ChloroKB, a Web-based application that enables the user to explore the chloroplast metabolic network in Arabidopsis.
Abstract
Higher plants, as autotrophic organisms, are effective sources of molecules. They hold great promise for metabolic engineering, but the behavior of plant metabolism at the network level is still incompletely described. Although structural models (stoichiometry matrices) and pathway databases are extremely useful, they cannot describe the complexity of the metabolic context, and new tools are required to visually represent integrated biocurated knowledge for use by both humans and computers. Here, we describe ChloroKB, a Web application (http://chlorokb.fr/) for visual exploration and analysis of the Arabidopsis (Arabidopsis thaliana) metabolic network in the chloroplast and related cellular pathways. The network was manually reconstructed through extensive biocuration to provide transparent traceability of experimental data. Proteins and metabolites were placed in their biological context (spatial distribution within cells, connectivity in the network, participation in supramolecular complexes, and regulatory interactions) using CellDesigner software. The network contains 1,147 reviewed proteins (559 localized exclusively in plastids, 68 in at least one additional compartment, and 520 outside the plastid), 122 proteins awaiting biochemical/genetic characterization, and 228 proteins for which genes have not yet been identified. The visual presentation is intuitive and browsing is fluid, providing instant access to the graphical representation of integrated processes and to a wealth of refined qualitative and quantitative data. ChloroKB will be a significant support for structural and quantitative kinetic modeling, for biological reasoning, when comparing novel data with established knowledge, for computer analyses, and for educational purposes. ChloroKB will be enhanced by continuous updates following contributions from plant researchers.
Higher plant metabolism is exceedingly complex due to the significantly different metabolic capacities displayed by various tissues and to the presence of the plastidial compartment, which can differentiate into different types (Jarvis and López-Juez, 2013) and interact with extraplastidial pathways (Rolland et al., 2012; Sweetlove and Fernie, 2013). This compartment is specific to photosynthetic eukaryotes and reflects plants’ evolutionary history. Another aspect of this history is the existence of branched and parallel metabolic pathways with many different protein isoforms (more than 10 in extreme cases), multiple spatial localizations, specific expression patterns, and generally varied kinetic or regulatory properties. This complexity often hinders our understanding of higher plant metabolism at the system level and is a hurdle to molecular engineering (Farré et al., 2015). Detailed kinetic models that simulate and predict the dynamic responses of metabolic networks are needed, which, to provide confident predictions, must be built from vast amounts of data collated from various sources. These data must first be assembled by performing a systematic inventory of the metabolic pathways related to a particular genome (i.e. genome-scale metabolic reconstructions; Seaver et al., 2012). Unfortunately, current databases specific for plant metabolic pathways remain limited when it comes to capturing all the essential details required for metabolic reconstructions and modeling (Seaver et al., 2012; Stobbe et al., 2014). Another major issue is that no current metabolic database refers to a precise metabolic state or to a given tissue; they all represent a global metabolic network where all possible reactions are combined, even though they may not take place in all tissues or every cell type (Stobbe et al., 2014). Finally, biological networks are often only partially described, and clear information is required on how these databases deal with incomplete knowledge, uncertainties, and conflicts (Stobbe et al., 2014).
Despite these limitations, attempts have been made to collate the information contained in these databases, for example by producing genome-scale structural models of Arabidopsis (Arabidopsis thaliana) in the form of stoichiometry matrices (Poolman et al., 2009; de Oliveira Dal’Molin et al., 2010; Mintz-Oron et al., 2012) from the annotations in the AraCyc database (Mueller et al., 2003) and KEGG (Kanehisa et al., 2016). Although these matrices provide a comprehensive model of a metabolic system, they fail to cover all the biological details (Dauga, 2015). Indeed, experimental evidence, along with tools for hypothesis generation and modeling, are needed to extend our knowledge about given tissues/states of a biological system. As part of this requirement, when seeking to exploit the vast amount of heterogenous data available in databases and the literature, biocuration (Howe et al., 2008; Burge et al., 2012; Dauga, 2015), which consists of coherently collecting, organizing, and structuring data from published articles, databases, and expert knowledge to make biological information accessible to both humans and computers, has become an essential aspect of the initial steps of modern biological discovery and biomedical research.
In this article, we have built on our experience in dynamic modeling (Curien et al., 2009, 2014) to present ChloroKB (the Arabidopsis Chloroplast Knowledge Base), a network-oriented reconstruction of the metabolism of Arabidopsis that takes spatial distribution within the cell into account. ChloroKB focuses on the following main points: (1) levels of biological information and how molecular actors are represented in their biological context; (2) extensive biocuration with explicit tracking of sources of experimental data (transparency); and (3) interactive and intuitive visual browsing. ChloroKB (http://chlorokb.fr) is a Web-based application combining the advantages of a visual graphical representation of a manually reconstructed metabolism with the features of a curated knowledge base. In its current form, ChloroKB represents 1,147 proteins in 125 interconnected metabolic maps.
RESULTS AND DISCUSSION
ChloroKB: Design Principles, Content, and Functionalities
A Multilevel Representation of the Arabidopsis Metabolic Network
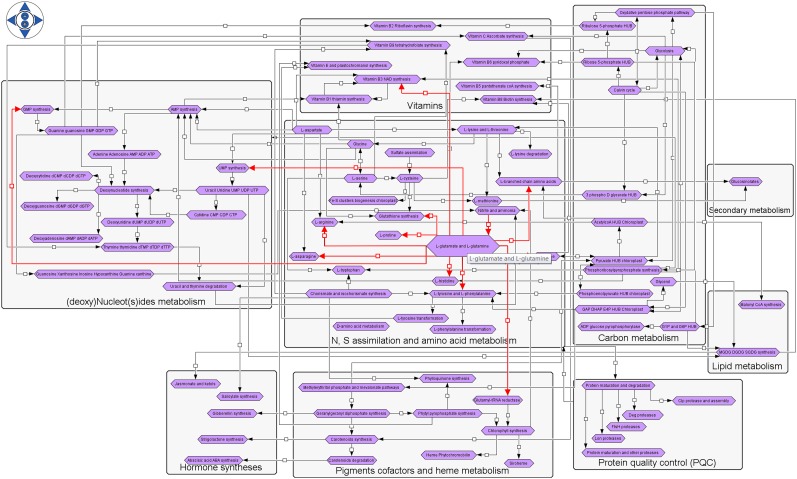
Various levels of graphical representation were considered during the development of ChloroKB to account for the different layers of organization within cells with the aim of allowing end users to visually explore networks at different scales. The first level, displayed from the home page, corresponds to a graphical and interactive representation of the overall reconstructed network organized by category of metabolic compounds and biochemical processes. Protein degradation and the metabolic processes related to the following compounds are represented: carbon precursors, amino acids, vitamins/cofactors, nucleosides and nucleotides, hormones, and pigments. The interconnections between the different pathways also are represented (Fig. 1). At this level, a comprehensive view of the network showing the numerous and complex dependencies between the different metabolic pathways making it up (in terms of inflow and outflow) is provided. This view allows users to track the origin and fate of a given compound by mousing over a pathway of interest (represented by a purple hexagonal box) to highlight the incoming/outgoing arrows (in red) and, thus, immediately view all the biochemically connected pathways. Mousing over arrows (or small squares) reveals the pathways they link in red. Each purple hexagonal icon can be clicked to display the metabolic map it represents; these maps correspond to the second level of knowledge representation.
