Numerous kaurene synthase-like cyclases have been identified in Isodon rubescens, providing new perspectives into understanding the evolution of diterpenoid chemical diversity in Lamiaceae.
Abstract
Ent-kaurene diterpenoids are the largest group of known Isodon diterpenoids. Among them, oridonin is accumulated in the leaves, and is the most frequently studied compound because of its antitumor and antibacterial activities. We have identified five copalyl diphosphate synthase (CPS) and six kaurene synthase-like (KSL) genes by transcriptome profiling of Isodon rubescens leaves. An in vitro assay assigns ten of them to five different diterpene biosynthesis pathways, except IrCPS3 that has a mutation in the catalytic motif. The Lamiaceae-specific clade genes (IrCPS1 and IrCPS2) synthesize the intermediate copalyl diphosphate (normal-CPP), while IrCPS4 and IrCPS5 synthesize the intermediate ent-copalyl diphosphate (ent-CPP). IrKSL2, IrKSL4, and IrKSL5 react with ent-CPP to produce an ent-isopimaradiene-like compound, ent-atiserene and ent-kaurene, respectively. Correspondingly, the Lamiaceae-specific clade genes IrKSL1 or IrKSL3 combined with normal-CPP led to the formation of miltiradiene. The compound then underwent aromatization and oxidization with a cytochrome P450 forming two related compounds, abietatriene and ferruginol, which were detected in the root bark. IrKSL6 reacts with normal-CPP to produce isopimaradiene. IrKSL3 and IrKSL6 have the γβα tridomain structure, as these proteins tend to possess the bidomain structure of IrKSL1, highlighting the evolutionary history of KSL gene domain loss and further elucidating chemical diversity evolution from a macroevolutionary stance in Lamiaceae.
The mint family (Lamiaceae) is the sixth largest family of flowering plants and one of the most economically valuable natural products for foods and toiletries. The Lamiaceae family produces a unique class of terpenoids called diterpenoids. The most common diterpenoids in Lamiaceae are abietanes, labdanes, neoclerodanes, primaranes, and ent-kauranes. These are taxa-specific compounds that have been found in six of the seven subfamilies (Harley et al., 2004). The diterpenoid skeleton, abietane, has the highest occurrence and is the most widespread among Lamiaceae (Vestri Alvarenga et al., 2001), but is characteristic of the Nepetoideae subfamily. Ent-kauranes are characteristic of the Isodon genus (Sun et al., 2006) and labdanes are characteristic of many genera in the subfamily Lamioideae (Harley et al., 2004). These diterpenoids fall into the labdane-related superfamily, which evolved from the same machinery involved in gibberellin (GA) metabolism (Peters, 2010). Biosynthesis is initiated by two sequential diterpene synthases (diTPS) that catalyze cyclization and/or rearrangement of the general acyclic precursor (E,E,E)-geranylgeranyl diphosphate (GGPP) to form different requisite skeletal structures (Zi et al., 2014). The first step is a protonation-initiated cyclization of GGPP by the class II diTPS copalyl diphosphate synthase (CPS) that leads to the formation of a bicyclic labdadienyl/copalyl diphosphate (CPP) with a specific stereochemistry (ent, syn, normal / (+)). CPP is further cyclized and/or rearranged in a diphosphate ionization-initiated reaction catalyzed by a class I diTPS, kaurene synthase-like cyclase (KSL), which is usually specific to a particular CPP stereoisomer.
The diterpenoid biosynthesis has been explored in nine species in Lamiaceae, including Salvia miltiorrhiza (Gao et al., 2009; Cui et al., 2015; Su et al., 2016), S. sclarea (Caniard et al., 2012; Schalk et al., 2012), Isodon eriocalyx (Li et al., 2012), Coleus forskohlii (Pateraki et al., 2014), Rosmarinus officinalis (Brückner et al., 2014), S. fruticosa (Božić et al., 2015), S. pomifera (Trikka et al., 2015), and S. divinorum (Pelot et al., 2017) from the biggest subfamily Nepetoideae, and Marrubium vulgare (Zerbe et al., 2014) from the subfamily Lamioideae. These enzymes have the ability to produce abietane, labdane, clerodane, and ent-kaurane diterpenes via an enzymatic hydrolysis-catalyzed reaction (Gao et al., 2009; Cui et al., 2015; Caniard et al., 2012; Schalk et al., 2012; Li et al., 2012; Pateraki et al., 2014; Zerbe et al., 2014; Brückner et al., 2014; Božić et al., 2015; Trikka et al., 2015; Pelot et al., 2017). Based on the stereochemistry of the intermediate that is initially cyclized by CPS, these enzymes can be divided into two groups. One group produces an intermediate of normal CPP, which has been found to be the precursor of a large array of diterpenoids with high economic and medicinal value, including tanshinone (Gao et al., 2009; Cui et al., 2015), sclareol (Caniard et al., 2012; Schalk et al., 2012), and forskolin (Pateraki et al., 2014). The other group produces the stereoisomeric intermediate of ent-CPP that is involved in GA biosynthesis as well as the synthesis of labdane or ent-kaurane in S. miltiorrhiza and I. eriocalyx (Li et al., 2012; Cui et al., 2015). Phylogenetic analysis has shown that these enzymes are grouped in accordance with their functions except the newly identified SdCPS1 and SdKSL1 from S. divinorum (Pelot et al., 2017), that is, those involved in normal-CPP biosynthesis form a sister group to those involved in ent-CPP biosynthesis, and positive selection has played an important role in this functional divergence (Cui et al., 2015). A similar divergence has also been observed in class I KSL enzymes that are responsible for the subsequent transformation of normal-CPP synthase products. Many of these enzymes have lost the characteristic internal/γ domain that is responsible for metabolite specialization, a deletion event found in KSLs in Lamiaceae (Hillwig et al., 2011; Caniard et al., 2012; Schalk et al., 2012; Brückner et al., 2014; Pateraki et al., 2014; Zerbe et al., 2014; Cui et al., 2015; Božić et al., 2015; Trikka et al., 2015). A recent work has shown the plasticity of class I KSLs, such as SsScS from S. sclarea, that can react with any available CPS product (Andersen-Ranberg et al., 2016; Jia et al., 2016). These studies demonstrate the value of Lamiaceae as a useful system to study diterpenoid chemical diversity evolution. While most recent works focus on specific biosynthesis pathways in given species, little is known about the diversified diterpenoids’ evolutionary origin.
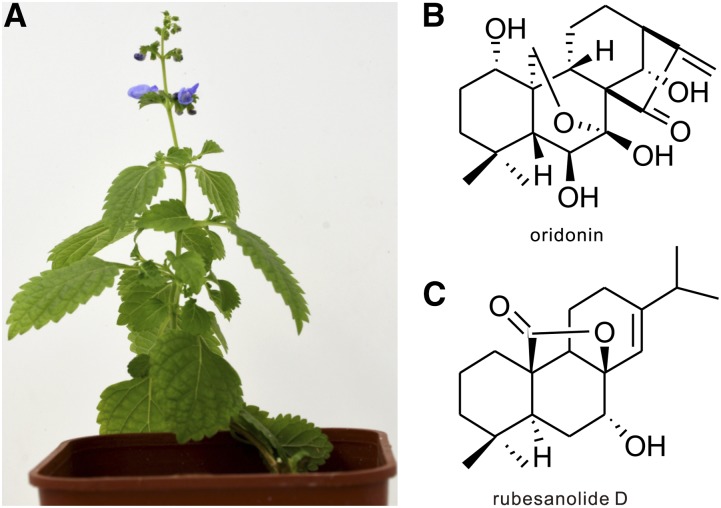
Here we report a large diTPS gene family from Isodon rubescens (tribe, Ocimeae; subfamily, Nepetoideae; Fig. 1A) composed of many ent-kaurane diterpenoids (Sun et al., 2006). One of these, oridonin (Fig. 1B), is found in the leaves of many Isodon plants and is considered to be a potential cancer chemoprevention agent with broad spectrum antitumor and antibacterial activities in vitro and in vivo (Zhou et al., 2007). In contrast to the numerous ent-kaurane diterpenoids, rubesanolides A-E, with only five abietane-related compounds, has been isolated from the leaves of I. rubescens (Zou et al., 2011, 2012). Rubesanolide D (Fig. 1C) has shown inhibition activity against biofilm formation of the dental bacterium Streptococcus mutans (Zou et al., 2012). Although seven diTPS genes have been reported from an I. rubescens transcriptome sequencing project (Zerbe et al., 2013), none of them were identified. Here we report 11 diTPS genes from a transcriptome sequencing project. We identified IrCPS4 that is predicted to be involved in oridonin biosynthesis, three KSL genes involved in ent-CPP interactions, and a normal-CPP mediated miltiradiene and isopimaradiene biosynthesis pathway. The three KSL genes bearing different domain structures are involved in the normal-CPP mediated biosynthesis, revealing information regarding the evolutionary origin of the γ-domain loss and further shedding light on the macroevolution of diterpenoid chemical diversity in Lamiaceae.
