A series of metabolic and transport genes involved in the nicotine pathway form a regulon under the control of jasmonate-responsive transcription factors in tobacco.
Abstract
In tobacco (Nicotiana tabacum), nicotine is the predominant alkaloid. It is produced in the roots and accumulated mainly in the leaves. Jasmonates play a central signaling role in damage-induced nicotine formation. The genome sequence of tobacco provides us an almost complete inventory of structural and regulatory genes involved in nicotine pathway. Phylogenetic and expression analyses revealed a series of structural genes of the nicotine pathway, forming a regulon, under the control of jasmonate-responsive ETHYLENE RESPONSE FACTOR (ERF) transcription factors. The duplication of NAD and polyamine metabolic pathways and the subsequent recruitment of duplicated primary metabolic genes into the nicotine biosynthesis regulon were suggested to be the drivers for pyridine and pyrrolidine ring formation steps early in the pathway. Transcriptional regulation by ERF and cooperatively acting MYC2 transcription factors are corroborated by the frequent occurrence of cognate cis-regulatory elements of the factors in the promoter regions of the downstream structural genes. The allotetraploid tobacco has homologous clusters of ERF genes on different chromosomes, which are possibly derived from two ancestral diploids and include either nicotine-controlling ERF189 or ERF199. A large chromosomal deletion was found within one allele of the nicotine-controlling NICOTINE2 locus, which is part of one of the ERF gene clusters, and which has been used to breed tobacco cultivars with a low-nicotine content.
In plants, a large number of structurally diverse specialized metabolites are produced through long, multistep, and often branched pathways (Arimura and Maffei, 2017). The proper functioning of such pathways, allowing massive metabolic flows leading to complex products from simple precursors, largely relies on the concerted expression of a large set of metabolic and transport genes, or structural genes, in different developmental and environmental contexts. The transcription factors regulating these pathways play a critical role in such coordination, which often occurs at the transcription level. The regulatory transcription factors and downstream structural genes, which form regulatory networks of multiple genes, or regulons, have begun to be explored intensively through molecular and genomics studies (De Geyter et al., 2012; Patra et al., 2013).
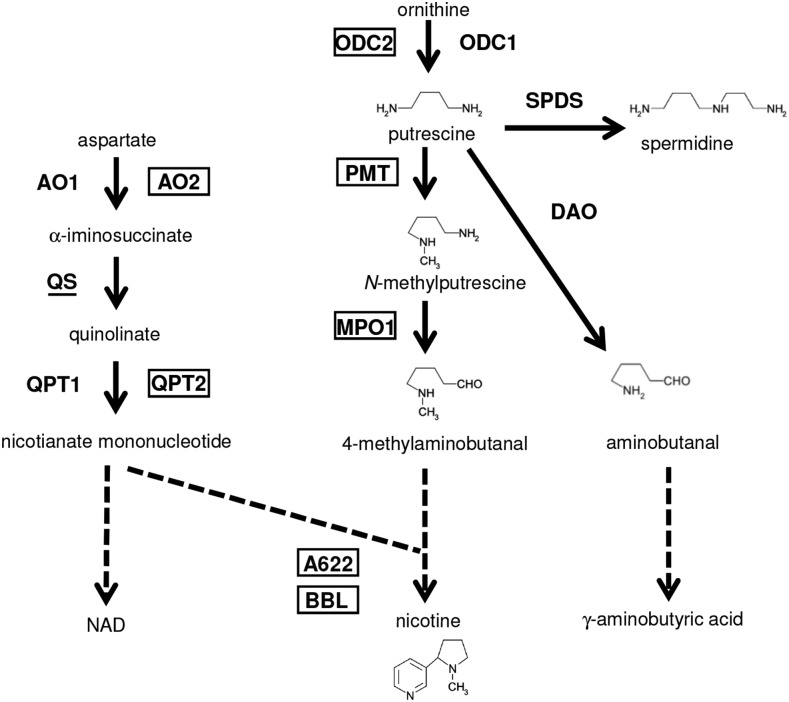
Nicotiana tabacum, hereafter called tobacco, is cultivated as an economically important crop around the globe (Davis and Nielsen, 1999). The main cultivated species, tobacco, is a natural allotetraploid possibly derived through the hybridization between two ancestral diploids that are closely related to current N. sylvestris and N. tomentosiformis (Murad et al., 2002; Sierro et al., 2013; Wang and Bennetzen, 2015). In tobacco, nicotine is an abundant predominant alkaloid produced in the roots and accumulating mainly in the leaves (Shoji and Hashimoto, 2011a; Dewey and Xie, 2013). As a defense toxin, nicotine production is drastically increased in response to damage caused by grazing herbivores (Baldwin, 1989), and jasmonates play a central signaling role in the damage-induced nicotine biosynthesis (Baldwin et al., 1994; Shoji et al., 2000, 2008). Nicotine has heterocyclic pyridine and pyrrolidine rings (Shoji and Hashimoto, 2011a; Dewey and Xie, 2013); the pyrrolidine ring is formed through consecutive reactions catalyzed by Orn decarboxylase (ODC; Imanishi et al., 1998; DeBoer et al., 2011a), putrescine N-methyltransferase (PMT; Hibi et al., 1994), and N-methylputrescine oxidase (MPO; Heim et al., 2007; Katoh et al., 2007), whereas enzymes involved in early steps of NAD synthesis, Asp oxidase (AO), quinolinate synthase (QS), and quinolinate phosphoribosyl transferase (QPT) are responsible for the formation of the pyridine ring (Sinclair et al., 2000; Katoh et al., 2006; Fig. 1). It has been proposed that PMT and MPO have evolved from spermidine synthase (SPDS) and diamine oxidase (DAO), two homologous enzymes with different catalytic activities, both of which accept putrescine as a substrate and thus are involved in polyamine metabolism (Hibi et al., 1994; Hashimoto et al., 1998; Junker et al., 2013; Naconsie et al., 2014). Two orphan oxidoreductases of different families, A622 (Hibi et al., 1994; DeBoer et al., 2009; Kajikawa et al., 2009) and berberine bridge enzyme-like (BBL) proteins (Kajikawa et al., 2011), are required for later steps including coupling of the two rings (Fig. 1), but the exact biochemical reactions catalyzed by these enzymes have yet to be determined. In tobacco roots, a pair of tonoplast-localized multidrug and toxic compound extrusion (MATE) family transporters, MATE1 and MATE2 (Shoji et al., 2009), mediate vacuolar sequestration of nicotine, whereas nicotine and vitamin B6, both with a pyridine ring, are imported into the cells by a purine permease-like transporter, nicotine uptake permease 1 (NUP1), localized at plasma membranes (Hildreth et al., 2011; Kato et al., 2014). In addition to the transport function, NUP1 was proposed to be involved in the regulation of root growth and nicotine biosynthesis, and thus may have a regulatory role as well (Hildreth et al., 2011; Kato et al., 2015).
Figure 1.
Pathways of nicotine, NAD, and polyamine metabolism in tobacco. Each defined enzymatic step is represented by an arrow and enzyme name, whereas undefined single or multiple step processes are represented by broken arrows. Boxes denote enzymes hypothesized to be involved predominantly in the nicotine biosynthesis pathway, whereas enzymes predominantly associated with related primary metabolic pathways are not framed. QS is underlined as it contributes to both nicotine and NAD pathways.
Nicotine contents in tobacco plants are genetically controlled by two distinct loci NICOTINE1 (NIC1) and NIC2, and their mutant alleles nic1 and nic2 have been used to breed a low-nicotine tobacco cultivars (Legg and Collins, 1971; Chaplin, 1975; Hibi et al., 1994). A small group of genes encoding closely related ETHYLENE RESPONSE FACTOR (ERF) transcription factors, which are in a clade within group IXa subfamily (Nakano et al., 2006), are clustered at NIC2 locus, and at least seven such genes (ERF17, ERF104ΔC, ERF115, ERF168, ERF179, ERF189, and ERF221), called NIC2-locus ERFs, were found to be deleted in the nic2 mutant (Shoji et al., 2010). In tobacco, all the NIC2-locus ERFs and their homologs are induced by jasmonates (Shoji et al., 2010), whereas salt stress induces the expression of most of the ERFs but not ERF189 and its closest homolog ERF199 (Shoji and Hashimoto, 2015). As master transcription factors regulating the pathway, jasmonate-inducible ERF189 and ERF199 directly up-regulate a nearly complete set of genes involved in nicotine biosynthesis and transport (all the genes mentioned above except NUP1) by recognizing GC-rich P box elements, which resemble, but differ from a typical GCC box, in the promoters of the downstream genes (Shoji and Hashimoto, 2011b, 2011c, 2012, 2013; Shoji et al., 2010, 2013). Interestingly, ORCA3 from Catharanthus roseus (van der Fits and Memelink, 2000) and JRE4/GAME9 from tomato (Solanum lycopersicum) and potato (Solanum tuberosum; Cárdenas et al., 2016; Thagun et al., 2016) are homologs of tobacco ERF189, and also regulate jasmonate-inducible defense metabolism, a part of indole alkaloid pathway and a nearly complete pathway for steroidal glycoalkaloid biosynthesis, respectively. Moreover, JRE4/GAME9 is in a cluster of related ERF genes in tomato and potato genomes, just like the NIC2-locus genes in tobacco (Cárdenas et al., 2016; Thagun et al., 2016). Through interaction with the nicotine-regulating ERF factors, a bHLH-family transcription factor MYC2, a key component in conserved jasmonate signaling (Goossens et al., 2016), positively regulates the nicotine pathway genes by directly binding to G box elements found in their promoters, as well as in way of the ERF genes (DeBoer et al., 2011b; Shoji and Hashimoto, 2011c; Zhang et al., 2012).
