Transcriptional control of brassinosteroid biosynthesis genes engages a transcription factor cascade in Arabidopsis.
Abstract
Brassinosteroids (BRs) are essential phytohormones regulating various developmental and physiological processes during normal growth and development. cog1-3D (cogwheel1-3D) was identified as an activation-tagged genetic modifier of bri1-5, an intermediate BR receptor mutant in Arabidopsis (Arabidopsis thaliana). COG1 encodes a Dof-type transcription factor found previously to act as a negative regulator of the phytochrome signaling pathway. cog1-3D single mutants show an elongated hypocotyl phenotype under light conditions. A loss-of-function mutant or inducible expression of a dominant negative form of COG1 in the wild type results in an opposite phenotype. A BR profile assay indicated that BR levels are elevated in cog1-3D seedlings. Quantitative reverse transcription-polymerase chain reaction analyses showed that several key BR biosynthetic genes are significantly up-regulated in cog1-3D compared with those of the wild type. Two basic helix-loop-helix transcription factors, PIF4 and PIF5, were found to be transcriptionally up-regulated in cog1-3D. Genetic analysis indicated that PIF4 and PIF5 were required for COG1 to promote BR biosynthesis and hypocotyl elongation. Chromatin immunoprecipitation and electrophoretic mobility shift assays indicated that COG1 binds to the promoter regions of PIF4 and PIF5, and PIF4 and PIF5 bind to the promoter regions of key BR biosynthetic genes, such as DWF4 and BR6ox2, to directly promote their expression. These results demonstrated that COG1 regulates BR biosynthesis via up-regulating the transcription of PIF4 and PIF5.
Brassinosteroids (BRs) are a class of naturally occurring hormone mediating multiple developmental and physiological processes during normal growth and development, such as photomorphogenesis, vascular differentiation, senescence, cell elongation, and cell division and differentiation (Clouse and Sasse, 1998). Within the last four decades, BR biosynthesis, catabolism, and signal transduction pathways have been largely elucidated. It is known that brassinolide (BL), the final product of the BR biosynthetic pathway and the most active BR, can directly interact with the extracellular domain of a Leu-rich repeat receptor-like kinase (LRR-RLK), BRASSINOSTEROID-INSENSITIVE1 (BRI1; Kinoshita et al., 2005; Hothorn et al., 2011; She et al., 2011). BL binding to BRI1 causes the release of BRI1 KINASE INHIBITOR1 from BRI1, allowing BRI1-BL to associate with a second LRR-RLK named BRI1-ASSOCIATED RECEPTOR KINASE1 (BAK1; Li et al., 2002; Nam and Li, 2002; Wang and Chory, 2006; Jaillais et al., 2011). Structure analyses revealed that BL is directly involved in the interaction of the BRI1-BL complex with BAK1, which is consistent with genetic data showing that BAK1 is essential for the early events of the BR signaling pathway (Gou et al., 2012; He et al., 2013; Santiago et al., 2013; Sun et al., 2013). The perception of BL at the cell surface by BRI1/BAK1 initiates a reversible phosphorylation and dephosphorylation cascade, leading to the dephosphorylation and inactivation of an important negative regulator, BRASSINOSTEROID-INSENSITIVE2 (BIN2), a GSK3-like kinase (Li and Nam, 2002; Mora-García et al., 2004; Kim et al., 2009, 2011).Without early BR signaling, BIN2 phosphorylates BRASSINAZOLE-RESISTANT1 (BZR1) and BRI1-EMS-SUPPRESSOR1 (BES1) to trigger their destruction (Wang et al., 2002b; Yin et al., 2002). Recent studies revealed that BIN2 has additional substrates that modulate downstream BR signaling. One of them is PHYTOCHROME INTERACTING FACTOR4 (PIF4), a basic helix-loop-helix transcription factor mainly regulating cell elongation (Castillon et al., 2007; de Lucas et al., 2008; Oh et al., 2012). BIN2 phosphorylates PIF4, leading this transcriptional regulator to a proteasome-mediated degradation process, which plays a prevalent role in timing hypocotyl elongation to late night (Bernardo-García et al., 2014). When BR signaling is induced, BIN2 is inactivated, and BZR1/BES1 and PIF4 are rapidly dephosphorylated and subsequently move into the nucleus to form a transcription factor complex synergistically activating a common set of BR-regulated genes involved in diverse processes of plant growth and development (He et al., 2002; Wang et al., 2002b; Yin et al., 2002; Sun et al., 2010; Yu et al., 2011; Oh et al., 2012).
The main BR biosynthetic pathway has also largely been elucidated (Zhao and Li, 2012). Campesterol is thought to be the first precursor entering into the specific BR biosynthetic pathway. Campesterol is converted to campestanol through a C-22 oxidation pathway. Campestanol is then converted to castasterone (CS) via an early or a late C-6 oxidation pathway (Suzuki et al., 1994a, 1994b; Fujioka et al., 1995; Choi et al., 1996, 1997). CS is ultimately converted to BL (Yokota et al., 1990; Suzuki et al., 1993). Both CS and BL are active BRs, with CS showing only about 10% of the activity of BL (Kinoshita et al., 2005). Until now, more than 70 BRs have been identified in plants (Fujioka and Yokota, 2003; Bajguz, 2007). Catalytic activities of several BR biosynthetic enzymes, such as DEETIOLATED2 (DET2), CONSTITUTIVE PHOTOMORPHOGENESIS AND DWARFISM (CPD), ROTUNDIFOLIA3 (ROT3), CYP90D1, DWARF4 (DWF4), BRASSINOSTEROID-6-OXIDASE1 (BR6ox1), and BR6ox2, have been elucidated. For example, DET2 catalyzes 5α reduction steps of the early BR biosynthetic pathway (Li et al., 1996; Fujioka et al., 1997, 2002; Noguchi et al., 1999b). CPD is involved in the C-3 oxidation steps of BR biosynthesis (Ohnishi et al., 2012). DWF4 encodes a C-22 hydroxylase that catalyzes the C-22 hydroxylation of multiple BR biosynthesis intermediates (Choe et al., 1998; Fujita et al., 2006). ROT3, also known as CYP90C1, and its homolog CYP90D1 are essential enzymes for the C-23 hydroxylation steps (Ohnishi et al., 2006). BR6ox1 and BR6ox2 catalyze multiple C-6 oxidation reactions connecting the early and late C-6 oxidation pathways. In addition, BR6ox2 participates in a final biosynthetic step, converting CS to BL (Bishop et al., 1999; Shimada et al., 2001; Kim et al., 2005).
Mutants impaired in BR biosynthesis show a severely retarded growth phenotype, whereas application of an excessive amount of BRs also inhibits root growth, suggesting that BR homeostasis is critical for optimal growth and development (Chory et al., 1991; Szekeres et al., 1996; Choe et al., 1998; Clouse and Sasse, 1998). Plants developed several strategies to tightly control endogenous BR levels. For example, the expression of many BR biosynthetic genes is negatively feedback controlled by the BR signaling pathway (Mathur et al., 1998; Bancoş et al., 2002). Activated BZR1 and BES1 can repress the expression of CPD and DWF4 by directly binding to their promoters when BR signaling is strong enough to maintain normal plant growth and development (He et al., 2005; Sun et al., 2010; Yu et al., 2011). Plants also can accelerate the BR biosynthetic rate to quickly produce more BRs when needed for certain developmental stages or environmental conditions. TEOSINTE BRANCHED1/CYCLOIDEA/PROLIFERATING CELL FACTOR1 (TCP1) and CESTA (CES) can up-regulate the expression of DWF4 and CPD by associating with the conserved motifs in their respective promoters (Guo et al., 2010; Poppenberger et al., 2011; Gao et al., 2015). Auxin or high temperature also can induce DWF4 expression (Chung et al., 2011; Maharjan and Choe, 2011). However, the detailed mechanisms controlling BR biosynthesis by environmental cues are poorly understood.
To identify additional regulators mediating BR biosynthesis, we carried out a large-scale genetic screen in an intermediate mutant of the BR receptor named bri1-5. A number of bri1-5 genetic modifiers have been isolated (Li et al., 2001, 2002; Zhou et al., 2004; Yuan et al., 2007; Guo et al., 2010). One of these mutants is cog1-3D (cogwheel1-3D). COG1 encodes a Dof-type transcription factor that is involved in phytochrome signaling and seed tolerance to deterioration (Park et al., 2003; Bueso et al., 2016). In this report, we provide strong evidence to show that COG1 can directly up-regulate the expression of PIF4 and PIF5 by associating with their promoter regions. PIF4 and PIF5 bind to the promoters of DWF4 and BR6ox2 to directly enhance their expression. Our analyses revealed that PIF4 and PIF5 are key regulators mediating BR biosynthesis, demonstrating that light signaling is critical to BR homeostasis.
RESULTS
cog1-3D Partially Suppresses the Short-Hypocotyl Phenotype of bri1-5
To identify additional components regulating BR signaling or homeostasis, we generated a large-scale activation-tagging pool in a weak bri1 allele, bri1-5, using a pBIB-BASTA-AT2 vector as described previously (Gou and Li, 2012). From about 120,000 transgenic plants, an extragenic suppressor was identified with significantly elongated hypocotyls compared with bri1-5 (Fig. 1, A and B). Scanning electron microscopy analysis indicated that the longer hypocotyls of the double mutants are caused mainly by elongated cells (Fig. 1, C and D). Genetic analysis indicated that the mutant phenotype is controlled by a dominant gene that cosegregates with the herbicide-resistant gene from the T-DNA of the activation-tagging construct.
Figure 1.
cog1-3D partially suppresses the short hypocotyl phenotype of bri1-5. A, Phenotypes of WS2, bri1-5, and bri1-5 cog1-3D seedlings. B, Hypocotyl lengths of the seedlings shown in A. C, Scanning electron micrographs showing that cog1-3D partially suppresses the short hypocotyl cell length phenotype of bri1-5. Middle parts of the hypocotyls were used for the scanning analyses. Bars = 150 μm. D, Cell lengths of bri1-5 and bri1-5 cog1-3D hypocotyls shown in C.
