Plants lacking the guard cell-expressed, proline-rich secreted protein FOCL1 are drought tolerant because they fail to form a stomatal cuticular ledge and produce stomatal pores that are covered by a continuous cuticle.
Abstract
Stomata are formed by a pair of guard cells which have thickened, elastic cell walls to withstand the large increases in turgor pressure that have to be generated to open the pore that they surround. We have characterized FOCL1, a guard cell-expressed, secreted protein with homology to Hyp-rich cell wall proteins. FOCL1-GFP localizes to the guard cell outer cuticular ledge and plants lacking FOCL1 produce stomata without a cuticular ledge. Instead the majority of stomatal pores are entirely covered over by a continuous fusion of the cuticle, and consequently plants have decreased levels of transpiration and display drought tolerance. The focl1 guard cells are larger and less able to reduce the aperture of their stomatal pore in response to closure signals suggesting that the flexibility of guard cell walls is impaired. FOCL1 is also expressed in lateral root initials where it aids lateral root emergence. We propose that FOCL1 acts in these highly specialized cells of the stomata and root to impart cell wall strength at high turgor and/or to facilitate interactions between the cell wall and the cuticle.
Plant cell walls typically consist of a network of cellulose, hemicellulose, pectin, and lignin, but also contain many structural proteins of unknown function such as Hyp-rich glycoproteins (HRGPs; Lamport et al., 2011). This group of proteins includes Pro-rich proteins (PRPs), arabinogalactan proteins (AGPs), and extensins. HRGPs are sequentially posttranslationally modified by Pro 4-hydroxylases, converting Pro residues to Hyp, and then by O-glycosyltransferases adding sugar moieties to Hyp residues. These posttranslational modifications are thought to contribute to the structural and possibly to the intercellular communication properties of the cell wall. Extensins are the best characterized of the plant HRGPs and these are commonly arabinosylated by the HPAT family of O-glycosyltransferases before being arabinogalactosylated (Velasquez et al., 2011; Ogawa-Ohnishi et al., 2013). Extensins are cross-linked at Tyr residues by peroxidases and processed by proteases which insolubilize and lock the extensins into the cell wall structure (Helm et al., 2008; Lamport et al., 2011). Extensins were originally isolated from elongating coleoptiles over 50 years ago (Lamport, 1963) and proposed to be involved in cell wall extensibility, but this has never been functionally confirmed (Lamport et al., 2011). Nonetheless, roles for extensins have been observed in Arabidopsis (Arabidopsis thaliana) embryo and root development (Cannon et al., 2008; Velasquez et al., 2011); embryos lacking EXT4 are defective with irregular cell size and shape and root hairs lacking EXT6-7, EXT10, and EXT12 show reduced root hair elongation. Similar root hair phenotypes are seen in plants lacking Pro 4-hydroxylase activity due to reduced Pro hydroxylation and O-arabinosylation of extensins, suggesting that these posttranslationally modified proteins influence root hair growth (Velasquez et al., 2011). There are 51 genes annotated as encoding extensins or extensin-like proteins in the Arabidopsis genome (Showalter et al., 2010), and it appears likely from their specific expression patterns that they are involved in a range of growth, developmental, and stress responses (Merkouropoulos and Shirsat, 2003), although plants manipulated to produce abnormally high levels of EXT1 appear to develop normally with the exception of having thicker stems (Roberts and Shirsat, 2006). Physiological roles in aerial tissues remain elusive and the failure to identify a function for extensins and indeed other HRGPs in shoots is likely to be due to redundancy within this large gene family, a common problem in plant cell wall protein studies.
The studies of extensins in root hairs described above indicate that it is possible to gain information about their function in a specialized and well-studied cell type. We therefore decided that because of the unique properties of guard cell walls and the tractability of measuring stomatal development and function, it might be possible to identify the function of a cell wall protein that is predominantly expressed in guard cells. Pairs of guard cells surround and adjust the aperture of stomatal pores in response to environmental signals that trigger changes to the turgor pressure of the cells (Kollist et al., 2014). Large turgor changes within guard cells occur over short time scales (typically minutes), with turgor increases causing stomatal opening, and decreases causing closure. Thus, in comparison to other cell types, guard cells require particularly strong and elastic cell walls. However, there is currently no genetic evidence of a role for cell wall HRGPs in stomatal function, although individual polysaccharide moieties of the mature guard cell wall are known to be important for pore aperture control as removal of the arabinan component of the guard cell wall or modifying pectin methyl esterification impairs stomatal opening and closing (Jones et al., 2003; Amsbury et al., 2016).
During leaf epidermal development the division of guard mother cells forms pairs of guard cells. Stomatal pores subsequently form between each guard cell pair, but little is known of the processes regulating guard cell wall maturation and stomatal pore formation. Microscopy observations show that the cell walls between adjacent guard cells (which are destined to line each stomatal pore) thicken and separate. The exterior surface of the leaf becomes coated with a waterproof layer of cuticle and an extended ledge or lip forms around each stomatal pore, which is known as the outer cuticular ledge (OCL). The exact functions of this cuticular ledge are unknown, but it has been proposed to prevent water loss by sealing the pore when the stomate is closed, to prevent water droplets entering when the pore is open, and to tilt its orientation to help open and close the stomatal pore (Fricker and Wilmer, 1996; Zhao and Sack, 1999; Kozma and Jenks, 2007). No specific proteins have yet been localized to the OCL. We report here that Arabidopsis plants lacking an OCL-localized gene product annotated (by TAIR; www.arabidopsis.org) as an “extensin-like protein” have larger stomata, show defects in stomatal closure, and most notably possess a malformed outer cuticular ledge that forms a fused cuticular layer over the stomatal pores. Hence, we named this protein FUSED OUTER CUTICULAR LEDGE1 (FOCL1). In addition to its roles in stomata, we also report that FOCL1 influences lateral root emergence. Our results therefore provide a link between a secreted Pro-rich protein and its function in the cell walls of specific plant cell types.
RESULTS
FOCL1 Has Features of Hyp-Rich Cell Wall Glycoproteins
The predicted amino acid sequence encoded by Arabidopsis gene At2g16630, named here as FOCL1, contains a putative signal sequence suggesting that it is secreted, and a Pro-rich domain with several motifs typical of HPRGs, including eight Pro-Val motifs, which are normally hydroxylated, and four repeated triple Pro residues that are most likely posttranslationally modified (Kieliszewski and Lamport, 1994; Menke et al., 2000; Fig. 1A; Supplemental Fig. S1). Nonetheless, the FOCL1 protein is not a classical extensin as it lacks the characteristic conserved Ser-poly-Pro repeats and the YXY or VYX domains required for Tyr intermolecular cross-linking (Kieliszewski and Lamport, 1994). However, FOCL1 may be intermolecularly bonded by Tyr residues in a different sequence context, as seen for the HRGP, PRP10 (Chen et al., 2015). FOCL1 orthologs occur across a wide range of plant species, but the closest homolog of FOCL1 in Arabidopsis encodes a protein of unknown function with only 24% identity. At2g20515 has similarity with the C terminus of FOCL1 but lacks the N-terminal and central Pro-rich regions of FOCL1 (Supplemental Fig. S1).
Figure 1.
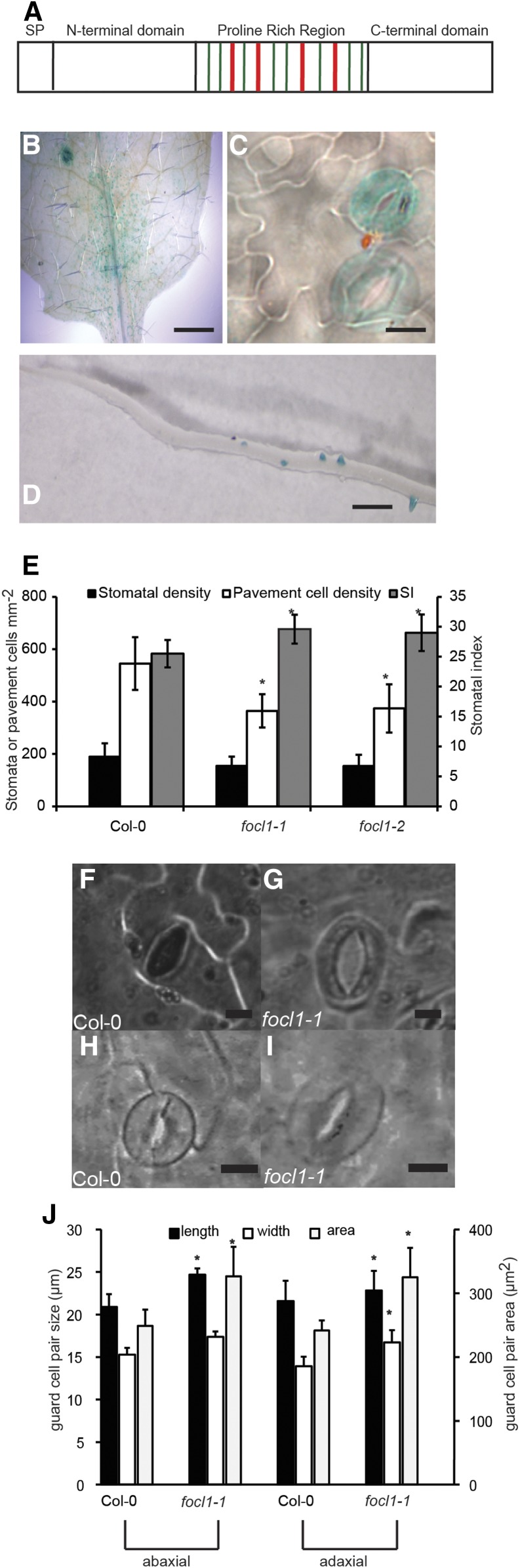
FOCL1 encodes a Pro-rich protein, is expressed in guard cells and roots, and affects stomatal index and stomatal complex size. A, Domain structure of the FOCL1 protein to illustrate positions of Pro, Val, and triple Pro motifs typical of HPRGs. Potentially hydroxylated prolines are indicated in green (PV context) or red (PPP context). SP, Signal peptide. B to D, Histochemical staining of 2-week-old Arabidopsis seedlings expressing pFOCL:GUS. B, Immature leaf; C, developing epidermis; D, developing lateral root. E, Stomatal index and pavement cell density of abaxial surfaces of fully expanded leaves. n = 7 to 9 plants; means of three areas from one leaf of each plant were compared. Representative experiment of three independent experiments is shown. F and G, Images of epidermal imprints of adaxial leaf surfaces. H and I, Cleared tissue of mature leaves. Bars = 500 μm in B, 10 μm in C, 250 μm in D, 5 μm in F and G, and 10 μm in H and I. J, Stomatal length and width and area. n = 4 to 7 plants; means of measurements from at least 10 stomates from one leaf of each plant were compared; asterisk indicates significant statistical difference from Col-0, P < 0.05. Error bars = sd.
