Abstract
Eukaryotic chromatin structure limits the initiation of DNA replication spatially to chromosomal origin zones and temporally to the ordered firing of origins during S phase. Here, we show that the level of histone H4 acetylation correlates with the frequency of replication initiation as measured by the abundance of short nascent DNA strands within the human c-myc and lamin B2 origins, but less well with the frequency of initiation across the β-globin locus. Treatment of HeLa cells with trichostatin A (TSA) reversibly increased the acetylation level of histone H4 globally and at these initiation sites. At all three origins, TSA treatment transiently promoted a more dispersive pattern of initiations, decreasing the abundance of nascent DNA at previously preferred initiation sites while increasing the nascent strand abundance at lower frequency genomic initiation sites. When cells arrested in late G1 were released into TSA, they completed S phase more rapidly than untreated cells, possibly due to the earlier initiation from late-firing origins, as exemplified by the β-globin origin. Thus, TSA may modulate replication origin activity through its effects on chromatin structure, by changing the selection of initiation sites, and by advancing the time at which DNA synthesis can begin at some initiation sites.
INTRODUCTION
The packaging of eukaryotic genomes into nucleosomes and higher order chromatin structures limits the access of replication factors to DNA. During the development of Xenopus and Drosophila embryos, correlated with the onset of zygotic transcription and global chromatin remodeling, there are dramatic changes in replication origin usage from apparently random initiations to more specific zones of initiation (1–5). The effect of chromosomal position on origin activity in Drosophila (6) and the observation that euchromatic domains of metazoan genomes generally replicate early in S phase whereas heterochromatic regions replicate late in S phase (7,8) argue for a regulatory role of chromatin structure in DNA replication.
The reversible acetylation of the N-terminal tails of histones is a prominent chromatin modification that is thought to alter the degree of chromatin compaction (9). A role for reversible histone acetylation in transcriptional regulation is well recognized, such that in general HAT binding and acetylation of histones are correlated with gene activation while HDAC binding and deacetylation are associated with repression (10). Several chromatin remodeling activities [FACT, CHRAC, histone acetyltransferases (HATs), histone deacetylases (HDACs)] have also been implicated in modulating DNA replication origin activity (11–15). Supporting a role for histone acetylation in the initiation phase of DNA replication, the pre-replication complex (pre-RC) components ORC1 and MCM2 have been shown to interact with the HAT HBO1 (11,14). Furthermore Cdc45, which associates with origins during initiation, co-purifies with the HDAC Rpd3 in budding yeast (16). Deletion of Rpd3 from the budding yeast genome allows many late-firing replication origins to initiate DNA synthesis earlier in S phase (17,18), and treatment of human cells with the HDAC inhibitor trichostatin A (TSA) causes earlier replication in S phase of normally late-replicating imprinted genes (19).
The initiation of DNA replication in eukaryotes is regulated spatially along chromosomes at sites known as replication origins (20–23) and temporally during S phase through the coordinated activation of individual replicons (24–26). In contrast to the simple model whereby DNA replication initiation occurs at sites precisely marked by the origin recognition complex (ORC) in the yeast Saccharomyces cerevisiae (27), DNA synthesis in higher eukaryotes often begins at a number of potential sites within a broad domain termed an initiation zone (21,28). Examples of initiation zones include those found at the hamster DHFR (29–32) and rhodopsin (33) loci, the c-myc locus in human (34–37), chicken and mouse cells (38), and the β-globin locus in human (39–41), mouse (42) and chicken (43) cells. Within a population of cells, there are sites within these loci that are used for replication initiation with greater frequency than other sites; however, it is currently unclear how this initiation site preference is determined. Although there is conservation of ORC and other replication initiation proteins from yeast to man, it has been difficult to identify sequence elements within mammalian replication origins that specify ORC binding (44,45). For example, it has been suggested that the initiation events spread over tens of kilobases at the hamster DHFR locus do not depend on unique sequences (46), but result from stochastic initiation at widely distributed complexes of MCM proteins (47), the putative replicative helicase (48–50).
In the present work, we examined the acetylation level of histone H4 at previously described initiation sites within the β-globin, lamin B2 and c-myc replication origin loci in human cells and used the HDAC inhibitor TSA to investigate if changes in histone acetylation levels at these sites correlate with changes in replication activity. We find that the pattern of initiation site selection within replication origin loci in human cells is altered upon treatment with TSA, becoming more dispersive. Thus, preferred initiation sites become less dominant while infrequently used initiation sites in the genome become more active after treatment with TSA. We show also that the β-globin origin can be induced to initiate DNA synthesis earlier in S phase after treatment with TSA. The results indicate that TSA can affect both the choice of initiation sites used within replication origin loci and globally throughout the genome as well as the time during S phase at which origins begin DNA synthesis.
MATERIALS AND METHODS
Cell culture and drug treatments
HeLa cells were maintained in DMEM with 10% newborn calf serum (Gibco) and 50 μg of gentamicin/ml in a humidified 5% CO2 atmosphere at 37°C. Synchronization of cells in late G1 phase was achieved by treating subconfluent HeLa cells with 200 μM mimosine (Sigma) for 24 h. Alternatively, cells were blocked at the G1/S boundary by addition of 1 μg/ml aphidicolin for 24 h. Treatment of cells with the histone deacetylase inhibitor trichostatin A (TSA; Sigma) was at a final concentration of 100 or 300 ng/ml, as indicated.
Flow cytometry
For DNA content analyses, 106 cells were trypsinized, washed in PBS and fixed overnight at −20°C in 70% ethanol. Cells were rehydrated in phosphate-buffered saline (PBS), treated with 100 U of RNase for 20 min at 37°C to digest RNA, and stained with 50 μg of propidium iodide/ml (Sigma). DNA content was analyzed on a Becton Dickenson FACScan flow cytometer. Measurement of nucleotide incorporation involved incubating cells in the presence of 50 μM bromodeoxyuridine (BrdU) for 1 h prior to trypsinization and overnight fixation in 70% ethanol. Cells were washed in PBS and stained with Alexa Fluor 488-conjugated anti-BrdU antibody (Molecular Probes) according to the manufacturer's recommendation. The percentage of cells incorporating BrdU was measured on a Becton Dickenson FACScan flow cytometer.
Western blotting
Whole-cell lysates were prepared from TSA-treated or untreated HeLa cells and examined by SDS–PAGE and western blotting with an anti-actin antibody (Upstate Biotechnology) or an antibody against acetylated-histone H4 (Upstate Biotechnology), which recognizes histone H4 acetylated at lysines 5, 8, 12 or 16.
Chromatin immunoprecipitation (ChIP)
TSA-treated or untreated cells were cross-linked according to the protocol of Ritzi et al. (51) with the following modifications. Cross-linked chromatin was resuspended in TE buffer (10 mM Tris–HCl, pH 7.4, 1 mM EDTA) and sonicated (Branson sonifier cell disrupter 200, 50% duty cycle, 10 s pulses, 7 pulses with 1 min intervals) on ice. The chromatin was digested with 0.1 U micrococcal nuclease (Sigma) per 100 μg of nucleoprotein at 37°C for 5 min to yield fragments <500 bp. Nucleoprotein (250 μg) for each chromatin immunoprecipitation was diluted with 11× NET (550 mM Tris–HCl, pH 7.4, 1.65 M NaCl, 5.5 mM EDTA, 5.5% NP-40) to a final concentration of 1× NET. Acetylated-histone H4 antibody or normal rabbit antiserum (Upstate Biotech) was used for immunoprecipitation. Washing of the antibody complex and purification of co-precipitated DNA was carried out according to Schepers et al. (52).
Nascent DNA
Nascent DNA was isolated by denaturing gel electrophoresis (39,53). Briefly, TSA-treated or untreated cells were trypsinized and washed twice in ice-cold PBS, pelleted by centrifugation, and resuspended in a small volume of ice-cold PBS with 10% glycerol. The cell suspensions were loaded into wells of a 1.25% alkaline low-melting point agarose gel (SeaPlaque GTG) prechilled to 4°C. Cells were lysed in the well for 10 min before the DNA marker was loaded and the current applied. The gel was run for 12 h at 40 V, neutralized in 1× Tris–acetate–EDTA (TAE) buffer and stained with ethidium bromide. The 1–2 kb nascent DNA size fraction was excised and purified (Qiagen). The total amount of 1–2 kb nascent DNA isolated was quantitated by OliGreen fluorescence (Molecular Probes) and ranged from 18 to 25 fg per cell in different preparations, consistent with previous determinations (53). This method eliminates all handling of DNA prior to size fractionation and avoids the variability inherent in other methods of nascent DNA isolation. Nascent strand abundances are normalized per nanogram of total nascent DNA, determined by OliGreen staining (Molecular Probes), inasmuch as an internal control STS whose replication is unaffected by TSA is not known. As reported by others (54), this normalization leads to somewhat higher SD values than we have reported previously (53,55). Nevertheless, SD values are typically <15–20% of the mean. Since the 95% prediction interval is <40% of the indicated mean, we consider decreases in nascent strand abundance of ≥40% to be significant.
