Abstract
Therapeutic application of the recently discovered small interfering RNA (siRNA) gene silencing phenomenon will be dependent on improvements in molecule bio-stability, specificity and delivery. To address these issues, we have systematically modified siRNA with the synthetic RNA-like high affinity nucleotide analogue, Locked Nucleic Acid (LNA). Here, we show that incorporation of LNA substantially enhances serum half-life of siRNA's, which is a key requirement for therapeutic use. Moreover, we provide evidence that LNA is compatible with the intracellular siRNA machinery and can be used to reduce undesired, sequence-related off-target effects. LNA-modified siRNAs targeting the emerging disease SARS, show improved efficiency over unmodified siRNA on certain RNA motifs. The results from this study emphasize LNA's promise in converting siRNA from a functional genomics technology to a therapeutic platform.
INTRODUCTION
Double-stranded small interfering RNA (siRNA) molecules have drawn much attention since it was unambiguously shown that they mediate potent gene knock-down in a variety of mammalian cells (1). This work followed the discovery of the phenomenon of RNA interference (RNAi) in Caenorhabditis elegans (2) and the demonstration of siRNAs as possible mediators of gene regulation in other eukaryotes (3–5).
SiRNA works through Watson–Crick base-pairing of an RNA guide sequence to the target RNA followed by specific degradation or translational block of the target [reviewed in (6,7)]. As such, siRNA technology offers the means to rationally design gene-specific inhibitors and in recent years such molecules have found widespread use as tools in functional genomic studies in mammalian cells in vitro.
However, application of siRNAs in vivo and their possible use as therapeutics still face several critical hurdles that have not yet been comprehensively addressed. For instance, siRNA delivery, bio-stability, pharmacokinetics and specificity, including off-target effects, will be major topics of further investigation.
Many of these issues are not new to oligonucleotide-based technologies being developed as drug platforms, such as antisense, aptamers and ribozymes. Here, critical advances have come from the development of nucleotide analogues with improved properties over natural nucleotides and recently several of these such as phosphorothioates (8,9), 2′-O-Me (10,11), 2′-O-allyl (10) and 2′-deoxy-fluorouridine (8,9) have been examined as a means to improve the prospect for siRNA therapy. Briefly, these studies have demonstrated that siRNAs can accommodate quite a number of modifications at both base-paired and non-base-paired positions without significant loss of activity. Moreover, some of the modified siRNAs were found to exhibit enhanced serum stability (11) and longer duration of action (10). Modification of the 5′ end of the antisense strand with 2′-O-allyl (10) or chemical blocking of the 5′-hydroxyl group (11) resulted in a dramatic loss in activity consistent with the proposed in vivo requirement for 5′ end phosphorylation. Also, more substantial modifications, such as total modification by 2′-O-Me (8) or PS modifications of every second or all internucleoside linkages (8,9) increased cytotoxic effects and resulted in a significant decrease or complete loss of activity.
Locked nucleic acid (LNA) is a family of conformationally locked nucleotide analogues which, amongst other benefits, imposes truly unprecedented affinity and very high nuclease resistance to DNA and RNA oligonucleotides (12–16). When used in antisense constructs, LNA has been reported to combine substantially increased potency in vitro and in vivo with minimal toxicity (12–17). Also, the commonly used LNA contains a methylene bridge connecting the 2′-oxygen with the 4′-carbon of the ribose ring. This bridge locks the ribose ring in the 3′-endo conformation characteristic of RNA (18–21). As such, LNA is a prime candidate for introducing critical new features into siRNAs without perturbing the overall A-form helical structure they require for activity (22).
Recently, Braasch et al. (8) provided the first evidence that LNA can be used to increase the thermal stability of siRNA molecules without affecting their function. In this report, we expand on these early findings by systematically pursuing the construction and biological testing of LNA-modified siRNA molecules, hereafter termed siLNA, aiming at in vivo applications. We show that LNA is substantially compatible with the siRNA machinery, and that siLNAs exhibit greatly improved bio-stability and shows enhanced inhibition at certain RNA targets. We further show that LNA can be used to reduce sequence-related off-target effects by either lowering incorporation of the siRNA sense-strand and/or by reducing the ability of inappropriately loaded sense-strands to cleave the target RNA.
MATERIALS AND METHODS
Oligonucleotides and plasmids
All siRNA and siLNA oligonucleotides used in this study are listed in Table 1. LNA containing oligonucleotides were synthesized by Santaris A/S (Hørsholm, Denmark), siRNA was ordered from MedProbe (Lund, Sweden) and DNA oligonucleotides from Invitrogen (Paisely, UK). Target sequences have been described elsewhere [firefly luciferase (1), Renilla (23), NPY (24), SARS 1–3 (25)]. The different siRNA sequences were used as unrelated controls in non-overlapping systems.
Table 1.
Sequences of siRNA and siLNA used in the study
Top strand depicts the sense strand in the 5′–3′ direction (same as the target sequence). Bottom strand depicts the antisense strand in the 3′–5′ direction (complementary to the target). LNA, uppercase; RNA, lower case; DNA, italic lowercase. All LNA-C monomers were methyl cytosines.
The plasmids used were pGL3-Control coding for firefly luciferase and pRL-TK coding for Renilla luciferase (Promega, Madison WI, USA). pS3Xs and pS3Xas with SARS 3 target in the sense or antisense direction, respectively, were constructed by ligation of a double-stranded DNA oligonucleotide corresponding to the SARS 3 target site with Xba I overhangs into the Xba I site in the 3′UTR of the firefly luciferase in the pGL3-plasmid. The sense or antisense direction of the insert was confirmed after ligation by PCR and sequencing.
