DCL4 inhibited intercellular VIGS, whereas DCL2 along with DCL2-processed/dependent siRNAs were involved in non-cell-autonomous VIRS in Nicotiana benthamiana.
Abstract
RNA silencing is an innate antiviral mechanism conserved in organisms across kingdoms. Such a cellular defense involves DICER or DICER-LIKEs (DCLs) that process plant virus RNAs into viral small interfering RNAs (vsiRNAs). Plants encode four DCLs that play diverse roles in cell-autonomous intracellular virus-induced RNA silencing (known as VIGS) against viral invasion. VIGS can spread between cells. However, the genetic basis and involvement of vsiRNAs in non-cell-autonomous intercellular VIGS remains poorly understood. Using GFP as a reporter gene together with a suite of DCL RNAi transgenic lines, here we show that despite the well-established activities of DCLs in intracellular VIGS and vsiRNA biogenesis, DCL4 acts to inhibit intercellular VIGS whereas DCL2 is required (likely along with DCL2-processed/dependent vsiRNAs and their precursor RNAs) for efficient intercellular VIGS trafficking from epidermal to adjacent cells. DCL4 imposed an epistatic effect on DCL2 to impede cell-to-cell spread of VIGS. Our results reveal previously unknown functions for DCL2 and DCL4 that may form a dual defensive frontline for intra- and intercellular silencing to double-protect cells from virus infection in Nicotiana benthamiana.
RNA silencing targets endogenous cellular nucleic acids and exogenous invasive pathogenic RNAs or DNAs for homologous RNA-dependent degradation, translation repression, or RNA-directed DNA methylation (RdDM) in eukaryotic organisms (Baulcombe, 2004; Sarkies and Miska, 2014). In plants, RNA silencing forms an innate defense against virus infection (Aliyari and Ding, 2009; Csorba et al., 2015). Such an antiviral mechanism involves DICER-LIKE (DCL) RNase type III enzymes. Most plants encode four DCLs of which DCL1 is responsible for production of microRNA, whereas DCL2, DCL3, and DCL4 are responsible for biogenesis of 22-, 24-, and 21-nucleotide small interfering RNA (siRNA), respectively (Mukherjee et al., 2013). DCL2 and DCL4 possess partially redundant functions in the production of transacting siRNA, but DCL2 acts predominantly to manufacture various-sized secondary siRNAs (Chen et al., 2010; Henderson et al., 2006; Xie et al., 2005). Unlike animal viruses, plant viruses have not yet been found to encode any microRNA or specific site that can be targeted by host cellular microRNAs. However, artificial microRNAs can inhibit plant virus invasion (Qu et al., 2007). In Arabidopsis (Arabidopsis thaliana), DCLs can process plant virus RNAs into viral small interfering RNAs (vsiRNAs) within individual cells. For instance, DCL4 and DCL4-processed 21-nucleotide vsiRNAs are involved in virus-induced RNA silencing (also known as VIGS), a kind of posttranscriptional gene silencing (PTGS; Bouché et al., 2006; Garcia-Ruiz et al., 2010; Qu et al., 2008). DCL2 and its cognate 22-nucleotide vsiRNAs may also affect VIGS in plant cells when DCL4 is absent or defective (Andika et al., 2015; Wang et al., 2011; Zhang et al., 2012). On the other hand, DCL3 and 24-nucleotide vsiRNAs are associated with RdDM and transcriptional gene silencing (TGS) in the protection of plant cells from DNA virus infection (Aregger et al., 2012; Blevins et al., 2006). In Arabidopsis, DCL4 and DCL2 also play hierarchical and redundant roles in intracellular antiviral silencing (Bouché et al., 2006; Garcia-Ruiz et al., 2010; Wang et al., 2011). Recently, a combined activity of DCL2 and DCL3 has been reported to be critical in defending plants from viroid infection (Katsarou et al., 2016). DCL1 can negatively regulate the DCL4-initiated antiviral RNA silencing pathway (Qu et al., 2008). However, the roles of the different DCLs in promoting intercellular VIGS for plant systemic acquired resistance to virus infection are unclear.
In response to virus infection, intracellular VIGS in the initial virus-infected cells triggers intercellular silencing in adjacent cells, which spreads systemically to remote tissues. This is known as non-cell-autonomous VIGS. Non-cell-autonomous VIGS combats incoming viruses and protects recipient cells from further viral invasion (Schwach et al., 2005). In Arabidopsis, spread of the phloem-originating PTGS from companion cells to nearby cells requires DCL4 and DCL4-processed 21-nucleotide siRNA signals (Dunoyer et al., 2005). However, whether 21-nucleotide siRNAs represent the bona fide silencing signals that are transportable among plant cells is highly controversial (Berg, 2016). On the other hand, DCL2 can stimulate transitive PTGS and biogenesis of secondary siRNAs (Mlotshwa et al., 2008). DCL2 can also restore silencing in the Arabidopsis dcl4 mutant that is deficient in cell-to-cell spread of transgene-mediated PTGS (Parent et al., 2015). Moreover, intercellular and systemic PTGS involve many cellular factors including RDR6, which has been shown to be required for efficient cell-to-cell movement of VIGS (Melnyk et al., 2011; Qin et al., 2012; Searle et al., 2010; Smith et al., 2007). Nonetheless, the genetic basis and the requirement of vsiRNAs for cell-to-cell and systemic spread of antiviral VIGS remain to be elucidated.
We previously developed a Turnip Crinkle Virus (TCV)-based local silencing assay to investigate intra- and intercellular VIGS in Nicotiana benthamiana (Qin et al., 2012; Ryabov et al., 2004; Shi et al., 2009; Zhou et al., 2008). TCV belongs to Carmovirus with a single positive-stranded RNA genome (Carrington et al., 1989). It encodes five proteins, namely, the RNA-dependent RNA polymerases P28 and its read-through P88; movement proteins P8 and P9; and coat protein (CP) P38 (Carrington et al., 1989; Hacker et al., 1992; Li et al., 1998). CP is a strong viral suppressor of RNA silencing (VSR; Chattopadhyay et al., 2015; Mérai et al., 2006; Pérez-Cañamás and Hernández, 2015; Qu et al., 2003; Thomas et al., 2003; Zhang et al., 2012). It is also required for cell-to-cell movement of TCV in N. benthamiana (Cohen et al., 2000; Li et al., 2009). TCV/GFP∆CP in which CP is replaced with the 714-nucleotide GFP sequence (dubbed “TcvGFP” hereafter) is movement deficient. This movement-deficient virus is still infectious but the virus remains restricted to the infected cell (Ryabov et al., 2004). Cell-to-cell spread of TCV/GFP∆CP can be complemented by heterologous silencing suppressors (Shi et al., 2009). However, in the absence of the strong VSR CP, the movement-deficient TCV/GFP∆CP can initiate intracellular VIGS that efficiently spreads to neighboring epidermal and mesophyll cells (Qin et al., 2012; Ryabov et al., 2004; Shi et al., 2009; Zhou et al., 2008). Using this intra- and intercellular VIGS assay together with a suite of transgenic DCL RNAi lines, we have examined how the different DCLs affect viral siRNA biogenesis and intra- and intercellular VIGS in N. benthamiana. Our findings lead us to propose a model where intra- and intercellular VIGS comprise two separate components of an integrated viral defense strategy in which DCL2 and DCL4 play different roles.
RESULTS
DCL RNAi Does Not Affect Cell-to-Cell Mobility of TCV/GFP∆CP
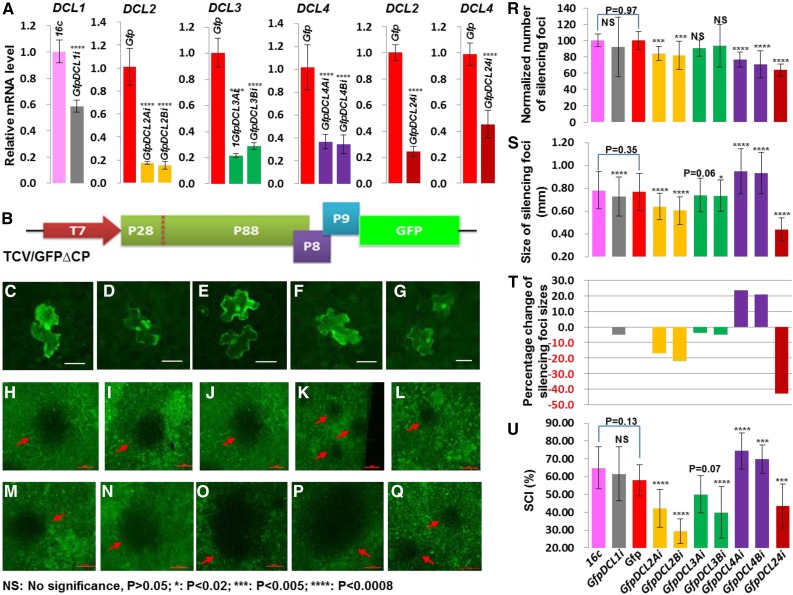
To dissect the genetic requirements and silencing signals involved in non-cell-autonomous intercellular VIGS (Fig. 1) in N. benthamiana (Nb), we utilized a suite of DCL RNAi transgenic Nb lines including DCL1i; DCL2Ai and DCL2Bi; DCL3Ai and DCL3Bi; DCL4Ai and DCL4Bi; and one double RNAi line DCL24i (Supplemental Table S1). We also used GFP transgenic lines 16cGFP; GfpDCL1i; and lines Gfp; GfpDCL2Ai and GfpDCL2Bi; GfpDCL3Ai and GfpDCL3Bi; GfpDCL4Ai and GfpDCL4Bi, which were derived from crosses between 16cGFP and Nb or DCL RNAi lines, respectively, as well as a triple cross line GfpDCL24i (Supplemental Table S1). We performed qRT-PCR assays and revealed that DCL2, DCL3, and DCL4 transcript levels were down-regulated by 60 to 80% in each of the two independent RNAi lines; but only ∼40% reduction was achieved for DCL1 in DCL1i (Fig. 1A). We then analyzed the impact of DCL RNAi on cell-to-cell mobility of TCV/GFP∆CP (Fig. 1, B–G). The upper epidermises of leaves in each DCL RNAi plant at the six-leaf stage were inoculated with TCV/GFP∆CP. As observed under the fluorescent microscope, strong GFP green fluorescence appeared only in single epidermal cells in leaves of the wild-type Nb control (Fig. 1C) and all DCL RNAi plants (Fig. 1, D–G). These results demonstrate that presence of TCV/GFP∆CP was limited to individual virus-infected epidermal cell and that DCL RNAi did not affect the movement-deficiency of TCV/GFP∆CP.
