Abstract
In Caenorhabditis elegans, the activity of transposable elements is repressed in the germline. One of the mechanisms involved in this repression is RNA interference (RNAi), a process in which dsRNA targets cleavage of mRNAs in a sequence-specific manner. The first gene found to be involved in RNAi and transposon silencing in C.elegans is mut-7, a gene encoding a putative exoribonuclease. Here, we show that the MUT-7 protein resides in complexes of ∼250 kDa in the nucleus and in the cytosol. In addition, we find that upon triggering of RNAi the cytosolic MUT-7 complex increases in size. This increase is independent of the presence of target RNA, but does depend on the presence of RDE-1 and RDE-4, two proteins involved in small interfering RNA (siRNA) production. Finally, using a yeast two-hybrid screen, we identified RDE-2/MUT-8 as one of the other components of this complex. This protein is encoded by the rde-2/mut-8 locus, previously implicated in RNAi and transposon silencing. Using genetic complementation analysis, we show that the interaction between these two proteins is required for efficient RNAi in vivo. Together these data support a role for the MUT-7/RDE-2 complex downstream of siRNA formation, but upstream of siRNA mediated target RNA recognition, possibly indicating a role in the siRNA amplification step.
INTRODUCTION
Transposons are segments of DNA that are capable of moving from one place in the host genome to another, thereby potentially threatening the stability of these genomes. The Caenorhabditis elegans genome contains numerous copies of transposable elements from various families; these are active in somatic tissues. Interestingly, transposition of these elements is undetectable in the germline of the canonical Bristol N2 strain (1). This observation led to the hypothesis that a host mechanism is responsible for the active silencing of transposons in the germline of C.elegans.
Indeed, forward genetic screens designed to identify mutants defective in this process resulted in strains with transposon activation in the germline (2,3). These mutants have been named ‘mut’ for their mutator phenotype. Surprisingly, although the screens were aimed at activating Tc1, the activation of transposition applies to multiple types of transposons, implying that a general silencing mechanism is lost (3). A large portion of the mutants obtained turned out to be defective in RNA interference (RNAi) as well (3). Vice versa, screens aimed at isolating mutants defective in RNAi, named ‘rde’ for their RNAi-deficiency, resulted in strains displaying transposon activation (4). These findings, combined with the fact that the mechanism of RNAi is conserved from fungi to human, suggest that RNAi represents an ancient defense mechanism to protect the genome against invading molecular parasites, like transposons and viruses (5).
RNAi refers to the process by which dsRNA induces sequence-specific mRNA degradation (6). The dsRNA is diced into small interfering RNAs (siRNAs) of ∼21–23 nt by a complex containing the RNase III enzyme DICER and the dsRNA-binding protein R2D2 (7–10). The latter facilitates incorporation of these siRNAs into an RNA-induced silencing complex (RISC), which cleaves homologous mRNAs. This cleavage reaction is most probably carried out by the Piwi domain of an Argonaute family protein (11,12), members of which have been associated with RNAi and RISC in several systems (4,11–17). In plants, fungi and C.elegans, RNA-directed RNA polymerases (RdRPs) are required for an efficient RNAi response (18–20). It has been suggested that antisense siRNAs anneal to their target mRNA and can trigger RdRP activity. This way dsRNA would be produced that again may serve as a substrate for the DICER enzyme (18). In agreement with this hypothesis, small antisense RNAs are able to trigger RNAi in a DICER-dependent fashion in C.elegans (21). This does not only require the DICER enzyme, but also depends on the mutator genes mut-14 and mut-7. The mut-14 gene encodes a member of the DEAD box helicase family (21), and mut-7 encodes a protein with a 3′ to 5′ exonuclease domain with homology to Escherichia coli RNase D (3).
A gene encoding a protein with an Rnase D-like domain, WEX, has been knocked-out in Arabidopsis thaliana by reverse genetics, and plants lacking a functional copy of this gene are defective in post-transcriptional gene silencing (22), consistent with the RNAi resistance phenotype in C.elegans. Interestingly, the human genome contains an ortholog of the mut-7 gene as well (AK094438.1), although this gene has not been characterized and a possible role in RNAi has yet to be established.
Screens aimed at identifying RNAi-deficient mutants in C.elegans resulted in four complementation groups; rde-1, rde-2, rde-3 and rde-4 (4). The rde-1 gene encodes a member of the Argonaute family (4). The rde-4 gene encodes a protein containing a dsRNA-binding domain, homologous to R2D2 (10,23). A complex containing RDE-1, RDE-4, the C.elegans DICER ortholog (DCR-1) and a DEAD box helicase (DRH-1) associates with long dsRNA and is suggested to execute the first step in RNAi to generate siRNAs (23). rde-2 and rde-3 have so far not been identified, but genetic analysis suggests that the mut-7 and rde-2 genes act downstream of rde-1 and rde-4 (21,24). Here, we show that the MUT-7 complex acts downstream of siRNA production, but upstream of target RNA recognition. In addition, we identified RDE-2 as another component of this complex.
MATERIALS AND METHODS
Strains
The Bristol strain N2 was used as standard wild-type strain. Alleles used are mut-7(pk720), mut-7(pk719), mut-7(pk204), mut-7(ne311), rde-2(pk735), rde-2(pk707), rde-2(pk715), rde-2(pk716), rde-2(pk718), rde-2(pk1657) and rde-2(ne221). ne221 was mapped by classical mapping strategies between dpy-14 and unc-13. pk716 was mapped between dpy-5 and pkP1062.
