An integrative glycobiology approach provided new insight into multiple modes of cell wall remodeling during pea border cell formation and mechanisms of their release.
Abstract
The adhesion of plant cells is vital for support and protection of the plant body and is maintained by a variety of molecular associations between cell wall components. In some specialized cases, though, plant cells are programmed to detach, and root cap-derived border cells are examples of this. Border cells (in some species known as border-like cells) provide an expendable barrier between roots and the environment. Their maturation and release is an important but poorly characterized cell separation event. To gain a deeper insight into the complex cellular dynamics underlying this process, we undertook a systematic, detailed analysis of pea (Pisum sativum) root tip cell walls. Our study included immunocarbohydrate microarray profiling, monosaccharide composition determination, Fourier-transformed infrared microspectroscopy, quantitative reverse transcription-PCR of cell wall biosynthetic genes, analysis of hydrolytic activities, transmission electron microscopy, and immunolocalization of cell wall components. Using this integrated glycobiology approach, we identified multiple novel modes of cell wall structural and compositional rearrangement during root cap growth and the release of border cells. Our findings provide a new level of detail about border cell maturation and enable us to develop a model of the separation process. We propose that loss of adhesion by the dissolution of homogalacturonan in the middle lamellae is augmented by an active biophysical process of cell curvature driven by the polarized distribution of xyloglucan and extensin epitopes.
The root cap is a specialized protective and sensing organ covering the root meristem (Arnaud et al., 2010), and many plants produce large numbers of sloughed off root cap cells, usually referred to as border cells or border-like cells in some plants, including Brassicacea species (Driouich et al., 2007, 2010). Following division of initials, root cap cells grow and are eventually positioned on the root surface; they are released subsequently into the rhizosphere as border cells (Driouich et al., 2007; Rost, 2011; Fig. 1). The outer cell layers of the root cap also secrete polysaccharide-rich mucilage, and the combined action of sloughing and mucilage secretion enables roots to penetrate the soil and ensure the replacement of cells mechanically challenged during growth (Driouich et al., 2007; McKenzie et al., 2013). Border cells also form a dynamic first-contact barrier against pathogens (Hawes et al., 2000; Cannesan et al., 2012; Driouich et al., 2013), and in some cases, border cells may attract or repel bacteria or form papillae in response to fungi (Hawes and Pueppke, 1986; Sherwood, 1987). Border cells, then, are a highly specialized component of the root system with functions and cellular properties distinct from other root cells. One important aspect of this specialization is the regulation of border cell release, but the underlying cellular mechanisms governing this are largely unknown.
Figure 1.
Developmental origin and morphology of pea border cells. A, Schematic showing the relationship of border cells to gross root apex anatomy. Border cells originate from the root cap initials and progress toward the root surface, eventually to be released as single cells. in, Initials; QC, quiescent center; RC, root cap. B, Low and higher magnification images of whole pea roots. The image at right shows a closeup of the boxed area in the image at left. C, Micrograph of the border cells isolated by agitation of root tips in liquid. D and E, Transverse (D) and longitudinal (E) sections of resin-embedded pea root apex stained using Toluidine Blue. A released border cell is indicated by the arrowheads. F, Intact single border cell labeled by Toluidine Blue. Note the curved shape of the cell. Bars = 1 mm (B), 30 μm (C–E), and 10 μm (F).
The adhesion and detachment of plant cells is chiefly mediated by the middle lamellae, the major component of which is the pectic polysaccharide homogalacturonan (HG). Compromised synthesis of HG results in altered border-like cell organization in Arabidopsis (Arabidopsis thaliana) quasimodo mutants (Bouton et al., 2002; Mouille et al., 2007; Durand et al., 2009; Mravec et al., 2014). Moreover, the postsynthetic demethyl esterification of HG by pectin methyl esterases (PMEs) is required for cell separation in pea (Pisum sativum) and Arabidopsis (Stephenson and Hawes, 1994; Wen et al., 1999; Mravec et al., 2014). Some studies also have implicated other polysaccharides in border cell processing. For example, the fucosylation of xyloglucan (XyG) is required for proper border cell morphology and their release in pea (Wen et al., 2008).
Consistent with their specialization, significant and characteristic modulations in gene expression are associated with border cell differentiation (Brigham et al., 1995), and some regulatory genes have been identified in Arabidopsis. Root cap-specific NAC transcription factors are involved in the process of maturation and separation of border-like cells, and they appear to control the expression of some cell wall-modifying enzymes, including the cellulase CEL5 (del Campillo et al., 2004). Moreover, triple mutants in which these NACs are defective or absent exhibit retention of the mature layers of root caps (Bennett et al., 2010).
Although Arabidopsis is a well-established model for plant molecular genetics, the very small size of the root presents significant difficulties for detailing the cell biology of border cells. In this study, we used pea roots as our model system (Fig. 1). Relatively large amounts of nearly pure border cells can be isolated by agitating root apices in liquid followed by gentle centrifugation (Fig. 1). This simple but effective procedure enables sufficient amounts of border cells to be isolated for glycomic, transcriptomic, and proteomic investigation (Brigham et al., 1995). We undertook a detailed analysis of changes in cell walls during pea root cap maturation and border cell formation using multiple approaches for profiling polysaccharides and selected associated genes. The aim was to obtain new insight into roles that specific glycan epitopes play in the specialized biology of border cells and to inform new thinking on how the release of border cells is established. An integrated glycobiology approach helped us to identify previously unknown modes of cell wall dynamics and to propose some operational mechanisms in this special type of cell separation process.
RESULTS
Carbohydrate Microarray Profiling Provides an Overview of Cell Wall Modulation in Pea Border Cells
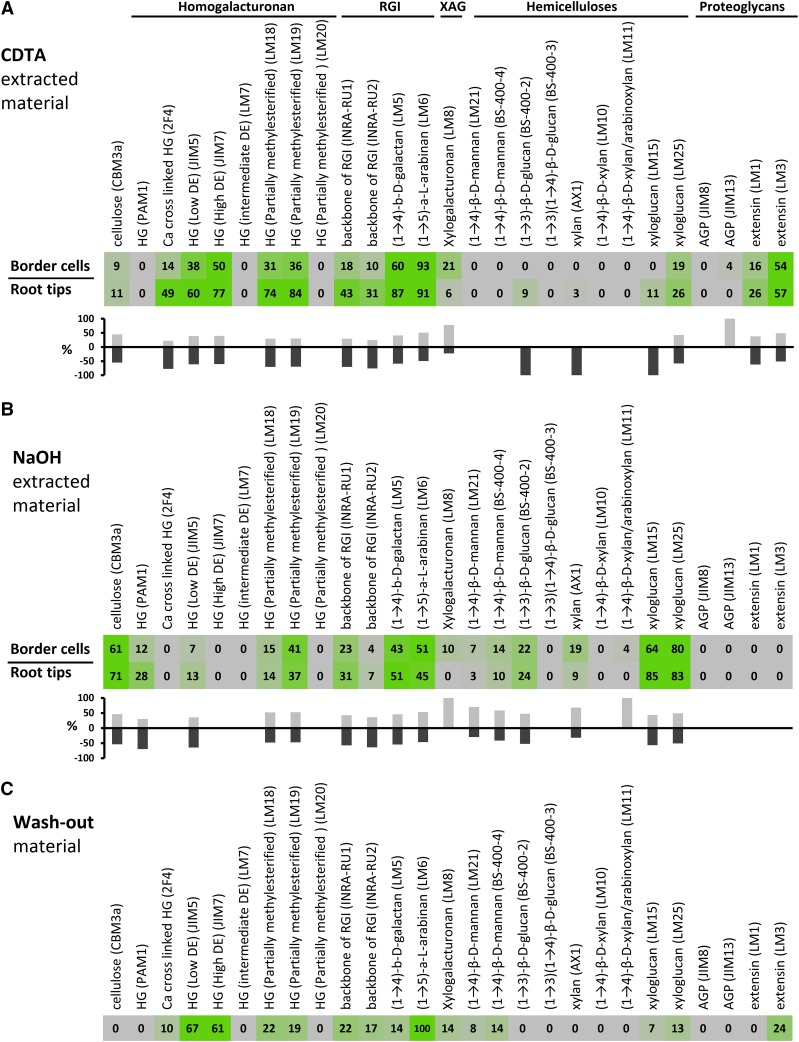
Using a variety of techniques, our initial aims were to describe in detail the compositional differences between border cells and roots and to identify which cell wall components are released to the external environment. Using a simple standard procedure, we were able to isolate approximately 0.1 mg of relatively pure border cells per one root tip (Fig. 1C). This enabled us to obtain enough alcohol-insoluble residue (AIR) to perform comprehensive microarray polymer profiling (CoMPP). CoMPP is a semiquantitative high-throughput method whereby cell wall polysaccharide components are extracted and then printed as microarrays onto membranes and probed with panels of monoclonal antibodies (mAbs) and/or carbohydrate-binding modules (Moller et al., 2007). CoMPP provides information about the relative abundance of polysaccharide epitopes and is useful for obtaining a comparative overview of glycans across sample sets. In this case, CoMPP was used to assess differences in epitope levels from the same weight of AIR from isolated border cells and washed 2- to 3-mm-long root apices. In addition to these two samples, we also isolated the liquid wash-out containing the soluble mucilage produced by root cap cells, and this was printed directly without any further processing.
Two extraction solvents were used sequentially, first 1,2-diaminocyclohexanetetraacetic acid (CDTA) and then NaOH. By disrupting calcium cross-links, CDTA releases pectic polysaccharides from cell walls and also promotes the removal of other polymer classes held in the wall by the pectic matrix. NaOH solubilizes polymers that are more firmly held in the wall, predominantly hemicelluloses. However, because cell wall polysaccharides form a coextensive network with multiple intramolecular associations and bond types, there is considerable overlap in the materials released by CDTA and NaOH. We analyzed the CDTA-extracted (Fig. 2A) and NaOH-extracted (Fig. 2B) material and liquid root tip wash-out (Fig. 2C) separately using 27 mAbs. The data are presented as heat maps in which mean spot signals are proportional to color intensity (Fig. 2). To aid the interpretation of differences between border cell versus root cap material, the same data also are presented in the form of bar graphs showing signal strength percentages (Fig. 2, A and B).
