Folate regulates DNA methylation to affect Arabidopsis flowering time.
Abstract
Folates, termed from tetrahydrofolate (THF) and its derivatives, function as coenzymes in one-carbon transfer reactions and play a central role in synthesis of nucleotides and amino acids. Dysfunction of cellular folate metabolism leads to serious defects in plant development; however, the molecular mechanisms of folate-mediated cellular modifications and physiological responses in plants are still largely unclear. Here, we reported that THF controls flowering time by adjusting DNA methylation-regulated gene expression in Arabidopsis (Arabidopsis thaliana). Wild-type seedlings supplied with THF as well as the high endogenous THF content mutant dihydrofolate synthetase folypoly-Glu synthetase homolog B exhibited significant up-regulation of the flowering repressor of Flowering Wageningen and thereby delaying floral transition in a dose-dependent manner. Genome-wide transcripts and DNA methylation profiling revealed that THF reduces DNA methylation so as to manipulate gene expression activity. Moreover, in accompaniment with elevated cellular ratios between monoglutamylated and polyglutamylated folates under increased THF levels, the content of S-adenosylhomo-Cys, a competitive inhibitor of methyltransferases, was obviously higher, indicating that enhanced THF accumulation may disturb cellular homeostasis of the concerted reactions between folate polyglutamylation and folate-dependent DNA methylation. In addition, we found that the loss-of-function mutant of CG DNA methyltransferase MET1 displayed much less responsiveness to THF-associated flowering time alteration. Taken together, our studies revealed a novel regulatory role of THF on epigenetic silencing, which will shed lights on the understanding of interrelations in folate homeostasis, epigenetic variation, and flowering control in plants.
In living organisms, tetrahydrofolate (THF) and its derivatives, collectively termed folates, are a group of essential B-complex vitamins that have long been recognized as necessary nutrients to support the normal cell differentiation and growth, critical coenzymes in one-carbon transfers to aid the production of nucleotide synthesis and amino acid metabolism, and methyl donors to be required for methylation reactions of important biological substances such as DNA, proteins, and lipids (Lucock, 2000; Hanson and Roje, 2001; Rébeillé et al., 1997; Cossins and Chen, 1997; Crider et al., 2012; Fitzpatrick et al., 2012). To accumulate sufficient amounts of folates and maintain their cellular homeostasis are extremely important for a broad range of metabolic functions and physiological processes, whereas inadequate folate uptakes usually cause various developmental defects and severe health problems in humans, including birth defects, vascular diseases, subfertility, and even increased risks of cancers (Lucock, 2000; Molloy, 2012; De-Regil et al., 2010; Rose, 1966; Choi and Friso, 2005). Unlike mammals, which depend entirely on the dietary supply of folates, plants own a complete folate biosynthetic machinery and are able to produce these essential vitamins de novo from pterin, para-aminobenzoic acid and Glu precursors (Hanson and Roje, 2001; Hanson and Gregory, 2011; Scott et al., 2000; Roje, 2007). Because of the significance of folates in human nutritional health, a great deal of effort has been invested in recent years with regard to folate synthesis and metabolism in plants (Fitzpatrick et al., 2012; Hanson and Gregory, 2011; Roje, 2007; Blancquaert et al., 2014), and there is growing concern about the molecular regulation of these important vitamins in normal genome function and related biological processes (Zhang et al., 2012; Zhou et al., 2013; Mehrshahi et al., 2010).
To enhance the stability, affinity, and specificity in functions of folates in living cells, most of them are attached with a polyglutamyl tail, whose reaction is catalyzed by folylpoly-Glu synthetases (FPGSs; Hanson and Gregory, 2011; Ravanel et al., 2001), and the increasing evidence in plants has suggested that folate polyglutamylation plays a pivotal role in determining the extent of cellular folate content and homeostasis (Zhou et al., 2013; Mehrshahi et al., 2010; Akhtar et al., 2010). Three FPGS isoforms with distinct subcellular localizations including plastid, mitochondria, and cytosol were discovered in Arabidopsis (Arabidopsis thaliana), named as AtDFB (dihydrofolate synthetase folypoly-Glu synthetase homolog B), AtDFC, and AtDFD, respectively (Mehrshahi et al., 2010; Ravanel et al., 2001). Previous studies have suggested that these FPGS homologs played key roles in regulation of folate abundance and homeostasis and participated in various developmental and metabolic processes in plants such as embryo and seed development, seedling establishment, root growth and morphogenesis, floral organ formation, and nitrogen utilization (Mehrshahi et al., 2010; Meng et al., 2014; Srivastava et al., 2011; Blancquaert et al., 2014; Jiang et al., 2013; Reyes-Hernández et al., 2014). However, little is known about the regulatory mechanisms of folate homeostasis-mediated cellular and molecular modifications on plant development as well as the underlying physiological relevance.
The appropriate timing of the developmental switch from the vegetative to reproductive phase is extremely important for maximizing reproductive success and propagation of plant species in natural ecological system, which is under the strict control of integrated genetic networks determined by various external and intrinsic regulatory factors (Aidyn et al., 2002; Simpson and Dean, 2002). With the discoveries of multiple genes involved in flowering control, the molecular genetic dissection to the functions of these genes have led to the identification of several signaling transduction pathways in plants (Aidyn et al., 2002; Simpson and Dean, 2002). Of these elucidated regulatory mechanisms from different genetic pathways, chromatin modeling-associated gene expression activity through altered DNA methylation patterns have been paid more attention in recent years, and a number of genes involved in floral transition have been defined that their functions are largely correlative with epigenetic modifications (Kakutani, 1997; Finnegan et al., 2000). One such gene, Flowering Wageningen (FWA), which has been extensively studied in detail since it was originally characterized from epigenetic mutants, displayed a heritable late-flowering phenotype (Kakutani, 1997; Soppe et al., 2000). To initiate normal flowering process, FWA expression is required to be silenced by methyltransferase1 (MET1)-mediated cytosine methylation to release its interfered effects on the function of key flower-promoting gene FLOWERING LOCUS T (FT; Ikeda et al., 2007), failure in FWA DNA methylation results of ectopically expressed FWA and changed flowering behaviors. However, the environmental or endogenous stimulating factors to trigger MET1-caused DNA methylation, and the underlying regulatory mechanism and physiological relevance have remained largely unknown.
