LEAFY and APETALA1/CAULIFLOWER have partially antagonistic activities in the control of floral initiation.
Abstract
The transcription factors LEAFY (LFY) and APETALA1 (AP1), together with the AP1 paralog CAULIFLOWER (CAL), control the onset of flower development in a partially redundant manner. This redundancy is thought to be mediated, at least in part, through the regulation of a shared set of target genes. However, whether these genes are independently or cooperatively regulated by LFY and AP1/CAL is currently unknown. To better understand the regulatory relationship between LFY and AP1/CAL and to obtain deeper insights into the control of floral initiation, we monitored the activity of LFY in the absence of AP1/CAL function. We found that the regulation of several known LFY target genes is unaffected by AP1/CAL perturbation, while others appear to require AP1/CAL activity. Furthermore, we obtained evidence that LFY and AP1/CAL control the expression of some genes in an antagonistic manner. Notably, these include key regulators of floral initiation such as TERMINAL FLOWER1 (TFL1), which had been previously reported to be directly repressed by both LFY and AP1. We show here that TFL1 expression is suppressed by AP1 but promoted by LFY. We further demonstrate that LFY has an inhibitory effect on flower formation in the absence of AP1/CAL activity. We propose that LFY and AP1/CAL act as part of an incoherent feed-forward loop, a network motif where two interconnected pathways or transcription factors act in opposite directions on a target gene, to control the establishment of a stable developmental program for the formation of flowers.
The onset of flowering is a key process during the life cycle of angiosperms and is controlled by a complex network of signaling pathways, which integrate information stemming from both environmental and developmental cues (Fornara et al., 2010). A detailed understanding of floral initiation is not only important for plant reproductive biology but is also helpful as a knowledge base for generating improved crop plants with higher yields.
The morphological changes that go along with the switch from the vegetative to the reproductive phase of development culminate in the initiation of floral primordia by the inflorescence meristem. These primordia are programmed to undergo floral organogenesis by the activities of so-called floral meristem identity genes of which LEAFY (LFY), APETALA1 (AP1), and the AP1 paralog CAULIFLOWER (CAL) are arguably most important in the model plant Arabidopsis (Arabidopsis thaliana; Ó’Maoiléidigh et al., 2014). These genes have been shown to activate key regulators of flower development and to suppress the expression of floral repressors, that is of genes that prevent flowering when the conditions for reproduction are not favorable (Busch et al., 1999; Kaufmann et al., 2010; Moyroud et al., 2011; Pajoro et al., 2014; Parcy et al., 1998; Wellmer et al., 2006; Winter et al., 2011). Thus, they appear to constitute a hub in the gene regulatory network that controls flowering.
LFY encodes a plant-specific transcription factor, whose structure has recently been elucidated in atomic detail (Hamès et al., 2008; Sayou et al., 2016). It is expressed most strongly on the flanks of the inflorescence meristem in floral anlagen (i.e. in incipient floral primordia) as well as in floral primordia at the earliest stages of development (Weigel et al., 1992). In contrast, LFY is not expressed in the inflorescence meristem proper where shoot identity genes such as TERMINAL FLOWER1 (TFL1; Bradley et al., 1997) prevent cells from adopting a floral fate (Baumann et al., 2015).
AP1 and CAL encode closely related MADS domain transcription factors (Kempin et al., 1995; Mandel et al., 1992), which act largely redundantly in the control of floral meristem identity specification. This is best demonstrated by ap1 cal double-mutant plants, which exhibit a severe delay in flower formation and undergo a massive overproliferation of inflorescence-like meristems (Bowman et al., 1993; Ferrándiz et al., 2000). AP1/CAL are expressed exclusively in floral primordia (Kempin et al., 1995; Mandel et al., 1992); thus, their expression commences later than that of LFY.
Over the years, the regulatory relationship between LFY and AP1/CAL has become a case example for gene interactions in the control of developmental transitions. LFY has been shown to directly activate AP1 and CAL expression in floral primordia (Wagner et al., 1999; William et al., 2004). AP1 (and probably CAL) then acts on LFY to reinforce its expression (Kaufmann et al., 2010; Liljegren et al., 1999). Thus, LFY and AP1/CAL are part of a positive feedback loop, which ensures that these floral meristem identity factors are expressed at high levels in early-stage floral primordia. Genetic evidence suggests that AP1/CAL and LFY then act in part redundantly to confer floral meristem identity fate. Specifically, it was shown that flower formation in lfy ap1 double mutants is much more severely affected than in either of the single mutants (Weigel et al., 1992). Also, the characterization of the gene expression programs acting downstream of LFY and AP1 through genomic technologies (Kaufmann et al., 2010; Moyroud et al., 2011; Pajoro et al., 2014; Wagner et al., 2004; Wellmer et al., 2006; William et al., 2004; Winter et al., 2011) revealed that LFY and AP1 share many target genes and often bind to adjacent sites in the Arabidopsis genome (Winter et al., 2011). The idea of a shared set of target genes has been confirmed by the results of a recent meta-analysis of available genome-wide data sets for the two transcription factors (Winter et al., 2015). It was shown that they jointly control the expression of ∼200 genes, which include many known regulators of flower development. These shared targets likely provide the molecular basis for the partial redundancy between LFY and AP1/CAL in the control of floral initiation. However, whether LFY and AP1/CAL provide independent inputs for the regulation of common target genes or instead function in a cooperative manner is currently unknown.
In this study, we investigated the regulatory relationship between LFY and AP1/CAL by determining the effects on gene expression that a specific activation of LFY has in the absence of AP1/CAL function. We found that while LFY can regulate some known targets independently of AP1/CAL, other genes appear to require the input of both classes of factors. We also found evidence for an antagonistic regulation of certain targets by LFY and AP1/CAL. Notably, these comprise several known regulators of floral induction, including TFL1. We show that LFY activity leads to an inhibition of flower formation in the absence of AP1/CAL function and propose that the antagonism between LFY and AP1/CAL contributes to a tight control of the onset of flower formation.
RESULTS
The Transcriptional Program Controlled by LFY in the Absence of AP1/CAL Function
To test the effects of LFY on gene expression in the absence of AP1/CAL function, we used a previously described transgenic line (Wagner et al., 1999), which constitutively expresses a fusion between LFY and the hormone-binding domain of the rat glucocorticoid receptor (LFY-GR) from the Cauliflower Mosaic Virus 35S promoter (p35S; line denoted: p35S:LFY-GR). It has been shown that the activation of LFY-GR through treatment of plants with the steroid hormone dexamethasone rescues the floral phenotypes of lfy null mutants, indicating that the fusion protein is fully functional (Wagner et al., 1999). We introgressed the p35S:LFY-GR transgene into an ap1-1 cal-1 double-mutant background, which is characterized by a considerable delay in flower formation and the accumulation of inflorescence-like meristems (Bowman et al., 1993). Using this line, we collected inflorescence tissue at different time points (0, 2, 4, 8, and 12 h) after activation of the LFY-GR fusion protein, as well as from mock-treated control plants (Fig. 1A). Next, we determined, through whole-genome microarray analysis (Supplemental Fig. S1), gene expression changes that occurred as a consequence of LFY-GR activation and identified 669 differentially expressed genes (DEGs) across all time points (Supplemental Data Set 1). As expected, the number of DEGs identified at the different time points increased overall over the course of the experiment (Supplemental Table S1). Similar to what has been described previously for AP1 (Kaufmann et al., 2010; Wellmer et al., 2006), we detected a preponderance of gene repression after LFY-GR activation in ap1 cal inflorescences (Supplemental Table S1; Fig. 1B). For the majority of genes (507 of 669), differential expression was observed in more than one time point (but excluding the 0-d time point), indicating that once a gene exhibited up- or down-regulation in response to LFY-GR activation, it was often differentially expressed also in subsequent time points. A Gene Ontology analysis of the DEGs revealed an enrichment of terms related to, for example, the regulation of transcription, meristem and flower development, and the response to different phytohormones (Fig. 1C), largely in agreement with what has been described previously for the gene regulatory program that acts downstream of LFY (Winter et al., 2011). Thus, in the absence of AP1/CAL function, LFY’s regulatory activity appears to be overall similar to that in a wild-type background.