Extract preparation
Nematodes were bleached and eggs were hatched o/n. Synchronized L1 larvae were grown in liquid culture and animals were collected as young adults/adults by sedimentation on ice, and purified by flotation on 30% sucrose. After washing the animals with extract buffer (10 mM HEPES, pH 7.1; 5 mM MgCl2, 2 mM DTT, 10% glycerol and protease inhibitors) (Complete protease inhibitor tablets; Roche), they were resuspended in 0.5 vol extract buffer. The suspension was dripped into N2 (l), and the resulting balls were ground in a mortar. The powder was thawed on ice and sheared using a douncer (30 strokes, pistol B). Crude extracts were subsequently centrifuged for 10 min at 18 000 g to obtain a soluble fraction (S18) and a pellet (P18). The supernatant was centrifuged at >100 000 g for 2 h to obtain a non-ribosome associated cytosolic fraction (S100) and a ribosome pellet fraction (P100). To obtain the nuclear fraction, the P18 was washed twice with extract buffer and subsequently resuspended in extract buffer with 500 mM NaCl, at 4°C for 2 h. This was again separated by centrifuging at 18 000 g for 10 min in a soluble nuclear fraction and a pellet, which was extracted once more with extract buffer.
Yeast two-hybrid screens
Yeast strain AH109 (Clontech), containing the reporter genes HIS3, ADE2 and LacZ, all depending on GAL4 for transcriptional activation was used to screen for MUT-7 interacting clones. The full-length mut-7 cDNA was cloned in frame with the GAL4 DNA-binding domain of the pGBKT7 vector (Clontech). This construct was used to screen a mixed-stage C.elegans poly(A) cDNA library cloned in the pACT vector (25,26). Two-hybrid screens to identify RDE-2 interacting clones were performed by cloning the full-length rde-2 gene into pDEST32 using the Gateway cloning system (Invitrogen). For these screens, we used yeast strain MaV203 containing the reporter genes HIS3, URA3 and LacZ, all depending on GAL4 for transcriptional activation. This construct was used to screen a mixed-stage C.elegans poly(A) cDNA library cloned in the pPC97 vector (27).
The resulting colonies were resuspended in appropriate selection medium and patched onto appropriate selection plates followed by a β-galactosidase assay.
Interaction domains were determined by cloning different parts of mut-7 and rde-2 into pDEST32 and pDEST22, respectively, using the Gateway system (Invitrogen). Different parts of mut-7 (amino acids 1–910, 643–787, 643–910 and 787–910) and constructs containing a stop codon at amino acids 811 or 812 were tested against the full-length rde-2 gene as prey. Different parts of rde-2 (amino acids 1–144, 1–286, 1–441, 1–585, 144–286, 144–441, 144–585, 286–441, 286–585 and 441–585) were tested against the full-length mut-7 gene as bait.
Antibodies
The C-terminal part of mut-7 (amino acids 727–910) was cloned into pET23 (Novagen) and expressed in E.coli BL21. Recombinant proteins were purified using Ni-NTA-agarose beads (Qiagen). Protein was purified under denaturing conditions, and was refolded by dialysis to phosphate-buffered saline (PBS). This protein was used for immunization of rabbits. RDE-2 antibodies were raised by injecting rabbits with the synthetic peptide CLPPLSSNQYFMNVRK. Antisera were subsequently purified against the synthetic peptide (Eurogentec).
Immunoprecipitation
Extracts were incubated with purified RDE-2 antibodies and protein A/G agarose beads (Santa Cruz Biotechnology) overnight in IP-buffer (2× buffer; PBS, 5 mM MgCl2, 1% NP-40). Beads were subsequently washed extensively with IP-buffer and proteins were eluted in sample buffer.
Re-localization assay
mut-7 and rde-2 cDNAs were cloned into the NheI site of pPD122.36 (A. Fire vector kit). To create a green fluorescent protein (GFP)-tagged MUT-7 or RDE-2 protein, the TMD (transmembrane domain) encoding cDNA was excised using KpnI. To create a vector solely expressing MUT-7 or RDE-2, both the TMD and GFP expressing cDNAs were excised using KpnI and NheI. The TMD-tagged MUT-7 or RDE-2 expressing vector was created by cloning back the TMD expressing cDNA in the vector solely expressing MUT-7 or RDE-2 using KpnI.
Transgenic worms were generated using standard micro-injection techniques.
Size-fractionation
Extracts were size-fractionated using a superdex200HR 10/30 column (Amersham), using buffer containing 10 mM HEPES, pH 7.0; 150 mM NaCl, 5 mM MgCl2, 2 mM DTT. Fractions of 250 μl were collected. Size markers used are Thyroglobulin (669 kDa), Ferritin (440 kDa), Catalase (232 kDa), Aldolase (158 kDa) and Albumin (67 kDa).
RESULTS
Subcellular localization of MUT-7
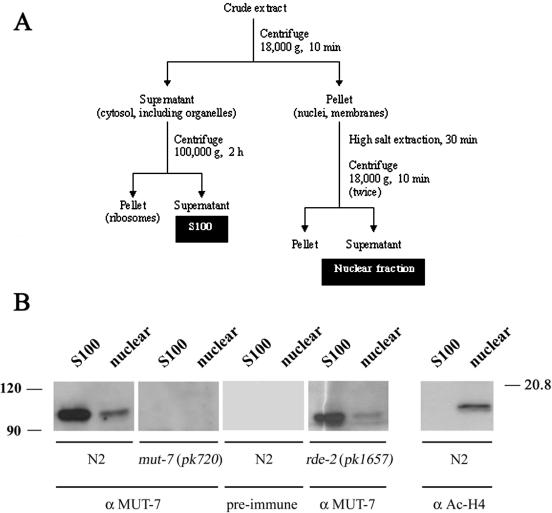
To biochemically characterize MUT-7 and study its involvement in RNAi and transposon silencing, we raised antibodies against a C-terminal fragment of the MUT-7 protein expressed in E.coli. Previous genetic characterization of strains containing mutations in the mut-7 gene indicated that the MUT-7 protein probably acts in the germline of C.elegans. Extracts were therefore prepared from young adult/adult worms, because these stages contain a fully developed reproductive system. To determine the subcellular localization of MUT-7, both cytosolic (S100) and nuclear extracts were prepared (Figure 1A). Separation of cytosol and nucleus was assayed by immunoblotting using a nuclear specific antibody (anti-acetylated histone H4, Figure 1B).
