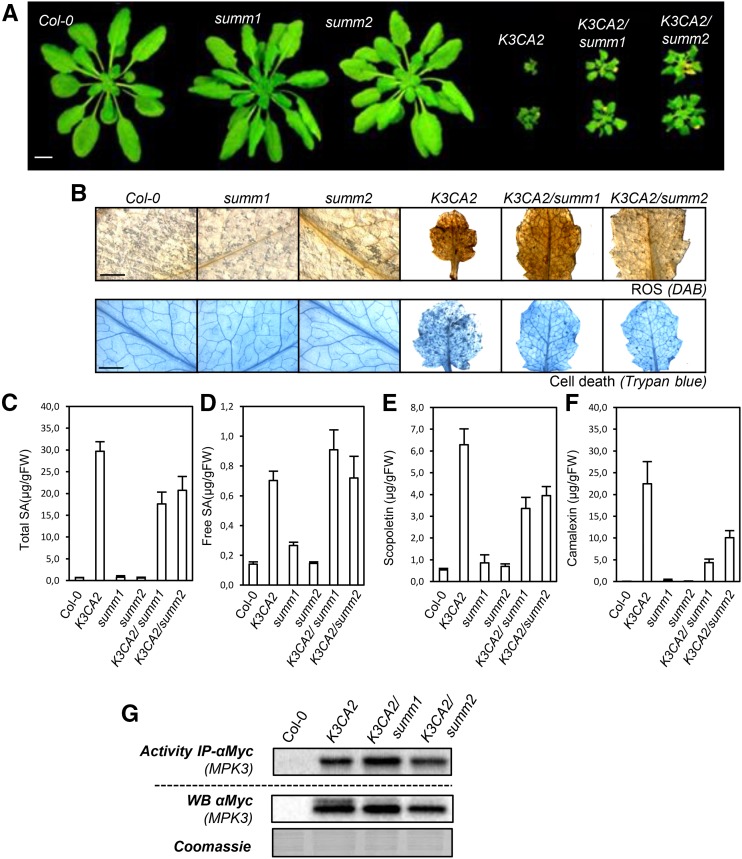
The stress-activated MPK3 and the SUPPRESSOR OF MKK1 MKK2 1/2 module control a similar set of responses, which include accumulation of salicylic acid, reactive oxygen species, and phytoalexins and modulation of defense genes.
Abstract
Mitogen-activated protein kinases (MAPKs) are important regulators of plant immunity. Most of the knowledge about the function of these pathways is derived from loss-of-function approaches. Using a gain-of-function approach, we investigated the responses controlled by a constitutively active (CA) MPK3 in Arabidopsis thaliana. CA-MPK3 plants are dwarfed and display a massive derepression of defense genes associated with spontaneous cell death as well as the accumulation of reactive oxygen species, phytoalexins, and the stress-related hormones ethylene and salicylic acid (SA). Remarkably CA-MPK3/sid2 and CA-MPK3/ein2-50 lines, which are impaired in SA synthesis and ethylene signaling, respectively, retain most of the CA-MPK3-associated phenotypes, indicating that the constitutive activity of MPK3 can bypass SA and ethylene signaling to activate defense responses. A comparative analysis of the molecular phenotypes of CA-MPK3 and mpk4 autoimmunity suggested convergence between the MPK3- and MPK4-guarding modules. In support of this model, CA-MPK3 crosses with summ1 and summ2, two known suppressors of mpk4, resulted in a partial reversion of the CA-MPK3 phenotypes. Overall, our data unravel a novel mechanism by which the MAPK signaling network contributes to a robust defense-response system.
To survive under different stresses during their life cycle, plants have developed various strategies. Their ability to adapt to changing environments largely relies on sensing and signaling processes that are based on manifold receptors and complex signaling networks. Mitogen-activated protein kinase (MAPK) cascades were shown to be key actors in plant signal transduction in response to various biotic and abiotic stresses (Colcombet and Hirt, 2008). The core of the MAPK pathways is formed by three types of kinases: MAPKK kinases (MAP3Ks), MAPK kinases (MAP2Ks), and MAP kinases (MAPKs), which phosphorylate and activate each other in a linear pathway. MAPKs, which are Ser/Thr kinases, are known to phosphorylate a wide range of substrates, including other kinases and transcription factors, leading to major stress-related cellular modifications. These three sets of protein kinases in MAPK pathways are encoded by multigene families (Ichimura et al., 2002). There are more than 60 MAP3Ks, 10 MAP2Ks, and 20 MAPKs in Arabidopsis thaliana, which define functional modules for multiple processes, including development and stress responses.
Interestingly, a given stress does not activate a single MAPK module. For example, the microbe-associated molecular pattern (MAMP) flg22, a derived peptide from Pseudomonas aeruginosa flagellin that reveals the presence of the pathogen to the plant, triggers the activation of several MAPKs: MPK3, MPK4, MPK6, MPK11, MPK1, and MPK13 (Meng and Zhang, 2013; Nitta et al., 2014). This activation occurs within minutes after flg22 perception by its cognate receptor complex FLS2-BAK1, peaking around 15 min and getting back to its basal level after 1 h (Nuhse et al., 2000; Asai et al., 2002). Upstream activators of the MAPKs also have been identified in some cases. The MAP3K MEKK1 activates the two MAP2Ks MKK1 and MKK2, which converge to activate MPK4 (Ichimura et al., 2006; Nakagami et al., 2006; Gao et al., 2008; Qiu et al., 2008). MPK3 and MPK6 are both activated by MKK4 and MKK5, but their upstream MAP3K(s) have not yet been unambiguously identified (Asai et al., 2002).
Whereas the MKK4/5-MPK3/6 module is thought to positively regulate immunity, the MEKK1-MKK1/2-MPK4 module was first described as a negative regulator (Petersen et al., 2000; Brodersen et al., 2006). Indeed, mekk1, mkk1/mkk2, and mpk4 loss-of-function mutants display constitutively activated defense responses in the absence of pathogens, spontaneous cell death, and the accumulation of reactive oxygen species (ROS) and salicylic acid (SA; Petersen et al., 2000; Gao et al., 2008), resulting in growth defects and dwarfed plants (Pitzschke et al., 2009; Frei dit Frey et al., 2014). Their altered developmental phenotypes can be partially suppressed by mutations/transgenes reducing SA levels, such as sid2 and NahG, as well as by growth at high temperature (Brodersen et al., 2006; Su et al., 2007). Recently, two allelic series of loss-of-function mutations able to fully suppress autoimmune-related growth defects of mekk1, mkk1/mkk2, and mpk4 were identified in a forward genetic screen (Kong et al., 2012; Zhang et al., 2012). summ1 (suppressor of mkk1 mkk2 1) is mutated in MEKK2, the closest homolog of MEKK1, and summ2 is mutated in a nucleotide-binding leucine-rich repeat (NB-LRR) protein. These results suggest that the MEKK1-MKK1/2-MPK4 module is guarded by the NB-LRR protein SUMM2 to avoid its manipulation by pathogen effectors. In accordance with the current model, the MEKK1-MKK1/2-MPK4 module would then be a positive regulator of stress responses.
Despite being activated by stresses with similar kinetics, three iconic MAPKs, MPK3, MPK4, and MPK6, are not functionally fully redundant. A growing number of publications describe MAPK substrates that may be targeted specifically by a single or a combination of several MAPKs (for review, see Bigeard et al., 2015). mpk3 and mpk6 single mutants show normal development, whereas mpk3mpk6 double mutants are barely viable (Wang et al., 2007). This strong developmental redundancy makes the investigation of the stress-related roles of these kinases complicated. The fact that some substrates, such as ACS2/6, ERF6, and WRKY22/28/33, are targeted by both MPK3 and MPK6 indicates that these MAPKs have at least partially similar roles in plant defense regulation (for review, see Bigeard et al., 2015). Interestingly, the phenotypical characterization of single mutants showed specific functions. For example, mpk3 mutants are susceptible to the necrotrophic pathogen Botrytis cinerea, whereas mpk6 is not (Ren et al., 2008). Both mpk3 and mpk6 mutants are impaired in flg22-triggered stomatal closure, suggesting that they have collaborative functions in the regulation of guard cell volume rather than redundant functions (Montillet et al., 2013). mpk3 but not mpk6 overproduces RBOHD-dependent ROS burst upon MAMP perception, suggesting an MPK3-specific negative role in MAMP-triggered immunity (MTI) responses (Ranf et al., 2011). Recently, an extensive transcriptomic analysis of mapk mutants upon flg22 treatment underlined the specificities of each MAPK in gene expression reprogramming during MTI (Frei dit Frey et al., 2014).
Our laboratory reported the identification of mutations that render Arabidopsis MAPKs constitutively active (CA; Berriri et al., 2012). Our previous work focused on plants expressing CA-MPK4 and demonstrated that these point mutations do not alter substrate specificity, making them a powerful tool to provide complementary evidence for loss-of-function-based analysis of MAPKs. In this study, we used the same approach to tackle MPK3 function. Our detailed characterization of CA-MPK3 lines brings new insights to the role of MPK3 in innate immunity.
RESULTS
The Dual Mutation D193G/E197A Results in Constitutive Activation of MPK3
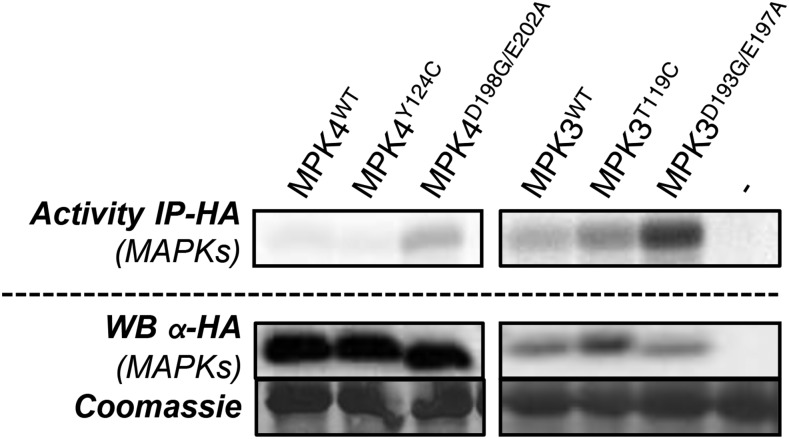
We recently reported the identification of two sets of mutations, Y144C and D218G/E222A, which trigger constitutive autoactivity to recombinant Arabidopsis MPK6 (Berriri et al., 2012). In vitro expression of CA MAPKs suggested that CA mutations may be transferrable to other plant MAPKs but that all mutations do not systematically activate MAPKs in planta. To select for mutations that trigger the strongest in vivo autoactivity in the stress-related MPK3, we expressed human influenza hemagglutinin (HA)-tagged MPK3WT, MPK3T119C, and MPK3D193G/E197A, as well as MPK4WT, MPK4Y124C, and MPK4D198G/E202A as controls, using the mesophyll protoplast expression system. We assayed MAPK activity after HA immunoprecipitation for their ability to phosphorylate myelin basic protein (MBP) and found that MPK3D193G/E197A and, to a lesser extent, MPK3T119C were more active than MPK3WT (Fig. 1).
