Abstract
Background
Type 2 diabetes (T2D) is a worldwide epidemic with considerable health and economic consequences. Sulfonylureas are widely used drugs for the treatment of patients with T2D. KCNJ11 and ABCC8 encode the Kir6.2 (pore-forming subunit) and SUR1 (regulatory subunit that binds to sulfonylurea) of pancreatic β cell KATP channel respectively with a critical role in insulin secretion and glucose homeostasis. TCF7L2 encodes a transcription factor expressed in pancreatic β cells that regulates insulin production and processing. Because mutations of these genes could affect insulin secretion stimulated by sulfonylureas, the aim of this study is to assess associations between molecular variants of KCNJ11, ABCC8 and TCF7L2 genes and response to sulfonylurea treatment and to predict their potential functional effects.
Methods
Based on a comprehensive literature search, we found 13 pharmacogenetic studies showing that single nucleotide polymorphisms (SNPs) located in KCNJ11: rs5219 (E23K), ABCC8: rs757110 (A1369S), rs1799854 (intron 15, exon 16 -3C/T), rs1799859 (R1273R), and TCF7L2: rs7903146 (intron 4) were significantly associated with responses to sulfonylureas. For in silico bioinformatics analysis, SIFT, PolyPhen-2, PANTHER, MutPred, and SNPs3D were applied for functional predictions of 36 coding (KCNJ11: 10, ABCC8: 24, and TCF7L2: 2; all are missense), and HaploReg v4.1, RegulomeDB, and Ensembl’s VEP were used to predict functions of 7 non-coding (KCNJ11: 1, ABCC8: 1, and TCF7L2: 5) SNPs, respectively.
Results
Based on various in silico tools, 8 KCNJ11 missense SNPs, 23 ABCC8 missense SNPs, and 2 TCF7L2 missense SNPs could affect protein functions. Of them, previous studies showed that mutant alleles of 4 KCNJ11 missense SNPs and 5 ABCC8 missense SNPs can be successfully rescued by sulfonylurea treatments. Further, 3 TCF7L2 non-coding SNPs (rs7903146, rs11196205 and rs12255372), can change motif(s) based on HaploReg v4.1 and are predicted as risk factors by Ensembl’s VEP.
Conclusions
Our study indicates that a personalized medicine approach by tailoring sulfonylurea therapy of T2D patients according to their genotypes of KCNJ11, ABCC8, and TCF7L2 could attain an optimal treatment efficacy.
Keywords: Sulfonylurea, Type 2 diabetes, Pharmacogenetics, ABCC8, KCNJ11, TCF7L2, Single nucleotide polymorphism, Bioinformatics, In silico
Background
The prevalence of diabetes is increasing at a fast rate, which was 6.4% (285 million) among adults aged 20–79 years in 2010, and will increase to 7.7% (438 million) by 2030 [1]. Among all diabetic cases, approximately 90% are patients with type 2 diabetes (T2D), which is associated with a number of microvascular complications including retinopathy, nephropathy, neuropathy, as well as macrovascular complications [2]. T2D is caused by a plethora of lifestyle and genetic factors [3, 4]. Current therapies for T2D include life-style modifications and use of oral antidiabetic drugs, with sulfonylurea being one of the most frequently used one [5]. There are a number of different sulfonylurea treatments for T2D patients, among which the commonly used ones are gliclazide, glibenclamide, glimepiride and glipizide [6].
Sulfonylurea promotes insulin secretion from the pancreatic β cells of the pancreas in a glucose-independent manner by binding to ATP-sensitive K+ (KATP) channel on the cell membrane of pancreatic β cells. KATP channel is a hetero-octamer comprising the inward-rectifier potassium ion channels Kir6.x (i.e., Kir6.1 and Kir6.2) that form the pore, and sulfonylurea receptors (SUR; i.e., SUR1, SUR2A, and SUR2B) that regulate the opening and closing of its associated Kir6.x potassium channel, as SUR is sensitive to ATP and ADP levels. The binding of sulfonylureas to the corresponding receptors could lead to an efflux of intracellular potassium, hyperpolarization of the β cell membrane, and the opening of voltage-gated calcium channels, which result in an increased secretion of insulin to circulation (Fig. 1).
Fig. 1.
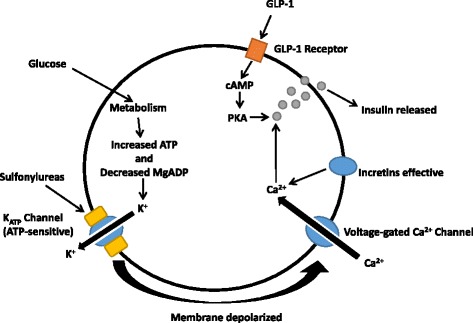
A schematic representation of the pancreatic β cell illustrating the molecular model for insulin secretion mediated by KATP channel comprising KCNJ11 and ABCC8 subunits in sulfonylurea treatment
The pancreatic β cell KATP channel consists of four pore-forming subunits of the inwardly rectifying potassium channel Kir6.2 and four regulatory subunits of the SUR1 [7–9]. When blood glucose concentrations rise, an increase in glucose metabolism results in a change of ADP/ATP ratio, which leads to a closing of KATP channel. The respective genes encoding Kir6.2 and SUR1, i.e., KCNJ11 and ABCC8, are located next to each other on human chromosome 11p15.15. Mutations in KCNJ11 or ABCC8 genes could decrease or abolish the metabolic sensitivity of β cell KATP channel function, leading to a constant depolarization of the cell membrane and a persistent insulin secretion even at very low plasma glucose concentrations [10]. E.g., single nucleotide polymorphism (SNP) E23K (i.e., rs5219) of KCNJ11 gene is associated with T2D risk (reviewed in [11]), is shown to result in a decrease or loss of sensitivity of KATP channel to the inhibitory effect of ATP [12] and/or an enhancement of activation by free fatty acids [13]. Further, mutations in ABCC8 gene could cause hyperinsulinemic hypoglycemia [10]. The β cell KATP channel can be pharmacologically regulated by sulfonylureas, which function by binding to and closing the KATP channel [14] that leads to membrane depolarization, which subsequently results in an activation of voltage-dependent calcium channels causing an influx of calcium, which then triggers insulin granule exocytosis.
TCF7L2 encodes a member of the T-cell factor (TCF) transcription factor that plays a critical role in Wnt signaling pathway [15], which is shown to be involved in β cell dysfunction in T2D [16]. TCF7L2 is a member of the TCF-lymphocyte enhancer factor (LEF) protein family [17], and the bipartite transcription factor β-catenin/TCF-LEF serves as an effector of cAMP-dependent protein kinase A (PKA) signaling to mediate the physiological effects of peptide hormones including glucagon-like peptide-1 (GLP-1), which utilizes cAMP as a second messenger [18, 19]. TCF7L2 gene SNPs are strongly associated with a higher risk of T2D development [15], which could be mediated by their influences on blood glucose homeostasis [20].
Sulfonylureas show considerable inter-individual variations in the hypoglycemic response, with approximately 10–20% of patients having a less than 20 mg/dl reduction in fasting plasma glucose (FPG) following the initiation of sulfonylurea therapy (called primary sulfonylurea failure) [21]. Further, about 50–60% of patients will initially have a greater than 30 mg/dl reduction in FPG, but will fail to reach the desired glycemic treatment goals [21]. In contrast, some T2D patients could have higher risks of mild or severe hypoglycemia in response to sulfonylurea treatment [22–24]. Molecular variants of sulfonylurea drug target genes KCNJ11, ABCC8, and TCF7L2 could lead to different responses to sulfonylurea therapy in T2D patients. Therefore, their impacts need to be carefully evaluated. The primary objective of this study is to predict functional effects of 36 coding (KCNJ11: 10, ABCC8: 24, and TCF7L2: 2, and all missense) and 7 non-coding (KCNJ11: 1, ABCC8: 1, and TCF7L2: 5) SNPs that were identified from published literatures and MutDB database (http://www.mutdb.org/) by applying a spectrum of in silico bioinformatics tools. Each Kir6.2 subunit has two transmembrane domains called M1 and M2, and the pore-forming domain is located between them [25]. The locations of 10 missense SNPs (including the well-studied E23K) in the KCNJ11 protein that comprises 390 amino acids [26] are shown in Fig. 2, respectively. Each SUR1 subunit has three transmembrane domains, i.e., TMD0, TMD1, and TMD3, and two nucleotide binding domains, i.e., NBD1 and NBD2. Between TMD0 and TMD1, there is a cytosolic loop called CL3 [27]. The locations of 24 missense SNPs (including the well-studied A1369S) in the ABCC8 protein that comprises 1581 amino acids [28] are shown in Fig. 3. The human TCF7L2 gene consists of 17 exons, five of which are alternatively spliced (i.e., exons 4, 13, 14, 15, and 16) and exhibits tissue-specific expression [29]. The differential splicing of TCF7L2 potentially gives rise to three groups of protein isoforms (i.e., short-, medium-, and large-length isoforms) with highly differential functional properties. These three groups depend on the predicted stop codon usages, which are located in exons 15, 16, 17 [30]. To date, TCF7L2 intronic SNP, rs7903146, represents the most significant risk variant for T2D [31]. However, four other non-coding SNPs, i.e., rs7901695, rs7895340, rs11196205 and rs12255372, have also been significantly associated with an increased risk of T2D [32] and have been widely studied. The locations of these 5 non-coding SNPs in the gene structure of TCF7L2 (including the well-studied intronic SNP rs7903146) are illustrated in Fig. 4.
Fig. 2.
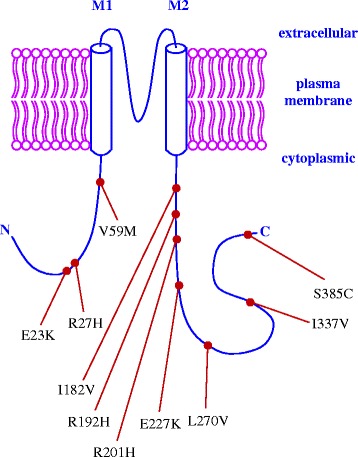
A schematic representation of 10 KCNJ11 missense SNPs in the protein product. Each Kir6.2 subunit (i.e., KCNJ11 protein product) contains two transmembrane domains, M1 and M2. Between M1 and M2, there is a pore-forming loop that creates the core of the K+ channel
Fig. 3.
