Abstract
The advancement in site directed spin labeling of proteins has enabled EPR studies to expand into newer research areas within the umbrella of protein-membrane interactions. Recently, membrane remodeling by amyloidogenic and non-amyloidogenic proteins has gained a substantial interest in relation to driving and controlling vital cellular processes such as endocytosis, exocytosis, shaping of organelles like endoplasmic reticulum, Golgi and mitochondria, intracellular vesicular trafficking, formation of filopedia and multivesicular bodies, mitochondrial fusion and fission, and synaptic vesicle fusion and recycling in neurotransmission. Misregulation in any of these processes due to an aberrant protein (mutation or misfolding) or alteration of lipid metabolism can be detrimental to the cell and cause disease. Dissection of the structural basis of membrane remodeling by proteins is thus quite necessary for an understanding of the underlying mechanisms, but it remains a formidable task due to the difficulties of various common biophysical tools in monitoring the dynamic process of membrane binding and bending by proteins. This is largely since membranes generally complicate protein structure analysis and this problem is amplified for structural analysis in the presence of different types of membrane curvatures. Recent EPR studies on membrane remodeling by proteins show that a significant structural information can be generated to delineate the role of different protein modules, domains and individual amino acids in the generation of membrane curvature. These studies also show how EPR can complement the data obtained by high resolution techniques such as X-ray and NMR. This perspective covers the application of EPR in recent studies for understanding membrane remodeling by amyloidogenic and non-amyloidogenic proteins that is useful for researchers interested in using or complimenting EPR to gain better understanding of membrane remodeling. We also discuss how a single protein can generate different type of membrane curvatures using specific conformations for specific membrane structures and how EPR is a versatile tool well-suited to analyze subtle alterations in structures under such modifying conditions which otherwise would have been difficult using other biophysical tools.
Graphical abstract
A. Introduction
Protein-membrane interactions are pivotal to many cellular processes [1–4]. Understanding the molecular mechanisms of these interactions is, therefore, of great importance. In the recent past, many proteins have been identified that can remodel membranes by their interplay with various cellular membranes [4, 5]. These include protein families like BIN/Amphiphysin/Rvs (BAR) [6–8], Eps 15-homology [9, 10], epsin [11], synaptotagmins [12] and synucleins [13–15].
Membrane remodeling proteins are involved in a plethora of cellular events within the cell like endocytosis, exocytosis, shaping of organelles such as endoplasmic reticulum, Golgi and mitochondria, intracellular vesicular trafficking, formation of filopedia and multivesicular bodies, mitochondrial fusion and fission, and synaptic vesicle fusion and recycling in neurotransmission. In these highly dynamic processes, interplay between proteins and lipids give rise to highly curved membrane structures which are not thermodynamically favored, unless stabilized by the curvature stabilizing proteins and lipids.
Processes requiring generation of membrane curvature are crucial to the survival and normal functional activity of the cell. For example, endocytosis is an energy-requiring process that is constantly used by cells to selectively move materials to the inside of the cell by forming an invagination in plasma membrane and then pinching-off into the cell internal. It is used for movement of solutes, microorganisms, cell debris, signaling molecules and fluid. The mechanism(s) underlying endocytosis is not completely understood due to the intrinsic complexity of the process requiring involvement of multiple protein and lipid players. It is a multistep process involving membrane bending, invagination, enlargement of the invaginated area, constriction of the plasma membrane, fusion of the membrane leaflets followed by excision or separation of the newly formed vesicle from the plasma membrane [4]. Different proteins are involved in the different steps and dysfunction of any one of these steps can lead to failure of the endocytic process. In addition, aberrant control of membrane curvature can compromise the activity of membrane remodeling proteins in various other membrane remodeling processes, leading to a diseased state. For example, some point mutations in the BAR (Bin1/amphiphysin/RVS167) domain-containing amphiphysin inhibit its ability to tubulate membranes in vitro and cause centronuclear myopathy. This disease causes muscle weakness and skeletal muscle atrophy and is characterized by a pronounced disruption of the membranous T tubule network. [16]. Mutation in dynamin-2 (DNM2), another membrane remodeling protein, also causes centronuclear myopathy further illustrating the importance of membrane curvature generation in the pathogenesis of this disease [17].
Various mechanisms have been proposed to explain how proteins can bend membranes (Fig. 1) [18–20]. One of these mechanisms is scaffolding where proteins directly impart their own intrinsic curvature onto the membrane. For example, amphiphysin contains a ‘banana-shaped’ BAR domain that directly molds the membrane into a curved shape. The specific alignment of multiple such BAR domains can generate a unidirectional membrane curvature that stabilizes membrane tubules [21, 22]. Another mechanism is wedging, where the asymmetric insertion of protein regions, often amphipathic helices, pushes the headgroups apart in a manner akin to the generation of membrane curvature by lipids with spontaneous positive curvature. Many proteins, including epsin, α-synuclein, IAPP and some BAR-domain containing proteins, called N-BAR proteins have such helical wedges. Non-helical wedges can also be found and are present in some BAR proteins such as pacsin [23]. In addition to pushing headgroups apart and affecting the spontaneous curvature of the membrane, wedges also add mass to the leaflet they insert into. This asymmetric addition of material to one leaflet could further promote membrane curvature by the bilayer couple mechanism. This mechanism arises from the expansion of the total area of one membrane leaflet compared to the other [4, 5, 18, 19, 24]. Using these and/or quite possibly other mechanisms such as molecular crowding of protein on membrane, partitioning of shaped transmembrane domains, and cytoskeleton-mediated ‘pulling and pushing’ of membrane, a variety of membrane morphologies like small vesicles, tubular structures, membrane protrusions and lipoprotein particles are generated and have been observed in vivo [4, 5, 18, 19, 24, 25]. Interestingly, a single protein, for example α-synuclein, can also generate different lipid morphologies in vitro, even by just modulating the protein-to-lipid ratio (Fig. 2) [13, 14]. Understanding the structure of a protein in membrane-bound form is difficult using techniques like X-ray crystallography and NMR spectroscopy and it becomes almost impossible to study a dynamic process like membrane remodeling. The complexity is further increased as the structure of a protein on different membrane morphologies is investigated considering that it is difficult to mimic the different membrane morphologies in crystals or NMR experiments.
FIGURE 1.
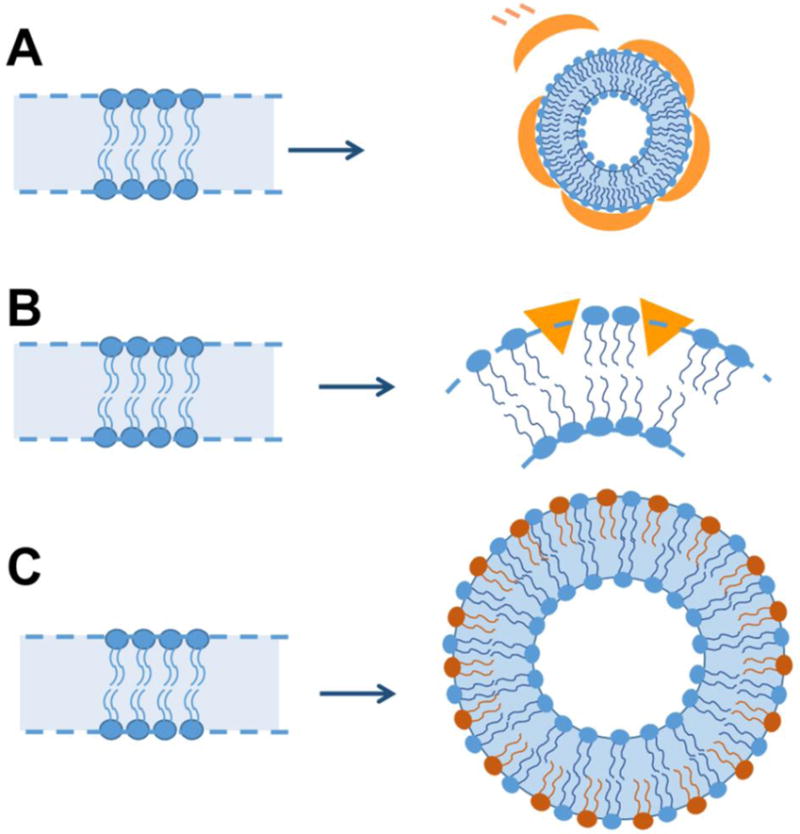
Mechanisms of membrane bending. A) Scaffolding mechanism - Membranes are bent by the scaffolding of intrinsically curved protein domains (Orange curved structures) on the membrane surface; B) Wedging mechanism - Membrane bending occurs by insertion of protein (Orange triangles) on the membrane surface that acts like a wedge pushing apart the phospholipid headgroups; and C) Bilayer couple mechanism – By asymmetric addition of material on one side of the bilayer, for example, small unilamellar vesicles are able to able to retain high curvature due to presence of ~twice the amount of lipid molecules in outer monolayer as compared to inner monolayer.
FIGURE 2.
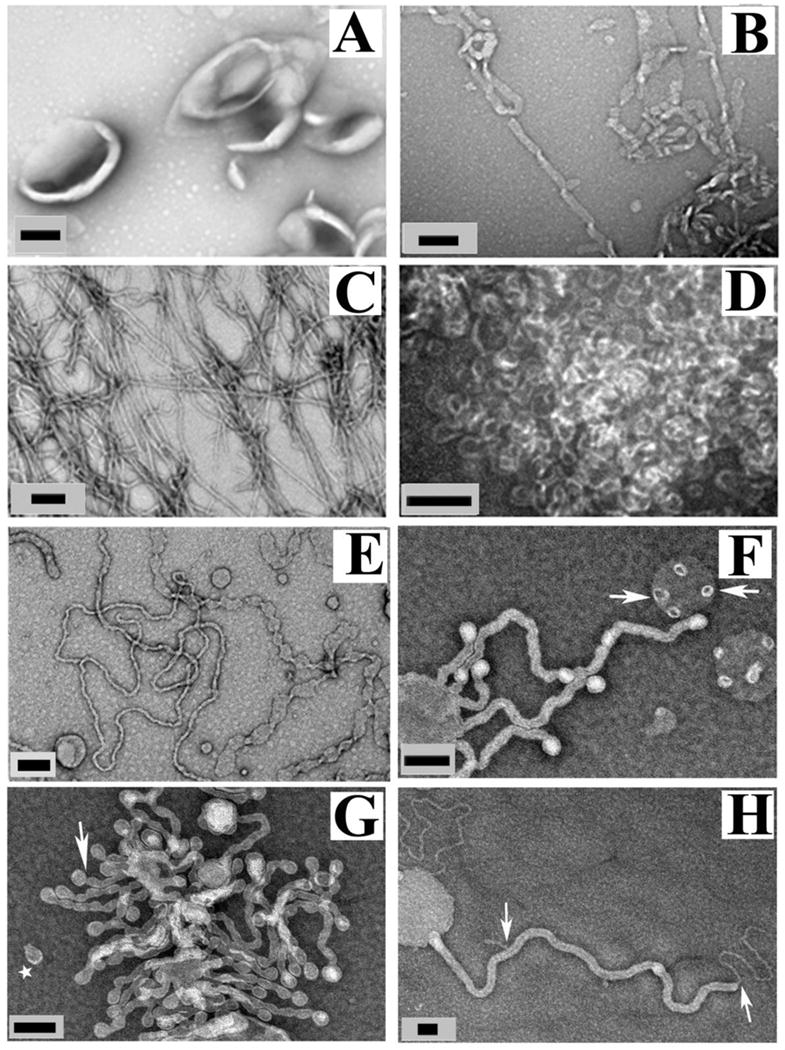
α-Synuclein induces different membrane morphologies based on lipid composition and amount of protein. A, negative stain electron micrograph of POPG vesicles without α-synuclein. B–D, α-synuclein with POPG lipid vesicles at varying protein-to-lipid (P/L) ratio of 1:40 (B), 1:20 (C), and 1:10 (D). E–G, Membrane composition affects the morphology of lipid vesicles in presence of α-synuclein. POPG/POPC (1:1) vesicles incubated with α-synuclein at 1:20 P/L. Arrows indicate “budding vesicles,” and star indicates “budded vesicle”. H, POPG/POPC (1:4) vesicles incubated with α-synuclein at 1:10 P/L molar ratio. Arrows show a smaller tube coming out from a larger tube. Black scale bar is 100 nm. Figure adapted and modified from (14).