Figure 1.
Subcellular localization of MUT-7. (A) Fractionation scheme indicating subsequent steps in preparation of a cytosolic extract (S100), and a nuclear extract. (B) Proteins in cytosolic (S100) and nuclear fractions from wild-type (N2), mut-7 deletion worms (pk720) or rde-2 deletion worms (pk1657) were separated by SDS–PAGE and analyzed on immunoblot, using MUT-7 antiserum, pre-immune serum or acetylated histon H4 antiserum. Molecular weight markers are indicated in kDa.
As shown in Figure 1B, immune serum specifically recognizes a protein of the predicted size (∼105 kDa) on immunoblots in the S100 and nuclear extracts of wild-type worms. This protein was not detected in the different subcellular fractions from mut-7 deletion worms or when pre-immune serum was used. This demonstrates that the immune serum specifically recognizes the MUT-7 protein and that MUT-7 is expressed in adult worms, and localizes to both the nucleus and the cytosol.
Size-fractionation of MUT-7 containing complexes
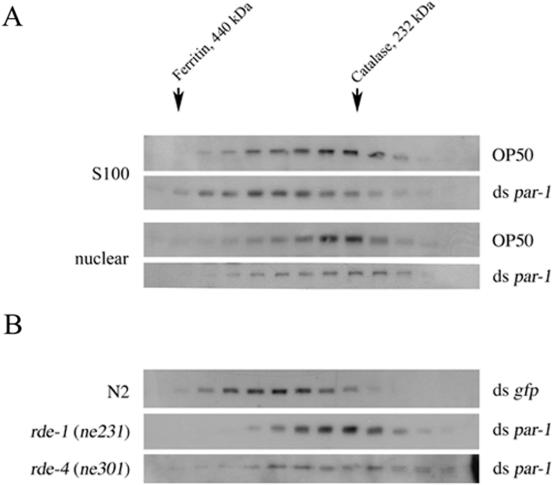
To establish whether the MUT-7 protein acts alone or is associated in a complex, we fractionated the S100 and nuclear extracts from over a size-exclusion column. As is shown in Figure 2A, both the nuclear and the cytosolic MUT-7 protein peak around the catalase marker protein of 232 kDa. These data show that the MUT-7 protein either oligomerizes or associates with other components to form complexes of ∼250 kDa in the cytosol and nucleus.
Figure 2.
Size-fractionation of MUT-7 containing complexes. (A) Cytosolic (S100) and nuclear extracts of N2 worms fed on OP50 bacteria or bacteria expressing par-1 dsRNA were fractionated over a size-exclusion column. The various fractions were analyzed by SDS–PAGE and immunoblotting using MUT-7 antiserum. Size-marker proteins are indicated (440 kDa, ferritin; 232 kDa, catalase). (B) As in (A), only N2 animals were fed on E.coli expressing gfp dsRNA.
RNAi effects on MUT-7 complexes
As mut-7 is required for the effects of dsRNA directed against germline-expressed genes, we analyzed whether treating animals with dsRNA had an effect on complexes containing MUT-7. Induction of RNAi by feeding C.elegans on E.coli expressing dsRNA for the par-1 gene neither affects the expression levels of the MUT-7 protein compared with animals fed on non-dsRNA expressing E.coli, nor does it affect the distribution between cytosol and nucleus (data not shown). However, size-fractionation of the S100 extract of animals fed on E.coli expressing dsRNA shows that the MUT-7 containing complex shifts in molecular weight from ∼250 to ∼350 kDa, indicating that additional factors are recruited (Figure 2A). Treatment of the ∼350 kDa complex with micrococcal nuclease had no effect on the size, suggesting that the observed shift is not caused by a nucleic acid species (data not shown). This shift is not seen in the nuclear fraction.
We have previously reported that mut-7 is required for the in vivo detection of siRNAs in C.elegans (21). Interestingly, also target RNA is required for siRNA accumulation (A. Grishok and C. Mello, unpublished data). To establish whether the observed shift in the molecular weight of the MUT-7 complex would depend on the presence of a target mRNA, wild-type animals were fed on bacteria expressing ds gfp RNA. Interestingly, in the absence of gfp mRNA, the increase in molecular weight of the MUT-7 containing complex is still observed (Figure 2B), implicating a function upstream of target RNA recognition.
Genetic experiments have placed mut-7 downstream of the rde-1 and rde-4 genes (21) and suggested that mut-7 is required for the execution step of RNAi and not initiation (24). To determine whether the molecular weight shift of the MUT-7 complex after feeding of dsRNA depends on the rde-1 and rde-4 genes, we fed rde-1(ne231) and rde-4(ne301) animals on bacteria expressing par-1 dsRNA. Neither rde-1(ne231) nor rde-4(ne301) animals show the molecular weight shift of the MUT-7 complex in the cytosol after feeding of dsRNA (Figure 2B). These data are consistent with the genetic data, placing MUT-7 downstream of RDE-1 and RDE-4, suggesting that MUT-7 functions after siRNA generation.