Figure 1.
Reconstructed network visible on the ChloroKB home page. The network includes N and S assimilation, synthesis of carbon precursors, amino acids, nucleotides, vitamins, cofactors, pigments, some lipids, hormones, glucosinolates, and plastidial protein degradation apparatus along with their major connections. Each purple icon represents a metabolic map that can be accessed by clicking on the icon.
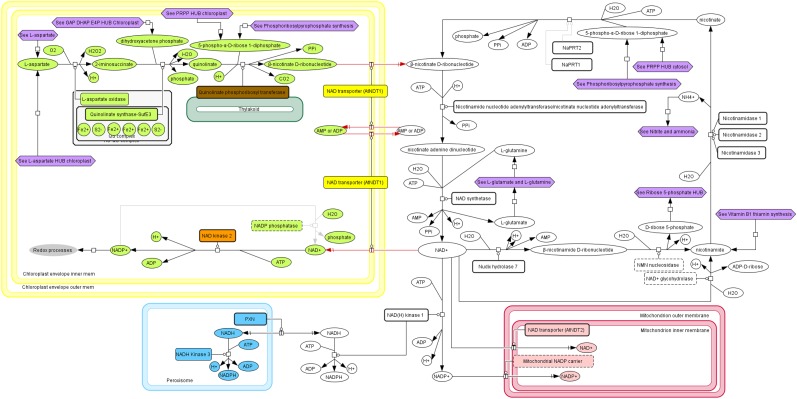
The second level is a graphical representation of a metabolic process, or map, which provides a detailed biochemical view and indicates the subcellular compartment where the reaction occurs. Although centered on plastidial processes, the reconstruction was extended to extrachloroplastic processes, since many metabolic pathways have multiple connections and share molecular components. In some cases (e.g. vitamin B3 synthesis; Fig. 2), metabolism is multicompartmentalized, while in others, different isoforms are distributed over several compartments. An extreme example of this multilocalization is the two isopentenyl diphosphate isomerases in the methylerythritol and mevalonate pathways (localized in four distinct compartments; Supplemental Fig. S3). By including cytosolic, mitochondrial, peroxisomal, and secretory pathways, ChloroKB offers a comprehensive view of metabolic processes at the level of the cell as a whole.
Figure 2.
How ChloroKB represents a multicompartmentalized process (vitamin B3 NAD synthesis). The chloroplast is represented surrounded by its double membrane, or envelope (in yellow), the peroxisome is in cyan, and the mitochondrion is in red. Cytosolic proteins and metabolites are represented in white.
Network and map reconstruction boundaries were determined based on (1) metabolite input and output, (2) a tradeoff between integration and readability, and (3) a reconstruction rule stating that all protein isoforms should be represented on a single map. According to this last rule, whether protein isoforms (isoenzymes) are coded by a single gene that undergoes alternative transcription (e.g. glutathione reductase 2) or whether they are produced by different genes (e.g. Asp kinase 1–Asp kinase 3), they are all represented on a single map. We tried to optimize integration (e.g. representing amino acid synthesis and degradation on the same page), but in some cases, compromises had to be found and some processes had to be fragmented. Connections between maps are represented explicitly by purple hexagonal boxes, except for the most common metabolites, ATP, ADP, AMP, NAD(P), NAD(P)+, inorganic pyrophosphate, and inorganic phosphate.
In addition to this pathway-oriented representation, hub maps also were constructed. Hub maps are centered on metabolites synthesized and converted by several different reactions (more than three in ChloroKB). This level of vision provides information on all the inputs and outputs related to a central metabolite and could help with metabolic engineering approaches. The current version of ChloroKB contains 14 hub maps centered on the following metabolites: 3-phospho-d-glycerate, acetate, acetyl-CoA, chorismate, pyruvate, phosphoenolpyruvate, glyceraldehyde 3-phosphate, dihydroxyacetone phosphate and erythrose 4-phosphate (GAP DHAP E4P HUB), fumarate, Glc-1-P and Glc-6-P (G1P and G6P HUB), l-Asp, phosphoribosyl pyrophosphate (PRPP HUB), and ribose 5-phosphate and ribulose 5-phosphate. The hub representation used in ChloroKB provides a convenient way to represent regulatory controls. Indeed, enzymes using hub metabolites are frequently the targets of specific regulatory interactions (allosteric or not) or regulatory modifications (oxidation, reduction, and phosphorylation); some examples of this type of regulation are presented below (see Chorismate HUB). We expect that these hub maps and descriptions of short-term regulatory processes will be especially useful to modelers, as intermediate metabolic steps, which generally do not play any regulatory roles, can be ignored in order to focus on important branching points.
Isoforms were explicitly considered in every process; therefore, the reconstruction can be considered almost complete at the cellular level for most biochemical pathways, such as amino acid synthesis; nucleotide synthesis, conversion, and degradation; and pigment and vitamin synthesis. Some reconstructed processes are unique to ChloroKB: (1) the protein degradation and protein quality control pathways in the chloroplast (six maps); (2) the contextualization of sulfate and nitrate assimilation in the cell; (3) thiosulfate metabolism; (4) nucleotide and deoxynucleotide metabolism at the cellular level, including interconversions and degradation; (5) photorespiration and how it relates to amino acid metabolism; (6) Fe-S cluster assembly; (7) jasmonate and ketol metabolism; (8) the pipecolate synthesis and degradation pathways, which were recently (partially) characterized; and (9) carotenoid synthesis and degradation with subplastidial localization of the proteins.
Through these maps and the links between them, ChloroKB offers users the possibility to navigate through a network starting from a global view and gradually zooming into a very detailed representation of molecular processes and their regulation in a few clicks.
Key Features of ChloroKB and Network Overview
In its current version, ChloroKB includes 125 maps (111 reviewed and 14 unreviewed; see “Biocuration of ChloroKB: Principles and Reviewing Process”) with subcellular compartments, 1,147 reviewed proteins, 228 unknown proteins (i.e. proteins assumed to be present but for which genes have yet to be identified), 724 molecular complexes, 1,700 simple molecules, 17 ions, and 81 chemical inhibitors. A total of 2,141 PubMed references and 91 additional references (not available through PubMed) were included for reviewed proteins, complexes, and metabolites. Quantitative data are available for 311 metabolites along with information relating to the experimental conditions used to determine this information.
