Disruption of the flavone synthase II gene in rice results in an altered cell wall lignin incorporating naringenin as a novel flavonoid component and improves biomass saccharification efficiency.
Abstract
Lignin, a ubiquitous phenylpropanoid polymer in vascular plant cell walls, is derived primarily from oxidative couplings of monolignols (p-hydroxycinnamyl alcohols). It was discovered recently that a wide range of grasses, including cereals, utilize a member of the flavonoids, tricin (3′,5′-dimethoxyflavone), as a natural comonomer with monolignols for cell wall lignification. Previously, we established that cytochrome P450 93G1 is a flavone synthase II (OsFNSII) indispensable for the biosynthesis of soluble tricin-derived metabolites in rice (Oryza sativa). Here, our tricin-deficient fnsII mutant was analyzed further with an emphasis on its cell wall structure and properties. The mutant is similar in growth to wild-type control plants with normal vascular morphology. Chemical and nuclear magnetic resonance structural analyses demonstrated that the mutant lignin is completely devoid of tricin, indicating that FNSII activity is essential for the deposition of tricin-bound lignin in rice cell walls. The mutant also showed substantially reduced lignin content with decreased syringyl/guaiacyl lignin unit composition. Interestingly, the loss of tricin in the mutant lignin appears to be partially compensated by incorporating naringenin, which is a preferred substrate of OsFNSII. The fnsII mutant was further revealed to have enhanced enzymatic saccharification efficiency, suggesting that the cell wall recalcitrance of grass biomass may be reduced through the manipulation of the flavonoid monomer supply for lignification.
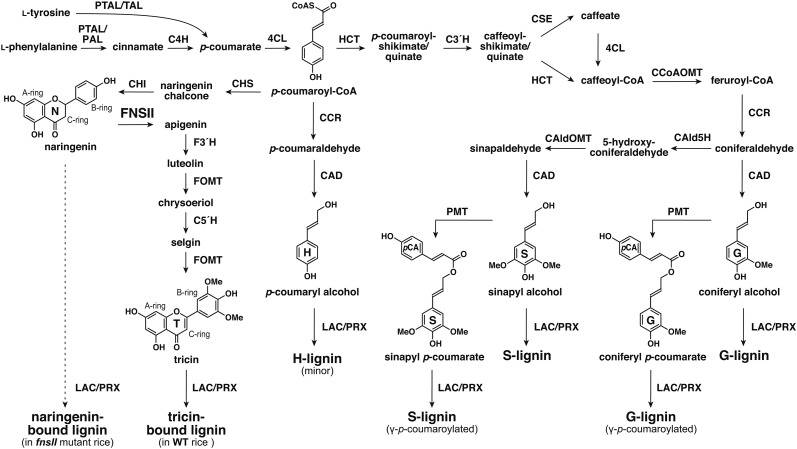
Phenylpropanoids are natural phenolic compounds widespread in plants, and they contribute to many aspects of plant development and responses toward biotic and abiotic stimuli. The phenylpropanoid pathway starts from l-Phe and/or l-Tyr that split(s) off from primary metabolism. Nonoxidative deaminations and successive hydroxylation and/or ligation with CoA produce p-coumaroyl-CoA, which serves as a common intermediate for many classes of phenylpropanoids (Fig. 1). Branching off from p-coumaroyl-CoA, flavonoids and monolignols are the two major downstream metabolite classes generated separately from the pathway (Dixon et al., 2002; Vogt, 2010; Barros et al., 2016).
Figure 1.
Proposed lignin biosynthetic pathway in grasses. PTAL, Phe and Tyr ammonia lyase; TAL, Tyr ammonia lyase; PAL, Phe ammonia lyase; C4H, cinnamate 4-hydroxylase; 4CL, 4-coumarate CoA ligase; HCT, p-hydroxycinnamoyl-CoA:quinate/shikimate p-hydroxycinnamoyltransferase; C3′H, p-coumaroyl ester 3-hydroxylase; CSE, caffeoyl shikimate esterase; CCR, cinnamoyl-CoA reductase; CCoAOMT, caffeoyl-CoA O-methyltransferase; CAld5H, coniferaldehyde 5-hydroxylase; CAldOMT, 5-hydroxyconiferaldehyde O-methyltransferase; CAD, cinnamyl alcohol dehydrogenase; PMT, p-coumaroyl-CoA:monolignol transferase; CHS, chalcone synthase; CHI, chalcone isomerase; FNSII, flavone synthase II; F3′H, flavonoid 3′-hydroxylase; FOMT, flavonoid O-methyltransferase; C5′H, crysoeriol 5′-hydroxylase; LAC, laccase; PRX, peroxidase.
Flavonoids are a large class of secondary metabolites widespread in vascular plants and certain bryophytes. The structures of flavonoids are highly diverse, and different classes are assigned based on the modification of the C6-C3-C6 backbone. Flavonoids display various physiological functions as antioxidants (Agati et al., 2012), phytoalexins (Koes et al., 1994; Du et al., 2010b), signaling molecules (Hassan and Mathesius, 2012), or pigments (Goto and Kondo, 1991). In the monocot family Poaceae, which are the grasses including the cereals, one of the predominant forms of flavonoids is tricin, a 3′,5′-dimethoxyflavone, commonly found as O-linked conjugates in vegetative tissues (Zhou and Ibrahim, 2010; Dong et al., 2014; Li et al., 2016). The biosynthesis of flavonoids is achieved by a combination of the phenylpropanoid pathway and the polyketide pathway. The sequential condensation of p-coumaroyl-CoA with three malonyl-CoAs is catalyzed by chalcone synthase (CHS) and followed by isomerization by chalcone isomerase (CHI) to form naringenin, a flavanone that is the precursor for the biosynthesis of all the other classes of flavonoids. To produce tricin conjugates, naringenin is converted into apigenin by FNSII, and sequential hydroxylations and O-methylations at the flavone B-ring furnish tricin, which is then further converted into the downstream tricin derivatives (Fig. 1).
Lignin, on the other hand, is an abundant phenylpropanoid polymer derived from oxidative couplings of monolignols (i.e. p-hydroxycinnamyl alcohols) and is one of the major cell wall components in vascular plants. By filling up spaces between cell wall polysaccharides (cellulose and hemicelluloses), lignin confers increased mechanical strength, imperviousness, and resistance to pathogens (Boerjan et al., 2003; Bonawitz and Chapple, 2010; Umezawa, 2010). Lignin biosynthesis and bioengineering have long been a major research focus, particularly because of its economic importance associated with agroindustrial utilizations of biomass. Lignin has traditionally been viewed as an impediment to chemical pulping, forage digestion by livestock, and cellulosic bioethanol production, but it is increasingly viewed as a potent source for producing aromatic commodities from biomass. Accordingly, the phenylpropanoid pathway responsible for synthesizing monolignols that build up lignin polymers has been one of the major targets in cell wall bioengineering studies (Ragauskas et al., 2014; Beckham et al., 2016; Rinaldi et al., 2016).
The biosynthesis of monolignols from p-coumaroyl-CoA involves aromatic hydroxylations and O-methylations as well as successive side chain reductions to generate the three canonical monolignols differing in their degree of aromatic methoxylation (Fig. 1; Boerjan et al., 2003; Bonawitz and Chapple, 2010; Umezawa, 2010). In angiosperms (i.e. in both dicots and monocots), lignins are majorly composed of guaiacyl (G) and syringyl (S) units derived from combinatorial radical couplings, initiated by laccases and/or peroxidases, of two monolignols, coniferyl and sinapyl alcohols, respectively, with a lower amount of p-hydroxyphenyl (H) units from p-coumaryl alcohol (Boerjan et al., 2003; Bonawitz and Chapple, 2010; Umezawa, 2010). While sharing this typical lignin trait with dicots, lignins in the major monocot family Poaceae (grasses including cereals) are partially acylated at the γ-position with p-coumarate. It has been established that such lignin acylations arise from lignification with γ-p-coumaroylated monolignols generated by a grass-specific acyltransferase, p-coumaroyl-CoA:monolignol transferase (PMT; Fig. 1; Petrik et al., 2014). Furthermore, it was demonstrated recently that various commelinid monocots, including Poaceae species, also incorporate a small amount of γ-feruloylated monolignols for lignification (Karlen et al., 2016).
Flavonoids are known to couple with monolignols, forming extractable flavonolignans, flavonolignols, and their O-glycosides. For example, silymarin extracted from milk thistle (Silybum marianum) seeds contains flavonolignans derived from the coupling of taxifolin and coniferyl alcohol (Kim et al., 2003; Wang et al., 2010). Hydnocarpin and 5′-methoxyhydnocarpin, coupling products of luteolin with coniferyl or sinapyl alcohol, were identified in Hydnocarpus wightiana (Parthasarathy et al., 1979), Onopordon corymbosum (Cardona et al., 1990), and Hymeneae palustris (Pettit et al., 2003). Other naturally occurring flavonolignans and flavonolignols include pseudotsuganol, hydnowightin, neohydnocarpin, palstatin, sinaiticin, and silandrin (Sharma et al., 1979; Foo and Karchesy, 1989; Pettit et al., 2003; Nyiredy et al., 2008). In monocots, the widespread nature and the high structural diversity of tricin-type flavonolignans and their related derivatives are well documented (Zhou and Ibrahim, 2010; Yang et al., 2014; Lan et al., 2016a; Li et al., 2016). More strikingly, after resolving the unknown signals in the NMR spectra of polymeric lignins isolated from wheat (Triticum aestivum) cell walls, tricin was recently discovered as an integrated component of lignins (Del Río et al., 2012). Subsequently, extensive surveys have revealed that tricin-bound lignins exist abundantly, particularly in the monocot family Poaceae, which comprises grasses including cereals. They also have been found in some commelinid monocot families outside Poaceae, such as Arecaceae (palms) and Bromeliaceae (pineapples and relatives), the noncommelinid family Orchidaceae (the orchids), particularly in the genus Vanilla, and also in certain dicots (Rencoret et al., 2013; Wen et al., 2013; Del Río et al., 2015; Lan et al., 2015, 2016a, 2016b; Koshiba et al., 2017).
Tricin, as an authentic lignin monomer in grasses, incorporates into the lignin polymers via combinatorial radical couplings, as in the way lignification takes place solely with monolignols in dicots and gymnosperms. Lacking the abilities to either undergo radical dehydrodimerization or to start the polymer chain elongations from the phloroglucinol A ring, tricin always occurs at one terminus of a lignin polymer chain and was proposed to function as a nucleation site for lignification (Lan et al., 2015). The discovery of the tricin-bound lignins, illustrating the plasticity of lignification and its strong interconnection with flavonoid biosynthesis, sheds new light on the studies of lignin biosynthesis and bioengineering. Currently, however, it remains largely unknown how tricin-bound lignins are biosynthesized and function in grass cell walls. Given that many of the grass biomass crops (e.g. sorghum [Sorghum bicolor], sugarcane [Saccharum officinarum], switchgrass [Panicum virgatum], and bamboo [Bambusa oldhamii]) produce substantial amounts of tricin-bound lignins (Lan et al., 2016b), it is also intriguing to investigate how tricin-bound lignins affect the utilization properties of cell walls.
