Abstract
PARN, a poly(A)-specific ribonuclease, binds the 5′ cap-structure of mRNA and initiates deadenylation-dependent decay. Eukaryotic initiation factor 4E (eIF4E) also binds to the cap structure, an interaction that is critical for initiating cap-dependent translation. The stability of various mRNA transcripts in human cell lines is reduced under conditions of serum starvation as determined by both functional and chemical half-lives. Serum starvation also leads to enhanced cap association by PARN. In contrast, the 5′ cap occupancy by eIF4E decreases under serum-deprivation, as does the translation of reporter transcripts. Further, we show that PARN is a phosphoprotein and that this modification can be modulated by serum status. Taken together, these data are consistent with a natural competition existing at the 5′ cap structure between PARN and eIF4E that may be regulated by changes in post-translational modifications. These phosphorylation-induced changes in the interplay of PARN and eIF4E may determine whether the mRNA is translated or decayed.
INTRODUCTION
Post-transcriptional regulation is increasingly recognized as a critical control point in gene expression. Modulation of mRNA stability can affect the steady-state levels of transcripts without any change in transcription rate (1,2), permitting rapid responses to metabolic or cell cycle events. Additionally, a number of factors have been identified, which can alter the levels of translation, such as mRNA secondary and tertiary structure, association of 3′ untranslated region (UTR) and 5′UTR-RNA binding proteins, and the phosphorylation of critical proteins (3–6).
The major decay pathways for mRNA species in eukaryotes initiate with deadenylation, followed either by decapping and 5′–3′ exonuclease digestion, or by 3′–5′ exonuclease decay (7–9). Recent reports have indicated that this initial, often rate-limiting step can be performed in mammalian cells by a poly(A)-specific ribonuclease (PARN) (10), an enzyme with affinity for the 5′ cap structure (11–13). PARN is the best characterized deadenylase in mammalian cells (13) and is an enzyme with homology to the RNase D family (14,15). It is not certain whether PARN is the major deadenylase, as other enzymes with deadenylating activity have been identified. However, deadenylation of poly(A) tails by PARN produces characteristic decay intermediates of ∼30 nt that correspond to the sequential removal of poly(A) binding protein (PABP) molecules, a pattern of deadenylation that has been observed in cytoplasmic extracts (10). Data indicate that PARN is constitutively expressed in most mammalian tissues, located in both the nucleus and cytoplasm (14,16) and is predicted to contain an RNA recognition motif (RRM) and an R3H domain. The R3H domain is a conserved sequence motif, identified in >100 proteins, which is thought to be involved in polynucleotide binding, including double and single-stranded DNA and RNA (http://bioinfo.weizmann.ac.il/cards-bin/carddisp?PARN&search=parn&suff=txt).
The two termini of most mammalian mRNAs are characterstically modified with a m7G(5′)ppp(5′)N cap at the 5′-end and a poly(A) tail at the 3′-end. These modifications occur co-transcriptionally and are believed to function in protecting against transcript decay, as well as stimulating translation. The 5′ cap structure is the recognition element for translation initiation factor eIF4E. Various reports suggest that the binding affinity of this protein for the cap may be enhanced by its interaction with eIF4G, creating a functional eIF4F complex (17,18), the formation of which is essential for appropriate translation of the vast majority of eukaryotic mRNAs. Furthermore, 5′ and 3′ termini communicate to facilitate the efficient translation initiation through interaction of PABP with the translation initiation factor eIF4G (19). During the process of mRNA decay, PARN has been shown to interact with 5′ cap in a manner that increases the processivity of poly(A) tail shortening (8–10). Therefore, it appears that for both the process of degradation and that of translation, the 5′ and 3′ termini of mRNA must be brought into close proximity to effectively perform either process. This suggests that the 5′–3′ interactions may be a pivotal control point that determines the mRNA fate.
Previous studies have indicated that it is unlikely that eIF4E and PARN can bind simultaneously to the 5′ cap (11,12). It is therefore an intriguing possibility that competition for cap-binding between PARN and eIF4E could regulate the fate of mRNA species in the cell, promoting degradation or translation, respectively. To investigate this hypothesis, we needed to subject cells to conditions that would be predicted to perturb this competition. There have been sporadic reports that depletion or withdrawal of nutrients (amino acids or serum) from cultured cells can induce rapid degradation of various mRNAs (20–26). Our experimental approach, therefore, focuses on serum-deprivation and investigates the associated effects on cap occupancy and transcript fate.
In this study, we report a significant increase in PARN association with the 5′ cap under conditions of serum starvation. This PARN–cap interaction is directly associated with an increase in the observed mRNA decay rate under serum-starvation conditions. Changes in the phosphorylation state of eIF4E and 4E-BP-1, which would reduce the cap occupancy by the eIF–4F complex, occur concomitantly under serum-starvation conditions. PARN was also shown to be a phosphoprotein whose post-translational modification is also affected under serum-starvation conditions. These data suggest that phosphorylation changes that occur during serum starvation may be responsible for the observed increased mRNA decay rates by promoting cap–PARN interactions.
MATERIALS AND METHODS
Tissue culture
Human liver Hep G2 cells were cultured (37°C, 5% CO2) in Eagle's modified essential medium supplemented with 10% (v/v) fetal calf serum, non-essential amino acids and 2 mM l-glutamine. Serum starvation was induced by culturing cells in serum-free medium for 15 h prior to experimentation.
Half-life determination of endogenous transcripts by northern analysis
Time courses were performed after transcription inhibition by actinomycin D (act D; 5 μg/ml−1 final), a treatment that has been previously shown to inhibit >94% of cellular transcription in Hep G2 cells (27). Total RNA was extracted from 4 × 106 cells using 1 ml of TRIzol reagent (Invitrogen), every 24 h over a 96 h time-course. Northern blot analysis and probe production for β-actin and 18S rRNA were as described previously (27). For glutamate dehydrogenase (GDH), the template was generated by PCR using forward primer 5′-GATGCATCATGGCTGA-3′, reverse primer 5′-AGCAAGCAACTGACTGCTCT-3′ and the following profile: 95°C for 4 min, 35 cycles of 95°C for 1 min, 55°C for 1 min and 72°C for 3 min with a final extension of 72°C for 8 min to produce a 335 bp fragment. All probes were radiolabelled using random hexamer primers and [α-32P]dCTP (GE Healthcare, Amersham Biosciences), as described previously (27). Resulting bands were visualized (PhosphorImager, Molecular Dynamics), quantified (ImageQuant, GE Healthcare, Amersham Biosciences) and β-actin and GDH values were corrected by comparison to the corresponding 18S rRNA value.
DNA Templates for in vitro transcription
Templates were generated by PCR using a forward primer that contains a phage SP6 RNA polymerase consensus promoter sequence (primer 1, 5′-CAAGCTATTTAGGTGACAC-3′) and reverse primers that introduce A40 tails on the resulting RNA (primer 2, 5′-T40CAGGTAAACTGG-3′, primer 3, 5′-T40ACAATAGCTAAGAATTTC-3′). PCR was performed with final concentrations of 1.5 mM MgCl2, 200 μM dNTPs, 50 pmol of each primer, 1× reaction buffer IV (AB Gene), and 2 U of DNA polymerase (AB Gene). DNA templates for these reactions were (i) pGEM–Luc (Promega) for luciferase (in conjunction with primers 1 and 3 at 50°C); (ii) pGEM–Luc with the 3′UTR substituted from cytochrome c oxidase subunit IV (COX 4) (28) generated as described in (29), amplified with primers 1 and 2 at 59°C for Luc–COX; (iii) Clones 84 and 12 were selected for their increased stability as described in (29). Both contain the COX 4 3′UTR with differing 50 nt insertions and were amplified with primers 1 and 2 at 59°C; and lastly (iv) Luc-5′ insertion was generated to introduce a stem loop into the 5′ UTR upstream of the luciferase open reading frame by HindIII and BamH1 excision from pJ6Ω, (30), ligation into HindIII and BamH1 digested pGEM–Luc and amplified with primers 1 and 3 at 50°C. All PCR reactions were hot start initiated with the following profile: 95°C for 4 min, then 35 cycles of 95°C for 1 min, hybridization for 1 min at specified temperatures, 72°C for 3.5 min, and a final 72°C for an 8 min step.