FOCL1 also shows conservation with an atypical AGP known as AGP31, both possessing distinctive tandem Pro-rich PKVPVISPDPPA/TTLPP domains (Showalter et al., 2010; Liu and Mehdy, 2007; Supplemental Fig. S2). As Pro residues of the AGP31 Pro-rich domain are known to be hydroxylated and glycosylated (Hijazi et al., 2012), it is likely that this is also the case for the conserved domain in FOCL1. However, FOCL1 and AGP31 have lower Pro content than many HRGPs and are therefore unlikely to have very high levels of posttranslational glycosylation. Thus, FOCL1 resembles a hydroxylated Pro-rich, structural cell wall protein, but it is neither a classical extensin nor a typical AGP.
BLAST analysis revealed homology of the FOCL1 N terminus with “Pollen Ole e 1 allergen and extensin family” proteins, but these lack the C-terminal domain and Pro-rich region present in FOCL1 (data not shown), suggesting that FOCL1 might be a chimeric protein, with a Pollen Ole e 1 extensin-like domain at the N terminus, a Pro-rich AGP31-like tandem repeat in the middle of its sequence, and an At2g20515-like domain at the C terminus.
FOCL1 Is Expressed in Guard Cells and Lateral Root Primordia
Published transcriptome data indicate that FOCL1 is strongly expressed in guard cell protoplasts and in roots and that expression levels are lower in root than in shoot tissue (Zimmermann et al., 2005; Winter et al., 2007; Yang et al., 2008). We examined FOCL1 expression patterns using plants expressing the β-glucuronidase gene under the control of the DNA region upstream of the FOCL1 coding sequence (pFOCL1:GUS). GUS expression was predominantly observed in immature and mature guard cells (Fig. 1, B and C). Staining was not present in guard cell precursors (such as guard mother cells), suggesting that FOCL1 is not directly involved in the formation or patterning of stomata during shoot development. Staining was also seen in emerged lateral roots (Fig. 1D) and developing primordia. Together, these results suggested that FOCL1 is an HRGP that could potentially function in the cell walls during guard cell maturation and function and during lateral root development.
Plants Lacking FOCL1 Have Large Stomata
Two independent Arabidopsis lines with T-DNA insertions 200 bp apart in the third exon of the FOCL1 gene were isolated and named focl1-1 and focl1-2 (Supplemental Fig. S3A). Expression of the FOCL1 transcript was not detectable by RT-PCR of homozygous focl1-1 plants with primers spanning the insertion site (Supplemental Fig. S3B), but a product was seen in focl1-2 with primers upstream of the insertion site, suggesting a truncated protein could be produced. focl1-1 and focl1-2 plants were both smaller than the wild type, with reduced rosette width at bolting. Growth of focl1-1 plants was more severely affected than focl1-2 plants, and these were smaller and paler than focl1-2 (Supplemental Fig. S4). As we had observed strong expression of FOCL1 in guard cells, we examined the leaf surfaces of these plants using epidermal imprints. Both focl1-1 and focl1-2 showed significant increases in abaxial stomatal index in the experiment shown in Figure 1E due to a significant decrease in the number of pavement cells. However, we observed no consistent alteration in stomatal density in replicated experiments, no clustering of stomata, and no arrested precursor cells as often seen in stomatal developmental mutants (e.g. Hunt and Gray, 2009). Instead, we observed an unusual phenotype; in both imprints and in cleared images of whole leaves, focl1 stomata were obviously larger than normal and had a pore that appeared to be different from the wild type (Fig. 1, F–I). Measurement of stomatal dimensions confirmed significant increases in width and length of focl1-1 guard cell pairs; on the abaxial and adaxial leaf surfaces, focl1-1 stomata were 31% and 34% larger than wild-type stomata when their area was calculated as an ellipse (Fig. 1J). To confirm that both the reduced rosette growth and larger stomata were caused by lack of FOCL1, focl1-1 and focl1-2 mutations were complemented by transformation with a genomic fragment containing the wild-type FOCL gene with an N-terminally fused GFP (focl1-1pFOCL1:GFP-FOCL1) or C terminally fused MYC tag (focl1-2pFOCL1:FOCL-MYC1). This GFP-FOCL1 protein rescued rosette growth and returned stomatal complex sizes to wild-type values in both mutant backgrounds (Supplemental Figs. S4, A and B, and S5, A and B).