Figure 1.
Activity of MPK3 and MPK4 wild-type and CA plants expressed in mesophyll protoplasts. The kinase activity shows HA immunoprecipitation (IP) of HA-tagged MAPK (wild type and mutant) expressed in Columbia-0 (Col-0) mesophyll protoplasts. Western blots (WB) show protein levels.
Plants Expressing MPK3D193G/E197A Show Higher MPK3 Activity and Dwarfism Associated with ROS Accumulation and Cell Death
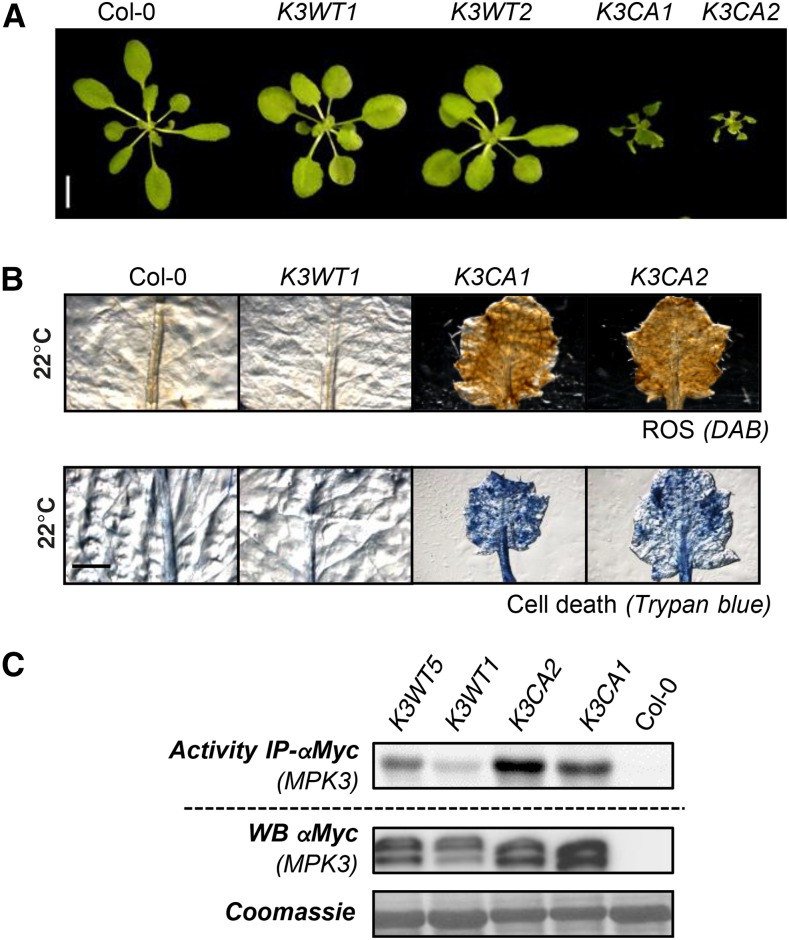
To investigate MPK3 function further, we created Arabidopsis transgenic lines expressing MPK3D193G/E197A. For this purpose, the mpk3-1 knockout mutant was transformed with vectors allowing the expression of the c-myc-tagged genomic sequence of MPK3WT and the mutated version MPK3D193G/E197A. These lines will be further referred to as K3WT and K3CA, respectively. Under standard growth conditions (soil, 22°C, long days), all K3CA T1 plants showed a dwarfed morphology associated with leaf serration and reduced fertility, whereas K3WT T1 plants did not show any noticeable morphological phenotype when compared with Col-0 (n > 20 for both genotypes). This phenotype was maintained in homozygous lines carrying a single insertion (Fig. 2A). Trypan Blue and 3,3′-diaminobenzidine (DAB) staining showed that this dwarfism is associated with spontaneous cell death and constitutive ROS accumulation (Fig. 2B). To characterize MPK3 protein levels in the different genetic backgrounds, we performed western-blot analysis of 15-d-old plantlets grown in vitro (Supplemental Fig. S1A). Unluckily, MPK3 antibody, which is raised against the 11 C-terminal residues of MPK3, does not detect the C-terminally c-myc-tagged MPK3 fusion proteins, making the comparison of MPK3 protein level between Col-0 and transgenic lines unachievable. Western immunoblots using c-myc antibodies showed that the level of MPK3-myc varies from one transgenic line to another, as expected when T-DNA insertion occurs randomly in the genome. As an alternative to direct MPK3 protein detection, we performed reverse transcription-quantitative PCR (RT-qPCR) analyses and found that the level of expression of MPK3-myc transcripts in transgenic lines, which also varies from one line to another, is overall similar to the level of MPK3 transcripts in Col-0 (Supplemental Fig. S1B). c-myc-based immunoprecipitation from K3WT and K3CA plantlets grown in vitro followed by kinase assays showed that K3CA lines exhibit a higher MPK3 activity than K3WT lines (Supplemental Fig. S1C). Similar results were obtained when the c-myc-based immunoprecipitation was performed from rosettes of pot-grown plants showing the strong dwarf phenotype (Fig. 2C). Importantly, the MPK3 kinase activities, as well as the dwarf phenotype of the K3CA plants, were specific for the CA mutation and apparently independent of MPK3 protein abundance levels (Fig. 2; Supplemental Fig. S1). Remarkably, K3CA plantlets grown in vitro showed no obvious phenotype while pot-grown plants showed a dwarfed morphology, suggesting that, in addition to the constitutive MPK3 activity, environmental factors are required for its full establishment.
Figure 2.
Phenotypes of plants expressing c-myc-tagged K3WT and K3CA. A, Representative photographs of 30-d-old rosettes of the indicated genotypes. Bar = 1 cm. B, Trypan Blue and DAB staining of representative leaves from the indicated genotypes. Bar = 2 mm. C, Kinase activity of MPK3-myc after immunoprecipitation (IP) using anti-c-myc antibody from the indicated genotypes. Western blots (WB) show protein levels.
K3CA Plants Still Respond to flg22 and Do Not Affect MPK4/6 Activities
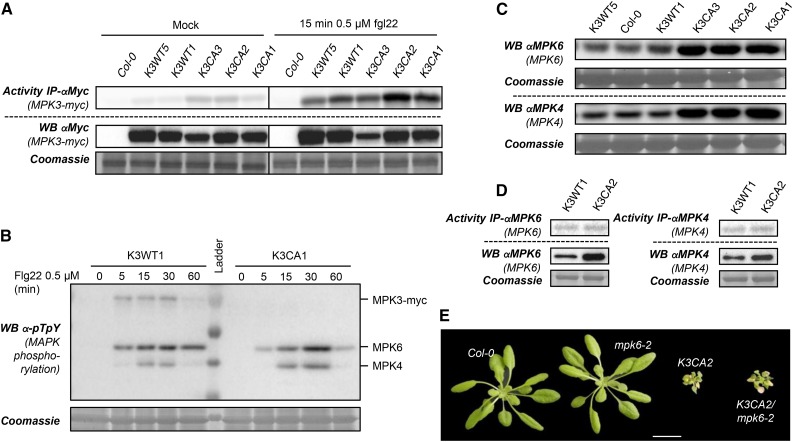
To determine whether the constitutive activity of K3CA plants interferes with MTI signaling, we challenged in vitro seedlings with the model MAMP flg22. As shown in Figure 3A, MPK3 activities in K3CA lines still increased drastically in response to flg22 compared with K3WT lines, demonstrating that the constitutive activity of MPK3 does not lead to a desensitized MPK3 pathway. It also indicates that the MPK3 activity increase triggered by CA mutations is mild compared with the MPK3 activity after MAMP treatment and that CA mutations, which are located in the activation loop close to the TEY activation motif, do not affect its phosphorylation by upstream MAP2Ks. As cross talk and compensatory mechanisms between MTI-related MAPK modules exist (Frei dit Frey et al., 2014), we were curious to see whether the constitutive activity of MPK3 might provoke changes in MPK6 and MPK4 activation. For this purpose, we used western immunoblots to monitor flg22-dependent activation loop phosphorylation of MPK4 and MPK6 in representative K3WT and K3CA lines (Fig. 3B). We did not observe any differences between these lines, as MPK4 and MPK6 had the typical kinetics of phosphorylation, with a rapid increase, a plateau at 15 to 30 min, and a decrease after 1 h (Fig. 3B). MPK4 and MPK6 protein levels also were not affected in in vitro-grown plantlets of K3CA lines compared with Col-0 and K3WT lines (Supplemental Fig. S1A). On the other hand, in the case of dwarfed plants grown on soil, we monitored an increase in the amount of MPK4/6 proteins in K3CA lines compared with Col-0 and K3WT lines (Fig. 3C). To exclude that K3CA dwarfism could be due to a higher accumulation of MPK4/6, which would trigger stronger MPK4/6 activities, we measured their activity after immunoprecipitation but did not see any increases in the K3CA line (Fig. 3D). Finally, we crossed K3CA2 plants with mpk6-2 but found that a lack of MPK6 does not suppress K3CA dwarfism (Fig. 3E). These results suggest that MPK6 likely does not mediate K3CA dwarfism.
Figure 3.
Characterization of MAPK protein levels and PAMP-triggered immunity in K3WT and K3CA lines. A, Kinase activity of MPK3-myc after immunoprecipitation (IP) using anti-c-myc antibody from in vitro plantlets of the indicated genotypes treated with flg22 for 15 min. Western blots (WB) show protein expression levels. B, Kinase phosphorylation in the MAPK activation loop monitored by western blot using anti-pTpY antibody from K3WT1 and K3CA1 transgenic lines treated with 1 µm flg22 for the indicated times. Note that the anti-pTpY antibody does not detect the phosphorylated MPK3 activation loop when it is mutated (D193G/E197A). C, Western blot analyses of MPK4/6 levels using MAPK-specific antibodies in rosettes of pot-grown plants of the indicated backgrounds. D, Kinase activity of MPK4 and MPK6 after immunoprecipitation using specific antibody from rosettes of the indicated genotypes. Western blots show protein expression levels. E, Representative photographs of 1-month-old K3CA2/mpk6-2 plants compared with parental lines and Col-0. Plants were cultivated at 22°C on soil in a culture chamber. Bar = 2 cm.