Quantitative PCR
Short (1–2 kb) nascent DNA and chromatin immunoprecipitated DNA was measured by quantitative real-time PCR (Q-PCR) on either an Applied Biosystems GeneAmp 5700 or Prism 7000 sequence detection system with amplification monitored by SYBR Green fluorescence as described previously (55). Copy numbers were determined by comparison to standard curves generated for each primer set by amplification of sheared, genomic DNA. The nascent strand abundance measurements (copies/ng nascent DNA) are presented as the means (and SD values) of triplicate analysis on at least three independent preparations of nascent DNA. The abundance of DNA precipitated by either anti-AcH4 or normal rabbit serum was determined in duplicate or triplicate Q-PCRs.
PCR primers
The primer sequences defining each sequence-tagged site (STS) are available upon request. The position of each STS in the corresponding GenBank sequence is as follows: STS-myc1 (3896–3964, AF176208), STS-myc2 (1829–1891, HUMMYCC), STS-myc3 (4488–4552, HUMMYCC), STS-myc4 (7866–7946, HUMMYCC), STS-L1 (2908–2995, M94363), STS-L2 (4039–4124, M94363), STS-L3 (4821–4896, M94363), STS-BG1 (33029–33107, U01317), STS-BG2 (41168–41250, U01317), STS-BG3 (54653–54728, U01317) and STS-BG4 (62073–62147, U01317). The sequences of the 10mer primers employed in Figure 8 were 5′-GTGCAATGAG-3′ and 5′-GGAAGACAAC-3′, with 45 PCR cycles at 94°C for 30 s, 35°C for 60 s and 72°C for 2 min.
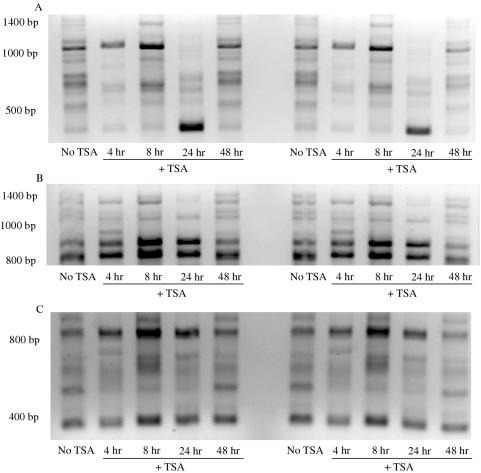
Figure 8.
TSA alters nascent strand abundance at uncharacterized sites. Nascent DNA isolated from cells treated with 100 ng/ml TSA for 4 h (A and B) or 8 h (C) was amplified by PCR with different 10mer primers using equal starting amounts of DNA. Products were purified, run on 2% agarose gels and stained with EtBr (inverse image). Samples from independent PCRs are shown to confirm the reproducibility of the assay.
RESULTS
Histone acetylation and initiation activity at replication origins
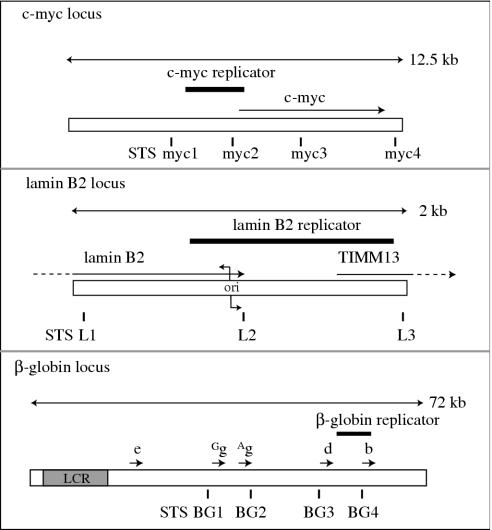
There has been only limited data directly implicating histone acetylation in controlling the initiation of DNA replication in human cells. Therefore, we first examined whether initiation site selection at previously described replication origins is correlated with the acetylation level of histones at these sites. The human β-globin, lamin B2 and c-myc loci contain regions characterized as bona fide genetic replicators (54–56), and maps of these origin loci are shown in Figure 1 with the locations of sequence-tagged sites (STSs) defined by specific primer sequences. The same primer sets were used to test for both the acetylation level of histone H4 and for replication initiation activity (nascent strand abundance). Chromatin immunoprecipitation was used to enrich for DNA sequences bound to acetylated histone H4, followed by analysis using quantitative real-time PCR (Q-PCR). Origin activity was determined with a Q-PCR-based nascent strand abundance assay using DNA 1–2 kb in size isolated by alkaline gel electrophoresis. DNA of this size fraction has been shown to comprise new, semiconservatively replicated DNA derived from replication origins and excludes Okazaki fragments (36,57,58). Replication initiation activity at many of these sites has been described previously using PCR-based assays and by other methods (34–36,39–41,43,56,59). The distances between adjacent sequence-tagged sites within the c-myc or β-globin loci range from 3 to 14 kb and hence reflect separate initiation events. The sequence-tagged sites at the lamin B2 locus span a region of only 2 kb that is centered at STS-L2, the location of which is ∼110 bp downstream of the transition point of bidirectional replication (ori) previously mapped at the nucleotide level (56,59). The STS-L1 and STS-L3 primers are located 1000 bp upstream and downstream of STS-L2, respectively, and thus may report on initiations at STS-L2 as well as at flanking sites.
Figure 1.
Maps of human replication origin loci. The human c-myc, lamin B2 and β-globin loci are labeled with the locations of sequence-tagged sites (STSs) analyzed by quantitative real time PCR (Q-PCR) in this work. Single-headed arrows above the open bars represent the indicated genes. Regions comprising the c-myc, β-globin and lamin B2 replicator elements are shown as solid boxes. LCR, β-globin locus control region. Bent arrows, lamin B2 replication initiation site.
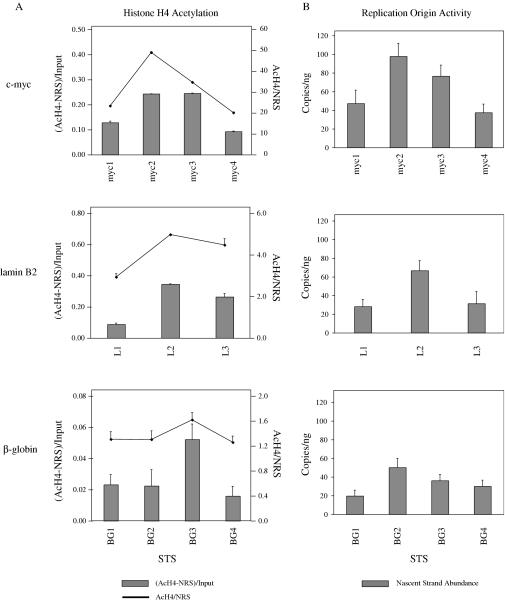
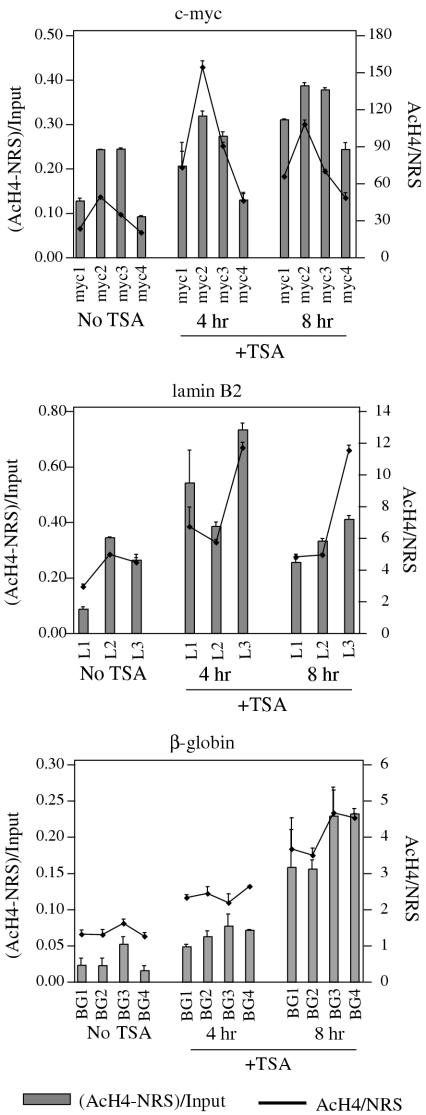
Comparison of the amount of cross-linked DNA immunoprecipitated with an antibody against acetylated-histone H4 (AcH4) to that precipitated by normal rabbit sera (NRS) yields an enrichment factor that can be used to define the relative acetylation level of histones at the corresponding STS. The level of histone acetylation was thus examined at several sites spanning the human β-globin, lamin B2 and c-myc replication origin loci (Figure 2A) and is displayed in two formats [(AcH4-NRS)/Input and AcH4/NRS] that show similar trends in AcH4 levels within each locus (Figure 2A). Consistent with the presence of the β-globin locus in heterochromatin, the AcH4 enrichment values [(AcH4-NRS)/Input] at β-globin sites were low (maximum 0.05), whereas the c-myc (maximum 0.25) and lamin B2 (maximum 0.35) loci showed higher AcH4 values, as expected of early replicating, transcribed chromatin.