We reported previously that OsFNSII is essential for the biosynthesis of extractable tricin metabolites (i.e. tricin O-glycosides and O-flavonolignans) in rice (Oryza sativa) seedlings (Lam et al., 2014). OsFNSII, which catalyzes the direct conversion of flavanones to flavones, is a cytochrome P450 enzyme (CYP93G1) belonging to the grass-specific 93G subfamily. In this study, we address the involvement of OsFNSII in lignification and examine cell wall properties upon tricin deficiency. A T-DNA insertional rice fnsII mutant was subjected to a series of analyses for the assessment of growth phenotypes, gene expression, as well as lignin structure. A series of chemical analyses demonstrated that the mutant produced cell walls with reduced lignin levels and decreased S/G lignin unit composition. NMR characterizations revealed the complete depletion of tricin along with the incorporation of naringenin, a flavanone substrate of OsFNSII, as a new component in cell wall lignin. Importantly, such lignin alterations resulted in enhanced cell wall digestibility without negative impact on growth and development. Together, our work establishes the essential role of OsFNSII in tricin lignification in the cell wall and suggests that grass biomass utilization may be enhanced by manipulation of the flavone biosynthesis pathway.
RESULTS
Expression of Flavonoid and Monolignol Biosynthetic Genes in Wild-Type Rice Plants
At the onset of this study, we performed in silico gene expression analysis of flavonoid and monolignol biosynthetic genes in wild-type rice (japonica cv Nipponbare; Sato et al., 2013). As with other putative/known tricin biosynthetic genes, such as OsCHS1 (Shih et al., 2008; Hong et al., 2012), OsCHI (Shih et al., 2008), and OsC5′H (CYP75B4; Lam et al., 2015), OsFNSII (CYP93G1; Lam et al., 2014) was expressed most prominently in culm at reproductive and ripening stages, where cell wall lignification typically occurs. We confirmed the concurrent expression of putative/known monolignol biosynthetic genes, including OsCAD2 (Zhang et al., 2006; Koshiba et al., 2013b), OsCAldOMT1 (Koshiba et al., 2013a), and OsPMT (Petrik et al., 2014), as well as the common phenylpropanoid genes OsPAL1/2 (Cass et al., 2015) and Os4CL3 (Gui et al., 2011; Supplemental Fig. S1). In addition, OsFNSII, along with its downstream OsC5′H (Fig. 1), was expressed in leaf at the vegetative stage and also in lemma and palea at the later stage of flower development, and several monolignol biosynthetic genes displayed similar spatial and temporal expression patterns (Supplemental Fig. S1). These data support our contention that OsFNSII is involved not only in the biosynthesis of soluble tricin metabolites (e.g. tricin O-glycosides and O-flavonolignans; Lam et al., 2014) but also of tricin monomer for lignification in the major rice vegetative tissues, as demonstrated further below.
Phenotype of OsFNSII-Knockout Mutant Rice
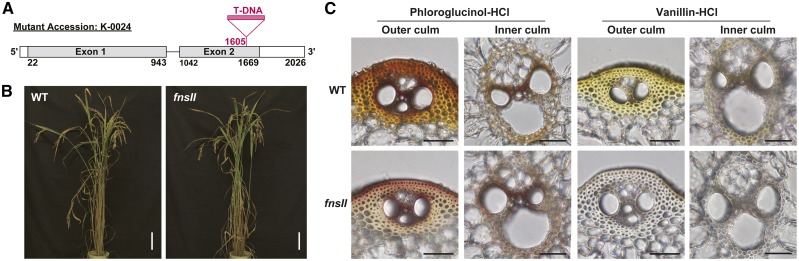
To further examine the involvement of OsFNSII in cell wall lignification, we reinvestigated a loss-of-function mutant rice (japonica cv Kitaake) that we characterized previously (Lam et al., 2014); this mutant has a T-DNA insertion in the second exon of the OsFNSII locus (Fig. 2A). Gene expression analysis on a homozygous mutant line (fnsII) using a quantitative real-time PCR approach suggested that, overall, with the exception of a slightly depressed OsC5′H expression, there are no significant changes in the major flavonoid and monolignol biosynthetic gene expression compared with wild-type plants (Supplemental Fig. S2). The mutant plants grew to maturity without displaying significant morphological changes compared with the wild-type controls (Fig. 2B). Although a slight reduction in plant height was observed, fnsII plants overall displayed a similar growth performance comparable with wild-type plants in terms of their culm length, tillering, fertility, and biomass production, at least under the growth conditions used (Table I).
Figure 2.
Gene structure, phenotype, and vasculature of FNSII-knockout mutant rice (fnsII) compared with a wild-type (WT) rice. A, Gene structure of OsFNSII (CYP93G1) in the T-DNA insertional mutant fnsII used in this study. B, Morphological phenotypes of wild-type and fnsII mutant plants at the harvest stage (45 d after heading). Bars = 10 cm. C, Histochemical analysis of culm cell walls in wild-type and fnsII mutant plants at the heading stage. Transverse cross sections of culms were stained by phloroglucinol-HCl and vanillin-HCl reagents for lignin and flavonoids, respectively. Bars = 40 μm.
Table I. Growth phenotypes, biomass yield, and fertility rate of wide-type and fnsII mutant plants.
Values are means ± sd (n = 5), and the asterisk indicates a significant difference from the wide type (Student’s t test: P < 0.05). CWR, Cell wall residue.
| Trait | Wild Type | fnsII |
|---|---|---|
| Plant height (cm)a | 115.0 ± 4.7 | 105.1 ± 6.0* |
| Culm length (cm)b | 86.3 ± 5.3 | 79.7 ± 7.5 |
| Ear length (cm) | 16.0 ± 2.5 | 15.3 ± 1.1 |
| Tiller number | 10.4 ± 1.7 | 12.2 ± 3.4 |
| Ear number | 14.6 ± 2.0 | 14.8 ± 2.6 |
| Dry mass of culm (g) | 4.8 ± 1.4 | 3.7 ± 0.7 |
| Dry mass of sheath (g) | 3.2 ± 0.5 | 3.6 ± 1.3 |
| Dry mass of leaf (g) | 3.0 ± 0.4 | 3.8 ± 1.1 |
| CWR yield of culm (%) | 63.2 ± 6.5 | 62.8 ± 2.5 |
| CWR yield of sheath (%) | 84.7 ± 0.7 | 80.2 ± 6.4 |
| CWR yield of leaf (%) | 73.6 ± 0.99 | 70.1 ± 4.66 |
| No. of panicles | 15.0 ± 2.2 | 15.4 ± 2.7 |
| Average mass per panicle (g) | 1.1 ± 0.2 | 1.2 ± 0.2 |
| Fertility rate (%) | 85.3 ± 3.9 | 83.6 ± 2.7 |
Length from the cotyledonary node to the tip of the top leaf. bLength from the cotyledonary node to the panicle base.
Histochemical Analysis of OsFNSII-Knockout Mutant Rice Cell Walls
Transverse sections from developing culms of fnsII mutant and wild-type plants were subjected to histochemical analyses using lignin and flavonoid staining reagents (Fig. 2C). As with wild-type plants, fnsII mutants developed morphologically normal vascular tissues with thick secondary walls in the cortical sclerenchyma fibers and vascular bundles. The fnsII cell walls exhibited positive colorations with phloroglucinol-HCl (Wiesner reagent), which is known to react with cinnamaldehyde end groups in the monolignol-derived lignin polymers. The staining of fnsII mutant cell walls, however, apparently was less intense than that of wild-type cell walls, indicating a decreased lignin content and/or a considerable alteration in lignin structure. In parallel, the sections were treated with vanillin-HCl, a well-known staining reagent for general flavonoid compounds (Gardner, 1975). The wild-type sections displayed a yellowish positive staining in the cortical sclerenchyma fibers and vascular bundle cell walls, suggesting a substantial amount of flavonoid, presumably tricin, bound to the cell walls. In contrast, no obvious flavonoid staining was observed for the fnsII mutant cell walls, suggesting a considerable depletion of flavonoids in the cell walls (Fig. 2C). These histochemical data collectively suggest that OsFNSII disruption does not lead to defects in vascular morphology but to potential reduction and/or alteration of flavonoid-bound lignins in cell walls.
Chemical Analysis of OsFNSII-Knockout Mutant Rice Cell Walls
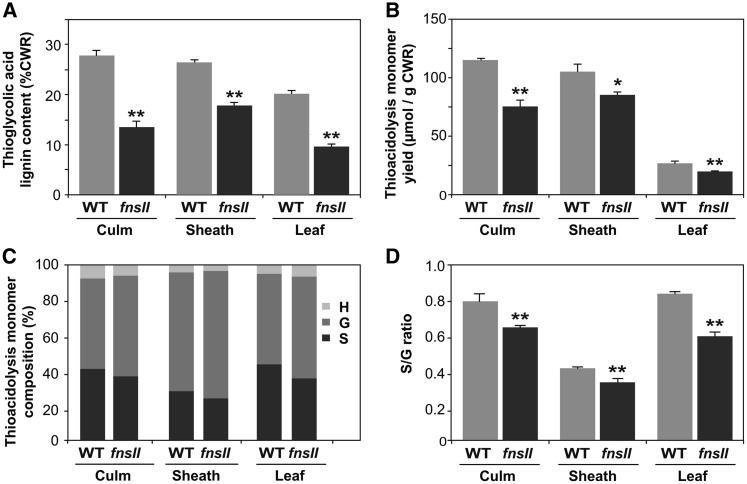
To investigate the cell wall chemotype of the fnsII mutant, we first performed a series of chemical analyses on extractive-free CWRs prepared from senesced culm, sheath, and leaf tissues; no significant differences were found in the yield of CWR per dry plant tissue between wild-type and fnsII mutant plants (Table I). Lignin content determined by thioglycolic acid assay was reduced remarkably, by 34% to 58%, in fnsII mutant cell walls compared with wild-type cell walls (Fig. 3A). This is in line with our earlier observation in the histochemical analysis (Fig. 2C). We also employed thioacidolysis to quantify lignin monomers released from monolignol-derived β-O-4 lignin substructures (Lapierre et al., 1986; Yamamura et al., 2012; Yue et al., 2012). The mutant cell walls released significantly less, by 17% to 33%, lignin monomers than wild-type cell walls upon thioacidolysis degradation (Fig. 3B), further confirming that OsFNSII disruption reduces the generation of lignins from monolignols. However, when the thioacidolysis monomer yield was expressed relative to lignin content, an opposite trend was observed in most of the samples (Supplemental Fig. S3). The total thioacidolysis-released H + G + S monomers and G monomers per thioglycolic lignin were significantly higher in all the tissues tested. Also, significant increases in S-type monomers in culm and leaf and H-type monomers in leaf tissues were observed. Intriguingly, the fnsII mutant cell walls appeared to show a trend of decreased S/G monomer ratio in all the tissues tested (Fig. 3, C and D). Taken together, our lignin analysis suggested that OsFNSII disruption somehow affects the content and composition of lignins derived from typical monolignols.
Figure 3.
