Abstract
Tyrosyl-DNA phosphodiesterase (TDP1) is a DNA repair enzyme that removes peptide fragments linked through tyrosine to the 3′ end of DNA, and can also remove 3′-phosphoglycolates (PGs) formed by free radical-mediated DNA cleavage. To assess whether TDP1 is primarily responsible for PG removal during in vitro end joining of DNA double-strand breaks (DSBs), whole-cell extracts were prepared from lymphoblastoid cells derived either from spinocerebellar ataxia with axonal neuropathy (SCAN1) patients, who have an inactivating mutation in the active site of TDP1, or from closely matched normal controls. Whereas extracts from normal cells catalyzed conversion of 3′-PG termini, both on single-strand oligomers and on 3′ overhangs of DSBs, to 3′-phosphate termini, extracts of SCAN1 cells did not process either substrate. Addition of recombinant TDP1 to SCAN1 extracts restored 3′-PG removal, allowing subsequent gap filling on the aligned DSB ends. Two of three SCAN1 lines examined were slightly more radiosensitive than normal cells, but only for fractionated radiation in plateau phase. The results suggest that the TDP1 mutation in SCAN1 abolishes the 3′-PG processing activity of the enzyme, and that there are no other enzymes in cell extracts capable of processing protruding 3′-PG termini. However, the lack of severe radiosensitivity suggests that there must be alternative, TDP1-independent pathways for repair of 3′-PG DSBs.
INTRODUCTION
When transient DNA strand breaks formed by DNA topoisomerase I fail to religate, the topoisomerase becomes irreversibly attached to the 3′ DNA terminus via a tyrosyl linkage (1,2). This linkage must then be cleaved by tyrosyl-DNA phosphodiesterase (TDP1) in order to allow repair of the break (3,4). DNA double-strand breaks (DSBs) induced by radiation and radiomimetic drugs typically bear 3′-phosphoglycolate (PG) and 3′-phosphate termini that must likewise be removed prior to any gap filling by DNA polymerase and rejoining of the break by DNA ligase (5–7). The human apurinic/apyrimidinic endonuclease Ape1 is capable of removing PGs from blunt and recessed 3′ ends, albeit inefficiently (8), and polynucleotide kinase/phosphatase (PNKP) can remove 3′-phosphates from blunt, recessed and protruding 3′ ends (9,10). However, PGs on protruding 3′ termini are refractory to both these enzymes, and TDP1 is the only enzyme known to be capable of processing such lesions, converting them to 3′-phosphates susceptible to removal by PNKP (8,11). Although previous data suggested that most of the processing of protruding 3′-PG termini in human cell extracts was indeed attributable to TDP1 (11), the possibility of alternative PG-processing enzymes or pathways was not excluded.
The rare hereditary disorder spinocerebellar ataxia with axonal neuropathy (SCAN1) has been linked to a homozygous mutation in the active site of TDP1 (12). SCAN1 patients exhibit adolescent-onset ataxia and peripheral neuropathy, accompanied by cerebellar atrophy as detected by magnetic resonance imaging (12). SCAN1 thus joins ataxia telangiectasia, xeroderma pigmentosum and Cockayne syndrome as DNA repair disorders with prominent neurological pathology (13–15).
In order to assess the biological significance of 3′-PG processing by TDP1, radiosensitivity of TDP1-mutant SCAN1 cells was determined, and processing of 3′-PG termini was examined in both whole-cell and nuclear extracts of these cells. While biochemical assays indicated a severe deficit in 3′-PG processing in SCAN1 cells, the cells showed only slight radiosensitivity.
MATERIALS AND METHODS
Materials
Cell lines from SCAN1 patients and from unaffected members of the same family were generated by transfection of peripheral lymphocytes with Epstein–Barr virus (12). The cells were maintained in suspension in upright T-75 flasks at a density of 105–106/ml in RPMI medium (Gibco) supplemented with 10% fetal bovine serum. Whole-cell and nuclear extracts were prepared from ∼5 × 108 cells, as described previously (16–18). A 3′-PG 14mer was prepared by bleomycin cleavage of a 5′-end-labeled 17mer, and an internally labeled 3′-PG-terminated plasmid was generated by ligating this 14mer and an unlabeled 3′-PG 13mer into the two 3′-resected ends of plasmid pSV56, as described previously (19). In order to overexpress TDP1 in human cells, the 1.8 kb BamHI/Bsu36I fragment (with the Bsu36I site converted to a blunt end by fill-in with T4 DNA polymerase) was isolated from pHN1894S (4) (generous gift from Howard Nash and Jeff Pouliot, NIMH) and cloned into the BglII and EcoRV sites of the mammalian expression vector pFLAG-CMV-2 (Sigma–Aldrich). This vector (pFLAG-TDP1), containing the complete TDP1 coding sequence, was transfected into human 293 cells using Superfect (Qiagen), and FLAG-TDP1 was purified on FLAG affinity beads, as described below.
Radiosensitivity
The cells were grown to plateau phase (∼2 × 106/ml) in 24-well plates and irradiated with 0.4–1.6 Gy 137Cs γ-rays (or mock irradiated) each day for 5 days. The cells were then diluted to 105/ml and the concentration of viable (trypan blue-excluding) cells was monitored for 19 days.
PG processing
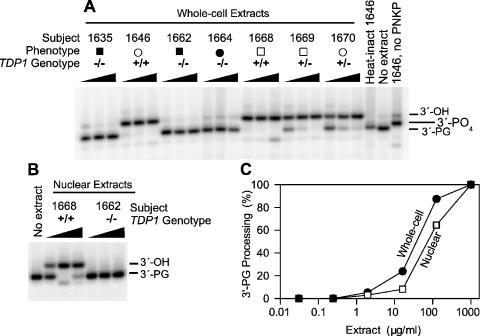
To assess processing of 3′-PG termini on the oligomeric substrate, 20 fmol of the substrate was incubated at 37°C for 2 h in 5 μl of 60 mM potassium acetate, 50 mM triethanolamine–HCl, pH 7.4, 2 mM ATP, 5 mM EDTA, 1 mM DTT and 100 μg/ml BSA, containing various amounts of whole-cell or nuclear extract (2 μl maximum). Samples were heated to 90°C for 5 min to inactivate proteins in the extract, and then 1.5 μl of 50 mM MgCl2 and 0.5 μl of 40 μg/ml PNKP (generous gift of Michael Weinfeld, Cross Cancer Institute) were added and the samples incubated for 1 h at 37°C. An equal volume of formamide containing 20 mM EDTA was added, and after denaturation for 1 min at 90°C the samples were subjected to denaturing electrophoresis on a 20% polyacrylamide sequencing gel. Post-treatment with PNKP converts any 3′-phosphate termini generated by TDP1 into 3′-OH termini, thus increasing the separation between the substrate and product bands on the gel. Initial screening of SCAN1 and normal cell lines (Figure 2) was performed in a double-blind fashion.
Figure 2.
