Abstract
Bisulfite genomic sequencing is the most widely used technique to analyze the 5-methylation of cytosines, the prevalent covalent DNA modification in mammals. The process is based on the selective transformation of unmethylated cytosines to uridines. Then, the investigated genomic regions are PCR amplified, subcloned and sequenced. During sequencing, the initially unmethylated cytosines are detected as thymines. The efficacy of bisulfite PCR is generally low; mispriming and non-specific amplification often occurs due to the T richness of the target sequences. In order to ameliorate the efficiency of PCR, we developed a new primer-design software called BiSearch, available on the World Wide Web. It has the unique property of analyzing the primer pairs for mispriming sites on the bisulfite-treated genome and determines potential non-specific amplification products with a new search algorithm. The options of primer-design and analysis for mispriming sites can be used sequentially or separately, both on bisulfite-treated and untreated sequences. In silico and in vitro tests of the software suggest that new PCR strategies may increase the efficiency of the amplification.
INTRODUCTION
In mammals, the prevalent covalent DNA modification is the 5′-methylation of cytosines in CpG dinucleotides (1). DNA methylation has fundamental physiological and pathological effects (2–4). It influences the functional state of the genome by determining the chromatin structure and the way the genome is packaged in the nucleus. It is implicated in the regulation of all the processes depending on chromatin structure, like differential gene expression and genomic imprinting, X-chromosome inactivation, recombination or replication.
Several methods have been developed to study DNA methylation based on methylation-sensitive restriction enzyme digestion (5–7), bisulfite modification (8,9) and immunolabeling (10). Some more recently developed techniques use a combination of these methods (11) or with other approaches such as the microarray technique (12), mass spectrometry (13,14), quantitative PCR (15) or DHPLC (16).
The most widely used method to study DNA methylation remains the bisulfite genomic sequencing technique (8,9), which gives information about the methylation profile of every single CpG site in a given sequence. This technique takes the advantage of the selective and complete transformation of unmethylated cytosines to uracil by sodium bisulfite. The chemically converted cytosines are amplified by PCR as thymines. After subcloning of the PCR product, the sequencing reveals the initial methylation profile of the region of interest. A technical advantage of the method resides in the use of PCR, which allows the analysis of samples with very low genomic DNA content, such as few haploid cells. However, the PCR amplification step is often the most difficult one in the whole process.
The challenge resides in the specific amplification of the bisulfite-treated DNA. The high redundancy of the target sequence due to the original GC richness creates long stretches of thymines, which are often difficult to read for the polymerases. Moreover, the DNA fragmentation during the treatment (17,18) leads to an empirical upper size limit of the PCR amplicon ∼400–500 bp. Indeed, only short amplicons are amplified and the need for nested primers and a second round of PCR is often necessary (19).
Therefore, a good primer design is crucial, since the dimer formations are greatly facilitated by the T and A richness of the sense and antisense oligos, respectively. Moreover, the primers designed for the bisulfite-treated template frequently amplify non-specific PCR products because of the mispriming of the highly redundant genome generated by the treatment. Although several primer-design softwares exist for the amplification of bisulfite-treated DNA (20–22), the problem remains unresolved. Some only transform the target sequences according to the bisulfite treatment and renders them suitable for primer-design by the conventional algorithms. MethPrimer and the recently published PerlPrimer both carry out the bisulfite transformation of the target sequences and design primers based on CpG island prediction (23).
However, an in silico test of the PCR primers for potential non-specific amplification of the bisulfite-treated genome would greatly facilitate in overcoming the difficulties of the technique. Here, we propose BiSearch for primer design to amplify normal or bisulfite-treated DNA and, if needed, for methylation-specific PCR (MSP) (24). The option of similarity search with the selected primer pairs on a bisulfite-treated genome is the most important feature of this algorithm. The search result is analyzed in an output file for the potential non-specific PCR products. The BiSearch software can be used for academic researchers at the http://bisearch.enzim.hu site.