K3CA Lines Show Induction of Stress-Related Genes
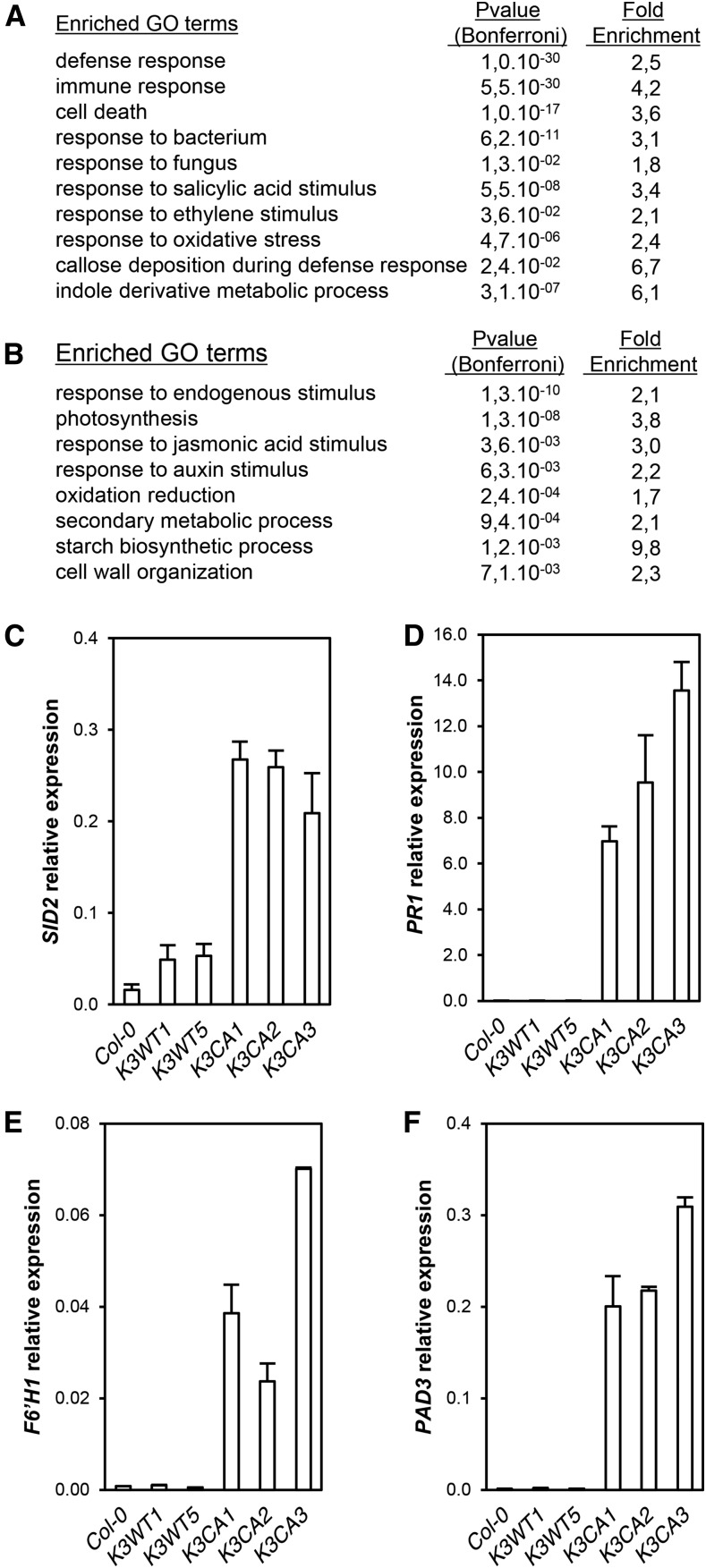
In order to identify the genes misregulated in K3CA lines, we performed a global transcriptome analysis. Plants of K3CA and K3WT lines, three independent lines for each, were grown on soil for 30 d. RNA was extracted and hybridization was performed on microarrays according to an experimental design limiting the positional effect of T-DNA insertions (Supplemental Fig. S2). A total of 1,848 up-regulated and 1,483 down-regulated CATMA probes, corresponding to 1,769 and 1,436 unique genes, were found in K3CA lines compared with K3WT lines, respectively (adjusted with a false discovery rate P < 0.05; Supplemental Tables S1 and S2). To identify the physiological processes that are altered in K3CA lines, we performed a Gene Ontology (GO) analysis of the differentially expressed genes. Notably, the set of up-regulated genes showed a significant enrichment of GO categories related to responses to pathogens, defense and immunity, and cell death as well as responses to oxidative stress and to the stress-related hormones ethylene and SA (Fig. 4A; Supplemental Table S3). Among the down-regulated genes, we found an enrichment in the categories of metabolism (photosynthesis, starch, and secondary metabolism) as well as the hormones auxin and jasmonic acid (Fig. 4B; Supplemental Table S3). The transcriptomic data obtained were confirmed by RT-qPCR using a set of relevant genes. Notably, the defense-related genes SID2 (SALICYLIC ACID INDUCTION-DEFICIENT2), PR1 (PATHOGENESIS-RELATED GENE1), PAD3 (PHYTOALEXIN DEFICIENT3), and F6ʹH1 (FERULOYL-COA 6ʹ-HYDROXYLASE1) were all up-regulated in K3CA lines (Fig. 4, C–F). Comparing our transcriptomic data with previously published ones revealed that about 40% and 30% of the genes up-regulated upon MAMP perception (30-min flg22 treatment) and bacterial infection (2 h post Pseudomonas syringae inoculation), respectively, were also up-regulated in K3CA lines (Supplemental Fig. S3, A–E). This result indicates that K3CA lines display active defenses due to MPK3 activation. It also suggests that MPK3 activity is important for the transcriptional reprograming during the first steps of pathogen perception.
Figure 4.
Misregulated genes in K3CA lines. A, Top enriched GO terms among the up-regulated genes. B, Top enriched GO terms among the down-regulated genes. C to F, SID2, PR1, F6ʹH1, and PAD3 transcript levels in K3CA and K3WT lines. Data are means ± sd of three technical repetitions.
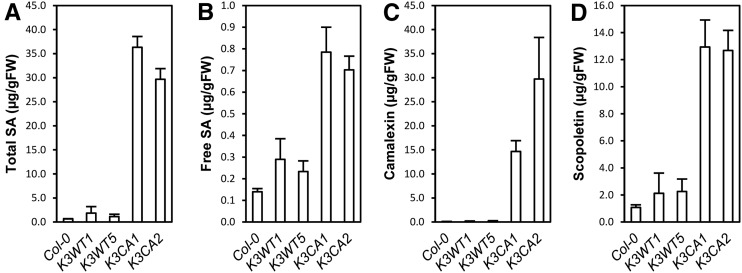
K3CA Plants Produce Ethylene and Accumulate SA, Camalexin, and Scopoletin
To understand the extent to which the transcriptomic data translate into a physiological phenotype, we quantified the levels of some defense-related hormones in K3CA plants. Ethylene production by 1-aminocyclopropane-1-carboxylic acid synthases (ACS) was reported to be up-regulated by the expression of a Nicotiana tabacum CA MEK2, which activates MPK3 and MPK6 (Liu and Zhang, 2004). Ethylene measurements were carried out using a photoacoustic detector. Surprisingly, it revealed only a minor increase of ethylene production in K3CA compared with K3WT lines (Supplemental Fig. S4A). In contrast, we found that total and free SA accumulated 10 to 20 and 2 to 3 times, respectively, in K3CA compared with K3WT and Col-0 plants (Fig. 5, A and B). Finally, camalexin and scopoletin, two major phytoalexins, were about 160 and 7 times more abundant, respectively, in K3CA than in K3WT and Col-0 plants (Fig. 5, C and D). Altogether, these findings established that MPK3 activity results in altered accumulation of defense compounds like defense hormones and phytoalexins.
Figure 5.
Characterization of SA and phytoalexin contents in K3CA lines. Total SA (A), free SA (B), camalexin (C), and scopoletin (D) contents were measured by HPLC in the indicated lines. Average contents of four independent biological replicates are represented. Error bars show se. FW, Fresh weight.