In this study, we reported that tetrahydrofolate, the biologically active form of folates (Ravanel et al., 2001; Beh, 2000), plays a pivotal role in determination of DNA methylation patterning in Arabidopsis genome. Seedlings supplied with THF, as well as the high endogenous THF content mutant Atdfb, showed dramatically induced effects on the release of MET1-mediated chromatin silencing and gene expression activity in a genome-wide scale, including the flowering regulation gene FWA. With elevated folate conditions, FWA constantly displayed transcription activity leading to retarded floral transition in a dose-dependent manner. Moreover, the loss of function of MET1 dramatically impaired THF-modulated flowering responses. Our studies uncover a fundamental role of folate homeostasis on epigenetically controlling gene expression to initiate different plant developmental events, which will shed light on how plants integrate epigenetic modifications, cellular functions, and physiological changes into a highly coordinated system to respond various environmental and intrinsic stimuli.
RESULTS
THF Regulates Floral Transition
To investigate the role of folate in plant development, we checked the effects of THF on seedling growth in Arabidopsis. As shown in Figure 1A, wild-type Arabidopsis plants grown on 0.5× Murashige and Skoog (MS) media with elevated THF concentrations displayed dramatically enhanced vegetative growth and postponed flowering time in a dose-dependent manner (Fig. 1, A–C), suggesting a potential regulatory role of THF in floral transition. To further confirm our hypothesis, we examined the flowering phenotype from the loss-of-function mutants of folylpoly-Glu synthetase (FPGS), the key enzyme to catalyze THF catabolism by polyglutamylation. It has been identified that there are three FPGS homologs in the Arabidopsis genome with different subcellular localizations, named as ATDFB/FPGS1, ATDFC/FPGS2, and ATDFD/FPGS3, respectively. Mutations in any AtDF genes led to disturbed folate homeostasis in the relative compartments of the cell (Mehrshahi et al., 2010; Srivastava et al., 2011). Interestingly, in all three tested Atdf mutants, only Atdfb (dihydrofolate synthetase folypoly-Glu synthetase homolog B), the one localized to plastids, was defective in normal floral transition process but not the others (Supplemental Figs. S1 and S2A). Our results showed that the flowering time of Atdfb mutants was later than that of wild-type plants, which is consistent with the higher endogenous THF level compared to those in wild-type control (Fig. 1, D and E; Supplemental Fig. S2, A–C); in addition, these mutants exhibited more serious responses under increased THF treatments (Fig. 1, F–H). These results indicated that AtDFB may play potential roles in the THF-associated developmental processes in Arabidopsis. To gain the insight of which components in the floral transition are affected by THF, we checked the expression of major flowering regulation genes in the elucidated genetic pathways, including CONSTANS, FT, FLOWERING LOUCUS C, and SUPPRESSOR OF OVEREXPRESSION OF CO1. However, none of them expressed differentially between wild type and Atdfb-3 seedlings (Supplemental Fig. S2D).
Figure 1.
THF regulates floral transition in Arabidopsis. A, Phenotypes of wild-type (WT) plants grown on 0.5× MS media supplemented with 0, 50, 100, and 150 µm THF. B and C, The effect of THF on flowering times. Days to bolting (B) and the rosette leaf number (C) were examined under the indicated THF concentrations from experiments as performed in A. D, Atdfb mutants flower late. Three-week-old wild-type and Atdfb mutant plants grown were shown. E, THF levels in wild type and Atdfb-3 mutant. The endogenous THF content was measured from wild-type and Atdfb-3 mutant plants grown in soil. F, Phenotypes of Atdfb-3 mutant grown on 0.5× MS media supplemented with 0, 50, 100, and 150 µm THF. G and H, Responses of Atdfb mutants to increased THF treatments. Experiments similar to those in B and C were performed from wild-type and Atdfb mutant plants under indicated THF concentrations. In A, D, and F, representative images are shown. In B, C, G, and H, data shown are mean ± sd, n ≥ 30 plants, P < 0.01. All plant materials were cultured in long-day conditions (16 h light/8 h dark) for 3 weeks before being analyzed.
THF Enhances FWA Expression to Postpone Flowering Time
To explore the possible mechanisms of high THF-resulted late flowering time, a transcriptional array analysis was performed by using wild-type seedlings and atdfb-3 mutant (Supplemental Table S1). Our data revealed that the transcription level of FWA, a previously reported flowering repression gene, was largely enhanced in Atdfb mutants, and the highly expressed FWA levels were also quantified by quantitative real-time PCR (qRT-PCR) analysis (Fig. 2, A and B). To further confirm the highly induced FWA transcription is specially related to the loss of function from AtDFB, we examined FWA transcripts in the other Atdf mutants, including atdfc and atdfd mutant lines. No apparent difference was found in the expression level of FWA in these mutants (Supplemental Fig. S3A), which is consistent with the flowering phenotypes observed in different Atdf mutants (Supplemental Fig. S2A).
Figure 2.
Elevated THF levels lead to ectopical expression of FWA and delayed floral transition. A and B, qRT-PCR analysis of FWA expression in the wild type (WT) and Atdfb mutants. Two-week-old seedlings grown on 0.5× MS media (A) or supplemented with 75 µm THF (B) were collected for the expression analysis. C, Examination of DNA methylation status in FWA promoter. FWA promoter sequences were generated by PCR amplification from the wild type and Atdfb-3 mutant treated with 0 or 75 µm THF, then digested by methylation-sensitive enzyme Hha1. D, Flowering phenotypes of the wild type, Atdfb-3, and Atdfb-3/fwa double mutant plants in long-day conditions for 35 d. Representative images are shown. E, Analysis of flowering time in the wild type, Atdfb-3, and Atdfb-3/fwa double mutant. F, FWA transcription levels in the wild type, Atdfb-3, and Atdfb-3/fwa mutant, respectively. For A, B, E, and F, data shown are means ± sd, P < 0.01.
Since the previous study reported FWA the ectopical expression-caused flowering defect is accompanied by the loss of DNA methylation in the direct repeats of FWA gene 5′ region (Soppe et al., 2000), we then examined the DNA methylation status of FWA promoter sequence using HhaI-based cytosine methylation assay (Woo et al., 2008), and our result dramatically showed that the 5′ region sequences of FWA gene amplified from wild-type seedlings exposed to THF and atdfb-3 mutant were more sensitive to the digestion of Hha1 enzyme (Fig. 2C), suggesting an association between the DNA hypomethylation of FWA promoter sequence and altered THF levels in these plant samples.