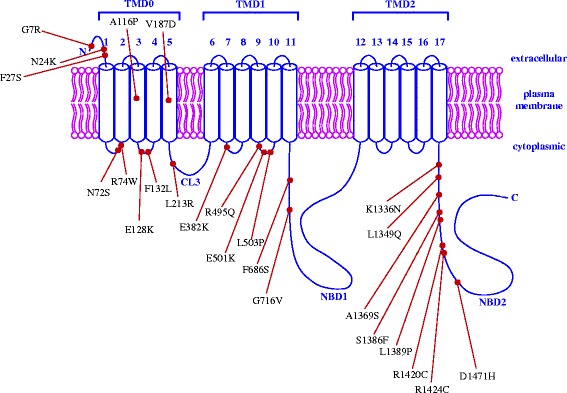
A schematic representation of 24 ABCC8 missense SNPs in the protein product. Each SUR1 subunit (i.e., ABCC8 protein product) contains 17 transmembrane helices, which are arranged in three transmembrane domains, i.e., TMD0, TMD1, and TMD2, respectively. The large cytosolic loop between TMD0 and TMD1 is called cytosolic loops 3 (CL3). The large cytosolic domains following TMD1 and TMD2 contain nucleotide-binding domain 1 (NBD1) and NBD2, respectively
Fig. 4.
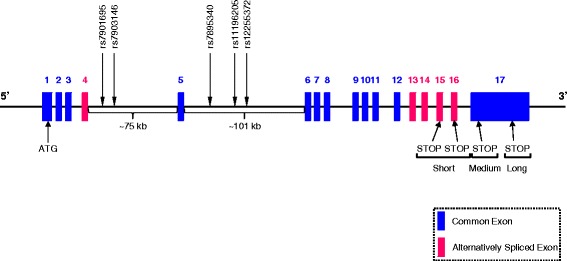
A schematic representation of 5 TCF7L2 non-coding SNPs in the gene structure. The start and stop codons are indicated by “ATG” (in exon 1) and “STOP” (in exons 15, 16, and 17), respectively. Because of alternative splicing, 3 groups of protein isoforms (i.e., short-, medium-, and large-length isoforms) can be generated by using different stop codons, which are indicated by “Short”, “Medium”, and “Long”, respectively
Methods
Literature search strategy
Comprehensive electronic literature searches of databases including PubMed, Google Scholar, Cochrane Library, Excerpta Medica Database (EMBASE) were performed up to June 1, 2016 using the following keywords: sulfonylurea, type 2 diabetes, KCNJ11, ABCC8, and TCF7L2. A manual search of the references cited in initially identified articles was also performed. Furthermore, we searched all relevant references of three comprehensive review articles [5, 33, 34]. The search was restricted to English language articles.
Inclusion and exclusion criteria
Randomized controlled trials and observational studies were eligible for inclusion in the current study. In vitro studies, animal studies, letters, reviews, and unrelated articles and duplicates were excluded from this study.
Data extraction
From each included study, the following data were extracted: first author, publication year, SNP name, gene name, National Center for Biotechnology Information (NCBI) dbSNP (http://www.ncbi.nlm.nih.gov/snp/) ID, study design, study subjects, control source, length of follow-up, and results.
In silico bioinformatics analysis
Computational predictions of functional impacts of non-synonymous SNPs (nsSNPs)
Five in silico tools were applied: (i) SIFT [35] (http://sift.jcvi.org/), (ii) PolyPhen-2 [36] (http://genetics.bwh.harvard.edu/pph2/), (iii) PANTHER [37] (http://www.pantherdb.org/tools/csnpScore.do), (iv) MutPred [38]) (http://mutpred.mutdb.org/), and (v) SNPs3D [39] (http://www.snps3d.org/).
Computational predictions of functional impacts of non-coding SNPs
Three in silico tools were applied: (i) HaploReg v4.1 [40, 41] (http://www.broadinstitute.org/mammals/haploreg/haploreg.php), (ii) RegulomeDB [42] (http://regulomedb.org/), and (iii) Ensembl’s VEP [43] (http://www.ensembl.org/Homo_sapiens/Tools/VEP?db=core).
Results
A total of 17 articles corresponding to 17 independent studies were qualified and subsequently included for evaluating the relationships between KCNJ11, ABCC8 and TCF7L2 SNPs and response to sulfonylurea in patients with T2D. The detailed characteristics of these 17 studies [44–60] were presented in Table 1. Of them, 13 studies gave positive results, which showed that 1 SNP located in KCNJ11 rs5219 (E23K) [45–47, 49, 50, 52], 3 SNPs located in ABCC8: rs757110 (A1369S) [46, 56], rs1799854 (intron 15, exon 16 -3C/T) [48, 53, 55], rs1799859 (R1273R) [48, 55], and 1 SNP located in TCF7L2: rs7903146 (intron 4) [58–60], were significantly associated with responses to sulfonylureas. It is noteworthy that no uniform definition of response to sulfonylurea therapy was used across these 17 independent studies. Javorsky et al. (2012) [50] defined response to sulfonylurea as change in HbA1c level to sulfonylurea at 6-month therapy. Feng et al. (2008) [46] defined response to sulfonylurea as percent decrease in FPG and also FPG at day 57 as < 7.8 mmol/l as well as percent decrease in HbA1c after 8-week sulfonylurea therapy, and Holstein et al. (2009) [47] defined this drug response phenotype as sulfonylurea-induced hypoglycemia, which refers to a symptomatic event requiring treatment with intravenous glucose that was confirmed by a blood glucose measurement of <50 mg/dl. Meirhaeghe et al. (2001) [53] defined response to sulfonylurea as post-treatment fasting insulin, FPG, fasting plasma total cholesterol, and fasting plasma triglyceride concentrations, and Schroner et al. (2011) [59] defined this drug response phenotype as change of HbA1c (%) and changes of FPG for 3-month treatment and 6-month treatment, respectively. Pearson et al. (2007) [58] defined response to sulfonylurea as failure to reach a target HbA1c < 7% within 1-year treatment and minimum HbA1c achieved within 1-year treatment, and they also considered time taken on sulfonylurea treatment to achieve target HbA1c < 7% as a drug response phenotype. In addition, Sesti et al. (2006) [45] defined secondary sulfonylurea failure as FPG greater than 300 mg/dl despite sulfonylurea-metformin combined therapy and appropriate diet, in the absence of other conditions causing hyperglycemia, but Holstein et al. (2011) [60] defined secondary sulfonylurea failure as the addition of insulin after at least 6-month sulfonylurea therapy and corresponding HbA1c ≥ 7%. In the following, we first summarize major results of SNPs’ effects on sulfonylurea responses in a gene-by-gene manner, and then, we present functional prediction results for nsSNPs and non-coding SNPs by respective online bioinformatics tools.
Table 1.
Characteristics of included studies (N = 17)*
| Study ID | Author | Year | Gene Symbol | SNP Name | dbSNP ID | Study Design | Study Subjects | Control Source | Length of Follow-up | Results | Association |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Study 1 | Gloyn et al. [44] | 2001 | KCNJ11 | E23K | rs5219 | RCT | 363 Caucasian T2D and 307 normoglycemic control subjects | UKPDS | 1 year | Variant allele did not significantly affect the response to SU therapy significantly | No |
| Study 2 | Sesti et al. [45] | 2006 | KCNJ11 | E23K | rs5219 | RCT | 525 Caucasian T2D patients with secondary SU failure | Hospital-based | NA | Secondary SU failure, K allele vs E allele (OR = 1.45; 95% CI 1.01–2.09; P = 0.04). Adjustment for age, gender, fasting glycemia, glycosylated hemoglobin, age at diagnosis, and duration of diabetes in a logistic regression analysis did not change this association (OR = 1.69; 95% CI: 1.02–2.78; P = 0.04) | Yes |
| Study 3 | Feng et al. [46] | 2008 | KCNJ11 | E23K | rs5219 | RCT | 1268 Chinese T2D patients treated with 8-week gliclazide | Hospital-based | 8 weeks | E23K variant of the KCNJ11 gene was significantly associated with decreases in FPG (P = 0.002). | Yes |
| Study 4 | Holstein et al. [47] | 2009 | KCNJ11 | E23K | rs5219 | Case–control | 43 T2D patients treated with glimepiride or glibenclamide | Hospital-based | NA | E23K variant was significantly associated with increased HbA1c levels (adjusted P = 0.04) independent of age, sex, body mass index, diabetes duration and SU dose. | Yes |
| Study 5 | Nikolac et al. [48] | 2009 | KCNJ11 | E23K | rs5219 | Cross-sectional | 228 Caucasian T2D patients with SU therapy | Hospital-based | NA | For KCNI11 E23K polymorphism, for different genotype groups, there were no significant differences of FPG, PPG, and HbA1c concentrations (P = 0.143, 0.675, and 0.824, respectively) | No |
| Study 6 | El-sisi et al. [49] | 2011 | KCNJ11 | E23K | rs5219 | Case–control | 50 Egyptian T2D patients with secondary SU failure | Hospital-based | NA | Secondary SU failure, EK + KK vs. EE (RR = 1.65; 95% CI: 1.04–2.6; P = 0.04). | Yes |
| Study 7 | Javorsky et al. [50] | 2012 | KCNJ11 | E23K | rs5219 | RCT | 55 T2D patients with 6-month treatment of gliclazide | Hospital-based | 6 months | For ΔHbA1c EK + KK vs. EE (1.15 ± 0.09 vs. 0.80 ± 0.13, P = 0.036) | Yes |
| Study 7 | Javorsky et al. [50] | 2012 | KCNJ11 | E23K | rs5219 | RCT | 28 T2D patients with 6-month treatment of glimepiride | Hospital-based | 6 months | For ΔHbA1c EK + KK vs. EE (1.10 ± 0.12 vs. 1.00 ± 0.19 P = 0.676) | No |
| Study 7 | Javorsky et al. [50] | 2012 | KCNJ11 | E23K | rs5219 | RCT | 14 T2D patients with 6-month treatment of glibenclamide | Hospital-based | 6 months | For ΔHbA1c EK + KK vs. EE (1.05 ± 0.11 vs. 0.98 ± 0.09 P = 0.633) | No |
| Study 8 | Ragia et al. [51] | 2012 | KCNJ11 | E23K | rs5219 | Case–control | 92 T2D patients (80 glimepiride/12 gliclazide) who had experienced at least one drug-associated hypoglycemic event, while 84 T2D patients (74 glimepiride/10 gliclazide) who had never experienced a hypoglycemic event | Hospital-based | NA | KCNJ11 E23K genotype and allele frequencies were not different between hypoglycemic and non-hypoglycemic T2D patients (P = 0.35 and 0.47, respectively). In logistic regression models before and after adjustment for other risk factors (age, body mass index, sulfonylurea mean daily dose, duration of T2D, renal function and CYP2C9 genotype), KCNJ11 E23K polymorphism did not affect hypoglycemia risk | No |
| Study 9 | Li et al. [52] | 2014 | KCNJ11 | E23K | rs5219 | RCT | 108 Chinese T2D patients treated with gliclazide for 16 weeks | Hospital-based | 16 weeks | Patients with the KK genotype had larger augmentations in changes (Δ) in acute insulin response (P = 0.049) and D body mass index (P = 0.003); Patients with the EK genotype had a lower variance in changes in fasting insulin levels (P = 0.049) and homeostasis model assessment of β cell function (P = 0.021) than those with the KK genotype | Yes |
| Study 1 | Gloyn et al. [44] | 2001 | KCNJ11 | L270V | rs1800467 | RCT | 363 Caucasian T2D patients | UKPDS | 1 year | Variant allele did not significantly affect the response to SU therapy significantly | No |