The reconstructed network provides general information on how metabolic pathways are organized in Arabidopsis. A numerical representation of subcellular localizations including multiple targeting is illustrated (Fig. 3; Supplemental Tables S1 and S2). Thus, a total of 559 proteins are targeted exclusively to the plastid (of which 85 are present in the envelope and 54 in the thylakoid). An additional 68 proteins (6% of the total) are targeted to the plastid and one or more other compartments. Dual targeting to the plastid and mitochondrion represents 50% of these cases, whereas dual targeting to the cytosol (13%) or peroxisome (6%) is less frequent. Cases of multiple targeting are scarce (e.g. five cases of targeting to the plastid, mitochondrion, and cytosol and a single case of targeting to four different compartments).
Figure 3.
Overview of the subcellular distribution of proteins represented in the reconstructed network. Proteins with multiple localizations were counted only once and are represented with additional color boxes indicating alternative subcellular localization. This counting does not refer to a specific cell type; hence, we use the term plastid rather than chloroplast. For more detailed counting, see Supplemental Table S1, and for gene/localization correspondence, see Supplemental Table S2.
In addition to plastidial proteins, the reconstruction in ChloroKB contains 278 purely cytosolic proteins, 88 proteins that are localized exclusively in the mitochondrion, and 27 proteins localized in the peroxisome. Other locations and additional cases of dual targeting between these compartments are shown in Figure 3.
Plastid metabolism is highly connected to extraplastidial metabolism (substrate/product relationships, pathways, or steps catalyzed in parallel in different compartments), and overall, 54% of the proteins involved in the metabolism of the different metabolites represented in ChloroKB (Fig. 1) are localized in the plastid compartment. The correspondence between the subcellular (and subplastidial) location and the protein’s gene identifier is provided in Supplemental Table S2.
Although an impressive number of biochemical and genetic analyses have been published since 2000, when the Arabidopsis genome sequence was published (Arabidopsis Genome Initiative, 2000), the reconstruction shows that 122 proteins (i.e. 10% of the proteins with a reviewed status) still require biochemical characterization and that subcellular localization remains unknown for 83 proteins. In addition, the coding gene has yet to be identified for 228 proteins.
Querying and Browsing ChloroKB
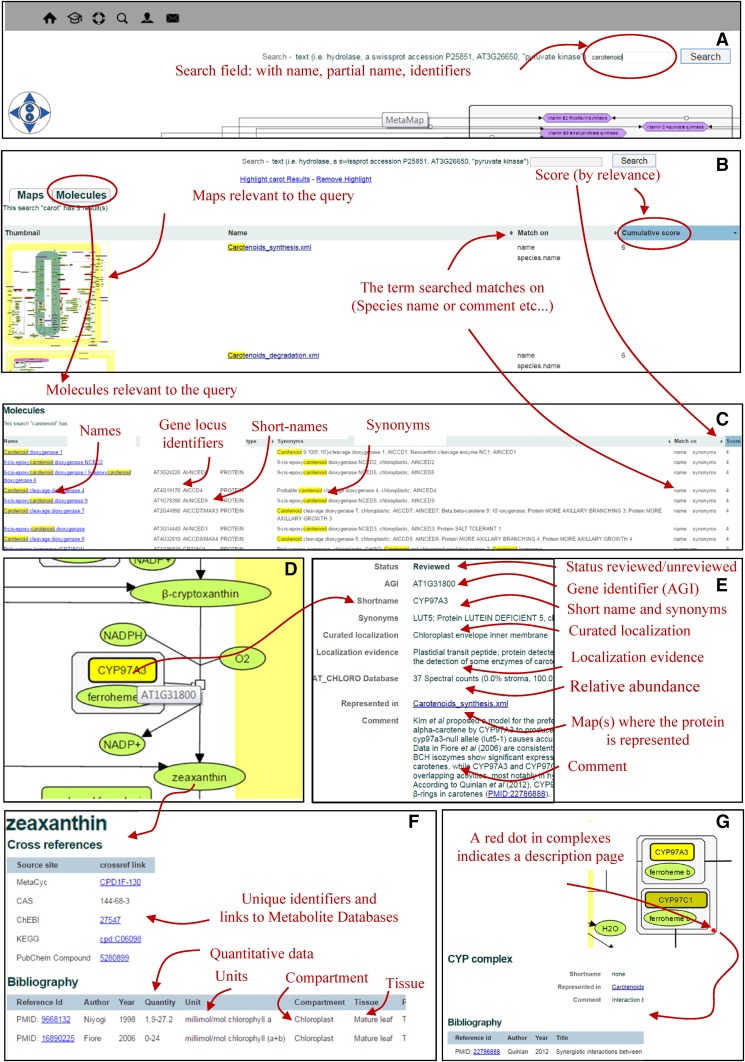
A search engine has been developed to query ChloroKB using name, keywords, locus identifier, or accession number (Fig. 4). Exact and partial text-matching capabilities have been implemented (Fig. 4A). Search results (maps [Fig. 4B] and molecules [Fig. 4C]) are ranked based on a scoring system that attributes a weight to preselected fields. A higher final score indicates that the query occurs more frequently in the result. To facilitate navigation and access to biological data, the same information can be retrieved in several different ways. For example, querying a protein name in the search field will return all the maps where the protein is represented (Fig. 4B). This information also appears in the protein description page, where all the maps on which the protein is represented are listed (Fig. 4E); the same principle was applied to metabolites. The fate of a metabolite of interest also can be tracked using ChloroKB, and additional fates of molecules (unmapped in the current version of ChloroKB) are indicated. This is a major advantage over other pathway databases and avoids the need to perform a refined analysis of the reactions involving the metabolite and time-consuming identification of their subcellular localization by literature mining (e.g. compare chorismate synthesis, the Calvin cycle, the oxidative pentose phosphate pathway, or carotenoid synthesis in ChloroKB and in other pathway databases).
Figure 4.
Querying and browsing in ChloroKB.
Browsing was made graphically fluid and interactive by applying the SVG format to pathways (see “Materials and Methods”). Clicking on a molecule of interest (Fig. 4, D–F) or clicking inside complexes displaying a red dot (Fig. 4G) opens a description page for that molecule or complex (Fig. 4E for a protein, Fig. 4F for a metabolite, and Fig. 4G for a complex), where related information such as localization, reaction, 3D structures, links to other maps, quantitative data, cross-references, bibliography, and comments can be found.
Biocuration of ChloroKB: Principles and Reviewing Process
A particular effort was devoted to ensuring the quality and reliability of the information presented in ChloroKB through biocuration. This step in development of the application involved analysis, interpretation, and integration of biological information from published work, and users can have high confidence in the information provided. All maps, proteins (including their subcellular locations; see below), and metabolites were biocurated, and cross-references to other resources and comments were added during this process (e.g. to highlight conflicting results or confusing protein nomenclature).