Figure 2.
Relative levels of (A) histone H4 acetylation and (B) initiation activity at sites spanning human replication origin loci. The level of histone H4 acetylation was determined by chromatin immunoprecipitation with an anti-acetylated-histone H4 (AcH4) antibody and Q-PCR. The data are calculated and presented in two formats. The first takes the difference between DNA precipitated by anti-AcH4 antibody and normal rabbit serum as a fraction of total input cross-linked DNA [(AcH4-NRS)/Input)]. The second method is simply a ratio of the amount of DNA precipitated by anti-AcH4 antibody to that precipitated by normal rabbit sera (AcH4/NRS). Replication activity was determined by nascent strand abundance assay and is presented as an average (and SD) from at least three independent nascent DNA isolations analyzed by Q-PCR in triplicate.
Replication origin activity (nascent strand abundance) at each STS is represented as copies per nanogram of 1–2 kb nascent DNA (Figure 2B). The c-myc locus showed a histone acetylation pattern that correlated well with initiation efficiency, with STS-myc2 showing both the highest AcH4 level and the greatest nascent strand abundance. STS-myc2 lies within a 2.4 kb fragment of the c-myc locus shown to possess replicator activity when placed at an ectopic site in the HeLa genome (35,53,55). Furthermore, ChIP analysis reveals that the 2.4 kb c-myc replicator contains preferential binding sites for ORC, Cdc6 and MCM proteins that neighbor, but are offset from sites with the highest levels of acetylated histone H4 [(60), M. Ghosh and M. Leffak, unpublished data]. Similarly the lamin B2 locus shows its highest AcH4 level at STS-L2, close to the transition point for bidirectional DNA replication initiation (59) and the site of ORC binding (61). In contrast, the late-replicating β-globin locus does not show a correlation between basal AcH4 level and replication origin activity, with STS-BG3 showing the highest AcH4 level but STS-BG2 displaying the greatest nascent strand abundance, consistent with previous reports (39–41,62). Conversely STS-BG4, which corresponds to the characterized replicator element (54,63), did not show the highest relative AcH4 level among the sites examined in the β-globin locus. While these data suggest that the elevated histone acetylation at the early firing c-myc and lamin B2 initiation sites may be related to activation of initiation complexes at sites of DNA synthesis, as has been suggested in yeast (17,18), it is nonetheless unlikely to be the sole determinant for pre-RC binding, based on observations that multiple cis-acting elements contribute to replicator function in mammalian cells (54–56,64). In S.cerevisiae, the initiation of replication occurs directly at sites where ORC is bound, and although ORC is conserved in higher eukaryotes (65), the demonstration that replication can initiate at multiple sites spanning large domains in mammalian cells suggests that other factors may select specific sites of initiation within a zone determined by ORC.
TSA treatment increases bulk histone acetylation and alters cell cycle progression
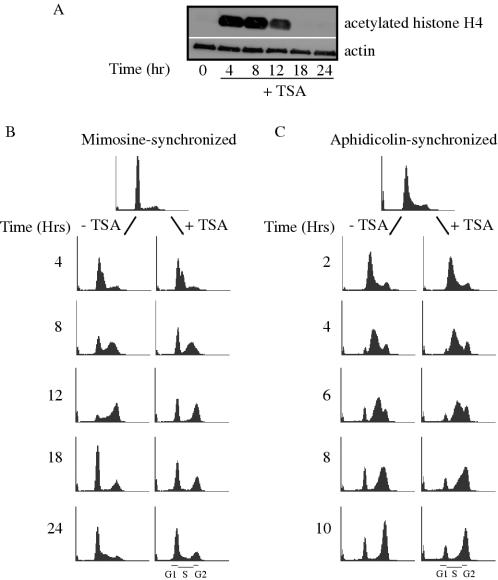
To investigate the role of histone acetylation in the initiation of DNA replication further, we made use of trichostatin A, a reversible inhibitor of histone deacetylases that has been shown to increase the acetylation level of histones within the mouse β-globin locus (66), and to affect the replication timing of imprinted genes in human cells (19). To confirm that TSA is able to increase the acetylation level of histone H4 in HeLa cells, logarithmically growing cells were treated with 100 ng/ml TSA for 4–24 h and harvested for protein analysis by western blotting. As can be observed in Figure 3A, TSA treatment greatly increased the acetylation level of histone H4 between 4 and 12 h of treatment. The AcH4 level returned to background by 18 h after the addition of TSA, consistent with breakdown of the drug, since either higher doses or additional TSA treatments could maintain this and other effects for a longer period of time (data not shown).
Figure 3.
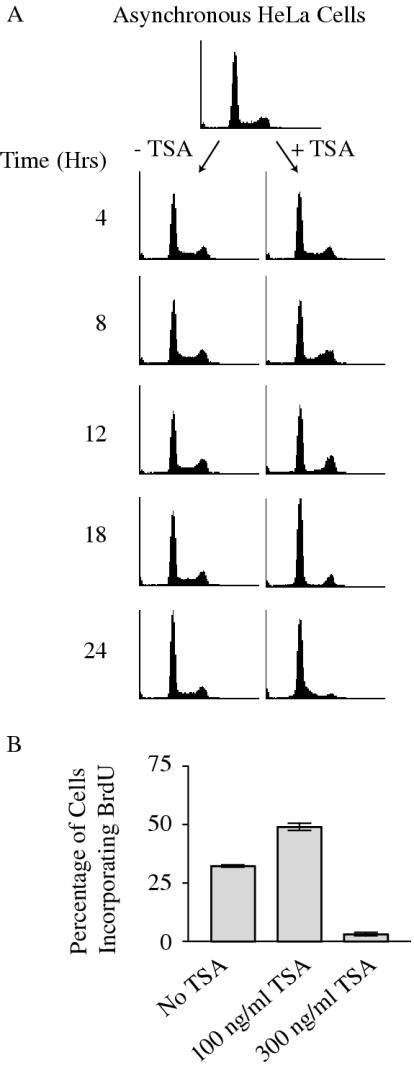
Single dose TSA treatment induces hyperacetylation of bulk histones. (A) HeLa cells treated with 100 ng/ml TSA for the indicated number of hours were lysed and analyzed by SDS–PAGE and western blotting. (B) HeLa cells were synchronized for 18 h with 0.2 mM mimosine before release into fresh medium in the absence or presence of 100 ng/ml TSA. Cells were harvested for analysis by flow cytometry. (C) HeLa cells were synchronized with 1 μg/ml aphidicolin for 24 h before release into fresh medium in the absence or presence of 100 ng/ml TSA.
Deletion of the Rpd3 histone deacetylase gene from the S.cerevisiae genome advances the timing of initiation from late-firing replication origins (17,18) and promoted quicker completion of DNA synthesis in rpd3Δ (17) or rpd3Δ, clb5Δ cells (17,18). To test whether a similar scenario exists in human cells, we synchronized HeLa cells with mimosine, a plant amino acid that arrests cells in late G1 phase before the establishment of active DNA replication forks (67–70). Mimosine-synchronized cells were released from the block into medium containing or lacking TSA. At various times after release, cells were harvested for examination of DNA content by flow cytometry. Similar to the effect of Rpd3 deletion in yeast (17), inhibition of HDACs with TSA in human cells promoted the progression of cells through S phase, such that by 12 h after release <30% of cells treated with TSA remained in S phase, whereas >80% of untreated cells remained in S phase (Figure 3B). That the cells completing S phase more rapidly indeed received TSA is supported by the observation that these cells arrested at G2/M and took longer to re-populate G1 than untreated cells, consistent with previous observations (66,71,72). In contrast to cells released from a mimosine block, cells released from an aphidicolin block into TSA did not complete S phase more quickly than control cells (Figure 3C). Since aphidicolin arrests cells after the establishment of replication forks, these data imply that TSA exerts an effect on replication late in G1 phase or early in S phase, at or near the time that replication complexes are activated.