Figure 1.
The major medicinally active diterpenoids in I. rubescens. A, The plant of I. rubescens. B, The typical ent-kaurene diterpenoid of oridonin. C, The typical abietane diterpenoid of rubesanolide D.
RESULTS
Transcriptome Sequencing and De Novo Assembly
A nonnormalized cDNA library was prepared for transcriptome sequencing using the Illumina HiSeq 2500 platform from leaf tissues of I. rubescens containing the highest oridonin content (Chinese Pharmacopoeia Committee 2015). A total of 27,248,946 reads were obtained. The Trinity de novo assembler (Grabherr et al., 2011) found 60,483 contigs with an average of 886 bp; contig length varied from 201 to 12,118 bp (Supplemental Table S1).
Annotation of diTPS Family in I. rubescens
All contigs were aligned to the GenBank nonredundant protein sequence database using BLASTX with e-values < 1e−5. Twenty-nine unigenes were annotated as copalyl diphosphate synthase or kaurene synthase-like cyclase genes (Supplemental Table S2). After manual assembly using known diTPS sequences from I. eriocalyx, S. miltiorrhiza, and a previous reported I. rubescens transcriptome sequencing data set (Zerbe et al., 2013), we found five putative diTPS with full-length cDNA, annotated as IrCPS1, IrCPS3, IrCPS4, IrKSL1, and IrKSL3 due to their homology to diTPS in S. miltiorrhiza (Supplemental Fig. S1). Together with 5′- and 3′-RACE, six other full-length diTPS were identified and annotated as IrCPS2, IrCPS5, IrKSL2, IrKSL4, IrKSL5, and IrKSL6. In total, we established five CPS and six KSL genes from I. rubescens (Supplemental Table S3) that comprise 28 unigenes (does not include c40624_g1_i1). Among these 11 genes, three of them, IrCPS1, IrKSL1, and IrKSL3, have corresponding full length sequence in the reported data set (Zerbe et al., 2013) with 98% to ∼99% identities. Four of them, IrCPS3, IrCPS4, IrCPS5, and IrKSL6 have corresponding sequences with 99% identity, and the other four genes have sequences with identities <93% (Supplemental Table S4).
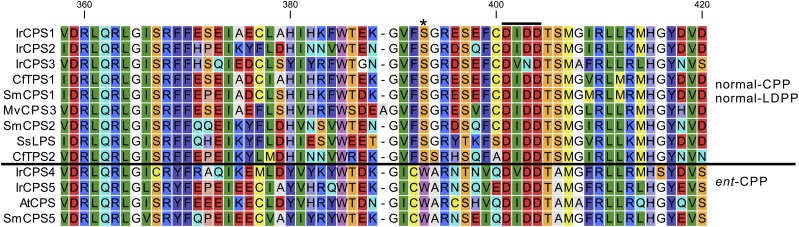
The deduced amino acid sequences of the four putative CPS genes (IrCPS1, IrCPS2, IrCPS4, and IrCPS5) contain the “(D,E)XDD” motif typical of CPS enzymes (Prisic et al., 2004), while the six putative KSL genes have the “DDXXD” motif that is characteristic of KSL enzymes (Christianson, 2006; Supplemental Fig. S1). In IrCPS3, the second Asp of the DXDD motif (corresponding to D379 in AtCPS from Arabidopsis [Arabidopsis thaliana]) is substituted by Asn (Fig. 2), The Asp amino acid has been reported to be essential in forming hydrogen-bonded “proton wires” during catalysis (Köksal et al., 2014).
Figure 2.
Alignment of the conserved DXDD motif and the positive selection site (Ser-362/Trp-364) of CPS from I. rubescens and other characterized CPSs. The DXDD motif is labeled above the alignment with a solid line, and the conserved Ser or Trp position is marked with an asterisk. The second Asp of the DXDD motif is substituted by Asn in IrCPS3. The enzymes are separated by the horizontal line according to normal-CPP/LDPP or ent-CPP stereochemistry. The CPS in the upper section (including IrCPS1 and IrCPS2 from this study) were characterized as being involved in normal-CPP/LDPP formation while those in the lower section (including IrCPS4 and IrCPS5 from this study) were characterized to be involved in ent-CPP production. CfTPS1 and CfTPS2 are from C. forskohlii, SmCPS1, SmCPS2, SmCPS5 from S. miltiorrhiza, MvCPS3 from M. vulgare, SsLPS from S. sclarea, and AtCPS from Arabidopsis.
The positive selection site Ser-362 (SmCPS1 in normal-CPP synthase)/Trp-364 (SmCPS5 in ent-CPP synthase) has shown to be essential for formation of normal-CPP versus ent-CPP (Cui et al., 2015). In I. rubescens, IrCPS1 and IrCPS2 have a Ser (S) at the 373 and 367 positions, as do the other six CPS from Lamiaceae (Fig. 2). This is necessary for proper enzymatic function to produce normal-CPP or normal-LDPP using GGPP as a substrate (Schalk et al., 2012; Pateraki et al., 2014; Zerbe et al., 2014; Cui et al., 2015). The other two genes, IrCPS4 and IrCPS5, have a Trp (W) in this position (Fig. 2), similar to SmCPS5 from S. miltiorrhiza and AtCPS, whose product is ent-CPP when using GGPP as a substrate (Cui et al., 2015).
Phylogenetic Relationship of diTPS within Lamiaceae
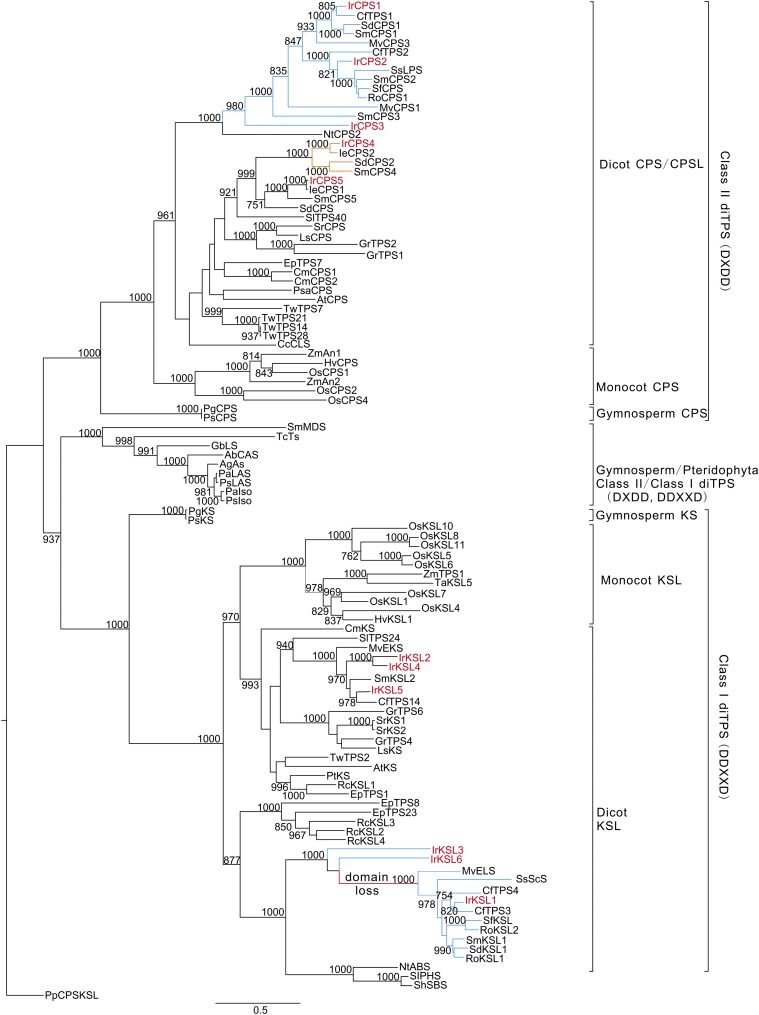
A maximum-likelihood tree was constructed with 96 diTPS enzymes with known functions (Supplemental Table S5). The diTPS from I. rubescens cluster with other genes from Lamiaceae, forming four clades. Two clades contain CPS and KSL that are involved in ent-CPP or its hydroxylated LDPP-mediated biosynthesis while the other two clades contain CPS and KSL that are involved in specialized normal-CPP mediated biosynthesis (Fig. 3).
Figure 3.