Figure 1.
Different roles of DCLs in the cell-to-cell spread of VIGS. A, Down-regulation of DCL expression by RNAi. Young leaves were collected from DCL RNAi plants at 7 dpi, and the level of DCL RNAs was analyzed by qRT-PCR. B, Schematic of the intracellular RNA silencing trigger TCV/GFP∆CP. The T7 promoter, viral RNA-dependent RNA polymerases (P28 and P88), movement proteins (P8 and P9), and GFP are indicated. C to G, Restricted localization of TCV/GFP∆CP in single epidermal cell of Nb (C), DCL1i (D), DCL2Ai (E), DCL3Bi (F), and DCL4Ai (G) plants. H to Q, Intercellular GFP silencing foci (dark patches indicated by red arrows). Photographs of silencing foci on leaves of 16cGFP (H), GfpDCL1i (I), Gfp (J), GfpDCL2Ai (K), and GfpDCL2Bi (L), GFPDCL3Ai (M), and GfpDCL3Bi (N), GfpDCL4Ai (O), and GfpDCL4Bi (P), and a triple-cross line GfpDCL24i (Q), were taken under a fluorescent microscope at 7 dpi. Bar = 500 μm. R, Normalized number of GFP silencing foci per upper epidermis. Silencing foci were counted at 7 dpi from 3 to 21 different plant leaves inoculated with TCV/GFP∆CP. S and T, Average size (diameters; S) and percentage change (T) of silencing foci. Eighty to 560 silencing foci on different upper epidermises were randomly selected and measured. U, SCI calculated as a percentage of number of silencing foci on lower epidermis out of the number of silencing foci on upper epidermis. Student’s t tests were performed for qRT-PCR and silencing data (mean ± sd) and P values are indicated (asterisks).
DCL4 RNAi Enhances, Whereas DCL2 RNAi Reduces, Cell-to-Cell Spread of VIGS
To test whether intra- and intercellular VIGS is affected by the down-regulation of individual DCL genes, we used GFP as a reporter and mechanically inoculated the movement-deficient TCV/GFP∆CP onto young leaves of 16cGFP (Fig. 1H), GfpDCL1i (Fig. 1I), Gfp (Fig. 1J), GfpDCL2Ai and GfpDCL2Bi (Fig. 1, K and L), GfpDCL3Ai and GfpDCL3Bi (Fig. 1, M and N), and GfpDCL4Ai and GfpDCL4Bi (Fig. 1, O, and P) plants. We then counted the number of GFP silencing foci on both upper and lower epidermises of the inoculated leaves, and measured sizes in diameter of 80 to 560 randomly selected silencing foci on the upper epidermises (Fig. 1, H–U; Supplemental Table S2). We used the number and size of silencing foci as well as the silencing cell-to-cell-spread index (dubbed “SCI” hereafter) to assess the influence of DCL RNAi on intra- and intercellular VIGS (Supplemental Text S1). Compared to 16cGFP and Gfp controls (Fig. 1, H and J), DCL2 RNAi caused 17% to 22% decrease in the average sizes of silencing foci (Fig. 1, K, L, S, and T; Supplemental Table S2). SCI was reduced from ∼58% in Gfp plants to 29% to 42% in GfpDCL2Ai and GfpDCL2Bi plants (Fig. 1U; Supplemental Table S2). DCL2 RNAi also caused a reduction in the number of silencing foci per leaf (Fig. 1R; Supplemental Table S2). RNAi knockdown of DCL1 or DCL3 did not affect the number of silencing foci and only reduced cell-to-cell movement of VIGS to a small extent, as evidenced by 3 to 5% decreases in silencing foci sizes and/or some reductions in SCI (Fig. 1, H–J, M, N, and R–U; Supplemental Table S2). This suggests that DCL3 and/or DCL1 may not contribute significantly to intercellular VIGS. However, a possible role cannot be ruled out completely due to discrepancy between the two GfpDCL3i lines and because we only have data from a single GfpDCL1i line. As with DCL2 RNAi, DCL4 RNAi caused a reduction in the number of silencing foci in GfpDCL4Ai and GfpDCL4Bi plants (Fig. 1R; Supplemental Table S2). This is consistent with the predominant role that DCL4 plays in intracellular VIGS. To our surprise, the average sizes of silencing foci increased by >20% (Fig. 1, O, P, S, and T; Supplemental Table S2). The SCI also raised from ∼58% in the Gfp controls to 70 to 75% in the two GfpDCL4 RNAi lines (Fig. 1U; Supplemental Table S2). These results demonstrate that DCL4 RNAi reduced intracellular silencing, but enhanced intercellular spread of VIGS. Taken together, our findings show that DCL4 RNAi enhances but DCL2 RNAi reduces cell-to-cell spread of VIGS in Nb.
DCL4 Interferes with DCL2 to Control Intercellular VIGS
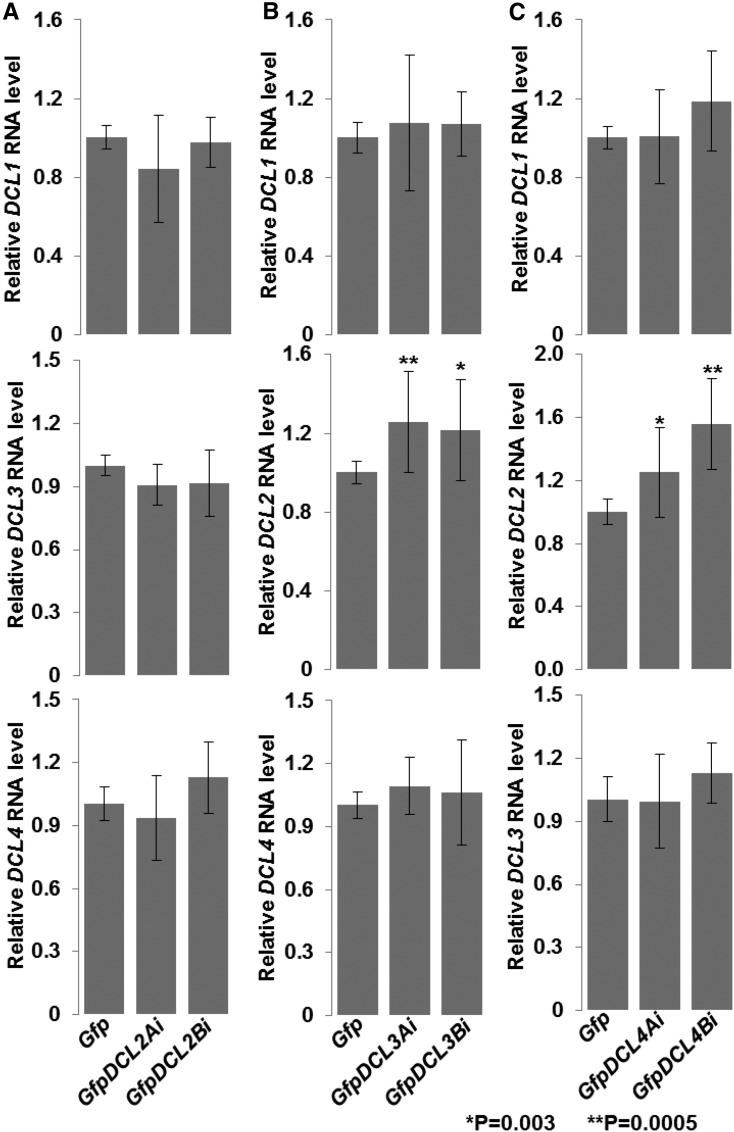
To investigate whether DCL4 and DCL2 would affect each other to influence cell-to-cell spread of VIGS in Nb, we inoculated the triple-cross GfpDCL24i plant with TCV/GFP∆CP. We found a marked reduction in the number of GFP silencing foci (Fig. 1R; Supplemental Table S2), consistent with the reduction in the number of silencing foci observed in GfpDCL2 RNAi and GfpDCL4 RNAi lines. However, in the triple-cross plants, the average sizes of silencing foci decreased by more than 40%, and SCI also fell from 58 to 44% when compared to the Gfp control (Fig. 1, Q, S, and T; Supplemental Table S2). These results demonstrate that simultaneous RNAi of DCL2 and DCL4 reduced both intra- and intercellular VIGS, similar to what is seen in GfpDCL2 RNAi lines, but to a greater extent. The inhibition of intercellular spread of VIGS in the triple-cross GfpDCL24i line is opposite to the increase in intercellular VIGS seen in the GfpDCL4 RNAi lines. This implies that DCL4 imposed an epistatic effect on DCL2 to affect intercellular VIGS. This conclusion is supported by data from qRT-PCR assays (Fig. 2). DCL2 RNAi had no obvious impact on mRNA levels of DCL1, DCL3, and DCL4 (Fig. 2A). However, DCL4 RNAi led to a 20 to 40% increase in DCL2 expression but had no substantial influence on the transcript levels of DCL1 or DCL3; DCL3 RNAi also enhanced the level of DCL2 transcripts, but did not affect expression of DCL1 or DCL4 (Fig. 2, B and C). Together with the specific RNAi effects on each DCL (Supplemental Data Set S1), these data reveal that DCL4 is involved in a negative regulation of DCL2 expression and as a consequence affecting the intercellular spread of VIGS in Nb.