Site directed spin labeling along with EPR has proven to be a useful tool for investigating protein structure regardless of membrane morphology. EPR is not limited by the size or shape of the lipid membranes to which proteins bind. The requirement of micro-molar concentration of proteins in microliter volumes - further eases the experimental measurements.
Site directed spin labeling of proteins followed by EPR studies have made significant advancements towards understanding of protein structure – both in solution as well as in the membrane bound form [26–29]. These approaches including measurements of mobility, polarity, membrane depth and distance distribution of the spin labels can be applied for exploring membrane remodeling by proteins and the application of EPR methods to membrane remodeling is described below.
B. Side directed spin labeling (SDSL)-EPR approach
SDSL is based on the incorporation of a spin labeled amino acid into a protein. For spin labeling of proteins, one or more cysteine residues are typically introduced into a protein and subsequently labeled using thiol reactive spin labels. The most commonly used and best characterized spin label for investigating protein structure is [1-oxy-2,2,5,5-tetramethylpyrroline-3-methyl]-methane-thiosulfonate (MTSL), typically referred to as R1.
EPR of spin labeled proteins can be used to obtain mobility, accessibility and distance information [29–32], all of which provides detailed structural information. The line shape of R1 obtained in continuous wave (CW) EPR is related to the local structure and dynamics at the labeled site. Sharp and narrow lines reflect a highly mobile R1 whereas a broad line (in the absence of spin-spin coupling, see below) is due to low mobility. The inverse of the central line width (ΔH0−1) or scaled mobility (Ms) are convenient and quantitative measures of line shapes [33]. Line shape analysis using these measures makes it possible to determine whether a labeling site is buried, on the surface of an ordered structure or in a loop/unfolded region [34]. Often times, protein regions binding to the membrane undergo a structural change from a disordered to an ordered structure and such changes can readily be resolved by line shape analysis [26, 30, 31, 33].
Accessibility measurements are very useful for membrane proteins in order to determine how deeply a given amino acid is buried in the membrane [31]. The immersion depth is measured by monitoring accessibility of R1 to polar (O2) and hydrophobic paramagnetic colliders (NiEDDA), which affect the relaxation properties of R1 in a concentration-dependent manner. These accessibility-based immersion depth measurements are based upon the differential partitioning of the colliders into the membrane; while O2 preferentially partitions into the membrane, NiEDDA is preferentially excluded from the membrane. The log of the ratio of the respective accessibilities was shown to be proportional to membrane immersion depth [31] and has been used in a large number of studies, many of which are also detailed below.
When two spin labels are introduced in a protein within 20 Å, or when two labels on different protein subunits come into proximity, line broadening due to spin-spin interaction can occur. Typically, these interactions are caused by dipolar interactions and cause a characteristic broadening that can be used to determine inter-spin distances [33]. Such distances provide important structural restraints that can be used for generating structural models. While dipolar interactions are generally dominant, an additional exchange interaction can also sometimes be observed. This interaction requires significantly strong orbital overlap between the free electrons on different spin labels and is, thus, limited to spin labels with short covalent connection or labels near physical contact. When multiple such labels are in contact with each other, as often seen in amyloid fibrils, single line EPR spectra can result [35–37]
C. Amyloidogenic proteins and membrane remodeling
Numerous studies have focused on the membrane binding of amyloidogenic proteins and correlated this with the ensuing cellular membrane disruption commonly observed in protein misfolding diseases such as Parkinson’s disease (misfolded α-synuclein), Alzheimer’s disease (misfolded amyloidβ and tau) and Type 2 diabetes (misfolded IAPP) [38]. The mechanisms of membrane damage however have remained contentious [38]. Different mechanisms have been proposed to explain the membrane disruption by α-synuclein and other amyloid proteins including pore formation, carpet mechanism, and lipid incorporation into aggregated (oligomers/fibrils) proteins [38–45]. Pore formation is further differentiated into barrel-stave models and toroidal pore models depending upon the arrangement of proteins and lipids within the pores on the membrane. Recent work performed by us and others on misfolded α-synuclein and IAPP revealed that many of the previously reported disruptions of membrane integrity were actually caused by uncontrolled membrane curvature induction, which is likely to be a mechanism leading to disruption of cellular membranes [13–15, 41].
1) α-Synuclein
Overview of α-synuclein membrane interaction
α-Synuclein is a presynaptic protein implicated in the pathogenesis of Parkinson’s disease [46, 47]. Membrane interaction of this protein is believed to be important for its physiological and pathological roles [39, 48, 49]. In physiology, α-synuclein seems to be involved in neurotransmitter release by affecting synaptic vesicle fusion and recycling, promote endocytosis, affect vesicular trafficking, and play a role in lipid/fatty acid transport [39, 49]. With respect to pathology, there is evidence for α-synuclein induced Golgi fragmentation, damage to lysosomes, and disruption of mitochondrial, endoplasmic reticulum and nuclear membranes [14, 49]. Due to the significance of membrane binding in the structure and function of α-synuclein, a large number of studies have focused on delineating the structure of membrane-bound α-synuclein (Fig. 3) [13, 50–61]. The solution structure of SDS micelle-bound α-synuclein solved using NMR showed a bent helical structure with two antiparallel helices (3–37 and 45–92) [52]. EPR experiments further confirmed the presence of a two-helical structure on lysophospholipid and SLAS (sodium lauroyl sarcosinate) micelles [51, 56]. To understand the structure of α-synuclein on SLAS, pulsed EPR and NMR spectroscopy were combined by using 18 EPR-derived interelectron spin label distance distributions with NMR-based secondary structure definitions and bond vector restraints [51]. However, as described in more detail below, using continuous wave EPR and four-pulse DEER measurements in combination with computational modelling it was shown that α-synuclein forms an extended amphipathic helix on intact phospholipid vesicles [53, 54]. Another study, using pulsed EPR measurements, carried out on bicelles, vesicles and rod-like micelles reinforced the presence of an extended helix [57]. Still, under some conditions EPR studies also detected the presence of a broken helix on lipid membranes [58, 59, 62]. The co-existence of broken or extended helix conformation is posited due to a small energy difference between the two conformations [50, 54, 60, 61]. Recent reports showed that α-synuclein could also exist as a tetramer in the native conformation, adding to the list of already debated conformations [63, 64]. This led to some important questions like – Which one of these is the physiological membrane bound structure of α-synuclein? Can α-synuclein exist in different membrane bound conformations? How could α-synuclein in a defined lipid environment exist in multiple conformations? As described below, a key feature controlling the α-synuclein structure is the state and morphology of the lipids or detergents. The following sections outline the structural information for the different α-synuclein conformations with particular emphasis on the role EPR played in these endeavors.
FIGURE 3.
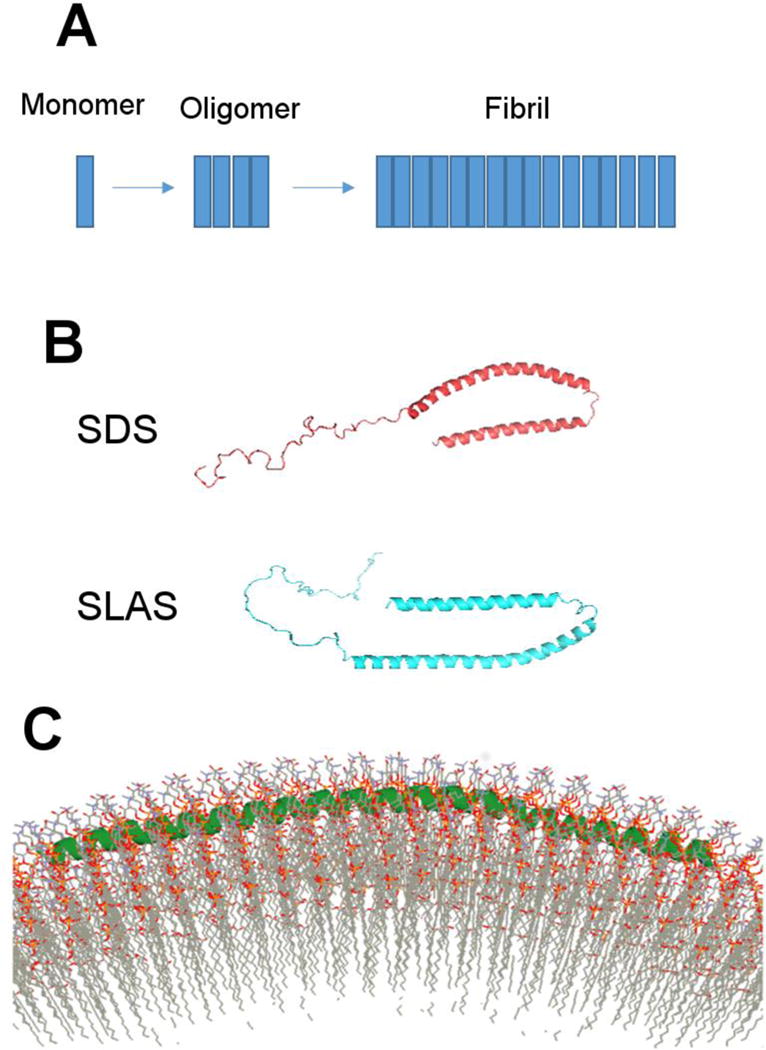
Multiple conformations of α-synuclein. A) Monomeric α-synuclein is mostly unstructured prior to aggregation. The largely unstructured protein attains β-sheet conformation in the oligomeric and fibrillar form; B) α-synuclein has bent-helical conformation on detergent micelles (~5 nm diameter). The structure varies slightly depending on whether α-synuclein is bound to SDS or SLAS micelle; and C) Extended helical form of α-synuclein from EPR experiments and simulated annealing molecular dynamics when bound to small unilamellar vesicles that is ~30 nm diameter. Panel C adapted from (54).