Genomics has greatly facilitated our understanding on specialized metabolism in plants. The genome sequence of tobacco (Sierro et al., 2014) allows us to characterize a whole range of features of the entire suite of genes involved in nicotine biosynthesis and related NAD and polyamine metabolism, such as fine molecular phylogenies, genomic arrangements, cis-element distributions in the promoters, and expression of individual genes. Here, we discuss the evolution of this specialized metabolic pathway specific to the Nicotiana lineage, with particular focus on gene regulatory aspects.
RESULTS
Genes in the Tobacco Genome Involved in Nicotine and Related Pathways
The genes encoding metabolic enzymes and transporters involved in nicotine and related primary metabolism (Fig. 1) were retrieved from tobacco genome sequence of TN90 cultivar (Sierro et al., 2014) using the BLAST functionality at the SOL Genomic Network (https://solgenomics.net/tools/blast/; Supplemental Table S1). To investigate the phylogenetic relationships, the tobacco enzymes, except A622 and BBL, were aligned with their homologs from N. sylvestris, N. tomentosiformis, tomato, pepper (Capsicum annuum), and Arabidopsis (Arabidopsis thaliana; Supplemental Fig. S1). In the phylogenetic trees, most proteins have two copies in tobacco, which group with their counterparts in ancestral diploids, forming a homeologous group of orthologs. It is possible to divide the proteins in each tree into multiple such orthologous groups. According to the groupings and ancestral origins of their products, the tobacco genes were, to our knowledge, newly named in this study (Supplemental Table S1; Supplemental Fig. S1), if not identical to already named ones. For A622 (Kajikawa et al., 2009), MATE (Shoji et al., 2009), and NUP (Hildreth et al., 2011) genes, orthologous genes (greater than 98% identity at the nucleotide level) were found in the genome. ODC and PMT genes were given the names, due to no clear matching to the reported ones (Riechers and Timko, 1999; Xu et al., 2004), and so were BBLd.2 and BBLe with their relations to other Nicotiana BBLs (Kajikawa et al., 2011; Supplemental Fig. S2).
Tobacco QPT1.2 and QPT2.2 of distinct groups, both from N. tomentosiformis, are ∼75 kb apart on Super Scaffold (SS) 1382, suggesting a relatively recent duplication giving rise to these two genes (Shoji and Hashimoto, 2011b). A two-gene cluster of nonhomologous A622L and MATE2 was found on chromosome 12; these genes from N. tomentosiformis are ∼128 kb away from each other on SS753. Other than those, no clustering was found at the SS level for the listed genes (Supplemental Table S1). Because of their uncertain placements in the genome, we could not confirm the genomic clustering of N. sylvestris-derived counterparts of the clustered genes.
Expression of Nicotine and Related Primary Metabolic Genes
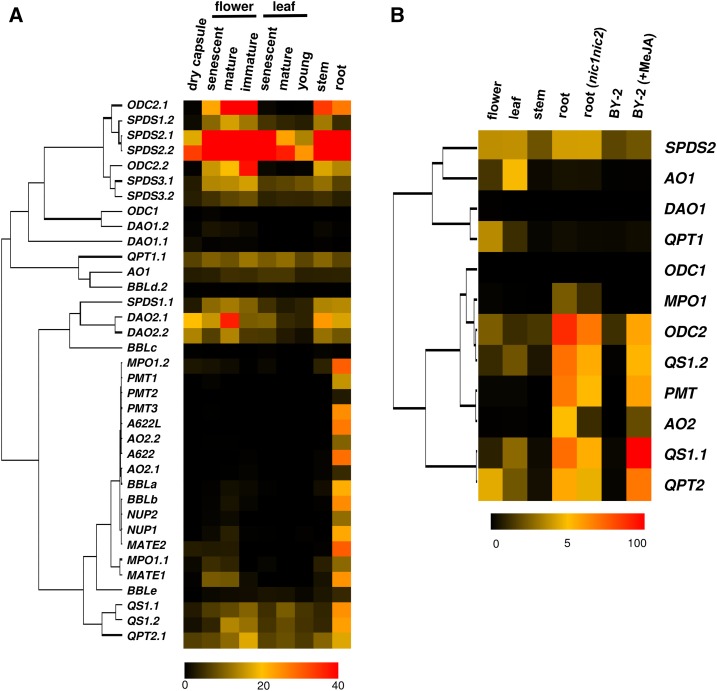
Transcript levels were estimated by RNA-seq analysis, which yields expression levels in fragments per kilobase of exon per million mapped sequence reads (FPKM), for each gene assigned on the genome. Based on the FPKM values in various tobacco tissues, all of the metabolic and transport genes were clustered (Fig. 2A); QPT1.2 and QPT2.2 were excluded because of apparently duplicate mapping of the reads. There is a discrete cluster of 19 genes, including nearly all genes in the nicotine biosynthesis regulon (Fig. 2A). The genes in this cluster are expressed preferentially in the nicotine-producing roots. Such characteristic expression largely restricted to the roots is not evident for genes that are possibly involved in related primary pathway rather than the alkaloid production and form other clusters (Fig. 2A). Although included in the regulon, ODC2.1 and ODC2.2 with substantial expression also in stems and flowers, and BBLc and BBLd.2 with very low FPKM values, are exceptional in this clustering.
Figure 2.
Heat map visualization of expression data of metabolic genes involved in nicotine and related primary metabolism in tobacco. A, Heat map visualizing FKPM values obtained by RNA-seq analysis. B, Heat map visualizing transcript levels estimated by qRT-PCR analysis. Transcript levels in various organs of tobacco plants and cultured BY-2 cells elicited or not with MeJA were analyzed. Using average values of three biological replicates, the levels are calculated relative to those of EF1α, and are shown relative to the highest one (set to 100) in the data set. Clustering was done with Cluster 3.0 (http://bonsai.hgc.jp/∼mdehoon/software/cluster/software.htm) and heat maps with a tree were drawn with Java TreeView (http://jtreeview.sourceforge.net/).
To complement the RNA-seq results and further address the differential regulation, we examined the expression patterns with quantitative reverse transcription (qRT)-PCR of the metabolic genes involved in early parts of the pathways, which overlap or work in parallel with related primary pathways (Fig. 1). Primer pairs were designed to specifically amplify each gene or group of orthologs (Supplemental Table S2). Transcript levels in organs from tobacco plants, including roots of nic1nic2 mutant with a low-nicotine trait (Legg and Collins, 1971), and cultured BY-2 cells elicited with methyl jasmonate (MeJA), were measured and represented relative to those of a housekeeping gene EF1α (Fig. 2B). As reflected in hierarchal clustering, the expression patterns were again clearly distinguished into two major groups of genes, one possibly forming nicotine biosynthesis regulon and one presumably devoted to parallel primary pathways (Fig. 1). ODC1 clusters with the genes of the former group, but only at a low level of statistical significance, which is likely to be due to its very low expression levels. Thus ODC1 was assigned to the latter group. The genes of the former, including ODC2, PMT, MPO1, AO2, QS1, and QPT2, strongly or often nearly exclusively express in the roots of wild-type tobacco and their levels are decreased to 18% to 68% levels in nic1nic2 mutant roots, reflecting the regulation by NIC loci and therefore by NIC2-locus ERFs. The genes of this group are markedly up-regulated by MeJA in the cultured cells, leading to alkaloid induction, except for MPO1 as has been reported in Shoji and Hashimoto (2008). In contrast to the genes involved in nicotine formation, expression of ODC1, SPDS2, DAO1, AO1, and QPT1 are not restricted to nicotine-producing tissues; they occur more ubiquitously, although their expression levels are generally lower (except SPDS2), which possibly reflects their contribution to primary pathways.
cis-Elements Predicted in Promoter Regions
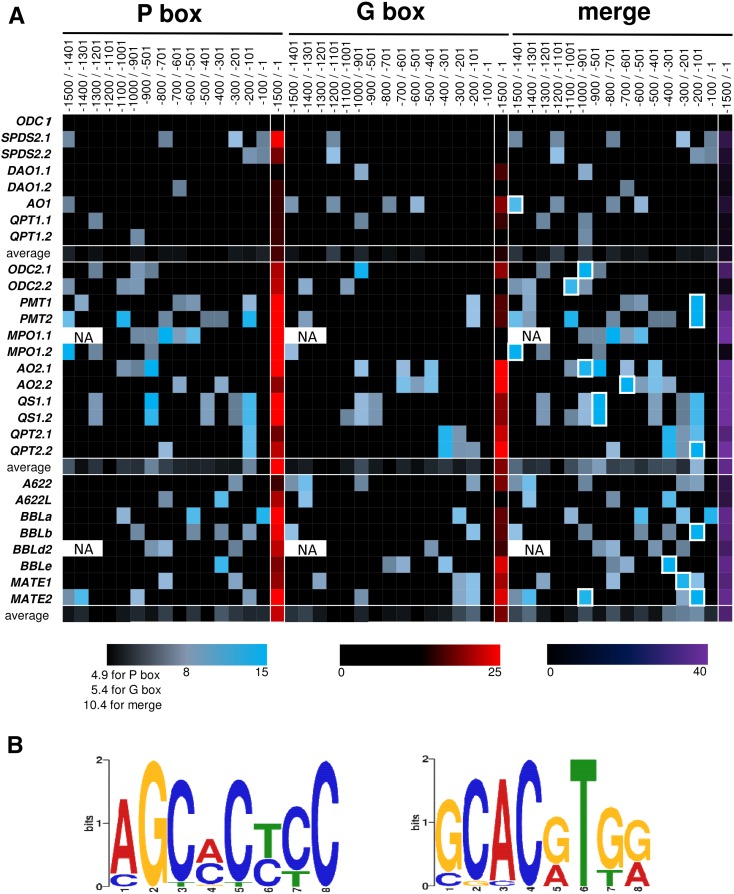
The availability of genomic sequences prompted us to predict ERF189-binding P box and MYC2-binding G box elements in the promoter regions of the genes involved in nicotine and related pathways. We examined whether such elements were enriched in nicotine pathway genes. Using weighted matrices (Shoji and Hashimoto 2011b, 2011c) representing P and G boxes, we computationally searched for the binding elements with cutoff scores of 5.0 for 10-mer P box and of 5.5 for 8-mer G box in 5′-flanking regions (−1,500 to −1; counted from the first ATG). The scores of the predicted elements in each 100-bp bin along the entire promoter sequences are visualized in a heat map (Fig. 3A). For both elements in the primary metabolic genes, these average score for each bin are relatively low (upper block), whereas for the nicotine pathway genes there are a number of bins with high scores (lower two blocks). Bins scoring highly for both P and G boxes (squared with white lines in the right column) are present in 14 out of the 20 gene promoters of nicotine pathway, implying that these elements are crucial to the expression of these genes.