In vitro transcription
All in vitro transcription was by high yield bacteriophage SP6 RNA polymerase in the absence or presence of m7G(5′)ppp(5′)G cap analogue (Epicentre). RNA was analysed for integrity and concentration by denaturing gel electrophoresis and rabbit reticulocyte lysate (nuclease treated, Promega) as described earlier (29).
Functional half-life calculation of luciferase mRNA and protein
In vitro transcribed, capped RNAs were electroporated into 5 × 106 Hep G2 cells as described previously (29). Briefly, cells were incubated in 0.5 ml of phosphate-buffered saline (PBS)/1.25% (v/v) dimethyl sulfoxide on ice before electroporation with 5–8 μg of RNA in a Biorad GenePulser II at 400 V/250 μF (0.2 cm pathlength cuvettes). Aliquots of cells (0.5 × 106) were lysed (cell culture lysis reagent, Promega) at various time points post electroporation and the luciferase activity was measured (luciferase assay reagent, Promega). Relative light units were plotted as a function of maximal activity against the time from which functional half-lives were derived (31). The half-life of the luciferase protein was determined by cycloheximide addition (100 μg.ml−1 final) 90 min post electroporation and the lysates were analysed at the times indicated.
Chemical half-life calculation of electroporated mRNA by quantitative RT–PCR
Total RNA was extracted (TRIzol, Invitrogen) from 4 × 106 cells at various time points post electroporation. Reverse transcriptions were performed on 10 μg of RNA using a Superscript first strand synthesis system (Invitrogen) after priming with random hexamers. Real-time PCR (LightCycler, Roche) was performed using Taqman (PE Applied Biosystems) probe technology. Probes and primers were designed using Primer Express Software (PE Applied Biosystems) for both luciferase and cytochrome c oxidase 7a-L (COX 7a-L) as a reference gene. Primer/probe details for luciferase are forward 5′-CCTTGTCGTATCCCTGGAAGA-3′, reverse 5′-GGGCGCACCTCTTTCGA-3′, probe 5′-TTGCAACCGCTTCCCCGACTTCT-3′ (71 bp amplicon) and for Cox 7a-L forward primer 5′-GCGGTTGCTGGTCAGTAACA-3′, reverse primer 5′-GCTTATCGTCCTCTGCCCAAT-3′ and probe 5′-TGCGGAATCTGCTGGCTCTTCGTC-3′ (63 bp amplicon).
Luciferase and cytochrome c oxidase amplicons were generated from the same cDNA sample using the LightCycler–FastStart DNA Master Hybridization Probes kit (Roche). Reactions were performed with final concentrations of 0.5 μM primers, 0.1 μM probe and 5 mM MgCl2 in 20 μl volume, and the following profile; 95°C for 10 min, 45 cycles of 95°C for 5 s and 60°C for 20 s. Quantification was by the relative standard curve method (User Bulletin 2, ABI 7700 Sequence Detection System). Standard curves were created for both target and reference using cDNA from T = 0 time point. For each time point, the amount of target and reference was calculated from linear regression of the appropriate standard curve, and target values were divided by their corresponding references. The values obtained for each transcript were plotted as percentage luciferase mRNA remaining versus time, and half-lives calculated from the slope of best fit. Analyses were performed on a minimum of three independent experiments with each RNA extraction reverse transcribed three times per time point (n = 9). For each time course, the mean mRNA remaining was plotted against time.
Radiolabel transfer, UV-irradiation and immunoprecipitation
Uncapped in vitro transcribed RNAs were labelled exclusively at the α-phosphate of the 5′ cap structure using recombinant guanylyl transferase and [α−32P]GTP (3000 Ci mmol−1, GE Healthcare, Amersham Biosciences) in the presence of 0.67 mM S-adenosyl-l-methionine (GE Healthcare, Amersham Biosciences), 50 mM Tris, pH 7.9, 1 mM MgCl2, 5 mM KCl and 5 mM DTT for 30 min at 37°C. The transcript was phenol extracted, precipitated and a fraction analysed on 5% (w/v) polyacrylamide/8 M urea gel.
The S100 extracts were prepared as in (32). For S10 fractions, cells were lysed in cold extraction buffer [20 mM HEPES–KOH, pH 7.5, 50 mM KCl, 25 mM β-glycerophosphate, 10% (v/v) glycerol, 0.1 mM EDTA and 0.5% (v/v) NP-40] and passed 7 times through a 21 gauge needle. The lysate was centrifuged at 10 000 g for 10 min at 4°C and the supernatant retained. Both S100 and S10 preparations were supplemented throughout with protease inhibitors (1 μg.ml−1 antipain, 1 μg.ml−1 leupeptin, 1 mM p-aminobenzamidine, 1 mM phenylmethylsulphonyl fluoride (PMSF), 1× Roche EDTA-free protease inhibitor cocktail and 0.2 mM EDTA) and phosphatase inhibitors (1 mM sodium fluoride (NaF), 25 mM β-glycerophosphate and 10 mM sodium orthovanadate).
For immunoprecipitations (IP), cap-labelled mRNA (20–50 fmol) was incubated with either 16 μg of S100 (PARN IP) or 40 μg of S10 (eIF4E IP) supplemented with 1.96 mM EDTA, 2.3% (w/v) polyvinyl alcohol, 0.87 mM ATP, 17.5 mM phosphocreatine for 5 min at 30°C. Reactions were UV-irradiated on ice (10 min for PARN, 32 min for eIF4E) in a Spectrolinker (Spectronics), before RNase A/T1 (0.25 U/10 U Ambion) digestion for 30 min at 30°C. Subsequent steps were performed as described previously (12). Briefly, 400 μl of NET2 buffer [50 mM Tris, pH 7.6, 150 mM NaCl and 0.01% (v/v) NP-40] was added post RNase digestion and reactions incubated with antisera for 1 h, rotating at 4°C. Interactions were collected using PANSORBIN (Calbiochem) (1 h, rotating at 4°C), washed and separated by denaturing gel electrophoresis (SDS–PAGE) [10% (w/v) for PARN, 15% (w/v) for eIF4E]. Products were visualized by ImageQuant analysis (GE Healthcare, Amersham Biosciences). Antisera against eIF4E from three different sources were tested for immunoprecipitation. The Santa Cruz preparation (Santa Cruz Biotechnology, Inc.) was the most effective and was therefore used for these experiments.
m7GTP-Sepharose 4B chromatography
This was modified from Martin et al. (33). Cell extracts were prepared as described for isoelectric focussing (IEF). Aliquots (100 μg) of post 600 g supernatant were incubated with 30 μl m7GTP-Sepharose (6 nmol ligand, GE Healthcare, Amersham Biosciences) for 15 min at 4°C. Beads were washed (20 mM HEPES, pH 7.9, 20% glycerol, 100 mM KCl, 0.2 mM EDTA, 1 mM PMSF and 1 mM DTT, supplemented with protease and phosphatase inhibitors as described above) before elution of associated proteins with SDS sample buffer and separation by denaturing gel electrophoresis (10% w/v). Immunoblot analysis was performed as described below with antibodies to PARN and eIF4E.
Isoelectric focussing
Cell extracts were prepared in S10 extraction buffer, supplemented with phosphatase and protease inhibitors (as above), sheared through a 21 gauge needle and centrifuged at 600 g. The supernatant was incubated overnight at 4°C with m7GTP-Sepharose 4B (GE Healthcare, Amersham Biosciences); the beads were collected by centrifugation, washed, resuspended in IEF buffer [3% (v/v) ampholines, 5% (w/v) CHAPS, 0.9 M β-mercaptoethanol and 6 M urea], electrophoresed through a 3.5–10 pH gradient IEF gel (34), transferred and western blotted with eIF4E antibodies.