To clone the gene responsible for the elongated hypocotyl phenotype, we backcrossed the double mutant with WS2, and the bri1-5 mutation was segregated out. The single mutants exhibit significantly longer hypocotyls than those of WS2 (Fig. 2A). Thermal asymmetry interlaced PCR was used to amplify the genomic sequences flanking the T-DNA insertion site (Liu and Whittier, 1995). Sequence analysis indicated that the T-DNA was inserted 1,400 bp upstream of the translation initiation codon of COG1 (At1g29160; Fig. 2B). The suppressor was named cog1-3D, as two dominant alleles, cog1-D and cog1-2D, were reported previously (Park et al., 2003; Bueso et al., 2016). To further determine whether COG1 is the gene responsible for the longer hypocotyl phenotype, transgenic plants harboring 35S:FLAG-COG1 were generated. Among 50 total transgenic plants obtained, over 90% of them showed elongated hypocotyl phenotypes, similar to that of the cog1-3D single mutant (Fig. 2, A and C). Real-time reverse transcription (RT)-PCR analysis confirmed that COG1 is indeed overexpressed in both the activation-tagged mutants and the 35S:FLAG-COG1 transgenic lines (Fig. 2D). These results confirmed that elevated expression of COG1 is the main cause of the longer hypocotyl phenotypes of the activation-tagged line.
Figure 2.
COG1 plays an important role in regulating hypocotyl elongation. A, Phenotypes of WS2, cog1-3D, and 35S:FLAG-COG1 seedlings. B, In cog1-3D, a T-DNA fragment from the activation-tagging construct pBIB-BASTA-AT2 was inserted at 1,400 bp upstream of the start codon of COG1 (At1g29160). The cog1-6 mutant contains a deletion of a single G at position 85 bp, resulting in a premature stop codon after amino acid 51. C, Hypocotyl measurements of the seedlings shown in A. D, Real-time RT-PCR assay indicated that the expression of COG1 was elevated in both cog1-3D and 35S:FLAG-COG1 seedlings. E, Phenotypes of Columbia-0 (Col-0), cog1-6, and cog1-D seedlings grown under long-day conditions. F, Hypocotyl lengths of the seedlings shown in E. G, Induced expression of a dominant negative chimeric gene, COG1-SRDX, inhibits hypocotyl elongation. Wild-type and GVG:COG1-SRDX transgenic seedlings were germinated on one-half-strength MS medium plates for 2 d and then transferred to one-half-strength MS medium supplemented with mock solution (dimethyl sulfoxide [DMSO]; top) or 10 μm DEX (bottom) for an additional 5 d. H, Hypocotyl lengths of the seedlings shown in G.
Loss of Function by Ethyl Methanesulfonate Point Mutation or Inducible Expression of the COG1-SRDX Chimeric Gene Results in a Short-Hypocotyl Phenotype
To further determine whether the function of COG1 is required for hypocotyl elongation, loss-of-function mutants of COG1 were searched from various resources. No T-DNA insertion alleles were identified from available databases. A cog1-6 mutant was identified as an intragenic suppressor of cog1-D, the first activation-tagged allele of COG1 (Park et al., 2003), from a large-scale ethyl methanesulfonate-mutagenized screen. The cog1-6 mutant contains a deletion of a single G at position 85, resulting in a premature stop codon (Fig. 2B). The cog1-6 mutation caused a significant inhibition of the long-hypocotyl phenotype of cog1-D and a slightly reduced hypocotyl length compared with that of wild-type seedlings, especially under red light (Fig. 2, E and F; Supplemental Fig, S1).
The subtle developmental phenotypes observed in cog1-6 plants suggested that homologs of COG1 may play redundant roles with COG1. As a matter of fact, phylogenetic analysis revealed that a number of proteins are closely related to COG1 (Yanagisawa, 2002). Therefore, we employed a dominant negative technology, named chimeric repressor gene silencing, to further examine the roles of COG1 and its homologs. This technology has been used successfully to determine the roles of numerous functionally redundant transcription factors (Hiratsu et al., 2003; Guo et al., 2010; Poppenberger et al., 2011). COG1 was fused with an EAR repressor domain (SRDX), and the chimeric version should effectively repress the expression of the target genes of COG1 and its redundant proteins. Since constitutive expression of COG1-SRDX driven by the 35S promoter resulted in a seedling lethality phenotype (Supplemental Fig. S2), we made a dexamethasone (DEX)-inducible COG1-SRDX construct (GVG:COG1-SRDX) and generated corresponding transgenic plants (Aoyama and Chua, 1997). Homozygous lines for the transgene were selected and examined for the hypocotyl growth phenotype with or without DEX treatment. When 2-d-old seedlings from one-half-strength Murashige and Skoog (MS) medium were transferred to the medium containing DEX and grown for an additional 5 d, the GVG:COG1-SRDX transgenic seedlings showed significantly shortened hypocotyls due to reduced cell length (Fig. 2, G and H; Supplemental Fig. S3), which is similar to the cog1-6 mutant and the COG1 antisense seedlings (Fig. 2, E and F; Park et al., 2003). Without DEX treatment, the transgenic plants show no defective phenotypes compared with the wild type (Fig. 2, G and H; Supplemental Fig. S3). Nontransgenic plants grown on one-half-strength MS medium with or without DEX treatment did not show any altered phenotypes, confirming that the shortened hypocotyl phenotype was indeed caused by induced expression of COG1-SRDX. These results demonstrated that COG1 plays an important role in hypocotyl elongation.
The cog1-3D Seedlings Are Insensitive to Exogenously Applied BRs Due to Elevated Endogenous BR Levels
COG1 partially suppressing the short-hypocotyl phenotype of bri1-5 suggests that COG1 is involved in BR-related pathways. To investigate how COG1 regulates BR-related pathways, we first examined the effect of 24-epibrassinolide (24-epiBL) on the hypocotyl growth of cog1-3D. The hypocotyl growth of WS2 is promoted by exogenously applied different concentrations of 24-epiBL ranging from 1 to 1,000 nm (Fig. 3A). The hypocotyl growth of cog1-3D mutants, however, is insensitive to exogenously supplied different concentrations of 24-epiBL (Fig. 3A).
Figure 3.
The cog1-3D seedlings showed reduced sensitivity to BR due to increased endogenous BR amounts. A, Hypocotyl growth analyses of 7-d-old cog1-3D and wild-type seedlings grown on one-half-strength MS agar plates containing different concentrations of 24-epiBL. B, The shortened hypocotyl phenotype of COG1-SRDX can be rescued by exogenous 24-epiBL application. WS2 and GVG:COG1-SRDX seedlings were grown on one-half-strength MS medium with or without 0.5 μm DEX or with 0.5 μm DEX and 100 nm 24-epiBL for 6 d. C, Endogenous BR levels in cog1-3D and wild-type plants. The data shown are averages and sd from three independent replicates. FW, Fresh weight.
Either blocking the BR signaling pathway or increasing endogenous levels of BRs can result in plant insensitivity to exogenous BRs. The fact that cog1-3D shows an elongated but not a shortened hypocotyl phenotype suggests the BR signaling is at least not blocked in cog1-3D. The phosphorylation status of BES1 was investigated with a specific anti-BES1 antibody in wild-type and cog1-3D seedlings after treatment with or without 1 μm 24-epiBL (Yin et al., 2002). Interestingly, the levels of both phosphorylated and unphosphorylated BES1 in cog1-3D mutants were higher than those of wild-type seedlings without BR treatment (Supplemental Fig. S4A). Upon BR treatment, unphosphorylated BES1 was increased dramatically, and phosphorylated BES1 disappeared in both wild-type and mutant seedlings. The amount of unphosphorylated BES1 in cog1-3D also was greater than that of wild-type seedlings (Supplemental Fig. S4A). Consistently, the expression of a BR signaling marker gene, SAUR-AC1, was increased significantly in cog1-3D plants (Supplemental Fig. S4B). These results confirmed that BR signaling is enhanced in cog1-3D plants.
Next, we investigated whether the endogenous levels of BRs are elevated in cog1-3D. If COG1 promotes BR biosynthesis, the shortened hypocotyl phenotype of COG1-SRDX should result from the reduced BR biosynthesis and the phenotype should be rescued by exogenously applied BRs. Indeed, the shortened hypocotyl phenotype of GVG:COG1-SRDX plants grown on medium containing DEX was rescued by supplying 100 nm 24-epiBL in the medium (Fig. 3B). In addition, blocking BR biosynthesis using a specific inhibitor, brassinazole (BRZ), should shorten the hypocotyls of cog1-3D plants (Asami et al., 2000). Indeed, the cog1-3D seedlings exhibited a hypocotyl length indistinguishable from that of wild-type controls when grown on one-half-strength MS medium agar plates supplied with 1 μm BRZ (Supplemental Fig. S5). These results support the hypothesis that COG1 promotes hypocotyl elongation by increasing endogenous BR levels.
BR profile analysis was performed using WS2 and cog1-3D seedlings. It is apparent that BL, CS, and several of their precursors are significantly elevated in cog1-3D seedlings (Fig. 3C). For example, dolichosterone, teasterone and typhasterol are increased by approximately 2-fold in cog1-3D seedlings compared with the wild type; CS is elevated by about 5-fold in cog1-3D; BL is increased by 3-fold in cog1-3D seedlings (Fig. 3C). These results suggested that COG1 possibly enhances BR biosynthesis by affecting multiple reactions in the biosynthesis pathway. Alternatively, it is also possible that BR catabolism is decreased in cog1-3D.
COG1 Up-Regulates the Expression of BR Biosynthetic Genes
To examine whether BR biosynthesis is enhanced or BR catabolism is decreased in cog1-3D, we compared the expression levels of all known key genes involved in both BR biosynthesis and catabolism from cog1-3D and wild-type seedlings. The transcript levels of DET2, CYP90D1, and BR6ox1 in cog1-3D showed no significant differences from those of the wild type (Supplemental Fig. S6), whereas the expression levels of CPD, DWF4, ROT3, and BR6ox2 are significantly up-regulated in cog1-3D and 35S:FLAG-COG1 compared with wild-type seedlings (Fig. 4A). Consistently, inducible expression of COG1-SRDX resulted in the down-regulation of these four genes (Fig. 4B). All the catabolism genes are not significantly down-regulated in cog1-3D compared with those in wild-type seedlings. Several of the catabolic genes are actually up-regulated in cog1-3D (Supplemental Fig. S7). These results suggested that BR accumulation in cog1-3D is caused mainly by enhanced BR biosynthesis rather than by decreased BR catabolism.