Maps included in ChloroKB were labeled with a status (reviewed or unreviewed) to indicate whether they have been revised by at least one independent expert (not involved in the map-building stage); an unreviewed status indicates that the map is currently undergoing revision.
At the protein level, a reviewed status indicates that the data associated with a given protein were verified for consistency and enriched through literature mining or relevant information obtained from another plant model, by adding details on reactions (e.g. catalysis, transport, or regulation), and including additional experimental evidence. Many reactions that are not currently available in pathway databases were included in ChloroKB based on literature searches. An unreviewed status on a protein description page means that the data and annotations associated with this category of protein were automatically collected from public resources (e.g. UniProt); otherwise, this means that the review is in progress.
The information related to subcellular and subplastidial localization received its own qualification. Thus, in ChloroKB, a protein’s subcellular localization was defined in one of three ways: (1) proteins for which the subcellular location was experimentally demonstrated (by mass spectrometry-based proteomics, GFP fusion analysis, western blot, immunogold labeling, activity measurement, etc.) were attributed a curated localization indicating the appropriate subcellular compartment (one or more); (2) proteins that have not been experimentally localized were labeled with an unknown localization; and (3) unreviewed proteins were labeled pending in this field. Because some subcellular localizations could be ambiguous or because information may be incomplete, a localization evidence comment also was included to justify the curated status of the localization. To facilitate alternative uses of this information, such as flux-balance analysis, subcellular localization also was indicated in the reaction field (see AT3G24170, http://chlorokb.fr/protein/AT3G24170).
Transport processes (uniport, cotransport, countertransport, and broad specific transport) can be complex. In ChloroKB, dedicated maps were built to describe some transport processes (Supplemental Fig. S4). Nominal cellular compartments (localization) were specified in the reaction fields on the description pages for each transporter included in the knowledge base.
Dealing with Incomplete and Partial Knowledge
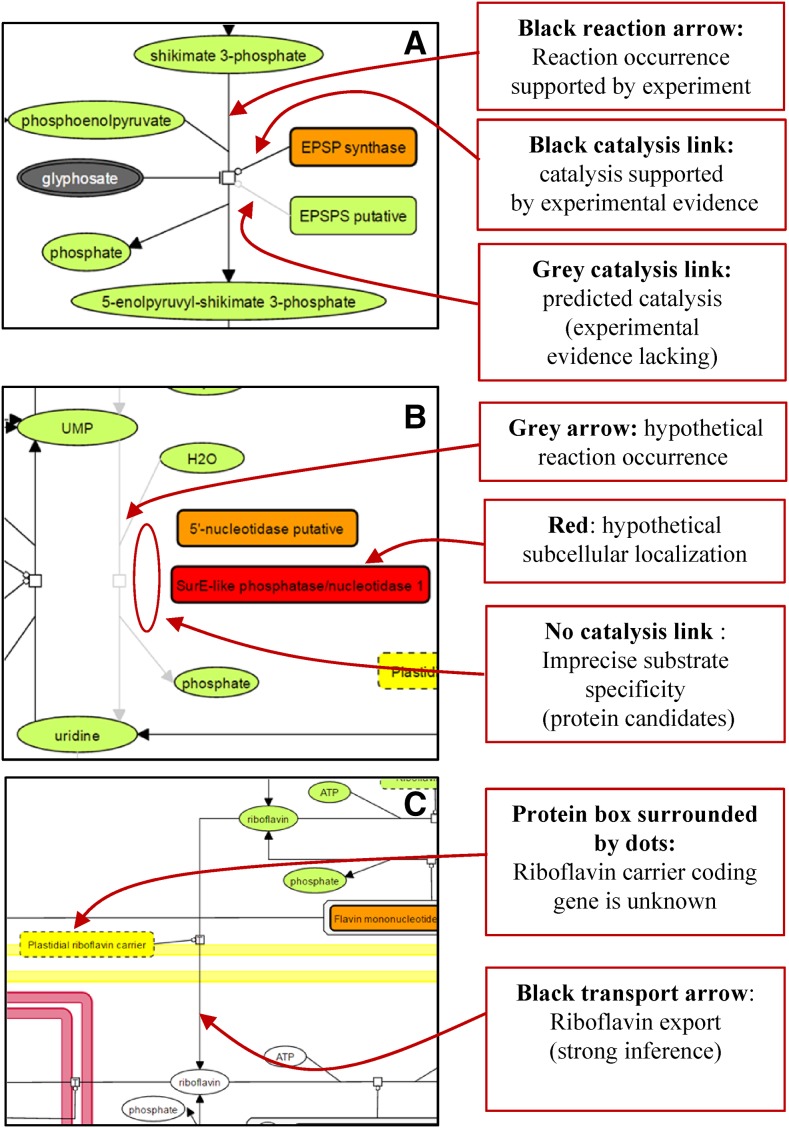
Biochemical networks necessarily contain some uncharacterized elements, and genome-scale metabolic reconstruction must be able to deal with incompleteness and manage uncertainties and/or unknown molecules (Fig. 5). As mentioned below (see “Data Representation and Graphical Annotation in ChloroKB”), this was dealt with in ChloroKB by defining a color code for reactions and catalysis. Examples are shown in Figure 5A, which illustrates 5-enolpyruvylshikimate-3-phosphate synthase activity (the target of glyphosate). This enzymatic activity is encoded by two genes in Arabidopsis, but only one has been characterized so far. In another example (Fig. 5B), the localization of the enzymes performing UMP dephosphorylation to produce uridine has not yet been documented. Indeed, several protein candidates (5′-nucleotidases) could catalyze this reaction. One 5′-nucleotidase was detected in the stroma (orange symbol in Fig. 5B), but its substrate specificity cannot be predicted from its sequence; therefore, no symbol for catalysis connects the protein to the reaction. The presence of this protein in the stroma simply indicates that it is a candidate for catalysis of the dephosphorylation reaction. Another candidate enzyme for this reaction is SurE-like phosphatase/nucleotidase 1 (Fig. 5B); in addition to catalyzing an undefined reaction, its subcellular localization is currently unknown, although it does possess an N-terminal extension that could be a plastid-targeting sequence. The red color of this enzyme indicates that its localization has not been experimentally determined (it also could be targeted to another compartment).
Figure 5.
How incomplete knowledge, prediction, and hypotheses are indicated in ChloroKB. The codes used to represent reactions or catalytic processes differ depending on whether they are supported by experimental data or represent hypothetical reactions (or transports) or predictions based on sequence similarity. Unknown subcellular localizations also can be distinguished from experimentally established localizations. See legends in the figure.