Figure 3.
ERF189-binding P box and MYC2-binding G box elements predicted in 5′-flanking regions from −1,500 to −1 (numbered from the first ATG) of metabolic and transport genes involved in nicotine and related primary metabolism. A, Heat map visualizing distributions of the elements predicted with Regulatory Sequence Analysis Tools (http://rsat.ulb.ac.be/rsat/); elements with scores greater than 5.5 for P box and 5.0 for G box were included. Colors reflect scores of the elements (or sums of those when multiples are predicted) in each bin. At borders between bins, elements were assigned into those proximal to the first ATG. Sums of scores for both boxes are in the right column (merge), where the bins including both P box and G box are squared with white lines. The values are averaged for a gene set in each block. Genomic sequence of a region from −1,500 to −1,200 is not available for MPO1.1 and BBLd.2. PMT3 and BBLc were excluded, because 5′-flanking sequences available were too short (<200 bp). B, Sequence logos representing conservations of sets of sequences related to P box (left) and G box (right) retrieved by MEME analysis from the promoter regions of the genes included in the lower two blocks of (A). NA, Not available.
To retrieve the cis-elements shared among the coregulated genes in a nontargeted way, Multiple EM for Motif Elicitation (MEME) analysis (http://meme-suite.org/; Bailey et al., 2006) was conducted using the 5′-flanking sequences (−1,500 to −1) of 20 genes, included in the lower two blocks in Figure 3A, as queries. Highly scoring sequences related to P box (rank, 2; e-value, 9.5e-004; log likelihood ratio, 203) and those to G box (rank, 4; e-value, 2.9e+0.00; log likelihood ratio, 188) were found in all 20 and 19 promoters, respectively. Logo graphics (Crooks et al., 2004) representing conservation among the retrieved sequences are shown (Fig. 3B).
Regulation of BBL Gene Promoter in Tobacco
In concert with other genes involved in nicotine biosynthesis, BBL genes are strongly expressed in the roots and induced by jasmonates in tobacco (Kajikawa et al., 2011). To gain insight into the transcriptional regulation of BBL genes, promoter reporter analyses were performed for NsBBLa from N. sylvestris (Supplemental Fig. S2). Because the genomic sequence was not available when we began the experiment, the region from −1.126 to −1 bp (counted from the first ATG) of NsBBLa was obtained with TAIL-PCR; the sequence was later confirmed 99% identical to that in the genome database and 97% to that of BBLa gene in tobacco genome. Four P box (scores 5.8 to 6.9) and two G box (scores 9.1 and 5.5) elements were predicted within the promoter sequence. To generate transgenic plants and hairy roots, a GUS reporter gene driven by the promoter was introduced into tobacco via Agrobacterium tumefaciens-mediated transformation.
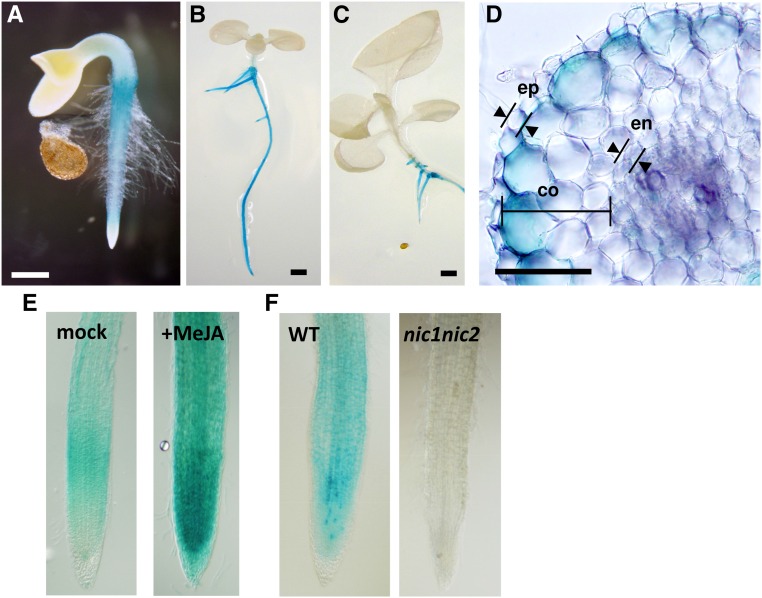
Expression patterns of the BBL promoter in transgenic seedlings and hairy roots were analyzed by visualizing GUS activities histochemically (Fig. 4). In transgenic seedlings at 2, 5, and 14 d after germination, GUS staining was visible in the roots, but not in any aerial tissues (Fig. 4, A–C). Strong staining was also observed in transgenic hairy roots (Fig. 4, D–F). In apical regions of the hairy roots, promoter activity was detected in differentiated cells as well as in actively dividing and elongating cells, but was absent in the root cap and in epidermal cells (Fig. 4, E and F). A cross section of the stained hairy roots in a differentiated zone was made to identify cell types associated with the activities; the promoter was expressed most strongly in the outmost cortex layer and moderately in the endodermis and parenchyma cells in the stele, but not in the epidermis (Fig. 4D). This expression pattern of the BBL promoter is quite similar to those reported for other nicotine pathway genes, such as PMT (Shoji et al., 2000), QPT2 (Shoji and Hashimoto 2011a), A622 (Shoji et al., 2002), and MATE1 (Shoji et al., 2009).
Figure 4.
Histochemical GUS staining of tobacco seedlings and hairy roots transformed with the reporter gene driven by NsBBLa promoter. The GUS reporter gene was driven by a promoter region from −1,162 to −1 (numbered from first ATG) of NsBBLa gene from N. sylvestris. Two- (A), five- (B), and fourteen- (C) day-old seedlings. D, A cross section of a transgenic hairy root in the differentiation zone. E, Hairy roots (wild-type line no. 8) were treated with 100 μm MeJA for 24 h. F, Hairy roots of wild type (line no. 13) and of nic1nic2 mutant (line no. 6). Bars = 0.5 mm in A, 2 mm in B and C, and 0.1 mm in D. co, Cortex; en, endodermis; ep, epidermis; WT, wild type.
To examine the jasmonate-mediated response of the BBL promoter, seven transgenic lines were treated with MeJA for 24 h, and the GUS activities in crude root extracts were measured fluorometrically. In five lines, significant increases of the activity (2.3 to 5.1 folds) relative to mock-treated controls were observed (Supplemental Fig. S3A). Indeed, GUS staining intensities were increased in transgenic hairy roots treated with MeJA, compared to the controls (Fig. 4E). To demonstrate the regulation of the BBL promoter by NIC loci, multiple lines of transgenic hairy roots with the promoter reporter were generated in wild-type and nic1nic2 mutant genotypes. Higher GUS activities in the wild-type background were clearly observed in histochemical (Fig. 4F) and fluorometric (Supplemental Fig. S3B) assays.
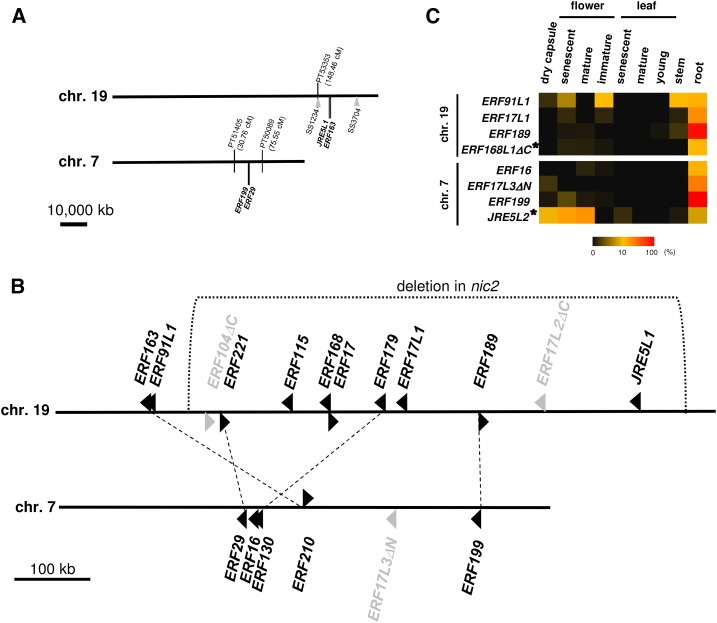
Gene Clusters of NIC2-Locus ERFs and Their Homologs
Clustering of ERF genes at NIC2 locus was presumed in a previous study (Shoji et al., 2010). To understand the genomic organization of the NIC2-locus gene cluster and its possible counterpart from another ancestral diploid, NIC2-locus ERF genes and their homologs were retrieved from the tobacco genome sequence. Twenty-two ERF genes were retrieved in total, including four genes on unplaced SSs (Supplemental Table S3). Two clusters of multiple ERF genes of the relevant group are found in the genome: one 12-gene cluster (spanning ∼660 kb, flanked with genetic maker PT53353) from N. tomentosiformis on chromosome (chr.) 19 and one six-gene cluster (∼320 kb, flanked with PT51405 and PT50089) from N. sylvestris on chr. 7 (Fig. 5; Bindler et al., 2011). A phylogenetic tree of the retrieved ERFs and their homologs can be found in Supplemental Figure S4. Although most ERFs retrieved are practically identical (>98% at nucleotide level) to the queries, ERF91Ls, ERF17Ls, and JRE5Ls were, to our knowledge, newly defined according to their phylogenies (Supplemental Fig. S4), and the ERF91L genes differ by an N-terminal 64 amino acid extensions, but are nearly identical to ERF91 (not in the tree). Also, the ERF17L genes are not orthologous to ERF17 or JRE4 from tomato, the nearest neighbor in the tree. Some ERFs are truncated and do not include full-length DNA-binding domains, and therefore may be nonfunctional as transcription factors; these are denoted with ΔN or ΔC. As reflected in disproportional gene numbers between the clusters, only five out of ten functional NIC2-locus ERFs in the T-genome have clear counterparts in the S-genome; there are at least four pairs of such ERFs in the clusters (Fig. 5).