Immunoprecipitation of 32P orthophosphate labelled PARN
Dynal protein G beads (Dynal ASA) were pre-blocked with 0.1 mg.ml−1 BSA in NET2 buffer (12) for 1 h at room temperature, then incubated in 500 μl of NET2 for a further hour with antisera to PARN (1:100 dilution of antisera kindly donated by E. Wahle) or without (described as 2° only). The beads were washed in NET2 buffer, before the addition of lysate. Cells were cultured in a 25 cm2 flask, washed twice in supplemented KRH buffer (121 mM NaCl, 4.9 mM KCl, 1.2 mM MgSO4, 0.33 mM CaCl2, 12 mM HEPES, pH 7.4, 50 μM H2NaPO4, 25 mM glucose, 2 mM l-glutamine and 1× non-essential amino acids) and incubated for 1 h at 37°C in 2 ml of supplemented KRH lacking serum with 500 μCi of radiolabelled orthophosphate (PBS11, GE Healthcare, Amersham Biosciences). Cells were washed in cold TBS, harvested, resuspended in 500 μl of cold IP buffer (150 mM NaCl, 1 mM EGTA, 1 mM EDTA, 1.0% (v/v) Triton X-100, 20 mM Tris, pH 7.5, 1 mM PMSF, 1 mM DTT, 10 mM β-glycero-phosphate, 10 mM NaF and Roche complete mini EDTA-free protease inhibitors) and lysed (7 passes through a 21 gauge needle). The lysate was cleared by centrifugation, equal amounts of protein were added to the anti-PARN treated and 2° only Dynal beads, supplemented to 400 μl with NET2 buffer and incubated at room temperature for 1 h. Beads were washed (5 × 1 ml NET2) before elution of complexes in sample loading buffer [20% (v/v) glycerol, 4% (w/v) SDS, 250 mM Tris, pH 6.8 and 100 mM DTT]. Samples were electrophoresed through 10% (w/v) SDS–PAGE and phosphorylated PARN, visualized by PhosphorImager and ImageQuant analysis.
Phosphoprotein analysis
Phosphorylated and non-phosphorylated fractions of total cell lysate were prepared following the manufacturer's recommendations from both serum-starved and stimulated cells using the Qiagen PhosphoPurification System (http://www.qiagen.com/catalog/auto/cget.asp?p=PhosphoProtein_purification_kit). As recommended, final volumes were concentrated with the nanosep columns provided and equivalent amounts from both phosphorylated and non-phosphorylated fractions of starved and stimulated samples were loaded onto 10% (w/v) and 15% (w/v) denaturing polyacrylamide gels.
Western analysis
Proteins were separated by SDS–PAGE or IEF and immobilized by wet (100 V, 1 h) or semi-dry (15 V, 1 h) transfer, respectively onto PVDF (Immobilon-P, Millipore Corporation) membrane in 25 mM Tris, 192 mM glycine, 0.02% (w/v) SDS and 15% (v/v) methanol. Proteins of interest were bound by rabbit polyclonal antisera [PARN (kindly provided by the Wormington and Wahle Laboratories), PABP (a gift of the Morley Laboratory)], or antibodies against eIF4E, 4E-BP1, and S6 ribosomal protein (NEB), eIF4E and eIF4G (Santa Cruz), followed by HRP-conjugated secondaries and visualized with ECL-plus reagents (GE Healthcare, Amersham Biosciences).
RESULTS
Endogenous transcripts are less stable under serum starvation
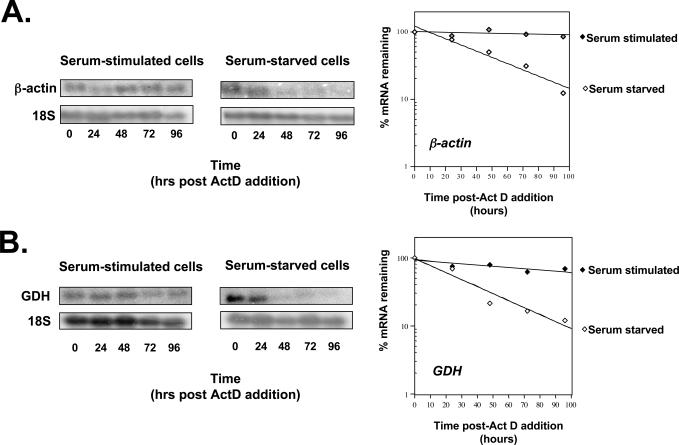
As a prerequisite to challenging the hypothesis described in the Introduction, conditions needed to be identified that would perturb the stability and/or translation of transcripts, allowing us to measure any concomitant alteration in cap occupancy. Several reports (20–26) had suggested that nutrient depletion or withdrawal could result in an increased mRNA turnover. We therefore initiated our studies by determining the turnover rates of transcripts encoded by two housekeeping genes in human Hep G2 and HeLa cells cultured under serum-starved or stimulated conditions. β-actin is a major cytoarchitectural component, while GDH catalyzes the oxidative deamination of glutamate in the mitochondrion. Both functions are considered vital, and both proteins are expressed at fairly constant levels. RNA was extracted from these cells grown under serum-stimulation or starvation conditions at various time points following transcription inhibition by act D and the stability of both β-actin and GDH transcripts was assessed by northern analyses (Figure 1). Under serum-stimulated conditions, β-actin mRNA was, as expected, relatively stable with a half-life of >96 h. In serum-deprived cells, however, its half-life was significantly reduced to 40 h (Figure 1A). A similar trend was seen with GDH transcripts. In serum-stimulated Hep G2 cells the mRNA also had a half-life >96 h, whereas in serum-starved cells this was reduced to 27 h (Figure 1B). In order to discern whether the reduction in the amount of mRNA found in serum-deprived cells at the later time points post-act D addition was due to increased cell death over the control cells, the viability of the cells in both conditions was assessed. Cell viability was found to be comparable at all time points post act-D addition, eliminating this possibility (data not shown). Therefore we conclude that serum starvation caused an increase in decay rates of β-actin and GDH mRNAs.
Figure 1.
The stability of endogenous mRNAs is reduced in serum-starved cells. Hep G2 cells were grown in Eagle's MEM supplemented with 10% fetal calf serum (serum-stimulated) or cultured in serum-free media for 15 h (serum starved). Transcription was inhibited by the addition of act D and total RNA was isolated at the times indicated. Northern blot analysis was performed with probes against β-actin mRNA and 18S rRNA (A) or GDH mRNA and 18S rRNA (B). Transcript half-lives were determined from the graphs on the right of the figure. Black diamonds represent serum-stimulated values; open diamonds represent serum-starved samples.
Serum starvation also decreases half-lives of exogenous transcripts
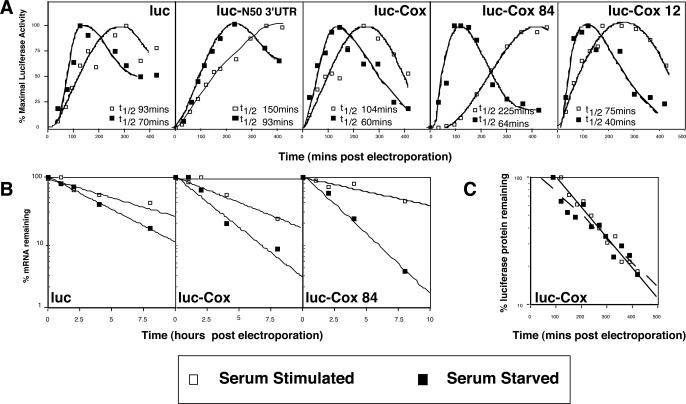
By inhibiting transcription with the intercalator act D, we were able to show that there is a more rapid turnover of certain endogenous transcripts in serum-starved conditions. It has been shown, however, that observations with different transcription inhibitors must be carefully interpreted. Harrold et al. estimated an mRNA half-life using different analytical approaches. They describe how transcription inhibitors act in different ways and gave different values for the decay rate of the same transcript. For example, 5,6-dichloro-1-beta-D-ribobenzimidazole (DRB) blocked RNA polymerase II transcription by >90% but only blocks the accumulation of non-RNA polymerase II RNA species by ∼11% (35). DRB also gave different levels of transcription inhibition in different cell types (35). In contrast, act D intercalates into the DNA inhibiting essentially all transcription. Moreover, act D prevents transcripts from properly attaching to the ribosome and thereby affecting translation as well as transcription. They also observed that the rate of transcript decay was increased in act D treated cells relative to that observed with DRB (35). Thus, although the utility of this approach for estimating mRNA half-lives may be considered debatable, we were using the same inhibitor in the same cell type and felt that this observation was a real and physiological effect. We wished, however, to confirm our observations in Figure 1 by an independent method. By electroporating cells with a reporter mRNA that cannot be generated by the cell, rates of transcript decay can be measured directly, obviating the need for any transcriptional inhibitors. A set of luciferase transcripts that varied only within a 50 nt cassette positioned in the 3′UTR (29), was electroporated into both serum-stimulated and serum-starved cells. Functional half-lives were determined for luciferase transcripts following electroporation in both starved and stimulated cells. The functional half-life is defined as the half-maximal value of luciferase activity (31) and the data are depicted in Figure 2. The trace from the luciferase transcript population in the starved cells was shifted to the left, indicating that it was substantially less stable (95 min) than in stimulated cells (150 min) (Figure 2A, leftmost panel). In order to determine whether this constituted a generic phenomenon or whether it was restricted to a subpopulation, a number of previously derived luciferase clones, which differed in the 3′ UTR, were utilized (29). Four additional luc-variant mRNAs were assayed to determine their stabilities in serum-starved and stimulated Hep G2 (Figure 2A) or HeLa cell lines (data not shown). In all cases, half-lives were reduced when cells were serum-deprived, irrespective of 5′ or 3′UTR composition. In addition, the maximal amount of translation product was also reduced (as would be predicted if less eIF4E was binding the cap).
