Guanine nucleotide exchange factor RopGEF2, which is directly activated by phytochrome phyB, suppresses red light-induced stomatal opening by activating ROP7 and ROP2.
Abstract
Circadian rhythm of stomatal aperture is mainly regulated by light/darkness. Blue and red light induce stomatal opening through different mechanisms that are mediated by special receptors. ROP2, a member of Rho GTPase family in Arabidopsis (Arabidopsis thaliana), has been found to negatively regulate light-induced stomatal opening. However, the upstream guanine nucleotide exchange factor (GEF) RopGEFs have not been revealed, and it is unclear which photoreceptor is required for the action of RopGEFs-ROPs. Here, we showed that RopGEF2 acted as a negative regulator in phytochrome B (phyB)-mediated red light-induced stomatal opening. Meanwhile, ROP7, another member of ROP family, acting redundantly with ROP2, was regulated by RopGEF2 in this process. RopGEF2 interacted with ROP7 and ROP2 and enhanced their intrinsic nucleotide exchange rates. Furthermore, the direct interactions between phyB and RopGEF2 were detected in vitro and in plants, and phyB enhanced the GEF activity of RopGEF2 toward both ROP7 and ROP2 under light. In addition, RopGEF4 functioned redundantly with RopGEF2 in red light-induced stomatal opening by activating both ROP7 and ROP2, and RopGEF2/RopGEF4 acted genetically downstream of phyB; however, the GEF activity of RopGEF4 was not directly enhanced by phyB. These results revealed that red light-activated phyB enhances the GEF activities of RopGEF2 and RopGEF4 directly or indirectly, and then activate both ROP7 and ROP2 in guard cells. The negative mechanism triggered by phyB prevents the excessive stomatal opening under red light.
Stomatal movements regulate gas exchange between the atmosphere and plants, optimizing the transpirational water loss and photosynthetic assimilation of CO2. In addition to responding to multiple environmental stimuli, stomatal movements also exhibit a daily circadian rhythm, with open stomata during the daytime and closed stomata at night. Both blue light and red light induce stomatal opening. Blue light is perceived by the phototropin receptors phot1 and phot2 (Kinoshita et al., 2001), causing the osmotic swelling of guard cells by activating H+-ATPases in the plasma membrane and resulting in the opening of the stomata (Kinoshita and Shimazaki, 2002). Red light acts as both an environmental signal and an energy source that induces stomatal opening. However, the molecular mechanisms underlying the stomatal response to red light has remained controversial for many years. It was proposed originally that red light induces mesophyll photosynthetic consumption of CO2 inside the leaf (Ci), which consequently opens stomata (Roelfsema et al., 2002), whereas several lines of evidence showed that stomata response to light even when the Ci was held constant (Messinger et al., 2006; Lawson et al., 2008). A close correlation between red light-dependent mesophyll photosynthesis and stomatal conductance has been well documented: metabolites or signals from mesophyll may coordinate the mesophyll and stomatal behavior. For example, sugar or malate, two photosynthetic products from mesophyll, may be transported to guard cells to affect stomatal movements (Stadler et al., 2003; Weise et al., 2008; Lee et al., 2008; Araújo et al., 2011); and a vapor phase signal (Sibbernsen and Mott, 2010; Mott et al., 2014) or an aqueous signal (Fujita et al., 2013) generated from mesophyll were transferred to epidermis. Photoreceptor phyB was found to mediate red light-enhanced photosynthesis (Guo et al., 2016). Recently, HIGH TEMPERATURE1 was found to mediate red light-induced stomatal opening that is both dependent on and independent of Ci (Matrosova et al., 2015), and it has been discussed that guard cells have direct responses to red light (Mott, 2009). Notably, red light receptor phyB has been revealed to participate in red light-induced stomatal opening, and Constitutive Photomorphogenic1 and the phytochrome interacting factors PIF3 and PIF4 act downstream of phyB (Mao et al., 2005; Wang et al., 2010). However, the molecular mechanisms linking phyB and stomatal opening have not been well understood.
Plants possess a distinct small GTPase family of ROPs (Yang, 2002; Gu et al., 2004). There are 11 ROP genes in Arabidopsis (Arabidopsis thaliana) that act as vital molecular switches in a number of cellular processes, including polar growth of pollen tubes (Gu et al., 2003), cell elongation during organogenesis (Fu et al., 2002; Brembu et al., 2005), interdigitated growth of pavement cells (Xu et al., 2010; Lin et al., 2015), and polar auxin transport (Nagawa et al., 2012; Chen et al., 2012; Lin et al., 2012; Huang et al., 2014). ROP2, ROP6, and ROP11 act as negative regulators of abscisic acid (ABA)-induced stomatal closure, seed germination, seedling growth, and gene expression (Lemichez et al., 2001; Hwang et al., 2011; Li and Liu, 2012). ROPs exert their functions in an active GTP-binding form. ROPs-specific guanine nucleotide exchange factors RopGEFs convert the GDP-bound forms of ROPs into the GTP-bound forms, thereby activating ROPs in plants. One DOCK family protein, SPIKE1, was revealed in Arabidopsis to control cell morphogenesis, and activation of ROPs is essential for the function of SPIKE1 (Qiu et al., 2002; Basu et al., 2008; Zhang et al., 2010; Ren et al., 2016). The genome of Arabidopsis also has a new RopGEFs family containing either a conserved central domain named “domain of unknown function 315” (DUF315) or a plant-specific ROP nucleotide exchanger (PRONE) domain, which has RopGEF catalytic activity toward ROPs (Berken et al., 2005; Gu et al., 2006; Shichrur and Yalovsky, 2006). The variable C- or N-terminal regions in RopGEFs are regulatory motifs of the GEF catalytic domains (Gu et al., 2006; Shichrur and Yalovsky, 2006). There are 14 RopGEF members and 11 ROPs in Arabidopsis, suggesting that certain physiological processes need the activation of distinct RopGEFs and ROPs: RopGEF1 activates ROP1 to regulate the polar growth of pollen tubes (Gu et al., 2006; Chang et al., 2013); RopGEF1 and RopGEF10 regulate root hair growth by activating ROP2 and ROP6 (Huang et al., 2013). RopGEF1 and RopGEF4 function as specific regulators of ROP11 in ABA-induced stomatal closure (Li and Liu, 2012; Li et al., 2016).
Several lines of evidence indicate that RopGEFs act as links connecting receptors or receptor kinases with ROP signaling. For example, the interactions between RopGEF12 and receptor kinase PRK2a (Zhang and McCormick, 2007), KPP (a RopGEF homolog) and the pollen-specific receptor kinases LePRK1 and LePRK2 (Kaothien et al., 2005), and RopGEF1 and the receptor-like kinase AtPRK2 (Chang et al., 2013) transmit the receptor kinases signals to the ROPs-mediated growth of pollen tube. The interactions between PIRF1/RopGEF11 and phytochrome light receptors or FERONIA receptor-like kinase connect receptor/receptor-like kinase signals and ROP response during root or root hair development (Shin et al., 2010; Duan et al., 2010).
ROP2 has been found to act as a negative regulator in light-induced stomatal opening. Stomata of CA-rop2 open slower and show reduced final aperture than wild type, whereas stomata of rop2 mutant and DN-rop2 open quicker and had larger final apertures than wild type in response to light (Jeon et al., 2008). The ROP-interactive CRIB-containing proteins (RIC7) have been revealed to act downstream of ROP2 in this process, and ROP2-RIC7 regulates stomatal opening by inhibiting the exocyst subunit Exo70B1 (Hong et al., 2016). However, the upstream RopGEFs involved in ROP-mediated light-induced stomatal opening have not been identified, and it is unclear which photoreceptor is required for the action of RopGEFs-ROPs. In this research, our genetic, physiological, and biochemical evidence supports that RopGEF2 functions redundantly with RopGEF4 to play a negative role in red light-induced stomatal opening by activating ROP2 and ROP7, another member of ROP family. Photoreceptor phyB is required for the action of RopGEF2/4-ROP7/2, and the GEF activity of RopGEF2 is directly enhanced by phyB in light, whereas the GEF activity of RopGEF4 is not directly enhanced by phyB.
RESULTS
ROP7 Negatively Regulates White Light-Induced Stomatal Opening
Our promoter:reporter analysis showed that the ROP7pro:GUS is expressed in guard cells (Fig. 1A). Therefore, we investigated whether ROP7 participates in white light-induced stomatal opening. The stomatal density and size of rop7 mutant showed no obvious difference to that of wild type (Supplemental Fig. S1, A and B). We then tested the stomatal opening of the T-DNA insertion rop7 mutant and CA-rop7 (constitutively active form of rop7; Brembu et al., 2005) line after white light illumination. The results showed that the stomatal apertures of the wild type, the rop7 mutant, and the CA-rop7 line increased gradually by the extension of light illumination. However, the rop7 mutant had a significantly larger stomatal aperture than those of wild type, and the apertures of CA-rop7 stomata were significantly smaller than those of wild type with 1, 2, or 3 h light treatment (Fig. 1B). The differences in stomatal apertures between wild type and mutants reached maximum with 2 h light treatment, so the stomatal apertures were measured hereafter in this article with this condition. Furthermore, the stomatal apertures of the two independent complemented lines of rop7 were restored to the wild-type level after 2 h light illumination (Fig. 1C). The differences of stomatal apertures between wild type and mutants were consistent with the stomatal conductance. The larger stomatal aperture in the rop7 mutant led to a higher stomatal conductance, and the CA-rop7 line exhibited a lower stomatal conductance (Fig. 1D). The excessive stomatal opening (faster opening of stomata with larger maximum value of aperture) of the rop7 mutant suggested that ROP7 is another negative regulator in white light-induced stomatal opening.
Figure 1.
ROP7 acts as a negative regulator of white light-induced stomatal opening. A, The GUS signal in epidermal guard cells of the transgenic plants expressing ROP7pro:GUS. B and C, The stomatal opening of the rop7 mutant and CA-rop7 line (B) and the complemented lines (Com-6 and Com-7; C) with white light treatment. Epidermal peels with closed stomata were illuminated with white light (150 μmol m−2 s−1) for 1, 2, or 3 h. The error bars represent the means ± se from three biological replicates (Student’s t test, n = 150, **P < 0.01). D, Stomatal conductance of CA-rop7 line, rop7 mutant, and two complemented lines of rop7 under white light. Leaves of 4- to 6-week-old plants were measured after 2 h white light illumination (150 μmol m−2 s−1). The error bars represent the means ± se from three biological replicates (Student’s t test, n = 15, **P < 0.01).