Phylogeny of I. rubescens diterpene synthases. The maximum likelihood tree illustrates the phylogenetic relationship of I. rubescens diterpene synthases with 96 representative characterized diTPS (Supplemental Table S5). Numbers on branches indicate the bootstrap percentage values calculated from 1000 bootstrap replicates. Physcomitrella patens CPS/kaurene was used as an outgroup. Blue lines show diTPS genes from Lamiaceae involved in normal-CPP/LDPP-mediated diterpenoid metabolism and red lines show the loss of the N-terminal γ domain found in eudicot and monocot KSLs. Yellow lines show genes from Lamiaceae involved in the specialized ent-CPP/LDPP-related diterpenoid metabolism. Red-marked enzymes show diTPS from I. rubescens in this study.
IrCPS1, IrCPS2, and IrCPS3 are found in the Lamiaceae-specialized CPS clade (Fig. 3), which also contains CfTPS1/CfTPS2 from C. forskohlii (Pateraki et al., 2014), SmCPS1/SmCPS2 from S. miltiorrhiza (Cui et al., 2015), MvCPS1/MvCPS3 from M. vulgare (Zerbe et al., 2014), RoCPS1 from R. officinalis (Brückner et al., 2014), SsLPS from S. sclarea (Schalk et al., 2012), SfCPS from S. fruticosa (Božić et al., 2015) and SdCPS1 from S. divinorum (Pelot et al., 2017). The enzymes mentioned above have catalytic activity with GGPP to form normal-CPP and the stereochemically related normal-LDPP (Schalk et al., 2012; Brückner et al., 2014; Pateraki et al., 2014; Zerbe et al., 2014; Cui et al., 2015; Božić et al., 2015) with the exception of SdCPS1 (Pelot et al., 2017). IrCPS4 and IrCPS5 are found in the ent-CPP synthase clade. IrCPS4 is closely related to an ent-CPS from I. eriocalyx (IeCPS2), which is predicted to be involved in the biosynthesis of Isodon ent-kaurene diterpenoids (Li et al., 2012). IrCPS5 is closely related to IeCPS1 and SmCPS5 that are involved in the GA biosynthetic pathway (Fig. 3).
IrKSL1 is grouped in the specialized Lamiaceae KSL clade, having the less common βα bidomain architecture through the loss of the γ domain at the N terminus (Fig. 3). These enzymes combine with the Lamiaceae-specialized CPS clade to form intermediates like miltiradiene, (13R)-manoyl oxide, and sclareol that are precursors to a large array of pharmaceutic compounds (Schalk et al., 2012; Brückner et al., 2014; Pateraki et al., 2014; Zerbe et al., 2014; Cui et al., 2015; Božić et al., 2015). It is interesting to note that IrKSL3 and IrKSL6 form a monophyletic clade with IrKSL1 despite the fact that they have the common γβα tridomain structures. IrKSL2, IrKSL4, and IrKSL5 belong to the KSLs that are involved in the ent-CPP mediated biosynthetic pathway that catalyzes reactions with ent-CPP and ent-LDPP (Fig. 3).
Functional Analysis of ent-CPP-Mediated Diterpene Synthesis
Consistent with its closer relationship to other dicot CPS and KSL genes involved in ent-CPP mediated GA or other related biosynthesis, IrCPS4 and IrCPS5 were found to produce ent-CPP. We first incubated IrCPS4 and IrCPS5 with GGPP alone for comparison with a known enzyme SmCPS5 (ent-CPP synthase) from S. miltiorrhiza. IrCPS4 and IrCPS5 yielded a diterpene with identical retention time and mass spectrum to the product of SmCPS5 after dephosphorylation, (Supplemental Fig. S2, A and B) illustrating that the primary product of IrCPS4 and IrCPS5 is copalyl diphosphate. The stereochemistry of IrCPS4 and IrCPS5 was further investigated by reaction assays of each protein with SmKSL1 (specific to normal-CPP) and AtKS (specific to ent-CPP). Reactions with IrCPS4 or IrCPS5 coupled with AtKS resulted in the formation of ent-kaurene, while no product was obtained with SmKSL1 (Supplemental Fig. S2A). This illustrates that IrCPS4 and IrCPS5 are ent-copalyl diphosphate synthase and are predicted to be involved in oridonin or GA biosynthesis in I. rubescens.
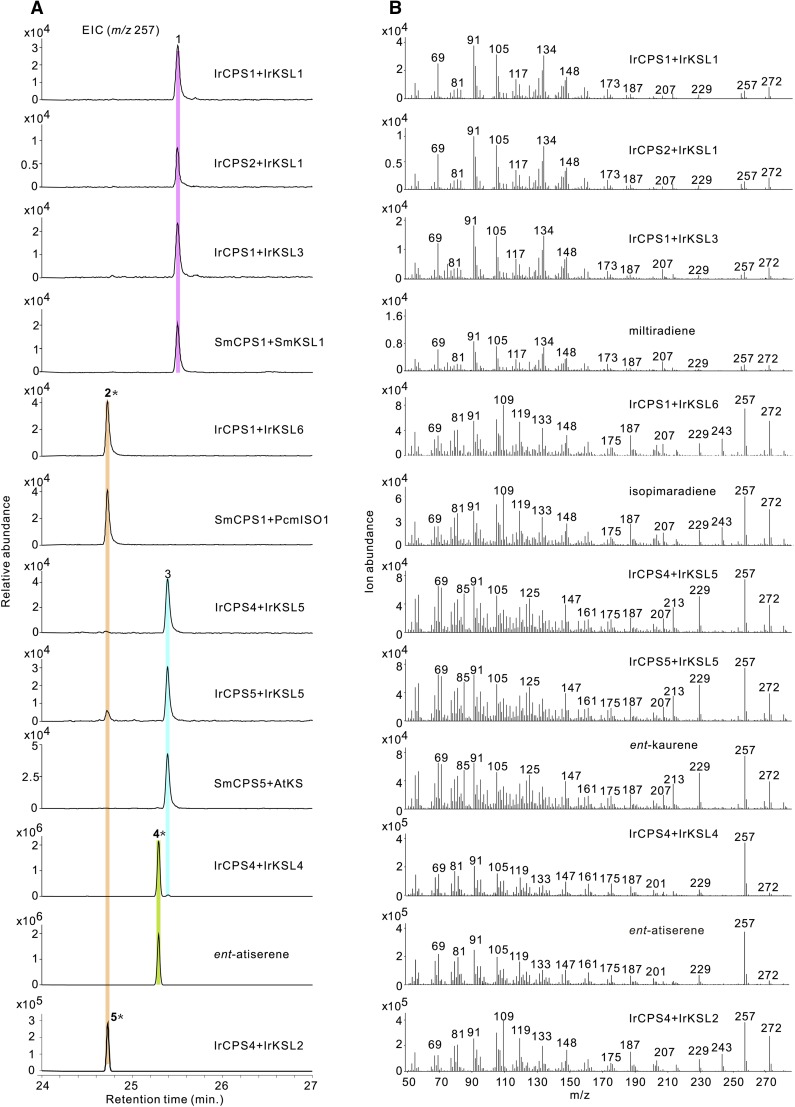
The catalytic activity of IrKSL2, IrKSL4, and IrKSL5 were each analyzed by incubation with IrCPS4 that was identified to produce ent-CPP. All enzymes were found to react with ent-CPP. Specifically, IrKSL2 generated a product with identical EI mass spectrum to isopimaradiene (isopimara-7,15-diene; Hall et al., 2013), IrKSL4 generated a main product of ent-atiserene (95%), and a minor product of ent-kaurene (5%), while IrKSL5 produced ent-kaurene (Fig. 4). At the same time, these enzymes also produced the same product when combined with IrCPS5 (Supplemental Fig. S3).
Figure 4.
GC-MS analysis of in vitro assays with I. rubescens diTPS on a Cyclodex-β GC column. A, Extracted ion chromatograms of m/z 257 of in vitro assays with IrCPS coupled with different IrKSL. The characterized SmCPS5 (ent-CPP synthase from S. miltiorrhiza), SmCPS1 (normal-CPP synthase from S. miltiorrhiza), SmKSL1 (miltiradiene synthase from S. miltiorrhiza), AtKS (ent-kaurene synthase from Arabidopsis), and PcmISO1 (pimaradiene from P. banksiana) were used as positive controls, along with the corresponding authentic standard ent-atiserene. (1) Miltiradiene, (2) isopimaradiene, (3) ent-kaurene, (4) ent-atiserene, and (5) ent-isopimaradiene-like compound. Asterisk indicates the unexpected enzyme products identified in this study. B, Corresponding mass spectra of recombinant enzyme assay products and authentic standard. EIC, Extracted ion chromatograms.
Many enzymes require copalyl diphosphate (normal-CPP) as a substrate including Isopimara-7,15-diene synthase from Picea abies (Martin et al., 2004) and the later identified PbmISO1 and PcmISO1 from Pinus banksiana and P. contorta (Hall et al., 2013). Thus, the stereochemistry of the IrKSL2 product, isopimaradiene, was further compared with PcmISO1. As expected, isopimaradiene was produced when combining PcmISO1 with SmCPS1 (normal-CPP) and has identical retention time with the product of IrCPS4 and IrKSL2 (Fig. 4). No product was observed when combining PcmISO1 with IrCPS4. Therefore, the product of IrCPS4 and IrKSL2 appears to have a different stereochemistry compared to isopimaradiene, however, its absolute configuration needs further identification.