Chemical lignin analysis of cell walls from wild-type (WT) and FNSII-knockout mutant (fnsII) rice plants. A, Lignin content determined by thioglycolic acid assay. B to D, Lignin composition analysis by thioacidolysis. Shown are total monomer yield per CWR (B) and relative abundances (C and D) of H-, G-, and S-type trithioethylpropane monomers released from H-, G-, and S-type lignins. Values are means ± sd from individually analyzed plants (n = 3), and asterisks indicate significant differences between wild-type and fnsII mutant plants (Student’s t test: *, P < 0.05 and **, P < 0.01).
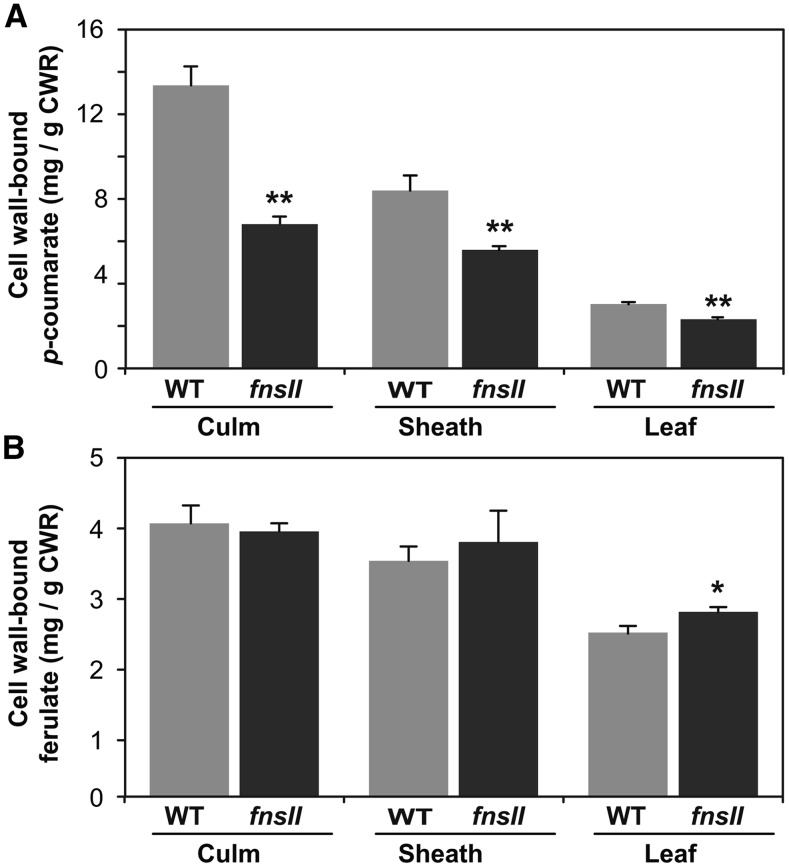
Cell wall-bound p-coumarates (pCAs) and ferulates (FAs) were quantified as the corresponding free acids released under mild alkaline hydrolysis of CWRs. The fnsII mutant cell walls displayed significantly reduced pCA levels (25%–48% less compared with wild-type controls), particularly in culm and sheath tissues (Fig. 4A), whereas FA levels were not affected significantly in all the vegetative tissues investigated (Fig. 4B). Given that a majority of pCA is bound to lignins whereas FA is bound mainly to hemicelluloses (arabinoxylans) in typical grass cell walls (Ralph, 2010), it is plausible that the reduced pCA levels in culm and sheath were associated with the reduced levels of lignins derived from monolignols (Fig. 3). This was further supported by comparing the pCA content per thioglycolic lignin between wild-type and fnsII mutant plants (Supplemental Fig. S4): there was no substantial difference in the content of pCA per lignin in the culm and sheath tissues. We also analyzed cell wall sugar composition via a combination of trifluoroacetic acid and sulfuric acid-catalyzed cell wall hydrolysis reactions (see “Materials and Methods”). Overall, wild-type and fnsII mutant cell walls displayed similar sugar profiles, suggesting that OsFNSII disruption does not affect the composition of cell wall polysaccharides; as is typical in grass cell walls, crystalline cellulose and arabinoxylans constitute a major part of cell wall polysaccharides in both wild-type and fnsII mutant tissues (Supplemental Fig. S5).
Figure 4.
Cell wall-bound p-coumarates (A) and ferulates (B) released from wild-type (WT) and FNSII-knockout mutant (fnsII) cell walls via mild alkaline hydrolysis. Values are means ± sd from individually analyzed plants (n = 3), and asterisks indicate significant differences between wild-type and fnsII mutant plants (Student’s t test: *, P < 0.05 and **, P < 0.01).
Two-Dimensional NMR Analysis of OsFNSII-Knockout Mutant Rice Cell Walls
To further investigate the impact of the OsFNSII-knockout mutation on cell wall structure, we performed two-dimensional NMR analysis on the cell walls isolated from fnsII and wild-type culm tissues. We first analyzed whole cell wall materials by simple swelling of CWRs in dimethyl sulfoxide (DMSO)-d6/pyridine-d5 after fine ball milling. This approach provides a global picture of the chemical composition and structure of cell wall lignins as well as polysaccharides (Kim and Ralph, 2010; Mansfield et al., 2012). For a more in-depth analysis, we analyzed lignin-enriched cell walls prepared from CWRs following the enzymatic removal of polysaccharides (Tobimatsu et al., 2013; Zhao et al., 2013).
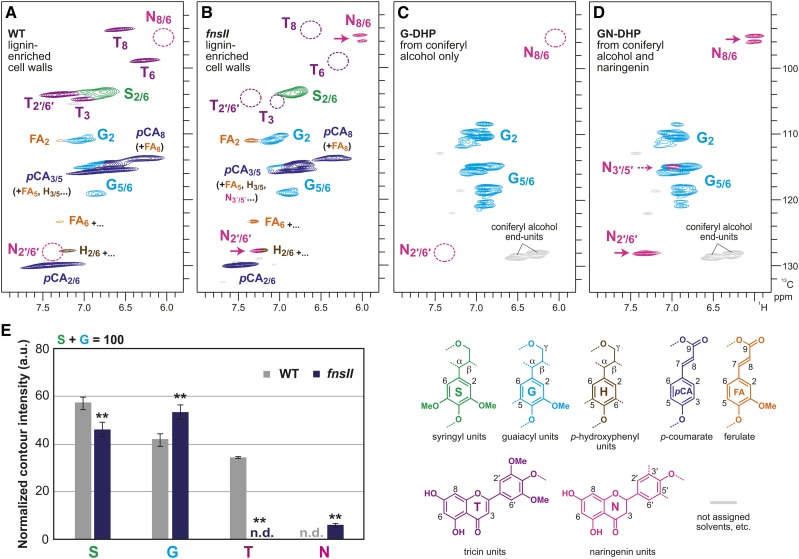
The aromatic subregions of the short-range 1H-13C correlation (heteronuclear single-quantum coherence [HSQC]) NMR spectra displayed typical lignin aromatic signals from G and S units (G and S) as well as those from H units (H), albeit at low levels (Fig. 5; Supplemental Fig. S6A). Volume integrations of these contour signals estimated 46% to 58% and 42% to 54% of S and G lignins, respectively (Fig. 5E). In line with our observation in thioacidolysis (Fig. 3D), S lignin signals were clearly depleted over G lignin signals in the fnsII mutant cell wall spectra. Besides the typical aromatic signals from the monolignol-derived lignins, the HSQC spectra of wild-type cell walls displayed the characteristic set of aromatic signals from lignin-bound tricin units (T); the chemical shifts of all the C-H correlations from the flavone aromatic system (T3, T8/6, and T2′/6′) are in total agreement with literature data (Del Río et al., 2012; Lan et al., 2015; Koshiba et al., 2017). In contrast, all these tricin signals were strikingly depleted to undetectable levels (less than 1%) in the spectra of fnsII mutant cell walls (Fig. 5, B and E). This clearly suggests that disruption of OsFNSII expression results in a strongly reduced incorporation of tricin into the lignin polymer.
Figure 5.
Aromatic subregions of short-range 1H-13C correlation (HSQC) NMR spectra of cell wall lignins from culm tissues of wild-type (WT) and FNSII-knockout mutant (fnsII) rice plants and in vitro synthetic lignin polymers (DHPs). A and B, Lignin-enriched cell walls of wild-type and fnsII mutant plants prepared by enzymatic removal of wall polysaccharides with crude cellulases. Contour coloration matches that of the lignin substructure units shown. C and D, DHPs prepared from coniferyl alcohol only (G-DHP) and from coniferyl alcohol along with naringenin (GN-DHP). Contour coloration matches that of the lignin substructure units shown. E, Normalized contour intensity of the major lignin and flavonoid aromatic signals appearing in the spectra of lignin-enriched cell walls. Values are means ± sd from individually analyzed plants (n = 3) and are expressed as percentages of the total S and G lignin units. Asterisks indicate significant differences between wild-type and fnsII mutant plants (Student’s t test: **, P < 0.01). n.d., Not detected; a.u., arbitrary unit.
In addition, a new set of aromatic signals appeared at δC/δH 95 to 96.5/6.2 in the fnsII mutant spectra. Based on the location of FNSII in the tricin biosynthetic pathway, we hypothesized that the new flavonoid-bound lignins could have been derived from incorporating naringenin intermediate into the lignin polymers (Fig. 1). To test this hypothesis, we prepared synthetic lignin polymers (GN-DHP) via in vitro peroxidase-catalyzed copolymerization of naringenin and coniferyl alcohol. A close comparison of the NMR spectra of the mutant cell walls and GN-DHP firmly established the incorporation of naringenin into the lignin polymers (Fig. 5, B and D). The resolved and diagnostic signals appearing at δC/δH 95 to 96.5/6.2 were assigned to C8-H8 and C6-H6 correlations of the naringenin flavanone aromatic system (N8/6). Although the signals from the naringenin B-ring most likely overlapped with G and H lignin aromatic signals (C2′-H6′ correlations [N2′/6′] at δC/δH ∼128/∼7.4; C3′-H5′ correlations [N3′/5′] at δC/δH ∼115/∼7), characteristic methylene signals from the naringenin C-ring (N3) also were well resolved and clearly seen at δC/δH 78.5/5.5 in the aliphatic subregions of the mutant and naringenin-incorporated GN-DHP spectra (Fig. 6; Supplemental Fig. S6B).
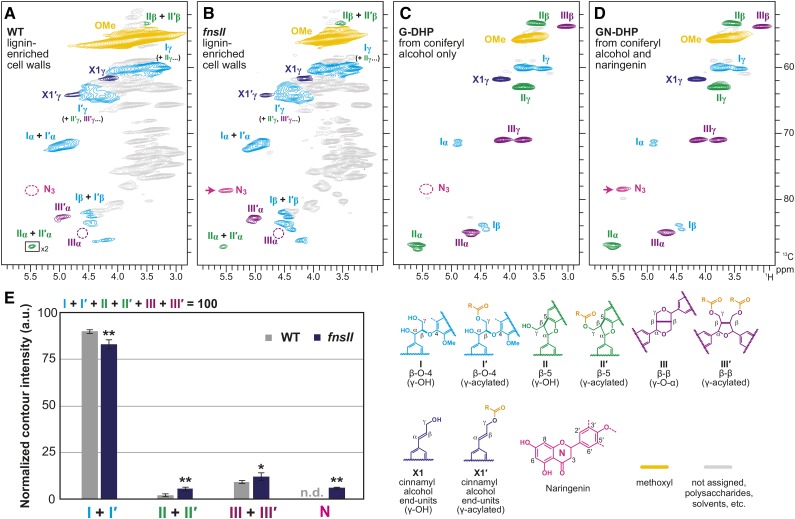
Figure 6.