Processing of a 3′-PG 14mer by extracts of lymphoblastoid cells from SCAN1 patients (closed symbols) and from unaffected members (open symbols) of the same family, including TDP1+/– carriers (squares, males; circles, females). The substrate was treated with 0.3, 1 or 3 mg/ml of each whole-cell extract (A), or with 0.14, 1.4 or 5 mg/ml of each nuclear extract (B) for 2 h in the presence of EDTA. Following heat denaturation of cellular proteins, the substrate was treated with PNKP to convert 3′-phosphate to 3′-OH termini. Titration with various concentrations of extract (C) showed that whole-cell extracts (closed circles) contained about twice as much PG-processing activity as nuclear extracts (open squares).
To assess processing of protruding 3′-PG termini on the plasmid substrate, the substrate was first incubated in whole-cell or nuclear extracts for various times. For whole-cell extracts, samples contained 10 ng substrate and 60 μg extract in the incubation buffer described above but with 0.5 mM magnesium acetate instead of EDTA, plus 50 μM dATP, dGTP, dCTP and ddTTP. Typically, 9 μl of extract was added to a total reaction volume of 20 μl. Potassium and magnesium were added to give the final concentrations indicated, after compensation for potassium and EDTA in extract storage buffer. For nuclear extracts, a similar buffer but with 1 mM magnesium acetate and 1 mM ATP was used. Following incubation at 37°C for various times, the samples were diluted into 0.14 ml of 1% SDS, 1 mg/ml proteinase K, 0.3 M NaCl, 20 mM Tris–HCl pH 7.6 and 10 mM EDTA, and heated to 65°C for 3 h. Following extraction with phenol and then with chloroform, the nucleic acids were ethanol-precipitated and the DNA cut with AvaI (40 U in 50 μl, 3 h at 37°C) to release the labeled oligomer. Samples were ethanol-precipitated in the presence of 2 μg tRNA and subjected to electrophoresis as above.
Phosphorylation in vivo
Human 293 cells were grown to 50% confluence in 60 mm dishes and transfected with pFLAG-TDP1 as follows. A mixture of 1.27 ml serum-free medium and 255 μl Superfect (Qiagen) was preincubated with 42 μg plasmid at room temperature for 10 min and then diluted into 8.5 ml complete medium. Medium in each dish was replaced with 1 ml of this transfection medium, and the dishes were rocked at 3 cycles/min at 37°C for 3 h. Following addition of 5 ml complete medium and incubation without rocking for 2 days, the medium was replaced with 2.5 ml phosphate-free medium containing 0.5 mCi [32P]phosphoric acid (Perkin-Elmer, 6000 Ci/mmol). After 30 min incubation, the cells were exposed to 0 or 20 Gy 60Co γ-rays (dose rate 4 Gy/min) and incubated for various times. The medium was removed and cells were scraped into 0.8 ml ice-cold phosphate-buffered saline, pelleted (3000 r.p.m., 4 min) and lysed by resuspending in 0.2 ml lysis buffer [10 mM HEPES-KOH, pH 7.8, 60 mM KCl, 1 mM EDTA, 0.5 mM phenylmethlysulfonyl fluoride (PMSF) and 0.5% NP-40], supplemented with protease inhibitors (Sigma–Aldrich P-8465) and phosphatase inhibitors (Sigma–Aldrich P-2850 and P-5726), each at 100:1 dilution. After 5 min on ice, the nuclei were pelleted and the supernatant (cytoplasmic fraction) was decanted. The nuclei were resuspended in 0.2 ml Dignam nuclear extraction buffer (20 mM Tris–HCl, pH 7.9, 0.2 mM EDTA, 0.42 M NaCl, 15 mM MgCl2, 25% glycerol, plus inhibitors as above), held on ice for 30 min and then centrifuged at 8000 g for 5 min. The supernatant (nuclear fraction) was decanted. Each cytoplasmic or nuclear fraction was diluted with an equal volume of 10 mM Tris–HCl, pH 7.6, 1 mM EDTA (TE), and prewashed FLAG affinity beads (30 μl bed volume) were added. FLAG-tagged TDP1 in the lysates was adsorbed to the beads (3 h at 8°C with rotation), and the beads were washed three times with inhibitor-free TE. Aliquots of the beads (∼5 μl bed volume for each sample) were treated with protein phosphatase 1 (PP1) (1 U at 30°C for 2 h), washed and then treated with YOP phosphatase (25 U at 30°C for 2 h), in 40 μl of the buffers provided by the vendor (New England Biolabs). In each case, the beads were washed once with the appropriate reaction buffer before each reaction, and control samples were incubated in buffer alone. Following phosphatase treatment, the beads were boiled for 5 min in 62.5 mM Tris–HCl, pH 6.8, 0.1 M 2-mercaptoethanol, 10% glycerol and 2% SDS, and the denatured proteins analyzed on 10% SDS–polyacrylamide gels. Proteins were electroblotted onto nitrocellulose and probed with anti-FLAG antibody (1000:1 dilution; Sigma–Aldrich F-3165), using a chemiluminescent goat anti-rabbit secondary antibody (Pierce). The blots were then subjected to phosphorimaging for 1–2 days to assess 32P incorporation into FLAG-TDP1.
For preparation of FLAG-TDP1 for reconstitution experiments, the above procedure was scaled up to five 150 mm dishes. After adsorption and washing as above, FLAG-TDP1 was eluted from FLAG affinity beads (200 μl bed volume) by rotating the beads for 16 h in 2.5 bed volumes TE containing 0.5 mM DTT, 0.5 mM PMSF and 160 μg/ml of the peptide DYKDDDDK (FLAG peptide, Sigma–Aldrich F-3290). An equal volume of glycerol was added and the enzyme was stored at −20°C.
RESULTS
Lack of 3′-prime-PG processing in SCAN1 extracts
To examine PG processing activities in human normal and SCAN1 cells, immortal lymphoblastoid cell lines were generated from SCAN1 patients and from unaffected members of the same family, including known heterozygous SCAN1 carriers. Whole-cell and nuclear extracts were prepared, and a 3′-PG-terminated 14mer (Figure 1A) was incubated in various dilutions of extract (Figure 2). These reactions contained EDTA, which prevents degradation of the oligomer by potent single-strand nucleases in the extracts, but also blocks PNKP-mediated conversion of 3′-phosphate termini generated by TDP1 to 3′-OH termini. Therefore, to increase the resolution of the assay, samples were treated with purified PNKP after heat-denaturation of cellular proteins.
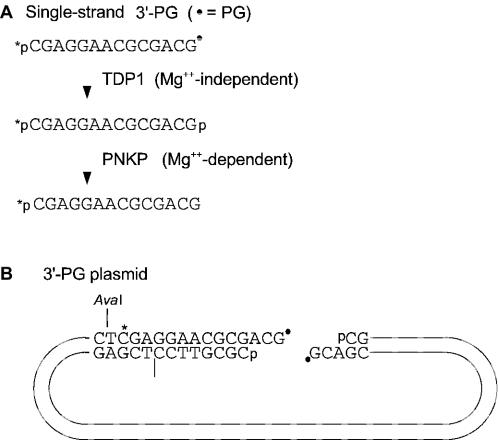
Figure 1.