Figure 2.
CoMPP of the pea border cell walls and wash-out material. Heat maps show mean spot signals from CoMPP data of border cells, root tips, and root wash-out material. A, Data for the CDTA fraction obtained from border cells and root tips. B, Data for the NaOH fraction obtained from border cell and root tips. C, Data for root wash-out material. The values are mean spot signals from two biological replicates, and color intensity is correlated with signal strength. Antibody names and the epitopes recognized are indicated. The bar graphs below the heat maps in A and B indicate relative differences, expressed as percentages, between the signal intensities of border cells and root tips.
CoMPP analysis indicated that all the epitopes present in root tips also could be detected in border cells, but there were some considerable differences in relative abundance and extractability. There were generally reduced levels of pectic epitopes in border cells compared with root tips, and this was most apparent in the CDTA-extracted border cell material (Fig. 2A). For example, the relative abundances of rhamnogalacturonan I (RGI) backbone epitopes recognized by the mAbs INRA-RUI and INRA-RU2 (Ralet et al., 2010) were reduced by 58% and 68%, respectively, in CDTA fractions, while the relative level of binding of all HG-directed mAbs was reduced by between 32% and 57% (Fig. 2A). Especially low binding was observed in border cells in the case of mAb 2F4 (a probe that recognizes HG in calcium complexes), with a reduction by 72% compared with root tips. One notable exception to this reduced pectin trend in border cells was the substantially higher levels (71%) of xylogalacturonan (XAG; recognized by mAb LM8) in border cells compared with root tips. This finding is consistent with previous work implicating XAG in cell separation in pea testae, root caps, and other angiosperm systems (Willats et al., 2004). For some epitopes, the signals obtained from root tips and border cells were very similar, and this was the case for (1→5)-α-l-arabinan and extensins recognized by the mAbs LM6 and LM3, respectively.
As expected, the NaOH fractions from both border cells and root tips were generally richer in hemicellulosic epitopes, notably those associated with XyG, xylan, and (1→3)-β-d-glucan compared with the CDTA fractions (Fig. 2B). It is also worth noting that, in general, there was less variance between the signals from border cells and root tips in the NaOH material compared with the CDTA material. This probably reflects the fact that nonhemicellulosic matrix polysaccharides are generally more subject to dynamic modulation in rapidly growing tissues. However, there was an increase in border cells in the signals obtained for the xylan and mannan epitopes recognized by the mAbs AX1 and LM21, respectively, although the signals were weak in both sample types. Some XAG was extracted from border cells using NaOH but not from root tips (Fig. 2B). CoMPP profiling of wash-out mucilage material provided an overview of which polysaccharides were released to the environment. These data showed that released material was compositionally quite different from both the root tips and border cells (Fig. 2C). It was relatively poor in hemicelluloses and proteoglycans but enriched in pectic polysaccharides, especially arabinan and HG. Taken together, these data indicate that there are substantial differences in the polysaccharide components of root tips versus isolated border cells. It is reasonable to propose that these differences reflect the specialized functional requirements of border cells.
Monosaccharide Composition Analysis and Fourier-Transformed Infrared Spectroscopy Confirm Quantitative Differences in Cell Wall Components in Border Cells and Root Tips
We extended our investigation of variations in cell wall components in root tips versus border cells by monosaccharide composition analysis of AIR using hydrolysis with trifluoroacetic acid (TFA) and high-performance anion-exchange chromatography with pulsed amperometric detection (HPAEC-PAD; Fig. 3A). CoMPP and monosaccharide composition analysis are complementary approaches that provide distinct outputs. Unlike CoMPP, monosaccharide composition analysis provides information about amounts of specific sugars between samples; but unlike the epitopes, recognized by mAbs in CoMPP, those sugars cannot be assigned directly to polysaccharides. Also, whereas CoMPP provides information about extractable polymers, monosaccharide composition analysis involves the hydrolysis of cell walls and, therefore, in theory, provides information about sugar levels per se. Even though underrepresentation of uronic acids has been reported (Manns et al., 2014) for similar analyses, significant relative differences were found in the monosaccharide levels of the two sample types. The lower level of GalUA in border cells (Fig. 3A) compared with root tips is consistent with the reduced binding in CoMPP analysis of pectin-directed mAbs in the CDTA fraction from border cells compared with the CDTA fraction from root tips. It is problematic to distinguish Xyl from Man by HPAEC-PAD, but the elevated Man/Xyl levels in border cells may correlate with the much higher CoMPP signals obtained in border cells for the Xyl-containing XAG. Arabinose levels were significantly higher in border cells than in root tips (Fig. 3A). However, in CoMPP analysis, signals from the anti-arabinan mAb LM6 were similar in border cells and root tips in CDTA fractions (Fig. 2A), although they were slightly higher in NaOH-extracted material from border cells compared with root tips (Fig. 2B). These marginal differences in arabinan suggest that the increased arabinose in border cells may be associated with another polysaccharide. One candidate is arabinoxylan, since binding of the xylan/arabinoxylan mAb AX1 was elevated in NaOH-extracted material from border cells compared with root tips (Fig. 2B).
Figure 3.
Biochemical analyses of differences between cell walls of root tips and border cells. A, Monosaccharide compositional analysis by HPAEC-PAD of AIR derived from border cells and root tips. Monosaccharide levels are shown as mole percentages (n = 3, error bars denote sd; *, P < 0.05, Student’s t test). B, FT-IR microspectroscopy showing differences in cell wall chemistry of border cells and root tips. Statistical comparison of border cell and root tip spectra was performed using Student’s t test. The regions with significant differences are marked with brackets and asterisks. Note the shape of the shoulder between 1,720 and 1,740 wave numbers (arrowhead), indicating a difference in the degree of esterification.
Another notable difference between root tips and border cells was the significantly reduced rhamnose levels in border cells, and this was consistent with the lower binding of the mAbs INRA-RU1 and INRA-RU2 that recognize the RGI backbone in border cells (Fig. 2A). Glc levels were significantly higher in root tips (Fig. 3A). This increase may be partially attributable to a higher level of XyG in root tips, since we observed slightly higher signals from two anti-XyG mAbs (LM15 and LM25) in CoMPP (Fig. 2, A and B). However, cellulose is generally the largest source of Glc in cell walls. Due to its insolubility in CoMPP extractions and only partial hydrolysis with TFA, we could not draw conclusions about the absolute levels of cellulose. On the other hand, imaging with the cellulose-specific dye Pontamine Scarlet 4B indicated thickened, cellulose-rich walls in the outer, but still attached, cell layers of root tips (see below). This observation at least partially supported the notion that Glc levels in root tips are indeed most likely due to higher cellulose levels.
The role of HG in cell adhesion is not simply related to HG levels per se but also to the degree and pattern of methyl esterification. Some information was provided by CoMPP using mAbs with specificities for HG with differing methyl esterification levels. But we also used Fourier-transformed infrared (FT-IR) spectroscopy to explore HG esterification levels further (Fig. 3B; Mouille et al., 2003; Wolf et al., 2012). Statistical analysis of the obtained spectra from AIR samples showed a significant difference within the region 1,400 to 1,720 cm−1 suggesting differences in the abundance of carboxylic and ester groups. In addition, the shape of the shoulder between 1,720 and 1,740 cm−1 specifically points to changes in the amount of ester bonds. Finally, we also observed significant differences in the so-called fingerprint area between 960 and 1,160 cm−1 that are suggestive of overall structural alterations of the cell wall polysaccharide fraction (Fig. 3B).
Quantitative RT-PCR Revealed Continuous Transcription of Cell Wall Biosynthetic Genes in Border Cells
The biochemical analyses showed that border cells possess complex cell walls, and we wanted to investigate whether border cells themselves contribute to the synthesis of their own cell walls.
Accordingly, we conducted quantitative reverse transcription (qRT)-PCR on selected genes involved in cell wall biosynthesis (Fig. 4). From available pea ESTs, we identified in silico orthologs of Arabidopsis cellulose synthase (PsCESA), galacturonosyl transferase8/quasimodo1 (PsGAUT), as well as galactan (PsGALS), arabinan (PsARA), and XAG (PsXAGS) synthases (Supplemental Table S1). We isolated RNA from border cells and performed qRT-PCR using the UBIQUITIN-CONJUGATING ENZYME gene as a reference. The most striking difference in gene expression levels between root tips and border cells was for PsXAGS and PsGAUT1. The much higher expression of PsXAGS is consistent with increased binding of the anti-XAG mAb LM8 to border cell material in CoMPP (Fig. 2A) and the higher Man/Xyl level in border cells (Fig. 3A). Similarly, the reduced expression of PsGAUT1 in border cells is in agreement with the decreased binding of the anti-HG mAbs to border cell material in CoMPP (Fig. 2A) and with the lower level of GalUA in border cells (Fig. 3A).
Figure 4.
Expression of cell wall biosynthetic genes in border cells. The bar graph shows relative expression levels derived from qRT-PCR data of the pea orthologs of five polysaccharide synthase genes. Note the significant down-regulation of PsGAUT and up-regulation of PsXAGS genes (n = 4, error bars denote se; *, P < 0.05, Student’s t test).
Interestingly, the expression level of PsCESA did not differ significantly between border cells and root tips, despite the relatively much higher levels of TFA-released Glc in root tips compared with border cells (Fig. 3A).
Although relatively limited in scope, the qRT-PCR results nonetheless yielded important information. Some cell wall biosynthetic genes are clearly still being expressed in border cells, despite their separation from the plant body. Also, at least for pectic domains, the differences in polysaccharide epitope levels correlated with the transcriptional activity of related genes.