Figure 2.
The stability of electroporated mRNAs is also significantly decreased in serum-starved cells. (A) Five reporter mRNAs containing the luciferase open reading frame (luc) or luc plus additional sequences in the 3′ UTR derived from a random 50mer (luc–N50–3′UTR), COX 4 (luc–cox), or two variants of the COX 4 3′ UTR (luc–cox84 and luc–cox12) were individually electroporated into serum-starved or serum-stimulated Hep G2 cells. Cells were lysed at the indicated times and luciferase activity was assessed. Relative luciferase units were plotted as a percentage of maximal activity to approximate functional mRNA half-lives. (B) Levels of three electroporated mRNAs (luc, luc–cox and luc–cox84) were directly determined by real time PCR and plotted versus time post electroporation on semilog plots to generate decay curves. (C) The half-life of the luciferase protein under these experimental conditions was assessed by treating cells with cycloheximide 90 min post electroporation and analysing luciferase enzyme activity at the times indicated. In all three panels, black diamonds represent serum-starved values; open squares represent serum-stimulated samples.
To measure transcript stabilities directly, chemical half-lives were derived using real time RT–PCR. Quantification of mRNAs encoding reporter (luciferase) and an endogenous control transcript (cytochrome c oxidase subunit VIIa, COX 7a-L mRNA) were performed from a single cDNA pool generated by random hexamer primed reverse transcription. Luciferase transcript levels, as well as two variants containing Cox4 3′ UTR sequences, were compared to the control at time points post electroporation and expressed as percentage luciferase mRNA that remained. As seen in Figure 2B, the half-lives of these mRNAs were significantly decreased under serum-starvation conditions. The absolute values obtained for chemical versus functional half-lives differed, as reported previously (31). Importantly, the relative hierarchy of stability and ratio of starved to stimulated half-life was similar. The most stable transcript in control conditions, Luc–Cox84 (6.9 ± 0.1 hours), had been identified previously as containing an element within the 3′UTR capable of enhancing stability (29). Despite this, it could not abrogate the loss of stability under serum starvation (1.8 ± 0.01 h). Similarly, Luc transcript stability decreased from 4.7 ± 0.1 to 3.4 ± 0.3 h and Luc–COX from 4.6 ± 0.6 to 2.2 ± 0.2 h.
Finally, to confirm that changes in the mRNA functional half-life were not attributable to differences in degradation rates of reporter protein, cycloheximide was added ∼90 min post electroporation, and luciferase activity monitored over time (Figure 2C). Only a minor difference in protein decay rates was observed (e.g. half-lives of 156 min in serum-starved Hep G2 cells versus 134 min in serum-stimulated cells). Similar data was also obtained in HeLa cells as well (protein half-lives of 200 min in serum-starved cells versus 210 min in serum-stimulated cells). Therefore, we conclude that the half-lives of mRNAs, whether the transcripts are endogenously produced or introduced into the cells, are significantly reduced under serum-starved conditions.
Occupancy of the 5′ cap structure by PARN and eIF4E is differentially affected by serum starvation
To determine whether the decrease in mRNA stability and translation could be due to a modulation in cap occupancy, various luciferase mRNAs (Figure 3A) were used to assess the interactions between PARN and the 5′ cap structure. Individual transcripts, radiolabelled exclusively at the 5′ cap, were incubated with S100 extracts, irradiated with UV light to cross link closely associated proteins, and treated with RNase to effectively transfer short cap-labelled oligomers to the proteins. Reaction mixtures were immunoprecipitated with anti-PARN antibodies and separated by denaturing gel electrophoresis (12). Under serum starvation, luciferase transcripts demonstrated a 3.03-fold increase in occupancy of the cap structure over stimulated cells, consistent with the observed loss of stability of the transcript in this condition (Figure 3B, top panel, lanes 1 and 2). Similarly, Luc–COX transcripts (substituted 3′ UTR sequence) also demonstrated increased (2.17-fold) PARN association (Figure 3B, top panel, lanes 5 and 6). A luciferase transcript with an insertion of a stem loop into the 5′ UTR (Figure 3B, top panel, lanes 3 and 4) also showed enhanced binding (2.63-fold). Since protein–RNA binding reactions were performed in the presence of EDTA, no significant degradation of the input RNA was noted under any conditions (data not shown). Therefore, we conclude that PARN protein in extracts from serum-starved cells is significantly better at interacting with the 5′ cap of an mRNA substrate.
Figure 3.
Serum starvation differentially affects the interaction of PARN and eIF4E with the mRNA 5′ cap. (A) Diagrammatic representation of the luciferase mRNA and its two variants used in this study. SL–Luc had a stem loop structure inserted into its 5′ UTR. Luc–Cox contained the COX4 3′ UTR attached to the luciferase opening reading frame. All constructs were capped and contained a 40 base adenosine 3′ tail. (B) The RNAs outlined in panel A were labelled exclusively at their 5′ cap structure and incubated in cell extracts derived from serum-stimulated (stimulated lanes) or serum-starved cells (starved lanes). Reactions mixtures were irradiated with UV light to covalently cross-link closely associated proteins and treated with RNase. Samples were then immunoprecipitated using α-PARN (top half) or α-eIF4E (bottom half) antisera. Immunoprecipitated proteins that were cross linked to the radioactive cap were separated on polyacrylamide gels containing SDS, visualized by phosphorimaging and analysed using ImageQuant software. (C) Post 600 g supernatants prepared from cells grown under serum-starvation conditions (starved lanes) or in the presence of 10% serum (stimulated lanes) were incubated with m7GTP-Sepharose beads. Beads were washed extensively and bound proteins were electrophoresed on a 10% polyacrylamide gel with SDS. Western blots were performed to assess the levels of bound PARN (top half) and eIF4E (bottom half) using specific antisera.
As there is an increase in PARN/cap interaction in cells deprived of serum, does this increase occur at the expense of other cap-binding proteins? The eIF4E is a cap-binding protein and, as a crucial member of a functional eIF4F complex, is required for cap-dependent protein translation. Perturbations in the cap–eIF4E interactions were, therefore, investigated using essentially the same strategy as for PARN with the same three transcripts. Although the same panel of radiolabelled transcripts was used, these transcripts were incubated with an S10 preparation to allow for binding of the eIF4F complex to the 40S ribosomal subunit. Cap-labelled RNA fragments that were cross-linked to eIF4E were immunoprecipitated with anti-eIF4E antisera, and the isolated complexes separated by denaturing gel electrophoresis. For each of these three transcripts, the interaction was stronger (2.17, 3.33 and 2.27-fold, respectively) for serum-stimulated (Figure 3B, lower panel, lanes 2, 4 and 6), than serum-deprived cells (Figure 3B, lower panel, lanes 1, 3 and 5). Therefore, it appears that one contributing factor to the increase in cap occupancy observed in serum-starved extracts may be a decrease in cap binding activity of the endogenous eIF4E fraction.
In order to further support these observations through an independent assay, cap binding was also assessed by interaction with immobilized cap analogue (m7GTP-Sepharose 4B). Cell lysates were prepared and aliquots (100 μg) were incubated with m7GTP-Sepharose 4B. Interacting proteins were separated by 10% (w/v) SDS–PAGE and analysed by western blotting with antibodies to PARN or eIF4E. As seen in Figure 3C, an increased interaction was observed with PARN in starved compared to stimulated cell extracts (63% ± 4.4 of starved sample), while association with eIF4E was decreased (49.5% ± 3.2 of stimulated sample). It must be stressed that each of the samples loaded on to the gel represented in Figure 3C lane 1 or 2 constitute the entire complement of protein capable of binding to the 7MeGTP beads. The signals derived from the subsequent western blot that was probed specifically for PARN and eIF4e strongly reinforces that these interactions appear reciprocal. These data, therefore, suggest that there is an decreased interaction of eIF4E with 5′ cap structures of transcripts in extracts from serum-starved cells, giving greater accessibility to PARN. The reduction in eIF4E interaction may also, in part, be responsible for the diminished levels of translation under these conditions.