Figure 5.
Clusters of NIC2-locus ERF genes and their homologs on tobacco chromosomes. A, Positions of clustered ERFs and flanking genetic markers on chromosomes. Positions of ERFs at ends of the clusters and the nearest genetic markers anchored on genome sequence are shown. Positions of genomic deletions on chr. 19 found in cv LA Burley 21 (Supplemental Table S5) are shown with gray arrowheads and corresponding SS numbers. B, Graphic views of the gene clusters. A large chromosomal region (∼650 kb) indicated was found to be deleted in nic2 mutant (Supplemental Fig. S5). Arrowheads indicate positions and orientations of predicted open reading frames of ERFs. The ERF genes, denoted with Δ and shown in gray, encode possibly nonfunctional transcription factors without full-length DNA-binding domains. Functional ERFs of same ortholog groups (Supplemental Fig. S4) are paired with dotted lines. C, Heat map visualizing FPKM values of ERF genes. ERF genes on unplaced SSs (but associated with chr. 7 or chr. 19; Supplemental Fig. S2) are marked with asterisks.
To delimit the genomic region deleted in nic2 mutant, genomic PCR analysis (Supplemental Fig. S5) was performed with primers designed to detect the sequences at various positions around the NIC2-locus gene cluster (Supplemental Table S4). Genomic DNAs from various NIC genotypes in two cultivars, Burley 21 and NC95, were used for the analysis. Some of the amplified fragments were sequenced to confirm the specific amplifications (Supplemental Fig. S5), which are sometimes unsuccessful possibly due to highly repetitive nature of tobacco genome (Sierro et al., 2014). A large chromosomal region (∼650 kb) including 10 out of 12 clustered ERF genes is missing in nic2 genetic background, whereas the remaining two genes, ERF163 and ERF91L1, are retained in the genotype (Fig. 5; Supplemental Fig. S5), in agreement with the previous data (Shoji et al., 2010). ERF91L1 was considered an equivalent of former ERF91. These results were also in line with expression data in nic2 mutant of the clustered ERFs and a gene that encodes COP9 signalosome subunit 7 and resides between ERF91L1 and the putative breakpoint of the deletion. Expression levels of the genes within the deleted region are very low, or below reliable detection limits, in nic2 mutant (Supplemental Fig. S6). As expression levels of ERF168L1ΔC and ERF104ΔC are also below the detection limits (Supplemental Fig. S6), these genes on unplaced SS3881 were assumed to reside somewhere in the deleted part of the cluster.
Because genes were predicted with transcriptomics data, only a small fraction of the high-expressed ERF genes are found in this database (Supplemental Table S3), and can be assigned FPKM data; eight ERF genes were annotated, including only one orthologous pair of functional genes, ERF189 and ERF199. FPKM data indicate that ERF189 and ERF199 are expressed almost exclusively in the roots, whereas a similar root-specific expression (albeit at a lower level) was observed for ERF16 and nonfunctional ERFs, ERF17L3ΔC and ERF168L1ΔC (Fig. 5C). Expression of JRE5L2 and ERF91L1 is not restricted to the roots, but is also apparent in flowers and other tissues (Fig. 5C).
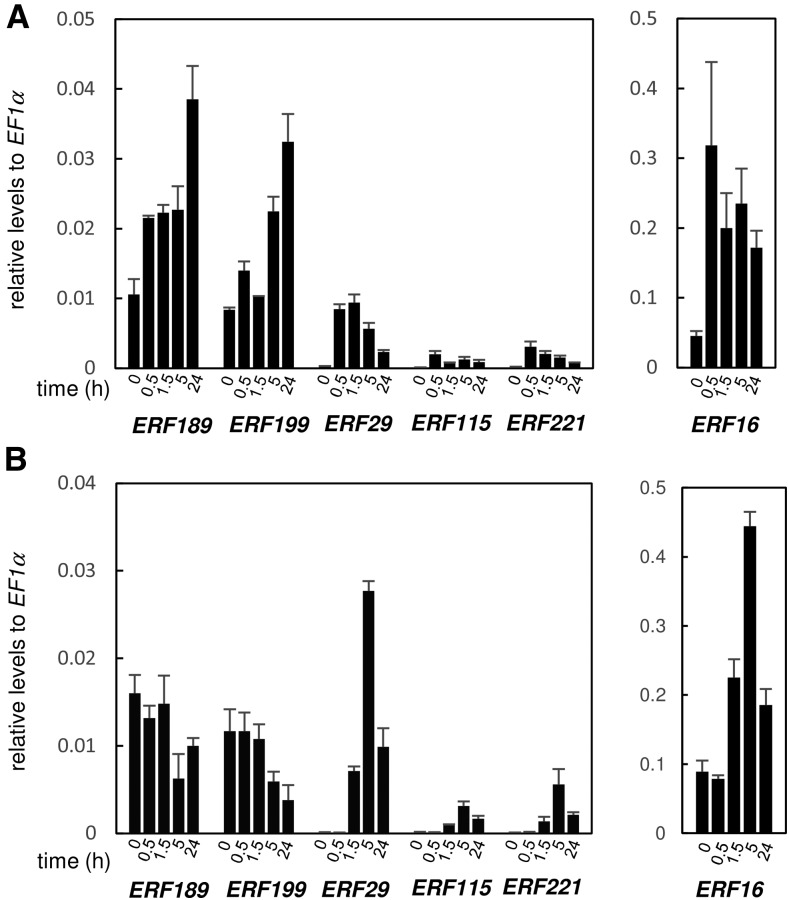
Responses of ERF189 and Its Close Homologs to Jasmonates and Salt Stress
The availability of the genome sequence allowed us to detect individual ERF genes with qRT-PCR by using carefully designed primers (Supplemental Table S2). We analyzed the responses to jasmonates and salt stress of ERF189 and ERF199, which were proposed to regulate the nicotine pathway (Shoji et al., 2010; Shoji and Hashimoto, 2015), and their close homologs, ERF16, ERF29, ERF115, and ERF221, which had been detected collectively in the previous studies, in tobacco hairy roots; the cultured roots were treated with MeJA at 100 μm and NaCl at 300 mm. A pair of ERF189 and ERF199 are gradually induced by MeJA, whereas NaCl treatment has negative impact on their expression (Fig. 6), as reported previously (Shoji et al., 2010; Shoji and Hashimoto 2015). ERF16 is from N. sylvestris and has no obvious counterpart from N. tomentosiformis (Supplemental Fig. S4). ERF16 is induced by both MeJA and NaCl (Fig. 6) and has the highest expression levels among the examined genes (according to qRT-PCR data; Figs. 5C and 6); it boasts a 7-fold induction at 0.5 h by MeJA and 5-fold induction at 5 h by NaCl. The expression levels of ERF29, ERF115, and ERF221, which are highly similar to each other at the sequence level (Supplemental Fig. S4), have very low expression relative to ERF16, ERF189, and ERF199 (Fig. 6), and thus may be not annotated as genes in the database. MeJA treatment caused rapid (reaching maximums within 0.5 to 1.5 h) and substantial induction (15 to 33 times) of ERF29, ERF115, and ERF221, whereas their expression also rapidly rose and reached 59- to 226-fold levels after 5 h upon NaCl treatment (Fig. 6); such large fold changes may reflect their low basal expression.
Figure 6.
Response of ERF189 and related genes to jasmonate and salt stress in tobacco hairy roots. Transcript levels were analyzed with qRT-PCR in tobacco hairy roots treated with 100 μm MeJA (A) or 300 mm NaCl (B) for 0, 0.5, 1.5, 5, or 24 h. The error bars indicate sds over three biological replicates. The levels are expressed relative to those of EF1α. Note that different vertical scales are adapted for ERF16 and for others.
Genomic Deletions Found in the nic1nic2 Mutant of Burley 21 Cultivar
To identify the genomic deletions in a nic1nic2 double mutant other than the ERF gene cluster (Fig. 5), genomic PCR analysis was carried out for the genes strongly down-regulated in the low-nicotine mutant (Supplemental Fig. S7); the genes highly ranked in a microarray-based screen (Shoji et al., 2010) along with ERF189 (rank, 8; signal ratio of nic1nic2 mutant to wild type, 0.025) were analyzed. We found that 13 genes have been deleted (rank, 1 to 38; ratio, 0.013 to 0.133), including presumed genes residing on the same SSs with the confirmed ones, on six SSs associated with at least four regions on three chromosomes in the nic1nic2 double mutant of cv Burley 21, or cv LA Burley 21 (Legg and Collins, 1971; Supplemental Table S5). The presumptive deletions encompass a relatively large chromosomal region, i.e. ∼1,730 kb deletion at a one end of chr. 17 (Supplemental Table S5). The deletions were found only in LA Burley 21 but not in other lines, such as the nic1 and nic2 single mutants of the same Burley 21 cultivar and the nic1nic2 double mutant of cv NC95 (Chaplin, 1975; Supplemental Fig. S7). As these are specific to a single line of LA Burley 21, the correlations of these deletions with NIC genotypes and thus nicotine biosynthesis are unlikely. The deletions found at two distinct positions on chr. 19 are far away and thus are apparently different from the deletion at NIC2 locus (Fig. 5A).