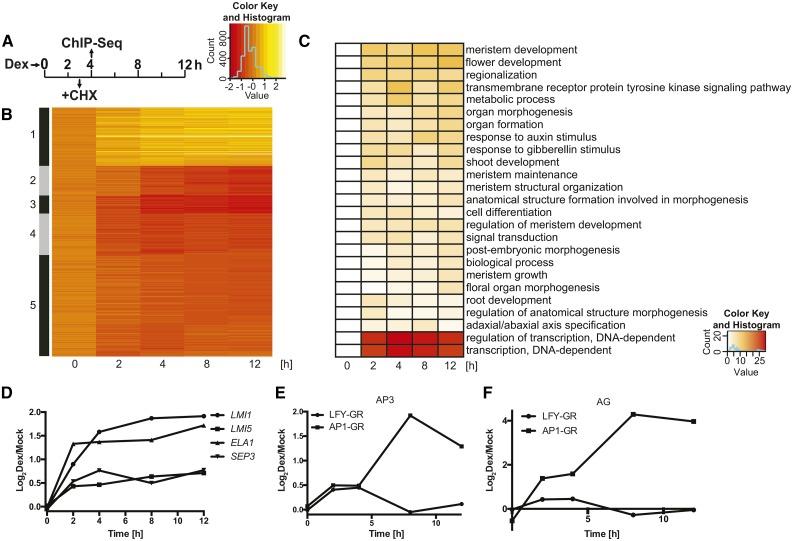
Figure 1.
Genes controlled by LFY in the absence of AP1/CAL function. A, Experimental set-up of the genome-wide analyses using inflorescence-like meristems from p35S:LFY-GR ap1 cal plants. CHX, cycloheximide; Dex, dexamethasone. B, K-means clustering (k = 5) of 669 DEGs identified in the p35S:LFY-GR ap1 cal time course experiment. Log2-transformed expression ratios (dexamethasone/mock) for the different time points (as indicated) were used for the analysis. C, Gene Ontology terms enriched among the DEGs at different time points (as indicated). Negative decadal logarithms of P values are shown. D, Response of selected LFY targets (as indicated) to an activation of LFY-GR in ap1 cal inflorescences. E and F, Response of AP3 (E) and AG (F) to an activation of LFY-GR and AP1-GR, respectively, in ap1 cal inflorescences. Data for AP1-GR were taken from Kaufmann et al. (2010). In D to F, log2-transformed fold-change values (dexamethasone/mock) from microarray experiments are shown.
To determine which of the DEGs may be directly regulated by LFY in ap1 cal inflorescence-like meristems, we activated LFY-GR in ap1 cal inflorescences in the presence of the translational inhibitor cycloheximide and conducted microarray experiments to identify gene expression changes that occur in the absence of protein biosynthesis. Because cycloheximide is a cellular toxin, we limited this analysis to a 3-h time point after LFY-GR activation to curb nonspecific effects (Fig. 1A). We found that 87 of the 116 (∼75%) DEGs identified in this experiment were also present among the 669 DEGs identified in the time course experiment and, as expected, these genes were responding at early time points after LFY-GR activation (Supplemental Data Set 2; Supplemental Fig. S2). We also compared the list of DEGs from the time course experiment to the results from published genome-wide localization studies for LFY in a wild-type background (Moyroud et al., 2011; Winter et al., 2011), as well as to the data from a chromatin immunoprecipitation-sequencing (ChIP-seq) experiment we conducted using LFY-specific antibodies we generated and inflorescence tissue from dexamethasone-treated p35S:LFY-GR ap1 cal plants. In total, we obtained evidence for LFY binding to 190 of the 669 DEGs (∼30%) identified in the time-course experiment (Supplemental Data Set 3), implying that many of the DEGs are indeed direct LFY targets. In agreement with this idea, we found among these genes well-characterized LFY targets such as LATE MERISTEM IDENTITY1 (LMI1; Saddic et al., 2006), LMI5 (William et al., 2004), SEPALLATA3 (Winter et al., 2011), and EUI-LIKE P450 A1 (Yamaguchi et al., 2014; Supplemental Fig. S3). The expression of these genes was up-regulated in response to LFY-GR activation in agreement with the previous reports (Fig. 1D). In contrast, other previously identified direct targets, such as the floral organ identity genes AP3 and AGAMOUS (AG), which are activated by LFY during early flower development (Parcy et al., 1998; Winter et al., 2011), showed no significant expression differences at any of the time points taken. This is in marked contrast to the strong and rapid induction of these genes after activation of an AP1-GR fusion protein in ap1 cal inflorescences (Fig. 1, E and F; Kaufmann et al., 2010). Because we detected in our ChIP-seq experiment LFY binding to the regulatory regions of these genes (as well as to those of the floral organ identity genes AP1 and PISTILLATA [PI]; Supplemental Fig. S4), it appears that the lack of a pronounced transcriptional response for AP3 and AG in the p35S:LFY-GR ap1 cal time course experiment was due to an absence of functional AP1.
Regulatory Interplay between LFY and AP1
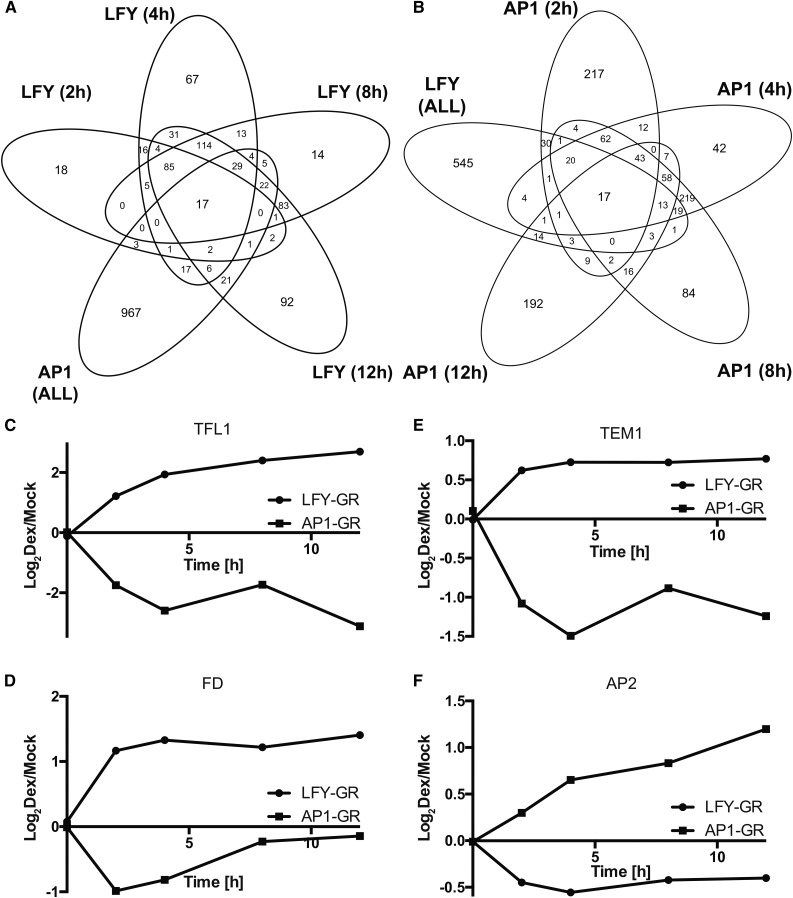
A recent meta-analysis of available data sets from transcriptomics experiments and genome-wide localization studies for LFY and AP1 showed that the two transcription factors share many (∼200) direct target genes (Winter et al., 2015). While the directionality of expression changes was identical for most of these genes, in some cases, opposite transcriptional responses to LFY or AP1 activation were detected. Because the expression profiling experiments used for this analysis were conducted with different plant material (inflorescences-like meristems of ap1 cal double mutants in the case of AP1; 9-d-old wild-type seedlings in the case of LFY), stage- and/or tissue-specific differences in the activities of the transcription factors could account for these observations. Yet, it is also possible that AP1 and LFY exhibit at least in some cases bona fide antagonistic activities. Given that the available genetic evidence suggest that the two transcription factors promote the initiation of flower development in a partially redundant manner, we considered this possibility unlikely. However, when we compared the results from the p35S:LFY-GR ap1 cal time course experiment with a data set stemming from an equivalent experiment (i.e. p35S:AP1-GR ap1 cal), we conducted previously for the analysis of AP1 function (Kaufmann et al., 2010), we not only found, as expected, a large overlap between the DEGs identified in the two experiments (143 genes; expected by chance: ∼33; Fig. 2, A and B; Supplemental Data Set 1), but also that the directionality of the observed expression changes appeared to be reversed for several genes (we identified 24 possible cases among the 143 DEGs shared between the data sets; Supplemental Data Set 1). Notably, the genes that showed opposite transcriptional responses included key regulators involved in the control of floral initiation such as TFL1, FLOWERING LOCUS D (FD; Abe et al., 2005; Wigge et al., 2005), TEMPRANILLO1 (Castillejo and Pelaz, 2008), and AP2 (Jofuku et al., 1994). The first three of these genes were down-regulated in response to AP1-GR activation but up-regulated by LFY-GR (Fig. 2, C–E). Conversely, AP2 expression was promoted by AP1-GR and repressed by LFY-GR activation (Fig. 2F). Because these experiments were conducted using the same tissue, identical time points after activation of the fusion proteins, as well as similar analysis procedures, it appears that AP1 and LFY have indeed antagonistic activities in the regulation of some of their target genes.