Structure of vesicle-bound α-synuclein from EPR spectroscopy
The ~14 nm long amphipathic helix formed by α-synuclein is one of the longest continuous helices on membranes, located at the lipid headgroup region per EPR studies [53, 54]. The evidence for existence of an extended helix was obtained by a combination of site-directed spin labeling in conjunction with EPR and simulated annealing molecular dynamics on small unilamellar vesicles composed of 30% 1-palmitoyl-2-oleoyl-sn-glycero-3-(phospho-L-serine)/70% 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine (wt/wt) [54]. This study used continuous wave (CW) and pulsed EPR data from a large number of singly and doubly spin-labeled α-synuclein encompassing the residues 9–89 responsible for membrane interaction of α-synuclein. CW EPR provided information for local secondary structure from mobility and accessibility information. The spectral changes in the absence and presence of membranes revealed which regions experience a change in mobility. The sharp and narrowly spaced lines for singly labeled α-synuclein derivatives in solution were characteristic of loop and unfolded regions. In the N-terminal region of the protein, these lines became broadened upon membrane binding with line shapes typically seen on the surface of ordered structures. The C-terminal residues did not exhibit any significant changes in spin label mobility implying that they did not partake in the membrane interaction of α-synuclein. Although there was some ordering in the N-terminal region, there was a negligible amount of strong immobilization as evidenced by the absence of outer peaks in the CW spectra of membrane bound α-synuclein [53, 54]. This indicated the absence of significant tertiary or quaternary interactions of α-synuclein on membranes contrasting with tightly-packed membrane proteins or deeply buried residues within globular proteins. The secondary structure and topology of membrane-bound α-synuclein was then further refined using O2 (∏O2) and NiEDDA (∏NiEDDA) accessibilities [31, 65–68]. The paramagnetic collider O2 readily partitions into the more hydrophobic lipid phase and hence has a larger effect on spin labels buried inside the lipid membrane as opposed to the surface exposed spin labels. In contrast, NiEDDA more efficiently collides with membrane surface exposed spin labels. The depth parameter (ϕ) obtained as ln (∏O2/∏NiEDDA) can then, after calibration, be used to quantify the immersion depth of the labeled site. For detailed methodology, see [26–28, 30, 31]. The depth parameter revealed a period oscillation consistent with the formation of an extended helical structure exposed to the membrane on one face and exposed to the solvent on the other [54]. To further refine the local structure, long distances were mapped using four-pulse DEER (Double electron-electron resonance) measurements that provided the information for three-dimensional arrangement of amino acid residues within the membrane bound protein [54]. Four-pulse DEER measurements can be used to obtain distances up to ~80 Å for ideal systems but for protein samples the range is often limited to ~50 to 60 Å [29, 56, 57, 69–75]. The distances can also be used to predict the presence of a specific type of secondary structural element. For example, for a continuous α-helix between labels at position 11R1 and 26R1, the distance will be approximately 15 × 1.5 Å (helix rise per amino acid) = 22.5 Å. This could, thus, be used for rapid testing of presence or absence of α-helical stretch. It should be mentioned that the R1 labels used for these types of distance measurements in amphipathic helices should face the same direction in order to minimize effects from side chain orientation. Either, both the labels should be facing the solvent environment or into the membrane. Eventually, the immersion depth data and long range distances from pulsed EPR were used as experimental constraints to refine the structure of α-synuclein using simulated annealing molecular dynamics [54]. The resulting structural model of α-synuclein uncovered an extended and slightly curved α-helix with a periodicity of 3.67 aa per turn and having a superhelical twist similar to right-handed coiled coil proteins [54]. This α-helix follows the curvature of the vesicle surface and the helix center lies just below the phosphate headgroup region of the phospholipid membrane. The positively charged, conserved lysine residues project perpendicularly to the membrane surface conveniently interacting with the negatively charged phosphate groups.
As described above, the collision gradient approach using accessibility of spin label to NiEDDA and O2 is well suited to determine the immersion depth and topology of membrane associated proteins. This method, however, has reduced and in some cases barely detectable resolution for the protein segments lying 5 Å or more above the phosphate headgroup due to the absence of concentration gradient of the paramagnetic colliders in this region. Overhauser dynamic nuclear polarization (ODNP)-enhanced NMR relaxometry that probes the local translational diffusion of hydration water can be used to measure gradient of water diffusion up to 20–30 Å away from the lipid phosphate on membrane. This technique is based on calculating the efficiency of dipolar relaxation between the electron spin of a nitroxide radical present on proteins and the nuclear spin of surrounding water protons. The combinatorial approach using EPR with 1H Overhauser dynamic nuclear polarization (ODNP)-enhanced NMR relaxometry enabled to measure membrane immersion depth along with the orientation of amino acid residues 20–30 Å away from the membrane surface [76]. The measurements revealed the formation of an extended α-synuclein helix on the membrane and the presence of a larger loop than an idealized α-helix for residues 90–96 of bound αS, that forms the transition segment linking the N- terminal α-helix and random C-terminus [76].
Membrane remodeling by α-synuclein
The unique length and wedge-like position of the α-synuclein helix suggested a possible role of α-synuclein in membrane remodeling. To answer the abovementioned questions and test the ability of α-synuclein to remodel membranes, we used a combinatorial approach of electron microscopy and EPR. These studies showed that α-synuclein can induce membrane curvature and remodel lipid membranes into different morphologies such as bilayer tubes [13, 14], micellar tubes [13], small vesicles [14] and lipoprotein nanoparticles [55, 77] (Fig. 2). α-Synuclein remodeled lipid vesicles with a wide variety of phospholipids lipids including 1-palmitoyl-2-oleoyl-sn-glycero-3-[phospho-RAC-(1-glycerol)] (POPG), 1-palmitoyl-2-oleoyl-sn-glycero-3-phospho-L-serine (POPS) and 1,2-dioleoyl-sn-glycero-3-phospho-L-serine (DOPS). The membrane remodeling ability of α-synuclein has since been confirmed by several other groups [15, 78–82]. α-Synuclein is present at a high concentration (~ 40 μM) in neuronal cells and any alteration in the concentration of α-synuclein can alter the membrane remodeling ability with physiological consequences. The various lipoprotein morphologies formed by the interaction of α-synuclein with the lipid membranes depended highly on the protein-to-lipid molar ratios (Fig. 2) [14]. Importantly, the presence of negatively charged phospholipids strongly promoted membrane remodeling. Because of the difficulty associated with understanding the structure of a protein bound to a specific curved membrane, we used EPR. To evaluate the conformation of α-synuclein on these various lipid assemblies, we developed a purification protocol to separate tubes, lipoprotein nanoparticles and unbound protein [13, 55]. The respective structure of α-synuclein on membrane tubes (formed from multilamellar POPG vesicles) and lipoprotein nanoparticles (generated from small unilamellar vesicles of POPG) was studied using both CW EPR and pulsed EPR. To achieve this objective, spin-labeled α-synuclein was incubated with lipid membranes at different protein-to-lipid molar ratios and then ultracentrifugation was carried out to separate the membrane bound α-synuclein from unbound protein.
i) α-synuclein structure on tubes
The conformational change in α-synuclein associated with tubulation was compared to previously obtained data on lipid vesicle-bound protein [13]. A total of eight spin labeled sites (31R1, 37R1, 41R1, 44R1, 48R1, 52R1, 70R1 and 76R1) within the membrane binding region of α-synuclein (~1-95 amino acid residues) were investigated (Fig. 4). The corresponding X-band EPR spectra showed a transition from a highly dynamic structure in solution to an ordered structure on the tubulated membrane with spectra similar to those previously observed for vesicle-bound α-synuclein [53, 54].
FIGURE 4.
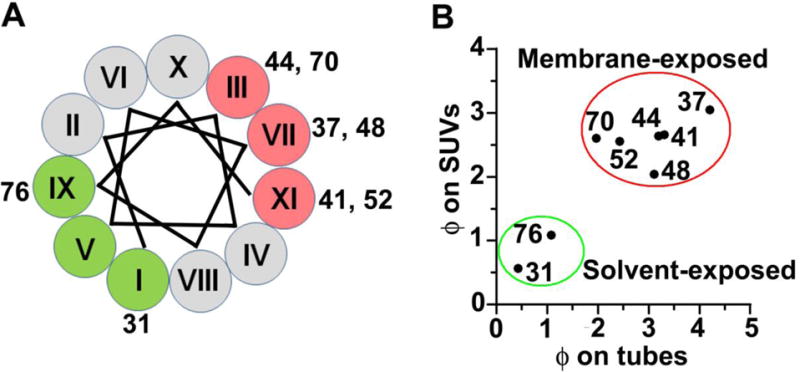
Identification of secondary structure and topology of α-synuclein using spin-label accessibilities to O2 and NiEDDA by continuous wave-EPR analysis. A, helical wheel depicting the solvent-exposed residues (green) and lipid-exposed (red) residues on membrane tubes. The roman numerals mark the positions of amino acids in the 11-amino acid repeat of α-synuclein. B, A similar distribution of Ф on small unilamellar vesicles (SUVs) to Ф on tubes means residues are at similar membrane depth on both SUVs and membrane tubes. Figure adapted from (13).
The membrane bound protein was then subjected to accessibility measurements toparamagnetic colliders and the data were compared to those obtained for vesicle-bound α-synuclein [13, 54]. In the vesicle-bound state, residues 37R1, 41R1, 44R1, 48R1, 52R1 and 70R1 were observed to face into the outer leaflet of lipid vesicles while residues 31R1 and 76R1 faced towards the solvent [53, 54]. A similar behavior was also observed for tube-bound protein. Based on the ϕ-values the residues can be grouped into two sets (green and red circles in Fig. 4B). Positions 31 and 76 displayed in green circles with reduced ϕ-values correspond to the solvent- exposed surface and are in positions IX and I on the helical wheel (Fig. 4B, colored in green). Positions 37, 41, 44, 48, 52, and 70 displayed in red circles with larger ϕ-values (Fig. 4B, red circle) correspond to the membrane-inserted residues and are in positions III, VII and IX on the helical wheel. Residues 41 and 44 displayed membrane insertion further suggesting an extended α-helical conformation on tubes [53, 54].
Measuring intramolecular distances between two amino acid residues provide valuable information regarding the conformational state of a protein. Distance measurements were carried out between key residues on α-synuclein to distinguish which conformation – broken helix or extended helix - is populated on membrane tubes [13] (Fig. 5). Three doubly spin labeled α-synuclein proteins were used for this purpose, 11R1/26R1, 22R1/52R1 and 63R1/81R1, encompassing the membrane binding region of α-synuclein [13]. The distances obtained on membranes tubes are almost same to that obtained on lipid vesicles, which indicated an extended helical conformation on membranes tubes. Specifically, the distance 50 Å for 22R1/50R1 pair is different than 23.3 Å observed with SDS micelles [13, 52]. An antiparallel bent helix that brings the residues 22R1 and 52 R1 closer to each other would provide a shorter distance unlike an extended helix. Thus, distances obtained through four-pulse DEER measurement can provide rapid information for the possibility of existence or non-existence of a α-helix unlike nitroxide accessibility scan using NiEDDA and O2 that requires more laborious exercise.
FIGURE 5.
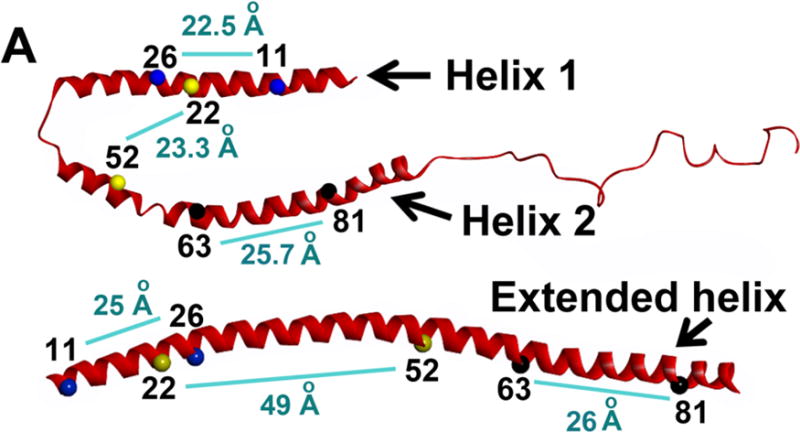
Four-pulse DEER experiment to distinguish bent-helical and extended conformation. The distances between residues connected by light blue line are displayed for bent-helix (SDS-bound structure of α-synuclein from NMR experiment, top) and extended helix (SUV-bound structure of α-synuclein from 4-pulse DEER experiment, bottom). These two α-synuclein conformation can be distinguished by measuring distances between spin labels at the indicated positions using 4-pulse DEER experiments. Figure adapted from (55).
ii) α-synuclein structure on lipid nanoparticles
Using the aforementioned purification protocol, it became possible to also investigate the structure of α-synuclein associated with lipid nanoparticles. Using four-pulse DEER experiment, we measured distances between spin labels in the same R1 pairs previously tested on membrane tubes (11R1/26R1, 22R1/52R1 and 63R1/81R1) [55]. The distance distribution for 11R1/26R1 was similar to that on membrane tubes suggesting a helical conformation in this stretch. But 22R1/52R1 displayed a shorter distance, 27 Å, like that obtained on SLAS micelles implying a break in the extended helical structure as opposed to the membrane tubes. 63R1/81R1 pair gave a distance matching that on tubes but with a broader distribution, suggesting that there could be multiple conformations of α-synuclein on lipoprotein nanoparticles. In the same study, it was discovered that nanoparticles generated by α-synuclein contain helical oligomers including trimers and tetramers. These nanoparticles closely resemble the trimeric and tetrameric α-synuclein suggested to be the native form present in cells [63, 64]. Our study raises the possibility that these trimers and tetramers of α-synuclein are possibly formed by interaction with lipids or fatty acids. α-Synuclein has strong affinity to lipids and during our study it was found that as α-synuclein is passed through a chromatographic column previously exposed to lipids, it transforms from unstructured to helical conformation. It is, therefore, of utmost importance to ensure that the experiments with α-synuclein are carefully performed to avoid misinterpretations.
