Abstract
Sac7d, a small, abundant, sequence-general DNA-binding protein from the hyperthermophilic archaeon Sulfolobus acidocaldarius, causes a single-step sharp kink in DNA (∼60°) via the intercalation of both Val26 and Met29. These two amino acids were systematically changed in size to probe their effects on DNA kinking. Eight crystal structures of five Sac7d mutant–DNA complexes have been analyzed. The DNA-binding pattern of the V26A and M29A single mutants is similar to that of the wild-type, whereas the V26A/M29A protein binds DNA without side chain intercalation, resulting in a smaller overall bending (∼50°). The M29F mutant inserts the Phe29 side chain orthogonally to the C2pG3 step without stacking with base pairs, inducing a sharp kink (∼80°). In the V26F/M29F-GCGATCGC complex, Phe26 intercalates deeply into DNA bases by stacking with the G3 base, whereas Phe29 is stacked on the G15 deoxyribose, in a way similar to those used by the TATA box-binding proteins. All mutants have reduced DNA-stabilizing ability, as indicated by their lower Tm values. The DNA kink patterns caused by different combinations of hydrophobic side chains may be relevant in understanding the manner by which other minor groove-binding proteins interact with DNA.
INTRODUCTION
The role of intrinsic DNA curvature in biologically important functions has been well documented. Curved DNA fragments are often found near functionally important sites, such as promoters and origins of replication (1–5). In eukaryotic cells, the genomic DNA is packed by histones into nucleosomes, which in turn form higher-order structures of chromatin. The structural organization of DNA in prokaryotes and archaeabacteria is somewhat less well understood. Sso7d and Sac7d are two small (∼7 kDa), abundant and basic chromosomal proteins from the hyperthermophilic archaeabacteria Sulfolobus solfataricus and S.acidocaldarius, respectively. Sac7d and Sso7d have unfolding temperatures >90°C (0.3 M KCl, pH 7.5,) and both are acid stable with Tm's of >60°C at pH 0. Both proteins bind to DNA without marked sequence preference and increase the Tm of DNA by more than 40°C (6–9). The extreme thermal, acid and chemical stability associated with these proteins makes them an attractive system for a variety of studies.
The crystal structures of several Sac7d/Sso7d-DNA complexes have revealed an unexpected DNA-binding mode (10–14). Binding of Sac7d/Sso7d causes a sharp kink in DNA and introduces significant DNA unwinding. They may play a role in the packing of DNA to organize chromatin structures in these archaea, which lack histones. Furthermore, they offer insights into the possible role of several classes of the minor groove DNA-binding proteins, including TBP (15–17), PurR (18), LacI (19), IHF (20), HU (21), HMG1 (22), SRY (23), LEF-1 (24), Sox2 (25), HMG-D (26) and NHP6A (27,28). The structures of these protein–DNA complexes have revealed a common theme whereby a protein inserts one or more hydrophobic side chains into DNA from the minor groove, unstacking two contiguous base pairs and thus producing noticeable kinks of the DNA duplex to varying degrees at one or more sites. However, these proteins possess very different global folds and they use different strategies for DNA recognition and binding (29–31). Interestingly, both Sac7d and TBP use hydrophobic amino acids on a β-sheet for the intercalation. These proteins also remodel the DNA conformation in distinct ways. The 60° single-step kink in the Sac7d/Sso7d-DNA complexes caused by Val26 and Met29 is the largest known value in a protein–DNA complex to date.
In these Sac7d/Sso7d-DNA complexes, the protein structure is similar to that of the unbound protein, consisting of an incomplete β-barrel with a triple-stranded β-sheet orthogonal to a β-hairpin. The small β-barrel is capped by an amphiphilic C-terminal α-helix. The triple-stranded β-sheet is placed across the DNA minor groove with the intercalation of the Val26 and Met29 side chains into DNA. The remarkable ability of a small, but stable, protein to sharply kink DNA may have implications in protein design. Here, we investigate the interaction and binding property of different Sac7d mutants of Val26 and Met29 with DNA. These two hydrophobic side chains were systematically changed to either smaller or larger sizes. Our results will help understand the molecular basis of protein-induced DNA kinks. New proteins may be engineered to modulate the extent of DNA curvature.
MATERIALS AND METHODS
Purification of Sac7d and crystallization of Sac7d-DNA complexes
The recombinant Sac7d mutant proteins were prepared and purified according to the published procedure (6). The purified protein was dialyzed against de-ionized water and stored as a lyophilized powder. These mutant–DNA complexes have been crystallized using the sitting drop vapor diffusion method at room temperature under similar condition as for wild-type Sac7d, except that the protein/DNA ratios were <1. The detailed crystallization conditions are summarized in Table 1S.
Data collection
Protein–DNA complex crystals were mounted on nylon loops and immediately frozen in liquid nitrogen. Data were collected at −150°C and processed by HKL/HKL2000 (32) program suite. Synchrotron data were collected at the BL17B2 beam line at the National Synchrotron Radiation Research Center (NSRRC) in Taiwan and BL18B beam line of Photon Factory (PF) (KEK, Tsukuba, Japan). In-house data were collected using MSC MicroMax 002 with a Rigaku R-Axis IV++ image-plate detector system. The crystal data and refinement statistics are given in Table 1. Datasets for crystals of the Sac7d M29F mutant in complex with the GCGATCGC, and the bromo-dU derivative, GCGAUbrCGC were both collected to 2.5 and 1.9 Å resolutions, respectively. The crystals have the same unit cells and space group for these two complexes, and therefore only the higher resolution data set for the M29F-GCGAUbrCGC complex is given in Table 1. For the two monoclinic crystal forms, the M29F-GCGAUbrCGC complex contains two molecules in an asymmetric unit, whereas the other M29F-GTAATTAC complex has only one.
Table 1.