MATERIALS AND METHODS
Bisulfite treatment
After DNA extraction from HEK and HepG2 cells, bisulfite treatment was performed according to the standard protocol (8,9). Briefly, DNA was denatured by 0.3 M NaOH treatment. Bisulfite solution was added for overnight treatment and the samples were heated at 55°C. After desalting, the treatment was completed by desulphonation and neutralization. Samples were ethanol precipitated and ready to use for PCR amplification.
PCR
The following primer pairs were designed by BiSearch to amplify a fragment of the bisulfite-treated tyrosine hydroxylase (TH, D00269) gene from genomic template:
THbs114 GAGGTTTTGGTGTTTATTAAA;
THbas308 TAAAATCTAATTACCTTCACTCC;
THbs566 TGTTAAGTAGGTAGAGGTTATTAT; and
THbas827 AACCTAAAAAAAACACACAC.
The AmpliTaqGold amplification system (Applied Biosystems) was used for the PCR of bisulfite-treated DNA with standard cycling conditions after an initial cycle of 10 min at 95°C: 30 cycles with denaturation at 95°C for 30 s, annealing at 56°C for 30 s and elongation at 72° for 90 s. A second PCR program was also performed, which differed from the first one by the annealing temperature lowered to 49°C (as indicated in the Results section).
RESULTS
Description of BiSearch
BiSearch software is composed of two basic algorithms. They can be used stepwise or separately. The first one is a primer design algorithm and the second one is a search with the selected primers through genomic sequences to find potential non-specific PCR products. We describe the main features of these algorithms in the following sections.
Primer design algorithm. The primer design software can propose primer pairs to amplify either bisulfite-treated (methylated or unmethylated target molecules) or untreated sequences.
Target sequence. The sequence to be amplified is entered in a plain text format. In case of primer design for PCR of bisulfite-treated target molecules, the conversion of the original sequence is carried out by the BiSearch software. In this case, every cytosine is converted to thymine with the memorization of the original CpG sites. As a result of the bisulfite treatment, the two DNA strands lose their original complementarities. For PCR amplification of the original sense or antisense strands, different primer pairs should be designed. The software proposes both options and converts the sequence according to the user's choice. Finally, if for diagnostic or other reasons only methylated alleles should be amplified (MSP), the conversion takes place only at the non-CpG cytosines as if all the CpG dinucleotides were methylated.
Primer selection. The primer design algorithm is a modification of the algorithm proposed by Kämpke et al. (25). Briefly, the program calculates various properties of the primer pairs picked up from the sense and antisense chains presented by the user. The primer pairs are scored according to these properties using simple weighted sums. All parameters used in the primer design are adjustable by the user. A modifiable optimum and acceptable range are set for all of them. The distance from the optimum is calculated and scored with different weights for each parameter.
The major criterion for primer pair design is the ability of duplex formation of the selected oligonucleotide with one specific site of the target sequence. Primers with one or more mismatches are not allowed by the software. Additional criteria are the low self (end) annealing and duplex formation abilities of the two primers, melting temperature analysis, GC content, primer and product length.
Melting temperature in °C is calculated according to Wetmur and Sninsky (26), by the following formula:
where T0 = 298.2 K, R = 1.987 (cal/mol K) the molar gas constant, ΔHp and ΔGp are the enthalpy and free energy of the primer p, p = (p1,…,pn) which are computed according to the nearest neighbor schemes, i.e. and , where enthalpy and free energy of a string consisting of two bases is given in Table 1 (26) and ΔHe = 5 kcal/mol, ΔGe = 1 kcal/mol and ΔGi = −2.2 kcal/mol. The dimensionless concentration of the primer in the PCR mixture is Cp and Csalt is the molar salt concentration. We use the GC content in the scoring of the primers, which measures the percentage of G and C of the primer. The self annealing, self end-annealing, pair annealing and pair end-annealing are calculated in the same way as described in (25). Finally, a score is calculated for primer pairs by using weighted sum of the distance of these calculated values of properties from their optimal values. To find the primer pairs with the best score, we use simple brute force searching, because the typical computational time is less than a minute. The left and right primers are searched in the sequence region defined by the user and by taking into account the maximal product length desired.