Substrates with 3′-PG termini. (A) Oligomeric substrate. The 3′-PG terminus can be converted to a 3′-phosphate by TDP1 and thence to a 3′-OH by PNKP. (B) The 5.5 kb 3′-PG plasmid substrate is internally labeled, and 3′-terminal processing can be monitored by cleaving the substrate with AvaI and analyzing the labeled fragment by denaturing gel electrophoresis.
Under these conditions, 3′-PG termini were quantitatively removed by incubation in 0.3 mg/ml whole-cell extract, but there was little if any removal by SCAN1 extracts even at 3 mg/ml (Figure 2A). Heterozygous TDP1+/– carriers appeared to have slightly less PG-processing activity, as PG removal was complete at 1 mg/ml but not at 0.3 mg/ml extract. As expected, a sample treated with extract but not subsequently treated with PNKP showed predominant accumulation of 3′-phosphate termini.
Although there was, in samples treated with SCAN1 whole-cell extracts, a trace of a band comigrating with the 3′-OH oligomer, the intensity of this band showed no apparent dependence on extract concentration. Moreover, no band was seen at the 3′-OH position in samples treated with a SCAN1 nuclear extract (Figure 2B), even though normal nuclear extracts contained nearly as much PG-processing activity as normal whole-cell extracts (Figure 2C). Thus, while the origin of the apparent 3′-OH band generated in SCAN1 whole-cell extracts is uncertain, it probably does not represent PG processing by residual TDP1 activity in the SCAN1 extracts. Based on titrations with normal and SCAN1 nuclear extracts (Figure 2B), it is estimated that, in the presence of EDTA, PG-processing activity is at least 100-fold lower in SCAN1 cells than in normal cells. Thus, as expected, the SCAN1 mutation in the active site of TDP1 (12) apparently abrogates its activity completely, at least for a PG substrate.
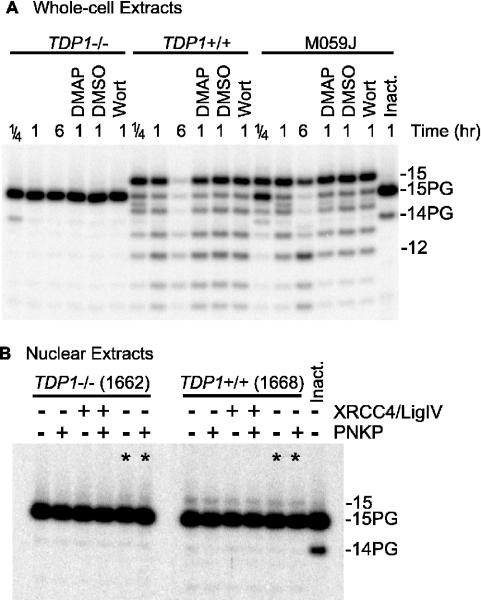
These results also indicate that, in the absence of divalent cations, nuclear and whole-cell extracts contain no enzymes capable of processing 3′-PG termini, other than TDP1. However, these data do not exclude the possibility of other Mg++-dependent PG-processing enzymes. When the 3′-PG oligomer was incubated in either normal or SCAN1 nuclear extract in the presence of Mg++, the 32P-label was completely converted to material that migrated at the position expected for a single nucleotide, suggesting that it was completely degraded by nucleases in the extracts. In SCAN1 extracts, the kinetics of degradation for unmodified oligomers suggested that both 3′→5′ and 5′→3′ exonucleolytic activities were present (data not shown). However, in similar experiments with the 3′-PG oligomer, neither full-length 3′-OH oligomer nor any intermediate-length fragments were visible; all label appeared as either completely intact or completely degraded substrate, suggesting predominantly 5′→3′ digestion, perhaps by exonuclease I (20). This degradation precluded use of the 3′-PG oligomer for assessing Mg++-dependent PG-processing activities. Therefore, PG processing was instead examined using a plasmid substrate (see Figure 1B) bearing an internal label near one 3′ end (Figure 3). In normal whole-cell extracts, in the presence of Mg++, conversion of the protruding 3′-PG terminus of this substrate to a 3′-OH was nearly complete within 15 min (Figure 3A). While there was also significant 3′→5′ resection, especially after 6 h, the plasmid substrate was subject to much less degradation than was the oligomer. The DNA-dependent protein kinase (DNA-PK) inhibitors wortmannin and dimethylaminopurine had no apparent effect on PG processing, and similar PG processing kinetics were seen in extracts of DNA-PK-deficient M059J cells (21), implying that PG processing did not require DNA-PK. Again, however, there was no detectable processing of the PG-terminated substrate in SCAN1 whole-cell extracts, indicating that, at least under these in vitro conditions, all PG processing was attributable to TDP1, and there were no Mg++-dependent activities in the extracts capable of processing protruding 3′-PG termini. These data also show that the PG terminus prevented 3′→5′ resection of the substrate. This resection may be attributable to DNase III, which is the predominant 3′→5′ exonuclease in mammalian cell extracts (22,23), and can digest 3′-OH- but not 3′-PG-terminated substrates (24).
Figure 3.
Processing of a plasmid bearing partially complementary PG-terminated 3-base 3′ overhangs by normal and SCAN1 extracts. The substrate was treated for the indicated times with 3 mg/ml whole-cell (A) or 1.8 mg/ml nuclear extract (B), deproteinized, treated (or not) with PNKP, cut with AvaI and subjected to denaturing gel electrophoresis. Some samples also contained 1 mM dimethylaminopurine (DMAP), 10 μM wortmannin (Wort) and/or 2% dimethyl sulfoxide (DMSO, solvent for wortmannin). The lanes marked (asterisk) in (B) indicate samples that were treated in the reaction buffer normally used for whole-cell extract. In (B), all samples treated with the normal extract showed 1.8–2.1% conversion to 3′-OH, while those treated with the SCAN1 extract showed no detectable conversion. A trace of 14-PG fragment seen in some samples is due to residual single-strand oligomer that was not incorporated into the plasmid.
Unexpectedly and in contrast to the 3′-PG oligomer, the plasmid substrate was subject to much less 3′-PG processing in nuclear than in whole-cell extracts, with only ∼2% conversion to 3′-OH after 6 h incubation in nuclear extracts from normal cells (Figure 3B). After 24 h incubation (data not shown), most of the 3′-PG bands disappeared, but little or no 3′-OH 14mer was detected, suggesting that the substrate was eventually either degraded or rendered insoluble. The disparity in the extent of PG processing between whole-cell and nuclear extracts was not due to the small differences between the reaction buffers, as the same result was obtained using the buffer normally used for whole-cell extracts (lanes marked ‘asterisk’ in Figure 3B); nor was it due to a lack of XRCC4/DNA ligase IV complex [a component of the end-joining repairosome that is often present in suboptimal amounts in nuclear extracts (17)], as addition of exogenous XRCC4 and ligase IV had no effect. The inefficiency of PG processing in nuclear extracts may be due to sequestration of DNA ends by DNA-PK, which is present in higher concentrations in the nucleus than in the cytoplasm (25). In any case, the small degree of PG processing seen in normal extracts was completely absent in SCAN1 nuclear extracts, consistent with its being catalyzed by TDP1 plus PNKP. When the normal nuclear extract was supplemented with 25 ng of additional, recombinant TDP1, conversion to 3′-OH was increased to ∼10%, and nearly all the conversion occurred within the first 15 min (data not shown). This result is consistent with the view that in nuclear extracts the 3′-PG terminus is briefly accessible to TDP1 before becoming sequestered in a DNA end-binding complex.