If the gene coding for a protein has not been identified but the existence of a catalytic or transport step is strongly supported by indirect experimental evidence or by logical reasoning, then the reaction/transport arrow symbol is represented in black on ChloroKB maps. An example of this situation is illustrated by riboflavin, which is synthesized exclusively within the chloroplast, and indirect evidence strongly suggests export to the cytosol (Sandoval et al., 2008; Fig. 5C). Another example is argininosuccinate lyase; this enzyme has never been characterized in Arabidopsis, but since l-Arg is synthesized in the chloroplast and a single gene has been identified, we chose to represent this reaction in black.
This encoding scheme allows users to distinguish between objectively verifiable knowledge and predictions (i.e. automatically inferred information) and suggestions due to extrapolation. Explanations for the color coding applied to different elements can be found in the associated text-based information (e.g. the comments section in the description page contains the following statement: “Awaiting biochemical/genetic characterization”).
Dealing with Conflicting Annotations
The traceability of protein localization predictions and the full description of available experimental evidence are novel aspects in ChloroKB. When a conflict occurred with an annotation (e.g. subcellular location, function, or name), curation mainly involved careful inspection of the literature and tracing of the source (Bernard et al., 2014).
If a conflicting subcellular localization arose, data from alternative literature sources were sought to determine the protein localization. An example is dihydroorotase, which is thought to be located in the chloroplast stroma. The localization evidence field indicates the following: “Presence of a transit peptide; GFP fusion indicates a cytosolic localization (PMID:22474184); however, immunological evidence in Pisum sativum (PMID:16669783) and biochemical evidence in Arabidopsis (PMID:16664505) indicated a plastidial localization, and in proteomics experiments, this protein was detected in the stroma (AT_CHLORO Database). This protein is therefore assumed to be chloroplastic.”
In all cases where confusion might arise, an explanation has been provided in the comment field of the protein description. All references cited on description pages correspond to data for the unique gene identifier (even if the names used by authors in their publications differ from those used in ChloroKB). Alternative protein names found in the literature were listed in the synonyms field in the protein description. In some cases, sequence analyses (BLAST search) had to be performed to verify the correspondence between the protein/gene considered in an article and the AGI identifier reported in the protein description.
When experimental data from different studies produced conflicting conclusions, this also was indicated in the comment field.
Using ChloroKB to Investigate the Metabolic Network of Arabidopsis
By combining data from independent sources in their original biological context, novel hints can be gleaned to help investigate a biochemical pathway. In this section, we will use the example of chorismate metabolism to demonstrate how ChloroKB can be applied practically to query the Arabidopsis metabolic network.
A search for chorismate in ChloroKB returns 13 maps (chorismate HUB, chorismate and isochorismate synthesis, chorismate mutases, Calvin cycle, etc.) in which the term chorismate appears either in the name of the map or as one of the components represented on the map. Results related to molecule descriptions (proteins and metabolites) for the query chorismate also are displayed. The chorismate and isochorismate biosynthesis map (Fig. 6) shows the catalytic steps in the conversion of phosphoenolpyruvate and erythrose 4-phosphate (purple hexagons at the top of the figure) to chorismate and the different uses to which chorismate is put by the cell (purple hexagons at the bottom of the figure). The connections with lignin precursor synthesis in the cytosol are indicated in pale gray (zoom displayed in Fig. 6). These connections are hypothetical and poorly experimentally documented in Arabidopsis; this uncertainty is graphically represented by the display of unknown proteins (boxes surrounded by dotted lines) and gray lines for reaction arrows and catalysis symbols. Much better characterized are the fates of chorismate and isochorismate. The chorismate and isochorismate biosynthesis maps (Fig. 6) indicate six different fates: they are precursors for the synthesis of a stress hormone (salicylate), a cofactor (phylloquinone), a vitamin (folate), and three amino acids (l-Trp, l-Tyr, and l-Phe). Each of these downstream pathways can be accessed by clicking on the relevant purple hexagonal icons. Alternatively, further details on chorismate (a hub metabolite) can be obtained by clicking on the See chorismate HUB icon (Fig. 7A). This hub map (Fig. 7B) shows all the enzymes competing for chorismate and offers a comprehensive view of what is currently known about short-term (allosteric) controls affecting these enzymes. For instance, both anthranilate synthase α-subunits bind l-Trp (an inhibitor); clicking on the red dot provides access to text-based information and a reference. More details are available for chorismate mutases (using the See chorismate mutases icon). The 3D structures of these enzymes and their biochemical and regulatory properties are known; therefore, a molecular model can be proposed (for details, see Fig. 7C).
Figure 6.
Exploration of chorismate and isochorismate metabolism. The so-called shikimate pathway, with the input carbon precursors (erythrose 4-phosphate and phosphoenolpyruvate) and the links toward erythrose 4-phosphate and phosphoenolpyruvate metabolisms (purple hexagons). The six outputs from chorismate and isochorismate (salicylate, phylloquinone, vitamin B9, and the three aromatic amino acids) are represented at the bottom. The transformation of erythrose 4-phosphate and phosphoenolpyruvate into chorismate has been characterized extensively, and all the catalytic conversions (arrows) are thus represented in black. In contrast, the connections between shikimate and shikimate/l-quinate metabolism in the cytosol are only poorly described, as represented by the pale gray color of the arrows. Shikimate O-hydroxycinnamoyltransferase in the cytosol uses shikimate (or l-quinate) as a substrate; therefore, shikimate and/or l-quinate must be exported from the chloroplast. This export step has yet to be characterized at the molecular level, and the nature of the molecule exported (shikimate, 3-dehydroshimikate, or quinate) is currently unclear. This uncertainty is indicated by the gray color of the symbols.
Figure 7.
Chorismate commitment to aromatic amino acid synthesis: passing from a high-level view of metabolism to detailed representation of regulatory processes. A, Zoom on the chorismate and isochorismate map showing the link toward the chorismate HUB. B, The chorismate HUB map is at the crossroads for 11 metabolic maps. Note the minus and plus signs above the metabolites in the protein complexes. The minus sign appended to l-Trp in the anthranilate synthase subunit 1 complex indicates that binding of l-Trp to this subunit in the complex inhibits the protein’s activity. The red dot indicates that text-based information is available for the complex. C, Detailed mechanistic description of how the activity of chorismate mutase1 (CM1) and CM3 are controlled, based on 3D data. The model shows that CM3 is activated only by l-Trp and that CM1 and CM2 are inhibited by l-Tyr and l-Phe and activated by l-Trp. l-Trp is a nonessential activator; therefore, CM1 and CM2 are active in the absence of l-Trp (a catalysis symbol links the protein and the reaction arrow). The presence of l-Trp renders the two enzymes more active, as symbolized by the thicker catalysis symbol connecting the protein and the reaction. Binding of l-Trp, l-Tyr, and l-Phe to CM1 is mutually exclusive (the protein has a single regulatory site); this is represented by three different complexes, each binding one of the three amino acids. When in complex with l-Tyr or l-Phe, the CM1 dimer is inactive, as represented by the absence of the catalysis symbol connecting these complexes to the reaction. CM2 is unregulated, and its function in the cytosol is unclear. Note that the box representing CM3 is surrounded by a thin line to indicate that it was not experimentally detected in leaf.