Summary of different α-synuclein structures and implications
Altogether, the combined CW EPR and pulsed EPR data showed that an extended helix was favored on lipid vesicles and tubes (micellar tubes and bilayer tubes) while a broken helix was favored on detergent micelles such as SDS and SLAS as well as on lipoprotein nanoparticles generated after membrane remodeling by α-synuclein. Overall, there seemed to be a clear indication that the energetic differences between the structures are relatively small. Also certain geometric conditions facilitate the formation of an extended structure. The extended helical structures can easily be accommodated on long tubes or on vesicles. In contrast, an extended helical structure (~14 nm in length) does not easily fit onto relatively small detergent micelles (~ 4 nm diameter SDS micelles) or nanoparticles (~10 nm diameter) In contrast, a ‘kink’ near the middle of the helix reduces the length and allows for a better fit [54].
Another important lesson learned from the EPR studies was that structural studies performed on membrane curvature inducing proteins like α-synuclein need to be carefully carried out. In particular, the resulting membrane morphologies need to be verified under the conditions of the experiments, as the conformation of protein can vary with conditions depending on how membrane morphology is altered.
After the discovery of the membrane curvature inducing potential of α-synuclein, in an in vivo study, overexpression of α-synuclein induced remodeling of membranous organelle system into highly curved structures at presynaptic termini in transgenic mice [83]. Curvature induction by α-synuclein -may play an important role in development of Parkinson’s disease demanding further future investigation. The studies discussed above are an example of how EPR measurements helped to provide new testable hypotheses for biological research.
2. Islet Amyloid Polypeptide-IAPP
Overview of IAPP membrane interaction
IAPP is an amyloidogenic peptide that is co-secreted with insulin and that has been linked to the development of type II diabetes mellitus [84, 85]. The 37 residue IAPP forms the main constituent of pancreatic amyloid that is characteristic of type II diabetes. IAPP amyloid is formed by misfolding of monomeric IAPP into oligomeric structures and then conversion of these oligomers into fibrils. Both oligomers and fibrils are suggested to be mediators of cellular toxicity [86, 87]. As in the case of α-synuclein, membrane damage has been considered one of the potential pathogenic features of IAPP [40]. Membranes have been shown to mediate the aggregation of IAPP and in turn IAPP can cause disruption of cellular membranes, a process that could cause toxicity [40, 41, 86, 88–90]. Different mechanisms of membrane damage have been proposed for IAPP [38]. As mentioned above for α-synuclein, many of these mechanisms, like pore formation and carpet mechanism, are commonly described for amyloidogenic proteins and antimicrobial peptides. An additional mechanisms describes the extraction of membrane lipids during the growth of fibrillar IAPP and thereby destabilizing the membrane [43]. However, IAPP analogues or rat IAPP that does not readily form fibrils are also able to disrupt membranes suggesting that multiple modes of membrane disruption might be present [91]. The existence of IAPP in different structural forms like monomer, oligomer and fibril complicates the investigation for understanding the mechanism of cellular toxicity. In solution, as a monomer it exhibits a random coil structure with some residual helicity that is transformed to a largely helical form after interacting with membranes [40, 89, 90]. The helical form can convert into largely β-sheet form that is observed in oligomeric and fibrillar IAPP. Unstructured monomeric IAPP can also directly transform into a β-sheet oligomer or fibril without the helical IAPP intermediate [92–95].
Understanding the details of different structural forms of IAPP is key to resolving the mechanism of cellular toxicity. A methodology combining site directed spin labeling with CW and pulsed EPR (analogous to that described for membrane-bound α-synuclein above) was used to determine the structure of fibrilar IAPP [94]. Initially, singly labeled IAPP peptides in fibrillar form were monitored by CW EPR. The change in inverse line width (ΔHo−1), a measure of spin label mobility, was monitored as the monomeric IAPP assembled into fibrillar form. Residues from 14–19 and 31–36 displayed ΔHo−1 that alternates with a periodicity of two. This periodicity is generally observed for β-sheet region where one face is buried more than the other. The data is in good agreement with a previous solid state NMR model where the abovementioned region forms β-sheet in IAPP fibrils [92]. Next, four-pulse DEER measurements were carried out with doubly labeled IAPP peptides to generate the three-dimensional arrangement of these two β-sheet regions with respect to each other and the intermolecular arrangement of different IAPP molecules in the fibril. The EPR data was then used as constraint to develop IAPP fibril model by simulated annealing molecular dynamics [94]. The resulting fibril model displayed a left-handed helical structure with an overall twisted morphology that was observed by electron microscopy [94].
One of the main challenges for understanding the structure of membrane-bound IAPP was to identify the conditions that stabilize its membrane bound form as the high aggregation propensity of IAPP often causes rapid misfolding of membrane bound IAPP. It was observed that under weakly stable α-helical conditions, membranes accelerate the aggregation and β-sheet formation of IAPP [90, 96]. Membranes likely enhance misfolding by the reduced dimensionality of diffusion and the increased local peptide concentration on the membrane surface [90, 96]. This acceleration towards aggregation was inhibited by targeting the helical form of IAPP using small molecules that bind to the residues in IAPP within the region with high helical propensity [97]. The resultant was decreased cellular toxicity of IAPP further underlining the importance of investigating the membrane interaction of IAPP. Low ionic strength and high negative charge of lipid membranes were found to kinetically trap the membrane bound form for hours [89, 90]. The structural evaluation of IAPP was, thus, realized by stabilizing the α-helical conformation on membrane.
Structure of vesicle-bound IAPP from EPR spectroscopy
To understand the mechanism of membrane disruption and to understand the mechanism of membrane-mediated misfolding, it is pertinent to identify the conformation of IAPP on the membranes. The initial membrane-bound structure of IAPP was investigated by CW EPR studies [89]. IAPP was bound to 100 nm unilamellar vesicles composed of 80% POPS and 20% 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine (POPC) (mol/mol). Based on accessibility and mobility analysis analogous to those described above, residues 9–20 were found to be in a helical conformation in the presence of membranes. This finding was in good agreement with prior circular dichroism spectra of IAPP obtained immediately after the addition lipid membranes indicating the presence of an α-helical conformation about ~15–19 amino acid in length [90]. In a later NMR study, the solution structure of IAPP on SDS micelles revealed a ‘kinked-helix’ formed by residues 7–17 and 21–28 [98]. While there is significant overlap between the helical regions, the differences again suggest that protein structures can be strongly influenced on the specific lipid or detergent context.
Membrane remodeling by IAPP
The presence of a helix at the interfacial region of membrane called for an investigation into whether IAPP can also induce membrane curvature through ‘helix-wedging’ mechanism like as described above for α-synuclein. To test this notion, IAPP was incubated with negatively charged lipid membranes and using various assays it was shown that IAPP is a highly efficient inducer of membrane curvature and it could be one of the mechanisms that IAPP utilizes to inflict membrane disruption [41]. Giant unilamellar vesicles with different compositions of POPS, POPC, POPG or DOPG lipid vesicles were used in this study. The first evidence of membrane curvature induction by IAPP came through phospholipid clearance assay and fluorescence microscopy data wherein large, negatively charged phospholipid vesicles were transformed into smaller structures [41]. Circular dichroism data revealed the membrane remodeling occurring through formation of helical conformation. The remodeling coincided with significant membrane disruption culminating in the formation of lipid tubules and smaller vesicles [41]. Another report also details the ability of IAPP to for vesicles and tubes from supported lipid membranes [99]. The ability to remodel membranes is very similar to that observed with α-synuclein. However, compared to α-synuclein, the amount of vesiculation was more pronounced than tubulation. One possibility for this difference could be the length of the helix. In case of α-synuclein, the length of the helix is ~14 nm while for IAPP it is ~2 nm. It is possible for a longer helix to stabilize long anisotropic curvature in tubular membrane structures as observed with α-synuclein. But a shorter helix, as in IAPP, may not be able provide stability to the anisotropic curvature in tubules. This instability could result in preferential formation of smaller vesicles with isotropic curvatures.
Another significant observation is that in presence of weakly negatively charged membranes, IAPP takes the role of membrane curvature sensor as opposed to an inducer of membrane curvature [41]. Whether it is the induction or sensing of membrane curvature, this could directly influence the function of IAPP in physiology as well as pathology. It could also impact the subcellular location of IAPP as demonstrated by preferential localization of IAPP on mitochondrial cristae that are highly curved membrane structures [41].
D. Non-amyloidogenic Proteins and membrane remodeling
Induction of membrane curvature is important in many cellular events such as endocytosis, exocytosis and cell division and in the maintenance of different membrane shapes, for example, as observed in Golgi complex and endoplasmic reticulum. Protein-membrane interactions play a major role in these events. Many proteins involved in these events have large size, contain multiple domains and form oligomers which makes structural and functional characterization an extremely formidable task. How EPR studies have supported investigation into the structural characterization and understanding of the mechanistic role of N-BAR proteins (amphiphysin and endophilin) and EHD2 (Eps15-homology domain-containing protein 2), a protein involved in induction of membrane curvature, is discussed below.
1) N-BAR Proteins (Endophilin and Amphiphysin)
One of the proteins which is involved in recycling of vesicles is endophilin that, like amphiphysin, forms part of the Bin/Amphiphysin/Rvs (BAR) superfamily. Endophilins are BAR domain containing proteins that function in clathrin-mediated endocytosis of synaptic vesicles [7, 22, 100, 101]. Mutation in this protein is associated with impaired synaptic vesicle recycling [102, 103].
Endophilin and amphiphysin possess an N-terminal helix followed by BAR domain linked to an SH3 domain by a variable linker region [104, 105]. The BAR domain forms a crescent-shaped dimer [100, 106, 107] which senses, induces and stabilizes membrane curvature. The SH3 domain is a protein-protein recognition module that also modulates the activity of endophilin. Evidence suggests a role of endophilin in synaptic endocytosis by interacting with synaptojanin and dynamin through its SH3 domain [108–110] and by inducing membrane curvature [100, 107, 111]. In vivo, endophilin localizes to the neck region of endocytotic vesicles at the synapses and in vitro, transforms large vesicles into small highly curved vesicles and lipid tubes [112–114].
While the crystal structures of N-NBAR proteins clearly show a bent structure (Fig. 6A) that is likely to act via scaffolding, there are other regions that are important for the control of membrane curvature that are typically not resolved in the crystal structures. For example, the N-terminal regions of N-BAR proteins are known to contain important regions for membrane interaction. Sequence analysis suggested that these regions might form amphipathic helices in the presence of membranes. Using EPR, we were able to directly demonstrate that the N-termini of endophilin and amphiphysin indeed insert into the membrane where they aid in curvature induction and stabilization [100]. Proteins bound to large unilamellar vesicles prepared from brain lipid extracts were studied. Using accessibility measurements, the depth parameter, Ф, was determined and the helices were shown to directly penetrate into the membrane where could act as wedges. As described below, the precise location of the helices, however, depended on whether the proteins were bound to tubes or vesicles suggesting that these regions act differently in stabilizing vesicles and tubules. There is an additional helical insert, called ‘helical 1 insert (H1I)’ in endophilin that is not present in amphiphysin. This insert was predicted to interact with membranes and this interaction was again directly demonstrated using EPR spectroscopy [101]. The same study also showed that the helices run perpendicular to the BAR domain, a position that is optimized for wedging [101]. This result was obtained from EPR distance and depth measurement coupled with simulated annealing-based structural refinement used for the structural analysis of membrane-bound α-synuclein [101].