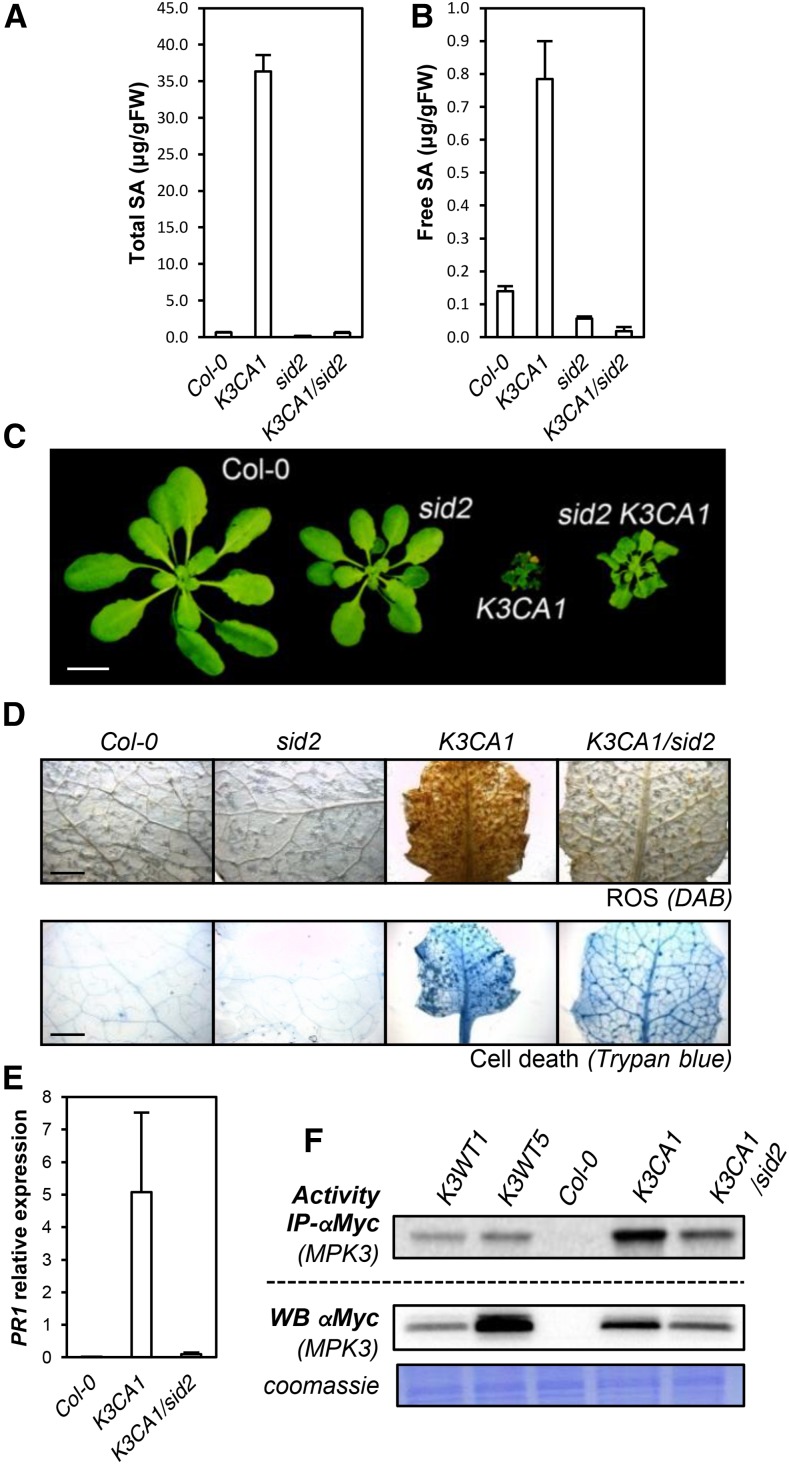
K3CA Phenotypes Are Mostly Dependent of SA Signaling But Independent of Ethylene Signaling
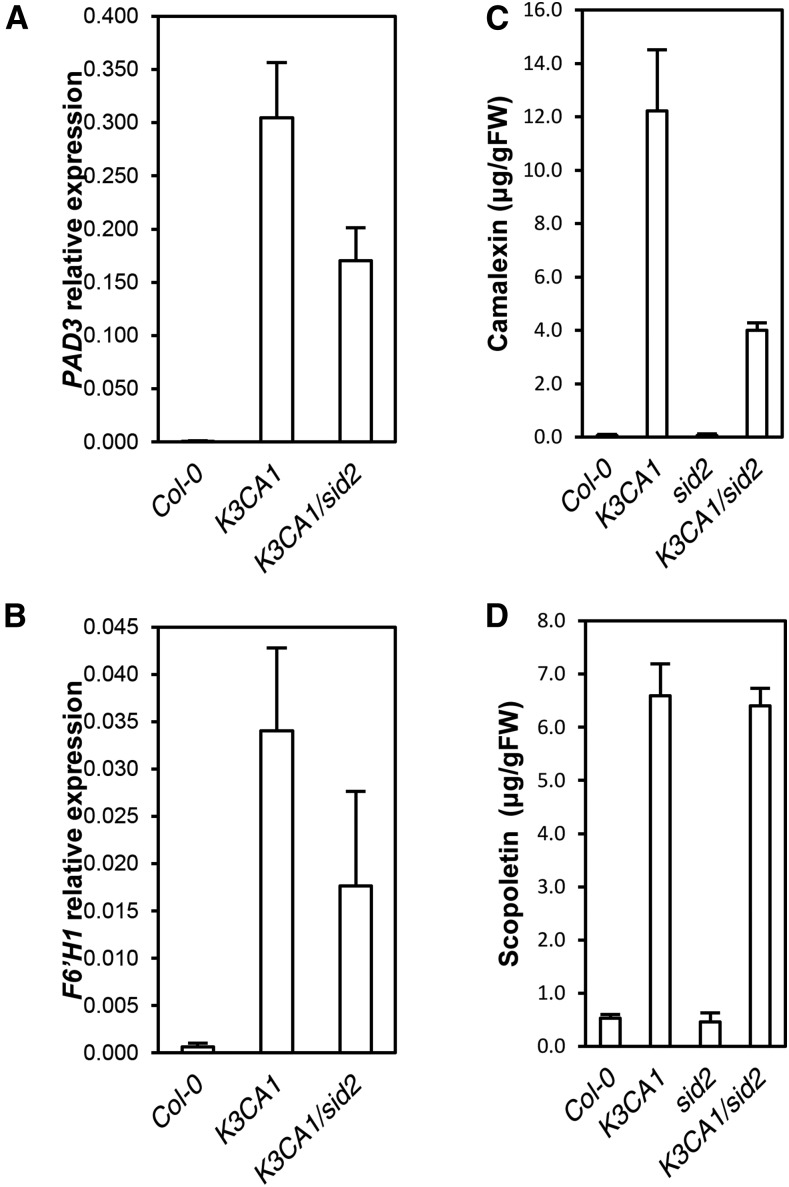
To further investigate the role of MPK3 in ethylene and SA production and the extent to which they contribute to K3CA phenotypes, we crossed K3CA1 plants with the ein2-50 (ethylene insensitive2) mutant impaired in ethylene-mediated signaling (Xu et al., 2008) and with sid2-2, a mutant impaired in SA accumulation during pathogen infection (Nawrath and Métraux, 1999). We did not observe any suppression of K3CA dwarfism by ein2-50 (Supplemental Fig. S4B), meaning that ethylene signaling is not of major importance for establishing the K3CA phenotype. In contrast, sid2-2 abolished SA accumulation in K3CA plants (Fig. 6, A and B) and led to a partial reversion of the dwarfed phenotype (Fig. 6C). Although K3CA1/sid2-2 plants were 3 to 5 times bigger than K3CA1 plants, they still remained affected in their development. In line with this observation, spontaneous ROS accumulation and cell death in K3CA1 also were only partially suppressed by sid2-2 (Fig. 6D), as was the up-regulation of PR1, which dropped about 100 times in K3CA/sid2 compared with K3CA but still remained severalfold higher compared with Col-0 (Fig. 6E). Additionally, although the decrease of PAD3 expression in K3CA/sid2 compared with K3CA correlated with the decrease in camalexin contents, the decrease of F6ʹH1 apparently had no effect on the level of scopoletin (Fig. 7). These results indicate that scopoletin accumulation, unlike camalexin, is SA independent. Interestingly, the partial suppression of the K3CA phenotypes in K3CA1/sid2-2 plants correlated with the mild decrease in MPK3 activity (Fig. 6F), suggesting that sustainable MPK3 activity is ensured by a positive feedback loop involving SA. Overall, our results show that MPK3 function in plant immunity is dependent on SA signaling.
Figure 6.
SA-related phenotypes of K3CA lines. A and B, Total (A) and free (B) SA contents measured by HPLC in the indicated genetic backgrounds. Data are means ± sd of four biological repetitions. FW, Fresh weight. C, Representative photographs of 1-month-old K3CA1/sid2-2 plants compared with parental lines and Col-0. Plants were cultivated at 22°C on soil in a culture chamber. Bar = 1 cm. D, Representative photographs of K3CA1/sid2-2 leaves compared with parental lines and Col-0 stained with DAB and Trypan Blue to visualize ROS and cell death. Bars = 2 mm. E, PR1 transcript levels in the indicated backgrounds. Data are means ± sd of four biological repetitions. F, Kinase activity toward MBP of MPK3-myc immunoprecipitated (IP) from the indicated lines. Western blots (WB) show MPK3-myc levels.
Figure 7.
SA-related phytoalexin phenotypes of K3CA lines. A and B, PAD3 (A) and F6ʹH1 (B) transcript levels in the indicated backgrounds. Data are means ± sd of four biological repetitions. C and D, Camalexin (C) and scopoletin (D) contents measured by HPLC in the indicated genetic backgrounds. Data are means ± sd of four biological repetitions.
Comparison of K3CA and mpk4 Phenotypes Shows That the Two Mutants Are Closely Related
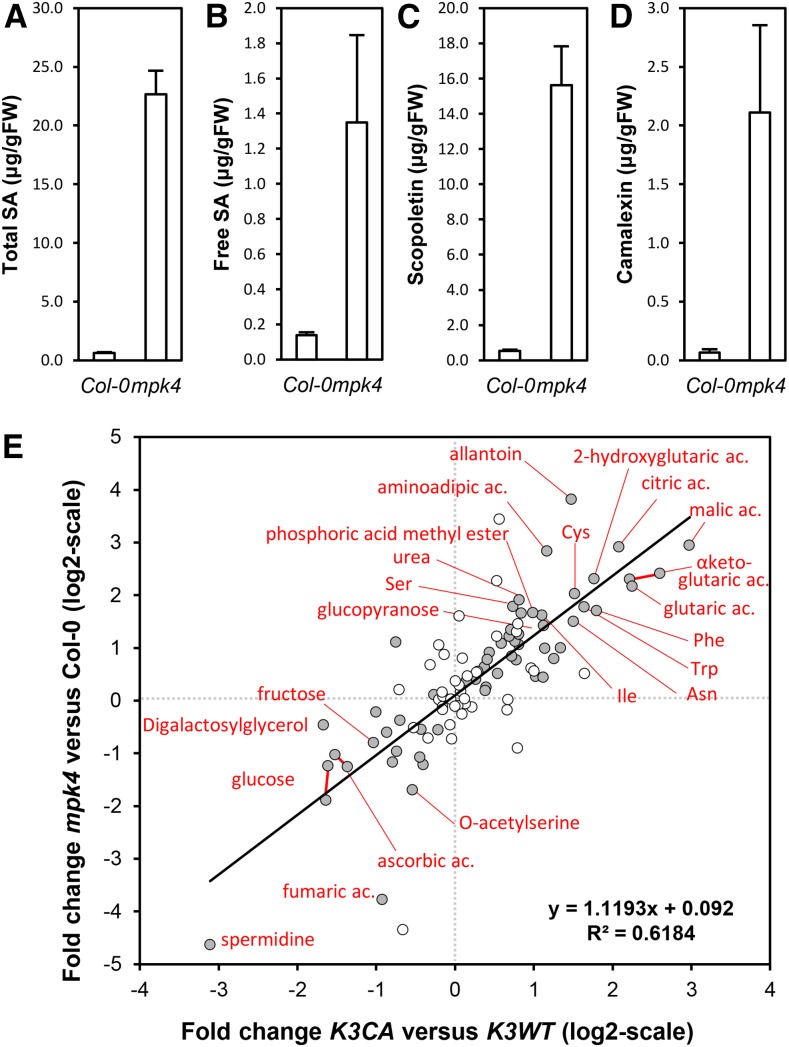
The similarity in the dwarf, cell death, and enhanced SA level phenotypes of the autoimmune mpk4 mutant and the K3CA lines prompted us to characterize these lines in close detail. The mpk4 phenotype is similarly related to high levels of the defense hormone SA and is suppressed by growth under high temperature. We grew K3CA plants at 28°C and observed an identical suppression of the autoimmune features (Supplemental Fig. S5, A and B). A comparison of the transcriptomes between K3CA lines grown on soil and mpk4 seedlings grown in vitro (Frei dit Frey et al., 2014) revealed 35% and 10% overlaps among the up- and down-regulated genes, respectively (Supplemental Fig. S3B). Interestingly, besides SA, mpk4 accumulates both scopoletin and camalexin (Fig. 8, A–D). To obtain an overview of the metabolic changes, we performed gas chromatography/mass spectrometry-time of flight profiling of Col-0, K3CA, K3WT, and mpk4 plants. We also included the mips1 mutant, which was shown previously to have a SA-related autoimmune phenotype (Meng et al., 2009). Overall, 99 peaks were identified corresponding to 91 metabolites (Supplemental Table S4). K3CA lines showed 26 and 11 metabolites to be significantly increased and reduced, respectively, compared with K3WT (P < 0.05). A more careful look to these metabolites showed that 11 amino acids and several molecules of the Krebs cycle, including citric acid, α-ketoglutaric acid, and malic acid, accumulated in K3CA plants. Among the reduced metabolites, there were important sugars, such as Glc and Fru, as well as spermidine. Interestingly, we observed a significant correlation between metabolite contents in K3CA and mpk4, which is another confirmation of the similarities of the responses activated in these two mutant lines (Fig. 8). No correlation was found between K3CA and mips1 (Supplemental Fig. S6), suggesting that the metabolome changes in these autoimmune mutants are not due to the high SA levels and that K3CA plants are phenotypically more similar to mpk4 than to mips1 plants.