Steady-state levels of proteins that interact at transcript termini do not differ between serum-starved and stimulated conditions, but differences in post-translational modifications are evident
The changes in relative cap occupancy between PARN and eIF4E noted above could be due to changes either in overall protein levels or in the overall activity of the available proteins. To determine whether the increase in PARN-cap occupancy in serum-starved cells was due to an increase in overall levels of this protein, western analyses were performed. As seen in Figure 4, the steady-state level of PARN was unchanged between serum-starved and stimulated cells in both S10 and S100 extracts. No difference in the level of the cap-binding protein eIF4E, the least abundant of the initiation factors (36), could be detected. The amounts of translationally related proteins that compete for a binding site on eIF4E, such as eIF4G or 4E-BP1 (17,37), were also assessed. Western analyses shown in Figure 4 revealed no detectable differences in the levels of these proteins between the serum-starved and stimulated cells. Finally, as a decrease in PABP coating the poly(A) tails could increase the accessibility for PARN and potentially enhance mRNA degradation, steady-state levels of PABP were compared in serum-starved versus stimulated cells. A slight decrease in serum-starved cells was noted (68% ± 8 n = 7). This observation is consistent with the repressed translation of PABP seen under conditions of growth arrest in mouse fibroblasts (38).
Figure 4.
Serum starvation does not significantly alter the levels of PARN or translation initiation factors associated directly or indirectly with the cap. Hep G2 cells were grown in Eagle's MEM supplemented with 10% fetal calf serum (stimulated lanes) or cultured in serum-free media for 15 h (starved lanes). Total protein from S10 or S100 fractions was prepared and electrophoresed on polyacrylamide gels containing SDS. Western blotting was performed with the indicated antisera to detect PARN, eIF4E, eIF4G, PABP and 4E-BP1. Proteins were visualized with ECL reagents.
Serum-stimulation has been shown to induce phosphorylation of eIF4G, 4E-BP1 and eIF4E (39–41). To confirm this in our system, the levels of phosphorylation of various proteins were assessed. Samples of total protein were separated by isoelectric focussing and eIF-4E levels were assessed by western blotting. As seen in Figure 5A, eIF4E from serum-starved cells showed a decrease in overall phosphorylation compared to serum-stimulated cells. Next, cell lysates were separated on an SDS–PAGE gel and western blots were probed with anti-4E-BP1 antisera that recognized pSer65 of 4E-BP1. As seen in Figure 5B, the proportion of phosphorylated 4E-BP1 was decreased in serum-starved cells. Similar results were obtained using 5 or 10 μg of the extract, confirming the quantitative nature of our assays. We conclude that the phosphorylation of eIF4E and 4E-BP1 is decreased under serum-starvation conditions. Based on the known properties of the dephosphorylated isoforms of these proteins (1), this is likely to be responsible for the decrease in cap occupancy that was observed for eIF4E in extracts prepared from serum-deprived cells.
Figure 5.
Phosphorylation of eIF4E and 4E-BP1 is impaired in serum-starved cells. Hep G2 cells were grown in Eagle's MEM supplemented with 10% fetal calf serum (stimulated lanes) or cultured in serum-free media for 15 h (starved lanes). (A) Cap binding proteins were purified on m7GTP-Sepharose 4B beads and electrophoresed through a 3.5–10 pH gradient isoelectric focussing gel. Western blotting was performed using α-eIF4E antisera. (B) Proteins from S10 or S100 cellular fractions prepared from Hep G2 cells grown under the indicated conditions were electrophoresed on a 15% polyacrylamide gel containing SDS. Phosphorylated 4E-BP1 binding protein was specifically detected by western blotting with antisera to the ser65 phosphorylated isoform.
PARN is a phosphoprotein with different phosphorylation profiles under serum-starved and stimulated conditions
As the increased cap occupancy by PARN in serum-starved cells could not be explained by increased steady-state levels, we evaluated differences in post-translational modifications. Using the NetPhos 2.0 Prediction programme (http://www.cbs.dtu.dk/services/NetPhos/), multiple serine, threonine and tyrosine residues within PARN were predicted to be potential sites for phosphorylation. To determine whether PARN was indeed phosphorylated in vivo, label transfer experiments were performed. Cells were cultured in the presence of radiolabelled orthophosphate, lysed, immunoprecipitated with α-PARN antisera and complexes were analysed by SDS–PAGE. Phosphorylation of PARN was clearly evident (Figure 6A), indicating that it is indeed a phosphoprotein. To establish a physiological relevance to the phosphorylation of PARN, preparations of starved and stimulated cells were subjected to separation using the Qiagen PhosphoProtein Purification System. Equal proportions of phosphoproteins and non-phosphorylated fractions were separated by SDS–PAGE. Data resulting from western analysis not only suggest that PARN is indeed a phosphoprotein, but also indicate that the ratio of non- to phosphorylated protein is modulated by serum. Starved cells demonstrate a greater proportion of phosphorylated PARN, in contrast to stimulated conditions (Figure 6B). To confirm the quality of the separation of phosphorylated isoforms, immunodetection was performed with a number of antisera specific for phosphoproteins, as well as proteins known not to contain phosphate (Figure 6B). Antibodies that recognize phosphoserines at position 65 of 4E-BP1 and 235/236 of the S6 ribosomal protein only detected protein in the phosphorylated fractions as expected (Figure 6B). In contrast, GDH, which is not known to be phosphorylated, was found exclusively in lanes corresponding to non-phosphorylated fractions (Figure 6B). Thus, it appears that PARN is a phosphoprotein and that this modification appears to be modulated by physiological growth conditions.
Figure 6.
PARN is a phosphoprotein. (A). Hep G2 cells were cultured in the presence of 32P orthophosphate for 60 min. Lysates were prepared and immunoprecipitated using α-PARN sera (α-PARN lane) or protein G alone (control lane). Immunoprecipitated proteins were analysed on a 10% polyacrylamide gel containing SDS and detected by phosphorimaging. (B) Total cell lysates from Hep G2 cells grown under serum-stimulated (lanes 1 and 2) or starved (lanes 3 and 4) conditions were separated into phosphorylated (lanes 1 and 3) and non-phosphorylated (lanes 2 and 4) fractions using the Qiagen phosphoprotein purification system. Fractions were electrophoresed on polyacrylamide gels containing SDS and the levels of the indicated proteins [PARN, known phosphoproteins (4E-BP1 and S6 ribosomal protein) and unphosphorylated GDH] were detected by Western blotting. The ratio of phosphorylated PARN appears to be modulated by serum status.
DISCUSSION
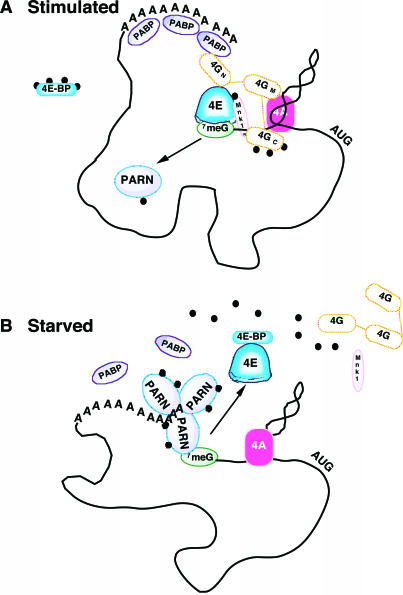
We have shown that serum deprivation of cultured cells causes a more rapid turnover of various endogenous and electroporated transcripts. In these serum-starved culture conditions, transcript populations that were more rapidly degraded also had more PARN bound to the 5′ cap. An increase in PARN cap-occupancy has been shown previously to reflect increased deadenylation activity (12). Conversely, cap-occupancy by eIF4E was reduced, consistent with the observed reduction in translation of these reporter mRNA species (despite equivalent electroporation efficiency), suggesting the loss of a functional eIF4F. Our data also demonstrate that PARN is a phosphoprotein, with this modification being modulated by serum conditions. PARN phosphorylation was increased in serum-deprived cells, whereas the phosphorylation of both eIF4E and its cognate binding protein 4E-BP1 was diminished under the same conditions. Taken together, these data are consistent with the equilibrium between mRNA deadenylation and translation, being modulated by a competition for the 5′ terminal cap between PARN and eIF4E, a competition that may be regulated by phosphorylation of these factors. A model outlining these observations is presented in Figure 7.