Aliphatic subregions of short-range 1H-13C correlation (HSQC) NMR spectra of cell wall lignins from culm tissues of wild-type (WT) and FNSII-knockout mutant (fnsII) rice plants and in vitro synthetic lignin polymers (DHPs). A and B, Lignin-enriched cell walls of wild-type and fnsII mutant plants prepared by enzymatic removal of wall polysaccharides with crude cellulases. Boxes labeled ×2 indicate regions that are vertically scaled 2-fold. Contour coloration matches that of the lignin substructure units shown. C and D, DHPs prepared from coniferyl alcohol only (G-DHP) and from coniferyl alcohol along with naringenin (GN-DHP). Contour coloration matches that of the lignin substructure units shown. E, Normalized contour intensity of the major lignin side chain and naringenin signals appearing in the spectra of lignin-enriched cell walls. Values are means ± sd from individually analyzed plants (n = 3) and are expressed as percentages of the total of I, I′, II, II′, III, and III′ side chain structures. Asterisks indicate significant differences between wild-type and fnsII mutant plants (Student’s t test: *, P < 0.05 and **, P < 0.01). n.d., Not detected; a.u., arbitrary unit.
The aliphatic subregions of the HSQC spectra also provide information on the major intermonomeric linkages in the lignin polymers (Fig. 6; Supplemental Fig. S6B). Typical lignin linkage signals from β-O-4 (I), β-5 (II), and β-β (III) units as well as those from the corresponding γ-acylated units (I′, II′, and III′) were visible in both wild-type and fnsII mutant cell wall spectra. Volume integrations of the relatively well-resolved Cα-Hα contours appearing in the lignin-enriched cell wall spectra allowed us to estimate the distributions of these lignin intermonomeric linkages (Fig. 6E). Our data suggested that the mutant lignins were depleted significantly in β-aryl ethers (I + I′) and augmented in phenylcoumarans (II + II′) and β-β (III + III′) units compared with wild-type lignins. As discussed further below, such shifts in the lignin linkage pattern might be a consequence of the reduction and partial replacement of tricin units by naringenin units. We also analyzed the profiles of cell wall polysaccharides based on the sugar anomeric correlations appearing in the whole cell wall spectra (Brennan et al., 2012; Kim and Ralph, 2014). Overall, distributions of the sugar correlations were similar between the wild-type and mutant spectra (Supplemental Fig. S6C), which is totally in line with the chemical data (Supplemental Fig. S5).
Digestibility of OsFNSII-Knockout Mutant Rice Cell Walls
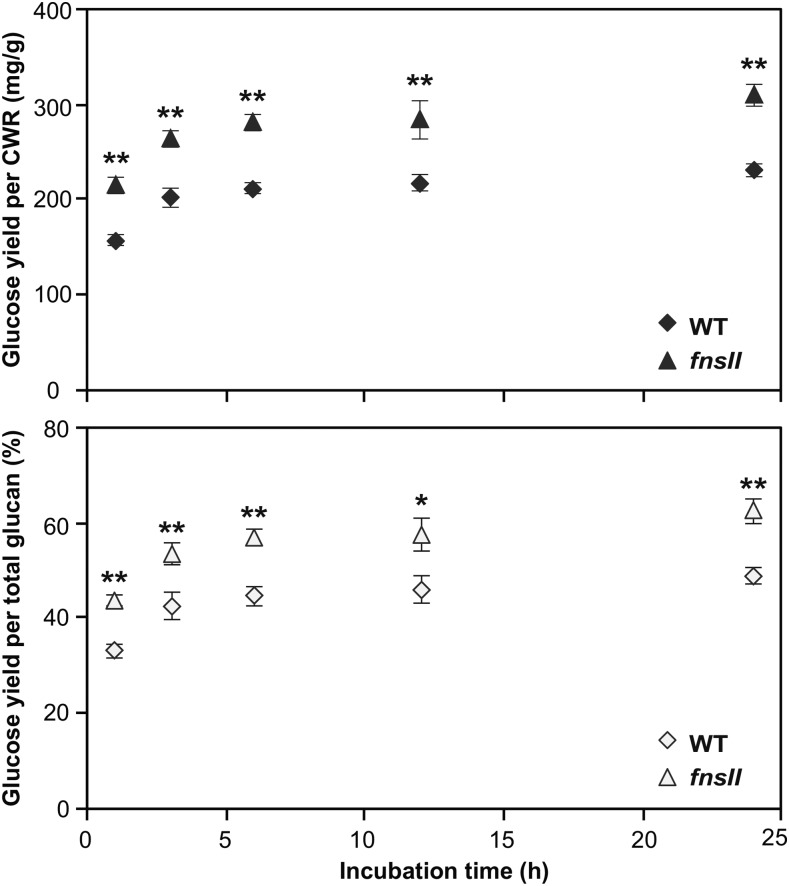
Lastly, to determine the effect of truncation of the tricin biosynthetic pathway on cell wall digestibility, we evaluated the enzymatic saccharification efficiency of the rice cell walls. Pulverized and destarched culm CWRs were digested, without any pretreatment, using a cocktail of commercially available cellulolytic enzymes (Hattori et al., 2012). Typical enzymatic hydrolysis profiles were obtained for both wild-type and mutant cell walls. Saccharification was rapid during the first 6 h of hydrolysis, and continued incubation released comparatively small amounts of additional Glc. As illustrated in Figure 7, it was clearly observed that the mutant cell walls yielded more Glc than the wild-type controls at all the incubation times examined. The enhancement of saccharification efficiency was 30% to 37% when expressed as Glc yield per cell wall and 25% to 32% when expressed as Glc yield per total glucan.
Figure 7.
Enzymatic saccharification of cell walls from culm tissues of wild-type (WT) and FNSII-knockout mutant (fnsII) rice plants. The saccharification efficiency is expressed as Glc yield per CWR (top) or as Glc yield per total glucan (bottom). Values are means ± sd from individually analyzed plants (n = 3), and asterisks indicate significant differences between wild-type and fnsII mutant plants (Student’s t test: *, P < 0.05 and **, P < 0.01).
DISCUSSION
FNSII Mutant Rice Produces Cell Wall Lignins Devoid of Tricin
The tricin biosynthetic pathway in rice was completely elucidated recently with the identification of a series of previously uncharacterized flavone enzymes (Kim et al., 2006; Lam et al., 2014, 2015). Among them, OsFNSII (CYP93G1) represents a branch-point enzyme for the entry to tricin biosynthesis in rice (Lam et al., 2014). Grass FNSIIs classified in the CYP93G subfamily were likely to have evolved independently from dicot FNSIIs, which belong to the 93B subfamily (Supplemental Fig. S7). Recombinant OsFNSII converts naringenin and eriodictyol into apigenin and luteolin, respectively. In addition, Arabidopsis overexpressing OsFNSII produces apigenin, luteolin, and chrysoeriol O-glycosides, which are normally not produced in the tissues examined. Furthermore, the accumulation of extractable flavones, including tricin O-glycosides and O-flavonolignans, was compromised in the fnsII mutant. Hence, OsFNSII is indispensable for the production of extractable tricin metabolites in rice (Lam et al., 2014).
This study provides compelling evidence that OsFNSII also is responsible for generating tricin monomer for cell wall lignification in rice. Our NMR analysis clearly demonstrated that the fnsII mutant produces cell wall lignins devoid of tricin residues. The tricin aromatic signals appearing in the HSQC spectra of the mutant cell walls are below detection limits (less than 1%), while those signals account for about 34% relative to the total of G and S lignin signals in the wild-type cell walls (Fig. 5E). It should be noted here that, as reported recently (Lan et al., 2016b), such HSQC NMR-based estimates of tricin concentrations most likely are excessive; tricin is mostly in lignins as the polymers’ terminal units, and typical HSQC experiments overquantify such more mobile terminal units compared with rigid internal units (Mansfield et al., 2012; Tobimatsu et al., 2013; Okamura et al., 2016). In fact, a recent study reported that tricin concentrations in grass lignins determined by a more reliable chemical method are typically 1% to 3% (Lan et al., 2016b). Given that all the tricin signals are below the detection limit in our HSQC NMR analysis for the fnsII mutant, it is conceivable that the actual concentration of lignin-bound tricin in this mutant is practically zero.
Interestingly, FNSII mutation also impacted the lignification of typical monolignols. The thioglycolic acid lignin assay estimated 34% to 58% lignin reductions in fnsII mutant cell walls compared with wild-type controls (Fig. 3A). In addition, we observed 18% to 35% reductions in the total yields of monolignol monomers released upon thioacidolysis (Fig. 3B), suggesting that the apparent lignin reduction in the fnsII mutant was caused not only by the loss of tricin units but also by the depletion in the lignin units derived from the canonical monolignols. This also is corroborated by the lower intensity of phloroglucinol-HCl lignin staining in vascular tissues (Fig. 2C) as well as by the reductions in lignin-bound pCA levels (Fig. 4A). On the other hand, when the yield of thioacidolysis-released monolignol monomers was expressed relative to the thioglycolic lignin content, an increase was observed in the fnsII mutant (Supplemental Fig. S3), implying that the mutant lignin is less condensed. Apparently, this is contradictory to what was observed in our NMR analysis: the fnsII mutant contained less noncondensed β-aryl ethers and more phenylcoumaran and β-β units than the wild-type control (Fig. 6E). It could be due partly to the fact that, unlike NMR, which provides structural information on the entire lignin, thioacidolysis analyzes only a fraction of the polymer containing cleavable β-aryl ethers; it is also reported that the acylation of lignin in grasses impedes the efficient cleavage of β-aryl ethers; thus, the lignin monomer yield determined for grass samples under typical thioacidolysis conditions could be substantially underestimated (Grabber et al., 1996; Yue et al., 2012).
In addition to the reduced lignin levels, we also observed significantly decreased S/G lignin unit ratios in all the mutant tissues as determined by both thioacidolysis (Fig. 3D) and NMR (Fig. 5E). It has been reported that disruptions in the monolignol biosynthetic pathway redirect the metabolic flux in the phenylpropanoid pathway and occasionally affect accumulations of flavonoids (Abdulrazzak et al., 2006; Besseau et al., 2007; Fornalé et al., 2010; Li et al., 2010; Vanholme et al., 2012; Fornalé et al., 2015). Therefore, it is conceivable that a blockage in a flavonoid pathway may, in turn, affect the generation of monolignols and their lignin polymers. Very recently, it was reported that a maize (Zea mays) mutant defective in CHS (Fig. 1) produces tricin-depleted cell walls with a substantially increased total lignin content (Eloy et al., 2017), which is apparently in contrast to our FNSII rice mutant, with lignins depleted in both tricin- and monolignol-derived units. As CHS is the entry enzyme for the flavonoid pathway branching off from the general phenylpropanoid pathway (Fig. 1), the down-regulation of CHS can redirect the carbon flux from the biosynthesis of flavonoids to canonical monolignols, which consequently results in plants with increased lignin levels. Such a scenario, however, may not prevail in our rice fnsII mutant, because FNSII functions downstream of the flavonoid pathways (Fig. 1). In fact, as discussed further below, fnsII mutant rice abnormally accumulates narigenin-incorporated lignins as well as other narigenin-derived flavone and flavanone metabolites, as we reported previously (Lam et al., 2014). These data suggest that the carbon flux redirected from the biosynthesis of tricin is at least partially compensated within the flavonoid pathway. Although further studies are required, the reduction of lignin content in the rice fnsII mutant may suggests a feedback system that controls the relative carbon flux between flavonoid and monolignol biosynthetic pathways. It also should be noted here that, unlike the case of CHS-defective maize (Eloy et al., 2017), CHS suppression in some dicot species resulted in no alterations or, as in our FNSII rice mutant, reductions in lignin levels (Li et al., 2010; Zuk et al., 2016). Therefore, cross-interactions between the flavonoid and monolignol pathway metabolisms also may be dependent on different metabolic plasticity in different plant species.
