An APC8-like protein regulates leaf petiole angle by modulating establishment of pulvinus in soybean.
Abstract
Leaf petiole angle (LPA) is an important plant architectural trait that affects canopy coverage, photosynthetic efficiency, and ultimately productivity in many legume crops. However, the genetic basis underlying this trait remains unclear. Here, we report the identification, isolation, and functional characterization of Glycine max Increased Leaf Petiole Angle1 (GmILPA1), a gene encoding an APC8-like protein, which is a subunit of the anaphase-promoting complex/cyclosome in soybean (Glycine max). A gamma ray-induced deletion of a fragment involving the fourth exon of GmILPA1 and its flanking sequences led to extension of the third exon and formation of, to our knowledge, a novel 3′UTR from intronic and intergenic sequences. Such changes are responsible for enlarged LPAs that are associated with reduced motor cell proliferation in the Gmilpa1 mutant. GmILPA1 is mainly expressed in the basal cells of leaf primordia and appears to function by promoting cell growth and division of the pulvinus that is critical for its establishment. GmILPA1 directly interacts with GmAPC13a as part of the putative anaphase-promoting complex. GmILPA1 exhibits variable expression levels among varieties with different degrees of LPAs, and expression levels are correlated with the degrees of the LPAs. Together, these observations revealed a genetic mechanism modulating the plant petiole angle that could pave the way for modifying soybean plant architecture with optimized petiole angles for enhanced yield potential.
Plant architecture is defined as the three-dimensional organization of the plant body. For aboveground plant parts, this includes stem height, branching pattern, and the shape and position of leaves and reproductive organs (Reinhardt and Kuhlemeier, 2002). Among these factors, leaf angle is a key determinant of canopy coverage that directly affects plant light reception, photosynthetic efficiency, and ultimately, productivity. For instance, maize (Zea mays) cultivars with more upright leaf angles above the uppermost ear (i.e., compact plant architecture) produce more grains per unit land area solely by permitting denser planting (Lu et al., 2007). As such, leaf angle in cereals is of agronomic importance and vital consideration for plant breeders.
Soybean (Glycine max) is one of the most important oilseed crops that provides edible oil for humans and is a major renewable feedstock for biodiesel production around the world. With the rapid increase in human consumption and industrial use, the demand for soybean yield has increased substantially and will continue to increase. As such, increasing soybean yield potential has become a long-term breeding objective. Soybean yield potential can be enhanced by creating ideal plant architecture that is mainly determined by leaf and stem composition. Leaf petiole angle (LPA), the degree of inclination between the leaf petiole and stem, is of particular importance in determining the plant architecture in soybean and many other legumes (Rodrigues and Machado, 2008; Zhou et al., 2012).
LPA is mainly controlled by the structure of a motor organ, termed the “pulvinus” (Volkov et al., 2010; Song et al., 2014), which is a jointlike thickening at the base of a leaf petiole, leaf, or leaflet. Typically, a pulvinus consists of a core of vascular tissues surrounded by a flexible, bulky cylinder of thin-walled parenchyma cells (Satter et al., 1990). The outer cells of the parenchyma, termed the “motor cells”, are responsible for nyctinastic and thigmonastic movement through water-driven volume changes (Moran, 2007; Rodrigues and Machado, 2008; Cortizo and Laufs, 2012). A loss-of-function mutation at the Elongated Petiolule1 or Petiolule-Like Pulvinus locus caused pulvini to change into petiolules with defects in leaf movement in Medicago truncatula (Chen et al., 2012; Zhou et al., 2012), indicating that leaf movability is associated with pulvinus development. In this study, we report the discovery and functional characterization of GmILPA1, a gene controlling LPA in soybean.
RESULTS
Characterization of the Soybean Gmilpa1 Mutant
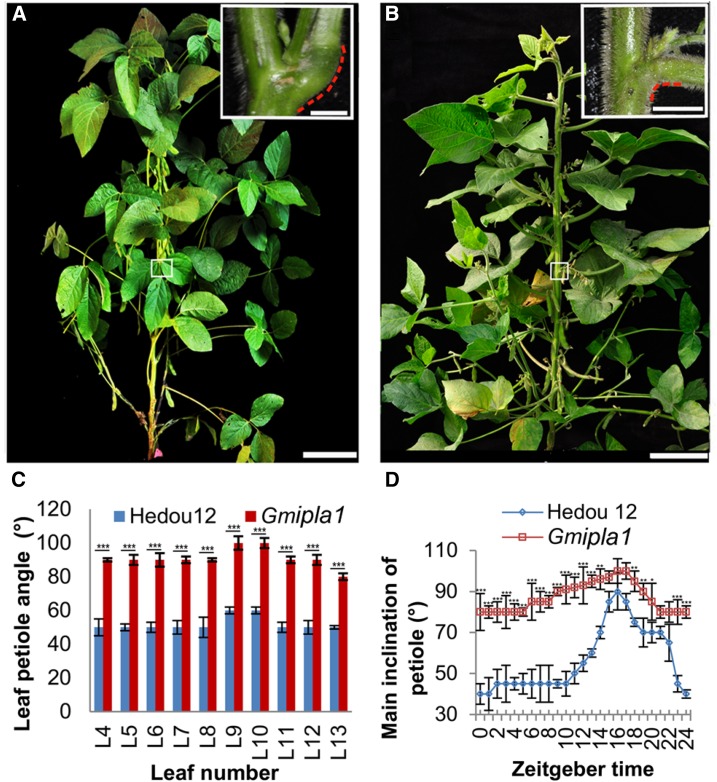
To understand the molecular basis of soybean LPA, we identified a soybean mutant with increased LPA, designated Gmilpa1, in a gamma ray-induced mutant population derived from the Chinese soybean cultivar, Hedou 12 (Song et al., 2015). The Gmilpa1 mutant had a larger LPA and a shorter petiole compared to the wild type. The pulvinus of the Gmilpa1 mutant was significantly smaller than that of the wild type (Fig. 1, A and B). The LPAs of trifoliate leaves in the Gmilpa1 mutant were larger than those in Hedou 12 (Fig. 1C). The nyctinastic movability of pulvini was diminished in the Gmilpa1 mutant compared to Hedou 12 (Fig. 1D).
Figure 1.