Figure 8.
Comparison of metabolite contents of mpk4 versus K3CA plants. A to D, Total SA (A), free SA (B), scopoletin (C), and camalexin (D) contents of mpk4 measured by HPLC. Data are means ± sd of four biological repetitions. FW, Fresh weight. E, Metabolite analysis of K3CA and mpk4 plants. Means of fold changes (log2) of K3CA/K3WT (x axis) are plotted against means of fold changes (log2) of mpk4/Col-0 (y axis). Gray dots show metabolites that are significantly misaccumulated in K3CA plants (Wilcoxon, Mann-Whitney test with P < 0.05). For clarity, compound names are indicated for fold changes (log2) higher than 1.5 or lower than −1.5.
Mutations That Suppress mpk4 Are Modifiers of the K3CA Phenotype
Several mutations have been reported to fully suppress mpk4 dwarfism (Kong et al., 2012; Zhang et al., 2012). In order to test whether these mutations also could suppress K3CA dwarfism, we crossed K3CA2 with summ1-1 and summ2-8. Both K3CA2/summ1-1 and K3CA2/summ2-8 were 2 to 3 times bigger than K3CA2 plants but still much smaller than control plants (K3WT and Col-0; Fig. 9A). Cell death and ROS accumulation were partially reduced in K3CA2/summ2-8 and K3CA2/summ1-1 (Fig. 9B). Whereas total SA levels also were partially reduced, free SA accumulation was not impacted by the summ1 or summ2 mutation (Fig. 9, C and D). Consistently, the transcript levels of the PR1 gene, which is a marker of SA signaling, were not significantly affected in K3CA2/summ1 and K3CA2/summ2 compared with control lines (Supplemental Fig. S7). In contrast, camalexin and scopoletin levels were both reduced in K3CA2/summ1-1 and K3CA2/summ2-8 compared with K3CA2, with a stronger reduction in camalexin, as were the biosynthetic marker genes of these pathways (Fig. 9, E and F; Supplemental Fig. S7). Besides, unlike what was observed in K3CA/sid2, the partial suppression of the K3CA phenotypes in K3CA1/summ1 and K3CA/summ2 plants did not appear to be linked to a decrease in MPK3 kinase activity or protein level (Fig. 9G). Altogether, these results indicate a bifurcation of the innate immunity pathway at the level of MPK3 for scopoletin and camalexin biosynthesis on the one hand and the MPK4-dependent SUMM1 and SUMM2 regulation on the other hand.
Figure 9.
Phenotypes of K3CA2 crossed with summ mutants. A, Representative photographs of K3CA2/summ1-1 and K3CA2/summ2-8 plants compared with parental lines and Col-0. Plants were cultivated at 22°C on soil in a culture chamber. Bar = 1 cm. B, Representative photographs of K3CA-2/summ1-1 and K3CA-2/summ2-8 leaves compared with parental lines and Col-0 stained with DAB and Trypan Blue to visualize ROS and cell death. Bars = 2 mm. C to F, Free (C) and total (D) SA, scopoletin (E), and camalexin (F) contents measured by HPLC in the indicated genetic backgrounds. Data are means ± sd of four biological repetitions. FW, Fresh weight. G, Kinase activity of MPK3-myc immunoprecipitated (IP) from the indicated genetic background. Western blots (WB) show protein expression levels.
DISCUSSION
MAPKs play central roles in MTI (Bigeard et al., 2015). Recent data suggest that they also have an important function during effector-triggered immunity (ETI; Tsuda et al., 2013). The main difference between these two processes lies in their kinetics of activation. MPK3 and MPK6 are rapidly and transiently activated upon MAMP perception, whereas this activation takes longer and is sustained for hours after the detection of a pathogenic effector (Tsuda et al., 2013). MPK4 also is activated transiently by MAMPs, but only indirect data suggest its activation during ETI (Berriri et al., 2012). These data fit the idea that MTI and ETI correspond to similar responses, differing mainly by their temporal pattern and amplitude of activation. The identification of particular roles of the MAPKs in the context of MTI and ETI is complex. A widely used strategy consists of using loss-of-function or gain-of-function protein kinase mutants. The gain-of-function strategy has been used extensively to investigate the role of MAP2Ks, for which replacing the two phosphorylated residues in the activation loop by phospho-mimicking residues usually triggers constitutive activity. We recently applied a gain-of-function strategy to the MAPK level by identifying mutations triggering constitutive activation (Berriri et al., 2012). As an alternative to inducible lines, which result in high protein accumulation and, thereby, possible nonspecific events, we created Arabidopsis lines expressing CA MAPKs under the control of their native promoter using genomic loci.
Plants Expressing CA-MPK3 Show Up-Regulation of SA-Dependent and Independent Defense Responses
Plants expressing CA-MPK3 showed autoimmune phenotypes characterized by dwarfism associated with spontaneous cell death, accumulations of ROS and SA, and modification of metabolism to produce the phytoalexins scopoletin and camalexin. MPK3 and MPK6 are thought to play redundant roles during immunity, as they share several features, notably their kinetics of activation and some of their substrates. For this purpose, it would have been interesting to compare the phenotypes of K3CA and K6CA plants. However, so far, we have been unable to create lines expressing CA-MPK6, possibly because, in our case, a constitutive activation of MPK6 in planta is deleterious. The enhanced production of camalexin by CA-MPK3 plants agrees with the findings that, upon B. cinerea infection, MPK3 and MPK6 have been proposed to phosphorylate the transcription factor WRKY33, which binds camalexin biosynthetic gene promoters and triggers camalexin accumulation (Ren et al., 2008; Mao et al., 2011). Moreover, our work confirms the report that Arabidopsis plants expressing dexamethasone-inducible CA-MKK5DD or NtMEK2DD, MAP2Ks upstream of MPK3 and MPK6, accumulate intermediate levels of camalexin (Ren et al., 2008; Lassowskat et al., 2014). We also found that CA-MPK3 is able to induce the accumulation of the phytoalexin scopoletin, which is known to have a role in defense in tobacco (Chong et al., 2002) and in iron uptake in Arabidopsis (Schmidt et al., 2014). K3CA plants accumulate intermediate compounds of the Krebs cycle, such as citric, malic, and α-ketoglutaric acids, suggesting an enhanced turnover of the tricarboxylic acid cycle. In agreement, sugars such as Glc and Fru were underaccumulated in K3CA lines. We also noticed a significant increase in several amino acids, which could be the consequence of cell death-related protein degradation.
K3CA plants produce slightly more ethylene than wild-type plants. However, ein2 was unable to suppress the K3CA dwarf phenotype, suggesting that ethylene is not a major player in MPK3 responses. This contradicts data reporting that ethylene is an important actor in the MKK5-dependent cell death (Liu et al., 2008) and the fact that ACS2/6 are MPK3/6 substrates (Liu and Zhang, 2004; Han et al., 2010). Although these contrasting results may depend on the use of different experimental systems, different outcomes also may stem from different expression and activation levels. In our work, MPK3 was expressed at endogenous levels and moderately activated by CA mutations. In contrast to ethylene, K3CA plants strongly accumulated SA, and consistently, mutation of the SID2 gene partially suppressed the morphological defects in CA-MPK3 plants, such as ROS accumulation and cell death. SA overaccumulation often is observed in autoimmune mutants and, therefore, is believed to be a crucial factor for inducing cell death (Bruggeman et al., 2015), as most of the mutant phenotypes are indeed suppressed by inhibiting SA biosynthesis. For example, the cell death phenotypes of mpk4 and mips1 are suppressed by the expression of the bacterial SA hydrolase nahG or by the introgression of a sid2 mutation (Brodersen et al., 2006; Meng et al., 2009). Nevertheless, not all MPK3 functions depend on SID2-triggered SA accumulation. First, while totally abolishing SA accumulation, sid2 did not fully revert K3CA-dependent dwarfism. Additionally, only camalexin accumulation was partially reduced by sid2, whereas scopoletin was unchanged. SA-dependent genes, such as PR1, which are highly expressed in K3CA lines, are only partially reduced by sid2. This result is in agreement with other studies. For example, cytokinin treatment induces scopoletin synthesis independently of SA (Grosskinsky et al., 2011), and the auxin 2,4-D but not SA or methyl jasmonate treatment triggered scopoletin synthesis in Arabidopsis (Kai et al., 2006). In line with a previous report (Tsuda et al., 2013), this result also illustrates the robustness and complexity of the defense signaling pathways modulated by a sustained activity of MPK3 that is both SA dependent and independent.