Functional Analysis of Normal-CPP-Mediated Diterpene Synthesis
Subsequently, we analyzed the function of the Lamiaceae-specialized clade gene products IrCPS1, IrCPS2,IrKSL1, IrKSL3, and IrKSL6 in vitro. IrCPS1 and IrCPS2, in combination with IrKSL1 or IrKSL3, formed miltiradiene as identified by comparison to the product of the combined activity of normal-CPP synthase (SmCPS1) and miltiradiene synthase (SmKSL1) from S. miltiorrhiza (Fig. 4). No product was observed when combining IrCPS1 and IrCPS2 with AtKS (specific to ent-CPP) providing further evidence the product of IrCPS1 and IrCPS2 was normal-CPP.
An unexpected result was observed when combining IrCPS1 or IrCPS2 with IrKSL6. The product has the same retention time and EI mass spectrum as the product of PcmISO1 and SmCPS1 (normal-CPP; Fig. 4; Supplemental Fig. S3). Thus, IrKSL6 can react with normal-CPP to produce isopimaradiene in I. rubescens.
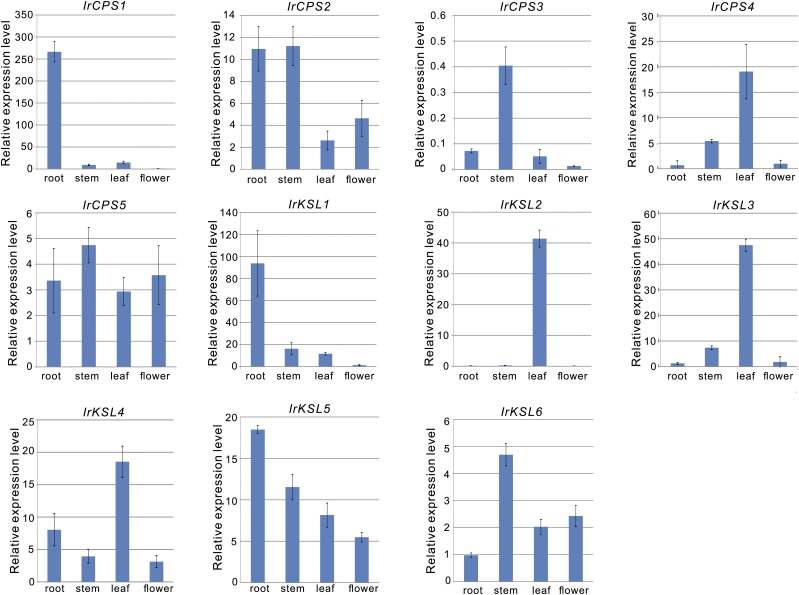
Expression Analysis of the Identified diTPS
To understand the physiological roles of the candidate diTPS in I. rubescens, we used quantitative reverse transcription PCR (qRT-PCR) to evaluate mRNA transcript levels in various organs, including root, stem, leaf, and flower. Transcripts of IrCPS1 and IrKSL1 were detected at extremely high levels in the root (Fig. 5), a similar finding to the expression patterns of SmCPS1 and SmKSL1 in S. miltiorrhiza (Cui et al., 2015). IrCPS2, IrCPS5, IrKLS5, and IrKSL6 were found to have relatively ubiquitous expression in all sampled organ tissues. IrCPS4, IrKSL2, IrKSL3, and IrKSL4 were highly expressed in the leaf tissues. IrCPS3 showed relative lower transcription levels in all organs, which was also similar to expression levels in SmCPS3 of S. miltiorrhiza (Cui et al., 2015; Fig. 5).
Figure 5.
The mRNA expression levels of the 11 candidate diTPS in root, stem, leaf, and flower tissues from I. rubescens. The expression level was normalized to that of actin. The error bars show the SDs from mean value (n = 3 experiments).
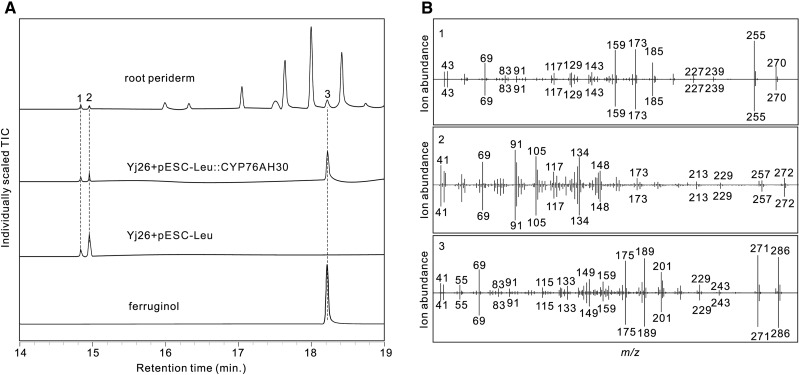
To confirm that abietane diterpenoids (like miltiradiene produced by IrCPS1 and IrKSL1/KSL3) exist in I. rubescens, different organs including root (periderm and cortex), stem, leaf, and flower were extracted with hexane and analyzed by GC-MS. Miltiradiene, together with abietatriene and ferruginol (two compounds related to miltiradiene biosynthesis), were detected in the periderm of the root (Fig. 6). No abietane diterpenoids were found in other organs (Supplemental Fig. S4).
Figure 6.
GC-MS detection of abietane diterpenoids in the periderm of the root and identification of CYP76AH30 in I. rubescens on a TR-5ms capillary column. A, Total ion chromatograms of abietane diterpenoids in the hexane extract of the root periderm and the ferruginol production from strain YJ26 transformed with the plasmid pESC-Leu::CYP76AH30. Strain YJ26 with the plasmid pESC-Leu was used as a control, along with the authentic standard of ferruginol. B, Mass spectra of (1) abietatriene, (2) miltiradiene, and (3) ferruginol from the root periderm (upper halves) and yeast (lower halves). TIC, Total ion chromatograms.
Oridonin is found in plant leaves and is a principle constituent of I. rubescens. This compound accumulates in the leaves at concentrations as high as 0.25% of the dry weight (Chinese Pharmacopoeia Committee, 2015). Thus, the highly expressed levels of IrCPS4 in the leaves illustrate that it may be responsible for oridonin biosynthesis in I. rubescens. Other diterpenes, including ent-atiserene, ent-isopimaradiene like compound and isopimaradiene were not found in the organs studied.
Identification of CYP76AH30 as Ferruginol Synthase in I. rubescens
The conversion of miltiradiene to ferruginol have been elucidated in S. miltiorrhiza (Guo et al., 2013), R. officinalis (Zi and Peters, 2013; Ignea et al., 2016), and S. fruticosa (Božić et al., 2015; Scheler et al., 2016), where a CYP76AH subfamily member plays an important role. It is interesting to note that CYP76AH1 from S. miltiorrhiza is characterized as ferruginol synthase, while CYP76AH22-24 from R. officinalis and S. fruticosa has the ability to produce both ferruginol and 11-hydroxyferruginol. Mutagenesis analysis revealed that reciprocal activity changes from ferruginol synthase to hydroxyferruginol synthase only depend on three amino acids (and vice versa; Scheler et al., 2016). When using CYP76AH1 from S. miltiorrhiza as a query sequence, one full-length homolog called CYP76AH30 was found in our transcriptome data. It shares 80.5% amino acid sequence identity with CYP76AH1, and 76.1% to 77.6% identities with CYP76AH22-24. The three residues, 301E, 303N and 479L of CYP76AH30 are different from both CYP76AH1 and CYP76AH22-24 (Supplemental Fig. S5).
The role of the CYP76AH30 protein was identified by transforming the yeast strain, YJ26 (Zhou et al., 2012) with a plasmid expressing the protein. This yeast strain harbors modules that express a GGPP synthase and farnesyl diphosphate synthase fusion, a SmCPS1 and SmKSL1 fusion, and a truncated hydroxy-3-methylglutaryl coenzymes A reductase (Zhou et al., 2012; Guo et al., 2013). The pESC yeast epitope tagging vector harboring CYP76AH30 (pESC-Leu::CYP76AH30) under the control of the GAL1 promoter was transformed into YJ26. After 48h of fermentation, we detected ferruginol from the strain transformed with CYP76AH30 (Fig. 6). Thus, our results show that CYP76AH30 is a ferruginol synthase and responsible for ferruginol biosynthesis in I. rubescens.