Figure 2.
Comparison of gene expression programs acting downstream of LFY and AP1. A and B, Overlap between genes differentially expressed after AP1-GR and LFY-GR activation in ap1 cal inflorescences. Overlaps for different time points (as indicated) are shown in A for the LFY-GR and in B for the AP1-GR time course experiments. C to F, Antagonistic regulation of selected genes after activation of AP1-GR and LFY-GR in ap1 cal inflorescences. Log2-transformed expression ratios (dexamethasone/mock) for the different time points (as indicated) are shown. Data for AP1-GR were taken from Kaufmann et al. (2010). C, TFL1. D, FD. E, TEMPRANILLO1. F, AP2.
LFY Activates TFL1 Expression in Absence of AP1/CAL
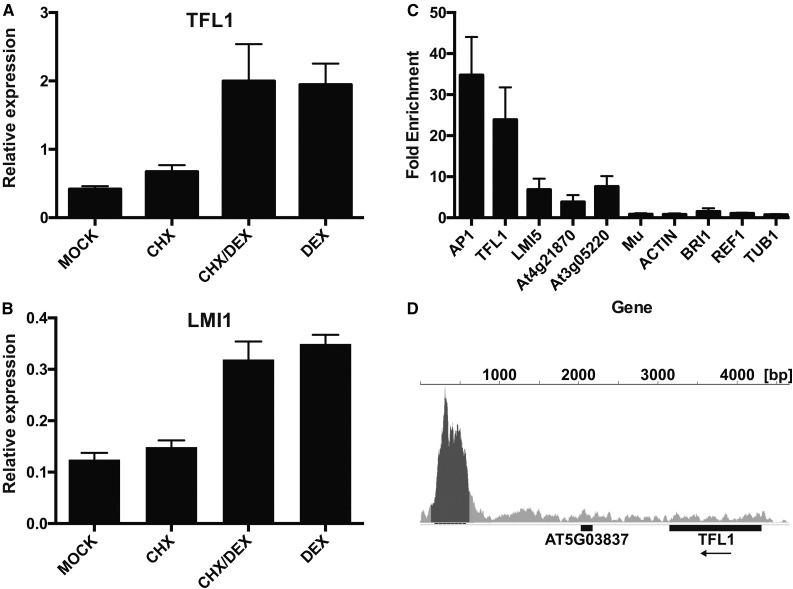
Of the genes with opposite transcriptional responses, TFL1 showed the strongest difference in expression after LFY-GR and AP1-GR activation, respectively. This result is contrary to the established view that TFL1 is directly repressed by both LFY and AP1 (Kaufmann et al., 2010; Liljegren et al., 1999; Winter et al., 2011). We therefore focused on this gene to validate our observations and to understand the function of the presumed antagonistic regulation by LFY and AP1. To confirm the microarray data, we used quantitative reverse transcription PCR (qRT-PCR) and detected up-regulation of TFL1 3 h after dexamethasone treatment of p35S:LFY-GR ap1 cal plants (Fig. 3A). We next asked whether the effect of LFY-GR activation on TFL1 expression is caused by direct or indirect regulation. When we activated LFY-GR in the presence of cycloheximide, we found that TFL1 expression (as well as expression of the known direct LFY target LMI1, which we used as a control; Fig. 3B) increased to an extent similar to that in plants in which protein biosynthesis had not been inhibited (Fig. 3A), suggesting that the effect is not mediated by intermediary proteins. Also, ChIP-qPCR assays (Fig. 3C) and the results of our ChIP-seq analysis (Fig. 3D) showed that in ap1 cal inflorescences, LFY-GR binds to a conserved region ∼2.8 to 3.3 kb downstream of TFL1, which is known to be bound by LFY in a wild-type background (Moyroud et al., 2011; Winter et al., 2011) and to be important for proper TFL1 regulation in the inflorescence (Serrano-Mislata et al., 2016). Taken together, these results strongly suggest that the induction of TFL1 by LFY-GR in ap1 cal is mediated by direct regulation.
Figure 3.
Direct activation of TFL1 by LFY. A and B, Relative expression of TFL1 (A) and LMI1 (B) in p35S:LFY-GR ap1 cal inflorescences after mock treatment (mock), treatments with cycloheximide (CHX) or dexamethasone (DEX) alone, or after a combined treatment with both cycloheximide and dexamethasone (CHX/DEX). C, Results of ChIP-qPCR experiments. Binding of LFY to selected known target genes (on the left, as indicated) and negative control regions (on the right; MU through TUB1) was tested after activation of LFY-GR in ap1 cal inflorescences. Error bars indicate ses of four biologically independent measurements. D, Results of ChIP-Seq experiments. Binding of LFY to the TFL1 locus was tested after activation of LFY-GR in ap1 cal inflorescences. A peak was detected ∼2.8 to 3.3 kb downstream of the transcribed region of the gene.
An ectopic activation of TFL1 expression has been reported previously in lines in which a fusion between LFY and the VP16 transcriptional activation domain was expressed from the LFY promoter (Parcy et al., 2002). In fact, this TFL1 overexpression was found to be partially responsible for the floral reversion phenotype observed in pLFY:LFY-VP16 plants, especially in an ag mutant background (Parcy et al., 2002). Because LFY normally promotes flowering (Weigel et al., 1992) and TFL1 represses it (Baumann et al., 2015), this apparent positive effect of LFY-VP16 on TFL1 expression was deemed to be antimorphic (Parcy et al., 2002). Under this scenario, the fusion of the strong activation domain VP16 to LFY would have converted the transcription factor from being a repressor of TFL1 expression into an activator. At the same time, it has been shown that LFY-VP16 can activate the expression of the floral homeotic gene AG independently of other factors, suggesting that the fusion protein can also function as a hypermorph relative to LFY alone (Parcy et al., 2002). Thus, LFY-VP16 may be able to act as a hypermorph for some target genes and as an antimorph for others.
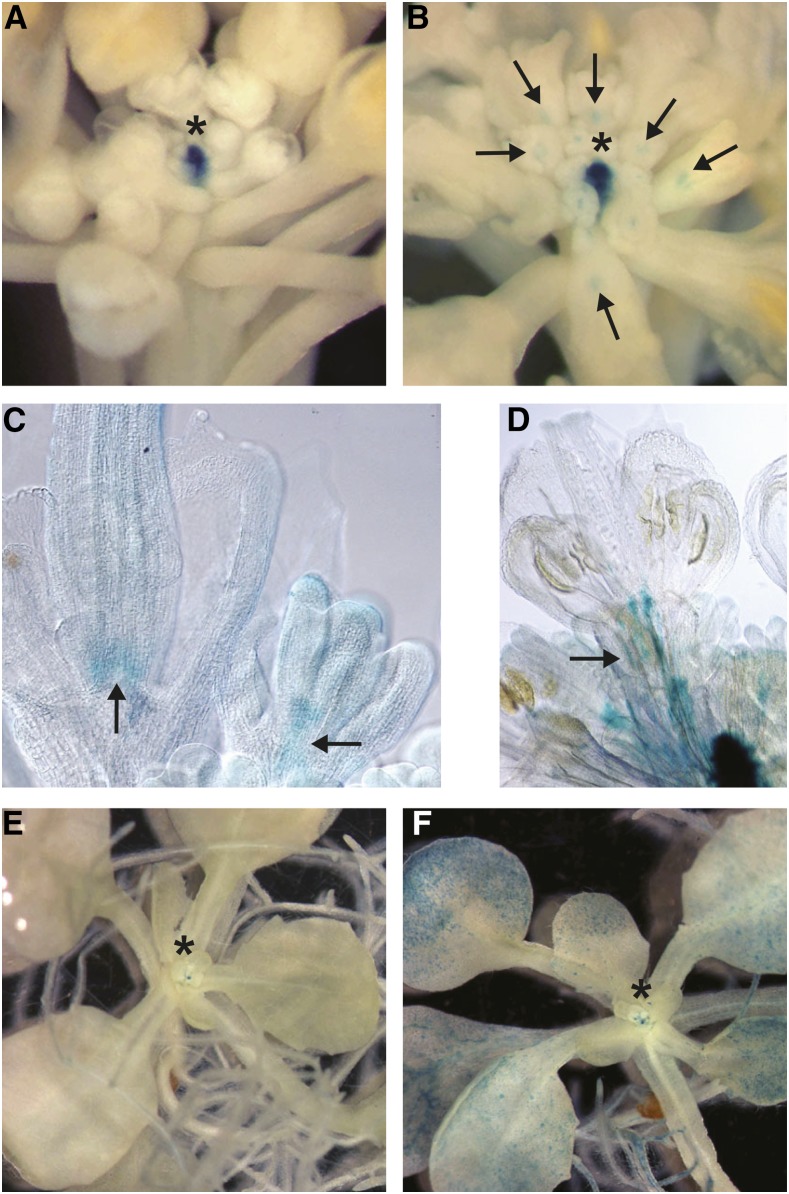
While this scenario remains a possibility, the results of the experiments outlined above strongly suggested that LFY’s normal function is to promote TFL1 expression, at least in the absence of AP1/CAL activity. Therefore, the observed ectopic expression of TFL1 in response to LFY-VP16 may not be a consequence of the fusion protein behaving as an antimorph, as previously suggested. To further investigate this, we first confirmed ectopic TFL1 expression in response to LFY-VP16 activity. To this end, we used a pTFL1:GUS reporter construct (Serrano-Mislata et al., 2016) to determine TFL1 promoter activity in lines expressing LFY-VP16 from the LFY promoter. Largely in agreement with the previous report (Parcy et al., 2002), we found pTFL1:GUS activity not only in the shoot meristem as in the wild type (Fig. 4A), but also in flowers, especially at the base of gynoecia (Fig. 4, B–D) where the LFY promoter is active at later floral stages (Weigel et al., 1992). Because the ectopic pTFL1:GUS expression in these lines could be an indirect effect of prolonged LFY-VP16 activity, we next tested whether an inducible expression of LFY-VP16 would lead to similar effects. For this, we analyzed pTFL1:GUS activity in plants expressing LFY-VP16 under control of a heat shock promoter (Benlloch et al., 2011). We found that pTFL1:GUS was broadly activated in wild-type seedlings that had been subjected to heat shock treatment but not in plants kept under normal growth conditions (Fig. 4, E and F). Taken together, these results, as well as the results of the experiments with the LFY-GR fusion protein, strongly suggest that LFY can promote TFL1 expression.