Figure 2.
Regulation of DCL2 expression by DCL3 and DCL4. A to C, Effects of RNAi of DCL2 (A), DCL3 (B), and DCL4 (C) on DCL gene expression. Young leaf tissues were collected at 6 to 8 leaf stage from four different plants of each transgenic line as indicated. RNA transcripts were analyzed by qRT-PCR. Four technical replicates for qRT-PCR assays were performed on each cDNA of four biological duplicates (n = 4; leaf tissues from four different transgenic plants). Student’s t tests were performed for data (mean ± sd) and P values are indicated (asterisks). DCL2i does not affect expression of DCL1, DCL3, or DCL4 (A). However, DCL3i (B) or DCL4i (C) resulted in increased mRNA levels of DCL2.
DCLs Play Differential Roles in vsiRNA Biogenesis
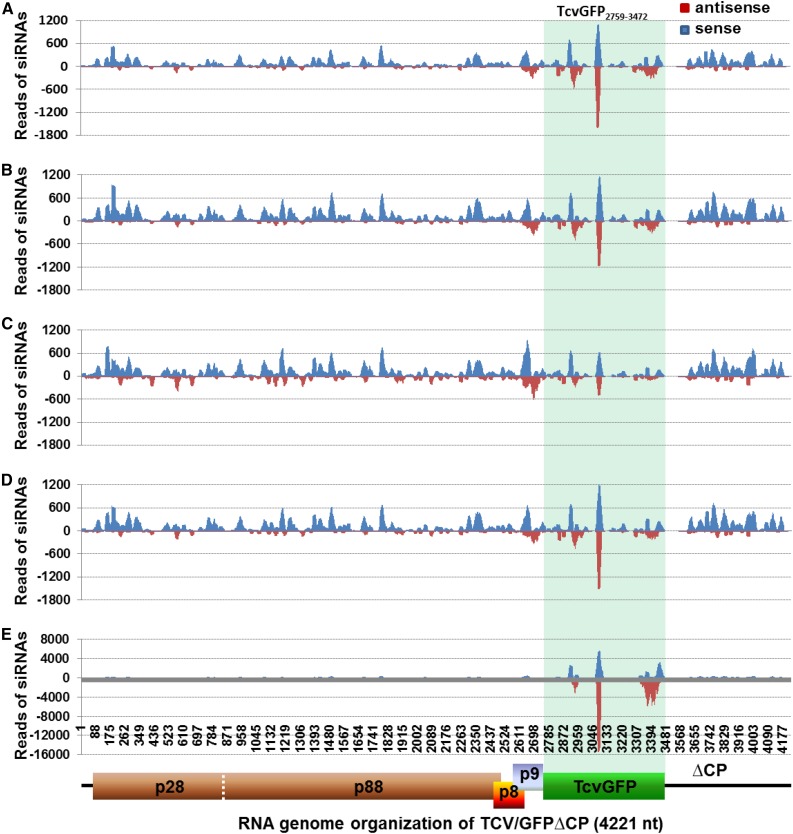
To further understand how DCLs contribute to intra- and intercellular VIGS, we performed next-generation sequencing of sRNA libraries for mock- or TCV/GFP∆CP-inoculated Gfp, GfpDCL1i, GfpDCL2Ai, GfpDCL3Bi, and GfpDCL4Ai (Supplemental Text S2; Supplemental Data Sets S1–S3; Supplemental Fig. S1). We then mapped vsiRNAs and TcvGFP siRNAs onto the sequence of TCV/GFP∆CP (Fig. 3; Supplemental Fig. S2) and TCV∆CP (Supplemental Figs. S3 and S4). We found abundant vsiRNAs in TCV/GFP∆CP-inoculated RNAi lines (Fig. 3, A–E; Supplemental Fig. S3, A–E), compared to their mock controls (Supplemental Figs. S2, A–E, and S4, A–E; Supplemental Table S3). This is consistent with induction of effective VIGS in these plants (Fig. 1, H–U; Supplemental Table S2). More vsiRNAs were recorded in GfpDCL1i, GfpDCL2Ai, and GfpDCL3Bi plants (Fig. 3, B–D; Supplemental Fig. S3, B–D) than in Gfp controls (Fig. 3A; Supplemental Fig. S3A; Supplemental Table S3). However, the reads of vsiRNAs, particularly in the sense polarity, decreased in GfpDCL4Ai plants (Fig. 3E; Supplemental Fig. S3E; Supplemental Table S3) despite a marked increase in the overall number of siRNAs (vsiRNAs and TcvGFP-siRNAs) mapped to TCV/GFP∆CP (Supplemental Table S3). These results are consistent with the reduced level of recombinant viral RNAs in TCV/GFP∆CP-inoculated DCL RNAi plants, compared to the non-RNAi controls (Supplemental Fig. S5). On the other hand, the distribution of vsiRNAs across TCV/GFP∆CP (Fig. 3, A–E) or TCV∆CP (Supplemental Fig. S3, A–E) was identical among all virus-inoculated RNAi and control plants. Taken together, these data demonstrate that DCL4 is able to efficiently target viral RNAs for the production of vsiRNAs during cell-autonomous VIGS. Our results also reveal that DCL2 is required for cell-to-cell spread of VIGS, and DCL2 could target viral RNA and TcvGFP mRNA for degradation in Nb.
Figure 3.
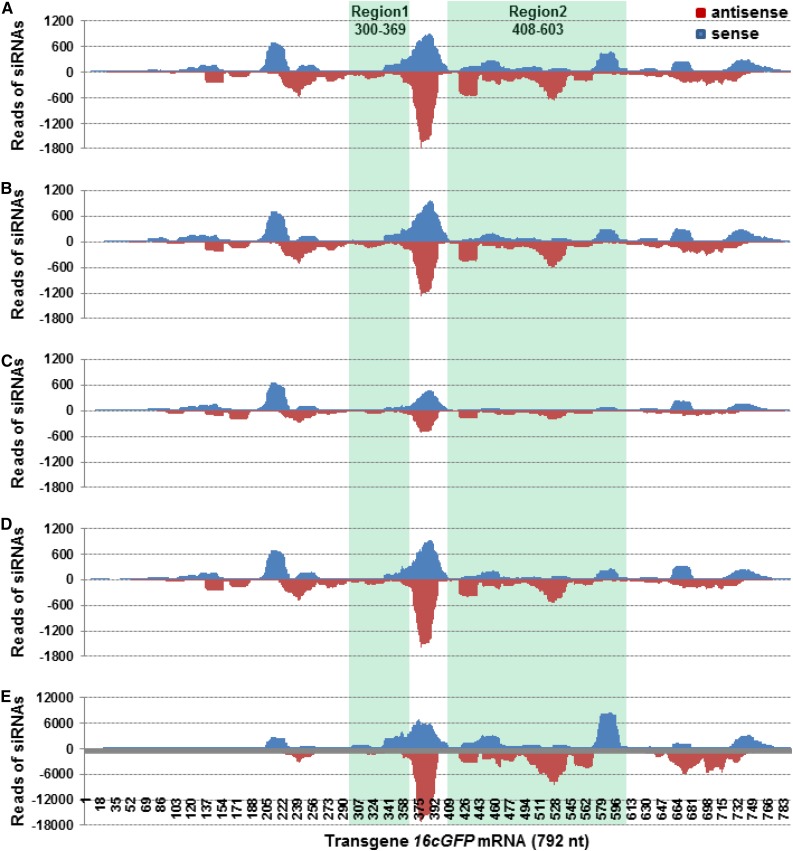
Distribution of 20- to 25-nucleotide vsiRNAs and siRNATcvGFP across the TCV/GFP∆CP RNA. A, Gfp. B, GfpDCL1i. C, GfpDCL2Ai. D, GfpDCL3Bi. E, GfpDCL4Ai. The sRNA libraries were generated from sRNA samples extracted from TCV/GFP∆CP-inoculated leaves. Blue and red bars represent siRNAs aligned to the sense (positive) and antisense (negative) strands of TCV/GFP∆CP viral RNA and TcvGFP mRNA (highlighted), respectively. The TCV/GFP∆CP genome organization is indicated.