TSA is known to cause a wide range of cellular effects, including cell cycle arrest, differentiation and cell death (71). We used flow cytometry to examine the DNA content of an asynchronous population of HeLa cells treated with 100 ng/ml TSA. Consistent with previous reports using higher drug doses (73,74), 100 ng/ml TSA arrested cells at both the G1 and G2 phases of the cell cycle, most notably after 18 h of treatment (Figure 4A). There did not appear to be an S-phase checkpoint induced, as cells that were in the early stages of S phase during the addition of TSA continued to replicate DNA, including from the late-firing β-globin origin (see below), and instead arrested at G2/M. Consistent with the transient hyperacetylation of histone H4 in bulk chromatin (Figure 3), by 24 h after TSA addition, cells successfully moved through the G2 block and mitosis and had begun to enter another S phase. These cells were actively synthesizing DNA as determined by the incorporation of the nucleotide analog BrdU (Figure 4B). Furthermore, addition of higher concentrations of TSA, or re-addition of TSA at later time points blocked both the recovery from the G2 arrest and subsequent ability to incorporate nucleotides (Figure 4B; data not shown), consistent with the ability of cells to degrade the drug.
Figure 4.
Low-dose TSA treatment causes cell cycle blocks. (A) Asynchronously growing HeLa cells were left untreated or were treated with 100 ng/ml TSA for the indicated number of hours and analyzed for DNA content by flow cytometry. (B) Asynchronously growing control cells (untreated) or treated with 100 ng/ml or 300 ng/ml TSA for 24 h were labeled with 50 μM BrdU for 1 h and analyzed for BrdU incorporation by flow cytometry.
TSA increases the acetylation level of histones at replication origins
The acetylation level of histone H4 in bulk chromatin is increased after treatment of cells with TSA (Figure 3). We were interested to determine whether increased acetylation also occurs to histones within replication origin loci, and used ChIP to examine the abundance of AcH4 at each replication origin STS in untreated HeLa cells or in cells treated with TSA (Figure 5). Consistent with the effect of TSA on bulk histones, increased AcH4 levels were observed at virtually every site examined, although differences were observed in the magnitude and timing of the change. For example, the largest changes observed at the c-myc locus were <3-fold, while increases as large as 14-fold were seen at the β-globin locus, possibly due to the higher basal AcH4 level at the c-myc locus and the heterochromatic nature of the β-globin domain. The largest increases in AcH4 levels at the β-globin locus occurred only upon longer treatment with TSA, consistent with the role of HDACs in forming the repressive chromatin structure found at late-replicating loci and the role of HATs, which are expected to be essential to replicate through repressive chromatin (75,76), but may be recruited to such sites only later during S phase. Thus, the enrichment for cells in the later stages of S phase 8 h after treatment with TSA (Figure 4A) may account for the coordinate appearance of AcH4 at the β-globin origin. Of interest is the observation that the greatest increase in histone H4 acetylation occurs at BG4, which corresponds to the genetically characterized β-globin replicator (63).
Figure 5.
TSA treatment causes hyperacetylation of histones at replication origin loci in human cells. ChIP was used to enrich for DNA sequences bound to chromatin with acetylated-histone H4 from either untreated HeLa cells or cells treated with 100 ng/ml TSA. Isolated DNA was quantitated in Q-PCR and AcH4 level was calculated as [(AcH4-NRS)/Input] and (AcH4/NRS).
TSA treatment alters replication origin activity
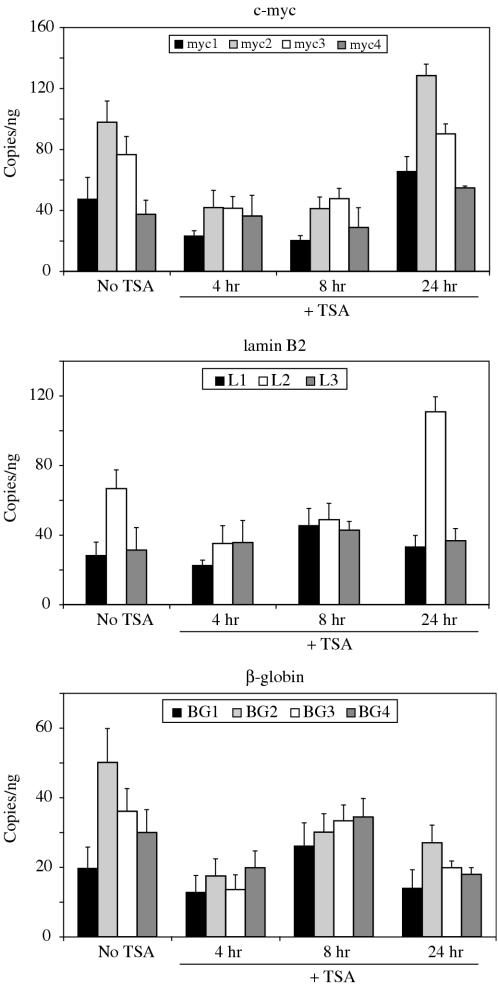
Since TSA treatment tended to even out the level of histone acetylation across each replicator locus, we wished to determine whether TSA treatment had a similar effect on replication initiation activity. Therefore, we used nascent strand abundance assays to investigate whether initiations at sequence tagged sites within origin loci changed upon treatment of cells with TSA. The data are displayed in Figure 6 as the nascent strand copy number at each STS per nanogram of nascent DNA. Treatment with 100 ng/ml TSA for 4 or 8 h led to both qualitative and quantitative changes in the pattern of initiations within the c-myc, lamin B2 and β-globin origins. This treatment decreased the abundance of 1–2 kb nascent strands at the most prominent sites within the c-myc locus (Figure 6). Interestingly, STS-myc2, which shows the highest level of nascent strands under normal conditions, appeared to be most affected by TSA treatment, with its initiation site usage decreasing to a greater extent than that for other sites within the locus. Sites located downstream of the replicator, including STS-myc3 and STS-myc4 were therefore used with greater frequency relative to STS-myc2 after treatment with TSA.
Figure 6.
TSA treatment alters the pattern of replication origin activity in HeLa cells. Asynchronously growing HeLa cells were treated for 4, 8 or 24 h with 100 ng/ml TSA and loaded on alkaline gels for isolation of 12 kb nascent DNA. Purified nascent DNA was quantitated by OliGreen fluorescence and quantitated by real-time PCR. The data presented are the average (and SD values) of triplicate analysis on at least three independent preparations of nascent DNA.
Replication initiation at the lamin B2 locus appeared to be less narrowly focused and to occur over a broader region after treatment with TSA, such that STS-L2 no longer showed the 2- to 3-fold higher nascent strand abundance compared to STS-L1 and STS-L3 (Figure 6). The delocalized initiation at the lamin B2 locus correlated with the greater increases in AcH4 levels at sites flanking STS-L2. At the β-globin locus, we further observed a change in the pattern of replication initiation (Figure 6), such that STS-BG2 was no longer used preferentially after treatment with TSA for 4 or 8 h. Although, the activity at each STS within the β-globin locus decreased after 4 h of TSA treatment relative to untreated cells, the nascent strand abundance at each STS increased during the next 4 h (8 h time point), consistent with a higher proportion of cells in the later stages of S phase at that time (Figure 4A). After 24 h of TSA treatment, a time by which AcH4 levels returned to background and cells recovered from the G2 arrest, the relative nascent strand abundance returned to pretreatment levels, such that at the c-myc locus, we observed STS-myc2 with the greatest abundance, followed by STS-myc3, STS-myc1 and STS-myc4 (Figure 6). The relative distributions of nascent strand abundance were also similar to pretreatment levels at the lamin B2 and β-globin loci. Differences in the absolute nascent strand abundances between control cells and cells 24 h post-TSA treatment may be explained by the enrichment of cells in very early S phase 24 h after TSA treatment (Figure 4).
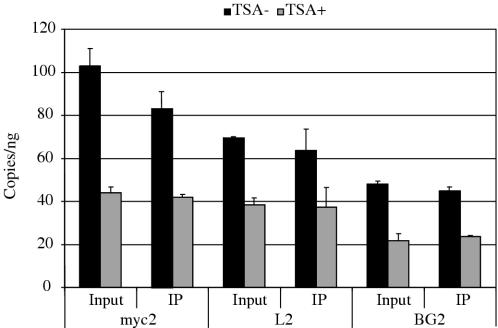
To confirm that it was newly synthesized DNA whose abundance was changed by TSA treatment, control or TSA treated cells were pulse-labeled with BrUdR and the putative short nascent strands isolated by alkaline electrophoresis were immunoprecipitated with anti-BrUdR antibody. The abundance of c-myc, lamin B2 and β-globin sequences in the input and immunoprecipitated DNA were compared after Q-PCR (Figure 7). The results showed that 80–100% of the input DNA at each STS comprised nascent DNA that could be immunoprecipitated by the anti-BrUdR antibody. The BrUdR-labeled DNA also recapitulated the TSA-induced decreased nascent strand abundance at the c-myc (myc2), lamin B2 (L2) and β-globin (BG2) sites.