Phenotype of Hedou 12 and the Gmilpa1 mutant. A, Hedou 12; B, the Gmilpa1 mutant at the R5 stage (reproductive stage with approximately 3.2-mm-long seeds in the pod at one of the four uppermost nodes on the main stem). Scale bars, 15 cm. Scale bars in top-right boxes, 1 cm. C, LPAs of the fourth to 13th leaf of Hedou 12 and the Gmilpa1 plants at the R5 stage. LPAs are the means ± ses of the means from 12 different plants. ***P < 0.001 (t test). D, Nyctinastic movement of pulvini in Hedou 12 and the Gmilpa1 mutant. Angles are the means ± ses of the means from 12 different plants. ***P < 0.001, **P < 0.01, *P < 0.05 (t test).
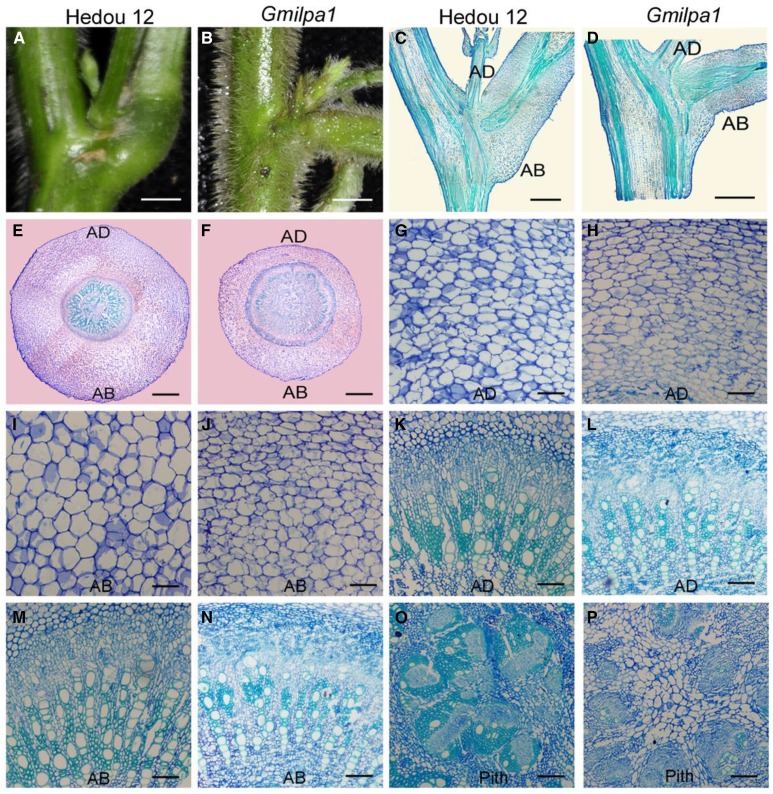
We studied the function of the GmILPA1 locus in soybean pulvini development by comparing the anatomical structure of pulvini in Hedou 12 and the Gmilpa1 mutant. The pulvinus of the Gmilpa1 mutant was smaller than that of Hedou 12, and the former did not have the kneelike structure on the abaxial side, which was present in the latter (Fig. 2, A to D). The ratio of the cortex on the abaxial side and the adaxial side at the joint was 2.14:1 in Hedou 12, and this was significantly lower in the Gmilpa1 mutant (1.54:1, n = 15, P < 0.05, t test). The cortex of the Gmilpa1 mutant was thinner than that of Hedou 12. In particular, there were fewer and smaller motor cells in the Gmilpa1 mutant than in Hedou 12 (Fig. 2, E to J). By contrast, the vascular cylinders of the Gmilpa1 mutant, particularly the phloem, cambium, and pith, were larger than those of Hedou 12 (Fig. 2, E, F, and K to P). These observations indicate that the GmILPA1 locus was largely responsible for shaping the pulvinus structure.
Figure 2.
Structural comparison of pulvini of 6-week-old Hedou 12 and the Gmilpa1 mutant. A and B, Pulvini of Hedou 12 (A) and the Gmilpa1 mutant (B), respectively. Scale bars, 1 cm. C and D, Longitudinal section of pulvini in Hedou 12 (C) and the Gmilpa1 mutant (D), respectively. Scale bars, 1 cm. E and F, Transection of pulvini in Hedou 12 (E) and the Gmilpa1 mutant (F), respectively. Scale bars, 400 μm. G to P, Partial magnification of transection of pulvini in Hedou 12 (G, K, and O) and the Gmilpa1 mutant (H, L, and P). Scale bars, 50 μm. G and H, The motor cells of Hedou 12 (G) and the Gmilpa1 mutant (H) on the adaxial side. I and J, The motor cells of Hedou 12 (I) and the Gmilpa1 mutant (J) on the abaxial side. K and L, The vascular cylinders of Hedou 12 (K) and the Gmilpa1 mutant (L) on the adaxial side. M and N, The vascular cylinders of Hedou 12 (M) and the Gmilpa1 mutant (N) on the abaxial side. O and P, The piths of Hedou 12 (O) and the Gmilpa1 mutant (P).
Cell differentiation and proliferation were also altered in the Gmilpa1 mutant compared with Hedou 12. In the Gmilpa1 mutant, there were fewer xylem fiber cells (Fig. 2, K to N), and these often clustered around the tracheary element cells far from the pith. By contrast, there were more xylem parenchyma cells among the tracheary elements in the Gmilpa1 mutant compared with Hedou 12 (Fig. 2, K to N), which was more prominent in the pith of the Gmilpa1 mutant (Fig. 2, O and P) than in Hedou 12. These observations indicate that the GmILPA1 locus clearly plays an important role in regulating cell differentiation and proliferation and shaping the structure of the pulvinus.
Molecular Mapping of the GmILPA1 Locus
To identify the mutation responsible for the observed phenotypic changes, a cross between Williams 82 and the Gmilpa1 mutant was made, and an F2 population comprising 891 individual F2 plants was obtained. Among these F2 plants, 670 showed the wild-type phenotypes and 221 exhibited the mutant phenotypes, fitting a 3:1 ratio (χ2 test, P = 0.89). This indicates that the phenotypic changes were controlled by a single locus, designated GmILPA1, and that the wild-type phenotypes were dominant over the mutant phenotypes.