Figure 7.
Schematic representation of changes in RNA–protein interactions and protein–protein assemblies on transcripts as a consequence of serum starvation or stimulation. This simplified diagram represents some of the known interactions at the 5′-terminus during cap-dependent translation of mRNA under serum-stimulated culture conditions. (A) When cells are stimulated by serum, there is a greater proportion of mRNA with eIF4E at the 5′ cap, resulting in a functional eIF4F complex that interacts with PABP at the 3′ terminus. This interaction protects both the 5′ cap and poly(A) tail from interaction with PARN. Serum stimulation causes phosphorylation (indicated by black dots) of eIF4E, its cognate binding proteins and eIF4G. The latter acts as a binding site for Mnk1, the kinase responsible for phosphorylation of eIF4E. This reflects our data in stimulated cells that show increased eIF4E-cap binding and higher levels of cap-dependent translation. (B) In contrast, our data show that cells deprived of serum favour interactions with PARN at the 5′ cap and this protein may be hyperphosphorylated, increasing the association with mRNA. This interaction appears to be at the expense of eIF4E binding and is consistent with the diminished cap-dependent translation in serum-deprived cells described here. A model of PARN cap-binding kinetics has been proposed by Virtanen et al. in which a single PARN molecule binds the cap, while a trimer generates the poly(A)-binding site and an active deadenylation complex (10). The competition between eIF4E and PARN may be regulated by further physiological changes and could represent a key mechanism in the balance between translation and degradation of transcripts.
Although these data are internally consistent, it must be considered that there are other proteins that can bind to the 5′ cap, in addition to PARN and eIF4E. The binding of YB-1 protein, for example, has been reported to cause the stabilization of mRNA (42) and may compete with eIF4E. However, YB-1, which can be localized to the nucleus or cytoplasm (43), has sequence selectivity for certain mRNAs (44) and the reduced stability of mRNA in serum-deprived cells suggests that YB-1 is not bound at high levels to the 5′ cap in this condition.
To further refute the possibility that we were observing an artefact, the stability of endogenous transcripts was also measured and was shown to decrease in response to serum-deprivation. β-actin and GDH transcripts had shorter half-lives in serum-starved Hep G2 cells. This is consistent with earlier work by Levy and Hug who noted that β-actin transcripts were less stable in quiescent- compared to proliferating Hep G2 and MCF-7 cells (20) and in a rat liver cell line deprived of amino acids (21), under conditions wherein other mRNA species were also shown to be more rapidly degraded. There are many other reports of serum or amino acid deprivation causing increased rates of decay for numerous endogenous transcripts in immortalized or primary cell lines (22–26). These experiments, however, did not address the mechanisms of increased mRNA decay. Noe et al. assessed transcript stability using a tetracycline responsive dihydrofolate reductase (DHFR) minigene introduced into CHO cells (45). Not only was the half-life of the DHFR mRNA decreased under serum starvation but this was accompanied by poly(A) tail shortening. Taken together with our data presented here, we propose that the decreased half-life observed in the DHFR minigene (and a subset of endogenous transcripts) under conditions of nutrient deprivation is a consequence of increased interaction of PARN, thereby enhancing deadenylation-dependent decay.
Gallie and Traugh previously postulated that cap-associated factors may be serum sensitive and that serum may affect the function of the cap and the activity of factors associated with it (46). The authors electroporated transcripts with and without a 5′ cap and/or poly(A) to investigate the synergism of cap and poly(A) tail on mRNA stability in response to insulin. Prior to insulin stimulation, cells were serum-deprived for 90 min and a small but consistent decrease in mRNA stability along with a decreased rate of translation, was observed. The effects in starved cells were more subtle than those described here, but this may relate to the duration of serum deprivation. They concluded that the accelerated mRNA decay was due to a modification of eIF4F in light of the rapid change in phosphorylation of eIF4E on serum addition. At the time of their work, PARN had not yet been identified. However, their observations and conclusions support the data presented here. Serum status does affect the complement of proteins bound to the 5′ cap such that deprivation increases the proportion of PARN bound and decreases association of eIF4E, leading to a reduction of cap-dependent translation. In contrast, serum stimulation favours cap interaction with eIF4E over PARN and enhances cap-dependent translation.
Our experiments show that more PARN is bound to the 5′ cap on serum withdrawal, yet there is no obvious increase in the steady-state level of the protein. Körner and et al. (14) have demonstrated that PARN is distributed between the nuclear and cytoplasmic compartments. This partitioning plays a crucial role in Xenopus oocytes, such that PARN is retained in the nucleus and is not released into the cytoplasm until meiosis. Thus, any alterations in this nuclear–cytoplasmic partitioning could provide a potential mechanism by which changes in PARN deadenylation are regulated. We compared PARN in the nuclear and cytoplasm under both growth conditions but found no difference in distribution (data not shown).
Although there were no changes in steady-state amounts or cellular distribution of either PARN or eIF4E, we were able to show that the phosphorylation state of both factors is modulated by serum status. Previous research has also shown that hypophosphorylated 4E-BP1, which is prevalent in serum-deprived cells, has an increased affinity for eIF4E, resulting in a decreased interaction of eIF4E with eIF4G, a concomitant decrease in eIF4F complex assembly and cap-dependent translation (47). Moreover, the affinity of eIF4E for the 5′ cap is reported to be increased as a result of the interaction between the eIF4F complex and PABP (48,49), an interaction that also increases the affinity of PABP for the poly(A) tail. This interaction decreases in serum-deprived cells (50), exposing both the 5′ cap and the poly(A) tail to PARN. Similarly, PGK1 mRNA stability is decreased in yeast expressing mutant forms of eIF4E and eIF4G1 (51), consistent with the release of these factors exposing transcripts to enhanced degradation. Binding of 4E-BP1 to eIF4E has also been shown to block phosphorylation of eIF4E (52). Although these results are consistent with our observations of phosphorylation and decreased translation under serum starvation, there are conflicting data as to whether phosphorylation of eIF4E modulates cap-binding (53–55) or control of translation (56,57). Decreased cap interaction by eIF4E for whatever reason, however, would allow increased access for PARN.
In conclusion, it is clear that various mechanisms operate post-transcriptionally to regulate both the steady-state levels of mRNA and their translation. Availability of mRNA template will dictate the levels of translation, as will events such as phosphorylation, which will affect the assembly and function of eIF4F. A wealth of previously published observations, some of which have been referenced here, describe decreases in either transcript levels and/or translation under nutrient deprivation and, conversely, enhancement when stimulated with nutrients, serum or hormones. Taken together with our data presented here, we suggest that a major post-transcriptional mechanism that controls the expression of at least a subset of transcripts is dependent upon the competition of PARN and eIF4E for the 5′ cap, as presented in Figure 7. Under conditions of nutrient deprivation, the cap interactions of eIF4E decrease in favour of PARN, potentially driven by the changes in phosphorylation states of these proteins themselves or their cognate binding proteins, shifting the balance away from cap-mediated translation, and critically increasing the deadenylation-dependent decay of transcripts.
Acknowledgments
Z.C-L wishes to thank Elmar Wahle (Universitaet Halle) and Michael Wormington (University of Virginia, USA) for the generous gift of PARN antisera, Simon Morley (Department of Biochemistry, University of Sussex) for kindly providing guanylyl transferase and PABP antisera. Z.C-L. and R.L. thank The Wellcome Trust for continued support and funding, the University of Newcastle upon Tyne for funding a Luccock Scholarship and the Newcastle Hospitals Special Trustees. J.W. was supported by a grant from the National Institutes of Health (GM072481). Funding to pay the Open Access publication charges for this article was provided by The Luccock Scholarship.