Antagonistic Influences of DCL4 and DCL2 on Accumulation of siRNAs Associated with Intercellular VIGS
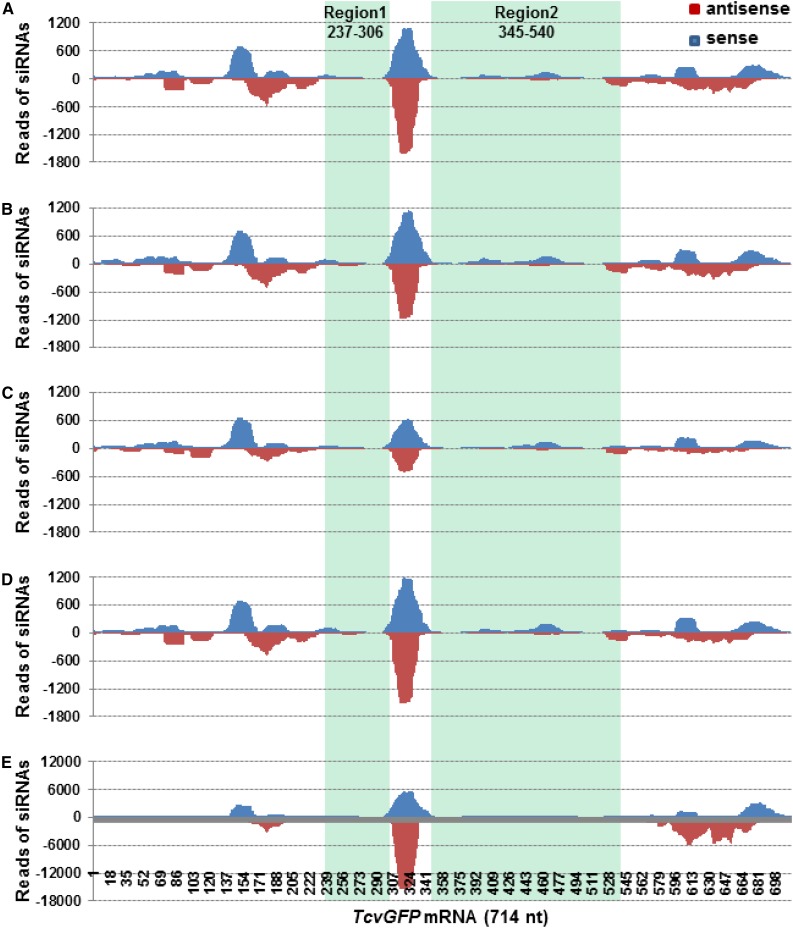
In contrast to the situation with vsiRNAs, GfpDCL RNAi lines differed in the generation of TcvGFP or transgene 16cGFP siRNAs (dubbed “siRNATcvGFP” and “siRNA16cGFP”) that are associated with intra- and intercellular VIGS. Note that the 714-nucleotide TcvGFP (Ryabov et al., 2004) and 792-nucleotide 16cGFP (Haseloff et al., 1997; Ruiz et al., 1998) mRNAs are not identical. Sequences between nucleotides 237 to 306 and 345 to 540 in TcvGFP (designated TcvGFP237-306 and TcvGFP345-540) differ from the corresponding regions 300 to 369 and 408 to 603 in 16cGFP (designated 16cGFP300-369 and 16cGFP408-603; Supplemental Fig. S6). Compared to the Gfp control and GfpDCL1i and GfpDCL3Bi plants, the levels of siRNATcvGFP and siRNA16cGFP were reduced in GfpDCL2Ai, but significantly increased in GfpDCL4Ai (Supplemental Table S3). We then mapped the siRNAs onto TcvGFP (Fig. 4; Supplemental Fig. S7) and 16cGFP mRNA (Fig. 5; Supplemental Fig. S8). The distribution of sense and antisense GFP siRNAs to regions that are identical in TcvGFP and 16cGFP was essentially the same in Gfp and in each of the DCL RNAi lines, but the levels of siRNATcvGFP and siRNA16cGFP were lower in GfpDCL2Ai, and much higher in GfpDCL4Ai, compared to Gfp, GfpDCL1i, and GfpDCL3Bi (Figs. 4, A–E, and 5, A–E; Supplemental Table S3). Moreover, in GfpDCL4Ai the level of siRNA16cGFP (2.5 million reads) was approximately double compared to the abundance of siRNATcvGFP (1.28 million reads; Supplemental Table S3). Such substantial differences between siRNATcvGFP and siRNA16cGFP levels suggest that the transgene 16cGFP mRNA was targeted and diced by intra- and intercellular VIGS to a greater extent than TcvGFP transcripts. In contrast, different profiles were observed for siRNATcvVGFP and siRNA16cGFP corresponding to the two less-similar regions (Region 1: TcvGFP237-306 and 16cGFP300-369; Region 2: TcvGFP345-540 and 16cGFP408-603; Figs. 4, A–E, and 5, A–E). TcvGFP237-306 and TcvGFP345-540 siRNAs were of low abundance and generally of sense polarity in the control and all RNAi lines (Fig. 4, A–E). However, higher levels of 16cGFP300-369 and 16cGFP408-603 siRNAs were observed, a significant amount of which was antisense. As with the other GFP siRNAs, the levels of 16cGFP300-369 and 16cGFP408-603 siRNAs were much higher in GfpDCL4Ai and lower in GfpDCL2Ai, compared to Gfp, GfpDCL1i, and GfpDCL3Bi (Fig. 5, A–E).
Figure 4.
Distribution of 20- to 25-nucleotide GFP siRNAs across the 714-nucleotide TcvGFP mRNA. A, Gfp. B, GfpDCL1i. C, GfpDCL2Ai. D, GfpDCL3Bi. E, GfpDCL4Ai. The sRNA libraries were generated from sRNA samples extracted from TCV/GFP∆CP-inoculated leaves. Blue and red bars represent siRNAs aligned to the sense (positive) and antisense (negative) strands of TcvGFP mRNA, respectively. The two regions (Region 1 and Region 2) having less sequence similarity with that of the transgene 16cGFP mRNA (see Fig. 5), as well as nucleotide coordinates, are indicated.
Figure 5.
Distribution of 20- to 25-nucleotide GFP siRNAs across the 792-nucleotide transgene 16cGFP mRNA. A, Gfp. B, GfpDCL1i. C, GfpDCL2Ai. D, GfpDCL3Bi. E, GfpDCL4Ai. The sRNA libraries were generated from sRNA samples extracted from TCV/GFP∆CP-inoculated leaves. Blue and red bars represent siRNAs aligned to the sense (positive) and antisense (negative) strands of 16cGFP mRNA, respectively. The two regions (Region 1 and Region 2) having less sequence similarity with that of the transgene TcvGFP mRNA (see Fig. 4) as well as nucleotide coordinates are indicated.
These results demonstrate that DCL4 and DCL2 antagonistically affected the accumulation of siRNAs associated with intercellular VIGS. The reduction of siRNATcvGFP and siRNA16cGFP in GfpDCL2Ai or the massive accumulation of these siRNAs in GfpDCL4Ai is likely to be due to the respective loss- or gain-of-function of DCL2-dependent production of primary or secondary siRNAs in these RNAi lines.
Potential DCL2-Processed/Dependent siRNA Signals for Intercellular VIGS
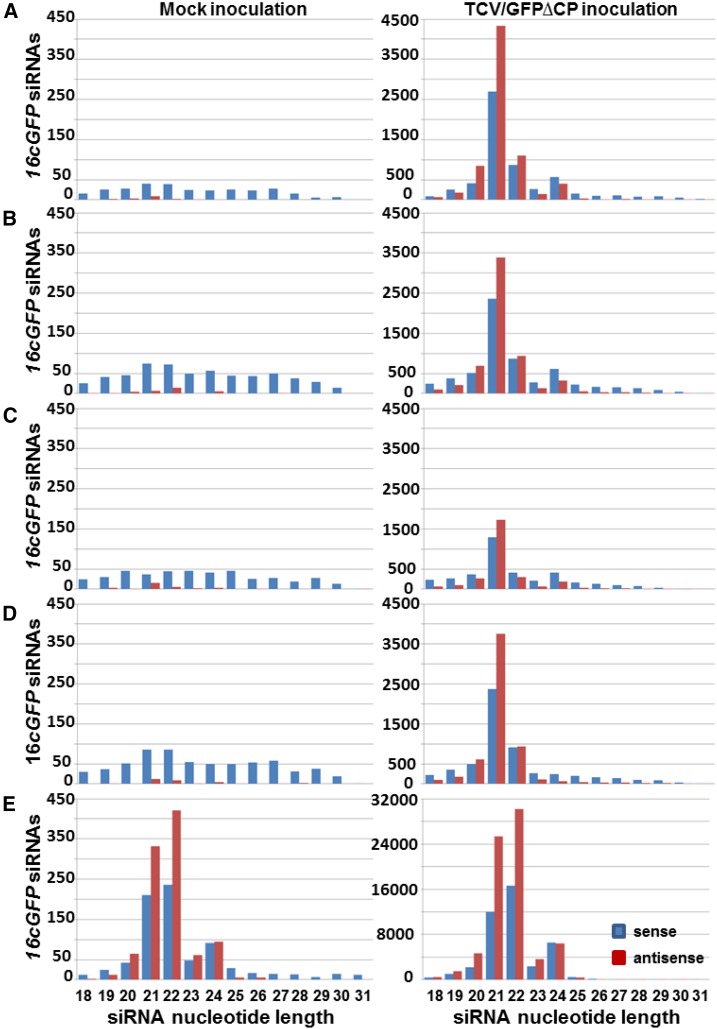
In Nb, the DCL2-processed siRNAs (Figs. 4C and 5C) and/or DCL2-dependent siRNAs (produced by DCL2-activated pathways; Figs. 4E and 5E) are likely to be involved in the intercellular spread of epidermal cell-originating VIGS. Consistent with this idea, an elevated level of 22-nucleotide siRNAs was only found in TCV/GFP∆CP-inoculated GfpDCL4Ai and GfpDCL4Bi plants that exhibited increased intercellular VIGS (Fig. 1; Supplemental Fig. S9). To examine this correlation further, we analyzed the size profiles of sense and antisense siRNA16cGFP (Fig. 6). The 21-, 22-, and 24-nucleotide siRNA16cGFP displayed similar size-profiles between Gfp and GfpDCL1i (Fig. 6, A and B, right panel). There was an obvious reduction in 24-nucleotide siRNA16cGFP in GfpDCL3Bi (Fig. 6D, right panel). However, among Gfp, GfpDCL1i, and GfpDCL3Bi, the 21-nucleotide siRNA16cGFP was always dominant whereas the levels of 22-nucleotide siRNAs remained similar (Fig. 6, A, B, and D, right panel; Supplemental Table S4). These findings further indicate that DCL1, DCL3, and DCL3-processed 24-nucleotide siRNAs may not significantly contribute to cell-to-cell spread of VIGS, consistent with the results of the local silencing assays (Fig. 1, H–U).
Figure 6.
Size profiles of transgene GFP siRNA16cGFP. A, Gfp. B, GfpDCL1i. C, GfpDCL2Ai. D, GfpDCL3Bi. E, GfpDCL4Ai. The sRNA libraries were generated from sRNA samples extracted from leaves with mock (left) or TCV/GFP∆CP (right) inoculation. Blue and red bars represent siRNAs aligned to the sense and antisense strands of the transgene 16cGFP mRNA, respectively.