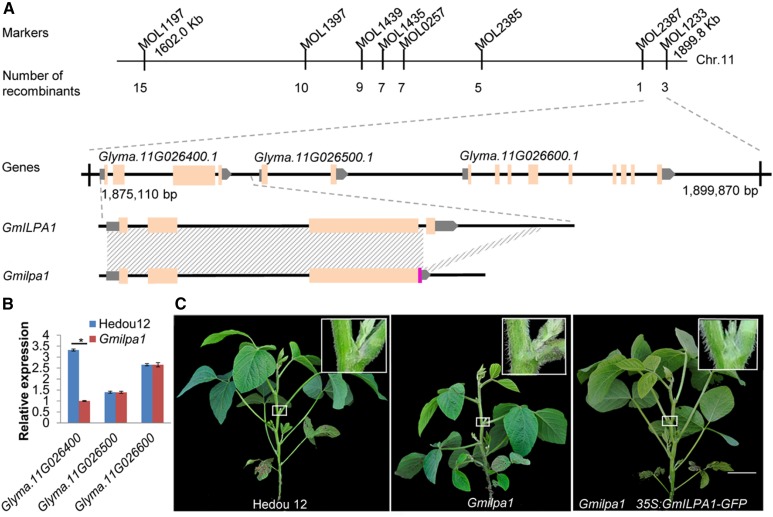
Using 165 INDEL markers developed earlier (Song et al., 2015), the GmILPA1 locus was mapped to a genomic region between MOL1197 and MOL1233 on chromosome 11, which is 189 kb according to the soybean reference genome sequence (Glycine max Wm82.a2.v1; Schmutz et al., 2010; Fig. 3A). Subsequently, MOL1197 and MOL1233 were used to search for recombinants between these two markers and the GmILPA1 locus from 1023 F2:3 individuals derived from the heterozygous F2 plants. We identified 15 recombinants that were phenotyped and genotyped with six fine mapping markers (MOL1397, MOL1439, MOL1435, MOL0257, MOL2385, MOL2387; Supplemental Table S1) between MOL1197 and MOL1233 and pinpointed the GmILPA1 locus to a 24.7-kb region between MOL2387 and MOL1233, which harbors three genes (Glyma.11G026400.1, Glyma.11G026500.1, and Glyma.11G026600.1) according to the Williams 82 reference genome (Glycine max Wm82.a2.v1; Fig. 3A).
Figure 3.
Map-based cloning of the GmILPA1 locus. A, Physical locations of markers defining the GmILPA1 region, the deletion region in Glyma.11G026400.1, and a new transcript identified in the Gmilpa1 mutant. Shading indicates identical sequences. The chromosomal positions of MOL1197 and MOL1233 are 1602.0 kb and 1890.8 kb, respectively (Glycine max Wm82.a2.v1). B, Expression levels of Glyma.11G026400.1, Glyma.11G026500.1, and Glyma.11G026600.1 in the fifth pulvini at the V5 stage of Hedou 12 and the Gmilpa1 mutant. Expression levels are presented as the means ± ses of the means from four biological replicates. *P < 0.05 (t test). C, Complementation of the Gmilpa1 mutant. Phenotypes of Hedou 12, the Gmilpa1 mutant, and T2 plants with the GmILPA1 transgene. Scale bar, 10 cm. Boxes in the top-right corner illustrate part of plants magnified 10×.
Sequence Comparison between the Wild Type and Gmilpa1 Mutant
In an attempt to identify the candidate gene for the GmILPA1 locus, the three genes and their flanking sequences in the defined GmILPA1 region in Hedou 12 (wild type) and the Gmilpa1 mutant were amplified and sequenced. A 1149-bp deletion that involved the 23-bp end sequence of the third intron and the entire fourth exon of Glyma.11G026400.1 and its 788-bp downstream flanking sequence was identified in the Gmilpa1 mutant (Fig. 3A and Supplemental Fig. S1A). No sequence changes were detected in the other two genes between the wild type and the Gmilpa1 mutant.
The transcripts of Glyma.11G026400.1 in Hedou 12 and the Gmilpa1 mutant were analyzed. We found that the transcript of Gm11G026400.1 in Hedou 12 was the same as that predicted by the reference genome. The transcript of Glyma.11G026400.1 in the Gmilpa1 mutant was altered by the deletion, including an extension of the third exon and the formation of, to our knowledge, a novel 3′UTR from 65-bp intronic and 77-bp intergenic sequences (Fig. 3A and Supplemental Fig. S1B). No transcript changes were detected in the other two genes between the wild type and the Gmilpa1 mutant.
The expression patterns of Glyma.11G026400.1, Glyma.11G026500.1, and Glyma.11G026600.1 in the fifth pulvini at the V5 stage (five trifoliate leaves unrolled) in Hedou 12 and the Gmilpa1 mutant were analyzed. Glyma.11G026400.1 was expressed at a significantly higher level in Hedou 12 than in the Gmilpa1 mutant. By contrast, no obvious difference in the expression level of Glyma.11G026500.1 and Glyma.11G026600.1 was detected between Hedou 12 and the Gmilpa1 mutant (Fig. 3B). These observations suggest that Glyma.11G026400.1 was most likely to be the candidate gene for the GmILPA1 locus.
Validation of the GmILPA1 Candidate by Complementation
To validate the candidacy of Glyma.11G026400.1 for the GmILPA1 locus, a construct that harbors the coding sequence (CDS) of Glyma.11G026400.1 amplified from Hedou 12 and the CDS of the green florescence protein (GFP) gene, coupled with the cauliflower mosaic virus 35S promoter, was made and introduced into the Gmilpa1 mutant using Agrobacterium tumefaciens-mediated transformation. Four independent transformation events carrying the 35S:GmILPA1:GFP expression cassette were obtained in the Gmilpa1 background (Supplemental Fig. S2A). In all four transformation events, transgenic T2 progeny lines with the expression of the GFP protein were observed to restore the wild-type LPA (Fig. 3C and Supplemental Fig. S2, A to D), indicating that Glyma.11G026400.1 was the GmILPA1 locus controlling LPA in soybean. We investigated the anatomical structure of pulvini in the four transformation events and found that all transgenic T2 progeny had the wild-type anatomical pulvinus structure. The cortex and vascular cylinders were the same as those of the wild-type plants (Supplemental Fig. S2E). These findings indicate that GmILPA1 can complement the Gmilpa1 mutant completely while under the control of the 35S promoter.