REFERENCES
- 1.Guhaniyogi J., Brewer G. Regulation of mRNA stability in mammalian cells. Gene. 2001;265:11–23. doi: 10.1016/s0378-1119(01)00350-x. [DOI] [PubMed] [Google Scholar]
- 2.Khodursky A.B., Bernstein J.A. Life after transcription—revisiting the fate of messenger mRNA. Trends Genet. 2003;19:113–115. doi: 10.1016/S0168-9525(02)00047-1. [DOI] [PubMed] [Google Scholar]
- 3.Gingras A.-C., Raught B., Sonenberg N. eIF4 initiation factors: effectors of mRNA recruitment to ribosomes and regulators of translation. Annu. Rev. Biochem. 1999;68:913–963. doi: 10.1146/annurev.biochem.68.1.913. [DOI] [PubMed] [Google Scholar]
- 4.Jackson R.J., Wickens M. Translational controls impinging on the 5′-untranslated region and initiation factor proteins. Curr. Opin. Genet. Dev. 1997;7:233–241. doi: 10.1016/s0959-437x(97)80133-5. [DOI] [PubMed] [Google Scholar]
- 5.Mitchell P., Tollervey D. mRNA stability in eukaryotes. Curr. Opin. Genet. Dev. 2000;10:193–198. doi: 10.1016/s0959-437x(00)00063-0. [DOI] [PubMed] [Google Scholar]
- 6.Stover P.J. New paradigms for nutrient control of genome translation. Nutr. Rev. 2003;61:427–431. doi: 10.1301/nr.2003.dec.427-431. [DOI] [PubMed] [Google Scholar]
- 7.Beelman C.A., Parker R. Degradation of mRNA in eukaryotes. Cell. 1995;81:179–183. doi: 10.1016/0092-8674(95)90326-7. [DOI] [PubMed] [Google Scholar]
- 8.Couttet P., Fromont-Racine M., Steel D., Pictet R., Grange T. Messenger RNA deadenylylation precedes decapping in mammalian cells. Proc. Natl Acad. Sci. USA. 1997;94:5628–5633. doi: 10.1073/pnas.94.11.5628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wilusz C.J., Gao M., Jones C.L., Wilusz J., Peltz S.W. Poly(A)-binding proteins regulate both yeast deadenylation and decapping in yeast cytoplasmic extracts. RNA. 2001;7:1416–1424. [PMC free article] [PubMed] [Google Scholar]
- 10.Korner C.G., Wahle E. Poly(A) tail shortening by a mammalian poly(A)-specific 3′-exoribonuclease. J. Biol. Chem. 1997;272:10448–10456. doi: 10.1074/jbc.272.16.10448. [DOI] [PubMed] [Google Scholar]
- 11.Dehlin E., Wormington M., Korner C.G., Wahle E. Cap-dependent deadenylation of mRNA. EMBO J. 2000;19:1079–1086. doi: 10.1093/emboj/19.5.1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gao M., Fritz D.T., Ford L.P., Wilusz J. Interaction between a poly(A)-specific ribonuclease and the 5′ cap influences mRNA deadenylation rates in vitro. Mol. Cell. 2000;5:479–488. doi: 10.1016/s1097-2765(00)80442-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Martinez J., Ren Y., Nilsson P., Ehrenberg M., Virtanen A. The mRNA cap structure stimulates rate of poly(A) removal and amplifies processivity of degradation. J. Biol. Chem. 2001;276:27923–27929. doi: 10.1074/jbc.M102270200. [DOI] [PubMed] [Google Scholar]
- 14.Korner C., Wormington M., Muckenthaler M., Schneider C., Dehlin E., Wahle E. The deadenylating nuclease (DAN) is involved in poly(A) tail removal during meiotic maturation of Xenopus oocytes. EMBO J. 1998;17:5427–5437. doi: 10.1093/emboj/17.18.5427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mian I.S. Comparative sequence analysis of ribonucleases HII, III, II PH and D. Nucleic Acids Res. 1997;25:3187–3195. doi: 10.1093/nar/25.16.3187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chiba Y., Johnson M.A., Lidder P., Vogel J.T., van Erp H., Green P.J. AtPARN is an essential poly(A) ribonuclease in Arabidopsis. Gene. 2004;328:95–102. doi: 10.1016/j.gene.2003.11.028. [DOI] [PubMed] [Google Scholar]
- 17.Haghighat A., Sonenberg N. eIF4G dramatically enhances the binding of eIF4E to the mRNA 5′-cap structure. J. Biol. Chem. 1997;272:21677–21680. doi: 10.1074/jbc.272.35.21677. [DOI] [PubMed] [Google Scholar]
- 18.Gross J.D., Moerke N.J., von der Haar T., Lugovskoy A.A., Sachs A.B., McCarthy J.E., Wagner G. Ribosome loading onto the mRNA cap is driven by conformational coupling between eIF4G and eIF4E. Cell. 2003;115:739–750. doi: 10.1016/s0092-8674(03)00975-9. [DOI] [PubMed] [Google Scholar]
- 19.Wells S.E., Hillner P.E., Vale R.D., Sachs A.B. Circularization of mRNA by eukaryotic translation initiation factors. Mol. Cell. 1998;2:135–140. doi: 10.1016/s1097-2765(00)80122-7. [DOI] [PubMed] [Google Scholar]
- 20.Levy J.R., Hug V. Regulation of insulin receptor gene expression. Cell cycle-mediated effects on insulin receptor mRNA stability. J. Biol. Chem. 1992;267:25289–25295. [PubMed] [Google Scholar]
- 21.Shay N.F., Nick H.S., Kilberg M.S. Molecular cloning of an amino acid-regulated mRNA (amino acid starvation-induced) in rat hepatoma cells. J. Biol. Chem. 1990;265:17844–17848. [PubMed] [Google Scholar]
- 22.Thissen J.P., Pucilowska J.B., Underwood L.E. Differential regulation of insulin-like growth factor I (IGF-I) and IGF binding protein-1 messenger ribonucleic acids by amino acid availability and growth hormone in rat hepatocyte primary culture. Endocrinology. 1994;134:1570–1576. doi: 10.1210/endo.134.3.7509741. [DOI] [PubMed] [Google Scholar]
- 23.Krupsky M., Kuang P.-P., Goldstein R.H. Regulation of type I collagen mRNA by amino acid deprivation in human lung fibroblasts. J. Biol. Chem. 1997;272:13864–13868. doi: 10.1074/jbc.272.21.13864. [DOI] [PubMed] [Google Scholar]
- 24.Wang H., Caldwell M., Lin S., Furneaux H., Gorospe M. HuR regulates cyclin A and cyclin B1 mRNA stability during cell proliferation. EMBO J. 2000;19:2340–2350. doi: 10.1093/emboj/19.10.2340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Malcolm T., Ettehadieh E., Sadowski I. Mitogen-responsive expression of RhoB is regulated by RNA stability. Oncogene. 2003;22:6142–6150. doi: 10.1038/sj.onc.1206638. [DOI] [PubMed] [Google Scholar]
- 26.Gallouzi I.E., Parker F., Chebli K., Maurier F., Labourier E., Barlat I., Capony J.P., Tocque B., Tazi J. A novel phosphorylation-dependent RNase activity of GAP-SH3 binding protein: a potential link between signal transduction and RNA stability. Mol. Cell. Biol. 1998;18:3956–3965. doi: 10.1128/mcb.18.7.3956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chrzanowska-Lightowlers Z.M.A., Preiss T., Lightowlers R.N. Inhibition of mitochondrial protein synthesis promotes increased stability of nuclear-encoded respiratory gene transcripts. J. Biol. Chem. 1994;269:27322–27328. [PubMed] [Google Scholar]
- 28.Zeviani M., Nakagawa M., Herbert J., Lomax M.I., Grossman L.I., Sherbany A.A., Miranda A.F., DiMauro S., Schon E.A. Isolation of a cDNA clone encoding subunit IV of human cytochrome c oxidase. Gene. 1987;55:205–217. doi: 10.1016/0378-1119(87)90281-2. [DOI] [PubMed] [Google Scholar]
- 29.Chrzanowska-Lightowlers Z.M.A., Lightowlers R.N. Fending off decay: a combinatorial approach in intact cells for identifying mRNA stability elements. RNA. 2001;7:435–444. doi: 10.1017/s1355838201001339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Veyrune J.-L., Carillo S., Vie A., Blanchard J.-M. c-fos mRNA instability determinants present within both the coding and the 3′ non coding region link the degradation of this mRNA to its translation. Oncogene. 1995;11:2127–2134. [PubMed] [Google Scholar]
- 31.Gallie D.R. The cap and poly(A) tail function synergistically to regulate mRNA translational efficiency. Genes Dev. 1991;5:2108–2116. doi: 10.1101/gad.5.11.2108. [DOI] [PubMed] [Google Scholar]