Crystal and refinement data of Sac7d mutant–DNA complexes
| V26A | V26A | M29A | M29A | V26A/M29A | M29F | M29F | V26F/M29Fa | |
|---|---|---|---|---|---|---|---|---|
| GCGATCGC | GTAATTAC | GCGATCGC | GTAATTAC | GCGATCGC | GCGAUbrCGC | GTAATTAC | GCGATCGC | |
| Crystal data | ||||||||
| a (Å) | 35.39 | 36.11 | 34.51 | 35.92 | 37.78 | 38.22 | 31.24 | 36.90 |
| b (Å) | 48.23 | 50.44 | 49.52 | 47.97 | 47.28 | 47.89 | 50.05 | 47.18 |
| c (Å) | 69.58 | 78.15 | 76.57 | 77.78 | 52.65 | 52.49 | 35.77 | 60.26 |
| β (degree) | 102.7 | 108.9 | ||||||
| Space group | P212121 | P212121 | P212121 | P212121 | P212121 | P21 | P21 | P212121 |
| Light source | Micro Max 002 | MicroMax 002 | MicroMax 002 | NSRRC BL17B2 | NSRRC BL17B2 | MicroMax 002 | MicroMax 002 | BL17B2 NSRRC |
| Resolution (Å) | 2.20 | 2.20 | 1.80 | 1.60 | 1.45 | 1.90 | 1.70 | 1.50 |
| No. of reflections | 5996 | 7491 | 12317 | 17958 | 17344 | 14849 | 11165 | 16734 |
| Rmerge (%) | 4.3 | 4.2 | 4.5 | 4.9 | 4.7 | 4.9 | 2.7 | 6.0 |
| Rmerge at the highest shell (%) | 26.1 (2.28–2.20 Å) | 40.3 (2.28–2.20 Å) | 26.4 (1.86–1.80 Å) | 49.9 (1.66–1.60 Å) | 39.5 (1.50–1.45 Å) | 26.9 (1.97–1.90 Å) | 6.8 (1.76–1.70 Å) | 38.9 (1.55–1.50 Å) |
| Completeness (%) | 94.9 | 97.1 | 96.6 | 98.0 | 99.4 | 98.6 | 94.4 | 95.7 |
| Completeness at the highest shell (%) | 77.7 (2.28–2.20 Å) | 88.8 (2.28–2.20 Å) | 94.3 (1.86–1.80 Å) | 96.8 (1.66–1.60 Å) | 99.1 (1.50–1.45 Å) | 93.4 (1.97–1.90 Å) | 85.4 (1.76–1.70 Å) | 94.2 (1.55–1.50 Å) |
| Refinement data | ||||||||
| No. of reflections | 5781 | 7147 | 11945 | 17132 | 16568 | 14194 | 10915 | 16272 |
| Rworking/Rfree (5% data) | 0.228/0.286 | 0.225/0.255 | 0.227/0.274 | 0.210/0.238 | 0.214/0.255 | 0.220/0.285 | 0.176/0.226 | 0.214/0.230 |
| r.m.s.d. bond distance (Å) | 0.010 | 0.016 | 0.014 | 0.017 | 0.016 | 0.016 | 0.014 | 0.015 |
| r.m.s.d. bond angle (deg.) | 1.54 | 1.58 | 1.84 | 1.79 | 1.90 | 1.78 | 1.83 | 1.72 |
| No. of atoms (protein/DNA) | 531/322 | 531/322 | 530/322 | 513/322 | 520/322 | 1029/644 | 519/322 | 532/322 |
| No. of waters | 58 | 95 | 144 | 140 | 207 | 156 | 235 | 191 |
| Accession number | 1WTW | 1WTX | 1WTR | 1WTV | 1XYI | 1WTP | 1WTQ | 1WTO |
aMAD data were collected at the beam line BL18B of PF (KEK).
Phasing and structure refinements
The structures of single mutants (V26A, M29A and M29F) complexed with DNA (GCGATCGC or GTAATTAC) were solved by the molecular replacement method using the program AMORE (33) in the CCP4 (34) suite or the CNS-SOLVE (35) program. The phase angles for V26A/M29A–GCGATCGC complex were first determined by SAD experiment on its iodine derivative V26A/M29A-GCGATiCGC at 2.0 Å and further improved by MAD experiment on its bromine derivative V26A/M29A-GCGATbrCGC at 1.9 Å. The phases for V26F/M29F-GCGATCGC complex structure were determined by MAD experiment on its bromine derivative V26A/M29A-GCGAUbrCGC at 2.5 Å. The wt-Sac7d protein and DNA models were manually fitted into the SAD/MAD maps calculated using program SOLVE/RESOLVE (36,37) with solvent flattening. Several distinct changes in the mutant–DNA complex structures were made using the program O (38). All refinements were carried out against |Fo| > 0σ data, using the simulated annealing and individual temperature factor refinements procedures incorporated in the program CNS. Free R factor was used to monitor the refinement. The stereochemical quality of the refined structures was evaluated using the program PROCHECK (39). DNA conformations were analyzed using the program X3DNA (40). Figures were prepared using the programs MOLSCRIPT (41), Raster-3D (42) and GRASP (43). Figure 2S was generated using the program NUCPLOT (44). The atomic coordinates of the eight Sac7d mutant–DNA complexes have been deposited in the RCSB Protein Data Bank. The accession numbers are listed in Table 1.
RESULTS
Overall structure of the complex
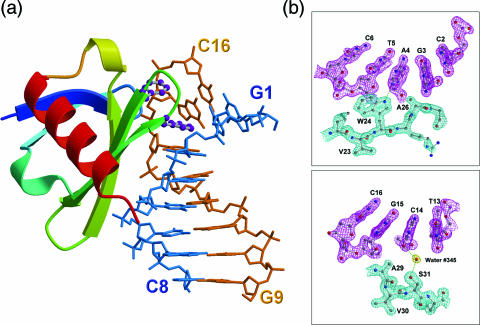
The crystal structures of eight Sac7d mutant–DNA complexes have been determined and refined at high resolution (Table 1). All φ/ψ angles and other conformational parameters in these structures fall within the acceptable regions. Figure 1a and b shows the overall structure of the V26F/M29F–GCGATCGC complex and some representative (2Fo − Fc) electron density maps of the V26A/M29A–DNA complex, respectively.