Sequence comparison and phylogenetic analysis demonstrated that GmILPA1 encoded an APC8-like protein, a subunit of the anaphase-promoting complex/cyclosome (APC/C; Supplemental Fig. S3). The Arabidopsis APC8 gene was previously identified by isolation of the apc8 mutant, which exhibited curly leaves, abnormal reproductive development, bushy inflorescences, and shorter siliques (Zheng et al., 2011). We examined the phenotypes of the apc8 mutant and the wild-type Arabidopsis (Arabidopsis thaliana) plants and found increased silique inclinations in the apc8 mutant, but the difference in LPA was hardly detected because the bushy structure of the apc8 mutant and Arabidopsis have no pulvini as exists in soybean. We introduced the 35S:GmILPA1:GFP expression cassette into the Arabidopsis apc8 mutant. Normal silique inclinations were observed in the GmILPA1 transgenic plants with the apc8 background, although these transgenic plants maintained other mutant phenotypes such as bushy inflorescences and shorter siliques (Supplemental Fig. S4), suggesting that GmILPA1 in soybean and APC8 in Arabidopsis may have functionally diverged after their split from a common ancestor.
Expression Pattern of GmILPA1
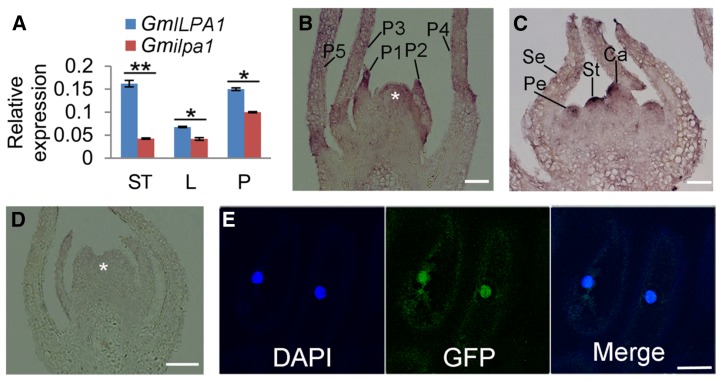
To further understand the function of GmILPA1, the expressions of GmILPA1 and Gmilpa1 among different tissues were examined using quantitative real-time PCR (RT-PCR). The expression levels of Gmilpa1 in the Gmilpa1 mutant were lower than those of GmILPA1 in Hedou 12 in apical stem tips, leaves, and pulvini at the V5 stage (Fig. 4A). RNA in situ hybridization revealed that the GmILPA1 transcripts were predominantly abundant at the base of leaf primordia, where the pulvinus is formed. In particular, the GmILPA1 transcripts were present in the basal region of the developing leaf primordia until the P3 stage, when pulvini were first apparent (Fig. 4B), suggesting that GmILPA1 might function at the base of leaf primordia at an early development stage of pulvinus initiation. In addition, GmILPA1 was also expressed in the petal, stamen, and carpel primordium (Fig. 4C). By contrast, no signal was detected by the sense probe (Fig. 4D). Expression of the GmILPA1-GFP fusion gene under the control of the 35S promoter in the epidermal cells of onion (Allium cepa L.) demonstrated that the fusion protein was localized in both the nucleus and cytoplasm (Fig. 4E).
Figure 4.
Expression and subcellular localization analysis of GmILPA1. A, Expression of GmILPA1 and Gmilpa1 in apical stem tips, leaves, and pulvini in Hedou 12 and the Gmilpa1 mutant. Expression levels are presented as the means ± ses of the means from four biological replicates. **P < 0.01 (t test). B to D, RNA in situ hybridization of GmILPA1 performed using RNA probes in anti-sense (B and C) and sense (D) directions. B and D, The shoot tips were embedded in paraffin at the VE developmental stage (12 d after planting). Asterisks denote the shoot apical meristem. Scale bars, 50 μm. C, The shoot tips were embedded in paraffin at the R1 stage (one open flower at any node). Scale bars, 50 μm. E, Subcellular localization of the GmILPA1-GFP fusion protein in onion epidermal cells under the control of the 35S promoter observed under dark field for green fluorescence (middle). The nuclei were counterstained with 4,6-diamidino-2-phenylindole. Scale bar, 50 μm. Ca, carpel primordium; DAPI, 4,6-diamidino-2-phenylindole; L, leaves; LP, leaf primordium; P, pulvini; Pe, petal primordium; Se, sepal primordium; St, stamen; ST, stem tips.
GmILPA1 Directly Interacts with GmAPC13a
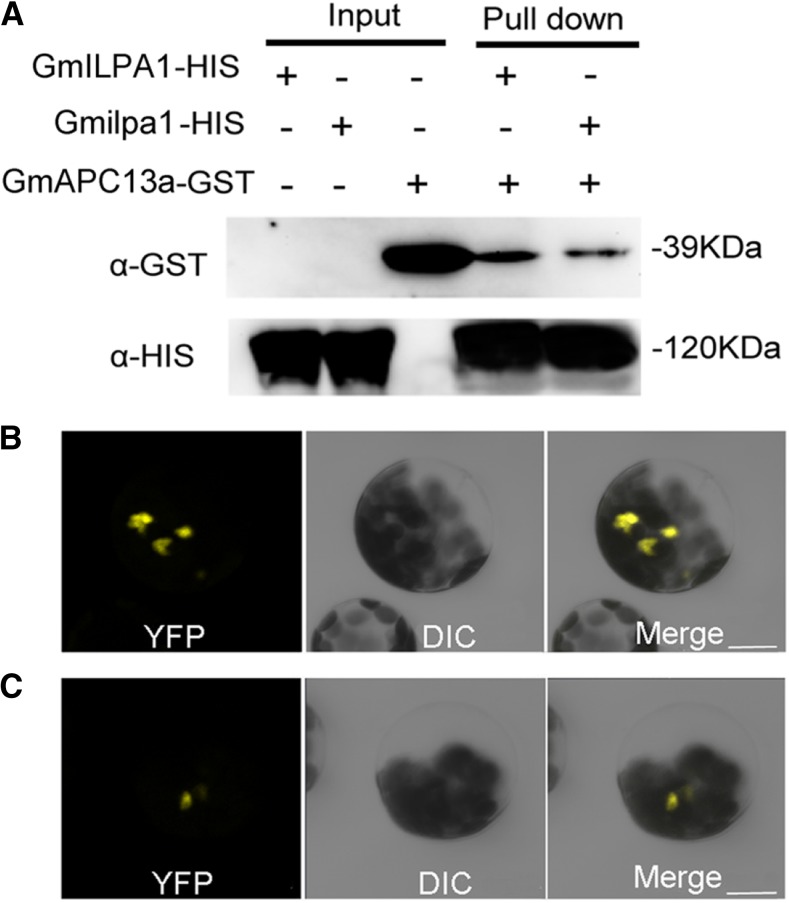