RopGEF2 Physically Interacts with ROP7 and Accelerates Its Intrinsic Nucleotide Exchange Rate
To identify the potential RopGEFs that activate ROP7 in light-induced stomatal opening, we performed a yeast two-hybrid assay by using ROP7 as bait. We cloned the coding region of all the RopGEFs except for RopGEF11 (we failed to amplify the coding region of RopGEF11 by PCR) and integrated it into the vector pGADT7. The results showed that ROP7 interacted with RopGEF1, 2, 3, 4, 5, and 7 in yeast (Fig. 2A). We next determined which ropgef mutants exhibited similar stomatal responses as rop7 with white light treatment. The stomatal analysis showed that ropgef1 and ropgef3 mutants exhibited the similar stomatal apertures as wild type, whereas ropgef2-1 and ropgef4-1 mutants showed larger stomatal apertures than that of wild type with white light illumination (Supplemental Fig. S2, A and B). The ropgef2, ropgef4, and ropgef2 ropgef4 mutants used in this research showed no obvious difference in stomatal density and size to wild type (Supplemental Fig. S1, A and B). We failed to obtain the null mutants of ropgef5 and ropgef7. Considering the interaction between ROP7 and RopGEF2 or RopGEF4, and the similar stomatal phenotypes of ropgef2 and ropgef4 to rop7, we next mainly investigated the role of RopGEF2 and RopGEF4 in light-induced stomatal opening. To further confirm the physical interaction between ROP7 and RopGEF2, we performed an in vitro pull-down assay and found that maltose binding protein (MBP)-fused RopGEF2 (MBP-RopGEF2) interacted with nucleotide-free and GDP-bound forms of the GST-ROP7 fusion protein, respectively (Fig. 2B). The interactions of RopGEF2 with nucleotide-free, GDP-ROP7 are consistent with the reported crystal structures of ROP-GDP-PRONE ternary and ROP-PRONE binary complexes (Thomas et al., 2007, 2009). This result provided further evidence that ROP7 directly interacts with RopGEF2.
Figure 2.
RopGEF2 physically interacts with ROP7 and enhances the guanine nucleotide exchange activity of ROP7. A, Analysis of the ROP7 interaction with RopGEFs in yeast. Yeast strains were grown on synthetic complete media without Trp and Leu (SC-LT) or without Trp, Leu, Ade, and His (SC-AHLT) for 4 to 5 d. B, The interaction between RopGEF2 and ROP7 was determined by an in vitro pull-down assay. C and D, The guanine nucleotide exchange activities of the small G protein positive control CDC42 (C) and ROP7 (D) increased with the elevation of the CDC42 or ROP7 concentrations. E, The guanine nucleotide exchange activities of ROP7 and CDC42 were enhanced by hDbs, a positive control of guanine nucleotide exchange factors. F, The guanine nucleotide exchange activity of ROP7 was enhanced by DUF315-GEF2.
RopGEFs catalyze nucleotide replacement on or disassociation from ROPs. The PRONE/DUF315 domains of RopGEFs exhibit GEF activities toward ROPs, and the activities are regulated by the variable N- and C-terminal regions (Gu et al., 2006; Berken et al., 2005). Therefore, we examined the GEF activity of the DUF315 domains of RopGEF2 (hereafter designated as DUF315-GEF2) toward ROP7 using the method reported by Gu et al. (2006). ROP7 was preloaded with unlabeled-GDP. To start the nucleotide exchange reaction, fluorescently labeled n-methylanthraniloyl (mant)-GTP was added to the reaction buffer and the fluorescent values were recorded every 0.2 s for 600 s by a spectrofluorometer. To investigate the effect of RopGEFs on the guanine nucleotide exchange rate of ROPs, the DUF315 domain of RopGEF proteins was added to the reaction buffer containing the ROP protein, unlabeled-GDP, and mant-GTP before the recording of fluorescent intensities. The results showed that the intrinsic guanine nucleotide exchange rates of ROP7 increased with the extension of the reaction time and that the reaction rates increased with elevated concentrations of ROP7 (Fig. 2D). Furthermore, the rate of mant-GTP incorporation into ROP7 was similar to its incorporation into the same concentration of CDC42, a positive control of small G protein (Fig. 2, C and D). The hDbs protein is an established guanine nucleotide exchange factor toward CDC42 (Murga-Zamalloa et al., 2010); therefore, we used hDbs here as a positive control of guanine nucleotide exchange factor. The incorporation rate of mant-GTP into CDC42 with hDbs treatment increased significantly relative to the incorporation rate into CDC42 alone, and the incorporation rate into ROP7 with hDbs treatment also increased greatly relative to the incorporation rate into ROP7 alone (Fig. 2E). We next analyzed the GEF activities of DUF315-GEF2 toward ROP7 and found that the GEF activity toward ROP7 increased with the elevation of DUF315-GEF2 concentrations (Fig. 2F). As a control, RopGEF2 or RopGEF4 alone without ROP GTPase did not induce fluorescent increase (Supplemental Fig. S3). These results clearly demonstrated that ROP7 has intrinsic guanine nucleotide exchange activity, and RopGEF2 enhances the activity of ROP7.
ropgef2 Mutants Exhibit Similar Stomatal Response as rop7 under White Light
Because RopGEF2 interacted with ROP7 and accelerated its intrinsic guanine nucleotide exchange rate, we next investigated whether RopGEF2 plays a similar role as ROP7 in light-induced stomatal opening. We first confirmed the guard cell expression of RopGEF2 by examining GUS expression in the RopGEF2pro:GUS transgenic lines and found that RopGEF2 was expressed in guard cells (Fig. 3C). We identified two T-DNA insertion ropgef2 mutants (Fig. 3A) and examined the expression of RopGEF2 in the mutants by quantitative RT-PCR. The results showed that the expression of RopGEF2 was greatly reduced in ropgef2-1 and ropgef2-2 mutants (Fig. 3B). We then analyzed the stomatal opening of ropgef2-1 and ropgef2-2 with white light treatment and found that, after 2 h light illumination, ropgef2-1 and ropgef2-2 exhibited larger stomatal apertures than the wild type (Fig. 3D). To obtain the complemented lines, RopGEF2 cDNA driven by its native promoter was introduced into ropgef2-1 mutant. The stomatal apertures of two independent complemented lines were nearly identical to those of the wild type (Fig. 3E), indicating that the stomatal phenotypes of the ropgef2 mutants were caused by the mutations in RopGEF2. In addition, we examined the stomatal conductance by a portable photosynthesis system and found that the stomatal conductance of ropgef2-1 was significantly higher than that of wild type, and the two complemented lines showed similar stomatal conductance to wild type (Fig. 3F). These results clearly demonstrated that ropgef2 mutants have similar stomatal response to rop7 under white light.
Figure 3.
RopGEF2 plays a negative role in white light-induced stomatal opening as ROP7. A, Structures of RopGEF2 gene and the T-DNA insertion sites of ropgef2-1 and ropgef2-2 mutants. B, Expression of RopGEF2 in ropgef2-1 and ropgef2-2 mutants was greatly lower than that of the wild type. The error bars represent the means ± se from three biological replicates (Student’s t test, n = 3, **P < 0.01). C, The GUS signals detected in the epidermis of the RopGEF2pro:GUS lines. D and E, Stomatal aperture in ropgef2-1 and ropgef2-2 mutants (D) and the complemented ropgef2-1 lines (Com-10 and Com-11; E) under white light. Epidermal peels with closed stomata were illuminated with white light (150 μmol m−2 s−1) for 2 h. The error bars represent the means ± se from three biological replicates (Student’s t test, n = 150, **P < 0.01). F, The stomatal conductance of ropgef2-1 and complemented lines. The intact leaves of 4- to 6-week-old plants were measured after 2 h white light illumination (150 μmol m−2 s−1). The error bars represent the means ± se from three biological replicates (Student’s t test, n = 15, **P < 0.01).
RopGEF2-ROP7 Plays a Negative Role in Red Light-Induced Stomatal Opening and Acts Genetically Downstream of phyB
Both blue and red light induce stomatal opening, we next determined whether RopGEF2-ROP7 participates in blue or red light signaling pathway. We analyzed the stomatal responses with blue or red light illumination and found that stomatal apertures of the rop7 and ropgef2-1 mutants were not different from the wild type under blue light (Fig. 4A), whereas the stomatal apertures of rop7 and ropgef2-1 mutants were significantly larger than those of the wild type under red light, and the complemented lines exhibited similar stomatal apertures to wild type (Fig. 4, B and C). These results demonstrated that RopGEF2-ROP7 play a negative role in red light-induced stomatal opening.
Figure 4.
RopGEF2 and ROP7 play a negative role in phyB-mediated, red light-induced stomatal opening. A, The ropgef2-1 and rop7 mutants showed similar stomatal apertures to wild type under blue light. B and C, The ropgef2-1 (B) and rop7 (C) mutants showed larger stomatal apertures than that of wild type under red light, and the complemented lines exhibited similar stomatal apertures to wild type. D and E, The phyB ropgef2-1 (D) and phyB rop7 (E) double mutants showed similar stomatal apertures to ropgef2-1 or rop7 single mutants under white light. Epidermal peels with closed stomata were illuminated with blue light (10 μmol m−2 s−1 in A), red light (50 μmol m−2 s−1 in B and C), or white light (150 μmol m−2 s−1 in D and E) for 2 h. The error bars represent the means ± se from three biological replicates (Student’s t test, n = 150, **P < 0.01).
Evidence shows that phyB is involved in red light-induced stomatal opening (Wang et al., 2010), therefore, we next asked whether RopGEF2-ROP7 act genetically downstream of phyB. The stomatal size of phyB was similar to that of wild type, whereas the stomatal density of phyB was lower than that of wild type (Supplemental Fig. S1, A and B), as reported previously (Kang et al., 2009; Casson and Hetherington, 2014), illustrating that the stomatal aperture of phyB can reflect stomatal opening. We obtained phyB ropgef2-1 and phyB rop7 double mutants by genetic crossing and analyzed the stomatal response of the double mutants. The results showed that the stomatal apertures of the phyB ropgef2-1 and phyB rop7 double mutants under white light were similar to the ropgef2-1 or rop7 single mutant, respectively, but quite different from that of the phyB mutant (Fig. 4, D and E), indicating that RopGEF2-ROP7 act genetically downstream of phyB.