Figure 1.
Sac7d-DNA interface. (a) Ribbon diagram of Sac7d V26F/M29F-GCGATCGC complex with two intercalating phenylalanine residues depicted as ball and stick. The aromatic ring of Phe26 residue stacks with the G3 base, whereas the phenyl ring of Phe29 is stacked on the deoxyribose of G15. (b) The (2Fo − Fc) Fourier electron density maps of the V26AM29A–GCGATCGC complex (contoured at 1.5σ level) in the regions at the protein–DNA interface (upper panel). The indole NH group of Trp24 forms hydrogen bond (3.0 Å) to the base N3 of A4. Adjacent to the intercalating site, Ser 31 (OG) forms a water-mediated hydrogen bond with C14 O2 (lower panel).
In these complexes, the five mutant protein structures are similar to wt-Sac7d, except for the loops 9–11, 36–39 and 48–52. Using backbone atoms from residues 10–60, the root-mean-square deviation (r.m.s.d.) values between the mutant- and wt-Sac7d proteins range between 0.24 and 0.86 Å. For the M29F–GCGATCGC complex, the asymmetric unit consists of two crystallographically independent and structurally similar complexes (the r.m.s.d. value between α-carbons of the two M29F–DNA complexes is 0.46 Å). The DNA structures of these two complexes are almost identical at the intercalation site, but different away from the binding site due to crystal packing.
These five mutants all bind to the minor groove of the DNA duplex and produce different kinks in DNA. Conformational changes occur in the DNA duplexes to match the protein surface. Numerous van der Waals contacts and hydrogen bonds occur between DNA, protein and associated water molecules. However, significant differences are found. The V26A and M29A single mutants are much similar to the wild-type protein. In the M29F–GCGATCGC complex, the Phe29 side chain of M29F mutant penetrates into the C2pG3 step without stacking with one of the base pairs. Together with the side chain of Val26 positioned vertically, they induce the largest sharp kink (∼80°) and disrupt the stacking of two adjacent base pairs.
For the V26F/M29F and V26A/M29A double mutants, the crystal structures reveal unprecedented changes. The Phe26 side chain in the V26F/M29F–GCGATCGC complex intercalates deeply into the gap of DNA bases by π–π stacking interactions with G3 base. Whereas the side chain of Phe29 is stacked on the deoxyribose of G15. In contrast, the V26A/M29A protein binds at the G3pA4 step and bends the DNA helix smoothly over several base pairs with a smaller overall kink angle (∼50°). Its crystal packing pattern is very similar to the Sso7d–DNA complex crystal structure (Sso7d-GCGAACGC/GCGTTCGC or Sso7d-GTGATCGC), but not to any of the other Sac7d/mutants–DNA complexes.
DNA deformation
In previous studies of the Sac7d/Sso7d–DNA complex structures, we observed that binding of Sac7d/Sso7d caused a sharp kink in DNA octamers (10,11). The helical structure of DNA is significantly altered by a side chain intercalation. In order to accommodate intercalating side chains, the minor groove is widened, and the helical twist locally is reduced, accompanying with a significant positive roll at one or more base steps.
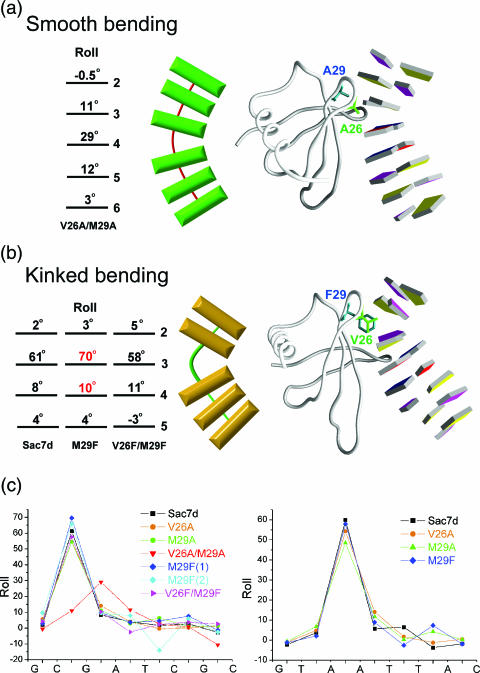
The Sac7d mutants induce two kinking modes: ‘smooth bending’ and ‘kinked bending’ (Figure 2a and b). The DNA conformation in the V26A/M29A–GCGATCGC complex is quite different from those in other Sac7d/Sso7d–DNA complexes. The DNA bending angles are 11°, 29° and 12° from the C2pG3, G3pA4 and A4pT5 steps, respectively, totaling 52°. The two small alanines cause a smaller, gradual ‘smooth bending’ of DNA by partial unstacking and rolling of several consecutive base-pair steps.
Figure 2.
Two modes of DNA bending. (a) The structure of V26A/M29A–DNA complex (right) and schematic diagram (left) stress gradual curvature by smooth bending. (b) The structure of M29F–DNA complex shows localization of curvature by kinked bending from minor groove of DNA. The contributions of each base-pair-step curvature to overall helix bending are listed with corresponding base-pair numbers. (c) Local base-pair step parameter, roll, for all eight Sac7d mutant–DNA binary structures, calculated using the X3DNA program. Note the largest roll at certain kinked step (G3pA4 in V26A/M29A, C2pG3 or A3pA4 in the others).
The binding sites of the DNA bound to wt-Sac7d (or its mutants) are located at different places in the complexes: at the C2pG3 step for GCGATCGC and at the central region of the A3pA4 step for GTAATTAC, respectively. The base at the 3′ end of the intercalating site is always a purine (G3 in GCGATCGC and A4 in GTAATTAC). Interestingly, Trp24 Nɛ1 always forms a hydrogen bond to its purine N3 atom. In eight Sac7d mutant–DNA complexes, the DNA-binding patterns and unit cells are quite different from each other, but the Trp24 residue plays a similar role like that in the wt-Sac7d-DNA complex. The binding site of the V26A/M29A in GCGATCGC shifts 1 bp, from the G2pC3 to the G3pA4 step and its Trp24 forms a hydrogen bond (3.0 Å) between indole NH group and the A4N3, a purine base at the 3′ side of the intercalating site (Figure 1b).