APC8 probably interacts with APC13 in Arabidopsis to promote the stable association of the APC/C complex (Schreiber et al., 2011). According to the Glyma.Wm82.a2.v1 reference genome, 2 GmAPC13-like genes, Glyma.19G223500.1 (GmAPC13a) and Glyma.03G226500.1 (GmAPC13b), were found. Phylogenetic analysis demonstrated that GmAPC13a was closer to Arabidopsis APC13 than GmAPC13b (Supplemental Fig. S5A). Furthermore, the expression patterns of GmAPC13a and GmAPC13b in apical stem tips, leaves, and pulvini in Hedou 12 and the Gmilpa1 mutant were also analyzed. GmAPC13a showed higher expression levels than GmAPC13b in these tissues of the wild type (Supplemental Fig. S5B). A coexpression pattern was found between GmAPC13a and GmILPA1; the expression levels of GmAPC13a and GmILPA1 decreased in the Gmilpa1 mutant compared to Hedou 12 (Fig. 4A and Supplemental Fig. S5B). By contrast, GmAPC13b showed the same expression level in the Gmilpa1 mutant and Hedou 12 (Supplemental Fig. S5B). Thus, we chose GmAPC13a to determine whether GmILPA1 could interact with GmAPC13 in soybean. We created the GmILPA1-HIS, Gmilpa1-HIS, and GmAPC13a-GST fusion proteins and performed pull-down assays to test whether GmILPA1 interacts with GmAPC13a in soybean. As shown in Figure 5A, the GmILPA1-HIS fusion protein was able to capture the GmAPC13a-GST fusion protein, indicating an in vitro interaction between GmILPA1 and GmAPC13a. Compared with the GmILPA1-HIS fusion protein, the capacity of the Gmilpa1-HIS fusion protein for capturing the GmAPC13a-GST fusion protein was significantly decreased (n = 3, P < 0.05, t test). A significant decrease was also detected using bimolecular fluorescence complementation (BiFC) assays (n = 4, P < 0.05, t test; Fig. 5, B and C). These observations suggest that the function of the Gmilpa1 protein may be substantially compromised through the reduction in its ability to interact with GmAPC13a. Together, these observations suggest that GmILPA1 may function through direct interaction with GmAPC13a in modulating LPA in soybean.
Figure 5.
Interaction between GmILPA1/Gmilpa1 and GmAPC13a. A, Interaction between GmILPA1/Gmilpa1 and GmAPC13a detected by HIS-antibody and GST-antibody in the pull-down assays. The “+” and “-” indicate reactions with or without tagged proteins, respectively. B and C, Interaction among GmILPA1 (B), Gmilpa1 (C), and GmAPC13a revealed by BiFC. Scale bars, 10 μm. α-GST, GST-antibody; α-HIS, HIS-antibody.
Expression Levels of GmILPA1 among Soybean Varieties
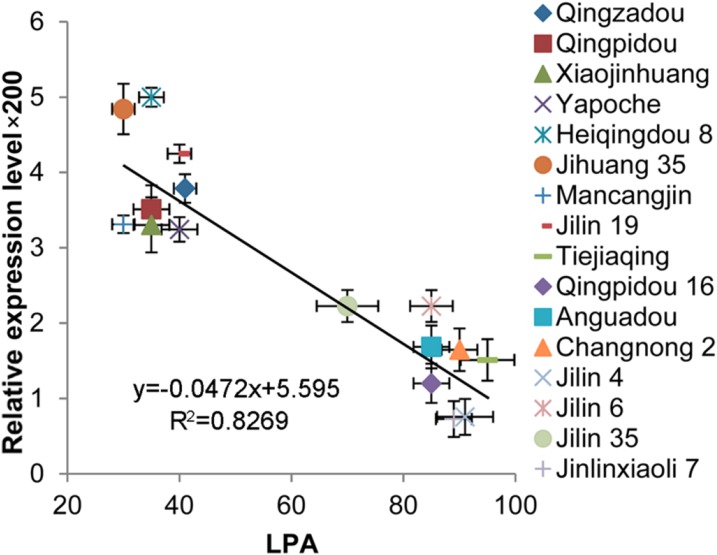
The relationship between LPA and the expression pattern of GmILPA1 in 16 representative soybean elite cultivars was analyzed. These cultivars were classified into two categories—small LPA cultivars and large LPA cultivars. Small LPA cultivars included Qingzadou, Qingpidou, Xiaojinhuang, Yapoche, Heiqingdou 8, Jihuang 35, Mancangjin, and Jilin 19, and these cultivars had LPAs less than 40°. Large LPA cultivars included Tiejiaqing, Qingpidou 16, Anguadou, Changnong 2, Jilin 4, Jinlin 6, Jinlin 35, and Jilinxiaoli 7; the LPAs of these cultivars exceeded 70° (Fig. 6). Overall, the expression levels of GmILPA1 in cultivars with small LPAs were higher than the cultivars with large LPAs, indicating the association between LPAs and the expression levels of GmILPA1. GmILPA1 was expressed at a higher level in the cultivars with smaller LPAs than in the cultivars with larger LPAs. The relationship between GmILPA1 and LPA was further demonstrated by the observed significant correlation of GmILPA1 expression level and LPA (R2 = 0.8269; Fig. 6). This is further evidence that GmILPA1 may function in an expression-dependent manner to regulate LPA.
Figure 6.
Association analysis of GmILPA1 expression and LPAs in 16 soybean cultivars. LPAs are the means ± ses from six different plants. Expression levels of GmILPA1, analyzed using young leaves at the V5 stage, are the means ± ses from six different plants. Horizontal and vertical bars are the ses of LPA and GmILPA1 expression level × 200, respectively. A significant correlation between the expression levels of GmILPA1 (y) and LPA (x) was found; the trend line was y = −0.0472x + 5.595 (R2 = 0.8269).