TM measurements
Melting curves were recorded with a Perkin Elmer UV/Vis spectrophotometer lambda 40 attached to a PTP-6 Peltier System. The siRNA/siLNA were dissolved in an RNase-free buffer (10 mM phosphate buffer, 100 mM NaCl, 0.1 mM EDTA pH 7) to a final concentration of 1.5 μM and measured in 1 cm path-length cells. Samples were denatured at 95°C for 3 min and slowly cooled to 20°C prior to measurements. Melting curves were recorded at 260 nm using a heating rate of 1°C/min, a slit of 2 nm and a response of 0.2 s. Tm values were obtained from the maxima of the first derivatives of the melting curves.
Cell cultures and virus
Cell lines used were human HEK 293, rat PC12 and monkey Vero. HEK 293 cells were maintained in DMEM supplemented with 10% foetal bovine serum, penicillin, streptomycin and glutamine. PC12 were maintained in DMEM supplemented with 10% horse serum, 5% foetal bovine serum, penicillin, streptomycin and glutamine. Vero cells were maintained in Eagle's MEM supplemented with 5% foetal bovine serum, penicillin, streptomycin and glutamine (Invitrogen). SARS-CoV, Frankfurt 1 isolate (GenBank AY291315) was amplified as previously described (25).
Transfection and inhibition experiments
Inhibition of firefly luciferase was performed in HEK293 cells by co-transfection of the target plasmid. HEK 293 cells were seeded in 500 μl antibiotic-free medium in 24-well plates the day before transfection to allow adherence and reach confluence of 70–90% at the time of transfection. The standard co-transfection mix was prepared for triplicate samples by adding 510 ng pGL3-Control, 51 ng pRL-TK and 340 ng siRNA to 150 μl Opti-MEM I (Invitrogen) and 3 μl LipofectAMINE 2000 (Invitrogen) to another 150 μl Opti-MEM I. The two solutions were mixed and incubated at room temperature for 20–30 min before 100 μl of the mix was added to each of three wells. The final volume of medium plus transfection mix was 600 μl and the final siRNA concentration was 13 nM. The cells were incubated with the transfection mix for 4 h and the medium was then replaced with new fully supplemented culturing medium. The cells were harvested 24 h later and luciferase activity was measured. The opposite amounts of plasmids were used in the Renilla luciferase assays.
The dose–response studies were performed analogously using a final siRNA concentration of 13 nM. The effective firefly luciferase siRNA was serially diluted with the unrelated siRNA targeting neuropeptide Y (NPY), reducing the effective amount of siRNA while keeping the total siRNA concentration constant. The plasmids pS3Xs and pS3Xas were used instead of pGL3-Control when assaying for the effects of the sense and antisense strand of SARS 3 siRNA and siLNA.
SiRNA and siLNA inhibition experiments with the endogenous target NPY were performed in PC12 cells as described above but without adding a target plasmid. The final siRNA concentration was 100 nM. mRNA was extracted 24 h post-transfection.
Inhibition of SARS-CoV-induced cytotoxicity followed a similar procedure. SARS-CoV infections and siRNA transfections of Vero cells were performed as described previously (25). Vero cells were seeded in 96-well plates, transfected with 85 nM siRNA using Lipofectamine 2000 and thereafter infected with 6000 TCID50 of SARS-CoV. The cytotoxicity was quantified with an LDH Cytotoxicity Detection Kit (Roche, Penzberg, Germany) 50 h later.
Luciferase activity
Luciferase activity was assessed according to the Dual-Luciferase Reporter Assay protocol (Promega) using a NovoSTAR 96-well format luminometer with substrate dispenser (BMG Labtechnologies, Offenburg, Germany). A 10 μl sample were placed in each well of a 96-well plate subsequent to which 50 μl Luciferace Assay Reagent II (substrate for firefly luciferase) was added to each well by the luminometer and the firefly activity was measured. Then 50 μl Stop and Glow (stop solution for firefly luciferase and substrate for Renilla luciferase) were added and Renilla luciferase activity measured. The mean of the luciferase activities measured for 10 s (100 readings) each were used to calculate ratios between firefly and Renilla luciferase.
Quantitative real-time PCR
Total RNA was isolated with RNeasy Mini and treated with RNase-Free DNase according to the manufacturer's protocol (Qiagen, Hilden, Germany). An amount of 400–800 ng of DNase-treated total RNA was used as template for first strand DNA synthesis according to the manufacturer's protocol (Applied Biosystems, Stockholm, Sweden). An aliquot (one-twentieth) of the cDNA reaction were analyzed by quantitative Real-Time PCR on an ABI PRISM 7000 (Applied Biosystems). Gene-specific primers and probes for the target genes NPY and cyclophilin A (24,26) were mixed separately with TaqMan Universal Mastermix (Applied Biosystems) and added to the cDNA to be analyzed. Samples were run in triplicate and the data obtained were analyzed with ABI Prism SDS Software (Applied Biosystems).