Table 1.
Nearest neighbor thermodynamic parameters used for primer design
| −ΔH(pi, pi+1) | −ΔG(pi, pi+1) | |
|---|---|---|
| AA or TT | 9.1 | 1.55 |
| AT | 8.6 | 1.25 |
| TA | 6.0 | 0.85 |
| CA or TG | 5.8 | 1.15 |
| GT or AC | 6.5 | 1.40 |
| CT or AG | 7.8 | 1.45 |
| GA or TC | 5.6 | 1.15 |
| CG | 11.9 | 3.05 |
| GC | 11.1 | 2.70 |
| GG or CC | 11.0 | 2.30 |
ΔH(pi, pi+1) and ΔG(pi, pi+1) are the enthalpy and free energy in kcal/mol required to disrupt the hydrogen bonds between the complementary bases of a paired chain, where pi and pi+1 are bases of primer p at the positions i and i + 1.
When a primer pair for the bisulfite-treated sequence is designed, sometimes CpG dinucleotides are found in the corresponding original sequence. In these cases the software proposes both the unmethylated and the methylated versions of the nucleotide at these sites, i.e. Y (C or T) for the sense strand and R (A or G) for the antisense strand. In order to avoid PCR bias, the software accepts only those primers where the maximal Tm difference between the ‘methylated’ and ‘unmethylated’ primer sequence is lower than 2.5°C.
In contrast, for primers designed for MSP, the inclusion of CpG dinucleotides in the primers is obligatory. Moreover, at least one of the primers has to contain the C of a methylated CpG at the 3′ end of the oligo. The program calculates the Tm difference between the methylated and a potential unmethylated sequence. In order to increase the specificity of the MSP reaction, the minimum acceptable difference was set at 8°C and only the oligonucleotide complementary to the methylated sequence is proposed.
The results are visualized in a table. For each primer pair, the sequences and the calculated individual parameter scores are shown. The sequence of the PCR product is also presented with the regions corresponding to the oligos underlined. The CpG dinucleotides are highlighted and their total number in the amplicon is indicated. If the primers are designed for a bisulfite-treated target sequence, all the other cytosines are converted to thymine.
The listed best primer pairs are significantly different from each other because the selected primers are filtered for resemblance. The software considers two primers significantly different from each other when the distance between their respective 5′ or 3′ ends is >3 nt. A primer pair is significantly different from another pair when at least one of the two primers is significantly different from the corresponding primer of the other pair. The software proposes only the best-scored primer pair from not significantly different pairs.
Post-design primer analysis. Although this option of the software may be applied to both bisulfite-treated and normal genomic sequences, we focus on the description of the first one because of its unique feature and methodological importance. A major difficulty of the PCR amplification of bisulfite-treated genomic DNA is the high redundancy of the target sequences. In fact, most of the sequences of interest are originally GC rich and they become extremely T rich after the chemical modification. Therefore, the primer pairs designed to a specific sequence may often amplify other non-specific products as well. In order to ameliorate the primer design and PCR efficiencies, we set up a novel option to test the binding capacity of the oligos to the original or bisulfite-treated genome. This option is not available for MSP.
Four genomic databases downloaded from the ensembl webserver are currently available to the user as a template: Homo sapiens, Mus musculus, Pan troglodytes and Rattus norvegicus. The bisulfite-converted versions are available for all of them at the BiSearch web site. A simple similarity search between the primers and the bisulfite-treated genome of interest is realized in a compressed file format. Both the sense and antisense oligonucleotides are searched on four strands. In fact, as mentioned previously, as a result of the bisulfite treatment the double-stranded DNA composed of two complementary strands is transformed to two single-stranded DNA molecules, which are not complementary anymore. Each primer is searched on the two non-complementary genomes created by the bisulfite transformation of the original sense and antisense strands. Moreover, the primers are also searched on the two additional virtual sequence strands. In fact, the complementary strands of the single-stranded DNA molecules are synthesized only during the PCR reaction by the reverse primers. In order to avoid the accumulation of potential non-specific PCR products, these initially nonexistent complementary strands have to be checked for similarity as well. Thus, both the primers are checked for similarity on four strands for every initially unique genome.