Screening of a C.elegans cDNA library for proteins interacting with MUT-7
To identify proteins associating with MUT-7, we screened over 5 million cDNA clones for their potential to interact with the full-length MUT-7 protein using the yeast two-hybrid system.
Most of the clones specifically interacting with MUT-7 correspond to a protein encoded by the gene F21C3.4. The identified clones ranged from full-length cDNAs to cDNAs encoding the last three-quarters of the protein (amino acids 163–585). All clones identified, including the EST yk818b11 from the C.elegans EST database, lack nucleotides 637–657 (amino acids 213–219) as compared with the annotated F21C3.4 gene in Wormbase (www.wormbase.org), and some clones contain a larger deletion from nucleotides 613–657 (amino acids 205–219), indicating that this RNA is to some extent alternatively spliced.
Two-hybrid screens using F21C3.4 as bait identified, amongst others, MUT-7 (amino acids 611–910) and the C-terminal part of F21C3.4 itself (amino acids 140–585 and 286–585), possibly indicating dimerization of this protein (data not shown).
F21C3.4 encodes a hypothetical protein of 66 kDa (CE27134) that shows strong homology to a predicted protein, CBG21903, in the closely related nematode C.briggsae, but shows no significant homology to proteins in other species.
Interaction domains of MUT-7 and F21C3.4
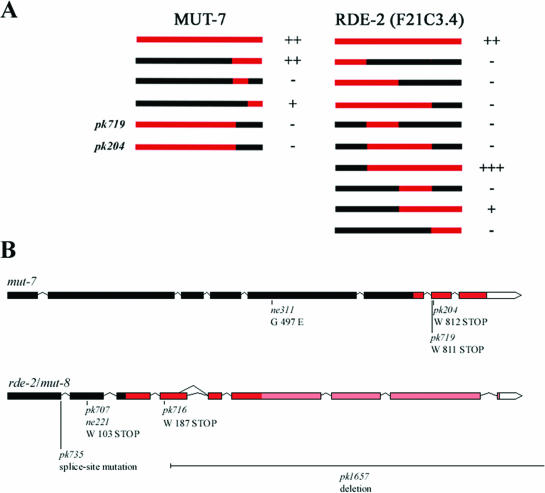
To characterize the interaction between MUT-7 and F21C3.4 in more detail, we mapped the interaction domains of both proteins. Parts of the mut-7 cDNA and the full-length mut-7 cDNA were tested against full-length and parts of F21C3.4. As indicated in Figure 3A, the C-terminal part of MUT-7 (amino acids 787–910) and most of F21C3.4 (amino acids 144–585, strong; amino acids 286–585, weak) are required for the interaction. The N-terminal part of F21C3.4 does not interact with MUT-7, but seems to inhibit the interaction between the two proteins, as deleting this region strongly increases the strength of the interaction. Interestingly, both nonsense alleles of mut-7 that were identified in a genetic screen, pk719 and pk204 (3), contain premature stop codons that cause a loss-of-interaction between MUT-7 and F21C3.4 (Figure 3A).
Figure 3.
Mapping of MUT-7 and RDE-2 interaction domains. (A) Different parts of the MUT-7 protein, indicated in red, were used as bait in a two-hybrid assay and tested against the full-length RDE-2 protein as prey for interaction. Different parts of the RDE-2 protein, indicated in red, were used as prey in the two-hybrid system and tested against the full-length MUT-7 protein as bait for interaction. Two mutant alleles of mut-7, indicated as pk719 and pk204, were tested as well. − to +++ indicate strength of interaction. (B) Mutant alleles of the mut-7 and rde-2/mut-8 genes. Exons are depicted as boxes, and introns as lines. Untranslated regions (UTRs) are depicted as white boxes. An alternative splice-site in rde-2 is depicted. Indicated are the different mutant alleles and the nature of the mutations. Alleles pk715 and pk718 are identical to pk716 and are probably not independent as they were identified from the same screen. This in contrast to the identical rde-2/mut-8 alleles pk707 and ne221 that were identified from different screens. Strongest binding domains are indicated in red, and the minimal binding domain of RDE-2 in light red.
F21C3.4 corresponds to rde-2/mut-8
Forward genetic screens aimed at identifying genes involved in RNAi or transposon silencing produced several complementation groups that are involved in these processes. Some of the genes mutated in these strains have been identified, but for others the causal mutation has not been identified yet.
F21C3.4 maps on chromosome I, at map position +1.87, corresponding to the region where the mutants rde-2(ne221), rde-3(ne298) (4) and mut-8(pk716) (28) have been mapped (this study, see Materials and Methods). We therefore sequenced F21C3.4 in these mutants and in remaining unmapped alleles obtained from the mutator screen (3), revealing that the alleles of rde-2 and mut-8 contained lesions within this gene, as depicted in Figure 3B.
pk735 contains a mutation in the splice donor site of the first intron. Aberrant splicing of the first intron would lead to a premature stop codon in the first intron. Alleles pk707 and ne221 contain nonsense mutations in the second exon, and allele pk716 contains a nonsense mutation in the fourth exon. Complementation tests using RNAi as a read-out show that pk716, pk735 and ne221 are indeed allelic (data not shown). In addition to these alleles, we isolated a deletion allele of rde-2 from a chemical deletion library (pk1657, Figure 3B) (29). This allele, removing the last three-quarters of the gene, is viable and is RNAi resistant (Table 1), showing that rde-2 is a non-essential gene, required for an effective RNAi response.
Table 1.