Serum stability
Duplexes of siRNA and siLNA (7 μM) were incubated at 37°C in 10% fetal bovine serum (Invitrogen) diluted in phosphate-buffered saline, 100% human or 100% mouse serum. Aliquots of 5 μl were withdrawn at different time points and immediately frozen in 15 μl 1.5× TBE-loading buffer. Samples were subjected to electrophoresis in 15% polyacrylamide–TBE under non-denaturing conditions and visualized by staining with SYBR gold and quantified by Typhoon 9400 hardware and Imagequant software (Amersham Biosciences, Uppsala, Sweden).
RESULTS
Serum stability of siLNA
LNA confers both unprecedented affinity as well as very high nuclease resistance on oligonucleotides (12–15,17). A priori, this suggests that LNA may be used to increase the functional half-life of siRNA in vivo by two different mechanisms, e.g. by enhancing the resistance of the constituent RNA strands against degradation by single-stranded RNases and by stabilizing the siRNA duplex structure that is critical for activity. To investigate how these mechanisms influence overall biostability, we assessed the integrity in fetal bovine serum, as well as human and mouse serum, of either unmodified siRNA or siLNAs that were modified to enhance exonuclease resistance (siLNA5) or further modified to also increase duplex stability (siLNA7). Briefly, siLNA5 has LNA modifications at the 3′ ends and exhibits a duplex stability similar to the unmodified siRNA1 (Tm = 69.7°C for siLNA5 and 70.5°C for siRNA1) whereas siLNA7 has six additional duplex stabilizing sense strand modifications at base-paired positions which increases its Tm to >90°C. As shown in Figure 1a and b, unmodified siRNA (siRNA1) was markedly degraded after 6 h during which it produces a smear of faster migrating species. A similar diffuse band was not observed with the 3′ end protected siLNA5, which in contrast to the unmodified siRNA showed only weak signs of degradation after 24 h. The more modified siLNA7 had a striking stability and showed no signs of degradation even at 48 h. When incubated in either undiluted human or mouse serum (Figure 1c and d), the unmodified siRNA1 were fully degraded within 6 h. An increased degradation rate was also observed with the siLNA5 where little full-length product remained at 24 h. In contrast, siLNA7 remained intact for the full 48 h of the assay in both human and mouse serum.
Figure 1.
Serum stability of siRNA and siLNA. The different siLNA/siRNA were incubated in 10% foetal bovine serum at 37°C and withdrawn at indicated time points. (a) The oligos were separated by PAGE and visualized with SYBR gold. ‘ds’ depicts double-stranded siRNA marker and ‘ss’ single-stranded. (b) Quantification of the bands. The mean and SD values are from three independent experiments. (c) Same layout as with foetal bovine serum but with 100% human serum or (d) 100% mouse serum.
Compatibility of LNA with the siRNA machinery
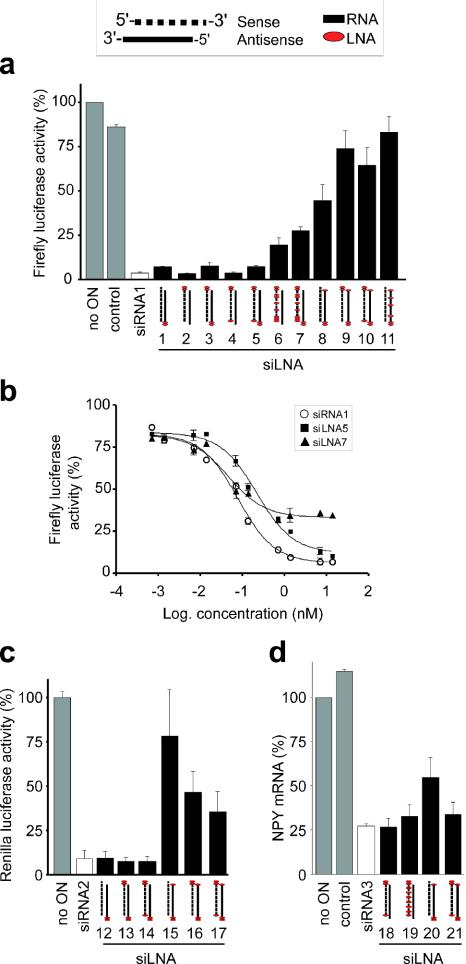
To verify experimentally the compatibility of LNA with the siRNA machinery, a range of different LNA-modified siRNAs (Table 1) were analysed for their ability to selectively inhibit firefly luciferase in cultured cells expressing both firefly and Renilla luciferase. As shown in Figure 2a, the firefly siRNA (siRNA1) effectively and selectively reduced firefly luciferase activity whereas an unrelated siRNA control was essentially without effect. Introduction of LNA modifications in the 3′ overhangs in either or both strands of the firefly siRNA revealed no loss of inhibitory effect (siLNA1–3). One LNA in the 5′ end of the sense strand was fully compatible with activity (siLNA4 and 5), while an LNA at the 5′ end of the antisense strand (siLNA8–11) dramatically impaired the inhibitory effect. To exclude the possibility that this impairment was due to a lack of a 5′ phosphate, which has been shown to be crucial for siRNA function (27), the 5′ end of the antisense strand in siLNA8 and 11 was phosphorylated in vitro. However, this procedure did not recover any of the lost effect (data not shown). SiLNAs wherein the sense strand was modified in the 3′ overhangs and at as many as seven base-paired positions (siLNA6 and 7) retained significant inhibitory activity whereas siLNAs comprising various combinations of either fully modified sense or antisense strands did not have any inhibitory effect (data not shown).