| Study 10 | Meirhaeghe et al. [53] | 2001 | ABCC8 | Intron 15, exon 16 -3C/T | rs1799854 | Cross-sectional | 70 T2D patients with SU therapy | 3 large representative French samples (in Lille, Strasbourg, and Toulouse) participating in the risk factor surveys of the WHO-MONICA | NA | For T2D patients treated with SU agents, those subjects bearing at least one -3C allele and had fasting plasma TG concentrations 35% lower than TT homozygotes [2.20 mmol/L (1.14–4.14) for TT vs. 1.43 mmol/L (0.81–2.52) for TC + CC; P = 0.026] | Yes |
| Study 11 | Zychma et al. [54] | 2002 | ABCC8 | Intron 15, exon 16 -3C/T | rs1799854 | Case–control | 68 Caucasian T2D patients who required insulin treatment and had known diabetes duration ≤ 5 years, compared to 99 Caucasian T2D patients receiving SU alone or in combination with metformin or acarbose with known diabetes duration ≥ 15 years | Hospital-based | NA | There was no significant impact of ABCC8 exon 16 -3C/T polymorphism on the early ineffectiveness of SU treatment (P = 0.4126 based on a Chi-square test) | No |
| Study 5 | Nikolac et al. [48] | 2009 | ABCC8 | Intron 15, exon 16 -3C/T | rs1799854 | Cross-sectional | 228 Caucasian T2D patients with SU therapy | Hospital-based | NA | CC genotype of the ABCC8 exon 16 polymorphism had significantly lower HbA1c concentration compared to the patients with T genotype [6.9 (6.2–7.7) mmol/L vs. 8.1 (6.7–8.8) mmol/L; P = 0.009] | Yes |
| Study 12 | Nikolac et al. [55] | 2012 | ABCC8 | Intron 15, exon 16 -3C/T | rs1799854 | Cross-sectional | 251 Caucasian T2D patients with SU therapy | Hospital-based | NA | Polymorphic allele carriers of the ABCC8 intron 15 -3C/T (which is 3 bp ahead of exon 16) polymorphism were more frequent in the subgroup of patients with the TG concentration increase after 6 months (P for genotype and allelic differences: 0.024 and 0.015, respectively) | Yes |
| Study 13 | Zhang et al. [56] | 2007 | ABCC8 | A1369S | rs757110 | RCT | 115 T2D patients with gliclazide treatment for 8 weeks | Hospital-based | 8 weeks | For ΔHbA1c TG + GG vs. TT (1.60 ± 1.39 vs. 0.76 ± 1.70, P = 0.044) | Yes |
| Study 3 | Feng et al. [46] | 2008 | ABCC8 | A1369S | rs757110 | RCT | 1268 Chinese T2D patients treated with 8-week gliclazide | Hospital-based | 8 weeks | Compared with TT genotype, subjects with the GG genotype had a 7.7% greater decrease in FPG (P < 0.001), an 11.9% greater decrease in 2-h plasma glucose (P = 0.003), and a 3.5% greater decrease in HbA1c (P = 0.06) | Yes |
| Study 14 | Sato et al. [57] | 2010 | ABCC8 | A1369S | rs757110 | Case–control | 32 patients with T2D admitted to hospital with severe hypoglycemia and 125 consecutive T2D outpatients without severe hypoglycemia, and all of the patients were taking glimepiride or glibenclamide | Hospital-based | NA | There were no significant differences in ABCC8 A1369S genotype distribution between patients with or without severe hypoglycemia (P = 0.26). Moreover, the A1369 allele tended to be less frequent in the hypoglycemic group (31 vs. 43%; OR = 1.65; 95% CI: 0.92–2.96; P = 0.09) | No |
| Study 5 | Nikolac et al. [48] | 2009 | ABCC8 | R1273R** | rs1799859 | Cross-sectional | 228 Caucasian T2D patients with SU therapy | Hospital-based | NA | GG genotype of the ABCC8 exon 31 polymorphism had significantly higher HbA1c concentration compared to the AA genotype [7.8 (6.9–8.8) mmol/L vs. 6.3 (5.7–6.8) mmol/L; P < 0.001] | Yes |
| Study 12 | Nikolac et al. [55] | 2012 | ABCC8 | R1273R** | rs1799859 | Cross-sectional | 251 Caucasian T2D patients with SU therapy | Hospital-based | NA | Wile-type G allele carriers had a significantly higher TG concentration when compared with the carriers of two variant A alleles (P = 0.023) | Yes |
| Study 15 | Pearson et al. [58] | 2007 | TCF7L2 | NA | rs7903146 | RCT | 901 T2D patients with SU treatment | GoDARTS | 12 months | Carriers of the risk allele were less likely to respond to SUs with an OR for failure of 1.95 (95% CI: 1.23–3.06; P = 0.005), comparing rs12255372 TT vs. GG. Including the baseline HbA1c strengthened this association (OR = 2.16, 95% CI: 1.21–3.86, P = 0.009) | Yes |
| Study 16 | Schroner et al. [59] | 2011 | TCF7L2 | NA | rs7903146 | RCT | 87 T2D patients with 6-month SU treatment in addition to metformin | Hospital-based | 6 months | Reduction in HbA1c: CC vs. CT + TT is 1.16 ± 0.07 vs. 0.86 ± 0.07%, P = 0.003 Reduction in FPG: 1.57 ± 0.12 vs. 1.14 ± 0.14 mmol/L, P = 0.031) | Yes |
| Study 17 | Holstein et al. [60] | 2011 | TCF7L2 | NA | rs7903146 | RCT | 189 T2D patients with 6-month SU treatment | Hospital-based | 6 months | T allele was significantly more frequent in the group of patients who failed to respond to SU (i.e., those with HbA1c ≥ 7%) (36%) than in the control (i.e., those with HbA1c < 7%) group (26%) (OR = 1.57, 95% CI: 1.01-2.45, P = 0.046) | Yes |
*Studies are grouped by different genes. For each gene, studies are first sorted by SNP Name, then by Year, and then by Author, in ascending orders. Abbreviations: CI confidence interval, FPG fasting plasma glucose, Go-DARTS Genetics of Diabetes Audit and Research Study in Tayside Scotland, HbA1c glycated hemoglobin A1c, OR odds ratio, RCT randomized clinical trial, SNP single nucleotide polymorphism, SU sulfonylurea, SUR sulfonylurea receptor, T2D type 2 diabetes, TG triglyceride, UKPDS United Kingdom Prospective Diabetes Study, WHO-MONICA World Health Organization-Multinational MONItoring of trends and determinants of CArdiovascular diseases, NA not available
**Because R1273R is a synonymous SNP, it is not included in functional prediction
KCNJ11
The most widely studied genetic polymorphism of KCNJ11 for sulfonylurea response is E23K (i.e., rs5219) located in exon 1 [33]. However, functional effects of KCNJ11 E23K polymorphism on the secretion and sensitivity of insulin in humans remain contentious [5]. Recent larger studies demonstrated that a significant reduction of insulin secretion, lower levels of insulin, and an improvement of insulin sensitivity were related to E23K variant in KCNJ11 gene [61]. Moreover, E23K variant was associated with T2D development, which means that the K allele carriers had an increased risk of T2D [44, 62, 63]. Furthermore, some studies also found that the K allele carriers had better therapeutic response to gliclazide in comparison with the EE homozygous wild-type group [50], as well as an increased risk of sulfonylurea treatment failure [45, 49]. In addition, E23K variant was significantly associated with an increase of glycated hemoglobin A1c (HbA1c) level [47] and fasting glucose level that patients with the KK homozygous variant genotype had lower fasting glucose levels than those with the EE/EK heterozygous genotype [52]. Importantly, recent evidence demonstrated that patients with KCNJ11 variants responded more efficiently to sulfonylurea than insulin [64–66]. Another KCNJ11 polymorphism that was associated with sulfonylurea treatment responses is rs5210 which is located in 3’- untranslated region (UTR). A study conducted in two independent cohorts of Chinese T2D patients (cohort 1: n = 661, cohort 2: n = 607) treated with gliclazide demonstrated that KCNJ11 rs5210 was positively associated with gliclazide response in cohort 1 study [46].
ABCC8
The most widely studied genetic polymorphism of ABCC8 for sulfonylurea response is S1369A (i.e., rs757110) located in exon 33 [67]. This genetic variant was demonstrated to influence antidiabetic efficacy of sulfonylurea treatment in Chinese [46, 56], as well as an increased sensitivity to gliclazide [56]. More importantly, KCNJ11 E23K and ABCC8 S1369A, two common KATP channel mutations that were in strong linkage disequilibrium, form a haplotype that appears to be associated with an increased T2D risk [68]. Additional ABCC8 gene polymorphisms including rs1799854 (intron 15, exon 16 -3C/T) and rs1799859 (exon 31) had been shown to be associated with sulfonylurea treatment efficacy in Caucasians [48, 55].
TCF7L2
Previous studies have shown that several non-coding genetic variants of TCF7L2 are associated with T2D risk in populations of diverse ancestries from countries encompassing United Kingdom [69], the Netherlands [70], Finland [32], Sweden [71], France [72], United States [73], India [74], and Japan [75] populations. Among these T2D-associated TCF7L2 variants, rs7903146 (intron 4) showed the strongest association with T2D [76]. Significant reductions in HbA1c and fasting plasma glucose levels following a combined sulfonylurea and metformin treatment between T2D patients with CC genotype and those with CT/TT genotype were associated with TCF7L2 rs7903146 variant allele [59]. Moreover, the rs12255372 variant, together with the rs7903146 variant, was shown to be associated with a significantly more frequent treatment failure [58–60]. It shall be noted that although in previous literatures, e.g., as in [32, 77], TCF7L2 rs7901695 and rs7903146 are indicated to be in intron 3, and rs7895340, rs11196205 and rs12255372 are indicated to be in intron 4, this is because exon 4, which is a variable exon, is often named as “3a” [78]. Because of a high incorporation in pancreatic β cells [79], exon 4 shall be included in the gene structure, such that rs7901695 and rs7903146 shall be indicated as located in intron 4, and rs7895340, rs11196205, and rs12255372 in intron 5, respectively, e.g., as in [80]. For the linear ordering of these 5 non-coding SNPs, according to the most updated (i.e., as of April 18, 2017) NCBI dbSNP, the chromosomal coordinates for rs7901695, rs7903146, rs7895340, rs11196205 and rs12255372 are 112994329, 112998590, 113041766, 113047288, and 113049143, respectively, on human chromosome 10 based on GRCh38.p7 assembly. Therefore, the linear ordering shall be rs7901695-rs7903146-rs7895340-rs11196205-rs12255372, as shown in Fig. 4 (all drawings in Figs. 1, 2, 3, and 4 are not to their exact scales and are for illustration purposes), which is agreement with that of [77].