DISCUSSION
GmILPA1 Might Be a Partial Functional Equivalent of the Arabidopsis APC8
Several complementary lines of evidence indicate that GmILPA1 might be only a partial functional equivalent of APC8 in Arabidopsis, although GmILPA1 is homologous to APC8. GmILPA1 in soybean is expressed at the base of leaf primordia to promote the establishment of pulvini, which do not exist in Arabidopsis. Sequence analysis showed that GmILPA1 arose from a recent duplication (5.41 mya; Supplemental Fig. S3), which occurred after the tetraploidization event (16 mya; Schlueter et al., 2004; Pfeil et al., 2005) in the ancestor of soybean. In legume plants, there are multiple copies of APC8-like genes, whereas in Arabidopsis, there is only a single APC8 gene. In soybean, there are four APC8-like genes that arose in different evolutionary time frames (Supplemental Fig. S3). These observations indicate that GmILPA1 might only possess a partial function of APC8 in regulating cell growth and division of the pulvinus, which is critical for pulvinus establishment in soybean. Overexpression of GmILPA1 could not completely complement the Arabidopsis apc8 mutant, especially in phenotypes, such as those with bushy inflorescences and shorter siliques (Supplemental Fig. S4). The expressional and functional differences between Arabidopsis APC8 and GmILPA1 could result from changes in other interacting genes. However, we used transgenic lines with the GFP reporter for better functional characterization of this gene. The partial complementation in the Arabidopsis apc8 mutant could be partially caused by fusion of GmILPA1 with the GFP tag, although complete complementation was achieved in the soybean Gmilpa1 mutant. It is worth noting that the complementation experiments in both Arabidopsis and soybean were performed with the constitutive 35S promoter. It is interpretable for no ectopic effects of these transgenic plants because GmILPA1 is only one subunit of the APC complex; the Arabidopsis APC/C consists of at least 11 core subunits (Capron et al., 2003; Foe and Toczyski, 2011; Schreiber et al., 2011) and the functional APC might depend on all parts of complex. However, it is not known whether the native GmILPA1 promoter would provide similar or identical complementation results.
GmILPA1 Appears to Function Via the APC Complex
The cell cycle plays a crucial role in regulating the growth and development of plants. In Arabidopsis, the APC/C is a 1.5-megadalton E3 ligase assembled from 13 different APC subunits that include APC8 and APC13 (Foe and Toczyski, 2011; Schreiber et al., 2011), acting complementarily to accomplish basic cell-cycle control and promote cell proliferation (Pines, 2011). In this study, we found a direct interaction between GmILPA1 and GmAPC13a via in vivo and in vitro experiments. In addition, GmILPA1 was coexpressed with GmAPC13a in different tissues between the wild type and the Gmilpa1 mutant. GmILPA1 was localized in the nuclei and cytoplasm of soybean, similar to the subcellular localization pattern of APC8 in Arabidopsis (Zheng et al., 2011). We speculate that GmILPA1 functions by modulating cell proliferation in concert with other subunits of the GmAPC complex in soybean.
Nyctinastic Movement Is Also Regulated by GmILPA1
Legumes both track and avoid the sun, thereby exhibiting nyctinastic movements. Pulvini show nyctinastic and thigmonastic movement through water-driven volume changes in their motor cells (Chen et al., 2012; Song et al., 2014). Two functionally different parts of motor cells, the adaxial flexor and the abaxial extensor, undergo rhythmic swelling and shrinking, causing volume changes of motor cells, and inducing petiole rising and falling (Volkov et al., 2010). The genetic mechanisms leading to pulvinar nyctinastic movement are not well known. Our data showed that the nyctinastic motions of pulvini and leaflets were both diminished in the Gmilpa1 mutant compared to Hedou 12 (Fig. 1D and Supplemental Fig. S6). Furthermore, the amount and size of abaxial and adaxial motor cells were reduced in Gmilpa1 pulvini (Fig. 2, C to J). Similar changes were also observed in the leaflet pulvini in the Gmilpa1 mutant and Hedou 12 (Supplemental Fig. S7). The defects in nyctinastic mobility in the Gmilpa1 mutant were attributed to developmental defects of the pulvinus. It would be useful to examine the relationship between pulvinar identity genes and GmILPA1 in pulvinus development, and to determine how the circadian system affects the role of GmILPA1 in the regulation of nyctinastic movement.
Our data suggest that the GmILPA1 gene may be a candidate for improvement of soybean architecture. In addition, the Gmilpa1 mutant had more flowers or beginning pods (pod was approximately 4.7-mm long at one of the four uppermost nodes) at the uppermost nodes rather than full pods (pod was approximately 19-mm long at one of the four uppermost nodes) compared to the wild type at the R5 stage (Fig. 1, A and B). The delayed podding in the Gmilpa1 mutant (Fig. 1, A and B) and the specific expression pattern of GmILPA1 in the floral meristem (Fig. 4C) indicate that GmILPA1 may also be involved in floral development or pod formation in soybean. Further research is needed to clarify the mechanism by which GmILPA1 regulates floral development. Given such a long period of divergence of soybean and Arabidopsis from a common ancestor, the formation of leaf angle or LPA by APC8-like genes in Arabidopsis and soybean suggests the potential application of this, to our knowledge, novel regulatory mechanism for leaf angle modification in legumes. If successful, this could optimize plant architecture for enhanced yield and greater adaptability to environmental stress.
MATERIALS AND METHODS
Plant Materials
Hedou 12 and Williams 82 were obtained from the Chinese Academy of Agricultural Sciences, and the Gmilpa1 mutant was created in our laboratory (Cheng et al., 2016). Wild-type Arabidopsis (Arabidopsis thaliana) and the apc8 mutant (Zheng et al., 2011) were obtained from Dr. Binglian Zheng at Fudan University. Arabidopsis seeds were surface sterilized with 75% ethyl alcohol. The sterilized seeds were stratified at 4°C for 3 d and transferred to half-strength Murashige & Skoog (1/2 MS) medium and soil for further growth at 22°C under conditions of 16 h of 150 μE/m2/s light, 8 h of dark, and 75% humidity.
DNA Extraction and Genetic Mapping
Genomic DNA was extracted using the DNeasy Plant Mini Kit (Qiagen). The anchor markers used for primary mapping were collected from Song et al. (2015). The fine mapping makers of the GmILPA1 locus were developed using INDELs between Hedou 12 and Williams 82 that had been analyzed previously (Song et al., 2015). The upstream and downstream sequences of the INDEL loci were collected from Phytozome for PCR primer design. The primer size ranged between 18 and 30 bp, and the GC content was set between 40 and 60%. The maximum Tm difference between the forward and reverse primers was set to 3°C. The size of the PCR products ranged from 100 to 600 bp depending on the primer design and polymorphism readability after polyacrylamide gel electrophoresis. The primers used are listed in Supplemental Table S1.