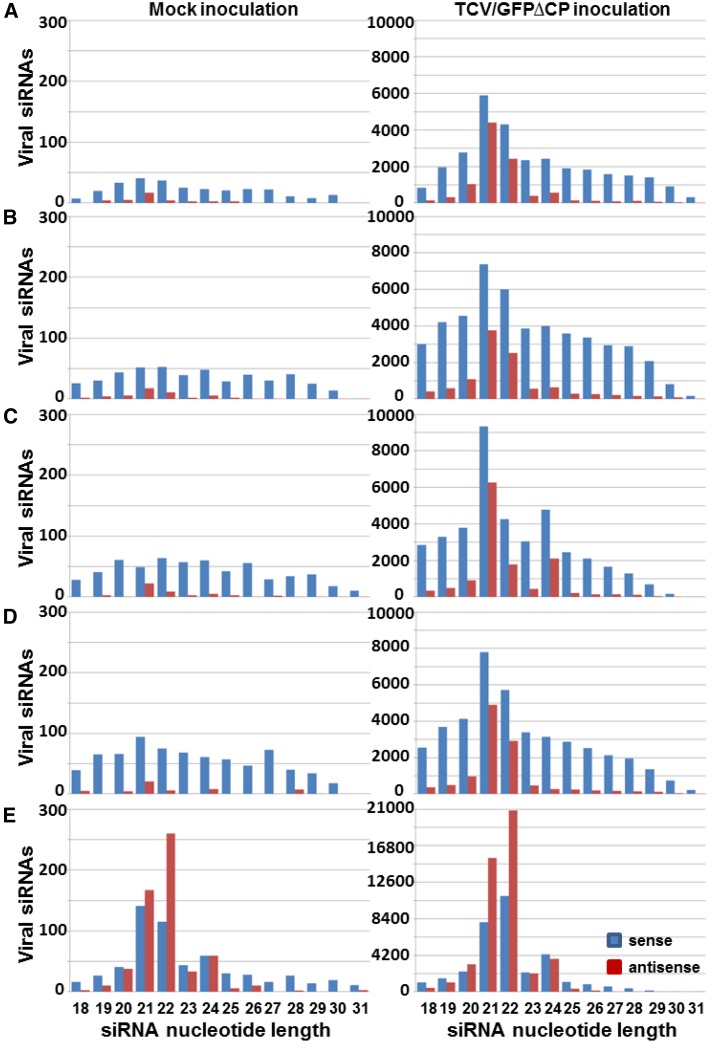
RNAi of DCL2 or DCL4 imposed contrasting effects on the accumulation of 21-, 22-, and 24-nucleotide siRNA16cGFP. Compared to Gfp, GfpDCL1i, and GfpDCL3Bi (Fig. 6, A, B, and D, right panel), the absolute reads of 21-, 22-, and 24-nucleotide siRNA16cGFP were reduced in GfpDCL2Ai (Fig. 6C, right panel), and were markedly increased in GfpDCL4Ai (Fig. 6E, right panel). Nonetheless, the percentage of the 22-nucleotide siRNA16cGFP decreased in GfpDCL2Ai, whereas the relative abundance of 21-nucleotide siRNA16cGFP was reduced in GfpDCL4Ai (Supplemental Table S4). These are in accordance with the respective roles of DCL4 and DCL2 in 21- and 22-nucleotide siRNA biosynthesis. The levels of DCL2-processed 22-nucleotide siRNA16cGFP and DCL2-dependent siRNA16cGFP were particularly low in GfpDCL2Ai (Fig. 6C, right panel), but copious in GfpDCL4Ai (Fig. 5E, right panel; Supplemental Table S4), consistent with the observed attenuation or enhancement of intercellular VIGS in the RNAi plants, respectively (Fig. 1). We also analyzed the size profiles of sense and antisense vsiRNA (Fig. 7). Distributions of 18-to 31-nucleotide vsiRNAs were not obviously altered among Gfp, GfpDCL1i, GfpDCL2Ai, and GfpDCL3Bi (Fig. 7, A–D, right panel). In these RNAi lines, the majority of vsiRNAs were 21 nucleotides in length (Fig. 7; Supplemental Table S4). However, in GfpDCL4Ai, vsiRNAs shifted their sizes largely to 22 nucleotides, although there were also marked increases in 21- and 24-nucleotide vsiRNAs (Fig. 7E, right panel). Notably, there was an approximate 10% reduction of 22-nucleotide vsiRNAs in GfpDCL2Ai compared to the Gfp control (Supplemental Table S4). Taken together, our data show that DCL4 plays a major, and DCL2 a minor, role in producing 21- or 22-nucleotide vsiRNAs for intracellular VIGS, whereas DCL2 is required to generate and perceive DCL2-processed/dependent mobile signals for intercellular VIGS. These conclusions are further supported by similar results that were generated from six extra sRNA libraries for the Gfp control and two different RNAi lines GfpDCL2Bi and GfpDCL4Bi (Supplemental Fig. S10, A–E).
Figure 7.
Size profiles of TCV/GFP∆CP viral siRNAs. A, Gfp. B, GfpDCL1i. C, GfpDCL2Ai. D, GfpDCL3Bi. E, GfpDCL4Ai. The sRNA libraries were generated from sRNA samples extracted from leaves with mock (left) or TCV/GFP∆CP (right) inoculation. Blue and red bars represent siRNAs aligned to the sense and antisense strands of TCV/GFP∆CP RNA, respectively.
DISCUSSION
In plants, DCLs play diverse roles in sense- and hairpin-RNA-mediated PTGS and TGS (Parent et al., 2015; Mlotshwa et al., 2008; Xie et al., 2004; 2005). DCL4, DCL2, and their cognate 21- and 22-nucleotide vsiRNAs are involved in cell-autonomous VIGS but their antiviral functioning roles are debated (Bouché et al., 2006; Fusaro et al., 2006; Garcia-Ruiz et al., 2010; Qu et al., 2008).
In this study, we reveal several interesting findings, as follows: (1) RNAi of the four DCL genes does not affect cell-to-cell movement deficiency of the immobile virus TCV/GFP∆CP (Fig. 1). This is consistent with our previous report that compromising of silencing machinery alone was not sufficient to promote virus movement (Shi et al., 2009). These findings ensure that any intercellular VIGS that we observe in our assays do not result from cell-to-cell movement of the recombinant viral RNA, and also argue against the idea that an altered ability to establish intra- and intercellular VIGS in these DCL RNAi lines may enable TCV-GFPΔCP to move locally or systemically more than in wild-type Nb plants.
(2) DCL4 inhibited non-cell-autonomous intercellular VIGS, whereas it acted as a major trigger for intracellular VIGS (Fig. 3), consistent with its critical role in cell-autonomous silencing and vsiRNA biogenesis (Wang et al., 2011; Xie et al., 2005). Our findings are also in agreement with previous reports that dcl4 mutations enhance transitivity of cell-autonomous PTGS and can rescue phloem-originating PTGS in Arabidopsis (Mlotshwa et al., 2008; Parent et al., 2015). The fact that DCL4 attenuates intercellular VIGS implies that DCL4-processed 21-nucleotide vsiRNAs are unlikely to be involved in cell-to-cell spread of VIGS in Nb.
(3) DCL2, probably along with DCL2-processed/dependent siRNAs and their precursor RNAs, is involved in intercellular VIGS. DCL2 was also able to target and degrade viral RNAs in plant cells but this activity was largely redundant when functional DCL4 was present (Fig. 3). These findings suggest that DCL2 could influence intracellular VIGS in Nb, although DCL2 is thought to be dispensable for antiviral silencing in Arabidopsis (Wang et al., 2011). Neither DCL1 nor DCL3 affected vsiRNA production or intra- and intercellular VIGS. Intriguingly, DCL2 played a key role in spreading VIGS from individual epidermal cells to adjacent epidermal and mesophyll cells—a formerly unidentified function in silencing-based antiviral defense.
(4) Silencing machinery degraded TcvGFP mRNA and the resultant siRNATcvGFP targeted identical regions in the transgene 16cGFP mRNA and generated siRNA16cGFP for intracellular VIGS in TCV/GFP∆CP-infected epidermal cells (Figs. 4 to 7). Such siRNATcvGFP and siRNA16cGFP in sense and antisense polarities then led to biogenesis of siRNAs associated with different parts across the 16cGFP and TcvGFP RNA sequences for intra- and intercellular VIGS. Our results thus imply that initial signals for intercellular VIGS might consist of sense and antisense siRNATcvGFP and siRNA16cGFP. Production of such signals in incipient epidermal cells (i.e. the TCV/GFP∆CP-infected cells) and subsequent induction of 16cGFP silencing in neighboring recipient cells (i.e. TCV/GFP∆CP noninfected cells) were influenced positively by DCL2, but negatively by DCL4 (Figs. 1 and 2). However, in contrast to complete loss-of-function genetic mutants, RNAi lines are partial loss of function. It is also possible that TCV/GFP∆CP infection could alter the expression of the DCL genes targeted by RNAi. Sequenced small RNAs were from all of the cells in the inoculated leaves, including the inducing and recipient cells. Considering these factors, it remains possible that long dsRNA precursors of DCL2 (or DCL4) could move between cells or long distance for induction of non-cell-autonomous VIGS.
Collectively, our results suggest that DCL4 and DCL2 play major but distinct roles in intra-/intercellular VIGS. Involvement of DCL2 and DCL2-processed/dependent siRNAs as well as their precursor RNAs in intercellular VIGS is consistent with the fact that DCL2 and DCL2-processed 22-nucleotide siRNA can effectively trigger biogenesis of secondary siRNAs in plants (Chen et al., 2010), and restore intercellular PTGS induced by sense- and hairpin-transgene RNAs in the Arabidopsis dcl4 mutant (Mlotshwa et al., 2008; Parent et al., 2015). It should be pointed out that silencing spread in our system is more complex than other systems because it is dependent on the expression of both p8 and p9 proteins of TCV (Zhou et al., 2008). Therefore, we are cautious to expand our findings to other examples of cell-to-cell spread of RNA silencing, such as the controversial Arabidopsis model in which, DCL4 and DCL4-processed 21-nucleotide siRNAs are thought to be directly involved in short-range cell-to-cell spread of phloem-originating PTGS.