Figure 4.
COG1 up-regulates the expression of BR biosynthetic genes, and COG1 regulation of hypocotyl elongation is dependent on BR biosynthesis. A, Expression levels of CPD, DWF4, ROT3, and BR6ox2 in WS2, cog1-3D, and 35S:FLAG-COG1 plants were detected by quantitative reverse transcription (qRT)-PCR. B, Expression levels of CPD, DWF4, ROT3, and BR6ox2 in WS2 and GVG:COG1-SRDX transgenic plants with or without DEX treatment. WS2 and GVG:COG1-SRDX seedlings were treated with mock solution (DMSO) or 10 μm DEX for 24 h before harvest for RNA extraction and qRT-PCR analyses. C, cog1-3D showed no or less effect on hypocotyl elongation in various BR biosynthetic mutants. One-week-old seedlings of WS2, cog1-3D, det2, det2 cog1-3D, cpd, cpd cog1-3D, dwf4, dwf4 cog1-3D, br6ox1 br6ox2, and br6ox1 br6ox2 cog1-3D are shown. D, Hypocotyl lengths of the seedlings shown in C.
To further investigate whether COG1 promoting hypocotyl elongation relies on BR biosynthesis, cog1-3D plants were crossed with various mutants deficient in BR biosynthesis. The homozygous det2 cog1-3D, cpd cog1-3D, dwf4 cog1-3D, and br6ox1 br6ox2 cog1-3D mutants were obtained and examined for their hypocotyl growth phenotypes. All these double or triple mutants show hypocotyl elongation phenotypes indistinguishable from the corresponding single or double mutants (Fig. 4, C and D). Furthermore, deficiency in BR signal transduction also completely inhibits the hypocotyl elongation of cog1-3D (Supplemental Fig. S8). These results clearly indicated that COG1 regulation of hypocotyl elongation depends on BR biosynthesis and signaling.
PIF4 and PIF5 Are Up-Regulated by COG1 and Are Required for COG1-Mediated BR Biosynthesis and Hypocotyl Elongation
Although the expression of CPD, DWF4, ROT3, and BR6ox2 is transcriptionally regulated by COG1, we failed to detect the direct association of COG1 with their promoters by chromatin immunoprecipitation (ChIP) analyses, indicating that COG1 regulates the expression of these genes possibly via other unknown transcription factors. An RNA sequencing analysis was performed to compare the expression profiles of WS2 and cog1-3D. We identified a total of 443 genes displaying statistically significant 2-fold expression changes in cog1-3D compared with wild-type seedlings, including 37 transcription factors (Supplemental Table S1). PIF4 and PIF5 are among those 37 transcription factors. PIF4 and PIF5 play a pivotal role in cell elongation by directly elevating the expression of genes involved in cell wall loosening (Huq and Quail, 2002; de Lucas et al., 2008; Oh et al., 2012; Supplemental Fig. S9). To investigate whether PIF4 and PIF5 are involved in COG1-mediated hypocotyl elongation, the expression of PIF4 and PIF5 in cog1-3D and wild-type plants was confirmed using real-time RT-PCR. Both PIF4 and PIF5 are up-regulated in cog1-3D as well as 35S:FLAG-COG1 seedlings compared with wild-type plants (Fig. 5A). To investigate whether the protein levels of PIF4 and PIF5 also are accumulated, we performed western-blot analysis. Consistently, the protein level of PIF5 is increased significantly in cog1-3D and 35S:FLAG-COG1 seedlings (Supplemental Fig. S10). PIF4 levels in cog1-3D and 35S:FLAG-COG1 were not examined due to unavailable specific antibody against PIF4. Inducible expression of COG1-SRDX, on the other hand, resulted in significantly decreased expression of PIF4 and PIF5 (Fig. 5B).
Figure 5.
PIF4 and PIF5 are essential for COG1 to promote BR biosynthesis and hypocotyl elongation. A, Expression levels of PIF4 and PIF5 in WS2, cog1-3D, and 35S: FLAG-COG1 seedlings. One-week-old WS2, cog1-3D, and 35S: FLAG-COG1 seedlings were harvested for RNA extraction and qRT-PCR analyses. B, Expression levels of PIF4 and PIF5 in WS2 and GVG:COG1-SRDX transgenic plants with or without DEX treatment. WS2 and GVG:COG1-SRDX seedlings were treated with either mock solution (DMSO) or 10 μm DEX for 24 h before harvesting for RNA extraction and qRT-PCR analyses. C and D, Phenotypes and hypocotyl measurements of WS2, cog1-3D, pif4, pif4 cog1-3D, pif5, pif5 cog1-3D, pif4 pif5, and pif4 pif5 cog1-3D seedlings. The pif4 pif5 double mutant was crossed with cog1-3D to yield a pif4 pif5 cog1-3D triple mutant that was then backcrossed with WS2 for at least five generations. E, qRT-PCR analysis of CPD, DWF4, ROT3, and BR6ox2 in WS2, pif4 pif5, and pif4 pif5 cog1-3D seedlings.
To further determine whether PIF4 and PIF5 are essential for COG1-mediated cell elongation, cog1-3D plants were crossed with pif4 and pif5 single and pif4 pif5 double mutants. The pif4 cog1-3D and pif5 cog1-3D seedlings exhibited partially inhibited hypocotyl elongation compared with the cog1-3D single mutant (Fig. 5, C and D), whereas pif4 pif5 cog1-3D triple mutant seedlings showed hypocotyl length almost identical to that of the pif4 pif5 double mutants (Fig. 5, C and D). Plants grown under continuous red light showed similar results (Supplemental Fig. S11). In addition, the expression levels of CPD, DWF4, ROT3, and BR6ox2 in pif4 pif5 cog1-3D triple mutants showed no significant differences from pif4 pif5 double mutants (Fig. 5E). These results suggested that the function of COG1 in regulating BR biosynthesis and hypocotyl elongation is dependent on PIF4 and PIF5.
COG1 Binds Directly to the Promoters of PIF4 and PIF5
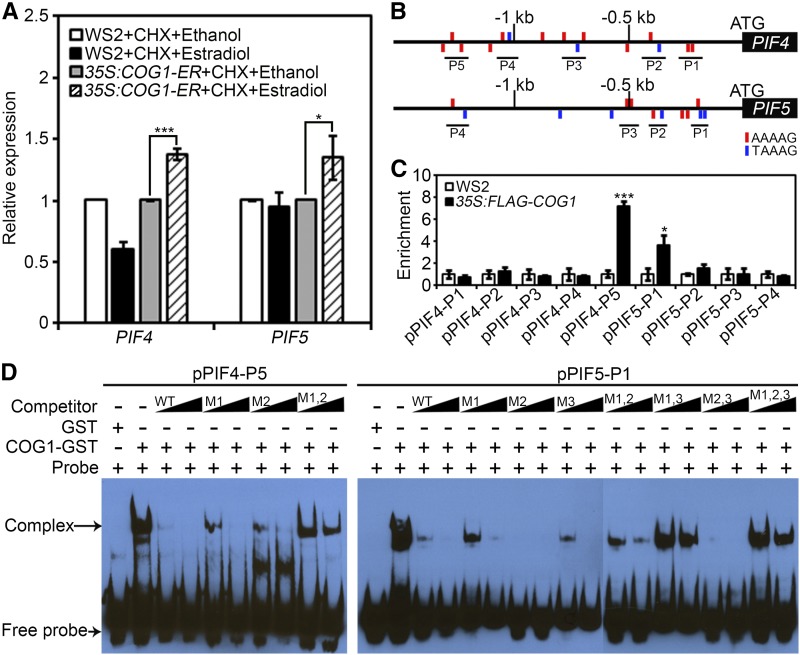
Since PIF4 and PIF5 are transcriptionally regulated by COG1 in plants and the function of COG1 is dependent on PIF4 and PIF5 (Fig. 5), we hypothesized that COG1 may directly regulate the expression of PIF4 and PIF5. To test this hypothesis, COG1 was fused with a hormone-binding domain of a human estrogen receptor (ER) at its C terminus, and transgenic plants harboring the chimeric gene driven by a 35S promoter (35S:COG1-ER) were generated. Six-day-old transgenic seedlings were pretreated with 10 μm cycloheximide (CHX) for 2 h to prevent the biosynthesis of new proteins. Estradiol was then added to the liquid culture to induce preexisting COG1 to enter the nucleus. The expression of PIF4 and PIF5 was rapidly activated following estradiol induction, demonstrating that COG1 can directly regulate the transcription of PIF4 and PIF5 (Fig. 6A). Previous studies demonstrated that Dof-type transcription factors bind to the 5′-A/TAAAG-3′ core sequences or their reversibly complementary sequences 5′-CTTTA/T-3′ in the promoters of their target genes to regulate gene expression in Arabidopsis (Arabidopsis thaliana; Yanagisawa, 2004). Sequence scanning revealed that there are several putative Dof DNA-binding motifs in the promoter regions of PIF4 and PIF5 (Fig. 6B). To determine whether COG1 associates with these motifs in the PIF4 and PIF5 promoters, 35S:FLAG-COG1 transgenic plants were used for ChIP assays. After immunoprecipitation of protein-DNA complexes from 7-d-old seedlings using an antibody against the FLAG epitope, enriched DNA fragments were amplified by qRT-PCR using primers that annealed near the Dof-binding motifs in the promoters of PIF4 and PIF5. Our results showed that at least one fragment in the PIF4 or PIF5 promoter region was indeed enriched in the ChIP assays, suggesting that COG1 associates directly with the promoters of PIF4 and PIF5 (Fig. 6C).
Figure 6.