In addition, we generated Atdfb-3/fwa double mutant to confirm the close relevance between altered flowing times and THF-caused FWA up-regulation. Not surprisingly, the late flowering defect showed in Atdfb-3 mutant was largely recovered in Atdfb-3/fwa double mutant, as that observed in wild-type plants (Fig. 2, D and E). Also, qRT-PCR analysis showed FWA transcription levels were comparable between wild-type and Atdfb-3/fwa double-mutant plants (Fig. 2F). Together, the data shown above clearly illustrate that THF-delayed floral transition is the consequence of ectopically expressed FWA due to the loss of DNA methylation.
THF-Modulated Gene Expression Activity Is Related to Reduced DNA Methylation Levels
To further investigate whether THF acts as a general role in epigenetically controlling gene expression, we performed a global transcriptional analysis to compare genes that are differentially expressed between wild-type and Atdfb mutant seedlings. Remarkably, a large number of THF up-regulated genes showed obviously changed expression patterns closely relating to the DNA methylation process (Fig. 3A; Supplemental Table S1; changes > 1.5 folds, P < 0.05); for example, At1g67105 which was identified from the HDA6-involved MET1 DNA methylation pathway (To et al., 2011), exhibited around 16-fold overexpression in Atdfb-3 mutant. The consistency of the microarray data for these highly differentially expressed genes was verified by qRT-PCR analysis (Supplemental Fig. S3B). These results shown above indicated that THF may play a general function in correlating with epigenetic modulation. To test this hypothesis, we detected the THF-modulated epigenetically controlling gene expression in further detail by scanning the DNA methylation status of the highly up-regulated genes. As shown in Figure 3B, the generated SD5 fragment in At1g67105 from Atdfb-3 mutants was completely insensitive to the digestion of McrBC, a specific enzyme to recognize methylated cytosines in DNA sequences (Fig. 3B), implying the dramatically reduced DNA methylation levels in these regions. Next, we further verified the THF effect on epigenetic silencing in vitro by culturing wild type with 75 µm THF. Our results clearly showed that the sensitivity of SD5 fragment from relative genomic loci to MrcBC was largely decreased in THF-treated wild-type seedlings, which bears a strong resemblance to that of the Atdfb-3 mutant (Fig. 3C). Moreover, as expected, the relative gene transcriptional activity was highly enhanced in THF-treated wild type and Atdfb-3 mutant (Fig. 3D). These data suggested that altered gene expression induced by THF closely correlates with the modulation of DNA methylation.
Figure 3.
Global analysis of THF-modulated gene expression profiles and DNA methylation patterns in selected genes. A, Analysis of transcriptional array for differentially expressed genes between wild type and Atdfb-3 mutant. B and C, Analysis of DNA methylation patterns. The THF up-regulated gene At1g67105 was used for McrBC sensitivity-based DNA methylation analysis between wild-type and Atdfb-3 seedlings grown on 0.5× MS media (B) or treated with 75 µm THF (C). Subregion of SD5 fragment from At1g67105 was amplified by PCR reactions. D, qRT-PCR confirmation of THF-affected gene expression. Fourteen-day-old wild-type and atdfb-3 seedlings treated 0 µm or 75 µm THF were collected for detecting At1g67105 expression levels. Data shown are means ± sd, P < 0.01.
THF Exhibits a Genome-Wide Regulation on DNA Methylation Patterning
To further explore the global effect of THF on DNA methylation, we conducted whole-genome bisulfite sequencing under in vivo and in vitro elevated THF conditions (Fig. 4, A–C; Supplemental Tables S2 and S3). Based on the generated high-coverage genome-wide methylome maps, we obtained in total 51,702,398, 52,273,244, and 49,911,116 uniquely mapped ∼90-nucleotide reads for Atdfb-3 mutant, wild-type, and THF-treated wild-type plants, respectively (Supplemental Table S2). The differentially methylated regions of genome map coverage were 30.04, 29.14, and 28.44 Mb in different samples (Fig. 4B). Compared to those in the control plants, we discovered that 4179 regions for CpG, 1637 regions for CpHpG, and 52 regions for CpHpH were shown altered DNA methylation levels in Atdfb-3 mutant with a significant change of more than 1.5-fold (Supplemental Tables S4–S6; P < 0.05), suggesting that the mainly THF-influenced DNA methylation regions were related to CG and CHG methylation contexts, which took up to 72.2% and 27.9%, respectively (Fig. 4A).
Figure 4.
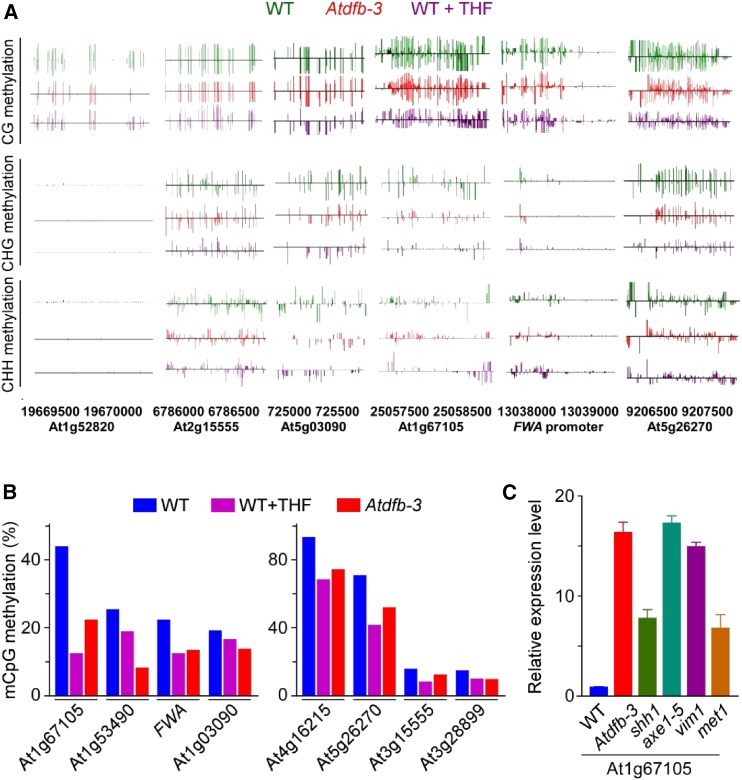
Genome-wide analyses of THF-modulated DNA methylation patterning. A, Differentially methylated genes between wild-type and Atdfb-3 seedlings based on whole-genome bisulfite sequencing. B, Analysis for differentially methylated region genome coverage. C, Analysis of total DNA methylation levels. D, Analysis of CG, CHG, and CHH methylation (where H = A, T, or C) from different elements in whole-genome genes. DNA methylation analysis was performed from 3′UTR, 5′UTR, CDS, and intron regions. E, Analysis of CG, CHG, and CHH methylation to elements of whole-genome nongenes including noncoding RNA (ncRNA), pseudogene, and transposon elements. For B to E analyses, sequences derived from the wild type treated with or without 75 µm THF and Atdfb-3 mutant were used.