DISCUSSION
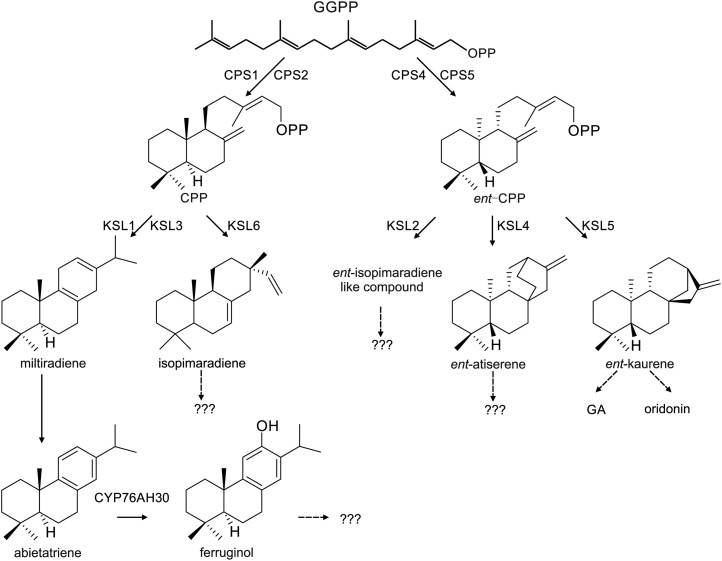
Here, we identified a large diTPS family in I. rubescens containing five CPS and six KSL genes. Ten of these genes were functionally characterized in vitro, representing the most extensive diTPS gene family found in Lamiaceae. Three of these genes are predicted to be involved in ent-kaurene diterpenoids, such as oridonin and the requisite of GA metabolism, while two other genes were also identified as KSLs functioning with ent-CPP as substrates. We also showed that there is a normal-CPP mediated biosynthesis pathway that exists in I. rubescens and that three KSL genes with different domain architectures are involved in this pathway, highlighting the role of KSL domain loss in the evolution of chemical diversification in diterpenoid biosynthesis.
I. rubescens Contains a Functionally Diverse diTPS Family
The intricate nature of diterpene metabolism (Fig. 7) is more complex than previously reported in I. rubescens (Sun et al., 2006). Despite containing four shared functional CPS genes, I. rubescens has four additional KSL genes compared to the two functional KSL genes identified in S. miltiorrhiza (Cui et al., 2015).
Figure 7.
The proposed diterpenoid biosynthesis in I. rubescens. Two sequential diterpene synthases CPS and KSL, and corresponding reactions are indicated, along with the possible downstream natural products. Dashed arrows indicate multiple enzymatic reactions.
Similar to S. miltiorrhiza, the two CPS (IrCPS1 and IrCPS2) and one KSL (IrKSL1) gene products of I. rubescens are closely related to the Lamiaceae specialized CPS and KSL, which have the same enzymatic activity as GGPP to produce normal-CPP and miltiradiene (Fig. 4). The transcription pattern of IrCPS1 and IrKSL1 was similar to those involved in tanshinone biosynthesis in S. miltiorrhiza and was mainly transcribed in the root periderm (Cui et al., 2015). Our in vitro results show the identification of miltiradiene together with abietatriene and ferruginol in the root periderm and further highlight the role of IrCPS1 in the miltiradiene mediated biosynthesis (Fig. 7). In I. rubescens, IrKSL3 also has the ability to react with normal-CPP to produce miltiradiene. IrKSL3 is highly expressed in leaves, suggesting its role in the biosynthesis of abietane diterpenoids such as rubesanolides A-E. However, miltiradiene was not detected in the aerial parts and its metabolic fate in planta calls for further study.
Phylogenetic and sequence analysis suggested that two genes, IrCPS4 and IrCPS5, had the ability to produce ent-CPP, the precursor of ent-kaurene diterpenoids. Unlike the relatively ubiquitous expression patterns of IrCPS55, IrCPS4 exhibited high transcription levels in leaf tissues (Fig. 5), suggesting its role in specialized diterpene metabolism, such as oridonin biosynthesis. The close phylogenetic relationship of IrCPS4 with IeCPS2 from I. eriocalyx, SmCPS4 from S. miltiorrhiza and the newly identified clerodienyl diphosphate synthase SdCPS2 from S. divinorum (Pelot et al., 2017) highlights a Lamiaceae-specific lineage (Fig. 3), responsible for specialized ent-kaurene or labdane diterpenoid formation in Lamiaceae plants. The role of IrCPS4 in oridonin biosynthesis should further be confirmed by RNA interference or virus-induced gene silencing in planta.
Three other KSL genes with unexpected enzymatic activities were identified in I. rubescens. The IrKSL2 gene product was found to be an ent-isopimaradiene-like molecule. Other ent-isopimaradiene-like compound derivatives have been isolated from Calceolaria foliosa (Chamy et al., 1989) and I. lophanthoides subsp. Gerardiana (Sun et al., 2006). The IrKSL4 gene product was found to be an ent-atiserene compound. A similar ent-atiserene gene product has been identified as a minor product (∼4%) of the RcKSL4 protein from castor bean (Ricinus communis; Jackson et al., 2014). Likewise, the product of IrKSL6, isopimaradiene, has not been found in Isodon species previously. Diterpenoid gene products from the leaves of I. rubescens revealed in this study provide a foundation for future in planta studies in the chemistry and metabolic fate of these molecules.
Macroevolutionary of Diterpenoids in Lamiaceae
Because secondary metabolites play vital roles in defense or as signaling compounds, their occurrence reflects adaptations and life strategies embedded in a given species. The identification of diTPS genes in S. miltiorrhiza and I. rubescens offer an opportunity to elucidate diterpenoid biosynthesis on a macroevolutionary level. The similar number and function of CPS enzyme in S. miltiorrhiza and I. rubescens shows that these genes may originate from a common ancestral origin. The resulting speciation of S. miltiorrhiza and I. rubescens led to the different evolution patterns of diterpenoids. As a result, we speculate that because the fleshy root of S. miltiorrhiza is often under herbivory attack (Chen et al., 2008) and survival has been based on the steady adaptation of evolving molecules to increase the complexity of their chemical arsenal. This is evident in the root production of miltiradiene, and the array of tanshinone compounds (Cui et al., 2015). The undershrub of I. rubescens is also attacked by herbivores and the increasing complexity of the leaf chemical arsenal, including oridonin and other ent-kaurene-derived compounds, is further elucidated in the growing number of diterpene biosynthetic pathways able to produce different complex chemical mixtures (Fig. 7). Diterpenoid pathways in Lamiaceae appear to have several instances of multifunctional ancestral pathways including the production of ent-kaurene generated by the same biosynthetic pathway as the universal hormone GA (Zi et al., 2014) and the production of miltiradiene involved in the more chemically elaborate mixture of normal-CPP mediated diterpenoids (Gao et al., 2009; Zerbe et al., 2014; Brückner et al., 2014; Pateraki et al., 2014).
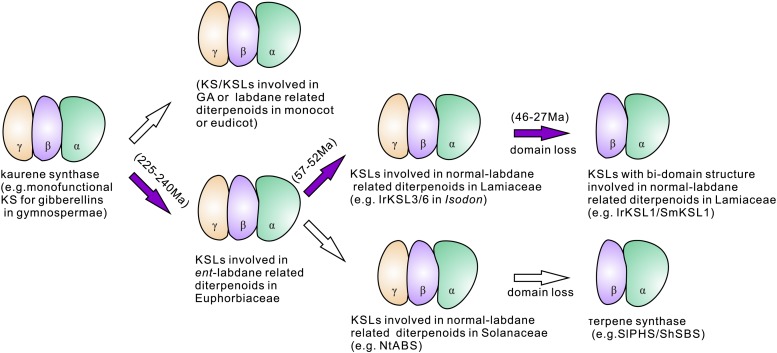
The Evolution of Domain Loss in Lamiaceae
To our knowledge, among KSL enzymes retaining the tridomain (γβα) structure in Lamiaceae, IrKSL3, and IrKSL6 have the ability to react with normal-CPP (Fig. 4). It’s most likely that the tridomain enzymes (IrKSL3 and IrKSL6) belong to an ancestral KSL clade that has the loss of γ domain, a widespread occurrence in the Lamiaceae (Gao et al., 2009; Caniard et al., 2012; Li et al., 2012; Schalk et al., 2012; Brückner et al., 2014; Pateraki et al., 2014; Božić et al., 2015; Cui et al., 2015; Trikka et al., 2015; Su et al., 2016). Therefore, these sequences from Isodon represent a step in the evolution of these Lamiaceae KSL with a domain loss. It is possible that those bidomain KSLs evolved independently in Lamiaceae on at least three separate occasions (Fig. 8). One instance is the evolution of the tridomain KSLs from an ancestral KSL that produced ent-kaurene for GA biosynthesis and arose from an early gene duplication and neofunctionalization during Angiosperm evolution, before the dicot-monocot split (Brückner et al., 2014; Figs. 3 and 8) at ∼240 to 225 million years ago (Ma; Zeng et al., 2014). The second possibility is the emergence of a three-domain KSL described here (IrKSL3 and IrKSL6) in Lamiaceae along with the radiation of the subfamily Nepetoideae at the Paleocene/Eocene boundary (57–52 Ma; Drew and Sytsma, 2012). The third option is the occurrence of βα bidomain KSLs along with the rise of the tribe Menthae in Subfamily Nepetoideae during the mid-Eocene (46 Ma; Drew and Sytsma, 2012). Because the genus Isodon arose 27 Ma (Yu et al., 2014), the existence of both tridomain (i.e. IrKSL3 and IrKSL6) and bidomain (i.e. IrKSL1) KSLs in I. rubescens may demonstrate that the bidomain KSLs originated between 46 Ma to 27 Ma.