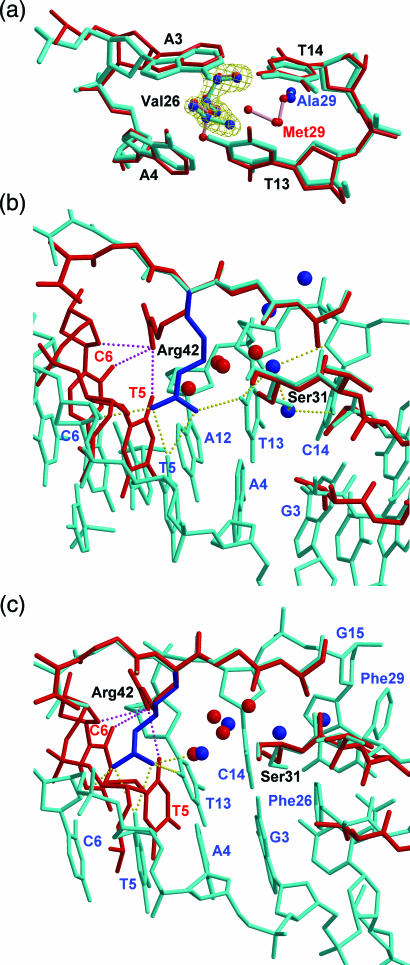
The DNA kink angles induced by the same single mutant protein (V26A, M29A or M29F) are all bigger at the C2pG3 than those at the A3pA4 step, especially in the M29F-DNA complexes. The mutant M29F uses the same conformations of Val26 and Phe29 to introduce the largest kink of ∼80° for the GCGATCGC, but a smaller kink of ∼68° for the GTAATTAC, close to that in the wt-Sac7d–DNA complex (Figure 2c). The Val26 side chain in the M29A–GTAATTAC complex penetrates horizontally into the ApA step, whereas in other Sac7d–DNA complexes, the valyl group is vertically intercalated (Figure 3a). The horizontal intercalation of Val26 causes a smaller roll angle (∼48°) than that in the vertical intercalation (∼68°). The M29A protein lacks the bulky side chain to force apart the central ApA base step that makes this unexpected conformation change of the Val side chain and causes the smallest overall bend angle (∼59°) among the ‘kinked bending’ DNA.
Figure 3.
Variations in Sac7d-DNA interactions. (a) The local structures of the M29A– (cyan) and wt-Sac7d– (red) in complex with GTAATTAC are superimposed at the intercalation site. In the M29A structure the isopropyl group of Val 26 penetrates horizontally into DNA base step, but in the wt-Sac7d structure (and other mutants), it intercalates vertically. The electron density map (contoured at 1.5σ level) of the side chain Val26 in M29A is shown in yellow. (b) V26A/M29A–GCGATCGC and (c) V26F/M29F–GCGATCGC complexes, colored in cyan, were superimposed on the wild-type Sac7d–DNA structures, colored in red. The side chain of Arg42 is flipped to opposite side and forms new hydrogen bonds (dashed lines), and the side chain of Ser31 is also rotated in both double mutant complexes.
Analyses of the three helical parameters, roll (Figure 2c), twist and rise (Figure 1S), reveal proteins with the larger side chains (Met or Phe) that cause bigger rolls of DNA than those with the smaller side chains (Ala or Val). Smaller twist angles and larger rises of DNA are also caused by the proteins with larger side chains. Crystal structures reveal a more obvious effect on the DNA deformation by changing the size of the bulky Met29 than that of Val26. Biochemical studies also show the mutations at these sites will lead to decreased binding affinities to DNA (Tables 2S and 3S). Moreover, it appears that mutation effects on DNA-kinking and unwinding is directly proportional to how much these two residues are changed.
Upon binding to the Sac7d mutants, all DNA minor grooves are widened significantly and many nucleotides surrounding the wedged site adopt the less common C3′-endo (N-type) sugar pucker that is more akin to an A-DNA. The ‘partial B-to-A like’ transformation has been repetitively observed in many minor groove binding protein–DNA complexes (45). DNA kinking is coupled to the intercalation of hydrophobic amino acids, and the kinked DNA configurations are further stabilized by direct contact between hydrophobic side chains and sugar. This may be due to that A-like nucleotides provide larger hydrophobic surfaces for interactions than B-like nucleotides (46).
Contacts between Sac7d mutants and DNA
In previous structures of the Sac7d/Sso7d–DNA complexes, four adaptor molecules were always found at the interface between DNA and protein (10,11). Arg42 is the only positive residue in the Sac7d–DNA interface. In all previous Sac7d/Sso7d–DNA structures, its guanidinium group forms hydrogen bonds with O2 atom of thymine, the third base after the intercalation site. The conformation of Arg42 in single-mutant complexes is the same with that in wt-Sac7d. However, in the V26A/M29A–GCGATCGC complex, the Arg42 adopts a very different conformation and occupies the space for those four-water adapters (Figure 3b and c). The guanidinium group now forms hydrogen bonds with both strands of the DNA. In the V26F/M29F–GCGATCGC structure, the guanidinium group also forms hydrogen bonds with two thymine O2 atoms, except Arg42 does not occupy the space for waters. These hydrogen bond(s) between NH2 of Arg42 to O2 of thymine determine the polarity of the Sac7d/Sso7–DNA binding, when DNA is not self-complementary.
The side chain of Ser31 in the V26A/M29A–DNA complex has the opposite orientation to that in the wild-type complex and it is almost in the plane of the G3-C14 base pair. The water hydrogen-bonded to the OG of Ser31 serves as an arm, extending into the gap between the DNA bases (Figure 1b). Interestingly, in both double mutant structures Ser31 has the similar water-mediated interactions with the DNA bases (Figure 2S).