COG1 interacts directly with the promoters of PIF4 and PIF5. A, Expression levels of PIF4 and PIF5 in WS2 and 35S:COG1-ER transgenic plants with or without estradiol treatment in the presence of CHX. WS2 and 35S:COG1-ER seedlings were pretreated with 10 μm CHX for 2 h, then either mock solution (ethanol) or 10 μm estradiol was added to the liquid solution for another 2 h before harvesting for RNA extraction and qRT-PCR analyses. B, PIF4 and PIF5 promoter regions contain multiple Dof motifs (red or blue upright lines). P1 to P5 indicate the DNA fragments amplified in ChIP-PCR assays. C, ChIP-quantitative PCR assays showing that COG1 associates with the promoters of PIF4 and PIF5 in vivo. One-week-old WS2 and 35S:FLAG-COG1 seedlings were harvested for ChIP experiments. qRT-PCR was used to quantify enriched DNA fragments in the PIF4 and PIF5 promoters. D, EMSA showing that COG1 binds to the Dof motifs in the promoter regions of PIF4 and PIF5 in vitro. The enriched DNA fragments in the promoters of PIF4 and PIF5 were incubated with GST-COG1 recombinant proteins as indicated. GST proteins were used as negative controls. Competition for COG1 binding was performed with 100× and 200× unlabeled probes containing the enriched DNA fragments (wild type [WT]) or mutated DNA fragments (M).
To further confirm the direct binding of COG1 to the PIF4 and PIF5 promoters, we performed an electrophoretic mobility shift assay (EMSA) using purified GST-COG1 fusion protein from Escherichia coli. Our results showed that GST-COG1 bound to the biotin-labeled Dof-binding motif-containing DNA fragments in the promoters of PIF4 and PIF5, as suggested by the ChIP assay (Fig. 6D). Furthermore, the binding could be competed off by adding an excessive amount of unlabeled same DNA fragments. However, the unlabeled DNA probes containing mutated Dof-binding motifs failed to compete off the binding of COG1 to the biotin-labeled wild-type DNA fragments (Fig. 6D). Control experiments indicated that GST protein alone failed to bind to the labeled DNA probes (Fig. 6D). Taken together, we conclude that COG1 can bind directly to the promoter region of PIF4 and PIF5 and directly enhance their expression.
PIF4 and PIF5 Directly Regulate the Expression of DWF4 and BR6ox2
Two pieces of evidence suggest that PIF4 and PIF5 might directly regulate the expression of BR biosynthetic genes. First, COG1 regulation of the expression of BR biosynthetic genes is dependent on PIF4 and PIF5 (Fig. 5E). Second, the transcript levels of BR6ox2 are reduced significantly in pif4 pif5 double mutants compared with those in the wild type (Fig. 5E). Given the fact that PIFs bind to the G-box motifs in their target gene promoters, we searched for G-box motifs in the promoters of CPD, DWF4, ROT3, and BR6ox2. G-box motifs were found in the promoters of CPD, DWF4, and BR6ox2 but not in the promoter of ROT3 (Fig. 7A; Supplemental Fig. S12A). To test whether PIF4 and PIF5 bind to the G-box-containing regions of these three BR biosynthetic genes, we performed ChIP assays using the 35S:PIF4-FLAG and 35S:PIF5-FLAG transgenic plants and anti-FLAG antibody. PCR analyses indicated that PIF4-FLAG specifically associates with the G-box-containing promoter regions of CPD, DWF4, and BR6ox2 (Fig. 7B; Supplemental Fig. S12B). PIF5-FLAG associates with the G-box-containing promoter region of DWF4 but not with the promoter regions of CPD and BR6ox2 (Fig. 7B). EMSA experiments using PIF4 and PIF5 proteins expressed in vitro further confirmed that PIF4 directly bound to the G-box-containing DNA fragments in the promoter regions of DWF4 and BR6ox2 (Fig. 7, C and D). PIF5 specifically bound to the G-box-containing DNA fragments in the promoter region of DWF4 (Fig. 7E). The binding could be effectively competed off by the addition of an excessive amount of unlabeled G-box-containing DNA probes (Fig. 7, C–E). As a control, DNA probes containing a mutated G-box motif (CACGGG) failed to compete off the binding of PIF4 or PIF5 to the biotin-labeled DNA fragments (Fig. 7, C–E). Moreover, we examined whether PIF4 and PIF5 regulate the expression of DWF4 and BR6ox2 in planta. qRT-PCR analysis showed that the expression of DWF4 and BR6ox2 is significantly up-regulated in both 35S:PIF4 and 35S:PIF5 transgenic plants relative to wild-type seedlings (Fig. 7F). PIF4 was taken as an example to further confirm that this regulation is via a direct process. Consistently, the expression of DWF4 and BR6ox2 can be rapidly up-regulated in 35S:PIF4-ER transgenic plants upon the treatment with estradiol in the presence of CHX (Fig. 7G). As a result, the final product of the BR biosynthetic pathway, BL, is increased significantly in 35S:PIF4 and 35S:PIF5 transgenic plants (Fig. 7H). Together, these results corroborated that PIF4 and PIF5 redundantly and directly regulate the expression of DWF4 and BR6ox2 to promote BR biosynthesis and hypocotyl elongation.
Figure 7.
PIF4 and PIF5 bind directly to the promoter regions of BR biosynthetic genes and activate their expression. A, Schematic diagrams of the promoter regions of two BR biosynthetic genes, DWF4 and BR6ox2. Red upright lines represent G-box DNA motifs. P1 and P2 are the DNA fragments for ChIP-PCR amplification. B, ChIP analysis showing that PIF4-FLAG and PIF5-FLAG interact with the G-box-containing regions within the DWF4 and BR6ox2 promoters upon precipitation with anti-FLAG antibody. One-week-old WS2, 35S:PIF4-FLAG, and 35S:PIF5-FLAG seedlings were harvested for ChIP experiments. qRT-PCR was used to quantify the enriched DNA fragments in the DWF4 and BR6ox2 promoters. C and D, EMSA indicates that PIF4 binds to the G-box motifs within the promoter regions of DWF4 and BR6ox2. The biotin-labeled probes containing the G-box elements were incubated with TNT-expressed PIF4 protein, which was used for gel-shift analyses. The probes incubated with the TNT expression system without the PIF4 template were used as negative controls. Nonlabeled probes containing G-box (wild type [WT]) or mutated G-box (M) with 10- or 100-fold molar concentrations over the biotin-labeled probe were used as cold competitors. E, EMSA shows that PIF5 binds to the G-box motifs in the promoter of DWF4. The fragments containing the G-box motifs present in the promoters of DWF4 were incubated with TNT-expressed PIF5 protein, which was used subsequently for EMSA analyses. Competition for PIF5 binding was performed using 10- and 100-fold unlabeled probes containing G-box or mutated G-box. F, DWF4 and BR6ox2 expression levels in 1-week-old WS2, 35S:PIF4-FLAG, and 35S:PIF5-FLAG seedlings. G, Expression levels of DWF4 and BR6ox2 in WS2 and 35S:PIF4-ER transgenic plants with or without estradiol treatment in the presence of CHX. WS2 and 35S:PIF4-ER seedlings were pretreated with 10 μm CHX for 2 h, then either mock solution (ethanol) or 10 μm estradiol was added to the liquid solution for another 2 h before harvesting for RNA extraction and qRT-PCR analyses. H, Endogenous BL levels in WS2, 35S:PIF4-FLAG, and 35S:PIF5-FLAG seedlings. The data shown are averages and sd from three independent replicates. FW, Fresh weight.
DISCUSSION
Regarding BR related research, the least characterized aspect is probably the mechanisms controlling BR homeostasis. Consistent with other growth-promoting phytohormones, such as auxin, GA, and cytokinin, both deficiency and excessive amounts of BR are harmful to plant growth and development. Unlike other phytohormones, BRs cannot be transported in a long-distance manner. Therefore, controlling BR homeostasis in a certain tissue or a cell is extremely important to their biological functions. In this study, we identified a dominant genetic suppressor, cog1-3D, of a weak BR receptor mutant named bri1-5. cog1-3D and cog1-3D bri1-5 showed significantly longer hypocotyl phenotypes relative to their corresponding backgrounds, WS2 and bri1-5, respectively (Figs. 1A and 2A). BR profile analyses revealed that multiple BR intermediates, including two biologically active forms, CS and BL, are drastically elevated in cog1-3D in comparison with those in wild-type seedlings (Fig. 3C). Transcription assays ruled out the possibility that it was caused by decreasing BR catabolism. Rather, it appeared that the accumulation of BRs is most likely caused by the enhanced BR biosynthesis. Consistently, several key BR biosynthesis genes are up-regulated in cog1-3D (Fig. 4A). Detailed analyses further suggested that cog1-3D up-regulation of the expression of BR biosynthetic genes is largely dependent on the function of PIF4 and PIF5. In the pif4 pif5 background, cog1-3D cannot promote the expression of BR biosynthetic genes and, as a result, fails to enhance hypocotyl growth (Fig. 5, C–E). EMSA experiments clearly indicated that COG1 binds directly to the promoter regions of PIF4 and PIF5 to regulate their expression. PIF4 and PIF5 associate with G-box motifs in the promoters of DWF4 and BR6ox2 (Figs. 6 and 7). Our results obtained from genetic, physiological, and biochemical analyses provide strong evidence to support that COG1 modulates BR biosynthesis and hypocotyl growth via PIF4 and PIF5 in Arabidopsis.
How can we explain the role of COG1 in promoting hypocotyl elongation in the wild type or a weak allele of the BRI1 mutant but not in BR-deficient mutants or a BRI1 null mutant? Our current observations suggest that COG1 activates the expression of PIF4 and PIF5, which subsequently enhance BR biosynthesis and signal transduction to promote hypocotyl growth. In the wild-type background, BRI1- and BAK1-mediated BR signaling results in the dephosphorylation and activation of a BZR1-PIF4 transcription factor complex that regulates the expression of a large number of genes involved in cell elongation to modulate hypocotyl growth (Wang et al., 2002b; Oh et al., 2012; Bernardo-García et al., 2014). COG1 up-regulates the expression of PIF4 and PIF5, enhancing the function of the BZR1-PIF4 complex on hypocotyl elongation. In addition, PIF4 and PIF5 also promote BR biosynthesis, leading to elevated BR signaling, thus further enhancing the function of the BZR1-PIF4 complex (Fig. 8A). In the bri1-5 background, however, BR signaling is partially impaired. Since BRs are largely accumulated in bri1-5 (Noguchi et al., 1999a), the elevation of BR biosynthesis alone cannot explain the hypocotyl growth seen in cog1-3D bri1-5. It is likely that the elevated PIF4 promotes the complex formation with a leaky amount of nucleus-localized functional BZR1 to regulate downstream gene expression and slightly enhance hypocotyl growth in bri1-5 (Fig. 8B). In BR signaling or biosynthesis null mutants, BZR1 and PIF4 are phosphorylated by BIN2, leading to their degradation (He et al., 2002; Bernardo-García et al., 2014). Thus, COG1-mediated up-regulation of PIF4 cannot promote the formation of the BZR1-PIF4 complex and fails to enhance hypocotyl growth (Fig. 8C). These results indicated that the function of COG1 in regulating hypocotyl growth is dependent on both PIF4 and BRs.