Figure 4.
Ectopic expression of TFL1 in LFY-VP16 lines. A, pTFL1:GUS activity in a wild-type inflorescence. Staining was restricted to the shoot apical meristem. B to D, pTFL1:GUS activity in pLFY:LFY-VP16 inflorescences. B, Staining was observed in the center of the shoot apical meristem as well as in flowers. C and D, Staining at the base of the gynoecium (C) and along pedicels (D) in cleared flowers. Arrows point to ectopic pTFL1:GUS activity in flowers. E and F, Ectopic activation of pTFL1:GUS in plants expressing LFY-VP16 from a heat shock promoter. E, Seedlings grown at ambient temperature with pTFL1:GUS activity in the shoot apical meristem. F, Heat-shocked seedling with additional staining in cotyledons, leaves, and roots. Asterisks in panels A, B, E, and F mark the position of the shoot apical meristem.
LFY Inhibits Flower Formation in the Absence of AP1/CAL
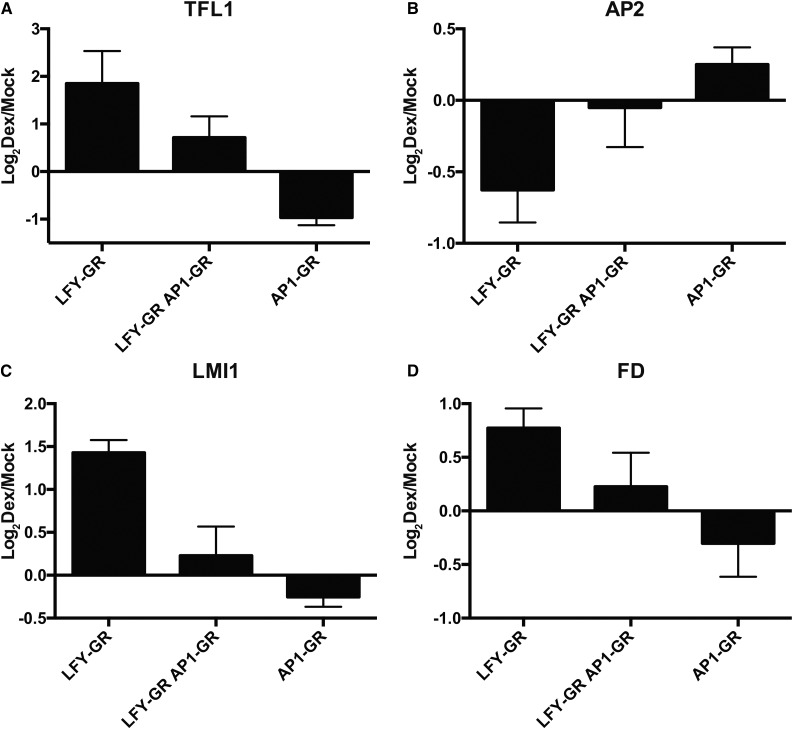
The results from the experiments outlined above implied that AP1 and LFY control the expression of TFL1 and of several other regulators of floral initiation in an antagonistic manner. To directly test this, we generated a line that carried p35S:LFY-GR as well as an pAP1:AP1-GR construct (Ó’Maoiléidigh et al., 2013) in an ap1 cal mutant background. Using this line, we compared the effects of an activation of both fusion proteins on gene expression to those detected after activating either AP1-GR or LFY-GR alone. We found that the simultaneous activation of AP1-GR or LFY-GR resulted in expression levels for TFL1, FD, and AP2 that were between those observed for either AP1-GR or LFY-GR alone (Fig. 5). These results imply that LFY and AP1 indeed provide opposite and possibly independent inputs for the regulation of several key regulators of flowering.
Figure 5.
Antagonistic activities of LFY and AP1. A to D, Effects of LFY-GR and AP1-GR activation, or of a simultaneous activation of both fusion proteins in ap1 cal inflorescences on the expression of selected genes (as indicated). Tissue was collected 3 h after treating plants with a dexamethasone-containing solution. Log2-transformed fold-change values (dexamethasone/mock) from qRT-PCR experiments are shown. Error bars indicate ses from four independent measurements.
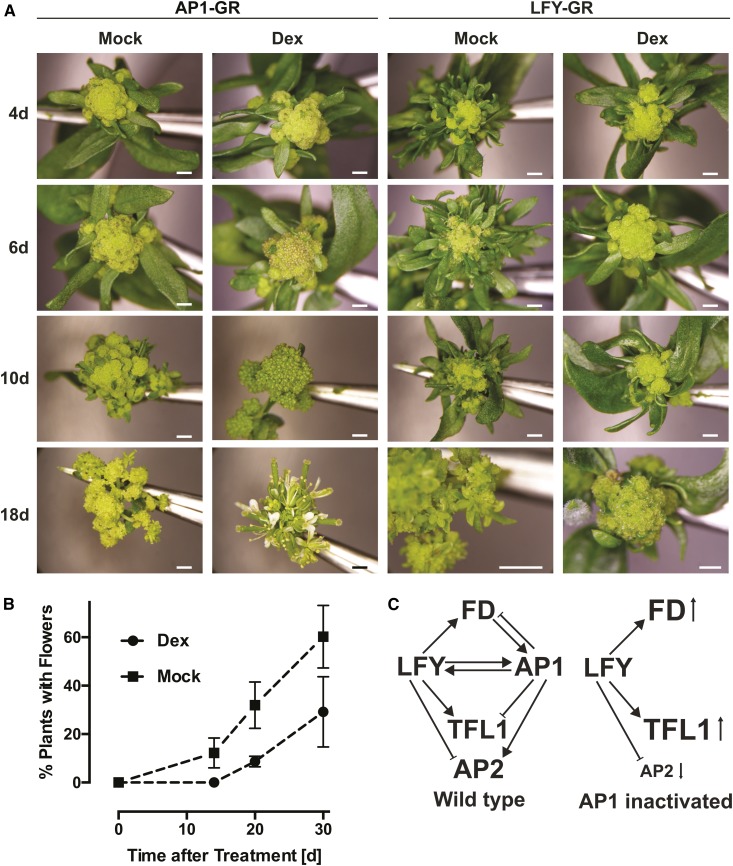
The LFY-dependent up-regulation of genes such as TFL1 and FD, which are repressed in emerging floral primordia, as well as the down-regulation of AP2, which is required for normal flower development, suggested that an activation of LFY-GR might further delay flower formation in ap1 cal double-mutant plants. To test this, we activated LFY-GR in the ap1 cal background and determined the time of flower formation relative to mock-treated control plants of the same genotype. As an additional control, we conducted the equivalent experiment with p35S:AP1-GR ap1 cal plants. While activation of AP1-GR led, as previously reported (Wellmer et al., 2006), to an immediate and synchronized onset of flower formation in all plants of the treatment population, neither dexamethasone nor mock-treated p35S:LFY-GR ap1 cal plants exhibited an instantaneous and coordinated flowering response (Fig. 6A). Most of the latter plants did eventually form flowers, but only after a very long delay following the treatment. Notably, the percentage of plants that made flowers at different time points after the treatment was considerably higher in the mock-treated than in the dexamethasone-treated p35S:LFY-GR ap1 cal plants (Fig. 6B). Thus, activation of LFY-GR in an ap1 cal double-mutant background led to a significant inhibition of flower formation relative to the one observed in ap1 cal plants. To further investigate whether this inhibition depends on a perturbation of AP1/CAL function, we germinated and grew p35S:LFY-GR and p35S:LFY-GR ap1 cal plants on dexamethasone-containing medium and found that flowering occurred earlier in the former plants than in the latter (Supplemental Fig. S5). We also tested whether the inhibitory effect on flower formation we observed after LFY-GR activation in ap1 cal plants could be abolished or diminished by a simultaneous activation of AP1. To this end, we employed a line that expressed both LFY-GR and a fusion between AP1 and the hormone binding domain of the androgen receptor (AP1-AR; Ó’Maoiléidigh et al., 2015) in an ap1 cal double-mutant background. As expected, we found that ap1 cal plants, in which AP1 and LFY had been activated at the same time, responded with an immediate onset of flower formation (Supplemental Fig. S6). Thus, an activation of AP1 can abrogate the inhibitory effect of LFY-GR on flowering.