Structural Cell Wall Modulations during Border Cell Maturation
The analyses described so far only considered differences in cell walls in fully detached border cells versus cell walls within roots. But we were also interested to track subtler modulations in cell wall structure and composition during border cell development. To do this, we used a combination of fluorescence immunocytochemistry, transmission electron microscopy (TEM), and immunogold TEM. Figure 5A shows a cross section of part of a pea root stained with the cellulose-specific dye Pontamine Scarlet 4B (Anderson et al., 2010). We observed that the cells toward the central region of the root tip had relatively thin walls but that cell walls became progressively thicker near the root surface. Loss of cell adhesion and the formation of intercellular spaces appeared to be correlated with cell wall thickening, but when cells became fully detached as border cells, their walls were notably thinner again.
Figure 5.
Cell wall morphology during border cell maturation. A, Resin section of part of a pea root apex stained with the cellulose-specific dye Pontamine Scarlet 4B (red signal) spanning the stages of border cell maturation and release. Blue signal emanates from autofluorescence under UV light. We identified four distinct regions of cell maturation: region 1, close to the root axis where the cells are fully adhered; region 2, where cells starts to separate with intercellular space formation and cell walls become thicker; region 3, the most outer layer of the root cap where border cells detached; and region 4, where border cells are released. B to E, Transmission electron micrographs of the regions of the pea root cap indicated in A. B, Cell walls (marked cw1 and cw2) of two cells (c1 and c2) in the central region of the root cap. Note the characteristic appearance of the middle lamella (indicated by the arrow). C, Two cells (c1 and c2) in the process of separation. Note the dissolution of the cell walls (cw1 and cw2) in separate layers (indicated by the arrow). D, The outer cell wall (cw) of a cell still attached to the root body has a diffuse, fibrillary appearance. E, Part of a fully detached border cell with a notably thin cell wall (cw). Numerous Golgi apparatus are visible (indicated by the arrowheads). Bars = 10 μm (A) and 500 nm (B–E).
To investigate these structural changes in more detail, we used TEM to examine the cell walls of cells from four regions of the root, spanning from near the root center, across the region of loosening attachment, and finally border cells themselves (Fig. 5, B–E). Material of medium electron density, characteristic of middle lamellae, was present between cells in region 1 (near the root center), and their relatively thin cell walls were typical of root parenchyma tissue (Fig. 5B). In region 2, (Fig. 5C), where cells were beginning to separate, much thicker cell walls were clearly visible, consistent with the Pontamine Scarlet 4B staining seen in Figure 5A. The TEM imaging of cell walls in region 2 revealed a laminate appearance, and in some cases, some delaminated cell wall material appeared to be partially detached from the cell walls proper (Fig. 5C). The outer cell walls in cells in region 3, from which border cells had released (Fig. 5D), had a diffuse fibrous appearance, with decreasing electron density away from the cell surface, and this was in marked contrast to the cell walls of the detached border cells, which were thin and electron dense (Fig. 5E). These observations indicate that the outermost portions of cell walls disintegrate, presumably releasing material into the environment.
Modulations in Specific Polysaccharide Epitopes across the Root Diameter and in Border Cells
TEM observations provided information about the quite significant alterations in cell wall morphology that accompany changes in adhesion and, finally, cell separation required for border cell release. Next, we examined the progressive modulations in specific polysaccharide epitopes associated with these dynamic cellular events. Transverse sections through root caps were labeled with a subset of the mAbs used in CoMPP, and an overview of the labeling profiles is shown in Supplemental Figures S1 and S2. For each mAb, the binding profile across the root was analyzed using software that scanned fluorescence intensity across the root from the center outward to the periphery. These scans and associated images show the large variations in the localization of different polysaccharide epitopes across the root diameter and in border cells. Whereas some epitopes were abundant in most cells (e.g. the anti-XyG mAb CCRCM1; Supplemental Fig. S2B), others were restricted to a few cell layers (e.g. the XAG recognized by LM8; Supplemental Fig. S1D), and some epitopes were abundant in border cells (e.g. the galactan epitopes recognized by LM5; Supplemental Fig. S1E). This further supported the notion that different cell wall components have different dynamics during root cap growth, and this might relate to the functional mechanisms of the release. To gain more insight into their possible roles, we decided to closely study their cellular localization.
Spatial Regulation of HG and RGI Epitopes
The HG domain of pectin has a widespread and well-characterized role in cell adhesion and is clearly of interest in relation to border cell detachment. We used three probes to localize HG with differing degrees of methyl esterification, the mAbs JIM5 and JIM7 and the recently developed oligosaccharide-based probe COS488 (Mravec et al., 2014; Fig. 6, A–F). JIM5 and JIM7 differ in their binding specificities, in that JIM5 preferentially binds HG with a lower degree of esterification (DE) while JIM7 preferentially binds HG with a higher DE (Willats et al., 2000). CoMPP analysis indicated that the partially methyl-esterified epitopes recognized by JIM5 and JIM7 were somewhat reduced in abundance in border cells compared with root caps, and the binding of mAb 2F4, which recognizes calcium cross-linked blocks of HG, was reduced considerably in border cells. The JIM7 signal was more ubiquitous than JIM5, which was more restricted to the central cylinder (Fig. 6, B and C). JIM5 preferentially binds to a partially esterified low-DE epitope rather than the completely deesterified regions (or blocks) of HG involved in calcium cross-linking-mediated cell adhesion (Willats et al., 1999). However, COS488 does bind exclusively to nonesterified HG (Mravec et al., 2014). Due to its small size, this probe has a higher penetrative capacity and is less prone to masking by other polysaccharides compared with the larger mAbs (Hervé et al., 2011; Mravec et al., 2014). In the center of the root cap, COS488 labeling showed a labeling pattern characteristic of middle lamellae, including at expanded triangular cell junctions (Fig. 6, D and E). Toward the surface, middle lamellae labeling became progressively more diffuse, but COS488 strongly labeled the thicker cell walls of detaching cells (Fig. 6F). Diffuse labeling also was observed on the outermost face of root cap cells, and this was presumably material left behind at the point of separation and is consistent with the TEM image in Figure 5D. COS488 also bound strongly to the thin walls of fully detached border cells (Fig. 6F). The highly penetrative efficiency of the COS488 probe enabled us to visualize this spatial HG dynamic also in vivo. The root tips could be immersed directly in the COS488-containing medium solution and observed without prior washing steps. These observations supported data from resin sections, and it was possible to observe both the diffused extracellular material and the detached cell walls labeled with COS488 (Supplemental Fig. S3).
Figure 6.
Distribution of pectin epitopes as detected in semithin resin sections of a pea root apex. All images are overlays showing emission from the β-glucan-specific dye Calcofluor White (blue signal) and emission from antibody labeling or staining. A, Control labeling with no primary antibody added. B and C, Immunolocalization (red signal) of partially methyl esterified HG with different esterification levels using the mAbs JIM5 (B) and JIM7 (C). D to F, Detection of deesterified HG by COS488 probe (green signal). Calcofluor White (blue signal) and propidium iodide (red signal) were used as counterstains. D, Low-magnification scan for overview. E, Portion of the root near the central cylinder. F, Region of the root periphery. Note that, in E, cell walls are thin and COS488 labels the middle lamellae and intercellular junctions (arrowhead). Note, in F, the diffuse nature of COS488 labeling, indicating the dissolution of middle lamellae (arrowhead). The cell walls (indicated by the arrow) of detached cells are notably thinner. G to I, Immunolocalization of RGI-related epitopes using mAbs: LM5 recognizing galactan (G), LM6 recognizing arabinan (H), and INRA-RU1 recognizing the RGI backbone (I). Note the cell wall localization of galactan but the extracellular localization of arabinan and RGI backbone epitopes in the shed material (indicated by arrowheads). Bars = 30 μm.
The branched side chain of pectic polymers associated with RGI significantly impacts pectin functionality, and we studied the distribution of RGI-related epitopes during border cell maturation (Fig. 6, G–I; Supplemental Fig. S2, E–G). Both galactan (recognized by the mAb LM5) and arabinan (recognized by the mAb LM6) epitopes appeared relatively more abundant toward the surface of the root cap; however, distinctly different localization patterns for LM5 and LM6 during the detachment phase were apparent (Fig. 6, G and H). While the galactan signal was confined to cell walls, the arabinan signal was confined mainly to extracellular material apparently detached from, or loosely attached to, walls (Fig. 6, G and H). This was also the case for the RGI backbone-specific mAb INRA-RU1. In cells in the central cylinder, RGI backbone labeling was present in the cell walls, whereas, after detachment, the labeling was confined almost exclusively to the extracellular material (Fig. 6I).