Figure 8.
Proposed process for the evolution of bidomain terpene synthases in Lamiaceae. The purple arrows show the proposed evolutionary steps from the ancient tridomain kaurene synthase to the bidomain KSLs in Lamiaceae.
It is interesting to note that the recent emergence enzymes MvELS in M. vulgare (12.6 Ma; Roy and Lindqvist, 2015) and SsScS in S. sclarea (10 Ma; Drew and Sytsma, 2012) exhibit more diversified enzymatic activity than other KSLs (Zerbe et al., 2014; Schalk et al., 2012; Andersen-Ranberg et al., 2016; Jia et al., 2016). This alludes to the fast evolution of KSL enzymes in Lamiaceae. To date, all of the identified βα bidomain KSL enzymes have dual substrate activities (Caniard et al., 2012; Brückner et al., 2014; Zerbe et al., 2014; Andersen-Ranberg et al., 2016; Jia et al., 2016), highlighting their roles in contributing to the diversity of diterpenoids in Lamiaceae. The proposed intermediate steps in KSL protein evolution studied here can be uncovered when additional genera are explored at the sequence level. Next generation sequencing technologies allow this at an accelerated pace and should provide valuable information on the evolution of enzymes (not just diterpene synthases) within or across plant families.
MATERIALS AND METHODS
Plant Materials
Seeds of Isodon rubescens were from Henan Province, given by Prof. Chen Suiqing. Plants were grown in a controlled environment chamber, maintained at 25°C under a 16-h-light/8-h-dark photoperiod.
RNA Extraction, cDNA Library Preparation, and Transcript Analyses by RNA-Seq
The leaves of five-month-old plants were used for RNA-seq analysis. RNA was extracted (HuaYueYang Biotechology) and shipped to the GENEWIZ Company (www.genewiz.com) for library construction and RAN-seq. The RNA quality was checked using an Agilent 2100 with Agilent Eukaryote Total RNA Nano Kit. The RNA-seq library was prepared using the TruSeq RNA Sample Prep Kit for Illumina starting with 1 μg of total RNA. The library was purified on Beckman AMPure XP beads. The barcoded RNA-seq library was assessed by qRT-PCR using the Library Quantification kit. The size range of the final cDNA libraries was determined on an Agilent bioanalyzer DNA7500 DNA chip (Agilent Technologies). The cDNA libraries were sequenced on one lane for 151 cycles from each end of the cDNA fragments on a HiSeq 2500 using a TruSeq SBS sequencing kit v3-HS (Illumina). The sequence images were transformed to bcl files with the Real Time Analysis 1.17.21.2 Illumina software, which were multiplexed to fastq files with CASAVA version 1.8.2. The quality-scores line in fastq files processed with Casava1.8.2 use an ASCII offset of 33 for presentation in the Sanger format.
Transcriptome Assembly and Annotation
The Trinity package version trinityrnaseq-r2013-02-25 (Grabherr et al., 2011) was utilized to assemble the transcriptome from the RNA-seq data. Before assembly, adaptor sequences were trimmed from both ends of the sequence and so were any low quality bases (<30 phred score). Sequences of 50 nucleotides and higher were utilized for the Trinity assembly, which was performed according to guidelines in the published protocols (Grabherr et al., 2011), and on the Trinity Web site (https://github.com/trinity-rnaseq/trinityrnaseq/wiki). Individual isotigs were annotated by searching the NCBI nonredundant protein sequence database using BLASTX software with default parameters. Isotig functions were assigned based on the annotation associated with the top hit that satisfied the following criteria: (1) ≥30% sequence identity; (2) ≥30% alignment coverage of either the query or subject sequences; and (3) with BLAST e-values < E-5. Manually assemblies were carried out with Bioedit and DNAMAN.
Cloning of the Full-Length diTPS Genes
The total RNA used for RNA-seq was further used to verify the full length of diTPS obtained from transcriptome assembly and clone the other diTPS by RACE. 5′-RACE and 3′-RACE were performed using the SMARTer RACE DNA amplification kit (Clontech Laboratories) according to the manufacturer’s protocol. All the primers for RACE-PCR were shown in Supplemental Table S6. The full lengths of diTPS genes were then cloned into pMD19-T (TaKaRa) vector for sequencing.
DiTPS Gene Expression Analysis
I. rubescens flowering plants were used to determine diTPS expression levels. Samples were harvested and immediately frozen in liquid nitrogen. RNA was extracted (HuaYueYang Biotechology) and RNA integrity and quality checked by denaturing gel electrophoresis. One to approximately five micrograms of total RNA was reverse transcribed into cDNA using the PrimerScript RT reagent kit with gDNA eraser (TaKaRa), according to the manufacturer’s instructions. The synthesized cDNA was then diluted 10-fold and 1 μL of this diluted template with 0.4 μL of ROX Reference Dye II (TaKaRa) were used for subsequent qRT-PCR analysis with a total PCR reaction volume of 20 μL. The primers sequences are shown in Supplemental Table S6. qRT-PCR was performed with a SYBR Green kit (TaKaRa) on ABI7500 real-time PCR detection system. All reactions were performed using the following PCR conditions: initial denaturation step of 95°C for 10 min, followed by 40 cycles for 95°C for 5 s, 60°C for 34 s, with a final melting stage from 60°C to 95°C. Primer specificity was assessed by agarose gel and melting curve analysis. The results were normalized with housekeeping gene actin. Relative expression levels were calculated as the mean of three technical replicates of three biological replicates.
Phylogenetic Analysis
The previously identified 96 diTPSs (Supplemental Table S5) were used to construct the phylogenetic tree follow the method described by Cui et al. (2015).
In Vitro Assays
For in vitro functional assays, the full coding sequence of diTPS obtained from RNA-seq data (IrCPS1, IrCPS3, IrCPS4, and IrKSL1) was proved by PCR and further synthesized with codon optimization for Escherichia coli (GENEWIZ Biotech) and cloned into pET32 plasmid (Merck). The full coding sequence of other seven diTPS obtained from RACE (IrCPS2, IrCPS3, IrCPS5, IrKSL2, IrKSL3, IrKSL4, IrKSL5, and IrKSL6) were digested with specific restriction enzyme site (Supplemental Table S6) and subcloned into the expression plasmid pET32a. The recombinant diTPS proteins were expressed in E. coli Tuner, affinity-purified and assayed with GGPP as the substrate.
SmCPS1 (GenBank: KC814639), SmCPS5 (KC814642), SmKSL1 (ABV08817) from Salvia miltiorrhiza, AtKS (AAC39443) from Arabidopsis (Arabidopsis thaliana), PcmISO1 (JQ240314) from Pinus banksiana were used in this study. The expression and purification of the recombinant proteins were performed as described by Cui et al. (2015). In short, the constructs were transformed into Tuner (DE3) or TransB (DE3) competent cells. Three to five positive colonies were cultured in LB medium with 50 mg·L−1 carbenicillin, and 0.1 to ∼0.4 mm isopropyl β-d-thiogalactopyranoside was added to induce the expression of the protein. Subsequently, cell pellets were collected and resuspended in assay buffer (50 mm P, pH 7.4, 10% glycerol, 2 mm DTT, and 10 mm MgCl2) and sonicated six times for 10 s, on ice. Lysate from the samples was centrifuged at 12,000g for 20 min at 4°C. The proteins were purified with the nickel-nitrilotriacetic acid agarose beads followed the previously described method from Cui et al. (2015).
The conversion of GGPP to CPP was carried out by incubating 200 μL of the purified enzymes with 20 to 50 μm GGPP (Sigma-Aldrich) for 2 to 4 h at 30°C. Assay mixtures were hydrolyzed (dephosphorylated) with 75 U bacterial alkaline phosphatase at pH 8 for 16 h at 37°C to produce hexane-soluble products. GGPP was converted to kaurene, miltiradiene, and other diterpenes by mixing 250 μL of purified recombinant CPS and KSL proteins with 20 to 50 μm GGPP, and incubated for 2 h to 4 h at 30°C. Assay mixtures were extracted three times with an equal volume of hexane. The hexane fractions were pooled, evaporated under N and resuspended in 50 μL of hexane or derivatized with 80 μL N-methyl-N-(trimethylsilyl) trifluoroacetamide at 80°C for 40 min, and then analyzed by GC-MS.