RopGEF4 Functions Redundantly with RopGEF2 in Red Light-Induced Stomatal Opening
Because both the stomata of ropgef2 and ropgef4 mutants had similar response to white light (Supplemental Fig. S2B), we asked whether RopGEF4 and RopGEF2 function redundantly in red light-induced stomatal opening. We first analyzed the stomatal response of ropgef4 single mutants. RopGEF4 was found to be expressed in guard cells in the RopGEF4pro:GUS transgenic lines (Supplemental Fig. S4C). We also identified two T-DNA insertion mutants, and the expression of RopGEF4 was greatly reduced in ropgef4-1 and ropgef4-2 mutants (Supplemental Fig. S4, A and B). The stomatal opening analysis showed that ropgef4-1 and ropgef4-2 mutants exhibited larger stomatal aperture as ropgef2 mutants with white light illumination (Supplemental Fig. S4D), and the phenotype of ropgef4-1 could be rescued by RopGEF4 cDNA driven by its native promoter (Supplemental Fig. S4E). Moreover, the stomatal conductance of ropgef4-1 mutant was also higher than that of wild type under white light (Supplemental Fig. S4G). These results indicated that ropgef4 mutants have similar stomatal response as ropgef2 mutants.
We next examined whether RopGEF4 and RopGEF2 function redundantly in light-induced stomatal opening by generating a ropgef2-1 ropgef4-1 double mutant. The stomatal aperture of ropgef2-1 ropgef4-1 double mutant was much larger than that of ropgef2-1 and ropgef4-1 single mutants (Supplemental Fig. S4F), and the stomatal conductance of the ropgef2-1 ropgef4-1 was also higher than that of each single mutant (Supplemental Fig. S4H), indicating that RopGEF2 and RopGEF4 function redundantly in light-induced stomatal opening. Moreover, the larger stomatal aperture of ropgef4-1 mutant could be observed under red light, not blue light, further supporting that RopGEF4 plays a negative role in red light-induced stomatal opening (Supplemental Fig. S5, A and B). We also obtained the phyB ropgef2-1 ropgef4-1 triple mutant by genetic crossing and found that the triple mutant showed similar stomatal aperture as the ropgef2-1 ropgef4-1 double mutant, but quite different from the phyB mutant, suggesting that RopGEF2/RopGEF4 act genetically downstream of phyB (Supplemental Fig. S5C). Taken together, these results indicated that RopGEF2 and RopGEF4 act redundantly in red light-induced stomatal opening and are situated genetically downstream of phyB.
White Light-Induced Translocation of ROP7 from Soluble Part to Membrane of Cells Was Greatly Reduced in ropgef2-1, ropgef2-1 ropgef4-1, and phyB Mutants
The ROPs target to plasma membrane (PM) upon posttranslational lipid modification, or coupled with their activation (Lavy et al., 2002; Sorek et al., 2007, 2010; Yalovsky 2015). ROP7 and CA-rop7 were found to be localized in the plasma membrane when interacted with the downstream effector AtSCAR2 (Uhrig et al., 2007). Therefore, we investigated whether light induces the translocation of EGFP-ROP7 from cytosol to the membrane. The 35Spro:EGFP-ROP7 lines used for ROP7 translocation analysis exhibited smaller stomatal apertures than that of 35Spro:EGFP line after white light illumination, which were similar to that of CA-rop7, indicating that EGFP-ROP7 is a functional protein (Supplemental Fig. S6). We checked the distribution of ROP7 in membrane and soluble part of cells in35Spro:EGFP-ROP7 transgenic lines by western blotting and found that about one-third of the total EGFP-ROP7 protein was detected in cell membrane in wild type after the seedling was grown in darkness for 8 h. The content of EGFP-ROP7 in membrane increased after 2 h light treatment, consequently the ratio between the membrane and the soluble part (M/S) significantly increased (Supplemental Fig. S7B), indicating that translocation of EGFP-ROP7 from the soluble part to the membrane, which may be PM according to Uhrig et al. (2007), was triggered by white light. However, the EGFP signal was not detectable in cell membrane of 35Spro:EGFP line both in darkness and light (Supplemental Fig. S7A). To determine whether the translocation of ROP7 was regulated by RopGEF2 and RopGEF4, we measured the EGFP-ROP7 distribution in ropgef2-1 and ropgef2-1 ropgef4-1 mutants before and after light treatment and found that the proportion of EGFP-ROP7 in the membrane of ropgef2-1 in darkness was significantly lower than that of wild type. Although the proportion of EGFP-ROP7 in cell membrane of ropgef2-1 increased after light illumination, the M/S value was greatly lower than that of wild type, indicating that the translocation of EGFP-ROP7 from the soluble part to the membrane was partially mediated by RopGEF2. In the ropgef2-1 ropgef4-1 double mutant, EGFP-ROP7 was not detectable in the cell membrane in either the darkness or light, indicating that the translocation of ROP7 to membrane was severely inhibited (Supplemental Fig. S7B). These results demonstrated that RopGEF4 functions redundantly with RopGEF2 in mediating white light-induced ROP7 translocation to membrane, and the translocation of ROP7 may be due to its activation.
Because phyB acts upstream of RopGEF2/4-ROP7 in light-induced stomatal opening, we also checked the translocation of EGFP-ROP7 in phyB mutant before and after white light treatment. The results showed that, similar to the ropgef2-1 ropgef4-1 double mutant, translocation of EGFP-ROP7 in the phyB mutant was also severely inhibited (Supplemental Fig. S7B), indicating that the translocation of ROP7 to membrane was mediated by phyB, which further supported that RopGEF2/RopGEF4-ROP7 act downstream of phyB.
ROP2 Functions Redundantly with ROP7 in Red Light-Induced Stomatal Opening, and the Activities of ROP7 and ROP2 Were Enhanced by Both RopGEF2 and RopGEF4
Both ROP2 and ROP7 act as negative regulators in light-induced stomatal movements, therefore, we investigated whether ROP2 and ROP7 function redundantly in light-induced stomatal opening. To test this possibility, we generated the rop2 rop7 double mutant by genetic crossing and found that after 2 h white light illumination, both rop2 and rop7 had larger stomatal apertures than the wild type, whereas the rop2 rop7 double mutant had a much larger aperture than the rop2 and rop7 single mutants. The differences between the rop2 rop7 double mutant and the rop2 or rop7 single mutants were significant, indicating that ROP2 and ROP7 function redundantly in this process (Fig. 5A). In consistence with the stomatal apertures, the stomatal conductance of rop2 rop7 double mutant was also significantly higher than that of rop2 and rop7 single mutants under white light (Supplemental Fig. S8). We then examined the stomatal responses of the rop2 mutant to red or blue light and found that the difference of stomatal aperture between rop2 and wild type was significant under red light, whereas the difference became insignificant under blue light, supporting that ROP2 also plays a negative role in red light-induced stomatal opening, and acts redundantly with ROP7 in this process (Fig. 5B).
Figure 5.
ROP2 functions redundantly with ROP7 in regulation of red light-induced stomatal opening. A, The rop2 rop7 double mutant showed larger stomatal aperture than rop2 and rop7 single mutants. B, The rop2 mutant showed a larger stomatal aperture than the wild type with red light treatment and similar stomatal aperture to the wild type with blue light treatment. Epidermal peels with closed stomata were illuminated with white light (150 μmol m−2 s−1) in A, and red light (50 μmol m−2 s−1) or blue light (10 μmol m−2 s−1) in B for 2 h. The bars represent the means ± se from three biological replicates (Student’s t test, n = 150, *P < 0.05 and **P < 0.01). C, RopGEF2 physically interacted with ROP7 in the in vitro pull-down assay. D, The guanine nucleotide exchange activity of ROP2 increased with the increasing concentrations of ROP2 protein. E, The guanine nucleotide exchange activity of ROP2 was enhanced by hDbs, a positive control of guanine nucleotide exchange factors. F, The guanine nucleotide exchange activity of ROP2 was enhanced by DUF315-GEF2.
To explore whether RopGEF2 interacts with ROP2, we performed an in vitro pull-down assay and found that MBP-RopGEF2 interacted with nucleotide-free and GDP-loaded GST-ROP2 (Fig. 5C), as with ROP7 (Fig. 2B). These results indicated that both ROP2 and ROP7 directly interact with RopGEF2. We next investigated whether the DUF315 domains of RopGEF2 activates ROP2. First, ROP2 showed an intrinsic guanine nucleotide exchange activity, which increased with the extension of the reaction time and elevated concentrations of ROP2 (Fig. 5D). In addition, hDbs effectually accelerated the incorporation rate of mant-GTP into ROP2 (Fig. 5E). DUF315-GEF2 also exhibited GEF activity toward ROP2, and the GEF activities increased with the elevation of protein concentrations (Fig. 5F), indicating that ROP2 is activated by RopGEF2, as is ROP7 (Fig. 2F). Similarly, the DUF315 domain of RopGEF4 also exhibited GEF activity toward both ROP7 and ROP2 (Supplemental Fig. S9, A and B). These results clearly demonstrated that the intrinsic guanine nucleotide exchange activities of both ROP7 and ROP2 were enhanced by RopGEF2 and RopGEF4. Taken together, these results suggested that ROP2 and ROP7 function redundantly in red light-induced stomatal opening, and their intrinsic guanine nucleotide exchange activities were enhanced by both RopGEF2 and RopGEF4, implying that RopGEF2/4-ROP7/2 play a negative role in red light-induced stomatal opening.