Figure 7.
TSA decreases the abundance of BrUdR labeled nascent strands at primary initiation sites. Cells were untreated or treated with 100 ng/ml TSA for 4 h prior to a 30 min BrUdR pulse. Short DNA fragments (1–2 kb) were isolated by alkaline electrophoresis and the abundance of sequences at STS-myc2, -L2 and -BG2 was quantitated by Q-PCR in aliquots of the total short DNA fraction (input) or the DNA immunoprecipitated by anti-BrUdR antibody.
TSA increases nascent strand abundance at uncharacterized sites
The absolute decreases in nascent strand abundance per nanogram of nascent DNA at most sites after 4 h of TSA treatment suggested that other previously inefficient sites of initiation were used more frequently after treatment with TSA. To test this idea, short 10mer primers designed for random amplification of genomic DNA were used to amplify anonymous sites in nascent DNA harvested from either untreated cells or cells treated with TSA. These primers generated a set of amplification products 400–1500 bp in size that could be visualized when electrophoresed through agarose and stained with ethidium bromide (Figure 8). Comparing the intensity of the PCR products from reactions using equivalent amounts of template nascent DNA from either TSA-treated or untreated cells revealed amplified products that increased, decreased or did not change in abundance upon treatment with TSA. These observations were seen with different 10mer primers (Figure 8A and B) and after either 4 h (Figure 8A and B) or 8 h (Figure 8C) of TSA treatment. The uncharacterized sites that increase in abundance upon treatment with TSA may compensate for the decreases observed at STSs assayed in Figure 6, and suggests that TSA treatment may lead to a less specific pattern of initiations throughout the genome.
TSA causes earlier initiation at a late-firing origin
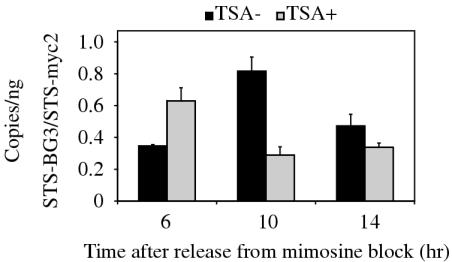
To investigate whether the time of initiation from a late-firing replication origin could be altered by TSA, we examined the nascent strand abundance at STS-BG3 which consistently showed the highest level of AcH4 in the β-globin locus. Nascent DNA was isolated from mimosine-synchronized cells released into medium containing or lacking TSA, and amplified by Q-PCR as in Figure 6 to determine nascent strand abundance. In cells lacking TSA, we observed maximal abundance at the β-globin STS-BG3 at 10 h after release from mimosine arrest (Figure 9), consistent with the location of the majority of cells in late S phase at that time (Figure 3b). In contrast, cells treated with TSA showed maximal abundance at STS-BG3 only 6 h after release from mimosine, suggesting accelerated firing of this origin in cells which completed S phase more rapidly. We observed maximal abundance of the early replicating STS-myc2 after 3 h of release from mimosine in both TSA-treated and untreated cells (data not shown), although the magnitude of the abundance for STS-myc2 and STS-BG3 was decreased in TSA-treated cells (Figure 9), consistent with previous observations (Figure 6). Similar selective acceleration of late origin firing was observed in rpd3Δ yeast mutants (18).
Figure 9.
Cells treated with TSA initiate synthesis from the late-firing β-globin origin earlier in S phase. Mimosine-synchronized cells were released into fresh medium or medium containing 100 ng/ml TSA for the indicated number of hours and loaded on alkaline gels for isolation of 1–2 kb nascent DNA. The nascent DNA was analyzed by Q-PCR in triplicate to determine the abundance of nascent strands at STS-myc2 and STS-BG3. The data are from two independent nascent DNA preparations and are normalized to STS-myc2.
DISCUSSION
Histone acetylation at replication initiation sites
To test the relationship between replication initiation and chromatin structure, we used a nascent strand abundance assay and chromatin immunoprecipitation with anti-acetylhistone H4 antibody, respectively, to compare the replication origin activity and levels of chromatin acetylation at the human β-globin, lamin B2 and c-myc loci. The level of histone H4 acetylation at initiation sites within the early replicating c-myc and lamin B2 loci correlated with the abundance of 1–2 kb nascent DNA strands at these origins, and the site within the lamin B2 and c-myc loci that showed the highest basal AcH4 level coincided with sites of known pre-RC formation or replicator activity (53,55,59,61,63). These data are consistent with models in which histone acetylation either contributes to, or results from, an open chromatin structure that facilitates replication initiation. Nevertheless, there are clearly factors other than the basal level of chromosomal protein acetylation that determine origin activity, since the HeLa β-globin locus did not show the same correlation between the basal AcH4 level and initiation pattern. In contrast, in the present work TSA treatment resulted in a high degree of AcH4 at STS-BG4 in the genetically characterized β-globin replicator (63). A recent examination of the chicken β-globin locus further suggested that there may exist a hierarchy of epigenetic states relevant to replication initiation and that even within the same locus different initiation sites may not show the same chromatin modifications (43). Thus, in erythroid precursor cells where the β-globin genes are not expressed, the β-globin domain was nonetheless in a decondensed state and replicated early in S phase. The relative nascent strand abundance at three initiation sites within CpG-enriched regions exhibited an inverse correlation with the extent of CpG methylation, and hyperacetylated histone H3 was found at only one of the three initiation sites.
HDAC inhibition as a modifier of chromatin structure
Increasing the acetylation level of histones is expected to loosen chromatin structure, making the DNA more accessible to protein factors (9). To test whether acetylation contributes to replication site selection, we treated HeLa cells with the HDAC inhibitor TSA to increase the acetylation level of histone H4 both at the bulk chromatin level and at specific chromosomal sites within replication initiation zones. The extent of the increase in AcH4 level varied among the sites investigated, suggesting that there may be differences in HDAC or HAT localization or activity among the sites, as has been proposed previously (66,77). Targeting of HDACs to the late-replicating heterochromatic β-globin locus, or delayed recruitment of HATs may be responsible for the increase in AcH4 levels only after longer periods of TSA treatment, at a time when the majority of cells were later in S phase, consistent with a link between HAT recruitment and DNA replication.
Overall, the TSA-dependent increase in AcH4 varied inversely with the basal level of acetylated histone H4, such that the highly acetylated c-myc locus showed the smallest relative changes while the heterochromatic β-globin locus showed the greatest changes. Within the early replicating c-myc and lamin B2 loci sites flanking the primary initiation sites showed the greatest relative increases in AcH4 with TSA treatment, supporting the view that differential HDAC accessibility may restrict origin activation.
TSA treatment led to decreased abundance of short nascent strands at the primary initiation sites in the early replicating c-myc (myc1, myc2), and lamin B2 (L2) loci. However, there were no consistent increases or decreases at the secondary sites examined (myc1, myc4, L1, L3). A unifying, though speculative model is that chromatin acetylation induces a broadening of the zone over which multiple initiations can occur and the activation of cryptic origins, consistent with the increased nascent strand abundance at several anonymous genomic sites (Figure 8). Despite the earlier replication of the β-globin locus in the presence of TSA, some of these cryptic initiations may fire before those in the β-globin locus leading to its decreased nascent strand abundance per nanogram of nascent DNA early during TSA treatment. Due to the very limited knowledge regarding sites of pre-RC assembly in human cells, it is currently unclear whether TSA plays a role in regulating pre-RC formation or whether its primary target is instead pre-RC activation. In S.cerevisiae, deletion of the HDAC Rpd3 allows the recruitment of Cdc45 and polymerase ε to late-firing origins earlier in S phase (17,18). Binding of these factors define activated origins, suggesting that the primary effect of the Rpd3 HDAC in yeast is not at the level of pre-RC formation.
We cannot rule out that TSA is indirectly influencing initiation by acting on targets other than histones (78) or that changes in nascent strand abundance are not due to alterations in the rate of nascent strand maturation. Supporting the view that nascent strand abundance reflects the efficiency of initiation, we note the changes in signals at the relatively closely spaced lamin B2 and c-myc STSs, and the observation that TSA stimulates only a subset of initiation sites and accelerates the replication of the β-globin locus relative to that of the c-myc locus. Therefore, some structural feature distinguishes initiation sites that are stimulated in response to HDAC inhibition from those that are not. A similar conclusion could be drawn from the data of Pappas et al. who found that elimination of SIR2 HDAC activity differentially restored origin activity within a panel of ARS elements in a S.cerevisiae cdc6-4 mutant temperature-sensitive for pre-RC formation (79), and of Aparicio et al. (18) who showed that the increased abundance of short nascent strands early in S phase in rpd3Δ yeast was due to premature initiation from late firing origins.