Figure 8.
A model for COG1 promoting hypocotyl elongation. A, In the wild-type background, COG1 activates the expression of PIF4, which not only enhances the function of the BZR1-PIF4 complex on hypocotyl elongation but also promotes BR biosynthesis, further enhancing BR signaling and, thus, the function of BZR1-PIF4 on cell elongation. B, In the bri1-5 background, the elevation of BR biosynthesis has no effect on hypocotyl growth, since BR signaling is partially impaired. However, the elevated PIF4 by COG1 promotes the complex formation of BZR1-PIF4 and, thus, downstream gene expression to partially enhance hypocotyl growth in bri1-5. C, In BR signaling or biosynthesis null mutants, BZR1 and PIF4 are phosphorylated by BIN2, leading to their degradation. Thus, the COG1-mediated up-regulation of PIF4 fails to enhance hypocotyl growth.
PIF4 and PIF5 were found initially to interact directly with PHYB and act as its downstream components to mediate phytochrome signaling (Huq and Quail, 2002; Shen et al., 2007). Recent studies suggested that they are the main integrators in light and hormonal signaling pathways to mediate hypocotyl elongation (de Lucas et al., 2008; Bai et al., 2012; Gallego-Bartolomé et al., 2012; Oh et al., 2012, 2014; Bernardo-García et al., 2014). PIF4 interacts directly with BZR1 and ARF6 to coregulate a large number of target genes critical for cell expansion, providing evidence for the cross talk among BR, auxin, and phytochrome during hypocotyl growth (Oh et al., 2014). Furthermore, GA promotes cell elongation largely by releasing the DELLA-mediated repression of PIF4, BZR1, and ARF6 (Bai et al., 2012; Gallego-Bartolomé et al., 2012). In addition to DELLA proteins, the activity and levels of PIF4 are regulated by several other mechanisms. Interaction with the Pfr form of PHYB induces the phosphorylation and degradation of PIF4 (Huq and Quail, 2002; Shen et al., 2007). A recent study found that BIN2 phosphorylates PIF4 to bring it to a degradation pathway (Bernardo-García et al., 2014). Additionally, the expression of PIF4 can be induced in response to far-red and red light (Huq and Quail, 2002). Temperature and the circadian clock also control the transcription levels of PIFs (Leivar and Quail, 2011). However, the transcription factors that directly regulate the expression of PIF4 are largely unknown. A previous study revealed that COG1 expression could be induced by far-red and red light, which is similar to that of PIF4 (Huq and Quail, 2002; Park et al., 2003). In this study, we demonstrated that COG1 binds directly to the promoter regions of PIF4 and PIF5 to regulate their expression (Fig. 6).
Biosynthetic pathways are frequently feedback regulated by their end products or their signaling pathway either positively or negatively. For example, ethylene can both positively and negatively regulate its biosynthesis by up-regulating the expression of ACS2 and ACS4 and down-regulating the expression of ACS6 (Wang et al., 2002a). Abscisic acid induces the expression of AtNCED3 to positively regulate its biosynthesis under dehydration stress (Yang and Tan, 2014). BR signaling is utilized to create a feedback inhibitory regulation loop to control the expression of BR biosynthetic genes (He et al., 2005; Sun et al., 2010; Yu et al., 2011). Together with previous studies, our results support a positive feedback regulatory loop of BR biosynthesis by PIF4-mediated signaling (Fig. 8). In the absence of BR, BIN2 phosphorylates PIF4, leading to its degradation process and the inhibition of hypocotyl elongation (Bernardo-García et al., 2014). In the presence of BR, activated BRI1 and BAK1 receptor kinase lead to the inactivation of BIN2. PIF4 then can interact with BZR1 to regulate the expression of their target genes and promote hypocotyl growth (Oh et al., 2012; Bernardo-García et al., 2014). On the other hand, PIF4 activates the expression of DWF4 and BR6ox2 to produce more BRs and further promote hypocotyl elongation. Meanwhile, BZR1 can inhibit the expression of BR biosynthetic genes when BRs are excessive to plant growth and development (He et al., 2005). Such positive and negative feedback loops ensure optimal BR effects on plant growth and development. How plants reset the equilibrium of BR in response to developmental and environmental signals is a question to be answered in the near future.
Understanding the mechanisms controlling BR homeostasis can help us to develop strategies to optimize plant growth and development via genetic engineering in the future. BR profile analysis showed that the substrates of DWF4 are always plentiful in plants. However, the products of the DWF4-catalyzed reactions are considerably low or even undetectable, suggesting that DWF4 catalyzes a flux-determining step in BR biosynthesis (Guo et al., 2010). BR6ox2 was identified as a bifunctional enzyme that mediates the conversion of CS to BL, the final and rate-limiting step in BR biosynthesis, as well as BR C-6 oxidation (Kim et al., 2005). Controlling the expression of DWF4 and/or BR6ox2 could be an effective approach to alter BR production in response to external and internal cues. TCP1 was identified as a transcription factor promoting the expression of DWF4 (Guo et al., 2010). Auxin and high temperature also can induce the expression of DWF4 via mechanisms yet to be determined (Chung et al., 2011; Maharjan and Choe, 2011). However, transcriptional regulation of BR6ox2 is not described in the literature. Our data revealed that PIF4 functions as a positive regulator enhancing the expression of DWF4 and BR6ox2, leading to an increase of bioactive levels of BRs in cog1-3D (Figs. 3C and 7). PIF5, a homolog of PIF4, also can bind to the promoter of DWF4 to activate its expression (Fig. 7).
Interestingly, CPD also is up-regulated in COG1- and PIF4-overexpressing plants, and the ChIP assay showed that PIF4 associates with the promoter of CPD (Fig. 4A; Supplemental Fig. S12C). However, we failed to detect the direct binding of PIF4 to the CPD promoter, indicating the existence of other transcription factors that regulate CPD expression together with PIF4. The expression levels of CPD and DWF4 are not altered significantly in pif4 pif5 double mutants compared with the wild type (Fig. 5E), which should be caused by the functional redundancy of other transcription factors, such as CES and TCP1, that regulate the expression of these two BR biosynthetic genes. And there might be more transcription factors regulating the level of these two BR biosynthetic genes. Teasterone, an earlier BR intermediate converted from cathasterone by ROT3, also is increased in cog1-3D (Fig. 3C). However, ROT3 is not regulated directly by PIF4 and PIF5, suggesting that COG1 may regulate ROT3 expression through other transcription factors. Typhasterol and CS are both elevated in cog1-3D, indicating that COG1 regulates genes essential for C-3 dehydrogenation, C-3 reduction, and C-2 hydroxylation. It is known that OsD2 and OsD11 are responsible for the C-3 dehydrogenation and C-3 reduction reactions in rice (Oryza sativa), respectively (Hong et al., 2003; Tanabe et al., 2005). C-2 hydroxylation of BRs is mediated by DDWF1 in pea (Pisum sativum; Kang et al., 2001). However, the enzymes catalyzing these reactions in Arabidopsis are not identified. It is possible that homologs of these three cytochrome P450s that are up-regulated in cog1-3D are the potential enzymes responsible for the corresponding reactions in Arabidopsis.
MATERIALS AND METHODS
Plant Materials
The Arabidopsis (Arabidopsis thaliana) bri1-5 cog1-3D double mutant was obtained from a large-scale activation-tagging screen for bri1-5 genetic modifiers as described previously (Li et al., 2001, 2002; Yuan et al., 2007; Guo et al., 2010). The cog1-3D single mutant was generated by backcrossing bri1-5 cog1-3D with WS2. Other plant materials, including bri1-5, bri1-4, det2, cpd, dwf4, and cog1-D, were described previously (Noguchi et al., 1999a; Park et al., 2003; Guo et al., 2010). The cog1-6 mutant was identified from an ethyl methanesulfonate-mutagenized cog1-D seed pool. The br6ox1 (Salk_148384), br6ox2 (Salk_148384), pif4 (Salk_140393), and pif5 (Salk_087012) T-DNA insertion lines were obtained from the Arabidopsis Biological Resource Center. The bri1-5, bri1-4, det2, and dwf4 mutants are in the WS2 background. The cpd, br6ox1/2, pif4, and pif5 mutants are in the Columbia-0 background. The double or triple mutants were generated by genetic crossing using the related single mutants. The cpd cog1-3D, br6ox1/2 cog1-3D, and pif4 pif5 cog1-3D mutants were then backcrossed with the WS2 ecotype for at least five times.
Identification of the cog1-3D Locus
The flanking genomic sequence of the inserted T-DNA of pBIB-BASTA-AT2 was amplified using thermal asymmetric interlaced PCR as described previously (Liu and Whittier, 1995; Yuan et al., 2007; Guo et al., 2010). The T-DNA insertion site was determined by sequencing the flanking genomic DNA.
Constructs and Plant Transformation
Gateway technology was employed to clone the coding sequences of COG1, PIF4, and PIF5 (Gou et al., 2010). The amplified COG1 coding sequence was cloned into a pEarleyGate 202 destination vector (Earley et al., 2006). The PIF4 and PIF5 coding sequences were cloned into a pBIB-35S-GWR-FLAG vector (Gou et al., 2010). For the construction of 35S:COG1-ER and 35S:PIF4-ER, the coding sequences of these two genes were introduced in a pBIB-35S-GWR-ER vector. For the construction of 35S:COG1-SRDX and GVG:COG1-SRDX, COG1-SRDX was PCR amplified and introduced in a pBIB-35S-GWR vector and a Gateway-compatible pTA7002 vector (Aoyama and Chua, 1997; Gou et al., 2010). These expression constructs were then transformed into Agrobacterium tumefaciens strain GV3101 for transformation of Arabidopsis plants by the floral dip method (Clough and Bent, 1998). The primers used are listed in Supplemental Table S2.