RopGEF2 Interacts with phyB in Light and Darkness, and the GEF Activity of RopGEF2 Is Enhanced by phyB in Light
Because RopGEF2/RopGEF4 acts genetically downstream of phyB, we asked whether phyB directly interacts with RopGEF2/RopGEF4 and affects their GEF activities. To test this possibility, we first performed yeast two-hybrid assay with RopGEF2 fused to the GAL4 activation domain and phyB fused to the GAL4 binding domain. Phycocyanobilin (PCB) was supplemented to the SC-HLT media as a phytochromobilin analog to reconstitute the photoactive phyB (Pfr form; Luo et al., 2014). The results showed that phyB interacted with RopGEF2 in light and darkness in yeast (Fig. 6A). We then performed an in vitro pull-down assay with MBP-RopGEF2 and phyB-GFP from the 35Spro:phyB-GFP plants grown in light or darkness and found that MBP-RopGEF2 directly interacted with phyB-GFP in light and darkness (Fig. 6B). Furthermore, we examined the interaction between phyB and RopGEF2 in Arabidopsis using the RopGEF2Pro:EGFP-RopGEF2 transgenic lines. The coimmunoprecipitation (co-IP) results showed that immunoprecipitation of EGFP-RopGEF2 pulled down endogenous phyB in both light and darkness (Fig. 6C). These results provided convincing evidence that phyB directly interacts with RopGEF2 in both light and darkness, suggesting that RopGEF2 interacts with both Pfr and Pr forms of phyB.
Figure 6.
phyB physically interacts with RopGEF2 in light and darkness, and enhances the GEF activity of RopGEF2 toward both ROP7 and ROP2 in light. A and B, The interaction between phyB and RopGEF2 was detected in yeast and by the in vitro pull-down assay in light (L) and darkness (D). Yeast strains were grown on synthetic complete media without Trp and Leu (SC-LT) or without Trp, Leu, and His (SC-HLT, containing 25 μm PCB) for 4 to 5 d; phyB-GFP was obtained from a 35Spro:phyB-GFP seedling grown in light or darkness. C, phyB interacted with RopGEF2 in vivo. Four-day-old RopGEF2pro:EGFP-RopGEF2 and 35Spro:EGFP seedling grown in light (L) or darkness (D) were subjected to a co-IP assay with anti-phyB antibody. The immunoprecipitates were detected with anti-phyB and anti-GFP antibodies, respectively. D and E, The full-length RopGEF2 showed GEF activities toward ROP7 (D) and ROP2 (E), and the GEF activities of RopGEF2 increased toward ROP7 (D) and ROP2 (E) after being incubated with phyB-GFP from a 35Spro:phyB-GFP seedling grown in light (phyB-light), while phyB-GFP (phyB-light) did not directly enhance the guanine nucleotide exchange activities of ROP7 (D) and ROP2 (E) without RopGEF2. F and G, phyB-GFP from a 35Spro:phyB-GFP seedling grown in darkness (phyB-dark) did not enhance the GEF activities of RopGEF2 toward ROP7 (F) and ROP2 (G).
The direct interaction between phyB and RopGEF2 suggested that phyB might affect the GEF activity of RopGEF2. It has been reported that the variable C- and N-terminal regions of RopGEF1 regulates its GEF activity by an autoinhibitory mechanism (Gu et al., 2006; Shichrur and Yalovsky, 2006), and that phyB might affect the GEF activity by regulating the C- or N-terminal inhibitory domains of RopGEF2. To test this possibility, we obtained the full-length RopGEF2 protein from Escherichia coli and found that the intrinsic guanine nucleotide exchange rates of both ROP7 and ROP2 were accelerated by full-length RopGEF2 (Fig. 6, D and E). However, the GEF activity of the full-length RopGEF2 was lower than the DUF315 domain of the protein (DUF315-GEF2 in Figs. 2F and 5F) at the same concentrations, suggesting that the variable regions of RopGEF2 are inhibitory to the GEF activity of DUF315 domain. Next, we checked whether phyB enhanced the GEF activity of RopGEF2. The fused phyB-GFP protein was immunoprecipitated by GFP antibody from the 35Spro:phyB-GFP transgenic lines grown in light or darkness. The full-length RopGEF2 protein was incubated with phyB-GFP, and then centrifuged. The supernatant containing RopGEF2 was added to the reaction mixture and started to record the change of fluorescent intensity. The RopGEF2, which had been incubated with phyB-GFP in light, showed higher GEF activities toward both ROP7 and ROP2 than that of RopGEF2 alone (Fig. 6, D and E), whereas RopGEF2, which had been incubated with phyB-GFP in darkness, exhibited similar GEF activities toward both ROP7 and ROP2 as RopGEF2 alone (Fig. 6, F and G). phyB-GFP in light did not accelerate the intrinsic guanine nucleotide exchange rates of ROP7 and ROP2 without RopGEF2 (Fig. 6, D and E). These results demonstrated that only photoactive phyB enhances the GEF activity of RopGEF2. Taken together, these results demonstrated that although phyB directly interacts with RopGEF2 in vivo and in vitro under light and darkness, only photoactive phyB contains the ability to enhance the GEF activity of RopGEF2.
phyB Shows No Direct Interaction with RopGEF4 and Does Not Affect the GEF Activity of RopGEF4
The functional redundancy between RopGEF4 and RopGEF2 in red light-induced stomatal opening prompted us to test whether phyB also directly interacted with RopGEF4 and enhanced the GEF activity of RopGEF4. The results from yeast two-hybrid and in vitro pull-down assays showed that phyB did not directly interact with RopGEF4 in light and darkness (Supplemental Fig. S10, A and B). We also detected whether the GEF activity of RopGEF4 was enhanced by phyB and found that the full-length RopGEF4 contained GEF activity toward both ROP7 and ROP2 as RopGEF2; however, the GEF activity was not enhanced by phyB with light irradiation (Supplemental Fig. S10, C and D). These results demonstrated that RopGEF4 does not directly interact with phyB, and the GEF activity of RopGEF4 is not directly enhanced by phyB.
DISCUSSION
Light is an important environmental stimulus that regulates stomatal movements. Red light-induced stomatal opening has a close relationship with mesophyll photosynthesis (Schwartz and Zeiger, 1984; Tominaga et al., 2001). It has been shown that red light receptor phyB mediates red light-enhanced photosynthesis, which will increase the ATP level and osmotic substances for stomatal opening (Guo et al., 2016). In addition, guard cell-specific transcription factor MYB60 has been revealed to play a positive role in light-induced stomatal opening (Cominelli et al., 2005). The expression of MYB60 was inhibited by phyB mutation, and enhanced by phyB overexpression (Wang et al., 2010). These reports supported that phyB activates some processes to enhance stomatal opening. Small G protein ROP2 has been reported to be a negative regulator of light-induced stomatal opening, which prevents the excessive stomatal opening (Jeon et al., 2008). However, the upstream RopGEFs that activate ROPs have not been identified, and it is unclear which photoreceptor is required for the action of RopGEFs-ROPs in this process. In this research, our genetic, physiological, and biochemical evidence supports that RopGEF2-ROP7/ROP2 directly activated by phyB plays a negative role in red light-induced stomatal opening. Photoactive phyB directly interacts with RopGEF2 and enhances the GEF activity of RopGEF2 toward both ROP7 and ROP2. At the same time, RopGEF4 acts redundantly with RopGEF2 by activating ROP7 and ROP2, whereas the GEF activity of RopGEF4 is not directly activated by phyB. Data from the previous reports and this article supported that the red light receptor phyB not only activates some positive processes to enhance stomatal opening, but also triggers the negative mechanism to prevent the excessive stomatal opening.
ROP7 and ROP2 Have Overlapping Functions as Negative Regulators in White Light-Induced Stomatal Opening
The negative regulatory mechanism in light-induced stomatal opening prevents the excessive opening of the stomata and consequently prevents excessive water loss. So far, only ROP2 has been found to act as a negative regulator in light-induced stomatal opening (Jeon et al., 2008). Mutation in ROP7, another member of the ROP family, led to an enhanced stomatal opening and higher stomatal conductance in response to white light; by contrast, the line expressing the constitutively active form of rop7 (CA-rop7) showed reduced stomatal apertures and lower stomatal conductance under white light (Fig. 1, B–D). Moreover, white light induced translocation of EGFP-ROP7 from the soluble to the membrane, and the translocation depended on RopGEF2/RopGEF4 (Supplemental Fig. S7B), suggesting that the translocation of ROP7 to membrane may be due to its activation. The translocation of active ROP7 to membrane is similar to the translocation of ROP2 in guard cells of Vicia faba: the constitutive active form of ROP2 (CA-rop2) was observed in PM, and dominant negative form of ROP2 (DN-rop2) was found in cytosol; furthermore, light induced the translocation of ROP2 from cytosol to PM (Jeon et al., 2008). These results clearly demonstrated that, similar to the role of ROP2, ROP7 acts as a negative regulator in white light-induced stomatal opening. The similar functions of ROP7 and ROP2 in white light-induced stomatal opening suggest that the two genes are likely to have functional redundancy. The rop2 rop7 double mutant exhibited much larger stomatal apertures and higher stomatal conductance than the rop2 or rop7 single mutants under white light (Fig. 5A; Supplemental Fig. S8), supporting that ROP7 and ROP2 have overlapping function in regulation of white light-induced stomatal opening.
Both RopGEF2 and RopGEF4 Act Upstream of ROP7 and ROP2 by Activating Their Guanine Nucleotide Exchange Activities
The nucleotide exchange activities of ROPs are enhanced by RopGEFs, which have been revealed to play roles in regulation of pollen growth (Gu et al., 2006; Zhang and McCormick, 2007), root hair growth (Won et al., 2009; Duan et al., 2010), and root development (Shin et al., 2010). Notably, RopGEF1 and RopGEF4 have been found to play negative roles in ABA-induced stomatal closure by activating ROP11 (Li and Liu, 2012; Li et al., 2016). However, the RopGEFs involved in light-induced stomatal opening have not been reported. In this study, we searched for the RopGEFs that participate in light-induced stomatal opening by using ROP7 as bait. Although ROP7 interacts with RopGEF1, 2, 3, 4, 5, and 7 in yeast (Fig. 2A), only ropgef2 and ropgef4 mutants showed similar stomatal responses to white light as rop7 (Fig. 3, D and F; Supplemental Fig. S4, D and G). Furthermore, the ropgef2-1 ropgef4-1 double mutant showed much larger stomatal apertures and higher stomatal conductance than the ropgef2-1 and ropgef4-1 single mutants (Supplemental Fig. S4, F and H), and also showed the more severe inhibition of light-induced EGFP-ROP7 translocation to membrane in the ropgef2-1 ropgef4-1 double mutant than that in ropgef2-1 single mutant (Supplemental Fig. S7B), supporting that RopGEF2 and RopGEF4 act redundantly as negative regulators in white light-induced stomatal opening.