The high resolution structures of the Sac7d mutant–GCGATCGC complexes afford a good opportunity to study the protein–DNA–water interactions in detail. As before (10,11), four recurring well-ordered water molecules are observed in the buried cavity located between the protein and DNA surfaces near the intercalation site. They fill up the cavity and enable close packing of protein–DNA interface. The four-water arrangements in the three single mutant complexes are similar to those in the wt-Sac7d-DNA complexes, whereas those in two double mutants are quite different. The buried cavity between the V26F/M29F mutant and DNA becomes narrower than that in the wt-Sac7d-DNA complexes due to the phenylalanine/base stacking interaction between Phe26 and the G3 base. The four bridging water molecules rearrange in the longitudinal direction along the helix axis to match the shapes of the protein–DNA interface. In the V26A/M29A–GCGATCGC complex, the space for these four-water adapters is occupied by the Arg 42. The four bridging water molecules rearrange to locate closer to the G3–C14 base pair. As noted above, it supports the idea that these interfacial water molecules may act as ‘modulators’ for different Sac7d mutants binding to DNA of varying sequences, a property required for sequence-general DNA-binding proteins.
DISCUSSION
The interactions of the minor groove intercalating proteins with DNA are primarily hydrophobic through amino acids, such as Val, Ile, Leu or Met, or the faces of aromatic amino acids (Phe or Tyr). Table 2 shows the comparison of the intercalation geometry of the minor groove binding protein–DNA complexes.
Table 2.
Comparisons of the base-pair steps at the place of insertion, the intercalating protein side chains and the DNA kink angle of minor groove binding protein–DNA complexes
| Protein | Amino acid | Step | Kink angle (°) | Reference | |
|---|---|---|---|---|---|
| Wt-Sac7d/Sso7d | V, M | CpG | 62 (∼70)a | β-sheet | (10,11) |
| ApA | 60 (∼70)a | ||||
| Sac7d mutants | |||||
| V26A/M29A | A, A | GpA | 29 (∼52)a | β-sheet | This work |
| V26F/M29F | F, F | CpG | 58 (∼68)a | This work | |
| V26A | A, M | CpG | 55 (∼69)a | This work | |
| ApA | 54 (∼68)a | ||||
| M29A | V, A | CpG | 55 (∼66)a | This work | |
| ApA | 49 (∼60)a | ||||
| M29F | V, F | CpG | 70 (∼80)a | This work | |
| ApA | 58 (∼68)a | ||||
| aTBP | F, F | TpA | 45 | β-sheet | (15) |
| F, F | ApG | 45 | |||
| yTBP, hTBP | F, F | TpA | 45 | β-sheet | (16,17) |
| F, F | ApA | 45 | |||
| PurR | L, (L′) | CpG | 45 | hinge α-helix | (18) |
| LacI | L | CpG | 45 | (leucine levers) | (19) |
| IHF | P | ApA | 59 (>160)a | β-ribbon arms | (20) |
| HU | P | TpT | 57 (∼105–140)a | (21) | |
| HMG group | L-shaped 3-helix | ||||
| Specific | |||||
| HMG1 | F | Pt-GpG | 61 | (22) | |
| SRYb | I, (F) | ApA | 65 (∼75)a | (23) | |
| LEF-1b | M, (F) | ApA | 52 (∼90)a | (24) | |
| POU/HMG/DNA (Oct1/Sox2/FGF4) | M, (F) | ApA | 45 (∼90)a | (25) | |
| Non-specific | |||||
| HMG-D | M | TpA | 49 | (26) | |
| A | ApT | 21 | |||
| V, T | GpA | 32 (∼111)a | |||
| yNHP6Ab | M, (Y) | TpG | 25 | (28) | |
| F | TpT | 20 (∼70)a | |||
aOverall bending angle of DNA in parenthesis.
bNMR structure.
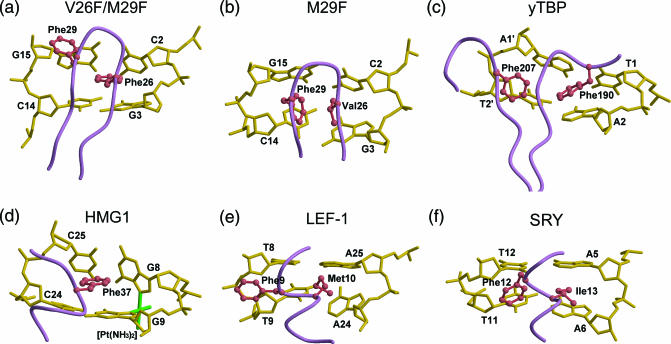
Two types of hydrophobic intercalating interactions are observed in the M29F– and V26F/M29F–DNA complexes. The Phe29 side chain of the M29F protein, together with the side chain of residue Val26, penetrates perpendicularly to the C2-G15/G3-C14 base pair planes, but the Phe26 side chain of the V26F/M29F protein acts as a parallel base-stacking intercalator (Figure 4a and b). Remarkably, the phenylalanine residues in other minor groove DNA-binding proteins, such as TBP, HMG1, SRY, LEF-1 and Sox2, make contact with DNA in a similar manner.
Figure 4.
Local structures of the minor groove-binding proteins. (a) Sac7d V26F/M29F, (b) Sac7d M29F, (c) yTBP, (d) HMG1, (e) LEF-1 and (f) SRY in complex with DNA are displayed near the intercalation sites. The polypeptide backbones are shown in purple, the side chains in coral and the DNA in gold.
The structures of the TBP–DNA complexes (15–17) reveal two pairs of phenylalanine residues partially intercalating into base steps, creating two ∼45° kinks at either end of the TATA element (Figure 4c). Interestingly, the Sac7d V26F/M29F mutant also uses a pair of the Phe side chains to stabilize its kink site in the minor groove in a similar way. Phe26 is parallel to the DNA G3 base at a distance 4.3 Å, and Phe29 is stacked against the deoxyribose ring of G15.