Next, we monitored the genome-wide methylation status among wild type, atdfb-3, and wild type incubated with THF, and we found that the total CG methylation was largely reduced both in THF-treated wild type and Atdfb-3 mutant in contrast to that in wild-type control plants. Analyses to various gene regions including 3′UTR, 5′UTR, CDS, and introns, as well as different nongene regions, including noncoding RNA, pseudogenes, and transposon elements displayed that the methylation level in these areas were strikingly impaired in THF-treated wild type and Atdfb-3 mutant (Fig. 4, D and E), indicating that a general controlling mechanism of THF on DNA methylation repression was existed in plants.
To uncover THF-modulated DNA methylation in detail, we selected some of these highly induced genes and further investigated their methylation status. As shown in Figure 5, DNA methylation was significantly suppressed in both THF-treated wild type and Atdfb-3 mutant relative to that of wild type in their genomic loci (Fig. 5, A and B), which was consistent with the changed sensitivity of these genes to DNA methylation-detecting enzyme McrBC (Fig. 3, B and C). Since it has been well-documented that MET1, the plant homolog of mammalian DNA (cytosine-5)-methyltransferase1, undertakes important roles not only in maintaining CG methylation but also cooperating with HDA6 to regulate CHG methylation (To et al., 2011), we hypothesized THF-modulated epigenetic silencing might be mainly implicated in MET1-mediated DNA methylation pathway. To test this hypothesis, we examined the transcriptional levels of THF up-regulated genes such as At1g67105 in various loss-of-function mutants covering different regulatory pathways of DNA methylation. As expected, the transcript of At1g67105 was more abundant in those mutants of MET1-centered methylation process such as met1, vim1, shh1, and axe1-5 (Fig. 5C). In contrast, the transcription level of these genes shows almost no change in DNA demethylation-related mutants ros1, ros3, and siRNA-mediated CHH DNA methylation-related mutants ktf1, nprd4, rdr2, sdc, nrpd1b, drm3, and kyp (Supplemental Fig. S4), suggesting that THF effects mainly function in MET1-mediated DNA methylation processes in these genes.
Figure 5.
Analysis of DNA methylation status in different regions of THF up-regulated genes. A, Genome browser views of DNA methylation in different regions of THF up-regulated genes. B, Analysis of CG methylation from THF up-regulated genes. C, Expression of THF up-regulated genes in mutants related to CG and CHG methylation. Data shown are means ± sd, P < 0.01. In A and B, plants from the wild type treated with 0 or 75 µm THF, and Atdfb-3 mutants were used for the analysis of differentially methylated modification.
To further verify THF-reduced DNA methylation through the MET1-regulated epigenetic modification pathway, we subsequently checked flowering phenotypes of the MET1 loss-of-function mutant under increased THF conditions. As shown in Figure 6, met1-3 exhibited largely insensitive responses to THF-modulated floral transition (Fig. 6, A–C); moreover, consistent with the late flowering behaviors of met1-3 mutant, the transcriptional activity of FWA was constantly higher in met1-3 mutant compared to that in wild-type plants (Fig. 6, D and E). These results have confirmed the close correlation between THF-modulated flowering control and MET1-mediated DNA methylation process.
Figure 6.
THF-regulated floral transition was caused by impaired folate-dependent epigenetic regulation due to disturbed folate polyglutamylation homeostasis. A, Flowering phenotypes of wild-type and met1-3 plants under THF treatment. Wild-type and met1-3 plants were grown on 0.5× MS media supplemented with indicated THF concentrations for 40 d prior to be pictured. Representative images are shown. B and C, Effects of THF on flowering times. Days to bolting (B) and the rosette leaf number (C) were examined under the indicated THF concentrations from experiments as performed in A. D, qRT-PCR analysis of FWA expression in the wild type and met1-3 mutants. E, FWA expression ratio (met1-3/WT) under THF treatment (75 µm). Two-week-old seedlings grown on 0.5× MS media or supplemented with 75 µm THF were collected for the expression analysis. F and G, Monoglutamated or polyglutamated THF levels (F) and the ratio between polyglutamylated and monoglutamylated folates (G) were analyzed in the wild type and Atdfb-3 mutant. H, Relative SAH levels in wild-type and Atdfb-3 plants. I, SAHH1 transcription level in the wild type and Atdfb-3 mutant. Fourteen-day-old seedlings supplied with 0 or 75 µm THF were collected for liquid chromatography-mass spectrometry measurement of folate glutamylation profiles and methyl-donor metabolites or qRT-PCR analysis. Data shown are means ± sd, P < 0.05.