FOCL1 Is Involved in the Formation of Stomatal Pore Outer Cuticular Ledges
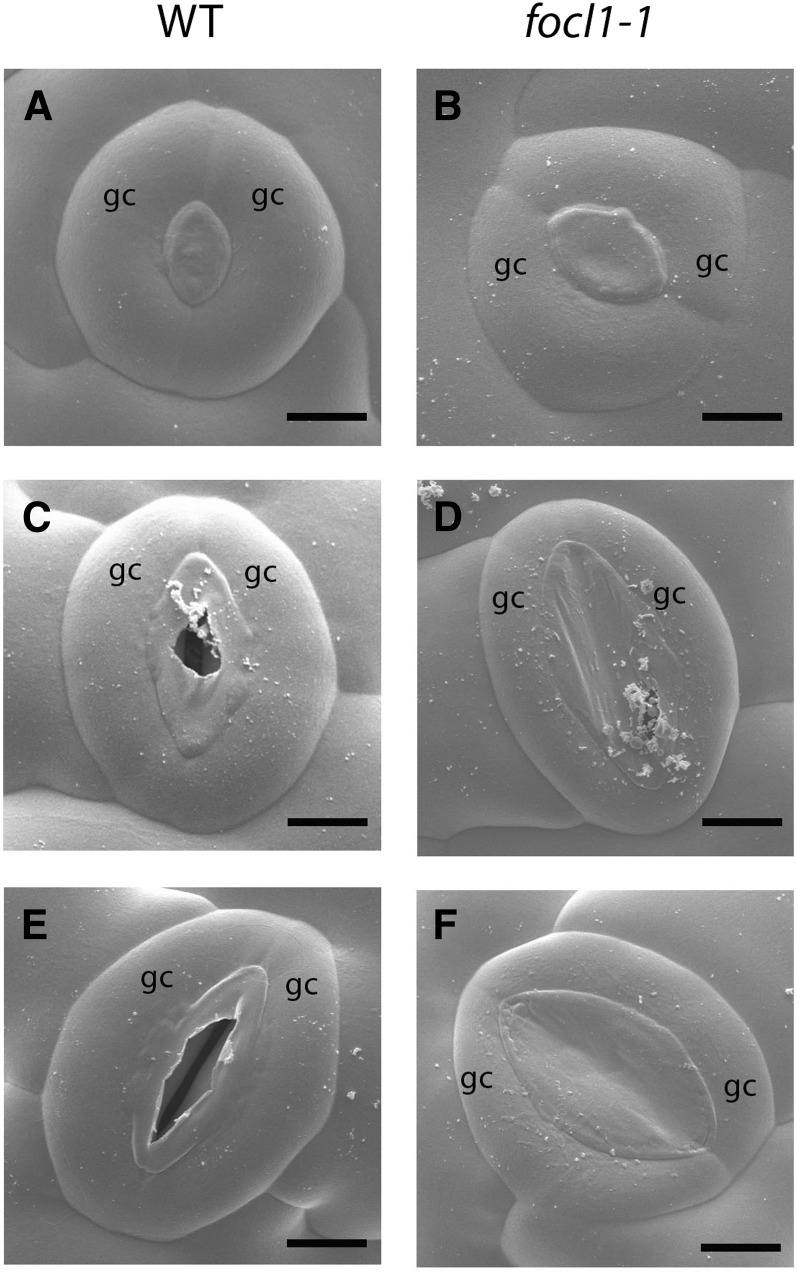
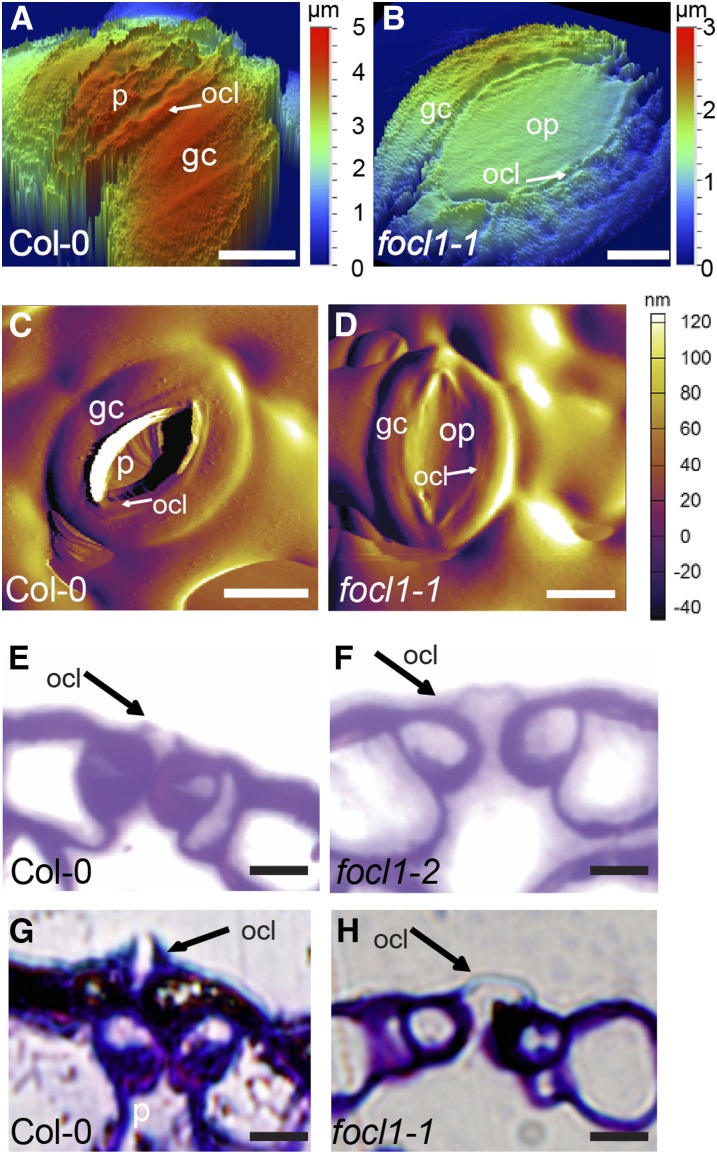
To investigate focl1 stomatal morphology in detail, we examined leaf surfaces using cryo-scanning electron microscopy (cryo-SEM) on 3-week-old plants. This revealed that in immature “rounded” stomates, the pore is covered by a cuticular layer, which appears to tear to form the outer cuticular ledge and to reveal and surround the pore as the guard cells lengthen and mature. In contrast, focl1 stomatal pores remained covered over or occluded by what appears to be an extension of the cuticle and do not form an outer cuticular ledge around the pore (Fig. 2, A–F). Further SEM analysis showed that even after fixation and dehydration, the majority (∼90%) of focl1 pores on mature leaves and stems remain occluded (Supplemental Fig. S6) with a minority of stomata forming a slit-like opening (Fig. 2; Supplemental Fig. S7). To confirm that the numerous occluded stomatal pores were not an artifact of electron microscopy, we imaged the epidermal surface topography of several stomates from fresh leaf tissue using both vertical scanning interferometry (VSI; Fig. 3, A and B) and atomic force microscopy (AFM; Fig. 3, C and D; Supplemental Fig. S8). These two techniques, which physically probe the surface of an object to measure height differences, both confirmed that focl1 stomatal pores are covered by what appears to be a continuous layer of cuticle. Furthermore, light microscopy of stained cross sections of stomata also revealed a continuous “fused” cuticular ledge formed between the edges of the two guard cells surrounding the pore (Fig. 3, E–H).
Figure 2.
FOCL1 is required for formation of the stomatal outer cuticular ledge. Cryo-SEM images of wild-type Col-0 (A, C, and E) and focl1-1 (B, D, and F) stomata at different stages of development reveal the occlusion of mature focl1-1 stomata by a membranous cuticle. A and B, Both wild-type and focl1-1 developing stomates have a plug of material in the pore between guard cells. C, In larger wild-type stomata, this material appears to be torn apart to reveal the stomatal pore, whereas in focl1-1 stomata (D), the pore remains generally occluded, although some tearing to reveal a subtending pore is visible. E, In mature wild-type stomata, a cuticular ridge bordering the central pore is formed. F, In mature focl1-1 stomata, the central pore can remain totally blocked by the membranous cuticular material. gc, Guard cell. Bars = 4 µm in A and B, 5 µm in C and D, and 6 µm in E and F.
Figure 3.
focl1 mutants have fused outer cuticular ledges. A and B, Abaxial surfaces of wild-type Col-0 and focl1 stomates imaged by VSI. Depth is indicated in nanometers. C and D, AFM deflection images of stomates. E and F, Transverse sections of stem epidermis stained with Toluidine blue. Position of outer cuticular ledges (ocl) indicated by arrows. G and H, Adaxial leaf epidermis. Bars = 5 μm in A, B, E, and F and 10 μm in C, D, G, and H. p, Stomatal pore; ocl, outer cuticular ledge; gc, guard cell; op, occluded pore.
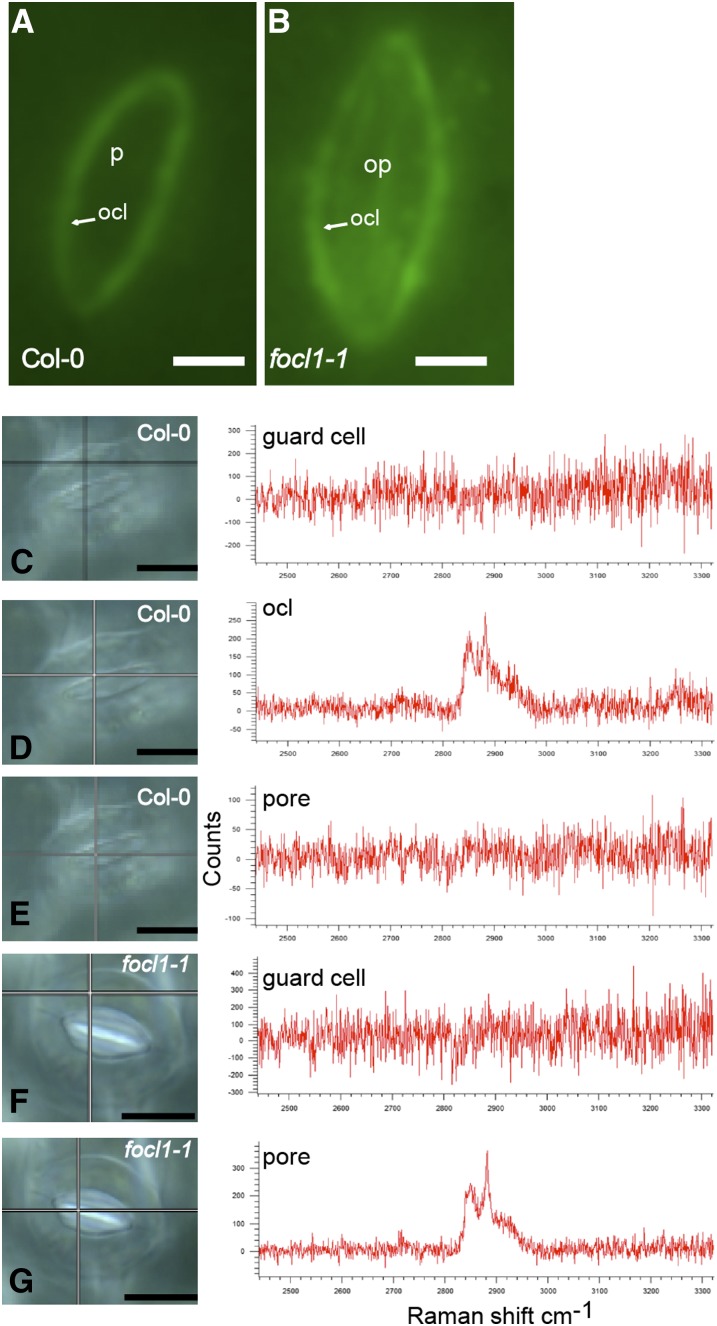
Staining with the lipophilic stain Nile red revealed a sharp discrete cuticular ledge surrounding the outer edge of wild-type stomatal pores, attached to the guard cells (Fig. 4A). In focl1 stomates, this staining was more diffuse and spread across the whole pore area (Fig. 4B). To further investigate the chemical nature of this lipophilic material covering the stomatal pores, we used Raman microscopy. The wild-type and focl1-1 guard cells produced similar Raman spectra when central regions of the cells distant from the ledge were analyzed (Fig. 4, C and F). Peaks of wavelength 2,840 and 2,880, indicative of epicuticular waxes (Greene and Bain, 2005), were observed in the cuticular ledge region of wild-type guard cells (Fig. 4D). Similar peaks in the spectra were observed after analysis of spots in the middle of the occluded focl1-1 pore (Fig. 4G), whereas the spectrum over the wild-type pore aperture area did not show peaks at these wavelengths (Fig. 4E). Thus, it appears that guard cells lacking FOCL1 are able to produce epicuticular wax material but are unable to properly form a cuticular ledge around their stomatal pores; consequently, the cuticle forms a continuous layer across the pore.