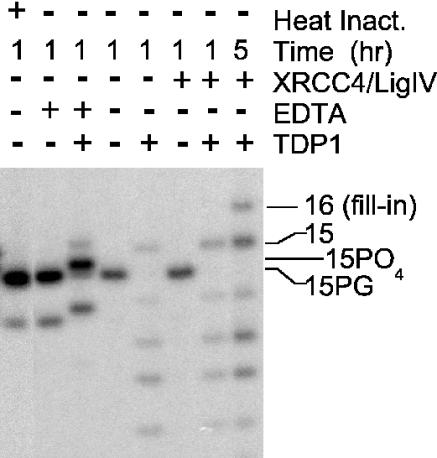
When SCAN1 whole-cell extracts were complemented with purified recombinant human TDP1, end processing was qualitatively similar to the processing seen in normal whole-cell extracts, with complete conversion of 3′-PG to 3′-OH termini (or to 3′-phosphate termini in the presence of EDTA) (Figure 4). Moreover, when the extracts were also supplemented with XRCC4/ligase IV complex, which is often rate-limiting in cell extracts (17), the substrate was extended by 1 nt, consistent with gap filling on DSB ends aligned by annealing of the self-complementary -CG at the termini (ddTTP is incorporated, preventing subsequent ligation; see Figure 1). Thus, except for XRCC4/ligase IV, all factors required for this alignment-based gap filling, which is the hallmark of the non-homologous end-joining repair pathway (26,27), were apparently present and active in these extracts.
Figure 4.
Reconstitution of 3′-PG processing and subsequent gap filling in SCAN1 extracts by recombinant TDP1. The plasmid substrate was treated with 3 mg/ml SCAN1 whole-cell extract (subject 1635) and analyzed as in Figure 3. In some cases, 25 ng of recombinant TDP1 and/or 200 ng of XRCC4/DNA ligase IV complex (Trevigen) were added. Some samples contained 5 mM EDTA instead of 0.5 mM MgCl2, as indicated.
Phosphorylation of TDP1 in vivo and stimulation by ionizing radiation
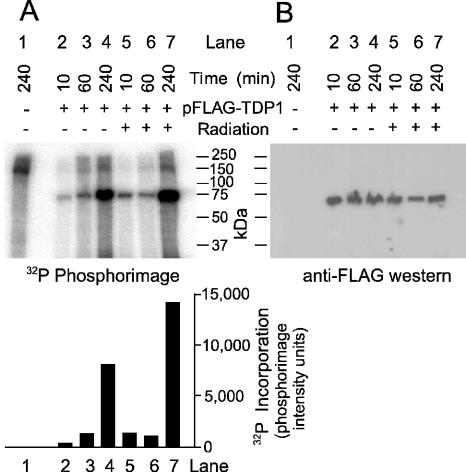
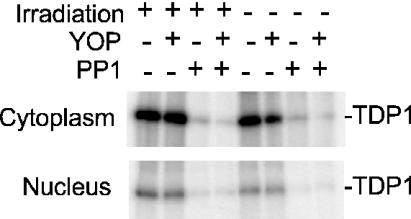
DNA DSBs induced by ionizing radiation initiate an extensive cascade of phosphorylation events, beginning with ATM and DNA-PK, and culminating in repair, cell cycle arrest and/or apoptosis (28–31). Many DNA repair enzymes are targets for phosphorylation as part of this cascade. To determine whether TDP1 is such a target, FLAG-TDP1 was overexpressed from a transfected plasmid vector in 293 cells in the presence of [32P]phosphate, and then affinity-purified from cell extracts. A prominent 32P-labeled ∼70 kDa band, comigrating with the FLAG-tagged protein detected on western blots, was found in extracts from transfected but not from untransfected cells, suggesting that TDP1 was indeed phosphorylated in vivo (Figure 5). For samples harvested 15 and 240 min after irradiation, phosphorylation was significantly stimulated by radiation. [The lack of apparent stimulation in the irradiated, 60 min sample may be attributable to low transfection efficiency or poor protein recovery in this sample, as suggested by a relatively weak FLAG-TDP1 band on the western blot (Figure 5B)]. In a second experiment wherein cytoplasmic and nuclear fractions were analyzed separately, radiation-induced stimulation of phosphorylation was detected in both fractions at all time points (Table 1). The average radiation-induced increase in [32P]phosphate incorporation into TDP1 for all nine samples from both experiments was 2.1 ± 0.3 fold (mean ± SE). These results suggest that TDP1 is a target for cell signaling in response to radiation. Treatment of the 32P-labeled FLAG-TDP1 with the serine/threonine-specific PP1 eliminated 32P-labeling almost completely, while the tyrosine-specific YOP phosphatase had little if any effect (Figure 6). Thus, TDP1 appears to be phosphorylated predominantly on serine or threonine residues. Although phosphorylation was stimulated only slightly by irradiation, the baseline phosphorylation level may have been elevated due to radiation from the 32P-radiolabel.
Figure 5.
Phosphorylation of TDP1. Cells irradiated with 0 or 20 Gy were incubated in the presence of [32P]orthophosphate for the indicated times. Affinity-purified FLAG-TDP1 was subjected to denaturing gel electrophoresis followed by phosphorimaging (A) or western blotting with FLAG antibody (B).
Table 1.
Radiation-induced enhancement of TDP1 phosphorylation (fold)
| Time | Experiment 1 | Experiment 2 | |
|---|---|---|---|
| Combined | Cytoplasm | Nucleus | |
| 20 min | 3.8 | 1.4 | 1.9 |
| 1 h | 1.1 | 1.6 | 1.3 |
| 4 h | 1.8 | 3.3 | 2.8 |
Figure 6.
Dephosphorylation of TDP1 by PP1. Affinity-purified phosphate-labeled TDP1 from cytoplasmic or nuclear extracts of cells treated with 0 or 20 Gy ionizing radiation was exposed to PP1 and/or YOP phosphatase.
Radiosensitivity of SCAN1 cells
If 3′-PG processing is inefficient or absent in SCAN1 cells in vivo, then SCAN1 cells should be more sensitive than normal cells to ionizing radiation, an agent that induces a high proportion of breaks with 3′-PG termini (7). Initial screening suggested that SCAN1 cells were no more sensitive than normal cells to a single dose of either radiation or hydrogen peroxide (data not shown). However, since DSB repair by non-homologous end-joining is primarily responsible for split-dose recovery (32), dose fractionation can accentuate the radiosensitizing effects of repair deficiencies.