Elevated THF Levels Disturbed Concerted Reactions between Folate Polyglutamylation and Folate-Dependent DNA Methylation
Since FPGS1/AtDFB-mediated folate polyglutamylation was involved in both plant development and chromatin silencing control (Zhou et al., 2013; Meng et al., 2014; Srivastava et al., 2011), we questioned whether increased THF levels affect cellular homeostasis of folate polyglutamylation. Thus, we quantified cellular levels of monoglutamtlated and polyglutamylated folates in THF-treated wild type and Atdfb-3 mutant. As shown in Figure 6, the contents of folate mono-Glus as well as the ratio between monoglutamylated and polyglutamylated folate types were drastically higher in THF-treated wild-type seedlings and Atdfb-3 mutant in contrast to untreated control plants (Fig. 6, F and 6G), suggesting that an out-of-balance situation between different folate types leads to the disturbed homeostasis of cellular folate polyglutamylation. Because folate mono-Glus are generally poor substrates for folate-dependent enzymes such as Met synthases (Cherest et al., 2000; Ravanel et al., 2004), we therefore investigated the influence of abnormal accumulation of folate mono-Glus on epigenetic silencing control by measuring contents of DNA methylation-related metabolites including Met, S-adenosyl-Met (SAM), S-adenosylhomo-Cys (SAH), and homo-Cys. Our results revealed that the level of SAH, a competitive inhibitor of methyltransferases, was abnormally higher in THF-treated wild-type seedlings and Atdfb mutant (Fig. 6H). The high SAH content shown in Atdfb-3 mutant was also in consistent with the previous finding (Zhou et al., 2013). In agreement with the increased SAH content, we also found the gene expression of SAHH1, the critical regulators required for removing SAH to avoid its inhibitory effect on SAM regeneration and SAM-dependent methyl transfer (Loenen, 2006), was enhanced in elevated THF conditions (Fig. 6I). Meanwhile, previous metabolite profiling data showed that the endogenous adenosine accumulated dramatically in Atdfb mutant (Srivastava et al., 2011). Together, our results shown above suggest that folate-dependent epigenetic regulation is largely impaired accompanying high THF-caused abnormality in plant folate mono-Glu homeostasis.
DISCUSSION
Tetrahydrofolate and its derivatives have long been recognized as one of the essential B vitamins in maintaining normal life because of their nutritional significance as well as crucial roles in one-carbon metabolism and epigenetic modification in living organisms (Hanson and Roje, 2001; Crider et al., 2012; Fitzpatrick et al., 2012; Lucock, 2000; Groth et al., 2016; Dinh et al., 2013). Thus, exploring folate homeostasis-associated biochemical reactions, cellular modifications, and underlying regulatory mechanisms are critical for people to improve the understanding of the important physiological functions of folates in plant development control and stress-evoked plant adaptive responses. In this study, we have demonstrated an important role of THF-regulated DNA methylation reduction and its effects on FWA-associated alteration in flowering behaviors through modulating folate poly-Glu homeostasis, which interferes in the concerted reactions of folate polyglutamylation and the folate-dependent methylation process. Since epigenetic regulation-associated developmental and physiological changes responding to endogenous and environmental signals play a significant role in plant fitness and stress adaptations (Chinnusamy and Zhu, 2009; Gutzat and Mittelsten Scheid, 2012; Becker and Weigel, 2012), our present study may uncover an essential role of folates in stress-stimulated plant quick responses against unfavorable environmental conditions, and the folate-centered epigenetic modifications may represent a flexible short-term strategy of stress adaptations in plants. In addition, as important regulators of cellular metabolism and enzymatic reactions, it is not surprising that folates may undertake dual functions in controlling normal developmental processes and mediating adaptive responses to stresses in plant cells through simultaneously cooperating cellular metabolisms. However, the detailed integration and complexity of involved regulatory mechanisms underlying signaling pathways and components are extraordinarily challenging and need to be further investigated.
Previous studies showed AtDFB mutation led to root growth defects (Mehrshahi et al., 2010; Srivastava et al., 2011), which were recovered by exogenously supplied 5-formyltetrahydrofolate (5-CHO-THF), a stable folate form, for complementing folate deficiency in atdfb mutants (Zhou et al., 2013; Srivastava et al., 2011). To examine whether 5-CHO-THF application rescues THF-resulted deregulation in FWA methylation status, we treated atdfb-3 and wild-type plants with 5-CHO-THF and some methyl-donor metabolites as well. Unfortunately, neither FWA epigenetic status nor flowering behaviors of atdfb-3 mutant could be changed with tested treatments (Supplemental Fig. S5, A–C), implying that complemented folate abundance by providing 5-CHO-THF may not be sufficient to alleviate the out-of-balance situation between different folate types and the disturbed homeostasis of cellular folate polyglutamylation. Moreover, the DNA methylation level and expression of THF accumulation up-regulated genes such as At1g67105 was not affected apparently with applied methyl-donor metabolites (Supplemental Fig. S5D). Interestingly, Zhou et al. (2013) recently reported that 5-CHO-THF application rescued ATDFB loss-of-function-derived defects in root development as well as DNA methylation and H3K9 dimethylation in ros1 mutant background. To combine the findings shown in our present study, it suggests that folate-associated epigenetic regulations might be varied from different mutant backgrounds, plant tissues, and targeted cellular compartments; thus, further intensive research to clarify the detailed DNA methylation control of folates in root development will enrich the molecular mechanisms of folate-centered epigenetic modification in plants. In addition, as roles of 5-CHO-THF in plants have still not been well understood, we could not completely exclude the possibility that 5-CHO-THF rescued root growth in atdfb mutant was generated from its nutritional effect due to the excess of folates (Srivastava et al., 2011).
Taking into account the key role of DNA methylation in nutritional control of reproductive status and the FWA contribution as a possible epigenetic flowering-time modifier in plant reproductive strategy (Kucharski et al., 2008; Fujimoto et al., 2008), our finding that THF-caused ectopic expression of FWA and altered floral transition due to impaired DNA methylation may imply a potential role of folate in natural variation, which might be ecologically important for genetic diversity in plant genome evolution (Becker and Weigel, 2012; Bossdorf et al., 2008). Interestingly, studies also showed that the epigenetic regulation of flowering from prolonged low-temperature treatments (Finnegan et al., 1998; Dennis and Peacock, 2007), indicating that epigenetic controls of reproductive status in plants may be involved in multiple signaling regulations, and the concerted integration between distinct signaling pathways and various target components eventually trigger the phenotypic changes in flowering behaviors.