DISCUSSION
A Regulon for Nicotine Biosynthesis Pathway
Concerted and substantial expression of structural genes involved in certain pathways is required for massive metabolic flows allowing production and accumulation of specialized metabolites. Such coordination of multiple genes is often extended into preceding primary pathways that supply precursors to downstream metabolism (van der Fits and Memelink, 2000; Cárdenas et al., 2016; Thagun et al., 2016; van Moerkercke et al., 2015). In tobacco, a series of metabolic and transport genes involved in a pathway leading to nicotine from Orn and Asp express nearly exclusively in the roots and jasmonate-elicited cultured cells (Fig. 2; Shoji and Hashimoto, 2011a). Such coexpression of these genes largely depends on pathway-controlling ERF transcription factors (Shoji and Hashimoto, 2013). The organization of these genes into such a regulon allows the coordination of distinct portions of the pathway from early ring formation steps to late steps, including ring coupling and nicotine transport (Fig. 1).
Detailed phylogenies (Supplemental Fig. S1) and expression profiling (Fig. 2) clearly distinguished the genes involved in the regulon, as shown in Figure 1. Genes involved in each step specific to the nicotine pathway, such as PMT, MPO, A622, BBL, and MATE, which generally belong to a single orthologous group (Supplemental Figs. S1 and S2), are commonly regulated as members of the regulon (Fig. 2). In contrast to the nicotine-specific genes, ODC, AO, QS, and QPT genes are involved in early steps that overlap with the polyamine or NAD pathway (Fig. 1). To satisfy the metabolic demands of different downstream pathways, two types of genes (genes from two groups of orthologs), are present for every overlapping step, except QS (Fig. 1; Supplemental Fig. S1); ODC2, AO2, and QPT2 are involved in the nicotine biosynthesis regulon, whereas ODC1, AO1, and QPT1 are not subjected to the regulation by the ERFs and jasmonates and are possibly devoted to the primary housekeeping pathways (Figs. 1 and 2). Of course, because such functional differentiations among the genes were presumed from their biased, but not mutually exclusive, expression patterns, transgenic or mutational approaches may be required to validate the contributions of each gene to specific metabolic pathways. Although no differences other than the transcriptional regulation between the gene types have been suggested to date, slight differences in the protein sequences (Supplemental Fig. S1) could give rise to functional differentiations, thus allowing for distinct metabolic roles (Schenck et al., 2015).
As reflected by the existence of two types of the genes for the overlapping steps, pyridine and pyrrolidine formation branches of nicotine pathway are presumed to have evolved from universal polyamine and NAD pathways through gene duplication followed with subfunctionalization or neofunctionalization of the duplicates (Fig. 1; Shoji and Hashimoto, 2011b; Naconsie et al., 2014). The Orn-derived pyrrolidine ring is utilized as a common building block for formation of tropane, nortropane, and nicotine alkaloids in a number of species of Solanaceae and other families (Shoji and Hashimoto, 2011a). The pyrrolidine formation branch, composed with ODC, PMT, and MPO, may have arisen before the diversification of the plants producing alkaloids containing the Orn-derived moiety. The assumption is supported with the fact that tomato and pepper have at least one gene of ODC2, PMT, and MPO1 (Supplemental Fig. S1; Stenzel et al., 2006). Whereas ODC2, which acts in parallel with ODC1, has retained its original catalytic activity, PMT and MPO1, which have been derived from SPDS and DAO respectively, have acquired novel activities through neofunctionalization (Junker et al., 2013; Naconsie et al., 2014), thus contributing to the innovation of the pyrrolidine-forming extension (Fig. 1). In contrast to the relatively ancient diversification of the pyrrolidine branch from polyamine metabolism, establishment of paralleled routes including AO, QS, and QPT for pyridine formation (Fig. 1), which supply the ring to NAD and nicotine production in tobacco, is presumed to have occurred around the time of the diversification of the Nicotiana lineage, given the specificity of AO2 and QPT2 to the lineage (Supplemental Fig. S1; Shoji and Hashimoto, 2011b; Ryan et al., 2012). This scenario is also in agreement with existence of a gene cluster of QPT1.1 and QPT2.1 in the tobacco genome (Supplemental Table S1) that may have arisen by a relatively recent gene duplication event. It remains to be addressed how and when the ERF factors, which play a central regulatory role in the nicotine biosynthesis regulon in present-day tobacco, have evolved to regulate distinct portions of the pathways that may have developed independently at different points in time.
Clusters of genes encoding nonhomologous proteins involved in specialized pathways has been widely recognized in plants (Nützmann and Osbourn, 2014), and alkaloid pathways follow suit (Winzer et al., 2012). In the tobacco genome, a cluster of A622L and MATE2 genes, which encode enzymes (DeBoer et al., 2009; Kajikawa et al., 2009) and transporters (Shoji et al., 2009), and which are both involved in late steps of the pathway, is situated on chromosome 12 (Supplemental Fig. S1). Localization of the cluster opens up the possibility of identifying genes for as yet undefined late steps based on their proximities to the cluster. Apart from this cluster, we could not find any clustering of the other nonhomologous genes; of course, before concluding, exact placements of unplaced SSs should be determined (Supplemental Fig. S1). The dispersion of these structural genes throughout the genome (Supplemental Table S1) implies that regulation of this functionality is dependent on promoter-binding transcription factors, rather than the chromatin-level regulation proposed for clustered genes (Wegel et al., 2009).
A number of P and G box elements, which are targeted by ERF189 and MYC2 transcription factors, respectively, were computationally predicted in the promoter regions of the genes (Fig. 3A). Occurrences of the predicted elements are more frequent among nicotine pathway genes regulated by those transcription factors, than they are among primary metabolic genes (Fig. 3A). This finding was complemented by the identification of P and G boxes as motifs conserved among the regulated promoters (Fig. 3B). These results support the notion that downstream structural genes have been recruited into the regulons under the control of the transcription factors through generating cognate cis-elements in their promoters (Shoji and Hashimoto, 2011b; Moghe and Last, 2015). Proximities of the two distinct boxes within the promoters (Fig. 3A; Shoji et al., 2010; Shoji and Hashimoto, 2011b, 2011c) imply the importance of such arrangements of functional cis-elements that possibly support cooperative action of the two transcription factors. As observed previously, both elements are likely to be present in similarly situated proximal promoter regions (Shoji et al., 2010; Shoji and Hashimoto, 2011b; Thagun et al., 2016); this is observed for genes such as PMT, QPT2, BBL, and MATE (Fig. 3A). However, this seems not always to be the case; the likely elements, though none of them have been validated experimentally, are also present in relatively distal promoter regions of some genes (Fig. 3A; Xu et al., 2004). It remains to be addressed whether such different placements of cis-elements in the promoters account for slight but significant differences in the expression patterns of the generally coregulated genes involved in the regulon, such as no elicitation of MPO1 in cultured cells and relatively relaxed suppression of ODC2 and MPO1 in nic1nic2 mutant (Fig. 3A; Shoji and Hashimoto, 2008).
Promoter of the BBLa gene follow the pattern of those of other genes involved in the regulon (Shoji et al., 2000, 2002, 2009; Shoji and Hashimoto 2011b) in terms of cell-type specificity (Fig. 4), response to jasmonates (Fig. 4E; Supplemental Fig. S3A), and suppression in the nic1nic2 mutant (Fig. 4F; Supplemental Fig. S3B). By recognizing resident P box elements, pathway-controlling ERF factors, with the help of MYC2, may mainly contribute to transcriptional regulation underlying such common characteristic expression. The functional importance of P and G box elements for regulation has been demonstrated by loss-of-function experiments with mutated promoters (Shoji et al., 2010; Shoji and Hashimoto, 2011b, 2011c). As yet, no gain-of-function analyses have been performed for these elements (Shoji et al., 2000; Xu and Timko, 2004). Compared to regulation by jasmonates and NIC genotypes, it has remained less clear how much the ERF factors contribute to the tissue-specific expression of the promoters. In tomato, expression of JRE4 is well correlated with those of downstream structural genes during developmental progressions (Cárdenas et al., 2016; Thagun et al., 2016). Root-specific expression of ERF189 and ERF199 indicated by RNA-seq (Fig. 5C) gives a first important cue to this issue. Cell-type specificity, in addition to root specificity, is considered characteristic to the regulon, as reflected by the epidermal expression of the NUP1 promoter contrasted with those of the ERF-regulated promoters (Kato et al., 2014).
Evolution of Clustered Transcription Factor Genes
The genetic locus NIC2 controlling nicotine content in tobacco (Legg and Collins, 1971; Hibi et al., 1994; Shoji et al., 2010) was finally elucidated; complete sequence of the tobacco genome (Sierro et al., 2014) allowed us to figure out the structure of ERF gene clusters at the NIC2 locus that originated from N. tomentosiformis and its counterpart from N. sylvestris (Fig. 5, A and B). A similar cluster of five ERF genes is present in the tomato genome (Cárdenas et al., 2016; Thagun et al., 2016). Whereas ERF and COP9 signalosome genes conserved between distantly related tomato and tobacco are similarly situated around the peripheral area of the clusters, central regions are occupied by ERF genes specific to each lineage, such as ERF189 and JRE4 (Fig. 5B; Supplemental Figs. S4 and S6). The structural arrangement of the clusters suggests more dynamic rearrangement of the divergent central area, which may favor formation of the ERFs devoted to lineage-specific alkaloid regulation. Tobacco has two sequences with similarities to JRE6 from tomato (not shown in Supplemental Table S3 and Supplemental Fig. S4 because of incomplete reading frames). In tomato, apart from the ERF gene cluster on chr. 1, JRE6 is present as a singleton on chr. 5 (Thagun et al., 2016). The tobacco JRE6-like sequences may be situated somewhere around clusters, because they reside on unplaced SSs associated with same chromosomes. The possible association of the JRE6-like sequences with the clusters in tobacco implies the involvement of JRE6 orthologs in ancient clusters and consequent relocation of them to other chromosomes during formation of the tomato cluster.