In the HMG1/cisplatin-modified DNA complex, the DNA is kinked, induced by the platinum crosslink that destacks the G8–G9 purine ring and exposes their hydrophobic surfaces in the minor groove (22). Like the Phe intercalators seen in TBP or Sac7d V26F/M29F complex with DNA, the phenyl ring of Phe32 at the amino terminus of helix II serves as a wedge and intercalates into the hydrophobic cleft and stacks onto the G9 base at a distance 3.5 Å (Figure 4d).
SRY (23) and LEF-1 (24) are members of the high-mobility group of proteins that bind DNA sequence-specifically and are expressed in a few cell types. LEF-1 uses a methionine (Met10) residue for intercalation at the ApA step. Additional stabilizing contacts at the binding site are made by Phe9 and Met13 to nearby bases. The aromatic ring of Phe9 stacks against the hydrophobic surface of the T8 deoxyribose (Figure 4e). The side chain of Ile in SRY, corresponding to the Met of LEF-1, also penetrates partly into the central ApA step of the DNA octamer. The wedged Phe12 is positioned orthogonally to the base edges of the minor groove and packed against the bases at the TpT step (Figure 4f).
In the Oct1/Sox2/FGF4 ternary complex (25), the HMG domain of Sox2 severely bends the DNA toward the major groove with an approximate overall bending angle of 90°. The hydrophobic side chains of Phe10 and Met11, same as that of LEF-1, are inserted between the ApA step and causing a ∼45° kink angle of the DNA axis. But the Sox2/DNA interaction pattern is more closely related to that of the SRY/DNA complex. The Phe side chains in these two proteins, instead of stacking on DNA deoxyribose, contact with the DNA bases at the TpT step in a similar way.
As noted above, a bulky hydrophobic side chain (Phe or Met) is favored at the site of insertion into the base stack. Together with another aromatic residue (Phe or Tyr), they support the penetrating side chain to pry open the minor groove through extensive van der Waals contacts with deoxyribose. In contrast, the phenylalanine side chain together with another smaller residue (Val) could intercalate into DNA perpendicularly as observed in the Sac7d M29F–DNA complexes. Despite the different modes of intercalation in DNA, the proteins bind exclusively in the minor groove and induce the helix unwinding, minor groove expansion and DNA bending. Although accomplished through very different 3D folds and structure motifs, the minor groove DNA-binding proteins that have the same combinations of hydrophobic intercalating side chains seem to interact with DNA in a similar way.
All Sac7d mutants form less extensive contacts with the backbone of the DNA due to the fact that only 6 (Lys3, 7, 9, 22, 28 and 65) of the 14 lysines are potentially involved in the hydrogen bonding interactions with phosphates. Instead, the complexes are stabilized by base-specific hydrogen bonds (Trp24 Nɛ1 with adenine or guanine-N3 and Arg42 NH1 or NH2 with thymine-O2), water-mediated contacts with DNA bases and hydrophobic interactions of residues 26 and 29 with DNA. The Tm of DNA in the M29F–DNA complex is lower than that in the V26F/M29F–DNA complex by about 10°C (Table 2S). This may be due to the absence of the phenylalanine/base stacking interactions. It illustrates the importance of the stacking forces between hydrophobic side chains and DNA. Any loss of the DNA base–base stacking energy would be compensated by the extensive van der Waals interactions between DNA and phenylalanine rings. In all Sac7d mutant–DNA complexes, we observed base-specific hydrogen bonds and water adapters in a manner similar to one another, whereas the hydrophobic interactions at the intercalation site provide major contribution to the difference of binding affinity between Sac7d mutants and DNA. The DNA-binding affinities of the non-sequence-specific HMG-D mutants suggest that intercalation by the primary residue Met13 is important for high affinity DNA binding (47). The M13F mutant has affinity most similar to wt-HMG-D, whereas the other mutants with small residues have decreasing affinities (Kdmut/Kdwt: 0.9 for M13F; 5.8 for M13A). This property emphasizes the importance of hydrophobic interactions in stabilizing these protein–DNA complexes.
In conclusion, DNA bending has long been recognized as an important component of biological activity. Intercalation represents an important mechanism by which protein-induced DNA bending can occur. The crystal structures of Sac7d mutants bound with DNA help elucidate the molecular basis of how different binding combinations of hydrophobic side chains are able to modulate the extent of DNA curvature.
SUPPLEMENTARY MATERIAL
Supplementary Material is available at NAR Online.
Supplementary Material
Acknowledgments
This research was supported by grants from Academia Sinica, the National Research Program for Genomic Medicine (NRPGM) and the National Science Council (NSC) for X-ray Core (NSC-93-3112-B-001-011-Y) to A.H.-J.W. We thank Dr S. Su for his early contribution during this work. The Open Access publication charges for this article were waived by Oxford University Press.