FNSII Mutant Rice Incorporates Naringenin as a Novel Lignin Component
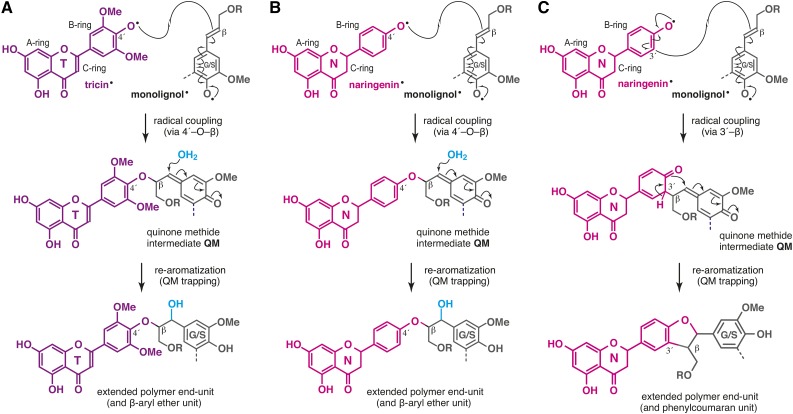
An intriguing discovery in this study was that the loss of tricin for lignification in the fnsII mutant was partially compensated by incorporating naringenin, a flavanone substrate of FNSII, as a new component of lignin polymer units (Fig. 1). In line with this, we previously reported the overaccumulation of soluble naringenin metabolites in the fnsII mutant seedlings (Lam et al., 2014). The successful generation of synthetic lignin polymer (GN-DHP) from naringenin and coniferyl alcohol in vitro indicates that naringenin is compatible in lignin polymerization; naringenin has the ability to be radicalized by peroxidases, cross-coupled with monolignols, and integrally incorporated into the lignin polymers. Our NMR analysis also demonstrated that the lignin-linked naringenin residues still contain the intact phloroglucinol A-rings (Figs. 5 and 6). This suggests that reactions of the p-hydroxyphenyl B-ring far exceed A-ring reactions during lignin polymerization with naringenin. Previous studies examining chemical and enzymatic oxidations of tricin (Lan et al., 2015) and analogous flavonoids (Itoh et al., 2007; Elumalai et al., 2012; Grabber et al., 2012) also have reported predominant reactions of cinnamoyl B-rings over phloroglucinol-type A-rings. Furthermore, these NMR data also can be interpreted that the newly incorporated naringenin units are linked majorly as the terminal units of the lignin polymer chains, as proposed for the canonical tricin units (Lan et al., 2015).
Tricin bearing the 3′,5′-dimethoxyl-p-hydroxyphenyl B-ring incorporates into lignin exclusively via 4′-O-β-type coupling, which ultimately creates β-aryl ether units in the lignin polymer chains (Fig. 8A; Lan et al., 2015). On the other hand, naringenin with a nonsubstituted p-hydroxyphenyl B-ring logically can couple with monolignols not only via 4′-O-β-type coupling for β-aryl ether units (Fig. 8B) but also via 3′-β-type coupling, yielding additional phenylcoumaran units at the lignin terminus (Fig. 8C). Therefore, our observation that fnsII mutant lignins had notably increased phenylcoumaran units (about 3-fold increase, based on HSQC signal integrations; Fig. 6E) could be explained partially by the replacement of the tricin lignin monomer by naringenin.
Figure 8.
Generation of flavonoid-bound lignin units upon lignification. A. The 4′-O-β pathway for β-aryl units via cross-coupling of tricin and monolignols upon lignification in wild-type rice cell walls. B and C, The 4′-O-β (B) and 3′-β (C) pathways for β-aryl ether and phenylcoumaran units via cross-coupling of naringenin and monolignols upon lignification in fnsII mutant rice cell walls.
As envisioned by the histochemical analysis with the vanillin-HCl reagent (Fig. 2C), the incorporation of naringenin into fnsII mutant lignins was unlikely to reach the level of tricin incorporation in wild-type lignins. In our HSQC analysis, whereas tricin signals account for ∼35% relative to the total G and S lignin signals in the wild-type cell wall spectra, naringenin signals reached only about 6% in the fnsII mutant spectra (Fig. 5E). Our previous metabolite study also suggested a relatively lower level of soluble naringenin metabolites in fnsII mutant seedlings compared with soluble tricin metabolites in wild-type seedlings (Lam et al., 2014). Meanwhile, OsFNSII disruption also may increase carbon flow to the production of flavone C-glycosides through CYP93G2, which utilize naringenin as a substrate (Du et al., 2010a; Lam et al., 2014).
Extensive studies on the biosynthesis and bioengineering of lignin have revealed the plasticity of lignification in planta. Manipulation of the canonical monolignol pathway had led to compositional alterations in the polymer due to the incorporation of nontraditional lignin monomers, such as caffeyl alcohol in a CCoAOMT-deficient plant (Wagner et al., 2011), 5-hydroxyconiferyl alcohol in CAldOMT-deficient plants (Jouanin et al., 2000; Ralph et al., 2001; Vanholme et al., 2010; Weng et al., 2010; Koshiba et al., 2013a), ferulic acid in CCR-deficient plants (Ralph et al., 2008; Wagner et al., 2013), and p-hydroxycinnamaldehydes in CAD-deficient plants (Kim et al., 2000; Marita et al., 2003; Sibout et al., 2005; Bouvier d’Yvoire et al., 2013; Koshiba et al., 2013b; Zhao et al., 2013; Anderson et al., 2015). Such malleability of lignification also is exemplified by the fact that numerous angiosperm plants produce seed coat-specific lignins derived from caffeyl and 5-hydroxyconiferyl alcohols (Chen et al., 2012, 2013; Tobimatsu et al., 2013). Our discovery that FNSII deficiency in rice results in the incorporation of naringenin into lignin further illustrates the substantial flexibility in the construction of lignin polymers in planta.
FNSII Mutant Rice Is Viable and Produces Biomass with an Improved Digestibility
As the quantity and quality of lignin affect many aspects of lignocellulosic biomass utilization, the regulation of lignin biosynthesis has been a primary target for cell wall bioengineering (Ragauskas et al., 2014; Beckham et al., 2016; Rinaldi et al., 2016). During biofuels production, lignin is a major recalcitrant barrier to the enzymatic saccharification of cell wall polysaccharides. Reduction of lignin content and/or alteration of lignin composition can improve the efficiency of enzymatic cell wall hydrolysis and downstream microbial fermentations (Chen and Dixon, 2007). However, such lignin modifications often result in developmental abnormalities, such as collapsed xylem, stunted growth, and infertility (Bonawitz and Chapple, 2013). Importantly, despite a considerably reduced lignin content and altered flavonoid-bound lignins, the fnsII mutant develops apparently intact vascular tissues (Fig. 2C) and displays overall normal plant growth, biomass production, and fertility, all comparable with the wild-type controls (Table I). Likewise, the recently reported tricin-depleted maize chs mutant displayed no growth defects (Eloy et al., 2017). Although a more comprehensive analysis of plant growth performance under various stress conditions should be examined in the future, it is suggested that the absence of integrated tricin in lignins is unlikely to be a major detrimental factor for growth and development, at least in rice and maize. At the same time, the fnsII mutant exhibits a remarkably enhanced cell wall digestibility (Fig. 7). Considering that tricin actually takes up small portions of rice cell walls (Lan et al., 2016b), the improved enzymatic saccharification efficiency of the rice fnsII mutant could be attributed mainly to the reduced lignin levels. On the contrary, the tricin-depleted maize chs mutant showed a substantially reduced saccharification efficiency, which, in turn, was attributed to the increased lignin levels (Eloy et al., 2017). Taken together, lignin content, rather than an absence or modification of lignin-bound tricin units, is likely a major factor affecting the saccharification efficiency observed for the tricin-truncated mutant plants.
Overall, we envision that genetic manipulations of tricin biosynthesis could be an alternative strategy to engineer grass cell walls for efficient biomass conversion processes without severely compromising plant fitness. Given that the CYP93G members (FNSIIs) are highly conserved in Poaceae (Lam et al., 2014; Supplemental Fig. S7), there is a strong potential to extend the application to bioenergy grass crops such as sorghum, sugarcane, switchgrass, and bamboo. Meanwhile, further generation of transgenic rice plants with altered flavonoid compositions in lignin will facilitate the elucidation of the physiology functions and phylogeny of tricin-bound lignins in grasses.
MATERIALS AND METHODS
Plant Materials
The rice (Oryza sativa) T-DNA insertion mutant of CYP93G1 (accession K-00244; cv Kitaake) was obtained originally from the Crop Biotech Institute of Kyung Hee University. Rice seeds were surface sterilized, germinated, and grown in a phytotoron under a 12-h photoperiod and an ∼30°C day/∼24°C night temperature regime. Wild-type and fnsII homozygous mutant plants were isolated by a genomic PCR approach as described previously (Lam et al., 2014), and primers used for genotyping are listed in Supplemental Table S1. Mature plants (45 d after the heading) were used for phenotypic characterization, harvested, and dried in a temperature-controlled room (27°C for 30 d) prior to cell wall characterization.
Gene Expression Analysis
Total RNA was extracted individually from lignifying culms of rice plants at the heading stage as described previously (Koshiba et al., 2013b) and reverse transcribed into cDNA using random hexamer (Invitrogen) as a primer. The gene expression assay used the 7300 Real-Time PCR System (Applied Biosystems) and primer sets listed in Supplemental Table S1. A ubiquitin gene (OsUBQ5; AK061988) was used as an internal control. Microarray-based gene expression data for in silico gene expression analysis (Supplemental Fig. S1) were retrieved from the Rice Expression Profile Database (Sato et al., 2013).
Histochemical Analysis
Fresh hand-cut specimens (∼8 mm) were excised from culms at the heading stage, fixed in formaldehyde:propionic acid:ethanol at a ratio of 3.7:5:50 (v/v/v), treated with ethanol:acetic acid at a ratio of 6:1 (v/v) to remove extractives, and agarose embedded. Sections were sliced at 100 μm thickness using a DTK-2000 microslicer (Dosaka EM). For lignin staining using the phloroglucinol-HCl method, sections were incubated in 1% (w/v) phloroglucinol in ethanol for 10 min and acidified in 17.5 n HCl for 10 min. For flavonoid staining using the vanillin-HCl method, sections were incubated in 1% (w/v) vanillin in ethanol for 10 min followed by incubation in 17.5 n HCl for 10 min. The sections treated were then observed with an Olympus BX51 microscope (Olympus Optical).