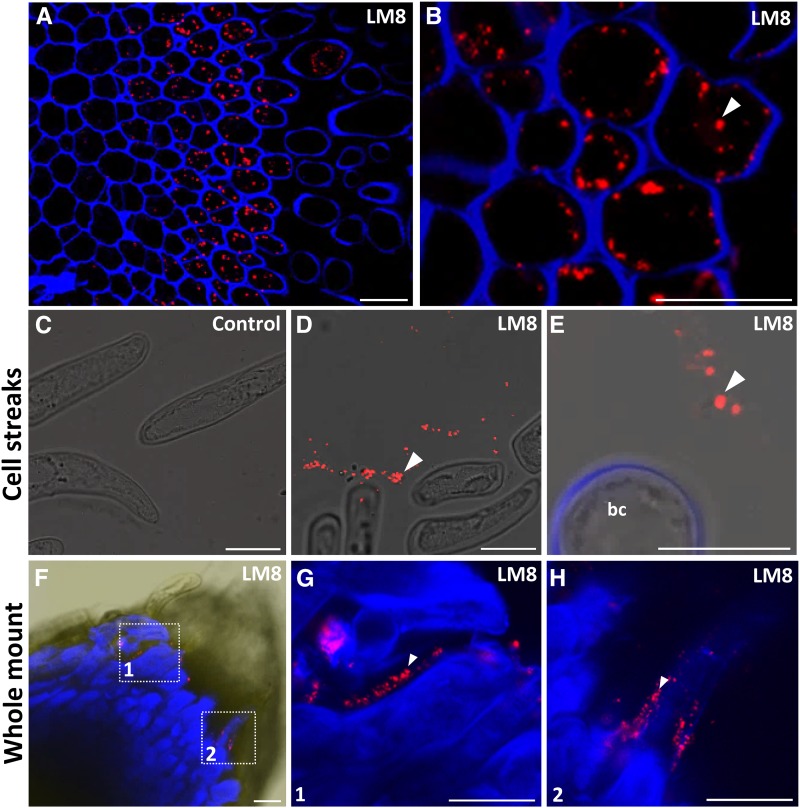
XAG Is Present in Intracellular and Extracellular Bodies
XAG is an HG subdomain substituted with Xyl and has been shown to be a polysaccharide associated with cell separation in several cell systems (Willats et al., 2004). The XAG epitope recognized by the mAb LM8 was restricted to a few distal cell layers toward the surface of pea root tips (Fig. 7A). This was a similar labeling pattern to that reported previously in pea roots (Willats et al., 2004), but in contrast to this earlier work, we observed that the signal was not present in cell walls but rather appeared as punctate distribution inside cells (Fig. 7, A and B). XAG certainly occurs as a mucilage layer on root surfaces in several species, including pea, and it is tempting to speculate that the intracellular vesicles are a component of the secretory machinery. To investigate this further, we attempted to visualize the LM8 epitope in material released from the root surface. To do this, we applied roots onto slides coated with Vectabond, a coating that immobilizes macromolecules by passive adsorption (Fig. 7, C–E). We observed large numbers of extracellular vesicle-like structures attached to slides (Fig. 7, C–E), and the size of these structures, between 0.5 and 1.5 μm, correlated with that of the intracellular bodies observed in sectioned resin-embedded material (Fig. 7, A and B). One explanation for these observations is that the extracellular bodies were secreted from roots, and this is consistent with the widely observed presence of the LM8 epitope on the surface of roots in mucilage. But we also considered that these structures may have simply been released from border cells ruptured during immobilization onto the Vectabond-coated microscope slides. We did not note any ruptured cells, but to further investigate this possibility, we also performed a whole-mount surface labeling of intact root apices (Fig. 7, F–H). Again, we observed LM8-labeled structures associated with the surface of separating cells and of similar size to the intracellular structures.
Figure 7.
Immunolocalization of XAG in the pea root apex. A, Immunolocalization of XAG in resin sections of pea root apex using the mAb LM8. Note that labeling is intracellular, punctate, and restricted to the last root cap layers and is absent from released border cells. B, Higher magnification image of the root cap outer layer showing LM8 labeling of intracellular punctate structures indicated by the arrowhead. C to E, LM8 labeling of border cells immobilized on the Vectabond-coated slides. C, Control labeling with no primary antibody added. D, LM8 labels distinct clouds of dot-like structures proximal to the border cells (arrowhead). E, Higher magnification image showing the LM8-labeled dots (arrowhead) and border cell (bc). Note that the dots are of a similar size (0.5–1.5 μm) to those observed in the resin sections shown in A and B. F, Whole-mount labeling of intact pea root tips with LM8. G and H, Higher magnification images of regions 1 and 2 marked in F showing the presence of the LM8-labeled dots (arrowheads) on the surface of the detaching cells. Images are overlays of the emission from Calcofluor White (blue signal) and LM8 labeling (red signal). Bars = 30 μm (A and F) and 10 μm (B–E, G, and H).
The most straightforward explanation for these findings is that the vesicle-like structures are involved in the export of XAG to the exterior of roots. Although intriguing, our work provides no evidence of any specific putative secretory mechanism(s) that may be involved. However, it is worth noting that a recent review highlighted the current paucity of knowledge relating to unconventional modes of protein secretion in plants, and this also may apply to polysaccharides (Robinson et al., 2016). Nevertheless, labeling data are consistent with the occurrence of XAG in the washed-out material and in border cell extractions (Fig. 2C). It is important to note that the XAG-containing bodies may remain associated with border cells during the isolation procedure and sediment together with border cells. This would explain the high abundance of XAG detected by CoMPP in border cell fractions.
XyG and Extensin Epitopes Display Polarized Distribution within Cell Wall Microdomains
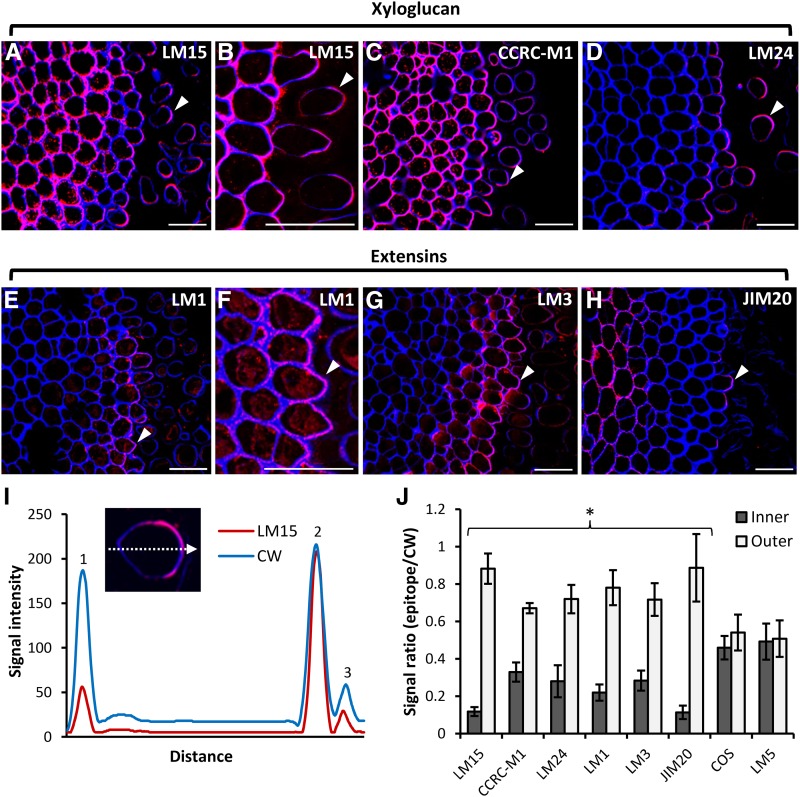
The longitudinal sections of pea apices (Fig. 1E) showed that cells approaching the root surface become more elongated, and we thought that this could be effected by some nonpectin components. XyG is one of the main hemicelluloses implicated in cell wall extensibility and cell elongation (Scheller and Ulvskov, 2010; Hayashi and Kaida, 2011; Park and Cosgrove, 2015), and we localized XyG in pea roots using three mAbs recognizing distinct XyG structural features: LM24, LM15, and CCRC-M1. LM24 and LM15 recognize the XLLG and XXXG motifs, respectively, and CCRC-M1 recognizes α-l-fucosylated XyG (Puhlmann et al., 1994; Pedersen et al., 2012). There were some clear differences in labeling patterns obtained for these three mAbs (Fig. 8, A–D). LM15 and CCRC-M1 both extensively labeled cell layers in the root cortex as well detached border cells (Fig. 8, A–C). In contrast, LM24 labeling was most abundant in the outermost cell and border cells (Fig. 8D).
Figure 8.
Polar localization of XyG and extensin epitopes. A to D, Immunolocalization of XyG epitopes in resin sections of a pea root apex using three XyG-specific mAbs, LM15 (A and B), CCRC-M1 (C), and LM24 (D). Note that all three mAbs exhibit up-regulated binding on the outward-facing (away from the root body) portion of cell walls of the peripheral root cap cells and detached border cells. E to H, Immunolocalization of extensin epitopes using the mAbs LM1 (E and F), LM3 (G), and JIM20 (H). Images are overlays of the emission from Calcofluor White (blue signal) and antibody labeling (red signal). Arrowheads indicate the most apparent cases of polar localization. I, Signal plot profile over a dashed arrow of a single cell (inset) stained with the mAb LM15 (red signal) and Calcofluor White (CW; blue signal). Numbered peaks indicate inner cell wall (1), outer cell wall (2), and shed material (3). Note the reduction of LM15 signal in the inner wall. J, Quantification of the ratios between the signal intensities of epitope labeling and Calcofluor White (used as a reference) recorded from the inner and outer cell walls. Note the reduction of XyG and extensin epitopes in the inner cell walls. No polarization of the COS and LM5 epitopes could be observed (n = 10, error bars denote se; *, P < 0.05, Student’s t test). Bars = 30 μm.
One striking feature of the binding of all three mAbs was that the labeling was unevenly distributed in both the outer layer of root cap cells and in border cells, such that the outer (i.e. facing away from the root body) portion of cell walls was more strongly labeled than the inner-facing walls (Fig. 8, A–D). We studied this in more detail using immunogold labeling with LM15 (Supplemental Fig. S4). We observed that, in cells of the root body, both adhered cell wall were labeled evenly, whereas for cells in the process of detaching, labeling was far more intense in the outer-facing wall, and this also was the case for detached border cells (Supplemental Fig. S4).