Figure 4.
A lipid rich cuticle extends across the pore of focl1 stomata. Wild-type Col-0 (A) and focl1-1 (B) stomatal surfaces imaged by fluorescent microscopy after staining with Nile red (which fluoresces green). C to G, Col-0 (C–E) and focl1-1 stomatal surfaces (F and G) were imaged using Raman spectroscopy over a range of wavelengths from ∼2,400 to 3,300 nm. Maps were obtained across stomata and point scans (indicated by cross-hairs) shown for different regions. Point scans taken from the guard cell surface (C and F), the wild-type cuticular ledge (D), and from the center of the pore region (E and H). Maps were taken from at least three independent biological samples, with similar results obtained in each case. Bars = 5 µm in A and B and 6 µm in C to G.
FOCL1 Protein Localizes to the OCL of Guard Cells
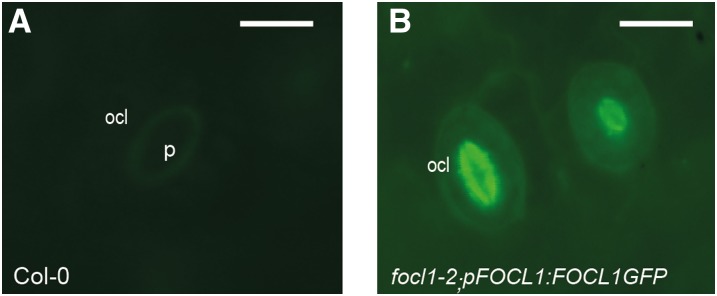
To investigate whether FOCL1 is a secreted cell wall protein as predicted by its sequence, and whether it could act in the formation of the guard cell cuticular ledge, we examined the subcellular localization of the FOCL1 protein. To do this, we analyzed the expression of a FOCL1-GFP fusion protein in vivo (in focl1-2 plants transformed with the promoter and coding region of FOCL1 in frame with a C-terminal GFP tag). The results shown in Figure 5, A and B, indicate that the fluorescent fusion protein accumulates specifically in the cuticular ledge of guard cells, further indicating that FOCL1 is secreted from guard cells and acts directly in the formation of the cuticular ledge.
Figure 5.
FOCL1-GFP localizes to the cuticular ledge. Seedlings of T2 lines of focl1-2 expressing pFOCL1:FOCL1-GFP were analyzed by epifluorescence microscopy. Wild-type Col-0 samples showed weak autofluorescence (A) compared to complemented focl1-2 plants (B) where FOCL1-GFP signal is largely restricted to the OCL in developing (right) and mature guard cell (left). Bar = 15 µm.
Lack of FOCL1 Impairs Stomatal Aperture Control and Transpiration
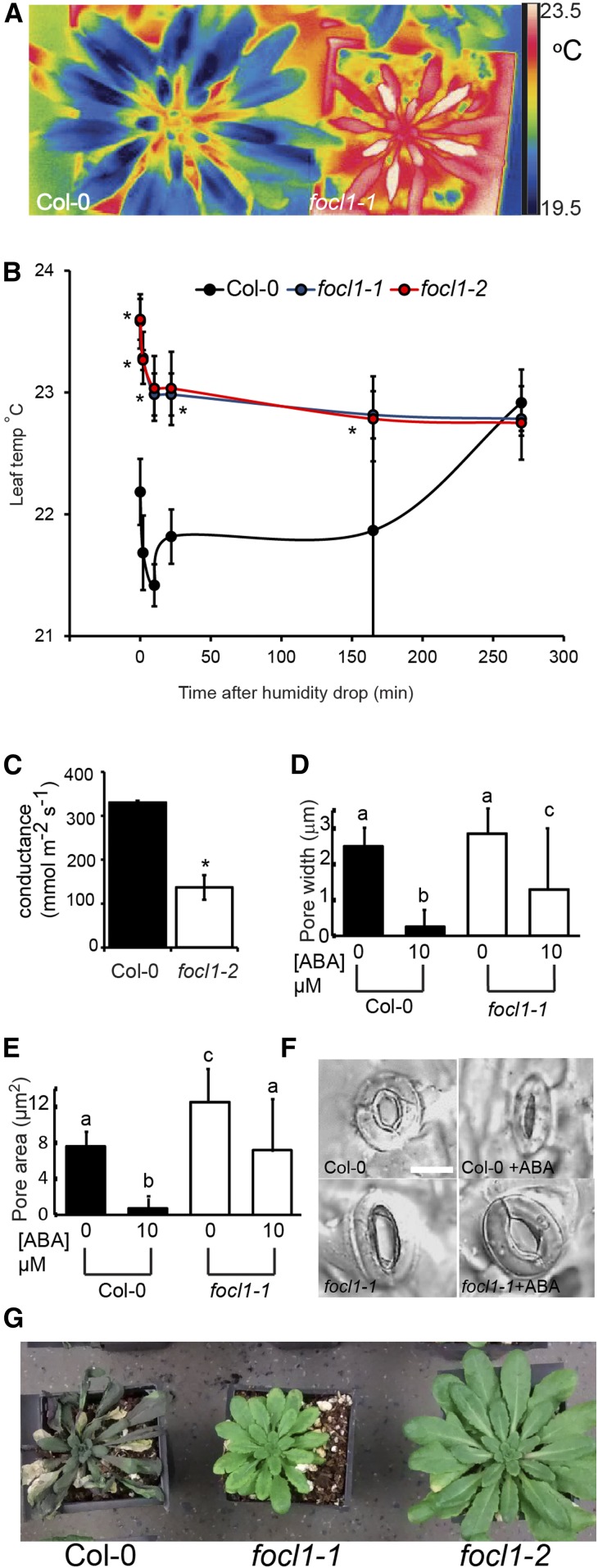
We tested whether the fused stomatal cuticle phenotype of focl1 mutants would affect the ability of plants to carry out gas exchange. To assess transpiration, plants were grown at high humidity and kept well-watered (in a propagator with a lid). Their leaf surface temperatures were monitored by infrared thermography, which is a proxy measure of transpiration rate. On average, mature leaves of FOCL1 mutants were ∼1°C warmer than control plants and remained hotter for at least 2.5 h after humidity was reduced (by removal of the propagator lid), suggesting a reduced level of transpiration and evaporative cooling in the focl1 plants (Fig. 6, A and B). The focl1 plants retained their warmer temperature throughout the experiment, while the wild-type plants slowly adjusted to the less humid environment by reducing their level of transpiration and eventually increasing their temperature to a similar level to that of the mutants (presumably by closing their stomatal pores). Leaf porometry measurements on well-watered unperturbed plants also confirmed a substantially reduced level of stomatal conductance from focl1 leaves (Fig. 6C), which is consistent with the observation that focl1 stomata are partially or completely occluded by a covering of cuticle (Figs. 2 and 3). We confirmed that the reduced transpiration phenotype was due to loss of FOCL1 by showing that leaf temperatures were returned to wild-type levels when focl1-1 or focl1-2 was complemented with the wild-type gene (in focl1-1pFOCL1:FOCL-MYC1, focl1-1pFOCL1:GFP-FOCL1, or focl1-2pFOCL1:FOCL-MYC1; Supplemental Fig. S9).
Figure 6.
focl1 mutants have impaired transpiration and stomatal aperture control. A, Infrared thermal images of representative mature Col-0 and focl1 plants taken at start of experiment. B, Time course of mean leaf temperature recorded by infrared thermography after reduction in humidity. focl1-1 and focl1-2 had similar temperatures throughout and are virtually indistinguishable on this graph. n = 6 plants of each genotype with measurements from three leaves of each plant. C, Leaf porometry measurements of Col-0 and focl1-2 stomatal conductance. n = 4 (one leaf from four plants of each genotype). Asterisk indicates significant statistical difference from Col-0, P < 0.01. D and E, Measurements of Col-0 and focl1-1 stomatal pore widths (D) and calculated pore areas (E) following incubation with 10 μm ABA. Bars with identical letters are not statistically different, P < 0.05. n = >100 stomata. Error bars = sd. Data from one independent experiment are shown; a replicated experiment showed similar results. F, Representative images of stomata from D and E. Bar = 10 μm. G, Representative images of 8-week-old plants under drought conditions after water was withheld for 7 d then rewatered for 3 d.