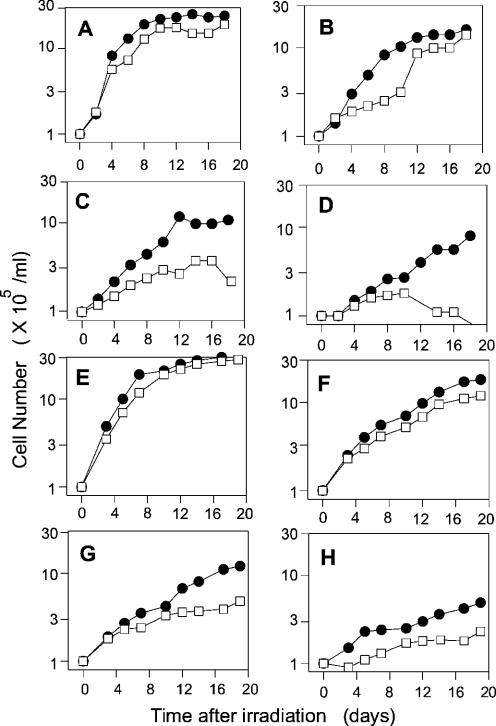
When plateau-phase cells were irradiated daily for five days, cell lines from two SCAN1 patients were reproducibly more radiosensitive than those from two unaffected individuals, as judged by the subsequent growth delay (Figure 7). For example, at 5 × 1.6 Gy, growth of normal cells resumed after 10 days, by which time the SCAN1 cells (subject 1664) had begun to die off (Figure 7D). Comparison of the response of SCAN1 cells to 5 × 0.8 Gy (Figure 7C and G) with the response of normal cells to 5 × 1.6 Gy (Figure 7D and H) suggested that in each case, the degree of radiosensitivity (dose-modifying factor) was ∼2-fold, although cells from SCAN1 subject 1664 (Figure 7D) appeared to be slightly more sensitive than those from subject 1662 (Figure 7H).
Figure 7.
Radiosensitivity of SCAN1 cells. Plateau-phase normal cells (closed circles, subject 1646) or SCAN1 cells (open squares, subject 1664) were irradiated daily for 5 days with 0 (A), 0.4 (B), 0.8 (C) or 1.6 (D) Gy of γ-rays, then diluted to 105/ml for assessment of cell growth. (E–H) Identical experiment with another normal and another SCAN1 cell line (subjects 1668 and 1662, respectively).
A third SCAN1 line (subject 1635) harboring the same mutant TDP1 allele and having the same apparent biochemical deficiency (Figure 2) did not show any radiosensitivity, consistently giving growth curves that were superimposable with those of normal cells (subject 1668) following treatment with either single-dose or fractionated radiation (data not shown). However, subjects 1662, 1664 and 1668 are more closely related to each other than to subject 1635, and thus may be more appropriate for comparison. Although growth assays may overlook small differences in radiosensitivity that might have been detected in clonogenic survival assays, we were unable to obtain reproducible survival data with any of these cell lines, by either agar cloning or limiting dilution assays, due to the low cloning efficiency (∼1%). Nevertheless, while the present results do not distinguish between growth delay and cell killing, they do show a difference between two of the three SCAN1 cell lines and normal cell lines in their response to radiation. Attempts to complement the apparent radiosensitivity of SCAN1 cells by expression of normal TDP1 are in progress, but are likewise complicated by the low cloning efficiency of these cells.
DISCUSSION
Because DNA ligases as well as gap-filling DNA polymerases require 3′-OH substrates, removal of 3′-PG termini is an essential early step in the repair of free radical-mediated DSBs. Candidates for carrying out such removal include dedicated 3′-terminal processing enzymes such as Ape1, PNKP and TDP1, as well as 3′ exonucleases such as DNase III, Mre11 and Wrn. However, based on the known specificities of these enzymes, none of them except TDP1 is expected to act on protruding 3′-PG termini of DSBs. Ape1 acts only on blunt and recessed 3′ ends (8), and neither PNKP nor DNase III has any activity toward 3′-PG termini (11,24). Although the activities of Mre11 and Wrn toward modified termini have not been rigorously examined, they both show a strong preference for recessed 3′ termini, and 3′ overhangs are extremely poor substrates for these enzymes even when they bear normal 3′-OH termini (33–35). The Saccharomyces cerevisiae Rad1/Rad10 endonuclease (and presumably its human ERCC1/XPF homolog) can remove a single 3′-PG (or 3′-OH) nucleotide from the 3′ end of a single-strand break in double-stranded DNA (36), but no such activity has been reported on PG-terminated 3′ overhangs. Similarly, while Artemis, a DNA-PK-activated endo/exonuclease, has been implicated in DSB repair (37), this enzyme appears to act only on 3′ overhangs longer than four bases (38).
Consistent with these predictions, in extracts of SCAN1 cells harboring mutant TDP1, there was no detectable processing of a PG terminus on a three-base 3′ overhang. Moreover, despite the fact that purified TDP1, whether produced in mammalian (data not shown) or bacterial (11) cells, is ∼100-fold less active toward 3′-PG than toward 3′-phosphotyrosyl substrates, the levels of TDP1 and PNKP in both whole-cell and nuclear extracts of normal cells are sufficient to rapidly convert any exposed 3′-PG termini to 3′-phosphate and then to 3′-OH termini (although in nuclear extracts DSB ends appear to be partially shielded from such processing). Both these results are consistent with the view that most if not all processing of protruding 3′-PG termini is dependent on TDP1.
Because a substantial proportion, perhaps as many as half, of DNA breaks induced by ionizing radiation bear 3′-PG termini (7), cells deficient in processing of PG-terminated DSBs are expected to be profoundly radiosensitive. However, in cell growth assays, only slight radiosensitivity was detected in two of three SCAN1 cell lines, and then only for fractionated radiation in plateau phase. These results suggest that most PG-terminated DSBs are still repaired in SCAN1 cells, presumably by TDP1-independent pathway(s). As noted above, blunt and recessed 3′-PG ends could be processed by Ape1, while 3′ overhangs longer than four bases could be processed by Artemis. Moreover, for DSBs with one 3′-PG and one 3′-phosphate terminus, it is possible that the strand with the 3′-phosphate could first be rejoined, and then the 3′-PG strand could be processed as if it were a single-strand break, with PG removal by Ape1. Thus, the fraction of radiation-induced DSBs with a strict requirement for TDP1 (perhaps only those with one- to four-base PG-terminated 3′ overhangs in both strands) may be quite small, possibly <10%. Finally, even for these breaks, there may be alternative, more complicated repair pathways that can substitute when simpler end-joining pathways fail, as suggested by the ‘repair foci’ of DSB repair factors that can be detected cytologically long after the majority of DSBs have already been repaired (39,40). Failure of these complexes to form properly in cell extracts could explain the complete lack of apparent processing in extract-based assays. Whether protruding PG termini on DSBs are more persistent in SCAN1 than in normal cells in vivo is not known, and although a post-labeling assay for PG formation and repair in vivo has been described previously (41,42), this assay is relatively insensitive and does not distinguish between single- and double-strand break termini. Thus, further work will be required to resolve the apparent disparity between the profound repair deficiency seen in cell extracts and the relatively mild radiosensitivity of SCAN1 cells.