Plants Expressing CA-MPK3 Phenocopy Loss-of-Function Mutants of the MEKK1-MKK1/2-MPK4 Pathway
Overall, the K3CA phenotype strongly resembles to loss-of-function mutants of the MEKK1-MKK1/2-MPK4 module (Petersen et al., 2000; Ichimura et al., 2006; Nakagami et al., 2006; Gao et al., 2008; Qiu et al., 2008). Like mpk4 plants (Brodersen et al., 2006), K3CA lines accumulated SA, camalexin, and scopoletin as well as ROS, but not to the same extent. Both K3CA and mpk4 show spontaneous cell death and comparable transcriptomes and metabolomes. On the other hand, the SA-dependent cell death phenotype of mips1 (Meng et al., 2009) was not correlated with a similar metabolome, as found for mpk4 and K3CA lines. Altogether, these data indicate the specificity of the MAPK immune phenotype and how constitutive activation of MPK3 alone results in the deregulation of similar aspects of defense as mutations in the MEKK1-MKK1/2-MPK4 module.
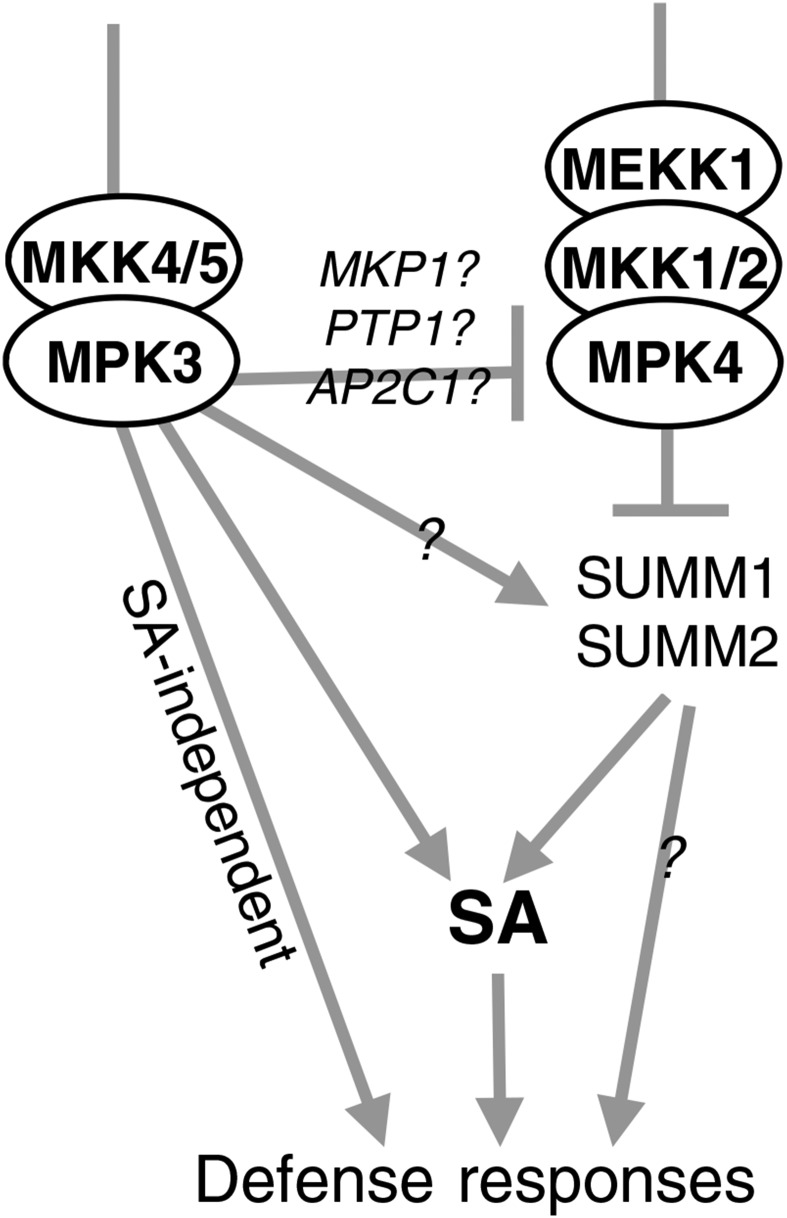
The phenotype of mutants impaired in the MEKK1-MKK1/2-MPK4 module has been attributed to the activation of a guarding pathway containing SUMM1/MEKK2 and SUMM2 (Kong et al., 2012; Zhang et al., 2012). Consequently, mutations impairing one of these two genes fully suppress mpk4 dwarfism as well as SA accumulation and the deregulation of defense marker genes (Kong et al., 2012; Zhang et al., 2012). The SUMM1/2 module was introgressed into K3CA plants to investigate whether it also regulates MPK3-related functions. The K3CA dwarfism, metabolite and ROS accumulation, as well as cell death phenotypes were partially reverted, but not to the same extent as in the summ1 and summ2 crosses with mpk4. This suggests that MPK3 regulates stress responses both dependently and independently of SUMM1/2 (Fig. 10). Our group recently showed that MPK3 also functions to restrict MAPK activation during MAMP signaling (Frei dit Frey et al., 2014). Indeed, in mpk3 plants, flg22-triggered MPK4 and MPK6 activation lasts longer than in wild-type plants. A possible mechanism is provided by various MAPK-triggered phosphatases that act on the immune MAPKs in a negative feedback loop. MAP Kinase Phosphatase1 (MKP1), Protein Tyr Phosphatase1, and AP2C1 all have been shown to reduce stress-triggered MAPK activation (Schweighofer et al., 2007; Bartels et al., 2009; Anderson et al., 2011). MKP1 itself is actually phosphorylated by MPK6, suggesting that it functions in negatively regulating MAPK kinetics during MAMP perception (Park et al., 2011). Therefore, it is possible that, in K3CA plants, the reduced basal activity of MPK4 is sufficient to trigger the activation of the guarding module SUMM1/2 and, thereby, leads to an autoimmune phenotype similar to that of mpk4. This could account for the partial reversion of the K3CA phenotype by summ1 and summ2. It is also plausible that MPK3 directly activates the SUMM1/2 module. Indeed, we observed an up-regulation of SUMM1 in K3CA plants that might be independent of MPK4 function. The fact that summ1 and summ2 do not fully suppress the K3CA phenotype also suggests that not all MPK3 responses depend on SUMM1/2. For example, the accumulation of camalexin and scopoletin is poorly suppressed by summ1/2 mutations in the K3CA lines. Since these two phytoalexins also accumulate in mpk4, it would be informative to know the extent to which their accumulation is suppressed in mpk4 by the summ1/2 mutations. Interestingly, SUMM2, an R gene of the CC-NB-LRR family, most likely triggers an NDR1-dependent signaling cascade leading to ETI (Aarts et al., 1998). It would be interesting to examine whether the K3CA phenotype also is suppressed by ndr1 and, thus, genetically define how the signaling cascades are connected. Furthermore, MPK3/6 kinases also have been unambiguously shown to be activated during the perception of the Xanthomonas effector AvrRPT2 by its cognate TIR-NB-LRR-type receptor RPS2 (Tsuda et al., 2013). One possibility is that the SUMM1/SUMM2-independent part of K3CA corresponds to the MPK3 function in ETI signaling. Overall, our study provides new insights into MAPK signaling in innate immunity but also raises a number of new questions that require future studies in this field.
Figure 10.
Working model for MPK3 function.
MATERIALS AND METHODS
Genetic Material and Growth Conditions
The Arabidopsis thaliana wild-type ecotype Col-0, mpk3-1 (Wang et al., 2007), mpk4-2 (Nakagami et al., 2006), mips1 (Meng et al., 2009), sid2-2 (Nawrath and Métraux, 1999), and summ2-8 and summ1-1 (Kong et al., 2012; Zhang et al., 2012) were used in this study.
To generate transgenic lines expressing CA-MPK3, two 3,056- and 723-bp PCR products, corresponding to MPK3 loci upstream and downstream of the stop codon, were amplified using the iProof (Bio-Rad) polymerase and Pr0112/Pr0113 and Pr0114/Pr0115 sets of primers (Supplemental Table S5) from Col-0 genomic DNA. Fragments were cloned in pGEMTeasy following the manufacturer’s protocol (Promega) and fully sequenced. The MPK3 locus was then reassembled in pGREEN0229 (Hellens et al., 2000) using appropriate restriction enzymes to create pGREEN-MPK3wt. A PCR fragment containing the PC2 tag (Bigeard et al., 2014) was then amplified using Pr0159/Pr0160 primers, digested with BglII, and inserted in the unique compatible BamHI site of pGREEN-MPK3wt to create pGREEN-MPK3wt-PC2. Finally, point mutations were created as described previously using Pr0317/Pr0318 primers to create the pMPK3::MPK3D193G/E197A-PC2 vector (Berriri et al., 2012). Whole constructs were sequenced before Agrobacterium tumefaciens transformation. mpk3-1 was then transformed by floral dipping (Clough and Bent, 1998). Positive transformants were selected on glufosinate. Based on the segregation of glufosinate resistance, lines carrying a single insertion were first selected and, in the next generation, plants carrying a homozygous T-DNA insertion were identified. With the exception of K3WT and K3CA transgenic lines and crosses, which were generated in a greenhouse, all described experiments were performed using plants grown in a growth cabinet (Percival) in long-day conditions, with a humidity of 70% at 22°C, if not mentioned otherwise.
Kinase Assay and Western Blot from Plants
For the protoplast experiment, appropriate wild-type and mutant open reading frames (MPK3, MPK3T119C, MPK3D193G/E197A, MPK4WT, MPK4Y124C, and MPK4D198G/E202A) in pDNR207 were recombined in pHaGWF7 using LR enzyme mix following the manufacturer’s protocol (Invitrogen). Protoplasts were prepared from Col-0 leaves and transformed following published protocols (Yoo et al., 2007).