RNA Isolation, Real-Time Quantitative PCR Analysis, and RACE Experiments
Total RNA was isolated from stem tips, leaves, and pulvini of soybean (Glycine max) using Trizol, according to the manufacturer’s instructions (Invitrogen Life Technologies). The RNA was treated with RNase-free DNase I (Cat. no. 2270; Takara) at 37°C for 30 min. RT-PCR was performed using the Prime-Script RT-PCR Kit (Cat. no. RR014; Takara), according to the manufacturer’s instructions. The expression of genes was analyzed by real-time quantitative RT-PCR, using SYBR premix Ex Taq (Cat. no. RR420; Takara) in the GeneAmp 5700 sequence Detection System (Applied Biosystems; http://www.appliedbiosystems.com) according to the manufacturer’s instructions. Four biological replicates were analyzed to quantify the levels of gene expression, and three technical replicates were performed to calculate the relative expression level using the 2−ΔΔCt method after normalization to Cons4 (ATP binding cassette transporter gene, Glyma.12G020500; Ping et al., 2014; Liu et al., 2016). The 3′ RACE experiments were conducted using an RLM-RACE kit (Invitrogen) following the manufacturer’s instructions.
Plasmid Construction and Transformation
The CDS of GmILPA1 (amplified from Hedou 12) was fused with the GFP-coding sequence and inserted into pCAMBIA3301 (CAMBIA) between the cauliflower mosaic virus 35S promoter and nopaline synthase terminator. The construct was introduced into the Gmilpa1 mutants by Agrobacterium-mediated transformation (Zhao et al., 2016), and Arabidopsis apc8 mutants were transformed using the floral dip method (Clough and Bent, 1998).
Multiple-sequence Alignment and Phylogenetic Analysis
We searched for homologous genes of APC8 from Phytozome (Phytozome V12.0; https://phytozome.jgi.doe.gov/pz/portal.html#) for the following genomes: Glycine max, Arabidopsis thaliana, Proteus vulgaris, Medicago truncatula, and Oryza sativa. The database was queried using AT3G48150 from Arabidopsis using the program BLASTP (https://blast.ncbi.nlm.nih.gov/Blast.cgi). All sequences were verified to contain six tetratricopeptide repeat domains using the SMART package (http://labix.org/smart/; Letunic et al., 2012). Alignment of the full-length APC8 protein sequences was performed, and the phylogenetic tree was constructed using the software MEGA 5.0 (www.megasoftware.net; Tamura et al., 2011). The neighbor-joining method was applied to construct trees as described in Zhao et al. (2016).
Histological Analysis
The pulvini of Hedou 12 and the Gmilpa1 mutant were fixed overnight at 4°C in an FAA buffer (3.7% (v/v) formaldehyde, 50% (v/v) ethanol, 5% (v/v) glacial acetic acid, 0.1% (v/v) Triton X-100) after two vacuum infiltrations of 30 min each. The tissues were successively incubated at room temperature for 1 h in 30% (v/v) ethanol, 50% (v/v) ethanol, 70% (v/v) ethanol, 85% (v/v) ethanol, 95% (v/v) ethanol, and 100% (v/v) ethanol (3×1 h), ethanol/xylene mix (75:25% (v/v), 1 h) and ethanol/xylene mix (50:50% (v/v), overnight). On d 3, the tissues were incubated at 60°C in a xylene/paraplast mix (1 V/1 V) overnight after been subjected to ethanol/xylene (25:75% (v/v), 1 h), 100% (v/v) xylene (3×1 h), and 100% (v/v) paraplast/xylene (25:75% (v/v), 1 h). The samples were then incubated at 60°C in 100% (v/v) paraplast for 2 d (with the solution changed twice per d). Tissues were embedded in 100% (v/v) paraplast, and sections were cut on a Model no. RM2245 microtome (Leica). The sections were then colored with toluidine blue and observed using a Model no. BX53M microscope (Olympus).
Subcellular Localization, Pull-down, and BiFC Assay
The construct used for transformation was expressed in onion (Allium cepa L.) epidermal cells as described in Sun et al. (2007). GFP fluorescence was observed using a confocal laser scanning microscope (Olympus). A HIS tag was fused to the C termini of GmILPA1 and Gmilpa1 in pColdTF, and a GST tag was fused to the C termini of GmAPC13a in pGEX-4T-3 for the pull-down assay. Protein expression, extraction, and immunoblot analysis were conducted as described in Cui et al. (2010) and Wang et al. (2013). GmILPA1 and Gmilpa1 were cloned into the pEarleyGate201-YN vector, and GmAPC13a was cloned into the pEarleyGate202-YC vector (Tian et al., 2011). These constructs were introduced into Arabidopsis (Col-0) mesophyll protoplasts (Wang et al., 2013). The localization of fluorescent proteins in protoplasts was visualized using a confocal microscope (Carl Zeiss).
RNA In Situ Hybridization
Tissue fixation for RNA in situ hybridization was performed following a protocol described in Feng et al. (2006). Tissues embedded in paraffin (Cat. no. P3683; Sigma-Aldrich) were sliced into 8-μm sections using a Model no. RM2245 microtome (Leica). The 329-bp 3′-region of GmILPA1 CDS was cloned into a pGEM-T easy vector (Promega) and used as the template to generate sense and anti-sense RNA probes. Digoxigenin-labeled RNA probes were prepared using a DIG RNA Labeling Kit (Cat. no. T7/SP6; 11175025910; Roche) according to the manufacturer’s instructions. Slides were observed under bright field using a Model no. BX53M microscope (Olympus).
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Deletion analysis in the Gmilpa1 mutant
Supplemental Figure S2. Identification of transgenic plants
Supplemental Figure S3. Phylogeny analysis of APC8-like proteins
Supplemental Figure S4. Phenotypes of Arabidopsis wild type (Col-0), the apc8 mutant, and the 35S:GmILPA1 T2 transgenic plants in the apc8 background
Supplemental Figure S5. Phylogeny analysis of APC13-like proteins and expression patterns of GmAPC13a and GmAPC13b
Supplemental Figure S6. Leaflet nyctinastic movement in Hedou 12 and the Gmilpa1 mutant
Supplemental Figure S7. Anatomical structure of the pulvinulus in Hedou 12 and the Gmilpa1 mutant
Supplemental Table S1. Primers used in this study
Supplementary Material
Acknowledgments
We thank Dr. Binglian Zheng from Fudan University for providing seeds of the Arabidopsis apc8 mutant.