Figure 2.
Effect of different LNA loads on the activity of siRNAs against exogenous (a) firefly luciferase gene, (c) Renilla luciferase or (d) endogenous NPY gene. (a) Firefly luciferase activity. HEK293 cells were co-transfected with firefly luciferase and plasmids, and siLNA (13 nM) and luciferase activity was assessed 24 h later. The firefly luciferase activity was normalized to the Renilla luciferase activity and the uninhibited activity (plasmids alone) was set to 100%. (b) Dose–response curves on selected siLNA targeting firefly luciferase. The total siLNA/siRNA concentration was kept constant at 13 nM; the ratio of effective and irrelevant siLNA/siRNA was varied. The graph shows the log concentration of the effective siLNA from one of two representative experiments, with mean and SD derived from duplicate samples. (c) Renilla luciferase activity was assessed as for firefly luciferase. The Renilla activity was normalized to the firefly luciferase activity. (d) NPY mRNA levels. Rat PC12 cells, endogenously expressing NPY, were transfected with siLNA (100 nM). NPY mRNA was measured 24 h later by quantitative PCR. The NPY mRNA levels were normalized to cyclophilin A. The uninhibited normalized NPY mRNA level was set to 100%. The mean and SD values in the case of luciferase are from two independent experiments performed in triplicate, and from two independent experiments performed in duplicate in the case of NPY.
Two of the firefly siLNAs, the lightly modified siLNA5 and the medium modified siLNA7, were further compared to the unmodified siRNA1 in a dose–response experiment (Figure 2b). SiLNA5 and siRNA1 had the same efficacy but a slight difference in potency (estimated EC50 siLNA5 0.22 nM, siRNA1 0.08 nM) whereas siLNA7 had a somewhat lower efficacy at the highest dose tested but similar potency (estimated EC50 0.05 nM).
Our finding that an LNA at the 5′ antisense position substantially impairs the function of the siRNA contrasts with the findings of Braasch et al. who recently published on the effect of several chemical modifications, including some LNA, on siRNA against human caveolin (8). To determine whether this discrepancy was due to the different choice of targets, we repeated the analysis, this time targeting Renilla luciferase. As shown in Figure 2c, we observe the same tendency as with the firefly target. SiLNA12 and 13, carrying modifications in the 3′ overhangs were as functional as unmodified Renilla siRNA (siRNA2). Again, the 5′ end of the sense strand could be modified without loss of activity (siLNA14), while an LNA in the 5′ antisense end significantly reduced the effect (siLNA15). Interestingly, in this case part of the activity that was lost as a result of the 5′ antisense LNA (siLNA15) could be recovered by simultaneously modifying the 3′ sense end (siLNA16) and even more so if modifications also included the 5′ sense end (siLNA17).
To substantiate the generality of our finding, we finally appraised siLNAs targeting an endogenous gene, neuropeptide Y (NPY) in PC12 cells. As shown in Figure 2d, the unmodified NPY siRNA (siRNA3) reduced the mRNA levels considerably whereas an unrelated control, siRNA against dopamine D2 receptor, did not. Sense strand LNA modifications were, as before, well tolerated with both lightly modified (siLNA18) and medium modified (siLNA19) displaying a similar inhibitory effect as unmodified siRNA. Again an LNA 5′ antisense modification substantially impaired activity (siLNA20), which, however, could be mostly recovered by simultaneous modification of the 5′ and 3′ sense end (siLNA21).
Positional effects of single LNA modifications
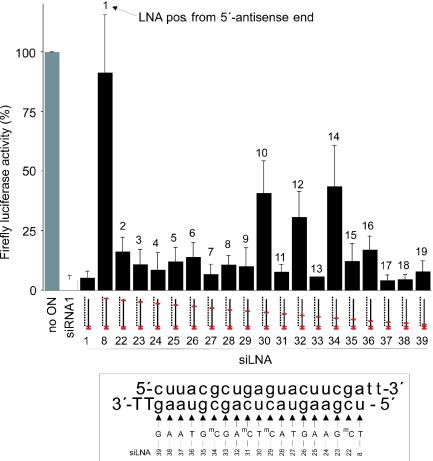
Next, we examined the effect of making single RNA to LNA exchanges at base-paired positions in the antisense strand of the firefly luciferase siLNA1. As shown in Figure 3, such exchanges were tolerated in most of the tested positions. Apart from the 5′ antisense end (siLNA8), the notable exceptions are positions 10 (siLNA30), 12 (siLNA32) and 14 (siLNA34), where introduction of LNA leads to a clear decrease of inhibitory activity. Although we cannot exclude that these modifications somehow prevent loading of the antisense strand into RISC, we believe this to be unlikely given the functionality of many significantly more modified siLNAs. Rather, as these positions are all close to the site where RNA target cleavage occurs [between pos. 10 and 11 of the siRNA strand counting from the 5′ end (28)], we suspect that the LNA modifications may exert a direct conformational or functional effect on the catalytic site.
Figure 3.
Effect on the activity of single RNA to LNA substitutions in the antisense strand. Mean and SD values are derived from two or more experiments.
The LNA substitutions at position 10 and 14 exchanged an RNA-U for an LNA-T and an RNA-C for an LNA-mC both of which lead to the introduction of an additional methyl-group on the nucleobase. In the A-form helix formed between the siRNA/RNA-target, these methyl groups will protrude into the major groove with potential effects on helical geometry or accessibility important for catalysis. To investigate this in more detail, we repeated the experiment with LNA-U in place of LNA-T at position 10. This new compound displayed activity similar to the unmodified siRNA (siRNA1), thus lending support to the importance of having native nucleobases close to the cleavage site (data not shown).