In silico bioinformatics analysis results
For KCNJ11, ABCC8 and TCF7L2 genes, functional prediction results for 36 nsSNPs by SIFT, PolyPhen-2, PANTHER, MutPred, and SNPs3D were presented in Table 2, and those prediction results for 7 non-coding SNPs by HaploReg v4.1, RegulomeDB and Ensembl’s VEP were presented in Table 3.
Table 2.
In silico predicted functional effects of 36 non-synonymous SNPs in the pharmacogenetics of sulfonylureas treatment by SIFT, PolyPhen-2, PANTHER, MutPred, and SNPs3D*
| SNP ID | Gene Symbol | SNP Name | dbSNP ID | SNP Location | Chromosome Location (GRCh38.p7) | SIFT Score/Prediction | PolyPhen-2 Score/Prediction | PANTHER subSPEC | PANTHER Pdeleterious | MutPred Pdeleterious | SNPs3D Score |
|---|---|---|---|---|---|---|---|---|---|---|---|
| SNP1 | KCNJ11 | E23K | rs5219 | Exon 1 | 11:17388025 | 1.00/Tolerated | 0.001/Benign | −0.69172 | 0.09044 | 0.35 | 2 |
| SNP2 | KCNJ11 | R27H | NA | Exon 1 | NA | 0.18/Tolerated | 0.006/Benign | −3.75303 | 0.67984 | 0.248 | NA |
| SNP3 | KCNJ11 | V59M | NA | Exon 1 | NA | 0.12/Tolerated | 0.999/Probably damaging | −2.72126 | 0.43076 | 0.855 | NA |
| SNP4 | KCNJ11 | I182V | NA | Exon 1 | NA | 0.98/Tolerated | 0.998/Probably damaging | −1.62168 | 0.20128 | 0.684 | NA |
| SNP5 | KCNJ11 | R192H | NA | Exon 1 | NA | 0.01/Affect Protein Function | 1.000/Probably damaging | −6.9765 | 0.98159 | 0.816 | NA |
| SNP6 | KCNJ11 | R201H | rs80356624 | Exon 1 | 11:17387490 | 0.00/Affect Protein Function | 1.000/Probably damaging | NA | NA | 0.981 | NA |
| SNP7 | KCNJ11 | E227K | NA | Exon 1 | NA | 0.00/Affect Protein Function | 1.000/Probably damaging | −7.17583 | 0.98487 | 0.94 | NA |
| SNP8 | KCNJ11 | L270V | rs1800467 | Exon 1 | 11:17387284 | 0.13/Tolerated | 0.003/Benign | −1.54301 | 0.18893 | 0.566 | 0.68 |
| SNP9 | KCNJ11 | I337V | rs5215 | Exon 1 | 11:17387083 | 0.73/Tolerated | 0.000/Benign | −0.89045 | 0.10817 | 0.462 | 0.94 |
| SNP10 | KCNJ11 | S385C | rs41282930 | NA | 11:17386938 | 0.02/Affect Protein Function | 0.380/Possibly damaging | NA | NA | 0.229 | NA |
| SNP11 | ABCC8 | G7R | NA | Exon 1 | NA | 0.00/Affect Protein Function | 1.000/Probably damaging | NA | NA | 0.863 | NA |
| SNP12 | ABCC8 | N24K | NA | Exon 1 | NA | 0.03/Affect Protein Function | 1.000/Probably damaging | NA | NA | 0.877 | NA |
| SNP13 | ABCC8 | F27S | NA | Exon 1 | NA | 0.00/Affect Protein Function | 0.884/Probably damaging | NA | NA | 0.858 | NA |
| SNP14 | ABCC8 | N72S | rs80356634 | Exon 2 | 11:17474961 | 0.12/Tolerated | 0.402/Possibly damaging | NA | NA | 0.802 | NA |
| SNP15 | ABCC8 | R74W | NA | Exon 2 | NA | 0.00/Affect Protein Function | 1.000/Probably damaging | NA | NA | 0.904 | NA |
| SNP16 | ABCC8 | A116P | NA | NA | NA | 0.12/Tolerated | 1.000/Probably damaging | NA | NA | 0.825 | NA |
| SNP17 | ABCC8 | E128K | NA | Exon 3 | NA | 0.02/Affect Protein Function | 1.000/Probably damaging | NA | NA | 0.829 | NA |
| SNP18 | ABCC8 | F132L | rs80356637 | Exon 3 | 11:17470119 | 0.16/Tolerated | 0.877/Possibly damaging | NA | NA | 0.847 | NA |
| SNP19 | ABCC8 | V187D | NA | Exon 4 | NA | 0.01/Affect Protein Function | 0.042/Benign | NA | NA | 0.857 | NA |
| SNP20 | ABCC8 | L213R | rs80356642 | Exon 5 | 11:17461767 | 0.41/Tolerated | 0.212/Possibly damaging | −3.12006 | 0.52998 | 0.786 | NA |
| SNP21 | ABCC8 | E382K | rs80356651 | NA | 11:17453151 | 0.27/Tolerated | 0.392/Possibly damaging | −1.96296 | 0.26172 | 0.872 | NA |
| SNP22 | ABCC8 | R495Q | NA | Exon 10 | NA | 0.00/Affect Protein Function | 1.000/Probably damaging | −8.28432 | 0.99496 | 0.906 | NA |
| SNP23 | ABCC8 | E501K | NA | Exon 10 | NA | 0.00/Affect Protein Function | 1.000/Probably damaging | −2.39817 | 0.35392 | 0.948 | NA |
| SNP24 | ABCC8 | L503P | NA | Exon 10 | NA | 0.00/Affect Protein Function | 1.000/Probably damaging | −4.69708 | 0.84515 | 0.964 | NA |
| SNP25 | ABCC8 | F686S | NA | Exon 15 | NA | 0.01/Affect Protein Function | 0.998/Probably damaging | −4.1174 | 0.75351 | 0.909 | NA |
| SNP26 | ABCC8 | G716V | rs72559723 | Exon 16 | 11:17427124 | 0.18/Tolerated | 1.000/Probably damaging | −8.97797 | 0.99747 | 0.974 | NA |
| SNP27 | ABCC8 | K1336N | NA | NA | NA | 0.25/Tolerated | 0.016/Benign | −1.84974 | 0.24044 | 0.693 | NA |
| SNP28 | ABCC8 | L1349Q | NA | Exon 33 | NA | 0.01/Affect Protein Function | 0.997/Probably damaging | −4.16681 | 0.76257 | 0.912 | NA |
| SNP29 | ABCC8 | A1369S | rs757110 | Exon 33 | 11:17396930 | 0.51/Tolerated | 0.000/Benign | −0.85285 | 0.1046 | 0.323 | 0.87 |
| SNP30 | ABCC8 | S1386F | NA | Exon 34 | NA | 0.00/Affect Protein Function | 1.000/Probably damaging | −4.60703 | 0.833 | 0.95 | NA |
| SNP31 | ABCC8 | L1389P | NA | Exon 34 | NA | 0.00/Affect Protein Function | 0.993/Probably damaging | −4.93634 | 0.87395 | 0.886 | NA |
| SNP32 | ABCC8 | R1420C | rs28938469 | Exon 35 | 11:17395659 | 0.00/Affect Protein Function | 1.000/Probably damaging | NA | NA | 0.863 | −1.19 |
| SNP33 | ABCC8 | I1424V | rs80356653 | Exon 35 | 11:17395647 | 0.00/Affect Protein Function | 0.988/Probably damaging | −2.02622 | 0.27413 | 0.882 | NA |
| SNP34 | ABCC8 | D1471H | NA | Exon 36 | NA | 0.00/Affect Protein Function | 0.994/Probably damaging | −4.60764 | 0.83308 | 0.913 | NA |
| SNP35 | TCF7L2 | P179H | rs3197486 | NA | 10:113141236 | 0.00/Affect Protein Function | 1.000/Probably damaging | −5.62868 | 0.93268 | 0.879 | 0.01 |
| SNP36 | TCF7L2 | K323N | rs2757884 | NA | 10:113151761 | 0.00/Affect Protein Function | 1.000/Probably damaging | −4.23841 | 0.77529 | 0.309 | −0.15 |
* Abbreviations: MutPred Mutation Prediction, PANTHER, Protein ANalysis THrough Evolutionary Relationships, PolyPhen-2 Polymorphism Phenotyping v2, SIFT Sorting Intolerant from Tolerant, SNP Single Nucleotide Polymorphism, subSPEC subStitution Position-specific Evolutionary Conservation, NA Not Available
Table 3.
In silico predicted functional effects of 7 non-coding SNPs in the pharmacogenetics of sulfonylureas treatment by Haploreg v4.1, RegulomeDB, and Ensembl’s VEP*
| SNP ID | Gene Symbol | dbSNP ID | SNP Location | Chromosome Location (GRCh38.p7) | HaploReg v4.1 Motifs changed by SNP |
RegulomeDB Score/Prediction | Ensembl’s VEP |
|---|---|---|---|---|---|---|---|
| SNP37 | KCNJ11 | rs5210 | 3’-UTR | 11:17386704 | None | 4/Minimal binding evidence | NA |
| SNP38 | ABCC8 | rs1799854 | Intron 15 | 11:17427157 | 4 altered motifs | 5/Minimal binding evidence | NA |
| SNP39 | TCF7L2 | rs7895340 | Intron 5 | 10:113041766 | Irf, PRDM1 | NA | NA |
| SNP40 | TCF7L2 | rs7901695 | Intron 4 | 10:112994329 | None | 5/Minimal binding evidence | NA |
| SNP41 | TCF7L2 | rs7903146 | Intron 4 | 10:112998590 | 7 altered motifs | 5/Minimal binding evidence | Risk factor |
| SNP42 | TCF7L2 | rs11196205 | Intron 5 | 10:113047288 | SMC3 | 5/Minimal binding evidence | Risk factor |
| SNP43 | TCF7L2 | rs12255372 | Intron 5 | 10:113049143 | 5 altered motifs | NA | Risk factor |
*Abbreviations: RegulomeDB Regulome Database, SNP Single Nucleotide Polymorphism, UTR Untranslated Region, VEP Variant Effect Predictor, NA Not Available
Analysis of functional effects of SNPs by SIFT
SIFT was used to predict the functional impact of an nsSNP on a protein molecule. An nsSNP with a SIFT score ≤ 0.05 is considered as having a deleterious effect on protein function [81]. A total of 22 nsSNPs were predicted to affect protein function (SIFT score range: 0.00-0.03) including 4 KCNJ11 missense SNPs (R192H, R201H, E227K, S385C), 16 ABCC8 missense SNPs (G7R, N24K, F27S, R74W, E128K, V187D, R495Q, E501K, L503P, F686S, L1349Q, S1386F, L1389P, R1420C, I1424V, D1471H), and 2 TCF7L2 missense SNPs (P179H, K323N), whereas the remaining 14 missense SNPs were predicted to be tolerated (SIFT score range: 0.12–1.00) (Table 2).