CONCLUSION
ChloroKB is a unique, extensively biocurated, and fully referenced visual interface, presenting a comprehensive view of chloroplast metabolism connected to whole-cell metabolism. Thanks to CellDesigner standard representations combined with the additional codes specifically developed for ChloroKB to represent biochemical complexity, as well as extensive biocuration, ChloroKB provides a complete overview of the current state of knowledge of complex molecular processes in their cellular context. Navigation between images and text-based information facilitates exploration of the knowledge accumulated over many years of research.
ChloroKB can be used for many different purposes: for hypothesis-driven analyses, to understand complex interconnected biochemical processes, and to identify missing information that requires additional biochemical characterization (Fig. 5). By allowing visualization of the network structure and facilitating access to relevant data, ChloroKB will be invaluable for efficient dynamic modeling. ChloroKB also might be useful in elucidating evolutionary issues. ChloroKB also provides information that can be useful when writing and reviewing articles, as it provides instant access to a wide range of up-to-date information on chloroplast components and their interactions. In addition to its use for teaching purposes, this visualization tool also will be a great help to nonexperts or young scientists who lack the years of experience and the knowledge acquired by senior scientists. As ChloroKB is based on the use of the freely accessible, easy-to-use software CellDesigner, the extent of the network can be improved continuously through community contributions; interested users and modelers are invited to use the icon Feedback and contribution to ChloroKB from the home page. In addition, a template file (XML format) containing all the graphical codes (as illustrated in Supplemental Fig. S2) is available upon request.
In the future, other metabolic or molecular processes will be added to ChloroKB, which will ultimately provide a complete representation of the molecular network in Arabidopsis chloroplasts and beyond. This work will facilitate the construction of equivalent models in crop plants or algae, using the maps as templates. In the long run, we hope that it will be possible to combine this metabolic network with a reconstruction of the gene network controlling metabolism, along the lines of the reconstructed genetic network controlling Arabidopsis flower development (Bouché et al., 2016).
MATERIALS AND METHODS
Data Representation and Graphical Annotation in ChloroKB
The biochemical context, Arabidopsis (Arabidopsis thaliana) chloroplast and other subcellular compartments, was reproduced using the CellDesigner formalism (Funahashi et al., 2008). Proteins, complexes, metabolites, ions, chemical inhibitors (called drugs in CellDesigner), their subcellular/subplastidial localization, and their various conversions (transport, catalysis, cleavage, regulation, posttranslational modifications, and assembly processes) were represented. CellDesigner supports graphical notation and listing of the symbols based on the Systems Biology Graphical Notation (Le Novère et al., 2009). This graphical notation was further extended to distinguish established knowledge from predictions and hypotheses (i.e. whether subcellular-subplastidial localization has been experimentally confirmed, whether protein function has been demonstrated, and whether a protein has been detected in the reference tissue). Results of biochemical, immunological, and/or proteomics-based studies were considered experimental evidence.
Graphically, proteins that were experimentally detected in leaf (the reference tissue in ChloroKB) are represented as a thick-bordered box, whereas proteins for which no experimental evidence for presence in leaf has yet been published are represented with a thin border. Each graphical box (protein) also was assigned a color referring to its curated subplastidial localization (orange = stroma, yellow = envelope, green = thylakoid, and purple = plastoglobule) or localization elsewhere in the cell (white = cytosol, pink = mitochondrion, and peach = endoplasmic reticulum/Golgi/vacuole). Proteins of unknown intraplastidial localization but which have been reported to be targeted to the plastid (e.g. GFP experiments and in vitro import assays) are indicated in pale green. Proteins for which localization is uncertain (i.e. for which no experimental evidence is available) are indicated in red. Graphical symbols representing metabolites follow the same color code, although only two colors (pale green and purple for the lumen) were retained for the chloroplast. Finally, noncovalent activation/inhibition of the protein in protein-metabolite complexes is systematically indicated, with a plus sign (for activators) or a minus sign (for inhibitors) appended to the metabolite in the protein-metabolite complexes (see description of chorismate metabolism, Fig. 4C). Covalent modifications by phosphorylation, Cys oxidation, and protein cleavage also are represented thanks to the graphical notation available in CellDesigner. The notion of unknown proteins (proteins for which a biological activity has been suggested or reported but for which the corresponding protein-coding gene has yet to be identified) also is represented on the maps.
Published data were biocurated extensively to distinguish between hypothetical and predicted reactions versus experimentally established protein activity. Conclusions of this curation are represented as follows. The arrow representing a reaction was colored black for experimentally demonstrated reactions (regardless of the catalytic protein); reactions that are strongly supported by indirect evidence also were indicated by black arrows. Hypothetical reactions were indicated by gray arrows, allowing users to readily distinguish between established knowledge and hypotheses. Predicted protein activity, based on a protein sequence similarity criterion but for which the protein/gene has not been experimentally characterized (e.g. biochemically, by functional complementation, by characterization of insertion lines, or by analysis of overexpression constructs), were connected to the reaction symbol by a gray catalysis symbol. Based on this representation, a rapid glance at a metabolic map can distinguish between experimentally characterized proteins and proteins awaiting characterization. Detailed legends can be displayed at any time by clicking in the top right region of any ChloroKB page (Supplemental Figs. S1 and S2).
Text-Based Information
The visual representation of the network components is associated with curated text-based information. This information can be accessed by clicking on the symbol representing a protein or metabolite in any metabolic map. A description page is available for all molecule types (proteins, unknown proteins, metabolites, ions, chemical inhibitors, and complexes). These pages list curated information (name, localization, function, reaction[s] catalyzed, and molecule[s] transported), hyperlinks to selected databases (see next paragraph), and PubMed references. Only bibliographic references reporting biochemical information (e.g. substrate specificity and kinetic data), experimental evidence for localization (e.g. antibody based, GFP fusion, activity measurements, and proteomics), or genetic data (i.e. insertion mutant phenotypic analyses) are included in ChloroKB; large-scale studies were voluntarily omitted. Semiquantitative data for plastidial proteins (i.e. spectral counting from the AT_CHLORO database; Ferro et al., 2010) and absolute quantification for metabolites also are reported in these description pages when available. For metabolites, literature references, original published units, tissues or subcellular compartments, experimental conditions, and comments are provided (see the ATP description page). Description pages also were produced for some protein-protein or protein-metabolite complexes; these are indicated by red dots. For regulatory proteins, the description page was used to briefly explain the properties of the complex.