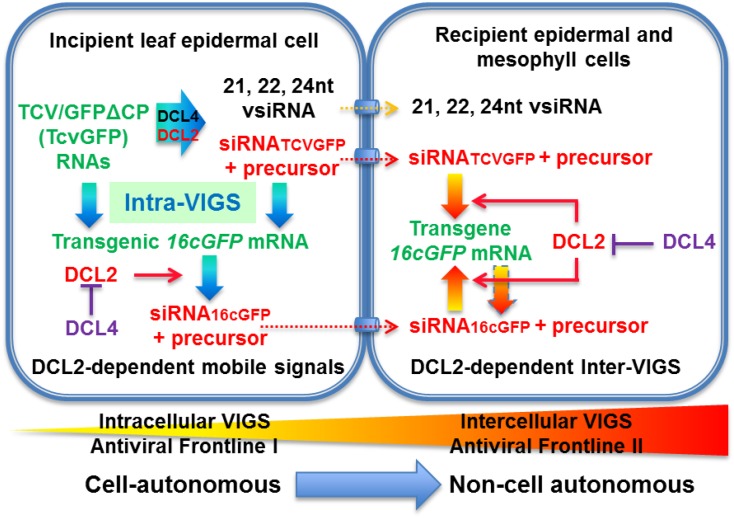
Nevertheless, our findings support a hypothesis that DCL4 is essential for cell-autonomous intracellular VIGS, but negatively regulates intercellular VIGS. This is likely to be achieved via DCL4-mediated epistatic interference over DCL2 because the latter is essential to promote cell-to-cell spread of VIGS. Indeed DCL4 can suppress the expression of DCL2 in Nb (Fig. 2). DCL2 is also required to generate and perceive mobile signals for systemic PTGS whereas DCL4 inhibits systemic PTGS (unpublished data). To put these findings in the context of RNA silencing-based defense, we propose two separate components of an integrated viral defense strategy in which DCL2 and DCL4 play different roles (Fig. 8). DCL4, the primary defender in the cell-autonomous intracellular VIGS, attacks viruses within the initially infected cells. Simultaneously it also inhibits non-cell-autonomous silencing. Thus, if this intracellular VIGS frontline in incipient cells was broken, for example through inhibiting DCL4 activity by VSR such as P1/HC-Pro and P38 (Csorba et al., 2015; Mlotshwa et al., 2008), intercellular VIGS would then be activated efficiently spreading to nearby recipient cells to form a second frontline against the virus. Non-cell-autonomous intercellular VIGS relies upon functional DCL2 and DCL2-processed/dependent siRNAs and their precursor RNAs. In this scenario, DCL2 is required to trigger the intercellular VIGS frontline and defend recipient cells from further virus invasion. DCL2 may also contribute to cell-autonomous VIGS, but DCL2 can only fulfill this activity when DCL4 is absent or dysfunctional. This explains why an increased intercellular VIGS was observed in DCL4 RNAi plants, but a decreased non-cell-autonomous VIGS was observed in DCL2 RNAi plants. Such a local dual-defense strategy would be more difficult for the virus to break down and may provide plants with an evolutionary advantage in their defense against viral pathogens (Supplemental Text S3).
Figure 8.
Cell- and non-cell autonomous VIGS in N. benthamiana. In incipient leaf epidermal cells (i.e. individual cells initially infected by TCV/GFP∆CP), DCL4 plays a critical role in biogenesis of vsiRNAs, siRNATCVGFP and transgene siRNA16cGFP. These siRNAs are associated with cell-autonomous intracellular VIGS to inhibit local virus infection. DCL4-processed siRNAs are unlikely involved in spread of VIGS from leaf epidermal cell to adjacent cells because DCL4 inhibits intercellular VIGS. In the incipient cell, DCL2 can also target and dice viral RNAs, TcvGFP and 16cGFP mRNA into siRNAs, but this activity is largely blocked by DCL4 (T sign). In contrast, the key functionality of DCL2 is to trigger efficient intercellular VIGS. This is likely achieved through its activities to produce DCL2-processed/dependent siRNAs (and/or their precursor long RNAs, highlighted red) in incipient cells and to perceive these mobile signals for non-cell-autonomous intercellular VIGS in recipient epidermal and mesophyll cells. Neither DCL1 nor DCL3 affects vsiRNA production or intra- and intercellular VIGS. Thus DCL4 and DCL2 play major but distinct roles in cell- and non-cell-autonomous VIGS that form a dual antiviral frontline in incipient and recipient cells. DCL4, the primary defender for the cell-autonomous intracellular VIGS, can attack viruses within the initially infected cells. However, if viruses break through this defense frontline, non-cell-autonomous intracellular VIGS can efficiently spread to nearby recipient cells. This is due to loss of the negative control of intercellular VIGS mediated by DCL4. Intercellular VIGS is dependent upon functional DCL2 and DCL2-processed/dependent siRNAs (and/or their precursor long RNAs), but it is negatively controlled by DCL4. RNAi of DCL4 results in fully functional DCL2 that enhances cell-to-cell spread of VIGS. The intercellular VIGS can then defend recipient cells from further virus infection. Such a dual-defense strategy can compensate each other to give host cells evolutionary advantage to battle against virus infection. This model is relevant to virus-VIGS interaction at the intra-/intercellular level, rather than to systemic virus infection. The potential spread of DCL2-processed/dependent siRNAs (and their precursor long RNAs, highlighted in red) to move from the incipient to recipient cell through plasmodesmata is indicated with dashed arrows and cylinder signs. Inter-VIGS, intercellular VIGS; intra-VIGS, intracellular VIGS.
MATERIALS AND METHODS
Plant Materials and Growth Conditions
Wild-type Nicotiana benthamiana and transgenic lines (Supplemental Table S1) were grown and maintained in insect-free growth rooms at 25°C with supplementary lighting to give a 16-h photoperiod.
Plasmid Constructs, Virus Inoculation, and Microscopy
Construction of TCV/GFPΔCP was described by Ryabov et al. (2004). The full-length GFP sequence was PCR-amplified using TCV/GFPΔCP as a DNA template and cloned into pMD18-T (Takara) to produce a pT7.GFP construct from which GFP RNA transcripts were produced by in vitro transcription using T7 RNA polymerase. Primers used for making this construct are listed in Supplemental Table S5. TCV/GFPΔCP RNA was generated by in vitro transcription and used to mechanically inoculate Nb, DCL RNAi, 16cGFP, Gfp, and GfpDCL RNAi plants as described by Ryabov et al. (2004). Inoculated leaves were collected and visualized under an Axiphot microscope (Carl Zeiss) as described by Ryabov et al. (2004).
Intra- and Intercellular VIGS Assays
We used a cell-specific, silencing suppression-free and movement-deficient Turnip Crinkle Virus (TCV/GFP∆CP)-based system to induce intracellular VIGS in a single epidermal cell, from which silencing spreads to form visible silencing foci covering 100 to 300 epidermal cells, equivalent to a circular area with a radius of 6 to 10 epidermal cells, on the leaf epidermis of transgenic 16cGFP plants (Qin et al., 2012; Ryabov et al., 2004; Zhou et al., 2008). Of important note, the precise location of a single epidermal cell that was initially infected with the movement-defective TCV/GFP∆CP could not be located before development of a visible silencing focus from the infected cell. Due to the compact TCV genome organization and viral gene expression strategy (Carrington et al., 1989; Cohen et al., 2000), it would be almost impossible to clone a second reporter gene, in addition to GFP, into TCV/GFP∆CP as an extra marker for measuring the initial infection of individual epidermal cell. Nevertheless, visible GFP silencing foci are a good indicator for induction and spread of TCV/GFP∆CP-induced intracellular VIGS. Upon mechanic inoculation, their appearance is a gradual process starting from the individual cell on the upper epidermises, which is initially infected by TCV/GFP∆CP. Intracellular GFP silencing is induced by TCV/GFP∆CP in the single epidermal cell, and subsequently moves horizontally and vertically to neighboring upper epidermal, mesophyll, and lower epidermal cells in a three-dimensional manner, i.e. occurrence of intercellular VIGS (Qin et al., 2012; Zhou et al., 2008).
To perform intra- and intercellular VIGS assays, a single young leaf (second from top) of each of four to six seedlings (six-leaf stage) of 16cGFP, Gfp, GfpDCL1i, GfpDCL2Ai, GfpDCL2Bi, GfpDCL3Ai, GfpDCL3Bi, GfpDCL4Ai, or GfpDCL4Bi lines were mechanically inoculated with an equal amount of RNA transcripts produced by in vitro transcription from 2.5 μg PacI-linearized TCV/GFPΔCP plasmid DNA, as described by Ryabov et al. (2004). Induction and spread of GFP silencing was routinely examined under long-wavelength UV light and recorded photographically using a D7000 Digital Camera (Nikon). Regions of leaf lamina in which silencing of GFP mRNA occurred show red chlorophyll fluorescence, whereas tissues expressing GFP show green fluorescence under long-wavelength UV light. Numbers and sizes of GFP silencing foci (dark patches) were counted, measured, and photographed under an Axiophot microscope (Carl Zeiss) using settings to visualize GFP green fluorescence, as described by Qin et al. (2012). Number of silencing foci on an individual leaf was normalized against the average number of silencing foci per leaf of the control plants (i.e. 16cGFP as control for GfpDCL1i, and Gfp as control for GfpDCL2Ai, GfpDCL2Bi, GfpDCL3Ai, GfpbDCL3Bi, GfpDCL4Ai, GfpDCL4Bi, and GfpDCL24i) to minimize disparities that could be caused by experimental variations such as leaf sizes among different plants and freshly generated inoculum RNA transcripts used in different experiments. Silencing cell-to-cell-spread index (SCI) was calculated as a percentage between the numbers of silencing foci counted on lower and upper (inoculated side) epidermises. Intra- and intercellular VIGS assays were performed for each of the transgenic lines in at least two separate experiments.