Analysis of functional effects of nsSNPs by PolyPhen-2
PolyPhen-2 calculates a naïve Bayes posterior probability for a given mutation that it will be benign (PolyPhen-2 score < 0.15), possibly damaging (PolyPhen-2 score is greater than or equal to 0.15 but is less than 0.85), or probably damaging (PolyPhen-2 score ≥ 0.85), respectively [82]. A total of 25 nsSNPs were predicted to be probably damaging to protein function (PolyPhen-2 score range: 0.877–1.000), which includes 5 KCNJ11 missense SNPs (V59M, I182V, R192H, R201H, E227K), 18 ABCC8 missense SNPs (G7R, N24K, F27S, R74W, A116P, E128K, F132L, R495Q, E501K, L503P, F686S, G716V, L1349Q, S1386F, L1389P, R1420C, I1424V, D1471H) and 2 TCF7L2 missense SNPs (P179H, K323N), and the remaining 11 SNPs were classified as benign (PolyPhen-2 score range: 0.000–0.402) (Table 2).
Analysis of functional effects of nsSNPs by PANTHER
PANTHER characterizes likely functional effect of amino acid variation by means of a hidden Markov model-based statistical modeling and evolutionary relationship. The SNP with subSPEC score ≤ −3 is considered as intolerant or deleterious, whereas SNP with subSPEC score > −3 is classified to be less deleterious [83]. A total of 14 amino acid substitutions were classified as intolerant (subSPEC score range: from−8.97797 to−3.12006) including 3 KCNJ11 missense SNPs (R27H, R192H, E227K), 9 ABCC8 missense SNPs (L213R, R495Q, L503P, F686S, G716V, L1349Q, S1386F, L1389P, D1471H) and 2 TCF7L2 missense SNPs (P179H, K323N), another 10 amino acid substitutions were classified as tolerated (subSPEC score range: from−2.72126 to−0.69172), and the remaining 12 amino acid substitutions did not have subSPEC scores (Table 2).
Analysis of functional effects of nsSNPs by MutPred
MutPred predicts molecular causes of disease or deleterious amino acid substitution. A total of 30 nsSNPs had p-values > 0.5, which were considered to be functional [84] (MutPred Pdeleterious range: 0.566-0.981), which included 6 KCNJ11 missense SNPs (V59M, I182V, R192H, R201H, E227K, L270V), 23 ABCC8 missense SNPs (G7R, N24K, F27S, N72S, R74W, A116P, E128K, F132L, V187D, L213R, E382K, R495Q, E501K, L503P, F686S, G716V, K1336N, L1349Q, S1386F, L1389P, R1420C, I1424V, D1471H) and 2 TCF7L2 missense SNPs (P179H, K323N) (Table 2).
Analysis of functional consequences of SNPs by SNPs3D
SNPs3D assigns molecular functional effects of nsSNPs based on structure and sequence analysis. Of the 36 nsSNPs, SNPs3D SVM score was available for only 7 nsSNPs (KCNJ11: 2, ABCC8: 3, and TCF7L2: 2). Of them, two nsSNPs, i.e., R1420C amino acid substitution of ABCC8 gene and K323N amino acid substitution of TCF7L2 gene, had SVM scores < 0, which were classified as deleterious substitutions [85] (Table 2).
Analysis of functional consequences of SNPs by HaploReg v4.1
HaploReg v4.1 is an online software for exploring annotations of the non-coding genome among those results of published genome-wide association studies or new sets of genetic variants, which help researchers to integrate DNA regulatory elements data with genetic variants to quickly formulate novel biological hypotheses [40, 41]. As predicted by HaploReg v4.1, rs1799854, rs7895340, rs7903146, rs11196205 and rs12255372 could change 4, 2 (i.e., Irf and PRDM1), 7, 1 (i.e., SMC3), and 5 DNA motifs for DNA-binding proteins, and could have regulatory effects on gene transcription. Neither rs5210 nor rs7901695 appear to change known motifs (Table 3).
Analysis of functional consequences of SNPs by RegulomeDB
RegulomeDB is a database that annotates SNPs with known and predicted regulatory elements in the intergenic regions of the human genome. Of the 7 non-coding SNPs, rs5210, rs1799854, rs7901695, rs7903146, and rs11196205 had RegulomeDB scores of 4, 5, 5, 5, and 5, respectively, which were all classified as having minimal binding evidence. Predictions were not available for either rs7895340 or rs12255372 (Table 3).
Analysis of functional consequences of SNPs by Ensembl’s VEP
The Ensembl’s VEP determines the effects of genetic variants on genes, transcripts, and protein sequences, as well as regulatory regions. Three non-coding SNPs of TCF7L2 gene, i.e., rs7903146, rs11196205 and rs12255372, were predicted as risk factors (Table 3).
Discussion
Sulfonylureas are a class of drugs that stimulates insulin secretion by closing KATP channels in pancreatic β cells. It has been estimated that 10–20% of individuals treated do not attain adequate glycemic control, and 5–10% initially responding to sulfonylurea subsequently lose the ability to maintain near-normal glycemic level [86]. This implies that genetic factors are linked with treatment efficacy of sulfonylureas. In our study, that includes 17 studies, two KCNJ11 SNPs — rs5219 (E23K) (exon 1) and rs5210 (3’-UTR), three ABCC8 SNPs — rs757110 (A1369S) (exon 33), rs1799854 (intron 15, exon 16 -3C/T), rs1799859 (R1273R) (exon 31), and two TCF7L2 SNPs rs7903146 (intron 4) and rs12255372 (intron 5) have been associated with response to sulfonylureas. Based on bioinformatics predictions for 36 selected coding SNPs (all are missense) for KCNJ11, ABCC8, and TCF7L2, by applying a set of computational tools — SIFT, PolyPhen-2, PANTHER, MutPred, and SNPs3D. Our bioinformatics prediction results demonstrated that 8 KCNJ11 missense SNPs (R27H, V59M, I182V, R192H, R201H, E227K, L270V, and S385C), 23 ABCC8 missense SNPs (G7R, N24K, F27S, N72S, R74W, A116P, E128K, F132L, V187D, L213R, E382K, R495Q, E501K, L503P, F686S, G716V, K1336N, L1349Q, S1386F, L1389P, R1420C, I1424V, D1471H), and 2 TCF7L2 missense SNPs (P179H, K323N) could affect protein functions with SIFT score ≤ 0.05, or PolyPhen-2 score ≥ 0.85, or PANTHER subSPEC score ≤ −3, or MutPred > 0.5, or SNPs3D score < 0. Of them, previous studies showed that mutant alleles of 4 KCNJ11 missense SNPs (R27H, V59M, R192H, and R201H) and 5 ABCC8 missense SNPs (G7R, N24K, F27S, R74W, and E128K) can be successfully rescued by sulfonylurea treatments. In addition, 3 TCF7L2 non-coding SNPs — rs7903146, rs11196205 and rs12255372 were predicted as risk factor based on Ensembl’s VEP, although their functional impacts in sulfonylurea results need to be elucidated by further experimental studies.
Conclusion
The ultimate goal of pharmacogenetics is the development of personalized medicine through individual genetic profiles which would accurately predict which individuals with a specific medical condition would respond to a specific medical therapy. Traditional medicine refers to the broad application of “standard of care” or “one-size-fits-all” treatments to all patients with a given diagnosis. In contrast, personalized medicine, often described as providing “the right drug for the right patient at the right dose and time” [87], tailors medical treatment according to each patient’s personal history, genetic profile and/or specific biomarkers [88, 89], Therefore, the full application of personalized medicine in health care will require significant changes in regulatory and reimbursement policies as well as legislative protections for privacy. The U.S. Food and Drug Administration has updated the labels of more than 120 drugs with recommendations for genetic testing prior to their use [90]. Currently, most genetic testing is based genotypic effects. Haplotypes of multiple linked genetic variants provide more precise information of their functional impacts than individual genetic markers [91, 92], which could also be potentially important for diagnosis and prognosis [93]. In future, regulatory authorities shall formulate clear guidelines for evaluating and approving personalized diagnostics and therapeutics and identify patients who can benefit from them.
Acknowledgements
Dr. Yu-Ping Wang and Dr. Tianhua Niu are supported in part by NIH 1R01GM109068-01A1. Dr. Tianhua Niu is also supported in part by a start-up fund of the Center for Bioinformatics and Genomics, Tulane University. Dr. Franck Mauvais-Jarvis is supported by NIH R01 DK074970, the American Diabetes Association (7-13-BS-101), and the Price-Goldsmith Endowed Chair in Nutrition.
Funding
Funded in part by NIH 1R01GM109068-01A1.
Availability of data and materials
Not applicable.
Authors’ contributions
TN conceived the idea for the project. TN and JS contributed to study design and conception. JS, YY and TN participated in data analysis and interpretation. JS, YY and TN drafted the manuscript. FMJ and YW revised it critically for intellectual content. All authors read and approved the final version of the manuscript.
Competing interests
The authors declare that they have no competing interests.
Consent for publication
Not applicable.