References
- Capron A, Serralbo O, Fülöp K, Frugier F, Parmentier Y, Dong A, Lecureuil A, Guerche P, Kondorosi E, Scheres B, Genschik P (2003) The Arabidopsis anaphase-promoting complex or cyclosome: molecular and genetic characterization of the APC2 subunit. Plant Cell 15: 2370–2382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Moreau C, Liu Y, Kawaguchi M, Hofer J, Ellis N, Chen R (2012) Conserved genetic determinant of motor organ identity in Medicago truncatula and related legumes. Proc Natl Acad Sci USA 109: 11723–11728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng W, Gao JS, Feng XX, Shao Q, Yang SX, Feng XZ (2016) Characterization of dwarf mutants and molecular mapping of a dwarf locus in soybean. J Integr Agric 15: 60345–60347 [Google Scholar]
- Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Cortizo M, Laufs P (2012) Genetic basis of the “sleeping leaves” revealed. Proc Natl Acad Sci USA 109: 11474–11475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui H, Wang Y, Xue L, Chu J, Yan C, Fu J, Chen M, Innes RW, Zhou JM (2010) Pseudomonas syringae effector protein AvrB perturbs Arabidopsis hormone signaling by activating MAP kinase 4. Cell Host Microbe 7: 164–175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng X, Zhao Z, Tian Z, Xu S, Luo Y, Cai Z, Wang Y, Yang J, Wang Z, Weng L, Chen J, Zheng L, et al. (2006) Control of petal shape and floral zygomorphy in Lotus japonicus. Proc Natl Acad Sci USA 103: 4970–4975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foe I, Toczyski D (2011) Structural biology: a new look for the APC. Nature 470: 182–183 [DOI] [PubMed] [Google Scholar]
- Pines J. (2011) Cubism and the cell cycle: the many faces of the APC/C. Nat Rev Mol Cell Biol 12: 427–438 [DOI] [PubMed] [Google Scholar]
- Letunic I, Doerks T, Bork P (2012) SMART 7: recent updates to the protein domain annotation resource. Nucleic Acids Res 40: D302–D305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Zhang D, Ping J, Li S, Chen Z, Ma J (2016) Innovation of a regulatory mechanism modulating semi-determinate stem growth through artificial selection in soybean. PLoS Genet 12: e1005818 10.1371/journal.pgen.1005818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu M, Zhou F, Xie CX, Li MS, Xu YB, Marilyn W, Zhang SH (2007) Construction of a SSR linkage map and mapping of quantitative trait loci (QTL) for leaf angle and leaf orientation with an elite maize hybrid. Yi Chuan 29: 1131–1138 [DOI] [PubMed] [Google Scholar]
- Moran N. (2007) Osmoregulation of leaf motor cells. FEBS Lett 581: 2337–2347 [DOI] [PubMed] [Google Scholar]
- Pfeil BE, Schlueter JA, Shoemaker RC, Doyle JJ (2005) Placing paleopolyploidy in relation to taxon divergence: a phylogenetic analysis in legumes using 39 gene families. Syst Biol 54: 441–454 [DOI] [PubMed] [Google Scholar]
- Ping J, Liu Y, Sun L, Zhao M, Li Y, She M, Sui Y, Lin F, Liu X, Tang Z, Nguyen H, Tian Z, et al. (2014) Dt2 is a gain-of-function MADS-domain factor gene that specifies semideterminacy in soybean. Plant Cell 26: 2831–2842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinhardt D, Kuhlemeier C (2002) Plant architecture. EMBO Rep 3: 846–851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodrigues TM, Machado SR (2008) Pulvinus functional traits in relation to leaf movements: a light and transmission electron microscopy study of the vascular system. Micron 39: 7–16 [DOI] [PubMed] [Google Scholar]
- Satter R, Vogelmann T, Gorton H (1990) The pulvinus: motor organ for leaf movement. Curr Topics Plant Physiol 3: 261–264 [Google Scholar]
- Schlueter JA, Dixon P, Granger C, Grant D, Clark L, Doyle JJ, Shoemaker RC (2004) Mining EST databases to resolve evolutionary events in major crop species. Genome 47: 868–876 [DOI] [PubMed] [Google Scholar]
- Schmutz J, Cannon SB, Schlueter J, Ma J, Mitros T, Nelson W, Hyten DL, Song Q, Thelen JJ, Cheng J, Xu D, Hellsten U, et al. (2010) Genome sequence of the palaeopolyploid soybean. Nature 463: 178–183 [DOI] [PubMed] [Google Scholar]
- Schreiber A, Stengel F, Zhang Z, Enchev RI, Kong EH, Morris EP, Robinson CV, da Fonseca PCA, Barford D (2011) Structural basis for the subunit assembly of the anaphase-promoting complex. Nature 470: 227–232 [DOI] [PubMed] [Google Scholar]
- Song K, Yeom E, Lee SJ (2014) Real-time imaging of pulvinus bending in Mimosa pudica. Sci Rep 4: 6466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song X, Wei H, Cheng W, Yang S, Zhao Y, Li X, Luo D, Zhang H, Feng X (2015) Development of INDEL markers for genetic mapping based on whole-genome re-sequencing in soybean. G3 (Bethesda) 5: 2793–2799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun W, Cao Z, Li Y, Zhao Y, Zhang H (2007) A simple and effective method for protein subcellular localization using Agrobacterium-mediated transformation of onion epidermal cells. Biologia 62: 529–532 [Google Scholar]
- Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S (2011) MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28: 2731–2739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian G, Lu Q, Zhang L, Kohalmi SE, Cui Y (2011) Detection of protein interactions in plant using a gateway compatible bimolecular fluorescence complementation (BiFC) system. J Vis Exp 55: e3473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volkov AG, Foster JC, Baker KD, Markin VS (2010) Mechanical and electrical anisotropy in Mimosa pudica pulvini. Plant Signal Behav 5: 1211–1221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Fan C, Zhang X, Zhu J, Fu YF (2013) BioVector, a flexible system for gene specific-expression in plants. BMC Plant Biol 13: 198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao B, Dai A, Wei H, Yang S, Wang B, Jiang N, Feng X (2016) Arabidopsis KLU homologue GmCYP78A72 regulates seed size in soybean. Plant Mol Biol 90: 33–47 [DOI] [PubMed] [Google Scholar]
- Zheng B, Chen X, McCormick S (2011) The anaphase-promoting complex is a dual integrator that regulates both MicroRNA-mediated transcriptional regulation of cyclin B1 and degradation of Cyclin B1 during Arabidopsis male gametophyte development. Plant Cell 23: 1033–1046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou C, Han L, Fu C, Chai M, Zhang W, Li G, Tang Y, Wang ZY (2012) Identification and characterization of petiolule-like pulvinus mutants with abolished nyctinastic leaf movement in the model legume Medicago truncatula. New Phytol 196: 92–100 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.