There is considerable structural divergence even between the two clusters originating from distinct Nicotiana diploids in tobacco (Fig. 5), suggesting relatively frequent loss and gain of ERF genes and their replacements within the clusters even after diversification of the two ancestral diploids. Tandem repeats of homologous sequences may have contributed to highly frequent unequal crossing-over events leading to such chromosomal changes around the regions. A number of genes encoding truncated ERFs (Fig. 5; Supplemental Table S3) may be vestiges of recombination events that have occurred in the intragenic regions. Phylogenetic analysis revealed the existence of only a very few homeologous pairs of functional ERF genes in the tobacco genome as well as a considerable number of remaining unpaired or possibly nonfunctional genes (Supplemental Fig. S4). For these, the biological relevance is unclear, considering evolutionary proximity of N. tomentosiformis and N. sylvestris. Phylogenetic (Supplemental Fig. S4) and gene expression (Figs. 5C and 6) evidence point to the functional importance of a pair of ERF189 and ERF199, one of the three Nicotiana-specific pairs (Fig. 5B; Supplemental Fig. S4). These have been previously proposed to function as a regulator of the nicotine pathway (Shoji et al., 2010; Shoji and Hashimoto, 2015). A pair of genes, ERF221 and ERF29, which is the nearest functional pair to ERF189 and ERF199 in the phylogenetic tree (Supplemental Fig. S4), is expressed basally at low levels and is induced not only by MeJA but also by NaCl (Figs. 5C and 6). It is intriguing to uncover how only a certain ERF gene (or a certain pair of genes in tobacco case), among the many clustered genes, has become the predominant regulatory gene (Figs. 5C and 6; Thagun et al., 2016), and functionally important for a specialized pathway in each plant lineage.
Genomic Deletions in Low-Nicotine Mutants
Molecular basis of nic2 mutation was described; the mutant allele has a large chromosomal deletion that encompasses ten ERF genes in the cluster at NIC2 locus on chr. 19, including ERF189 (Fig. 5B; Supplemental Fig. S5). Repetitive sequences present throughout the tobacco genome (Sierro et al., 2014) prevented further delimitation of the deleted region; regions of ∼13 and 20 kb in size remain to be elucidated (Supplemental Fig. S5). Whole-genome or targeted resequencing and cytogenetic analyses may help to precisely determine breakpoints of the deletion and to detect possible structural changes at chromosomal level, respectively. Absence of a major deletion in the counterpart locus on chr. 7 has yet to be confirmed in nic1 genetic background (T. Shoji, unpublished data).
We found a number of genomic deletions other than that at NIC2 locus in the nic1nic2 mutant of Burley 21 cultivar, or cv LA Burley 21, on at least four positions on three chromosomes (Supplemental Table S5; Supplemental Fig. S7). A lot of genes, the expression of which is severely suppressed in the nic1nic2 mutant (signal ratio of nic1nic2 to wild type < 0.113; Shoji et al., 2010), are missing in the mutant (Supplemental Table S5; Supplemental Fig. S7). These deletions are specific to LA Burley 21 and not found in other varieties, and may not be associated with the NIC genotypes and thus with nicotine biosynthesis. Note that the nic1nic2 double mutant was not generated by crossing the corresponding single mutants, but rather derived from an original Cuban cigar cultivar as its genotype (Legg and Collins, 1971). The presence of multiple deletions specific to a certain line implies that the nature of the tobacco genome is highly prone to structural variations.
MATERIALS AND METHODS
Plant Growth and Treatment
Sterilized seeds of Nicotiana tabacum (tobacco), N. sylvestris, and N. tomentosiformis were germinated and grown to seedlings on half-strength Gamborg B5 medium solidified with 0.3% (w/v) gellan gum and supplemented with 2% (w/v) Suc. The wild-type, nic1, nic2, and nic1nic2 (registered as cv LA Burley 21) genotypes in the Burley 21 cultivar (Legg and Collins, 1971) were obtained from the USDA, the wild-type and nic1nic2 (registered as LAFC53) genotypes in the NC95 cultivar (Chaplin, 1975) from John Hamill (Deakin University), and N. tabacum cv Petit Havana line SR1, N. sylvestris, and N. tomentosiformis from Japan Tobacco. Two-week-old seedlings were transferred onto soil in pots and grown to maturity in the greenhouse. Tobacco BY-2 cells were cultured as described by Nagata et al. (1992). To induce nicotine biosynthesis genes, 4-d-old BY-2 cells were rinsed five times to remove auxin, transferred to auxin-free medium supplemented with MeJA at 100 μm, and cultured for 24 h.
RNA-Seq Analysis
Sequencing data for RNA from dry capsule (SRX495517, SRX495518, and SRX495519), senescent flower (SRX495530 and SRX495531), mature flower (SRX495520, SRX495521, and SRX495522), immature flower (SRX495602, SRX495603, and SRX495605), senescent leaf (SRX495532, SRX495534, and SRX495535), mature leaf (SRX495523, SRX495524, and SRX495525), young leaf (SRX495606, SRX495607, and SRX495608), stem (SRX495598, SRX495600, and SRX495601), and root (SRX495526, SRX495527, and SRX495529) of N. tabacum TN90 were obtained from BioProject PRJNA208209 (https://www.ncbi.nlm.nih.gov/bioproject/; Sierro et al., 2014).
Reads were mapped to the genome of N. tabacum TN90 (ftp://ftp.solgenomics.net/genomes/Nicotiana_tabacum/assembly/Ntab-TN90_AYMY-SS.fa.gz) using the software HISAT2 (v2.0.5; https://ccb.jhu.edu/software/hisat2/; Kim et al., 2015) and filtered using the software SAMtools (v1.3.1; http://samtools.sourceforge.net/; Li et al., 2009) to retain properly paired read pairs not annotated as “secondary,” “QC failed,” “duplicate,” or “supplementary.”
For each N. tabacum TN90 gene model (ftp://ftp.solgenomics.net/genomes/Nicotiana_tabacum/annotation/Ntab-TN90_AYMY-SS_NGS_rnaseq.gff3), expression was calculated from the filtered mapped reads using the software StringTie (v1.3.1c; https://ccb.jhu.edu/software/stringtie/; Pertea et al., 2016). Gene level expression values were obtained by summing the FPKM of the gene’s isoforms.
qRT-PCR Analysis
Total RNA was isolated from samples using RNeasy kit (Qiagen) and then converted to first-strand cDNA using ReverTra Ace qPCR RT Master Mix (Toyobo). The cDNA templates were amplified using a LightCycler 96 (Roche) with SYBR Premix Ex Taq (Takara) according to Shoji et al. (2010). The primer sequences are given in Supplemental Table S2. EF1a was used as a reference gene. Each assay was repeated at least three times. Based on amplifications from equal molar quantities of cloned amplicons, amplifications from different primer pairs were normalized.
Computational Analyses
Full-length protein sequences were aligned with the software ClustalW (http://www.genome.jp/tools/clustalw/; Thompson et al., 1994), and using the alignments, phylogenetic trees were generated with the software MEGA6 (http://www.megasoftware.net/; Tamura et al., 2013) with the neighbor-joining algorithm.
The putative ERF189 and MYC2-binding elements in the 5′-flanking regions from −1,500 to −1 bp (numbered from the first ATG) of metabolic and transport genes (Supplemental Table S1) were searched and scored with Regulatory Sequence Analysis Tools (http://rsat.ulb.ac.be/rsat/; Turatsinze et al., 2008), using weight matrices for P box (Shoji and Hashimoto, 2011a) and G box (Shoji and Hashimoto, 2011b).
MEME analysis (v. 4, 11.2; http://meme-suite.org/; Bailey et al., 2006) was performed by selecting an option of zero or one occurrence per sequence and setting minimum width to 6 and maximum width to 15 and 12 when retrieving P box- and G box-related sequences, respectively.
Plant Transformation
The 5′-flanking region of NsBBLa was cloned from N. sylvestris genomic DNA by a TAIL-PCR method (Liu et al., 1995). A promoter region from −1,126 to −1 (numbered from the first ATG) of NsBBLa was placed upstream of a GUS coding sequence on pGBW3 using Gateway Technology (Thermo Fisher Scientific). To generate transgenic tobacco plants and hairy roots, leaf discs from N. tabacum cv NC95 of wild type and nic1nic2 mutant were infected with Agrobacterium tumefaciens strain EHA105 and A. rhizogenes strain ATCC15834 harboring the GUS reporter construct, respectively (Horsch et al., 1985). Wild-type tobacco hairy roots used in Figure 6 were induced in a similar fashion, but with N. tabacum cv Petit Havana line SR1 and the A. rhizogenes strain without the binary vector. The hairy root lines were subcultured every week in 125-mL glass flasks filled with 30 mL of liquid B5 medium supplemented with 2% (w/v) Suc with shaking at 100 rpm in the dark. MeJA and NaCl were directly added to 4-d-old cultures to final concentrations of 100 μm and 300 mm, respectively.
GUS Reporter Assays
The GUS activity was detected histochemically and cross sections of the hairy roots were prepared as described by Shoji et al. (2000). Images of the stained tissues and sections were captured with a model no. SZX12 (Olympus) or a model no. Eclipse E-1000 (Nikon) microscope, both equipped with a model no. DP-70 digital camera (Olympus).
Frozen hairy roots were homogenized in 50 mm potassium P buffer (pH 7.0), 10 mm EDTA (pH 8.0), 0.1% (v/v) Triton X-100, and 0.1% (w/v) Sarcosyl. After centrifugation, the supernatants were desalted through NAP-5 columns (GE Healthcare) with P-buffered saline. Protein concentrations were determined with a Coomassie Protein Assay Reagent (Thermo Fisher Scientific). The protein solutions was supplemented with 4-methylumbelliferyl-β-d-glucuronide at 150 μm and incubated at 37°C for the reactions. The amount of 4-methylumbeliferone formed was measured by using a model no. F-4500 fluorescence spectrophotometer (Hitachi).