We next explored whether the alterations in the morphology of focl1 stomata and their cuticles affected their ability to close their pores in response to environmental stimuli. To investigate the effect of the lack of FOCL1 on stomatal aperture control, we measured stomatal pores from isolated epidermal strips following incubation with 10 μm abscisic acid (ABA; a plant stress hormone that triggers stomatal closure). All pores in the field of view were measured as it was not possible to tell under light microscopy whether they were covered over or not. Although the focl1 stomata closed to some extent in response to ABA, they were unable to close as fully as wild-type stomata and the width and areas of their pore apertures remained significantly larger (Fig. 6, D–F). To take account of the increased stomatal complex size in focl1 in these experiments, we calculated the relative reductions in pore width and area; in the presence of ABA, wild-type stomatal pore width and area decreased by 90% and 91%, but focl1-1 stomatal pore width and area decreased by only 54% and 42%, respectively. Thus, it appears that loss of FOCL1 leads to impaired guard cell movement. However, despite their impaired ABA-inducible stomatal closure, focl1 plants wilted less readily than the wild type when water was withheld for 7 d, presumably because of their occluded stomata and reduced level of transpiration. In these experiments, both focl1 lines displayed drought tolerance, showing no visible signs of water stress, whereas the wild-type plants were unable to recover when rewatered (Fig. 6G).
FOCL1 Acts during Lateral Root Emergence and Influences Root Architecture
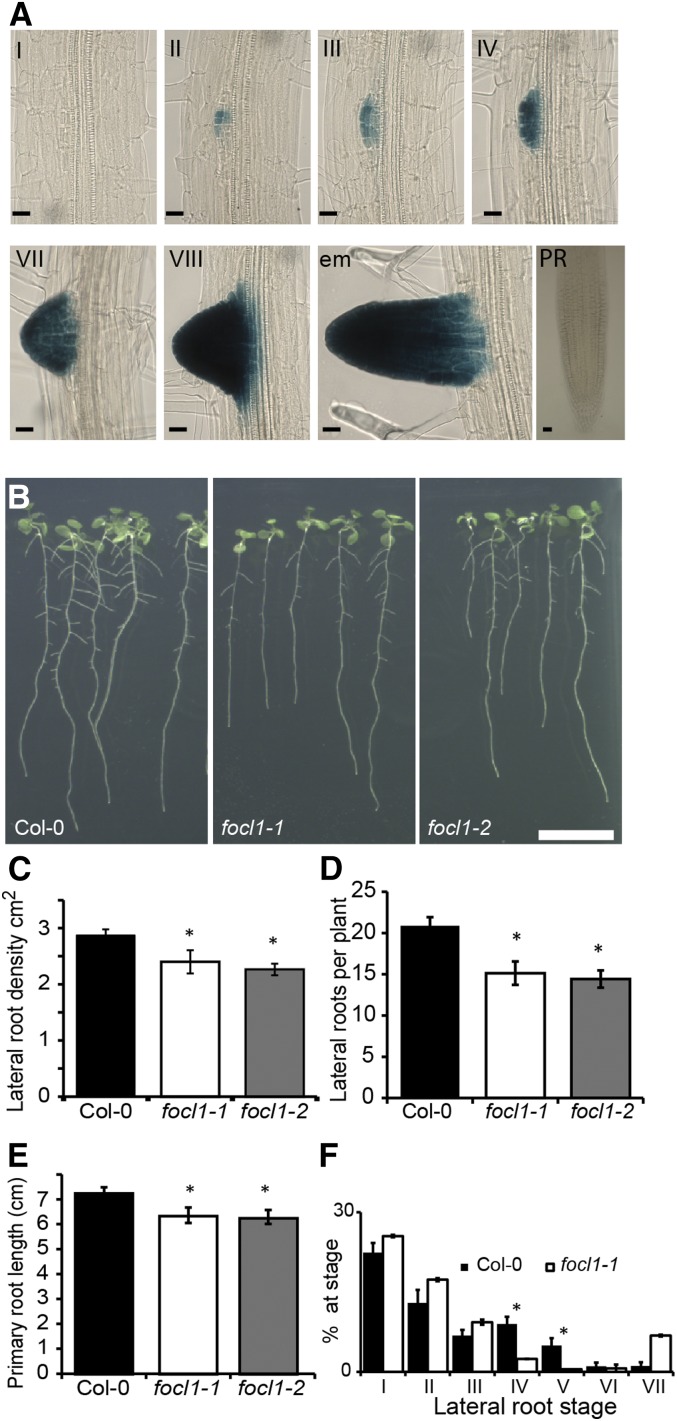
A detailed study of pFOCL1:GUS roots indicated that FOCL1 is expressed at a very early stage of lateral root development. Lateral roots originate from lateral root founder cells located opposite xylem pole pericycle cells. FOCL1 is expressed soon after division of the founder cells (Fig. 7A). GUS expression is first seen in stage II primordia (Péret et al., 2009) and then continues throughout the further stages of lateral root primordia development (stages III to VII) and emergence (Fig. 7A). FOCL1 expression appeared to be specifically associated with the developing and emerging lateral root primordia, and no staining was observed in the surrounding or the overlying cells of the parent root prior to emergence.
Figure 7.
FOCL1 affects root growth. A, GUS expression pattern in pFOCL1:GUS roots. Lateral root emergence stages are indicated with Roman numerals. PR, Primary root. Bars = 20 μm. B, Images of seedlings 8 d after transfer to light for root growth analysis. Bar = 1 cm. C to E, Measurements of roots 11 d after light transfer: lateral root density (C), number of lateral root branches per seedling (D), and primary root lengths (E). n = 12 seedlings. A typical experiment from three independent replicates is shown, each experiment showing a similar result (F) proportion of lateral roots at each stages of primordial development. Asterisk indicates statistically significant from Col-0, P < 0.05. n = 7 (Col-0) or 8 (focl1-1). Error bars = se.
As FOCL1 is expressed in early root development, we explored whether focl1-1 and focl1-2 mutants had defects in lateral root primordia development and emergence. Lateral root numbers, density, primary root lengths, and lateral root stages were measured in 11-d-old seedlings. As shown in Figure 7, C to E, there was a significant reduction in primary root length, lateral root number, and lateral root density in focl1 seedlings compared to the wild type. To further explore if this defect was due to defects in lateral root growth rate or in lateral root initiation and/or emergence, roots were cleared and all stages of lateral root primordia scored. The results shown in Figure 7F indicate that lateral root development was significantly delayed in focl1-1 at stages IV and V. These are the stages when a series of anticlinal and periclinal divisions produce a dome shape structure that protrudes through the cortex toward the epidermal layer prior to emergence. These data indicate that the FOCL1 protein is required for the growth of early lateral root primordia through the parent root.
DISCUSSION
FOCL1 Is a Putative Cell Wall Structural Protein
Plants produce many nonenzymatic proteins that are believed to influence the structure and mechanical properties of their cell walls. However, despite extensive study, the function of most of these proteins remains elusive. We characterized a putative Arabidopsis cell wall structural protein that is required for the correct functioning of guard cells and lateral root initials. The expression of FOCL1 in these discrete cell types of the epidermis and root suggests that this protein is required to create the particular cell wall properties associated with their specific functions. The FOCL1 protein has a predicted signal sequence and Pro-rich region typical of cell wall HRGPs (Kieliszewski and Lamport, 1994). The deduced protein sequence bears limited similarity to extensins except for several potentially hydroxylated Pro residues that are conserved with the Pro rich domain of AGP31 (Supplemental Fig. S2). Thus, FOCL1 is not an extensin and appears to be the only member of a distinct subgroup of Arabidopsis HRGPs. The Pro-rich sequence suggests that FOCL1 most likely interacts with other cell wall components through its primary structure or through specific posttranslational modifications of Hyp residues. Through these interactions it may guide the assembly of new cell wall material, or it may be involved in maintaining the structure and rigidity of the cell wall.
Role and Structure of the Stomatal OCL
The guard cell wall and its extracellular matrix have an important and specialized role in the functioning of stomata and in preventing plant desiccation (Jones et al., 2003). We show that FOCL1 is localized in the guard cell outer cuticular ledge and that plants lacking FOCL1 have their stomata occluded by a continuous layer of cuticle formed from a fused outer cuticular ledge. The retarded growth of these plants is most likely explained by reduced CO2 entry and carbon assimilation, although it is possible that the delayed development of their root initials may also contribute to poor seedling establishment. The timing of FOCL1 expression during guard cell maturation (Fig. 1) and the relatively normal structure of stomates beneath the focl1 cuticle suggest that OCL formation occurs after guard mother cell division and pore formation. This indicates that the focl1 guard cells may have a defect in the framework or assembly of the cell wall that normally sculpts the cuticular ledge into a distinct elliptical shape (Fig. 2). This defective cell wall is also likely to be the reason for the increased size of focl1 stomata; turgor pressure is probably exerting a force to inflate the guard cells that is normally restrained in wild-type guard cells by their more rigid cell wall framework. It is possible, but less likely, that larger stomata could be due to reduced intercellular CO2 concentration resulting from abrogated stomatal function. Low intercellular CO2 concentration has been associated with an increase in stomatal complex size, but this is normally linked to a decrease in stomatal density (Franks and Beerling, 2009), and focl1 showed an increase in stomatal index and no difference in density, suggesting that it is most likely due to an impairment in guard cell wall function. In line with this proposal, we also found that the focl1 stomata were impaired in their ability to close (Fig. 6). This is most likely due to a defect in the guard cell walls and may be indicative of a lack of elasticity in the rather large focl1 guard cells.