Figure 6.
Activation of LFY in ap1 cal mutants delays the onset of flowering. A, Response of p35S:AP1-GR ap1 cal (on the left) and p35S:LFY-GR ap1 cal (on the right) plants to treatments with a dexamethasone-containing (Dex) or a mock solution. Images of inflorescences 4, 6, 10, and 18 d after the treatment are shown. Note the complete lack of floral structures in dexamethasone-treated p35S:LFY-GR ap1 cal plants at the 18-d time point. Scale bars = 1 mm. B, Onset of flowering in dexamethasone- and mock-treated p35S:LFY-GR ap1 cal plants. Percentage of plants in a treatment population with visible floral structures are shown. Error bars indicate ses calculated from three independent measurements. C, Model for the activities of LFY and AP1 (and CAL) during floral initiation. Left: gene interactions in the wild type; right: gene interactions after inactivation of AP1/CAL. Font size changes for gene symbols and small arrows indicate up- and down-regulation, respectively. TFL1, LFY, and AP1/CAL appear to be part of an incoherent feed-forward loop, where LFY promotes expression of TFL1 but is also involved in activating the TFL1 repressors, AP1/CAL. At the same time, LFY and AP1/CAL are also part of a positive feedback loop and reinforce each other’s expression through direct regulation.
DISCUSSION
Our study aimed at understanding the regulatory interplay between LFY and AP1/CAL, which are master regulators of floral initiation, a process that is of exceptional importance for both plant reproductive biology and agriculture. By testing the effects of LFY activation on gene expression in the absence of AP1/CAL function, we found that several known LFY targets are unaffected by the perturbation of AP1/CAL function (i.e. they respond normally to an activation of LFY), while other genes appear to require the input from both regulators. We further showed that LFY and AP1/CAL regulate several genes antagonistically, including some known to be involved in controlling the initiation of flower development (Fig. 5). Our results further suggest that this antagonism leads to LFY suppressing flower development in the absence of AP1/CAL function (Fig. 6). Thus, in addition to being part of a positive feedback loop that reinforces their expression and ensures high levels of activity during the onset of flower formation (Kaufmann et al., 2010; Wagner et al., 1999), LFY and AP1/CAL appear to be part of an incoherent feed-forward loop, where LFY promotes expression of TFL1 but is also involved in activating the TFL1 repressors, AP1/CAL (Fig. 6C). In general, an incoherent feed-forward loop is a gene network motif where two interconnected signaling pathways or transcription factors act in opposite directions on a target gene. The motif is thought to facilitate the time- or dosage-dependent control of a regulatory output (Kim et al., 2008). Thus, by providing positive as well as negative inputs, LFY might ensure that flower development can commence only when AP1/CAL levels are sufficiently high to efficiently activate the expression of other key floral regulators and to override its own inhibitory activity. Hence, this incoherent feed-forward loop may contribute to the establishment of a stable developmental program required for the formation of flowers.
Using the shoot identity gene TFL1 as an example, we further characterized the antagonistic activity between LFY and AP1/CAL and found that TFL1 expression is promoted by LFY but repressed by AP1. The concomitant activation of both transcription factors led to expression levels that are between those obtained after activating either AP1 or LFY alone (Fig. 5), suggesting that they may provide independent negative and positive regulatory inputs, respectively. Our finding that TFL1 is up-regulated upon LFY activation in ap1 cal inflorescences goes against the established view that both LFY and AP1 repress TFL1. However, several previous observations support the idea of LFY actually promoting TFL1 expression. For example, it has been shown that the meristem over-proliferation phenotype of ap1 cal double mutants (or that of ap1 cal fruitful triple mutants) depends on the activity of both LFY and TFL1 (Ferrándiz et al., 2000). Also, the expression patterns of LFY and TFL1 were found to overlap in ap1 cal inflorescence-like meristems (Ferrándiz et al., 2000), suggesting that they are not mutually antagonistic. Further support comes from a recent study of functional elements within the TFL1 promoter, which showed that the removal of a section of the promoter located 3′ of the gene, which contains LFY binding sites, is required for the maintenance of TFL1 expression in the inflorescence meristem (Serrano-Mislata et al., 2016). Moreover, very recently, it was noted after reanalyzing published gene expression data (those from Winter et al., 2011) that an activation of LFY-GR in wild-type seedlings leads to an up-regulation of TFL1 (Denay et al., 2017. Thus, it appears likely that LFY activates TFL1 expression at least in the inflorescence meristem. Because LFY is not expressed in the inflorescence meristem proper (Weigel et al., 1992) it has been suggested that this process may require LFY protein movement (Serrano-Mislata et al., 2016), which has been previously shown to occur (Sessions et al., 2000).
Parcy et al. (2002) found that the expression of an activated form of LFY, LFY-VP16, from the LFY promoter causes ectopic TFL1 expression in flowers, a result we confirmed and expanded on in this study (Fig. 4). Because it has been shown that LFY-VP16-mediated TFL1 misexpression causes partial floral reversion, a phenotype also seen in lfy mutants, LFY-VP16 has been interpreted as being an antimorphic version of LFY (Parcy et al., 2002). However, our results showing that LFY-GR promotes TFL1 expression in ap1 cal inflorescences suggests that LFY-VP16 rather functions as a hypermorph in this case.
If LFY was a constitutive activator of TFL1, one would expect to find TFL1 expression in floral anlagen, where LFY, but not AP1/CAL, is expressed. However, we never observed TFL1 expression in this domain and there is no evidence from the available literature that this may occur. Thus, the control of TFL1 expression by LFY may involve additional components, which might not be present in incipient floral primordia. The requirement of additional promoting factors with tissue- and/or stage-specific expression could also explain why ectopic TFL1 promoter activity was observed in LFY-VP16-expressing plants (Fig. 4) but has not been reported for lines expressing LFY from the 35S promoter. Under this scenario, only an activated version of LFY would be able to overcome the requirement for cofactor activity. This would be similar to the previously reported cofactor-independent induction of the floral homeotic gene AG in vegetative tissues by LFY-VP16, which is not seen in p35S:LFY overexpression lines (Parcy et al., 1998).
Taken together, our results provide evidence for an unexpected role of LFY in the control of floral initiation and further highlight the exceptional complexity of the gene regulatory network that controls this essential process during plant development. A key question that remains to be answered is why LFY and AP1/CAL act sometimes antagonistically, but more often function in concert. It is possible that these different regulatory outputs depend on the presence or absence of additional factors at target gene promoters. A biochemical characterization of the regulatory complexes that control floral initiation may be required to unravel the molecular mechanisms underlying these activities.
MATERIALS AND METHODS
Plant Growth
Unless specified otherwise, plants were grown on a soil:vermiculite:perlite (5:3:2) mixture at 20°C under constant illumination with cool-white fluorescent light. For the pTFL1:GUS experiments, plants were grown on a mixture of sphagnum:perlite:vermiculite (2:1:1) at 21°C under long-day conditions (16 h light, 8 h darkness). Light was provided by cool-white fluorescent lamps at 150 μmol m−2 s−1.