Extensins also have been implicated in cell elongation (Velasquez et al., 2011; Taylor et al., 2012; Nguema-Ona et al., 2014). Although less pronounced than for XyG, we also observed a polarized distribution of extensin epitopes recognized by the mAbs LM1, LM3, and JIM20 (Fig. 8, E–J). LM1 and LM3 labeling was confined to the outer two to three cell layers of the root cap, and labeling within walls was of a fragmented nature (Fig. 8, E–G). In both cases, labeling was more abundant in the outer-facing wall of the outermost cells. JIM20 labeled cell walls in some inner cortical cells, and compared with LM1 and LM3, labeling was sparse in the outer cell layers. Nevertheless, where labeling was present, it was more abundant in the outermost portion of cell walls (Fig. 8H). We also quantified this phenomenon by means of calculating ratios between the reference signal Calcofluor White and the signal obtained from epitope detection. This quantification clearly demonstrated the polarized distribution of XyG and extensin epitopes but not epitopes associated with HG (detected by the COS probe) and galactan (detected by LM5; Fig. 8, I and J).
Extensive Cell Wall Degradation Activities Have Not Been Detected with Chromogenic Polymer Substrates
Our biochemical and in situ analyses showed a flux of cell wall components such that some appear to increase in abundance during maturation and some appear to decline. These observations suggested that cell wall functional fine-tuning associated with border cell release is achieved by coordinated cell wall remodeling rather than by extensive polysaccharide degradation. To test this assumption, we used a recently described set of chromogenic hydrogel substrates developed for screening hydrolytic cell wall-degrading enzyme activities (Kračun et al., 2015). We used a panel of seven chromogenic substrates (chromogenic polymer hydrogel [CPH]) representing major cell wall components and one insoluble chromogenic biomass substrate (pectin-rich orange [Citrus spp.] peel as a tested substrate for HG-degrading enzymes). As with the CoMPP work, we separately analyzed homogenates of border cells, root tips, as well as wash-out material, and using this approach, no appreciable enzyme activity was observed, at least not at detection levels reported for CPH substrates (Kračun et al., 2015) even after 24 h (Supplemental Fig. S5). Although this was a limited survey, these data nonetheless support the notion that extensive hydrolysis of major cell wall components is not the major determinant of the release of border cells.
The Role of Extensin Glycosylation and HG Deesterification in the Release of Border Cells
In this work, evidence obtained from a variety of techniques pointed to subtle and specific modulations of cells being instrumental in border cell release, and extensins and HG emerged as candidates in this processes. Very limited genetic resources are available for cell biology in pea, but we were able to use two chemical inhibitors to perturb HG and extensins: (−)-epigallocatechin gallate (EGCG) and 3,4-dehydro-l-proline (3,4-DHP).
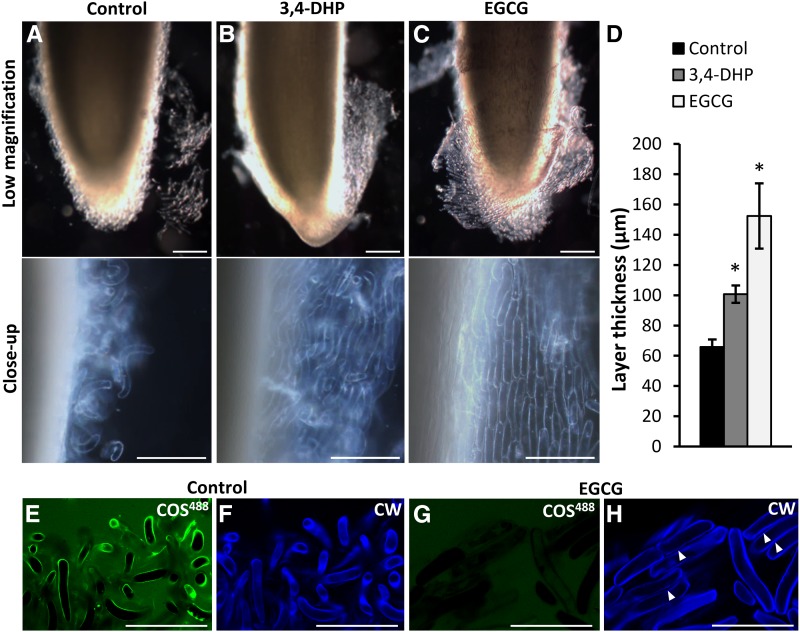
3,4-DHP is an inhibitor of Hyp biosynthesis that is necessary for extensin glycosylation (Cooper et al., 1994). Root apices were first agitated to remove loosely attached border cells and then incubated in growth medium supplemented with 200 μm 3,4-DHP (Fig. 9). 3,4-DHP treatment inhibited the release of border cells compared with untreated control roots (Fig. 9). In contrast to controls, the border cells of 3,4-DHP-treated apices formed nonseparated sheets or layers of adhered cells, and very few cells were fully detached.
Figure 9.
Effect on border cell release of the inhibition of extensin glycosylation and pectin methyl deesterification. A to C, Appearance of dissected pea apices after incubation overnight in Murashige and Skoog (MS) medium alone (A), MS medium supplemented with 200 μm extensin glycosylation inhibitor 3,4-DHP (B), or MS medium supplemented with 100 μm PME inhibitor EGCG (C). The top row shows low-magnification images, and the bottom row shows closeup images of the area of release. Note that border cell release is inhibited when 3,4-DHP or EGCG is present. D, Quantification of the thickness of the layer of loosely attached border cells (n = 12, error bars denote se; *, P < 0.05, Student’s t test). E to H, Effect of EGCG on HG deesterification studied by the COS488 probe. E and F, Border cells from a nontreated control. G and H, Cells from root tips grown in 100 μm EGCG. E and G, In vivo labeling of border cells with COS488 (green signal). F and H, Labeling of cell walls with Calcofluor White (CW; blue signal). Note the nonseparated files of cells indicated by arrowheads in H. Bars = 100 μm.
The pharmacological inhibition of PMEs and HG deesterification by EGCG (Lewis et al., 2008; Wolf et al., 2012; Mravec et al., 2014) had an even stronger effect. The EGCG-grown apices formed thick layers of straight cells in files, and the effect of EGCG was evident after 2 h from the point of EGCG application (Supplemental Fig. S6). Moreover, the inhibition of pectin demethyl esterification was demonstrated by labeling with COS488. The EGCG-treated border cells were completely devoid of COS488 labeling, and no COS488 labeled shed cell wall material could be observed (Fig. 9, E–H). Although we cannot rule out some direct effect of EGCG on cell wall components, including the blockage of COS488 binding, these results indicate that pectin methyl deesterification is not required for the formation of border cells per se but rather for their further spatial separation.
Border Cells Consistently Display a Curved Morphology That Is Affected by Cell Wall Components
The fact that both 3,4-DHP or EGCG treatments resulted in notably straighter border cells (Fig. 9, A–C) led us to speculate that curved and elongated shape is an additional aspect of their release.
To investigate these effects further, we undertook a detailed analysis of the length and degree of curvature of border cells (Fig. 10). Most border cells were between 100 and 200 µm long, but some were as short as 50 µm and some were longer than 300 µm (Fig. 10A). Similarly, most border cells were moderately curved (between 0° and 45°), but some displayed extreme, almost circular levels of curvature (Fig. 10B). Length and curvature were altered by EGCG or 3,4-DHP (Fig. 10B). In both treatments, considerably fewer cells could be observed in the pool of cells longer than 150 µm, and far fewer cells with extreme curvature (Fig. 10, A and B). These findings provided further indication for a cell wall-driven biophysical component to the curvature process, and to study the progression of curvature, we observed and recorded border cells as they detached from a section of root over a period of 2 h (Fig. 10, C and D). The real-time imaging revealed a predicable pattern of curvature and release events. At the beginning of the observation period, when cells were first detaching, we observed that cells were consistently curved away from the root surface, with the longer, convex portion of the cells closest to the root surface. This pattern continued throughout the observation period, and more cells were seen to start to curve, again in the same direction, until by 2 h, fully detached cells were visible. The consistent nature of the cell curvature is suggestive of a programmed developmental process, one that presumably facilitates cell detachment.
Figure 10.
Characterization of border cell shape and visualization of the detachment sequence. A and B, Quantification of the length (A) and degree of curvature (B) of isolated border cells released from roots incubated in control solution, 3,4-DHP, and EGCG (n = 300). Note the reductions of length and curvature in the cases of 3,4-DHP and EGCG incubation. The insets in B show examples of the different categories of curvature and how the curvature angle (χ) was measured. C, Simple setup for real-time observation of border cell release. A seedling was placed on a slide, and the root was submerged in a large drop of water contained in a PAP pen-marked area. D, Records of real-time observation of the process of detachment over a period of 2 h. The arrowheads indicate an instance of border cell release. Bar = 20 μm.
DISCUSSION
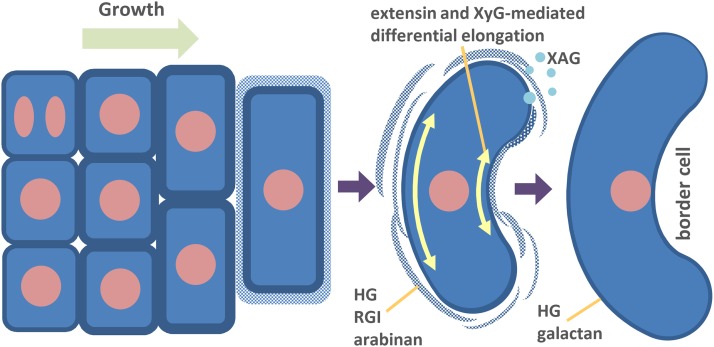
The adhesion of plant cells together via cell wall polysaccharides is crucial for the multicellularity of plants (Domozych and Domozych, 2014). The integrity of cell walls and their adhesion to neighboring walls are essential for the support and protection of the plant body (Bouton et al., 2002; Verger et al., 2016; Langhans et al., 2017). However, growth and development requires that cell walls also can be fined-tuned to allow extension or detachment in some specialized cases (Roberts et al., 2002; Lewis et al., 2006; Ogawa et al., 2009; Swain et al., 2011). Border cell release is one such example of this, and the systematic investigation we undertook demonstrates just how subtle and complex this fine-tuning is. By the time of their release, border cells become elongated and curved, and their cell walls become thinner than those of cells in the cell layer from which they detached (Fig. 11). Our work has provided new insight into how these substantial changes in cellular morphology are orchestrated by a series of multiple, complex, and spatially organized modulations of distinct cell wall epitopes that may play direct functional roles or may be associated with downstream events.