BiSearch utilizes a simple search method to analyze the potential amplification products of a designed primer pair. We use a simple matching string to fasten the searching. The matching criterion is defined by the user via a simple match string (00001111222…), which means the maximal number of mismatch from the 3′ end of the nucleotide. In this example, there is no mismatch for the first four nucleotides, one is allowed between the fifth and eight nucleotides, and a second one between the eight and twelve or two if no mismatches are present in the first eight positions. To check the matches of a given primer at a given genomic position, the algorithm compares the number of mismatches from the 3′ end of the primer with the corresponding position of the matching string. If the number of mismatches is below the limit on the whole primer sequence, then the hit is accepted as an annealing site.
Search for PCR products. All the forward and reverse primer annealing sites are collected, and their positions are compared. A sequence is accepted as a PCR target, when two properly localized primer binding sites are found on one of the bisulfite-treated and on its virtual complementary strand and are within <1 kb distance. All the potential PCR product sequences are shown with the details of their genomic location.
In silico test of BiSearch
Primer design and post-design analysis. We designed primer pairs for the amplification of a portion of the bisulfite-treated human tyrosine hydroxylase gene (TH, D00269). Default conditions were used (primer length between 15 and 35 bases, Tm between 50 and 60°C). The optimal GC content was set at 30% and the maximal product length was limited at 400 bp. An important feature of the program is the proposal of significantly different primer pairs for the amplification of various DNA fragments from the larger original input sequence. The score of the primer pair proposed as the first choice was 12.30. These oligonucleotides, called THbs566 and THbas827, amplify a PCR fragment of 262 bp. Another forward and reverse primer, called THbs114 and THbas308 respectively, was also selected. The score of this primer pair was worse: 14.18. THbs114 and THbas308 amplify a PCR fragment of 195 bp.
The selected primers were subjected to post-design sequence analysis. The search results for the first primer pair with the default parameters showed ∼40 hits on each original bisulfite-treated strand and 2500 hits per virtual strand. Moreover, one non-specific PCR fragment was also identified. This fragment was only 20 bp longer than the specific one (282 bp), but a few mismatches were present in both primers. It is important to note that when search parameters were modified in order to accept only perfect matches, the number of hits diminished by two orders of magnitude. The second primer pair gave much less genomic hits and only the specific PCR fragment was detected.
For untreated sequences, usually only a few and almost always <20 hits were found per primer with the default parameters, which allows a few mismatches at the 5′ end of the sequence. However, the primers designed for the amplification of treated DNA gave much more, often several thousands of hits with the same parameters, when searched on a bisulfite genomic database. Although this may be surprising at first glance, the extreme genomic redundancy after bisulfite treatment accounts for these results. The sense primers are composed of only A, G and T nucleotides, like the two bisulfite-treated original DNA strands. In general, no hits are found on the two virtual antisense strands with these primers, since they are composed of only T, A and C nucleotides. The antisense primers give hits only on the virtual strands with no match on the two existing bisulfite-treated strands. The number of hits on each relevant strand (originals and virtuals, respectively) follows a Poisson-distribution (data not shown).
In vitro test of BiSearch
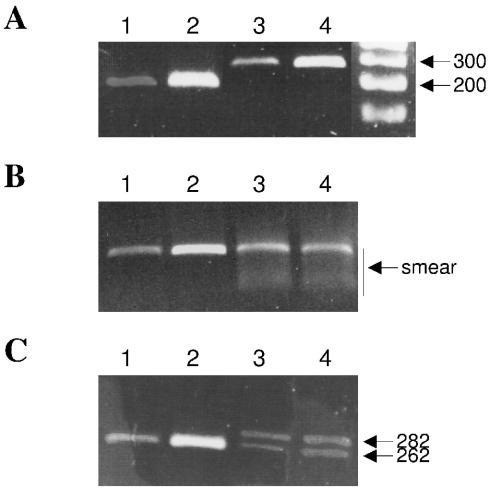
We tested the selected primers using the PCR on bisulfite-treated genomic DNA template. Standard reaction conditions were used with a hot start enzyme. The annealing temperature was set at the primers' Tm (not below). Both primer pairs amplified the specific PCR fragment (Figure 1A). No traces of contaminating PCR product or primer dimers were observed on the gel.