Plant Material
The p35S:LFY-GR ap1-1 cal-1, p35S:AP1-GR ap1-1 cal-1, pAP1:AP1-GR ap1-1 cal-1, and pAP1:AP1-AR ap1-1 cal-1 lines have been described previously (Ó’Maoiléidigh et al., 2015; Wellmer et al., 2006; Winter et al., 2011). To generate the p35S:LFY-GR pAP1:AP1-GR ap1-1 cal-1 line, the p35S:LFY-GR transgene was introgressed into pAP1:AP1-GR ap1-1 cal-1 plants. The presence of both transgenes was determined by genotyping using primer pairs KG2 and KG14 for the p35S:LFY-GR transgene and primers DM400 and DM494 for the pAP1:AP1-GR transgene. A similar strategy was followed to generate the p35S:LFY-GR pAP1:AP1-AR ap1-1 cal-1 line.
The TFL1 reporter line has been described previously (Serrano-Mislata et al., 2016). pTFL1:GUS contains the GUS gene flanked by the full-length TFL1 regulatory regions, 2.2 kb of the 5′ and 4.6 kb of the 3′ region. The pLFY:LFY-VP16 and pHS:LFY-VP16 lines were kindly provided by Dr. François Parcy and have been described previously (Benlloch et al., 2011; Parcy et al., 2002).
RNA Extraction
Total RNA was isolated from tissue samples using the Plant Total RNA kit (Sigma-Aldrich). For microarray experiments, quality of selected RNA samples was evaluated on a Bioanalyzer instrument using a RNA Nano 6000 kit (Agilent).
qRT-PCR Experiments
Total RNA extracts were treated with the DNA-free kit (Ambion) to remove genomic DNA contaminations. cDNA synthesis was performed using RNA preparations, oligo(dT) primers, Ribolock RNase inhibitor, and RevertAid H Minus reverse transcriptase (all reagents from Thermo-Fisher). Relative transcript abundance of selected genes (see Supplemental Table S3 for a list of genes and primers used) was determined using the Roche LightCycler 480 system and the LC480 SYBR Green I Master kit (Roche Applied Sciences). Measurements were taken for four biologically independent sets of samples. LightCycler melting curves were obtained for the reactions, revealing single peak melting curves for all amplification products. The amplification data were analyzed using the second derivative maximum method, and resulting Cp values were converted into relative expression values using the comparative Ct method (Livak and Schmittgen, 2001). Expression of one reference gene, REF1 (At1g13320), was used to normalize the data (Czechowski et al., 2005).
Tissue Collection for Gene Expression Profiling Experiments
For all experiments, we used ∼4-week-old p35S:LFY-GR ap1-1 cal-1 plants. For each sample, inflorescence tissue from ∼25 plants was collected using jeweler’s forceps as previously described (Wellmer et al., 2006). At least four biologically independent sets of samples were generated for each experiment. For the time course experiments, we treated inflorescences with a solution containing 10 μm dexamethasone (Sigma-Aldrich), 0.01% (v/v) ethanol, and 0.015% (v/v) Silwett l-77 (De Sangosse), or with an identical mock solution that lacked dexamethasone. Using plastic pipettes, the solutions were directly applied onto the inflorescences so that the cauliflower-like structures were completely drenched. We then collected inflorescence tissue after 2, 4, 8, and 12 h, as well as from untreated plants (0-h time point). For cycloheximide experiments, we treated inflorescences of p35S:LFY-GR ap1-1 cal-1 plants with a dexamethasone-containing or a mock solution (as described above) that contained in addition 10 μm cycloheximide (Sigma-Aldrich). Tissue was collected 3 h after the treatment. RNA was extracted from samples as described above.
Tissue Collection for Quantitative RT-PCR Experiments
For qRT-PCR experiments, tissue was collected from untreated control plants and from plants 3 h after treatment with either a dexamethasone-containing or a mock solution. For cycloheximide experiments, we treated inflorescences of p35S:LFY-GR ap1-1 cal-1 plants with a dexamethasone-containing or a mock solution (as described above) that contained 10 μm cycloheximide (Sigma-Aldrich).
Gene Expression Profiling Experiments
Gene expression profiling experiments using custom-designed Agilent Arabidopsis whole-genome microarrays were conducted as previously described (Kaufmann et al., 2010).
Data were analyzed using the limma (Linear models for Microarray Analysis) software package (Smyth, 2004). Background correction was done using the subtract method with an offset of 50. Within array normalization was performed using the loess method and 1,000 iterations, while between array normalization was done using the Aquantile method. To assess the reproducibility of the microarray experiments, correlograms were generated using the corrgram package in R. To identify DEGs, a P value cutoff of <0.01 (adjusted to control the false discovery rate using the Benjamini-Hochberg procedure) and a log2-transformed fold change value of >0.5 were applied. Gene Ontology analysis was performed using the online tool DAVID (Huang et al., 2007). Statistical significance calculations were performed with Fisher’s exact testing. Gene Ontology terms identified were deemed significant if the P value for enrichment was <0.01.
To identify candidates for an antagonistic response to LFY-GR and AP1-GR activation, expression data for DEGs from this study were compared to those from Kaufmann et al. (2010). Only genes with opposite directionality in expression changes over more than one time point were selected as candidates.
Generation of a Polyclonal Antiserum for LFY
To generate a LFY-specific antibody, we cloned the cDNA sequence coding for amino acid residues 223 to 420 of LFY (fragment noted LFY-C) into the Escherichia coli expression plasmid pET16b (Clontech). For that purpose, the cDNA fragment was amplified using primers LR177 and LR178 (Supplemental Table S3). The resulting PCR fragment and plasmid pET16b were then digested using NdeI/BamHI, and the fragments were ligated together to yield pET16b LFY-C (pLR48). BL21(DE3) pLysS RARE were transformed with pLR48 and freshly transformed colonies were resuspended in 500 mL Luria-Bertani medium. Cultures were grown at 37°C until an OD600 ∼0.6 was reached. Protein expression was induced by adding 0.5 mm isopropylthio-β-galactoside to the culture. After an overnight incubation at 19°C, cells were collected by centrifugation (5,000g for 10 min at 5°C) and kept frozen at −80°C until protein extraction and purification. Total proteins were extracted by resuspending the frozen cell pellets on ice in 15 mL resuspension buffer (20 mm Tris-HCl, pH 7.5, 0.5 m NaCl, 5 mm imidazole, 5 mm dithiothreitol [DTT], 5% glycerol, and 1:100 Complete Protease Inhibitor Cocktail [Roche]). Cells were lysed by sonication, while keeping the samples on ice for 5 min between sonication cycles. Cell extract was then centrifuged for 10 min at 18,000g at 4°C. Total solLBuble proteins (supernatant) were collected and used in the subsequent protein purification of His-tagged C-terminal domain of LFY. His-Select nickel-affinity gel (800 μL gel slurry; Sigma-Aldrich) was first equilibrated in resuspension buffer. The total soluble protein fraction was incubated with the equilibrated resin for 1 h at 4°C with rotation. Subsequent steps were carried out on a small chromatography column. The flow-through (unbound proteins) was discarded, and the resin was washed with 5 × 1 mL of Wash1 buffer (20 mm Tris-HCl, pH 7.5, 0.5 m NaCl, 5 mm imidazole, 5 mm DTT, and 5% glycerol), followed by washes with 5 × 1 mL of Wash2 buffer (20 mm Tris-HCl, pH 7.5, 0.5 m NaCl, 50 mm imidazole, 5 mm DTT, and 5% glycerol). His-tagged LFY-C was then eluted using 3× 0.5 mL elution buffer (20 mm Tris-HCl, pH 7.5, 0.5 m NaCl, 380 mm imidazole, 5 mm DTT, and 5% glycerol). Purified His-tagged LFY-C was dialyzed overnight against Dialysis buffer (20 mm Tris-HCl, pH 7.5, 0.5 m NaCl, and 5% glycerol) and used for immunizations and production of rabbit polyclonal antibodies against LFY (immunization was performed by Biogenes). Whole serum was used for ChIP experiments.