Figure 11.
Proposed model for the formation of border cells. By continuous growth, the root cap cells appear on the surface of the root tip and become elongated. During this progression, the cell walls undergo multilevel dynamics, which finally result in the dissolution of a part of the cell wall enriched in HG, RGI backbone, and arabinan. XAG is secreted into the environment via secretory vesicles or in the form of aggregates. The final getaway also could be augmented by extensin and XyG-mediated differential elongation and the formation of curvature. The intact, relatively thin cell walls of the released cells contain HG and galactan epitopes.
The low-magnification images in Supplemental Figures S1 and S2 provide a striking overview of the flux of different epitopes across the root during border cell maturation. However, the combination of immunocytochemical, TEM, gene expression, and biochemical analyses revealed developmentally regulated cell wall microdomain modulations in unprecedented detail and enable us to putatively assign roles to specific polysaccharides and to propose a model for border cell maturation and release (Fig. 11).
During the developmental progression of border cells, it is striking that root cell walls become thicker and enriched in pectic epitopes associated with the pectic side chains, arabinan, galactan, and the rhamnogalacturonan backbone bearing those side chains. There is evidence in various cell systems that both pectic arabinan and galatan can have roles in cell wall structural integrity, but in the case of galactan, it also could be important in plasticity and cell expansion (Willats et al., 1999; McCartney et al., 2003; Harholt et al., 2006; Verhertbruggen et al., 2013). The superabundance of RGI epitopes in the outermost cell layers is consistent with a requirement for cells to be prepared for the rapid elongation that characterizes the later stages of border cell formation but also with the degree of strength required when encountering an environment without supporting neighboring cells in a potentially mechanically challenging rhizosphere.
The newly developed probe COS488, which has a higher level of specificity for deesterified HG than previous probes, enabled us to monitor the dual functions of HG during border cell development. Deesterified HG has a structural role, again well documented in other cell systems, that involves cell wall stiffening via the calcium cross-linking of deesterified HG blocks (Willats et al., 2001; Daher and Braybrook, 2015). We observed that the walls of the outer layers of root cells were strongly labeled with COS488, as were the walls of detached border cells. As with RGI, this is likely a response to the structural requirements inherent in an isolated existence in the rhizosphere. But deesterified HG in middle lamellae also mediates cell adhesion, and the reduction or degradation of HG promotes cell separation. Our labeling with COS488, and the TEM work, revealed how border cell detachment is accompanied by the dissolution of HG between cells that becomes diffuse in nature.
One of the most novel and intriguing sets of observations we made relates to cell curvature, the polarized distribution of XyG and extensin epitopes, and the possible functional link between these cellular effects. Both XyG and extensin epitopes were up-regulated consistently on the outer face of cell walls of cells in the outermost root cell layer. This correlated with the cell curvature of separating cells away from the root surface. In other words, XyG epitopes and extensin were least abundant in the longer portions of cell walls. Both XyG and extensin are implicated in cell wall rigidification (Hayashi and Kaida, 2011; Velasquez et al., 2011; Hijazi et al., 2014), and conversely, the depolymerization or reduction in cross-linking of these polymers is associated with increased cell wall extension (Park and Cosgrove, 2015). It is tempting, therefore, to speculate that XyG and extensins are processed in outer root cell layers with the purpose of mechanically conditioning border cells such that they curve as they separate from roots. If this is the case, what is the purpose? Although we do not provide conclusive evidence, we propose that curvature is a programmed cell wall-determined biophysical effect that is synergistic with the well-understood effects of HG dissolution in middle lamellae. There is evidence from particle physics and biological systems that curvature can promote separation by reducing contact surface area (Derjaguin et al., 1975; Afferante et al., 2016). Indeed, the everyday process of pinching contact lenses to remove them is based on this principle. On the other hand, some cells are not curved but are still released to the environment, suggesting that this feature of border cells is supportive but not strictly required for separation. More functional analyses, including modeling, would be necessary to elucidate this interesting but previously neglected phenomenon.
In conclusion, this work has demonstrated the high degree of cellular and subcellular level sophistication inherent in the specialized process of border cell formation and has provided new insight into how modulations in cell wall components underpin a distinct process that promotes separation. It is worth noting that, although the release of border cells into soil is the final step in a complex series of biosynthesis events in planta, it is just the beginning of no doubt equally complex events that operate in the rhizosphere. From this point on, border cell wall components become substrates for carbohydrate-processing enzymes, and our findings also are of relevance for understanding this aspect of soil ecology.
MATERIALS AND METHODS
Isolation of Border Cells and CoMPP
Pea (Pisum sativum variety Norli) seeds were sterilized using 10% sodium hypochlorite for 5 to 10 min in a glass beaker and washed with sterile water several times. Seeds were allowed to soak in water for 4 h and then put in sterile water-wetted paper on the bottom of plastic boxes. They were kept in the dark at room temperature for 3 d. Only 2- to 3-cm-long radicles were used. The 1- to 2-mm-long root apices were dissected and immersed in 1 mL of PBS in 2-mL microtubes. Approximately 30 root tips were collected in one microtube. The microtubes were shaken in a ThermoMixer C (Eppendorf) set at 1,000 rpm speed and room temperature for 1 h. The liquid with cells was pipetted to a new microtube and centrifuged at 2,300g for 3 min. The supernatant was carefully removed, and the sediment was weighed. In total, 60 mg of border cells was collected. The samples were snap frozen in liquid nitrogen and ground with steel balls in a tissue lyser (Qiagen) for 1 min at 30 s−1 frequency. AIR was isolated by washing the homogenate with 70% ethanol followed by drying. To perform sequential extraction, 300 μL of CDTA was added, and the microtubes were shaken at 60°C for 1.5 h. The samples were centrifuged at 2,300g for 3 min, and the supernatant was removed and pipetted to a new microtube. NaOH at 4 m + 0.1% (w/v) NaHB was added to the sediment and shaken for the next 1.5 h. The samples were centrifuged at 2,300g for 3 min, and the supernatant was removed and pipetted to new microtubes. The printing on the nitrocellulose membrane was done on an ArrayJet printer in two replicates and four dilutions, probed, and quantified as described by Moller et al. (2007).
Monosaccharide Composition Analysis
One milligram of AIR was hydrolyzed in 2 m TFA for 1 h at 120°C and lyophilized. Monosaccharide composition determination was performed with HPAEC-PAD with monosaccharide standards of l-Fuc, l-Rha, l-Ara, d-Gal, d-Glc, d-Xyl, d-GalUA, and d-GlcA (Sigma-Aldrich) as described by Harholt et al. (2006).
FT-IR Spectroscopy
FT-IR spectroscopy was performed on a Nicolet iN10 infrared microscope system (Thermo Scientific) on AIR samples laid on gold-coated slides. The spectral data were averaged, base line corrected and normalized, and analyzed as described by Wolf et al. (2012).
Immunolocalization
Pea root apices 2 to 3 mm long were dissected and fixed in 4% paraformaldehyde in PBS for 1 h and washed with PBS. The samples were dehydrated in a methanol series (50%, 80%, and 100%, v/v) for 30 min then infiltrated with methanol:LR White Resin (1:1, v/v) for 24 h and twice in pure resin for 12 h. The resin was polymerized at 60°C overnight and cut on an ultramicrotome (Leica UC7) to generate 1-μm-thick sections. The sections were mounted on Superfrost slides that were blocked with 5% (w/v) milk in PBS for 30 min and incubated with primary antibodies at 1:10 dilution (except for INRA-RU1 and CCRC-M1, which were used at 1:100 dilution). The slides were washed three times with PBS for 10 min and probed with secondary antibodies. Anti-mouse and anti-rat antibodies, conjugated to AlexaFluor555 (Invitrogen), were used at 1:500 dilution. Both primary and secondary probing was done for 1 h. After the final three 10-min-long washing steps with PBS, sections were stained with Calcofluor White (Sigma-Aldrich) in PBS (0.1 mg L−1) for 10 min, washed with PBS, and mounted in CitiFluor medium (Agar Scientific). In the case of whole-mount labeling, the apices were dissected, fixed, and further processed on small 24-well plates. To obtain the border cell streaks, roots were dissected, and the tip was gently moved several times across the Vectabond-coated (Vector Laboratories) slides and left to dry for at least 30 min. In both cases, the labeling procedure was the same as for the resin sections.
Confocal and Light Microscopy
The scanning of fluorescently labeled sections was done on a Leica SP5 II confocal microscope equipped with UV diode (405 nm), argon (488 nm), and HeNe (543 nm) lasers. Light microscopy was done on an Olympus BX41 microscope equipped with a ColorView I camera and analySIS GetIT image-acquisition software using UPlaN 4× and 10× objectives and a phase-contrast Annulus Ph1. All recordings were performed at room temperature (20°C–23°C). The images were processed using GIMP2 software only to enhance the contrast and brightness. Measurements were performed by ImageJ software https://imagej.nih.gov/ij/index.html.
TEM
Five-millimeter-long segments of the pea root apices were excised and plunge frozen in liquid propane. The cells were then freeze substituted using a modified version of the protocol described previously by Domozych (1999). Briefly, the frozen apices were kept in 0.5% glutaraldehyde/acetone (w/v) for 24 h at −80°C. Solid osmium tetroxide was then added (final concentration, 0.5% osmium tetroxide), and the substitution was continued for an additional 24 h. The cells were gradually warmed to room temperature over an 8-h period, washed with acetone, and then infiltrated/embedded in London Resin. Polymerization was by UV light over 16 h. Fifty- to 70-nm sections were obtained using a Leica ultramicrotome and collected on formvar-coated nickel grids. The sections were viewed with a Zeiss Libra 120 transmission electron microscope at 120 kV.