In some cases, the proteins have not been characterized in Arabidopsis and much of what we know, such as kinetic properties or localization, is available only from orthologs expressed in other model plant systems (e.g. pea [Pisum sativum], spinach [Spinacia oleracea], and tobacco [Nicotiana tabacum]). This information is scattered throughout the literature but has been gathered together in ChloroKB to guide model building and to facilitate the characterization of Arabidopsis proteins. The organism from which evidence was obtained is listed as the plant species alongside the references cited in the description pages (see the Rubisco map). A similar approach was applied to quantitative data provided for metabolites when the information was unavailable for Arabidopsis but was available from other model species.
Cross-Referencing of Other Databases
Cross-references to related molecular biology databases is provided in description pages in ChloroKB. For proteins, these include TAIR (Huala et al., 2001; Berardini et al., 2015), UniProt (UniProt Consortium, 2015), KEGG (Kanehisa et al., 2016), ENZYME (Bairoch 2000), and two more specialized databases, AT_CHLORO (Ferro et al., 2010) for semiquantitative information on the relative abundance of chloroplast proteins and MASCP Gator (Joshi et al., 2011). MASCP Gator centralizes proteomics data from different Arabidopsis tissues and, thus, makes it possible to explore protein expression in different organs in addition to the mesophyllic leaf tissues used as a reference for ChloroKB. A link to SABIO-RK (Wittig et al., 2012) is provided for biochemical reaction kinetics (even though it only documents a very small number of Arabidopsis proteins) and to The Protein Databank (Berman et al., 2000) for protein 3D structures. For metabolites, links to ChEBI (Hastings et al., 2013), MetaCyc (Caspi et al., 2014), and PubChem (Kim et al., 2016) are provided. PubChem links are used for direct visualization of metabolites in ChloroKB. Links to MetaCyc reaction identifiers are provided for an atomic-scale representation of reactions.
If the 3D structure of the Arabidopsis protein is unavailable but it is known for the homologous enzyme from another plant model, the Protein Data Bank code is included in the comment field.
Cross-references to specialized metabolic pathway databases also were included directly on maps in the following three cases: the Aralip database (http://aralip.plantbiology.msu.edu/pathways/pathways; Beisson et al., 2003) in the malonyl-CoA synthesis and monogalactosyldiacylglycerol, digalactosyldiacylglycerol, and sulfoquinovosyldiacylglycerol synthesis maps; the oxylipin profiling database (http://www.oxylipins.uni-goettingen.de/) in the jasmonate and ketol maps; and AtIPD (http://www.atipd.ethz.ch/; Vranova et al., 2011) in the geranylgeranyl diphosphate map.
Software Architecture, Data Model, and Visualization
These structured heterogenous biological data can be exported as XML files for further computer manipulation (e.g. embedded into graphical format; see below). In terms of architecture, the ChloroKB application was built using a model view controller architecture. This architecture is based on the Grails framework (https://grails.org), a ready-to-use development environment for Web applications. The model view controller design split the software into three interconnected parts making it possible to use: (1) the view to display the Web layer; (2) the model to store the information from the database; and (3) the controller to manipulate the data. Data were organized and stored using MongoDB (https://www.mongodb.com), an open-source document-oriented database. This solution was selected because it provides high-performance data persistence, which increases data availability and automatic scalability. Data were stored in the form of documents, which are themselves gathered into collections. The data model currently consists of three main collections: (1) map, a collection of CellDesigner maps; (2) record, a collection containing descriptive data related to all molecules (proteins, metabolites, complexes, etc.); and (3) crossref_URL, a collection of all URLs linking ChloroKB information to other public resources. A detailed description of the ChloroKB data model can be found in Supplemental Data S1. For interactive and animated map visualization, we chose the scalable vector graphics (SVG) format, an XML-based vector image format for two-dimensional graphics. The SVG images and their behavior are defined in XML text files; therefore, they can be searched, indexed, scripted, and compressed. In association with javascript, this technological choice also allows extensive browsing options (zoom in/out, embedded hyperlinks, scrolling, mouse over, etc.).
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Graphical code and explanations.
Supplemental Figure S2. Contextualized graphical code.
Supplemental Figure S3. Spatial organization represented in ChloroKB.
Supplemental Figure S4. Transport reactions in ChloroKB.
Supplemental Table S1. Curated subcellular and suborganellar localization in ChloroKB.
Supplemental Table S2. Gene identifiers and corresponding curated subcellular localizations in ChloroKB.
Supplemental Table S3. Gene identifiers, protein names, and short names in ChloroKB.
Supplemental Data S1. ChloroKB data model.
Acknowledgments
We thank Jacques Joyard, who initiated this project, for continued support since the very beginning; the staff of the Plant and Cell Physiology laboratory and attendees of the ChloroKB’s day, who participated in testing ChloroKB; Florence Combes for technical assistance; Pierre Baldet, Elisa Dell’Aglio, Younès Dellero, Eric Maréchal, Renaud Dumas, and Águila Ruiz-Sola for reviewing specific maps; and Maighread Gallagher-Gambarelli for advice on English usage and editing suggestions.
Glossary
- SVG
scalable vector graphics
Footnotes
This work was supported by the French National Research Agency (grant no. ANR-10-LABEX-04 GRAL Labex, Grenoble Alliance for Integrated Structural Cell Biology).
Articles can be viewed without a subscription.