Similar to many DNA repair proteins (28,30,31,43,44), TDP1 is phosphorylated, and its phosphorylation is stimulated by ionizing radiation, providing indirect evidence of possible involvement in pathways for repair of radiation-induced damage. The kinase(s) involved have yet to be identified, but phosphorylation on serine/threonine residues as well as stimulation by ionizing radiation would be consistent with ATM and/or DNA-PK (28,30,31,43–45). Phosphorylation is clearly not required for the basic enzymatic function of TDP1, as recombinant proteins produced in bacteria or in human cells show qualitatively similar activity (H. Tatavarthi and L. F. Povirk, unpublished data). However, phosphorylation could modulate TDP1 activity or influence its interactions with other repair proteins.
Although not proven, it is generally assumed that SCAN1 pathology is a consequence of the failure of mutant TDP1 to efficiently repair topoisomerase I-associated DNA damage (12). This repair deficiency could indirectly confer sensitivity to oxidative DNA damage, as certain oxidative lesions tend to promote formation of topoisomerase I cleavable complexes (46), which upon replication can be converted to cytotoxic topoisomerase-terminated DSBs (47). Nevertheless, a role for PG-terminated DSBs in SCAN1 pathology cannot be excluded. The neuronal apoptosis seen in knockout mice lacking critical DNA end-joining proteins, such as XRCC4 and DNA ligase IV (48,49), suggest that in the absence of DSB repair, terminally differentiating neurons accumulate sufficient DSBs to produce significant neurological pathology. There is evidence that at least some of these ‘spontaneous’ DSBs reflect damage by oxygen free radicals associated with normal oxidative metabolism (50). Moreover, SCAN1 pathology is similar to that of both Friedreich ataxia and ataxia with oculomotor apraxia (AOA1) (51), both of which have been linked to oxidative damage. Friedreich ataxia is associated with a mutant frataxin protein, which leads to increased oxidative stress due to a defect in iron homeostasis (52–56). AOA1 cells are sensitive to hydrogen peroxide and harbor a mutation in aprataxin; while the exact function of the aprataxin protein is not known, it interacts with the single-strand break repair protein XRCC1. In S.cerevisiae of appropriate genetic backgrounds, TDP1 deficiency confers sensitivity to ionizing radiation as well as to bleomycin, a radiomimetic drug that specifically induces PG-terminated DSBs (57). Thus, while the exact sequence of events culminating in the SCAN1 phenotype remains to be elucidated, several lines of circumstantial evidence suggest a possible linkage among TDP1 deficiency, oxidative damage, DSBs and cerebellar ataxia. Even though lymphoblastoid cell lines derived from SCAN1 patients show at most only mild radiosensitivity, it is possible that endogenous tissues of SCAN1 patients are more sensitive to free radical damage than the cell lines. Because cultured cells are subjected to higher oxygen tensions than would occur in vivo, they may be more likely to adapt to TDP1 deficiency, for example, by upregulating alternative pathways for repair of DSBs and other oxidative damage.
Acknowledgments
We thank Michael Weinfeld for providing PNKP. This work was supported by Grants CA40615 and AG023783 from the National Institutes of Health, US DHHS. Funding to pay the Open Access publication charges for this article was provided by Grant CA40615.
REFERENCES
- 1.Hsiang Y.H., Hertzberg R., Hecht S., Liu L.F. Camptothecin induces protein-linked DNA breaks via mammalian DNA topoisomerase I. J. Biol. Chem. 1985;260:14873–14878. [PubMed] [Google Scholar]
- 2.Pommier Y. DNA topoisomerase I and II in cancer chemotherapy: update and perspectives. Cancer Chemother. Pharmacol. 1993;32:103–108. doi: 10.1007/BF00685611. [DOI] [PubMed] [Google Scholar]
- 3.Pouliot J.J., Yao K.C., Robertson C.A., Nash H.A. Yeast gene for a Tyr-DNA phosphodiesterase that repairs topoisomerase I complexes. Science. 1999;286:552–555. doi: 10.1126/science.286.5439.552. [DOI] [PubMed] [Google Scholar]
- 4.Interthal H., Pouliot J.J., Champoux J.J. The tyrosyl-DNA phosphodiesterase Tdp1 is a member of the phospholipase D superfamily. Proc. Natl Acad. Sci. USA. 2001;98:12009–12014. doi: 10.1073/pnas.211429198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hutchinson F. Chemical changes induced in DNA by ionizing radiation. Prog. Nucleic Acids Res. Mol. Biol. 1985;32:115–154. doi: 10.1016/s0079-6603(08)60347-5. [DOI] [PubMed] [Google Scholar]
- 6.Ward J.F. DNA damage produced by ionizing radiation in mammalian cells: identities, mechanisms of formation, and reparability. Prog. Nucleic Acid. Res. Mol. Biol. 1988;35:95–125. doi: 10.1016/s0079-6603(08)60611-x. [DOI] [PubMed] [Google Scholar]
- 7.Henner W.D., Rodriguez L.O., Hecht S.M., Haseltine W.A. gamma-Ray induced deoxyribonucleic acid strand breaks. 3′ glycolate termini. J. Biol. Chem. 1983;258:711–713. [PubMed] [Google Scholar]
- 8.Suh D., Wilson D.M., III, Povirk L.F. 3′-Phosphodiesterase activity of human apurinic/apyrimidinic endonuclease at DNA double-strand break ends. Nucleic Acids Res. 1997;25:2495–2500. doi: 10.1093/nar/25.12.2495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Karimi-Busheri F., Daly G., Robins P., Canas B., Pappin D.J., Sgouros J., Miller G.G., Fakhrai H., Davis E.M., Le Beau M.M., Weinfeld M. Molecular characterization of a human DNA kinase. J. Biol. Chem. 1999;274:24187–24194. doi: 10.1074/jbc.274.34.24187. [DOI] [PubMed] [Google Scholar]
- 10.Jilani A., Ramotar D., Slack C., Ong C., Yang X.M., Scherer S.W., Lasko D.D. Molecular cloning of the human gene, PNKP, encoding a polynucleotide kinase 3′-phosphatase and evidence for its role in repair of DNA strand breaks caused by oxidative damage. J. Biol. Chem. 1999;274:24176–24186. doi: 10.1074/jbc.274.34.24176. [DOI] [PubMed] [Google Scholar]