MATERIALS AND METHODS
Plant Materials and Growth Conditions
All Arabidopsis (Arabidopsis thaliana) genotypes used in the study are in the Columbia (Col-0) background. The Atdfb mutant lines (SALK_015472, SAIL_556_G08) and DNA methylation-related T-DNA insertion lines (met1-3: cs16394,shh1: SALK_073390, axe1-5:CS66153, vim1: SALK_050903C, rdr2: SALK_020850, ktf1: SALK_001254C, nrpd1b: CS66152, drm3: SALK_024820C, sdc: SALK_017605, kyp: SALK_105816C, nprd4: SALK_020157C, ros1: SALK_006944, ros3: SALK_022363C, fwa: SALK_064256C) were obtained from the Arabidopsis Biological Resource Center. Arabidopsis plants were grown in soil (Scotts Metro-Mix 200) or in petri dishes containing 0.5× strength of MS media (Sigma-Aldrich), 1.5% (w/v) Suc (Sigma-Aldrich), and 0.8% (w/v) agar (Sigma-Aldrich) in a controlled growth chamber at 22°C under white fluorescent light (100 μmol m−2 s−1) with a 16-h light/8-h dark photoperiod. For different chemical treatments, plants were grown in 0.5× MS supplied with THF (Schircks Laboratories), 5-CHO-THF (Schircks Laboratories), Met (Sigma-Aldrich), or SAM (Sigma-Aldrich), respectively, under indicated concentrations and conditions.
Analysis of Flowering Time
For soil-grown wild-type and mutant plants, the rosette leaf number and bolting days were scored as performed previously (Liu et al., 2013). For seedlings grown on 0.5× MS media in petri dishes, we scored the number of rosette leaves and the days at the stage of bolting when stems were about 3 mm as described previously (Yuan et al., 2016).
Transcriptional Analysis of Genes Involved in Flowering Time Control
The abundance of mRNAs including CONSTANS, FT, FLOWERING LOUCUS C, and SUPPRESSOR OF OVEREXPRESSION OF CO1 were analyzed from seedlings grown in soil under 16-h-light/8-h-dark cycles. Total mRNA was extracted by using Trizol reagent (Sigma-Aldrich), and first-strand cDNA was synthesized using a cDNA synthesis kit (Invitrogen). Primers used for the RT-PCR reactions were as described previously (He et al., 2004).
RT-qPCR Analysis
Two-week-old seedlings were collected and total RNA was extracted using Trizol reagent (Sigma-Aldrich). ReverTra Ace qPCR RT Master Mix with gDNA remover (FSQ301; Toyobo) was used for reverse transcription. RT-qPCR reactions were performed with SYBR Premix Ex Taq II (Tli RNaseH Plus; RR420A; Takara) on the Applied Biosystems 7500 Fast real-time PCR system using gene-specific primers (Supplemental Table S8; Woo et al., 2008; Yu et al., 2010). Three technical repeats were performed for each sample, and relative gene expression levels were calculated by normalization to the reference gene ACTIN2.
DNA Methylation Analysis
DNA methylation was analyzed by bisulfite sequencing, DNA gel blotting, and chop-PCR. For bisulfite sequencing, 2 mg of genomic DNA was treated and purified according to the protocol for the EpiTect bisulfite kit (Qiagen). The purified DNA was amplified, and the amplification product was cloned into the pMD18-T vector (Takara) for sequencing. The DNA methylation levels at CG, CHG, and CHH sites were separately calculated. For DNA gel blotting, genomic DNA was cleaved with the DNA methylation-sensitive endonucleases HpaII, MspI, and HaeIII at 37°C for 12 h and run on 1% agarose gels. The endonuclease McrBC cleaves DNA containing methylated cytosine but has no action on unmethylated DNA. For chop-PCR, genomic DNA was cleaved with McrBC, and the products were subjected to PCR and quantitative PCR.
Whole-Genome Bisulfite Sequencing
Raw sequencing data were mapped to TAIR10 reference genome modified by the single-nucleotide polymorphisms between Col-0. Only the sequences mapped to unique positions on the Arabidopsis genome were retained for DNA methylation analysis. Gene annotations were downloaded from The Arabidopsis Information Resource. One-kilobase upstream and downstream surrounding regions were included to calculate the DNA methylation levels of genes. The methylation levels of genes were estimated by pooling the read counts that show at least 5-fold coverage. CG, CHG, and CHH methylation was calculated. Annotated genes or transposons including 1-kb upstream and downstream flanking sequences were aligned to the modified TAIR10 reference genome. The average methylation level for each 100-bp interval was plotted. The lengths of genes or transposons were adjusted to a common sum. DNA methylation levels in 200-kb windows were plotted across each chromosome to indicate the genome-wide DNA methylation status. Samples from wild type, Atdfb-3, and wild type treated with THF were analyzed. The fold change of DNA methylation between wild type and Atdfb-3 in the 200-kb window was calculated and shown across each chromosome. To evaluate the correlation between differentially expressed gene DNA methylation, we determined the DNA methylation levels of different classes of differentially expressed genes using the method described previously (Zhou et al., 2013). The whole-genome bisulfite sequencing data have been deposited in NCBI database (http://www.ncbi.nlm.nih.gov/), and the related SRA accession number for the bisulfite sequencing clean data reported in this paper is SRP101407.
Microarray Analysis
Total RNA was extracted from 2-week-old Arabidopsis seedlings, and gene expression analysis was performed by the Beijing BioChain Institute Incorporation. Arabidopsis genome sequences and annotated gene models were downloaded from TAIR10 (http://www.Arabidopsis.org). The experiments were biologically repeated three times.
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Characterization of Atdfb homozygous mutants.
Supplemental Figure S2. Analysis of flowering phenotype and transcription of key flowering regulation genes in Atdfb loss-of-function mutants.
Supplemental Figure S3. qRT-PCR analysis of gene expression in Atdf mutants.
Supplemental Figure S4. Genome-wide analysis of differentially up-regulated genes in Atdfb-3 and various DNA methylation mutants.
Supplemental Figure S5. Application of methyl-donor metabolites cannot recover the MET1-mediated DNA methylation defect in Atdfb-3 mutant.
Supplemental Table S1. List of differentially expressed genes between Atdfb-3 and wild type (P < 0.05).
Supplemental Table S2. Whole-genome bisulfite sequencing analysis.
Supplemental Table S3. Average cytosine levels of CG, CHG, and CHH methylation.
Supplemental Table S4. Differentially CpG methylated regions in wild-type and Atdfb-3 genome.
Supplemental Table S5. Differentially CpHpG methylated regions in wild-type and Atdfb-3 genome.