- 32.Ford L.P., Wilusz J. An in vitro system using HeLa cytoplasmic extracts that reproduces regulated mRNA stability. Methods Enzymol. 1999;17:21–27. doi: 10.1006/meth.1998.0703. [DOI] [PubMed] [Google Scholar]
- 33.Martin M.E., Munoz F.M., Salinas M., Fando J.L. Ischaemia induces changes in the association of the binding protein 4E-BP1 and eukaryotic initiation factor (eIF) 4G to eIF4E in differentiated PC12 cells. Biochem. J. 2000;351:327–334. [PMC free article] [PubMed] [Google Scholar]
- 34.Scheper G.C., Morrice N.A., Kleijn M., Proud C.G. The mitogen-activated protein kinase signal-integrating kinase Mnk2 is a eukaryotic initiation factor 4E kinase with high levels of basal activity in mammalian cells. Mol. Cell. Biol. 2001;21:743–754. doi: 10.1128/MCB.21.3.743-754.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Harrold S., Genovese C., Kobrin B., Morrison S.L., Milcarek C. A comparison of apparent mRNA half-life using kinetic labeling techniques vs decay following administration of transcriptional inhibitors. Anal. Biochem. 1991;198:19–29. doi: 10.1016/0003-2697(91)90500-s. [DOI] [PubMed] [Google Scholar]
- 36.Duncan R., Milburn S.C., Hershey J.W.B. Regulated phosphorylation and low abundance of HeLa cell initiation factor 4F suggest a role in translational control. J. Biol. Chem. 1987;262:380–388. [PubMed] [Google Scholar]
- 37.Mader S., Lee H., Pause A., Sonenberg N. The translation initiation factor eIF-4E binds to a common motif shared by the translation factor eIF-4g and the translational repressors 4E-binding proteins. Mol. Cell. Biol. 1995;15:4990–4997. doi: 10.1128/mcb.15.9.4990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hornstein E., Git A., Braunstein I., Avni D., Meyuhas O. The expression of poly(A)-binding protein gene is translationally regulated in a growth-dependent fashion through a 5′-terminal oligopyrimidine tract motif. J. Biol. Chem. 1999;274:1708–1714. doi: 10.1074/jbc.274.3.1708. [DOI] [PubMed] [Google Scholar]
- 39.Flynn A., Proud C.G. Ser209, but not Ser53, is the major site of phosphorylation in initiation factor eIF4E in serum treated chinese hamster ovary cells. J. Biol. Chem. 1995;270:21684–21688. doi: 10.1074/jbc.270.37.21684. [DOI] [PubMed] [Google Scholar]
- 40.Gingras A.-C., Raught B., Gygi S.P., Niedzwiecka A., Miron M., Burley S.K., Polakiewicz R.D., Wyslouch-Cieszynska A., Aebersold R., Sonenberg N. Hierarchical phosphorylation of the translation inhibitor 4E-BP1. Genes Dev. 2001;15:2852–2864. doi: 10.1101/gad.912401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Raught B., Gingras A.C., Gygi S.P., Imataka H., Morino S., Gradi A., Aebersold R., Sonenberg N. Serum-stimulated, rapamycin-sensitive phosphorylation sites in the eukaryotic translation initiation factor 4GI. EMBO J. 2000;19:434–444. doi: 10.1093/emboj/19.3.434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Evdokimova V., Ruzanov P., Imataka H., Raught B., Svitkin Y.V., Ovkinov L., Sonenberg N. The major mRNA-associated protein YB-1 is a potent 5′ cap-dependent mRNA stabiliser. EMBO J. 2001;20:5491–5502. doi: 10.1093/emboj/20.19.5491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jurchott K., Bergmann S., Stein U., Walther W., Janz M., Manni I., Piaggio G., Fietze E., Dietel M., Royer H.D. YB-1 as a cell cycle-regulated transcription factor facilitating cyclin A and cyclin B1 gene expression. J. Biol. Chem. 2003;278:27988–27996. doi: 10.1074/jbc.M212966200. [DOI] [PubMed] [Google Scholar]
- 44.Kohno K., Izumi H., Uchiumi T., Ashizuka M., Kuwano M. The pleiotropic functions of the Y-box-binding protein, YB-1. BioEssays. 2003;25:691–698. doi: 10.1002/bies.10300. [DOI] [PubMed] [Google Scholar]
- 45.Noe V., Ciudad C.J., Chasin L.A. Effect of differential polyadenylation and cell growth phase on dihydrofolate reductase mRNA stability. J. Biol. Chem. 1999;274:27807–27814. doi: 10.1074/jbc.274.39.27807. [DOI] [PubMed] [Google Scholar]
- 46.Gallie D.R., Traugh J.A. Serum and insulin regulate cap function in 3T3-L1 cells. J. Biol. Chem. 1994;269:7174–7179. [PubMed] [Google Scholar]
- 47.Pyronnet S., Dostie J., Sonenberg N. Suppression of cap-dependent translation in mitosis. Genes Dev. 2001;15:2083–2093. doi: 10.1101/gad.889201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Borman A.M., Michel Y.M., Kean K.M. Biochemical characterisation of cap-poly(A) synergy in rabbit reticulocyte lysates: the eIF4G-PABP interaction increases the functional affinity of eIF4E for the capped mRNA 5′-end. Nucleic Acids Res. 2000;28:4068–4075. doi: 10.1093/nar/28.21.4068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.von Der Haar T., Ball P.D., McCarthy J.E. Stabilization of eukaryotic initiation factor 4E binding to the mRNA 5′-Cap by domains of eIF4G. J. Biol. Chem. 2000;275:30551–30555. doi: 10.1074/jbc.M004565200. [DOI] [PubMed] [Google Scholar]
- 50.Fraser C.S., Pain V.M., Morley S.J. The association of initiation factor 4F with poly(A)-binding protein is enhanced in serum-stimulated Xenopus kidney cells. J. Biol. Chem. 1999;274:196–204. doi: 10.1074/jbc.274.1.196. [DOI] [PubMed] [Google Scholar]
- 51.Schwartz D.C., Parker R. Mutations in translation initiation factors lead to increased rates of deadenylation and decapping of mRNas in Saccharomyces cerevisiae. Mol. Cell. Biol. 1999;19:5247–5256. doi: 10.1128/mcb.19.8.5247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wang X., Flynn A., Waskiewicz A.J., Webb B.L., Vries R.G., Baines I.A., Cooper J.A., Proud C.G. The phosphorylation of eukaryotic initiation factor eIF4E in response to phorbol esters, cell stresses, and cytokines is mediated by distinct MAP kinase pathways. J. Biol. Chem. 1998;273:9373–9377. doi: 10.1074/jbc.273.16.9373. [DOI] [PubMed] [Google Scholar]
- 53.Minich W.B., Balasta M.L., Goss D.J., Rhoads R.E. Chromatographic resolution of in vivo phosphorylated and nonphosphorylated eukaryotic translation initiation factor eIF-4E: increased cap affinity of the phosphorylated form. Proc. Natl Acad. Sci. USA. 1994;91:7668–7672. doi: 10.1073/pnas.91.16.7668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Scheper G.C., van Kollenburg B., Hu J., Luo Y., Goss D.J., Proud C.G. Phosphorylation of eukaryotic initiation factor 4E markedly reduces its affinity for capped mRNA. J. Biol. Chem. 2002;277:3303–3309. doi: 10.1074/jbc.M103607200. [DOI] [PubMed] [Google Scholar]
- 55.Zuberek J., Wyslough-Cieszynska A., A N., Dadlez M., Stepinski J., Augustyniak W., Gingras A.-C., Zhang Z., Burley S.K., Sonenberg N., et al. Phosphorylation of eIF4E attenuates its interaction with mRNA 5′ cap analogues by electrostatic repulsion: Intein-mediated protein ligation strategy to obtain phosphorylated protein. RNA. 2003;9:52–61. doi: 10.1261/rna.2133403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kleijn M., O V.H., Thomas A.A. Phosphorylation of eIF-4E and initiation of protein synthesis in P19 embryonal carcinoma cells. J. Cell. Biochem. 1995;59:443–452. doi: 10.1002/jcb.240590405. [DOI] [PubMed] [Google Scholar]
- 57.McKendrick L., Morley S.J., Pain V.M., Jagus R., Joshi B. Phosphorylation of eukaryotic initiation factor 4E (eIF4E) at Ser209 is not required for protein synthesis in vitro and in vivo. Eur. J. Biochem. 2001;268:5375–5385. doi: 10.1046/j.0014-2956.2001.02478.x. [DOI] [PubMed] [Google Scholar]