Chromatin Immunoprecipitation-Sequencing Experiments
For the identification of LFY-GR binding sites, we collected inflorescence tissue from ∼4-week-old 35S:LFY-GR ap1-1 cal-1 plants, 4 h after treatment with a solution containing 10 μm dexamethasone (Sigma-Aldrich), 0.01% (v/v) ethanol, and 0.015% (v/v) Silwet L-77 (De Sangosse). Approximately 500 μL of inflorescence tissue was vacuum infiltrated four times for 15 min in cross-linking solution (1% formaldehyde, 10 mm NaPi, pH 7.0, 50 mm NaCl, and 100 mm Suc) at room temperature. The cross-linking reaction was stopped by adding Gly to a final concentration of 0.1 m and vacuum infiltrating for 5 min. The tissue was washed three times with water before being ground to a fine powder in liquid nitrogen. Chromatin was isolated using Extraction buffers 1, 2, and 3 (Plant ChIP-Seq kit; Diagenode) and resuspended in nuclei lysis buffer (50 mm Tris-HCl, pH 8.1, 10 mm EDTA, and 1% SDS) and then sonicated to achieve an average fragment size of ∼200 to 500 bp. Cellular debris were removed by centrifugation. The chromatin solution was then diluted to 1 mL with 1× ChIP Dilution buffer (Plant ChIP-Seq kit) and precleared by incubating 60 μL of protein-A agarose beads (50% suspension in 1× ChIP dilution buffer; Santa Cruz Biotech) for 90 min at 4°C. Fifty microliters of the precleared chromatin solution was then removed and stored at −80°C to be used as input control. Immunoprecipitation was carried out using 15 μL of rabbit polyclonal antibodies raised against LFY-C overnight at 4°C, and 60 μL of protein-A agarose beads (50% suspension in 1× ChIP dilution buffer; Santa Cruz Biotech) was added to the chromatin-antibody solution and incubated for 2 h at 4°C. The beads were then washed five times using 1× ChIP Dilution buffer (Plant ChIP-Seq kit) before the immunocomplexes were eluted by adding 400 μL elution buffer 1 (Plant ChIP-Seq kit) and incubating at 65°C for 30 min with shaking at 1,300 rpm. Samples were spun to remove remaining agarose beads and 16 μL of elution buffer 2 (Plant ChIP-Seq kit) was added with samples being incubated overnight at 65°C with shaking at 1,300 rpm. The input control samples were treated similarly, with the addition of 400 μL of elution buffer 1 and 16 μL of elution buffer 2 (Plant ChIP-Seq kit), followed by overnight incubation at 65°C with shaking at 1,300 rpm.
DNA was purified and precipitated from these samples using buffers from the Plant ChIP-Seq kit according to the manufacturer’s instructions. DNA concentration was measured using the Qubit dsDNA HS assay kit (Thermo-Fisher). Sequencing of libraries was carried out by the Beijing Genomics Institute using an Illumina HiSeq 2000 instrument.
Burrows-Wheeler Aligner (Li and Durbin, 2009) was used to map raw data to the Arabidopsis reference genome (version: TAIR10), using a minimum quality score of 20. The resulting output file was converted into .bam format with samtools (Li et al., 2009). MACS (model-based Analysis for ChIP-Seq) version 1.4 (Zhang et al., 2008) was used for peak calling and Integrative Genomics Viewer (Robinson et al., 2011) for visualization. Peak annotation was performed using the annotatePeaks.pl script (http://homer.salk.edu).
ChIP-Quantitative PCR Experiments
Tissue was collected from ∼4-week-old p35S:LFY-GR ap1-1 cal-1 plants 4 h after dexamethasone treatment. ChIP was performed as described above and relative DNA abundance of selected genomic regions (see Supplemental Table S4 for a list of genes and the primers used) was then determined using the Roche LightCycler 480 system and the LC480 SYBR Green I Master kit (Roche Applied Sciences). Measurements were taken for four biologically independent sets of samples. LightCycler melting curves were obtained for the reactions, revealing single peak melting curves for all amplification products. The amplification data were analyzed using the second derivative maximum method, and resulting Cp values were converted into relative enrichment values using the comparative cycle threshold method where input DNA was used to normalize the data (Livak and Schmittgen, 2001). Five genomic regions (MU, ACTIN, BRI1, REF1, and TUB1; Supplemental Table S4) with no known LFY binding sites were used to normalize the data. Their Cp values were averaged for each sample.
Analysis of pTFL1:GUS Activity
For the reporter analysis of TFL1 in pLFY:LFY-VP16 inflorescence apices, pTFL1:GUS and pLFY:LFY-VP16 plants were crossed and a F3 homozygous line for both transgenes was selected for analysis. A minimum of three inflorescence tips of pTFL1:GUS and pTFL1:GUS; pLFY:LFY-VP16 plants after bolting were collected and stained for GUS activity as described in Serrano-Mislata et al., 2016. After staining, a few apices were cleared with a chloral hydrate solution (72% chloral hydrate and 11% glycerol) for 3 d before microscopy.
For the reporter analysis of TFL1 in pHS:LFY-VP16 seedlings, the pTFL1:GUS transgene was introgressed into a pHS:LFY-VP16 background. F1 seeds were grown on Murashige and Skoog petri dishes (2.2 g/L Murashige and Skoog medium, 20 g/L saccharose, 0.1 g/L MES, and 6 g/L agar, pH 5.9) for 14 d at 24°C under long-day conditions. Then, one-half of the plates were heat-shocked for 3 h at 37°C (on three consecutive days) and the other one-half were kept under normal growing conditions. One day after the last heat shock treatment, 8 to 10 seedlings per genotype and treatment were stained for GUS activity as described by Serrano-Mislata et al., 2016.
Accession Numbers
Microarray and ChIP-seq data sets have been deposited with the Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo/) under accession numbers GSE96799 and GSE96806, respectively.
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Microarray analysis of LFY-GR time course experiment.
Supplemental Figure S2. Activation of LFY-GR in the presence of cycloheximide.
Supplemental Figure S3. ChIP-seq results for known LFY target genes.
Supplemental Figure S4. ChIP-seq results for floral organ identity genes.
Supplemental Figure S5. Flowering after LFY-GR activation in wild-type and ap1 cal double-mutant plants.
Supplemental Figure S6. Activation of AP1 rescues the late flowering phenotype of ap1 cal plant after by LFY-GR activation.
Supplemental Table S1. DEGs identified in the p35S:LFY-GR ap1 cal time course experiment.
Supplemental Table S2. Primers used for PCR genotyping and the generation of constructs.
Supplemental Table S3. Primers used for qRT-PCR analysis.
Supplemental Table S4. Primers used for ChIP-qPCR analysis.
Supplemental Data Set 1. DEGs identified in LFY-GR ap1 cal time course experiment.
Supplemental Data Set 2. DEGs identified after activating LFY-GR in ap1 cal inflorescences in the presence of cycloheximide.
Supplemental Data Set 3. DEGs with LFY binding sites and results of ChIP-seq experiments.
Supplementary Material
Acknowledgments
We thank José Luis Riechmann for sharing the design of the Agilent microarrays and François Parcy for reagents.