Kinase assays and western-blot analyses were performed as described previously (Berriri et al., 2012). Briefly, for kinase assays, proteins were extracted in a nondenaturant buffer and normalized to 1 µg µL−1 using Bradford reagent. A total of 100 µg was then used for the immunoprecipitation/kinase assay on MBP and 20 µg for related western blots. In the figures 1, 2, 3, 6, and 9, the top image shows the radioactive labeling of MBP and the bottom one shows the blot demonstrating protein concentrations and its cognate Coomassie Blue staining of the membrane at the level of the large subunit of Rubisco. If samples were used only for western blots, proteins were extracted using Laemmli 2× buffer (v/w), and 10 µL was loaded on SDS gels.
DAB and Trypan Blue Staining
ROS staining was performed as described previously (Daudi and O’Brien, 2012). For Trypan Blue staining, plant leaves were boiled 1 min in Trypan Blue solution (3.3 mg mL−1 Trypan Blue, 33.3% acetic acid, 33.3% phenol, and 33.3% glycerol) diluted in 1 volume of 100% ethanol. Leaves were then bleached in choral hydrate (2.5 g mL−1) and conserved in 50% glycerol. Photographs were taken using a binocular loop Leica MZ16F apparatus.
Transcriptome and qPCR Experiments
The transcriptomic experiment was performed in Transcriptome Platform following an established protocol (Danquah et al., 2015). Rosettes of 1-month-old plants grown in Percival were collected. RNAs were extracted using the NucleoSpin RNA kit (Macherey-Nagel). cRNA were synthesized, labeled, and hybridized on CATMAv7 Agilent arrays (Two-Color Microarray-Based Gene Expression Analysis). Differentially expressed genes were identified using a previously described pipeline in which data normalization is done using the Loess procedure and differential expression assessed by Limma with an adjusted P ≤ 0.05 (Danquah et al., 2015). Data analysis (GO term enrichment) was carried out with DAVID Bioinformatics Resources (Huang et al., 2009).
For quantitative reverse transcription-PCR analysis, cDNA was synthetized from 1 µg of DNase-treated RNA using SuperScript II reverse transcriptase (Invitrogen) following the manufacturer’s protocol. For reverse transcription, oligo(dT) primers were used. The SYBR FAST Universal qPCR Kit (Kapa Biosystems) was used to prepare qPCR mix. Primers (Supplemental Table S4) were used at 100 nm final concentration. Biological triplicates were performed. The following PCR program was used: 95°C for 30 s and then 40 cycles of 95°C for 5 s and 60°C for 20 s; a dissociation step also was programmed to validate the PCR products. qPCR was carried out on a CFX384 Touch Real-Time PCR device (Bio-Rad) and analyzed with CFX Manager Software (Bio-Rad).
Metabolite Measurements
The experiment was performed in Platform Metabolism-Metabolome of the Institute of Plant Sciences Paris Saclay. To reduce the variability linked to the position of T-DNA insertion, samples were pooled from three independent K3CA and K3WT lines. Three biological replicates were done. Aerial parts were collected, and samples were lyophilized and extracted before being injected into the gas chromatography-time of flight apparatus according to the Platform Metabolism-Metabolome protocol (Tcherkez et al., 2012). SA, camalexin, and scopoletin assays were performed as described previously (Bruggeman et al., 2014).
For ethylene measurement, transgenic plants were grown in a 12-h photoperiod at 22°C on peat pellet substrate (Jiffy). At the start of the experiments, the peat pellets were treated once with NPK fertilizer (Wuxal liquid; Algukon). Light was cold white (80 µmol m−2 s−1). Seven- to 9-week-old plants with substrate were enclosed in a 150-mL cuvette with a flat glass lid. Cuvettes were flushed every 2 h for 20 min with synthetic air containing 350 µL L−1 CO2 (Air Liquide). Ethylene in the headspace was detected using a photoacoustic detector (ETD300; Sensor Sense; Schellingen et al., 2014). Measurements on plant-containing cuvettes were compared among them and with a cuvette containing only a peat pellet as a background control.
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL databases under the following accession numbers: At3g45640 (MPK3), At4g01370 (MPK4), At4g08480 (SUMM1), At1g12280 (SUMM2), At1g74710 (SID2), At5g03280 (EIN2), At3g26830 (PAD3), At3g13610 (F6ʹH1), and At2g14610 (PR1).
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Characterization of MAPKs in K3WT and K3CA in vitro-grown plantlets in resting conditions.
Supplemental Figure S2. Microarray experiment overview.
Supplemental Figure S3. Comparison of misregulated genes in K3CA plants with published transcriptome.
Supplemental Figure S4. Ethylene-related phenotype.
Supplemental Figure S5. The K3CA autoimmune phenotype is suppressed by high temperature.
Supplemental Figure S6. Comparison of metabolite contents of mips1 versus K3CA plants.
Supplemental Figure S7. Misregulated genes in K3CA crossed with mutants.
Supplemental Table S1. Transcriptome of K3WT and K3CA: list of 1,848 up-regulated genes in K3CA lines.
Supplemental Table S2. Transcriptome of K3WT and K3CA: list of 1,483 down-regulated genes in K3CA lines.
Supplemental Table S3. Transcriptome of K3WT and K3CA: GO categories for up- and down-regulated genes.
Supplemental Table S4. Metabolome of K3WT, K3CA, Col-0, mpk4, and mips1 plants.
Supplemental Table S5. Primers.
Acknowledgments
We thank Yuelin Zhang and Jane Parker for summ1/2 and sid2 mutants, respectively, Filip Vandenbussche for ethylene measurement, and Jean Bigeard for helpful discussions and critical reading of the article.
Glossary
- MAMP
microbe-associated molecular pattern
- ROS
reactive oxygen species
- SA
salicylic acid
- NB-LRR
nucleotide-binding leucine-rich repeat
- MTI
MAMP-triggered immunity
- HA
hemagglutinin
- CA
constitutively active
- Col-0
Columbia-0
- DAB
3,3′-diaminobenzidine
- qPCR
quantitative PCR
- GO
Gene Ontology
- ETI
effector-triggered immunity
Footnotes
This work was supported by the Institut National de Recherche Agronomique, the Agence Nationale de la Recherche, and LabEx Saclay Plant Sciences (grant no. ANR-10-LABX-0040-SPS), managed by the French National Research Agency under the Investments for the Future program (grant no. ANR-11-IDEX-0003-02), by the French Ministry of Research (Ph.D. fellowship to B.G.), and by the Research Foundation Flanders (grant no. FWO G.0656.13N to D.V.D.S.) and Ghent University (Bijzonder Onderzoeksfonds grant to D.V.D.S.).
References
- Aarts N, Metz M, Holub E, Staskawicz BJ, Daniels MJ, Parker JE (1998) Different requirements for EDS1 and NDR1 by disease resistance genes define at least two R gene-mediated signaling pathways in Arabidopsis. Proc Natl Acad Sci USA 95: 10306–10311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson JC, Bartels S, González Besteiro MA, Shahollari B, Ulm R, Peck SC (2011) Arabidopsis MAP Kinase Phosphatase 1 (AtMKP1) negatively regulates MPK6-mediated PAMP responses and resistance against bacteria. Plant J 67: 258–268 [DOI] [PubMed] [Google Scholar]
- Asai T, Tena G, Plotnikova J, Willmann MR, Chiu W, Gomez-Gomez L, Boller T, Ausubel FM, Sheen J (2002) MAP kinase signalling cascade in Arabidopsis innate immunity. Nature 415: 977–983 [DOI] [PubMed] [Google Scholar]
- Bartels S, Anderson JC, González Besteiro MA, Carreri A, Hirt H, Buchala A, Métraux JP, Peck SC, Ulm R (2009) MAP kinase phosphatase1 and protein tyrosine phosphatase1 are repressors of salicylic acid synthesis and SNC1-mediated responses in Arabidopsis. Plant Cell 21: 2884–2897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berriri S, Garcia AV, Frei dit Frey N, Rozhon W, Pateyron S, Leonhardt N, Montillet JL, Leung J, Hirt H, Colcombet J (2012) Constitutively active mitogen-activated protein kinase versions reveal functions of Arabidopsis MPK4 in pathogen defense signaling. Plant Cell 24: 4281–4293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bigeard J, Colcombet J, Hirt H (2015) Signaling mechanisms in pattern-triggered immunity (PTI). Mol Plant 8: 521–539 [DOI] [PubMed] [Google Scholar]
- Bigeard J, Pflieger D, Colcombet J, Gérard L, Mireau H, Hirt H (2014) Protein complexes characterization in Arabidopsis thaliana by tandem affinity purification coupled to mass spectrometry analysis. Methods Mol Biol 1171: 237–250 [DOI] [PubMed] [Google Scholar]
- Brodersen P, Petersen M, Nielsen HB, Zhu S, Newman MA, Shokat KM, Rietz S, Parker J, Mundy J (2006) Arabidopsis MAP kinase 4 regulates salicylic acid- and jasmonic acid/ethylene-dependent responses via EDS1 and PAD4. Plant J 47: 532–546 [DOI] [PubMed] [Google Scholar]
- Bruggeman Q, Garmier M, de Bont L, Soubigou-Taconnat L, Mazubert C, Benhamed M, Raynaud C, Bergounioux C, Delarue M (2014) The polyadenylation factor subunit CLEAVAGE AND POLYADENYLATION SPECIFICITY FACTOR30: a key factor of programmed cell death and a regulator of immunity in Arabidopsis. Plant Physiol 165: 732–746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruggeman Q, Raynaud C, Benhamed M, Delarue M (2015) To die or not to die? Lessons from lesion mimic mutants. Front Plant Sci 6: 24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chong J, Baltz R, Schmitt C, Beffa R, Fritig B, Saindrenan P (2002) Downregulation of a pathogen-responsive tobacco UDP-Glc:phenylpropanoid glucosyltransferase reduces scopoletin glucoside accumulation, enhances oxidative stress, and weakens virus resistance. Plant Cell 14: 1093–1107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Colcombet J, Hirt H (2008) Arabidopsis MAPKs: a complex signalling network involved in multiple biological processes. Biochem J 413: 217–226 [DOI] [PubMed] [Google Scholar]