Complementation analysis between various alleles of mut-7 and rde-2, using RNAi sensitivity as a read-out
| Genotype mut-7 | rde-2 | RNAi |
|---|---|---|
| +/+ | pk1657/pk1657 | − |
| pk204/+ | +/+ | + |
| ne311/+ | +/+ | + |
| +/+ | pk716/+ | + |
| pk204/+ | pk716/+ | + |
| ne311/+ | pk716/+ | − |
+, Fully sensitive; −, fully resistant.
RNAi sensitivity was assayed by scoring dead eggs after feeding animals on E.coli expressing pos-1 dsRNA.
In vivo interaction of MUT-7 and RDE-2
The observed interaction between MUT-7 and RDE-2 was identified in an artificial system and does not necessarily reflect the in vivo situation. To investigate whether these proteins interact in vivo we used three strategies.
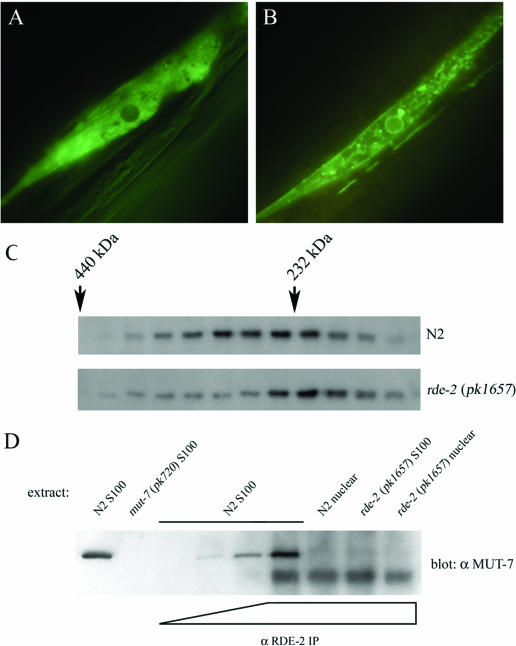
The first strategy is a re-localization experiment (D. Blanchard, H. Hutter, J. Fleenor, and A. Fire, unpublished data). For this, both proteins were co-expressed in body wall muscle cells under transcriptional control of the myo-3 promoter. To visualize the intracellular localization, one of both proteins was tagged with GFP at its N-terminus. Both RDE-2 (Figure 4A) and MUT-7 (data not shown) are predominantly present in the cytosol. Upon co-expression of GFP-tagged RDE-2 and a transmembrane domain-tagged MUT-7 protein, green fluorescence was no longer observed in the cytosol, but localized to membranous compartments (Figure 4B), indicating that both proteins are associated with one another under these conditions. Without a membrane tag on MUT-7, the RDE-2 fusion protein does not localize to the membrane. The same re-localization was observed in the inverse experiment (data not shown).
Figure 4.
In vivo interaction between MUT-7 and RDE-2. (A) A body wall muscle cell co-expressing MUT-7 and GFP-tagged RDE-2. (B) Co-expression of GFP-tagged RDE-2 and TMD-tagged MUT-7 under transcriptional control of the myo-3 promotor. (C) The S100 extracts prepared from wild-type (N2) and rde-2 deletion worms (pk1657) were size-fractionated and analyzed by immunoblotting using MUT-7 antiserum for detection. (D) Cytosolic (S100) and nuclear extracts of N2 and rde-2 deletion worms (pk1657) fed on OP50 bacteria were immunoprecipitated using increasing amounts of RDE-2 antiserum (0, 0.5, 2.0 and 10.0 μl). Samples were analyzed by SDS–PAGE and subsequent immunoblotting using MUT-7 antiserum. Cytosolic N2 and mut-7(pk720) extracts were loaded for MUT-7 antibody specificity.
Second, we size-fractionated S100 and nuclear extracts of rde-2(pk1657) deletion animals. If MUT-7 and RDE-2 indeed reside in the same complex, one would predict a drop in the molecular weight of the MUT-7 containing complexes compared with the wild-type situation. Indeed, as is shown in Figure 4C, the MUT-7 protein from rde-2(pk1657) S100 extracts peaks consistently at a lower molecular weight (∼200 kDa) than that of wild-type animals (∼250 kDa). However, this was not observed for the nuclear extract (data not shown), indicating that MUT-7 and RDE-2 are associated with each other in the cytosol, but not in the nucleus.
Finally, we performed a co-immunoprecipitation experiment. Antibodies were raised against a synthetic peptide corresponding to the C-terminal region of the RDE-2 protein.
Using these antibodies, we could specifically immunoprecipitate the MUT-7 protein from the S100 extract from wild-type worms, but not from extracts prepared from rde-2(pk1657) deletion worms (Figure 4D), although MUT-7 subcellular distribution and expression levels are not affected by the rde-2(pk1657) mutation (Figure 1B). We could not co-immunoprecipitate MUT-7 with RDE-2 antibodies from nuclear extract. Together these data show that the MUT-7 and RDE-2 proteins are associated with each other in the cytosol, but not in the nucleus.
Genetic interaction of mut-7 and rde-2
It is likely that the identified mutant alleles of mut-7 and rde-2 abolish the interaction between the two proteins. For the nonsense alleles of mut-7, we demonstrated this in the two-hybrid system (Figure 3A). For the identified alleles of rde-2 we did not do this, but all identified rde-2 mutations would delete the interaction domain with MUT-7 as it was identified in yeast. However, ne311, a newly identified allele of mut-7 resulting in RNAi resistance and transposon de-silencing, contains a missense mutation in the catalytic ribonuclease domain of MUT-7 (Figure 3B), outside the identified interaction domain. If ne311 would indeed create a catalytically less active allele of MUT-7, this allele may behave in a dominant negative fashion.