Replication origin activity and timing
TSA significantly decreased the abundance of short, 1–2 kb nascent DNA strands at all of the most frequently used initiation sites of the β-globin, lamin B2 and c-myc origins. The decreased nascent strand abundances are not likely due to formation of double-strand breaks that could otherwise alter the pool of 1–2 kb nascent DNA, since although high concentrations of TSA can activate the ATM kinase, this activation is not mediated by double-strand break formation (80). Consistent with this interpretation, flow cytometry detected no increase in apoptotic (‘sub-G1’) cells after TSA treatment, nor did cells arrest in S phase. Moreover, BrUdR incorporation showed that >80% of the short DNAs isolated by alkaline electrophoresis comprised bona fide nascent DNAs.
Although the earlier initiation of late-firing replication origins (Figure 9) may have contributed to the ability of TSA-treated cells to progress through S phase (Figure 3B), it may not be the only factor responsible for this observation. The experiment of Figure 3C, wherein cells were synchronized with aphidicolin and released into medium lacking or containing TSA, addresses the question of whether faster replication fork progression or nascent strand maturation rate contributed to this phenomenon. Aphidicolin inhibits the replicative DNA polymerases and synchronizes cells at the G1/S transition (81), after the establishment of replication forks and replication bubbles. We observed no measurable difference in S phase progression or completion between untreated and TSA-treated cells released from aphidicolin arrest. Aphidicolin has been reported to synchronize cells at a point in the cell cycle ∼2 h after that of mimosine (70), suggesting that the ability of cells released from mimosine into TSA to complete S phase more rapidly may be related to the rate of activation of replication complexes, or the activation of additional replication origins since mimosine arrests cells prior to the establishment of replication forks (67–70). Consistent with the latter view, quantitation of total 1–2 kb nascent DNA strands from cells released from a mimosine block into medium lacking or containing TSA for 3, 6 or 10 h showed no significant difference (data not shown), although there are clearly fewer TSA-treated cells that enter S phase (Figure 3B). Thus the initiation of replication at additional origins probably contributes to faster S phase completion.
The observed decreases in nascent strand abundances at the c-myc, lamin B2 and β-globin sites per nanogram of nascent DNA necessitates that there be increases in the abundance of 1–2 kb nascent DNA elsewhere in the genome. Indeed random amplification of nascent DNA demonstrated that there are other initiation sites, as yet uncharacterized, that become capable of initiation or are used with greater frequency upon treatment with TSA. The relative decrease of replication initiation at the primary c-myc, lamin B2 and β-globin origin sites, as well as at several anonymous site, suggests that TSA is causing earlier initiations to occur at previously unused neighboring sites. This explanation is consistent with the TSA-induced increase in short nascent strands per S phase cell, the induction of new nascent strands that compensate on a weight basis for the decrease observed at the downregulated initiation sites, and the more rapid completion of S phase. Interestingly, the activation of quiescent origin sites by TSA appears to be the reverse of what occurs during cellular differentiation, where changes in chromatin structure associated with tissue-specific gene expression result in a more restrictive pattern of initiations (1–5).
Acknowledgments
Funding to pay the Open Access publication charges for this article was provided by the OhioLINK Library and Information Network.
REFERENCES
- 1.Shinomiya T., Ina S. Analysis of chromosomal replicons in early embryos of Drosophila melanogaster by two-dimensional gel electrophoresis. Nucleic Acids Res. 1991;19:3935–3941. doi: 10.1093/nar/19.14.3935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sasaki T., Sawado T., Yamaguchi M., Shinomiya T. Specification of regions of DNA replication initiation during embryogenesis in the 65-kilobase DNApolalpha-dE2F locus of Drosophila melanogaster. Mol. Cell. Biol. 1999;19:547–555. doi: 10.1128/mcb.19.1.547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Maric C., Levacher B., Hyrien O. Developmental regulation of replication fork pausing in Xenopus laevis ribosomal RNA genes. J. Mol. Biol. 1999;291:775–788. doi: 10.1006/jmbi.1999.3017. [DOI] [PubMed] [Google Scholar]
- 4.Hyrien O., Mechali M. Chromosomal replication initiates and terminates at random sequences but at regular intervals in the ribosomal DNA of Xenopus early embryos. EMBO J. 1993;12:4511–4520. doi: 10.1002/j.1460-2075.1993.tb06140.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hyrien O., Maric C., Mechali M. Transition in specification of embryonic metazoan DNA replication origins. Science. 1995;270:994–997. doi: 10.1126/science.270.5238.994. [DOI] [PubMed] [Google Scholar]
- 6.Lu L., Tower J. A transcriptional insulator element, the su(Hw) binding site, protects a chromosomal DNA replication origin from position effects. Mol. Cell. Biol. 1997;17:2202–2206. doi: 10.1128/mcb.17.4.2202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schubeler D., Scalzo D., Kooperberg C., van Steensel B., Delrow J., Groudine M. Genome-wide DNA replication profile for Drosophila melanogaster: a link between transcription and replication timing. Nature Genet. 2002;32:438–442. doi: 10.1038/ng1005. [DOI] [PubMed] [Google Scholar]
- 8.Brewer B.J., Lockshon D., Fangman W.L. The arrest of replication forks in the rDNA of yeast occurs independently of transcription. Cell. 1992;71:267–276. doi: 10.1016/0092-8674(92)90355-g. [DOI] [PubMed] [Google Scholar]
- 9.Lowell J.E., Pillus L. Telomere tales: chromatin, telomerase and telomere function in Saccharomyces cerevisiae. Cell. Mol. Life Sci. 1998;54:32–49. doi: 10.1007/s000180050123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kornberg R.D., Lorch Y. Twenty-five years of the nucleosome, fundamental particle of the eukaryote chromosome. Cell. 1999;98:285–294. doi: 10.1016/s0092-8674(00)81958-3. [DOI] [PubMed] [Google Scholar]
- 11.Burke T.W., Cook J.G., Asano M., Nevins J.R. Replication factors MCM2 and ORC1 interact with the histone acetyltransferase HBO1. J. Biol. Chem. 2001;276:15397–15408. doi: 10.1074/jbc.M011556200. [DOI] [PubMed] [Google Scholar]
- 12.Alexiadis V., Varga-Weisz P.D., Bonte E., Becker P.B., Gruss C. In vitro chromatin remodelling by chromatin accessibility complex (CHRAC) at the SV40 origin of DNA replication. EMBO J. 1998;17:3428–3438. doi: 10.1093/emboj/17.12.3428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Alexiadis V., Halmer L., Gruss C. Influence of core histone acetylation on SV40 minichromosome replication in vitro. Chromosoma. 1997;105:324–331. doi: 10.1007/BF02529747. [DOI] [PubMed] [Google Scholar]
- 14.Iizuka M., Stillman B. Histone acetyltransferase HBO1 interacts with the ORC1 subunit of the human initiator protein. J. Biol. Chem. 1999;274:23027–23034. doi: 10.1074/jbc.274.33.23027. [DOI] [PubMed] [Google Scholar]
- 15.Formosa T. Changing the DNA landscape: putting a SPN on chromatin. Curr. Top. Microbiol. Immunol. 2003;274:171–201. doi: 10.1007/978-3-642-55747-7_7. [DOI] [PubMed] [Google Scholar]
- 16.Gavin A.C., Bosche M., Krause R., Grandi P., Marzioch M., Bauer A., Schultz J., Rick J.M., Michon A.M., Cruciat C.M., et al. Functional organization of the yeast proteome by systematic analysis of protein complexes. Nature. 2002;415:141–147. doi: 10.1038/415141a. [DOI] [PubMed] [Google Scholar]
- 17.Vogelauer M., Rubbi L., Lucas I., Brewer B.J., Grunstein M. Histone acetylation regulates the time of replication origin firing. Mol. Cell. 2002;10:1223–1233. doi: 10.1016/s1097-2765(02)00702-5. [DOI] [PubMed] [Google Scholar]