Cell Wall Preparations
Extractive-free CWRs for chemical analysis and NMR were prepared as described previously (Yamamura et al., 2012). Briefly, dried rice plant tissues were pulverized with a TissueLyser (Qiagen), extracted sequentially with methanol, hexane, and distilled water, and then freeze dried to give CWRs. For NMR analysis, CWRs (∼300 mg) were further ball milled using the Planetary Micro Mill Pulverisette 7 (Fritsch) with ZrO2 vessels containing ZrO2 ball bearings (600 rpm, 12 cycles of 10 min at 5-min intervals; Mansfield et al., 2012; Tobimatsu et al., 2013). For whole cell wall NMR analysis, 60 mg of the ball-milled CWRs was directly swelled in 600 μL of DMSO-d6/pyridine-d5 (4:1, v/v). In parallel, ∼240 mg of the ball-milled CWRs was further digested with crude cellulases (Cellulysin; Calbiochem) according to methods described previously (Tobimatsu et al., 2013). The obtained lignin-enriched CWRs (∼40–60 mg) were dissolved in 600 µL of DMSO-d6/pyridine-d5 (4:1, v/v) and subjected to NMR analysis.
Chemical Analysis
Lignin content was estimated by the thioglycolic acid method (Suzuki et al., 2009). Analytical thioacidolysis was performed according to the method described previously (Yamamura et al., 2012), and the released lignin monomers were derivatized with N,O-bis(trimethylsilyl)acetamide and quantified by gas chromatography-mass spectrometry using 4,4′-ethylenebisphenol as an internal standard (Yue et al., 2012). Cell wall-bound pCA and FA were quantified using the methods described by Yamamura et al. (2011). The monosaccharide composition of the cell wall polysaccharides, excluding crystalline cellulose, was determined by hydrolyzing CWRs with trifluoroacetic acid and analyzing the released monosaccharides as alditol acetates by gas chromatography-mass spectrometry with inositol acetate as an internal standard (Chen et al., 2012). The crystalline cellulose content of the residue was determined by washing it with the Updegraff reagent (Updegraff, 1969) followed by a complete hydrolysis with 72% sulfuric acid (Hattori et al., 2012) and Glc quantified by the Glc CII test kit (Wako Pure Chemical Industries).
Generation of Synthetic Lignin Polymers
DHPs from coniferyl alcohol and naringenin were generated by the so-called bulk polymerization method (Tobimatsu et al., 2008, 2011). Briefly, 100 mL of acetone:sodium phosphate buffer (0.1 m, pH 6.5; 1:9, v/v) containing 0.5 mmol of coniferyl alcohol (for G-DHP) or 0.425 mmol of coniferyl alcohol together with 0.075 mmol of naringenin (for GN-DHP), along with 100 mL of hydrogen peroxide solution (0.6 mmol), were added separately to 25 mL of sodium phosphate buffer (pH 6.5) containing 5 mg of horseradish peroxidase (type IV; Sigma-Aldrich) over 1 h at room temperature. The solution was further stirred for 14 h, and the precipitates formed were collected by centrifugation (13,640g, 15 min), washed with distilled water (50 mL × 4), and lyophilized to afford G-DHP (∼53 mg, 59% weight yield) or GN-DHP (∼37 mg, 38% weight yield) as colorless powders. The DHPs (∼30 mg) were dissolved in 600 μL of DMSO-d6/pyridine-d5 (4:1, v/v) for NMR analysis.
Two-Dimensional NMR Analysis
NMR spectra were acquired on the Avance III 800US system (Bruker Biospin) equipped with a cryogenically cooled 5-mm TCI gradient probe. Adiabatic HSQC NMR experiments were carried out using standard implementation (hsqcgcep.3) with parameters described in the literature (Mansfield et al., 2012). Data processing and analysis used Bruker TopSpin 3.1 software (Bruker Biospin), and the central DMSO solvent peaks (δC/δH, 39.5/2.49 ppm) were used as an internal reference. HSQC plots were obtained with typical matched Gaussian apodization in F2 and squared cosine-bell apodization and one level of linear prediction (32 coefficients) in F1. For volume integration, linear prediction was turned off and no correction factors were used. For the integration of lignin and flavonoid aromatic signals (Fig. 5), C2-H2 correlations from G units (G) and C2-H2/C6-H6 correlations from S units (S), C2′-H2′/C6′-H6′ correlations from tricin (T), and C8-H8/C6-H6 correlations from naringenin (N) residues were used, and the S, T, and N integrals were logically halved. For integrations of lignin intermonomeric linkages (Fig. 6), well-resolved Cα-Hα contours from I, I′, II, II′, III, and III′ units and C3-H3 contours from N were integrated, and III, III′, and N integrals were logically halved. The relative contour intensities listed in Figures 5E and 6E are derived from three biological replicates and expressed on G + S = 100 and I + I′ + II + II′ + III + III′ = 100 bases, respectively.
Determination of Enzymatic Saccharification Efficiency
Enzymatic saccharification efficiency was determined essentially by the method described by Hattori et al. (2012). Briefly, CWRs were destarched and subjected to enzymatic hydrolysis with a cellulolytic enzyme cocktail composed of Celluclast 1.5 L, Novozyme 188, and Ultraflo L (Novozymes) in a sodium citrate buffer (pH 4.8). The Glc concentration at each incubation time point was determined by the Glc CII test kit (Wako Pure Chemical Industries). Cellulose content for the calculation of cellulose-to-Glc conversion was determined independently by the hydrolysis of destarched CWRs with sulfuric acid (Hattori et al., 2012).
Phylogenetic Analysis
The unrooted phylogenetic tree was constructed by the neighbor-joining method using MEGA6 (Tamura et al., 2013) with default parameters. Bootstrapping with 1,000 replications was performed.
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession number AK100972 (OsFNSII, LOC_Os04g01140). Accession numbers for the sequences used in the phylogenetic analysis are shown in the tree or in the legend of Supplemental Figure S7.
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Gene expression data of flavonoid and monolignol biosynthetic genes in wild-type rice plants.
Supplemental Figure S2. Relative expression levels of flavonoid and monolignol biosynthetic genes in fnsII mutant culms.
Supplemental Figure S3. Thioacidolysis yield per thioglycolic lignin content in wild-type and fnsII mutant rice tissues.
Supplemental Figure S4. Cell wall-bound p-coumarate content per thioglycolic lignin content in wild-type and fnsII mutant rice tissues.
Supplemental Figure S5. Sugar composition in wild-type and fnsII mutant rice tissues.
Supplemental Figure S6. HSQC NMR spectra of whole culm cell walls from wild-type and fnsII mutant rice.
Supplemental Figure S7. Phylogenetic tree of CYP93 proteins.
Supplemental Table S1. Primers used in this study.
Supplementary Material
Acknowledgments
We thank Naoyuki Matsumoto, Keiko Tsuchida, and Megumi Ozaki for assisting in the analysis of rice cell walls; Dr. Hironori Kaji and Ayaka Maeno for assistance in NMR analysis; Drs. Arata Yoshinaga, Keiji Takabe, Tomoya Imai, and Junji Sugiyama for assistance and helpful suggestions for histochemical analysis. A part of this study was conducted using the facilities in the DASH/FBAS at the Research Institute for Sustainable Humanosphere, Kyoto University, and the NMR spectrometer in the JURC at the Institute for Chemical Research, Kyoto University.
Glossary
- G
guaiacyl
- S
syringyl
- H
hydroxyphenyl
- CWR
cell wall residue
- FA
ferulate
- DMSO
dimethyl sulfoxide
- HSQC
heteronuclear single-quantum coherence
Footnotes
This work was supported by the Research Grants Council of Hong Kong, China (grant nos. 17114514 and 17123315), the Japan Society for the Promotion of Science (Grants-in-Aid for Scientific Research nos. 16K14958 and 16H06198), the Japan Science and Technology Agency/Japan International Cooperation Agency (Science and Technology Research Partnership for Sustainable Development), and the Research Institute for Sustainable Humanosphere, Kyoto University (grant no. 2016-5-2-1).
Articles can be viewed without a subscription.
