and efficient gene transfer into mammalian and non-mammalian cells. Proc. Natl Acad. Sci. USA. 1993;90:8033–8037. doi: 10.1073/pnas.90.17.8033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yee J.K., Miyanohara A., Laforte P., Bouic K., Burns J.C., Friedman T. A general method for the generation of high-titer, pantropic retroviral vectors: highly efficient infection of primary hepatocytes. Proc. Natl Acad. Sci. USA. 1994;91:9564–9568. doi: 10.1073/pnas.91.20.9564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sakai A., Hirabayasi Y., Aizawa S., Tanaka M., Ida S., Oka S. Investigation of a new p24 antigen detection system by the chemiluminescence-enzyme-immunoassay. J. Jap. Assoc. Infect. Dis. 1999;73:205–212. doi: 10.11150/kansenshogakuzasshi1970.73.205. [DOI] [PubMed] [Google Scholar]
- 38.Kuwasaki T., Hosono K., Takai K., Ushijima K., Nakashima H., Saito T., Yamamoto N., Takaku H. Hairpin antisense oligonucleotides containing 2′-methoxynucleosides with base-pairing in the stem region at the 3′-end: penetration, localization, and anti-HIV activity. Biochem. Biophys. Res. Commun. 1996;228:623–631. doi: 10.1006/bbrc.1996.1707. [DOI] [PubMed] [Google Scholar]
- 39.Clever J., Mirandar D., Jr, Parslow T.G. RNA structure and packaging signals in the 5′ leader region of the human immunodeficiency virus type 1 genome. J. Virol. 2002;76:12381–12387. doi: 10.1128/JVI.76.23.12381-12387.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.McBride M.S., Panganiban A.T. Position dependence of functional hairpins important for human immunodeficiency virus type 1 RNA encapsidation in vivo. J. Virol. 1997;71:2050–2058. doi: 10.1128/jvi.71.3.2050-2058.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sullenger B.A., Gallardo H.F., Ungers G.E., Gilboa E. Overexpression of TAR sequences renders cells resistant to human immunodeficiency virus replication. Cell. 1990;63:601–608. doi: 10.1016/0092-8674(90)90455-n. [DOI] [PubMed] [Google Scholar]
- 42.Sullenger B.A., Lee T.C., Smith C.A., Ungers G.E., Gilboa E. Expression of chimeric tRNA-driven antisense transcripts renders NIH 3T3 cells highly resistant to Moloney murine leukemia virus replication. Mol. Cell. Biol. 1990;10:6512–6523. doi: 10.1128/mcb.10.12.6512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhang Y.A., Nemunaitis J., Scanlon K.J., Tong A.W. Anti-tumorigenic effect of K-ras ribozyme against human lung cancer cell line heterotransplants in nude mice. Gene Ther. 2000;7:2041–2050. doi: 10.1038/sj.gt.3301331. [DOI] [PubMed] [Google Scholar]
- 44.Habu Y., Miyano-Kurosaki N., Nagawa T., Matsumoto N., Takeuchi H., Takaku H. Inhibition of HIV-1 replication by an HIV-1 dependent ribozyme expression vector with the Cre/loxP (ON/OFF) system. Antivir. Chem. Chemother. 2002;13:273–281. doi: 10.1177/095632020201300502. [DOI] [PubMed] [Google Scholar]
- 45.Mulligan R.C. The science of gene therapy. Science. 1993;260:926–932. doi: 10.1126/science.8493530. [DOI] [PubMed] [Google Scholar]
- 46.Miller A.D., Rosman G.J. Improved retroviral vectors for gene transfer. BioTechniques. 1989;7:980–987. [PMC free article] [PubMed] [Google Scholar]