Figure 1.
PCR amplification of bisulfite-treated human genomic DNA with primer pairs designed by the BiSearch software. The primers were used to amplify fragments of the human tyrosine hydroxylase gene. (A) Specific fragments of 195 and 262 bp were amplified at 56°C by primer pairs THbs113/THbas308 (lanes 1 and 2) and THbs565/THbas827 (lanes 3 and 4), respectively. The THbs565/THbas827 primer pair was also tested at 49°C annealing temperature. (B) Samples in lanes 1 and 2 are the same as in lanes 3 and 4 on panel A. The two other PCR products were obtained at the lower annealing temperature. The smears reveal an inefficient reaction. (C) The same gel after longer migration resolved the PCR products of lanes 3 and 4 into two distinct bands corresponding to the predicted lengths by BiSearch search.
In some experimental protocols, low annealing temperature is proposed for the amplification of bisulfite-treated DNA (19). We also tested the second primer pair in a PCR experiment with identical conditions than the first time, except for lowering the annealing temperature by 7°C. The resulting PCR product contained a low-molecular weight smear, suggesting an inefficient amplification and a band around the predicted amplicon length (Figure 1B). A longer migration on a 2% agarose gel also revealed that two distinct PCR products were present in both reaction tubes at the end of the second PCR (Figure 1C). These bands correspond in length to the predicted ones by the software (262 and 282 bp, respectively). These results validate the BiSearch software and underscore the utility of the special post-design primer analysis option.
DISCUSSION
DNA methylation has a major role in the regulation of chromatin structure and function and is implicated in the pathogenesis of several hereditary and malignant disorders (2–4). The pattern and level of CpG methylation is intensively studied by the bisulfite genomic sequencing method (8,9). After the bisulfite conversion of unmethylated cytosines, PCR amplification precedes the determination of methylation pattern. Due to the chemical reaction, the target for the amplification is a highly U/T rich molecule embedded in a similarly U/T rich background. These conditions lower the efficiency of the PCR since mispriming often occurs.
The in silico analysis of primers for annealing to various genomic loci and for the potential non-specific PCR products can greatly facilitate the PCR end-point assays and increase the efficiency of the reaction. In this paper, we present the first and unique tool capable of doing such an analysis: BiSearch, the combined new primer-design and post-design analysis software. The software is able to design primers for the PCR amplification of both bisulfite-treated and untreated sequences. The functions may be used sequentially or separately.
The primer-design function of BiSearch was successfully utilized in an unrelated study of transcriptional gene regulation when we analyzed two CpG islands on the human chromosome 16. One of the alternative well-known primer-design algorithms was unable to propose primer pairs for the amplification of the bisulfite-treated sequence, while BiSearch was able to propose several pairs and the selection of specific ones was possible after the Primer Genome Search. The DNA methylation analyses of both CpG islands were carried out successfully (Arányi et al., manuscript in preparation).
The primer selection by BiSearch is achieved according to the consideration of different parameters adjusted by the user. An important feature of the program is the proposal of primer pairs significantly different from each other. This approach allows the user to decide the portion of a longer DNA sequence that is the best target for PCR amplification. The decision may depend on the scores of the designed primers and other experimental constraints.
These constraints may be independent of the PCR itself, but sometimes other factors, not included in the primer design calculation, may influence the efficiency of the amplification reaction. One of them is the non-specific annealing and mispriming of the primers. To circumvent this problem, the software proposes the unique post-design primer analysis option.