REFERENCES
- 1.Hagerman P.J. Sequence-directed curvature of DNA. Annu. Rev. Biochem. 1990;59:755–781. doi: 10.1146/annurev.bi.59.070190.003543. [DOI] [PubMed] [Google Scholar]
- 2.McFall S.M., Chugani S.A., Chakrabarty M.A. Transcriptional activation of the catechol and chlorocatechol operons: variations on a theme. Gene. 1998;223:257–267. doi: 10.1016/s0378-1119(98)00366-7. [DOI] [PubMed] [Google Scholar]
- 3.McGill G., Fisher D.E. DNA bending and the curious case of Fos/Jun. Chem. Biol. 1998;5:R29–R38. doi: 10.1016/s1074-5521(98)90139-9. [DOI] [PubMed] [Google Scholar]
- 4.Muller H.P., Varmus H.E. DNA bending creates favored sites for retroviral integration: an explanation for preferred insertion sites in nucleosomes. EMBO J. 1994;13:4704–4714. doi: 10.1002/j.1460-2075.1994.tb06794.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zahn K., Blattner F.R. Sequence-induced DNA curvature at the bacteriophage lambda origin of replication. Nature. 1985;317:451–453. doi: 10.1038/317451a0. [DOI] [PubMed] [Google Scholar]
- 6.McAfee J.G., Edmondson S.P., Datta P.K., Shriver J.W., Gupta R. Gene cloning, expression, and characterization of the Sac7 proteins from the hyperthermophile Sulfolobus acidocaldarius. Biochemistry. 1995;34:10063–10077. doi: 10.1021/bi00031a031. [DOI] [PubMed] [Google Scholar]
- 7.McAfee J.G., Edmondson S.P., Zegar I., Shriver J.W. Equilibrium DNA binding of Sac7d protein from the hyperthermophile Sulfolobus acidocaldarius: fluorescence and circular dichroism studies. Biochemistry. 1996;35:4034–4045. doi: 10.1021/bi952555q. [DOI] [PubMed] [Google Scholar]
- 8.McCrary B.S., Edmondson S.P., Shriver J.W. Hyperthermophile protein folding thermodynamics: differential scanning calorimetry and chemical denaturation of Sac7d. J. Mol. Biol. 1996;264:784–805. doi: 10.1006/jmbi.1996.0677. [DOI] [PubMed] [Google Scholar]
- 9.Lundback T., Hansson H., Knapp S., Ladenstein R., Hard T. Thermodynamic characterization of non-sequence-specific DNA-binding by the Sso7d protein from Sulfolobus solfataricus. J. Mol. Biol. 1998;276:775–784. doi: 10.1006/jmbi.1997.1558. [DOI] [PubMed] [Google Scholar]
- 10.Gao Y.G., Su S.Y., Robinson H., Padmanabhan S., Lim L., McCrary B.S., Edmondson S.P., Shriver J.W., Wang A.H.-J. The crystal structure of the hyperthermophile chromosomal protein Sso7d bound to DNA. Nature Struct. Biol. 1998;5:782–786. doi: 10.1038/1822. [DOI] [PubMed] [Google Scholar]
- 11.Robinson H., Gao Y.G., McCrary B.S., Edmondson S.P., Shriver J.W., Wang A.H.-J. The hyperthermophile chromosomal protein Sac7d sharply kinks DNA. Nature. 1998;392:202–205. doi: 10.1038/32455. [DOI] [PubMed] [Google Scholar]
- 12.Krueger J.K., McCrary B.S., Wang A.H.-J., Shriver J.W., Trewhella J., Edmondson S.P. The solution structure of the Sac7d/DNA complex: A small-angle X-ray scattering study. Biochemistry. 1999;38:10247–10255. doi: 10.1021/bi990782c. [DOI] [PubMed] [Google Scholar]
- 13.Su S.Y., Gao Y.G., Robinson H., Liaw Y.C., Edmondson S.P., Shriver J.W., Wang A.H.-J. Crystal structures of the chromosomal proteins Sso7d/Sac7d bound to DNA containing T-G mismatched base-pairs. J. Mol. Biol. 2000;303:395–403. doi: 10.1006/jmbi.2000.4112. [DOI] [PubMed] [Google Scholar]
- 14.Ko T.P., Chu H.M., Chen C.Y., Chou C.C., Wang A.H.-J. Structures of the hyperthermophilic chromosomal protein Sac7d in complex with DNA decamers. Acta Crystallogr. D Biol. Crystallogr. 2004;60:1381–1387. doi: 10.1107/S0907444904012065. [DOI] [PubMed] [Google Scholar]
- 15.Kim Y., Geiger J.H., Hahn S., Sigler P.B. Crystal structure of a yeast TBP/TATA-box complex. Nature. 1993a;365:512–520. doi: 10.1038/365512a0. [DOI] [PubMed] [Google Scholar]
- 16.Kim J.L., Nikolov D.B., Burley S.K. Co-crystal structure of TBP recognizing the minor groove of a TATA element. Nature. 1993b;365:520–527. doi: 10.1038/365520a0. [DOI] [PubMed] [Google Scholar]
- 17.Nikolov D.B., Chen H., Halay E.D., Hoffmann A., Roeder R.G., Burley S.K. Crystal structure of a human TATA box-binding protein/TATA element complex. Proc. Natl Acad. Sci. USA. 1996;93:4862–4867. doi: 10.1073/pnas.93.10.4862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schumacher M.A., Choi K.Y., Zalkin H., Brennan R.G. Crystal structure of LacI member, PurR, bound to DNA: minor groove binding by alpha helices. Science. 1994;266:763–770. doi: 10.1126/science.7973627. [DOI] [PubMed] [Google Scholar]
- 19.Lewis M., Chang G., Horton N.C., Kercher M.A., Pace H.C., Schumacher M.A., Brennan R.G., Lu P. Crystal structure of the lactose operon repressor and its complexes with DNA and inducer. Science. 1996;271:1247–1254. doi: 10.1126/science.271.5253.1247. [DOI] [PubMed] [Google Scholar]
- 20.Rice P.A., Yang S.W., Mizuuchi K., Nash H. Crystal structure of an IHF-DNA complex: a protein-induced DNA U-turn. Cell. 1996;87:1295–1306. doi: 10.1016/s0092-8674(00)81824-3. [DOI] [PubMed] [Google Scholar]
- 21.Swinger K.K., Lemberg K.M., Zhang Y., Rice P.A. Flexible DNA bending in HU-DNA cocrystal structures. EMBO J. 2003;22:3749–3760. doi: 10.1093/emboj/cdg351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ohndorf U.M., Rould M.A., He Q., Pabo C.O., Lippard S.J. Basis for recognition of cisplatin-modified DNA by high-mobility-group proteins. Nature. 1999;399:708–712. doi: 10.1038/21460. [DOI] [PubMed] [Google Scholar]
- 23.Werner M.H., Huth J.R., Gronenborn A.M., Clore G.M. Molecular basis of human 46X, Y sex reversal revealed from the three-dimensional solution structure of the human SRY-DNA complex. Cell. 1995;81:705–714. doi: 10.1016/0092-8674(95)90532-4. [DOI] [PubMed] [Google Scholar]