The replacement at position 12 (siRNA32) substituted an RNA-A for an LNA-A indicating that factors other than helical structure or accessibility are also important for proper catalytic activity. Given the increased affinity imposed by LNA, it seems likely that one such factor may be a changed thermodynamic fingerprint of the siRNA in the vicinity of the cleavage site, the importance of which has been indicated by recent reports (29,30).
Reducing ‘off-target’ effects
Much experimental data supports the notion that cells can incorporate both strands of an siRNA into the RISC complex but that preference is given to one of the two strands (3,27). An explanation for this strand-bias was provided by Schwarz et al. (31) and Khvorova et al. (29), who proposed that the strand that displays the weakest binding energy at its closing 5′ base-pair is incorporated preferentially. As a functional genomic tool and as a prospective therapeutic, the incorporation of the unwanted, non-target complementary, sense strand is a concern as it is a likely cause of ‘off-target’ effects (32) and may lower the potency of the siRNA by limiting incorporation of the intended antisense strand.
If relative binding energies at the ends of the siRNA duplex determine strand bias, it ought to be possible to favour incorporation of the antisense strand by selectively enhancing the affinity of the 5′ sense end with LNA. This intriguing possibility was already hinted at by the previous observations that activity loss due to LNA incorporation at the 5′ antisense end could be largely rescued by compensatory modifications in the 5′ end of the sense strand (which a priori would serve to restore the relative binding energies of the two ends of the siLNA).
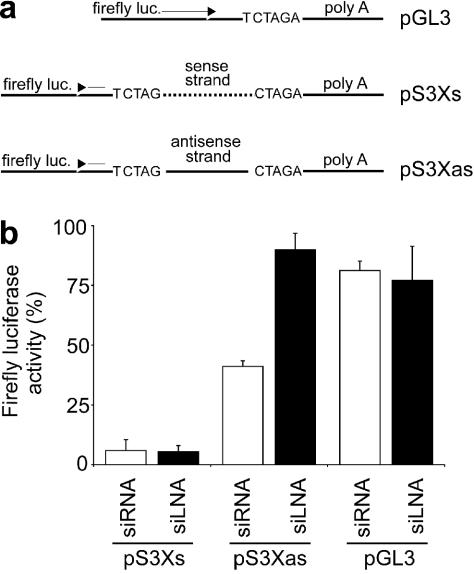
To examine in more detail the ability of LNA to direct strand loading, we constructed a plasmid system that made it possible to monitor the activity of both the sense and antisense strand. Briefly, a target region derived from the SARS virus to which a medium effective siRNA (SARS 3-siRNA) had previously been identified (25) was cloned into the 3′UTR of the firefly luciferase gene in both the sense (pS3Xs) and antisense (pS3Xas) orientation (Figure 4a). The SARS siRNA (Table 1) has identical closing base-pairs at both ends (A:U) making it likely that enough of both the antisense and sense strand would be incorporated into RISC to observe activity on the respective targets.
Figure 4.
Sense and antisense strand activity of siRNA and 5′ sense end modified siLNA. (a) Schematic representation of the SARS 3 target cloned in the sense (pS3Xs) or the antisense (pS3Xas) direction behind firefly luciferase. Also shown is the parental luciferase plasmid without the SARS 3 target (pGL3). (b) Activity of siRNA and siLNA against sense (pS3Xs), antisense (pS3Xas) or control target (pGL3-Control). Mean and SD values are from two experiments performed in duplicate.
As shown in Figure 4b, both SARS3-siRNA and SARS3-siLNA (modified at the 3′ overhangs and at the 5′ sense end) inhibited the sense target (pS3Xs) and to the same extent indicating that both siRNA and siLNA are effective in loading the antisense strand into RISC. However, when tested for sense strand activity, the outcome was different. Here, the siRNA showed clear downregulation of the target (pS3Xas), albeit the effect was less than that observed with the antisense strand. In contrast, no activity was observed with the siLNA sense strand strongly supporting the conclusion that the 5′ sense LNA modification has altered strand-bias in favour of incorporation of the antisense strand. Both siRNA and siLNA were also tested against the control plasmid pGL3 and found to have no effect on luciferase expression.