This algorithm of the software carries out a simple similarity search with the designed primer for highly homologous genomic sequences. The search is currently available on four different genomes each downloaded from the ensembl webserver (27). The genomic databases are extendable upon request. The search is also available on the bisulfite-treated counterpart of these genomes. Each bisulfite genomic database was generated by the BiSearch program. It consists of two sense strands corresponding to the initial complementary strands whose complementarities were lost upon treatment. The database also contains the two virtual complementary strands synthesized only during the PCR.
The similarity search of BiSearch software is fundamentally different from Blast searches (28), which calculate the probability of identity of two sequences. The Blast software searches with a minimal word size (11 nt by default) i.e. it takes into account only sequences that harbor a stretch of a given length completely identical to the query sequence. Moreover, because of the high redundancy of the bisulfite-treated genomic DNA and the relatively high memory usage of Blast, the search cannot be carried out on a usual PC because typical memory usage of a run is >2 Gb.
The BiSearch software compares base by base the primer sequence from the 3′ end toward the 5′ end with the target genome sequence. The acceptable total number of mismatches from the 3′ end of the primer can be defined by the user for each nucleotide position. This approach may accept sequences as similar if their 3′ end (more important for the PCR) is identical, while their 5′ end is divergent. The software considers all the sequences fitting the criteria as hits of equal validity. The extreme genomic redundancy after the bisulfite treatment often leads to several thousands of search hits when the default parameters are used. This is approximately hundred times more than in the case of the classic genomic primers. In the next step, the program analyses the genomic sequences homologous to the primers for potential amplification and shows as PCR products, those regions that are <1 kb long, flanked by the designed primer(s) in a correct orientation on the complementary strands. Non-specific PCR products are frequently found by the program just as empirically observed without the use of this new in silico tool.
The use of this new primer-design software and the coupled post-design primer analysis reveals potential new strategies for the PCR amplification of bisulfite-treated target sequences. First, GC rich gene regulatory sequences studied for DNA methylation are often contaminated by repeat sequences. Even if some repeat elements are variable and allow specific amplification of the original genomic fragment, the bisulfite treatment strongly reduces the sequence diversity and renders the PCR, inefficient and non-specific. We thus suggest a repeat analysis by the Repeatmasker software prior to primer design (29).
Second, based on empirical considerations, primers designed previously for bisulfite-treated DNA were between 25 and 35 bases long. In the present study, two primer pairs were designed by the BiSearch software and tested both in silico and in vitro. The minimal and maximal acceptable primer length was set at 15 and 35 bases, respectively. All the four selected primers were between 20 and 24 bases long and amplified the specific fragment (see Figure 1A), suggesting that the use of longer oligonucleotides may be unnecessary.
Third, when setting the parameters for primer design, the GC content may be crucial. The original sequence before bisulfite treatment usually contains at least 60% of G+C, since the investigated regions are often CpG islands (30). Assuming that half of the G+C content is G, the remaining G+C content after bisufite treatment is still 30%. In order to avoid long stretches of T or A in the primer sequence, which can diminish the efficiency of amplification reaction, it is useful to set the optimal primer G+C content at 30%.
Fourth, generally suggested PCR conditions used low annealing temperatures (19). However, not surprisingly, our results showed that higher annealing temperatures resulted in more specific PCR conditions (see Figure 1C). Moreover, as discussed above, the post-design primer analysis with default parameters often shows several thousands of matches when the primers are searched on the bisulfite-treated genome. When the parameters are changed and only complete matches are accepted, the high number of hits decreases by hundred times. This clearly indicates that the use of more elevated annealing temperatures during the PCR should result in a more specific and efficient amplification. In conclusion, the use of the combined BiSearch software facilitates primer selection and indicates new strategies to increase the efficiency of PCR on bisulfite-treated genomic target sequences.
Acknowledgments
The authors thank Gábor Tusnády for useful comments. This work was financed by PXE International, Inc., GVOP-3.1.1-2004-05-0143/3.0 and OTKA T34131, D42207. G.E.T. was supported by the Boólyai János Scholarship. Funding to pay the Open Access publication charges for this article was provided by GVOP-3.1.1-2004-05-0293/3.0.