RNA Extraction and Northern Hybridization
For quantitative real-time PCR (qRT-PCR), total RNAs were extracted from leaf tissues using the RNAprep Pure Plant Kit (Tiangen) as recommended by the manufacturer. For northern blot, total RNAs were extracted from leaf tissues with Trizol reagent (Invitrogen) as recommended by the manufacturer. To analyze siRNAs, low-molecular-mass small RNAs were enriched from total RNA as described by Hamilton and Baulcombe (1999). The enriched small RNAs (2.5 μg) were fractionated on an 18% denaturing polyacrylamide-7 m urea gel in 1 × Tris-borate-EDTA buffer. Small RNAs were transferred to Hybond-N+ membranes (Amersham Biosciences) by upward capillary transfer in 20× SSC buffer, then cross-linked to the membranes with a UVP CX 2000 UV Crosslinker for four times (upside, underside, upside, underside) at 120 mJ/cm2, 1 min each time. The membranes were hybridized with digoxigenin (Dig)-labeled GFP RNA probes prepared by in vitro transcription using the pT7.GFP and Dig RNA Labeling Kit (Roche) as recommended by the manufacturer. The hybridization chemiluminescence signals were detected with a ChemiDoc XRS+ imaging System (Bio-Rad).
qRT-PCR
TCV/GFP∆CP or mock-inoculated leaves of Nb, DCL RNAi, 16cGFP, Gfp, and GfpDCL RNAi plants were taken at 7 d postinoculation (dpi) in three repeated experiments for RNA extraction. The first-strand cDNA was synthesized using total RNAs treated with RNase-free DNase I as templates by the M-MLV Reverse Transcriptase (Promega). The qRT-PCR analyses of DCLs mRNA or TCV/GFP∆CP RNA levels were performed using specific primers (Supplemental Table S5) and the SYBR Green Mix. The amplification program for SYBR Green I was performed at 95°C for 10 s, 58°C for 30 s, and 72°C for 20 s on the CFX96 machine (Bio-Rad), following the manufacturer’s instructions. Quadruplicate quantitative assays (four technical replicates) were performed on cDNA of each of three to four biological duplicates (leaf tissues from three to four different treated plants). The relative RNA quantification was calculated using the expression 2−ΔΔCt and normalized to the amount of GAPDH (GenBank accession no. TC17509) as described by Qin et al. (2012).
Construction of sRNA Library and sRNA Sequencing
Fragments of 18- to 30-bases-long RNA were isolated from total RNA extracted from mock- or TCV/GFP∆CP-inoculated leaf tissues of three to four different plants collected at 7 dpi after being separated through 15% denaturing PAGE. Then sRNAs were excised from the gel and sequentially ligated to 3′- and 5′-adapters. After each ligation step, sRNAs were purified after 15% denaturing PAGE. The final purified ligation products were reversely transcribed into cDNA using reverse transcriptase (Finnzymes Oy). The first-strand cDNA was PCR-amplified using Phusion DNA Polymerase (Finnzymes Oy). The purified DNA fragments were used for clustering and sequencing by Illumina HiSeq 2000 (Illumina) at the Beijing Genomics Institute. It should be noted that a pool of leaves from three to four different plants was used for construction of each sRNA library. This avoided potential variations between individual treated plants, in particular these for TCV/GFP∆CP-based intra- and intercellular VIGS assays due to some variations of TCV/GFP∆CP replication in different plants.
Bioinformatics Analysis of sRNA Sequences
Illumina HighSeq 2000 sequencing produced 11 to 12 million reads per sRNA library. The reads were cropped to remove adapter sequences and were aligned to the reference sequences using the software Bowtie2 (Langmead and Salzberg, 2012; Ryabov et al., 2014). The reference sequences included TCV/GFP∆CP, viral TcvGFP, and the 16cGFP transgene (Haseloff et al., 1997; Ruiz et al., 1998; Ryabov et al., 2004); DCL1, DCL2, DCL3, and DCL4 gene sequences (Nakasugi et al., 2013); and the set of 50 tobacco microRNAs identified in Nicotiana plants (Nakasugi et al., 2014; Pandey et al., 2008). SAMtools pileup was used to produce the siRNA and miRNA coverage profiles. For correlation analyses for the small RNA libraries, we determined numbers of the miRNA hits corresponding to the previously identified set of 50 Nicotiana miRNAs (Nakasugi et al., 2014; Pandey et al., 2008). All analyzed small RNA libraries contained similar proportions of host-encoded miRNA reads (Supplemental Data Sets S1–S3), indicating equivalence and direct comparability of the sRNA data sets. Indeed outcomes of comparisons between normalized siRNAs generated from target sequences against the total sRNA reads for all the libraries (per 10 million sRNA reads) are consistent with that the reads of siRNAs were directly compared.
Statistical Analysis
Normalized number of RNA silencing foci per leaf, sizes of RNA silencing foci, SCI, and qRT-PCR data between control and various treatments, were analyzed by Student’s t tests using an online program (http://www.physics.csbsju.edu/stats/t-test.html). It is worthwhile noting that ∼4% or more change in the silencing foci sizes is of statistical significance due to the large numbers of samples (80–560) tested between wild-type controls and RNAi lines (Fig. 1; Supplemental Table S2).
Supplemental Data
The following supplemental materials are available.
Supplemental Text S1. Parameters for assessing intra- and intercellular VIGS.
Supplemental Text S2. DCLs play differential roles in vsiRNA biogenesis.
Supplemental Text S3. Local VIGS versus virus interaction at the intra-/intercellular level.
Supplemental Figure S1. Total small RNA profiles.
Supplemental Figure S2. Distribution of vsiRNAs and siRNATcvGFP across the TCV/GFP∆CP RNA.
Supplemental Figure S3. Distribution of vsiRNAs across the TCV∆CP RNA.
Supplemental Figure S4. Distribution of vsiRNAs across the TCV∆CP RNA.
Supplemental Figure S5. Impact of DCLi on TCV/GFP∆CP RNA replication.
Supplemental Figure S6. Comparisons between transgene 16cGFP and viral TcvGFP sequences.
Supplemental Figure S7. Distribution of 20- to 25-nucleotide GFP siRNAs across the 714-nucleotide TcvGFP mRNA.
Supplemental Figure S8. Distribution of 20- to 25-nucleotide GFP siRNAs across the 792-nucleotide transgene 16cGFP mRNA.
Supplemental Figure S9. Northern detection of TCV/GFPΔCP siRNAs.
Supplemental Figure S10. DCL2 and DCL2-dependent siRNAs for non-cell-autonomous intercellular VIGS.
Supplemental Table S1. DCL RNAi lines used in this study.
Supplemental Table S2. Impact of DCL RNAi on cell-to-cell spread of virus-induced RNA silencing.
Supplemental Table S3. Summary of total viral and/or GFP siRNAs in mock- or TCV/GFP∆CP-inoculated leaves.
Supplemental Table S4. Percentage of 16cGFP and TCV/GFPΔCP 21-, 22-, and 24-nucleotide siRNA.
Supplemental Table S5. Primers used in this study.
Supplemental Data Set S1. Summary of sRNA and miRNA reads.
Supplemental Data Set S2. Correlation analyses of miRNA profiles among 10 sRNA libraries.
Supplemental Data Set S3. Comparisons of miRNAs among 10 sRNA libraries.
Supplementary Material
Acknowledgments
We thank David Baulcombe for transgenic line 16cGFP and RDR6i seeds. Y.H. thanks Dr. Alison Tör for checking English grammar and style throughout the manuscript.
Footnotes
Articles can be viewed without a subscription.