Improving on low efficacy siRNAs
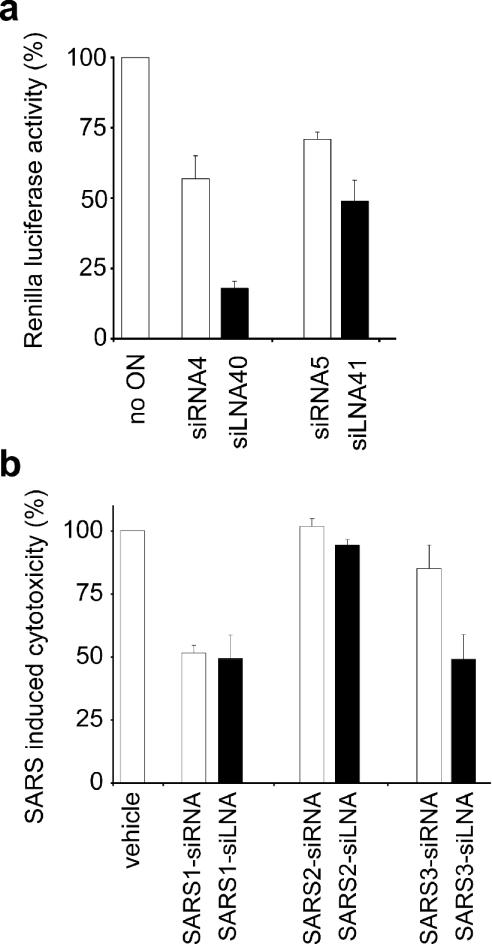
As described above, incorporation of sense strand may decrease the potency of siRNAs by simply lowering the number of RISC complexes loaded with the antisense strand. Having established that 5′ sense LNA appeared able to redirect strand loading, we next examined whether these modifications would also be able to improve the potency of two inefficient siRNAs targeting Renilla luciferase, siRNA4 and 5 (23). Both of these siRNAs have a strong G:C base-pair at their antisense end and a weak A:U base-pair at their 5′ sense end (Table 1) making it likely that at least part of the poor activity could be due to strand incorporation of the sense strand. As shown in Figure 5a, the activity of both siRNAs were improved by the 5′ sense LNA modification with siRNA4 reducing residual luciferase activity from 57 to 18% (siLNA 40) and siRNA5 from 71 to 49% (siLNA 41).
Figure 5.
LNA improvement of medium-efficient siRNAs. (a) Renilla luciferase activity. Effect of siRNA and siLNA depending on the target sequence (siRNA4 and 5 have different target sequences, accordingly also siLNA40 and 41). (b) Effect of siRNA and siLNA on SARS-induced cytotoxicity depending on the target sequence (SARS 1–3). Vero cells were transfected with 85 nM siRNA or siLNA and then infected with 6000 TCID50 SARS-CoV. SARS-induced cytotoxicity was assessed 50 h later. The untransfected but infected sample was set to 100% cytotoxicity. Mean and SD values in the Renilla case are from two experiments performed in triplicate, and in the SARS case from three experiments performed in quadruplicate.
To investigate if an LNA 5′ sense modification could rescue mediocre siRNAs against a therapeutically important target, we compared the ability of three siRNAs and their corresponding siLNAs to protect Vero cells from death induced by severe acute respiratory syndrome-associated coronavirus (SARS-CoV). Of the three siRNAs, the reasonably effective SARS-1 siRNA and the ineffective SARS-2 siRNA both have a strong G:C base-pairs at the 5′ sense end and a weak A:U base-pair at the 5′ antisense (Table 1). As such, further stabilization by LNA of their 5′ sense end is not expected to lead to improved activity and consistent with this notion none was observed (Figure 5b). In contrast, the modestly effective SARS-3 siRNA, which has A:U base-pairs at both ends showed the expected improvement when modified with LNA.
Decreasing or increasing the viral titres did not change the relative behaviour of the siRNAs and siLNAs although a generally greater reduction or increase in cytotoxicity was noted at the lower and higher titers, respectively (data not shown). Unrelated siRNA and siLNA controls (firefly luciferase and NPY) showed no inhibition of virus-induced cytotoxicity.
DISCUSSION
We have shown that the nucleotide analogue LNA is substantially compatible with the siRNA intracellular machinery, preserving molecule integrity whilst offering several improvements that are relevant to the development of siRNA technology for therapeutic use. Notably, LNA offers the means to improve dramatically the half-life of siRNAs through a combination of enhanced nuclease stability and stabilization of the duplex structure. As this property can be obtained with a modest number of LNA modifications that do not affect the ability of the siRNA to mediate target knock-down, we expect that siLNAs may exhibit significantly enhanced efficacy when administered in vivo compared to their unmodified counterparts.
Off-target effects brought about by inappropriate loading of siRNA sense strands constitute a major concern for the use of siRNAs as genomic tools and prospective drugs. Our data provide evidence that LNA can be used to minimize such effects, acting through two different mechanisms.
First, LNA substitution can alter strand-bias through selectively increasing the affinity of the closing base-pair at the 5′ end of the siRNA sense strand. We note that our analyses so far have been confined to the ultimate 5′ sense position and we cannot exclude the possibility that even greater strand-bias may be imposed by additional modifications to neighbouring positions.
Second, LNA may be incorporated into positions in the sense strand that, once loaded into the RISC complex, impair its ability to participate in target cleavage. We have identified these activity-impairing positions (pos. 10, 12 and 14) by systematically analysing a whole set of single LNA insertions in the antisense strand and found them to be in the vicinity of the target cleavage site. The evidence that similar substitutions applied to the sense strand will have a similar effect is indirect. Nevertheless, we see no reason to believe this will not be the case. If so, the combination of two or more activity-impairing LNA modifications may facilitate complete loss of activity of RISC complexes inappropriately loaded with sense strands.
The ability to influence strand loading by LNA modification at the 5′ sense end also provides an opportunity to improve the potency of ineffective siRNAs by further enhancing antisense strand incorporation into RISC. Although our data demonstrate that such enhanced loading is not a general phenomenon, it is an option to use LNA in this way where the choice of target sequence is restrained.