The DUF315 domains of RopGEFs exhibit GEF activities toward ROPs, and the activities are regulated by the variable N- and C-terminal regions (Gu et al., 2006; Shichrur and Yalovsky, 2006). The GEF activities of RopGEF2 and RopGEF4 toward ROP7 and ROP2 were supported by this evidence: ROP7 and ROP2 exhibited intrinsic guanine nucleotide exchange activities, which increased with the elevated concentration of the proteins (Figs. 2D and 5D). The guanine nucleotide exchange rates of ROP7 and ROP2 were greatly accelerated by the DUF315 domains of RopGEF2 (Figs. 2F and 5F) and RopGEF4 (Supplemental Fig. S9, A and B). However, the GEF activities of full-length RopGEF2/RopGEF4 toward both ROP7 and ROP2 were greatly lower than the GEF activities of DUF315 domain in RopGEF2/RopGEF4 at the same concentrations (Fig. 6, D and E; Supplemental Fig. S10, C and D), suggesting that the variable regions of RopGEF2/RopGEF4 are inhibitory to the GEF activity of DUF315 domains, and the GEF activities of full-length RopGEF2/RopGEF4 might be regulated by upstream molecules in vivo. These results clearly demonstrated that both RopGEF2 and RopGEF4 enhance the guanine nucleotide exchange activities of ROP7 and ROP2, and the GEF activities of RopGEF2 and RopGEF4 might be regulated by upstream mechanism in plants.
RopGEF2/RopGEF4-ROP7/ROP2 Participate in Red Light-Induced Stomatal Opening and Act Genetically Downstream of phyB
Both red and blue light induce stomatal opening through different mechanisms that are mediated by special receptors. The stomatal opening analysis with red or blue light irradiation showed that the larger stomatal apertures of rop7, rop2, ropgef2, and ropgef4 could be observed in red light, not blue light (Figs. 4, A–C, and 5B; Supplemental Fig. S5, A and B). In Arabidopsis, blue light-induced stomatal opening is mediated by phot1 and phot2 (Kinoshita et al., 2001), whereas red light-induced stomatal opening is mainly mediated by phyB (Wang et al., 2010). Because RopGEF2/RopGEF4-ROP7/ROP2 play a negative role in red light-induced stomatal opening, it is possible that they act genetically downstream of phyB. Stomatal opening responses of the phyB ropgef2-1 ropgef4-1 triple mutant was similar to the ropgef2-1 ropgef4-1 double mutant in white light, whereas it was quite different from the phyB single mutant, demonstrating that RopGEF2/RopGEF4 act genetically downstream of phyB (Supplemental Fig. S5C). These results provided convincing evidence that RopGEF2/RopGEF4-ROP7/ROP2 play a negative role in red light-induced stomatal opening and are situated genetically downstream of phyB.
phyB Physically Interacts with RopGEF2 in Both Light and Darkness and Enhances Its GEF Activities toward Both ROP7 and ROP2 in Light
Several RopGEFs directly interact with receptors or receptor-like kinases and transmit the signals to downstream ROPs (Kaothien et al., 2005; Zhang and McCormick, 2007; Chang et al., 2013). Notably, PIRF1/RopGEF11 interacted with phyA and phyB in the cytosol in darkness, and the Pr form of phyA enhances the guanine nucleotide exchange activity of RopGEF11 during root development (Shin et al., 2010). As a red light receptor, phyB is essential for light-regulated plant growth and development (Franklin and Quail, 2010). Several lines of evidence showed that phyB is also involved in the regulation of defense and stress responses (González et al., 2012; Liu et al., 2012; Moreno and Ballaré, 2014; Wang et al., 2016), likely to be related to its regulation of stomatal development (Kang et al., 2009; Liu et al., 2012; Casson and Hetherington, 2014) and movements (Wang et al., 2010; González et al., 2012). Because RopGEF2/RopGEF4 play a negative role in red light-induced stomatal opening, and act downstream of phyB, RopGEF2/RopGEF4 may act as a link between phyB and ROP7/ROP2. Our results showed that RopGEF2 directly interacted with phyB, which was detected by the yeast two-hybrid assay, in vitro pull-down assay, and in vivo co-IP in both light and darkness (Fig. 6, A–C), demonstrating that the interaction is independent of light. It is worth noting that the GEF activities of RopGEF2 toward both ROP7 and ROP2 were enhanced by phyB from plants grown in light, not in the dark (Fig. 6, D–G), suggesting that the enhancement on GEF activity of RopGEF2 depends on the activation of phyB by light. However, in the same experimental conditions, we could not detect the physical interaction between phyB and RopGEF4, and phyB did not directly enhance the GEF activity of RopGEF4 toward ROP7 and ROP2 in light. These results demonstrated that RopGEF2 and RopGEF4 act redundantly as negative regulators in red light-induced stomatal opening by activating ROP7 and ROP2, and RopGEF2 is directly activated by photoactive phyB, whereas RopGEF4 is not directly activated by active phyB.
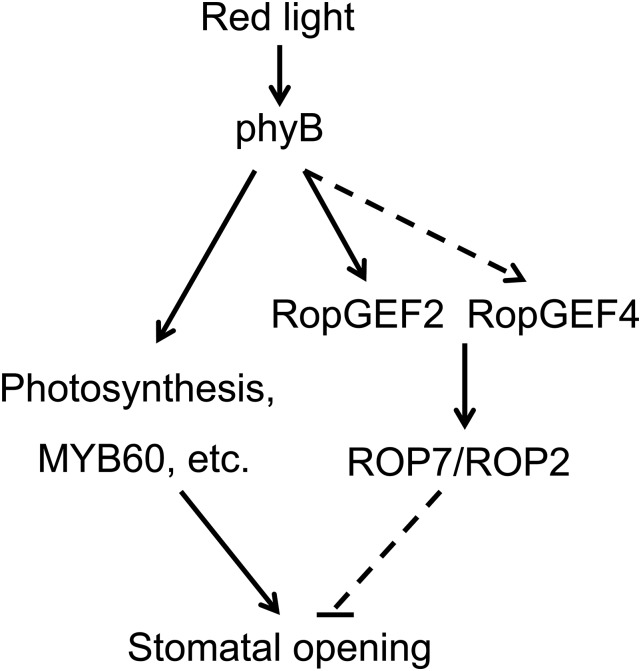
Upon red light irradiation, the Pr form of phyB was converted to the biologically active Pfr form, which will be translocated from the cytoplasm to the nucleus. However, the nucleus import of phyB needs transport facilitators (Pfeiffer et al., 2012). In the phyB-GFP line, the GFP signal in nucleus was not clear at darkness. After 2 h red light illumination, the nucleus fluorescent intensity increased, and the phyB-GFP signal was detectable in the periphery of the cells (Yamaguchi et al., 1999). Therefore, the photoactive Pfr form of phyB will stay in the cytoplasm for a period of time. In this research, the incubation time of RopGEF2 with phyB-GFP from plants grown under light was 10 min, and then the RopGEF2 in the supernatant has a higher GEF activity than RopGEF2 alone. We proposed that Pr form of phyB is activated by red light irradiation, and subsequently the Pfr form of phyB in cytoplasm enhances the GEF activity of RopGEF2, and RopGEF4 is activated by phyB indirectly. The Pfr form of phyB was then translocated to the nucleus, and active RopGEF2/RopGEF4 enhance the guanine nucleotide exchange activity of ROP7/ROP2, which will play a negative role to prevent the excessive opening of stomata (Fig. 7).
Figure 7.
A schematic drawing showing that RopGEF2/RopGEF4-ROP7/ROP2, activated by phyB, play a negative role in red light-induced stomatal opening.
The interaction between phyB and RopGEF2 is independent of light, whereas the enhancement of GEF activity of RopGEF2 by phyB is dependent on light, implying that the domains for interaction and activation in phyB may be different. Further research needs to determine which domain in phyB is responsible for its interaction with RopGEF2, and which domain is responsible for the activation on RopGEF2. Evidence shows that phosphorylation of phyB in Ser-86 inhibits its function by accelerating the dark reversion (Medzihradszky et al., 2013; Hajdu et al., 2015), therefore, it is interesting to investigate whether the phosphorylation of phyB affects its activation on the GEF activity of RopGEF2. ABA induced the degradation of RopGEF1 and RopGEF2 (Li et al., 2016; Zhao et al., 2015), and the negative mechanism of RopGEF2-ROP7/ROP2 triggered by phyB contributes to the circadian rhythm of stomatal movements. Further research needs to examine whether RopGEF2 undergoes degradation and de novo synthesis along with the daily rhythm of stomatal movements.
MATERIALS AND METHODS
Plant Materials and Growth Conditions
The plant materials used in this study were Arabidopsis (Arabidopsis thaliana) with the Col-0 background. rop2 (SALK_055328), ropgef2-1 (SALK_130229), rop7 (GK-212D04), ropgef2-2 (GK-061B10), ropgef4-1 (Salk_107520), ropgef4-2 (Sail_184_C08), ropgef1 (GK-586B11), and ropgef3 (SALK_021751) were T-DNA insertion mutants. The phyB-9 mutant had a premature stop codon at the 396th amino acid, and the phyB-9 mutant with Ler background was backcrossed with Col-0 wild type for three times (named “phyB mutant” in this article). The mutants were identified with the primers shown in Supplemental Table S1. To check the expression levels of RopGEF2 and RopGEF4 in the ropgef2 and ropgef4 mutants, total RNA from leaves of 3- to 4-week-old plants was isolated using TRIzol (Invitrogen), and cDNA was prepared using the PrimeScript RT reagent kit (Takara). The relative expression of the two genes in the corresponding mutants was performed using SYBR Premix ExTaq (Takara). The primers used for quantitative RT-PCR are shown in Supplemental Table S1. The quantitative RT-PCR was conducted in a Real-Time PCR System (ABI PRISM 7500; Applied Biosystems) according to the manufacturer’s recommendations. Each experiment was repeated three times. Seedlings were grown in a greenhouse under long-day conditions (16-h-light/8-h-dark cycle), with a photon flux density of 150 μmol m−2 s−1 and a temperature of 18°C to 22°C.