References
- Aliyari R, Ding SW (2009) RNA-based viral immunity initiated by the Dicer family of host immune receptors. Immunol Rev 227: 176–188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andika IB, Maruyama K, Sun L, Kondo H, Tamada T, Suzuki N (2015) Differential contributions of plant Dicer-like proteins to antiviral defences against potato virus X in leaves and roots. Plant J 81: 781–793 [DOI] [PubMed] [Google Scholar]
- Aregger M, Borah BK, Seguin J, Rajeswaran R, Gubaeva EG, Zvereva AS, Windels D, Vazquez F, Blevins T, Farinelli L, Pooggin MM (2012) Primary and secondary siRNAs in Geminivirus-induced gene silencing. PLoS Pathog 8: e1002941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baulcombe D. (2004) RNA silencing in plants. Nature 431: 356–363 [DOI] [PubMed] [Google Scholar]
- Berg JM. (2016) Retraction. Science 354: 190. [DOI] [PubMed] [Google Scholar]
- Blevins T, Rajeswaran R, Shivaprasad PV, Beknazariants D, Si-Ammour A, Park HS, Vazquez F, Robertson D, Meins F Jr., Hohn T, Pooggin MM (2006) Four plant Dicers mediate viral small RNA biogenesis and DNA virus induced silencing. Nucleic Acids Res 34: 6233–6246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouché N, Lauressergues D, Gasciolli V, Vaucheret H (2006) An antagonistic function for Arabidopsis DCL2 in development and a new function for DCL4 in generating viral siRNAs. EMBO J 25: 3347–3356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrington JC, Heaton LA, Zuidema D, Hillman BI, Morris TJ (1989) The genome structure of Turnip Crinkle Virus. Virology 170: 219–226 [DOI] [PubMed] [Google Scholar]
- Chattopadhyay M, Stupina VA, Gao F, Szarko CR, Kuhlmann MM, Yuan X, Shi K, Simon AE (2015) Requirement for host RNA-silencing components and the virus-silencing suppressor when second-site mutations compensate for structural defects in the 3′ untranslated region. J Virol 89: 11603–11618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen HM, Chen LT, Patel K, Li YH, Baulcombe DC, Wu SH (2010) 22-Nucleotide RNAs trigger secondary siRNA biogenesis in plants. Proc Natl Acad Sci USA 107: 15269–15274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen Y, Gisel A, Zambryski PC (2000) Cell-to-cell and systemic movement of recombinant green fluorescent protein-tagged Turnip Crinkle Viruses. Virology 273: 258–266 [DOI] [PubMed] [Google Scholar]
- Csorba T, Kontra L, Burgyán J (2015) Viral silencing suppressors: tools forged to fine-tune host-pathogen coexistence. Virology 479-480: 85–103 [DOI] [PubMed] [Google Scholar]
- Dunoyer P, Himber C, Voinnet O (2005) DICER-LIKE 4 is required for RNA interference and produces the 21-nucleotide small interfering RNA component of the plant cell-to-cell silencing signal. Nat Genet 37: 1356–1360 [DOI] [PubMed] [Google Scholar]
- Fusaro AF, Matthew L, Smith NA, Curtin SJ, Dedic-Hagan J, Ellacott GA, Watson JM, Wang MB, Brosnan C, Carroll BJ, Waterhouse PM (2006) RNA interference-inducing hairpin RNAs in plants act through the viral defence pathway. EMBO Rep 7: 1168–1175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Ruiz H, Takeda A, Chapman EJ, Sullivan CM, Fahlgren N, Brempelis KJ, Carrington JC (2010) Arabidopsis RNA-dependent RNA polymerases and dicer-like proteins in antiviral defense and small interfering RNA biogenesis during Turnip Mosaic Virus infection. Plant Cell 22: 481–496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hacker DL, Petty IT, Wei N, Morris TJ (1992) Turnip Crinkle Virus genes required for RNA replication and virus movement. Virology 186: 1–8 [DOI] [PubMed] [Google Scholar]
- Hamilton AJ, Baulcombe DC (1999) A species of small antisense RNA in posttranscriptional gene silencing in plants. Science 286: 950–952 [DOI] [PubMed] [Google Scholar]
- Haseloff J, Siemering KR, Prasher DC, Hodge S (1997) Removal of a cryptic intron and subcellular localization of green fluorescent protein are required to mark transgenic Arabidopsis plants brightly. Proc Natl Acad Sci USA 94: 2122–2127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henderson IR, Zhang X, Lu C, Johnson L, Meyers BC, Green PJ, Jacobsen SE (2006) Dissecting Arabidopsis thaliana DICER function in small RNA processing, gene silencing and DNA methylation patterning. Nat Genet 38: 721–725 [DOI] [PubMed] [Google Scholar]
- Langmead B, Salzberg SL (2012) Fast gapped-read alignment with Bowtie 2. Nat Methods 9: 357–359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katsarou K, Mavrothalassiti E, Dermauw W, van Leeuwen T, Kalantidis K (2016) Combined activity of DCL2 and DCL3 is crucial in the defense against Potato Spindle Tuber viroid. PLoS Pathog 12: e1005936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C, Zhang K, Zeng X, Jackson S, Zhou Y, Hong Y (2009) A cis element within flowering locus T mRNA determines its mobility and facilitates trafficking of heterologous viral RNA. J Virol 83: 3540–3548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li WZ, Qu F, Morris TJ (1998) Cell-to-cell movement of turnip crinkle virus is controlled by two small open reading frames that function in trans. Virology 244: 405–416 [DOI] [PubMed] [Google Scholar]
- Melnyk CW, Molnar A, Baulcombe DC (2011) Intercellular and systemic movement of RNA silencing signals. EMBO J 30: 3553–3563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mérai Z, Kerényi Z, Kertész S, Magna M, Lakatos L, Silhavy D (2006) Double-stranded RNA binding may be a general plant RNA viral strategy to suppress RNA silencing. J Virol 80: 5747–5756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mlotshwa S, Pruss GJ, Peragine A, Endres MW, Li J, Chen X, Poethig RS, Bowman LH, Vance V (2008) DICER-LIKE2 plays a primary role in transitive silencing of transgenes in Arabidopsis. PLoS One 3: e1755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukherjee K, Campos H, Kolaczkowski B (2013) Evolution of animal and plant dicers: early parallel duplications and recurrent adaptation of antiviral RNA binding in plants. Mol Biol Evol 30: 627–641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakasugi K, Crowhurst R, Bally J, Waterhouse P (2014) Combining transcriptome assemblies from multiple de novo assemblers in the allo-tetraploid plant Nicotiana benthamiana. PLoS One 9: e91776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakasugi K, Crowhurst RN, Bally J, Wood CC, Hellens RP, Waterhouse PM (2013) De novo transcriptome sequence assembly and analysis of RNA silencing genes of Nicotiana benthamiana. PLoS One 8: e59534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandey SP, Shahi P, Gase K, Baldwin IT (2008) Herbivory-induced changes in the small-RNA transcriptome and phytohormone signaling in Nicotiana attenuata. Proc Natl Acad Sci USA 105: 4559–4564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parent J-S, Bouteiller N, Elmayan T, Vaucheret H (2015) Respective contributions of Arabidopsis DCL2 and DCL4 to RNA silencing. Plant J 81: 223–232 [DOI] [PubMed] [Google Scholar]
- Pérez-Cañamás M, Hernández C (2015) Key importance of small RNA binding for the activity of a glycine-tryptophan (GW) motif-containing viral suppressor of RNA silencing. J Biol Chem 290: 3106–3120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin C, Shi N, Gu M, Zhang H, Li B, Shen J, Mohammed A, Ryabov E, Li C, Wang H, et al. (2012) Involvement of RDR6 in short-range intercellular RNA silencing in Nicotiana benthamiana. Sci Rep 2: 467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qu F, Ren T, Morris TJ (2003) The coat protein of Turnip Crinkle Virus suppresses posttranscriptional gene silencing at an early initiation step. J Virol 77: 511–522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qu F, Ye X, Morris TJ (2008) Arabidopsis DRB4, AGO1, AGO7, and RDR6 participate in a DCL4-initiated antiviral RNA silencing pathway negatively regulated by DCL1. Proc Natl Acad Sci USA 105: 14732–14737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qu J, Ye J, Fang R (2007) Artificial microRNA-mediated virus resistance in plants. J Virol 81: 6690–6699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz MT, Voinnet O, Baulcombe DC (1998) Initiation and maintenance of virus-induced gene silencing. Plant Cell 10: 937–946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryabov EV, van Wezel R, Walsh J, Hong Y (2004) Cell-to-cell, but not long-distance, spread of RNA silencing that is induced in individual epidermal cells. J Virol 78: 3149–3154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryabov EV, Wood GR, Fannon JM, Moore JD, Bull JC, Chandler D, Mead A, Burroughs N, Evans DJ (2014) A virulent strain of deformed wing virus (DWV) of honeybees (Apis mellifera) prevails after Varroa destructor-mediated, or in vitro, transmission. PLoS Pathog 10: e1004230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarkies P, Miska EA (2014) Small RNAs break out: the molecular cell biology of mobile small RNAs. Nat Rev Mol Cell Biol 15: 525–535 [DOI] [PubMed] [Google Scholar]
- Schwach F, Vaistij FE, Jones L, Baulcombe DC (2005) An RNA-dependent RNA polymerase prevents meristem invasion by Potato Virus X and is required for the activity but not the production of a systemic silencing signal. Plant Physiol 138: 1842–1852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Searle IR, Pontes O, Melnyk CW, Smith LM, Baulcombe DC (2010) JMJ14, a JmjC domain protein, is required for RNA silencing and cell-to-cell movement of an RNA silencing signal in Arabidopsis. Genes Dev 24: 986–991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y, Ryabov EV, van Wezel R, Li C, Jin M, Wang W, Fan Z, Hong Y (2009) Suppression of local RNA silencing is not sufficient to promote cell-to-cell movement of Turnip Crinkle Virus in Nicotiana benthamiana. Plant Signal Behav 4: 15–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith LM, Pontes O, Searle I, Yelina N, Yousafzai FK, Herr AJ, Pikaard CS, Baulcombe DC (2007) An SNF2 protein associated with nuclear RNA silencing and the spread of a silencing signal between cells in Arabidopsis. Plant Cell 19: 1507–1521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas CL, Leh V, Lederer C, Maule AJ (2003) Turnip crinkle virus coat protein mediates suppression of RNA silencing in Nicotiana benthamiana. Virology 306: 33–41 [DOI] [PubMed] [Google Scholar]
- Wang XB, Jovel J, Udomporn P, Wang Y, Wu Q, Li WX, Gasciolli V, Vaucheret H, Ding SW (2011) The 21-nucleotide, but not 22-nucleotide, viral secondary small interfering RNAs direct potent antiviral defense by two cooperative argonautes in Arabidopsis thaliana. Plant Cell 23: 1625–1638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Z, Allen E, Wilken A, Carrington JC (2005) DICER-LIKE 4 functions in trans-acting small interfering RNA biogenesis and vegetative phase change in Arabidopsis thaliana. Proc Natl Acad Sci USA 102: 12984–12989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Z, Johansen LK, Gustafson AM, Kasschau KD, Lellis AD, Zilberman D, Jacobsen SE, Carrington JC (2004) Genetic and functional diversification of small RNA pathways in plants. PLoS Biol 2: E104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Zhang X, Singh J, Li D, Qu F (2012) Temperature-dependent survival of Turnip Crinkle Virus-infected Arabidopsis plants relies on an RNA silencing-based defense that requires dcl2, AGO2, and HEN1. J Virol 86: 6847–6854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y, Ryabov E, Zhang X, Hong Y (2008) Influence of viral genes on the cell-to-cell spread of RNA silencing. J Exp Bot 59: 2803–2813 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.