- Danquah A, de Zélicourt A, Boudsocq M, Neubauer J, Frei Dit Frey N, Leonhardt N, Pateyron S, Gwinner F, Tamby JP, Ortiz-Masia D, et al. (2015) Identification and characterization of an ABA-activated MAP kinase cascade in Arabidopsis thaliana. Plant J 82: 232–244 [DOI] [PubMed] [Google Scholar]
- Daudi A, O’Brien J (2012) Detection of hydrogen peroxide by DAB staining in Arabidopsis leaves. Bio Protoc 2: e263. [PMC free article] [PubMed] [Google Scholar]
- Frei Dit Frey N, Garcia AV, Bigeard J, Zaag R, Bueso E, Garmier M, Pateyron S, de Tauzia-Moreau ML, Brunaud V, Balzergue S, et al. (2014) Functional analysis of Arabidopsis immune-related MAPKs uncovers a role for MPK3 as negative regulator of inducible defences. Genome Biol 15: R87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao M, Liu J, Bi D, Zhang Z, Cheng F, Chen S, Zhang Y (2008) MEKK1, MKK1/MKK2 and MPK4 function together in a mitogen-activated protein kinase cascade to regulate innate immunity in plants. Cell Res 18: 1190–1198 [DOI] [PubMed] [Google Scholar]
- Grosskinsky DK, Naseem M, Abdelmohsen UR, Plickert N, Engelke T, Griebel T, Zeier J, Novák O, Strnad M, Pfeifhofer H, et al. (2011) Cytokinins mediate resistance against Pseudomonas syringae in tobacco through increased antimicrobial phytoalexin synthesis independent of salicylic acid signaling. Plant Physiol 157: 815–830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han L, Li GJ, Yang KY, Mao G, Wang R, Liu Y, Zhang S (2010) Mitogen-activated protein kinase 3 and 6 regulate Botrytis cinerea-induced ethylene production in Arabidopsis. Plant J 64: 114–127 [DOI] [PubMed] [Google Scholar]
- Hellens RP, Edwards EA, Leyland NR, Bean S, Mullineaux PM (2000) pGreen: a versatile and flexible binary Ti vector for Agrobacterium-mediated plant transformation. Plant Mol Biol 42: 819–832 [DOI] [PubMed] [Google Scholar]
- Huang W, Sherman BT, Lempicki RA (2009) Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc 4: 44–57 [DOI] [PubMed] [Google Scholar]
- Ichimura K, Casais C, Peck SC, Shinozaki K, Shirasu K (2006) MEKK1 is required for MPK4 activation and regulates tissue-specific and temperature-dependent cell death in Arabidopsis. J Biol Chem 281: 36969–36976 [DOI] [PubMed] [Google Scholar]
- Ichimura K, Shinozaki K, Tena G, Sheen J, Henry Y, Champion A, Kreis M, Zhang S, Hirt H, Wilson C, et al. (2002) Mitogen-activated protein kinase cascades in plants: a new nomenclature. Trends Plant Sci 7: 301–308 [DOI] [PubMed] [Google Scholar]
- Kai K, Shimizu B, Mizutani M, Watanabe K, Sakata K (2006) Accumulation of coumarins in Arabidopsis thaliana. Phytochemistry 67: 379–386 [DOI] [PubMed] [Google Scholar]
- Kong Q, Qu N, Gao M, Zhang Z, Ding X, Yang F, Li Y, Dong OX, Chen S, Li X, et al. (2012) The MEKK1-MKK1/MKK2-MPK4 kinase cascade negatively regulates immunity mediated by a mitogen-activated protein kinase kinase kinase in Arabidopsis. Plant Cell 24: 2225–2236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lassowskat I, Böttcher C, Eschen-Lippold L, Scheel D, Lee J (2014) Sustained mitogen-activated protein kinase activation reprograms defense metabolism and phosphoprotein profile in Arabidopsis thaliana. Front Plant Sci 5: 554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H, Wang Y, Xu J, Su T, Liu G, Ren D (2008) Ethylene signaling is required for the acceleration of cell death induced by the activation of AtMEK5 in Arabidopsis. Cell Res 18: 422–432 [DOI] [PubMed] [Google Scholar]
- Liu Y, Zhang S (2004) Phosphorylation of 1-aminocyclopropane-1-carboxylic acid synthase by MPK6, a stress-responsive mitogen-activated protein kinase, induces ethylene biosynthesis in Arabidopsis. Plant Cell 16: 3386–3399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao G, Meng X, Liu Y, Zheng Z, Chen Z, Zhang S (2011) Phosphorylation of a WRKY transcription factor by two pathogen-responsive MAPKs drives phytoalexin biosynthesis in Arabidopsis. Plant Cell 23: 1639–1653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng PH, Raynaud C, Tcherkez G, Blanchet S, Massoud K, Domenichini S, Henry Y, Soubigou-Taconnat L, Lelarge-Trouverie C, Saindrenan P, et al. (2009) Crosstalks between myo-inositol metabolism, programmed cell death and basal immunity in Arabidopsis. PLoS ONE 4: e7364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng X, Zhang S (2013) MAPK cascades in plant disease resistance signaling. Annu Rev Phytopathol 51: 245–266 [DOI] [PubMed] [Google Scholar]
- Montillet J, Leonhardt N, Mondy S, Tranchimand S, Chevalier A, Castresana C, Hirt H, Laurie C (2013) An abscisic acid-independent oxylipin pathway controls stomatal closure and immune defense in Arabidopsis. PLoS Biol 11: e1001513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagami H, Soukupová H, Schikora A, Zárský V, Hirt H (2006) A mitogen-activated protein kinase kinase kinase mediates reactive oxygen species homeostasis in Arabidopsis. J Biol Chem 281: 38697–38704 [DOI] [PubMed] [Google Scholar]
- Nawrath C, Métraux JP (1999) Salicylic acid induction-deficient mutants of Arabidopsis express PR-2 and PR-5 and accumulate high levels of camalexin after pathogen inoculation. Plant Cell 11: 1393–1404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nitta Y, Ding P, Zhang Y (2014) Identification of additional MAP kinases activated upon PAMP treatment. Plant Signal Behav 9: e976155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nuhse T, Peck SC, Hirt H, Boller T (2000) Microbial elicitors induce activation and dual phosphorylation of the Arabidopsis thaliana MAPK 6. J Biol Chem 275: 7521–7526 [DOI] [PubMed] [Google Scholar]
- Park HC, Song EH, Nguyen XC, Lee K, Kim KE, Kim HS, Lee SM, Kim SH, Bae DW, Yun DJ, et al. (2011) Arabidopsis MAP kinase phosphatase 1 is phosphorylated and activated by its substrate AtMPK6. Plant Cell Rep 30: 1523–1531 [DOI] [PubMed] [Google Scholar]
- Petersen M, Brodersen P, Naested H, Andreasson E, Lindhart U, Johansen B, Nielsen HB, Lacy M, Austin MJ, Parker JE, et al. (2000) Arabidopsis map kinase 4 negatively regulates systemic acquired resistance. Cell 103: 1111–1120 [DOI] [PubMed] [Google Scholar]
- Pitzschke A, Djamei A, Bitton F, Hirt H (2009) A major role of the MEKK1-MKK1/2-MPK4 pathway in ROS signalling. Mol Plant 2: 120–137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu JL, Zhou L, Yun BW, Nielsen HB, Fiil BK, Petersen K, MacKinlay J, Loake GJ, Mundy J, Morris PC (2008) Arabidopsis mitogen-activated protein kinase kinases MKK1 and MKK2 have overlapping functions in defense signaling mediated by MEKK1, MPK4, and MKS1. Plant Physiol 148: 212–222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ranf S, Eschen-Lippold L, Pecher P, Lee J, Scheel D (2011) Interplay between calcium signalling and early signalling elements during defence responses to microbe- or damage-associated molecular patterns. Plant J 68: 100–113 [DOI] [PubMed] [Google Scholar]
- Ren D, Liu Y, Yang KY, Han L, Mao G, Glazebrook J, Zhang S (2008) A fungal-responsive MAPK cascade regulates phytoalexin biosynthesis in Arabidopsis. Proc Natl Acad Sci USA 105: 5638–5643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schellingen K, Van Der Straeten D, Vandenbussche F, Prinsen E, Remans T (2014) Cadmium-induced ethylene production and responses in Arabidopsis thaliana rely on ACS2 and ACS6 gene expression. BMC Plant Biol 14: 214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt H, Günther C, Weber M, Spörlein C, Loscher S, Böttcher C, Schobert R, Clemens S (2014) Metabolome analysis of Arabidopsis thaliana roots identifies a key metabolic pathway for iron acquisition. PLoS ONE 9: e102444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schweighofer A, Kazanaviciute V, Scheikl E, Teige M, Doczi R, Hirt H, Schwanninger M, Kant M, Schuurink R, Mauch F, et al. (2007) The PP2C-type phosphatase AP2C1, which negatively regulates MPK4 and MPK6, modulates innate immunity, jasmonic acid, and ethylene levels in Arabidopsis. Plant Cell 19: 2213–2224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su SH, Suarez-Rodriguez MC, Krysan P (2007) Genetic interaction and phenotypic analysis of the Arabidopsis MAP kinase pathway mutations mekk1 and mpk4 suggests signaling pathway complexity. FEBS Lett 581: 3171–3177 [DOI] [PubMed] [Google Scholar]
- Tcherkez G, Gúrard F, Gilard F, Lamothe M, Mauve C, Gout E, Bligny R (2012) Metabolomic characterisation of the functional division of nitrogen metabolism in variegated leaves. Funct Plant Biol 39: 959–967 [DOI] [PubMed] [Google Scholar]
- Tsuda K, Mine A, Bethke G, Igarashi D, Botanga CJ, Tsuda Y, Glazebrook J, Sato M, Katagiri F (2013) Dual regulation of gene expression mediated by extended MAPK activation and salicylic acid contributes to robust innate immunity in Arabidopsis thaliana. PLoS Genet 9: e1004015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Ngwenyama N, Liu Y, Walker JC, Zhang S (2007) Stomatal development and patterning are regulated by environmentally responsive mitogen-activated protein kinases in Arabidopsis. Plant Cell 19: 63–73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu SL, Rahman A, Baskin TI, Kieber JJ (2008) Two leucine-rich repeat receptor kinases mediate signaling, linking cell wall biosynthesis and ACC synthase in Arabidopsis. Plant Cell 20: 3065–3079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo SD, Cho YH, Sheen J (2007) Arabidopsis mesophyll protoplasts: a versatile cell system for transient gene expression analysis. Nat Protoc 2: 1565–1572 [DOI] [PubMed] [Google Scholar]
- Zhang Z, Wu Y, Gao M, Zhang J, Kong Q, Liu Y, Ba H, Zhou J, Zhang Y (2012) Disruption of PAMP-induced MAP kinase cascade by a Pseudomonas syringae effector activates plant immunity mediated by the NB-LRR protein SUMM2. Cell Host Microbe 11: 253–263 [DOI] [PubMed] [Google Scholar]