References
- Abdulrazzak N, Pollet B, Ehlting J, Larsen K, Asnaghi C, Ronseau S, Proux C, Erhardt M, Seltzer V, Renou JP, et al. (2006) A coumaroyl-ester-3-hydroxylase insertion mutant reveals the existence of nonredundant meta-hydroxylation pathways and essential roles for phenolic precursors in cell expansion and plant growth. Plant Physiol 140: 30–48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agati G, Azzarello E, Pollastri S, Tattini M (2012) Flavonoids as antioxidants in plants: location and functional significance. Plant Sci 196: 67–76 [DOI] [PubMed] [Google Scholar]
- Anderson NA, Tobimatsu Y, Ciesielski PN, Ximenes E, Ralph J, Donohoe BS, Ladisch M, Chapple C (2015) Manipulation of guaiacyl and syringyl monomer biosynthesis in an Arabidopsis cinnamyl alcohol dehydrogenase mutant results in atypical lignin biosynthesis and modified cell wall structure. Plant Cell 27: 2195–2209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barros J, Serrani-Yarce JC, Chen F, Baxter D, Venables BJ, Dixon RA (2016) Role of bifunctional ammonia-lyase in grass cell wall biosynthesis. Nat Plants 2: 16050. [DOI] [PubMed] [Google Scholar]
- Beckham GT, Johnson CW, Karp EM, Salvachúa D, Vardon DR (2016) Opportunities and challenges in biological lignin valorization. Curr Opin Biotechnol 42: 40–53 [DOI] [PubMed] [Google Scholar]
- Besseau S, Hoffmann L, Geoffroy P, Lapierre C, Pollet B, Legrand M (2007) Flavonoid accumulation in Arabidopsis repressed in lignin synthesis affects auxin transport and plant growth. Plant Cell 19: 148–162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boerjan W, Ralph J, Baucher M (2003) Lignin biosynthesis. Annu Rev Plant Biol 54: 519–546 [DOI] [PubMed] [Google Scholar]
- Bonawitz ND, Chapple C (2010) The genetics of lignin biosynthesis: connecting genotype to phenotype. Annu Rev Genet 44: 337–363 [DOI] [PubMed] [Google Scholar]
- Bonawitz ND, Chapple C (2013) Can genetic engineering of lignin deposition be accomplished without an unacceptable yield penalty? Curr Opin Biotechnol 24: 336–343 [DOI] [PubMed] [Google Scholar]
- Bouvier d’Yvoire M, Bouchabke-Coussa O, Voorend W, Antelme S, Cézard L, Legée F, Lebris P, Legay S, Whitehead C, McQueen-Mason SJ, et al. (2013) Disrupting the cinnamyl alcohol dehydrogenase 1 gene (BdCAD1) leads to altered lignification and improved saccharification in Brachypodium distachyon. Plant J 73: 496–508 [DOI] [PubMed] [Google Scholar]
- Brennan M, McLean JP, Altaner C, Ralph J, Harris PJ (2012) Cellulose microbril angles and cell-wall polymers in different wood types of Pinus radiata. Cellulose 19: 1385–1404 [Google Scholar]
- Cardona ML, Garcia B, Pedro JR, Sinisterra JF (1990) Flavonoids, flavonolignans and a phenylpropanoid from Onopordon corymbosum. Phytochemistry 29: 629–631 [Google Scholar]
- Cass CL, Peraldi A, Dowd PF, Mottiar Y, Santoro N, Karlen SD, Bukhman YV, Foster CE, Thrower N, Bruno LC, et al. (2015) Effects of PHENYLALANINE AMMONIA LYASE (PAL) knockdown on cell wall composition, biomass digestibility, and biotic and abiotic stress responses in Brachypodium. J Exp Bot 66: 4317–4335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen F, Dixon RA (2007) Lignin modification improves fermentable sugar yields for biofuel production. Nat Biotechnol 25: 759–761 [DOI] [PubMed] [Google Scholar]
- Chen F, Tobimatsu Y, Havkin-Frenkel D, Dixon RA, Ralph J (2012) A polymer of caffeyl alcohol in plant seeds. Proc Natl Acad Sci USA 109: 1772–1777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen F, Tobimatsu Y, Jackson L, Nakashima J, Ralph J, Dixon RA (2013) Novel seed coat lignins in the Cactaceae: structure, distribution and implications for the evolution of lignin diversity. Plant J 73: 201–211 [DOI] [PubMed] [Google Scholar]
- Del Rio JC, Lino AG, Colodette JL, Lima CF, Gutiérrez A, Martínez ÁT, Lu F, Ralph J, Rencoret J (2015) Differences in the chemical structure of the lignins from sugarcane bagasse and straw. Biomass Bioenergy 81: 322–338 [Google Scholar]
- Del Río JC, Rencoret J, Prinsen P, Martínez ÁT, Ralph J, Gutiérrez A (2012) Structural characterization of wheat straw lignin as revealed by analytical pyrolysis, 2D-NMR, and reductive cleavage methods. J Agric Food Chem 60: 5922–5935 [DOI] [PubMed] [Google Scholar]
- Dixon RA, Achnine L, Kota P, Liu CJ, Reddy MSS, Wang L (2002) The phenylpropanoid pathway and plant defence: a genomics perspective. Mol Plant Pathol 3: 371–390 [DOI] [PubMed] [Google Scholar]
- Dong X, Chen W, Wang W, Zhang H, Liu X, Luo J (2014) Comprehensive profiling and natural variation of flavonoids in rice. J Integr Plant Biol 56: 876–886 [DOI] [PubMed] [Google Scholar]
- Du Y, Chu H, Chu IK, Lo C (2010a) CYP93G2 is a flavanone 2-hydroxylase required for C-glycosylflavone biosynthesis in rice. Plant Physiol 154: 324–333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du Y, Chu H, Wang M, Chu IK, Lo C (2010b) Identification of flavone phytoalexins and a pathogen-inducible flavone synthase II gene (SbFNSII) in sorghum. J Exp Bot 61: 983–994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eloy NB, Voorend W, Lan W, Saleme MdLS, Cesarino I, Vanholme R, Smith RA, Goeminne G, Pallidis A, Morreel K, Nicomedes JJr, Ralph J, Boerjan W (2016) Silencing chalcone synthase impedes the incorporation of tricin in lignin and increases lignin content. Plant Physiol 173: 998–1016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elumalai S, Tobimatsu Y, Grabber JH, Pan X, Ralph J (2012) Epigallocatechin gallate incorporation into lignin enhances the alkaline delignification and enzymatic saccharification of cell walls. Biotechnol Biofuels 5: 59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foo LY, Karchesy J (1989) Pseudotsuganol, a biphenyl-linked pinoresinol-dihydroquercetin from Douglas-fir bark: isolation of the first true flavonolignan. J Chem Soc Chem Commun 217–219 [Google Scholar]
- Fornalé S, Rencoret J, Garcia-Calvo L, Capellades M, Encina A, Santiago R, Rigau J, Gutiérrez A, Del Río JC, Caparros-Ruiz D (2015) Cell wall modifications triggered by the down-regulation of coumarate 3-hydroxylase-1 in maize. Plant Sci 236: 272–282 [DOI] [PubMed] [Google Scholar]
- Fornalé S, Shi X, Chai C, Encina A, Irar S, Capellades M, Fuguet E, Torres JL, Rovira P, Puigdomènech P, et al. (2010) ZmMYB31 directly represses maize lignin genes and redirects the phenylpropanoid metabolic flux. Plant J 64: 633–644 [DOI] [PubMed] [Google Scholar]
- Gardner RO. (1975) Vanillin-hydrochloric acid as a histochemical test for tannin. Stain Technol 50: 315–317 [DOI] [PubMed] [Google Scholar]
- Goto T, Kondo T (1991) Structure and molecular stacking of anthocyanins: flower color variation. Angew Chem Int Ed Engl 30: 17–33 [Google Scholar]
- Grabber JH, Quideau S, Ralph J (1996) p-Coumaroylated syringyl units in maize lignin: implications for β-ether cleavage by thioacidolysis. Phytochemistry 43: 1189–1194 [Google Scholar]
- Grabber JH, Ress D, Ralph J (2012) Identifying new lignin bioengineering targets: impact of epicatechin, quercetin glycoside, and gallate derivatives on the lignification and fermentation of maize cell walls. J Agric Food Chem 60: 5152–5160 [DOI] [PubMed] [Google Scholar]
- Gui J, Shen J, Li L (2011) Functional characterization of evolutionarily divergent 4-coumarate:coenzyme A ligases in rice. Plant Physiol 157: 574–586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hassan S, Mathesius U (2012) The role of flavonoids in root-rhizosphere signalling: opportunities and challenges for improving plant-microbe interactions. J Exp Bot 63: 3429–3444 [DOI] [PubMed] [Google Scholar]
- Hattori T, Murakami S, Mukai M, Yamada T, Hirochika H, Ike M, Tokuyasu K, Suzuki S, Sakamoto M, Umezawa T (2012) Rapid analysis of transgenic rice straw using near-infrared spectroscopy. Plant Biotechnol 29: 359–366 [Google Scholar]
- Hong L, Qian Q, Tang D, Wang K, Li M, Cheng Z (2012) A mutation in the rice chalcone isomerase gene causes the golden hull and internode 1 phenotype. Planta 236: 141–151 [DOI] [PubMed] [Google Scholar]
- Itoh N, Katsube Y, Yamamoto K, Nakajima N, Yoshida K (2007) Laccase-catalyzed conversion of green tea catechins in the presence of gallic acid to epitheaflagallin and epitheaflagallin 3-O-gallate. Tetrahedron 63: 9488–9492 [Google Scholar]
- Jouanin L, Goujon T, de Nadaï V, Martin MT, Mila I, Vallet C, Pollet B, Yoshinaga A, Chabbert B, Petit-Conil M, et al. (2000) Lignification in transgenic poplars with extremely reduced caffeic acid O-methyltransferase activity. Plant Physiol 123: 1363–1374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karlen SD, Zhang C, Peck ML, Smith RA, Padmakshan D, Helmich KE, Free HCA, Lee S, Smith BG, Lu F, et al. (2016) Monolignol ferulate conjugates are naturally incorporated into plant lignins. Sci Adv 2: e1600393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim BG, Lee Y, Hur HG, Lim Y, Ahn JH (2006) Flavonoid 3′-O-methyltransferase from rice: cDNA cloning, characterization and functional expression. Phytochemistry 67: 387–394 [DOI] [PubMed] [Google Scholar]
- Kim H, Ralph J (2010) Solution-state 2D NMR of ball-milled plant cell wall gels in DMSO-d6/pyridine-d5. Org Biomol Chem 8: 576–591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H, Ralph J (2014) A gel-state 2D-NMR method for plant cell wall profiling and analysis: a model study with the amorphous cellulose and xylan from ball-milled cotton linters. RSC Advances 4: 7549–7560 [Google Scholar]
- Kim H, Ralph J, Yahiaoui N, Pean M, Boudet AM (2000) Cross-coupling of hydroxycinnamyl aldehydes into lignins. Org Lett 2: 2197–2200 [DOI] [PubMed] [Google Scholar]
- Kim NC, Graf TN, Sparacino CM, Wani MC, Wall ME (2003) Complete isolation and characterization of silybins and isosilybins from milk thistle (Silybum marianum). Org Biomol Chem 1: 1684–1689 [DOI] [PubMed] [Google Scholar]
- Koes RE, Quattrocchio F, Mol JNM (1994) The flavonoid biosynthetic pathway in plants: function and evolution. BioEssays 16: 123–132 [Google Scholar]
- Koshiba T, Hirose N, Mukai M, Yamamura M, Hattori T, Suzuki S, Sakamoto M, Umezawa T (2013a) Characterization of 5-hydroxyconiferaldehyde O-methyltransferase in Oryza sativa. Plant Biotechnol 30: 157–167 [Google Scholar]
- Koshiba T, Murakami S, Hattori T, Mukai M, Takahashi A, Miyao A, Hirochika H, Suzuki S, Sakamoto M, Umezawa T (2013b) CAD2 deficiency causes both brown midrib and gold hull and internode phenotypes in Oryza sativa L. cv. Nipponbare. Plant Biotechnol 30: 365–373 [Google Scholar]