Ethics approval and consent to participate
Not applicable.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Abbreviations
- ABCC8
ATP Binding Cassette Subfamily C Member 8
- ADP
Adenosine diphosphate
- ATP
Adenosine triphosphate
- cAMP
Cyclic adenosine monophosphate
- CL
cytosolic loop
- dbSNP
Single Nucleotide Polymorphism database
- EMBASE
Excerpta Medica Database
- GLP-1
Glucagon-like peptide-1
- HbA1c
Glycated hemoglobin A1c
- Go-DARTS
Genetics of Diabetes Audit and Research Study in Tayside Scotland
- KATP
ATP-sensitive K+ channel
- KCNJ11
Potassium channel, inwardly rectifying subfamily J, member 11
- Kir
Inwardly rectifying K(+)
- LEF
Lymphocyte enhancer factor
- MutPred
Mutation prediction
- NBD
nucleotide binding domain
- NCBI
National Center for Biotechnology Information
- nsSNP
Non-synonymous single nucleotide polymorphism
- PANTHER
Protein analysis through evolutionary relationships
- PKA
Protein kinase A
- PolyPhen-2
Polymorphism phenotyping v2
- RCT
Randomized clinical trial
- RegulomeDB
Regulome database
- SIFT
Sorting intolerant from tolerant
- SNP
Single nucleotide polymorphism
- SU
Sulfonylurea
- subSPEC
subStitution Position-specific Evolutionary Conservation
- SUR
Sulfonylurea receptor
- T2D
Type 2 diabetes
- TCF
T-cell factor
- TCF7L2
T-cell factor 7-like 2
- TMD
transmembrane domain
- UKPDS
United Kingdom Prospective Diabetes Study
- UTR
Untranslated region
- VEP
Variant effect predictor
- WHO-MONICA
World Health Organization-Multinational MONItoring of trends and determinants of CArdiovascular diseases
- Wnt
Wingless type
Contributor Information
Jingwen Song, Email: olivia22061@163.com.
Yunzhong Yang, Email: yyang16@tulane.edu.
Franck Mauvais-Jarvis, Email: fmauvais@tulane.edu.
Yu-Ping Wang, Email: wyp@tulane.edu.
Tianhua Niu, Phone: (504) 988-2058, Email: tniu@tulane.edu.
References
- 1.Federation ID . IDF Diabetes Atlas. Fourth 2009. [Google Scholar]
- 2.Cade WT. Diabetes-related microvascular and macrovascular diseases in the physical therapy setting. Phys Ther. 2008;88(11):1322–1335. doi: 10.2522/ptj.20080008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ripsin CM, Kang H, Urban RJ. Management of blood glucose in type 2 diabetes mellitus. Am Fam Physician. 2009;79(1):29–36. [PubMed] [Google Scholar]
- 4.Riserus U, Willett WC, Hu FB. Dietary fats and prevention of type 2 diabetes. Prog Lipid Res. 2009;48(1):44–51. doi: 10.1016/j.plipres.2008.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Semiz S, Dujic T, Causevic A. Pharmacogenetics and personalized treatment of type 2 diabetes. Biochem Med (Zagreb) 2013;23(2):154–171. doi: 10.11613/BM.2013.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pearson ER. Pharmacogenetics and future strategies in treating hyperglycaemia in diabetes. Front Biosci. 2009;14:4348–4362. doi: 10.2741/3532. [DOI] [PubMed] [Google Scholar]
- 7.Aguilar-Bryan L, Nichols CG, Wechsler SW, Clement JP, Boyd AE, 3rd, Gonzalez G, Herrera-Sosa H, Nguy K, Bryan J, Nelson DA. Cloning of the beta cell high-affinity sulfonylurea receptor: a regulator of insulin secretion. Science. 1995;268(5209):423–426. doi: 10.1126/science.7716547. [DOI] [PubMed] [Google Scholar]
- 8.Inagaki N, Gonoi T, Clement JP, Namba N, Inazawa J, Gonzalez G, Aguilar-Bryan L, Seino S, Bryan J. Reconstitution of IKATP: an inward rectifier subunit plus the sulfonylurea receptor. Science. 1995;270(5239):1166–1170. doi: 10.1126/science.270.5239.1166. [DOI] [PubMed] [Google Scholar]
- 9.Sakura H, Ammala C, Smith PA, Gribble FM, Ashcroft FM. Cloning and functional expression of the cDNA encoding a novel ATP-sensitive potassium channel subunit expressed in pancreatic beta-cells, brain, heart and skeletal muscle. FEBS Lett. 1995;377(3):338–344. doi: 10.1016/0014-5793(95)01369-5. [DOI] [PubMed] [Google Scholar]
- 10.Jahnavi S, Poovazhagi V, Kanthimathi S, Balamurugan K, Bodhini D, Yadav J, Jain V, Khadgawat R, Sikdar M, Bhavatharini A, et al. Novel ABCC8 (SUR1) gene mutations in Asian Indian children with congenital hyperinsulinemic hypoglycemia. Ann Hum Genet. 2014;78(5):311–319. doi: 10.1111/ahg.12070. [DOI] [PubMed] [Google Scholar]
- 11.Bonfanti DH, Alcazar LP, Arakaki PA, Martins LT, Agustini BC, de Moraes Rego FG, Frigeri HR. ATP-dependent potassium channels and type 2 diabetes mellitus. Clin Biochem. 2015;48(7–8):476–482. doi: 10.1016/j.clinbiochem.2014.12.026. [DOI] [PubMed] [Google Scholar]
- 12.Schwanstecher C, Meyer U, Schwanstecher M. K(IR)6.2 polymorphism predisposes to type 2 diabetes by inducing overactivity of pancreatic beta-cell ATP-sensitive K(+) channels. Diabetes. 2002;51(3):875–879. doi: 10.2337/diabetes.51.3.875. [DOI] [PubMed] [Google Scholar]
- 13.Riedel MJ, Boora P, Steckley D, de Vries G, Light PE. Kir6.2 polymorphisms sensitize beta-cell ATP-sensitive potassium channels to activation by acyl CoAs: a possible cellular mechanism for increased susceptibility to type 2 diabetes? Diabetes. 2003;52(10):2630–2635. doi: 10.2337/diabetes.52.10.2630. [DOI] [PubMed] [Google Scholar]
- 14.Ashcroft FM, Rorsman P. K(ATP) channels and islet hormone secretion: new insights and controversies. Nat Rev Endocrinol. 2013;9(11):660–669. doi: 10.1038/nrendo.2013.166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jin T, Liu L. The Wnt signaling pathway effector TCF7L2 and type 2 diabetes mellitus. Mol Endocrinol. 2008;22(11):2383–2392. doi: 10.1210/me.2008-0135. [DOI] [PubMed] [Google Scholar]
- 16.Ip W, Chiang YT, Jin T. The involvement of the wnt signaling pathway and TCF7L2 in diabetes mellitus: The current understanding, dispute, and perspective. Cell Biosci. 2012;2(1):28. doi: 10.1186/2045-3701-2-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.MacDonald BT, Tamai K, He X. Wnt/beta-catenin signaling: components, mechanisms, and diseases. Dev Cell. 2009;17(1):9–26. doi: 10.1016/j.devcel.2009.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liu Z, Habener JF. Glucagon-like peptide-1 activation of TCF7L2-dependent Wnt signaling enhances pancreatic beta cell proliferation. J Biol Chem. 2008;283(13):8723–8735. doi: 10.1074/jbc.M706105200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jin T, George Fantus I, Sun J. Wnt and beyond Wnt: multiple mechanisms control the transcriptional property of beta-catenin. Cell Signal. 2008;20(10):1697–1704. doi: 10.1016/j.cellsig.2008.04.014. [DOI] [PubMed] [Google Scholar]
- 20.Yi F, Brubaker PL, Jin T. TCF-4 mediates cell type-specific regulation of proglucagon gene expression by beta-catenin and glycogen synthase kinase-3beta. J Biol Chem. 2005;280(2):1457–1464. doi: 10.1074/jbc.M411487200. [DOI] [PubMed] [Google Scholar]
- 21.DeFronzo RA. Pharmacologic therapy for type 2 diabetes mellitus. Ann Intern Med. 1999;131(4):281–303. doi: 10.7326/0003-4819-131-4-199908170-00008. [DOI] [PubMed] [Google Scholar]
- 22.Zammitt NN, Frier BM. Hypoglycemia in type 2 diabetes: pathophysiology, frequency, and effects of different treatment modalities. Diabetes Care. 2005;28(12):2948–2961. doi: 10.2337/diacare.28.12.2948. [DOI] [PubMed] [Google Scholar]
- 23.Gangji AS, Cukierman T, Gerstein HC, Goldsmith CH, Clase CM. A systematic review and meta-analysis of hypoglycemia and cardiovascular events: a comparison of glyburide with other secretagogues and with insulin. Diabetes Care. 2007;30(2):389–394. doi: 10.2337/dc06-1789. [DOI] [PubMed] [Google Scholar]
- 24.Kalra S, Mukherjee JJ, Venkataraman S, Bantwal G, Shaikh S, Saboo B, Das AK, Ramachandran A. Hypoglycemia: The neglected complication. Indian J Endocr Metab. 2013;17(5):819–834. doi: 10.4103/2230-8210.117219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Capener CE, Shrivastava IH, Ranatunga KM, Forrest LR, Smith GR, Sansom MS. Homology modeling and molecular dynamics simulation studies of an inward rectifier potassium channel. Biophys J. 2000;78(6):2929–2942. doi: 10.1016/S0006-3495(00)76833-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Haghvirdizadeh P, Mohamed Z, Abdullah NA, Haghvirdizadeh P, Haerian MS, Haerian BS. KCNJ11: Genetic Polymorphisms and Risk of Diabetes Mellitus. J Diabetes Res. 2015;2015:908152. doi: 10.1155/2015/908152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mikhailov MV, Mikhailova EA, Ashcroft SJ. Molecular structure of the glibenclamide binding site of the beta-cell K(ATP) channel. FEBS Lett. 2001;499(1–2):154–160. doi: 10.1016/S0014-5793(01)02538-8. [DOI] [PubMed] [Google Scholar]
- 28.Park JS, Lee HJ, Park CH. A novel mutation of ABCC8 gene in a patient with diazoxide-unresponsive congenital hyperinsulinism. Korean J Pediatr. 2016;59(Suppl 1):S116–S120. doi: 10.3345/kjp.2016.59.11.S116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mondal AK, Das SK, Baldini G, Chu WS, Sharma NK, Hackney OG, Zhao J, Grant SF, Elbein SC. Genotype and tissue-specific effects on alternative splicing of the transcription factor 7-like 2 gene in humans. J Clin Endocrinol Metab. 2010;95(3):1450–1457. doi: 10.1210/jc.2009-2064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hansson O, Zhou Y, Renstrom E, Osmark P. Molecular function of TCF7L2: Consequences of TCF7L2 splicing for molecular function and risk for type 2 diabetes. Curr Diab Rep. 2010;10(6):444–451. doi: 10.1007/s11892-010-0149-8. [DOI] [PubMed] [Google Scholar]