Determination of Stomatal Density and Size
Fully expanded leaves from 3- to 4-week-old plants were collected to determine the stomatal density and size. The abaxial epidermal strips were peeled and placed on a slide. Images (×200 magnification) were photographed with a microscope (Eclipse 80i; Nikon). The stomatal density (stomatal number per area) and stomatal size (the length between the junctions of the two guard cells) was determined. Epidermal strips were peeled from 10 independent leaves, and three images were taken from one strip. Stomatal density was calculated from the data of 30 images, and stomatal size was the average value from 30 stomata. The data are presented as the means ± se (n = 30).
Stomatal Aperture Measurements under Various Light Treatments
Stomatal aperture assays were performed essentially as described in Li et al. (2009). Briefly, 3- to 4-week-old fully expanded rosette leaves were used for the stomatal assay. To close the stomata, the leaves were collected and incubated in MES buffer (30 mm KCl, 0.1 mm CaCl2, 10 mm MES, pH 6.1) in the dark for 1 h, and then the epidermis was peeled and illuminated with white (150 μmol m−2 s−1), red (50 μmol m−2 s−1), or blue light (10 μmol m−2 s−1) for the indicated time in each figure. The stomatal apertures were measured under a microscope. Fifty stomata were selected randomly for three independent replicates before or after light treatments. The data are presented as the means ± se (n = 150).
Measurements of Leaf Stomatal Conductance
The plants of each genotype were grown under long-day conditions (16 h light/8 h dark) for 4 to 6 weeks. Measurement of stomatal conductance of intact leaves was performed after 2 h white light illumination (150 μmol m−2 s−1) using a portable photosynthesis system (LI-6400XT; LI-COR). Five leaves were selected for three independent replicates. The data are presented as the means ± se (n = 15).
GUS Staining to Determine the Expression of ROP7, RopGEF2, and RopGEF4 in Guard Cells
To obtain the ROP7pro:GUS, RopGEF2pro:GUS, and RopGEF4pro:GUS constructs, the promoter regions of ROP7, RopGEF2, and RopGEF4 were amplified from genomic DNA using the primers shown in Supplemental Table S1. The promoter sequences of ROP7, ROPGEF2, and ROPGEF4 were introduced into the pCAMBIA1391 vector, which was then transformed into Arabidopsis using the floral dipping method (Clough and Bent, 1998). The transformants were selected from MS media containing hygromycin (50 mg·L−1) and transferred to soil for further growth and GUS staining. Abaxial epidermis was peeled from 3- or 4-week-old fully expanded rosette leaves, and the GUS activity was checked using the method from Jefferson (1987).
Obtaining the Complemented rop7, ropgef2, and ropgef4 Lines
To obtain the complemented lines of rop7, ropgef2, and ropgef4 mutants, we made the ROP7pro:ROP7, RopGEF2pro:RopGEF2, and RopGEF4pro:RopGEF4 constructs. The promoter regions of ROP7, RopGEF2, and RopGEF4 were obtained as described for the GUS expression constructs for the three genes. The coding regions of ROP7, RopGEF2, and RopGEF4 were amplified from wild-type cDNA with the primers shown in Supplemental Table S1. The promoter and coding region of each gene were integrated into the pCAMBIA1300 vector and introduced into the corresponding mutants. The homozygous hygromycin-resistant mutants were selected in the T3 generation.
Measuring ROP7 Translocation from Cytosol to the Membrane
The coding region of ROP7 was fused with the pEGAD vector. The EGFP-ROP7 construct driven by the CaMV 35S promoter was introduced into wild type, phyB, ropgef2-1, and ropgef2-1 ropgef4-1 mutants. To measure the translocation of EGFP-ROP7 from the soluble part to the membrane, 2-week-old seedlings were collected by the end of 8 h dark period and after 2 h light illumination, and the soluble part and membrane were separated essentially as described by Zhang et al. (2016). In brief, 0.5 mg seedlings was ground and dissolved in 500 μL lysis buffer (50 mm Tris, pH 7.5, 20 mm NaF, 150 mm NaCl, 5 mm DTT, 1 mm PMSF, 0.1% NP40, and 1× proteinase inhibitor cocktail), and centrifuged at 10,000g for 20 min, and then the supernatant was spun at 60,000g for 60 min. The microsome pellet (membrane proteins) was resuspended with 500 μL lysis buffer. The same volume of membrane proteins and soluble proteins was loaded for immunoblotting analysis.
Guanine Nucleotide Exchange Assay
To investigate the guanine nucleotide exchange activity of ROPs and the effect of RopGEFs on the activities of ROPs, the cDNA sequences of ROP7, ROP2, and the DUF315 domains or full length of RopGEF2 and RopGEF4 were amplified and integrated into the pMAL-c2X vector with a MBP tag and introduced into Escherichia coli BL21 cells. Protein expression, purification, and guanine nucleotide exchange assays were essentially carried out according to the methods described by Gu et al. (2006). The MBP-RopGEFs and MBP-ROPs fusion proteins were expressed in E. coli BL21 cells and the MBP tag was cleaved using Factor Xa (New England Biolabs). The guanine nucleotide exchange activity of different concentrations of ROPs or CDC42 (a positive control of ROPs; Cytoskeleton), and the effect of RopGEFs or hDbs (a positive control of GEFs; Cytoskeleton) on the activity of ROPs was measured using a spectrofluorometer (F-7000; Hitachi). To examine the effect of phyB on the GEF activities of RopGEF2 and RopGEF4, phyB-GFP protein immunoprecipitated by GFP antibody from the 35Spro:phyB-GFP line grown in light or darkness was mixed with RopGEF2 or RopGEF4 for 10 min, centrifuged, and then the supernatant containing RopGEF2 or ROpGEF4 was added to the reaction mixture. The changes in the fluorescent intensity were recorded every 0.2 s for 600 s at an excitation of 360 nm and an emission of 440 nm.
Yeast Two-Hybrid Analysis
Yeast two-hybrid analysis was performed according to the Yeast Protocols Handbook. ROP7 or phyB were fused to the GAL4 DNA binding domain (BD) in the bait vector pGBKT7. ROPGEFs were fused to the GAL4 DNA activating domain (AD) in the prey vector pGADT7. pGBKT7-ROP7 and pGADT7-RopGEFs, or pGBKT7-phyB and pGADT7-RopGEFs, were cotransformed into AH109 cells cultured on synthetic complete media lacking Leu and Trp (SC-LT) for 4 to 5 d at 30°C. The transformants were then spotted onto synthetic complete media lacking Leu, Trp, Ade, and His (SC-AHLT) or synthetic complete media lacking Leu, Trp, and His (SC-HLT; Clontech) and the colony growth was checked after 4 to 5 d. For the interaction between phyB and RopGEF2 or RopGEF4, the SC-HLT media was supplemented with 25 μm PCB. All the experiments were repeated at least three times.
In Vitro Protein Interaction Assays
To check the direct interaction between RopGEF2 and ROP7/ROP2, or phyB with RopGEF2/RopGEF4, the MBP-RopGEF2, MBP-RopGEF4, GST-ROP7, and GST-ROP2 fusion proteins were expressed in E. coli BL21 cells and purified. phyB-GFP was obtained from 35Spro:phyB-GFP line. GST-ROPs were preloaded with GDP in nucleotide loading buffer as described by Gu et al. (2006). Approximately 10 μg GST-ROPs, phyB-GFP, or MBP-RopGEFs was used in each experiment. GST-ROP bound to glutathione-conjugated agarose beads (Sangon) were incubated with MBP-RopGEF proteins in the reaction buffer. The protein complex was washed extensively to remove unbound proteins, and then the proteins were separated by SDS-PAGE. The anti-MBP antibody (New England Biolabs), anti-GST antibody (GenScript), and anti-phyB antibody (HangZhou HuaAn Biotechnology; a gift from Prof. Jian-ping Yang, Chinese Academy of Agricultural Sciences) were used to detect the equal loading or interactions.
Protein Extraction and in Vivo Co-IP
To obtain the RopGEF2pro:EGFP-RopGEF2 transgenic lines, the promoter and coding region of RopGEF2 was integrated into the pEGAD vector and introduced into the wild type. Total protein extracts were obtained with lysis buffer (50 mm Tris, pH7.5, 20 mm NaF, 150 mm NaCl, 5 mm DTT, 1 mm PMSF, 0.1% NP40, and 1× proteinase inhibitor cocktail) from 4-d-old RopGEF2pro:EGFP-RopGEF2 or 35Spro:EGFP transgenic lines grown in light or darkness (16 h light/8 h dark). For co-IP of RopGEF2 and phyB, 3 μg anti-GFP antibody (Roche) was added to 20 μL of protein A-sefinose beads (Sangon) incubated for 2 h at 4°C. Then, equal amounts of total protein in 1 mL lysis buffer were incubated with the mixture for 3 h at 4°C. All precipitates were washed three times with lysis buffer, suspended with 2× SDS sample buffer, and boiled for 10 min. All of these experiments were repeated three times.
Accession Numbers
Sequence data for the genes described in this article can be found in the Arabidopsis database under the following accession numbers: RopGEF1 (AT4G38430), RopGEF2 (AT1G01700), RopGEF3 (AT4G00460), RopGEF4 (AT2G45890), ROP7 (AT5G45970), and ROP2 (AT1G20090).
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. The stomatal density and size in rop7, phyB, ropgef2, ropgef4, and ropgef2 ropgef4 mutants.
Supplemental Figure S2. ropgef1and ropgef3 have similar stomatal apertures as wild type, whereas ropgef2 and ropgef4 showed larger stomatal apertures than wild type in response to white light.
Supplemental Figure S3. RopGEF2 or RopGEF4 alone (without ROP GTPase) does not induce the fluorescence increase.
Supplemental Figure S4. RopGEF4 and RopGEF2 have overlapping function in white light-induced stomatal opening.
Supplemental Figure S5. RopGEF4 plays a negative role in phyB-mediated red light-induced stomatal opening.