- 18.Aparicio J.G., Viggiani C.J., Gibson D.G., Aparicio O.M. The Rpd3-Sin3 histone deacetylase regulates replication timing and enables intra-S origin control in Saccharomyces cerevisiae. Mol. Cell. Biol. 2004;24:4769–4780. doi: 10.1128/MCB.24.11.4769-4780.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bickmore W.A., Carothers A.D. Factors affecting the timing and imprinting of replication on a mammalian chromosome. J. Cell. Sci. 1995;108:2801–2809. doi: 10.1242/jcs.108.8.2801. [DOI] [PubMed] [Google Scholar]
- 20.DePamphilis M.L. Replication origins in metazoan chromosomes: fact or fiction? Bioessays. 1999;21:5–16. doi: 10.1002/(SICI)1521-1878(199901)21:1<5::AID-BIES2>3.0.CO;2-6. [DOI] [PubMed] [Google Scholar]
- 21.Gilbert D.M. Making sense of eukaryotic DNA replication origins. Science. 2001;294:96–100. doi: 10.1126/science.1061724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hyrien O., Marheineke K., Goldar A. Paradoxes of eukaryotic DNA replication: MCM proteins and the random completion problem. Bioessays. 2003;25:116–125. doi: 10.1002/bies.10208. [DOI] [PubMed] [Google Scholar]
- 23.Todorovic V., Falaschi A., Giacca M. Replication origins of mammalian chromosomes: the happy few. Front. Biosci. 1999;4:D859–D868. doi: 10.2741/todorovic. [DOI] [PubMed] [Google Scholar]
- 24.Dimitrova D.S., Prokhorova T.A., Blow J.J., Todorov I.T., Gilbert D.M. Mammalian nuclei become licensed for DNA replication during late telophase. J. Cell. Sci. 2002;115:51–59. doi: 10.1242/jcs.115.1.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dimitrova D.S., Todorov I.T., Melendy T., Gilbert D.M. Mcm2, but not RPA, is a component of the mammalian early G1-phase prereplication complex. J. Cell. Biol. 1999;146:709–722. doi: 10.1083/jcb.146.4.709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cimbora D.M., Groudine M. The control of mammalian DNA replication: a brief history of space and timing. Cell. 2001;104:643–646. [PubMed] [Google Scholar]
- 27.Bielinsky A.K., Gerbi S.A. Chromosomal ARS1 has a single leading strand start site. Mol. Cell. 1999;3:477–486. doi: 10.1016/s1097-2765(00)80475-x. [DOI] [PubMed] [Google Scholar]
- 28.Benbow R.M., Zhao J., Larson D.D. On the nature of origins of DNA replication in eukaryotes. Bioessays. 1992;14:661–670. doi: 10.1002/bies.950141004. [DOI] [PubMed] [Google Scholar]
- 29.Vaughn J., Dijkwel P., Hamlin J.L. Replication initiates in a broad zone in the amplified CHO dihydrofolate reductase domain. Cell. 1990;61:1075–1087. doi: 10.1016/0092-8674(90)90071-l. [DOI] [PubMed] [Google Scholar]
- 30.Bianchi A., Wells R.D., Heintz N.H., Caddle M.S. Sequences near the origin of replication of the DHFR locus of Chinese hamster ovary cells adopt left-handed Z-DNA and triplex structures. J. Biol. Chem. 1990;265:21789–21796. [PubMed] [Google Scholar]
- 31.Dijkwel P.A., Vaughn J.P., Hamlin J.L. Replication initiation sites are distributed widely in the amplified CHO dihydrofolate reductase domain. Nucleic Acids Res. 1994;22:4989–4996. doi: 10.1093/nar/22.23.4989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dijkwel P.A., Wang S., Hamlin J.L. Initiation sites are distributed at frequent intervals in the Chinese hamster dihydrofolate reductase origin of replication but are used with very different efficiencies. Mol. Cell. Biol. 2002;22:3053–3065. doi: 10.1128/MCB.22.9.3053-3065.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dijkwel P.A., Mesner L.D., Levenson V.V., d'Anna J., Hamlin J.L. Dispersive initiation of replication in the Chinese hamster rhodopsin locus. Exp. Cell. Res. 2000;256:150–157. doi: 10.1006/excr.2000.4809. [DOI] [PubMed] [Google Scholar]
- 34.Tao L., Dong Z., Leffak M., Zannis-Hadjopoulos M., Price G. Major DNA replication initiation sites in the c-myc locus in human cells. J. Cell. Biochem. 2000;78:442–457. doi: 10.1002/1097-4644(20000901)78:3<442::aid-jcb9>3.0.co;2-1. [DOI] [PubMed] [Google Scholar]
- 35.Trivedi A., Waltz S.E., Kamath S., Leffak M. Multiple initiations in the c-myc replication origin independent of chromosomal location. DNA Cell. Biol. 1998;17:885–896. doi: 10.1089/dna.1998.17.885. [DOI] [PubMed] [Google Scholar]
- 36.Waltz S.E., Trivedi A.A., Leffak M. DNA replication initiates non-randomly at multiple sites near the c-myc gene in HeLa cells. Nucleic Acids Res. 1996;24:1887–1894. doi: 10.1093/nar/24.10.1887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nenguke T., Aladjem M.I., Gusella J.F., Wexler N.S., Arnheim N. Candidate DNA replication initiation regions at human trinucleotide repeat disease loci. Hum. Mol. Genet. 2003;12:1021–1028. doi: 10.1093/hmg/ddg111. [DOI] [PubMed] [Google Scholar]
- 38.Girard-Reydet C., Gregoire D., Vassetzky Y., Mechali M. DNA replication initiates at domains overlapping with nuclear matrix attachment regions in the xenopus and mouse c-myc promoter. Gene. 2004;332:129–138. doi: 10.1016/j.gene.2004.02.031. [DOI] [PubMed] [Google Scholar]
- 39.Kamath S., Leffak M. Multiple sites of replication initiation in the human β-globin gene locus. Nucleic Acids Res. 2001;29:809–817. doi: 10.1093/nar/29.3.809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dhar V., Mager D., Iqbal A., Schildkraut C.L. The coordinate replication of the human β-globin gene domain reflects its transcriptional activity and nuclease hypersensitivity. Mol. Cell. Biol. 1988;8:4958–4965. doi: 10.1128/mcb.8.11.4958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Epner E., Forrester W.C., Groudine M. Asynchronous DNA replication within the human β-globin gene locus. Proc. Natl Acad. Sci. USA. 1988;85:8081–8085. doi: 10.1073/pnas.85.21.8081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Aladjem M.I., Rodewald L.W., Lin C.M., Bowman S., Cimbora D.M., Brody L.L., Epner E.M., Groudine M., Wahl G.M. Replication initiation patterns in the β-globin loci of totipotent and differentiated murine cells: evidence for multiple initiation regions. Mol. Cell. Biol. 2002;22:442–452. doi: 10.1128/MCB.22.2.442-452.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Prioleau M.N., Gendron M.C., Hyrien O. Replication of the chicken beta-globin locus: early-firing origins at the 5′ HS4 insulator and the rho- and beta(A)-globin genes show opposite epigenetic modifications. Mol. Cell. Biol. 2003;23:3536–3549. doi: 10.1128/MCB.23.10.3536-3549.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bogan J.A., Natale D.A., Depamphilis M.L. Initiation of eukaryotic DNA replication: conservative or liberal? J. Cell. Physiol. 2000;184:139–150. doi: 10.1002/1097-4652(200008)184:2<139::AID-JCP1>3.0.CO;2-8. [DOI] [PubMed] [Google Scholar]
- 45.Vashee S., Cvetic C., Lu W., Simancek P., Kelly T.J., Walter J.C. Sequence-independent DNA binding and replication initiation by the human origin recognition complex. Genes Dev. 2003;17:1894–1908. doi: 10.1101/gad.1084203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mesner L.D., Li X., Dijkwel P.A., Hamlin J.L. The dihydrofolate reductase origin of replication does not contain any nonredundant genetic elements required for origin activity. Mol. Cell. Biol. 2003;23:804–814. doi: 10.1128/MCB.23.3.804-814.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Edwards M.C., Tutter A.V., Cvetic C., Gilbert C.H., Prokhorova T.A., Walter J.C. MCM2-7 complexes bind chromatin in a distributed pattern surrounding the origin recognition complex in Xenopus egg extracts. J. Biol. Chem. 2002;277:33049–33057. doi: 10.1074/jbc.M204438200. [DOI] [PubMed] [Google Scholar]
- 48.Ishimi Y. A DNA helicase activity is associated with an MCM4, -6, and -7 protein complex. J. Biol. Chem. 1997;272:24508–24513. doi: 10.1074/jbc.272.39.24508. [DOI] [PubMed] [Google Scholar]