Supplemental Table S6. Differentially CpHpH methylated regions in wild-type and Atdfb-3 genome.
Supplemental Table S7. Primers used in this study.
Supplementary Material
Glossary
- THF
tetrahydrofolate
Footnotes
This work was supported by grants from the Ministry of Science and Technology of China (2013CB967300) and National Natural Science Foundation of China (31530006) to Y.-k.H. (Capital Normal University, China).
References
- Aidyn M, Frédéric C George C (2002) Control of flowering time: Interacting pathways as a basis for diversity. Plant Cell (Suppl) 14: S111–S130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akhtar TA, Orsomando G, Mehrshahi P, Lara-Núñez A, Bennett MJ, Gregory JFIII III, Hanson AD (2010) A central role for gamma-glutamyl hydrolases in plant folate homeostasis. Plant J 64: 256–266 [DOI] [PubMed] [Google Scholar]
- Becker C, Weigel D (2012) Epigenetic variation: Origin and transgenerational inheritance. Curr Opin Plant Biol 15: 562–567 [DOI] [PubMed] [Google Scholar]
- Beh M. (2000) Tetrahydrofolate and tetrahydromethanopterin compared: Functionally distinct carriers in C-1 metabolism. Biochem J 350: 609–629 [PMC free article] [PubMed] [Google Scholar]
- Blancquaert D, De Steur H, Gellynck X, Van Der Straeten D (2014) Present and future of folate biofortification of crop plants. J Exp Bot 65: 895–906 [DOI] [PubMed] [Google Scholar]
- Bossdorf O, Richards CL, Pigliucci M (2008) Epigenetics for ecologists. Ecol Lett 11: 106–115 [DOI] [PubMed] [Google Scholar]
- Cherest H, Thomas D, Surdin-Kerjan Y (2000) Polyglutamylation of folate coenzymes is necessary for methionine biosynthesis and maintenance of intact mitochondrial genome in Saccharomyces cerevisiae. J Biol Chem 275: 14056–14063 [DOI] [PubMed] [Google Scholar]
- Chinnusamy V, Zhu JK (2009) Epigenetic regulation of stress responses in plants. Curr Opin Plant Biol 12: 133–139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi SW, Friso S (2005) Interactions between folate and aging for carcinogenesis. Clin Chem Lab Med 43: 1151–1157 [DOI] [PubMed] [Google Scholar]
- Cossins EA, Chen L (1997) Folates and one-carbon metabolism in plants and fungi. Phytochemistry 45: 437–452 [DOI] [PubMed] [Google Scholar]
- Crider KS, Yang TP, Berry RJ, Bailey LB (2012) Folate and DNA methylation: a review of molecular mechanisms and the evidence for folate’s role. Adv Nutr 3: 21–38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dennis ES, Peacock WJ (2007) Epigenetic regulation of flowering. Curr Opin Plant Biol 10: 520–527 [DOI] [PubMed] [Google Scholar]
- De-Regil LM, Fernández-Gaxiola AC, Dowswell T, Peña-Rosas JP (2010) Effects and safety of periconceptional folate supplementation for preventing birth defects. Cochrane Database Syst Rev 10: CD007950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dinh TT, O’Leary M, Won SY, Li S, Arroyo L, Liu X, Defries A, Zheng B, Cutler SR, Chen X (2013) Generation of a luciferase-based reporter for CHH and CG DNA methylation in Arabidopsis thaliana. Silence 4: 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finnegan EJ, Genger RK, Kovac K, Peacock WJ, Dennis ES (1998) DNA methylation and the promotion of flowering by vernalization. Proc Natl Acad Sci USA 95: 5824–5829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finnegan EJ, Peacock WJ, Dennis ES (2000) DNA methylation, a key regulator of plant development and other processes. Curr Opin Genet Dev 10: 217–223 [DOI] [PubMed] [Google Scholar]
- Fitzpatrick TB, Basset GJ, Borel P, Carrari F, DellaPenna D, Fraser PD, Hellmann H, Osorio S, Rothan C, Valpuesta V, et al. (2012) Vitamin deficiencies in humans: Can plant science help? Plant Cell 24: 395–414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujimoto R, Kinoshita Y, Kawabe A, Kinoshita T, Takashima K, Nordborg M, Nasrallah ME, Shimizu KK, Kudoh H, Kakutani T (2008) Evolution and control of imprinted FWA genes in the genus Arabidopsis. PLoS Genet 4: e1000048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groth M, Moissiard G, Wirtz M, Wang H, Garcia-Salinas C, Ramos-Parra PA, Bischof S, Feng S, Cokus SJ, John A, et al. (2016) MTHFD1 controls DNA methylation in Arabidopsis. Nat Commun 7: 11640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutzat R, Mittelsten Scheid O (2012) Epigenetic responses to stress: Triple defense? Curr Opin Plant Biol 15: 568–573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanson AD, Gregory JF III (2011) Folate biosynthesis, turnover, and transport in plants. Annu Rev Plant Biol 62: 105–125 [DOI] [PubMed] [Google Scholar]
- Hanson AD, Roje S (2001) One-carbon metabolism in higher plants. Annu Rev Plant Physiol Plant Mol Biol 52: 119–137 [DOI] [PubMed] [Google Scholar]
- He Y, Tang RH, Hao Y, Stevens RD, Cook CW, Ahn SM, Jing L, Yang Z, Chen L, Guo F, et al. (2004) Nitric oxide represses the Arabidopsis floral transition. Science 305: 1968–1971 [DOI] [PubMed] [Google Scholar]
- Ikeda Y, Kobayashi Y, Yamaguchi A, Abe M, Araki T (2007) Molecular basis of late-flowering phenotype caused by dominant epi-alleles of the FWA locus in Arabidopsis. Plant Cell Physiol 48: 205–220 [DOI] [PubMed] [Google Scholar]
- Kakutani T. (1997) Genetic characterization of late-flowering traits induced by DNA hypomethylation mutation in Arabidopsis thaliana. Plant J 12: 1447–1451 [DOI] [PubMed] [Google Scholar]
- Kucharski R, Maleszka J, Foret S, Maleszka R (2008) Nutritional control of reproductive status in honeybees via DNA methylation. Science 319: 1827–1830 [DOI] [PubMed] [Google Scholar]