FIGURE 6.
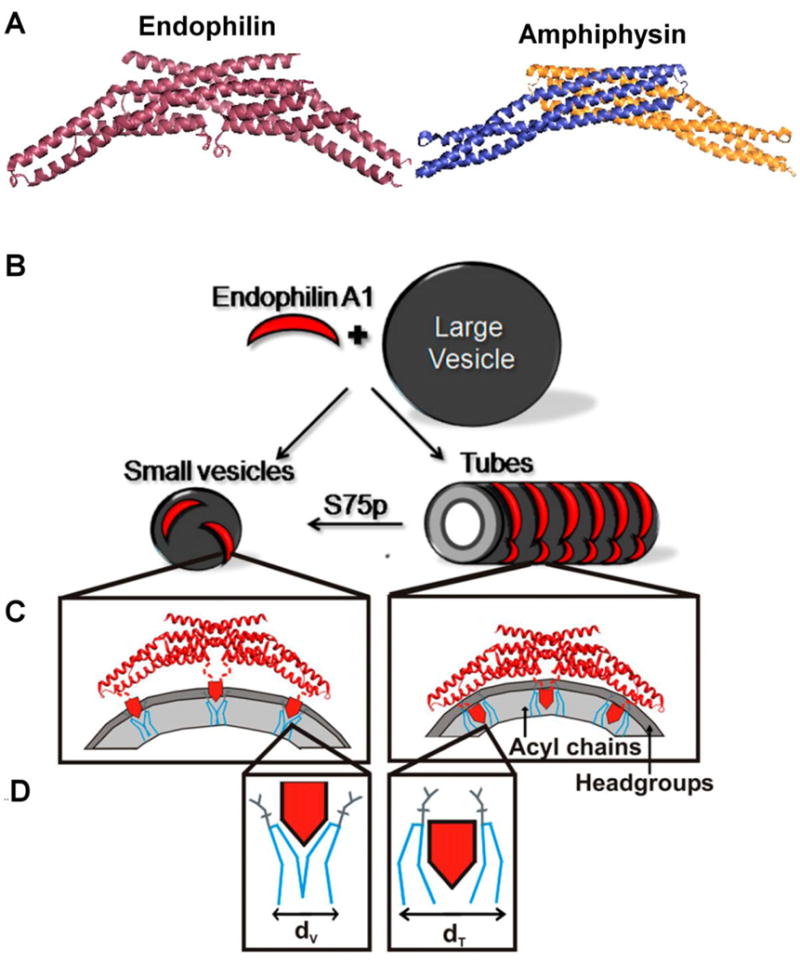
Endophilin A1 adopts different conformation on tubes and vesicles and the structure is modulated by phosphorylation. (A) Crystal structure of Endophilin A1 BAR domain (PDB ID-1ZWW) and Amphiphysin BAR domain (PDB ID-4ATM). The amphipathic helices were not resolved in the crystal structure. (B) Endophilin induces vesiculation or tubulation on binding to membranes. (C) Endophilin stabilizes curvature on highly-curved small vesicles (Left) using its amphipathic helices (red pentagons) by wedging into the headgroup region (dark gray) and forcing the adjacent lipid molecules to bend (D, blue lines). On tubes (B, Right) the amphipathic helices insert deeply into the acyl chains (C, Right), and relatively fills larger space for tubes (dt) than for vesicles (dv) in the acyl chain region (light gray). The deeper insertion of helices in tubes pull the BAR domain towards a closer proximity to the lipid headgroups (C, Right). Hence, a negative charge at S75 that disfavors deeper membrane insertion of helices, promotes vesiculation over tubulation. B–D panels adapted from (7).
A somewhat surprising result from the EPR studies was that the BAR domains of endophilin and amphiphysin seemed to be far away from membranes in the vesicle-bound states. However, there was strong evidence that the BAR domain plays an important role in membrane curvature induction [6, 100]. To resolve this seeming contradiction, it was necessary to further evaluate the structures of tube-bound endophilin [7] and amphiphysin [8] (Fig.6). Interestingly, some significant differences were observed for the tube-bound proteins compared to their vesicle-bound counterparts: (a) both the N-terminal helix (helix 0) and the central helical insert (H1I) were found to insert deeper into the tubes as compared to small vesicles; (b) the BAR domain of amphiphysin as well as endophilin made contacts with the membrane surface of tubes that were absent on vesicles. The presence of two different conformations on tubes and vesicles follows the theme of different structures seen for different membrane morphologies and it could mean important regulatory mechanisms can modify the conformation during different steps in endocytosis.
It is notable that a single mutation at position S75 can act like a conformational switch in endophilin [7]. The residue S75 is inserted into the membrane in tubes but is exposed to aqueous environment in vesicles. A physiologically significant S75D phosphomimetic mutation favors vesicle formation over tubulation, suggesting that post translational modifications could regulate what types of membrane morphologies can be generated.
2) EHD2 - Eps15-homology domain-containing protein 2
Introduction
EHD2 is a membrane remodeling protein involved in the regulation of caveolae formation by localizing to the neck of caveolae that are flask-shaped invaginations of plasma membrane and stabilizing the highly curved membrane surface [115]. EHD2 contains an N-terminal G domain with ATPase activity, a middle helical domain followed by C-terminal EH domain. According to X-ray structure analysis, EHD2 and dynamin have structurally related G domains [9]. Another shared property is the ability to tubulate vesicles with negatively charged phospholipids and oligomerization into ring-like structures around the membrane tubules [9, 116]. Despite the availability of X-ray crystal data, the question regarding the structural changes associated with membrane tubulation remained unanswered. EPR made two contributions toward understanding the EHD2 mechanism of action. First, accessibility measurements directly identified membrane contacts and second, the N-terminal membrane binding region of EHD2 was poorly resolved in the crystal structure, but could be located based upon distance measurements by EPR. The following sections describe both of these aspects.
Mapping of membrane-binding regions
Based on previous crystallography data and mutagenesis studies, it was suggested that residues within the tip region of the helical domain (320–340) could mediate membrane binding (Fig. 7). Using SDSL and EPR we tested whether this region was responsible for recruiting EHD2 to the membrane [10, 117]. First, the three native cysteines in EHD2 were mutated to serines. This cysteine-less mutant showed uncompromised ability to interact with membranes like the wild-type EHD2 protein. Second, six residues in the helical region were individually replaced with cysteines and spin labeled. The spin-labeled proteins were incubated with lipid vesicles prepared from bovine brain lipids (Folch). The spectra of spin label at Phe322 that is positioned at the tip of the helical region displayed ordering after binding to Folch lipid vesicles. Spin labels at positions 320, 321, 323, 324 and 328 also revealed ordering, thereby confirming the interaction of the amino acids at these positions with the membrane. Accessibilities of the spin labels to O2 and NiEDDA further supported the notion that this region of the helical domain inserts into the membrane [10]. A subsequent study [117] found that this membrane binding region also included additional amino acids in the tip region, as the helical domain can undergo a domain rotation that allows additional amino acids to come into contact with the membrane (Fig. 7).
FIGURE 7.
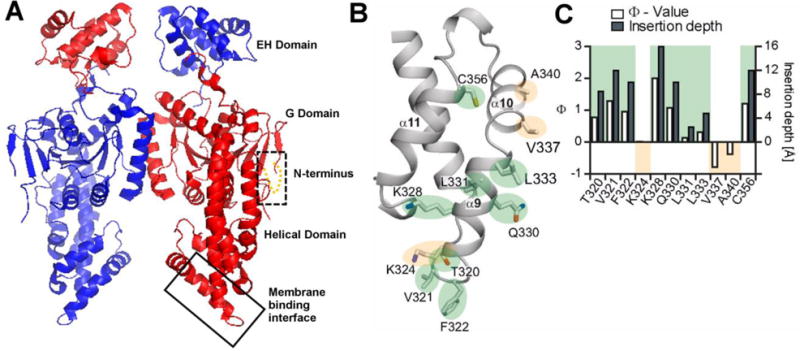
Crystal structure of EHD2 dimer (PDB ID:2QPT). A) EHD2 has a GTPase domain (G domain), followed by a helical domain and a Eps15-homology (EH) domain at the C-terminal end. The box with broken line shows the location of the N-terminus region that was originally unresolved in the crystal structure of EHD2 but was localized by EPR. The box with solid line indicates the region of the helical domain which was identified through EPR studies as the primary membrane binding site. B) Magnified primary membrane binding site (shown by solid box in A) with details of the membrane binding interface of EHD2. Membrane inserting residues are highlighted in green while the residues that do not insert into membrane are highlighted in yellow. C) Ф was calculated for EHD2 labeled positions 330, 331, 333, 337, 340, 356 in the presence of Folch-SUVs (open bars/left y-axis). Positive Φ values show membrane interaction. The membrane insertion depth of each residue was derived by calibrating with spin-labeled lipids (solid bars/right y axis). Panel B and C are from (117).
Trilateration method to locate N-terminal region only poorly resolved in X-ray structure
The previous X-ray data could not resolve the structural information for residues 1–18 although these residues were found to be highly conserved. Mobility analysis of spin labels at position 2–9, surprisingly revealed a predominantly ordered conformation even though the lack of assignment in the X-ray data had suggested a potentially flexible conformation [10]. EPR spectra also showed the presence of a small fraction of highly mobile component. The fraction of mobile component increased when hydrophobic residues Phe2, Trp4 and Leu5 were substituted with R1, indicating the importance of these amino acids in stabilizing the ordered conformation through the hydrophobic interaction.
To determine the location of the N-terminus, a trilateration method using DEER was used. EHD2 double mutant variants were generated with one spin label at position 5 and the other one at any one of the positions 28, 294, 303 and 313. The distances ranged from 27 to 48 Å pinning the position close to the G domain. This prediction was then confirmed by X-ray crystrallography of a Leu5Met mutant that was generated as a selenomethionine derivative.
Interestingly, this structure changed upon membrane interaction as the N-terminus switches out of its binding pocket in the G-domain and binds to the membrane. This conformational change was revealed by mobility and accessibility analysis of spin labeled EHD2 derivatives labeled individually at positions 2–9. All of the labels displayed a spectral change in the presence of lipid vesicles indicative of membrane interaction and depth measurements of labels at position 2–9 revealed membrane penetration at all sites except for positions 3 and 4. Altogether, the data indicated a shallow membrane insertion of the N-terminus. In contrast to the membrane binding region of the helical domain, this N-terminal binding region was not essential and is considered a secondary membrane binding site. For example, while the N-terminal region binds to the membrane, cryo-EM data showed that EHD2, devoid of the first 18 residues, can still efficiently induce membrane curvature. The N-terminal membrane binding region did however affect cellular localization. Overall, it is thought that the N terminus region constitutes a switch region controlling recruitment of EHD2 to specific membranes [10].
E. Conclusion
This review focused on different membrane remodeling proteins and how EPR can be a valuable tool to explore the structure and underlying mechanism for membrane curvature induction. A common theme in the aforementioned studies is the tremendous interplay between the precise structure of the lipids and the proteins. A large variety of membrane structures can be stabilized by the same protein under different conditions. Importantly, these different lipid structures are often associated, in turn, with different protein conformations. As described above, it is possible to bind α-synuclein to the same lipids under slightly different conditions and obtain either tubules or nanoparticles. This will result in very different conformations that either contain an extended or a bent helical structure. As described above, it appears that many of the reported differences in the α-synuclein literature have resulted from differences in the precise lipid state and membrane morphology. This makes it essential to verify the exact membrane structure when studying membrane remodeling proteins at the time of measurement. In our experience, the structural analysis of membrane remodeling proteins requires careful optimization with respect to finding the best conditions that favor tubulation, vesiculation or nanoparticle formation. Such optimization should also include kinetic stability studies to ensure that the lipid state remains unchanged during the measurements. A surprisingly large number of proteins and peptides have recently been reported to have membrane curvature effects. According to this notion, it might even be generally advisable to investigate whether a given protein can impact lipid structure, if one performs structural studies.