Identification of CYP76AH30 Using the Engineering Yeast
Full-length cDNA of CYP76AH30 were cloned from the leaves using primers shown in Supplemental Table S6. The open reading frame of CYP76AH30 was subcloned into the yeast epitope-tagged vector pESC-Leu using BamHI and SalI, yielding pESC-Leu::CYP76AH30. The plasmid was transformed into the miltiradiene production strain YJ26, using lithium acetate/single-stranded carrier DNA/polyethylene glycol transformation method (Gietz and Woods, 2002; Zhou et al., 2012). Transformants were selected on yeast nitrogen base without amino acids (YNB) medium containing 20 g L−1 Glc and grown at 30°C for 48 h. The recombinant yeast strains were grown in YNB medium containing 2% Glc (YNB/Glc) at 30°C, shaking at 250 rpm, for 48 h, then transferred to 50 ml YNB/Glc medium in 250 mL flasks and grown to an initial OD600 of 0.05, and cultivated an additional 12 to 16 h to reach logarithmic phase. Cells were centrifuged and washed twice with sterile water to remove any residual Glc. The cells were then resuspended in 50 mL YNB medium containing 2% Gal (YNB/gal) for induction, and grown for 30 to 72 h to produce diterpenoids. Yeast cultures were extracted three times by ultrasonication with an equal volume of hexane. After separation, the organic extract was concentrated under vacuum, and the residue resuspended in hexane for GC-MS analysis.
Diterpene Analysis Using GC-MS
Plant material was lyophilized for 48 h and 100 mg of the lyophilized powder extracted with 2 mL hexane. The extracts were sonicated twice for 15 min and centrifuged (3,000g) for 10 min. The supernatant was evaporated under nitrogen, resuspended in 50 μL of hexane and analyzed by GC-MS.
Two GC-MS systems were used in this study. Plant material and the entire in vitro assay product were first carried out using a Trace 1310 series GC with a TSQ8000 MS detector (Thermo Fisher Scientific). A 1-μL portion of extract was injected in splitless mode onto the column. Chromatographic separation was performed on a TR-5ms capillary column (30 m × 0.25 mm i.d.; DF = 0.25 μm; Thermo Fisher Scientific). Helium was used as the carrier gas at a constant flow rate of 1 mL/min through the column. The injector temperature was set at 280°C and the injector temperature was 280°C. The oven program was as follows: 50°C for 2 min, linear ramp at a rate of 20°C·min−1 to 200°C, followed with a linear ramp at a rate of 5°C·min−1 to 300°C, held at 300°C for 10 min. The transfer line temperature was 280°C.
The other GC-MS system is Agilent GC-MS 7890B connected to an Agilent 7000B triple quadruple MS with electron impact ionization. A 1-μL portion of the extract was injected in splitless mode onto the column. The column used was a chiral column Agilent Cyclodex-β (30 m × 0.25 mm i.d., 0.25-μm film thickness). Helium was used as the carrier gas for GC at a flow rate of 1.0 ml·min−1. The injector temperature was 250°C. The oven program was as follows: 50°C for 2 min, linear ramp at a rate of 8°C·min−1 to 230°C, then held at 230°C for 8 min. The transfer line temperature was 280°C.
Identification of ent-kaurene, miltiradiene, and isopimaradiene were achieved by comparison to the previously defined enzymes. Ent-atiserene and ferruginol were identified by comparison to authentic standard.
Accession Numbers
The Illumina-derived nucleotide sequences reported in this article have been submitted to the short read archive at NCBI under accession number GSE96954. Nucleotide sequences of characterized enzymes can be found in the GenBank/EMBL data libraries with the following accession numbers: IrCPS1 (KU180499), IrCPS2 (KU180500), IrCPS3 (KU180501), IrCPS4 (KU180502), IrCPS5 (KU180503), IrKSL1 (KU180504), IrKSL2 (KU180505), IrKSL3 (KU180506), IrKSL4 (KX580633), IrKSL5 (KX580634), IrKSL6 (KX580635), and Ir76AH30 (KX580636).
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Amino acid alignment of diterpene synthase in I. rubescens and S. miltiorrhiza.
Supplemental Figure S2. GC-MS analysis of in vitro assays with I. rubescens IrCPS4 and IrCPS5.
Supplemental Figure S3. GC-MS analysis of different clade of IrKSL with the corresponding IrCPS on a TR-5ms capillary column.
Supplemental Figure S4. GC-MS detection of abietane diterpenoids in different organs.
Supplemental Figure S5. Amino acid alignment of CYP76AH30 with CYP76AH1 and CYP76AH22-24.
Supplemental Table S1. The number and length of all of the unigenes obtained by RNA-seq.
Supplemental Table S2. Unigenes annotated as copalyl diphosphate synthase and kaurene synthase genes.
Supplemental Table S3. Information of diTPS gene family in I. rubescens.
Supplemental Table S4. BLAST result of genes identified from this article to the reference transcriptome sequencing data set.
Supplemental Table S5. List of plant diTPS involved in this article.
Supplemental Table S6. Primers used in this study.
Supplementary Material
Acknowledgments
We thank Professors Suiqing Chen and Yun Lei giving the seeds of I. rubescens. We thank Professor Reuben J. Peters for kindly providing the reference standard ent-antiserene.
Footnotes
Articles can be viewed without a subscription.
References
- Andersen-Ranberg J, Kongstad KT, Nielsen MT, Jensen NB, Pateraki I, Bach SS, Hamberger B, Zerbe P, Staerk D, Bohlmann J, Møller BL, Hamberger B (2016) Expanding the landscape of diterpene structural diversity through stereochemically controlled combinatorial biosynthesis. Angew Chem Int Ed Engl 55: 2142–2146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Božić D, Papaefthimiou D, Brückner K, de Vos RC, Tsoleridis CA, Katsarou D, Papanikolaou A, Pateraki I, Chatzopoulou FM, Dimitriadou E, et al. (2015) Towards elucidating carnosic acid biosynthesis in Lamiaceae: functional characterization of the three first steps of the pathway in Salvia fruticosa and Rosmarinus officinalis. PLoS One 10: e0124106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brückner K, Božić D, Manzano D, Papaefthimiou D, Pateraki I, Scheler U, Ferrer A, de Vos RC, Kanellis AK, Tissier A (2014) Characterization of two genes for the biosynthesis of abietane-type diterpenes in rosemary (Rosmarinus officinalis) glandular trichomes. Phytochemistry 101: 52–64 [DOI] [PubMed] [Google Scholar]
- Caniard A, Zerbe P, Legrand S, Cohade A, Valot N, Magnard JL, Bohlmann J, Legendre L (2012) Discovery and functional characterization of two diterpene synthases for sclareol biosynthesis in Salvia sclarea (L.) and their relevance for perfume manufacture. BMC Plant Biol 12: 119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chamy MC, Piovano M, Garbarino JA, Gambaro V, Miranda C (1989) Foliosate, a bis-diterpene and 9-epi-ent-7,15-isopimaradiene derivatives from Calceolaria foliosa. Phytochemistry 28: 571–574 [Google Scholar]
- Chen C, Guo XX, Shi YQ, Tang XH (2008) Primary study on the species and vertical distribution of underground pests of Salvia miltiorrhiza. J Anhui Agri Sci 36: 116–142 [Google Scholar]
- Chinese Pharmacopoeia Committee (2015) Pharmacopoeia of the People’s Republic of China. China Medical Science Press, Beijing, China [Google Scholar]
- Christianson DW. (2006) Structural biology and chemistry of the terpenoid cyclases. Chem Rev 106: 3412–3442 [DOI] [PubMed] [Google Scholar]
- Cui G, Duan L, Jin B, Qian J, Xue Z, Shen G, Snyder JH, Song J, Chen S, Huang L, Peters RJ, Qi X (2015) Functional divergence of diterpene syntheses in the medicinal plant Salvia miltiorrhiza. Plant Physiol 169: 1607–1618 (in Chinese) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drew BT, Sytsma KJ (2012) Phylogenetics, biogeography, and staminal evolution in the tribe Mentheae (Lamiaceae). Am J Bot 99: 933–953 [DOI] [PubMed] [Google Scholar]
- Gao W, Hillwig ML, Huang L, Cui G, Wang X, Kong J, Yang B, Peters RJ (2009) A functional genomics approach to tanshinone biosynthesis provides stereochemical insights. Org Lett 11: 5170–5173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gietz RD, Woods RA (2002) Transformation of yeast by lithium acetate/single-stranded carrier DNA/polyethylene glycol method. Methods Enzymol 350: 87–96 [DOI] [PubMed] [Google Scholar]