- 11.Inamdar K.V., Pouliot J.J., Zhou T., Lees-Miller S.P., Rasouli-Nia A., Povirk L.F. Conversion of phosphoglycolate to phosphate termini on 3′ overhangs of DNA double-strand breaks by the human tyrosyl-DNA phosphodiesterase hTdp1. J. Biol. Chem. 2002;276:24323–24330. doi: 10.1074/jbc.M204688200. [DOI] [PubMed] [Google Scholar]
- 12.Takashima H., Boerkoel C.F., John J., Saifi G.M., Salih M.A., Armstrong D., Mao Y., Quiocho F.A., Roa B.B., Nakagawa M., Stockton D.W., Lupski J.R. Mutation of TDP1, encoding a topoisomerase I-dependent DNA damage repair enzyme, in spinocerebellar ataxia with axonal neuropathy. Nature Genet. 2002;32:267–272. doi: 10.1038/ng987. [DOI] [PubMed] [Google Scholar]
- 13.Hoeijmakers J.H. Nucleotide excision repair. II: from yeast to mammals. Trends. Genet. 1993;9:211–217. doi: 10.1016/0168-9525(93)90121-w. [DOI] [PubMed] [Google Scholar]
- 14.Brooks P.J. DNA repair in neural cells: basic science and clinical implications. Mutat. Res. 2002;509:93–108. doi: 10.1016/s0027-5107(02)00222-1. [DOI] [PubMed] [Google Scholar]
- 15.Rolig R.L., McKinnon P.J. Linking DNA damage and neurodegeneration. Trends Neurosci. 2000;23:417–424. doi: 10.1016/s0166-2236(00)01625-8. [DOI] [PubMed] [Google Scholar]
- 16.Baumann P., West S.C. DNA end-joining catalyzed by human cell-free extracts. Proc. Natl Acad. Sci. USA. 1998;95:14066–14070. doi: 10.1073/pnas.95.24.14066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Huang J., Dynan W.S. Reconstitution of the mammalian DNA double-strand break end-joining reaction reveals a requirement for an Mre11/Rad50/NBS1-containing fraction. Nucleic Acids Res. 2002;30:667–674. doi: 10.1093/nar/30.3.667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lee J.W., Blanco L., Zhou T., Bebenek K., Garcia-Diaz M., Kunkel T.A., Wang Z., Povirk L.F. Implication of DNA polymerase lambda in alignment-based gap filling for nonhomologous DNA end joining in human nuclear extracts. J. Biol. Chem. 2004;279:805–811. doi: 10.1074/jbc.M307913200. [DOI] [PubMed] [Google Scholar]
- 19.Bennett R.A.O., Gu X.Y., Povirk L.F. Construction of a vector containing a site-specific DNA double-strand break with 3′-phosphoglycolate termini and analysis of the products of end-joining in CV-1 cells. Int. J. Radiat. Biol. 1996;70:623–636. doi: 10.1080/095530096144509. [DOI] [PubMed] [Google Scholar]
- 20.Lee B.I., Wilson D.M. The RAD2 domain of human exonuclease 1 exhibits 5′ to 3′ exonuclease and flap structure-specific endonuclease activities. J. Biol. Chem. 1999;274:37763–37769. doi: 10.1074/jbc.274.53.37763. [DOI] [PubMed] [Google Scholar]
- 21.Lees-Miller S.P., Godbout R., Chan D.W., Weinfeld M., Day R.S., III, Barron G.M., Allalunis-Turner J. Absence of p350 subunit of DNA-activated protein kinase from a radiosensitive human cell line. Science. 1995;267:1183–1185. doi: 10.1126/science.7855602. [DOI] [PubMed] [Google Scholar]
- 22.Hoss M., Robins P., Naven T.J., Pappin D.J., Sgouros J., Lindahl T. A human DNA editing enzyme homologous to the Escherichia coli DnaQ/MutD protein. EMBO J. 1999;18:3868–3875. doi: 10.1093/emboj/18.13.3868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mazur D.J., Perrino F.W. Identification and expression of the TREX1 and TREX2 cDNA sequences encoding mammalian 3′→5′ exonucleases. J. Biol. Chem. 1999;274:19655–19660. doi: 10.1074/jbc.274.28.19655. [DOI] [PubMed] [Google Scholar]
- 24.Inamdar K.V., Yu Y., Povirk L.F. Resistance of 3′-phosphoglycolate DNA ends to digestion by mammalian DNase III. Radiat. Res. 2002;157:306–311. doi: 10.1667/0033-7587(2002)157[0306:ropdet]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- 25.Anderson C.W., Lees-Miller S.P. The nuclear serine/threonine protein kinase DNA-PK. Crit. Rev. Eukaryot. Gene Expr. 1992;2:283–314. [PubMed] [Google Scholar]
- 26.Daza P., Reichenberger S., Göttlich B., Hagmann M., Feldmann E., Pfeiffer P. Mechanisms of nonhomologous DNA end-joining in frogs, mice and men. Biol. Chem. 1996;377:775–786. doi: 10.1515/bchm3.1996.377.12.775. [DOI] [PubMed] [Google Scholar]
- 27.Chen S., Inamdar K.V., Pfeiffer P., Feldmann E., Hannah M.F., Yu Y., Lee J.W., Zhou T., Lees-Miller S.P., Povirk L.F. Accurate in vitro end-joining of a DNA double-strand break with partially cohesive 3′-overhangs and 3′-phosphoglycolate termini: effect of Ku on repair fidelity. J. Biol. Chem. 2001;276:24323–24330. doi: 10.1074/jbc.M010544200. [DOI] [PubMed] [Google Scholar]
- 28.Valerie K., Povirk L.F. Regulation and mechanisms of mammalian double-strand break repair. Oncogene. 2003;22:5792–5812. doi: 10.1038/sj.onc.1206679. [DOI] [PubMed] [Google Scholar]
- 29.Jackson S.P. Sensing and repairing DNA double-strand breaks. Carcinogenesis. 2002;23:687–696. doi: 10.1093/carcin/23.5.687. [DOI] [PubMed] [Google Scholar]
- 30.Lees-Miller S.P. The DNA-dependent protein kinase, DNA-PK: 10 years and no ends in sight. Biochem. Cell Biol. 1996;74:503–512. doi: 10.1139/o96-054. [DOI] [PubMed] [Google Scholar]
- 31.Goodarzi A.A., Block W.D., Lees-Miller S.P. The role of ATM and ATR in DNA damage-induced cell cycle control. Prog. Cell Cycle Res. 2003;5:393–411. [PubMed] [Google Scholar]
- 32.Whitmore G.F., Varghese A.J., Gulyas S. Cell cycle responses of two X-ray sensitive mutants defective in DNA repair. Int. J. Radiat. Biol. 1989;56:657–665. doi: 10.1080/09553008914551881. [DOI] [PubMed] [Google Scholar]
- 33.Kamath-Loeb A.S., Shen J.C., Loeb L.A., Fry M. Werner syndrome protein. II. Characterization of the integral 3′→5′ DNA exonuclease. J. Biol. Chem. 1998;273:34145–3450. doi: 10.1074/jbc.273.51.34145. [DOI] [PubMed] [Google Scholar]
- 34.Paull T.T., Gellert M. The 3′ to 5′ exonuclease activity of Mre 11 facilitates repair of DNA double-strand breaks. Mol. Cell. 1998;1:969–979. doi: 10.1016/s1097-2765(00)80097-0. [DOI] [PubMed] [Google Scholar]