Consistent with the findings of Braasch et al. (8), the present data confirm that excessive LNA modifications, each of which are permissive when introduced as separate modifications, can reduce the knockdown efficacy of the siRNA. Based on the present data, we can only speculate as to the underlying causes which may include changes in siRNA structure that affect RISC loading, problems with unwinding the duplex due to excessive thermo-stability, changes in release kinetics after substrate cleavage, etc. Whatever the cause, as we have shown, the potential key therapeutic benefits of introducing LNA into siRNAs can all be achieved with relative few modifications that do not compromise siRNA activity. Other modifications than LNA has been shown to provide benefits to siRNA and could be conceivable when successfully combined with LNA.
In conclusion, the RNA-like character of LNA combined with its enhanced biophysical characteristics, e.g. increased nuclease resistance and affinity, enabled us to construct hybrid RNA–LNA molecules with new and favourable properties over unmodified siRNA. We anticipate that these new molecules, which we have termed siLNA, will impact positively on the use of RNAi technology in functional genomics and the broader perspective of translating the technology into a drug platform.
Acknowledgments
This study was supported by the PhD program in biotechnology with an industrial focus and the Foundation for Knowledge and Competence Development, which also provided the funding to pay the Open Access publication charges for this article.
REFERENCES
- 1.Elbashir S.M., Harborth J., Lendeckel W., Yalcin A., Weber K., Tuschl T. Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature. 2001;411:494–498. doi: 10.1038/35078107. [DOI] [PubMed] [Google Scholar]
- 2.Fire A., Xu S., Montgomery M.K., Kostas S.A., Driver S.E., Mello C.C. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature. 1998;391:806–811. doi: 10.1038/35888. [DOI] [PubMed] [Google Scholar]
- 3.Elbashir S.M., Lendeckel W., Tuschl T. RNA interference is mediated by 21- and 22-nucleotide RNAs. Genes Dev. 2001;15:188–200. doi: 10.1101/gad.862301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hamilton A.J., Baulcombe D.C. A species of small antisense RNA in posttranscriptional gene silencing in plants. Science. 1999;286:950–952. doi: 10.1126/science.286.5441.950. [DOI] [PubMed] [Google Scholar]
- 5.Zamore P.D., Tuschl T., Sharp P.A., Bartel D.P. RNAi: double-stranded RNA directs the ATP-dependent cleavage of mRNA at 21 to 23 nucleotide intervals. Cell. 2000;101:25–33. doi: 10.1016/S0092-8674(00)80620-0. [DOI] [PubMed] [Google Scholar]
- 6.Dorsett Y., Tuschl T. siRNAs: applications in functional genomics and potential as therapeutics. Nature Rev. Drug Discov. 2004;3:318–329. doi: 10.1038/nrd1345. [DOI] [PubMed] [Google Scholar]
- 7.Dykxhoorn M., Novina C., Sharp P.A. Killing the messenger: short RNAs that silence gene expression. Nature Rev. Mol. Cell Biol. 2003;4:457–467. doi: 10.1038/nrm1129. [DOI] [PubMed] [Google Scholar]
- 8.Braasch D.A., Jensen S., Liu Y., Kaur K., Arar K., White M.A., Corey D.R. RNA interference in mammalian cells by chemically-modified RNA. Biochemistry. 2003;42:7967–7975. doi: 10.1021/bi0343774. [DOI] [PubMed] [Google Scholar]
- 9.Harborth J., Elbashir S.M., Vandenburgh K., Manninga H., Scaringe S.A., Weber K., Tuschl T. Sequence, chemical, and structural variation of small interfering RNAs and short hairpin RNAs and the effect on mammalian gene silencing. Antisense Nucleic Acid Drug Dev. 2003;13:83–105. doi: 10.1089/108729003321629638. [DOI] [PubMed] [Google Scholar]
- 10.Amarzguioui M., Holen T., Babaie E., Prydz H. Tolerance for mutations and chemical modifications in a siRNA. Nucleic Acids Res. 2003;31:589–595. doi: 10.1093/nar/gkg147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Czauderna F., Fechtner M., Dames S., Aygun H., Klippel A., Pronk G.J., Giese K., Kaufmann J. Structural variations and stabilising modifications of synthetic siRNAs in mammalian cells. Nucleic Acids Res. 2003;31:2705–2716. doi: 10.1093/nar/gkg393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wahlestedt C., Salmi P., Good L., Kela J., Johnsson T., Hokfelt T., Broberger C., Porreca F., Lai J., Ren K., et al. Potent and nontoxic antisense oligonucleotides containing locked nucleic acids. Proc. Natl Acad. Sci. USA. 2000;97:5633–5638. doi: 10.1073/pnas.97.10.5633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Braasch D.A., Corey D.R. Locked nucleic acid (LNA): fine-tuning the recognition of DNA and RNA. Chem. Biol. 2001;8:1–7. doi: 10.1016/s1074-5521(00)00058-2. [DOI] [PubMed] [Google Scholar]