Real-Time PCR
Two 2-mm glass beads were added to frozen cells in 2-mL microtubes, and cells were disrupted by Tissue Lyzer II (Qiagen) for 2 min at a frequency 30 s−1. RNA was extracted with the Plant RNA Extraction Kit (Qiagen) and eluted in 30 µL. RNA concentration was measured on a Nanodrop 1000, and RNA quality was measured by agarose gel electrophoresis. Based on Nanodrop readings, 1.2 µg of RNA was DNase treated with DNA-free turbo (Ambion), and cDNA was synthesized with the iScript cDNA synthesis kit (Bio-Rad). For real-time PCR, cDNA of root samples was diluted 1:9. One microliter of diluted root tip cDNA or 1 µL of undiluted border cell cDNA was used for real-time PCR with LightCycler 480 SYBR Green I master (Roche) and reference dye from Brilliant II SYBR Green QPCR master mix (Agilent Technologies). A 25-µL real-time PCR with 0.015 µm reference dye and 0.4 µm primers was run in duplicate on Agilent MX3000P as follows: 15 min at 95°C, 40 cycles at 95°C for 10 s and 60°C for 30 s, with reading at the end of each cycle, followed by continuous readings from 60°C to 95°C for the dissociation curve.
Analysis of Hydrolytic Activities
The border cells, root tips, and wash-out material were isolated and homogenized as described for the CoMPP procedure in three replicates of 60 root tips. The wash-out material was used directly without any processing. The soluble extracts of border cells and root tips were obtained by diluting the homogenized powder with 1 mL of sterile water followed by centrifugation at 2,700g for 3 min. The supernatants were collected into new Eppendorf tubes and used directly for the analysis on two assay plates containing seven CPH substrates and one insoluble chromogenic biomass substrate from orange (Citrus spp.) peel obtained from GlycoSpot. After activation of the substrates, 50 µL of extracts together with 100 µL of sterile water were transferred to each well with a substrate; 150 µL of water was used as a negative control, and 150-µL water solutions of the respective enzymes at 1 unit mL−1 dilution were used as positive controls. The reaction plates were incubated for 1.5 and 24 h, respectively, at constant 150 rpm horizontal shaking at room temperature. The reaction solution was then transferred to a collection plate by centrifugation, and the absorbance was recorded at 517 nm (for red-colored substrates) and 595 nm (for blue-colored substrates) using a SpectraMax M5 plate reader (Molecular Devices).
Physiological Experiments
Whole roots were excised and incubated in liquid Arabidopsis (Arabidopsis thaliana) Murashige and Skoog medium (Duchefa; 1% saccharose, pH 5.7) supplemented with 1-Naphthaleneacetic acid (0.5 mg L−1) and kinetin (0.05 mg L−1). Stock solutions of 200 mm 3,4-DHP and 100 mm EGCG (both Sigma-Aldrich) were made in water and used at 200 µm (3,4-DHP) or 100 µm (EGCG) final concentration. The roots were incubated at room temperature in the dark for 18 h and analyzed mounted in 50% glycerol solution in a cavity containing glass slides by light microscopy. The real-time recording was performed on the whole pea seedling in which the loosely attached border cells were removed by gentle agitation. The seedling was placed on the slide having the root tip submerged in water or water supplemented with EGCG and recorded in 10-min intervals for 2 h.
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Overview of the differential distribution of pectin-related epitopes in pea apices.
Supplemental Figure S2. Overview of the differential distribution of extensin and XyG epitopes in pea apices.
Supplemental Figure S3. In vivo imaging of deesterified HG during border cell detachment.
Supplemental Figure S4. Immunogold labeling of XyG with the mAb LM15 in pea root cap cells.
Supplemental Figure S5. Setup and results of analysis of cell wall component-degrading enzymes using chromogenic substrates.
Supplemental Figure S6. Imaging of the EGCG effect on the release of border cells over the 2-h period.
Supplemental Table S1. List of pea orthologs of Arabidopsis cell wall biosynthetic genes identified in publicly available ESTs databases and used in this study.
Supplementary Material
Acknowledgments
We thank Martina Pičmanová for help with the isolation of border cells.
Glossary
- HG
homogalacturonan
- AIR
alcohol-insoluble residue
- CoMPP
comprehensive microarray polymer profiling
- mAb
monoclonal antibody
- CDTA
1,2-diaminocyclohexanetetraacetic acid
- RGI
rhamnogalacturonan I
- TFA
trifluoroacetic acid
- HPAEC-PAD
high-performance anion-exchange chromatography with pulsed amperometric detection
- FT-IR
Fourier-transformed infrared
- qRT
quantitative reverse transcription
- TEM
transmission electron microscopy
- DE
degree of esterification
- XAG
xylogalacturonan
- XyG
xyloglucan
- CPH
chromogenic polymer hydrogel
- EGCG
(−)-epigallocatechin gallate
- 3,4-DHP
3,4-dehydro-l-proline
Footnotes
This work was supported by the European Union FP7 Marie Curie action projects CeWalDyn (grant no. 329830) and ITN WallTraC (grant no. 263916), the Innovation Funds Denmark projects Bio-Value (grant no. 0603-00522B) and B21st (grant no. 001-2011-4), the Villum Foundation project PLANET (grant no. 00009283), and the U.S. National Science Foundation (grant nos. NSF-MCB 0919925 and NSF-DBI 0922805).
Articles can be viewed without a subscription.
References
- Afferante L, Heepe L, Casdorff K, Gorb SN, Carbone G (2016) A theoretical characterization of curvature controlled adhesive properties of bio-inspired membranes. Biomimetics 1: 3 [Google Scholar]
- Anderson CT, Carroll A, Akhmetova L, Somerville C (2010) Real-time imaging of cellulose reorientation during cell wall expansion in Arabidopsis roots. Plant Physiol 152: 787–796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnaud C, Bonnot C, Desnos T, Nussaume L (2010) The root cap at the forefront. C R Biol 333: 335–343 [DOI] [PubMed] [Google Scholar]
- Bennett T, van den Toorn A, Sanchez-Perez GF, Campilho A, Willemsen V, Snel B, Scheres B (2010) SOMBRERO, BEARSKIN1, and BEARSKIN2 regulate root cap maturation in Arabidopsis. Plant Cell 22: 640–654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouton S, Leboeuf E, Mouille G, Leydecker MT, Talbotec J, Granier F, Lahaye M, Höfte H, Truong HN (2002) QUASIMODO1 encodes a putative membrane-bound glycosyltransferase required for normal pectin synthesis and cell adhesion in Arabidopsis. Plant Cell 14: 2577–2590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brigham LA, Woo HH, Nicoll SM, Hawes MC (1995) Differential expression of proteins and mRNAs from border cells and root tips of pea. Plant Physiol 109: 457–463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cannesan MA, Durand C, Burel C, Gangneux C, Lerouge P, Ishii T, Laval K, Follet-Gueye ML, Driouich A, Vicré-Gibouin M (2012) Effect of arabinogalactan proteins from the root caps of pea and Brassica napus on Aphanomyces euteiches zoospore chemotaxis and germination. Plant Physiol 159: 1658–1670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper JB, Heuser JE, Varner JE (1994) 3,4-Dehydroproline inhibits cell wall assembly and cell division in tobacco protoplasts. Plant Physiol 104: 747–752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daher FB, Braybrook SA (2015) How to let go: pectin and plant cell adhesion. Front Plant Sci 6: 523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- del Campillo E, Abdel-Aziz A, Crawford D, Patterson SE (2004) Root cap specific expression of an endo-beta-1,4-D-glucanase (cellulase): a new marker to study root development in Arabidopsis. Plant Mol Biol 56: 309–323 [DOI] [PubMed] [Google Scholar]
- Derjaguin BV, Muller VM, Toporov YP (1975) Effect of contact deformations on the adhesion of particles. J Colloid Interface Sci 53: 314–326 [Google Scholar]
- Domozych DS. (1999) Disruption of the Golgi apparatus and secretory mechanism in the desmid, Closterium acerosum, by brefeldin A. J Exp Bot 50: 1323–1330 [Google Scholar]
- Domozych DS, Domozych CE (2014) Multicellularity in green algae: upsizing in a walled complex. Front Plant Sci 5: 649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Driouich A, Durand C, Cannesan MA, Percoco G, Vicré-Gibouin M (2010) Border cells versus border-like cells: are they alike? J Exp Bot 61: 3827–3831 [DOI] [PubMed] [Google Scholar]
- Driouich A, Durand C, Vicré-Gibouin M (2007) Formation and separation of root border cells. Trends Plant Sci 12: 14–19 [DOI] [PubMed] [Google Scholar]
- Driouich A, Follet-Gueye ML, Vicré-Gibouin M, Hawes M (2013) Root border cells and secretions as critical elements in plant host defense. Curr Opin Plant Biol 16: 489–495 [DOI] [PubMed] [Google Scholar]
- Durand C, Vicré-Gibouin M, Follet-Gueye ML, Duponchel L, Moreau M, Lerouge P, Driouich A (2009) The organization pattern of root border-like cells of Arabidopsis is dependent on cell wall homogalacturonan. Plant Physiol 150: 1411–1421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harholt J, Jensen JK, Sørensen SO, Orfila C, Pauly M, Scheller HV (2006) ARABINAN DEFICIENT 1 is a putative arabinosyltransferase involved in biosynthesis of pectic arabinan in Arabidopsis. Plant Physiol 140: 49–58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawes MC, Gunawardena U, Miyasaka S, Zhao X (2000) The role of root border cells in plant defense. Trends Plant Sci 5: 128–133 [DOI] [PubMed] [Google Scholar]