The stomatal OCL has been little studied, and FOCL1 is the only protein known to be localized to this structure. Mutant plants that are unable to synthesize cutin, such as lacs2, have diminished cuticular ledges and increased transpiration rates, indicating a probable role in preventing water loss (Li et al., 2007; Macgregor et al., 2008). In contrast, plants lacking FOCL1 have the opposite phenotype: an overgrowth of the cuticular ledges associated with reduced transpiration, suggesting that FOCL1 defines the extent of the OCL in guard cells. The OCL is an extension of the guard cell wall derived from the middle lamella, which contains unesterified pectins and glycans (Majewska-Sawka et al., 2002; Merced and Renzaglia, 2014; Wilson et al., 2015; Amsbury et al., 2016). Plant cuticles are anchored to cell walls by extended pectic lamellae and can be released by pectinase or cellulase treatment (Jeffree, 2006). As the Pro-rich region of FOCL1 is likely to be decorated with pectic side chains containing Gal and arabinose (Hijazi et al., 2012), it is possible that the posttranslationally modified FOCL1 protein normally interacts with pectin or cutin in the OCL where it is located (Fig. 5B). Thus, FOCL1 could be required to facilitate interactions between the guard cell wall and the cuticle that are necessary for OCL formation (Jeffree, 2006).
FOCL1 Is Involved in Lateral Root Development
Plants lacking FOCL1 show defects in primary root and lateral root development. However, in our experiments, pFOCL1:GUS staining was not consistently observed in the primary root (Fig. 7), and it is possible that reduced primary root growth is related to the smaller size of focl1 plants due to their covered-over stomata or that additional FOCL1 promoter regions are required for primary root expression. Nonetheless, the specific GUS expression pattern in developing and emerged lateral roots and lateral root defects in focl1 plants indicate that FOCL1 has a direct effect on lateral root development. The lateral root emergence process is thought to involve a separation of overlying cortex and epidermal cells along their middle lamella. Indeed, cell wall modifications have previously been shown to play a role in lateral root development (Swarup et al., 2008). Several genes encoding cell wall remodeling enzymes show specific expression in the cells overlaying new lateral root primordia and are induced by auxin, which plays a key role in initiation, emergence, and elongation of lateral roots (Swarup et al., 2008; Voss et al., 2015). It is unlikely that FOCL1 is directly involved in this cell separation process though as its expression is restricted to developing lateral root primordia and is never detected in the outer tissues. Interestingly, the reduced cutin levels in the lacs2 mutant cause both a defective OCL and increased lateral root formation (Macgregor et al., 2008), which may be related to an altered root cuticle or indirectly related to the increased transpiration in these mutants. Thus, the focl1 root phenotype, like the focl1 occluded stomata phenotype, might also result from a defective relationship between the cell wall and the cuticle.
Our experimental results indicate that FOCL1 is not required for lateral root initiation but is required for the development of lateral root primordia prior to emergence (Péret et al., 2009). During this period the lateral root initial cells of the pericyle divide periclinally and expand radially, while the endodermal cell layer overlaying the primordium separates to allow the lateral root to expand and protrude through into the cortical layer. The process by which the lateral root passes through these cell layers is poorly understood but is believed to involve both biomechanical forces and cell wall modifications (Geldner, 2013). Indeed, it has recently been suggested that a buildup in turgor pressure within the cells of the primordium through the regulation of water flux by aquaporin activity and auxin enables the lateral root to extend and force itself through the overlying cell layers (Péret et al., 2012). Thus, it appears possible that in lateral root primordia, and in guard cells, FOCL1 could provide the cell wall strength that allows cells to withstand the high turgor pressures required to expand and to fulfill their function. Alternatively, FOCL1 could be involved in guiding and directing newly synthesized components into the cell wall that are required for cellular expansion and function.
In conclusion, we propose that FOCL1 is specifically required for the function of lateral root tip cells and guard cells by playing a role in assembling or strengthening the cell wall and in anchoring it to the developing cuticle. As it appears that the same protein has been recruited to fulfill a function in the walls of cell types with two very different functions, focl1 mutants provide a new tool for the study of HRGPs. We hope that future studies of focl1 roots and stomata may reveal the precise role of a plant Pro-rich cell wall protein.
MATERIALS AND METHODS
Plant Materials
Arabidopsis (Arabidopsis thaliana) plants were grown on a 9-h-day (200 µmol m−2 s−1 light, 22°C), 15-h-night (16°C) cycle at 60% humidity. T-DNA insertion lines WiscDsLoxHs053_08G (focl1-1; Woody et al., 2007) and SK5131 (focl1-2; Robinson et al., 2009) were obtained from NASC, Nottingham, UK. Plants were confirmed as homozygous for the insertion by PCR verification with primers WiscDsLoxHs053_08G, 5′-GAGCCATCAGCTTGTTCTCAC-3′ and 5′-TGTTCATGTCCCTCTGGAATG-3′, or SK5131, 5′-GCTTCCACCATTGCCTCAAA-3′ and 5′-TGTTCATGTCCCTCTGGAATG-3′. To confirm lack of, or truncated, FOCL1 transcript RT-PCR was carried out. RNA was extracted with a Spectrum RNA kit (Sigma-Aldrich) and 2 μg converted to cDNA with Maxima H Minus reverse transcriptase (Thermo Fisher Scientific). cDNAs were diluted 5-fold and FOCL1 transcript amplified using primers foclf1 5′-GCTTCAGGTCCTGCACAGAAA-3′, foclr1 5′-TCTGCAGGTCCCGGAATTAG-3′, and foclr2 5′-ACAAAAAGAACTTGGCTGAACTGG-3′. ACT3 was amplified as loading control using 5′-CTCCGGCGACTTGACAGAGAAG-3′ and 5′-GGAGGATGGCATGAGGAAGAGA-3′.
Histochemical GUS Staining
pFOCL1:GUS gene construct was produced by PCR amplifying 2 kb upstream from the FOCL1 translation start site with primers 5′-TGTATGATAATTCGAGCTACGATTCTAGGCGCAAAAG-3′ and 5′-AGAAAGCTGGGTCGGAGCAATAAAGAAGAAGAAGAGAAAC-3′ and combined by Gibson cloning (Gibson et al., 2009) into pBGWFS7 (Karimi et al., 2002) containing the upstream region of EPF2 (Hunt and Gray, 2009), which was then removed by digestion with SacI and AscI. The plasmid was transformed into Agrobacterium tumefaciens GV3101 by freeze/thaw, and plants transformed by the floral dip method (Clough and Bent, 1998). Transformants were selected by spraying with Basta (Liberty, Agrevo). Histochemical staining for GUS activity was carried out on leaves of T1 seedlings in 50 mm potassium phosphate, 1 mM potassium ferrocyanide, 1 mm potassium ferricyanide, 0.2% Triton X-100, 2 mm 5-bromo-4-chloro-3-indolyl-β-d-GlcA, and 10 mm EDTA after vacuum infiltration at 37°C. Leaves were decolorized in 70% ethanol and cleared in 80% chloral hydrate and images captured with an Olympus BX51 microscope connected to a DP51 digital camera using Cell B software. Expression pattern shown was typical of several independently transformed lines. GUS staining in the roots was performed on 11-d-old roots as described previously (Lucas et al., 2011).
Genetic Complementation
pFOCL1:FOCL1-GFP was generated by amplifying genomic DNA with primers 5′-TGTATGATAATTCGAGCTACGATTCTAGGCGCAAAAG-3′ and 5′-AGAAAGCTGGGTCGGAGCAATAAAGAAGAAGAAGAGAAAC-3′ and combined via Gibson cloning into pMDC107 previously cut with XbaI and AscI. pFOCL1:FOCL1-MYC was generated by cutting pFOCL1:FOCL1-GFP with KpnI and SacI. The MYC tag from pCTAPa (Rubio et al., 2005) was amplified using the primers 5′-tggtacctaacagcgggttaattaac-3′ and 5′-tgaacgatcggggaaattcg-3′ and the product digested with KpnI and SacI and ligated into a similarly cut pFOCL1:FOCL-GFP to create pFOCL1:FOCL-MYC. The plasmid was transformed into A. tumefaciens strain GV3101 by freeze thaw by floral dip method and selected on 0.5× Murashige and Skoog plates containing 5 mg/L hygromycin.
pFOCL1:GFP-FOCL1 was generated by overlapping PCR using primers 5′-TAAAACGACGGCCAGTGCCAACGATTCTAGGCGCAAAAG-3′, 5′-CTTTACTCATGGTGGCTAAGCAGAGAAC-3′, 5′-TTACACCATTTTTGTATAGTTCTACCATGCC-3′, 5′-ACTATACAAAAATGGTGTAACCGGATATG-3′, and 5′-CGATCGGGGAAATTCGAGCTTTGCTGAGCGTTGATGTG-3′. The products were ligated into pJET1.2 by blunt ended cloning then excised using XhoI and XbaI. The digested product was ligated into pMDC99 cut with SalI and SpeI and transformed into focl1-1 as above.