- 31.Haddad SA, Palmer JR, Lunetta KL, Ng MC, Consortium M, Ruiz-Narvaez EA. A novel TCF7L2 type 2 diabetes SNP identified from fine mapping in African American women. PLoS One. 2017;12(3):e0172577. doi: 10.1371/journal.pone.0172577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Scott LJ, Bonnycastle LL, Willer CJ, Sprau AG, Jackson AU, Narisu N, Duren WL, Chines PS, Stringham HM, Erdos MR, et al. Association of transcription factor 7-like 2 (TCF7L2) variants with type 2 diabetes in a Finnish sample. Diabetes. 2006;55(9):2649–2653. doi: 10.2337/db06-0341. [DOI] [PubMed] [Google Scholar]
- 33.Aquilante CL. Sulfonylurea pharmacogenomics in Type 2 diabetes: the influence of drug target and diabetes risk polymorphisms. Expert Rev Cardiovasc Ther. 2010;8(3):359–372. doi: 10.1586/erc.09.154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Maruthur NM, Gribble MO, Bennett WL, Bolen S, Wilson LM, Balakrishnan P, Sahu A, Bass E, Kao WH, Clark JM. The pharmacogenetics of type 2 diabetes: a systematic review. Diabetes Care. 2014;37(3):876–886. doi: 10.2337/dc13-1276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ng PC, Henikoff S. Predicting deleterious amino acid substitutions. Genome Res. 2001;11(5):863–874. doi: 10.1101/gr.176601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Adzhubei IA, Schmidt S, Peshkin L, Ramensky VE, Gerasimova A, Bork P, Kondrashov AS, Sunyaev SR. A method and server for predicting damaging missense mutations. Nat Methods. 2010;7(4):248–249. doi: 10.1038/nmeth0410-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Thomas PD, Campbell MJ, Kejariwal A, Mi H, Karlak B, Daverman R, Diemer K, Muruganujan A, Narechania A. PANTHER: a library of protein families and subfamilies indexed by function. Genome Res. 2003;13(9):2129–2141. doi: 10.1101/gr.772403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li B, Krishnan VG, Mort ME, Xin F, Kamati KK, Cooper DN, Mooney SD, Radivojac P. Automated inference of molecular mechanisms of disease from amino acid substitutions. Bioinformatics. 2009;25(21):2744–2750. doi: 10.1093/bioinformatics/btp528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yue P, Melamud E, Moult J. SNPs3D: candidate gene and SNP selection for association studies. BMC Bioinformatics. 2006;7:166. doi: 10.1186/1471-2105-7-166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ward LD, Kellis M. HaploReg: a resource for exploring chromatin states, conservation, and regulatory motif alterations within sets of genetically linked variants. Nucleic Acids Res. 2012;40(Database issue):D930–934. doi: 10.1093/nar/gkr917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ward LD, Kellis M. HaploReg v4: systematic mining of putative causal variants, cell types, regulators and target genes for human complex traits and disease. Nucleic Acids Res. 2016;44(D1):D877–881. doi: 10.1093/nar/gkv1340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Boyle AP, Hong EL, Hariharan M, Cheng Y, Schaub MA, Kasowski M, Karczewski KJ, Park J, Hitz BC, Weng S, et al. Annotation of functional variation in personal genomes using RegulomeDB. Genome Res. 2012;22(9):1790–1797. doi: 10.1101/gr.137323.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.McLaren W, Pritchard B, Rios D, Chen Y, Flicek P, Cunningham F. Deriving the consequences of genomic variants with the Ensembl API and SNP Effect Predictor. Bioinformatics. 2010;26(16):2069–2070. doi: 10.1093/bioinformatics/btq330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gloyn AL, Hashim Y, Ashcroft SJ, Ashfield R, Wiltshire S, Turner RC, Study UKPD. Association studies of variants in promoter and coding regions of beta-cell ATP-sensitive K-channel genes SUR1 and Kir6.2 with Type 2 diabetes mellitus (UKPDS 53) Diabet Med. 2001;18(3):206–212. doi: 10.1046/j.1464-5491.2001.00449.x. [DOI] [PubMed] [Google Scholar]
- 45.Sesti G, Laratta E, Cardellini M, Andreozzi F, Del Guerra S, Irace C, Gnasso A, Grupillo M, Lauro R, Hribal ML, et al. The E23K variant of KCNJ11 encoding the pancreatic beta-cell adenosine 5’-triphosphate-sensitive potassium channel subunit Kir6.2 is associated with an increased risk of secondary failure to sulfonylurea in patients with type 2 diabetes. J Clin Endocrinol Metab. 2006;91(6):2334–2339. doi: 10.1210/jc.2005-2323. [DOI] [PubMed] [Google Scholar]
- 46.Feng Y, Mao G, Ren X, Xing H, Tang G, Li Q, Li X, Sun L, Yang J, Ma W, et al. Ser1369Ala variant in sulfonylurea receptor gene ABCC8 is associated with antidiabetic efficacy of gliclazide in Chinese type 2 diabetic patients. Diabetes Care. 2008;31(10):1939–1944. doi: 10.2337/dc07-2248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Holstein A, Hahn M, Stumvoll M, Kovacs P. The E23K variant of KCNJ11 and the risk for severe sulfonylurea-induced hypoglycemia in patients with type 2 diabetes. Horm Metab Res. 2009;41(5):387–390. doi: 10.1055/s-0029-1192019. [DOI] [PubMed] [Google Scholar]
- 48.Nikolac N, Simundic AM, Katalinic D, Topic E, Cipak A, Zjacic Rotkvic V. Metabolic control in type 2 diabetes is associated with sulfonylurea receptor-1 (SUR-1) but not with KCNJ11 polymorphisms. Arch Med Res. 2009;40(5):387–392. doi: 10.1016/j.arcmed.2009.06.006. [DOI] [PubMed] [Google Scholar]
- 49.El-Sisi AE, Hegazy SK, Metwally SS, Wafa AM, Dawood NA. Effect of genetic polymorphisms on the development of secondary failure to sulfonylurea in egyptian patients with type 2 diabetes. Ther Adv Endocrinol Metab. 2011;2(4):155–164. doi: 10.1177/2042018811415985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Javorsky M, Klimcakova L, Schroner Z, Zidzik J, Babjakova E, Fabianova M, Kozarova M, Tkacova R, Salagovic J, Tkac I. KCNJ11 gene E23K variant and therapeutic response to sulfonylureas. Eur J Intern Med. 2012;23(3):245–249. doi: 10.1016/j.ejim.2011.10.018. [DOI] [PubMed] [Google Scholar]
- 51.Ragia G, Tavridou A, Petridis I, Manolopoulos VG. Association of KCNJ11 E23K gene polymorphism with hypoglycemia in sulfonylurea-treated type 2 diabetic patients. Diabetes Res Clin Pract. 2012;98(1):119–124. doi: 10.1016/j.diabres.2012.04.017. [DOI] [PubMed] [Google Scholar]
- 52.Li Q, Chen M, Zhang R, Jiang F, Wang J, Zhou J, Bao Y, Hu C, Jia W. KCNJ11 E23K variant is associated with the therapeutic effect of sulphonylureas in Chinese type 2 diabetic patients. Clin Exp Pharmacol Physiol. 2014;41(10):748–754. doi: 10.1111/1440-1681.12280. [DOI] [PubMed] [Google Scholar]
- 53.Meirhaeghe A, Helbecque N, Cottel D, Arveiler D, Ruidavets JB, Haas B, Ferrieres J, Tauber JP, Bingham A, Amouyel P. Impact of sulfonylurea receptor 1 genetic variability on non-insulin-dependent diabetes mellitus prevalence and treatment: a population study. Am J Med Genet. 2001;101(1):4–8. doi: 10.1002/ajmg.1297. [DOI] [PubMed] [Google Scholar]
- 54.Zychma MJ, Gumprecht J, Strojek K, Grzeszczak W, Moczulski D, Trautsolt W, Karasek D. Sulfonylurea receptor gene 16–3 polymorphism - association with sulfonylurea or insulin treatment in type 2 diabetic subjects. Med Sci Monit. 2002;8(7):CR512–515. [PubMed] [Google Scholar]
- 55.Nikolac N, Simundic AM, Saracevic A, Katalinic D. ABCC8 polymorphisms are associated with triglyceride concentration in type 2 diabetics on sulfonylurea therapy. Genet Test Mol Biomarkers. 2012;16(8):924–930. doi: 10.1089/gtmb.2011.0337. [DOI] [PubMed] [Google Scholar]
- 56.Zhang H, Liu X, Kuang H, Yi R, Xing H. Association of sulfonylurea receptor 1 genotype with therapeutic response to gliclazide in type 2 diabetes. Diabetes Res Clin Pract. 2007;77(1):58–61. doi: 10.1016/j.diabres.2006.10.021. [DOI] [PubMed] [Google Scholar]
- 57.Sato R, Watanabe H, Genma R, Takeuchi M, Maekawa M, Nakamura H. ABCC8 polymorphism (Ser1369Ala): influence on severe hypoglycemia due to sulfonylureas. Pharmacogenomics. 2010;11(12):1743–1750. doi: 10.2217/pgs.10.135. [DOI] [PubMed] [Google Scholar]
- 58.Pearson ER, Donnelly LA, Kimber C, Whitley A, Doney AS, McCarthy MI, Hattersley AT, Morris AD, Palmer CN. Variation in TCF7L2 influences therapeutic response to sulfonylureas: a GoDARTs study. Diabetes. 2007;56(8):2178–2182. doi: 10.2337/db07-0440. [DOI] [PubMed] [Google Scholar]
- 59.Schroner Z, Javorsky M, Tkacova R, Klimcakova L, Dobrikova M, Habalova V, Kozarova M, Zidzik J, Rudikova M, Tkac I. Effect of sulphonylurea treatment on glycaemic control is related to TCF7L2 genotype in patients with type 2 diabetes. Diabetes Obes Metab. 2011;13(1):89–91. doi: 10.1111/j.1463-1326.2010.01324.x. [DOI] [PubMed] [Google Scholar]