- Jiang L, Liu Y, Sun H, Han Y, Li J, Li C, Guo W, Meng H, Li S, Fan Y, et al. (2013) The mitochondrial folylpolyglutamate synthetase gene is required for nitrogen utilization during early seedling development in arabidopsis. Plant Physiol 161: 971–989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu WZ, Kong DD, Gu XX, Gao HB, Wang JZ, Xia M, Gao Q, Tian LL, Xu ZH, Bao F, et al. (2013) Cytokinins can act as suppressors of nitric oxide in Arabidopsis. Proc Natl Acad Sci USA 110: 1548–1553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loenen WA. (2006) S-adenosylmethionine: Jack of all trades and master of everything? Biochem Soc Trans 34: 330–333 [DOI] [PubMed] [Google Scholar]
- Lucock M. (2000) Folic acid: Nutritional biochemistry, molecular biology, and role in disease processes. Mol Genet Metab 71: 121–138 [DOI] [PubMed] [Google Scholar]
- Mehrshahi P, Gonzalez-Jorge S, Akhtar TA, Ward JL, Santoyo-Castelazo A, Marcus SE, Lara-Núñez A, Ravanel S, Hawkins ND, Beale MH, et al. (2010) Functional analysis of folate polyglutamylation and its essential role in plant metabolism and development. Plant J 64: 267–279 [DOI] [PubMed] [Google Scholar]
- Meng H, Jiang L, Xu B, Guo W, Li J, Zhu X, Qi X, Duan L, Meng X, Fan Y, et al. (2014) Arabidopsis plastidial folylpolyglutamate synthetase is required for seed reserve accumulation and seedling establishment in darkness. PLoS One 9: e101905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molloy AM. (2012) Genetic aspects of folate metabolism. Subcell Biochem 56: 105–130 [DOI] [PubMed] [Google Scholar]
- Reyes-Hernández BJ, Srivastava AC, Ugartechea-Chirino Y, Shishkova S, Ramos-Parra PA, Lira-Ruan V, Díaz de la Garza RI, Dong G, Moon JC, Blancaflor EB, et al. (2014) The root indeterminacy-to-determinacy developmental switch is operated through a folate-dependent pathway in Arabidopsis thaliana. New Phytol 202: 1223–1236 [DOI] [PubMed] [Google Scholar]
- Ravanel S, Block MA, Rippert P, Jabrin S, Curien G, Rébeillé F, Douce R (2004) Methionine metabolism in plants: Chloroplasts are autonomous for de novo methionine synthesis and can import S-adenosylmethionine from the cytosol. J Biol Chem 279: 22548–22557 [DOI] [PubMed] [Google Scholar]
- Ravanel S, Cherest H, Jabrin S, Grunwald D, Surdin-Kerjan Y, Douce R, Rébeillé F (2001) Tetrahydrofolate biosynthesis in plants: Molecular and functional characterization of dihydrofolate synthetase and three isoforms of folylpolyglutamate synthetase in Arabidopsis thaliana. Proc Natl Acad Sci USA 98: 15360–15365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rébeillé F, Macherel D, Mouillon JM, Garin J, Douce R (1997) Folate biosynthesis in higher plants: Purification and molecular cloning of a bifunctional 6-hydroxymethyl-7,8-dihydropterin pyrophosphokinase/7,8-dihydropteroate synthase localized in mitochondria. EMBO J 16: 947–957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roje S. (2007) Vitamin B biosynthesis in plants. Phytochemistry 68: 1904–1921 [DOI] [PubMed] [Google Scholar]
- Rose DP. (1966) Folic acid deficiency in leukemia and lymphomas. J Clin Pathol 19: 29–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott J, Rébeillé F, Fletcher J (2000) Folic acid and folates: The feasibility for nutritional enhancement in plant foods. J Sci Food Agr 80: 795–824 [Google Scholar]
- Simpson GG, Dean C (2002) Arabidopsis, the Rosetta stone of flowering time? Science 296: 285–289 [DOI] [PubMed] [Google Scholar]
- Soppe WJ, Jacobsen SE, Alonso-Blanco C, Jackson JP, Kakutani T, Koornneef M, Peeters AJ (2000) The late flowering phenotype of fwa mutants is caused by gain-of-function epigenetic alleles of a homeodomain gene. Mol Cell 6: 791–802 [DOI] [PubMed] [Google Scholar]
- Srivastava AC, Ramos-Parra PA, Bedair M, Robledo-Hernández AL, Tang Y, Sumner LW, Díaz de la Garza RI, Blancaflor EB (2011) The folylpolyglutamate synthetase plastidial isoform is required for postembryonic root development in Arabidopsis. Plant Physiol 155: 1237–1251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- To TK, Kim JM, Matsui A, Kurihara Y, Morosawa T, Ishida J, Tanaka M, Endo T, Kakutani T, Toyoda T, et al. (2011) Arabidopsis HDA6 regulates locus-directed heterochromatin silencing in cooperation with MET1. PLoS Genet 7: e1002055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo HR, Dittmer TA, Richards EJ (2008) Three SRA-domain methylcytosine-binding proteins cooperate to maintain global CpG methylation and epigenetic silencing in Arabidopsis. PLoS Genet 4: e1000156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Z, Haberer G, Matthes M, Rattei T, Mayer KF, Gierl A, Torres-Ruiz RA (2010) Impact of natural genetic variation on the transcriptome of autotetraploid Arabidopsis thaliana. Proc Natl Acad Sci USA 107: 17809–17814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan S, Zhang ZW, Zheng C, Zhao ZY, Wang Y, Feng LY, Niu G, Wang CQ, Wang JH, Feng H, et al. (2016) Arabidopsis cryptochrome 1 functions in nitrogen regulation of flowering. Proc Natl Acad Sci USA 113: 7661–7666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Deng X, Miki D, Cutler S, La H, Hou YJ, Oh J, Zhu JK (2012) Sulfamethazine suppresses epigenetic silencing in Arabidopsis by impairing folate synthesis. Plant Cell 24: 1230–1241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou HR, Zhang FF, Ma ZY, Huang HW, Jiang L, Cai T, Zhu JK, Zhang C, He XJ (2013) Folate polyglutamylation is involved in chromatin silencing by maintaining global DNA methylation and histone H3K9 dimethylation in Arabidopsis. Plant Cell 25: 2545–2559 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.