Supplemental Figure S6. The 35Spro:EGFP-ROP7 lines used for ROP7 translocation experiments exhibit a smaller stomatal aperture than that of the 35Spro:EGFP line after light illumination.
Supplemental Figure S7. The light-induced translocation of ROP7 from the soluble part to the membrane was greatly reduced in ropgef2-1, ropgef2-1 ropgef4-1, and phyB mutants.
Supplemental Figure S8. rop2 and rop7 have higher stomatal conductance than that of wild type, and the rop2 rop7 double mutant has much higher stomatal conductance than that of rop2 and rop7 single mutants.
Supplemental Figure S9. RopGEF4 enhances the guanine nucleotide exchange activity of both ROP7 and ROP2.
Supplemental Figure S10. RopGEF4 does not directly interact with phyB.
Supplemental Table S1. A list of primers used in this study.
Supplementary Material
Acknowledgments
We thank the ABRC for providing T-DNA insertion lines, Professor Hong-Quan Yang (Shanghai Jiaotong University) for providing the pGBKT7-phyB construct, Professor Jian-Ping Yang (Chinese Academy of Agricultural Sciences) for providing the phyB antibody, Professor Akira Nagatani (Kyoto University) for providing the 35Spro:phyB-GFP transgenic line, and Professor Li-Geng Ma (Capital Normal University) for providing the seeds of the phyB mutant.
References
- Araújo WL, Nunes-Nesi A, Osorio S, Usadel B, Fuentes D, Nagy R, Balbo I, Lehmann M, Studart-Witkowski C, Tohge T, et al. (2011) Antisense inhibition of the iron-sulphur subunit of succinate dehydrogenase enhances photosynthesis and growth in tomato via an organic acid-mediated effect on stomatal aperture. Plant Cell 23: 600–627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basu D, Le J, Zakharova T, Mallery EL, Szymanski DB (2008) A SPIKE1 signaling complex controls actin-dependent cell morphogenesis through the heteromeric WAVE and ARP2/3 complexes. Proc Natl Acad Sci USA 105: 4044–4049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berken A, Thomas C, Wittinghofer A (2005) A new family of RhoGEFs activates the Rop molecular switch in plants. Nature 436: 1176–1180 [DOI] [PubMed] [Google Scholar]
- Brembu T, Winge P, Bones AM (2005) The small GTPase AtRAC2/ROP7 is specifically expressed during late stages of xylem differentiation in Arabidopsis. J Exp Bot 56: 2465–2476 [DOI] [PubMed] [Google Scholar]
- Casson SA, Hetherington AM (2014) Phytochrome B is required for light-mediated systemic control of stomatal development. Curr Biol 24: 1216–1221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang F, Gu Y, Ma H, Yang Z (2013) AtPRK2 promotes ROP1 activation via RopGEFs in the control of polarized pollen tube growth. Mol Plant 6: 1187–1201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Naramoto S, Robert S, Tejos R, Löfke C, Lin D, Yang Z, Friml J (2012) ABP1 and ROP6 GTPase signaling regulate clathrin-mediated endocytosis in Arabidopsis roots. Curr Biol 22: 1326–1332 [DOI] [PubMed] [Google Scholar]
- Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Cominelli E, Galbiati M, Vavasseur A, Conti L, Sala T, Vuylsteke M, Leonhardt N, Dellaporta SL, Tonelli C (2005) A guard-cell-specific MYB transcription factor regulates stomatal movements and plant drought tolerance. Curr Biol 15: 1196–1200 [DOI] [PubMed] [Google Scholar]
- Duan Q, Kita D, Li C, Cheung AY, Wu HM (2010) FERONIA receptor-like kinase regulates RHO GTPase signaling of root hair development. Proc Natl Acad Sci USA 107: 17821–17826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franklin KA, Quail PH (2010) Phytochrome functions in Arabidopsis development. J Exp Bot 61: 11–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu Y, Li H, Yang Z (2002) The ROP2 GTPase controls the formation of cortical fine F-actin and the early phase of directional cell expansion during Arabidopsis organogenesis. Plant Cell 14: 777–794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita T, Noguchi K, Terashima I (2013) Apoplastic mesophyll signals induce rapid stomatal responses to CO2 in Commelina communis. New Phytol 199: 395–406 [DOI] [PubMed] [Google Scholar]
- González CV, Ibarra SE, Piccoli PN, Botto JF, Boccalandro HE (2012) Phytochrome B increases drought tolerance by enhancing ABA sensitivity in Arabidopsis thaliana. Plant Cell Environ 35: 1958–1968 [DOI] [PubMed] [Google Scholar]
- Gu Y, Li S, Lord EM, Yang Z (2006) Members of a novel class of Arabidopsis Rho guanine nucleotide exchange factors control Rho GTPase-dependent polar growth. Plant Cell 18: 366–381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu Y, Vernoud V, Fu Y, Yang Z (2003) ROP GTPase regulation of pollen tube growth through the dynamics of tip-localized F-actin. J Exp Bot 54: 93–101 [DOI] [PubMed] [Google Scholar]
- Gu Y, Wang Z, Yang Z (2004) ROP/RAC GTPase: an old new master regulator for plant signaling. Curr Opin Plant Biol 7: 527–536 [DOI] [PubMed] [Google Scholar]
- Guo Z, Wang F, Xiang X, Ahammed GJ, Wang M, Onac E, Zhou J, Xia X, Shi K, Yin X, et al. (2016) Systemic induction of photosynthesis via illumination of the shoot apex is mediated sequentially by phytochrome B, auxin and hydrogen peroxide in tomato. Plant Physiol 172: 1259–1272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hajdu A, Ádám É, Sheerin DJ, Dobos O, Bernula P, Hiltbrunner A, Kozma-Bognár L, Nagy F (2015) High-level expression and phosphorylation of phytochrome B modulates flowering time in Arabidopsis. Plant J 83: 794–805 [DOI] [PubMed] [Google Scholar]
- Hong D, Jeon BW, Kim SY, Hwang JU, Lee Y (2016) The ROP2-RIC7 pathway negatively regulates light-induced stomatal opening by inhibiting exocyst subunit Exo70B1 in Arabidopsis. New Phytol 209: 624–635 [DOI] [PubMed] [Google Scholar]
- Huang GQ, Li E, Ge FR, Li S, Wang Q, Zhang CQ, Zhang Y (2013) Arabidopsis RopGEF4 and RopGEF10 are important for FERONIA-mediated developmental but not environmental regulation of root hair growth. New Phytol 200: 1089–1101 [DOI] [PubMed] [Google Scholar]
- Huang JB, Liu H, Chen M, Li X, Wang M, Yang Y, Wang C, Huang J, Liu G, Liu Y, et al. (2014) ROP3 GTPase contributes to polar auxin transport and auxin responses and is important for embryogenesis and seedling growth in Arabidopsis. Plant Cell 26: 3501–3518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang JU, Jeon BW, Hong D, Lee Y (2011) Active ROP2 GTPase inhibits ABA- and CO2-induced stomatal closure. Plant Cell Environ 34: 2172–2182 [DOI] [PubMed] [Google Scholar]
- Jefferson R. (1987) Assaying chimeric genes in plants: the GUS gene fusion system. Plant Mol Biol Report 5: 387–405 [Google Scholar]
- Jeon BW, Hwang JU, Hwang Y, Song WY, Fu Y, Gu Y, Bao F, Cho D, Kwak JM, Yang Z, Lee Y (2008) The Arabidopsis small G protein ROP2 is activated by light in guard cells and inhibits light-induced stomatal opening. Plant Cell 20: 75–87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang CY, Lian HL, Wang FF, Huang JR, Yang HQ (2009) Cryptochromes, phytochromes, and COP1 regulate light-controlled stomatal development in Arabidopsis. Plant Cell 21: 2624–2641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaothien P, Ok SH, Shuai B, Wengier D, Cotter R, Kelley D, Kiriakopolos S, Muschietti J, McCormick S (2005) Kinase partner protein interacts with the LePRK1 and LePRK2 receptor kinases and plays a role in polarized pollen tube growth. Plant J 42: 492–503 [DOI] [PubMed] [Google Scholar]
- Kinoshita T, Doi M, Suetsugu N, Kagawa T, Wada M, Shimazaki K (2001) Phot1 and phot2 mediate blue light regulation of stomatal opening. Nature 414: 656–660 [DOI] [PubMed] [Google Scholar]
- Kinoshita T, Shimazaki K (2002) Biochemical evidence for the requirement of 14-3-3 protein binding in activation of the guard-cell plasma membrane H+-ATPase by blue light. Plant Cell Physiol 43: 1359–1365 [DOI] [PubMed] [Google Scholar]
- Lavy M, Bracha-Drori K, Sternberg H, Yalovsky S (2002) A cell-specific, prenylation-independent mechanism regulates targeting of type II RACs. Plant Cell 14: 2431–2450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawson T, Lefebvre S, Baker NR, Morison JI, Raines CA (2008) Reductions in mesophyll and guard cell photosynthesis impact on the control of stomatal responses to light and CO2. J Exp Bot 59: 3609–3619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee M, Choi Y, Burla B, Kim YY, Jeon B, Maeshima M, Yoo JY, Martinoia E, Lee Y (2008) The ABC transporter AtABCB14 is a malate importer and modulates stomatal response to CO2. Nat Cell Biol 10: 1217–1223 [DOI] [PubMed] [Google Scholar]
- Lemichez E, Wu Y, Sanchez JP, Mettouchi A, Mathur J, Chua NH (2001) Inactivation of AtRac1 by abscisic acid is essential for stomatal closure. Genes Dev 15: 1808–1816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li JH, Liu YQ, Lü P, Lin HF, Bai Y, Wang XC, Chen YL (2009) A signaling pathway linking nitric oxide production to heterotrimeric G protein and hydrogen peroxide regulates extracellular calmodulin induction of stomatal closure in Arabidopsis. Plant Physiol 150: 114–124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Liu D (2012) ROPGEF1 and ROPGEF4 are functional regulators of ROP11 GTPase in ABA-mediated stomatal closure in Arabidopsis. FEBS Lett 586: 1253–1258 [DOI] [PubMed] [Google Scholar]