Stomatal Density, Size, and Aperture Measurements
Stomatal density was taken from fully mature leaf surfaces (three areas per leaf) using nail varnish imprints from dental resin impressions (Impression plus; TryCare) and mounted directly onto slides. Images were recorded using an Olympus DX51 light microscope. To analyze stomatal complex size, images from imprints (three areas per leaf and at least 10 stomata per plant) were measured using Line tool in Image J. Stomatal complex size was calculated using the formula area = πab, where a is the guard cell pair short radius and b the long radius.
The control of stomatal apertures was analyzed using leaf abaxial epidermis (Webb and Hetherington, 1997). Strips of epidermis were taken from leaves of 5- to 6-week-old plants (three to five\ leaves of each genotype) using tweezers and then floated on resting buffer (10 mm MES, pH 6.2) for 10 min. Strips were transferred to opening buffer (10 mm MES and 50 mm KCl, pH 6.2) in the light (300 µmol m−2 s−1), aerated with CO2-free air, and maintained at 20°C for 2 h. To investigate the effect of ABA on stomatal aperture, opening buffer was supplemented with 10 µm ABA. Pore widths and lengths were recorded from at least 100 stomata for each treatment. Pore area was calculated as above.
Microscopy and Cell Surface Analyses
For cryo-SEM, excised leaves were placed flat on a brass stub, stuck down with cryo glue consisting of a 3:1 mixture of Tissue-Tec (Scigen Scientific) and Aquadag colloidal graphite (Agar Scientific), and plunge frozen in liquid nitrogen with vacuum applied. Cryo fracture leaf samples were placed vertically in recessed stubs held by cryo glue. Frozen samples were transferred under vacuum to the prep chamber of a PT3010T cryo-apparatus (Quorum Technologies) maintained at −145°C. Surface ice was removed using a sublimation protocol consisting of −90°C for 3 min. For cryo fracture, no sublimation was carried out, and instead a level semirotary cryo knife was used to randomly fracture the leaf. All samples were sputter coated with platinum to a thickness of 5 nm. Samples were then transferred and maintained cold, under vacuum into the chamber of a Zeiss EVO HD15 SEM fitted with a cryo-stage. SEM images were captured using a gun voltage of 6 kV, I probe size of 460 pA, a SE detector, and a working distance of 5 to 6 mm.
Scanning electron microscopy specimens were fixed overnight in 3% glutaraldehyde, 0.1 m sodium cacodylate buffer, washed in 0.1 m sodium cacodylate buffer, and secondary fixed in 2% aqueous osmium tetroxide 1 h before dehydrating through 50 to 100% ethanol series 30 min each and drying over anhydrous copper sulfate. Specimens were critically point dried using CO2 as the transitional fluid then mounted with sticky tabs on 12.5-mm diameter stubs and coated in an Edwards S150B sputter coater with ∼25 to 30 nm of gold. Specimens were viewed using a Philips SEM XL-20 at accelerating voltage of 20 kV. For AFM, 28-d-old leaves were excised and fixed to glass slides using Provil Novo before submerging under a drop of water and imaging with an Asylum MFP-3D (Oxford Instruments) using contact mode. Height and deflection images were obtained with triangular silicon nitride probes (Bruker SNL10; nominal spring constant 0.35 N/m) using Asylum instrumentation software by scanning at 2 Hz on contact mode with set point 1 V.
VSI was carried out on abaxial surfaces of fully expanded leaves, with leaf held flat by pressing on to double-sided tape using a Wyko NT9100 surface Profiler and images were analyzed on Vision 4.10. For light microscopy, stem samples (∼1-cm lengths from the bases of branches of mature plants) were fixed in 4% (w/v) formaldehyde in PEM buffer (0.1 m PIPES, 2 mm EGTA, and 1 mm MgSO4, adjusted to pH7) by vacuum infiltration and then dehydrated in an ethanol series (30 min each at 30, 50, 70, and 100% ethanol) and infiltrated with LR White Resin (London Resin Company) diluted in ethanol (45 min each at 10, 20, 30, 50, 70, and 90% resin then 3 × 8 h+ at 100%). Samples were stood vertically in gelatin capsules filled with resin and polymerized >5 d at 37°C. Three-micrometer sections were cut using a Reichert-Jung Ultracut E ultramicrotome, stained with Toluidine Blue, and visualized using an Olympus BX51 microscope, and images were captured using Cell B software. Epidermal peels were stained by adding a drop of 1 ng/µL Nile red in 50% DMSO and imaged by fluorescence microscopy with an Olympus DX51 microscope using 460- to 490-nm excitation, 510- to 550-nm emission, and 505-nm dichroic mirror. FOCL-GFP images were captured as above, with a 1-s exposure time.
Raman Spectroscopy
Raman microscopy was performed using a Renishaw InVia system fitted with a 532-nm laser and a 2,400 lines/mm grating. Fresh leaf sample blocks (5 × 5 mm) were attached to aluminum slides using carbon tape and Raman 2D mapping was carried out using a 100× objective with a 1 s/pixel exposure time, 3× accumulation. Spectral range was set at 2439 to 3324 (center 2900) Raman shift (cm−1). Data were analyzed using Renishaw WiRE software, with scans being obtained across stomatal regions of interest from at least three independent biological samples.
Transpiration Measurements
Transpiration rates were measured using a porometer (Decagon Devices) with three measurements taken per plant from four plants of each genotype. Only focl1-2 was studied as focl1-1 leaves were too small to insert into the porometer chamber. Infrared thermography was used as a proxy measure of evaporative cooling from transpiration. Eight-week-old plants were kept under a propagator lid for 24 h before analysis. The lid was removed 4 h into the photoperiod and images captured with a FLIR SC660 thermal imaging camera and analyzed using ThermaCAM Researcher Professional 2.9. For each image, the mean temperature from spot readings from the center of three fully expanded leaves from six plants of each genotype was calculated and a mean temperature per plant used for statistical analyses.
Root Growth Analysis
Seedlings were grown vertically on 0.5× Murashige and Skoog plates and number of emerged lateral roots and primary root lengths were recorded at 9, 10, and 11 d. Roots were then cleared (Péret et al., 2012) and mounted in 50% glycerol and stages of lateral root primordia were determined using a Leica DMRB microscope.
Statistical Analysis
Unpaired t tests were performed using Microsoft Excel.
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession number NP_179254.2.
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Alignment of FOCL1 amino acid sequence with orthologs from wheat and Physcomitrella patens and with closest Arabidopsis homolog, At2g20515.
Supplemental Figure S2. Alignment of deduced amino acid sequences of FOCL1 and AGP31.
Supplemental Figure S3. Insertion positions and expression of focl1-1 and focl1-2.
Supplemental Figure S4. Rosette widths of focl1-1 and focl1-2.
Supplemental Figure S5. Complementation of focl1 restores stomatal complex size to the wild type.
Supplemental Figure S6. Wide view of abaxial epidermis of mature leaves of Col-0 and focl1-2.
Supplemental Figure S7. SEM of focl1-1 stomate showing partial opening.
Supplemental Figure S8. Deflection images for two stomata from Col-0 and focl1-1.
Supplemental Figure S9. Complementation of focl1-1 and focl1-2 restores leaf temperature to the wild type.
Supplementary Material
Acknowledgments
We thank Dr. Joe Quirk, Chris Hill, and Dr. Ray Weightman (SLCU Cambridge) for assistance with VSI, SEM, cryo-SEM, and Raman spectroscopy.