- 60.Holstein A, Hahn M, Korner A, Stumvoll M, Kovacs P. TCF7L2 and therapeutic response to sulfonylureas in patients with type 2 diabetes. BMC Med Genet. 2011;12:30. doi: 10.1186/1471-2350-12-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Villareal DT, Koster JC, Robertson H, Akrouh A, Miyake K, Bell GI, Patterson BW, Nichols CG, Polonsky KS. Kir6.2 variant E23K increases ATP-sensitive K+ channel activity and is associated with impaired insulin release and enhanced insulin sensitivity in adults with normal glucose tolerance. Diabetes. 2009;58(8):1869–1878. doi: 10.2337/db09-0025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Vaxillaire M, Veslot J, Dina C, Proenca C, Cauchi S, Charpentier G, Tichet J, Fumeron F, Marre M, Meyre D, et al. Impact of common type 2 diabetes risk polymorphisms in the DESIR prospective study. Diabetes. 2008;57(1):244–254. doi: 10.2337/db07-0615. [DOI] [PubMed] [Google Scholar]
- 63.Zhou D, Zhang D, Liu Y, Zhao T, Chen Z, Liu Z, Yu L, Zhang Z, Xu H, He L. The E23K variation in the KCNJ11 gene is associated with type 2 diabetes in Chinese and East Asian population. J Hum Genet. 2009;54(7):433–435. doi: 10.1038/jhg.2009.54. [DOI] [PubMed] [Google Scholar]
- 64.Pearson ER, Flechtner I, Njolstad PR, Malecki MT, Flanagan SE, Larkin B, Ashcroft FM, Klimes I, Codner E, Iotova V, et al. Switching from insulin to oral sulfonylureas in patients with diabetes due to Kir6.2 mutations. N Engl J Med. 2006;355(5):467–477. doi: 10.1056/NEJMoa061759. [DOI] [PubMed] [Google Scholar]
- 65.Siklar Z, Ellard S, Okulu E, Berberoglu M, Young E, Savas Erdeve S, Mungan IA, Hacihamdioglu B, Erdeve O, Arsan S, et al. Transient neonatal diabetes with two novel mutations in the KCNJ11 gene and response to sulfonylurea treatment in a preterm infant. J Pediatr Endocrinol Metab. 2011;24(11–12):1077–1080. doi: 10.1515/jpem.2011.250. [DOI] [PubMed] [Google Scholar]
- 66.Dupont J, Pereira C, Medeira A, Duarte R, Ellard S, Sampaio L. Permanent neonatal diabetes mellitus due to KCNJ11 mutation in a Portuguese family: transition from insulin to oral sulfonylureas. J Pediatr Endocrinol Metab. 2012;25(3–4):367–370. doi: 10.1515/jpem-2011-0191. [DOI] [PubMed] [Google Scholar]
- 67.Klen J, Dolzan V, Janez A. CYP2C9, KCNJ11 and ABCC8 polymorphisms and the response to sulphonylurea treatment in type 2 diabetes patients. Eur J Clin Pharmacol. 2014;70(4):421–428. doi: 10.1007/s00228-014-1641-x. [DOI] [PubMed] [Google Scholar]
- 68.Fatehi M, Raja M, Carter C, Soliman D, Holt A, Light PE. The ATP-sensitive K(+) channel ABCC8 S1369A type 2 diabetes risk variant increases MgATPase activity. Diabetes. 2012;61(1):241–249. doi: 10.2337/db11-0371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Groves CJ, Zeggini E, Minton J, Frayling TM, Weedon MN, Rayner NW, Hitman GA, Walker M, Wiltshire S, Hattersley AT, et al. Association analysis of 6,736 U.K. subjects provides replication and confirms TCF7L2 as a type 2 diabetes susceptibility gene with a substantial effect on individual risk. Diabetes. 2006;55(9):2640–2644. doi: 10.2337/db06-0355. [DOI] [PubMed] [Google Scholar]
- 70.van Vliet-Ostaptchouk JV, Shiri-Sverdlov R, Zhernakova A, Strengman E, van Haeften TW, Hofker MH, Wijmenga C. Association of variants of transcription factor 7-like 2 (TCF7L2) with susceptibility to type 2 diabetes in the Dutch Breda cohort. Diabetologia. 2007;50(1):59–62. doi: 10.1007/s00125-006-0477-z. [DOI] [PubMed] [Google Scholar]
- 71.Mayans S, Lackovic K, Lindgren P, Ruikka K, Agren A, Eliasson M, Holmberg D. TCF7L2 polymorphisms are associated with type 2 diabetes in northern Sweden. Eur J Hum Genet. 2007;15(3):342–346. doi: 10.1038/sj.ejhg.5201773. [DOI] [PubMed] [Google Scholar]
- 72.Cauchi S, Meyre D, Dina C, Choquet H, Samson C, Gallina S, Balkau B, Charpentier G, Pattou F, Stetsyuk V, et al. Transcription factor TCF7L2 genetic study in the French population: expression in human beta-cells and adipose tissue and strong association with type 2 diabetes. Diabetes. 2006;55(10):2903–2908. doi: 10.2337/db06-0474. [DOI] [PubMed] [Google Scholar]
- 73.Zhang C, Qi L, Hunter DJ, Meigs JB, Manson JE, van Dam RM, Hu FB. Variant of transcription factor 7-like 2 (TCF7L2) gene and the risk of type 2 diabetes in large cohorts of U.S. women and men. Diabetes. 2006;55(9):2645–2648. doi: 10.2337/db06-0643. [DOI] [PubMed] [Google Scholar]
- 74.Chandak GR, Janipalli CS, Bhaskar S, Kulkarni SR, Mohankrishna P, Hattersley AT, Frayling TM, Yajnik CS. Common variants in the TCF7L2 gene are strongly associated with type 2 diabetes mellitus in the Indian population. Diabetologia. 2007;50(1):63–67. doi: 10.1007/s00125-006-0502-2. [DOI] [PubMed] [Google Scholar]
- 75.Hayashi T, Iwamoto Y, Kaku K, Hirose H, Maeda S. Replication study for the association of TCF7L2 with susceptibility to type 2 diabetes in a Japanese population. Diabetologia. 2007;50(5):980–984. doi: 10.1007/s00125-007-0618-z. [DOI] [PubMed] [Google Scholar]
- 76.Cauchi S, El Achhab Y, Choquet H, Dina C, Krempler F, Weitgasser R, Nejjari C, Patsch W, Chikri M, Meyre D, et al. TCF7L2 is reproducibly associated with type 2 diabetes in various ethnic groups: a global meta-analysis. J Mol Med. 2007;85(7):777–782. doi: 10.1007/s00109-007-0203-4. [DOI] [PubMed] [Google Scholar]
- 77.Sale MM, Smith SG, Mychaleckyj JC, Keene KL, Langefeld CD, Leak TS, Hicks PJ, Bowden DW, Rich SS, Freedman BI. Variants of the transcription factor 7-like 2 (TCF7L2) gene are associated with type 2 diabetes in an African-American population enriched for nephropathy. Diabetes. 2007;56(10):2638–2642. doi: 10.2337/db07-0012. [DOI] [PubMed] [Google Scholar]
- 78.Zhou Y, Park SY, Su J, Bailey K, Ottosson-Laakso E, Shcherbina L, Oskolkov N, Zhang E, Thevenin T, Fadista J, et al. TCF7L2 is a master regulator of insulin production and processing. Hum Mol Genet. 2014;23(24):6419–6431. doi: 10.1093/hmg/ddu359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Osmark P, Hansson O, Jonsson A, Ronn T, Groop L, Renstrom E. Unique splicing pattern of the TCF7L2 gene in human pancreatic islets. Diabetologia. 2009;52(5):850–854. doi: 10.1007/s00125-009-1293-z. [DOI] [PubMed] [Google Scholar]
- 80.Chiang YT, Ip W, Jin T. The role of the Wnt signaling pathway in incretin hormone production and function. Front Physiol. 2012;3:273. doi: 10.3389/fphys.2012.00273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Kumar P, Henikoff S, Ng PC. Predicting the effects of coding non-synonymous variants on protein function using the SIFT algorithm. Nat Protoc. 2009;4(7):1073–1081. doi: 10.1038/nprot.2009.86. [DOI] [PubMed] [Google Scholar]
- 82.Hicks S, Wheeler DA, Plon SE, Kimmel M. Prediction of missense mutation functionality depends on both the algorithm and sequence alignment employed. Hum Mutat. 2011;32(6):661–668. doi: 10.1002/humu.21490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Niu T, Liu N, Yu X, Zhao M, Choi HJ, Leo PJ, Brown MA, Zhang L, Pei YF, Shen H, et al. Identification of IDUA and WNT16 Phosphorylation-Related Non-Synonymous Polymorphisms for Bone Mineral Density in Meta-Analyses of Genome-Wide Association Studies. J Bone Miner Res. 2016;31(2):358–368. doi: 10.1002/jbmr.2687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Manickam M, Ravanan P, Singh P, Talwar P. In silico identification of genetic variants in glucocerebrosidase (GBA) gene involved in Gaucher’s disease using multiple software tools. Front Genet. 2014;5:148. doi: 10.3389/fgene.2014.00148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Wei Q, Wang L, Wang Q, Kruger WD, Dunbrack RL., Jr Testing computational prediction of missense mutation phenotypes: functional characterization of 204 mutations of human cystathionine beta synthase. Proteins. 2010;78(9):2058–2074. doi: 10.1002/prot.22722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Ortega VE, Meyers DA. Pharmacogenetics: implications of race and ethnicity on defining genetic profiles for personalized medicine. J Allergy Clin Immunol. 2014;133(1):16–26. doi: 10.1016/j.jaci.2013.10.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Sadee W, Dai Z. Pharmacogenetics/genomics and personalized medicine. Hum Mol Genet. 2005;14(2):R207-214. [DOI] [PubMed]
- 88.Dancey JE, Bedard PL, Onetto N, Hudson TJ. The genetic basis for cancer treatment decisions. Cell. 2012;148(3):409–420. doi: 10.1016/j.cell.2012.01.014. [DOI] [PubMed] [Google Scholar]
- 89.Evans WE, Relling MV. Moving towards individualized medicine with pharmacogenomics. Nature. 2004;429(6990):464–468. doi: 10.1038/nature02626. [DOI] [PubMed] [Google Scholar]
- 90.Table of Pharmacogenomic Biomarkers in Drug Labels. [http://www.fda.gov/Drugs/ScienceResearch/ResearchAreas/Pharmacogenetics/ucm083378.htm]. [DOI] [PubMed]
- 91.Niu T. Algorithms for inferring haplotypes. Genet Epidemiol. 2004;27(4):334–347. doi: 10.1002/gepi.20024. [DOI] [PubMed] [Google Scholar]
- 92.Niu T, Qin ZS, Xu X, Liu JS. Bayesian haplotype inference for multiple linked single-nucleotide polymorphisms. Am J Hum Genet. 2002;70(1):157–169. doi: 10.1086/338446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Lee JE, Choi JH, Lee JH, Lee MG. Gene SNPs and mutations in clinical genetic testing: haplotype-based testing and analysis. Mutat Res. 2005;573(1–2):195–204. doi: 10.1016/j.mrfmmm.2004.08.018. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Not applicable.