To test this, we performed complementation tests between various alleles of mut-7 and rde-2. Animals heterozygous for only mut-7(pk716), mut-7(pk204) or mut-7(ne311) are wild-type for RNAi, indicating that these alleles behave in a recessive fashion. However, in rde-2(pk716) heterozygous animals, a dominant effect of mut-7(ne311) is observed, but not of mut-7(pk204) (Table 1). Apparently, lowering the dosage of rde-2 by 50% sensitizes the animals for semi-dominant alleles of mut-7, showing that mut-7 and rde-2 interact genetically and suggests that interaction between the two wild-type proteins is required for RNAi.
DISCUSSION
MUT-7 interacts with the novel protein RDE-2
In this paper, we report the identity of rde-2 (F21C3.4). Independent screens identified several alleles of this gene (3,4) that was named rde-2 (4) and mut-8 (28). We show that the RDE-2 protein interacts with MUT-7 in the cytosol. Interestingly, mut-7(ne311) behaves dominantly in heterozygous rde-2(pk716) animals. We propose that the MUT-7 protein produced from the mut-7(ne311) allele still interacts with RDE-2, but the complex formed is not functional as MUT-7 is catalytically inactive. Thus, mutant MUT-7 sequesters 50% of the available RDE-2, which is already limited in heterozygous rde-2(pk716) animals. This would imply that 25% functional MUT-7/RDE-2 is not sufficient for efficient RNAi. This, and the fact that all identified nonsense mutations in the mut-7 and rde-2 genes delete the interaction domains between the two proteins, indicates that interaction between the two proteins is required for RNAi.
The observation that the RDE-2 protein can interact with itself in the two-hybrid system could indicate that RDE-2 functions as a dimer or even that the MUT-7/RDE-2 complex as a whole functions as a dimer. However, considering the size of the MUT-7/RDE-2 complex in the cytosol (∼250 kDa) and the size of the MUT-7 complex in the cytosol of rde-2(pk1657) animals (∼200 kDa), it seems likely that MUT-7 associates with an RDE-2 monomer, rather than a dimer.
Role of the MUT-7/RDE-2 complex in RNAi
Much has been learned about the RNAi pathway and the function of the proteins involved. In Drosophila a complex containing R2D2, a dsRNA-binding protein, assists Dicer2 in processing long dsRNA into siRNAs (7,10,30). R2D2 subsequently facilitates siRNA passage from Dicer2 to RISC (10), where it is presumably loaded onto Argonaute2 (Ago2). The siRNA directs the latter to the target RNA where Ago2 can exert its ‘slicing’ activity (11,12), thereby cleaving the target.
In C.elegans RDE-4, a dsRNA-binding protein, and DCR-1 have been shown to be in a complex together (23) and are required for generating siRNAs (7–9,31), analogs to R2D2 and Dicer2 in Drosophila. Furthermore, genetic analysis showed that rde-1, an AGO2 homolog, and rde-4 are essential for the generation of a heritable interfering agent, presumably siRNAs, but are dispensable for the interference thereafter. On the other hand, mut-7 and rde-2 are required downstream in the RNAi reaction, but are not needed for the initial siRNA generating response (24). Consistent with these results, we show that the response of the MUT-7/RDE-2 complex on dsRNA depends on rde-1 and rde-4.
Although MUT-7 is not required for siRNA production in vitro, feeding of dsRNA to mut-7 mutant animals does not result in detectable levels of siRNAs in vivo (21). Also, rde-2 mutant animals do not produce detectable levels of siRNAs in vivo (data not shown). This is not a general RNAi deficiency phenotype, since mut-14(pk738) animals do show significant levels of siRNAs in vivo (21). Detection of siRNAs in vivo, but not in vitro, also requires the presence of target RNA [A. Grishok, C. Mello, unpublished data; (21)]. This suggests that the siRNAs that are detected in vivo are mainly derived from the RdRP-mediated amplification cycle in C.elegans (18). Together these data hint at a role for the MUT-7/RDE-2 complex in the amplification step of the RNAi pathway in C.elegans.
Dual role for MUT-7
From our studies, it appears that MUT-7 is present in both cytosol and nucleus, but that the MUT-7 protein in the nucleus is embedded in a different complex compared with the cytosol. Nuclear MUT-7 does not associate with RDE-2 and the complex does not alter in molecular weight upon exposure of dsRNA. Previous reports have implicated RNAi-like processes in nuclear events (32–36). More specifically, in fission yeast heterochromatin formation requires components of the RNAi machinery, including an RdRP homolog, and deletion of these components results in loss of histone H3 lysine-9 methylation (37,38), and impairment of centromere function (33). The latter results in miss-segregation of chromosomes. Interestingly, both mut-7 and rde-2 mutants show a high incidence of males (him) phenotype (3,4), caused by non-disjunction of the X-chromosome. It will therefore be of interest to further study the nuclear MUT-7 protein, and its role in chromosome segregation.
Acknowledgments
We thank A. Bathoorn for technical assistance, M. Tijsterman for critical reading of the manuscript, R. Barstead and M. Vidal for kindly sharing the yeast two-hybrid libraries, G. Hannon for support in MUT-7 antiserum production and Yuji Kohara for EST yk818b11. This research was partly funded by a VENI fellowship from The Netherlands Organization for Scientific Research (NWO) to R.F.K., and a grant by the Research Institute of Diseases in the Elderly (RIDE) to R.H.A.P. Funding to pay the Open Access publication charges for this article was provided by The Netherlands Organization for Scientific Research (NWO).