REFERENCES
- 1.Gruenbaum Y., Stein R., Cedar H., Razin A. Methylation of CpG sequences in eukaryotic DNA. FEBS Lett. 1981;124:67–71. doi: 10.1016/0014-5793(81)80055-5. [DOI] [PubMed] [Google Scholar]
- 2.Laird P.W. The power and the promise of DNA methylation markers. Nature Rev. Cancer. 2003;3:253–266. doi: 10.1038/nrc1045. [DOI] [PubMed] [Google Scholar]
- 3.Robertson K.D., Wolffe A.P. DNA methylation in health and disease. Nature Rev. Genet. 2000;1:11–19. doi: 10.1038/35049533. [DOI] [PubMed] [Google Scholar]
- 4.Singal R., Ginder G.D. DNA methylation. Blood. 1999;93:4059–4070. [PubMed] [Google Scholar]
- 5.Bird A.P. Use of restriction enzymes to study eukaryotic DNA methylation: II. The symmetry of methylated sites supports semi-conservative copying of the methylation pattern. J. Mol. Biol. 1978;118:49–60. doi: 10.1016/0022-2836(78)90243-7. [DOI] [PubMed] [Google Scholar]
- 6.Bird A.P., Southern E.M. Use of restriction enzymes to study eukaryotic DNA methylation: I. The methylation pattern in ribosomal DNA from Xenopus laevis. J. Mol. Biol. 1978;118:27–47. doi: 10.1016/0022-2836(78)90242-5. [DOI] [PubMed] [Google Scholar]
- 7.Riggs A.D. X inactivation, differentiation, and DNA methylation. Cytogenet. Cell. Genet. 1975;14:9–25. doi: 10.1159/000130315. [DOI] [PubMed] [Google Scholar]
- 8.Frommer M., McDonald L.E., Millar D.S., Collis C.M., Watt F., Grigg G.W., Molloy P.L., Paul C.L. A genomic sequencing protocol that yields a positive display of 5-methylcytosine residues in individual DNA strands. Proc. Natl Acad. Sci. USA. 1992;89:1827–1831. doi: 10.1073/pnas.89.5.1827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Clark S.J., Harrison J., Paul C.L., Frommer M. High sensitivity mapping of methylated cytosines. Nucleic Acids Res. 1994;22:2990–2997. doi: 10.1093/nar/22.15.2990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Adouard V., Dante R., Niveleau A., Delain E., Revet B., Ehrlich M. The accessibility of 5-methylcytosine to specific antibodies in double-stranded DNA of Xanthomonas phage XP12. Eur. J. Biochem. 1985;152:115–121. doi: 10.1111/j.1432-1033.1985.tb09170.x. [DOI] [PubMed] [Google Scholar]
- 11.Xiong Z., Laird P.W. COBRA: a sensitive and quantitative DNA methylation assay. Nucleic Acids Res. 1997;25:2532–2534. doi: 10.1093/nar/25.12.2532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gitan R.S., Shi H., Chen C.M., Yan P.S., Huang T.H. Methylation-specific oligonucleotide microarray: a new potential for high-throughput methylation analysis. Genome Res. 2002;12:158–164. doi: 10.1101/gr.202801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tost J., Schatz P., Schuster M., Berlin K., Gut I.G. Analysis and accurate quantification of CpG methylation by MALDI mass spectrometry. Nucleic Acids Res. 2003;31:e50. doi: 10.1093/nar/gng050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Humeny A., Beck C., Becker C.M., Jeltsch A. Detection and analysis of enzymatic DNA methylation of oligonucleotide substrates by matrix-assisted laser desorption ionization time-of-flight mass spectrometry. Anal. Biochem. 2003;313:160–166. doi: 10.1016/s0003-2697(02)00568-7. [DOI] [PubMed] [Google Scholar]