Glossary
- HRGP
Hyp-rich glycoprotein
- OCL
outer cuticular ledge
- VSI
vertical scanning interferometry
- AFM
atomic force microscopy
- cryo-SEM
cryo-scanning electron microscopy
- ABA
abscisic acid
References
- Amsbury S, Hunt L, Elhaddad N, Baillie A, Lundgren M, Verhertbruggen Y, Scheller HV, Knox JP, Fleming AJ, Gray JE (2016) Stomatal function requires pectin de-methyl-esterification of the guard cell wall. Curr Biol 26: 2899–2906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cannon MC, Terneus K, Hall Q, Tan L, Wang Y, Wegenhart BL, Chen L, Lamport DTA, Chen Y, Kieliszewski MJ (2008) Self-assembly of the plant cell wall requires an extensin scaffold. Proc Natl Acad Sci USA 105: 2226–2231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Ye D, Held MA, Cannon MC, Ray T, Saha P, Frye AN, Mort AJ, Kieliszewski MJ (2015) Identification of the abundant hydroxyproline-rich glycoproteins in the root walls of wild-type Arabidopsis, an ext3 mutant line, and its phenotypic revertant. Plants (Basel) 4: 85–111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Franks PJ, Beerling DJ (2009) Maximum leaf conductance driven by CO2 effects on stomatal size and density over geologic time. Proc Natl Acad Sci USA 106: 10343–10347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fricker MD, Wilmer C (1996) Stomata, Ed 2 Chapman and Hall, London [Google Scholar]
- Geldner N. (2013) The endodermis. Annu Rev Plant Biol 64: 531–558 [DOI] [PubMed] [Google Scholar]
- Gibson DG, Young L, Chuang RY, Venter JC, Hutchison CA III, Smith HO (2009) Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat Methods 6: 343–345 [DOI] [PubMed] [Google Scholar]
- Greene PR, Bain CD (2005) Total internal reflection Raman spectroscopy of barley leaf epicuticular waxes in vivo. Colloids Surf B Biointerfaces 45: 174–180 [DOI] [PubMed] [Google Scholar]
- Helm M, Schmid M, Hierl G, Terneus K, Tan L, Lottspeich F, Kieliszewski MJ, Gietl C (2008) KDEL-tailed cysteine endopeptidases involved in programmed cell death, intercalation of new cells, and dismantling of extensin scaffolds. Am J Bot 95: 1049–1062 [DOI] [PubMed] [Google Scholar]
- Hijazi M, Durand J, Pichereaux C, Pont F, Jamet E, Albenne C (2012) Characterization of the arabinogalactan protein 31 (AGP31) of Arabidopsis thaliana: new advances on the Hyp-O-glycosylation of the Pro-rich domain. J Biol Chem 287: 9623–9632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunt L, Gray JE (2009) The signaling peptide EPF2 controls asymmetric cell divisions during stomatal development. Curr Biol 19: 864–869 [DOI] [PubMed] [Google Scholar]
- Jeffree CE. (2006) The fine structure of the plant cuticle. In Riederer M, Muller C, eds, Biology of the Plant Cuticle. Blackwell, Oxford, UK, pp 11–125 [Google Scholar]
- Jones L, Milne JL, Ashford D, McQueen-Mason SJ (2003) Cell wall arabinan is essential for guard cell function. Proc Natl Acad Sci USA 100: 11783–11788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karimi M, Inzé D, Depicker A (2002) GATEWAY vectors for Agrobacterium-mediated plant transformation. Trends Plant Sci 7: 193–195 [DOI] [PubMed] [Google Scholar]
- Kieliszewski MJ, Lamport DTA (1994) Extensin: repetitive motifs, functional sites, post-translational codes, and phylogeny. Plant J 5: 157–172 [DOI] [PubMed] [Google Scholar]
- Kollist H, Nuhkat M, Roelfsema MR (2014) Closing gaps: linking elements that control stomatal movement. New Phytol 203: 44–62 [DOI] [PubMed] [Google Scholar]
- Kozma DK, Jenks MA (2007) Eco-physiological and molecular-genetic determinants of plant cuticle function in drought and salt stress tolerance. In MA Jenks, PM Hasegawa, SM Jain, eds, Advances in Molecular Breeding Toward Drought and Salt Tolerant Crops. Springer, Dordrecht, The Netherlands, pp 91–120 [Google Scholar]
- Lamport DT, Kieliszewski MJ, Chen Y, Cannon MC (2011) Role of the extensin superfamily in primary cell wall architecture. Plant Physiol 156: 11–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamport DTA. (1963) Oxygen fixation into hydroxyproline of plant cell wall protein. J Biol Chem 238: 1438–1440 [PubMed] [Google Scholar]
- Li Y, Beisson F, Koo AJ, Molina I, Pollard M, Ohlrogge J (2007) Identification of acyltransferases required for cutin biosynthesis and production of cutin with suberin-like monomers. Proc Natl Acad Sci USA 104: 18339–18344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C, Mehdy MC (2007) A nonclassical arabinogalactan protein gene highly expressed in vascular tissues, AGP31, is transcriptionally repressed by methyl jasmonic acid in Arabidopsis. Plant Physiol 145: 863–874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucas M, Swarup R, Paponov IA, Swarup K, Casimiro I, Lake D, Peret B, Zappala S, Mairhofer S, Whitworth M, et al. (2011) Short-Root regulates primary, lateral, and adventitious root development in Arabidopsis. Plant Physiol 155: 384–398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macgregor DR, Deak KI, Ingram PA, Malamy JE (2008) Root system architecture in Arabidopsis grown in culture is regulated by sucrose uptake in the aerial tissues. Plant Cell 20: 2643–2660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majewska-Sawka A, Münster A, Rodríguez-García MI (2002) Guard cell wall: immunocytochemical detection of polysaccharide components. J Exp Bot 53: 1067–1079 [DOI] [PubMed] [Google Scholar]
- Menke U, Renault N, Mueller-Roeber B (2000) StGCPRP, a potato gene strongly expressed in stomatal guard cells, defines a novel type of repetitive proline-rich proteins. Plant Physiol 122: 677–686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merced A, Renzaglia K (2014) Developmental changes in guard cell wall structure and pectin composition in the moss Funaria: implications for function and evolution of stomata. Ann Bot (Lond) 114: 1001–1010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merkouropoulos G, Shirsat AH (2003) The unusual Arabidopsis extensin gene atExt1 is expressed throughout plant development and is induced by a variety of biotic and abiotic stresses. Planta 217: 356–366 [DOI] [PubMed] [Google Scholar]
- Ogawa-Ohnishi M, Matsushita W, Matsubayashi Y (2013) Identification of three hydroxyproline O-arabinosyltransferases in Arabidopsis thaliana. Nat Chem Biol 9: 726–730 [DOI] [PubMed] [Google Scholar]
- Péret B, De Rybel B, Casimiro I, Benková E, Swarup R, Laplaze L, Beeckman T, Bennett MJ (2009) Arabidopsis lateral root development: an emerging story. Trends Plant Sci 14: 399–408 [DOI] [PubMed] [Google Scholar]
- Péret B, Li G, Zhao J, Band LR, Voß U, Postaire O, Luu DT, Da Ines O, Casimiro I, Lucas M, et al. (2012) Auxin regulates aquaporin function to facilitate lateral root emergence. Nat Cell Biol 14: 991–998 [DOI] [PubMed] [Google Scholar]
- Roberts K, Shirsat AH (2006) Increased extensin levels in Arabidopsis affect inflorescence stem thickening and height. J Exp Bot 57: 537–545 [DOI] [PubMed] [Google Scholar]
- Robinson SJ, Tang LH, Mooney BA, McKay SJ, Clarke WE, Links MG, Karcz S, Regan S, Wu YY, Gruber MY, et al. (2009) An archived activation tagged population of Arabidopsis thaliana to facilitate forward genetics approaches. BMC Plant Biol 9: 101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubio V, Shen Y, Saijo Y, Liu Y, Gusmaroli G, Dinesh-Kumar SP, Deng XW (2005) An alternative tandem affinity purification strategy applied to Arabidopsis protein complex isolation. Plant J 41: 767–778 [DOI] [PubMed] [Google Scholar]
- Showalter AM, Keppler B, Lichtenberg J, Gu D, Welch LR (2010) A bioinformatics approach to the identification, classification, and analysis of hydroxyproline-rich glycoproteins. Plant Physiol 153: 485–513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swarup K, Benková E, Swarup R, Casimiro I, Péret B, Yang Y, Parry G, Nielsen E, De Smet I, Vanneste S, et al. (2008) The auxin influx carrier LAX3 promotes lateral root emergence. Nat Cell Biol 10: 946–954 [DOI] [PubMed] [Google Scholar]
- Velasquez SM, Ricardi MM, Dorosz JG, Fernandez PV, Nadra AD, Pol-Fachin L, Egelund J, Gille S, Harholt J, Ciancia M, et al. (2011) O-glycosylated cell wall proteins are essential in root hair growth. Science 332: 1401–1403 [DOI] [PubMed] [Google Scholar]
- Voss U, Wilson MH, Kenobi K, Gould PD, Robertson FC, Peer WA, Lucas M, Swarup K, Casimiro I, Holman TJ, et al. (2015) The circadian clock rephases during lateral root organ initiation in Arabidopsis thaliana Nat Commun 6: 7641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webb AA, Hetherington AM (1997) Convergence of the abscisic acid, CO2, and extracellular calcium signal transduction pathways in stomatal guard cells. Plant Physiol 114: 1557–1560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson MH, Holman TJ, Sørensen I, Cancho-Sanchez E, Wells DM, Swarup R, Knox JP, Willats WG, Ubeda-Tomás S, Holdsworth M, et al. (2015) Multi-omics analysis identifies genes mediating the extension of cell walls in the Arabidopsis thaliana root elongation zone. Front Cell Dev Biol 3: 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winter D, Vinegar B, Nahal H, Ammar R, Wilson GV, Provart NJ (2007) An “Electronic Fluorescent Pictograph” browser for exploring and analyzing large-scale biological data sets. PLoS One 2: e718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woody ST, Austin-Phillips S, Amasino RM, Krysan PJ (2007) The WiscDsLox T-DNA collection: an Arabidopsis community resource generated by using an improved high-throughput T-DNA sequencing pipeline. J Plant Res 120: 157–165 [DOI] [PubMed] [Google Scholar]
- Yang Y, Costa A, Leonhardt N, Siegel RS, Schroeder JI (2008) Isolation of a strong Arabidopsis guard cell promoter and its potential as a research tool. Plant Methods 4: 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao L, Sack FD (1999) Ultrastructure of stomatal development in Arabidopsis (Brassicaceae) leaves. Am J Bot 86: 929–939 [PubMed] [Google Scholar]
- Zimmermann P, Hennig L, Gruissem W (2005) Gene-expression analysis and network discovery using Genevestigator. Trends Plant Sci 10: 407–409 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.