Genomic PCR Analysis
Genomic DNA was isolated using the CTAB method (Murray and Thompson, 1980) and used for PCR with Ex Taq DNA polymerase (Takara). The thermal program details for each primer pair are available upon request.
Accession Numbers
The promoter sequence of NsBBLa genes from N. sylvestris can be found in the GenBank/EMBL/DDBJ database under accession number LC201808.
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Phylogenetic trees of enzyme proteins involved in nicotine and related primary metabolism.
Supplemental Figure S2. A phylogenetic tree of BBL proteins from tobacco and its ancestral Nicotiana diploids.
Supplemental Figure S3. GUS activities in tobacco hairy roots transformed with the reporter gene driven by NsBBLa promoter.
Supplemental Figure S4. A phylogenetic tree of NIC2-locus ERFs and related proteins.
Supplemental Figure S5. A chromosomal region deleted in nic2 mutant was delimitated with genomic PCR analysis.
Supplemental Figure S6. Transcript levels of genes around NIC2 locus in the roots of wild-type and nic2 mutant tobacco were analyzed by qRT-PCR.
Supplemental Figure S7. Genes deleted in LA Burley 21 of nic1nic2 genotype were found by genomic PCR analysis.
Supplemental Table S1. Metabolic enzyme and transporter genes involved in nicotine and related metabolism in tobacco.
Supplemental Table S2. Primer sequences for qRT-PCR analysis.
Supplemental Table S3. NIC2-locus ERF genes and their homologs in tobacco.
Supplemental Table S4. Primer sequences for genomic PCR analysis to detect the chromosomal deletion around NIC2 locus in the mutants.
Supplemental Table S5. Gene found to be deleted in LA Burley 21.
Supplemental Table S6. Primer sequences for genomic PCR analysis to detect genes deleted in LA Burley 21.
Supplementary Material
Acknowledgments
We thank Drs. Tsuyoshi Nakagawa (Shimane University) and John Hamill (Deakin University) for providing the pGBW3 vector and seeds of the NC95 cultivar, respectively. We are grateful to Drs. James Battey and David Page for critically reading and correcting the manuscript.
Footnotes
[CC-BY]: Article free via Creative Commons CC-BY 4.0 license.
References
- Arimura G, Maffei M, editors (2017) Plant Specialized Metabolism: Genomics, Biochemistry, and Biological Functions. CRC Press, New York [Google Scholar]
- Bailey TL, Williams N, Misleh C, Li WW (2006) MEME: discovering and analyzing DNA and protein sequence motifs. Nucleic Acids Res 34: W369–W73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baldwin IT. (1989) Mechanism of damage-induced alkaloid production in wild tobacco. J Chem Ecol 15: 1661–1680 [DOI] [PubMed] [Google Scholar]
- Baldwin IT, Schmelz EA, Ohnmeiss TE (1994) Wound-induced changes in root and shoot jasmonic acid pools correlate with induced nicotine synthesis in Nicotiana sylvestris spegazzini and comes. J Chem Ecol 20: 2139–2157 [DOI] [PubMed] [Google Scholar]
- Bindler G, Plieske J, Bakaher N, Gunduz I, Ivanov N, Van der Hoeven R, Ganal M, Donini P (2011) A high density genetic map of tobacco (Nicotiana tabacum L.) obtained from large scale microsatellite marker development. Theor Appl Genet 123: 219–230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cárdenas PD, Sonawane PD, Pollier J, Vanden Bossche R, Dewangan V, Weithorn E, Tal L, Meir S, Rogachev I, Malitsky S, et al. (2016) GAME9 regulates the biosynthesis of steroidal alkaloids and upstream isoprenoids in the plant mevalonate pathway. Nat Commun 7: 10654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaplin JF. (1975) Registration of LAFC53 tobacco germplasm. Crop Sci 15: 282 [Google Scholar]
- Crooks GE, Hon G, Chandonia JM, Brenner SE (2004) WebLogo: a sequence logo generator. Genome Res 14: 1188–1190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis DL, Nielsen MT, editors (1999) Tobacco: Production, Chemistry, and Technology. Blackwell Science, Oxford, UK [Google Scholar]
- DeBoer KD, Lye JC, Aitken CD, Su AK, Hamill JD (2009) The A622 gene in Nicotiana glauca (tree tobacco): evidence for a functional role in pyridine alkaloid synthesis. Plant Mol Biol 69: 299–312 [DOI] [PubMed] [Google Scholar]
- DeBoer KD, Dalton HL, Edward FJ, Hamill JD (2011a) RNAi-mediated down-regulation of ornithine decarboxylase (ODC) leads to reduced nicotine and increased anatabine levels in transgenic Nicotiana tabacum L. Phytochemistry 72: 344–355 [DOI] [PubMed] [Google Scholar]
- DeBoer K, Tilleman S, Pauwels L, Vanden Bossche R, De Sutter V, Vanderhaeghen R, Hilson P, Hamill JD, Goossens A (2011b) APETALA2/ETHYLENE RESPONSE FACTOR and basic helix-loop-helix tobacco transcription factors cooperatively mediate jasmonate-elicited nicotine biosynthesis. Plant J 66: 1053–1065 [DOI] [PubMed] [Google Scholar]
- De Geyter N, Gholami A, Goormachtig S, Goossens A (2012) Transcriptional machineries in jasmonate-elicited plant secondary metabolism. Trends Plant Sci 17: 349–359 [DOI] [PubMed] [Google Scholar]
- Dewey RE, Xie J (2013) Molecular genetics of alkaloid biosynthesis in Nicotiana tabacum. Phytochemistry 94: 10–27 [DOI] [PubMed] [Google Scholar]
- Goossens J, Fernández-Calvo P, Schweizer F, Goossens A (2016) Jasmonates: signal transduction components and their roles in environmental stress responses. Plant Mol Biol 91: 673–689 [DOI] [PubMed] [Google Scholar]
- Hashimoto T, Tamaki K, Suzuki K, Yamada Y (1998) Molecular cloning of plant spermidine synthases. Plant Cell Physiol 39: 73–79 [DOI] [PubMed] [Google Scholar]
- Heim WG, Sykes KA, Hildreth SB, Sun J, Lu RH, Jelesko JG (2007) Cloning and characterization of a Nicotiana tabacum methylputrescine oxidase transcript. Phytochemistry 68: 454–463 [DOI] [PubMed] [Google Scholar]
- Hibi N, Higashiguchi S, Hashimoto T, Yamada Y (1994) Gene expression in tobacco low-nicotine mutants. Plant Cell 6: 723–735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hildreth SB, Gehman EA, Yang H, Lu RH, Ritesh KC, Harich KC, Yu S, Lin J, Sandoe JL, Okumoto S, Murphy AS, Jelesko JG (2011) Tobacco nicotine uptake permease (NUP1) affects alkaloid metabolism. Proc Natl Acad Sci USA 108: 18179–18184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horsch RB, Fry JE, Hoffmann NL, Eichholtz D, Rogers SG, Fraley RT (1985) A simple and general method for transferring genes into plants. Science 227: 1229–1231 [DOI] [PubMed] [Google Scholar]
- Imanishi S, Hashizume K, Nakakita M, Kojima H, Matsubayashi Y, Hashimoto T, Sakagami Y, Yamada Y, Nakamura K (1998) Differential induction by methyl jasmonate of genes encoding ornithine decarboxylase and other enzymes involved in nicotine biosynthesis in tobacco cell cultures. Plant Mol Biol 38: 1101–1111 [DOI] [PubMed] [Google Scholar]
- Junker A, Fischer J, Sichhart Y, Brandt W, Dräger B (2013) Evolution of the key alkaloid enzyme putrescine N-methyltransferase from spermidine synthase. Front Plant Sci 4: 260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kajikawa M, Hirai N, Hashimoto T (2009) A PIP-family protein is required for biosynthesis of tobacco alkaloids. Plant Mol Biol 69: 287–298 [DOI] [PubMed] [Google Scholar]
- Kajikawa M, Shoji T, Kato A, Hashimoto T (2011) Vacuole-localized berberine bridge enzyme-like proteins are required for a late step of nicotine biosynthesis in tobacco. Plant Physiol 155: 2010–2022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato K, Shitan N, Shoji T, Hashimoto T (2015) Tobacco NUP1 transports both tobacco alkaloids and vitamin B6. Phytochemistry 113: 33–40 [DOI] [PubMed] [Google Scholar]
- Kato K, Shoji T, Hashimoto T (2014) Tobacco nicotine uptake permease regulates the expression of a key transcription factor gene in the nicotine biosynthesis pathway. Plant Physiol 166: 2195–2204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh A, Shoji T, Hashimoto T (2007) Molecular cloning of N-methylputrescine oxidase from tobacco. Plant Cell Physiol 48: 550–554 [DOI] [PubMed] [Google Scholar]
- Katoh A, Uenohara K, Akita M, Hashimoto T (2006) Early steps in the biosynthesis of NAD in Arabidopsis start with aspartate and occur in the plastid. Plant Physiol 141: 851–857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D, Langmead B, Salzberg SL (2015) HISAT: a fast spliced aligner with low memory requirements. Nat Methods 12: 357–360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Legg PG, Collins GB (1971) Inheritance of percent total alkaloids in Nicotiana tabacum L. II. Genetic effects of two loci in Burley 21 × LA Burley 21 populations. Can J Genet Cytol 13: 287–291 [Google Scholar]
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R; 1000 Genome Project Data Processing Subgroup (2009) The sequence alignment/map (SAM) format and SAMtools. Bioinformatics 25: 2078–2079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu YG, Mitsukawa N, Oosumi T, Whittier RF (1995) Efficient isolation and mapping of Arabidopsis thaliana T-DNA insert junctions by thermal asymmetric interlaced PCR. Plant J 8: 457–463 [DOI] [PubMed] [Google Scholar]