- 49.You Z., Ishimi Y., Mizuno T., Sugasawa K., Hanaoka F., Masai H. Thymine-rich single-stranded DNA activates Mcm4/6/7 helicase on Y-fork and bubble-like substrates. EMBO J. 2003;22:6148–6160. doi: 10.1093/emboj/cdg576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lee J.K., Hurwitz J. Processive DNA helicase activity of the minichromosome maintenance proteins 4, 6, and 7 complex requires forked DNA structures. Proc. Natl Acad. Sci. USA. 2001;98:54–59. doi: 10.1073/pnas.98.1.54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ritzi M., Tillack K., Gerhardt J., Ott E., Humme S., Kremmer E., Hammerschmidt W., Schepers A. Complex protein–DNA dynamics at the latent origin of DNA replication of Epstein–Barr virus. J. Cell. Sci. 2003;116:3971–3984. doi: 10.1242/jcs.00708. [DOI] [PubMed] [Google Scholar]
- 52.Schepers A., Ritzi M., Bousset K., Kremmer E., Yates J.L., Harwood J., Diffley J.F., Hammerschmidt W. Human origin recognition complex binds to the region of the latent origin of DNA replication of Epstein–Barr virus. EMBO J. 2001;20:4588–4602. doi: 10.1093/emboj/20.16.4588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Malott M., Leffak M. Activity of the c-myc replicator at an ectopic chromosomal location. Mol. Cell. Biol. 1999;19:5685–5695. doi: 10.1128/mcb.19.8.5685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wang L., Lin C.M., Brooks S., Cimbora D., Groudine M., Aladjem M.I. The human β-globin replication initiation region consists of two modular independent replicators. Mol. Cell. Biol. 2004;24:3373–3386. doi: 10.1128/MCB.24.8.3373-3386.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Liu G., Malott M., Leffak M. Multiple functional elements comprise a mammalian chromosomal replicator. Mol. Cell. Biol. 2003;23:1832–1842. doi: 10.1128/MCB.23.5.1832-1842.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Paixao S., Colaluca I.N., Cubells M., Peverali F.A., Destro A., Giadrossi S., Giacca M., Falaschi A., Riva S., Biamonti G. Modular structure of the human lamin B2 replicator. Mol. Cell. Biol. 2004;24:2958–2967. doi: 10.1128/MCB.24.7.2958-2967.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Giacca M., Pelizon C., Falaschi A. Mapping replication origins by quantifying relative abundance of nascent DNA strands using competitive polymerase chain reaction. Methods. 1997;13:301–312. doi: 10.1006/meth.1997.0529. [DOI] [PubMed] [Google Scholar]
- 58.Burhans W., Vassilev L., Caddle M., Heintz N., DePamphilis M. Identification of an origin of bidirectional DNA replication in mammalian chromosomes. Cell. 1990;62:955–965. doi: 10.1016/0092-8674(90)90270-o. [DOI] [PubMed] [Google Scholar]
- 59.Abdurashidova G., Deganuto M., Klima R., Riva S., Biamonti G., Giacca M., Falaschi A. Start sites of bidirectional DNA synthesis at the human lamin B2 origin. Science. 2000;287:2023–2026. doi: 10.1126/science.287.5460.2023. [DOI] [PubMed] [Google Scholar]
- 60.Kinoshita Y., Johnson E.M. Site-specific loading of an MCM protein complex in a DNA replication initiation zone upstream of the c-MYC gene in the HeLa cell cycle. J. Biol. Chem. 2004;279:35879–35889. doi: 10.1074/jbc.M401640200. [DOI] [PubMed] [Google Scholar]
- 61.Abdurashidova G., Danailov M.B., Ochem A., Triolo G., Djeliova V., Radulescu S., Vindigni A., Riva S., Falaschi A. Localization of proteins bound to a replication origin of human DNA along the cell cycle. EMBO J. 2003;22:4294–4303. doi: 10.1093/emboj/cdg404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Keller C., Ladenburger E.M., Kremer M., Knippers R. The origin recognition complex marks a replication origin in the human TOP1 gene promoter. J. Biol. Chem. 2002;277:31430–31440. doi: 10.1074/jbc.M202165200. [DOI] [PubMed] [Google Scholar]
- 63.Aladjem M.I., Rodewald L.W., Kolman J.L., Wahl G.M. Genetic dissection of a mammalian replicator in the human β-globin locus. Science. 1998;281:1005–1009. doi: 10.1126/science.281.5379.1005. [DOI] [PubMed] [Google Scholar]
- 64.Altman A.L., Fanning E. Defined sequence modules and an architectural element cooperate to promote initiation at an ectopic mammalian chromosomal replication origin. Mol. Cell. Biol. 2004;24:4138–4150. doi: 10.1128/MCB.24.10.4138-4150.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Bell S.P. The origin recognition complex: from simple origins to complex functions. Genes Dev. 2002;16:659–672. doi: 10.1101/gad.969602. [DOI] [PubMed] [Google Scholar]
- 66.Im H., Grass J.A., Christensen H.M., Perkins A., Bresnick E.H. Histone deacetylase-dependent establishment and maintenance of broad low-level histone acetylation within a tissue-specific chromatin domain. Biochemistry. 2002;41:15152–15160. doi: 10.1021/bi026786q. [DOI] [PubMed] [Google Scholar]
- 67.Krude T. Mimosine arrests proliferating human cells before onset of DNA replication in a dose-dependent manner. Exp. Cell. Res. 1999;247:148–159. doi: 10.1006/excr.1998.4342. [DOI] [PubMed] [Google Scholar]
- 68.Mosca P.J., Dijkwel P.A., Hamlin J.L. The plant amino acid mimosine may inhibit initiation at origins of replication in Chinese hamster cells [Erratum (1993). Mol. Cell. Biol., 13, 1981] Mol. Cell. Biol. 1992;12:4375–4383. doi: 10.1128/mcb.12.10.4375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Watson P.A., Hanauske-Abel H.H., Flint A., Lalande M. Mimosine reversibly arrests cell cycle progression at the G1–S phase border. Cytometry. 1991;12:242–246. doi: 10.1002/cyto.990120306. [DOI] [PubMed] [Google Scholar]
- 70.Lalande M. A reversible arrest point in the late G1 phase of the mammalian cell cycle. Exp. Cell. Res. 1990;186:332–339. doi: 10.1016/0014-4827(90)90313-y. [DOI] [PubMed] [Google Scholar]
- 71.Marks P.A., Richon V.M., Rifkind R.A. Histone deacetylase inhibitors: inducers of differentiation or apoptosis of transformed cells. J. Natl Cancer Inst. 2000;92:1210–1216. doi: 10.1093/jnci/92.15.1210. [DOI] [PubMed] [Google Scholar]
- 72.Noh E.J., Lee J.S. Functional interplay between modulation of histone deacetylase activity and its regulatory role in G2–M transition. Biochem. Biophys. Res. Commun. 2003;310:267–273. doi: 10.1016/j.bbrc.2003.09.013. [DOI] [PubMed] [Google Scholar]
- 73.Nair A.R., Boersma L.J., Schiltz L., Chaudhry M.A., Muschel R.J., Chaudry A. Paradoxical effects of trichostatin A: inhibition of NF-Y-associated histone acetyltransferase activity, phosphorylation of hGCN5 and downregulation of cyclin A and B1 mRNA. Cancer Lett. 2001;166:55–64. doi: 10.1016/s0304-3835(01)00418-9. [DOI] [PubMed] [Google Scholar]
- 74.Hoshikawa Y., Kwon H.J., Yoshida M., Horinouchi S., Beppu T. Trichostatin A induces morphological changes and gelsolin expression by inhibiting histone deacetylase in human carcinoma cell lines. Exp. Cell. Res. 1994;214:189–197. doi: 10.1006/excr.1994.1248. [DOI] [PubMed] [Google Scholar]
- 75.Hasan S., Hottiger M.O. Histone acetyl transferases: a role in DNA repair and DNA replication. J. Mol. Med. 2002;80:463–474. doi: 10.1007/s00109-002-0341-7. [DOI] [PubMed] [Google Scholar]
- 76.Chen H., Tini M., Evans R.M. HATs on and beyond chromatin. Curr. Opin. Cell. Biol. 2001;13:218–224. doi: 10.1016/s0955-0674(00)00200-3. [DOI] [PubMed] [Google Scholar]
- 77.Johnson C.A., O'Neill L.P., Mitchell A., Turner B.M. Distinctive patterns of histone H4 acetylation are associated with defined sequence elements within both heterochromatic and euchromatic regions of the human genome. Nucleic Acids Res. 1998;26:994–1001. doi: 10.1093/nar/26.4.994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Yoshida M., Shimazu T., Matsuyama A. Protein deacetylases: enzymes with functional diversity as novel therapeutic targets. Prog. Cell Cycle Res. 2003;5:269–278. [PubMed] [Google Scholar]
- 79.Pappas D.L., Jr, Frisch R., Weinreich M. The NAD+-dependent Sir2p histone deacetylase is a negative regulator of chromosomal DNA replication. Genes Dev. 2004;18:769–781. doi: 10.1101/gad.1173204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Bakkenist C.J., Kastan M.B. DNA damage activates ATM through intermolecular autophosphorylation and dimer dissociation. Nature. 2003;421:499–506. doi: 10.1038/nature01368. [DOI] [PubMed] [Google Scholar]
- 81.Spadari S., Focher F., Sala F., Ciarrocchi G., Koch G., Falaschi A., Pedrali-Noy G. Control of cell division by aphidicolin without adverse effects upon resting cells. Arzneimittelforschung. 1985;35:1108–1116. [PubMed] [Google Scholar]