Glossary
- ChIP-seq
chromatin immunoprecipitation-sequencing
- DEG
differentially expressed gene
- DTT
dithiothreitol
- qRT-PCR
quantitative reverse transcription PCR
References
- Abe M, Kobayashi Y, Yamamoto S, Daimon Y, Yamaguchi A, Ikeda Y, Ichinoki H, Notaguchi M, Goto K, Araki T (2005) FD, a bZIP protein mediating signals from the floral pathway integrator FT at the shoot apex. Science 309: 1052–1056 [DOI] [PubMed] [Google Scholar]
- Baumann K, Venail J, Berbel A, Domenech MJ, Money T, Conti L, Hanzawa Y, Madueno F, Bradley D (2015) Changing the spatial pattern of TFL1 expression reveals its key role in the shoot meristem in controlling Arabidopsis flowering architecture. J Exp Bot 66: 4769–4780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benlloch R, Kim MC, Sayou C, Thévenon E, Parcy F, Nilsson O (2011) Integrating long-day flowering signals: a LEAFY binding site is essential for proper photoperiodic activation of APETALA1. Plant J 67: 1094–1102 [DOI] [PubMed] [Google Scholar]
- Bowman JL, Alvarez J, Weigel D, Meyerowitz EM, Smyth DR (1993) Control of flower development in Arabidopsis thaliana by APETALA1 and interacting genes. Development 119: 721–743 [Google Scholar]
- Bradley D, Ratcliffe O, Vincent C, Carpenter R, Coen E (1997) Inflorescence commitment and architecture in Arabidopsis. Science 275: 80–83 [DOI] [PubMed] [Google Scholar]
- Busch MA, Bomblies K, Weigel D (1999) Activation of a floral homeotic gene in Arabidopsis. Science 285: 585–587 [DOI] [PubMed] [Google Scholar]
- Castillejo C, Pelaz S (2008) The balance between CONSTANS and TEMPRANILLO activities determines FT expression to trigger flowering. Curr Biol 18: 1338–1343 [DOI] [PubMed] [Google Scholar]
- Czechowski T, Stitt M, Altmann T, Udvardi MK, Scheible WR (2005) Genome-wide identification and testing of superior reference genes for transcript normalization in Arabidopsis. Plant Physiol 139: 5–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denay G, Chahtane H, Tichtinsky G, Parcy F (2017) A flower is born: an update on Arabidopsis floral meristem formation. Curr Opin Plant Biol 35: 15–22 [DOI] [PubMed] [Google Scholar]
- Ferrándiz C, Gu Q, Martienssen R, Yanofsky MF (2000) Redundant regulation of meristem identity and plant architecture by FRUITFULL, APETALA1 and CAULIFLOWER. Development 127: 725–734 [DOI] [PubMed] [Google Scholar]
- Fornara F, de Montaigu A, Coupland G (2010). SnapShot: control of flowering in Arabidopsis. Cell 141: e551–e552. [DOI] [PubMed]
- Hamès C, Ptchelkine D, Grimm C, Thevenon E, Moyroud E, Gérard F, Martiel JL, Benlloch R, Parcy F, Müller CW (2008) Structural basis for LEAFY floral switch function and similarity with helix-turn-helix proteins. EMBO J 27: 2628–2637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang DW, Sherman BT, Tan Q, Collins JR, Alvord WG, Roayaei J, Stephens R, Baseler MW, Lane HC, Lempicki RA (2007) The DAVID Gene Functional Classification Tool: a novel biological module-centric algorithm to functionally analyze large gene lists. Genome Biol 8: R183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jofuku KD, den Boer BG, Van Montagu M, Okamuro JK (1994) Control of Arabidopsis flower and seed development by the homeotic gene APETALA2. Plant Cell 6: 1211–1225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaufmann K, Wellmer F, Muiño JM, Ferrier T, Wuest SE, Kumar V, Serrano-Mislata A, Madueño F, Krajewski P, Meyerowitz EM, et al. (2010) Orchestration of floral initiation by APETALA1. Science 328: 85–89 [DOI] [PubMed] [Google Scholar]
- Kempin SA, Savidge B, Yanofsky MF (1995) Molecular basis of the cauliflower phenotype in Arabidopsis. Science 267: 522–525 [DOI] [PubMed] [Google Scholar]
- Kim D, Kwon YK, Cho KH (2008) The biphasic behavior of incoherent feed-forward loops in biomolecular regulatory networks. BioEssays 30: 1204–1211 [DOI] [PubMed] [Google Scholar]
- Li H, Durbin R (2009) Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25: 1754–1760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R; 1000 Genome Project Data Processing Subgroup (2009) The Sequence Alignment/Map format and SAMtools. Bioinformatics 25: 2078–2079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liljegren SJ, Gustafson-Brown C, Pinyopich A, Ditta GS, Yanofsky MF (1999) Interactions among APETALA1, LEAFY, and TERMINAL FLOWER1 specify meristem fate. Plant Cell 11: 1007–1018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods 25: 402–408 [DOI] [PubMed] [Google Scholar]
- Mandel MA, Gustafson-Brown C, Savidge B, Yanofsky MF (1992) Molecular characterization of the Arabidopsis floral homeotic gene APETALA1. Nature 360: 273–277 [DOI] [PubMed] [Google Scholar]
- Moyroud E, Minguet EG, Ott F, Yant L, Posé D, Monniaux M, Blanchet S, Bastien O, Thévenon E, Weigel D, et al. (2011) Prediction of regulatory interactions from genome sequences using a biophysical model for the Arabidopsis LEAFY transcription factor. Plant Cell 23: 1293–1306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- ÓMaoiléidigh DS, Wuest SE, Rae L, Raganelli A, Ryan PT, Kwasniewska K, Das P, Lohan AJ, Loftus B, Graciet E, et al. (2013) Control of reproductive floral organ identity specification in Arabidopsis by the C function regulator AGAMOUS. Plant Cell 25: 2482–2503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ó’Maoiléidigh DS, Graciet E, Wellmer F (2014) Gene networks controlling Arabidopsis thaliana flower development. New Phytol 201: 16–30 [DOI] [PubMed] [Google Scholar]
- Ó’Maoiléidigh DS, Thomson B, Raganelli A, Wuest SE, Ryan PT, Kwaśniewska K, Carles CC, Graciet E, Wellmer F (2015) Gene network analysis of Arabidopsis thaliana flower development through dynamic gene perturbations. Plant J 83: 344–358 [DOI] [PubMed] [Google Scholar]
- Pajoro A, Madrigal P, Muiño JM, Matus JT, Jin J, Mecchia MA, Debernardi JM, Palatnik JF, Balazadeh S, Arif M, et al. (2014) Dynamics of chromatin accessibility and gene regulation by MADS-domain transcription factors in flower development. Genome Biol 15: R41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parcy F, Bomblies K, Weigel D (2002) Interaction of LEAFY, AGAMOUS and TERMINAL FLOWER1 in maintaining floral meristem identity in Arabidopsis. Development 129: 2519–2527 [DOI] [PubMed] [Google Scholar]
- Parcy F, Nilsson O, Busch MA, Lee I, Weigel D (1998) A genetic framework for floral patterning. Nature 395: 561–566 [DOI] [PubMed] [Google Scholar]
- Robinson JT, Thorvaldsdóttir H, Winckler W, Guttman M, Lander ES, Getz G, Mesirov JP (2011) Integrative genomics viewer. Nat Biotechnol 29: 24–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saddic LA, Huvermann B, Bezhani S, Su Y, Winter CM, Kwon CS, Collum RP, Wagner D (2006) The LEAFY target LMI1 is a meristem identity regulator and acts together with LEAFY to regulate expression of CAULIFLOWER. Development 133: 1673–1682 [DOI] [PubMed] [Google Scholar]
- Sayou C, Nanao MH, Jamin M, Posé D, Thévenon E, Grégoire L, Tichtinsky G, Denay G, Ott F, Peirats Llobet M, et al. (2016) A SAM oligomerization domain shapes the genomic binding landscape of the LEAFY transcription factor. Nat Commun 7: 11222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serrano-Mislata A, Fernández-Nohales P, Doménech MJ, Hanzawa Y, Bradley D, Madueño F (2016) Separate elements of the TERMINAL FLOWER 1 cis-regulatory region integrate pathways to control flowering time and shoot meristem identity. Development 143: 3315–3327 [DOI] [PubMed] [Google Scholar]
- Sessions A, Yanofsky MF, Weigel D (2000) Cell-cell signaling and movement by the floral transcription factors LEAFY and APETALA1. Science 289: 779–782 [DOI] [PubMed] [Google Scholar]
- Smyth GK. (2004) Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Stat Appl Genet Mol Biol 3: Article3. [DOI] [PubMed]
- Wagner D, Sablowski RW, Meyerowitz EM (1999) Transcriptional activation of APETALA1 by LEAFY. Science 285: 582–584 [DOI] [PubMed] [Google Scholar]
- Wagner D, Wellmer F, Dilks K, William D, Smith MR, Kumar PP, Riechmann JL, Greenland AJ, Meyerowitz EM (2004) Floral induction in tissue culture: a system for the analysis of LEAFY-dependent gene regulation. Plant J 39: 273–282 [DOI] [PubMed] [Google Scholar]
- Weigel D, Alvarez J, Smyth DR, Yanofsky MF, Meyerowitz EM (1992) LEAFY controls floral meristem identity in Arabidopsis. Cell 69: 843–859 [DOI] [PubMed] [Google Scholar]
- Wellmer F, Alves-Ferreira M, Dubois A, Riechmann JL, Meyerowitz EM (2006) Genome-wide analysis of gene expression during early Arabidopsis flower development. PLoS Genet 2: e117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wigge PA, Kim MC, Jaeger KE, Busch W, Schmid M, Lohmann JU, Weigel D (2005) Integration of spatial and temporal information during floral induction in Arabidopsis. Science 309: 1056–1059 [DOI] [PubMed] [Google Scholar]
- William DA, Su Y, Smith MR, Lu M, Baldwin DA, Wagner D (2004) Genomic identification of direct target genes of LEAFY. Proc Natl Acad Sci USA 101: 1775–1780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winter CM, Austin RS, Blanvillain-Baufumé S, Reback MA, Monniaux M, Wu MF, Sang Y, Yamaguchi A, Yamaguchi N, Parker JE, et al. (2011) LEAFY target genes reveal floral regulatory logic, cis motifs, and a link to biotic stimulus response. Dev Cell 20: 430–443 [DOI] [PubMed] [Google Scholar]
- Winter CM, Yamaguchi N, Wu MF, Wagner D (2015) Transcriptional programs regulated by both LEAFY and APETALA1 at the time of flower formation. Physiol Plant 155: 55–73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi N, Winter CM, Wu MF, Kanno Y, Yamaguchi A, Seo M, Wagner D (2014) Gibberellin acts positively then negatively to control onset of flower formation in Arabidopsis. Science 344: 638–641 [DOI] [PubMed] [Google Scholar]
- Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, Bernstein BE, Nusbaum C, Myers RM, Brown M, Li W, et al. (2008) Model-based analysis of ChIP-Seq (MACS). Genome Biol 9: R137. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.