- 14.Kurreck J., Wyszko E., Gillen C., Erdmann V.A. Design of antisense oligonucleotides stabilized by locked nucleic acids. Nucleic Acids Res. 2002;30:1911–1918. doi: 10.1093/nar/30.9.1911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Crinelli R., Bianchi M., Gentilini L., Magnani M. Design and characterization of decoy oligonucleotides containing locked nucleic acids. Nucleic Acids Res. 2002;30:2435–2443. doi: 10.1093/nar/30.11.2435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bondensgaard K., Petersen M., Singh S.K., Rajwanshi V.K., Kumar R., Wengel J., Jacobsen J.P. Structural studies of LNA:RNA duplexes by NMR: conformations and implications for RNase H activity. Chemistry. 2000;6:2687–2695. doi: 10.1002/1521-3765(20000804)6:15<2687::aid-chem2687>3.0.co;2-u. [DOI] [PubMed] [Google Scholar]
- 17.Braasch D.A., Liu Y., Corey D.R. Antisense inhibition of gene expression in cells by oligonucleotides incorporating locked nucleic acids: effect of mRNA target sequence and chimera design. Nucleic Acids Res. 2002;30:5160–5167. doi: 10.1093/nar/gkf651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Obika S., Nanbu D., Hari Y., Andoh J., Morio K., Doi T., Imanishi T. Stability and structural features of the duplexes containing nucleoside analogues with a fixed N-type conformation, 2′-O,4′-C-methyleneribonucleosides. Tetrahedron Lett. 1998;39:5401–5404. [Google Scholar]
- 19.Koshkin A.A., Singh S.K., Nielsen P., Rajwanshi V.K., Kumar R., Meldgaard M., Olsen C.E., Wengel J. LNA (Locked Nucleic Acids): synthesis of the adenine, cytosine, guanine, 5-methylcytosine, thymine and uracil bicyclonucleoside monomers, oligomerisation, and unprecedented nucleic acid recognition. Tetrahedron. 1998;54:3607–3630. [Google Scholar]
- 20.Singh S.K., Nielsen P., Koshkin A.A., Wengel J. LNA (Locked nucleic acids): synthesis and high-affinity recognition. J. Chem. Commun. 1998:455–456. [Google Scholar]
- 21.Koshkin A.A., Nielsen P., Meldgaard M., Rajwanshi V.K., Singh S.K., Wengel J. LNA (Locked Nucleic Acid): an RNA mimic forming exceedingly stable LNA:LNA duplexes. J. Am. Chem. Soc. 1998;120:13252–13253. [Google Scholar]
- 22.Chiu Y.L., Rana T.M. RNAi in human cells: basic structural and functional features of small interfering RNA. Mol. Cell. 2002;10:549–561. doi: 10.1016/s1097-2765(02)00652-4. [DOI] [PubMed] [Google Scholar]
- 23.Xu Y., Zhang H.Y., Thormeyer D., Larsson O., Du Q., Elmen J., Wahlestedt C., Liang Z. Effective small interfering RNAs and phosphorothioate antisense DNAs have different preferences for target sites in the luciferase mRNAs. Biochem. Biophys. Res. Commun. 2003;306:712–717. doi: 10.1016/s0006-291x(03)01024-6. [DOI] [PubMed] [Google Scholar]
- 24.Thonberg H., Scheele C.C., Dahlgren C., Wahlestedt C. Characterization of RNA interference in rat PC12 cells: requirement of GERp95. Biochem. Biophys. Res. Commun. 2004;318:927–934. doi: 10.1016/j.bbrc.2004.04.119. [DOI] [PubMed] [Google Scholar]
- 25.Elme´n J., Wahlestedt C., Brytting M., Wahren B., Ljungberg K. SARS virus inhibited by siRNA. Preclinica. 2004;2:135–142. [Google Scholar]
- 26.Medhurst A.D., Harrison D.C., Read S.J., Campbell C.A., Robbins M.J., Pangalos M.N. The use of TaqMan RT-PCR assays for semiquantitative analysis of gene expression in CNS tissues and disease models. J. Neurosci. Methods. 2000;98:9–20. doi: 10.1016/s0165-0270(00)00178-3. [DOI] [PubMed] [Google Scholar]
- 27.Nykanen A., Haley B., Zamore P.D. ATP requirements and small interfering RNA structure in the RNA interference pathway. Cell. 2001;107:309–321. doi: 10.1016/s0092-8674(01)00547-5. [DOI] [PubMed] [Google Scholar]
- 28.Elbashir S.M., Martinez J., Patkaniowska A., Lendeckel W., Tuschl T. Functional anatomy of siRNAs for mediating efficient RNAi in Drosophila melanogaster embryo lysate. EMBO J. 2001;20:6877–6888. doi: 10.1093/emboj/20.23.6877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Khvorova A., Reynolds A., Jayasens S.D. Functional siRNAs and miRNAs exhibit strand bias. Cell. 2003;115:209–216. doi: 10.1016/s0092-8674(03)00801-8. [DOI] [PubMed] [Google Scholar]
- 30.Reynolds A., Leake D., Boese Q., Scaringe S., Marshall W.S., Khvorova A. Rational siRNA design for RNA interference. Nat. Biotechnol. 2004;22:326–330. doi: 10.1038/nbt936. [DOI] [PubMed] [Google Scholar]
- 31.Schwarz D.S., Hutvagner G., Du T., Xu Z., Aronin N., Zamore P.D. Asymmetry in the assembly of the RNAi enzyme complex. Cell. 2003;115:199–208. doi: 10.1016/s0092-8674(03)00759-1. [DOI] [PubMed] [Google Scholar]
- 32.Jackson A.L., Bartz S.R., Schelter J., Kobayashi S.V., Burchard J., Mao M., Li B., Cavet G., Linsley P.S. Expression profiling reveals off-target gene regulation by RNAi. Nat. Biotechnol. 2003;21:635–637. doi: 10.1038/nbt831. [DOI] [PubMed] [Google Scholar]