In relatively simple systems, like IAPP and α-synuclein, it was possible to obtain protein structure information from EPR data alone without the need for any prior structural information from NMR or crystallography. For more complex structures, like the EHD2 and BAR proteins discussed here, this would not have been feasible. However, the availability of high-resolution structural data from the soluble form of these proteins provided a structural framework to guide the EPR studies. Although analysis of the EHD2 and BAR protein crystal structures alone has already provided tremendous insights into potential mechanisms of action, the structural differences between the solution and membrane bound states of these proteins, as well as the lipid state dependence of the membrane bounds structure still make it essential to investigate what the exact membrane bound state looks like. The strength of EPR is that it can provide such information.
HIGHLIGHTS.
Membrane remodeling by proteins is vital for many cellular processes
Protein structure analysis on curved membranes problematic for most techniques
EPR-based structure analysis can be applied to all membrane and detergent systems
Amyloidogenic and non-amyloidogenic proteins induce variety of membrane curvatures
Different membrane structures often associated with different protein structures
Acknowledgments
This work was supported by NIH grant GM115736 and NS084345 (to R.L.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Farsad K, De Camilli P. Mechanisms of membrane deformation. Curr Opin Cell Biol. 2003;15:372–381. doi: 10.1016/s0955-0674(03)00073-5. [DOI] [PubMed] [Google Scholar]
- 2.Drin G, Antonny B. Amphipathic helices and membrane curvature. FEBS Lett. 2010;584:1840–1847. doi: 10.1016/j.febslet.2009.10.022. [DOI] [PubMed] [Google Scholar]
- 3.Doherty GJ, McMahon HT. Mechanisms of endocytosis. Annu Rev Biochem. 2009;78:857–902. doi: 10.1146/annurev.biochem.78.081307.110540. [DOI] [PubMed] [Google Scholar]
- 4.McMahon HT, Gallop JL. Membrane curvature and mechanisms of dynamic cell membrane remodelling. Nature. 2005;438:590–596. doi: 10.1038/nature04396. [DOI] [PubMed] [Google Scholar]
- 5.Jarsch IK, Daste F, Gallop JL. Membrane curvature in cell biology: An integration of molecular mechanisms. J Cell Biol. 2016;214:375–387. doi: 10.1083/jcb.201604003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Peter BJ, Kent HM, Mills IG, Vallis Y, Butler PJ, Evans PR, McMahon HT. BAR domains as sensors of membrane curvature: the amphiphysin BAR structure. Science. 2004;303:495–499. doi: 10.1126/science.1092586. [DOI] [PubMed] [Google Scholar]
- 7.Ambroso MR, Hegde BG, Langen R. Endophilin A1 induces different membrane shapes using a conformational switch that is regulated by phosphorylation. Proc Natl Acad Sci U S A. 2014;111:6982–6987. doi: 10.1073/pnas.1402233111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Isas JM, Ambroso MR, Hegde PB, Langen J, Langen R. Tubulation by amphiphysin requires concentration-dependent switching from wedging to scaffolding. Structure. 2015;23:873–881. doi: 10.1016/j.str.2015.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Daumke O, Lundmark R, Vallis Y, Martens S, Butler PJ, McMahon HT. Architectural and mechanistic insights into an EHD ATPase involved in membrane remodelling. Nature. 2007;449:923–927. doi: 10.1038/nature06173. [DOI] [PubMed] [Google Scholar]
- 10.Shah C, Hegde BG, Morén B, Behrmann E, Mielke T, Moenke G, Spahn CM, Lundmark R, Daumke O, Langen R. Structural insights into membrane interaction and caveolar targeting of dynamin-like EHD2. Structure. 2014;22:409–420. doi: 10.1016/j.str.2013.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lai CL, Jao CC, Lyman E, Gallop JL, Peter BJ, McMahon HT, Langen R, Voth GA. Membrane binding and self-association of the epsin N-terminal homology domain. J Mol Biol. 2012;423:800–817. doi: 10.1016/j.jmb.2012.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Martens S, Kozlov MM, McMahon HT. How synaptotagmin promotes membrane fusion. Science. 2007;316:1205–1208. doi: 10.1126/science.1142614. [DOI] [PubMed] [Google Scholar]
- 13.Mizuno N, Varkey J, Kegulian NC, Hegde BG, Cheng N, Langen R, Steven AC. Remodeling of lipid vesicles into cylindrical micelles by α-synuclein in an extended α-helical conformation. J Biol Chem. 2012;287:29301–29311. doi: 10.1074/jbc.M112.365817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Varkey J, Isas JM, Mizuno N, Jensen MB, Bhatia VK, Jao CC, Petrlova J, Voss JC, Stamou DG, Steven AC, Langen R. Membrane curvature induction and tubulation are common features of synucleins and apolipoproteins. J Biol Chem. 2010;285:32486–32493. doi: 10.1074/jbc.M110.139576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Westphal CH, Chandra SS. Monomeric synucleins generate membrane curvature. J Biol Chem. 2013;288:1829–1840. doi: 10.1074/jbc.M112.418871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nicot AS, Toussaint A, Tosch V, Kretz C, Wallgren-Pettersson C, Iwarsson E, Kingston H, Garnier JM, Biancalana V, Oldfors A, Mandel JL, Laporte J. Mutations in amphiphysin 2 (BIN1) disrupt interaction with dynamin 2 and cause autosomal recessive centronuclear myopathy. Nat Genet. 2007;39:1134–1139. doi: 10.1038/ng2086. [DOI] [PubMed] [Google Scholar]
- 17.Bitoun M, Maugenre S, Jeannet PY, Lacène E, Ferrer X, Laforêt P, Martin JJ, Laporte J, Lochmüller H, Beggs AH, Fardeau M, Eymard B, Romero NB, Guicheney P. Mutations in dynamin 2 cause dominant centronuclear myopathy. Nat Genet. 2005;37:1207–1209. doi: 10.1038/ng1657. [DOI] [PubMed] [Google Scholar]
- 18.Kozlov MM, Campelo F, Liska N, Chernomordik LV, Marrink SJ, McMahon HT. Mechanisms shaping cell membranes. Curr Opin Cell Biol. 2014;29:53–60. doi: 10.1016/j.ceb.2014.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zimmerberg J, Kozlov MM. How proteins produce cellular membrane curvature. Nat Rev Mol Cell Biol. 2006;7:9–19. doi: 10.1038/nrm1784. [DOI] [PubMed] [Google Scholar]
- 20.Baumgart T, Capraro BR, Zhu C, Das SL. Thermodynamics and mechanics of membrane curvature generation and sensing by proteins and lipids. Annu Rev Phys Chem. 2011;62:483–506. doi: 10.1146/annurev.physchem.012809.103450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Frost A, Perera R, Roux A, Spasov K, Destaing O, Egelman EH, De Camilli P, Unger VM. Structural basis of membrane invagination by F-BAR domains. Cell. 2008;132:807–817. doi: 10.1016/j.cell.2007.12.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mizuno N, Jao CC, Langen R, Steven AC. Multiple modes of endophilin-mediated conversion of lipid vesicles into coated tubes: implications for synaptic endocytosis. J Biol Chem. 2010;285:23351–23358. doi: 10.1074/jbc.M110.143776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang Q, Navarro MV, Peng G, Molinelli E, Goh SL, Judson BL, Rajashankar KR, Sondermann H. Molecular mechanism of membrane constriction and tubulation mediated by the F-BAR protein Pacsin/Syndapin. Proc Natl Acad Sci U S A. 2009;106:12700–12705. doi: 10.1073/pnas.0902974106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Graham TR, Kozlov MM. Interplay of proteins and lipids in generating membrane curvature. Curr Opin Cell Biol. 2010;22:430–436. doi: 10.1016/j.ceb.2010.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shibata Y, Hu J, Kozlov MM, Rapoport TA. Mechanisms shaping the membranes of cellular organelles. Annu Rev Cell Dev Biol. 2009;25:329–354. doi: 10.1146/annurev.cellbio.042308.113324. [DOI] [PubMed] [Google Scholar]
- 26.Ambroso MR, Haworth IS, Langen R. Structural Characterization of Membrane-Curving Proteins: Site-Directed Spin Labeling, EPR, and Computational Refinement. Methods Enzymol. 2015;564:259–288. doi: 10.1016/bs.mie.2015.07.002. [DOI] [PubMed] [Google Scholar]
- 27.Margittai M, Langen R. Spin labeling analysis of amyloids and other protein aggregates. Methods Enzymol. 2006;413:122–139. doi: 10.1016/S0076-6879(06)13007-4. [DOI] [PubMed] [Google Scholar]
- 28.Altenbach C, López CJ, Hideg K, Hubbell WL. Exploring Structure, Dynamics, and Topology of Nitroxide Spin-Labeled Proteins Using Continuous-Wave Electron Paramagnetic Resonance Spectroscopy. Methods Enzymol. 2015;564:59–100. doi: 10.1016/bs.mie.2015.08.006. [DOI] [PubMed] [Google Scholar]
- 29.Jeschke G, Polyhach Y. Distance measurements on spin-labelled biomacromolecules by pulsed electron paramagnetic resonance. Phys Chem Chem Phys. 2007;9:1895–1910. doi: 10.1039/b614920k. [DOI] [PubMed] [Google Scholar]
- 30.Hubbell WL, Gross A, Langen R, Lietzow MA. Recent advances in site-directed spin labeling of proteins. Curr Opin Struct Biol. 1998;8:649–656. doi: 10.1016/s0959-440x(98)80158-9. [DOI] [PubMed] [Google Scholar]
- 31.Altenbach C, Greenhalgh DA, Khorana HG, Hubbell WL. A collision gradient method to determine the immersion depth of nitroxides in lipid bilayers: application to spin-labeled mutants of bacteriorhodopsin. Proc Natl Acad Sci U S A. 1994;91:1667–1671. doi: 10.1073/pnas.91.5.1667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jeschke G. DEER distance measurements on proteins. Annu Rev Phys Chem. 2012;63:419–446. doi: 10.1146/annurev-physchem-032511-143716. [DOI] [PubMed] [Google Scholar]
- 33.Hubbell WL, Cafiso DS, Altenbach C. Identifying conformational changes with site-directed spin labeling. Nat Struct Biol. 2000;7:735–739. doi: 10.1038/78956. [DOI] [PubMed] [Google Scholar]
- 34.Hubbell WL, Mchaourab HS, Altenbach C, Lietzow MA. Watching proteins move using site-directed spin labeling. Structure. 1996;4:779–783. doi: 10.1016/s0969-2126(96)00085-8. [DOI] [PubMed] [Google Scholar]
- 35.Margittai M, Langen R. Template-assisted filament growth by parallel stacking of tau. Proc Natl Acad Sci U S A. 2004;101:10278–10283. doi: 10.1073/pnas.0401911101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ladner CL, Chen M, Smith DP, Platt GW, Radford SE, Langen R. Stacked sets of parallel, in-register beta-strands of beta2-microglobulin in amyloid fibrils revealed by site-directed spin labeling and chemical labeling. J Biol Chem. 2010;285:17137–17147. doi: 10.1074/jbc.M110.117234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Margittai M, Langen R. Fibrils with parallel in-register structure constitute a major class of amyloid fibrils: molecular insights from electron paramagnetic resonance spectroscopy. Q Rev Biophys. 2008;41:265–297. doi: 10.1017/S0033583508004733. [DOI] [PubMed] [Google Scholar]