- Hawes MC, Pueppke SG (1986) Sloughed peripheral root cap cells: yield from different species and callus formation from single cells. Am J Bot 73: 1466–1473 [Google Scholar]
- Hayashi T, Kaida R (2011) Functions of xyloglucan in plant cells. Mol Plant 4: 17–24 [DOI] [PubMed] [Google Scholar]
- Hervé C, Marcus SE, Knox JP (2011) Monoclonal antibodies, carbohydrate-binding modules, and the detection of polysaccharides in plant cell walls. Methods Mol Biol 715: 103–113 [DOI] [PubMed] [Google Scholar]
- Hijazi M, Velasquez SM, Jamet E, Estevez JM, Albenne C (2014) An update on post-translational modifications of hydroxyproline-rich glycoproteins: toward a model highlighting their contribution to plant cell wall architecture. Front Plant Sci 5: 395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kračun SK, Schückel J, Westereng B, Thygesen LG, Monrad RN, Eijsink VG, Willats WG (2015) A new generation of versatile chromogenic substrates for high-throughput analysis of biomass-degrading enzymes. Biotechnol Biofuels 8: 70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langhans M, Weber W, Babel L, Grunewald M, Meckel T (2017) The right motifs for plant cell adhesion: what makes an adhesive site? Protoplasma 254: 95–108 [DOI] [PubMed] [Google Scholar]
- Lewis KC, Selzer T, Shahar C, Udi Y, Tworowski D, Sagi I (2008) Inhibition of pectin methyl esterase activity by green tea catechins. Phytochemistry 69: 2586–2592 [DOI] [PubMed] [Google Scholar]
- Lewis MW, Leslie ME, Liljegren SJ (2006) Plant separation: 50 ways to leave your mother. Curr Opin Plant Biol 9: 59–65 [DOI] [PubMed] [Google Scholar]
- Manns D, Deutschle AL, Saakec B, Meyer AS (2014) Methodology for quantitative determination of the carbohydrate composition of brown seaweeds (Laminariaceae). RSC Advances 4: 25736–25746 [Google Scholar]
- McCartney L, Steele-King CG, Jordan E, Knox JP (2003) Cell wall pectic (1→4)-beta-d-galactan marks the acceleration of cell elongation in the Arabidopsis seedling root meristem. Plant J 33: 447–454 [DOI] [PubMed] [Google Scholar]
- McKenzie BM, Mullins CE, Tisdall JM, Bengough AG (2013) Root-soil friction: quantification provides evidence for measurable benefits for manipulation of root-tip traits. Plant Cell Environ 36: 1085–1092 [DOI] [PubMed] [Google Scholar]
- Moller I, Sørensen I, Bernal AJ, Blaukopf C, Lee K, Øbro J, Pettolino F, Roberts A, Mikkelsen JD, Knox JP, et al. (2007) High-throughput mapping of cell-wall polymers within and between plants using novel microarrays. Plant J 50: 1118–1128 [DOI] [PubMed] [Google Scholar]
- Mouille G, Ralet MC, Cavelier C, Eland C, Effroy D, Hématy K, McCartney L, Truong HN, Gaudon V, Thibault JF, et al. (2007) Homogalacturonan synthesis in Arabidopsis thaliana requires a Golgi-localized protein with a putative methyltransferase domain. Plant J 50: 605–614 [DOI] [PubMed] [Google Scholar]
- Mouille G, Robin S, Lecomte M, Pagant S, Höfte H (2003) Classification and identification of Arabidopsis cell wall mutants using Fourier-transform infrared (FT-IR) microspectroscopy. Plant J 35: 393–404 [DOI] [PubMed] [Google Scholar]
- Mravec J, Kračun SK, Rydahl MG, Westereng B, Miart F, Clausen MH, Fangel JU, Daugaard M, Van Cutsem P, De Fine Licht HH, et al. (2014) Tracking developmentally regulated post-synthetic processing of homogalacturonan and chitin using reciprocal oligosaccharide probes. Development 141: 4841–4850 [DOI] [PubMed] [Google Scholar]
- Nguema-Ona E, Vicré-Gibouin M, Gotté M, Plancot B, Lerouge P, Bardor M, Driouich A (2014) Cell wall O-glycoproteins and N-glycoproteins: aspects of biosynthesis and function. Front Plant Sci 5: 499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogawa M, Kay P, Wilson S, Swain SM (2009) ARABIDOPSIS DEHISCENCE ZONE POLYGALACTURONASE1 (ADPG1), ADPG2, and QUARTET2 are polygalacturonases required for cell separation during reproductive development in Arabidopsis. Plant Cell 21: 216–233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park YB, Cosgrove DJ (2015) Xyloglucan and its interactions with other components of the growing cell wall. Plant Cell Physiol 56: 180–194 [DOI] [PubMed] [Google Scholar]
- Pedersen HL, Fangel JU, McCleary B, Ruzanski C, Rydahl MG, Ralet MC, Farkas V, von Schantz L, Marcus SE, Andersen MC, et al. (2012) Versatile high resolution oligosaccharide microarrays for plant glycobiology and cell wall research. J Biol Chem 287: 39429–39438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puhlmann J, Bucheli E, Swain MJ, Dunning N, Albersheim P, Darvill AG, Hahn MG (1994) Generation of monoclonal antibodies against plant cell-wall polysaccharides. I. Characterization of a monoclonal antibody to a terminal alpha-(1→2)-linked fucosyl-containing epitope. Plant Physiol 104: 699–710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ralet MC, Tranquet O, Poulain D, Moïse A, Guillon F (2010) Monoclonal antibodies to rhamnogalacturonan I backbone. Planta 231: 1373–1383 [DOI] [PubMed] [Google Scholar]
- Roberts JA, Elliott KA, Gonzalez-Carranza ZH (2002) Abscission, dehiscence, and other cell separation processes. Annu Rev Plant Biol 53: 131–158 [DOI] [PubMed] [Google Scholar]
- Robinson DG, Ding Y, Jiang L (2016) Unconventional protein secretion in plants: a critical assessment. Protoplasma 253: 31–43 [DOI] [PubMed] [Google Scholar]
- Rost TL. (2011) The organization of roots of dicotyledonous plants and the positions of control points. Ann Bot (Lond) 107: 1213–1222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheller HV, Ulvskov P (2010) Hemicelluloses. Annu Rev Plant Biol 61: 263–289 [DOI] [PubMed] [Google Scholar]
- Sherwood RT. (1987) Papilla formation in corn root cap cells and leaves inoculated with Collectotrichum graminicola. Phytopathology 77: 930–934 [Google Scholar]
- Stephenson MB, Hawes MC (1994) Correlation of pectin methylesterase activity in root caps of pea with root border cell separation. Plant Physiol 106: 739–745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swain S, Kay P, Ogawa M (2011) Preventing unwanted breakups: using polygalacturonases to regulate cell separation. Plant Signal Behav 6: 93–97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor CM, Karunaratne CV, Xie N (2012) Glycosides of hydroxyproline: some recent, unusual discoveries. Glycobiology 22: 757–767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velasquez SM, Ricardi MM, Dorosz JG, Fernandez PV, Nadra AD, Pol-Fachin L, Egelund J, Gille S, Harholt J, Ciancia M, et al. (2011) O-Glycosylated cell wall proteins are essential in root hair growth. Science 332: 1401–1403 [DOI] [PubMed] [Google Scholar]
- Verger S, Chabout S, Gineau E, Mouille G (2016) Cell adhesion in plants is under the control of putative O-fucosyltransferases. Development 143: 2536–2540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verhertbruggen Y, Marcus SE, Chen J, Knox JP (2013) Cell wall pectic arabinans influence the mechanical properties of Arabidopsis thaliana inflorescence stems and their response to mechanical stress. Plant Cell Physiol 54: 1278–1288 [DOI] [PubMed] [Google Scholar]
- Wen F, Celoy RM, Nguyen T, Zeng W, Keegstra K, Immerzeel P, Pauly M, Hawes MC (2008) Inducible expression of Pisum sativum xyloglucan fucosyltransferase in the pea root cap meristem, and effects of antisense mRNA expression on root cap cell wall structural integrity. Plant Cell Rep 27: 1125–1135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen F, Zhu Y, Hawes MC (1999) Effect of pectin methylesterase gene expression on pea root development. Plant Cell 11: 1129–1140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willats WG, Limberg G, Buchholt HC, van Alebeek GJ, Benen J, Christensen TM, Visser J, Voragen A, Mikkelsen JD, Knox JP (2000) Analysis of pectic epitopes recognised by hybridoma and phage display monoclonal antibodies using defined oligosaccharides, polysaccharides, and enzymatic degradation. Carbohydr Res 327: 309–320 [DOI] [PubMed] [Google Scholar]
- Willats WG, McCartney L, Steele-King CG, Marcus SE, Mort A, Huisman M, van Alebeek GJ, Schols HA, Voragen AG, Le Goff A, et al. (2004) A xylogalacturonan epitope is specifically associated with plant cell detachment. Planta 218: 673–681 [DOI] [PubMed] [Google Scholar]
- Willats WGT, Orfila C, Limberg G, Buchholt HC, van Alebeek GJ, Voragen AG, Marcus SE, Christensen TM, Mikkelsen JD, Murray BS, et al. (2001) Modulation of the degree and pattern of methyl-esterification of pectic homogalacturonan in plant cell walls: implications for pectin methyl esterase action, matrix properties, and cell adhesion. J Biol Chem 276: 19404–19413 [DOI] [PubMed] [Google Scholar]
- Willats WGT, Steele-King CG, Marcus SE, Knox JP (1999) Side chains of pectic polysaccharides are regulated in relation to cell proliferation and cell differentiation. Plant J 20: 619–628 [DOI] [PubMed] [Google Scholar]
- Wolf S, Mravec J, Greiner S, Mouille G, Höfte H (2012) Plant cell wall homeostasis is mediated by brassinosteroid feedback signaling. Curr Biol 22: 1732–1737 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.