Hypocotyl Measurements
Seeds were surface sterilized for 15 min in 10% bleach, washed five times with sterilized water, and planted on one-half-strength MS agar plates containing 1% (w/v) Suc, with or without 24-epiBL, or BRZ. Plants were pretreated at 4°C for 3 d and then transferred to a growth chamber set at 22°C with a 16-h-light/8-h-dark photoperiod or in continuous red light for 6 d. The GVG:COG1-SRDX seeds were planted on one-half-strength MS agar plates containing 1% (w/v) Suc. Two days after germination, the seedlings were transferred to one-half-strength MS agar plates containing 1% (w/v) Suc and 10 μm DEX, growing for another 5 d. Hypocotyls of the seedlings were then scanned and measured as reported previously (Yuan et al., 2007). All measurements were repeated three times, and at least 20 seedlings were measured for each genotype. Student’s t test was performed (*, P < 0.05; **, P < 0.01; and ***, P < 0.001).
qRT-PCR Analyses
Long-day-grown seedlings were harvested directly or after treatment with DEX or estradiol and CHX at the same time of day and frozen in liquid nitrogen for RNA extraction. Total RNA was extracted using the RNAprep Pure Plant Kit (Tiangen Biotech). Five micrograms of total RNA was reverse transcribed into first-strand cDNAs using M-MLV reverse transcriptase (Thermo Fisher Scientific) according to the manufacturer’s instructions. The cDNA samples diluted 10-fold were used as PCR templates. qRT-PCR was performed using SYBR Premix Ex Taq II (TaKaRa) on a Step One Plus Real-Time PCR machine (Thermo Fisher Scientific). The relative expression level was calculated from three replicates using the ΔΔCt method after normalization to an ACT2 control. All the qRT-PCR analyses were performed for three biological replicates, yielding similar results. Data shown are means and se from three biological replicates. Student’s t test was performed (*, P < 0.05; **, P < 0.01; ***, P < 0.001; and ns, not significant). The primers used are listed in Supplemental Table S2.
BR Profile Assays
Samples were analyzed for BR contents as described previously with a few modifications (Swaczynova et al., 2007). Briefly, 50 mg of fresh Arabidopsis tissue samples was homogenized to a fine consistency using 3-mm zirconium oxide beads and an MM 301 vibration mill at a frequency of 30 Hz for 3 min (Retsch). The samples were then extracted overnight with stirring at 4°C using a benchtop laboratory rotator (Stuart SB3; Bibby Scientific) after the addition of 1 mL of ice-cold 60% acetonitrile and 10 pmol of [2H3]BL, [2H3]CS, [2H3]24-epiBL, [2H3]24-epicastasterone, [2H3]28-norbrassinolide, and [2H3]28-norcastasterone as internal standards (OlChemIm). The samples were further centrifuged, purified on polyamide SPE columns (Supelco), and then analyzed by ultra-high-performance liquid chromatography-tandem mass spectrometry (Micromass). The data were analyzed using Masslynx 4.1 software (Waters), and BR content was finally quantified by the standard isotope dilution method (Rittenberg and Foster, 1940).
ChIP-PCR Assay
Two grams of one-week-old WS2, 35S:FLAG-COG1, 35S:PIF4-FLAG, and 35S:PIF5-FLAG transgenic seedlings was harvested for ChIP experiments. Chromatin was isolated and sonicated to generate DNA fragments with an average size of 500 bp. The solubilized chromatin was immunoprecipitated by an agarose-conjugated anti-FLAG antibody (Sigma-Aldrich). The coimmunoprecipitated DNA was recovered and analyzed by quantitative PCR with SYBR Premix Ex Taq II (TaKaRa). Relative fold enrichment was calculated by normalizing the amount of a target DNA fragment against that of an ACT2 promoter fragment and then against the respective input DNA samples. Three independent biological repeats were performed, yielding similar results. Shown are representative data from one biological replicate. Student’s t test was performed (*, P < 0.05; **, P < 0.01; and ***, P < 0.001). The primers used are listed in Supplemental Table S2.
DNA Gel-Shift Assay
The full-length COG1 coding region was cloned into a pGEX-4T-3 vector (GE). The resulting constructs were sequenced to confirm the correct open reading frame and sequences and then transformed into the Escherichia coli Rosetta strain for recombinant protein expression. Bacteria were grown in 100 mL of Luria-Bertani medium supplemented with 100 μg mL−1 ampicillin and 30 μg mL−1 chloramphenicol, and the GST-COG1 recombinant proteins were purified with GST-Sefinose Resin (Sangon Biotech). PIF4 and PIF5 were synthesized using the TNT SP6 High-Yield Wheat Germ Protein Expression System (Promega). The probes were synthesized and labeled with biotin using the Biotin 3′ End DNA Labeling Kit (Thermo Fisher Scientific). Unlabeled competitor probes were generated from dimerized oligonucleotides. EMSA experiments were performed using the Chemiluminescent Nucleic Acid Detection Module (Thermo Fisher Scientific) according to the manufacturer’s instructions. Probe sequences are shown in Supplemental Table S2.
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL databases under the following accession numbers: COG1 (At1g29160), PIF4 (At2g43010), PIF5 (At3g59060), DET2 (At2g38050), CPD (At5g05690), DWF4 (At3g50660), BR6ox1 (At5g38970), BR6ox2 (At3g30180), ROT3 (At4g36380), and CYP90D1 (At3g13730).
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. The cog1-6 mutant was identified as an intragenic suppressor of cog1-D.
Supplemental Figure S2. Phenotypes of 35S:COG1-SRDX transgenic plants.
Supplemental Figure S3. Induced expression of COG1-SRDX leads to reduced cell length.
Supplemental Figure S4. BR signal transduction is elevated in the cog1-3D mutant.
Supplemental Figure S5. BRZ inhibits the hypocotyl growth of cog1-3D seedlings.
Supplemental Figure S6. Expression of DET2, CYP90D1, and BR6ox1 in cog1-3D.
Supplemental Figure S7. Expression of BR catabolism genes in cog1-3D.
Supplemental Figure S8. The bri1-4 mutation completely inhibits the long-hypocotyl phenotype of cog1-3D.
Supplemental Figure S9. PIF4 and PIF5 promote cell elongation.
Supplemental Figure S10. Protein levels of PIF5 in cog1-3D and 35S:FLAG-COG1 seedlings.
Supplemental Figure S11. COG1-mediated hypocotyl elongation is dependent on PIF4 and PIF5.
Supplemental Figure S12. PIF4 regulates the expression of CPD.
Supplemental Table S1. The expression of 37 transcription factors is significantly altered in cog1-3D compared with WS2.
Supplemental Table S2. List of primers and probes used in this study.
Supplementary Material
Acknowledgments
We thank Dr. Yanhai Yin (Iowa State University) and Dr. Haodong Chen (Peking University) for providing the BES1 and PIF5 antibodies, respectively.
Glossary
- BR
brassinosteroid
- BL
brassinolide
- CS
castasterone
- RT
reverse transcription
- DEX
dexamethasone
- MS
Murashige and Skoog
- 24-epiBL
24-epibrassinolide
- BRZ
brassinazole
- ChIP
chromatin immunoprecipitation
- CHX
cycloheximide
- qRT
quantitative reverse transcription
- EMSA
electrophoretic mobility shift assay
- DMSO
dimethyl sulfoxide
Footnotes
This work was supported by the National Natural Science Foundation of China (grant nos. 31530005, 31470380, and 91317311 to J.L.), the Ministry of Education (2011 Scholarship Award for Excellent Doctoral Student to Z.W.), the China Postdoctoral Science Foundation (grant no. 2016M602889 to Z.W.), the Ministry of Agriculture of the People’s Republic of China (grant no. 2016ZX08009-003-002 to K.H.), the Návrat program from the Ministry of Education, Youth, and Sports of the Czech Republic (grant no. LK21306 to O.N.), the Czech Science Foundation (grant no. GA14-34792S to O.N.), and the Institute for Basic Science (grant no. IBS-R013-D1 to H.G.N.).