- Moghe GD, Last RL (2015) Something old, something new: conserved enzymes and the evolution of novelty in plant specialized metabolism. Plant Physiol 169: 1512–1523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murad L, Lim KY, Christopodulou V, Matyasek R, Lichtenstein CP, Kovarik A, Leitch AR (2002) The origin of tobacco’s T genome is traced to a particular lineage within Nicotiana tomentosiformis (Solanaceae). Am J Bot 89: 921–928 [DOI] [PubMed] [Google Scholar]
- Murray MG, Thompson WF (1980) Rapid isolation of high molecular weight plant DNA. Nucleic Acids Res 8: 4321–4325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naconsie M, Kato K, Shoji T, Hashimoto T (2014) Molecular evolution of N-methylputrescine oxidase in tobacco. Plant Cell Physiol 55: 436–444 [DOI] [PubMed] [Google Scholar]
- Nagata T, Nemoto Y, Hasezawa S (1992) Tobacco BY-2 cells as the HeLa cells in the cell biology of higher plants. Int Rev Cytol 132: 1–30 [Google Scholar]
- Nakano T, Suzuki K, Fujimura T, Shinshi H (2006) Genome-wide analysis of the ERF gene family in Arabidopsis and rice. Plant Physiol 140: 411–432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nützmann HW, Osbourn A (2014) Gene clustering in plant specialized metabolism. Curr Opin Biotechnol 26: 91–99 [DOI] [PubMed] [Google Scholar]
- Patra B, Schluttenhofer C, Wu Y, Pattanaik S, Yuan L (2013) Transcriptional regulation of secondary metabolite biosynthesis in plants. Biochim Biophys Acta 1829: 1236–1247 [DOI] [PubMed] [Google Scholar]
- Pertea M, Kim D, Pertea GM, Leek JT, Salzberg SL (2016) Transcript-level expression analysis of RNA-seq experiments with HISAT, StringTie and Ballgown. Nat Protoc 11: 1650–1667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riechers DE, Timko MP (1999) Structure and expression of the gene family encoding putrescine N-methyltransferase in Nicotiana tabacum: new clues to the evolutionary origin of cultivated tobacco. Plant Mol Biol 41: 387–401 [DOI] [PubMed] [Google Scholar]
- Ryan SM, Cane KA, DeBoer KD, Sinclair SJ, Brimblecombe R, Hamill JD (2012) Structure and expression of the quinolinate phosphoribosyltransferase (QPT) gene family in Nicotiana. Plant Sci 188-189: 102–110 [DOI] [PubMed] [Google Scholar]
- Schenck CA, Chen S, Siehl DL, Maeda HA (2015) Non-plastidic, tyrosine-insensitive prephenate dehydrogenases from legumes. Nat Chem Biol 11: 52–57 [DOI] [PubMed] [Google Scholar]
- Shoji T, Hashimoto T (2008) Why does anatabine, but not nicotine, accumulate in jasmonate-elicited cultured tobacco BY-2 cells? Plant Cell Physiol 49: 1209–1216 [DOI] [PubMed] [Google Scholar]
- Shoji T, Hashimoto T (2011a) Nicotine biosynthesis. In Ashihara H, Crozier A, Komamine A, eds, Plant Metabolism and Biotechnology. John Wiley & Sons, New York, pp 191-216 [Google Scholar]
- Shoji T, Hashimoto T (2011b) Recruitment of a duplicated primary metabolism gene into the nicotine biosynthesis regulon in tobacco. Plant J 67: 949–959 [DOI] [PubMed] [Google Scholar]
- Shoji T, Hashimoto T (2011c) Tobacco MYC2 regulates jasmonate-inducible nicotine biosynthesis genes directly and by way of the NIC2-locus ERF genes. Plant Cell Physiol 52: 1117–1130 [DOI] [PubMed] [Google Scholar]
- Shoji T, Hashimoto T (2012) DNA-binding and transcriptional activation properties of tobacco NIC2-locus ERF189 and related transcription factors. Plant Biotechnol 29: 35–42 [Google Scholar]
- Shoji T, Hashimoto T (2013) Smoking out the masters; transcriptional regulators for nicotine biosynthesis. Plant Biotechnol 30: 217–224 [Google Scholar]
- Shoji T, Hashimoto T (2015) Stress-induced expression of NICOTINE2-locus genes and their homologs encoding Ethylene Response Factor transcription factors in tobacco. Phytochemistry 113: 41–49 [DOI] [PubMed] [Google Scholar]
- Shoji T, Inai K, Yazaki Y, Sato Y, Takase H, Shitan N, Yazaki K, Goto Y, Toyooka K, Matsuoka K, Hashimoto T (2009) Multidrug and toxic compound extrusion-type transporters implicated in vacuolar sequestration of nicotine in tobacco roots. Plant Physiol 149: 708–718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shoji T, Kajikawa M, Hashimoto T (2010) Clustered transcription factor genes regulate nicotine biosynthesis in tobacco. Plant Cell 22: 3390–3409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shoji T, Mishima M, Hashimoto T (2013) Divergent DNA-binding specificities of a group of ETHYLENE RESPONSE FACTOR transcription factors involved in plant defense. Plant Physiol 162: 977–990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shoji T, Ogawa T, Hashimoto T (2008) Jasmonate-induced nicotine formation in tobacco is mediated by tobacco COI1 and JAZ genes. Plant Cell Physiol 49: 1003–1012 [DOI] [PubMed] [Google Scholar]
- Shoji T, Winz R, Iwase T, Nakajima K, Yamada Y, Hashimoto T (2002) Expression patterns of two tobacco isoflavone reductase-like genes and their possible roles in secondary metabolism in tobacco. Plant Mol Biol 50: 427–440 [DOI] [PubMed] [Google Scholar]
- Shoji T, Yamada Y, Hashimoto T (2000) Jasmonate induction of putrescine N-methyltransferase genes in the root of Nicotiana sylvestris. Plant Cell Physiol 41: 831–839 [DOI] [PubMed] [Google Scholar]
- Sierro N, Battey JN, Ouadi S, Bakaher N, Bovet L, Willig A, Goepfert S, Peitsch MC, Ivanov NV (2014) The tobacco genome sequence and its comparison with those of tomato and potato. Nat Commun 5: 3833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sierro N, Battey JN, Ouadi S, Bovet L, Goepfert S, Bakaher N, Peitsch MC, Ivanov NV (2013) Reference genomes and transcriptomes of Nicotiana sylvestris and Nicotiana tomentosiformis. Genome Biol 14: R60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinclair SJ, Murphy KJ, Birch CD, Hamill JD (2000) Molecular characterization of quinolinate phosphoribosyltransferase (QPRtase) in Nicotiana. Plant Mol Biol 44: 603–617 [DOI] [PubMed] [Google Scholar]
- Stenzel O, Teuber M, Dräger B (2006) Putrescine N-methyltransferase in Solanum tuberosum L., a calystegine-forming plant. Planta 223: 200–212 [DOI] [PubMed] [Google Scholar]
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S (2013) MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol Biol Evol 30: 2725–2729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thagun C, Imanishi S, Kudo T, Nakabayashi R, Ohyama K, Mori T, Kawamoto K, Nakamura Y, Katayama M, Nonaka S, et al. (2016) Jasmonate-responsive ERF transcription factors regulate steroidal glycoalkaloid biosynthesis in tomato. Plant Cell Physiol 57: 961–975 [DOI] [PubMed] [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ (1994) CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res 22: 4673–4680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turatsinze JV, Thomas-Chollier M, Defrance M, van Helden J (2008) Using RSAT to scan genome sequences for transcription factor binding sites and cis-regulatory modules. Nat Protoc 3: 1578–1588 [DOI] [PubMed] [Google Scholar]
- van der Fits L, Memelink J (2000) ORCA3, a jasmonate-responsive transcriptional regulator of plant primary and secondary metabolism. Science 289: 295–297 [DOI] [PubMed] [Google Scholar]
- van Moerkercke A, Steensma P, Schweizer F, Pollier J, Gariboldi I, Payne R, Vanden Bossche R, Miettinen K, Espoz J, Purnama PC, et al. (2015) The bHLH transcription factor BIS1 controls the iridoid branch of the monoterpenoid indole alkaloid pathway in Catharanthus roseus. Proc Natl Acad Sci USA 112: 8130–8135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Bennetzen JL (2015) Current status and prospects for the study of Nicotiana genomics, genetics, and nicotine biosynthesis genes. Mol Genet Genomics 290: 11–21 [DOI] [PubMed] [Google Scholar]
- Wegel E, Koumproglou R, Shaw P, Osbourn A (2009) Cell type-specific chromatin decondensation of a metabolic gene cluster in oats. Plant Cell 21: 3926–3936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winzer T, Gazda V, He Z, Kaminski F, Kern M, Larson TR, Li Y, Meade F, Teodor R, Vaistij FE, et al. (2012) A Papaver somniferum 10-gene cluster for synthesis of the anticancer alkaloid noscapine. Science 336: 1704–1708 [DOI] [PubMed] [Google Scholar]
- Xu B, Sheehan MJ, Timko MP (2004) Differential induction of ornithine decarboxylase (ODC) gene family members in transgenic tobacco (Nicotiana tabacum L. cv. Bright Yellow 2) cell suspensions by methyl-jasmonate treatment. Plant Mol Biol 44: 101–116 [Google Scholar]
- Xu B, Timko M (2004) Methyl jasmonate induced expression of the tobacco putrescine N-methyltransferase genes requires both G-box and GCC-motif elements. Plant Mol Biol 55: 743–761 [DOI] [PubMed] [Google Scholar]
- Zhang HB, Bokowiec MT, Rushton PJ, Han SC, Timko MP (2012) Tobacco transcription factors NtMYC2a and NtMYC2b form nuclear complexes with the NtJAZ1 repressor and regulate multiple jasmonate-inducible steps in nicotine biosynthesis. Mol Plant 5: 73–84 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.