- 15.Zeschnigk M., Bohringer S., Price E.A., Onadim Z., Masshofer L., Lohmann D.R. A novel real-time PCR assay for quantitative analysis of methylated alleles (QAMA): analysis of the retinoblastoma locus. Nucleic Acids Res. 2004;32:e125. doi: 10.1093/nar/gnh122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Couvert P., Poirier K., Carrie A., Chalas C., Jouannet P., Beldjord C., Bienvenu T., Chelly J., Kerjean A. DHPLC-based method for DNA methylation analysis of differential methylated regions from imprinted genes. Biotechniques. 2003;34:356–362. doi: 10.2144/03342rr06. [DOI] [PubMed] [Google Scholar]
- 17.Grunau C., Clark S.J., Rosenthal A. Bisulfite genomic sequencing: systematic investigation of critical experimental parameters. Nucleic Acids Res. 2001;29:65. doi: 10.1093/nar/29.13.e65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Raizis A.M., Schmitt F., Jost J.P. A bisulfite method of 5-methylcytosine mapping that minimizes template degradation. Anal. Biochem. 1995;226:161–166. doi: 10.1006/abio.1995.1204. [DOI] [PubMed] [Google Scholar]
- 19.Warnecke P.M., Stirzaker C., Song J., Grunau C., Melki J.R., Clark S.J. Identification and resolution of artifacts in bisulfite sequencing. Methods. 2002;27:101–107. doi: 10.1016/s1046-2023(02)00060-9. [DOI] [PubMed] [Google Scholar]
- 20.Li L.C., Dahiya R. MethPrimer: designing primers for methylation PCRs. Bioinformatics. 2002;18:1427–1431. doi: 10.1093/bioinformatics/18.11.1427. [DOI] [PubMed] [Google Scholar]
- 21.Marshall O.J. PerlPrimer: cross-platform, graphical primer design for standard, bisulphite and real-time PCR. Bioinformatics. 2004;20:2471–2472. doi: 10.1093/bioinformatics/bth254. Available at ( http://perlprimer.sourceforge.net) [DOI] [PubMed] [Google Scholar]
- 22.Singal R., Grimes S.R. Microsoft Word macro for analysis of cytosine methylation by the bisulfite deamination reaction. Biotechniques. 2001;30:116–120. doi: 10.2144/01301bc02. [DOI] [PubMed] [Google Scholar]
- 23.Gardiner-Garden M., Frommer M. CpG islands in vertebrate genomes. J. Mol. Biol. 1987;196:261–282. doi: 10.1016/0022-2836(87)90689-9. [DOI] [PubMed] [Google Scholar]
- 24.Herman J.G., Graff J.R., Myohanen S., Nelkin B.D., Baylin S.B. Methylation-specific PCR: a novel PCR assay for methylation status of CpG islands. Proc. Natl Acad. Sci. USA. 1996;93:9821–9826. doi: 10.1073/pnas.93.18.9821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kampke T., Kieninger M., Mecklenburg M. Efficient primer design algorithms. Bioinformatics. 2001;17:214–225. doi: 10.1093/bioinformatics/17.3.214. [DOI] [PubMed] [Google Scholar]
- 26.Wetmur J., Sninsky J. Nucleic acid hybridization and unconventional bases. In: Innis M., Gelfand D., Sninsky J., editors. PCR strategies. Academic Press; 1995. pp. 69–83. [Google Scholar]
- 27.Birney E., Andrews D., Bevan P., Caccamo M., Cameron G., Chen Y., Clarke L., Coates G., Cox T., Cuff J., et al. Ensembl 2004. Nucleic Acids Res. 2004;32:D468–D470. doi: 10.1093/nar/gkh038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Altschul S.F., Gish W., Miller W., Myers E.W., Lipman D.J. Basic local alignment search tool. J. Mol. Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 29.Smit A., Green P. RepeatMasker. 1996. Available at ( http://www.repeatmasker.org)
- 30.Bird A., Taggart M., Frommer M., Miller O.J., Macleod D. A fraction of the mouse genome that is derived from islands of nonmethylated, CpG-rich DNA. Cell. 1985;40:91–99. doi: 10.1016/0092-8674(85)90312-5. [DOI] [PubMed] [Google Scholar]