References
- Aoyama T, Chua NH (1997) A glucocorticoid-mediated transcriptional induction system in transgenic plants. Plant J 11: 605–612 [DOI] [PubMed] [Google Scholar]
- Asami T, Min YK, Nagata N, Yamagishi K, Takatsuto S, Fujioka S, Murofushi N, Yamaguchi I, Yoshida S (2000) Characterization of brassinazole, a triazole-type brassinosteroid biosynthesis inhibitor. Plant Physiol 123: 93–100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai MY, Shang JX, Oh E, Fan M, Bai Y, Zentella R, Sun TP, Wang ZY (2012) Brassinosteroid, gibberellin and phytochrome impinge on a common transcription module in Arabidopsis. Nat Cell Biol 14: 810–817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bajguz A. (2007) Metabolism of brassinosteroids in plants. Plant Physiol Biochem 45: 95–107 [DOI] [PubMed] [Google Scholar]
- Bancoş S, Nomura T, Sato T, Molnár G, Bishop GJ, Koncz C, Yokota T, Nagy F, Szekeres M (2002) Regulation of transcript levels of the Arabidopsis cytochrome p450 genes involved in brassinosteroid biosynthesis. Plant Physiol 130: 504–513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernardo-García S, de Lucas M, Martínez C, Espinosa-Ruiz A, Davière JM, Prat S (2014) BR-dependent phosphorylation modulates PIF4 transcriptional activity and shapes diurnal hypocotyl growth. Genes Dev 28: 1681–1694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bishop GJ, Nomura T, Yokota T, Harrison K, Noguchi T, Fujioka S, Takatsuto S, Jones JD, Kamiya Y (1999) The tomato DWARF enzyme catalyses C-6 oxidation in brassinosteroid biosynthesis. Proc Natl Acad Sci USA 96: 1761–1766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bueso E, Muñoz-Bertomeu J, Campos F, Martínez C, Tello C, Martínez-Almonacid I, Ballester P, Simón-Moya M, Brunaud V, Yenush L, et al. (2016) Arabidopsis COGWHEEL1 links light perception and gibberellins with seed tolerance to deterioration. Plant J 87: 583–596 [DOI] [PubMed] [Google Scholar]
- Castillon A, Shen H, Huq E (2007) Phytochrome Interacting Factors: central players in phytochrome-mediated light signaling networks. Trends Plant Sci 12: 514–521 [DOI] [PubMed] [Google Scholar]
- Choe S, Dilkes BP, Fujioka S, Takatsuto S, Sakurai A, Feldmann KA (1998) The DWF4 gene of Arabidopsis encodes a cytochrome P450 that mediates multiple 22α-hydroxylation steps in brassinosteroid biosynthesis. Plant Cell 10: 231–243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi YH, Fujioka S, Harada A, Yokota T, Takatsuto S, Sakurai A (1996) A brassinolide biosynthetic pathway via 6-deoxocastasterone. Phytochemistry 43: 593–596 [Google Scholar]
- Choi YH, Fujioka S, Nomura T, Harada A, Yokota T, Takatsuto S, Sakurai A (1997) An alternative brassinolide biosynthetic pathway via late C-6 oxidation. Phytochemistry 44: 609–613 [Google Scholar]
- Chory J, Nagpal P, Peto CA (1991) Phenotypic and genetic analysis of det2, a new mutant that affects light-regulated seedling development in Arabidopsis. Plant Cell 3: 445–459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung Y, Maharjan PM, Lee O, Fujioka S, Jang S, Kim B, Takatsuto S, Tsujimoto M, Kim H, Cho S, et al. (2011) Auxin stimulates DWARF4 expression and brassinosteroid biosynthesis in Arabidopsis. Plant J 66: 624–638 [DOI] [PubMed] [Google Scholar]
- Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Clouse SD, Sasse JM (1998) Brassinosteroids: essential regulators of plant growth and development. Annu Rev Plant Physiol Plant Mol Biol 49: 427–451 [DOI] [PubMed] [Google Scholar]
- de Lucas M, Davière JM, Rodríguez-Falcón M, Pontin M, Iglesias-Pedraz JM, Lorrain S, Fankhauser C, Blázquez MA, Titarenko E, Prat S (2008) A molecular framework for light and gibberellin control of cell elongation. Nature 451: 480–484 [DOI] [PubMed] [Google Scholar]
- Earley KW, Haag JR, Pontes O, Opper K, Juehne T, Song K, Pikaard CS (2006) Gateway-compatible vectors for plant functional genomics and proteomics. Plant J 45: 616–629 [DOI] [PubMed] [Google Scholar]
- Fujioka S, Inoue T, Takatsuto S, Yanagisawa T, Yokota T, Sakurai A (1995) Identification of a new brassinosteroid, cathasterone, in cultured-cells of catharanthus-roseus as a biosynthetic precursor of teasterone. Biosci Biotechnol Biochem 59: 1543–1547 [Google Scholar]
- Fujioka S, Li J, Choi YH, Seto H, Takatsuto S, Noguchi T, Watanabe T, Kuriyama H, Yokota T, Chory J, et al. (1997) The Arabidopsis deetiolated2 mutant is blocked early in brassinosteroid biosynthesis. Plant Cell 9: 1951–1962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujioka S, Takatsuto S, Yoshida S (2002) An early C-22 oxidation branch in the brassinosteroid biosynthetic pathway. Plant Physiol 130: 930–939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujioka S, Yokota T (2003) Biosynthesis and metabolism of brassinosteroids. Annu Rev Plant Biol 54: 137–164 [DOI] [PubMed] [Google Scholar]
- Fujita S, Ohnishi T, Watanabe B, Yokota T, Takatsuto S, Fujioka S, Yoshida S, Sakata K, Mizutani M (2006) Arabidopsis CYP90B1 catalyses the early C-22 hydroxylation of C27, C28 and C29 sterols. Plant J 45: 765–774 [DOI] [PubMed] [Google Scholar]
- Gallego-Bartolomé J, Minguet EG, Grau-Enguix F, Abbas M, Locascio A, Thomas SG, Alabadí D, Blázquez MA (2012) Molecular mechanism for the interaction between gibberellin and brassinosteroid signaling pathways in Arabidopsis. Proc Natl Acad Sci USA 109: 13446–13451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Y, Zhang D, Li J (2015) TCP1 modulates DWF4 expression via directly interacting with the GGNCCC motifs in the promoter region of DWF4 in Arabidopsis thaliana. J Genet Genomics 42: 383–392 [DOI] [PubMed] [Google Scholar]
- Gou X, He K, Yang H, Yuan T, Lin H, Clouse SD, Li J (2010) Genome-wide cloning and sequence analysis of leucine-rich repeat receptor-like protein kinase genes in Arabidopsis thaliana. BMC Genomics 11: 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gou X, Li J (2012) Activation tagging. Methods Mol Biol 876: 117–133 [DOI] [PubMed] [Google Scholar]
- Gou X, Yin H, He K, Du J, Yi J, Xu S, Lin H, Clouse SD, Li J (2012) Genetic evidence for an indispensable role of somatic embryogenesis receptor kinases in brassinosteroid signaling. PLoS Genet 8: e1002452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Z, Fujioka S, Blancaflor EB, Miao S, Gou X, Li J (2010) TCP1 modulates brassinosteroid biosynthesis by regulating the expression of the key biosynthetic gene DWARF4 in Arabidopsis thaliana. Plant Cell 22: 1161–1173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He JX, Gendron JM, Sun Y, Gampala SS, Gendron N, Sun CQ, Wang ZY (2005) BZR1 is a transcriptional repressor with dual roles in brassinosteroid homeostasis and growth responses. Science 307: 1634–1638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He JX, Gendron JM, Yang Y, Li J, Wang ZY (2002) The GSK3-like kinase BIN2 phosphorylates and destabilizes BZR1, a positive regulator of the brassinosteroid signaling pathway in Arabidopsis. Proc Natl Acad Sci USA 99: 10185–10190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He K, Xu S, Li J (2013) BAK1 directly regulates brassinosteroid perception and BRI1 activation. J Integr Plant Biol 55: 1264–1270 [DOI] [PubMed] [Google Scholar]
- Hiratsu K, Matsui K, Koyama T, Ohme-Takagi M (2003) Dominant repression of target genes by chimeric repressors that include the EAR motif, a repression domain, in Arabidopsis. Plant J 34: 733–739 [DOI] [PubMed] [Google Scholar]
- Hong Z, Ueguchi-Tanaka M, Umemura K, Uozu S, Fujioka S, Takatsuto S, Yoshida S, Ashikari M, Kitano H, Matsuoka M (2003) A rice brassinosteroid-deficient mutant, ebisu dwarf (d2), is caused by a loss of function of a new member of cytochrome P450. Plant Cell 15: 2900–2910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hothorn M, Belkhadir Y, Dreux M, Dabi T, Noel JP, Wilson IA, Chory J (2011) Structural basis of steroid hormone perception by the receptor kinase BRI1. Nature 474: 467–471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huq E, Quail PH (2002) PIF4, a phytochrome-interacting bHLH factor, functions as a negative regulator of phytochrome B signaling in Arabidopsis. EMBO J 21: 2441–2450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaillais Y, Hothorn M, Belkhadir Y, Dabi T, Nimchuk ZL, Meyerowitz EM, Chory J (2011) Tyrosine phosphorylation controls brassinosteroid receptor activation by triggering membrane release of its kinase inhibitor. Genes Dev 25: 232–237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang JG, Yun J, Kim DH, Chung KS, Fujioka S, Kim JI, Dae HW, Yoshida S, Takatsuto S, Song PS, et al. (2001) Light and brassinosteroid signals are integrated via a dark-induced small G protein in etiolated seedling growth. Cell 105: 625–636 [DOI] [PubMed] [Google Scholar]
- Kim TW, Guan S, Burlingame AL, Wang ZY (2011) The CDG1 kinase mediates brassinosteroid signal transduction from BRI1 receptor kinase to BSU1 phosphatase and GSK3-like kinase BIN2. Mol Cell 43: 561–571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim TW, Guan S, Sun Y, Deng Z, Tang W, Shang JX, Sun Y, Burlingame AL, Wang ZY (2009) Brassinosteroid signal transduction from cell-surface receptor kinases to nuclear transcription factors. Nat Cell Biol 11: 1254–1260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim TW, Hwang JY, Kim YS, Joo SH, Chang SC, Lee JS, Takatsuto S, Kim SK (2005) Arabidopsis CYP85A2, a cytochrome P450, mediates the Baeyer-Villiger oxidation of castasterone to brassinolide in brassinosteroid biosynthesis. Plant Cell 17: 2397–2412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinoshita T, Caño-Delgado A, Seto H, Hiranuma S, Fujioka S, Yoshida S, Chory J (2005) Binding of brassinosteroids to the extracellular domain of plant receptor kinase BRI1. Nature 433: 167–171 [DOI] [PubMed] [Google Scholar]