- Li Z, Waadt R, Schroeder JI (2016) Release of GTP exchange factor mediated down-regulation of abscisic acid signal transduction through ABA-induced rapid degradation of RopGEFs. PLoS Biol 14: e1002461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin D, Nagawa S, Chen J, Cao L, Chen X, Xu T, Li H, Dhonukshe P, Yamamuro C, Friml J, et al. (2012) A ROP GTPase-dependent auxin signaling pathway regulates the subcellular distribution of PIN2 in Arabidopsis roots. Curr Biol 22: 1319–1325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin D, Ren H, Fu Y (2015) ROP GTPase-mediated auxin signaling regulates pavement cell interdigitation in Arabidopsis thaliana. J Integr Plant Biol 57: 31–39 [DOI] [PubMed] [Google Scholar]
- Liu J, Zhang F, Zhou J, Chen F, Wang B, Xie X (2012) Phytochrome B control of total leaf area and stomatal density affects drought tolerance in rice. Plant Mol Biol 78: 289–300 [DOI] [PubMed] [Google Scholar]
- Luo Q, Lian HL, He SB, Li L, Jia KP, Yang HQ (2014) COP1 and phyB physically interact with PIL1 to regulate its stability and photomorphogenic development in Arabidopsis. Plant Cell 26: 2441–2456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao J, Zhang YC, Sang Y, Li QH, Yang HQ (2005) A role for Arabidopsis cryptochromes and COP1 in the regulation of stomatal opening. Proc Natl Acad Sci USA 102: 12270–12275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matrosova A, Bogireddi H, Mateo-Peñas A, Hashimoto-Sugimoto M, Iba K, Schroeder JI, Israelsson-Nordström M (2015) The HT1 protein kinase is essential for red light-induced stomatal opening and genetically interacts with OST1 in red light and CO2-induced stomatal movement responses. New Phytol 208: 1126–1137 [DOI] [PubMed] [Google Scholar]
- Medzihradszky M, Bindics J, Ádám É, Viczián A, Klement É, Lorrain S, Gyula P, Mérai Z, Fankhauser C, Medzihradszky KF, et al. (2013) Phosphorylation of phytochrome B inhibits light-induced signaling via accelerated dark reversion in Arabidopsis. Plant Cell 25: 535–544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Messinger SM, Buckley TN, Mott KA (2006) Evidence for involvement of photosynthetic processes in the stomatal response to CO2. Plant Physiol 140: 771–778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno JE, Ballaré CL (2014) Phytochrome regulation of plant immunity in vegetation canopies. J Chem Ecol 40: 848–857 [DOI] [PubMed] [Google Scholar]
- Mott KA. (2009) Opinion: stomatal responses to light and CO2 depend on the mesophyll. Plant Cell Environ 32: 1479–1486 [DOI] [PubMed] [Google Scholar]
- Mott KA, Berg DG, Hunt SM, Peak D (2014) Is the signal from the mesophyll to the guard cells a vapour-phase ion? Plant Cell Environ 37: 1184–1191 [DOI] [PubMed] [Google Scholar]
- Murga-Zamalloa CA, Atkins SJ, Peranen J, Swaroop A, Khanna H (2010) Interaction of retinitis pigmentosa GTPase regulator (RPGR) with RAB8A GTPase: implications for cilia dysfunction and photoreceptor degeneration. Hum Mol Genet 19: 3591–3598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagawa S, Xu T, Lin D, Dhonukshe P, Zhang X, Friml J, Scheres B, Fu Y, Yang Z (2012) ROP GTPase-dependent actin microfilaments promote PIN1 polarization by localized inhibition of clathrin-dependent endocytosis. PLoS Biol 10: e1001299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeiffer A, Nagel MK, Popp C, Wüst F, Bindics J, Viczián A, Hiltbrunner A, Nagy F, Kunkel T, Schäfer E (2012) Interaction with plant transcription factors can mediate nuclear import of phytochrome B. Proc Natl Acad Sci USA 109: 5892–5897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu JL, Jilk R, Marks MD, Szymanski DB (2002) The Arabidopsis SPIKE1 gene is required for normal cell shape control and tissue development. Plant Cell 14: 101–118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren H, Dang X, Yang Y, Huang D, Liu M, Gao X, Lin D (2016) SPIKE1 activates ROP GTPase to modulate petal anisotropic growth in Arabidopsis. Plant Physiol 172: 358–371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roelfsema MRG, Hanstein S, Felle HH, Hedrich R (2002) CO2 provides an intermediate link in the red light response of guard cells. Plant J 32: 65–75 [DOI] [PubMed] [Google Scholar]
- Schwartz A, Zeiger E (1984) Metabolic energy for stomatal opening. Roles of photophosphorylation and oxidative phosphorylation. Planta 161: 129–136 [DOI] [PubMed] [Google Scholar]
- Shichrur K, Yalovsky S (2006) Turning ON the switch—RhoGEFs in plants. Trends Plant Sci 11: 57–59 [DOI] [PubMed] [Google Scholar]
- Shin DH, Cho MH, Kim TL, Yoo J, Kim JI, Han YJ, Song PS, Jeon JS, Bhoo SH, Hahn TR (2010) A small GTPase activator protein interacts with cytoplasmic phytochromes in regulating root development. J Biol Chem 285: 32151–32159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sibbernsen E, Mott KA (2010) Stomatal responses to flooding of the intercellular air spaces suggest a vapor-phase signal between the mesophyll and the guard cells. Plant Physiol 153: 1435–1442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorek N, Poraty L, Sternberg H, Bar E, Lewinsohn E, Yalovsky S (2007) Activation status-coupled transient S acylation determines membrane partitioning of a plant Rho-related GTPase. Mol Cell Biol 27: 2144–2154 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Sorek N, Segev O, Gutman O, Bar E, Richter S, Poraty L, Hirsch JA, Henis YI, Lewinsohn E, Jürgens G, Yalovsky S (2010) An S-acylation switch of conserved G domain cysteines is required for polarity signaling by ROP GTPases. Curr Biol 20: 914–920 [DOI] [PubMed] [Google Scholar]
- Stadler R, Büttner M, Ache P, Hedrich R, Ivashikina N, Melzer M, Shearson SM, Smith SM, Sauer N (2003) Diurnal and light-regulated expression of AtSTP1 in guard cells of Arabidopsis. Plant Physiol 133: 528–537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas C, Fricke I, Scrima A, Berken A, Wittinghofer A (2007) Structural evidence for a common intermediate in small G protein-GEF reactions. Mol Cell 25: 141–149 [DOI] [PubMed] [Google Scholar]
- Thomas C, Fricke I, Weyand M, Berken A (2009) 3D structure of a binary ROP-PRONE complex: the final intermediate for a complete set of molecular snapshots of the RopGEF reaction. Biol Chem 390: 427–435 [DOI] [PubMed] [Google Scholar]
- Tominaga M, Kinoshita T, Shimazaki K (2001) Guard-cell chloroplasts provide ATP required for H+ pumping in the plasma membrane and stomatal opening. Plant Cell Physiol 42: 795–802 [DOI] [PubMed] [Google Scholar]
- Uhrig JF, Mutondo M, Zimmermann I, Deeks MJ, Machesky LM, Thomas P, Uhrig S, Rambke C, Hussey PJ, Hülskamp M (2007) The role of Arabidopsis SCAR genes in ARP2-ARP3-dependent cell morphogenesis. Development 134: 967–977 [DOI] [PubMed] [Google Scholar]
- Wang F, Guo Z, Li H, Wang M, Onac E, Zhou J, Xia X, Shi K, Yu J, Zhou Y (2016) Phytochrome A and B function antagonistically to regulate cold tolerance via abscisic acid-dependent jasmonate signaling. Plant Physiol 170: 459–471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang FF, Lian HL, Kang CY, Yang HQ (2010) Phytochrome B is involved in mediating red light-induced stomatal opening in Arabidopsis thaliana. Mol Plant 3: 246–259 [DOI] [PubMed] [Google Scholar]
- Weise A, Lalonde S, Kühn C, Frommer WB, Ward JM (2008) Introns control expression of sucrose transporter LeSUT1 in trichomes, companion cells and in guard cells. Plant Mol Biol 68: 251–262 [DOI] [PubMed] [Google Scholar]
- Won SK, Lee YJ, Lee HY, Heo YK, Cho M, Cho HT (2009) Cis-element- and transcriptome-based screening of root hair-specific genes and their functional characterization in Arabidopsis. Plant Physiol 150: 1459–1473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu T, Wen M, Nagawa S, Fu Y, Chen JG, Wu MJ, Perrot-Rechenmann C, Friml J, Jones AM, Yang Z (2010) Cell surface- and rho GTPase-based auxin signaling controls cellular interdigitation in Arabidopsis. Cell 143: 99–110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yalovsky S. (2015) Protein lipid modifications and the regulation of ROP GTPase function. J Exp Bot 66: 1617–1624 [DOI] [PubMed] [Google Scholar]
- Yamaguchi R, Nakamura M, Mochizuki N, Kay SA, Nagatani A (1999) Light-dependent translocation of a phytochrome B-GFP fusion protein to the nucleus in transgenic Arabidopsis. J Cell Biol 145: 437–445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z. (2002) Small GTPases: versatile signaling switches in plants. Plant Cell (Suppl) 14: S375–S388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang B, Wang X, Zhao Z, Wang R, Huang X, Zhu Y, Yuan L, Wang Y, Xu X, Burlingame AL, et al. (2016) OsBRI1 activates BR signaling by preventing binding between the TPR and kinase domains of OsBSK3 via phosphorylation. Plant Physiol 170: 1149–1161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C, Kotchoni SO, Samuels AL, Szymanski DB (2010) SPIKE1 signals originate from and assemble specialized domains of the endoplasmic reticulum. Curr Biol 20: 2144–2149 [DOI] [PubMed] [Google Scholar]
- Zhang Y, McCormick S (2007) A distinct mechanism regulating a pollen-specific guanine nucleotide exchange factor for the small GTPase Rop in Arabidopsis thaliana. Proc Natl Acad Sci USA 104: 18830–18835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao S, Wu Y, He Y, Wang Y, Xiao J, Li L, Wang Y, Chen X, Xiong W, Wu Y (2015) RopGEF2 is involved in ABA-suppression of seed germination and post-germination growth of Arabidopsis. Plant J 84: 886–899 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.