- Koshiba T, Yamamoto N, Tobimatsu Y, Yamamura M, Suzuki S, Hattori T, Mukai M, Noda S, Shibata D, Sakamoto M, et al. (2017) MYB-mediated upregulation of lignin biosynthesis in Oryza sativa towards biomass refinery. Plant Biotechnol 34: 7–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam PY, Liu H, Lo C (2015) Completion of tricin biosynthesis pathway in rice: cytochrome P450 75B4 is a unique chrysoeriol 5′-hydroxylase. Plant Physiol 168: 1527–1536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam PY, Zhu FY, Chan WL, Liu H, Lo C (2014) Cytochrome P450 93G1 is a flavone synthase II that channels flavanones to the biosynthesis of tricin O-linked conjugates in rice. Plant Physiol 165: 1315–1327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lan W, Lu F, Regner M, Zhu Y, Rencoret J, Ralph SA, Zakai UI, Morreel K, Boerjan W, Ralph J (2015) Tricin, a flavonoid monomer in monocot lignification. Plant Physiol 167: 1284–1295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lan W, Morreel K, Lu F, Rencoret J, Del Río JC, Voorend W, Vermerris W, Boerjan W, Ralph J (2016a) Maize tricin-oligolignol metabolites and their implications for monocot lignification. Plant Physiol 171: 810–820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lan W, Rencoret J, Lu F, Karlen SD, Smith BG, Harris PJ, Del Río JC, Ralph J (2016b) Tricin-lignins: occurrence and quantitation of tricin in relation to phylogeny. Plant J 88: 1046–1057 [DOI] [PubMed] [Google Scholar]
- Lapierre C, Monties B, Roland C (1986) Preparative thioacidolysis of spruce lignin: isolation and identification of main monomeric products. Holzforschung 40: 47–50 [Google Scholar]
- Li M, Pu Y, Yoo CG, Ragauskas AJ (2016) The occurrence of tricin and its derivatives in plants. Green Chem 18: 1439–1454 [Google Scholar]
- Li X, Bonawitz ND, Weng JK, Chapple C (2010) The growth reduction associated with repressed lignin biosynthesis in Arabidopsis thaliana is independent of flavonoids. Plant Cell 22: 1620–1632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mansfield SD, Kim H, Lu F, Ralph J (2012) Whole plant cell wall characterization using solution-state 2D NMR. Nat Protoc 7: 1579–1589 [DOI] [PubMed] [Google Scholar]
- Marita JM, Vermerris W, Ralph J, Hatfield RD (2003) Variations in the cell wall composition of maize brown midrib mutants. J Agric Food Chem 51: 1313–1321 [DOI] [PubMed] [Google Scholar]
- Nyiredy S, Samu Z, Szücs Z, Gulácsi K, Kurtán T, Antus S (2008) New insight into the biosynthesis of flavanolignans in the white-flowered variant of Silybum marianum. J Chromatogr Sci 46: 93–96 [DOI] [PubMed] [Google Scholar]
- Okamura H, Nishimura H, Nagata T, Kigawa T, Watanabe T, Katahira M (2016) Accurate and molecular-size-tolerant NMR quantitation of diverse components in solution. Sci Rep 6: 21742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parthasarathy MR, Ranganathan KR, Sharma D (1979) 13C NMR of flavonolignans from Hydnocarpus wightiana. Phytochemistry 18: 506–508 [Google Scholar]
- Petrik DL, Karlen SD, Cass CL, Padmakshan D, Lu F, Liu S, Le Bris P, Antelme S, Santoro N, Wilkerson CG, et al. (2014) p-Coumaroyl-CoA:monolignol transferase (PMT) acts specifically in the lignin biosynthetic pathway in Brachypodium distachyon. Plant J 77: 713–726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pettit GR, Meng Y, Stevenson CA, Doubek DL, Knight JC, Cichacz Z, Pettit RK, Chapuis JC, Schmidt JM (2003) Isolation and structure of palstatin from the Amazon tree Hymeneae palustris. J Nat Prod 66: 259–262 [DOI] [PubMed] [Google Scholar]
- Ragauskas AJ, Beckham GT, Biddy MJ, Chandra R, Chen F, Davis MF, Davison BH, Dixon RA, Gilna P, Keller M, et al. (2014) Lignin valorization: improving lignin processing in the biorefinery. Science 344: 1246843. [DOI] [PubMed] [Google Scholar]
- Ralph J. (2010) Hydroxycinnamates in lignification. Phytochem Rev 9: 65–83 [Google Scholar]
- Ralph J, Kim H, Lu F, Grabber JH, Leplé JC, Berrio-Sierra J, Derikvand MM, Jouanin L, Boerjan W, Lapierre C (2008) Identification of the structure and origin of a thioacidolysis marker compound for ferulic acid incorporation into angiosperm lignins (and an indicator for cinnamoyl CoA reductase deficiency). Plant J 53: 368–379 [DOI] [PubMed] [Google Scholar]
- Ralph J, Lapierre C, Lu F, Marita JM, Pilate G, Van Doorsselaere J, Boerjan W, Jouanin L (2001) NMR evidence for benzodioxane structures resulting from incorporation of 5-hydroxyconiferyl alcohol into lignins of O-methyltransferase-deficient poplars. J Agric Food Chem 49: 86–91 [DOI] [PubMed] [Google Scholar]
- Rencoret J, Ralph J, Marques G, Gutiérrez A, Martínez Á, del Río JC (2013) Structural characterization of lignin isolated from coconut (Cocos nucifera) coir fibers. J Agric Food Chem 61: 2434–2445 [DOI] [PubMed] [Google Scholar]
- Rinaldi R, Jastrzebski R, Clough MT, Ralph J, Kennema M, Bruijnincx PCA, Weckhuysen BM (2016) Paving the way for lignin valorisation: recent advances in bioengineering, biorefining and catalysis. Angew Chem Int Ed Engl 55: 8164–8215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato Y, Takehisa H, Kamatsuki K, Minami H, Namiki N, Ikawa H, Ohyanagi H, Sugimoto K, Antonio BA, Nagamura Y (2013) RiceXPro version 3.0: expanding the informatics resource for rice transcriptome. Nucleic Acids Res 41: D1206–D1213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma DK, Ranganathan KR, Parthasarathy MR, Bhushan B, Seshadri TR (1979) Flavonolignans from Hydnocarpus wightiana. Planta Med 37: 79–83 [Google Scholar]
- Shih CH, Chu H, Tang LK, Sakamoto W, Maekawa M, Chu IK, Wang M, Lo C (2008) Functional characterization of key structural genes in rice flavonoid biosynthesis. Planta 228: 1043–1054 [DOI] [PubMed] [Google Scholar]
- Sibout R, Eudes A, Mouille G, Pollet B, Lapierre C, Jouanin L, Séguin A (2005) CINNAMYL ALCOHOL DEHYDROGENASE-C and -D are the primary genes involved in lignin biosynthesis in the floral stem of Arabidopsis. Plant Cell 17: 2059–2076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki S, Suzuki Y, Yamamoto N, Hattori T, Sakamoto M, Umezawa T (2009) High-throughput determination of thioglycolic acid lignin from rice. Plant Biotechnol 26: 337–340 [Google Scholar]
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S (2013) MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol 30: 2725–2729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tobimatsu Y, Chen F, Nakashima J, Escamilla-Treviño LL, Jackson L, Dixon RA, Ralph J (2013) Coexistence but independent biosynthesis of catechyl and guaiacyl/syringyl lignin polymers in seed coats. Plant Cell 25: 2587–2600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tobimatsu Y, Davidson CL, Grabber JH, Ralph J (2011) Fluorescence-tagged monolignols: synthesis, and application to studying in vitro lignification. Biomacromolecules 12: 1752–1761 [DOI] [PubMed] [Google Scholar]
- Tobimatsu Y, Takano T, Kamitakahara H, Nakatsubo F (2008) Studies on the dehydrogenative polymerizations of monolignol β-glycosides. Part 3. Horseradish peroxidase-catalyzed polymerizations of triandrin and isosyringin. J Wood Chem Technol 28: 69–83 [Google Scholar]
- Umezawa T. (2010) The cinnamate/monolignol pathway. Phytochem Rev 9: 1–17 [Google Scholar]
- Updegraff DM. (1969) Semimicro determination of cellulose in biological materials. Anal Biochem 32: 420–424 [DOI] [PubMed] [Google Scholar]
- Vanholme R, Ralph J, Akiyama T, Lu F, Pazo JR, Kim H, Christensen JH, Van Reusel B, Storme V, De Rycke R, et al. (2010) Engineering traditional monolignols out of lignin by concomitant up-regulation of F5H1 and down-regulation of COMT in Arabidopsis. Plant J 64: 885–897 [DOI] [PubMed] [Google Scholar]
- Vanholme R, Storme V, Vanholme B, Sundin L, Christensen JH, Goeminne G, Halpin C, Rohde A, Morreel K, Boerjan W (2012) A systems biology view of responses to lignin biosynthesis perturbations in Arabidopsis. Plant Cell 24: 3506–3529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogt T. (2010) Phenylpropanoid biosynthesis. Mol Plant 3: 2–20 [DOI] [PubMed] [Google Scholar]
- Wagner A, Tobimatsu Y, Goeminne G, Phillips L, Flint H, Steward D, Torr K, Donaldson L, Boerjan W, Ralph J (2013) Suppression of CCR impacts metabolite profile and cell wall composition in Pinus radiata tracheary elements. Plant Mol Biol 81: 105–117 [DOI] [PubMed] [Google Scholar]
- Wagner A, Tobimatsu Y, Phillips L, Flint H, Torr K, Donaldson L, Pears L, Ralph J (2011) CCoAOMT suppression modifies lignin composition in Pinus radiata. Plant J 67: 119–129 [DOI] [PubMed] [Google Scholar]
- Wang K, Zhang H, Shen L, Du Q, Li J (2010) Rapid separation and characterization of active flavonolignans of Silybum marianum by ultra-performance liquid chromatography coupled with electrospray tandem mass spectrometry. J Pharm Biomed Anal 53: 1053–1057 [DOI] [PubMed] [Google Scholar]
- Wen JL, Sun SL, Xue BL, Sun RC (2013) Quantitative structural characterization of the lignins from the stem and pith of bamboo (Phyllostachys pubescens). Holzforschung 67: 613–627 [Google Scholar]
- Weng JK, Mo H, Chapple C (2010) Over-expression of F5H in COMT-deficient Arabidopsis leads to enrichment of an unusual lignin and disruption of pollen wall formation. Plant J 64: 898–911 [DOI] [PubMed] [Google Scholar]
- Yamamura M, Hattori T, Suzuki S, Shibata D, Umezawa T (2012) Microscale thioacidolysis method for the rapid analysis of β-O-4 substructures in lignin. Plant Biotechnol 29: 419–423 [Google Scholar]
- Yamamura M, Wada S, Sakakibara N, Nakatsubo T, Suzuki S, Hattori T, Takeda M, Sakurai N, Suzuki H, Shibata D, et al. (2011) Occurrence of guaiacyl/p-hydroxyphenyl lignin in Arabidopsis thaliana T87 cells. Plant Biotechnol 28: 1–8 [Google Scholar]
- Yang Z, Nakabayashi R, Okazaki Y, Mori T, Takamatsu S, Kitanaka S, Kikuchi J, Saito K (2014) Toward better annotation in plant metabolomics: isolation and structure elucidation of 36 specialized metabolites from Oryza sativa (rice) by using MS/MS and NMR analyses. Metabolomics 10: 543–555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yue F, Lu F, Sun RC, Ralph J (2012) Syntheses of lignin-derived thioacidolysis monomers and their uses as quantitation standards. J Agric Food Chem 60: 922–928 [DOI] [PubMed] [Google Scholar]
- Zhang K, Qian Q, Huang Z, Wang Y, Li M, Hong L, Zeng D, Gu M, Chu C, Cheng Z (2006) GOLD HULL AND INTERNODE2 encodes a primarily multifunctional cinnamyl-alcohol dehydrogenase in rice. Plant Physiol 140: 972–983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Q, Tobimatsu Y, Zhou R, Pattathil S, Gallego-Giraldo L, Fu C, Jackson LA, Hahn MG, Kim H, Chen F, et al. (2013) Loss of function of cinnamyl alcohol dehydrogenase 1 leads to unconventional lignin and a temperature-sensitive growth defect in Medicago truncatula. Proc Natl Acad Sci USA 110: 13660–13665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou JM, Ibrahim RK (2010) Tricin: a potential multifunctional nutraceutical. Phytochem Rev 9: 413–424 [Google Scholar]
- Zuk M, Działo M, Richter D, Dymińska L, Matuła J, Kotecki A, Hanuza J, Szopa J (2016) Chalcone Synthase (CHS) gene suppression in flax leads to changes in wall synthesis and sensing genes, cell wall chemistry and stem morphology parameters. Front Plant Sci 7: 894. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.