- Leivar P, Quail PH (2011) PIFs: pivotal components in a cellular signaling hub. Trends Plant Sci 16: 19–28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Lease KA, Tax FE, Walker JC (2001) BRS1, a serine carboxypeptidase, regulates BRI1 signaling in Arabidopsis thaliana. Proc Natl Acad Sci USA 98: 5916–5921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Nagpal P, Vitart V, McMorris TC, Chory J (1996) A role for brassinosteroids in light-dependent development of Arabidopsis. Science 272: 398–401 [DOI] [PubMed] [Google Scholar]
- Li J, Nam KH (2002) Regulation of brassinosteroid signaling by a GSK3/SHAGGY-like kinase. Science 295: 1299–1301 [DOI] [PubMed] [Google Scholar]
- Li J, Wen J, Lease KA, Doke JT, Tax FE, Walker JC (2002) BAK1, an Arabidopsis LRR receptor-like protein kinase, interacts with BRI1 and modulates brassinosteroid signaling. Cell 110: 213–222 [DOI] [PubMed] [Google Scholar]
- Liu YG, Whittier RF (1995) Thermal asymmetric interlaced PCR: automatable amplification and sequencing of insert end fragments from P1 and YAC clones for chromosome walking. Genomics 25: 674–681 [DOI] [PubMed] [Google Scholar]
- Maharjan PM, Choe S (2011) High temperature stimulates DWARF4 (DWF4) expression to increase hypocotyl elongation in Arabidopsis. J Plant Biol 54: 425–429 [Google Scholar]
- Mathur J, Molnár G, Fujioka S, Takatsuto S, Sakurai A, Yokota T, Adam G, Voigt B, Nagy F, Maas C, et al. (1998) Transcription of the Arabidopsis CPD gene, encoding a steroidogenic cytochrome P450, is negatively controlled by brassinosteroids. Plant J 14: 593–602 [DOI] [PubMed] [Google Scholar]
- Mora-García S, Vert G, Yin Y, Caño-Delgado A, Cheong H, Chory J (2004) Nuclear protein phosphatases with Kelch-repeat domains modulate the response to brassinosteroids in Arabidopsis. Genes Dev 18: 448–460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nam KH, Li J (2002) BRI1/BAK1, a receptor kinase pair mediating brassinosteroid signaling. Cell 110: 203–212 [DOI] [PubMed] [Google Scholar]
- Noguchi T, Fujioka S, Choe S, Takatsuto S, Yoshida S, Yuan H, Feldmann KA, Tax FE (1999a) Brassinosteroid-insensitive dwarf mutants of Arabidopsis accumulate brassinosteroids. Plant Physiol 121: 743–752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noguchi T, Fujioka S, Takatsuto S, Sakurai A, Yoshida S, Li J, Chory J (1999b) Arabidopsis det2 is defective in the conversion of (24R)-24-methylcholest-4-En-3-one to (24R)-24-methyl-5α-cholestan-3-one in brassinosteroid biosynthesis. Plant Physiol 120: 833–840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh E, Zhu JY, Bai MY, Arenhart RA, Sun Y, Wang ZY (2014) Cell elongation is regulated through a central circuit of interacting transcription factors in the Arabidopsis hypocotyl. eLife 3: doi/10.7554/eLife.03031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh E, Zhu JY, Wang ZY (2012) Interaction between BZR1 and PIF4 integrates brassinosteroid and environmental responses. Nat Cell Biol 14: 802–809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohnishi T, Godza B, Watanabe B, Fujioka S, Hategan L, Ide K, Shibata K, Yokota T, Szekeres M, Mizutani M (2012) CYP90A1/CPD, a brassinosteroid biosynthetic cytochrome P450 of Arabidopsis, catalyzes C-3 oxidation. J Biol Chem 287: 31551–31560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohnishi T, Szatmari AM, Watanabe B, Fujita S, Bancos S, Koncz C, Lafos M, Shibata K, Yokota T, Sakata K, et al. (2006) C-23 hydroxylation by Arabidopsis CYP90C1 and CYP90D1 reveals a novel shortcut in brassinosteroid biosynthesis. Plant Cell 18: 3275–3288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park DH, Lim PO, Kim JS, Cho DS, Hong SH, Nam HG (2003) The Arabidopsis COG1 gene encodes a Dof domain transcription factor and negatively regulates phytochrome signaling. Plant J 34: 161–171 [DOI] [PubMed] [Google Scholar]
- Poppenberger B, Rozhon W, Khan M, Husar S, Adam G, Luschnig C, Fujioka S, Sieberer T (2011) CESTA, a positive regulator of brassinosteroid biosynthesis. EMBO J 30: 1149–1161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rittenberg D, Foster GL (1940) A new procedure for quantitative analysis by isotope dilution, with application to the determination of amino acids and fatty acids. J Biol Chem 133: 737–744 [Google Scholar]
- Santiago J, Henzler C, Hothorn M (2013) Molecular mechanism for plant steroid receptor activation by somatic embryogenesis co-receptor kinases. Science 341: 889–892 [DOI] [PubMed] [Google Scholar]
- She J, Han Z, Kim TW, Wang J, Cheng W, Chang J, Shi S, Wang J, Yang M, Wang ZY, et al. (2011) Structural insight into brassinosteroid perception by BRI1. Nature 474: 472–476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen Y, Khanna R, Carle CM, Quail PH (2007) Phytochrome induces rapid PIF5 phosphorylation and degradation in response to red-light activation. Plant Physiol 145: 1043–1051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimada Y, Fujioka S, Miyauchi N, Kushiro M, Takatsuto S, Nomura T, Yokota T, Kamiya Y, Bishop GJ, Yoshida S (2001) Brassinosteroid-6-oxidases from Arabidopsis and tomato catalyze multiple C-6 oxidations in brassinosteroid biosynthesis. Plant Physiol 126: 770–779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y, Fan XY, Cao DM, Tang W, He K, Zhu JY, He JX, Bai MY, Zhu S, Oh E, et al. (2010) Integration of brassinosteroid signal transduction with the transcription network for plant growth regulation in Arabidopsis. Dev Cell 19: 765–777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y, Han Z, Tang J, Hu Z, Chai C, Zhou B, Chai J (2013) Structure reveals that BAK1 as a co-receptor recognizes the BRI1-bound brassinolide. Cell Res 23: 1326–1329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki H, Fujioka S, Takatsuto S, Yokota T, Murofushi N, Sakurai A (1993) Biosynthesis of brassinolide from castasterone in cultured cells of Catharanthus roseus. J Plant Growth Regul 12: 101–106 [Google Scholar]
- Suzuki H, Fujioka S, Takatsuto S, Yokota T, Murofushi N, Sakurai A (1994a) Biosynthesis of brassinolide from teasterone via typhasterol and castasterone in cultured cells of Catharanthus roseus. J Plant Growth Regul 13: 21–26 [Google Scholar]
- Suzuki H, Inoue T, Fujioka S, Takatsuto S, Yanagisawa T, Yokota T, Murofushi N, Sakurai A (1994b) Possible involvement of 3-dehydroteasterone in the conversion of teasterone to typhasterol in cultured cells of Catharanthus roseus. Biosci Biotechnol Biochem 58: 1186–1188 [Google Scholar]
- Swaczynova J, Novak O, Hauserova E, Fuksova K, Sisa M, Kohout L, Strnad M (2007) New techniques for the estimation of naturally occurring brassinosteroids. J Plant Growth Regul 26: 1–14 [Google Scholar]
- Szekeres M, Németh K, Koncz-Kálmán Z, Mathur J, Kauschmann A, Altmann T, Rédei GP, Nagy F, Schell J, Koncz C (1996) Brassinosteroids rescue the deficiency of CYP90, a cytochrome P450, controlling cell elongation and de-etiolation in Arabidopsis. Cell 85: 171–182 [DOI] [PubMed] [Google Scholar]
- Tanabe S, Ashikari M, Fujioka S, Takatsuto S, Yoshida S, Yano M, Yoshimura A, Kitano H, Matsuoka M, Fujisawa Y, et al. (2005) A novel cytochrome P450 is implicated in brassinosteroid biosynthesis via the characterization of a rice dwarf mutant, dwarf11, with reduced seed length. Plant Cell 17: 776–790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang KL, Li H, Ecker JR (2002a) Ethylene biosynthesis and signaling networks. Plant Cell (Suppl) 14: S131–S151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Chory J (2006) Brassinosteroids regulate dissociation of BKI1, a negative regulator of BRI1 signaling, from the plasma membrane. Science 313: 1118–1122 [DOI] [PubMed] [Google Scholar]
- Wang ZY, Nakano T, Gendron J, He J, Chen M, Vafeados D, Yang Y, Fujioka S, Yoshida S, Asami T, et al. (2002b) Nuclear-localized BZR1 mediates brassinosteroid-induced growth and feedback suppression of brassinosteroid biosynthesis. Dev Cell 2: 505–513 [DOI] [PubMed] [Google Scholar]
- Yanagisawa S. (2002) The Dof family of plant transcription factors. Trends Plant Sci 7: 555–560 [DOI] [PubMed] [Google Scholar]
- Yanagisawa S. (2004) Dof domain proteins: plant-specific transcription factors associated with diverse phenomena unique to plants. Plant Cell Physiol 45: 386–391 [DOI] [PubMed] [Google Scholar]
- Yang YZ, Tan BC (2014) A distal ABA responsive element in AtNCED3 promoter is required for positive feedback regulation of ABA biosynthesis in Arabidopsis. PLoS ONE 9: e87283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin Y, Wang ZY, Mora-Garcia S, Li J, Yoshida S, Asami T, Chory J (2002) BES1 accumulates in the nucleus in response to brassinosteroids to regulate gene expression and promote stem elongation. Cell 109: 181–191 [DOI] [PubMed] [Google Scholar]
- Yokota T, Ogino Y, Takahashi N, Saimoto H, Fujioka S, Sakurai A (1990) Brassinolide is biosynthesized from castasterone in Catharanthus roseus crown gall cells. Agric Biol Chem 54: 1107–1108 [Google Scholar]
- Yu X, Li L, Zola J, Aluru M, Ye H, Foudree A, Guo H, Anderson S, Aluru S, Liu P, et al. (2011) A brassinosteroid transcriptional network revealed by genome-wide identification of BESI target genes in Arabidopsis thaliana. Plant J 65: 634–646 [DOI] [PubMed] [Google Scholar]
- Yuan T, Fujioka S, Takatsuto S, Matsumoto S, Gou X, He K, Russell SD, Li J (2007) BEN1, a gene encoding a dihydroflavonol 4-reductase (DFR)-like protein, regulates the levels of brassinosteroids in Arabidopsis thaliana. Plant J 51: 220–233 [DOI] [PubMed] [Google Scholar]
- Zhao B, Li J (2012) Regulation of brassinosteroid biosynthesis and inactivation. J Integr Plant Biol 54: 746–759 [DOI] [PubMed] [Google Scholar]
- Zhou A, Wang H, Walker JC, Li J (2004) BRL1, a leucine-rich repeat receptor-like protein kinase, is functionally redundant with BRI1 in regulating Arabidopsis brassinosteroid signaling. Plant J 40: 399–409 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.