- 35.Huang S., Beresten S., Li B., Oshima J., Ellis N.A., Campisi J. Characterization of the human and mouse WRN 3′→5′ exonuclease. Nucleic Acids Res. 2000;28:2396–2405. doi: 10.1093/nar/28.12.2396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Guzder S.N., Torres-Ramos C., Johnson R.E., Haracska L., Prakash L., Prakash S. Requirement of yeast Rad1-Rad10 nuclease for the removal of 3′-blocked termini from DNA strand breaks induced by reactive oxygen species. Genes Dev. 2004;18:2283–2291. doi: 10.1101/gad.1232804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Moshous D., Callebaut I., de Chasseval R., Corneo B., Cavazzana-Calvo M., Le Deist F., Tezcan I., Sanal O., Bertrand Y., Philippe N., Fischer A., de Villartay J.P. Artemis, a novel DNA double-strand break repair/V(D)J recombination protein, is mutated in human severe combined immune deficiency. Cell. 2001;105:177–186. doi: 10.1016/s0092-8674(01)00309-9. [DOI] [PubMed] [Google Scholar]
- 38.Ma Y., Pannicke U., Schwarz K., Lieber M.R. Hairpin opening and overhang processing by an Artemis/DNA-dependent protein kinase complex in nonhomologous end joining and V(D)J recombination. Cell. 2002;108:781–794. doi: 10.1016/s0092-8674(02)00671-2. [DOI] [PubMed] [Google Scholar]
- 39.Bradbury J.M., Jackson S.P., Fernandez-Capetillo O., Celeste A., Nussenzweig A. The complex matter of DNA double-strand break detection Focusing on foci: H2AX and the recruitment of DNA-damage response factors. Cell Cycle. 2003;2:426–427. [PubMed] [Google Scholar]
- 40.Bradbury J.M., Jackson S.P. The complex matter of DNA double-strand break detection. Biochem. Soc. Trans. 2003;31:40–44. doi: 10.1042/bst0310040. [DOI] [PubMed] [Google Scholar]
- 41.Weinfeld M., Soderlind K.J. 32P-postlabeling detection of radiation-induced DNA damage: identification and estimation of thymine glycols and phosphoglycolate termini. Biochemistry. 1991;30:1091–1097. doi: 10.1021/bi00218a031. [DOI] [PubMed] [Google Scholar]
- 42.Bertoncini C.R., Meneghini R. DNA strand breaks produced by oxidative stress in mammalian cells exhibit 3′-phosphoglycolate termini. Nucleic Acids Res. 1995;23:2995–3002. doi: 10.1093/nar/23.15.2995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Durocher D., Jackson S.P. DNA-PK, ATM and ATR as sensors of DNA damage: variations on a theme? Curr. Opin. Cell Biol. 2001;13:225–231. doi: 10.1016/s0955-0674(00)00201-5. [DOI] [PubMed] [Google Scholar]
- 44.Shiloh Y. ATM and related protein kinases: safeguarding genome integrity. Nature Rev. Cancer. 2003;3:155–168. doi: 10.1038/nrc1011. [DOI] [PubMed] [Google Scholar]
- 45.Jeggo P.A. DNA-PK: at the cross-roads of biochemistry and genetics. Mutat. Res. 1997;384:1–14. doi: 10.1016/s0921-8777(97)00009-8. [DOI] [PubMed] [Google Scholar]
- 46.Pourquier P., Ueng L.M., Fertala J., Wang D., Park H.J., Essigmann J.M., Bjornsti M.A., Pommier Y. Induction of reversible complexes between eukaryotic DNA topoisomerase I and DNA-containing oxidative base damages. 7, 8-dihydro-8-oxoguanine and 5-hydroxycytosine. J. Biol. Chem. 1999;274:8516–8523. doi: 10.1074/jbc.274.13.8516. [DOI] [PubMed] [Google Scholar]
- 47.Hsiang Y.H., Lihou M.G., Liu L.F. Arrest of replication forks by drug-stabilized topoisomerase I-DNA cleavable complexes as a mechanism of cell killing by camptothecin. Cancer Res. 1989;49:5077–5082. [PubMed] [Google Scholar]
- 48.Barnes D.E., Stamp G., Rosewell I., Denzel A., Lindahl T. Targeted disruption of the gene encoding DNA ligase IV leads to lethality in embryonic mice. Curr. Biol. 1998;8:1395–1398. doi: 10.1016/s0960-9822(98)00021-9. [DOI] [PubMed] [Google Scholar]
- 49.Gao Y., Sun Y., Frank K.M., Dikkes P., Fujiwara Y., Seidl K.J., Sekiguchi J.M., Rathbun G.A., Swat W., Wang J., et al. A critical role for DNA end-joining proteins in both lymphogenesis and neurogenesis. Cell. 1998;95:891–902. doi: 10.1016/s0092-8674(00)81714-6. [DOI] [PubMed] [Google Scholar]
- 50.Karanjawala Z.E., Hsieh C.L., Lieber M.R. Overexpression of Cu/Zn superoxide dismutase is lethal for mice lacking double-strand break repair. DNA Repair. 2003;2:285–294. doi: 10.1016/s1568-7864(02)00218-5. [DOI] [PubMed] [Google Scholar]
- 51.Caldecott K.W. DNA single-strand break repair and spinocerebellar ataxia. Cell. 2003;112:7–10. doi: 10.1016/s0092-8674(02)01247-3. [DOI] [PubMed] [Google Scholar]
- 52.De Michele G., Di Salle F., Filla A., D'Alessio G., Ambrosio G., Viscardi L., Scala R., Campanella G. Magnetic resonance imaging in “typical” and “late onset” Friedreich's disease and early onset cerebellar ataxia with retained tendon reflexes. Ital. J. Neurol. Sci. 1995;16:303–308. doi: 10.1007/BF02249105. [DOI] [PubMed] [Google Scholar]
- 53.Pandolfo M. The molecular basis of Friedreich ataxia. Adv. Exp. Med. Biol. 2002;516:99–118. doi: 10.1007/978-1-4615-0117-6_5. [DOI] [PubMed] [Google Scholar]
- 54.Cooper J.M., Bradley J.L. Friedreich's ataxia. Int. Rev. Neurobiol. 2002;53:147–173. doi: 10.1016/s0074-7742(02)53006-3. [DOI] [PubMed] [Google Scholar]
- 55.Schulz J.B., Dehmer T., Schols L., Mende H., Hardt C., Vorgerd M., Burk K., Matson W., Dichgans J., Beal M.F., Bogdanov M.B. Oxidative stress in patients with Friedreich ataxia. Neurology. 2003;55:1719–1721. doi: 10.1212/wnl.55.11.1719. [DOI] [PubMed] [Google Scholar]
- 56.Lodi R., Rajagopalan B., Bradley J.L., Taylor D.J., Crilley J.G., Hart P.E., Blamire A.M., Manners D., Styles P., Schapira A.H., Cooper J.M. Mitochondrial dysfunction in Friedreich's ataxia: from pathogenesis to treatment perspectives. Free Radic. Res. 2002;36:461–466. doi: 10.1080/10715760290021324. [DOI] [PubMed] [Google Scholar]
- 57.Liu C., Pouliot J.J., Nash H.A. Repair of topoisomerase I covalent complexes in the absence of the tyrosyl-DNA phosphodiesterase Tdp1. Proc. Natl Acad. Sci. USA. 2002;99:14970–14975. doi: 10.1073/pnas.182557199. [DOI] [PMC free article] [PubMed] [Google Scholar]