REFERENCES
- 1.Emmons S.W., Yesner L. High-frequency excision of transposable element Tc 1 in the nematode Caenorhabditis elegans is limited to somatic cells. Cell. 1984;36:599–605. doi: 10.1016/0092-8674(84)90339-8. [DOI] [PubMed] [Google Scholar]
- 2.Collins J., Saari B., Anderson P. Activation of a transposable element in the germ line but not the soma of Caenorhabditis elegans. Nature. 1987;328:726–728. doi: 10.1038/328726a0. [DOI] [PubMed] [Google Scholar]
- 3.Ketting R.F., Haverkamp T.H., van Luenen H.G., Plasterk R.H. Mut-7 of C.elegans, required for transposon silencing and RNA interference, is a homolog of Werner syndrome helicase and RNaseD. Cell. 1999;99:133–141. doi: 10.1016/s0092-8674(00)81645-1. [DOI] [PubMed] [Google Scholar]
- 4.Tabara H., Sarkissian M., Kelly W.G., Fleenor J., Grishok A., Timmons L., Fire A., Mello C.C. The rde-1 gene, RNA interference, and transposon silencing in C.elegans. Cell. 1999;99:123–132. doi: 10.1016/s0092-8674(00)81644-x. [DOI] [PubMed] [Google Scholar]
- 5.Plasterk R.H. RNA silencing: the genome's immune system. Science. 2002;296:1263–1265. doi: 10.1126/science.1072148. [DOI] [PubMed] [Google Scholar]
- 6.Fire A., Xu S., Montgomery M.K., Kostas S.A., Driver S.E., Mello C.C. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature. 1998;391:806–811. doi: 10.1038/35888. [DOI] [PubMed] [Google Scholar]
- 7.Bernstein E., Caudy A.A., Hammond S.M., Hannon G.J. Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature. 2001;409:363–366. doi: 10.1038/35053110. [DOI] [PubMed] [Google Scholar]
- 8.Ketting R.F., Fischer S.E., Bernstein E., Sijen T., Hannon G.J., Plasterk R.H. Dicer functions in RNA interference and in synthesis of small RNA involved in developmental timing in C.elegans. Genes Dev. 2001;15:2654–2659. doi: 10.1101/gad.927801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Knight S.W., Bass B.L. A role for the RNase III enzyme DCR-1 in RNA interference and germ line development in Caenorhabditis elegans. Science. 2001;293:2269–2271. doi: 10.1126/science.1062039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liu Q., Rand T.A., Kalidas S., Du F., Kim H.E., Smith D.P., Wang X. R2D2, a bridge between the initiation and effector steps of the Drosophila RNAi pathway. Science. 2003;301:1921–1925. doi: 10.1126/science.1088710. [DOI] [PubMed] [Google Scholar]
- 11.Liu J., Carmell M.A., Rivas F.V., Marsden C.G., Thomson J.M., Song J.J., Hammond S.M., Joshua-Tor L., Hannon G.J. Argonaute2 Is the Catalytic Engine of Mammalian RNAi. Science. 2004;305:1437–1441. doi: 10.1126/science.1102513. [DOI] [PubMed] [Google Scholar]
- 12.Song J.J., Smith S.K., Hannon G.J., Joshua-Tor L. Crystal structure of Argonaute and its implications for RISC slicer activity. Science. 2004;305:1434–1437. doi: 10.1126/science.1102514. [DOI] [PubMed] [Google Scholar]
- 13.Hammond S.M., Boettcher S., Caudy A.A., Kobayashi R., Hannon G.J. Argonaute2, a link between genetic and biochemical analyses of RNAi. Science. 2001;293:1146–1150. doi: 10.1126/science.1064023. [DOI] [PubMed] [Google Scholar]
- 14.Bohmert K., Camus I., Bellini C., Bouchez D., Caboche M., Benning C. AGO1 defines a novel locus of Arabidopsis controlling leaf development. EMBO J. 1998;17:170–180. doi: 10.1093/emboj/17.1.170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kataoka Y., Takeichi M., Uemura T. Developmental roles and molecular characterization of a Drosophila homologue of Arabidopsis Argonaute1, the founder of a novel gene superfamily. Genes Cells. 2001;6:313–325. doi: 10.1046/j.1365-2443.2001.00427.x. [DOI] [PubMed] [Google Scholar]
- 16.Martinez J., Patkaniowska A., Urlaub H., Luhrmann R., Tuschl T. Single-stranded antisense siRNAs guide target RNA cleavage in RNAi. Cell. 2002;110:563–574. doi: 10.1016/s0092-8674(02)00908-x. [DOI] [PubMed] [Google Scholar]
- 17.Catalanotto C., Azzalin G., Macino G., Cogoni C. Involvement of small RNAs and role of the qde genes in the gene silencing pathway in Neurospora. Genes Dev. 2002;16:790–795. doi: 10.1101/gad.222402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sijen T., Fleenor J., Simmer F., Thijssen K.L., Parrish S., Timmons L., Plasterk R.H., Fire A. On the role of RNA amplification in dsRNA-triggered gene silencing. Cell. 2001;107:465–476. doi: 10.1016/s0092-8674(01)00576-1. [DOI] [PubMed] [Google Scholar]
- 19.Cogoni C., Macino G. Gene silencing in Neurospora crassa requires a protein homologous to RNA-dependent RNA polymerase. Nature. 1999;399:166–169. doi: 10.1038/20215. [DOI] [PubMed] [Google Scholar]