- 38.Butterfield SM, Lashuel HA. Amyloidogenic protein-membrane interactions: mechanistic insight from model systems. Angew Chem Int Ed Engl. 2010;49:5628–5654. doi: 10.1002/anie.200906670. [DOI] [PubMed] [Google Scholar]
- 39.Snead D, Eliezer D. Alpha-synuclein function and dysfunction on cellular membranes. Exp Neurobiol. 2014;23:292–313. doi: 10.5607/en.2014.23.4.292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jayasinghe SA, Langen R. Membrane interaction of islet amyloid polypeptide. Biochim Biophys Acta. 2007;1768:2002–2009. doi: 10.1016/j.bbamem.2007.01.022. [DOI] [PubMed] [Google Scholar]
- 41.Kegulian NC, Sankhagowit S, Apostolidou M, Jayasinghe SA, Malmstadt N, Butler PC, Langen R. Membrane Curvature-sensing and Curvature-inducing Activity of Islet Amyloid Polypeptide and Its Implications for Membrane Disruption. J Biol Chem. 2015;290:25782–25793. doi: 10.1074/jbc.M115.659797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Engel MF. Membrane permeabilization by Islet Amyloid Polypeptide. Chem Phys Lipids. 2009;160:1–10. doi: 10.1016/j.chemphyslip.2009.03.008. [DOI] [PubMed] [Google Scholar]
- 43.Sparr E, Engel MF, Sakharov DV, Sprong M, Jacobs J, Kruijff B de, Höppener JW, Killian JA. Islet amyloid polypeptide-induced membrane leakage involves uptake of lipids by forming amyloid fibers. FEBS Lett. 2004;577:117–120. doi: 10.1016/j.febslet.2004.09.075. [DOI] [PubMed] [Google Scholar]
- 44.Cecchi C, Stefani M. The amyloid-cell membrane system. The interplay between the biophysical features of oligomers/fibrils and cell membrane defines amyloid toxicity. Biophys Chem. 2013;182:30–43. doi: 10.1016/j.bpc.2013.06.003. [DOI] [PubMed] [Google Scholar]
- 45.Andreasen M, Lorenzen N, Otzen D. Interactions between misfolded protein oligomers and membranes: A central topic in neurodegenerative diseases? Biochim Biophys Acta. 2015;1848:1897–1907. doi: 10.1016/j.bbamem.2015.01.018. [DOI] [PubMed] [Google Scholar]
- 46.Dettmer U, Selkoe D, Bartels T. New insights into cellular α-synuclein homeostasis in health and disease. Curr Opin Neurobiol. 2016;36:15–22. doi: 10.1016/j.conb.2015.07.007. [DOI] [PubMed] [Google Scholar]
- 47.Tofaris GK, Spillantini MG. Physiological and pathological properties of alpha-synuclein. Cell Mol Life Sci. 2007;64:2194–2201. doi: 10.1007/s00018-007-7217-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Auluck PK, Caraveo G, Lindquist S. α-Synuclein: membrane interactions and toxicity in Parkinson’s disease. Annu Rev Cell Dev Biol. 2010;26:211–233. doi: 10.1146/annurev.cellbio.042308.113313. [DOI] [PubMed] [Google Scholar]
- 49.Beyer K. Mechanistic aspects of Parkinson’s disease: alpha-synuclein and the biomembrane. Cell Biochem Biophys. 2007;47:285–299. doi: 10.1007/s12013-007-0014-9. [DOI] [PubMed] [Google Scholar]
- 50.Lokappa SB, Ulmer TS. Alpha-synuclein populates both elongated and broken helix states on small unilamellar vesicles. J Biol Chem. 2011;286:21450–21457. doi: 10.1074/jbc.M111.224055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rao JN, Jao CC, Hegde BG, Langen R, Ulmer TS. A combinatorial NMR and EPR approach for evaluating the structural ensemble of partially folded proteins. J Am Chem Soc. 2010;132:8657–8668. doi: 10.1021/ja100646t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ulmer TS, Bax A, Cole NB, Nussbaum RL. Structure and dynamics of micelle-bound human alpha-synuclein. J Biol Chem. 2005;280:9595–9603. doi: 10.1074/jbc.M411805200. [DOI] [PubMed] [Google Scholar]
- 53.Jao CC, Der-Sarkissian A, Chen J, Langen R. Structure of membrane-bound alpha-synuclein studied by site-directed spin labeling. Proc Natl Acad Sci U S A. 2004;101:8331–8336. doi: 10.1073/pnas.0400553101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Jao CC, Hegde BG, Chen J, Haworth IS, Langen R. Structure of membrane-bound alpha-synuclein from site-directed spin labeling and computational refinement. Proc Natl Acad Sci U S A. 2008;105:19666–19671. doi: 10.1073/pnas.0807826105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Varkey J, Mizuno N, Hegde BG, Cheng N, Steven AC, Langen R. α-Synuclein oligomers with broken helical conformation form lipoprotein nanoparticles. J Biol Chem. 2013;288:17620–17630. doi: 10.1074/jbc.M113.476697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Borbat P, Ramlall TF, Freed JH, Eliezer D. Inter-helix distances in lysophospholipid micelle-bound alpha-synuclein from pulsed ESR measurements. J Am Chem Soc. 2006;128:10004–10005. doi: 10.1021/ja063122l. [DOI] [PubMed] [Google Scholar]
- 57.Georgieva ER, Ramlall TF, Borbat PP, Freed JH, Eliezer D. Membrane-bound alpha-synuclein forms an extended helix: long-distance pulsed ESR measurements using vesicles, bicelles, and rodlike micelles. J Am Chem Soc. 2008;130:12856–12857. doi: 10.1021/ja804517m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Drescher M, Veldhuis G, van Rooijen BD, Milikisyants S, Subramaniam V, Huber M. Antiparallel arrangement of the helices of vesicle-bound alpha-synuclein. J Am Chem Soc. 2008;130:7796–7797. doi: 10.1021/ja801594s. [DOI] [PubMed] [Google Scholar]
- 59.Drescher M, van Rooijen BD, Veldhuis G, Subramaniam V, Huber M. A stable lipid-induced aggregate of alpha-synuclein. J Am Chem Soc. 2010;132:4080–4082. doi: 10.1021/ja909247j. [DOI] [PubMed] [Google Scholar]
- 60.Ferreon AC, Gambin Y, Lemke EA, Deniz AA. Interplay of alpha-synuclein binding and conformational switching probed by single-molecule fluorescence. Proc Natl Acad Sci U S A. 2009;106:5645–5650. doi: 10.1073/pnas.0809232106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Trexler AJ, Rhoades E. Alpha-synuclein binds large unilamellar vesicles as an extended helix. Biochemistry. 2009;48:2304–2306. doi: 10.1021/bi900114z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Bortolus M, Tombolato F, Tessari I, Bisaglia M, Mammi S, Bubacco L, Ferrarini A, Maniero AL. Broken helix in vesicle and micelle-bound alpha-synuclein: insights from site-directed spin labeling-EPR experiments and MD simulations. J Am Chem Soc. 2008;130:6690–6691. doi: 10.1021/ja8010429. [DOI] [PubMed] [Google Scholar]
- 63.Bartels T, Choi JG, Selkoe DJ. α-Synuclein occurs physiologically as a helically folded tetramer that resists aggregation. Nature. 2011;477:107–110. doi: 10.1038/nature10324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wang W, Perovic I, Chittuluru J, Kaganovich A, Nguyen LT, Liao J, Auclair JR, Johnson D, Landeru A, Simorellis AK, Ju S, Cookson MR, Asturias FJ, Agar JN, Webb BN, Kang C, Ringe D, Petsko GA, Pochapsky TC, Hoang QQ. A soluble α-synuclein construct forms a dynamic tetramer. Proc Natl Acad Sci U S A. 2011;108:17797–17802. doi: 10.1073/pnas.1113260108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Isas JM, Langen R, Haigler HT, Hubbell WL. Structure and dynamics of a helical hairpin and loop region in annexin 12: a site-directed spin labeling study. Biochemistry. 2002;41:1464–1473. doi: 10.1021/bi011856z. [DOI] [PubMed] [Google Scholar]
- 66.Altenbach C, Klein-Seetharaman J, Hwa J, Khorana HG, Hubbell WL. Structural features and light-dependent changes in the sequence 59–75 connecting helices I and II in rhodopsin: a site-directed spin-labeling study. Biochemistry. 1999;38:7945–7949. doi: 10.1021/bi990014l. [DOI] [PubMed] [Google Scholar]
- 67.Glasgow BJ, Gasymov OK, Abduragimov AR, Yusifov TN, Altenbach C, Hubbell WL. Side chain mobility and ligand interactions of the G strand of tear lipocalins by site-directed spin labeling. Biochemistry. 1999;38:13707–13716. doi: 10.1021/bi9913449. [DOI] [PubMed] [Google Scholar]
- 68.Oh KJ, Zhan H, Cui C, Altenbach C, Hubbell WL, Collier RJ. Conformation of the diphtheria toxin T domain in membranes: a site-directed spin-labeling study of the TH8 helix and TL5 loop. Biochemistry. 1999;38:10336–10343. doi: 10.1021/bi990520a. [DOI] [PubMed] [Google Scholar]
- 69.Pannier M, Veit S, Godt A, Jeschke G, Spiess HW. Dead-time free measurement of dipole-dipole interactions between electron spins. J Magn Reson. 2000;142:331–340. doi: 10.1006/jmre.1999.1944. [DOI] [PubMed] [Google Scholar]
- 70.Jeschke G. Distance measurements in the nanometer range by pulse EPR. Chemphyschem. 2002;3:927–932. doi: 10.1002/1439-7641(20021115)3:11<927::AID-CPHC927>3.0.CO;2-Q. [DOI] [PubMed] [Google Scholar]
- 71.Jeschke G, Bender A, Paulsen H, Zimmermann H, Godt A. Sensitivity enhancement in pulse EPR distance measurements. J Magn Reson. 2004;169:1–12. doi: 10.1016/j.jmr.2004.03.024. [DOI] [PubMed] [Google Scholar]
- 72.Chiang YW, Borbat PP, Freed JH. The determination of pair distance distributions by pulsed ESR using Tikhonov regularization. J Magn Reson. 2005;172:279–295. doi: 10.1016/j.jmr.2004.10.012. [DOI] [PubMed] [Google Scholar]
- 73.Altenbach C, Kusnetzow AK, Ernst OP, Hofmann KP, Hubbell WL. High-resolution distance mapping in rhodopsin reveals the pattern of helix movement due to activation. Proc Natl Acad Sci U S A. 2008;105:7439–7444. doi: 10.1073/pnas.0802515105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Banham JE, Timmel CR, Abbott RJ, Lea SM, Jeschke G. The characterization of weak protein-protein interactions: evidence from DEER for the trimerization of a von Willebrand Factor A domain in solution. Angew Chem Int Ed Engl. 2006;45:1058–1061. doi: 10.1002/anie.200503720. [DOI] [PubMed] [Google Scholar]