- Grabherr MG, Haas BJ, Yassour M, Levin JZ, Thompson DA, Amit I, Adiconis X, Fan L, Raychowdhury R, Zeng Q, et al. (2011) Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat Biotechnol 29: 644–652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo J, Zhou YJ, Hillwig ML, Shen Y, Yang L, Wang Y, Zhang X, Liu W, Peters RJ, Chen X, Zhao ZK, Huang L (2013) CYP76AH1 catalyzes turnover of miltiradiene in tanshinone biosynthesis and enables heterologous production of ferruginol in yeasts. Proc Natl Acad Sci USA 110: 12108–12113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall DE, Zerbe P, Jancsik S, Quesada AL, Dullat H, Madilao LL, Yuen M, Bohlmann J (2013) Evolution of conifer diterpene synthases: diterpene resin acid biosynthesis in lodgepole pine and jack pine involves monofunctional and bifunctional diterpene synthases. Plant Physiol 161: 600–616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harley RM, Atkins S, Budanstev AL, Cantino PD, Conn BJ, Grayer R, Harley MM (2004) Flowering plants, dicotyledons. In Kubitzki K., ed., The Families and Genera of Vascular Plants, Vol 6, Springer, Berlin, Germany, pp. 167–275 [Google Scholar]
- Hillwig ML, Xu M, Toyomasu T, Tiernan MS, Wei G, Cui G, Huang L, Peters RJ (2011) Domain loss has independently occurred multiple times in plant terpene synthase evolution. Plant J 68: 1051–1060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ignea C, Athanasakoglou A, Ioannou E, Georgantea P, Trikka FA, Loupassaki S, Roussis V, Makris AM, Kampranis SC (2016) Carnosic acid biosynthesis elucidated by a synthetic biology platform. Proc Natl Acad Sci USA 113: 3681–3686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson AJ, Hershey DM, Chesnut T, Xu M, Peters RJ (2014) Biochemical characterization of the castor bean ent-kaurene synthase(-like) family supports quantum chemical view of diterpene cyclization. Phytochemistry 103: 13–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia M, Potter KC, Peters RJ (2016) Extreme promiscuity of a bacterial and a plant diterpene synthase enables combinatorial biosynthesis. Metab Eng 37: 24–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Köksal M, Potter K, Peters RJ, Christianson DW (2014) 1.55 Å-resolution structure of ent-copalyl diphosphate synthase and exploration of general acid function by site-directed mutagenesis. Biochim Biophys Acta 1840: 184–190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li JL, Chen QQ, Jin QP, Gao J, Zhao PJ, Lu S, Zeng Y (2012) IeCPS2 is potentially involved in the biosynthesis of pharmacologically active Isodon diterpenoids rather than gibberellin. Phytochemistry 76: 32–39 [DOI] [PubMed] [Google Scholar]
- Martin DM, Fäldt J, Bohlmann J (2004) Functional characterization of nine Norway spruce TPS genes and evolution of gymnosperm terpene synthases of the TPS-d subfamily. Plant Physiol 135: 1908–1927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pateraki I, Andersen-Ranberg J, Hamberger B, Heskes AM, Martens HJ, Zerbe P, Bach SS, Møller BL, Bohlmann J, Hamberger B (2014) Manoyl oxide (13R), the biosynthetic precursor of forskolin, is synthesized in specialized root cork cells in Coleus forskohlii. Plant Physiol 164: 1222–1236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelot KA, Mitchell R, Kwon M, Hagelthorn DM, Wardman JF, Chiang A, Bohlmann J, Ro DK, Zerbe P (2017) Biosynthesis of the psychotropic plant diterpene salvinorin A: discovery and characterization of the Salvia divinorum clerodienyl diphosphate synthase. Plant J 89: 885–897 [DOI] [PubMed] [Google Scholar]
- Peters RJ. (2010) Two rings in them all: the labdane-related diterpenoids. Nat Prod Rep 27: 1521–1530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prisic S, Xu M, Wilderman PR, Peters RJ (2004) Rice contains two disparate ent-copalyl diphosphate synthases with distinct metabolic functions. Plant Physiol 136: 4228–4236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy T, Lindqvist C (2015) New insights into evolutionary relationships within the subfamily Lamioideae (Lamiaceae) based on pentatricopeptide repeat (PPR) nuclear DNA sequences. Am J Bot 102: 1721–1735 [DOI] [PubMed] [Google Scholar]
- Schalk M, Pastore L, Mirata MA, Khim S, Schouwey M, Deguerry F, Pineda V, Rocci L, Daviet L (2012) Toward a biosynthetic route to sclareol and amber odorants. J Am Chem Soc 134: 18900–18903 [DOI] [PubMed] [Google Scholar]
- Scheler U, Brandt W, Porzel A, Rothe K, Manzano D, Božić D, Papaefthimiou D, Balcke GU, Henning A, Lohse S, et al. (2016) Elucidation of the biosynthesis of carnosic acid and its reconstitution in yeast. Nat Commun 7: 12942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su P, Tong Y, Cheng Q, Hu Y, Zhang M, Yang J, Teng Z, Gao W, Huang L (2016) Functional characterization of ent-copalyl diphosphate synthase, kaurene synthase and kaurene oxidase in the Salvia miltiorrhiza gibberellin biosynthetic pathway. Sci Rep 6: 23057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun HD, Huang SX, Han QB (2006) Diterpenoids from Isodon species and their biological activities. Nat Prod Rep 23: 673–698 [DOI] [PubMed] [Google Scholar]
- Trikka FA, Nikolaidis A, Ignea C, Tsaballa A, Tziveleka LA, Ioannou E, Roussis V, Stea EA, Božić D, Argiriou A, et al. (2015) Combined metabolome and transcriptome profiling provides new insights into diterpene biosynthesis in S. pomifera glandular trichomes. BMC Genomics 16: 935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vestri Alvarenga SA, Pierre Gastmans J, do Vale Rodrigues G, Moreno PR, de Paulo Emerenciano V (2001) A computer-assisted approach for chemotaxonomic studies—diterpenes in Lamiaceae. Phytochemistry 56: 583–595 [DOI] [PubMed] [Google Scholar]
- Yu XQ, Maki M, Drew BT, Paton AJ, Li HW, Zhao JL, Conran JG, Li J (2014) Phylogeny and historical biogeography of Isodon (Lamiaceae): rapid radiation in south-west China and Miocene overland dispersal into Africa. Mol Phylogenet Evol 77: 183–194 [DOI] [PubMed] [Google Scholar]
- Zeng L, Zhang Q, Sun R, Kong H, Zhang N, Ma H (2014) Resolution of deep angiosperm phylogeny using conserved nuclear genes and estimates of early divergence times. Nat Commun 5: 4956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zerbe P, Chiang A, Dullat H, O’Neil-Johnson M, Starks C, Hamberger B, Bohlmann J (2014) Diterpene synthases of the biosynthetic system of medicinally active diterpenoids in Marrubium vulgare. Plant J 79: 914–927 [DOI] [PubMed] [Google Scholar]
- Zerbe P, Hamberger B, Yuen MM, Chiang A, Sandhu HK, Madilao LL, Nguyen A, Hamberger B, Bach SS, Bohlmann J (2013) Gene discovery of modular diterpene metabolism in nonmodel systems. Plant Physiol 162: 1073–1091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou GB, Chen SJ, Wang ZY, Chen Z (2007) Back to the future of oridonin: again, compound from medicinal herb shows potent antileukemia efficacies in vitro and in vivo. Cell Res 17: 274–276 [DOI] [PubMed] [Google Scholar]
- Zhou YJ, Gao W, Rong Q, Jin G, Chu H, Liu W, Yang W, Zhu Z, Li G, Zhu G, Huang L, Zhao ZK (2012) Modular pathway engineering of diterpenoid synthases and the mevalonic acid pathway for miltiradiene production. J Am Chem Soc 134: 3234–3241 [DOI] [PubMed] [Google Scholar]
- Zi J, Mafu S, Peters RJ (2014) To gibberellins and beyond! Surveying the evolution of (di)terpenoid metabolism. Annu Rev Plant Biol 65: 259–286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zi J, Peters RJ (2013) Characterization of CYP76AH4 clarifies phenolic diterpenoid biosynthesis in the Lamiaceae. Org Biomol Chem 11: 7650–7652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou J, Pan L, Li Q, Pu J, Yao P, Zhu M, Banas JA, Zhang H, Sun H (2012) Rubesanolides C-E: abietane diterpenoids isolated from Isodon rubescens and evaluation of their anti-biofilm activity. Org Biomol Chem 10: 5039–5044 [DOI] [PubMed] [Google Scholar]
- Zou J, Pan L, Li Q, Zhao J, Pu J, Yao P, Gong N, Lu Y, Kondratyuk TP, Pezzuto JM, et al. (2011) Rubesanolides A and B: diterpenoids from Isodon rubescens. Org Lett 13: 1406–1409 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.