References
- Arabidopsis Genome Initiative (2000) Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 408: 796–815 [DOI] [PubMed] [Google Scholar]
- Bairoch A. (2000) The ENZYME database in 2000. Nucleic Acids Res 28: 304–305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beisson F, Koo AJ, Ruuska S, Schwender J, Pollard M, Thelen JJ, Paddock T, Salas JJ, Savage L, Milcamps A, et al. (2003) Arabidopsis genes involved in acyl lipid metabolism: a 2003 census of the candidates, a study of the distribution of expressed sequence tags in organs, and a Web-based database. Plant Physiol 132: 681–697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berardini TZ, Reiser L, Li D, Mezheritsky Y, Muller R, Strait E, Huala E (2015) The Arabidopsis information resource: making and mining the “gold standard” annotated reference plant genome. Genesis 53: 474–485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, Shindyalov IN, Bourne PE (2000) The Protein Data Bank. Nucleic Acids Res 28: 235–242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernard T, Bridge A, Morgat A, Moretti S, Xenarios I, Pagni M (2014) Reconciliation of metabolites and biochemical reactions for metabolic networks. Brief Bioinform 15: 123–135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouché F, Lobet G, Tocquin P, Périlleux C (2016) FLOR-ID: an interactive database of flowering-time gene networks in Arabidopsis thaliana. Nucleic Acids Res 44: D1167–D1171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burge S, Attwood TK, Bateman A, Berardini TZ, Cherry M, O’Donovan C, Xenarios L, Gaudet P (2012) Biocurators and biocuration: surveying the 21st century challenges. Database (Oxford) 2012: bar059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caspi R, Altman T, Billington R, Dreher K, Foerster H, Fulcher CA, Holland TA, Keseler IM, Kothari A, Kubo A, et al. (2014) The MetaCyc database of metabolic pathways and enzymes and the BioCyc collection of pathway/genome databases. Nucleic Acids Res 42: D459–D471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curien G, Bastien O, Robert-Genthon M, Cornish-Bowden A, Cárdenas ML, Dumas R (2009) Understanding the regulation of aspartate metabolism using a model based on measured kinetic parameters. Mol Syst Biol 5: 271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curien G, Cárdenas ML, Cornish-Bowden A (2014) Analytical kinetic modeling: a practical procedure. Methods Mol Biol 1090: 261–280 [DOI] [PubMed] [Google Scholar]
- Dauga D. (2015) Biocuration: a new challenge for the tunicate community. Genesis 53: 132–142 [DOI] [PubMed] [Google Scholar]
- de Oliveira Dal’Molin CG, Quek LE, Palfreyman RW, Brumbley SM, Nielsen LK (2010) AraGEM, a genome-scale reconstruction of the primary metabolic network in Arabidopsis. Plant Physiol 152: 539–549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farré G, Twyman RM, Christou P, Capell T, Zhu C (2015) Knowledge-driven approaches for engineering complex metabolic pathways in plants. Curr Opin Biotechnol 32: 54–60 [DOI] [PubMed] [Google Scholar]
- Ferro M, Brugiere S, Salvi D, Seigneurin-Berny D, Court M, Moyet L, Ramus C, Miras S, Mellal M, Le Gall S, et al. (2010) AT_CHLORO, a comprehensive chloroplast proteome database with subplastidial localization and curated information on envelope proteins. Mol Cell Proteomics 9: 1063–1084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Funahashi A, Matsuoka Y, Jouraku A, Morohashi M, Kikuchi N, Kitano H (2008) CellDesigner 3.5: a versatile modeling tool for biochemical networks. Proc IEEE 96: 1254–1265 [Google Scholar]
- Hastings J, de Matos P, Dekker A, Ennis M, Harsha B, Kale N, Muthukrishnan V, Owen G, Turner S, Williams M, et al. (2013) The ChEBI reference database and ontology for biologically relevant chemistry: enhancements for 2013. Nucleic Acids Res 41: D456–D463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howe D, Costanzo M, Fey P, Gojobori T, Hannick L, Hide W, Hill DP, Kania R, Schaeffer M, St Pierre S, et al. (2008) Big data: the future of biocuration. Nature 455: 47–50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huala E, Dickerman AW, Garcia-Hernandez M, Weems D, Reiser L, LaFond F, Hanley D, Kiphart D, Zhuang M, Huang W, et al. (2001) The Arabidopsis Information Resource (TAIR): a comprehensive database and web-based information retrieval, analysis, and visualization system for a model plant. Nucleic Acids Res 29: 102–105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarvis P, López-Juez E (2013) Biogenesis and homeostasis of chloroplasts and other plastids. Nat Rev Mol Cell Biol 14: 787–802 [DOI] [PubMed] [Google Scholar]
- Joshi HJ, Hirsch-Hoffmann M, Baerenfaller K, Gruissem W, Baginsky S, Schmidt R, Schulze WX, Sun Q, van Wijk KJ, Egelhofer V, et al. (2011) MASCP Gator: an aggregation portal for the visualization of Arabidopsis proteomics data. Plant Physiol 155: 259–270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanehisa M, Sato Y, Kawashima M, Furumichi M, Tanabe M (2016) KEGG as a reference resource for gene and protein annotation. Nucleic Acids Res 44: D457–D462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S, Thiessen PA, Bolton EE, Chen J, Fu G, Gindulyte A, Han L, He J, He S, Shoemaker BA, et al. (2016) PubChem substance and compound databases. Nucleic Acids Res 44: D1202–D1213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Novère N, Hucka M, Mi H, Moodie S, Schreiber F, Sorokin A, Demir E, Wegner K, Aladjem MI, Wimalaratne SM, et al. (2009) The systems biology graphical notation. Nat Biotechnol 27: 735–741 [DOI] [PubMed] [Google Scholar]
- Mintz-Oron S, Meir S, Malitsky S, Ruppin E, Aharoni A, Shlomi T (2012) Reconstruction of Arabidopsis metabolic network models accounting for subcellular compartmentalization and tissue-specificity. Proc Natl Acad Sci USA 109: 339–344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller LA, Zhang PF, Rhee SY (2003) AraCyc: a biochemical pathway database for Arabidopsis. Plant Physiol 132: 453–460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poolman MG, Miguet L, Sweetlove LJ, Fell DA (2009) A genome-scale metabolic model of Arabidopsis and some of its properties. Plant Physiol 151: 1570–1581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rolland N, Curien G, Finazzi G, Kuntz M, Marechal E, Matringe M, Ravanel S, Seigneurin-Berny D (2012) The biosynthetic capacities of the plastids and integration between cytoplasmic and chloroplast processes. Annu Rev Genet 46: 233–264 [DOI] [PubMed] [Google Scholar]
- Sandoval FJ, Zhang Y, Roje S (2008) Flavin nucleotide metabolism in plants: monofunctional enzymes synthesize FAD in plastids. J Biol Chem 283: 30890–30900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seaver SMD, Henry CS, Hanson AD (2012) Frontiers in metabolic reconstruction and modeling of plant genomes. J Exp Bot 63: 2247–2258 [DOI] [PubMed] [Google Scholar]
- Stobbe MD, Jansen GA, Moerland PD, van Kampen AHC (2014) Knowledge representation in metabolic pathway databases. Brief Bioinform 15: 455–470 [DOI] [PubMed] [Google Scholar]
- Sweetlove LJ, Fernie AR (2013) The spatial organization of metabolism within the plant cell. Annu Rev Plant Biol 64: 723–746 [DOI] [PubMed] [Google Scholar]
- UniProt Consortium (2015) UniProt: a hub for protein information. Nucleic Acids Res 43: D204–D212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vranova E, Hirsch-Hoffmann M, Gruissem W (2011) AtIPD: a curated database of Arabidopsis isoprenoid pathway models and genes for isoprenoid network analysis. Plant Physiol 156: 1655–1660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wittig U, Kania R, Golebiewski M, Rey M, Shi L, Jong L, Algaa E, Weidemann A, Sauer-Danzwith H, Mir S, et al. (2012) SABIO-RK: database for biochemical reaction kinetics. Nucleic Acids Res 40: D790–D796 [DOI] [PMC free article] [PubMed] [Google Scholar]