- 24.Love J.J., Li X., Case D.A., Giese K., Grosschedl R., Wright P.E. Structural basis for DNA bending by the architectural transcription factor LEF-1. Nature. 1995;376:791–795. doi: 10.1038/376791a0. [DOI] [PubMed] [Google Scholar]
- 25.Remenyi A., Lins K., Nissen L.J., Reinbold R., Scholer H.R., Wilmanns M. Crystal structure of a POU/HMG/DNA ternary complex suggests differential assembly of Oct4 and Sox2 on two enhancers. Genes Dev. 2003;17:2048–2059. doi: 10.1101/gad.269303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Murphy F.H., Sweet R.M., Churchill M.E.A. The structure of a chromosomal high mobility group protein–DNA complex reveals sequence-neutral mechanisms important for non-sequence-specific DNA recognition. EMBO J. 1999;18:6610–6618. doi: 10.1093/emboj/18.23.6610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Allian F.H.-T., Yen Y.M., Masse J.E., Schultzu P., Dieckmann T., Johnson R.C., Fegon J. Solution structure of the HMG protein NHP6A and its interaction with DNA reveals the structural determinants for non-sequence-specific binding. EMBO J. 1999;18:2563–2579. doi: 10.1093/emboj/18.9.2563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Masse J.E., Wong B., Yen Y.M., Allian F.H.-T., Johnson R.C., Fegon J. The S.cerevisiae architectural HMGB protein NHP6A complexed with DNA: DNA and protein conformational changes upon binding. J. Mol. Biol. 2002;323:263–284. doi: 10.1016/s0022-2836(02)00938-5. [DOI] [PubMed] [Google Scholar]
- 29.Werner M.H., Gronenborn A.M., Clore G.M. Intercalation, DNA kinking, and the control of transcription. Science. 1996;271:778–784. doi: 10.1126/science.271.5250.778. [DOI] [PubMed] [Google Scholar]
- 30.Dickerson R.E. DNA bending: the prevalence of kinkiness and the virtues of normality. Nucleic Acids Res. 1998;26:1906–1926. doi: 10.1093/nar/26.8.1906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Werner M.H., Burley S.K. Architectural transcription factors: proteins that remodel DNA. Cell. 1996;88:733–736. doi: 10.1016/s0092-8674(00)81917-0. [DOI] [PubMed] [Google Scholar]
- 32.Otwinowski Z., Minor W. Processing of the X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 33.Navaza J. AmoRe: an automated package for molecular replacement. Acta Crystallogr. A. 1994;50:157–163. [Google Scholar]
- 34.Collaborative Computational Project Number 4. The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D Biol. Crystallogr. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 35.Brunger A.T., Adams P.D., Clore G.M., DeLano W.L., Gros P., Grosse-Kunstleve R.W., Jiang J.S., Kuszewski J., Nilges M., Pannu N.S., et al. Crystallography & NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr. D Biol. Crystallogr. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 36.Terwilliger T.C., Berendzen J. Automated MAD and MIR structure solution. Acta Crystallogr. D Biol. Crystallogr. 1999;55:849–861. doi: 10.1107/S0907444999000839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Terwilliger T.C. Map-likelihood phasing. Acta Crystallogr. D Biol. Crystallogr. 2001;57:1763–1775. doi: 10.1107/S0907444901013749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jones T.A., Zou J.Y., Cowan S.W., Kjeldgaard M. Improved methods for the building of protein models in electron density and the location of errors in these models. Acta Crystallogr. A. 1991;47:392–400. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- 39.Laskowski R.A., MacArthur M.W., Moss D.S., Thornton J.M. PROCHECK: a program to check the stereochemical quality of proteins structures. J. Appl. Crystallogr. 1993;26:283–291. [Google Scholar]
- 40.Lu X.J., Olson W.K. X3DNA: a software package for the analysis, rebuilding and visualization of three-dimensional nucleic acid structures. Nucleic Acids Res. 2003;31:5108–5121. doi: 10.1093/nar/gkg680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kraulis P.J. MOLSCRIPT: a program to produce both detailed and schematic plots of proteins structures. J. Appl. Crystallogr. 1991;24:946–950. [Google Scholar]
- 42.Merritt E.A., Murphy M.E.P. Raster3D version 2.0: a program for photorealistic molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 1994;50:869–873. doi: 10.1107/S0907444994006396. [DOI] [PubMed] [Google Scholar]
- 43.Nicholls A., Bharadwaj R., Honig B. GRASP: graphical representation and analysis of surface-properties. Biophys. J. 1993;64:166–170. [Google Scholar]
- 44.Luscombe N.M., Laskowski R.A., Thornton J.M. NUCPLOT: a program to generate schematic diagrams of protein–DNA interactions. Nucleic Acids Res. 1997;25:4940–4945. doi: 10.1093/nar/25.24.4940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dostal L., Chen C.Y., Wang A.H.-J., Welfle H. Partial B-to-A DNA transition upon minor groove binding of protein Sac7d monitored by Raman spectroscopy. Biochemistry. 2004;43:9600–9609. doi: 10.1021/bi049192r. [DOI] [PubMed] [Google Scholar]
- 46.Tolstorukov M.Y., Jernigan R.L., Zhurkin V.B. Protein–DNA hydrophobic recognition in the minor groove is facilitated by sugar switching. J. Mol. Biol. 2004;337:65–76. doi: 10.1016/j.jmb.2004.01.011. [DOI] [PubMed] [Google Scholar]
- 47.Klass J., Murphy F.V., IV, Fouts S., Serenil M., Changela A., Siple J., Churchill M.E.A. The role of intercalating residues in chromosomal high-mobility-group protein DNA binding, bending and specificity. Nucleic Acids Res. 2003;31:2852–2864. doi: 10.1093/nar/gkg389. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.