- 75.Hilger D, Jung H, Padan E, Wegener C, Vogel KP, Steinhoff HJ, Jeschke G. Assessing oligomerization of membrane proteins by four-pulse DEER: pH-dependent dimerization of NhaA Na+/H+ antiporter of E. coli. Biophys J. 2005;89:1328–1338. doi: 10.1529/biophysj.105.062232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Cheng CY, Varkey J, Ambroso MR, Langen R, Han S. Hydration dynamics as an intrinsic ruler for refining protein structure at lipid membrane interfaces. Proc Natl Acad Sci U S A. 2013;110:16838–16843. doi: 10.1073/pnas.1307678110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Eichmann C, Campioni S, Kowal J, Maslennikov I, Gerez J, Liu X, Verasdonck J, Nespovitaya N, Choe S, Meier BH, Picotti P, Rizo J, Stahlberg H, Riek R. Preparation and Characterization of Stable α-Synuclein Lipoprotein Particles. J Biol Chem. 2016;291:8516–8527. doi: 10.1074/jbc.M115.707968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Ouberai MM, Wang J, Swann MJ, Galvagnion C, Guilliams T, Dobson CM, Welland ME. α-Synuclein senses lipid packing defects and induces lateral expansion of lipids leading to membrane remodeling. J Biol Chem. 2013;288:20883–20895. doi: 10.1074/jbc.M113.478297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Jiang Z, de Messieres M, Lee JC. Membrane remodeling by α-synuclein and effects on amyloid formation. J Am Chem Soc. 2013;135:15970–15973. doi: 10.1021/ja405993r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Braun AR, Lacy MM, Ducas VC, Rhoades E, Sachs JN. α-Synuclein-induced membrane remodeling is driven by binding affinity, partition depth, and interleaflet order asymmetry. J Am Chem Soc. 2014;136:9962–9972. doi: 10.1021/ja5016958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Pandey AP, Haque F, Rochet JC, Hovis JS. α-Synuclein-induced tubule formation in lipid bilayers. J Phys Chem B. 2011;115:5886–5893. doi: 10.1021/jp1121917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Shi Z, Sachs JN, Rhoades E, Baumgart T. Biophysics of α-synuclein induced membrane remodelling. Phys Chem Chem Phys. 2015;17:15561–15568. doi: 10.1039/c4cp05883f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Boassa D, Berlanga ML, Yang MA, Terada M, Hu J, Bushong EA, Hwang M, Masliah E, George JM, Ellisman MH. Mapping the subcellular distribution of α-synuclein in neurons using genetically encoded probes for correlated light and electron microscopy: implications for Parkinson’s disease pathogenesis. J Neurosci. 2013;33:2605–2615. doi: 10.1523/JNEUROSCI.2898-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Butler PC, Chou J, Carter WB, Wang YN, Bu BH, Chang D, Chang JK, Rizza RA. Effects of meal ingestion on plasma amylin concentration in NIDDM and nondiabetic humans. Diabetes. 1990;39:752–756. doi: 10.2337/diab.39.6.752. [DOI] [PubMed] [Google Scholar]
- 85.Janson J, Soeller WC, Roche PC, Nelson RT, Torchia AJ, Kreutter DK, Butler PC. Spontaneous diabetes mellitus in transgenic mice expressing human islet amyloid polypeptide. Proc Natl Acad Sci U S A. 1996;93:7283–7288. doi: 10.1073/pnas.93.14.7283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Janson J, Ashley RH, Harrison D, McIntyre S, Butler PC. The mechanism of islet amyloid polypeptide toxicity is membrane disruption by intermediate-sized toxic amyloid particles. Diabetes. 1999;48:491–498. doi: 10.2337/diabetes.48.3.491. [DOI] [PubMed] [Google Scholar]
- 87.Lorenzo A, Razzaboni B, Weir GC, Yankner BA. Pancreatic islet cell toxicity of amylin associated with type-2 diabetes mellitus. Nature. 1994;368:756–760. doi: 10.1038/368756a0. [DOI] [PubMed] [Google Scholar]
- 88.Magzoub M, Miranker AD. Concentration-dependent transitions govern the subcellular localization of islet amyloid polypeptide. FASEB J. 2012;26:1228–1238. doi: 10.1096/fj.11-194613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Apostolidou M, Jayasinghe SA, Langen R. Structure of alpha-helical membrane-bound human islet amyloid polypeptide and its implications for membrane-mediated misfolding. J Biol Chem. 2008;283:17205–17210. doi: 10.1074/jbc.M801383200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Jayasinghe SA, Langen R. Lipid membranes modulate the structure of islet amyloid polypeptide. Biochemistry. 2005;44:12113–12119. doi: 10.1021/bi050840w. [DOI] [PubMed] [Google Scholar]
- 91.Last NB, Rhoades E, Miranker AD. Islet amyloid polypeptide demonstrates a persistent capacity to disrupt membrane integrity. Proc Natl Acad Sci U S A. 2011;108:9460–9465. doi: 10.1073/pnas.1102356108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Luca S, Yau WM, Leapman R, Tycko R. Peptide conformation and supramolecular organization in amylin fibrils: constraints from solid-state NMR. Biochemistry. 2007;46:13505–13522. doi: 10.1021/bi701427q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Sumner Makin O, Serpell LC. Structural characterisation of islet amyloid polypeptide fibrils. J Mol Biol. 2004;335:1279–1288. doi: 10.1016/j.jmb.2003.11.048. [DOI] [PubMed] [Google Scholar]
- 94.Bedrood S, Li Y, Isas JM, Hegde BG, Baxa U, Haworth IS, Langen R. Fibril structure of human islet amyloid polypeptide. J Biol Chem. 2012;287:5235–5241. doi: 10.1074/jbc.M111.327817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Jayasinghe SA, Langen R. Identifying structural features of fibrillar islet amyloid polypeptide using site-directed spin labeling. J Biol Chem. 2004;279:48420–48425. doi: 10.1074/jbc.M406853200. [DOI] [PubMed] [Google Scholar]
- 96.Williamson JA, Loria JP, Miranker AD. Helix stabilization precedes aqueous and bilayer-catalyzed fiber formation in islet amyloid polypeptide. J Mol Biol. 2009;393:383–396. doi: 10.1016/j.jmb.2009.07.077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Hebda JA, Saraogi I, Magzoub M, Hamilton AD, Miranker AD. A peptidomimetic approach to targeting pre-amyloidogenic states in type II diabetes. Chem Biol. 2009;16:943–950. doi: 10.1016/j.chembiol.2009.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Nanga RP, Brender JR, Vivekanandan S, Ramamoorthy A. Structure and membrane orientation of IAPP in its natively amidated form at physiological pH in a membrane environment. Biochim Biophys Acta. 2011;1808:2337–2342. doi: 10.1016/j.bbamem.2011.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Kinnunen PK, Domanov YA, Mattila JP, Varis T. Formation of lipid/peptide tubules by IAPP and temporin B on supported lipid membranes. Soft Matter. 2015;11:9188–9200. doi: 10.1039/b925228b. [DOI] [PubMed] [Google Scholar]
- 100.Gallop JL, Jao CC, Kent HM, Butler PJ, Evans PR, Langen R, McMahon HT. Mechanism of endophilin N-BAR domain-mediated membrane curvature. EMBO J. 2006;25:2898–2910. doi: 10.1038/sj.emboj.7601174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Jao CC, Hegde BG, Gallop JL, Hegde PB, McMahon HT, Haworth IS, Langen R. Roles of amphipathic helices and the bin/amphiphysin/rvs (BAR) domain of endophilin in membrane curvature generation. J Biol Chem. 2010;285:20164–20170. doi: 10.1074/jbc.M110.127811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Rikhy R, Kumar V, Mittal R, Krishnan KS. Endophilin is critically required for synapse formation and function in Drosophila melanogaster. J Neurosci. 2002;22:7478–7484. doi: 10.1523/JNEUROSCI.22-17-07478.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Guichet A, Wucherpfennig T, Dudu V, Etter S, Wilsch-Bräuniger M, Hellwig A, González-Gaitán M, Huttner WB, Schmidt AA. Essential role of endophilin A in synaptic vesicle budding at the Drosophila neuromuscular junction. EMBO J. 2002;21:1661–1672. doi: 10.1093/emboj/21.7.1661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Huttner WB, Schmidt A. Lipids, lipid modification and lipid-protein interaction in membrane budding and fission–insights from the roles of endophilin A1 and synaptophysin in synaptic vesicle endocytosis. Curr Opin Neurobiol. 2000;10:543–551. doi: 10.1016/s0959-4388(00)00126-4. [DOI] [PubMed] [Google Scholar]
- 105.Kjaerulff O, Brodin L, Jung A. The structure and function of endophilin proteins. Cell Biochem Biophys. 2011;60:137–154. doi: 10.1007/s12013-010-9137-5. [DOI] [PubMed] [Google Scholar]
- 106.Weissenhorn W. Crystal structure of the endophilin-A1 BAR domain. J Mol Biol. 2005;351:653–661. doi: 10.1016/j.jmb.2005.06.013. [DOI] [PubMed] [Google Scholar]
- 107.Masuda M, Takeda S, Sone M, Ohki T, Mori H, Kamioka Y, Mochizuki N. Endophilin BAR domain drives membrane curvature by two newly identified structure-based mechanisms. EMBO J. 2006;25:2889–2897. doi: 10.1038/sj.emboj.7601176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Sundborger A, Soderblom C, Vorontsova O, Evergren E, Hinshaw JE, Shupliakov O. An endophilin-dynamin complex promotes budding of clathrin-coated vesicles during synaptic vesicle recycling. J Cell Sci. 2011;124:133–143. doi: 10.1242/jcs.072686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Ringstad N, Gad H, Löw P, Paolo G Di, Brodin L, Shupliakov O, De Camilli P. Endophilin/SH3p4 is required for the transition from early to late stages in clathrin-mediated synaptic vesicle endocytosis. Neuron. 1999;24:143–154. doi: 10.1016/s0896-6273(00)80828-4. [DOI] [PubMed] [Google Scholar]
- 110.Schuske KR, Richmond JE, Matthies DS, Davis WS, Runz S, Rube DA, van der Bliek AM, Jorgensen EM. Endophilin is required for synaptic vesicle endocytosis by localizing synaptojanin. Neuron. 2003;40:749–762. doi: 10.1016/s0896-6273(03)00667-6. [DOI] [PubMed] [Google Scholar]
- 111.Farsad K, Ringstad N, Takei K, Floyd SR, Rose K, De Camilli P. Generation of high curvature membranes mediated by direct endophilin bilayer interactions. J Cell Biol. 2001;155:193–200. doi: 10.1083/jcb.200107075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Meinecke M, Boucrot E, Camdere G, Hon WC, Mittal R, McMahon HT. Cooperative recruitment of dynamin and BIN/amphiphysin/Rvs (BAR) domain-containing proteins leads to GTP-dependent membrane scission. J Biol Chem. 2013;288:6651–6661. doi: 10.1074/jbc.M112.444869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Milosevic I, Giovedi S, Lou X, Raimondi A, Collesi C, Shen H, Paradise S, O’Toole E, Ferguson S, Cremona O, De Camilli P. Recruitment of endophilin to clathrin-coated pit necks is required for efficient vesicle uncoating after fission. Neuron. 2011;72:587–601. doi: 10.1016/j.neuron.2011.08.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Ferguson SM, Ferguson S, Raimondi A, Paradise S, Shen H, Mesaki K, Ferguson A, Destaing O, Ko G, Takasaki J, Cremona O, O’Toole E, De Camilli P. Coordinated actions of actin and BAR proteins upstream of dynamin at endocytic clathrin-coated pits. Dev Cell. 2009;17:811–822. doi: 10.1016/j.devcel.2009.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Morén B, Shah C, Howes MT, Schieber NL, McMahon HT, Parton RG, Daumke O, Lundmark R. EHD2 regulates caveolar dynamics via ATP-driven targeting and oligomerization. Mol Biol Cell. 2012;23:1316–1329. doi: 10.1091/mbc.E11-09-0787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Pant S, Sharma M, Patel K, Caplan S, Carr CM, Grant BD. AMPH-1/Amphiphysin/Bin1 functions with RME-1/Ehd1 in endocytic recycling. Nat Cell Biol. 2009;11:1399–1410. doi: 10.1038/ncb1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Melo AA, Hegde BG, Shah C, Larsson E, Isas M, Kunz S, Lundmark R, Langen R, Daumke O. Structural insights into the activation mechanism of dynamin-like Eps15-homology domain proteins. Proceedings of the National Acedemy of Sciences USA. 2017 doi: 10.1073/pnas.1614075114. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]