- 20.Dalmay T., Hamilton A., Rudd S., Angell S., Baulcombe D.C. An RNA-dependent RNA polymerase gene in Arabidobsis is required for posttranscriptional gene silencing mediated by a transgene but not by a virus. Cell. 2000;101:543–553. doi: 10.1016/s0092-8674(00)80864-8. [DOI] [PubMed] [Google Scholar]
- 21.Tijsterman M., Ketting R.F., Okihara K.L., Sijen T., Plasterk R.H. RNA helicase MUT-14-dependent gene silencing triggered in C.elegans by short antisense RNAs. Science. 2002;295:694–697. doi: 10.1126/science.1067534. [DOI] [PubMed] [Google Scholar]
- 22.Glazov E., Phillips K., Budziszewski G.J., Meins F., Levin J.Z. A gene encoding an RNase D exonuclease-like protein is required for post-transcriptional silencing in Arabidopsis. Plant J. 2003;35:342–349. doi: 10.1046/j.1365-313x.2003.01810.x. [DOI] [PubMed] [Google Scholar]
- 23.Tabara H., Yigit E., Siomi H., Mello C.C. The dsRNA binding protein RDE-4 interacts with RDE-1, DCR-1, and a DExH-box helicase to direct RNAi in C.elegans. Cell. 2002;109:861–871. doi: 10.1016/s0092-8674(02)00793-6. [DOI] [PubMed] [Google Scholar]
- 24.Grishok A., Tabara H., Mello C.C. Genetic requirements for inheritance of RNAi in C.elegans. Science. 2000;287:2494–2497. doi: 10.1126/science.287.5462.2494. [DOI] [PubMed] [Google Scholar]
- 25.Durfee T., Becherer K., Chen P.L., Yeh S.H., Yang Y., Kilburn A.E., Lee W.H., Elledge S.J. The retinoblastoma protein associates with the protein phosphatase type 1 catalytic subunit. Genes Dev. 1993;7:555–569. doi: 10.1101/gad.7.4.555. [DOI] [PubMed] [Google Scholar]
- 26.Elledge S.J., Mulligan J.T., Ramer S.W., Spottswood M., Davis R.W. Lambda YES: a multifunctional cDNA expression vector for the isolation of genes by complementation of yeast and Escherichia coli mutations. Proc. Natl Acad. Sci. USA. 1991;88:1731–1735. doi: 10.1073/pnas.88.5.1731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Walhout A.J., Sordella R., Lu X., Hartley J.L., Temple G.F., Brasch M.A., Thierry-Mieg N., Vidal M. Protein interaction mapping in C.elegans using proteins involved in vulval development. Science. 2000;287:116–122. doi: 10.1126/science.287.5450.116. [DOI] [PubMed] [Google Scholar]
- 28.Ketting R.F., Plasterk R.H. A genetic link between co-suppression and RNA interference in C.elegans. Nature. 2000;404:296–298. doi: 10.1038/35005113. [DOI] [PubMed] [Google Scholar]
- 29.Jansen G., Hazendonk E., Thijssen K.L., Plasterk R.H. Reverse genetics by chemical mutagenesis in Caenorhabditis elegans. Nature Genet. 1997;17:119–121. doi: 10.1038/ng0997-119. [DOI] [PubMed] [Google Scholar]
- 30.Lee Y.S., Nakahara K., Pham J.W., Kim K., He Z., Sontheimer E.J., Carthew R.W. Distinct roles for Drosophila Dicer-1 and Dicer-2 in the siRNA/miRNA silencing pathways. Cell. 2004;117:69–81. doi: 10.1016/s0092-8674(04)00261-2. [DOI] [PubMed] [Google Scholar]
- 31.Parrish S.N., Fire A. Distinct roles for RDE-1 and RDE-4 during RNA interference in C.elegans. RNA. 2001;7:1397–1402. [PMC free article] [PubMed] [Google Scholar]
- 32.Mochizuki K., Fine N.A., Fujisawa T., Gorovsky M.A. Analysis of a piwi-related gene implicates small RNAs in genome rearrangement in tetrahymena. Cell. 2002;110:689–699. doi: 10.1016/s0092-8674(02)00909-1. [DOI] [PubMed] [Google Scholar]
- 33.Hall I.M., Noma K., Grewal S.I. RNA interference machinery regulates chromosome dynamics during mitosis and meiosis in fission yeast. Proc. Natl Acad. Sci. USA. 2003;100:193–198. doi: 10.1073/pnas.232688099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dudley N.R., Labbe J.C., Goldstein B. Using RNA interference to identify genes required for RNA interference. Proc. Natl Acad. Sci. USA. 2002;99:4191–4196. doi: 10.1073/pnas.062605199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pal-Bhadra M., Bhadra U., Birchler J.A. RNAi related mechanisms affect both transcriptional and posttranscriptional transgene silencing in Drosophila. Mol. Cell. 2002;9:315–327. doi: 10.1016/s1097-2765(02)00440-9. [DOI] [PubMed] [Google Scholar]
- 36.Zilberman D., Cao X., Jacobsen S.E. ARGONAUTE4 control of locus-specific siRNA accumulation and DNA and histone methylation. Science. 2003;299:716–719. doi: 10.1126/science.1079695. [DOI] [PubMed] [Google Scholar]
- 37.Hall I.M., Shankaranarayana G.D., Noma K., Ayoub N., Cohen A., Grewal S.I. Establishment and maintenance of a heterochromatin domain. Science. 2002;297:2232–2237. doi: 10.1126/science.1076466. [DOI] [PubMed] [Google Scholar]
- 38.Volpe T.A., Kidner C., Hall I.M., Teng G., Grewal S.I., Martienssen R.A. Regulation of heterochromatic silencing and histone H3 lysine-9 methylation by RNAi. Science. 2002;297:1833–1837. doi: 10.1126/science.1074973. [DOI] [PubMed] [Google Scholar]